Open Access Article
Open Access ArticleIntegrating metagenomics and untargeted metabolomics to analyze the relationship between microbial dynamics and non-volatile metabolomic profiles in plant-derived microbial fuel cells (MFCs)†
Kun Tanga,
Suxuan Lia,
Yiye Luoa,
Wenning Fengb,
Zhichao Zhanga,
Liusheng Wangb,
Hui Zhao*b,
Xiaoyu Chena,
Xiaohu Lic and
Zhiyong Wu*a
aFlavors and Fragrance Engineering & Technology Research Center of Henan Province, College of Tobacco Science, Henan Agricultural University, Zhengzhou, Henan 450046, P. R. China. E-mail: zhiyongwu@henau.edu.cn; Fax: +86-371-68555219; Tel: +86-151-37111794
bTechnology Center of China Tobacco Hebei Industrial Co., Ltd, Shijiazhuang, Hebei 050051, P. R. China. E-mail: zhaohui1014@126.com
cSchool of Materials Science & Engineering, Beihang University, Beijing 102206, P. R. China
First published on 10th June 2025
Abstract
Microbial Fuel Cells (MFC) are an emerging biomass energy technology that harnesses the power of electroactive bacteria living on a bacterial biofilm to convert biomass energy within waste materials into usable electricity. A pivotal aspect of MFC research involves understanding the behavior and underlying mechanisms of electroactive bacteria during extracellular electron transfer to the anode, which plays a crucial role in energy conversion. In this paper, four MFCs were operated at external resistances of 500 and 1000 ohms, and the changes in the biofilm's electroactive bacterial composition due to altered external resistances were indicated by the voltage and power differences. After stable power generation, total DNA was extracted from the biofilm for sequencing, and metabolites were tested. The expression trends of genes and the differences in final metabolites from the whole period indicate that electron transfer gene families are associated with Shewanella, Pseudomonas, Trichococcus, and Enterococcus, while tyrosine and purine metabolism showed significant differences in effective metabolite accumulation among communities with varying energy output efficiency. Omics techniques revealed, to some extent, the coordination mechanisms and bacterial interactions within biofilms during microbial community succession.
1. Introduction
Microbial Fuel Cells (MFCs) are an energy technology that harnesses renewable biomass to generate power, recovering energy from biomass waste and industrial wastewater using living microorganisms as electrode catalysts. Microorganisms use the anode as an electron acceptor to oxidize and degrade organic substrates, hence, MFCs are promising for energy-efficient and cost-saving waste treatment processes. The phenomenon of microbial electrochemistry was first observed between 1908 and 1911, when Potter demonstrated voltage generation during microbial metabolism using organic substrates as electron donors.1 In 1962, Davis and Yarbrough conceptualized converting this phenomenon into a biological fuel cell.2 Since then, MFCs technology has been actively developing. MFCs may become a future alternative “green” energy technology because they produce sustainable electricity from biodegradable organic compounds through microbial metabolism. In MFCs, microorganisms degrade organic compounds through anaerobic respiration and donate electrons to an external circuit, coupling the removal of organic matter with electricity generation. These systems have been proven effective in laboratory-scale settings, and research on scaling up designs for industrial and urban wastewater treatment is ongoing.3 Once integrated into waste treatment processes, MFCs technology is expected to have advantages over aerobic treatment systems due to lower biomass yield and reduced energy consumption. Furthermore, unlike most anaerobic digestion tanks that produce biogas, MFCs bioreactors can operate at ambient temperature and do not require the handling of final products produced within the cells.The expansion and development of MFCs have primarily focused on optimizing reactor design and identifying cost-effective materials, aimed at reducing costs and improving efficiency to meet the requirements for large-scale treatment.4 However, it is necessary to have a deeper understanding of the microbial composition and to maximize the range and rate of degradation of complex substrates affected by biodegradation to achieve optimal performance, which is highly dependent on the metabolic capabilities of the resident microorganisms. Therefore, it is essential to identify the active populations within mixed microbial communities to design microbial inoculants for large-scale MFCs.5 The most effective communities were expected to contain a large number of genes involved in biodegradation and extracellular electron transfer.
In the MFCs with natural microbial communities as anodes, the detected bacterial populations exhibit significant diversity, including members of the Proteobacteria, Firmicutes, and Bacteroides phyla. These microbial populations were all active in terms of the electricity generation capability of the MFCs. However, the formation of specific symbiotic combinations with characteristic power generation properties has still not been observed. Although studies have reported on the qualitative temporal changes in the structure of microbial communities in MFCs, the power output of mixed microbial community MFCs has not shown a clear association with any specific species richness. Rabaey et al.6 obtained cultures from batch-fed glucose mode MFCs that generated power using autocrine mediators such as pyocyanin produced by Pseudomonas aeruginosa. In this context, the increase in output rate of the mixed community MFC over time is considered to be due to the increase in abundance of these bacteria producing mediators, as the same as the enhancement of their self-mediated capabilities.
To date, these assumptions cannot be verified, as Aelterman et al.7 reported that the maximum power density of a continuous-flow MFC fed with acetate increased threefold, which was attributed to the shift from a community dominated by Proteobacteria (with a small proportion of Firmicutes and Actinobacteria) to one dominated by Brevibacillus sp., a member of the Gram-negative bacteria. In a subsequent study, it was demonstrated that metabolic products produced by Pseudomonas were capable of being produced by Brevibacillus.8 Therefore, the rate output of the mixed community MFC is once again considered to be due to the derivative effects of the community and metabolites. Previous analyses of MFCs have primarily focused on wastewater treatment MFCs. At present, integrated meta-genomics and metabolomics studies of mixed-culture based microbial fuel cells were relatively rare.9,10 Herein, we report on the metagenome and metabolome analysis of anodic communities from four MFCs based on plants as the degradation source and stock culture, intended to serve as part of a modular MFC system for demonstrating degradation characterization. Examination of the biochemical repair potential of the communities and their inocula revealed that despite the MFC bioreactors differing in design, inocula, and feedstock composition, they developed a shared core community. The MFC communities revealed changes in population structure that occurred during the operation of the bioreactors under real-world conditions, based on their respective inoculation scenarios.
2. Materials and methods
2.1 Microorganisms and culture media
Pandanus amaryllifolius Roxb. (obtained from Hainan Wanquan River Tropical Botanical Agriculture Co., Ltd, China) was used as the inoculum source for MFCs. The composition of the MFC medium (per liter): NH4Cl, 0.25 g; CaCl2: 0.25 g; MgSO4: 0.50 g; K2HPO4: 1.00 g; KH2PO4: 1.00 g; NaHCO3: 8 g; NaCl: 2.00 g; deionized water: 1000.00 mL; peptone: 1.00 g; yeast extract: 1.00 g; beef extract: 1.00 g; glucose: 1 g. All culture medium formulations were sterilized under high pressure at 115 °C for 30 minutes, except for vitamins and minerals which were filtered sterilized through a 0.2 μm pore size membrane (Nalgene SFCA membrane, USA).2.2 MFC operation
The leaves of Pandanus amaryllifolius Roxb. were grounded into powder and filtered through a 0.6 mm sieve (Endecotts Co., Ltd, UK) to remove large particles, and then stored at 4 °C. The powdered particles were then fed into a liquid medium at a 10% volume ratio to initiate four replicate MFCs. During the initial enrichment period (approximately a month), the MFCs were operating in batch mode. During this period, the anodic slurry was repeatedly replaced (3–4 times) by mixing with fresh N2-purging culture medium, and then the entire volume of the anodic slurry was replaced with fresh culture medium to continue operating the MFCs in batch mode until a reproducible voltage generation was observed. The MFCs was operating at room temperature (21–22 °C) and samples were taken for chemical and microbial community analysis.2.3 MFC configuration
Four sets of dual-chamber MFCs were constructed, each chamber with an effective working volume of 150 mL. The MFCs were acclimated simultaneously with external resistances of 500 and 1000 Ω for 80 days. The dual-chamber MFCs consist of an anodic chamber and a cover plate made of plexiglass, with platinum wire used as the external circuit wire between the cathode and the anode, with additional resistances (K1, K3) of 500 Ω and (K2, K4) of 1000 Ω. The anodic electrode was a carbon fiber brush (PRF Composite Materials, UK) placed inside the anodic chamber. The cathodic electrode was a square piece of graphite, and the cation exchange membrane (20 cm2, DuPont, USA) was pretreated by boiling continuously for 1 hour in a solution of 6% w/v H2O2, H2O, 0.5 M H2SO4, and H2O, and then stored in deionized water in the dark before assembly between the anodic and cathodic chambers. There were six small holes above both the anode and cathode chambers to facilitate anaerobic operations, with argon gas aeration.2.4 MFC electrochemical measurement
The output voltage on the external resistor was recorded at five-minute intervals by a battery monitoring system (Xinwei, China) and subsequently processed by the data acquisition software. The ohmic resistance of the MFC was measured using electrochemical impedance spectroscopy, polarization curves and power density curves were obtained by varying the external resistance through a variable resistance box (ZX21, Dongmao, China). After adjusting the external resistance (1500–10 Ω) and stabilizing it for 20 minutes during the polarization process, the output voltage was recorded after stabilizing it for 15 minutes with a digital multimeter (UNI-T UT33D). The current and power density were calculated according to Ohm's law.2.5 Untargeted metabolomics analysis preparation and extraction
The bacteria pellets (about 107 bacteria) were taken, mixed with 1000 μL of extraction solution (MeOH![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ACN
ACN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O, 2
H2O, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v)), the extraction solution contained deuterated internal standards, and the mixed solution were vortexed for 30 s. Add 2 homogenization beads and homogenize for 4 min (35 Hz), then transferred to an ice-water bath to sonicate for 5 min (repeat 3 times). The samples were then allowed to thaw at room temperature and vortexed for 30 s. This freeze–thaw cycle was repeated three times, then the samples were sonicated for 10 min in 4 °C water bath, and incubated for 1 h at 40 °C to precipitate proteins. The samples were centrifuged at 12
1 (v/v)), the extraction solution contained deuterated internal standards, and the mixed solution were vortexed for 30 s. Add 2 homogenization beads and homogenize for 4 min (35 Hz), then transferred to an ice-water bath to sonicate for 5 min (repeat 3 times). The samples were then allowed to thaw at room temperature and vortexed for 30 s. This freeze–thaw cycle was repeated three times, then the samples were sonicated for 10 min in 4 °C water bath, and incubated for 1 h at 40 °C to precipitate proteins. The samples were centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm (RCF = 13
000 rpm (RCF = 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 (×g), R = 8.6 cm) for 15 min at 4 °C. The supernatant was transferred to a fresh glass vial for analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatant of samples.
800 (×g), R = 8.6 cm) for 15 min at 4 °C. The supernatant was transferred to a fresh glass vial for analysis. The quality control (QC) sample was prepared by mixing an equal aliquot of the supernatant of samples.
2.6 UPLC conditions
For polar metabolites, LC-MS/MS analyses were performed using an UHPLC system (Vanquish, Thermo Fisher Scientific) with a Waters ACQUITY UPLC BEH Amide (2.1 mm × 50 mm, 1.7 μm) coupled with Orbitrap Exploris 120 mass spectrometer (Orbitrap MS, Thermo). The mobile phase consisted of 25 mmol L−1 ammonium acetate and 25 ammonia hydroxide in water (pH = 9.75) (A) and acetonitrile (B). The auto-sampler temperature was 4 °C, and the injection volume was 2 μL. The Orbitrap Exploris 120 mass spectrometer was used with its ability to acquire MS/MS spectra on information-dependent acquisition (IDA) mode in the control of the acquisition software (Xcalibur, Thermo). In this mode, the acquisition software continuously evaluated the full-scan MS spectrum. The ESl source conditions were set as following: sheath gas flow rate as 50Arb, Aux gas flow rate as 15 Arb, capillary temperature 320 °C, full MS resolution as 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, MS/MS resolution as 15
000, MS/MS resolution as 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, collision energy: SNCE 20/30/40, spray voltage as 3.8 kV (positive) or −3.4 kV (negative), respectively.
000, collision energy: SNCE 20/30/40, spray voltage as 3.8 kV (positive) or −3.4 kV (negative), respectively.
2.7 Metagenomics analysis
Following the method of Cai et al.,11 metagenomic analysis was conducted on the entire microbial community DNA of the electrochemically active biofilms (ECABs) to further study the impact of charge transfer resistance (CTR) on the metabolic pathways at the molecular level. Two ECAB samples (0.5 × 1 cm2) were collected from each reactor using aseptic scissors. According to the manufacturer's instruction, 0.2 g of sample was used to extract total genomic DNA with the he E.Z.N.A.® soil DNA Kit (Omega Bio-tek, Norcross, GA, USA). Concentration and purity of extracted DNA was determined with SynergyHTX and NanoDrop2000, respectively. DNA quality was checked on 1% agarose gel. DNA extract was fragmented to an average size of about 350 bp using Covaris M220 (Gene Company Limited, China) for paired-end library construction. Paired-end library was constructed using NEXTFLEX Rapid DNA-Seq (Bioo Scientific, Austin, TX, USA). Paired-end sequencing was performed on Illumina NovaSeq X Plus (Illumina Inc., San Diego, CA, USA) at Majorbio Bio-Pharm Technology Co., Ltd (Shanghai, China).2.8 Data processing and statistical analysis
The raw data of metagenomics were maintained using Microsoft Excel 2019, and figures were drawn using OriginPro (version 2022, OriginLab, USA), SIMCA (version 14.1, Umetrics, Sweden), and TBtools (version 1.1, China). The raw data of untargeted metabolomics were converted to the mzXML format using ProteoWizard and processed with an in-house program, which was developed using R package and XCMS, for peak detection, extraction, alignment, and integration. The R package and the BiotreeDB (version 3.0) were applied in metabolite identification.3. Results
3.1 Electronic performance
In typical microbial metabolism classification, whether the energy obtained by microorganisms came from photosynthesis or the chemical reactions of substances, oxidation-reduction was a core process.12 In the process of microorganisms obtaining energy, it can be divided into three forms according to the different final electron acceptors: fermentation, anaerobic respiration, and aerobic respiration.13 Based on this theory, we explain the electric energy function in the process of microorganisms attaching to the anode of an MFC and carrying out oxidation-reduction reactions by the electronic performance displayed during the electron transfer between microorganisms and extracellular electron acceptors or donors.As expected, the fresh culture medium act with a light green color, while after electricity production transformation, the entire anode turned yellow-green and eventually evolved into a dark green. During the microbial enrichment and domestication process in the fuel cell, the initial pH of the anode culture medium was 6.46, then began to decline to 6.28 after 15 days, stabilized at 6.20 after 30 days, and reached 6.24 after 45 days. To better study the dynamic changes of the MFCs, the voltage performance of the fuel cell at days 0, 15, 30, and 45 was analyzed. After approximately four weeks (3–4 cycles) of biological anode enrichment, a stable output voltage was obtained from the MFCs with external resistances of 500 Ω and 1000 Ω. After the inoculation source entered the two-chamber anode MFCs, the MFCs experienced a lag phase (stage I). Then, the voltage increased (Fig. 1). After four batches of cyclic periods, the voltage output began stable and reached 0.4 ± 0.1 V when the battery was operated under conditions of 500 Ω and 1000 Ω external resistance, and it can be found that the power generation time was about 100 hours. When the output voltage was stable, polarization curves were obtained by changing the external resistance between 100 and 1500 Ω (Fig. 2). The results demonstrated that the open-circuit voltages were 0.48 V (K2), 0.53 V (K3), and 0.65 V (K4) for the respective MFC configurations. The maximum power densities were 69.34 mW m−2 (K1), 164.33 mW m−2 (K2), 82.16 mW m−2 (K3), 199.44 mW m−2 (K4), respectively, and the voltage variation tended to decrease linearly, which was a typical ohmic polarization stage. The MFC mass transfer resistance will finally become the primary resistance to electron transfer, resulting in a significant decline in power density. Previous studies showed that external resistance may affect the thickness of the biofilm.14 The reduction of external resistance led to a greater difference between the anodic potential and the redox potential of the electron acceptor, resulting in the transfer of energy from microbial growth to extracellular polymer formation,15 thereby extending the domestication period. K1–K4 showed lower maximum voltage values after the initial two cycles, except for K2. The secondary metabolism after microbial activity and electron transfer often leads to changes in metabolites, which were considered to be the cause of changes in physicochemical parameters.16,17 The results of the assembled MFCs indicated that the microbial community culture of the MFCs can be divided into four stages (0 days, 15 days, 30 days, 45 days). Therefore, the structure and function of the microbial communities during these periods, as well as the succession of metabolites and functional genes were further explored to reveal the relationship between the microbial community and the electrochemical function of the MFCs.
3.2 Microbial anodic community dynamics in MFC
The metagenomic sequences were compared with the NCBI NR database to determine the dominant assumed taxa at each time point, and the initial community was classified by abundance, as shown in Table S1.† The community composition at the phyla and genus levels between different time points were shown in Fig. 3 and 4, respectively. Throughout the dynamic changes of the MFC microbial community, Proteobacteria, Firmicutes, and Bacteroidetes were the dominant phyla in the dynamic changes of the entire samples. Proteobacteria, Firmicutes, and Bacteroidetes were often reported to be found in the electrochemical functional microbiota of two-chamber MFCs.17,18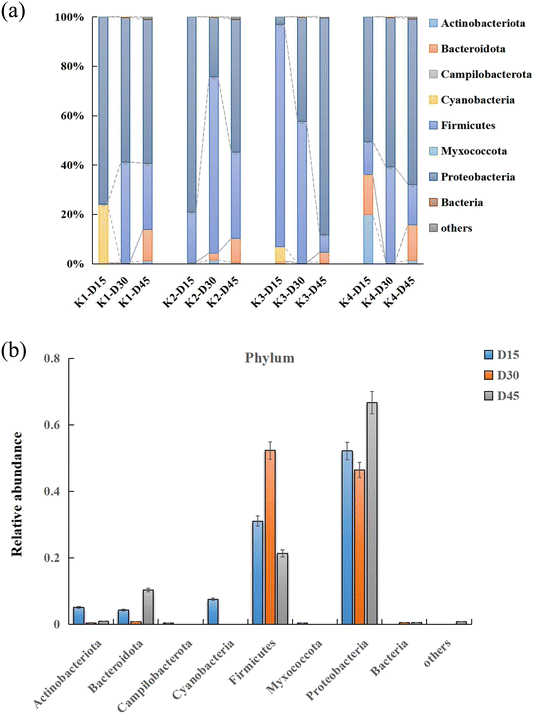 | ||
| Fig. 3 (a) The annotation diagrams of the top ten abundant MFC phylums in each treatment. (b) The overall phylums of MFC distinguished by different trends over time. | ||
 | ||
| Fig. 4 (a) The annotation diagrams of the top ten abundant MFC genuses in each treatment. (b) The overall genuses of MFC distinguished by different trends over time. | ||
During the initial stage of culture (0–15 days, stage I), the experimental treatment primarily involved strains of Escherichia coli, Enterococcus, and Acinetobacter. Escherichia coli, a member of the Enterobacteriaceae family and a facultative anaerobic fermentative Gram-negative bacterium, has been reported to generate electricity in microbial fuel cells (MFCs) by using oxidizable metabolic products such as H2 and H2S as redox mediators to transfer electrons. However, the limited electron transfer capacities of these metabolic products often resulted in lower electrical power output of the MFC.19 Acinetobacter, a non-fermentative Gram-negative bacterium also belonging to the Proteobacteria phylum, and Enterococcus, a facultative Gram-positive bacterium belonging to the Firmicutes phylum, coexist with the electroactive microorganisms in the MFCs system. These microorganisms metabolize organic matter and release electrons to the anode, which is the principle of MFCs. Some studies have shown that feeding Enterococcus can reduce the relative abundance of air-conditioned pathogenic bacteria Acinetobacter and Escherichia coli,20 which was consistent with the characteristics of the microbial community in the early stage of the electrochemical “domestication” process of MFCs. During the metabolism of Escherichia coli, succinate was an intermediate product in the tricarboxylic acid (TCA) cycle, and its accumulation was relatively low. Therefore, most studies adopted a two-stage fermentation model involving aerobic cultivation and anaerobic acid production. This means that the aerobic environment increases the concentration of bacteria, while the anaerobic stage produces succinate.21 Scientists have enabled these bacteria to generate electricity through the extracellular electron transfer (EET) process. During the dynamic equilibrium of the substrate biological oxidation process in the early stage, the electrons produced by microorganisms were difficult to pass through the cell membrane. However, under artificial intervention, the addition of electron mediators can facilitate the oxidation metabolism of some bacterial genera. The goal of this stage was to decompose and metabolize substrates in the entire MFCs, breaking down macromolecular nutrients such as proteins, polysaccharides, and lipids into small molecules like amino acids, monosaccharides, and fatty acids.
During the intermediate stage of an MFC (15–30 days, stage I), genera such as Citrobacter and Trichococcus began to thrive rapidly, accounting for 4.03–67.40% of the total MFC microbial population. Their relative abundance gradually declined in the later stage. It is speculated that Citrobacter transferred electrons through its self-secreted redox-active compounds during power generation,22,23 while Trichococcus has been confirmed to produce lactic acid, acetate, and ethanol from glucose. However, it is still unclear whether this Trichococcus genus possesses EET capabilities. Interestingly, previous studies speculated that Trichococcus might serve as a potential electric-syntrophy partner to other genuses.24
Similar to the microbiome characteristics of other reported MFCs, in the final stage of MFC domestication (30–45 days, stage II), as microbial and metabolite enrichment changed, we found that the Proteobacteria and Firmicutes gradually accounted for 90–100% of the entire bacterial community composition. This transition was also commonly observed in macroalgal biomass25 and domestic wastewater26 during the microbial community structure development process of MFCs. Previous studies demonstrated that the Proteobacteria and Firmicutes were defined as the EAB in MFCs reactors.27 The most commonly observed prokaryotic phylum and specie in each sample were Shewanella and Pseudomonas under Proteobacteria, and Trichococcus under Firmicutes (Fig. 4a and b). Shewanella was discovered in 1988 and can exist in various sedimentary water bodies, or in lactic acid environments, Fe(III) and Mn(IV) reduction environments, and sulfate environments. It has been confirmed to have a variety of extracellular electron transfer forms and possesses electrochemical functional characteristics. Shewanella has a complete set of extracellular electron transfer chains,28,29 which were mediated by direct electron transfer (DET) between cells and electrodes through the electron transfer channels formed by outer membrane cytochromes (Mtr and CymA systems), the other was mediated electron transfer (MET), which was enabled via redox-active molecules such as flavins secreted by Shewanella spp. Similarly, Pseudomonas species have been identified as utilizing MET signaling for transmission, which secreted different electronic mediators like phenazines as molecule-mediated transfer,28,30 and it seemed that the electrochemical activity was mainly acted upon and affected at different times in this experiment (Fig. 4a).
3.3 Gene classification analysis of MFC microbial activity
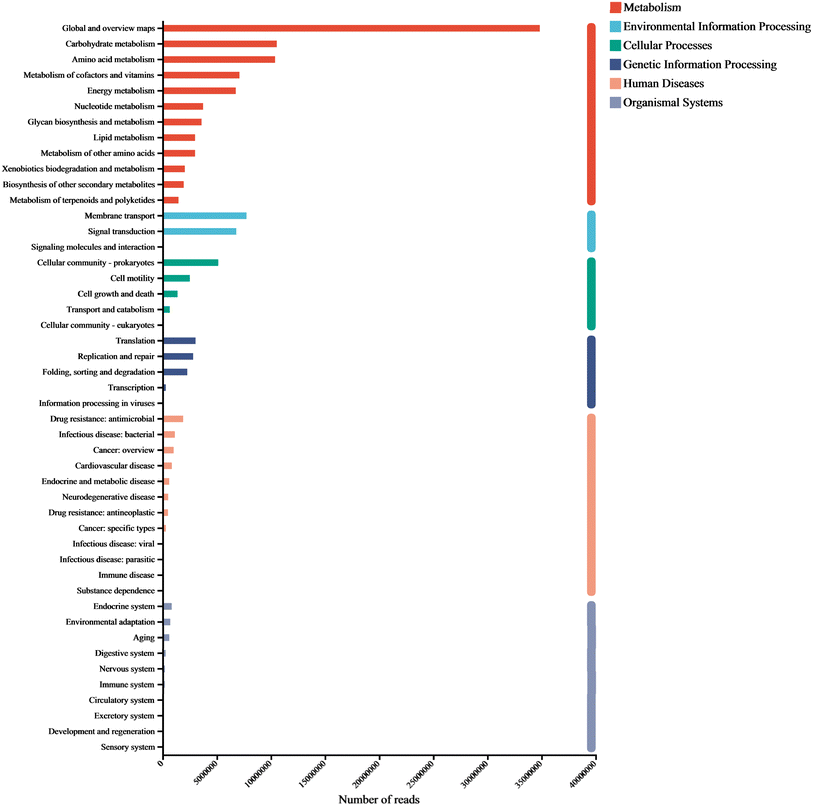
To explore the metabolic potential of the microbiome, gene annotation and classification were performed using the KEGG database. Fig. 5 illustrated the biochemical pathways of functional genes, including metabolism (67.66%), environmental information processing (11.14%), cellular processes (7.41%), genetic information processing (6.45%), human diseases (5.19%), and organismal systems (2.15%). As each category can be further divided into subcategories, a total of 47 subtypes were presented in the figure, with Global and overview maps, carbohydrate metabolism, and amino acid metabolism dominating (relative abundance >7%), followed by metabolism of cofactors and vitamins, energy metabolism, membrane transport, signal transduction, and cellular community -prokaryotes (relative abundance >4%).Carbohydrates is a major component of cell structures and energy supply, playing a key role in regulating microbial activity.8 We found that carbohydrate metabolism accounted for the largest proportion of microbial functional genes. In addition to carbohydrate metabolism, amino acid metabolism, membrane transport, and signal transduction are often used as signature pathways in MFC metabolism groups.31 As small molecular organic compounds, vitamins typically participate in metabolism in the form of cofactors. Moreover, cofactors provide redox carriers for biosynthetic and catabolic reactions and are crucial for the intracellular transfer of microbial energy, therefore, the regulation of cofactor abundance may be beneficial for the efficient production of target metabolites.32
The metabolism of amino acids is related to higher gene abundance in energy metabolism, nucleotide metabolism, and other biosynthetic pathways such as amino sugars. These genes affect the synthesis of polysaccharides (PS) and further impact the synthesis of extracellular polymeric substances (EPS).33 Further annotation analysis through the KEGG database indicates that these metagenomic genes are associated with many secondary metabolism pathways (Fig. 6 & 7). To analyze the key genes in the anaerobic respiratory pathway within the electrochemical pathway, we conducted a screening of gene clusters that have been reported.11 This was based on the functional abundance of species and electron transfer (c-Cyts, riboflavin, and conductive pili) in the samples. The results (Fig. S1†) corroborated that function 1 (riboflavin synthase, ribE, RIB5, K00793), function 2 (electron transfer flavoprotein beta subunit, fixA, etfB; K03521), function 3 (riboflavin kinase/FMN adenylyltransferase, ribF, K11753), function 4 (flagellin, fliC, K02406), function 5 (type IV pilus assembly protein PilA, PilA, K02650), function 6 (c-Cyts, torC, K03532), and function 7 (electron-transferring-flavoprotein dehydrogenase, ETFDH, K00311) were particularly enriched in the genera Shewanella and Pseudomonas. These gene families may play a crucial role in both direct and mediated EET processes, thereby inducing faster electron transfer rates and higher bioelectrochemical catalytic activity in MFCs.
 | ||
| Fig. 6 KEGG functional classification of microbial communities in different MFC stages, relative abundance of KEGG orthologous functions annotated at level 2. | ||
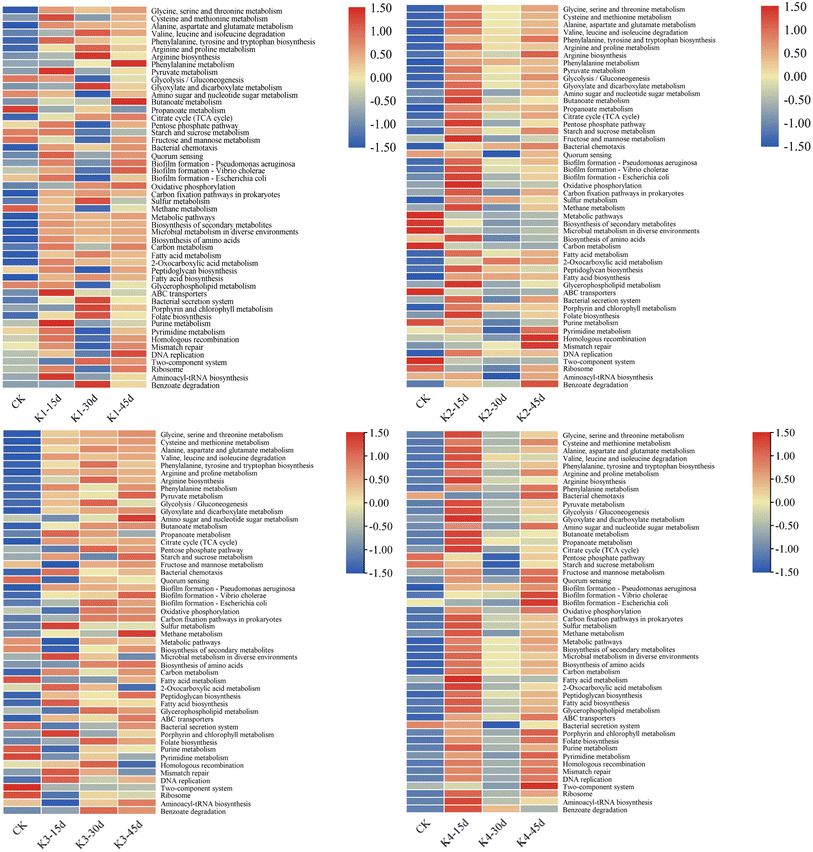 | ||
| Fig. 7 Heatmap predictions of the top 50 most abundant KEGG orthologous functions in the communities at different stages. | ||
3.4 Metabolic differences of microbial activity analysis in MFC
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 230 peaks were detected in both positive and negative ion modes. After removing data with missing values greater than 20% and QC-verified RSD values less than 30%, 12
230 peaks were detected in both positive and negative ion modes. After removing data with missing values greater than 20% and QC-verified RSD values less than 30%, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 954 peaks were retained. Among them, 1098 metabolites were annotated by the KEGG and HMDB databases. The KEGG database annotated a total of 211 metabolites, mainly belonging to categories such as alkaloids (30 types), fatty acids (29 types), shikimate and phenylpropanoids (24 types), organoheterocyclics (20 types), amino acids and peptides (17 types), as shown in Fig. 8. The HMDB database annotated a total of 387 metabolites, including organic heterocyclic compounds (99 types), organic acids and derivatives (48 types), benzene compounds (47 types), lipids and similar lipid molecules (27 types), and alkaloids (21 types). As shown in the figure, the organic heterocyclic compounds in the anodic solution were mainly purines, pyrazoles, and pyridine derivatives, while the organic acids and derivatives mainly include amino acids, peptides, and analogues such as biological amino acids, peptides, and analogues.
954 peaks were retained. Among them, 1098 metabolites were annotated by the KEGG and HMDB databases. The KEGG database annotated a total of 211 metabolites, mainly belonging to categories such as alkaloids (30 types), fatty acids (29 types), shikimate and phenylpropanoids (24 types), organoheterocyclics (20 types), amino acids and peptides (17 types), as shown in Fig. 8. The HMDB database annotated a total of 387 metabolites, including organic heterocyclic compounds (99 types), organic acids and derivatives (48 types), benzene compounds (47 types), lipids and similar lipid molecules (27 types), and alkaloids (21 types). As shown in the figure, the organic heterocyclic compounds in the anodic solution were mainly purines, pyrazoles, and pyridine derivatives, while the organic acids and derivatives mainly include amino acids, peptides, and analogues such as biological amino acids, peptides, and analogues.
As shown in Fig. 9, the annotated metabolites in the tyrosine pathway included tyrosine, 4-hydroxyphenylpyruvic acid, homogentisate, gentisate, and fumarate. When compared with K1 as the control treatment, fumarate was significantly upregulated in K2 and K4, whereas gentisate was significantly upregulated in K3 compared to K2 and K4. The analysis of the results indicated that the different degradation transformations of homogentisate in this pathway led to the final differences in metabolite levels. The precursor substance tyrosine in this pathway is involved in amino acid metabolism, undergoing deamination, hydration, and decarboxylation to produce the intermediate homogentisate, which is then further processed through oxidation-reduction and isomerohydrolysis to yield fumarate.34
Fumarate is a short-chain fatty acid that can be reduced by fumarate dehydrogenase using NADH as a cofactor in an MFC. The electrons pass through the anodic chamber to the cation exchange membrane, making the cathodic electrode an electron acceptor, ultimately affecting the levels of metabolism in the TCA cycle.35 Additionally, gentisate, which is synthesized through a two-step enzymatic reaction via a selective pathway, is mainly reported to affect the degradation of aromatic hydrocarbons, especially the benzoate or naphthalene pathway.36 This may explain the differences in the enrichment of terephthalic acid in K2 and K4. The results suggest that these two substances may act as intermediaries in the main metabolic difference pathways.
Metabolites annotated in the purine metabolism pathway included four parent metabolites: guanosine 5′-triphosphate (GTP), guanosine 5'-(tetrahydrogen triphosphate) (GMP), xanthosine-5′-monophosphate (XMP), and inosinemonophosphate (IMP), as well as two respective daughter metabolites. This pathway is part of the nucleotide metabolism, and by combining the enzyme and substance annotations we detected, we found that xanthine was significantly upregulated in K1 and K3. In the nucleotide reactions involving GMP and IMP, the phosphorylase activity led to a significant downregulation of the substances in K1 and K3, which is somewhat related to the ultimate fate of the substances as xanthine. Since the reverse catalytic enzymes of the substances were detected but the parent metabolites did not show significant differences, the results of the metabolic pathways indicate that the electron transport via mixed colonies involves multiple stages of metabolism, and in communities with large differences in energy output efficiency, there is effective accumulation of different metabolites. This is often associated with nucleotide, energy, amino acid, and carbon metabolism.
3.5 Discussion
The results of the detection of the oxidative phosphorylation pathway in this experiment (Fig. S5†) has shown that electrons are not simply transferred with the potential gradient between the electrode and the battery, nor are they transferred through the intracellular electron transport chain in the biochemical system. The presence of electron carriers is not only for transferring electrons from the electrode to the cell in a simple manner. The capture of electrons is usually accompanied by the co-transport of protons or cations37,38 which can be released and participate in intracellular reduction and energy metabolism. Some dehydrogenases route electrons into the quinone pool, whereas other types act as pumps of protons. Majority of dehydrogenases are only produced when required suggests that bacteria choose which enzymes from their DNA library to generate depending on the circumstances of their surroundings.39 Additionally, reducing agents such as NADH and FADH2, along with ATP, are vital for intracellular redox balance, the production and movement of metabolites, cellular response to stress, and the regulation of gene expression.40 Disturbances in intracellular ATP and NADH levels affect the entire cellular metabolism and redirect metabolic fluxes.41 For example, when Shewanella is inoculated into an MFC system, they can continuously obtain energy by degrading organic acids, thereby promoting cell growth and metabolism. And by releasing electrons to the anode to maintain redox balance, the anode replaces the insoluble metal as the final electron acceptor. For some electrochemically inactive bacteria in the system, most can interact with the electrode under the presence of electron shuttles, such as flavin, coenzyme Q, or its analogs, which can be directly integrated into the intrinsic electron transfer chain through membrane transport proteins or diffusion, and then participate in intracellular reduction and energy metabolism.42EPS components influence the structural and functional properties of biofilms. During biofilm development, EPS facilitates cell–cell recognition/co-adhesion and microbial aggregation, primarily mediated by mechanical sensing or specific adhesin (protein)–receptor (polysaccharide) interactions between species.49,50 EPS components can be categorized into two types:51 I. Cell surface-associated matrix proteins, primarily including surface appendages such as flagella, type IV pilus assembly protein PilA, and functional amyloid proteins. These appendages regulate bacterial adhesion, mechanical stability, and quorum sensing by influencing bacterial motility and attachment to solid surfaces. II. Extracellular components, including secreted polysaccharides, proteins, eDNA, and extracellular RNA (eRNA), which contribute to matrix scaffolding and functionality. In summary, the EPS matrix not only acts as a physical barrier against external substances but also modulate the diffusion of various molecules within the biofilm. This creates gradient distributions of nutrients and chemical components—such as oxygen, pH, signaling molecules, inorganic ions, and metabolites across the biofilm's spatial structure.
The formation of biofilms (BF) has been demonstrated to be regulated by multiple signaling molecules. BF-associated signaling molecules enable bacteria to sense the presence of neighboring bacteria and adjust their biological activities accordingly.52 These signaling molecules are metabolic byproducts autonomously produced during bacterial metabolism. They are not only regulate intracellular activities of individual bacteria, but also secret extracellular to coordinate intercellular behaviors. Depending on the regulatory modes of different signaling molecules53 (interspecies, intraspecies, or intra-/extracellular interactions), biofilm formation is primarily co-regulated by three key mechanisms: quorum sensing (QS), c-di-GMP signaling,54 and two-component systems.55 Quorum Sensing (QS) is defined as a process through which bacteria communicate and coordinate group behaviors using self-produced signaling molecules. This system encompasses a broad range of regulatory pathways. c-di-GMP Signaling regulates transitions in bacterial lifestyles via intracellular concentration gradients: Low c-di-GMP levels: Promote the expression of motility-related genes. High c-di-GMP levels: Enhance the production of adhesion factors and extracellular matrix components, facilitating bacterial attachment to cellular or biomaterial surfaces and promoting BF formation. The critical role of c-di-GMP in BF formation is achieved by regulating biofilm-associated factors such as Psl, Pel, alginate polysaccharides, the surface adhesin CdrA, and type IV pili synthesis. For example, In Pseudomonas aeruginosa, c-di-GMP promotes the production of multiple adhesins. The synthesis of type IV pilus depends on the regulation of c-di-GMP-binding domain proteins PilZ and FimX.
Two-Component systems, composed of a kinase receptor and a cytoplasmic response regulator: The sensor kinase detects external stimuli and undergoes autophosphorylation at a conserved histidine residue. The phosphoryl group is transferred to the response regulator, activating its ability to bind to gene promoters and modulate gene expression.56 Taking the GAC system as an example, this regulatory network is a complex multi kinase network that plays a major role in regulating bacterial planktonic growth mode and biofilm growth mode. The GAC system consists of the transmembrane sensor kinase GacS and its coupling regulator GacA.57 GasS has an autophosphorylated conserved histidine residue, and the activated phosphate group can be transferred to its coupling regulator GacA, affecting the transcription of two sRNAs, RsmZ and RsmY.58 In addition to regulating QS to affect the formation of BF, in vitro studies have found that it also participates in the transition between bacterial planktonic state and BF growth mode by regulating extracellular polysaccharides Pel, Psl, and type IV pilus.59
The two primary electron transfer mechanisms exhibited by Shewanella and Pseudomonas in MFCs were direct electron transfer (DET) and mediated electron transfer (MET).61 The efficiency of extracellular electron transfer (EET) mainly influenced by the transfer between cells and electrodes in DET, including interactions between organisms and electrodes, as well as the effects of electrode reactor materials. For MET, taking Shewanella as an example, Zang et al. reported four strategies to enhance the EET performance of Shewanella: increase in riboflavin, enhancement in intracellular reducing force, promotion of biofilm formation and expansion of the substrate spectrum.62
In an MFC, the anode serves as the final electron acceptor in the metabolic chain of bacteria. The electron transfer between the bacterial cell and the anode has two effects: on the one hand, the anode produces current by collecting electrons from the bacterial cell; on the other hand, for a complete bacterial respiratory process, electrons must be transferred to the anode. In the absence of any other electron acceptors, if the electrons produced by metabolism cannot be smoothly transferred to the anode, the growth of bacterial cells will be suppressed. Therefore, it can be inferred that the bacteria that remain active after the discharge of the fuel cell are those that can adapt to the fuel cell environment and are easy to transfer electrons to the electrode. The next generation of bacteria cultivated using these bacteria for inoculation should have similar properties, enabling them to better utilize the electrochemical electrode as the final electron acceptor in the respiratory chain. This electrochemical activation process of the bacteria is analogous to the natural selection principle in Darwin's theory of evolution.73
4. Conclusion
We conducted a comprehensive study for the first time on the biofilm in a mixed-culture microbial fuel cell (MFC) under various stages of development, focusing on its formation, electroactivity, and the expression of genes related to catabolic pathways and anaerobic respiration. The results from the metagenomics and metabolomics studies indicate that: (i) K2-1000 exhibited mature and high-power output at stage I, and through the study of microbial dynamics, genera such as Pseudomonas and Trichococcus were associated with electrochemical phenomena; (ii) dynamic analysis of the main categories of 10 microorganisms on the biocathode was conducted, and gene and metabolic function tracking was performed for four treatments; (iii) seven electron transport gene functions have suggested an association with the main genera at three periods.In this work, community and metabolic substances of MFC were studied using omics methods, which to some extent can reveal the basic characteristics of coordination and interaction among bacteria in the biofilm. We observed that external resistance is an important reference factor to study the impact of microbial community succession. Through electrochemical domestication, samples were taken at three time points during the stable period, and the dynamics of microorganisms and representative genera were analyzed. The study found that as domestication enters the stable phase, the functional expression of the main microbial communities is closely related to electrochemical phenomena. By studying functional genes, it was found that electron conductance gene families show correlation with genera such as Shewanella, Pseudomonas, Trichococcus, Enterococcus, and even Psychrobacter (K3, stage II). Metabolically, by studying the differences in KEGG metabolic pathways, tyrosine and purine metabolism showed the highest enrichment. In communities with significant differences in output energy efficiency, effective accumulation of different metabolites was observed.
Ethical statement
This article does not contain any studies with human or animal subjects.Data availability
The data supporting this article have been included as part of the ESI.† More raw data used for the preparation of the manuscript are available from the corresponding author.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
This research was supported by the grants from the National Key R&D Program of China (No. 2022YFB2902503, 2021YFB2900604), National Natural Science Foundation of China (No. 22002006), Henan Provincial Department of Science and Technology (242102110081) and China Tobacco Fund of Hebei (No. HBZY2024A079).References
- M. C. Potter, Electrical effects accompanying the decomposition of organic compounds, Proc. R. Soc. London, Ser. B, 1911, 84, 260–276, DOI:10.1098/rspb.1911.0073.
- J. B. Davis and H. F. Yarbrough Jr., Preliminary experiments on a microbial fuel cell, Science, 1962, 137, 615–616, DOI:10.1126/science.137.3530.615.
- M. N. Naseer, A. A. Zaidi, H. Khan, S. Kumar, M. T. bin Owais and J. Jaafar, et al., Mapping the field of microbial fuel cell: A quantitative literature review (1970–2020), Energy Rep., 2021, 7, 4126–4138 CrossRef . https:/doi.org/10.1016/j.egyr.2021.06.082.
- M. Al-Sahari, A. A. Al-Gheethi, R. M. S. R. Mohamed, G. Yashni, D. V. N. Vo and N. Ismail, Microbial fuel cell systems; developments, designs, efficiencies, and trends: A comparative study between the conventional and innovative systems, Chemosphere, 2022, 298, 134244, DOI:10.1016/j.chemosphere.2022.134244.
- J. Zhao, F. Li, Y. Cao, X. Zhang, T. Chen and H. Song, et al., Microbial extracellular electron transfer and strategies for engineering electroactive microorganisms, Biotechnol. Adv., 2021, 53, 107682, DOI:10.1016/j.biotechadv.2020.107682.
- K. Rabaey, N. Boon, S. D. Siciliano, M. Verhaege and W. Verstraete, Biofuel cells select for microbial consortia that self-mediate electron transfer, Appl. Environ. Microbiol., 2004, 70, 5373–5382, DOI:10.1128/AEM.70.9.5373-5382.2004.
- P. Aelterman, K. Rabaey, P. Clauwaert and W. Verstraete, Microbial fuel cells for wastewater treatment, Water Sci. Technol., 2006, 54, 9–15, DOI:10.2166/wst.2006.702.
- T. H. Pham, N. Boon, K. De Maeyer, M. Höfte, K. Rabaey and W. Verstraete, Use of Pseudomonas species producing phenazine-based metabolites in the anodes of microbial fuel cells to improve electricity generation, Appl. Microbiol. Biotechnol., 2008, 80, 985–993 CrossRef CAS PubMed . https://doi.org10.1007/s00253-008-1619-7.
- E. Hakalehto. Mixed Culture Cultivation in Microbial Bioprocesses. Mixed Cultures in Industrial Bioprocesses, Springer Cham, Switzerland, 2025, vol. 2, pp. 9–69. doi: DOI:10.1007/978-3-031-73526-4.
- M. Sharma, E. S. Salama, P. Zhang, L. Zhang, X. Xing and J. Yue, et al., Microalgae-assisted microbial fuel cells for electricity generation coupled with wastewater treatment: Biotechnological perspective, J. Water Process Eng., 2022, 49, 102966, DOI:10.1016/j.jwpe.2022.102966.
- T. Cai, Y. Zhang, N. Wang, Z. Zhang, X. Lu and G. Zhen, Electrochemically active microorganisms sense charge transfer resistance for regulating biofilm electroactivity, spatio-temporal distribution, and catabolic pathway, Chem. Eng. J., 2022, 442, 136248, DOI:10.1016/j.cej.2022.136248.
- D. Nath and M. M. Ghangrekar, Plant secondary metabolites induced electron flux in microbial fuel cell: Investigation from laboratory-to-field scale, Sci. Rep., 2020, 10, 17185, DOI:10.1038/s41598-020-74092-y.
- A. Dhanda, S. Das, B. K. Dubey and M. M. Ghangrekar, Anodic inoculum pre-treatment with green strategies for enhanced electron shuttling and suppressing methanogens in microbial fuel cell: A review, Bioresour. Technol. Rep., 2023, 24, 101593, DOI:10.1016/j.biteb.2023.101593.
- T. Liu, Y. Y. Yu, D. Li, H. Song, X. Yan and W. N. Chen, The effect of external resistance on biofilm formation and internal resistance in Shewanella inoculated microbial fuel cells, RSC Adv., 2016, 6, 20317–20323, 10.1039/C5RA26125B.
- W. F. Cai, J. F. Geng, K. B. Pu, Q. Ma, D. W. Jing and Y. H. Wang, et al., Investigation of a two-dimensional model on microbial fuel cell with different biofilm porosities and external resistances, Chem. Eng. J., 2018, 333, 572–582, DOI:10.1016/j.cej.2017.09.189.
- G. Zhang, Q. Zhao, Y. Jiao, K. Wang, D. J. Lee and N. Ren, Efficient electricity generation from sewage sludge using biocathode microbial fuel cell, Water Res., 2012, 46, 43–52, DOI:10.1016/j.watres.2011.10.036.
- H. Hassan, B. Jin, E. Donner, S. Vasileiadis, C. Saint and S. Dai, Microbial community and bioelectrochemical activities in MFC for degrading phenol and producing electricity: microbial consortia could make differences, Chem. Eng. J., 2018, 332, 647–657, DOI:10.1016/j.cej.2017.09.114.
- N. Zhao, Y. Jiang, M. Alvarado-Morales, L. Treu, I. Angelidaki and Y. Zhang, Electricity generation and microbial communities in microbial fuel cell powered by macroalgal biomass, Bioelectrochemistry, 2018, 123, 145–149, DOI:10.1016/j.bioelechem.2018.05.002.
- B. E. Logan, C. Murano, K. Scott, N. D. Gray and I. M. Head, Electricity generation from cysteine in a microbial fuel cell, Water Res., 2005, 39, 942–952, DOI:10.1016/j.watres.2004.11.019.
- S. Cheng, M. Chen, M. Gao, T. Qiu, S. Tian and S. Li, et al., Effects of Enterococcus faecalis administration on the community structure of airborne bacteria in weanling piglet and layer hen houses, J. Gen. Appl. Microbiol., 2021, 67, 162–169, DOI:10.2323/jgam.2020.11.001.
- J. Lobato, P. Cañizares, F. J. Fernández and M. A. Rodrigo, An evaluation of aerobic and anaerobic sludges as start-up material for microbial fuel cell systems, New Biotechnol., 2012, 29, 415–420, DOI:10.1016/j.nbt.2011.09.004.
- S. Xu and H. Liu, New exoelectrogen Citrobacter sp. SX-1 isolated from a microbial fuel cell, J. Appl. Microbiol., 2011, 111, 1108–1115, DOI:10.1111/j.1365-2672.2011.05129.x.
- J. Huang, N. Zhu, Y. Cao, Y. Peng, P. Wu and W. Dong, Exoelectrogenic Bacterium Phylogenetically Related to Citrobacter freundii, Isolated from Anodic Biofilm of a Microbial Fuel Cell, Appl. Biochem. Biotechnol., 2015, 175, 1879–1891, DOI:10.1007/s12010-014-1418-9.
- Z. Wang, T. Wang, B. Si, J. Watson and Y. Zhang, Accelerating anaerobic digestion for methane production: Potential role of direct interspecies electron transfer, Renewable Sustainable Energy Rev., 2021, 145, 111069, DOI:10.1016/j.rser.2021.111069.
- S. Ren, M. Usman, D. C. Tsang, S. O-Thong, I. Angelidaki and X. Zhu, et al., Hydrochar-facilitated anaerobic digestion: evidence for direct interspecies electron transfer mediated through surface oxygen-containing functional groups, Environ. Sci. Technol., 2020, 54, 5755–5766, DOI:10.1021/acs.est.0c00112.
- O. Lefebvre, T. T. Ha Nguyen, A. Al-Mamun, I. S. Chang and H. Y. Ng, T-RFLP reveals high β-Proteobacteria diversity in microbial fuel cells enriched with domestic wastewater, J. Appl. Microbiol., 2010, 109, 839–850, DOI:10.1111/j.1365-2672.2010.04735.x.
- B. E. Logan and J. M. Regan, Electricity-producing bacterial communities in microbial fuel cells, Trends Microbiol., 2006, 14, 512–518, DOI:10.1016/j.tim.2006.10.003.
- K. Rabaey, N. Boon, M. Höfte and W. Verstraete, Microbial phenazine production enhances electron transfer in biofuel cells, Environ. Sci. Technol., 2005, 39, 3401–3408, DOI:10.1021/es048563o.
- D. Saffarini, K. Brockman, A. Beliaev, R. Bouhenni and S. Shirodkar. Shewanella oneidensis and extracellular electron transfer to metal oxides. Bacteria-metal Interactions, Springer Cham, Switzerland, 2015, vol. 2, pp. 21–40, doi: DOI:10.1007/978-3-319-18570-5_2.
- Y. Yu, Y. Wu, B. Cao, Y. G. Gao and X. Yan, Adjustable bidirectional extracellular electron transfer between Comamonas testosteroni biofilms and electrode via distinct electron mediators, Electrochem. Commun., 2015, 59, 43–47 CrossRef CAS.
- C. Li, L. Hao, J. Cao, K. Zhou, F. Fang and Q. Feng, et al., Mechanism of Fe–C micro-electrolysis substrate to improve the performance of CW-MFC with different factors: insights of microbes and metabolic function, Chemosphere, 2022, 304, 135410, DOI:10.1016/j.chemosphere.2022.135410.
- D. Z. Khater, K. M. El-Khatib and R. Y. Hassan, Effect of vitamins and cell constructions on the activity of microbial fuel cell battery, J. Genet. Eng. Biotechnol., 2018, 16, 369–373, DOI:10.1016/j.jgeb.2018.02.011.
- Y. Wang, D. Li, X. Song, X. Cao, Z. Xu and W. Huang, Intensifying anoxic ammonium removal by manganese ores and granular active carbon fillings in constructed wetland-microbial fuel cells: Metagenomics reveals functional genes and microbial mechanisms, Bioresour. Technol., 2022, 352, 127114, DOI:10.1016/j.biortech.2022.127114.
- J. Y. Wang, X. T. Dai, Q. L. Gao, H. K. Chang, S. Zhang and C. L. Shan, et al., Tyrosine metabolic reprogramming coordinated with the tricarboxylic acid cycle to drive glioma immune evasion by regulating PD-L1 expression, Ibrain, 2023, 9, 133–147, DOI:10.1002/ibra.12107.
- X. Chen, X. Dong, Y. Wang, Z. Zhao and L. Liu, Mitochondrial engineering of the TCA cycle for fumarate production, Metab. Eng., 2015, 31, 62–73, DOI:10.1016/j.ymben.2015.02.002.
- P. S. Phale, A. Basu, P. D. Majhi, J. Deveryshetty, C. Vamsee-Krishna and R. Shrivastava, Metabolic diversity in bacterial degradation of aromatic compounds, OMICS: J. Integr. Biol., 2007, 11, 252–279, DOI:10.1089/omi.2007.0004.
- H. T. Pham Bioelectrochemical technologies: Current and potential applications in agriculture resource recovery. Recent Advancement in Microbial Biotechnology. Academic Press. 2021, pp. 209–308. doi: DOI:10.1016/B978-0-12-822098-6.00002-1.
- R. A. Rozendal, H. V. Hamelers and C. J. Buisman, Effects of membrane cation transport on pH and microbial fuel cell performance, Environ. Sci. Technol., 2006, 40, 5206–5211, DOI:10.1021/es060387r.
- S. S. S. Garimella, S. V. Rachakonda, S. S. Pratapa, G. D. Mannem and G. Mahidhara, From cells to power cells: harnessing bacterial electron transport for microbial fuel cells (MFCs), Ann. Microbiol., 2024, 74, 19, DOI:10.1186/s13213-024-01761-y.
- W. Xiao and J. Loscalzo, Metabolic responses to reductive stress, Antioxid. Redox Signaling, 2020, 32, 1330–1347, DOI:10.1089/ars.2019.7803.
- I. Mesquita, and F. Rodrigues. Cellular metabolism at a glance. Metabolic Interaction in Infection, Springer Cham, Switzerland, 2018, vol. 109, pp. 3–27. doi: DOI:10.1007/978-3-319-74932-7_1.
- E. Hakalehto, Advances in Biochemical Engineering/Biotechnology, Mixed Cultures in Industrial Bioprocesses., Springer Cham, Switzerland, 2025, vol. 189, pp. 9–69 doi: DOI:10.1007/978-3-031-73526-4.
- U. Schroder, Anodic electron transfer mechanisms in microbial fuel cells and their energy efficiency, Phys. Chem. Chem. Phys., 2007, 9, 2619–2629, 10.1039/b703627m.
- L. Karygianni, Z. Ren, H. Koo and T. Thurnheer, Biofilm matrixome: extracellular components in structured microbial communities, Trends Microbiol., 2020, 28, 668–681, DOI:10.1016/j.tim.2020.03.016.
- S. U. Gerbersdorf and S. Wieprecht, Biostabilization of cohesive sediments: revisiting the role of abiotic conditions, physiology and diversity of microbes, polymeric secretion, and biofilm architecture, Geobiology, 2015, 13, 68–97, DOI:10.1111/gbi.12115.
- H. L. Røder, N. M. Olsen, M. Whiteley and M. Burmølle, Unravelling interspecies interactions across heterogeneities in complex biofilm communities, Environ. Microbiol., 2020, 22, 5–16, DOI:10.1111/1462-2920.14834.
- N. N. Henriksen, M. F. Hansen, H. T. Kiesewalter, J. Russel, J. Nesme and K. R. Foster, et al., Biofilm cultivation facilitates coexistence and adaptive evolution in an industrial bacterial community, npj Biofilms Microbiomes, 2022, 8, 59, DOI:10.1038/s41522-022-00323-x.
- Z. Tsvetanova and H. Najdenski, Pathogenic Bacteria in Waters and Drinking Water Associated Biofilms, Ecological Engineering and Environment Protection, 2017, 1, 50–61, DOI:10.32006/eeep.2017.1.5061.
- S. Rath, S. Fatma and S. Das, Unraveling the multifaceted role of extracellular DNA (eDNA) of biofilm in bacterial physiology, biofilm formation, and matrixome architecture, Crit. Rev. Biochem. Mol. Biol., 2025, 1, 1–32, DOI:10.1080/10409238.2025.2497270.
- S. Di Cio and J. E. Gautrot, Cell sensing of physical properties at the nanoscale: Mechanisms and control of cell adhesion and phenotype, Acta Biomater., 2016, 30, 26–48, DOI:10.1016/j.actbio.2015.11.027.
- Y. Xu, H. Xu, J. Yan and G. Song, Mechanical force induced activation of adhesion G protein–coupled receptor, MBM, 2024, 2, 100078, DOI:10.1016/j.mbm.2024.100078.
- D. Singhi and P. Srivastava, Role of bacterial cytoskeleton and other apparatuses in cell communication, Front. Mol. Biosci., 2020, 7, 158, DOI:10.3389/fmolb.2020.00158.
- A. Iaconis, L. M. De Plano, A. Caccamo, D. Franco and S. Conoci, Anti-Biofilm Strategies: A Focused Review on Innovative Approaches, Microorganisms, 2024, 12, 639, DOI:10.3390/microorganisms12040639.
- N. Mangwani, H. R. Dash, A. Chauhan and S. Das, Bacterial quorum sensing: functional features and potential applications in biotechnology, J. Mol. Microbiol. Biotechnol., 2012, 22, 215–227, DOI:10.1159/000341847.
- M. Valentini and A. Filloux, Multiple roles of c-di-GMP signaling in bacterial pathogenesis, Annu. Rev. Microbiol., 2019, 73, 387–406, DOI:10.1146/annurev-micro-020518-115555.
- N. Papon and A. M. Stock, Two-component systems, Curr. Biol., 2019, 29, R724–R725, DOI:10.1016/j.cub.2019.06.010.
- J. Cheung and W. A. Hendrickson, Sensor domains of two-component regulatory systems, Curr. Opin. Microbiol., 2010, 13, 116–123, DOI:10.1016/j.mib.2010.01.016.
- S. Salar, S. Silletti and F. D. Schubot, Evidence of Bidirectional Transmembrane Signaling by the Sensor Histidine Kinase GacS from Pseudomonas aeruginosa, J. Biol. Chem., 2025, 301, 108521, DOI:10.1371/journal.pgen.1006032.
- A. Brencic, K. A. McFarland, H. R. McManus, S. Castang, I. Mogno and S. L. Dove, et al., The GacS/GacA signal transduction system of Pseudomonas aeruginosa acts exclusively through its control over the transcription of the RsmY and RsmZ regulatory small RNAs, Mol. Microbiol., 2009, 73, 434–445, DOI:10.1111/j.1365-2958.2009.06782.x.
- X. Wang, M. Liu, C. Yu, J. Li and X. Zhou, Biofilm formation: mechanistic insights and therapeutic targets, Mol. Biomed., 2023, 4, 49, DOI:10.1186/s43556-023-00164-w.
- M. Lukaszczyk, B. Pradhan and H. Remaut, The biosynthesis and structures of bacterial pili, Bacterial cell walls and membranes, 2019, 92, 369–413, DOI:10.1007/978-3-030-18768-2_12.
- A. Kumar, L. H. H. Hsu, P. Kavanagh, F. Barrière, P. N. Lens and L. Lapinsonnière, et al., The ins and outs of microorganism–electrode electron transfer reactions, Nat. Rev. Chem., 2017, 1, 0024, DOI:10.1038/s41570-017-0024.
- Y. Zang, B. Cao, H. Zhao, B. Xie, Y. Ge and H. Liu, et al., Mechanism and applications of bidirectional extracellular electron transfer of Shewanella, Environ. Sci.: Processes Impacts, 2023, 25, 1863–1877, 10.1039/d3em00224a.
- D. Min, L. Cheng, F. Zhang, X. N. Huang, D. B. Li and D. F. Liu, et al., Enhancing extracellular electron transfer of Shewanella oneidensis MR-1 through coupling improved flavin synthesis and metal-reducing conduit for pollutant degradation, Environ. Sci. Technol., 2017, 51, 5082–5089, DOI:10.1021/acs.est.6b04640.
- J. Lange, R. Takors and B. Blombach, Zero-growth bioprocesses: A challenge for microbial production strains and bioprocess engineering, Eng. Life Sci., 2017, 17, 27–35, DOI:10.1002/elsc.201600108.
- A. Hirose, A. Kouzuma and K. Watanabe, Towards development of electrogenetics using electrochemically active bacteria, Biotechnol. Adv., 2019, 37, 107351, DOI:10.1016/j.biotechadv.2019.02.007.
- A. Almatouq, A. O. Babatunde, M. Khajah, G. Webster and M. Alfodari, Microbial community structure of anode electrodes in microbial fuel cells and microbial electrolysis cells, J. Water Process Eng., 2020, 34, 101140, DOI:10.1016/j.jwpe.2020.101140.
- D. Nosek and A. Cydzik-Kwiatkowska, Microbial structure and energy generation in microbial fuel cells powered with waste anaerobic digestate, Energies, 2020, 13, 4712, DOI:10.3390/en13184712.
- N. Uria, I. Ferrera and J. Mas, Electrochemical performance and microbial community profiles in microbial fuel cells in relation to electron transfer mechanisms, BMC Microbiol., 2017, 17, 1–12, DOI:10.1186/s12866-017-1115-2.
- M. Degli Esposti. Phylogeny and Evolution of Bacteria and Mitochondria, CRC Press, Boca Raton, 2018. doi: DOI:10.1201/b22399.
- M. M. Curtis, Z. Hu, C. Klimko, S. Narayanan, R. Deberardinis and V. Sperandio, The gut commensal Bacteroides thetaiotaomicron exacerbates enteric infection through modification of the metabolic landscape, Cell Host Microbe, 2014, 16, 759–769, DOI:10.1016/j.chom.2014.11.005.
- T. Conway, and P. S. Cohen. Commensal and pathogenic Escherichia coli metabolism in the gut. Metabolism and Bacterial Pathogenesis, Wiley Online Library, America, 2015, vol. 3, pp. 343–362. doi: DOI:10.1128/9781555818883.ch16.
- E. Bornet and A. J. Westermann, The ambivalent role of Bacteroides in enteric infections, Trends Microbiol., 2022, 30, 104–108, DOI:10.1016/j.tim.2021.11.009.
Footnote |
| † Electronic supplementary information (ESI) available: The table of original microbiome of plant inoculants, the relative analysis of gene functional contribution, score scatter plot of OPLS-DA model, permutation plot test of OPLS-DA model, metabolites in heat map analysis, metabolic pathway prediction analysis, the detected analysis related to the oxidative phosphorylation pathway. See DOI: https://doi.org/10.1039/d5ra01080b |
| This journal is © The Royal Society of Chemistry 2025 |