Open Access Article
Open Access ArticleFluorescence dye-conjugated magnetic core–shell silica nanoparticles for enhanced nucleic acid visualization
Tammar Hussein Ali *ab,
Ammar Alhasana,
Hasanain Salah Naeema,
Israa Jabera,
Ruqaya Sabah Abdulhusseina,
Nihad A. M. Al-Rashedi
*ab,
Ammar Alhasana,
Hasanain Salah Naeema,
Israa Jabera,
Ruqaya Sabah Abdulhusseina,
Nihad A. M. Al-Rashedi c and
Siti Aishah Hasbullah
c and
Siti Aishah Hasbullah d
d
aCollege of Pharmacy, Al-Muthanna University, 66001 Samawah, Al Muthanna, Iraq. E-mail: tammar@mu.edu.iq; tammar86@gmail.com
bCollege of Pharmacy, National University of Science and Technology, Dhi Qar, 64001, Iraq
cDepartment of Biology, College of Science, Al Muthanna University, 66001 Samawah, Iraq
dDepartment of Chemical Sciences, Faculty of Science and Technology, Universiti Kebangsaan Malaysia, 43600 UKM Bangi, Selangor, Malaysia
First published on 11th July 2025
Abstract
Traditional fluorescent dyes employed for the detection of nucleic acids are associated with significant challenges and encounter environmental and economic obstacles, including high cost, toxicity, long staining times, and insufficient sensitivity. Thus, efforts have been devoted to replacing the prevalent ethidium bromide (EB) dye with safer and more stable dyes; however, some of the aforementioned drawbacks continue to hinder progress in this field. In this work, we developed a novel neutral fluorescent magnetic core–shell nanoparticle dye by synthesizing Fe2O3 nanoparticles and subsequently coating them with a silica shell. The silica coating not only stabilized the magnetic core but also facilitated the conjugation of the nanoparticles with 4-hydroxy coumarin (C4@NpFeSi), 7-hydroxy coumarin (C7@NpFeSi), and fluorescein (Flu@NpFeSi). The samples were comprehensively characterized via TEM, XRD, TGA, UV-vis absorption, and fluorescence spectroscopy, which confirmed their successful synthesis, and thus, the resulting particles could be utilized for visualizing nucleic acids in the solid phase. Fluorescence studies demonstrated that DNA-C7@NpFeSi exhibits an emission band centered at 458 nm (λex = 325 nm), which represents an increase in FL intensity by 2-fold in comparison with C7@NpFeSi. However, DNA-Flu@NpFeSi exhibited an emission peak at 650 nm (λex = 515 nm), which can be attributed to the intercalation binding between Flu dye and DNA protonation, increasing the fluorescence intensity by ∼10 fold compared to the free Flu@NpFeSi. Agarose gel electrophoresis confirmed effective DNA visualization with distinct bands resolved for 50 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp fragments crossing three distinct DNA ladders, indicating highly efficient magnetic separation. These results highlight that DNA-Flu@NpFeSi is an efficient alternative to ethidium bromide due to its high sensitivity, low toxicity, and cost-effectiveness.
000 bp fragments crossing three distinct DNA ladders, indicating highly efficient magnetic separation. These results highlight that DNA-Flu@NpFeSi is an efficient alternative to ethidium bromide due to its high sensitivity, low toxicity, and cost-effectiveness.
Introduction
The visualization of DNA plays a pivotal role in the early detection, diagnosis, and monitoring of diseases, particularly in identifying cancer biomarkers.1–3 In the context of cancer diagnosis, techniques with the ability to clearly visualize DNA fragments are essential for facilitating the identification of genetic mutations and abnormalities, thereby enabling precise diagnosis and guiding therapeutic interventions.4,5 Among the available methods for nucleic acid separation, agarose gel electrophoresis remains one of the most efficient and effective techniques for the separation and analysis of nucleic acids due to its ability to leverage an electric field to separate DNA fragments based on size.6,7 The global electrophoresis reagent market has grown considerably to a valuable size, with a compound annual growth rate (CAGR) of 4.47% forecasted for the period from 2024 to 2031, driven by increasing demand for genomics and clinical diagnosis.8 The COVID-19 pandemic further emphasized the significance of electrophoresis in biomedical research, where it played a critical role in developing effective therapies by enabling the precise labeling of DNA, RNA, or protein molecules. This, in turn, spurred interest in developing safer and more environmentally friendly electrophoretic reagents.9 This demand has driven researchers to investigate new dye formulations that are more rationally and scientifically sustainable from their inception. Consequently, the electrophoresis reagent market is projected to achieve a higher CAGR of 5.11% during the forecast period.10,11Historically, ethidium bromide (EB) has been the most common and widely used reagent of choice for DNA staining due to its high fluorescence upon intercalation with DNA.12,13 However, it suffers from several drawbacks, especially given that it poses significant health and environmental risks.14 It is known as an ionic compound, which leads to the formation of a strong complex with nucleic acid.15,16 More importantly, EB is considered mutagenic and genotoxic to humans, which causes various alterations by affecting the biological processes of DNA transcription and replication, and causing structural and functional alterations in cells.16 Consequently, there are significant safety concerns regarding EB waste disposal for laboratory staff, which is creating regulatory issues regarding its disposal into the environment due to it contaminating both soil and water.17,18 Furthermore, EB is expensive, and it presents significant economic drawbacks associated with hazardous waste disposal. For example, a case study reported that annual disposal costs of EB range from 5000 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 $ (USD), depending on the volume, packaging, and disposal procedures.19
000 $ (USD), depending on the volume, packaging, and disposal procedures.19
These drawbacks have driven research on safer and more efficient alternatives, where introducing nanotechnology offers innovative solutions for DNA labeling and visualization.20 Nanotechnology is a highly promising field, enabling the assembly, manipulation, and manufacturing of nanomaterials, thereby translating nanoscience theory into practical applications with the potential to stimulate scientific innovation, while greatly benefiting society.21–24 Nanoparticles have introduced innovative functionalities, increased efficiencies, and expanded capabilities in numerous medical applications, including medical sensing, cellular imaging, and DNA labeling.25–29 For instance, Zanoli et al. (2012) reported the use of functionalized gold nanoparticles for DNA labeling based on PCR amplification; however, their findings were based on the detection of specific DNA sequences and only a few systems were applied to real samples.30 Secondly, Prieto et al. (2014) described electrophoretic gels using methyl green (GM) for DNA labeling.31 Furthermore, Li and group (2024) used CRISPR-Cas12a complexes to detect colorectal cancer by identifying DNA methylation features at the aM level.32
In this context, fluorescent dyes continue to play a crucial role in DNA labeling due to their ability to provide high contrast imaging of nucleic acids.33,34 Among the fluorescent dyes, coumarin-based fluorescent probes have been reported to be employed for the fluorescent imaging of live cells due to their low cytotoxicity and reasonable cell permeability.35,36 Similarly, fluorescein dye is one of the first labels that has been widely employed in many different biological and biochemical applications.37 Recently, fluorescein has been used to assist with lesion localization and visualizing the extent of tumor resection in neurosurgical oncology due to its accumulation in areas of blood–brain barrier breakdown.38
Therefore, this study explored the development of fluorescent-magnetic nanoparticles as alternatives to ethidium bromide for DNA visualization. Specifically, coumarin and fluorescein conjugated with superparamagnetic iron oxide (Fe2O3) nanoparticles were synthesized and evaluated for DNA visualization. The resulting Flu@NpFeSi nanoparticles exhibited strong fluorescence, high stability, and effective superparamagnetic behavior in comparison with C7@NpFeSi and C4@NpFeSi, making them suitable and cost-effective for DNA detection. Overall, a comparison between these particles and the most commonly used dyes is presented in Table 1, revealing their optical properties and sensitivity with the detection method. These findings not only highlight the potential of the developed nanoparticles as an alternative to ethidium bromide but also pave the way for the next-generation development of DNA labeling and detection.
| Ethidium bromide78,79 | SYBR safe80–85 | SYBR green86,87 | SYBR gold88,89 | Crystal violet90–92 | YOYO-1 (ref. 93–95) | Hoechst 33258 (ref. 96–98) | EvaGreen99–101 | PicoGreen102–104 | AccuClear105–107 | AccuBlue105–107 | C4@NpFeSi | C7@NpFeSi | Flu@NpFeSi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Excitation | 300 nm | 280 nm | 495–500 nm | 496 nm | ∼590 nm | 491 nm | 360 nm | 500 nm | 500 nm | 468 nm | 500 nm | 325 nm | 325 nm | 515 nm |
| 502 nm | ||||||||||||||
| Emission | 605 nm | 530 nm | 520–522 nm | 539 nm | 640 nm | 509 nm | 460 nm | 530 nm | 525 nm | 507 nm | 530 nm | 456 nm | 456 nm | 650 nm |
| Sensitivity with DNA | 1 ng | 3 ng | Highly sensitive (>60 pg) | Ultra-sensitive (>25 pg) | 16 ng | Highly sensitive (pg) | 10 ng | 50 pg to 50 ng | 50 pg to 2 μg | 0.03–250 ng | 0.2–100 ng | Highly sensitive (pg) | 25 ng | 40 ng |
| Hazardous | Hazardous | Non-hazardous | Hazardous | Hazardous | Non-hazardous | Hazardous | Hazardous | Non-hazardous | Hazardous | Hazardous | Non-hazardous | Non-hazardous | Non-hazardous | Non-hazardous |
| Detection method | UV | UV or blue light | Fluorescence (blue light excitation) | UV, blue light, or laser scanners | UV | Fluorescence | Fluorescence | Fluorescence | Fluorescence | Fluorescence | Fluorescence | Fluorescence or UV | Fluorescence or UV | Fluorescence or UV |
| Colour | Orang | Green | Green | Gold | Non-fluorescent | Green | Blue | Green | Green | Green | Green | Orang | Orang | Blue |
Materials and characterization
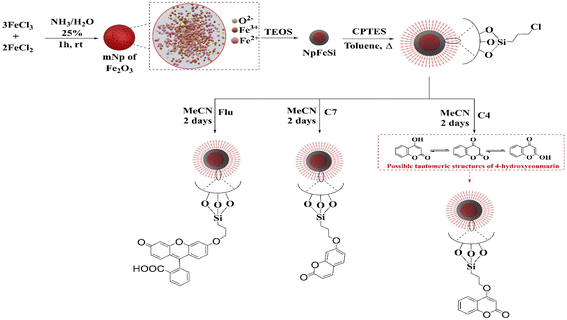
All chemicals were obtained from Merck and Sigma-Aldrich and used as-received without purification. The reactants used were iron(III) chloride anhydrous (99.9% w/w), iron(II) chloride tetrahydrate (98% w/w), tetraethoxysilane (98% w/w), (3-chloropropyl)trimethoxysilane (97% w/w), 7-hydroxycoumarin (99% w/w), 4-hydroxycoumarin (98% w/w), and fluorescein sodium salt. All other chemicals and solvents were purchased from various commercial sources.IR spectra were recorded on a PerkinElmer Tensor 27 ATR FT-IR spectrometer. Dried nanoparticles were subjected to thermal gravimetric and differential thermal analysis (TG-DTA) at a ramping rate of 20 °C min−1. Transmission electron microscopy (TEM) was performed on a Talos L120C operating at 120 kV with 0.2 nm resolution. Samples were prepared by dispersing the nanoparticles in ethanol and sonicating for 5 min before deposition on an amorphous carbon-coated 300 mesh copper grid, and then allowing the solvent to evaporate. The surface morphology of the samples was analyzed by field emission scanning electron microscopy (SEM) on an EI Quanta 400F field emission scanning electron microscope (FESEM) equipped with an Oxford-Instruments INCA 400 X-Max detector for energy-dispersive X-ray spectroscopy (EDX) measurement at ×300 magnification (spot size 1 mm × 1 mm) and an accelerating voltage of 20 kV. A few milligrams of dried sample were added to an FESEM cell, and subsequently scanned by the electronic microscope. The XRD patterns of the particles were recorded on a Bruker D8 Advance X-ray diffraction AXS using Cu Kα radiation (d = 1.54 Å) at a voltage of 40 kV and current of 40 mA at ambient temperature.
Synthesis of magnetic nanoparticle-supported dye (dye@NpFeSi)
The core–shell magnetic nanoparticles were prepared via the chemical co-precipitation of Fe3+ and Fe2+ ions at a molar ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and then functionalization by (3-chloropropyl)triethoxysilane (CPTES) according to our previously reported method.16,39–42 The resulting silyl chloride-functionalized magnetic nanoparticles (2.0 g) were dispersed in MeCN/MeOH (50/20 mL) and ultrasonicated for 20 min. A nucleophilic dye (1.0 g) (sodium fluorescein, 7-hydroxy coumarin, and 4-hydroxy coumarin) was added dropwise with triethylamine (0.5 mL) and the reaction mixture was stirred at 65 °C for 3 days. After cooling to room temperature, the product was isolated by centrifugation and magnetic decantation. It was washed with a mixture of water/ethanol (3 × 20 mL) to ensure removal of the unreacted dye, and then washed repeatedly with acetone (3 × 20 mL) and dried for 6 h at 45 °C to obtain dye@NpFeSi (∼2.4–2.6 g) as a brown solid.
1, and then functionalization by (3-chloropropyl)triethoxysilane (CPTES) according to our previously reported method.16,39–42 The resulting silyl chloride-functionalized magnetic nanoparticles (2.0 g) were dispersed in MeCN/MeOH (50/20 mL) and ultrasonicated for 20 min. A nucleophilic dye (1.0 g) (sodium fluorescein, 7-hydroxy coumarin, and 4-hydroxy coumarin) was added dropwise with triethylamine (0.5 mL) and the reaction mixture was stirred at 65 °C for 3 days. After cooling to room temperature, the product was isolated by centrifugation and magnetic decantation. It was washed with a mixture of water/ethanol (3 × 20 mL) to ensure removal of the unreacted dye, and then washed repeatedly with acetone (3 × 20 mL) and dried for 6 h at 45 °C to obtain dye@NpFeSi (∼2.4–2.6 g) as a brown solid.
DNA interaction by the gel electrophoresis method
TBE buffer solution (1×) was used to prepare the (2% w/v) agarose gel for gel electrophoresis. The agarose mixture was heated in a microwave until fully dissolved with occasional gentle swirling for mixing. The result solution was allowed to cool to approximately 50 °C (until it was safe to handle), and then poured into a gel tray. Five microliters of each Flu@NpFeSi, C7@NpFeSi, and C4@NpFeSi was individually added to separate agarose solutions. A comb was placed in the gel tray to create wells for the sample loading. The cooled agarose was carefully poured into the gel tray, ensuring even distribution and the probe comb placement. The gel was allowed to set for 15 min at room temperature. Then, the comb was carefully removed, and 5 μL of each type of 1 kb, 100 bp and 50 bp ladder (GoldBio, USA) (Fig. 1) was added to a separate well. The gel tray was placed in an electrophoresis chamber, which was filled with 1× TBE buffer. Electrophoresis was performed at a voltage of 110 V for 30 min. After that, to improve the band separation, the voltage was adjusted to 75 V for one hour. The resulting migration patterns were visualized under UV irradiation (G-BOX, SYNGENE).DNA release from nanoparticles
Particle purification was performed by adding 50 μL of AW1 elution buffer, which contained a high concentration of chaotropic salt, and incubation at 70 °C for 10 min to dissociate the bonded DNA from the magnetic nanoparticles. Subsequently, the mixture was vortexed until the pellet was fully resuspended visibly. Next, the sample was centrifuged for 30 s at approximately 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, and the supernatant was removed. The particles were washed twice using a buffer solution containing absolute ethanol and centrifuged for 1 min at 14
000 rpm, and the supernatant was removed. The particles were washed twice using a buffer solution containing absolute ethanol and centrifuged for 1 min at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Then, the solution was decanted, and the nanoparticles were collected and dried in an oven at 60 °C for 6 h.
000 rpm. Then, the solution was decanted, and the nanoparticles were collected and dried in an oven at 60 °C for 6 h.
Computational study
The Chimera and AutoDock Vina programs were used to perform the docking simulations. The interactions were visualized using the Discovery Studio software. The docking was carried out for the studied complexes formed between the synthesized particles and DNA. The protein structure (PDB: 6VOY) was retrieved from the Research Collaboratory for Structural Bioinformatics (RCSB) database. To ensure the reliability of the results, at least five independent docking runs were performed consecutively. Gasteiger charges and energy minimization was also considered in the calculations.Results and discussion
Nanoparticle characterization
According to Fig. 2, the XRD patterns of the produced powders indicate the existence of an amorphous core–shell quartz and hematite modified with C4, C7, and Flu compounds. Our prior investigation demonstrated that the precursor magnetic core–shell (NpFeSi and NpFeSiCl) consists of a mixture of Fe2O3 and SiO2.43 The peak width of the pattern peaks recorded in the three modified materials (C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi) did not exhibit any variations. According to the comparison of NpFeSi and NpFeSiCl, the only minor variations observed were in their crystallinity and particle sizes, which can be related to the variable quantities of reactant (C4, C7, and Flu) on the surface of NpFeSi. An interesting observation is that the phase particles of C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi exhibit diffraction peaks at 26.8°, 30.4°, 35.7°, 36.5°, 43.2°, 53.4°, 57.4°, 59.9°, and 62.9°. The Fe2O3 percentages of 70%, 3.0%, 5.0%, 1.0%, and 30% on the (110), (202), (122), (211) and (214) crystallographic planes, respectively, are consistent with a 62% rhombohedral phase, as shown by the PDF Reference Code 033-0664 (indexed in ICDDPDF2). Based on PDF Ref Cod: 033-1161 (indexed in ICDDPDF2), the diffractions of hexagonal phase SiO2 indicate the presence of (101), (202), (211), and (102) crystallographic planes with percentages of 100%, 4%, 1%, and 8%, respectively, and the overall match percentage is 42%. These values distinctly indicate the formation of Fe2O3 hematite core–shell silica nanoparticles, which reveals that the particles possess super-paramagnetic behavior, as previously reported.39 It is important to note that the magnetic properties of these particles have already been thoroughly investigated and documented in our earlier research.39,44The particles were subjected to TEM analysis for comprehensive scrutiny. The TEM images demonstrated the presence of a hematite core encased in a quartz shell, as shown in Fig. 3. The electron diffraction of the particles indicated that their outer shell (quartz) consisted of three distinct compositions, i.e., C4, C7, and Flu, validating the development of core–shell particles. The generation of particles featuring a high porosity core–shell structure was confirmed by the TEM images, which revealed different particle size distributions. The results obtained from the TEM images indicate that the average dimensions (area) of C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi were 165 nm, 42 nm, and 69 nm2, respectively. The decrease in the dimensions of C7@NpFeSi and Flu@NpFeSi can be attributed to the increased reaction between the C7 and Flu molecules with NpFeSi particles or/and their agglomeration, which could be due to the drying of the specimen on the grid for the TEM observations. The relative contribution of the magnetostatic interaction appeared to increase, leading to ordering and self-assembly.45,46 In agreement, the particles exhibited varying diameters of around 10, 7.5, and 9.2 nm along the longer axis for C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi, respectively. They presented details regarding the atomic structure and shape of the nanoparticles. The lattice fringes of the magnetic core could be distinctly observed. The estimated lattice d-spacing was ∼0.33 and 0.29 nm for C4@NpFeSi; ∼0.2 and 0.15 nm for C7@NpFeSi; and ∼0.25 and 0.16 nm for Flu@NpFeSi, corresponding to (101), (111), (202), (214), (110), (102), (211), and (122), respectively.43,47 The SAED pattern displays luminous rings indicating a substantial quantity of hematite nanocrystals (polycrystalline phase) and spot reflections associated with individual particles of the three synthesized core–shell (C4, C7, and Flu)@NpFeSi materials.
 | ||
| Fig. 3 TEM images of three core–shell nanoparticles C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi, with SAED of each NPs. | ||
Upon analyzing the literature studies with the aid of the FESEM images, it was noted that the morphology of the NpFeSi and NpFeSiCl structures exhibited a uniform phase and possessed irregular forms with rough surfaces.48–51 Fig. 4 exhibits the FESEM images of the NpFeSi-modified material at various magnification levels. The FESEM images demonstrate a significant change in the morphology of the NpFeSi surface after being treated with organic compounds. There was a discernible disparity in the degree of surface roughness and loading. The surface of C4@NpFeSi displayed a coarse texture with bright aggregations after modification with 4-hydroxycoumarin, as shown in Fig. 4(A). By changing the position of the hydroxyl group on the initial compound 7-hydroxycoumarin, a significant change in the structure of C7@NpFeSi took place, as shown in Fig. 4(B). The surface of C7@NpFeSi exhibited a smooth and undulating texture, with a greater degree of loading in comparison to C4@NpFeSi according to the TGA analysis. This observation is consistent with the visual attributes of the organic substances, which were revealed using FESEM.52–54
 | ||
| Fig. 4 FESEM images of three core–shell samples: (A) C4@NpFeSi, (B) C7@NpFeSi, and (C) Flu@NpFeSi material with EDX analysis. | ||
The rough and rocky texture observed in these locations can be attributed to the lower concentration of C7 reactants in this particular area. According to the fluorescence results, it can be suggested that they show consistent and regular distribution patterns on the Flu@NpFeSi surface, while maintaining the surface roughness, as shown in Fig. 4(C).
EDX analysis was performed to determine the elemental percentage composition in the (C4, C7, and Flu)@NpFeSi materials. The results demonstrate that the composition of the resultant materials differs among the reactants (C4, C7, and Flu). There was a noticeable alteration in the C4@NpFeSi substance, with a reduction in the carbon content of 34% and an increase in the chlorine content of 1.1%. In the case of C7@NpFeSi, its carbon content increased to 46%, while its chlorine content decreased to 0.6%. This is because the C7 compounds have a stronger interaction with the surface of NpFeSiCl, resulting in a reduction in the chlorine substitution by the C7 compounds. The elemental composition of Flu@NpFeSi in the EDX Table was approximately comparable to that of C7@NpFeSi. The Flu@NpFeSi composition was comprised of approximately 21% iron (Fe), 30% oxygen (O), 1.8% silicon (Si), 45.8% carbon (C), and 0.9% chlorine (Cl). The positioning of the hydroxyl group on Flu in relation to C7 and C4, as well as the molecular structure of Flu, are consistent with these values.
FT-IR investigation was performed to verify the existence of several functional groups after the modification processes on the surface of the NpFeSi-Cl material. Fig. 5 presents the FT-IR spectra of the NpFeSi-Cl, C4@NpFeSi, C7@NpFeSi, and Flu@NpFeSi materials. The spectrum of NpFeSi-Cl exhibited a distinct absorption peak at 3282 cm−1, which suggests the presence of stretching vibrations from the remaining O–H groups. Subsequently, a weak and sharp identifiable vibration at 2925 cm−1 appeared, which is ascribed to the sp3–C–H vibration of the hydrocarbon chain. The presence of an absorption peak at 1023 cm−1 indicates the occurrence of the asymmetric stretching vibration of Si–O–Si. The band at 1624 cm−1 can be attributed to the bending vibration of water molecules that are absorbed on the silica surface.55,56 The findings provide evidence of the creation of a silica matrix on the surface of NpFe, resulting in the synthesis of the NpFeSi material. Then, this material was further modified into NpFeSi-Cl through the immobilization process. The spectra of NpFeSi-Cl, when treated with C4, C7, and Flu molecules, exhibited a significant reduction in intensity. Although the C4 and C7 compounds share a similar structure, they displayed different spectra because of the differing positions of the hydroxyl group. The spectrum of C7@NpFeSi displays clear peaks at 1697, 1519, and 1126 cm−1, which correspond to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C
O, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, and C–O functional groups, respectively.57 By contrast, the spectrum of C4@NpFeSi displays a notable decrease in peak intensity, accompanied by a substantial change in peak positions in comparison to C7@NpFeSi due to the increased loading of C7 molecules compared to C3 molecules, as illustrated by the TGA results. The heightened resonance effect in the aromatic ring (C
C, and C–O functional groups, respectively.57 By contrast, the spectrum of C4@NpFeSi displays a notable decrease in peak intensity, accompanied by a substantial change in peak positions in comparison to C7@NpFeSi due to the increased loading of C7 molecules compared to C3 molecules, as illustrated by the TGA results. The heightened resonance effect in the aromatic ring (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C 1450 cm−1) can be attributed to the close proximity of the enol group to the carbonyl group (C
C 1450 cm−1) can be attributed to the close proximity of the enol group to the carbonyl group (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O 1603 cm−1); furthermore, the C4 reactant exhibits a low loading ratio on the NpFeSi surface. The spectrum of Flu@NpFeSi indicates the presence of hydroxyl and carbonyl groups, which correspond to the oscillation of the carboxylic acid group at wavenumbers of 3359 and 1636 cm−1, respectively.58 This indicates that the Flu molecules were successfully bound to the surface of the NpFeSi particles.
O 1603 cm−1); furthermore, the C4 reactant exhibits a low loading ratio on the NpFeSi surface. The spectrum of Flu@NpFeSi indicates the presence of hydroxyl and carbonyl groups, which correspond to the oscillation of the carboxylic acid group at wavenumbers of 3359 and 1636 cm−1, respectively.58 This indicates that the Flu molecules were successfully bound to the surface of the NpFeSi particles.
Thermogravimetric analysis (TGA) was employed to ascertain the composition and organic content of the synthesized magnetic nanocomposites, as shown in Fig. 6. A more straightforward study was facilitated by mathematical data processing utilizing differential thermogravimetry (DTG) spectra. All the thermodynamic curves of the three new compositions (C4, C7, and Flu)@NpFeSi exhibited three characteristic zones of weight loss. The weight loss in the first region below 200 °C can be attributed to the desorption of retained moisture and residual organic solvent.59 The following stage (the second region) of mass reduction at 200 °C is linked to the thermal oxidation and pyrolysis of the organic constituents.
The TGA-DTG curves of (C4, C7, and Flu)@NpFeSi exhibited a significant decomposition stage at 160 °C, 139 °C, and 119 °C, with the corresponding mass loss of 0.435 mg (4.344%), 0.248 mg (2.551%), and 0.256 mg (2.622%), respectively. The first region in the TGA-DTG curve corresponds to the varying percentages of water adsorbed inside the nanoparticle matrix of the three product materials ((C4, C7, and Flu)@NpFeSi). The water loss temperature was higher in C4@NpFeSi and progressively decreased in Flu@NpFeSi, which is attributed to the greater availability of reactive organic compounds on the nanoparticle surface in Flu@NpFeSi compared to C4@NpFeSi. The increase in the content of Flu molecules resulted in a reduction in the content of hydroxyl groups (–OH) on the surface of SiO2, thereby diminishing the percentage of water adsorbed within the nanoparticle. The mass loss in C7@NpFeSi was lower than that in Flu@NpFeSi due to the higher concentration of polar functional groups, particularly carboxylic group (–COOH), in the Flu structure relative to C7. The existence of these functional groups will enhance the adsorption of water molecules on the surface relative to the matrix itself. Consequently, the thermal measurement revealed a greater weight loss at a lower temperature for Flu@NpFeSi.
The second mass loss region was seen at 270 °C (0.621 mg, 6.21%), 269 °C (0.606 mg, 6.24%), and 264 °C (0.429 mg, 4.4%), corresponding to (C4, C7, and Flu)@NpFeSi, respectively, which can be ascribed to the decomposition of the chloropropyl group bonded to silica.
The third mass loss region was between approximately 300 °C to 600 °C and showed the characteristic weight loss at 330 °C (0.59 mg, 5.9%), 374 °C (0.774 mg, 7.97%), and 453 °C (1.288 mg, 13.2%), corresponding to (C4, C7, and Flu)@NpFeSi, respectively. The mass loss rates for C4@NpFeSi and C7@NpFeSi differed, exhibiting a variation in their TGA-DTG curves. The disparity in the TGA-DTG curves of C4@NpFeSi and C7@NpFeSi at approximately 300–400 °C arises from the availability of C7 molecules on the silica surface, while exhibiting identical decomposition profiles for both C4 and C7. Conversely, the Flu@NpFeSi particles exhibited a variation in their TGA-DTG curve throughout the third principal region. This region can be characterized as a non-sharp thermal decomposition region, which is attributed to the elevated molecular weight of Flu molecules, their abundance on the silica surface, and the presence of polar carboxylic groups that enhance the internal molecular forces. Consequently, the thermal stability of Flu@NpFeSi surpassed that of both C4@NpFeSi and C7@NpFeSi.
Particle synthesis
The co-precipitation method, initially delineated by Massart,60 is the most prevalent methodology, which has subsequently experienced extensive research and modification. This process yields magnetic nanoparticles with exact size and outstanding magnetic properties. The synthesis of the Fe2O3 hematite magnetic nanoparticles entails the co-precipitation of ferrous and ferric salts in an aqueous medium, utilizing a strong basic solution (aqueous ammonia at pH 9) at ambient temperatures.61,62 Scheme 1 illustrates the primary framework for the synthesis and modification of the core–shell magnetic nanoparticles. A modified Stöber technique was utilized to synthesize SiO2, which was then coated via TEOS hydrolysis. Through meticulous regulation of the conditions, the particles were protected from oxidation and aggregation, leading to improved surface functionalization.63,64 This process involved the treatment of CPTES for grafting an organic halide onto the nanoparticle surface, followed by the final coupling of the nanoparticles with the designated chemicals including 4-hydroxy coumarin (C4), 7-hydroxy coumarin (C7), and fluorescein (Flu). According to the results from the FESEM, FTIR, and TEM analyses, the loading percentage of the C7 molecule exceeded that of the C4 compound. This disparity can be ascribed to the steric barrier of the C4 molecule and its existence in three tautomeric keto–enol forms in aqueous solution.65,66 The position of the hydroxyl group relative to the carbonyl group on the C7 ring influences the resonance phenomena, and subsequently impacts the formation of tautomers. The deposition of Flu molecules on the surface of NpFeSi-Cl resulted in a significant change in surface morphology. The interactions between Flu molecules and alkyl halides occurred seamlessly, which was not affected by steric hindrance. However, the main novelty of this design lies in introducing a spacer between the particles and the fluorescent molecules to prevent their direct interaction, which can reduce the solid-state quenching due to the interaction with the particles. Additionally, this spacer provides flexibility to the fluorescent molecules for more effective interaction with biological molecules. Moreover, the ether linkage will allow additional hydrogen bond interactions, which serve as a directing and detecting agent for biological targets, thereby enhancing the fluorescent properties and the polarity. As a result, the design of the synthesized particles was intended for visualizing nucleic acids in the solid phase.Optical properties
Next, the light absorption properties of 4-hydroxy coumarin (coumarin-C7) conjugated with Si-Fe2O3 nanoparticles, both pre- and post-incorporated with DNA, were investigated. Fig. 8(a) shows the absorption spectra of aqueous C7@NpFeSi at different concentrations. The absorption peak appears at 325 nm. However, the UV-vis absorption of DNA-C7@NpFeSi nanoparticles was shown to have the same peak with a higher intensity in comparison to that for the highest concentration of C7@NpFeSi (0.1 mg mL−1), as shown in Fig. 8(b). The absorption results indicate that there is no possibility of intercalation interaction with DNA, and thus no red-shift was recorded. The observation of a higher intensity upon incorporation with DNA could be caused by the hydrogen interaction, which also increased the stability of the complex.69
Furthermore, the light absorption properties of fluorescein (Flu) conjugated with Si-Fe2O3 nanoparticles, both pre- and post-incorporated with DNA, were investigated. Fig. 9(a) shows the absorption spectra of aqueous Flu@NpFeSi. The absorption shoulder with a peak appears at 460 nm. The low intensity of this peak corresponds to the fluorescence quenching of fluorescein, owing to the presence of oxygen molecular, which is well known to be a good quencher, in addition to the weak interaction between the two electron clouds of perpendicular π–π system of phenyl and xanthene rings and/or interaction with hydroxyl group on the particle surface, and this result in agreement with previous research work.70,71 However, the UV-vis absorption peak of DNA-Flu@NpFeSi nanoparticles was shown to shift to 515 nm with a higher intensity, as shown in Fig. 9(b), due to the intercalation between Flu dye and DNA.69
However, the fluorescence emission of the C7@NpFeSi nanoparticles and DNA-C7@NpFeSi nanoparticles was investigated following UV-visible absorption measurement. The FL emission spectra of the C7@NpFeSi nanoparticles at the optimum excitation of 325 nm show an obvious emission band peak centered at 456 nm, as presented in Fig. 10(a), which is attributed to their large π–π conjugated system with electron-rich and charge transfer properties.72 Also, the DNA-C7@NpFeSi nanoparticles exhibited stable fluorescence at 325 nm excitation with an emission band centered at 458 nm. They display similar fluorescence properties compared to that of the C7@NpFeSi nanoparticles core but with a higher FL intensity, as shown in Fig. 10(b). The increase in the FL emission intensity of DNA-C7@NpFeSi is due to a process driven by collisions between DNA, which is already known as a fluorescence agent, and the C7@NpFeSi core, and by the strong intra/inter hydrogen interactions affinity of the carbonyl groups with the hydroxyl groups on the surface. Therefore, the synthesized DNA-C7@NpFeSi is found to be a potential candidate for assembly-based fluorescence enrichment.
 | ||
| Fig. 10 Fluorescence emission of the (a) C7@NpFeSi nanoparticle and the (b) DNA-C7@NpFeSi nanoparticle. | ||
Furthermore, the fluorescence emission spectrum of fluorescein conjugated with manganic nanoparticles (Flu@NpFeSi) was observed by employing their maximum absorbance wavelength as the excitation wavelength. Fig. 11(a) shows that once excited by light at a wavelength of 460 nm, the Flu@NpFeSi nanoparticles in a water medium emitted fluorescence light at a wavelength of 511 nm and 650 nm due to the fluorescein emission when conjugated with Fe2O3 nanoparticles. These results are attributed to the coexistence of different fluorescein ionic forms in the solution, which can produce dual emission peaks. This occurs due to light absorption, followed by proton transfer in the excitation state, leading to equilibrium between the protonated and deprotonated forms, which in agreement with previously reported studies in the literature.71,73
 | ||
| Fig. 11 Fluorescence emission of the (a) Flu@NpFeSi nanoparticle and the (b) DNA-Flu@NpFeSi nanoparticle. | ||
Compared to the DNA-Flu@NpFeSi nanoparticles, the FL emission spectra at an excitation of 515 nm show an emission band peaking at 650 nm, as shown in Fig. 11(b), which can be attributed to the intercalation binding between Flu dye and nucleotide. In addition, this is due to the possibility of the protonation of DNA that occurs because of the protonation of its purine bases. Subsequently, cytosine protonation is due to the proton-induced conformational transition within the guanine/cytosine pairs with the sequence proton transfer from (N-7) of guanine to (N-3) of cytosine and/or protonation of cytosine by transfer of the proton from the carboxylic acid part of the Flu dye to cytosine.74 The DNA-Flu@NpFeSi nanoparticles also exhibited stable fluorescence with a higher FL intensity. The ionic interaction increased the FL emission intensity of DNA-Flu@NpFeSi.69
Among the synthesized dye@NpFeSi nanoparticles, both C7@NpFeSi and Flu@NpFeSi exhibit similar excellent optical properties both pre- and post-interaction with DNA. The main difference lies in their wavelength range (color), with C7@NpFeSi displaying a blue color and Flu@NpFeSi showing a reddish-orange color.
Nanoparticle interaction with DNA
The agarose gel electrophoresis image illustrates effective DNA band separation using three distinct DNA ladders, i.e., 50 bp plus ladder, 100 bp plus ladder, and 1 kb DNA ladder, applied in lanes Flu@NpFeSi, C7@NpFeSi, and C4@NpFeSi, as shown in Fig. 12. The clear, well-defined bands ranging from 50 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp confirm the successful size resolution and high sample quality. The prominent bands at 500 bp and 1000 bp reflect higher DNA concentrations, validating both the accurate quantification and proper ladder migration. The consistent and uniform migration of bands across the gel indicates the optimal electrophoretic conditions. Initially, the gel was run at 110 V for 30 min, a standard setting for resolving DNA fragments within this size range and gel concentration. Subsequently, the voltage was reduced to 75 V and maintained for an additional hour to enhance the band separation. This approach followed best practices for optimizing the resolution in agarose gel electrophoresis.75 The resulting band clarity supports reliable verification of DNA markers and fragment sizing. The minor smears or faint bands suggest potential pipetting errors or DNA degradation, which can be addressed by improving the sample preparation. These observations are consistent with the common challenges in electrophoresis experiments, as noted in molecular biology protocols.76 These optimizations are crucial for reliable applications, including PCR product validation and detailed fragment analysis. However, the use of an external magnetic field facilitates the efficient extraction of DNA-dye@NpFeSi from the mixture, representing a key advantage in minimizing its environmental toxicity and reducing its disposal-related costs. Also, a reusability test was performed after releasing the DNA from the particle and activating it. A similar efficacy was recorded up to three repeated uses, as shown in Fig. 13.
000 bp confirm the successful size resolution and high sample quality. The prominent bands at 500 bp and 1000 bp reflect higher DNA concentrations, validating both the accurate quantification and proper ladder migration. The consistent and uniform migration of bands across the gel indicates the optimal electrophoretic conditions. Initially, the gel was run at 110 V for 30 min, a standard setting for resolving DNA fragments within this size range and gel concentration. Subsequently, the voltage was reduced to 75 V and maintained for an additional hour to enhance the band separation. This approach followed best practices for optimizing the resolution in agarose gel electrophoresis.75 The resulting band clarity supports reliable verification of DNA markers and fragment sizing. The minor smears or faint bands suggest potential pipetting errors or DNA degradation, which can be addressed by improving the sample preparation. These observations are consistent with the common challenges in electrophoresis experiments, as noted in molecular biology protocols.76 These optimizations are crucial for reliable applications, including PCR product validation and detailed fragment analysis. However, the use of an external magnetic field facilitates the efficient extraction of DNA-dye@NpFeSi from the mixture, representing a key advantage in minimizing its environmental toxicity and reducing its disposal-related costs. Also, a reusability test was performed after releasing the DNA from the particle and activating it. A similar efficacy was recorded up to three repeated uses, as shown in Fig. 13.
 | ||
| Fig. 12 Agarose gel electrophoresis performed using dye@NpFeSi mixed with 50 plus (1 μL), 100 plus (1.3 μL), and 1 kb (0.8 μL) DNA ladders. | ||
Overall, the visualization of nucleic acid bands using the dye@NpFeSi nanoparticle shows a promising result, enabling clear band separation at high and low concentrations. Among the synthesized nanoparticles, the most intense and brightest band was displayed with the Flu@NpFeSi nanoparticles.
A simulation program was applied to give a principal idea about the interaction. The cryo-EM structure of HTLV-1 instasome (6VOY) was the selected DNA model, and a simple structure of dye molecule with a trimethoxysilylpropoxyl spacer was used as the ligand to reduce the complexity of the nanoparticle. Fig. 14(A and B) show the C4 dye ligand interaction with the protein group around the nucleotides (DNA) only by hydrogen and hydrophobic bonds. Although the C7 dye ligand interacts with the protein by multiple bonds and one hydrogen bond with nucleotides via DT (D-threonine) on the open edge end of the DNA, as depicted in Fig. 14(C and D). A significantly different interaction by multiple hydrogen and hydrophobic bonds is shown with the Flu dye ligand and both nucleotides and protein. The main interaction was with two amino acids, DC (D-cytosine) and DT (D-threonine) of nucleotides, also on the open edge end of the DNA, but more deeply inside the helix structure as shown in Fig. 14(E and F). This interaction could be considered as an intercalator, which is in agreement with the previous study.77 The corresponding binding energies for the C4, C7, and Flu dyes were equal to −4.9, −6.1, and −6.3 kcal mol−1, respectively.
 | ||
| Fig. 14 Docking structure obtained from the interaction of the magnetic and DNA module complex. (A) and (B) C4@NpFeSi, (C) and (D) C7@NpFeSi, and (E) and (F) Flu@NpFeSi. | ||
Conclusion
The synthesis of a new fluorescence magnetic nanoparticle dye was facilitated by incorporating a covalently surface-modified magnetic core–shell silica particle with 4-hydroxy coumarin, 7-hydroxy coumarin, and fluorescein as a dye. The propyl group was used in the particle design as a spacer to increase the potential of interaction along the particle surface. This spacer separated the dye molecules from the particles, which could also reduce the solid-state quenching caused by the particle interaction. This combination was intended for visualizing nucleic acids in the solid phase. An interesting fluorescence result was recorded for Flu@NpFeSi after interacting with DNA. Excitation at 515 nm resulted in an emission band peaking at 650 nm, which could be attributed to the intercalation binding between the dye particles and DNA protonation. Agarose gel electrophoresis was applied to demonstrate the effectiveness of the dye in detecting DNA bands. The distinct bands, ranging from 50 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp, indicated successful fragment sizing and good sample quality. Furthermore, cost-effective and simple separation using the proper positioning of an external magnetic field minimized the environmental toxicity. The simulation study for C4@NpFeSi showed that it forms one hydrogen bond interaction with DNA, while C7@NpFeSi exhibits multiple bonds and one hydrogen bond with nucleotides via DT (D-threonine). A significantly different interaction via multiple hydrogen and hydrophobic bonds was shown with Flu@NpFeSi, where its main interaction is with two amino acids, DC (D-cytosine) and DT (D-threonine), of the nucleotides deep inside the helix structure of DNA. The corresponding binding energies for C4, C7, and Flu dyes were equal to −4.9, −6.1, and −6.3 kcal mol−1, respectively.
000 bp, indicated successful fragment sizing and good sample quality. Furthermore, cost-effective and simple separation using the proper positioning of an external magnetic field minimized the environmental toxicity. The simulation study for C4@NpFeSi showed that it forms one hydrogen bond interaction with DNA, while C7@NpFeSi exhibits multiple bonds and one hydrogen bond with nucleotides via DT (D-threonine). A significantly different interaction via multiple hydrogen and hydrophobic bonds was shown with Flu@NpFeSi, where its main interaction is with two amino acids, DC (D-cytosine) and DT (D-threonine), of the nucleotides deep inside the helix structure of DNA. The corresponding binding energies for C4, C7, and Flu dyes were equal to −4.9, −6.1, and −6.3 kcal mol−1, respectively.
Data availability
The data supporting this article have been included in the main manuscript.Author contributions
All authors contributed equally.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
The authors of this paper gratefully acknowledge financial support from the Iraq Ministry of Higher Education and Scientific Research, the Faculty of Pharmacy of Al Muthanna University and the Ministry of Higher Education, Malaysia under the fundamental research grant scheme (FRGS) with the grant number of (FRGS/1/2022/STG04/UKM/02/4). They would also like to thank the Department of Chemistry, College of Science, University of Malaya.References
- T. Wittenberger, S. Sleigh, D. Reisel, M. Zikan, B. Wahl, M. Alunni-Fabbroni, A. Jones, I. Evans, J. Koch, T. Paprotka, H. Lempiäinen, T. Rujan, B. Rack, D. Cibula and M. Widschwendter, Epigenomics, 2014, 6, 311–327 CrossRef CAS PubMed.
- F. Sabir, M. Zeeshan, U. Laraib, M. Barani, A. Rahdar, M. Cucchiarini and S. Pandey, Cancers, 2021, 13(14), 3396 CrossRef CAS PubMed.
- X. Wu, J. Li, A. Gassa, D. Buchner, H. Alakus, Q. Dong, N. Ren, M. Liu, M. Odenthal, D. Stippel, C. Bruns, Y. Zhao and R. Wahba, Int. J. Biol. Sci., 2020, 16, 1551–1562 CrossRef CAS PubMed.
- H. Satam, K. Joshi, U. Mangrolia, S. Waghoo, G. Zaidi, S. Rawool, R. P. Thakare, S. Banday, A. K. Mishra, G. Das and S. K. Malonia, Biology, 2023, 12(7), 997 CrossRef CAS PubMed.
- S. D. Dwivedi, S. D. Yadav, D. Sahu, D. Singh and M. R. Singh, Gene Rep., 2024, 36, 101963 CrossRef CAS.
- P. Y. Lee, J. Costumbrado, C.-Y. Hsu and Y. H. Kim, J. Visualized Exp., 2012, 20(62), 3923 Search PubMed.
- B. D. Hames, Gel Electrophor. Proteins, 2023, 1–52 Search PubMed.
- databridgemarket, Global Electrophoresis Reagents Market Size, Share, and Trends Analysis Report – Industry Overview and Forecast to 2032, 2023 Search PubMed.
- W. Song, Z. Fang, F. Ma, J. Li, Z. Huang, Y. Zhang, J. Li and K. Chen, Front. Microbiol., 2023, 14, 1217567 CrossRef PubMed.
- B. Data, Global Electrophoresis Reagents Market – Industry Trends and Forecast to 2031, 2023 Search PubMed.
- Mordor Intelligence, Electrophoresis Reagents Market Size & Share Analysis - Growth Trends & Forecasts (2024–2029), 2023 Search PubMed.
- N. Akbay, Z. Seferoğlu and E. Gök, J. Fluoresc., 2009, 19, 1045–1051 CrossRef CAS PubMed.
- G. Mazzini and M. Danova, Methods Mol. Biol., 2017, 1560, 239–259 CrossRef CAS PubMed.
- S. Singh, N. Singh and A. N. Singh, Int. J. Res. Anal. Rev., 2018, 5, 226–233 Search PubMed.
- M. J. Waring, J. Mol. Biol., 1965, 13, 269–282 CrossRef CAS PubMed.
- T. Hussein Ali, A. Mousa Mandal, A. Alhasan and W. Dehaen, J. Mol. Liq., 2022, 359, 119345 CrossRef CAS.
- O. Moradi, M. Norouzi, A. Fakhri and K. Naddafi, J. Environ. Health Sci. Eng., 2014, 12, 1–9 CrossRef PubMed.
- P. Singh, P. K. Narang and Akanksha, Environ. Ecol., 2024, 42, 851–854 CrossRef.
- P. N. Hengen, Trends Biochem. Sci., 1994, 19, 257–258 CrossRef CAS PubMed.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- S. Bayda, M. Adeel, T. Tuccinardi, M. Cordani and F. Rizzolio, Molecules, 2020, 25, 1–15 Search PubMed.
- A. Ahmed, A. Singh and S. Arya, J. Energy Storage, 2025, 105, 114783 CrossRef.
- A. Dubey, A. Singh, A. Ahmed, A. K. Sundramoorthy, S. P. Patole and S. Arya, Surf. Interfaces, 2024, 51, 104731 CrossRef CAS.
- A. Ahmed, S. Verma, P. Mahajan, A. K. Sundramoorthy and S. Arya, Sci. Rep., 2023, 13, 1–12 CrossRef PubMed.
- B. S. Gaylord, A. J. Heeger and G. C. Bazan, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 10954–10957 CrossRef CAS PubMed.
- M. Sun, B. Sun, Y. Liu, Q. D. Shen and S. Jiang, Sci. Rep., 2016, 6, 1–12 CrossRef PubMed.
- F. Feng, H. Wang, L. Han and S. Wang, J. Am. Chem. Soc., 2008, 130, 11338–11343 CrossRef CAS PubMed.
- A. Samanta and I. L. Medintz, Nanoscale, 2016, 8, 9037–9095 RSC.
- C. H. Liu, M. H. Tsao, S. L. Sahoo and W. C. Wu, RSC Adv., 2017, 7, 5937–5947 RSC.
- L. M. Zanoli, R. D'Agata and G. Spoto, Anal. Bioanal. Chem., 2012, 402, 1759–1771 CrossRef CAS PubMed.
- D. Prieto, G. Aparicio, P. E. Morande and F. R. Zolessi, Histochem. Cell Biol., 2014, 142, 335–345 CrossRef CAS PubMed.
- Y. Li, M. Cai, W. Zhang, Y. Liu, X. Yuan, N. Han, J. Li, S. Jin and C. Ding, Biosens. Bioelectron., 2024, 244, 115810 CrossRef CAS PubMed.
- Y. W. Jun, E. M. Harcourt, L. Xiao, D. L. Wilson and E. T. Kool, Nat. Commun., 2022, 13, 5043 CrossRef CAS PubMed.
- L. Wang, V. Reipa and J. Blasic, Bioconjug. Chem., 2004, 15, 409–412 CrossRef CAS PubMed.
- S. Das, H. K. Indurthi, P. Saha and D. K. Sharma, Dyes Pigm., 2024, 228, 112257 CrossRef CAS.
- R. Alabada, A. Ayub, Y. Ajaj, S. Ishwar, R. H. Alshammari, A. Abduldayeva, A. Imran, Z. Ahmad and R. M. K. Mohamed, J. Alloys Compd., 2024, 977, 173400 CrossRef CAS.
- J. B. Grimm, L. M. Heckman and L. D. Lavis, in Progress in Molecular Biology and Translational Science, Elsevier Inc., 1st edn, 2013, vol. 113, pp. 1–34 Search PubMed.
- X. Zhang, A. Habib, E. Jaman, A. N. Mallela, N. M. Amankulor and P. O. Zinn, J. Neurosurg. Sci., 2023, 67, 374–379 Search PubMed.
- T. H. Ali, A. M. Mandal, T. Heidelberg, R. S. Duali Hussen and E. W. Goh, RSC Adv., 2020, 10, 38818–38830 RSC.
- T. H. Ali, A. M. Mandal, T. Heidelberg and R. S. D. Hussen, RSC Adv., 2022, 12, 13566–13579 RSC.
- A. Halilu, T. H. Ali, A. Y. Atta, P. Sudarsanam, S. K. Bhargava and S. B. Abd Hamid, Energy Fuels, 2016, 30, 2216–2226 CrossRef CAS.
- A. Halilu, T. H. Ali, P. Sudarsanam and S. K. Bhargava, Symmetry, 2019, 11, 524 CrossRef CAS.
- T. H. Ali, M. Mandal and T. Heidelberg, RSC Adv., 2020, 10, 38818–38830 RSC.
- A. Halilu, T. H. Ali, P. Sudarsanam and S. K. Bhargava, Symmetry, 2019, 11, 1–11 CrossRef.
- Y. Takeno, Y. Murakami, T. Sato, T. Tanigaki, H. S. Park, D. Shindo, R. M. Ferguson and K. M. Krishnan, Appl. Phys. Lett., 2014, 105, 5 CrossRef PubMed.
- A. S. Najm, H. S. Naeem, K. O. Alabboodi, S. A. Hasbullah, H. A. Hasan, A. M. Holi, A. A. AL-Zahrani, K. Sopian, B. Bais, H. S. Majdi and A. J. Sultan, Sci. Rep., 2022, 12, 1–21 CrossRef PubMed.
- A. S. Najm, V. Selvanathan, T. M. Aljuwaya, L. S. Sabri, M. S. Jamal, A. A. Al-zahrani, A. M. Holi, I. Jaber, A. Al Ghamdi, M. T. Amin, K. Sopian, R. A. Ismail, H. Moria, B. Bais, H. S. Majdi, S. Chowdhury, A. J. Sultan and T. M. Alhuzaymi, APL Mater., 2023, 11, 1–23 Search PubMed.
- J. Liu, S. Dong, Q. He, S. Yang, M. Xie and P. Deng, Biomolecules, 2019, 9, 245–260 CrossRef CAS PubMed.
- S. Suresh, S. Karthikeyan and K. Jayamoorthy, J. Adv. Res., 2016, 7(5), 739–747 CrossRef CAS.
- A. S. Najm, H. S. Naeem, D. A. R. M. Alwarid, A. Aljuhani, S. A. Hasbullah, H. A. Hasan, K. Sopian, B. Bais, H. J. Al-Iessa, H. S. Majdi, A. J. Sultan and H. Moria, Coatings, 2022, 12, 1400 CrossRef CAS.
- M. M. Ba-abbad, A. Benamour, D. Ewis and A. W. Mohammad, JOM, 2022, 74, 3531–3539 CrossRef CAS.
- D. Mallah, B. Bi, F. Mirjalili and A. Bamoniri, Sci. Rep., 2024, 14, 1–18 CrossRef PubMed.
- A. S. Najm, A. Aljuhani, H. S. Naeem, K. Sopian, R. A. Ismail, A. M. Holi, L. S. Sabri, A. Abdullah AL-Zahrani, R. T. Rasheed and H. Moria, RSC Adv., 2022, 12, 29613–29626 RSC.
- H. Pang, Y. Sun, J. Zhou, M. Xie, H. Lin and Y. Yong, Crystals, 2020, 10, 1–18 CrossRef.
- S. C. Feifel and F. Lisdat, J. Nanobiotechnol., 2011, 9, 1–12 CrossRef PubMed.
- H. S. Naeem, I. Jaber, S. Sapari, F. I. A. Razak, B. M. Rudaini, N. H. A. Razak, S. N. Asmaa and S. A. Hasbullah, Sains Malays., 2024, 53, 1937–1952 CrossRef CAS.
- A. S. Najm, N. Ahmad, I. Jaber and N. Hisham, Inorg. Chim. Acta, 2022, 533, 120776 CrossRef CAS.
- P. Heksahidropirimidina, Sains Malays., 2024, 53, 839–850 CrossRef.
- T.-L. Hsieh, P.-S. Hung, C.-J. Wang, K.-C. Tso, H.-Y. Wang, C.-T. Cheng, Y.-C. Lin, R.-J. Chung, K.-H. Wei, P.-W. Wu and P.-C. Chen, Nanotechnology, 2020, 31, 1–38 CrossRef PubMed.
- R. Massart, IEEE Trans. Magn., 1981, 17, 1247–1248 CrossRef.
- A. Fadli and A. Adnan, Mater. Sci. Eng., 2019, 622, 012013 CAS.
- M. Rahmayanti, Nat. Sci. J. Sci. Technol., 2020, 54–58 CrossRef.
- C. Sun, J. S. H. Lee and M. Zhang, Adv. Drug Delivery Rev., 2008, 60, 1252–1265 CrossRef CAS PubMed.
- N. Geerts and E. Eiser, Soft Matter, 2010, 6, 4647–4660 RSC.
- C. A. Pranzoni, A. Pacciaroni and V. Sosa, Arch. Org. Chem., 2011, 7, 1–12 RSC.
- M. M. Abdou, R. A. El-saeed and S. Bondock, Recent Advances in 4-Hydroxycoumarin Chemistry. Part 1: Synthesis, King Saud University, 2015 Search PubMed.
- S. Singh, V. V Begoyan, M. Tanasova, K. Waters, M. Seel and R. Pandey, J. Phys. Org. Chem., 2018, 31(9), e3852 CrossRef.
- M. K. Verma, R. Kumar and M. Sharma, J. Mol. Struct., 2024, 1312, 138605 CrossRef CAS.
- H. Hilal and J. A. Taylor, Dyes Pigm., 2007, 75, 483–490 CrossRef CAS.
- X. F. Zhang, J. Zhang and L. Liu, J. Fluoresc., 2014, 24, 819–826 CrossRef CAS PubMed.
- J. Panchompoo, L. Aldous, M. Baker, M. I. Wallace and R. G. Compton, Analyst, 2012, 137, 2054–2062 RSC.
- H. Raunio, O. Pentikäinen and R. O. Juvonen, Int. J. Mol. Sci., 2020, 21, 1–17 Search PubMed.
- M. Mekawy, A. Saito, H. Shimizu and T. Tominaga, Nanomaterials, 2015, 5, 874–884 CrossRef CAS PubMed.
- T. I. Smol'janinova, V. A. Zhidkov and G. V Sokolov, Nucleic Acids Res., 1982, 10, 2121–2134 CrossRef PubMed.
- D. W. Sambrook and J. Russell, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York., 3rd edn, 2001 Search PubMed.
- J. Green and M. R. Ambrook, Molecular Cloning: A Laboratory Manual, Cold Spring Harbor Laboratory Press, New York, 2019 Search PubMed.
- C.-H. Liu, M.-H. Tsao, S. L. Sahoo and W.-C. Wu, RSC Adv., 2017, 7, 5937–5947 RSC.
- A. Gurunathan and M Subalakshmi, Decontaminating Ethidium Bromide: Advances in Adsorptive, Photocatalytic, and Biodegradation Approaches, 2025, p. 2025051106, DOI:10.20944/preprints202505.1106.v1.
- P.-H. Chang and B. Sarkar, J. Environ. Manage., 2021, 278, 111586 CrossRef CAS PubMed.
- N. Svanvik, G. Westman, D. Y. Wang and M. Kubista, Anal. Biochem., 2000, 281, 26 CrossRef CAS PubMed.
- J. Isacsson, H. Cao, L. Ohlsson, S. Nordgren, N. Svanvik, G. Westman, M. Kubista, R. Sjoback and U. Sehlstedt, Mol. Cell. Probes, 2000, 14, 321 CrossRef CAS PubMed.
- B. A. Armitage, Top. Curr. Chem., 2005, 253, 55 CrossRef CAS.
- W. E. Evenson, L. M. Boden, K. A. Muzikar and D. J. O'Leary, J. Org. Chem., 2012, 77, 10967–10971 CrossRef CAS PubMed.
- A. Biancardi, T. Biver, A. Marini, B. Mennucci and F. Secco, Phys. Chem. Chem. Phys., 2011, 13, 12595–12602 RSC.
- C. S. O'Neil, J. L. Beach and T. D. Gruber, Electrophoresis, 2018, 39, 1474–1477 CrossRef PubMed.
- H. Zipper, H. Brunner, J. Bernhagen and F. Vitzthum, Nucleic Acids Res., 2004, 32, e103 CrossRef PubMed.
- G. Cosa, K. S. Focsaneanu, J. R. McLean, J. P. McNamee and J. C. Scaiano, Photochem. Photobiol., 2001, 73, 585–599 CrossRef CAS PubMed.
- P. J. Kolbeck, W. Vanderlinden, G. Gemmecker, C. Gebhardt, M. Lehmann, A. Lak, T. Nicolaus, T. Cordes and J. Lipfert, Nucleic Acids Res., 2021, 49, 5143–5158 CrossRef CAS PubMed.
- R. S. Tuma, M. P. Beaudet, X. Jin, L. J. Jones, C.-Y. Cheung, S. Yue and V. L. Singer, Anal. Biochem., 1999, 268, 278–288 CrossRef CAS PubMed.
- M. J. Bartel, M. F. Picco and M. B. Wallace, Gastrointest. Endosc. Clin. N. Am., 2015, 25, 243–260 CrossRef PubMed.
- E. Q. Adams and L. Rosenstein, J. Am. Chem. Soc., 1914, 36, 1452–1473 CrossRef CAS.
- S. Mani and R. N. Bharagava, Rev. Environ. Contam. Toxicol., 2016, 237, 71–104 CAS.
- H. S. Rye, S. Yue, D. E. Wemmer, M. A. Quesada, R. P. Haugland, R. A. Mathies and A. N. Glazer, Nucleic Acids Res., 1992, 20, 2803–2812 CrossRef CAS PubMed.
- L. Wang, J. R. Pyle, K. A. Cimatu and J. Chen, J. Photochem. Photobiol., A, 2018, 367, 411–419 CrossRef CAS PubMed.
- J. R. Pyle and J. Chen, Beilstein J. Nanotechnol., 2017, 8, 2296–2306 CrossRef CAS PubMed.
- S. A. Latt, G. Stetten, L. A. Juergens, H. F. Willard and C. D. Scher, J. Histochem. Cytochem., 1975, 23, 493–505 CrossRef CAS PubMed.
- S. A. Latt and G. Stetten, J. Histochem. Cytochem., 1976, 24, 24–33 CrossRef CAS PubMed.
- J. Bucevičius, G. Lukinavičius and R. Gerasimaitė, Chemosensors, 2018, 6(2), 18 CrossRef.
- F. Sang and J. Ren, J. Sep. Sci., 2006, 29, 1275–1280 CrossRef CAS PubMed.
- J. Ihrig, R. Lill and U. Mühlenhoff, Anal. Biochem., 2006, 359, 265–267 CrossRef CAS PubMed.
- B. Bruijns, R. Tiggelaar and H. Gardeniers, Data Brief, 2017, 10, 132–143 CrossRef PubMed.
- V. L. Singer, L. J. Jones, S. T. Yue and R. P. Haugland, Anal. Biochem., 1997, 249, 228–238 CrossRef CAS PubMed.
- G. G. Schofield, Biotechniques, 2004, 37, 778–780 CrossRef CAS PubMed.
- A. I. Dragan, J. R. Casas-Finet, E. S. Bishop, R. J. Strouse, M. A. Schenerman and C. D. Geddes, Biophys. J., 2010, 99, 3010–3019 CrossRef CAS PubMed.
- B. B. Bruijns, R. M. Tiggelaar and J. G. E. Gardeniers, Anal. Biochem., 2016, 511, 74–79 CrossRef CAS PubMed.
- H.-J. He, E. V. Stein, D. Paul and K. D. Cole, Biotechniques, 2018, 64, 59–68 CrossRef CAS PubMed.
- S. Lager, M. C. de Goffau, U. Sovio, S. J. Peacock, J. Parkhill, D. S. Charnock-Jones and G. C. S. Smith, Microbiome, 2018, 6, 151 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2025 |