Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Synthesis of new Ag(I) and Cd(II) hydrozanomide-based complexes with antibacterial and anticancer properties†
Alina Climova *a,
Ekaterina Pivovarova
*a,
Ekaterina Pivovarova a,
Małgorzata Szczesioa,
Katarzyna Gobisb,
Agnieszka Korga-Plewkoc,
Magdalena Iwanc,
Edyta Kordialik-Bogackad,
Sylwia Ścieszkad,
Jaromir Marekef and
Agnieszka Czylkowska
a,
Małgorzata Szczesioa,
Katarzyna Gobisb,
Agnieszka Korga-Plewkoc,
Magdalena Iwanc,
Edyta Kordialik-Bogackad,
Sylwia Ścieszkad,
Jaromir Marekef and
Agnieszka Czylkowska *a
*a
aInstitute of General and Ecological Chemistry, Faculty of Chemistry, Lodz University of Technology, Zeromskiego 116, Lodz, 90-924, Poland. E-mail: alina.climova@dokt.p.lodz.pl; agnieszka.czylkowska@p.lodz.pl
bDepartment of Organic Chemistry, Faculty of Pharmacy, Medical University of Gdańsk, 107 Gen. Hallera Ave., Gdańsk, 80-416, Poland
cIndependent Medical Biology Unit, Faculty of Pharmacy, Medical University of Lublin, Jaczewskiego 8b, Lublin, 20-093, Poland
dInstitute of Fermentation Technology and Microbiology, Faculty of Biotechnology and Food Sciences, Lodz University of Technology, Wólczańska 171/173, Lodz, 90-924, Poland
eCore Facility Biomolecular Interactions and Crystallography, CEITEC MU, Masaryk University, Kamenice 5, Brno, 62500, Czech Republic
fDepartment of Chemistry, Faculty of Science, Masaryk University, Kamenice 5, Brno, 62500, Czech Republic
First published on 10th June 2025
Abstract
A series of new Ag(I) and Cd(II) hydrozanomide-based complexes have been prepared and studied. The synthesized compounds have undergone extensive spectroscopic and structural characterization, including F-AAS, FTIR, single crystal XRD and TGA analyses. Crystal structures of two representative complexes, 4 and 6, have revealed that the silver atom in complex 4 assumes a four-coordinate geometry with two ligands. Additionally, the stability of these complexes in DMSO has been determined using UV-vis spectroscopy. The biological activity of these metal complexes has been assessed, focusing on their antibacterial and anticancer properties against LN-229 and U87 cell lines. Among the tested compounds, silver-based complexes demonstrated significant antibacterial activity against a wide range of microorganisms. Moreover, cytotoxicity studies have shown that the metal complexes exhibit higher anticancer activity compared to their parent ligands. Notably, the Ag(I) complexes of L3, L4, and L5 have emerged as the most promising candidates for their selective toxicity against cancer cells, without harmful effects on normal human fibroblast cells. These findings highlight the potential of Ag(I) complexes as promising anticancer agents with selective toxicity and potent antibacterial properties.
Introduction
Coordination chemistry is a promising field that has several advantages in the development of new therapies for cancer treatment.1 Metal complexes, often derived from transition metals such as platinum, have shown promising anticancer properties. The most commonly used and effective ones are cisplatin, carboplatin, and oxaliplatin.2 The ability of metal ions to coordinate with biomolecules such as DNA underlies their effectiveness in inhibiting cell proliferation. The mechanism of action is attributed to their ability to penetrate the cell, bind to cellular DNA, forming cross-links that alter the DNA structure and disrupt its function.3,4 One of the major benefits of coordination compounds is their inherent tunability. This allows researchers to precisely adjust these compounds to specific biological targets, enhancing their selectivity and reducing side effects.5 Through precise compound design, researchers synthesize complexes that selectively interact with cancer cells, sparing healthy tissues and minimizing damage to normal cells.6–9 This targeted approach enhances therapeutic efficacy and minimizes the risk of adverse side effects. It also promotes safer and more efficient treatments, making coordination compounds an attractive option for the development of personalized medicine. The capacity to modify these compounds to optimize their pharmacokinetic and pharmacodynamic properties presents a significant opportunity to advance personalized treatment options. Although coordination compounds have promising therapeutic potential, many metal-based candidates, especially those already close to clinical application, have been discontinued due to unacceptable toxicity. For this reason, this area needs more extensive research and more expanded database.10Ongoing research efforts around the world are aiming to reduce the side effects of chemotherapy through the development of novel complexes containing dn-electron metals as alternatives to platinum-based drugs.11,12 Silver has a more effective antimicrobial effect than penicillin and other antibiotics, and also causes a similar effect on antibiotic-resistant strains of bacteria.13 Budagumpi's team studied the antimicrobial properties of benzimidazolium salts and their bis-N-heterocyclic carbene silver(I) complexes. Some of the tested compounds showed excellent antibacterial activity against E. coli, with a MIC value of 4 μg mL−1, which is comparable to the standard antibiotic ampicillin. One of the compounds demonstrated a MIC value of 2–4 μg mL−1 against S. aureus, E. coli, and P. aeruginosa, which is better than the ampicillin value.14,15 Ag ions have a significant ability to inactivate influenza viruses, some entero- and adenoviruses, as well as inhibit the activity of the AIDS virus.16 Moreover, silver complexes exhibited more potent cytotoxicity with the least toxicity compared to cisplatin.17 Cd(II) complexes have attracted attention due to the growing recognition of their role in biological processes and rich structural chemistry.18–21 Cadmium complexes showed anticancer activity against Mia Paca-2, BxPC-3 and Panc-1 pancreatic cancer cell lines with IC50 values up to 16.0 μM.22 In addition, it has been reported that complexes of this metal inhibit T-cell leukemia tumor growth in mice.23 Cadmium complexes have also attracted attention as potential antimicrobial agents.24,25 Cadmium can disrupt the functioning of bacterial cells by embedding into their membranes and disrupting their integrity, as well as inhibiting the activity of various bacterial enzymes, which disrupt their metabolic processes and lead to cell death.26,27
Previously, we synthesized new organic pyridine and pyrazine based-ligands and a series of neutral Cu(II) and Zn(II) complexes based on them.28,29 The present work is a continuation of our previous studies on the biological activity of metal complexes, and herein, we report the synthesis, spectroscopic and structural characterization of them. We investigated new Ag(I) and Cd(II) complexes based on the previously described ligands: L1 – N′-(benzylidene)-6-chloropyrazine-2-carbohydrazonamide, L2 – 6-chloro-N′-(4-nitrobenzylidene) picolinohydrazonamide, L3 – N′-(benzylidene)-4-chloropicolinohydrazonamide, L4 – (4-nitrobenzylidene)-6-(pyrrolidin-1-yl)picolinohydrazonamide, L5 – (benzylidene)-6-morpholinopyrazine-2-carbohydrazonamide and L6 – (benzylidene)-6-(pyrrolidin-1-yl)pyrazine-2-carbohydra-zonamide (Fig. 1).
All synthesized complexes were characterized by F-AAS, FTIR, UV-vis, and TGA analysis. Only for two coordination compounds (4 and 6) the single crystals were obtained. The crystal structures of complexes 4 and 6 were described, where Ag atom of complex 4 is four-coordinated by two ligand molecules. Additionally, antibacterial and anticancer activity of the obtained compounds were evaluated. Among all the tested compounds, silver complexes demonstrated the most promising results in almost all microorganisms used. Anticancer activity measurements revealed that synthesized metal complexes generally exhibited greater cytotoxic potential than their parent ligands, with varying sensitivity between different cell lines. The results were compared to the activity of the human skin fibroblast cell line BJ. The Ag complexes of L3, L4, and L5 were the most promising compounds, as they showed selective toxicity towards cancer cells while having no effect on normal cells.
Result and discussion
Synthesized complexes
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1632; δ(NH) 1550; β(CH) 1366; ν(C
N) 1632; δ(NH) 1550; β(CH) 1366; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1444; ν(N–N) 1169; γ(CH) 855.
C) 1444; ν(N–N) 1169; γ(CH) 855.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1626; δ(NH) 1558; β(CH) 1382; ν(C
N) 1626; δ(NH) 1558; β(CH) 1382; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1457; ν(N–N) 1160, 1130; γ(CH) 854.
C) 1457; ν(N–N) 1160, 1130; γ(CH) 854.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1622; δ(NH) 1560; β(CH) 1373; ν(C
N) 1622; δ(NH) 1560; β(CH) 1373; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1445; ν(N–N) 1175, 1120; γ(CH) 881.
C) 1445; ν(N–N) 1175, 1120; γ(CH) 881.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1632; δ(NH) 1593; β(CH) 1360; ν(C
N) 1632; δ(NH) 1593; β(CH) 1360; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1449; ν(N–N) 1165, 1006; γ(CH) 881.
C) 1449; ν(N–N) 1165, 1006; γ(CH) 881.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1629; δ(NH) 1573, 1525; β(CH) 1372; ν(C
N) 1629; δ(NH) 1573, 1525; β(CH) 1372; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1447; ν(N–N) 1117, 1069; γ(CH) 885.
C) 1447; ν(N–N) 1117, 1069; γ(CH) 885.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1629; δ(NH) 1527, 1565; β(CH) 1377; ν(C
N) 1629; δ(NH) 1527, 1565; β(CH) 1377; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1490, 1449; ν(N–N) 1147, 1024; γ(CH) 862.
C) 1490, 1449; ν(N–N) 1147, 1024; γ(CH) 862.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1624; δ(NH) 1557; β(CH) 1381; ν(C
N) 1624; δ(NH) 1557; β(CH) 1381; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1456; ν(N–N) 1160, 1105; γ(CH) 854.
C) 1456; ν(N–N) 1160, 1105; γ(CH) 854.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1632; δ(NH) 1538; β(CH) 1376; ν(C
N) 1632; δ(NH) 1538; β(CH) 1376; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1447; ν(N–N) 1186, 1123; γ(CH) 870.
C) 1447; ν(N–N) 1186, 1123; γ(CH) 870.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) 1630; δ(NH) 1558; β(CH) 1404; ν(C
N) 1630; δ(NH) 1558; β(CH) 1404; ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) 1457; ν(N–N) 1175, 1149; γ(CH) 857.
C) 1457; ν(N–N) 1175, 1149; γ(CH) 857.FTIR
FTIR spectra of free ligands and Ag(I) and Cd(II) coordination compounds are shown in Fig. S1.† The spectra of pure ligands have been described previously.28,29 The main frequencies and distribution are summarised in Table S1.†The stretching vibration mode of the –NH– group is observed in the range of 3470–3300 cm−1, composed of two peaks. After complexation, these vibration modes are shifted and decreased in intensity. For complex 5, there is an overlap of the ν(NH) vibrations due to the presence of water molecules. In the range of 3100–3000 cm−1, stretching vibrations of CH groups are observed for all complexes.
The sharp CN bands observed in the range of 1632–1614 cm−1 (ref. 30–33) for the ligands shifted after complexation and did not change their intensity. A more significant shift was observed for complexes 8 and 9, which is proposed to be due to complexation via nitrogen from the pyridine/pyrazine ring. Complexes 4, 6, 8, 9 showed more significant vibrations in the region of 1170–1070 cm−1 (ν(NN)), indicating participation of the imine group in complexation. Strong absorption peaks were observed in the pure ligand at 1475–1449 cm−1, corresponding to the ν(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) vibration mode. These peaks shifted towards higher wavenumbers after complex formation. At lower wavelengths, bands were observed at 1406–1361, 998–949 and 970–690 cm−1 that correspond to β(CH), δ(C
C) vibration mode. These peaks shifted towards higher wavenumbers after complex formation. At lower wavelengths, bands were observed at 1406–1361, 998–949 and 970–690 cm−1 that correspond to β(CH), δ(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), and γ(CH), respectively.
C), and γ(CH), respectively.
Based on the obtained data, coordination in complexes 1, 2, 3, 5, and 7 is proposed via the nitrogen atom of the pyridine/pyrazine ring. For the other complexes, the imine nitrogen atom is involved in coordination. In these cases, ligands act as bidentants, chelating metal centers. Complex 4 is an exception, as the ligand forms an infinite, one-dimensional polymer chain upon binding to the metal, which is considered in the crystal structure part. The proposed coordination in complex 6 has been confirmed by the crystal structure of the compound. Spectra of 1H and 13C (Fig. S2–S5†) were also obtained for complex 7, which also (together with EA and F-AAS) clarify the proposed complex structure.
NMR
The 1H NMR spectrum of the free ligand L2 (Fig. S2†) reveals several characteristic signals, including NH2 protons (at δ 7.16 and 7.48 ppm), pyridine protons (between δ 7.66 and 8.24), and a distinct azomethine proton at δ 8.57. Upon complexation with cadmium (complex 7), there are slight downfield shifts observed for the pyridine protons, indicating pyridine involvement in metal coordination (Fig. S3†). The 13C NMR (Fig. S4†) spectrum of L2 reveals characteristic signals from its pyridine and 4-nitrophenyl groups, connected through a hydrazone bond. Key signals are seen at δ 157.232 (C–Cl), 151.534 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N pyridine), and 142.103 ppm (C
N pyridine), and 142.103 ppm (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N hydrazone). Upon complexation with Cd(II), forming complex 7 (Cd(L2)2Cl2), there are specific downfield shifts in the pyridine ring carbon atoms (e.g., δ 157.232 → 157.52 ppm), confirming the coordination through the pyridine nitrogen atoms (Fig. S5†). These changes in chemical shifts reflect the decreased electron density around the coordination sites, as electrons are transferred to the Cd(II) center. The consistent spectral pattern in the aromatic/heterocyclic region (120–160 ppm) confirms that the structural framework of the ligand remains intact after coordination, without any degradation or rearrangement. This indicates successful synthesis of a stable and pure complex.
N hydrazone). Upon complexation with Cd(II), forming complex 7 (Cd(L2)2Cl2), there are specific downfield shifts in the pyridine ring carbon atoms (e.g., δ 157.232 → 157.52 ppm), confirming the coordination through the pyridine nitrogen atoms (Fig. S5†). These changes in chemical shifts reflect the decreased electron density around the coordination sites, as electrons are transferred to the Cd(II) center. The consistent spectral pattern in the aromatic/heterocyclic region (120–160 ppm) confirms that the structural framework of the ligand remains intact after coordination, without any degradation or rearrangement. This indicates successful synthesis of a stable and pure complex.
The tetrahedral geometry of the complex is supported by spectral changes in both 1H and 13C NMR spectra, where two L2 ligands are coordinated to cadmium via pyridine nitrogens, while two chloride ions complete the coordination sphere.
Crystal structure
TGA
Fig. S6–S8† present thermal decomposition patterns of the obtained Ag(I) and Cd(II) coordination compounds. Table 1 demonstrates the decomposition stages of the complexes with the identified temperature ranges. All investigated compounds were stable at room temperature and decomposed gradually during the heating process.| Compound | Stages | Temperature ranges (°C) | Final product | |||
|---|---|---|---|---|---|---|
| I | II | III | IV | |||
| Complex 1 | 4 | 118–156 | 156–205 | 205–642 | 642–820 | Ag |
| Complex 2 | 4 | 100–210 | 210–220 | 220–665 | 665–820 | Ag |
| Complex 3 | 4 | 110–155 | 155–255 | 255–675 | 675–820 | Ag |
| Complex 4 | 3 | 135–240 | 240–350 | 350–525 | — | Ag |
| Complex 5 | 4 | 35–175 | 175–250 | 250–600 | 600–710 | Ag |
| Complex 6 | 3 | 195–245 | 245–485 | 485–560 | — | Ag |
| Complex 7 | 3 | 220–350 | 350–660 | 660–745 | — | CdO |
| Complex 8 | 3 | 305–400 | 400–590 | 590–710 | — | CdO |
| Complex 9 | 3 | 260–360 | 360–680 | 680–760 | — | CdO |
The first three complexes are stable up to 100 °C and more and decomposed gradually in a similar way. They consist 4 main decomposition steps. The first stage of decomposition of the complex 1 proves the presence of H2O molecule in these compounds (−3.84%). Complex 2 and 3 showed a low mass loss (less than 1%) of water from the surface of the test compounds. As the temperature increases, all three complexes undergo gradual thermal decomposition combined with the destruction of the organic ligand. The last stage for all coordination compounds is the decomposition of the residual parts of the organic ligand, loss of nitrate anions and the formation of Ag34,35 (22.41% for complex 1, 21.1% for complex 2, 24.3% for complex 3).
Complex 5 is stable at room temperature, but above 35 °C the dehydration process begins, which is the first step of decomposition. In the second step of thermal decomposition, destruction of morpholine is observed with a mass loss of 17.7% (theor. 17.5%). The process was completed in the fourth step at 710 °C, where the final solid product of decomposition is metallic Ag (21.2%).
Complexes 4 and 6 are the most stable among all studied silver complexes. Thermal decomposition patterns of these complexes contain three decomposition steps each in a similar manner. All compounds decompose progressively. The decompositions started at 135 °C and 185 °C with mass losses of 13.2% and 15.7%, corresponding to the combustion of pyrrolidine (complex 4, 13.8% theor.) and pyrrole (complex 6, 15% theor.). The last third step for these two coordination compounds is the decomposition of the residual parts of the organic ligand, loss of nitrate anions and the formation of Ag (19% for complex 4 and 23.4% for complex 6).
Complexes 7, 8, 9 demonstrate very high stability up to 220 °C, 305 °C, 260 °C, respectively. Decomposition started with the partial loss of part of organic ligands for all three complexes. The second mass losses can be assigned to the release of the rest parts of the organic ligands to form CdO. The total mass loss observed experimentally during the 3rd step at 710 °C and higher (96.18% for complexes 7, 95.65% for complexes 8, 96.52% for complexes 9). This behaviour can be connected with the CdCl2 volatilization.36–38
Solution stability
The stability of metal complexes is a crucial property that provides insight into their interactions with biological molecules, as it directly impacts their reactivity, bioavailability, and effectiveness in therapeutic applications. Additionally, the solubility of these complexes in dimethyl sulfoxide (DMSO) is of considerable importance, as solubility data are essential for optimizing storage conditions and ensuring compatibility with biological assays.39 DMSO, a widely used solvent in drug discovery, serves as a medium that supports the stability and bioavailability of complexes under assay conditions, which is critical for maintaining the integrity of analytical protocols and enabling accurate assessments of biological activity. For the obtained complexes, the time-dependent UV-vis investigations in DMSO (Fig. 5 and S9†) were carried out from 0 min to 48 h (0 min, 30 min, 1 h, 2 h, 3 h, 24 h, 48 h).The most stable compounds were complexes 1 and 3, where the hypochromic effect appeared to the least extent, representing 0.84% and 2.31%, respectively. At the same time hypsochromic shifts are absent. Similarly, no distinctive changes were observed for complexes 2, 4, 5, 6, and 9, where Δε ranges from 5% to 9.8%. Complexes 7 and 8 showed the least stable behaviour, where the hypochromic effect was 27.7% and 36.3%, and the peak maxima shifted by 15 and 4 nm, respectively.
Antibacterial study
In this study, the antibacterial activity of the Ag(I) and Cd(II) complexes was investigated. The results were compared to the previous study, where antibacterial properties of their ligands were measured.29 Some of the results are presented in the Table 2, the whole data is available in the Table S8.† The inhibition zones around the wells (Fig. 6) were measured as well and presented in the Table S9.†| Microorganism | Complexes and ligands | Ref. drug | |||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 | L1 | 4 | L4 | 5 | L5 | 6 | L6 | ||
| Gram-positive bacteria | MIC (μg mL−1) | Van | |||||||
| S. aureus ATCC 25923 | <10 | 500 | <10 | >500 | 50 | >500 | 50 | 500 | 1 |
| S. epidermidis ATCC 12228 | 10 | 500 | 50 | >500 | <10 | >500 | <10 | 500 | 1 |
| E. faecalis ATCC 29212 | 50 | 250 | 50 | >500 | 50 | >500 | >500 | 500 | 1 |
| Gram-negative bacteria | MIC (μg mL−1) | Cip | |||||||
| S. typhimurium ATCC 14028 | <10 | >500 | 100 | >500 | 50 | >500 | 50 | 500 | 1 |
Complex 1 exhibited antibacterial activity against all tested microorganisms, showing MIC values <10 μg mL−1 for Staphylococcus aureus, Listeria monocytogenes, Escherichia coli and Salmonella typhimurium. Among all the tested complexes, complex 1 exhibited the strongest antibacterial activity. Complex 2 also exhibited activity against all tested microorganisms, except Staphylococcus epidermidis. The lowest MIC values (10 mg L−1) for complex 2 were obtained against Listeria monocytogenes and Escherichia coli. For complex 3 the results were similar, but MIC value of 10 μg mL−1 was also found for Bacillus subtilis. Complex 4 showed high antibacterial activity against Staphylococcus aureus, Listeria monocytogenes, Escherichia coli (MIC 10 or <10 μg mL−1). In the case of complex 5, it was determined that the complex showed high activity against all tested microorganisms except Escherichia coli. Among all tested microorganisms, complex 6 was not effective against Enterococcus faecalis and Escherichia coli. Its highest activity was against Staphylococcus epidermidis (MIC <10 μg mL−1). Compared to the silver complexes, the cadmium complexes exhibited lower antibacterial properties. However, in contrast to the corresponding ligands, complexes 8 and 9 exhibited moderate activity against both Gram-positive and Gram-negative bacteria. Among them, complex 9 exhibited the strongest antibacterial properties against Staphylococcus aureus, Enterococcus faecalis, Escherichia coli (10 μg mL−1). Complex 7 exhibited the lowest antibacterial activity among all tested compounds.
Anticancer study
The MTT assay results after 48 hours of incubation demonstrated variable cytotoxic effects of the tested compounds across three cell lines: glioblastoma U87, LN-229, and normal fibroblast BJ (Table 3). Compounds L1, L3, L5 exhibited limited cytotoxicity with IC50 values exceeding 50 μM. However, significant cytotoxic activity was observed in certain metal complexes. For instance, complex 1 showed potent cytotoxicity in both LN-229 (IC50 = 25.22 ± 3.75 μM) and BJ cells (IC50 = 20.48 ± 5.21 μM). L2 and its Ag derivative (complex 2) also demonstrated activity, particularly in LN-229 (IC50 = 40.77 ± 6.12 μM and 37.24 ± 6.55 μM, respectively) and BJ cells (IC50 = 35.48 ± 4.33 μM and 14.46 ± 2.82 μM, respectively). Complex 4 showed the most significant cytotoxicity in U87 cells (IC50 = 1.22 ± 0.34 μM), while complex 9 was highly active in BJ cells (IC50 = 1.05 ± 0.32 μM). Interestingly, some compounds exhibited selective cytotoxicity. As an example, complex 6 showed strong activity across all three cell lines, with IC50 values of 10.17 ± 2.98 μM in U87, 20.98 ± 3.07 μM in LN-229, and 10.62 ± 2.14 μM in BJ cells. Overall, metal complexes of Ag(I) and Cd(II) derivatives, generally exhibited greater cytotoxic potential compared to their parent ligands, with variations in cell line sensitivity. BJ fibroblasts were more susceptible, suggesting that metal complexation may enhance cytotoxic effects.| U87 | LN-229 | BJ | |
|---|---|---|---|
| L1 | >50 | >50 | >50 |
| Complex 1 | 8.15 ± 2.67 | 25.22 ± 3.75 | 20.48 ± 5.21 |
| L2 | >50 | 40.77 ± 6.12 | 35.48 ± 4.33 |
| Complex 2 | >50 | 37.24 ± 6.55 | 14.46 ± 2.82 |
| Complex 7 | >50 | >50 | 41.66 ± 4.18 |
| L3 | >50 | >50 | >50 |
| Complex 3 | 32.03 ± 7.08 | >50 | >50 |
| Complex 8 | 10.87 ± 3.80 | >50 | 25.84 ± 4.48 |
| L4 | >50 | 48.11 ± 6.09 | >50 |
| Complex 4 | 1.22 ± 0.34 | >50 | >50 |
| Complex 9 | >50 | 45.34 ± 2.18 | 1.05 ± 0.32 |
| L5 | >50 | >50 | >50 |
| Complex 5 | 49.67 ± 3.11 | >50 | >50 |
| L6 | >50 | >50 | 42.36 ± 6.17 |
| Complex 6 | 10.17 ± 2.98 | 20.98 ± 3.07 | 10.62 ± 2.14 |
| Cisplatin | 39.56 ± 1.13 | 29.09 ± 0.23 | >50 |
Ag(I) complexes of L3, L4 and L5 are the most promising compounds, that demonstrated selective toxicity towards cancer cells while showing no effect on normal cells (Fig. S10†).
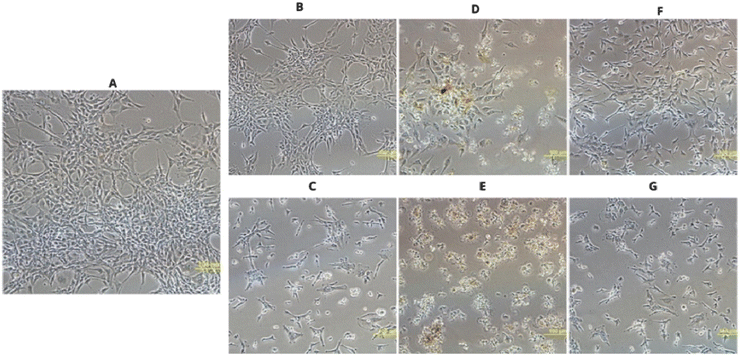
Microscopic observations confirmed the results of the MTT assay. Cells treated with the Ag-containing complexes shrank, which is a sign of toxic effects (Fig. 7C, E and G). Cells exposed to the complex 4 appeared more rounded and detached from the substrate (Fig. 7E), indicating that this compound has the strongest effect. Ligands L3, and L5 did not exhibit such effects, and the cell morphology remained similar to that of normal cells (Fig. 7B and F). Incubation with the L4 ligand caused slight, partial cell shrinkage (Fig. 7D).
Apoptosis detection revealed that approximately 50% of the cells incubated with the complexes 3 and 5 were in the early stage of apoptosis (Fig. 8C and G). In the case of the complex 4, a population of cells in the late stage of apoptosis was also present, confirming the stronger effect of this compound (Fig. 8E). Histograms for cultures treated with the ligands did not differ significantly from the histograms for control cells (Fig. 8B, D and F).
Cell cycle analysis showed that in cultures treated with Ag(I) complexes, the number of cells in the G1 and G2/M phases decreased, while the number of subG1 cells, which correspond to apoptotic cells, increased. This was most notable for complex 4, as shown in Fig. 9. No cell cycle arrest was observed, indicating that the complexes have a cytotoxic rather than a cytostatic effect.
Experimental part
The chemical compounds for the complexes and ligands syntheses were obtained from Sigma-Aldrich. The chemicals were used without additional purification.Synthesis of the ligands
The synthesis routes, structures and properties of the ligands, used for this work, were fully described in our previous publications28,29 and presented in ESI.†Synthesis of the complexes
A suitable amount of ligand was dissolved in methanol, with constant stirring at 90 °C. After the ligands have been dissolved, a metal salt (AgNO3 or CdCl2), dissolved in water, was added to each solution in an equimolar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The mixtures were left to stir at 90 °C for two hours. After that, they were cooled to room temperature and stored in the refrigerator to allow the formation of complexes. The resulting precipitates were filtered, washed with methanol, and dried at room temperature.
1. The mixtures were left to stir at 90 °C for two hours. After that, they were cooled to room temperature and stored in the refrigerator to allow the formation of complexes. The resulting precipitates were filtered, washed with methanol, and dried at room temperature.
X-ray crystallographic analysis of complex 4
Diffraction measurements were performed using a XtaLAB Synergy, Dualflex diffractometer, (Rigaku Oxford Diffraction, Poland–Japan–UK), with a Pilatus 300 K detector at low temperature (100.0(2) K) using MoKα radiation (0.71073 Å). Diffraction data were processed using CrysAlisPRO software (Rigaku Oxford Diffraction, CrysAlisPRO; Rigaku Oxford Diffraction Ltd: Yarnton, Oxfordshire, England). Solving and refinement of the crystal structure was performed with SHELX40,41 using full-matrix least-squares minimization on F2. All H atoms were geometrically optimized and allowed as riding atoms. ShelXle and Mercury softwares42,43 were used to visualize the molecular structure.CCDC 2403771† contains the supplementary crystallographic data for this paper. These data can be obtained free of charge from The Cambridge Crystallographic Data Centre via https://www.ccdc.cam.ac.uk/structures.
X-ray crystallographic analysis of complex 6
Single crystal of complex 6 was obtained by one-spot synthetic method at temperature 90 °C. X-ray diffraction data were collected at 119.98(13) K with a Rigaku Oxford Diffraction Synergy Custom system and a Rigaku HyPix-6000HE detector using Mo Kα radiation (λ = 0.710 73 Å) generated by a Rigaku MicroMax-007 HF DW. The data collection, reduction, and absorption corrections were performed using CrysAlisPro (version 1.171.43.112a).44 The crystal structure was solved and refined using SHELXT.41,45 Images of the structure were created using Diamond 5.0.2.46 The Crystallographic Information File for complex 6 has been deposited in the Cambridge Crystallographic Data Centre (CCDC deposition number 2388390†), from which it can be obtained free of charge using https://www.ccdc.cam.ac.uk/structures/.F-AAS
The content of solid metals were determined using F-AAS (Flame Atomic Absorption Spectroscopy) spectroscopy with a source of light and using air/acetylene flame. Analytical spectral lines were used for absorbance measurement where the limit of quantification was 0.04 mg L−1. Solid samples were decomposed using the Anton Paar Multiwave 3000 closed system instrument. Mineralization conditions: 240 °C under pressure 60 bar for 45 min.Elemental analysis
The contents of carbon, hydrogen and nitrogen were determined by a Vario micro company Elementar Analysensysteme GmbH.FTIR
FTIR spectra were recorded with an IRTracer-100 Shimadzu Spectrometer (4000–600 cm−1) with the accuracy of recording 1 cm−1 using KBr pellets.Thermogravimetric analysis
The thermal analyses were carried out with a Netzsch STA 449 F1 Jupiter thermoanalyzer (Netzsch-Geratebau GmbH, Selb, Germany) coupled with a Netzsch Aeolos Quadro QMS 403 mass spectrometer (Netzsch-Geratebau GmbH, Selb, Germany). The thermolysis of compounds in the air atmosphere was studied by TG-DTA techniques in the range of temperature 25–800 °C at a heating rate of 10 °C min−1; TGA-MS and DTA curves were recorded under the air atmosphere v = 20 mL min−1 using ceramic crucibles, and as a reference material ceramic crucible was used.Solution stability
The sample solutions (50 μL (1.0 × 10−3 M)) of the complexes were prepared in DMSO (Sigma Aldrich, ≥99.9%). The absorption spectra of the complexes were measured with increasing time (0 min, 30 min, 60 min, 2 h, 3 h, 24 h, 48 h) at room temperature. All measurements were performed on the Shimadzu UV-2401PC.Antibacterial study
24 hour bacterial cultures with a density of 1–2 × 108 CFU mL−1 were plated (0.1 mL) on Mueller–Hinton Agar (Merck, Germany). Subsequently, wells with a diameter of 10 mm were cut out from and filled with 100 μL of the tested compound at a concentration of 10–500 μg mL−1 (diluted in DMSO). After an 18-hour incubation at either 30 °C (for Bacillus ssp.) or 37 °C (for other tested microorganisms), the inhibition zones around the wells were measured, and the results were expressed in millimetres (Table S9†).
The microorganism cultures with a density of 1.5 × 108 CFU mL−1 (corresponding to a McFarland standard of 0.5) were plated on Mueller–Hinton Agar (Merck, Germany) following the procedures recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST).47 Wells measuring 10 mm in diameter were cut into the medium. The tested compounds were dissolved in a DMSO solution, which was recommended due to their chemical structures. Subsequently, 100 μL of prepared solutions containing the tested compounds at concentrations ranging from x μg mL−1 to x μg mL−1 were added to each well. The plates were incubated at 30 °C (for Bacillus subtilis and Bacillus cereus) or 37 °C (for other bacteria) for 18 hours. After incubation, the minimum inhibitory concentration (MIC value) was determined (the lowest concentration of the tested compounds that prevents bacterial growth) and expressed in μg mL−1. The experiment was conducted in three independent replicates. Ciprofloxacin (Sigma Aldrich, Saint Louis, MO, USA) was used as a reference antimicrobial agent against Gram-negative bacteria, while vancomycin (Sigma Aldrich, Saint Louis, MO, USA) was used as a reference antimicrobial agent against Gram-positive bacteria.
Anticancer study
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U mL−1), streptomycin (10 mg mL−1) and amphotericin B (250 μg mL−1). The cells were incubated at 37 °C, 5% CO2 atmosphere. The cells were maintained in the logarithmic growth phase by regular passage at 80% confluence.
000 U mL−1), streptomycin (10 mg mL−1) and amphotericin B (250 μg mL−1). The cells were incubated at 37 °C, 5% CO2 atmosphere. The cells were maintained in the logarithmic growth phase by regular passage at 80% confluence.Conclusion
In this study nine new Ag(I) and Cd(II) coordination compounds with the hydrozanomide-based ligands were synthesized and characterized as potential antibacterial and anticancer agents. The newly obtained complexes were characterized using numerous physicochemical and spectral methods of analysis. The crystal structure for complexes 4 and 6 was determined, where complex 4 is a coordination compound of cationic silver(I) with two ligands and one silver nitrate salt, forming a distorted tetrahedral arrangement for Ag(I). In the case of complex 6, the metal atoms link molecules of the ligand in the crystal structure into an one-dimensional chain, and the asymmetric cell contains one molecule of the ligand coordinated by 2 Ag atoms in two special positions with occupation ½ and one nitrate anion. In the case of the other coordination compounds, based on the difference in the FTIR spectra of the complexes and free ligands, the coordination between metal–organic ligand takes place. The thermal decomposition patterns of the Ag(I) and Cd(II) coordination compounds exhibit varying stability and decomposition behaviours. While all compounds are stable at room temperature, their decomposition occurs in multiple stages, involving the loss of water, organic ligands, and nitrate anions, ultimately forming metallic Ag or CdO. Complexes 4 and 6 exhibit the highest stability among the silver complexes, while complexes 7, 8, and 9 show exceptional thermal stability, with decomposition involving organic ligand loss and CdO formation, followed by potential CdCl2 volatilization at high temperatures. Besides that, a study has been carried out to examine the stability of complexes upon 48 h in a DMSO, where the most stable compounds were complexes 1 and 3.The results of the antibacterial activity of complexes were compared to the activity of the parent ligands, which was measured in the previous study. All tested compounds demonstrated high or moderate activity against the studied microorganisms, specifically against Staphylococcus aureus and Escherichia coli. Complexes 1 and 4 showed the most promising activity, where MIC values were 10 μg mL−1 or <10 μg mL−1 against some of the Gram-negative and Gram-positive bacteria. Based on the achieved results, it was found that despite the overall higher toxicity of cadmium compared to silver, the synthesised silver complexes showed more potential as antibacterial agents.
The IC50 results demonstrated, that certain metal complexes exhibited significant cytotoxic activity. Complexes 1, 6, 8 showed high toxicity values towards the tested glioblastoma cells. At the same time, they also showed high toxicity to normal BJ cells, which makes it difficult to consider them as potential drug candidates. The most promising compounds for further research are those that exhibit selective toxicity towards cancer cells without harming normal cells, as has been shown by complexes 3, 4 and 5. Under the toxic influence of these complexes, the cells shrank, which was confirmed by microscopic observations. Based on the cell apoptosis assay results, it can clearly be observed that in the case of complex 4, a population of cells at a late stage of apoptosis was present, indicating a stronger effect of this compound. These findings confirm, that complex 4 can be considered as a potential selective anticancer metallodrug candidate and can be subjected to further biological studies.
Data availability
Crystallographic data for complex 4 has been deposited at the https://www.ccdc.cam.ac.uk/structures with the deposition number CCDC 2403771; crystallographic data for complex 6 has been deposited at the https://www.ccdc.cam.ac.uk/structures with the deposition number CCDC 2388390; crystallographic data for L6 has been deposited at the https://www.ccdc.cam.ac.uk/structures under CCDC 2241574 and can be obtained from DOI: 10.5517/ccdc.csd.cc2f7jv2 (https://dx.doi.org/10.5517/ccdc.csd.cc2f7jv2); the data supporting this article have been included as part of the ESI.†Author contributions
Conceptualization, A. C. (Alina Climova), A. C. (Agnieszka Czylkowska).; methodology, A. C. (Alina Climova), E. P., M. S., K. G., A. K.-P., M. I., E. K.-B., S. S., J. M., A. C. (Agnieszka Czylkowska); formal analysis, A. C. (Alina Climova), E. P., M. S., K. G., A. K.-P., M. I., J. M., A. C. (Agnieszka Czylkowska); investigation A. C. (Alina Climova), E. P., M. S., K. G., M. I., J. M.; data curation, A. C. (Alina Climova), A. C. (Agnieszka Czylkowska); writing – original draft preparation, A. C. (Alina Climova), E. P., M. S., A. K.-P., M. I., J. M., A. C. (Agnieszka Czylkowska); writing – review and editing, A. C. (Alina Climova), E. P., A. C. (Agnieszka Czylkowska); visualization, A. C. (Alina Climova); supervision, A. C. (Agnieszka Czylkowska). All authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This article has been completed while the first two authors were the Doctoral Candidates in the Interdisciplinary Doctoral School at the Lodz University of Technology, Poland (A. C. (Alina Climova), E. P.).References
- V. K. Singh, V. K. Singh, A. Mishra, A. A. Singh, G. Prasad and A. K. Singh, Polyhedron, 2023, 241, 116485 CrossRef.
- C. Zhang, C. Xu, X. Gao and Q. Yao, Theranostics, 2022, 12, 2115–2132 Search PubMed.
- S. Dasari and P. Bernard Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 Search PubMed.
- P. B. Tchounwou, S. Dasari, F. K. Noubissi, P. Ray and S. Kumar, J. Exp. Pharmacol., 2021, 13, 303–328 Search PubMed.
- K. L. Haas and K. J. Franz, Chem. Rev., 2009, 109, 4921–4960 CrossRef CAS PubMed.
- D. Buac, S. Schmitt, G. Ventro, F. Rani Kona and Q. Ping Dou, Mini-Rev. Med. Chem., 2012, 12, 1193–1201 Search PubMed.
- D. van der Westhuizen, D. I. Bezuidenhout and O. Q. Munro, Dalton Trans., 2021, 50, 17413–17437 RSC.
- C. Santini, M. Pellei, V. Gandin, M. Porchia, F. Tisato and C. Marzano, Chem. Rev., 2014, 114, 815–862 CrossRef CAS PubMed.
- U. Ndagi, N. Mhlongo and M. Soliman, Drug Des., Dev. Ther., 2017, 11, 599–616 CrossRef CAS PubMed.
- U. Ndagi, N. Mhlongo and M. Soliman, Drug Des., Dev. Ther., 2017, 11, 599–616 CrossRef CAS PubMed.
- M. M. González-Ballesteros, C. Mejía and L. Ruiz-Azuara, FEBS Open Bio, 2022, 12, 880–899 CrossRef PubMed.
- E. Z. Jahromi, A. Divsalar, A. A. Saboury, S. Khaleghizadeh, H. Mansouri-Torshizi and I. Kostova, J. Iran. Chem. Soc., 2016, 13, 967–989 Search PubMed.
- A. B. G. Lansdown, J. Wound Care, 2002, 11, 125–130 CrossRef CAS PubMed.
- G. Achar, V. C. Ramya, K. Upendranath and S. Budagumpi, Appl. Organomet. Chem., 2017, 31, e3770 CrossRef.
- G. Achar, C. R. Shahini, S. A. Patil, J. G. Małecki, S.-H. Pan, A. Lan, X.-R. Chen and S. Budagumpi, J. Inorg. Biochem., 2018, 183, 43–57 Search PubMed.
- H. H. Lara, N. V. Ayala-Nuñez, L. Ixtepan-Turrent and C. Rodriguez-Padilla, J. Nanobiotechnol., 2010, 8, 1 Search PubMed.
- S. K. Raju, A. Karunakaran, S. Kumar, P. Sekar, M. Murugesan and M. Karthikeyan, Ger. J. Pharm. Biomater., 2022, 1, 06–28 Search PubMed.
- I. Majumder, P. Chakraborty, S. Dasgupta, C. Massera, D. Escudero and D. Das, Inorg. Chem., 2017, 56, 12893–12901 CrossRef CAS PubMed.
- A. Halder, S. Mazumdar, A. Das, P. Karmakar and D. Ghoshal, ChemistrySelect, 2017, 2, 11832–11839 Search PubMed.
- Y. M. Zhang, L. Y. Wang, B. L. Li, J. H. Yang and Y. Zhang, J. Mol. Struct., 2008, 875, 527–539 CrossRef CAS.
- S. A. Musavi, M. Montazerozohori, A. Masoudiasl, R. Naghiha, S. Joohari and A. Assoud, J. Mol. Struct., 2017, 1145, 65–75 CrossRef CAS.
- T. Karmakar, Y. Kuang, N. Neamati and J. B. Baruah, Polyhedron, 2013, 54, 285–293 CrossRef CAS.
- X. Zhou, Y. Koizumi, M. Zhang, M. Natsui, S. Koyota, M. Yamada, Y. Kondo, F. Hamada and T. Sugiyama, Cancer Sci., 2015, 106, 635–641 CrossRef CAS PubMed.
- F. B. Biswas, T. G. Roy, Md. A. Rahman and T. Bin Emran, Asian Pac. J. Trop. Med., 2014, 7, S534–S539 CrossRef CAS PubMed.
- M. S. Aljahdali and A. A. El-Sherif, Bioinorg. Chem. Appl., 2020, 2020, 1–17 CrossRef.
- A. Cuypers, M. Plusquin, T. Remans, M. Jozefczak, E. Keunen, H. Gielen, K. Opdenakker, A. R. Nair, E. Munters, T. J. Artois, T. Nawrot, J. Vangronsveld and K. Smeets, BioMetals, 2010, 23, 927–940 CrossRef CAS PubMed.
- A. Pal, S. Bhattacharjee, J. Saha, M. Sarkar and P. Mandal, Crit. Rev. Microbiol., 2022, 48, 327–355 CrossRef CAS PubMed.
- A. Climova, E. Pivovarova, M. Szczesio, K. Gobis, D. Ziembicka, A. Korga-Plewko, J. Kubik, M. Iwan, M. Antos-Bielska, M. Krzyżowska and A. Czylkowska, J. Inorg. Biochem., 2023, 240, 112108 CrossRef CAS PubMed.
- A. Climova, E. Pivovarova, M. Szczesio, A. Olczak, J. Wojciechowski, K. Gobis, D. Ziembicka, A. Korga-Plewko, J. Kubik, M. Iwan, I. Korona-Głowniak and A. Czylkowska, Polyhedron, 2023, 245, 116634 CrossRef CAS.
- D. Kwiatek, M. Kubicki, J. Belter, R. Jastrząb, H. Wiśniewska, S. Lis and Z. Hnatejko, Polyhedron, 2017, 33, 187–194 CrossRef.
- D. Sutradhar, H. Chowdhury, S. Banerjee, N. C. Saha and B. K. Ghosh, Inorg. Chim. Acta, 2019, 485, 86–97 CrossRef CAS.
- H. M. Abd El-Lateef and M. M. Khalaf, Appl. Organomet. Chem., 2025, 39, 78–97 Search PubMed.
- P. Mech-Warda, A. Giełdoń, A. Kawiak, N. Maciejewska, M. Olszewski, M. Makowski and A. Chylewska, Molecules, 2022, 27, 3704 CrossRef CAS PubMed.
- K. Gutmańska, P. Szweda, M. Daszkiewicz, T. Mazur, K. Szaciłowski, A. Ciborska and A. Dołęga, Appl. Organomet. Chem., 2023, 37, e7207 CrossRef.
- B. Cláudia, E. Almeida Neves and É. TG Cavalheiro, Thermochim. Acta, 2001, 370, 49–55 CrossRef.
- C.-G. Elinder, Cadmium and health. Cadmium: Uses, Occurrence, and Intake, 1986 Search PubMed.
- Z. Yolcu, Erzincan Üniv. Bilimleri Enst. Derg., 2020, 13, 25–32 Search PubMed.
- A. Di Santo, G. A. Echeverría, O. E. Piro, H. Pérez, A. Ben Altabef and D. M. Gil, J. Mol. Struct., 2017, 1134, 492–503 CrossRef CAS.
- K. V. Balakin, Y. A. Ivanenkov, A. V. Skorenko, Y. V. Nikolsky, N. P. Savchuk and A. A. Ivashchenko, SLAS Discovery, 2004, 9, 22–31 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. A, 2008, 64, 112–122 CrossRef CAS PubMed.
- G. M. Sheldrick, Acta Crystallogr., A: Found Adv., 2015, 71, 3–8 CrossRef PubMed.
- C. F. Macrae, I. Sovago, S. J. Cottrell, P. T. A. Galek, P. McCabe, E. Pidcock, M. Platings, G. P. Shields, J. S. Stevens, M. Towler and P. A. Wood, J. Appl. Crystallogr., 2020, 53, 226–235 CrossRef CAS PubMed.
- C. B. Hübschle, G. M. Sheldrick and B. Dittrich, J. Appl. Crystallogr., 2011, 44, 1281–1284 CrossRef PubMed.
- CAP: Rigaku Oxford Diffraction, CrysAlisPro Software System, Rigaku Corporation, Wrocław, Poland, 2024 Search PubMed.
- G. M. Sheldrick, Acta Crystallogr., Sect. C: Struct. Chem., 2015, 71, 3–8 Search PubMed.
- Diamond: Diamond – Crystal and Molecular Structure Visualization, Crystal Impact GbR, Bonn, Germany, 2024 Search PubMed.
- C. G. Giske, J. Turnidge, R. Cantón and G. Kahlmeter, J. Clin. Microbiol., 2022, 60, e00276 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. CCDC 2388390 and 2403771. For ESI and crystallographic data in CIF or other electronic format see DOI: https://doi.org/10.1039/d5ra02001h |
| This journal is © The Royal Society of Chemistry 2025 |