 Open Access Article
Open Access ArticleResearch progress on the preparation, performance, mechanisms, and applications of carbon nanomaterials against Gram-negative bacteria
Chengwei Zhang†
 b,
Ying Zheng†a,
Xiaoyan Lina and
Shaohuang Weng
b,
Ying Zheng†a,
Xiaoyan Lina and
Shaohuang Weng *a
*a
aDepartment of Pharmaceutical Analysis, School of Pharmacy, Fujian Medical University, Fuzhou 350122, P. R. China. E-mail: shweng@fjmu.edu.cn
bSchool of Basic Medical Sciences, Fujian Medical University, Fuzhou, 350122, China
First published on 30th June 2025
Abstract
The excessive and inappropriate use of antibiotics in clinical settings has led to bacterial mutations and the emergence of drug resistance. This escalating problem presents a major challenge in the treatment of infections, particularly those caused by Gram-negative and multidrug-resistant bacteria, for which effective therapeutic options are increasingly limited. As a result, there is an urgent need to develop novel antimicrobial agents. Recent studies have demonstrated that carbon nanomaterials, owing to their potent antibacterial activity and low susceptibility to resistance, are emerging as promising candidates in antimicrobial research. This review provides a comprehensive overview of recent advancements in the synthesis strategies, antibacterial efficacy against Gram-negative bacteria, underlying antimicrobial mechanisms, and biomedical applications of four main carbon nanomaterials (CNMs) of graphene quantum dots (GQDs), carbon dots (CDs), carbon nanotubes (CNTs), and graphene. The preparation methods and antibacterial mechanisms of CNMs with antibacterial properties are the main section. Additionally, the review addresses the current challenges hindering their clinical translation and explores potential future directions with a focus on efficacy and safety. By collating current findings, this review aims to establish a theoretical foundation for the development of innovative antimicrobial agents targeting Gram-negative bacteria.
1. Introduction
Since the discovery of penicillin by Alexander Fleming in 1929, the development of a vast array of antimicrobial agents has significantly advanced global public health.1 However, the widespread, excessive, and inappropriate use of antibiotics has led to the emergence of antibiotic-resistant bacterial strains.2 In response to this growing threat, the World Health Organization (WHO) published a list of priority antibiotic-resistant pathogens in 2017, classifying them into critical, high, and medium priority groups.3 Alarmingly, some of these pathogens have developed resistance to nearly all available antibiotics, posing a severe threat to human health and survival.4Owing to their unique structural characteristics, Gram-negative bacteria exhibit a higher propensity for developing antibiotic resistance compared to Gram-positive bacteria, contributing to increased morbidity and mortality rates worldwide.5 Notably, the majority of the WHO-designated priority resistant pathogens belong to the Gram-negative group. These bacteria possess a distinctive three-layered cell envelope, a key factor in their resistance mechanisms. The outermost layer, the outer membrane (OM), is a defining feature of Gram-negative bacteria and serves as a protective barrier against external threats.6 The inner leaflet of the OM consists of phospholipids, while the outer leaflet contains lipopolysaccharides (LPS), which are known to trigger endotoxin shock in humans.7 Additionally, the OM is embedded with outer membrane proteins (OMPs), which allow small molecules like amino acids and sugars to pass through. Beneath the OM lies the peptidoglycan cell wall, a rigid exoskeleton composed of repeating units of N-acetylglucosamine and N-acetylmuramic acid, which maintains cell shape and structural integrity.8 The innermost layer is the inner membrane (IM), a phospholipid bilayer that not only offers structural support but also plays a crucial role in molecular transport, biosynthesis, and cellular division by anchoring DNA and facilitating sister-chromatid separation.9 The OM plays a pivotal role in bacterial resistance to a wide range of antibiotics, including β-lactams, quinolones, and polymyxins. Structural modifications in the OM, such as alterations in porin hydrophobicity or mutations in membrane components, can significantly reduce antibiotic permeability and enhance resistance.10 In contrast, Gram-positive bacteria lack this protective OM, rendering them generally more susceptible to antibiotics.11–13 Consequently, treatment options for infections caused by multidrug-resistant Gram-negative bacteria are becoming increasingly limited.
Gram-negative bacteria are responsible for severe infections in humans, particularly in individuals with weakened or underdeveloped immune systems, such as neonates, the elderly, and patients undergoing surgery or cancer treatments. These infections are often associated with high morbidity and mortality rates.14 Antibiotic-resistant infections caused by Gram-negative bacteria have emerged as one of the most formidable challenges in clinical medicine.15 Notably, drug-resistant Gram-negative bacteria are the primary pathogens responsible for ventilator-associated pneumonia, catheter-associated bloodstream infections, and other intensive care unit (ICU)-acquired sepsis, including urinary tract infections.16 Alarmingly, recent WHO reports on novel antibiotic formulations indicate that of the 50 new antimicrobial agents currently under development, only a small fraction exhibit efficacy against Gram-negative bacteria. In response to the growing threat of antibiotic resistance, various strategies have been explored to combat Gram-negative and multidrug-resistant bacteria via diverse molecular mechanisms. These approaches include the development of novel antibiotics,17 derivatives of existing antibiotics,18,19 antibiotic adjuvants,20 antimicrobial peptides derived from mammals and other organisms21 and lipopeptides and its assembly.22–24 Among these approaches, antimicrobial peptides designed from venom peptides have demonstrated considerable promise due to their potent therapeutic properties.25,26
Nevertheless, despite initial efficacy, bacterial populations eventually develop resistance, reinforcing concerns about the so-called “post-antibiotic era”—a looming global health crisis that underscores the urgent need for innovative antimicrobial agents to combat drug-resistant Gram-negative bacteria.27 In recent decades, rapid advances in nanotechnology have provided promising alternatives for antimicrobial therapy.28 Among these, carbon nanomaterials (CNMs) have attracted significant attention due to their diverse synthesis methods, unique structural features, favourable physicochemical properties, and relatively high biocompatibility. These materials are primarily composed of carbon atoms, which can form single bonds through sp3 hybridization as well as stable double and triple bonds through sp2 and sp hybridization, enabling the formation of various allotropes with distinct structures and properties.29 Notable examples of CNMs include zero-dimensional graphene quantum dots, carbon quantum dots (CQDs), one-dimensional carbon nanotubes, and two-dimensional graphene. As research on CNMs has progressed, their intrinsic physicochemical properties, such as large surface area and nanoscale dimensions, have been found to confer unique antibacterial capabilities. These attributes enable CNMs to inhibit or eliminate microbial populations while minimizing the risk of bacterial resistance. Consequently, their application has broadened the scope of nanomaterial-based antimicrobial research, offering novel strategies for the development of effective antibacterial agents. Furthermore, CNMs are relatively easy to synthesize, require inexpensive and abundantly available raw materials, and hold significant potential for antimicrobial therapy.
Despite the growing body of research on the antibacterial properties of CNMs, considerable challenges remain in their clinical application, particularly in their use against Gram-negative bacteria. Given the continuous advancements in antimicrobial nanomaterials, it is essential to systematically review and evaluate the potential of carbon-based nanomaterials for addressing Gram-negative bacterial infections. This review focuses on the research progress of four key CNMs—graphene quantum dots (GQDs), carbon dots (CDs), carbon nanotubes (CNTs), and graphene—with respect to their antibacterial activity against Gram-negative bacteria. The discussion encompasses their synthesis methods, physicochemical properties, antibacterial mechanisms, and current biomedical applications. Finally, the challenges associated with the practical application of these CNMs in antimicrobial therapy are examined, along with potential future research directions.
2. Preparation of antibacterial carbon nanomaterials
The antibacterial properties of CNMs are intricately linked to their microstructure and surface chemical characteristics, which are primarily influenced by their preparation methods. These methods can generally be categorized into two main approaches: top-down and bottom-up.2.1 Top-down preparation methods
The top-down method involves the breakdown of larger carbon materials into nanoscale particles or structures through physical or chemical processes. The fundamental principle of this approach is to begin with bulk materials and progressively reduce their size via physical actions or chemical reactions, ultimately yielding nanostructures.30To facilitate a comprehensive comparison of various top-down synthesis techniques, Table 1 summarizes the characteristics, applicable material types, and technical advantages of several commonly used methods. The top-down methods for preparing antibacterial CNMs each exhibit distinct characteristics. For instance, laser ablation offers high precision and purity but comes at the cost of being energy-intensive and expensive. The arc discharge method, while particularly effective for producing CNTs with high crystallinity, is complex and requires substantial energy input. In contrast, electrochemical methods are notable for being environmentally friendly, cost-effective, and efficient, though there is a need for further improvements in QYs and purity. Ultrasonic synthesis, which is simple, non-toxic, and inexpensive, also faces challenges related to size control and yield consistency.59,60 Overall, each preparation method possesses distinct advantages in terms of product properties, controllability, and economic feasibility. The choice of a method should be based on the specific requirements of the intended application.
| Preparation method | Main features | Preparable nanostructures | Advantages | References |
|---|---|---|---|---|
| Laser ablation | High-energy laser pulses vaporize or decompose carbon materials, forming nanoscale particles | Carbon dots, carbon nanotubes, graphene quantum dots | High precision, high purity, good controllability | 39 and 54 |
| Electric arc discharge | A high current between graphite electrodes induces evaporation and re-condensation of carbon, forming nanostructures | Carbon nanotubes, carbon dots, graphene nanosheets | High efficiency, high product quality, simple equipment | 55 and 56 |
| Electrochemical method | Voltage applied to a carbon electrode in an electrolyte induces exfoliation, forming carbon nanostructures | Carbon dots, carbon nanotubes, graphene oxide | Environmentally friendly, efficient, easy to operate | 46 and 57 |
| Ultrasonic synthesis | Microbubbles generated by pressure waves collapse, creating shear forces that break down carbon materials into nanoscale sizes. Forces, breaking down large carbon-based materials into nanoscale sizes | Carbon dots, graphene quantum dots | Easy to operate, non-toxic | 53 and 58 |
2.2 Bottom-up synthesis methods
The bottom-up approach involves the synthesis of CNMs through the aggregation or chemical reactions of atoms, molecules, or smaller units. Compared to top-down methods, this approach is more effective in producing CNMs with uniform sizes and well-defined morphologies.61The selection of carbon precursors plays a crucial role in hydrothermal synthesis. These precursors range from natural biomaterials, such as apple juice, hemicellulose, bamboo leaves, cabbage, and black tea, to waste materials, including discarded polytetrafluoroethylene (PTFE) from syringes and hyaluronic acid.63–67 Under high-temperature and high-pressure conditions, these precursors undergo dehydration, polymerization, carbonization, and passivation, ultimately forming CQDs.68,69 This approach underscores the economic, environmentally friendly, and non-toxic nature of hydrothermal synthesis by enabling the repurposing of waste materials.70,71
Significant progress has been made in utilizing hydrothermal synthesis to produce antibacterial CDs. For instance, Ghataty et al.72 synthesized highly fluorescent bovine serum albumin (BSA)-derived carbon dots (B-CDs) using BSA as a precursor. These BCDs demonstrated strong antibacterial activity, effectively inhibiting the growth of various wound-infecting pathogens. Their antibacterial mechanism is likely associated with the presence of hydroxyl functional groups on the BCDs' surface, which disrupt the integrity of bacterial cell membranes. Similarly, Wei et al.73 synthesized nitrogen-doped carbon dots (N-CDs) using folic acid and acetamide through hydrothermal synthesis. These N-CDs exhibited selective recognition and inhibition of various microorganisms, demonstrating particularly strong antibacterial effects against E. coli. Their antibacterial activity is attributed to disruption of bacterial cell membranes, which compromises bacterial viability. Ma et al.74 employed a microwave-assisted glycolysis of polyethylene terephthalate (PET) waste, combined with hydrothermal synthesis, to prepare N-CDs. These N-CDs exhibited excellent fluorescence properties and broad-spectrum antibacterial activity against both Gram-positive and Gram-negative bacteria. Their antibacterial mechanism is primarily attributed to electrostatic interactions between the positively charged N-CDs surfaces and negatively charged bacterial cell membranes, ultimately leading to membrane rupture.
The hydrothermal synthesis method holds great promise for the production of antibacterial CDs. By carefully selecting carbon sources and doping elements, both antibacterial activity and fluorescence properties of CQDs can be tailored to meet specific application requirements. These antibacterial CDs exhibit significant potential in biomedicine, environmental protection, and water treatment. However, due to the high-temperature and high-pressure conditions involved in the process, strict safety measures must be implemented to ensure both effective and safety in large-scale applications.68,75
A key advantage of the microwave-assisted method is its compatibility with a wide range of carbon sources, including small organic molecules such as citric acid and urea, as well as natural biomaterials like orange peel powder and mango leaves.77–79 For example, Lee et al.77 synthesized nitrogen-doped CQDs with a high QY (22.26%) within 30 minutes using citric acid and urea as precursors. The resulting CQDs demonstrated excellent photostability and low cytotoxicity, making them particularly suitable for fluorescent labelling of bacterial cells. Similarly, Yalshetti et al.80 utilized microwave-assisted synthesis with hibiscus leaf extract as a carbon source to produce CQDs exhibiting antibacterial, anti-inflammatory, and wound-healing properties, thereby expanding their potential for biomedical applications.
Furthermore, microwave-assisted synthesis holds great promise for the development of functional CQDs tailored for specific applications. By precisely controlling synthesis conditions, CQDs can be optimized for use in metal ion detection, bioimaging, and drug delivery.68 For instance, Osman et al.81 synthesized nitrogen and sulphur co-doped carbon quantum dots (N–S/CQDs) from alfalfa biomass using a microwave-assisted synthesis, which demonstrated high sensitivity for detecting nitrofurazone in pharmaceutical samples. This process not only ensures rapid synthesis but also aligns with the principles of green chemistry by being environmentally friendly and cost-effective. Despite the significant advancements achieved with microwave-assisted synthesis, challenges remain in precisely controlling the size and uniformity of the CQDs. Future research should focus on elucidating the underlying mechanisms of microwave-driven synthesis and optimizing reaction conditions to achieve greater precision in CQDs size control.
Table 2 provides a comparative overview of the key characteristics, commonly used template materials, and advantages of various bottom-up synthesis methods. The bottom-up approach for the fabrication of antibacterial CNMs involves the spontaneous formation of nanostructures under controlled chemical reaction conditions. This method is highly versatile and adaptable, offering a wide range of applications. Among the various techniques, the hydrothermal and solvothermal methods are favoured for their environmental benefits, low cost, and tuneable reaction conditions. However, these methods present challenges in terms of controlling the size of the synthesized products and often require extended synthesis times. In contrast, microwave-assisted synthesis is efficient, scalable, and capable of producing high yields. However, it may be hindered by high energy consumption, which could result in increased operational costs. Pyrolysis and carbonization methods are simple, cost-effective, and compatible with a broad range of raw materials, but they require further optimization to achieve uniformity in product size and yield. The chemical vapor deposition method, while capable of producing high-quality CNMs, necessitates advanced equipment and incurs significant operational costs.59,60 Overall, while the bottom-up approach enables the controllable synthesis of various carbon nanostructures with relatively lower energy consumption, further research is required to enhance size control and reduce production costs.
| Preparation method | Main features | Prepared nanostructures | Advantages | References |
|---|---|---|---|---|
| Hydrothermal method | Precursors dissolved in water or organic solvents undergo high-temperature, high-pressure reactions | Carbon dots, graphene, carbon nanotubes | Cost-effective, highly controllable, non-toxic, environmentally friendly | 85–87 |
| Solvothermal method | High-temperature, high-pressure reactions in non-aqueous or organic solvents synthesize carbon nanomaterials | Carbon dots, graphene, porous carbon materials | Broad applicability, controllable conditions, cost-effective, eco-friendly | 88–90 |
| Microwave method | Microwave radiation uniformly heats organic matter, inducing carbonization for nanoscale carbon production | Carbon dots, graphene | High efficiency, controlled particle size, scalable, eco-friendly | 91 and 92 |
| Pyrolysis | Thermal decomposition of carbon sources in a vacuum or inert atmosphere | Carbon dots, graphene, carbon nanotubes, porous carbon materials | Cost-effective, versatile raw materials, flexible, easy operation | 93–96 |
| Carbonization | Organic materials decompose under high-temperature, anaerobic conditions to form carbon structures | Carbon dots, porous carbon materials | Cost-effective, free of harmful chemicals | 97 and 98 |
| Chemical vapor deposition | Gaseous precursors decompose at high temperatures, depositing carbon nanomaterials on a substrate | Carbon dots, graphene, carbon nanotubes | Scalable production, high quality, excellent controllability | 99 and 100 |
3. Structural properties of carbon nanomaterials and their antibacterial activity against Gram-negative bacteria
3.1 Graphene quantum dots
Graphene quantum dots are nanoscale fragments of graphene, typically measuring only a few nanometers in all three dimensions, classifying them as quasi-zero-dimensional materials.101 GQDs integrate the exceptional physicochemical properties of graphene with the distinctive characteristics of quantum dots. Unlike bulk graphene, GQDs exhibit pronounced quantum confinement effects and surface defects, endowing them with unique optical and electronic properties, such as wavelength-dependent fluorescence emission, high photostability, excellent biocompatibility, and ease of conjugation with biological molecules.102,103 These attributes render GQDs highly promising for a wide range of applications, including nonlinear optics, magnetic media, catalysis, pharmaceuticals, and functional materials.104,105 Recent advancements have further highlighted the potential of GQDs in antimicrobial applications, particularly against Gram-negative bacteria.The antibacterial activity of GQDs against Gram-negative bacteria underscores their potential as effective antimicrobial agents. One key factor influencing their antibacterial efficacy is functionalization, which enhances their physicochemical properties. Luo et al.106 synthesized adenosine-functionalized graphene quantum dots (A-GQDs) via a two-step microwave-assisted method, yielding materials with strong fluorescence characteristics. Notably, A-GQDs exhibited remarkable biocompatibility and two-photon fluorescence for cell imaging. Under white light illumination, A-GQDs displayed selective antibacterial activity, showing no effect on S. aureus but almost completely inhibiting the growth of E. coli, demonstrating their specificity against Gram-negative bacteria.
Moreover, multi-element-doped GQDs have exhibited substantial antibacterial effects against Gram-negative bacteria. Huang et al.107 developed halogen/nitrogen co-doped graphene quantum dots (X/N-PGQDs) as light-emitting diode-assisted bactericidal agents. These X/N-PGQDs demonstrated a MIC of less than 0.5 μg mL−1 against Gram-negative bacteria, indicating their potent antimicrobial efficacy. Additionally, GQDs-based composites have shown promise in combating Gram-negative bacteria. For instance, Yu et al.108 successfully conjugated AgNPs onto the surface of GQDs, forming a GQDs-AgNPs composite with broad-spectrum antibacterial properties. Under 450 nm excitation, this composite exhibited significantly enhanced bactericidal activity against Gram-negative bacteria compared to GQDs alone.
Research further suggests that integrating GQDs with other functional materials can significantly improve their antibacterial efficacy. Yin et al.109 synthesized GQDs using a bottom-up approach and combined them with alendronate (ALN) and AgNPs to create the composite material ALN-GQDs-Ag. This composite exhibited strong antibacterial activity against Streptococcus mutans biofilms, highlighting its potential for practical antimicrobial applications. Furthermore, the antibacterial performance of GQDs composites can be optimized by fine-tuning the GQDs-to-material ratio. For example, Yang et al.110 synthesized a novel photocatalyst, GQDs/NH2-MIL-125, using a solvothermal method. Their findings revealed that a 2% GQDs loading in the composite achieved 92% antibacterial activity against E. coli, underscoring the crucial role of GQDs concentration in optimizing antibacterial performance.
3.2 Carbon dots
Carbon dots are fluorescent carbon-based nanomaterials characterized by a core composed of sp2-hybridized carbon atoms and a surface composed of sp3-hybridized carbon atoms. These unique nanostructures exhibit exceptional electrical and chemical stability, low biotoxicity, environmental friendliness, and strong optical properties. Typically, CDs have a size of less than 10 nm, which enables them to closely resemble biological entities such as proteins, DNA, ion channels, and glomerular filtration barriers in the human body. Furthermore, the surface of CDs, depending on the synthesis method, is often functionalized with groups such as carboxyl, carbonyl, hydroxyl, and other functional moieties derived from precursor materials. These surface modifications contribute to their high-water solubility and biocompatibility.111 Consequently, CDs are increasingly favored for various biomedical applications, including drug delivery, gene therapy, photosensitization, and antibacterial therapies.112,113 Additionally, ongoing research continues to highlight their potential in multifunctional diagnostic platforms, cellular and bacterial bioimaging, and the advancement of nanomedicine.114–116In recent years, there has been growing interest in the antibacterial activity of CDs. As research has progressed, the mechanisms underlying their antibacterial properties have become a primary focus. Early studies emphasized light-activated antibacterial properties, while more recent advancements have successfully overcome the limitations imposed by lightdependence. Moreover, strategies for functional modification are being explored to address the emerging issue of antibiotic resistance. This evolving field has made significant strides, as evidenced by the recent advances summarized in the timeline of antibacterial CDs research (Fig. 1).
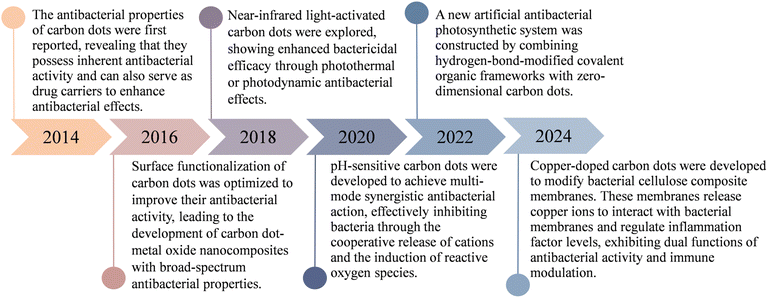 | ||
| Fig. 1 Research progress on antibacterial carbon dots.117–122 | ||
A pioneering study on the visible and natural light-activated antibacterial properties of CDs demonstrated that ethylenediamine (EDA)-functionalized carbon quantum dots (EDA-CQDs) could effectively inhibit E. coli both in suspension and on agar surfaces under visible or natural light.123 Experimental results revealed a significant reduction in bacterial colony counts on agar plates following treatment with light-activated EDA-CQDs, indicating a strong inhibitory effect on E. coli growth in culture media. Subsequently, Pandey et al.124 synthesized antibacterial CDs that exhibited antimicrobial activity independent of light activation. Using citric acid as the carbon source and β-alanine as the surface passivating agent, they successfully produced CDs via microwave-assisted synthesis. Their findings demonstrated that these CDs inhibited the growth of several Gram-negative bacterial strains, including E. coli, Salmonella, Pseudomonas, Agrobacterium, and Pectinobacterium. Notably, these CDs retained their antibacterial efficacy even after prolonged incubation in the absence of light, addressing the need for CDs with inherent antibacterial properties that do not rely on external light activation. Furthermore, unlike conventional antibiotics, these CDs exhibited a significantly lower tendency to induce bacterial resistance, offering a promising strategy to mitigate the global challenge of antibiotic resistance. Further advancing this field, Wu et al.125 synthesized levofloxacin-functionalized carbon dots (L-CDs) using a simple one-pot hydrothermal method. These L-CDs exhibited potent antibacterial activity, with a MIC of 0.125 μg mL−1 against E. coli, and demonstrated strong inhibitory effects against antibiotic-resistant bacterial strains. Moreover, L-CDs displayed excellent biocompatibility both in vitro and in vivo, effectively overcoming the limitations associated with traditional antibiotics in clinical applications. The antibacterial properties of CDs can be attributed not only to their nanoscale size and unique surface chemistry but also to the presence of various functional groups introduced during synthesis. To enhance both antibacterial efficacy and biocompatibility, CDs must be functionalized with specific chemical groups. These modifications play a critical role in determining the primary antibacterial mechanisms, thereby optimizing their effectiveness.126 Based on the distinct properties of different functional groups, researchers have developed a wide range of functionalized CDs to address the diverse challenges associated with combating Gram-negative bacterial infections. Table 3 provides a summary of the effects of various functional group modifications on the antibacterial performance of CDs.
| Type | Functional group formula | Antimicrobial mechanism | Antimicrobial effect | References |
|---|---|---|---|---|
| Quaternary ammonium group | –NR4+ | Positively charged nitrogen interacts with negatively charged bacterial surfaces, disrupting membranes and causing cell death | Fast sterilization, most effective at pH ≥ 9 | 127 and 128 |
| Quaternary phosphonate group | –PR4+ | Larger ionic radii and stronger polarization of phosphorus enable better adsorption to bacterial surfaces | Broad applicability, effective in pH range 2–12 | 129 and 130 |
| Guanidino group | –C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) NH)NH2 NH)NH2 |
Protonation of the guanidine group allows electrostatic interactions, rupturing bacterial membranes and leading to cell death | Highly effective against drug-resistant strains | 131 and 132 |
| N-Halamine group | –NX (X = Cl, Br) | Positively charged halogens transfer to bacterial receptors, inhibiting growth | Renewable, efficient, broad-spectrum sterilization | 133 and 134 |
3.3 Carbon nanotubes
Carbon nanotubes are one-dimensional nanomaterials composed of graphene sheets, where carbon atoms are arranged in a sp2 hybridized structure. These sheets are coiled around a central axis at a specific helical angle, forming seamless, hollow tubes. Depending on the number of graphene sheets, CNTs can be classified as single-walled carbon nanotubes (SWCNTs) or multi-walled carbon nanotubes (MWCNTs). Due to their exceptional tensile strength, mechanical durability, large surface area, light-weight, chemical stability, superior transmembrane ability, and remarkable electronic, optical, and magnetic properties,135–137 CNTs have significant potential for applications in drug delivery,138 biosensors,139 and tumour hyperthermia.140 Furthermore, the discovery of their antibacterial activity has sparked growing interest in their use for environmental and human health applications.141The antibacterial efficacy of CNTs, particularly against Gram-negative bacteria, is closely related to their physicochemical properties. The synthesis method significantly influences the types and concentrations of impurities in CNTs, which in turn affect their antibacterial performance. For example, Kang et al.142 investigated the interactions between purified and raw SWCNTs and the model strain E. coli K12. Their results demonstrated that highly purified SWCNTs exhibited stronger antibacterial activity than raw CNTs. Additionally, factors such as surface area, dispersibility, and other physicochemical characteristics influence the antibacterial activity of CNTs. CNTs with better dispersibility and larger surface areas are more effective in interacting with bacteria, enhancing direct contact and uptake. For instance, SWCNTs, with smaller diameters and lengths compared to MWCNTs, offer a larger surface area, making them more efficient in inhibiting E. coli growth.143 Functionalization plays a pivotal role in modulating the antibacterial properties of CNTs. Pasquini et al.144 examined nine different functionalized SWCNTs and their interactions with E. coli. Their findings revealed that functionalization could indirectly influence the antibacterial activity of CNTs by altering their physicochemical properties, with the effects varying depending on the functional group applied. Additionally, combining CNTs with other materials to create composite structures has proven to be an effective strategy for enhancing their antibacterial properties. For example, Khalil et al.56 synthesized AgNPs-CNTs using an arc discharge method. These composites exhibited significantly enhanced antibacterial performance compared to individual AgNPs alone.
Furthermore, optimizing the loading ratio of metal nanoparticles in CNT-based composites is essential for maximizing antibacterial activity. Assis et al.145 employed a microwave-assisted hydrothermal synthesis to load various concentrations (1, 5, and 10 wt%) of copper nanoparticles (CuNPs) onto MWCNTs. Their findings indicated that CNT-Cu 10, with a 10 wt% copper load, exhibited the highest antibacterial effect, with a MIC of 64 μg mL−1 against E. coli. Additionally, novel synthesis methods aimed at optimizing the structure and surface properties of CNTs have been shown to significantly improve their antibacterial performance. For instance, Yan et al.146 used a sacrificial template method to prepare oxygen-modified nitrogen-doped CNTs (O-NCNTs-6), which demonstrated excellent antibacterial properties, achieving inactivation rates of 96% and 93% for S. aureus and P. aeruginosa, respectively, when tested on stainless steel electrodes in PBS. Furthermore, external factors, such as the dispersing medium, bacterial species, and the interaction method between CNTs and bacteria, can also impact the antibacterial activity of CNTs against Gram-negative bacteria.
3.4 Graphene
Graphene is a two-dimensional carbon nanomaterial composed of a single layer of carbon atoms bonded via sp2 hybridization, forming a hexagonal crystal structure. It serves as the fundamental structural unit for other carbon allotropes, including fullerene (zero-dimensional), CNTs (one-dimensional), and graphite and diamond (three-dimensional). The thickness of a single graphene layer is approximately 0.35 nm, conferring exceptional stability and endowing the material with unique physical properties. These properties include outstanding thermal and mechanical performance, exceptional electronic conductivity, and remarkable optical characteristics.147,148 Additionally, graphene possesses a high surface area, excellent catalytic activity, and notable antibacterial properties, rendering it highly suitable for applications across diverse fields, including electronics, information technology, energy, physics, biomedicine, and materials science.149–151 Among these applications, graphene-based nanomaterials have garnered significant attention in the field of biomedicine. Graphene derivatives, such as GO, reduced graphene oxide (rGO), GQDs, and graphene-based nanocomposites, have been extensively explored for use in biosensors,152,153 bioimaging,154–156 drug delivery,157,158 and photothermal therapy159 among other applications. Furthermore, graphene nanomaterials have shown considerable promise as antimicrobial agents. To date, numerous studies have demonstrated the effective antibacterial activity of graphene-based nanomaterials against Gram-negative bacteria. The first reports on the antimicrobial properties of graphene nanomaterials specifically targeted Gram-negative bacteria. In 2010, Hu et al.160 were the first to report the antibacterial properties of graphene, discovering that GO and rGO could inhibit E. coli growth by over 90%. Since then, research has expanded to include a variety of graphene composites engineered to target Gram-negative bacteria. For instance, Liu et al.161 prepared GO coatings on polymer matrices and investigated their antibacterial activity against both E. coli and S. aureus. The results indicated that thinner GO coatings exhibited stronger antibacterial activity against E. coli, while thicker coatings were more effective against S. aureus. In another study, Matharu et al.162 successfully fabricated antibacterial polymer nanocomposite fibers containing GO nanosheets using a pressure-assisted spinning method. These GO-based nanocomposites demonstrated significant antibacterial potential, with bacterial reduction rates ranging from 46% to 85%, and the nanocomposite containing 8% GO showing the highest biological activity. This highlights the importance of combining novel fabrication techniques with material composites to enhance the antibacterial performance of graphene-based nanomaterials. Thangaraj et al.94 employed onion peel waste to prepare rGO through a one-step pyrolysis method. The study demonstrated that rGO-WiC, produced with the aid of a ferrocene catalyst, exhibited an inhibition zone of 566 mm2 against K. pneumoniae at a concentration of 200 μg mL−1, with the largest antibacterial effect observed against E. coli (inhibition zone area of 647 mm2). Notably, the antibacterial activity of rGO-WiC was found to be 38.5% greater than that of rGO produced without a catalyst. Moreover, optimizing the structure and functionalization of composite materials has proven to be an effective strategy for enhancing the antibacterial properties of graphene-based nanomaterials. For example, Guo et al.163 utilized GO as a template to develop a copper–iron sulfide nanocomposite (GO/CuFeSx NC) and tested various Cu/Fe ratios to identify the most effective antibacterial nanoenzyme combination. Under near-infrared (NIR) light stimulation, at a temperature of 45 °C, the GO/Cu3Fe1Sx nanocomposite at a concentration of 40 μg mL−1, combined with 100 μM hydrogen peroxide (H2O2), completely eradicated E. coli, demonstrating remarkable antibacterial activity.4. Antimicrobial mechanism of carbon nanomaterials
The antibacterial efficacy of CNMs against Gram-negative bacteria is primarily attributed to their distinctive physicochemical properties, including specific morphology, size, and surface functionalization. Extensive research has elucidated several widely recognized antibacterial mechanisms, including physical disruption of bacterial membranes, oxidative stress induction, photothermal and photodynamic effects, lipid extraction, inhibition of bacterial energy metabolism, suppression of nucleic acid synthesis, DNA damage, bacterial encapsulation and isolation, and catalytic sterilization (Fig. 2).4.1 Physical/mechanical disruption
CNMs of varying dimensionality can induce mechanical damage to bacterial outer membranes or cell walls, exhibiting both bactericidal and bacteriostatic effects. Zero-dimensional, one-dimensional, and two-dimensional CNMs interact with bacterial membranes in ways analogous to point, line, and planar interactions, respectively. The degree of mechanical disruption is closely associated with the nanomaterials' structural characteristics. Chatterjee et al.164 demonstrated that carboxylated nanodiamonds, upon adhesion to the E. coli cell wall, altered the structural integrity of membrane proteins, ultimately leading to membrane rupture and bacterial cell death. Similarly, Wang et al.165 synthesized CQDs from Artemisia argyi (A-CDs) using a simulated smoking method. Their findings revealed that A-CDs exhibited selective antibacterial activity by inhibiting enzyme functions essential for Gram-negative bacterial cell wall synthesis. Notably, A-CDs selectively compromised the cell walls of Gram-negative bacteria without affecting those of Gram-positive bacteria, highlighting their targeted specificity. Kang et al.142,143 investigated the interaction between CNTs and bacteria, demonstrating that direct contact with CNTs induced morphological deformations, membrane disruption, intracellular leakage, and ultimately, bacterial cell death. These findings underscore the potent antibacterial properties of CNTs. For graphene materials (GMs), Wang et al.166 employed molecular dynamics simulations to investigate graphene's cutting action, revealing that graphene sheets could disrupt phospholipid bilayers due to their sharp, sheet-like structures. Subsequent studies confirmed that when bacterial cells came into contact with GMs, the sharp edges of graphene sheets physically lacerated bacterial membranes, leading to cytoplasmic leakage and bacterial inactivation.167 Furthermore, the orientation and angle of graphene sheets during contact significantly influenced their bactericidal efficiency. Shangguan et al.168 reported that CDs synthesized from branched polyethylenimine (bPEI25000-CDs) exhibited notable antibacterial activity. Specifically, positively charged ALA-bPEI25000-CDs strongly interacted with bacterial cell membranes, leading to structural damage and cellular dysfunction. Scanning electron microscopy (SEM) images revealed that E. coli cells treated with these CDs displayed rough, deformed surfaces, ultimately resulting in membrane rupture. Similarly, Ruan et al.169 synthesized N–S/CQDs, which exhibited broad-spectrum antibacterial activity. This activity was closely linked to their nanoscale size, surface functional groups, and the ability to generate ROS. Mechanical disruption was identified as a key antibacterial mechanism, with N–S/CQDs effectively penetrating bacterial membranes, inducing structural damage, and inhibiting bacterial proliferation. The incorporation of amine functional groups conferred a positive charge to the N–S/CQDs, enhancing electrostatic interactions with negatively charged bacterial membranes, and thereby improving antibacterial efficacy. By fine-tuning the surface properties and dimensions of N–S/CQDs, their antibacterial performance was further optimized, offering new strategies for developing advanced antimicrobial nanomaterials. Zhao et al.170 observed pronounced morphological changes in S. aureus cells treated with vancomycin–malic acid–amines carbon dots (VMA-CDs), including pore formation and membrane rupture, indicating severe bacterial damage. These findings confirmed that VMA-CDs exert their antibacterial effects by disrupting bacterial membrane integrity. Furthermore, Wang et al.171 synthesized CDs from ornidazole via microwave radiation and investigated their selective antibacterial mechanisms against Porphyromonas gingivalis (P. gingivalis). The study revealed that CDs with specific molecular weights (1–10 kDa) exhibited strong antibacterial effects against P. gingivalis, while having minimal impact on Streptococcus mutans. This selectivity was attributed to the nanoscale dimensions of the CDs, which facilitated membrane penetration, membrane integrity disruption, and the release of antibacterial functional groups, such as nitro groups, effectively eliminating the target bacteria. Li et al.172 showed that carbon dots with specific surface functional groups could cause significant morphological changes in bacterial cells. Their study revealed that the positively charged carbon dots could strongly interact with bacterial cell membranes, leading to structural damage and cellular dysfunction. Scanning electron microscopy (SEM) images indicated rough and deformed surfaces of E. coli cells treated with these carbon dots, ultimately resulting in membrane rupture and cell death. Furthermore, it was observed that carbon dots with amine functional groups could enhance electrostatic interactions with negatively charged bacterial membranes, thereby improving antibacterial efficacy. By adjusting the surface properties and dimensions of carbon dots, their antibacterial performance could be further optimized, providing new strategies for developing advanced antimicrobial nanomaterials. Mortimer et al.173 indicated that CNTs cause mechanical damage to bacterial cells through direct contact, leading to membrane rupture and cytoplasmic leakage. The physical properties of carbon nanomaterials, such as their sharp edges and high surface area, enable them to interact closely with bacterial cell membranes and exert physical disruption effects. This not only damages the bacterial cell membrane but also compromises the integrity of the cell wall, thereby enhancing the antibacterial efficacy of carbon-based nanomaterials.4.2 Oxidation stress
The induction of oxidative stress by CNMs is widely recognized as an effective antibacterial mechanism. This process disrupts the oxidant–antioxidant balance within bacterial cells, leading to cellular damage. Oxidative stress can occur through two primary pathways: ROS-dependent and ROS-independent mechanisms. The ROS-dependent pathway is initiated by the excessive accumulation of intracellular ROS, such as superoxide anions (˙O2−), hydrogen peroxide, hydroxyl radicals (˙OH), and singlet oxygen. Numerous studies have demonstrated that elevated ROS levels can cause protein inactivation, lipid peroxidation, mitochondrial dysfunction, and membrane degradation, ultimately resulting in bacterial necrosis or apoptosis.174 In contrast, ROS-independent oxidative stress refers to the damage or oxidation of essential cellular structures or intracellular components without the direct involvement of ROS. This pathway represents an alternative oxidative stress response that does not rely on ROS generation. For instance, graphene and its derivatives can induce oxidative stress by oxidizing intracellular glutathione, thereby increasing ROS levels and promotes the oxidation of fatty acids. This oxidation leads to the formation of lipid peroxides, triggering free radical reactions, resulting in the destruction and lysis of biological membranes and induce bacterial cell death. Gurunathan et al.175 in their investigation of the antibacterial properties of graphene against P. aeruginosa, reported a 3.8-fold and 2.7-fold increase in intracellular ROS levels in bacteria treated with GO and rGO, respectively. These findings confirm that graphene exerts its antibacterial effects via ROS-dependent oxidative stress. Similarly, other studies have proposed that oxidative stress is a key mechanism underlying the antibacterial properties of CNTs.143 Vecitis et al.176 demonstrated that the interaction between MWCNTs and E. coli led to an increase in glutathione oxidation within bacterial cells. The degree of oxidation was directly proportional to the concentration of metallic SWNTs, indicating that enhanced oxidative stress was responsible for the observed antibacterial activity. This suggests that CNTs interfere with bacterial metabolism through oxidative stress, leading to intracellular dysfunction and effective bacterial inhibition. Further research by Bing et al.177 explored the effects of CDs on E. coli growth and their underlying antibacterial mechanisms. The study revealed significant variations in ROS levels induced by three types of CQDs (S-CQDs, C-CQDs, and G-CQDs), indicating that endogenous ROS levels play a crucial role in triggering bacterial apoptosis and determining the antibacterial efficacy of these nanomaterials. Recent studies have provided deeper insights into the oxidative stress mechanism of carbon dots. Cui et al.178 demonstrated that spermidine-capped carbon dots (S-PCDs) could effectively induce intracellular ROS generation in Staphylococcus aureus. Their experiments showed that S–P/CDs, at a concentration equal to the MIC, elevated ROS production to 5.25-fold that of the control group. As the S–P/CDs concentration increased, ROS levels rose further, reaching 6.52-fold the control at four times the MIC. The study revealed that S–P CDs, with their high positive charge (+47.06 mV), could adsorb onto the negatively charged bacterial surface via electrostatic interactions. This interaction not only facilitated the generation of ROS but also induced significant oxidative stress, leading to cell membrane damage and eventual bacterial cell death. The ROS generation pathways were found to involve the activation of oxidative stress-related genes such as dmpI, narJ, and narK. The upregulation of these genes promoted increased oxidative stress through non-photonic pathways within the bacterial cells, further exacerbating membrane damage and cell death. Similarly, Rosato et al.179 found that GQDs exposed to blue light exhibited significantly enhanced antibacterial activity against E. coli. In their study, GQDs were administered to E. coli suspensions and treated with blue light. The results showed that blue light stimulation enhanced GQDs' antibacterial activity in a time-dependent manner attributed to ROS production by GQDs under blue light irradiation. Sun et al.180 discovered that CDs can efficiently generate ROS under irradiation, which can disrupt bacterial cell membranes and proteins. They found that nitrogen-doped CDs exhibited higher ROS generation efficiency when stimulated by blue light, resulting in stronger antibacterial effects. The study also highlighted the importance of CD-bacterial cell envelope interactions in the antibacterial process. CDs can specifically target and bind to bacterial cell surfaces, enhancing ROS generation and improving antibacterial efficacy. Recently, Xia et al.181 prepared nitrogen-doped carbon quantum dots to inactivate antibiotic resistant bacteria through spontaneous generation of extracellular and intracellular ROS, which was supported by the significant regulation of genes related to oxidative stress.4.3 Photothermal effect
Under NIR irradiation, CNMs exhibit exceptional photothermal conversion efficiency, rendering them highly effective in antibacterial applications. Various photothermal CNMs have been developed, characterized by strong NIR absorption and the ability to efficiently convert light energy into heat. The localized heat generation can effectively inhibit bacterial growth and prevent biofilm formation. In 2007, Kim et al.182 utilized E. coli as a model organism to explore the potential of CNTs in photothermal antibacterial therapy. This study was among the first to propose that CNMs exert antibacterial effects via photothermal mechanisms. The findings demonstrated that CNTs could self-assemble into clusters on bacterial surfaces and exhibit responsiveness to NIR laser stimulation. Upon NIR excitation, CNTs generated localized high temperatures, significantly reducing E. coli viability.Beyond CNTs, other CNMs, such as GO and rGO, also exhibit photothermal antibacterial properties. However, rGO demonstrates superior photothermal antibacterial efficiency compared to GO, primarily due to its higher intrinsic thermal conductivity and enhanced NIR absorption. Tan et al.183 investigated the antibacterial activity of rGO/Ag nanocomposites against E. coli and K. pneumoniae. Their results indicated that rGO/Ag nanocomposites exhibited enhanced synergistic antibacterial activity through photothermal effects. Under NIR irradiation, these nanocomposites not only generated heat but also induced membrane disruption and ROS production, thereby significantly improving antibacterial efficacy.
Additionally, in the case of CDs, modifying the core structure through transition metal doping or forming composites with metal nanoparticles has been identified as an effective strategy to enhance their photothermal antibacterial performance.184
4.4 Photodynamic effect
The principle of photodynamic therapy (PDT) was first discovered by Oscar Raab in Munich in 1900 through a serendipitous observation. PDT is based on three essential components: oxygen, a photosensitizer, and visible light (typically laser light). Upon irradiation with light of a specific wavelength, the photosensitizer becomes excited, initiating a photochemical reaction. The photosensitizer transfers energy to surrounding oxygen molecules, resulting in the generation of highly reactive singlet oxygen. This singlet oxygen subsequently interacts with nearby biological macromolecules through oxidative reactions, leading to cytotoxicity and ultimately bacterial cell death. CNMs have demonstrated efficacy as photosensitizers in PDT, exhibiting antibacterial activity through the light-dependent generation of ROS. Ristic et al.185 synthesized GQDs via electrochemical reactions and observed ROS production upon light excitation. These GQDs exhibited inhibitory effects against methicillin-resistant Staphylococcus aureus (MRSA) and E. coli, confirming their potential as PDT agents for antibacterial applications. In 2016, Kuo et al.186 utilized nitrogen-doped graphene quantum dots (N-GQDs) as photosensitizers to eliminate E. coli through PDT. Their findings revealed that GQDs with higher nitrogen doping levels (5.1%) exhibited significantly enhanced PDT effects and antibacterial activity compared to those with lower nitrogen content (2.9%). Building upon this, Kuo et al.187 further synthesized nitrogen-doped, amine-functionalized graphene quantum dots (AN-GQDs). These AN-GQDs demonstrated a higher capacity for ROS generation compared to unmodified GQDs, effectively combating multidrug-resistant bacteria through PDT. Zühlke et al.188 explored the use of amorphous carbon nanodots (CNDs) for photodynamic sterilization. By controlling the irradiation wavelength to exceed 290 nm, they minimized damage to bacterial outer membranes while enhancing immune responses. Due to their small particle size, CNDs were able to penetrate bacterial membranes and generate ROS internally under ultraviolet (UV) light, thus accelerating bacterial inactivation. Experimental results showed that photodynamic inactivation using UV light in conjunction with CNDs was ten times more effective than UV treatment alone. Bacterial membrane integrity was further confirmed through aggregation studies and atomic force microscopy (AFM). In a study by Kadyan et al.189 the unique properties of GQDs under UV and visible light irradiation were investigated. The results indicated that nitrogen-doped GQDs acted as efficient photosensitizers, generating various ROS, including H2O2, ˙OH, ˙O2−, and singlet oxygen. These ROS interacted with water and dissolved oxygen (O2), inducing oxidative stress within bacterial cells and leading to cytotoxic effects and bacterial death. Rosato et al.179 demonstrated that CNMs like GQDs exhibit excellent photothermal conversion efficiency under NIR irradiation, inhibiting bacterial growth and biofilm formation. Temperature increases enhanced bacterial membrane permeability, facilitating antimicrobial agent penetration. Simultaneously, the photodynamic effect of CNMs can generate ROS under light irradiation. For example, GQDs and CDs produce singlet oxygen and superoxide anions under specific wavelengths of light, which have strong oxidizing properties, causing oxidative damage to bacterial cells.4.5 Phospholipids extraction
CNMs can also interact with bacterial cells through lipid extraction mechanisms. Bacterial membranes, particularly those of Gram-negative bacteria, are composed of both an outer and an inner lipid bilayer, which are essential for functions such as nutrient uptake and the expulsion of toxic substances. Disruption of the structural and functional integrity of these membranes results in bacterial inhibition or cell death.190 In contrast to mechanical damage, which physically disrupts the bacterial cell wall or outer membrane, CNMs with abundant surface-active sites can penetrate the bacterial lipid bilayers. This penetration facilitates the extraction of phospholipids from the lipid bilayer onto the surface of the nanomaterials, thereby compromising membrane integrity and ultimately leading to bacterial death. In 2013, Tu et al.191 conducted both experimental and theoretical studies to elucidate the lipid extraction effect of GO nanosheets on bacterial membranes. Their findings revealed that GO nanosheets disrupted the bacterial membrane by directly extracting significant quantities of phospholipid molecules, leading to bacterial death. Molecular dynamics simulations further demonstrated that the two-dimensional planar structure of GO interacts with phospholipids through van der Waals forces and strong hydrophobic interactions, facilitating the adsorption of phospholipids onto the GO surface. Liu and Yi et al.192,193 further confirmed GO–phospholipid bilayer interactions, noting that lipid extraction is size-dependent, with larger GO sheets exhibiting stronger effects. Other studies identified lipid extraction as a primary mechanism for CNT-induced lipid bilayer damage.1944.6 Inhibition of bacterial energy metabolism
Bacteria rely on metabolic processes to convert nutrients into energy and to eliminate metabolic waste, essential for growth and survival. Unlike eukaryotic cells, bacteria lack mitochondria. ATPases involved in energy metabolism are embedded in the bacterial cell membrane. Many antibacterial agents exert their effects by inhibiting the synthesis of peptidoglycan (PG), a critical component of the bacterial cell wall, thereby disrupting bacterial energy metabolism. Recent studies show that CNMs can induce bacterial cell death by suppressing energy metabolism. Mashino et al.195 reported that fullerene derivatives accept electrons from the bacterial respiratory chain, reacting with oxygen to generate H2O2. This reaction inhibits oxygen uptake in the E. coli respiratory chain, suggesting that antibacterial activity of fullerene derivatives is mediated by the disruption of bacterial energy metabolism. In addition, the ability of GQDs to inhibit PG synthesis has been extensively studied. Xin et al.28 demonstrated that D-glutamic acid-functionalized GQDs (DGGs) can penetrate bacterial cell membranes and specifically target MurD ligase, an enzyme critical for PG biosynthesis. By inhibiting MurD's catalytic activity, DGGs disrupt cell wall synthesis, leading to membrane damage, cytoplasmic leakage, and ultimately bacterial death. DGGs exhibited high antibacterial activity and selective toxicity in vitro. In contrast, unmodified GQDs and L-glutamic acid-functionalized GQDs, although capable of penetrating bacterial membranes, showed weaker interactions with MurD ligase and did not exhibit significant antibacterial activity.4.7 Inhibition of nucleic acid synthesis and DNA damage
To effectively inhibit bacterial growth and reproduction at the molecular level, various antibacterial agents have been developed that target nucleic acid synthesis and DNA damage mechanisms. In the context of CNMs, it has been demonstrated that these materials can directly interact with DNA, thereby exerting antibacterial effects. Wu et al.196 demonstrated that GO can form complexes with DNA through coordination interactions, which protect the DNA from enzymatic degradation. However, in the presence of copper ions, GO can disrupt the DNA structure, leading to damage.197 Subsequent studies have confirmed that CNMs exhibit antibacterial activity by inhibiting nucleic acid synthesis, binding to DNA molecules, impeding DNA replication, and inducing DNA damage. Li et al.46 synthesized positively charged CQDs using a one-step electrochemical method with vitamin C as the precursor. These CQDs exhibited broad-spectrum antibacterial activity. Experimental results revealed that the positively charged CQDs were capable of diffusing into bacterial cells, disrupting the cell wall, and non-covalently binding to bacterial DNA and RNA. This interaction resulted in the loosening of the DNA structure, leading to the unwinding of the double helix, fragmentation of the genetic material, and subsequent cell inactivation, ultimately resulting in bacterial cell death even at very low concentrations. Moreover, studies have shown that spermidine-functionalized carbon quantum dots (Spd-CQDs), which exhibit a high positive surface charge (zeta potential of up to 60.6 mV), can interact multivalently with negatively charged bacterial membranes. This interaction causes severe cytoplasmic damage and leakage. Notably, at low concentrations, Spd-CQDs can bind to nucleic acids such as plasmid DNA and small interfering RNA (siRNA), disrupting gene expression regulation and exerting antibacterial effects.198 Nangan et al.67 synthesized fluorescent carbon dots (S-CDs) via a hydrothermal method using waste PTFE from discarded syringes as the precursor material. These S-CDs exhibited significant antibacterial activity by adhering to bacterial cell walls, penetrating the membrane, and binding to DNA. This interaction led to DNA unwinding, compromising bacterial structural integrity, and enhancing antibacterial efficacy. Furthermore, Yao et al.199 highlighted that quantum dots can damage DNA either through their intrinsic nanoscale morphology or via the release of metal ions. Their antibacterial activity is also attributed to the generation of ROS and the inhibition of DNA damage repair mechanisms.4.8 Wrapping isolation
In addition to the previously discussed antibacterial mechanisms, certain CNMs with large surface areas can inhibit bacterial proliferation by enveloping the bacteria, thereby isolating them from their environment. Researchers have found that GMs, with their hexagonal two-dimensional crystal structure, can act as a physical barrier, effectively separating bacteria from their surroundings and preventing the proliferation of Gram-negative bacteria. Although encapsulated bacteria retain their original shape, the separation from the external environment restricts the exchange of substances and information between bacterial cells. This disruption reduces their intracellular metabolic activity and vitality, ultimately inhibiting bacterial growth. Furthermore, the encapsulation effect of GMs can also induce damage to the bacterial cell membrane.200 Chen et al.201 proposed that GO exerts its antibacterial effects by wrapping and entangling microorganisms, which leads to depolarization of the bacterial cell membrane or electrolyte leakage. Furthermore, it was observed that graphene nanoplatelets (GNPs) larger than 5.2 nm can adhere to bacterial membranes due to the strong hydrophobic interactions between graphene and the lipid bilayer. Larger and better-dispersed GO nanoplatelets can more efficiently envelop bacteria, blocking active sites on the bacterial membrane, thereby inhibiting bacterial growth and reducing their survival potential.202 Studies on the antibacterial properties of CNTs have also demonstrated a similar encapsulation effect. Chen et al.203 confirmed that the antibacterial mechanism of CNTs is associated with both diameter-dependent perforation and length-dependent encapsulation of bacterial cell walls and membranes. Longer MWCNTs, by increasing the contact area with the bacterial cell wall, are more likely to encapsulate bacteria, thereby exhibiting enhanced antibacterial activity.4.9 Catalytic sterilization
Certain CNMs have been found to exhibit enzyme-like catalytic activities, enabling the conversion of H2O2 into highly reactive ˙OH. As a potent oxidizing agent, ˙OH induces oxidative stress responses that disrupt bacterial cell membranes, inactivate proteins, and cause DNA damage, ultimately interfering with bacterial growth and metabolic processes. This catalysis-based antibacterial mechanism presents new opportunities for the application of CNMs in antimicrobial research. Qi et al.204 synthesized iron-doped carbon dot nanozymes (Fe-CDs) using Eucommia ulmoides (a traditional Chinese medicinal herb) as a biomass-derived carbon source via a hydrothermal method. The peroxidase-like (POD) activity of Fe-CDs was confirmed by monitoring the oxidation of 3,3′,5,5′-tetramethylbenzidine (TMB). Furthermore, when Fe-CDs were combined with H2O2, a significant reduction in the characteristic absorption intensity of methylene blue (MB) at 665 nm was observed, further verifying their ability to catalyze the conversion of H2O2 into ˙OH. Antibacterial experiments demonstrated that the combination of Fe-CDs and H2O2 produced a significantly larger inhibition zone compared to either agent alone, achieving a bactericidal rate of 94.82%. In another study, Wang et al.205 developed copper-doped carbon dots (Cu-CDs) with exceptional POD activity using cherry blossom biomass via a hydrothermal process. UV-vis spectroscopy was employed to evaluate the oxidation capacity of different CDs on TMB solutions. The results demonstrated that Cu-CDs efficiently catalyzed the decomposition of H2O2 into ˙OH, achieving bactericidal rates exceeding 90% against both E. coli and S. aureus. These findings underscore the potent catalytic activity of metal-doped CDs in antimicrobial applications. Additionally, Zhang et al.206 engineered multifunctional glucose oxidase/carbon dots@copper metal–organic framework nanofibers (GOx/CDs@MOF NFs). Their study revealed that glucose oxidase (GOx) catalyzed the conversion of physiological glucose into gluconic acid and H2O2, concurrently lowering the pH of the system. This acidic environment effectively enhanced the POD activity of the Cu-MOF, further validating the potential of CNMs to improve antibacterial efficacy through catalytic mechanisms.5. Antimicrobial applications of carbon nanomaterials in medicine
In summary, CNMs, due to their remarkable antibacterial properties, hold significant promise in the field of medical antibacterial applications. Their potential is increasingly recognized across a wide range of medical domains, including oral health, antimicrobial dressings, tissue engineering, medical sutures, pneumonia treatments, and urinary catheters. As research in this area progresses, these materials are expected to play an increasingly pivotal role in combating bacterial infections, providing innovative solutions for both infection prevention and treatment within clinical settings.5.1 Dentistry
In dental medicine, P. gingivalis, a non-glycolytic, Gram-negative anaerobic bacterium, is a primary pathogen responsible for periodontitis. CDs have emerged as promising antibacterial agents, interacting with the negatively charged surface structures of P. gingivalis via their positively charged surfaces. This interaction aids in controlling infections and contributes to the treatment of dental caries and periodontal diseases. Liu et al.207 were the first to investigate the antibacterial effects of fluorescent CDs on P. gingivalis, an obligate anaerobe commonly found in the oral cavity. Using metronidazole as a precursor, they synthesized fluorescent CDs via a one-step hydrothermal method. Their findings revealed that at a concentration of 1.25 μg mL−1, the fluorescent CDs achieved an inhibition rate of 71.7% against P. gingivalis, with the antibacterial effect increasing proportionally with higher concentrations of CDs. This discovery holds considerable promise for the diagnosis, treatment, and prevention of dental caries and periodontal diseases, paving the way in further exploration into the application of fluorescent CDs for targeting oral bacteria. Liang et al.208 successfully synthesized therapeutic CQDs (T-CQDs and M-CQDs) using sulfonyl metronidazole and metronidazole as precursors through a hydrothermal process. These CQDs demonstrated strong inhibitory effects against P. gingivalis, with TCDs exhibiting the additional ability to penetrate bacterial biofilms, enhancing their efficacy in suppressing the growth of P. gingivalis. This advancement represents a novel strategy for improving periodontitis treatment outcomes, offering new therapeutic options for managing oral bacterial infections.5.2 Antibacterial accessories
In the field of biomedical materials, the development of biomaterials with antimicrobial properties plays a critical role in medical applications, particularly in preventing microbial infections associated with medical devices and wound care. Recent studies have demonstrated the potential of polymer-functionalized CNTs as effective antimicrobial agents for use in biomedical coatings and wound dressings. Simmons et al.209 fabricated SWCNTs coatings functionalized with polyvinylpyrrolidone-iodine (PVP-I), which exhibited significant antibacterial activity against E. coli. When incorporated into conductive nanofiber bandages, these materials demonstrated excellent flexibility and antimicrobial efficacy, effectively reducing bacterial colonization at wound sites and accelerating the healing process. Furthermore, CNTs functionalized with amino acids such as arginine and lysine, as well as those modified with antimicrobial peptides, have shown exceptional antibacterial performance, underscoring their potential for development as advanced antimicrobial biomaterials.210–212 Beyond CNTs, other CNMs have also exhibited promising antibacterial capabilities. For instance, Wang et al.213 developed an antibacterial composite film by incorporating lanthanum-doped nitrogen and phosphorus co-doped carbon quantum dots (La@N–P-CQDs) into a polyvinyl alcohol (PVA) matrix. This composite film achieved an inhibition rate exceeding 99% against E. coli. In addition to its strong antibacterial properties, the PVA/La@N–P-CQDs composite film exhibited excellent hydrophilicity, biocompatibility, and fluorescence intensity, making it a multifunctional material with potential applications in biomedical settings. Notably, in vivo studies demonstrated its ability to significantly enhance wound healing, highlighting its potential as an effective antimicrobial adjunct in clinical medicine.5.3 Tissue engineering
Infectious bone defects present a significant challenge in the treatment of orthopedic infectious diseases, particularly those resulting from severe open fractures caused by high-impact trauma. These injuries are often accompanied by varying degrees of wound contamination, extensive soft tissue damage, compromised blood supply, and substantial bone loss. As a result, there is an urgent need for the development of bone graft materials that not only possess robust antibacterial properties but also enable controlled drug release and demonstrate excellent osteogenic activity, thereby addressing both infection and bone defect repair following open fractures.214 Graphene-based nanocomposites have shown considerable promise in promoting the growth of human and mammalian cells, positioning them as potential candidates for use as scaffolding materials in tissue engineering applications. Mazaheri et al.215 fabricated graphene oxide–chitosan (GO–CS) composites as antibacterial scaffolds to support stem cell proliferation. Similarly, Faghihi et al.216 employed GO nanosheets as reinforcing agents in polyacrylic acid/gelatin composite hydrogels, creating antibacterial scaffolds with enhanced suitability for tissue engineering applications. Building on these advancements, Weng et al.217 incorporated antibiotics into graphene-based three-dimensional composite scaffolds, creating localized, controlled-release systems that combine both bone-filling and antibacterial functions. This innovative strategy leverages the superior properties of graphene to enhance local drug concentrations while minimizing systemic side effects, offering a promising solution for treating infectious bone defects associated with open fractures.5.4 Medical sutures
Medical sutures play a critical role in wound closure by approximating tissue edges, stabilizing the wound, and facilitating natural healing. To minimize adverse reactions, suture materials must exhibit excellent biocompatibility and biodegradability. However, despite advancements in sterile techniques, suture materials remain susceptible to microbial adhesion, potentially leading to post-surgical infections. Therefore, the ideal suture material should possess intrinsic antibacterial properties to mitigate the risk of infection. Recent advancements in nanotechnology have highlighted the potential of carbon-based nanomaterials, such as graphene and CNTs, in enhancing the antibacterial properties of sutures, thereby improving wound infection control. For instance, Ma et al.218 incorporated mechanically exfoliated graphene into a PVA matrix to fabricate PVA/GO nanocomposite fibres using gel spinning and stretching techniques. While PVA fibres alone lack antibacterial properties, the incorporation of GO significantly enhanced the antimicrobial activity of the composite fibres. In wound healing experiments, nanocomposite fibres containing 0.3 wt% GO exhibited a marked reduction in healing time compared to conventional surgical sutures, suggesting their potential as a novel alternative for surgical applications. In another study, de Moura et al.219 developed two antibacterial nanocomposites by combining GNPs and MWCNTs with a poly(lactic acid) (PLA) and thermoplastic polyurethane (TPU) blend at a 60/40 ratio. The resulting composites, designated as 60/40/GNP2 and 60/40/MWCNTs/GNP1, demonstrated significant antibacterial activity in agar diffusion assays, with distinct inhibition zones, indicating their efficacy in preventing bacterial colonization. Furthermore, recent research suggests that integrating natural polymers with GO could provide an effective strategy for developing next-generation antimicrobial suture materials. Gayathri et al.220 fabricated suture materials using cellulose, chitosan (CS), and GO. Their agar diffusion and simulated body fluid (SBF) studies confirmed that these sutures exhibited strong antibacterial activity, particularly against E. coli, further underscoring their potential as advanced antimicrobial suture materials.5.5 Pneumonia
Bacterial pneumonia remains a prevalent respiratory infection in modern medicine, with its treatment increasingly complicated by the rise of multidrug-resistant bacterial strains.221 The widespread use of antibiotics has contributed to the development and dissemination of resistant pathogens, posing a significant threat to global public health. Consequently, traditional antibiotic therapies are often ineffective against these resistant bacteria, highlighting the urgent need for novel antimicrobial strategies and materials. Serrano-Aroca et al.222 identified carbon-based nanomaterials as promising candidates for the treatment of bacterial pneumonia. These materials exhibit potent antibacterial activity primarily through physical mechanisms, such as disruption of bacterial cell walls and the generation of ROS. Such mechanisms are particularly valuable for targeting pneumonia-causing pathogens, offering substantial clinical potential. Notably, carbon nanomaterials can impair bacterial cell wall integrity and disrupt biofilm structures, thus increasing bacterial susceptibility to antibiotics. This characteristic is especially significant for treating biofilm-associated pneumonia infections, which are often more resistant to conventional therapies. Furthermore, the biocompatibility and low toxicity of carbon nanomaterials make them highly attractive for clinical applications. For instance, combining carbon nanomaterials with antibiotics has been shown to enhance antibacterial efficacy, particularly against drug-resistant bacterial strains responsible for pneumonia. Research conducted by Wu et al.223 supports these findings, demonstrating that carbon nanomaterials have promising effects in controlling pneumonia infections caused by resistant bacteria. When used in conjunction with antibiotics, carbon nanomaterials significantly improve treatment outcomes. These results suggest that carbon nanomaterials can serve both as standalone therapeutic agents and as adjuncts to traditional antibiotics, offering a synergistic effect that enhances treatment efficacy against resistant pathogens. This interaction not only improves overall treatment effectiveness but also provides a valuable complement to conventional antibiotics, introducing a novel approach to combating bacterial pneumonia. Despite these promising results, clinical data remain limited, and large-scale clinical trials are essential to further assess the safety and efficacy of carbon nanomaterials in real-world medical settings.5.6 Medical catheter
Catheter-associated urinary tract infections (CAUTIs) represent a major category of hospital-acquired infections, accounting for approximately 40% of such cases. These infections significantly impact patients' quality of life and pose considerable risks to their health and safety. Consequently, enhancing the antibacterial properties of urinary catheters is of paramount clinical importance for the prevention of urinary tract infections. CNMs, including CNTs, graphene-based materials, and CDs, possess unique physicochemical properties and inherent antibacterial activity, positioning them as promising candidates for the development of antimicrobial catheters. Zhang et al.224 demonstrated that CNMs exhibit robust antibacterial properties with a reduced propensity for inducing bacterial resistance, contributing to their growing use in antimicrobial applications. In particular, the integration of CNMs into medical catheters has shown considerable potential, as these materials can disrupt bacterial outer membranes through both physical and chemical interactions, thereby slowing the development of microbial resistance. The distinctive properties of CNMs make them invaluable for the design of antibacterial composite materials, with significant promise for reducing the incidence of CAUTIs. Teixeira-Santos et al.225 further explored the potential of nitrogen-functionalized graphene composites for urinary catheter applications. Their study revealed that nitrogen-functionalized graphene nanoplatelets (N-GNPs), when incorporated into a polydimethylsiloxane (PDMS) matrix to form N-GNP/PDMS composites, significantly inhibited the formation of both single-species and multi-species biofilms, particularly against S. aureus. Compared to pure PDMS, the N-GNP/PDMS composite exhibited increased surface roughness and hydrophobicity, which effectively reduced the colonization of S. aureus, P. aeruginosa, and K. pneumoniae, while also diminishing S. aureus biofilm formation. In multi-species biofilms, a reduction in total cell counts was also observed. Furthermore, N-GNPs were found to alter membrane permeability in S. aureus and promote the generation of ROS, while primarily affecting cellular metabolism in Gram-negative bacteria. Overall, the N-GNP/PDMS composite demonstrated significant potential as a urinary catheter coating material by effectively inhibiting biofilm development. Given the escalating issue of antibiotic resistance, the development of novel antimicrobial materials is becoming increasingly critical. The discovery of N-GNP/PDMS composites provides a promising direction for the design of urinary catheters and could play a pivotal role in reducing the risk of catheter-associated infections.6. The toxicity about antibacterial CNMs
6.1 Toxicity assessment and metabolism of CNMs
The increasing applications of carbon nanomaterials in biomedicine necessitate a comprehensive understanding of their toxicity profiles. Li et al.226 indicated those various carbon-based nanomaterials, including carbon black nanoparticles, nanodiamonds, fullerenes, and graphene quantum dots, exhibit diverse toxicological behaviours depending on their physicochemical properties. Researchers employ a range of in vitro and in vivo assays to evaluate their cytotoxic effects, encompassing MTT assays, flow cytometry, and confocal microscopy, which provide insights into cell viability, cell cycle progression, apoptosis, and autophagy. Comparative analyses between different carbon nanomaterials reveal that dimensionality significantly influences toxicity. For instance, Raja et al.227 discovered that zero-dimensional carbon dots demonstrate dose-dependent cytotoxicity in human lung epithelial cells (A549), with IC50 values ranging from 200 to 500 μg mL−1, whereas one-dimensional carbon nanotubes exhibit distinct biological interactions. These disparities underscore the importance of considering material dimensions in toxicity assessments. Furthermore, surface functionalization emerges as a critical determinant of biocompatibility. Ecological risk assessments further refine safety thresholds, with Kim et al.228 proposing 10 μg L−1 as an environmental safety limit for aquatic systems based on Daphnia magna immobilization tests.CNMs usually have high biological stability, which makes them relatively difficult to degrade in vivo.229 CNMs typically exhibit high biological stability, making them relatively difficult to degrade in vivo. Although there is currently a lack of direct research in the metabolic behaviour of CNMs against Gram negative bacteria, the in vivo metabolism or distribution of biological CNMs continues to attract significant interest. Kim et al.228 quantified this concern through comparative pharmacokinetic studies, revealing that carbon nanotubes exhibit 30% slower clearance rates than gold nanoparticles in murine models. Correspondingly, researchers are trying to overcome non-biodegradability and stability challenges. Surface modification and functionalization strategies effectively address biodegradability limitations. Li et al.226 demonstrated that carboxyl group incorporation enhances both water dispersibility and enzymatic degradation of carbon nanotubes, significantly reducing accumulation risks. Chen et al.230 developed poly(amidoamine) dendrimer-crosslinked nanotubes that maintain antimicrobial efficacy while improving colloidal stability. Emerging research directions focus on intrinsically biodegradable designs that retain antimicrobial activity.
6.2 Balancing efficacy and biosafety
While carbon nanomaterials effectively combat Gram-negative bacteria via membrane disruption and oxidative stress pathways, their potential cytotoxicity toward eukaryotic cells requires meticulous evaluation. Nasirzadeh et al.231 observed a 60% viability loss in HEK293 kidney cells at 50 μg mL−1 pristine graphene oxide exposure, attributing this toxicity to lysosomal membrane destabilization. This finding underscores the urgency of developing targeted antimicrobial strategies. Advanced material design approaches demonstrate promising solutions to this biosafety challenge. Li et al.226 engineered sulfur-doped reduced graphene oxide nanosheets that achieved a 5-log reduction in Pseudomonas aeruginosa at 120 μg mL−1 while maintaining 92% viability in L929 fibroblasts. Chiou et al.232 and Zhang et al.233 designed and prepared biomass-derived CNMs with excellent antibacterial activities and balanced the enhanced biocompatibility and reduced hemolysis at antimicrobial concentrations several times higher than the effective dose. Thus, the preparation of CNMs from biomass or the surface bioactivity modification are important strategies to improve both antibacterial and biosafety properties of CNMs.7. Summary and outlook
The emergence of multidrug-resistant (MDR) and extensively drug-resistant (XDR) bacteria has made the development of new antimicrobial agents an urgent priority. Gram-negative bacteria, due to their unique biological structure, have long posed significant challenges in the development of effective antimicrobial agents. Unfortunately, in recent years, the field of antimicrobial drug development targeting Gram-negative bacteria has remained largely confined to existing classes of antibiotics. There remains a notable scarcity of new antimicrobial agents with novel mechanisms of action in clinical development. Carbon nanomaterials, with their inherent antimicrobial activity and distinctive physical antibacterial properties, offer new promise in an era characterized by widespread antibiotic misuse. If effectively applied in infection treatment, these materials may provide an innovative approach to combating bacterial resistance. However, several unresolved issues remain regarding the clinical application of carbon nanomaterials in antibacterial therapy. These challenges and development directions include: (1) the potential toxicity assessment of CNMs on human cells and tissues is incomplete. It is necessary to clarify the structure of CNMs and their interactions with biological targets, as well as their antibacterial molecular mechanisms. Furthermore, by precisely controlling parameters such as material size, shape, and surface chemical properties, and developing purification and large-scale preparation methods, some CNMs with tuneable antibacterial activity and minimal toxicity can be achieved. (2) There is insufficient understanding of the precise localization of CNMs in bacteria, cells, and organisms, and it is necessary to conduct safety evaluations and in vivo distribution research on antibacterial CNMs. Animal models and evaluation indicators need to be further studied for in vivo safety research to understand the in vivo long-term safety and stability of CNMs in vivo. The imaging, in vivo metabolism, and pharmacokinetics study methods of CNMs are needed to determine the optimal dosage regimen. (3) There is still a lack of consistency in the structure and properties of carbon dots across different production batches. Quality control standards, safety standards, usage strategies, and specialized guidelines and regulations need to be developed as a regulatory framework for fair use of antibacterial CNMs. (4) The combination of CNMs with other antibacterial drugs. For example, deepening the combination strategy of CNMs with traditional antibiotics, antimicrobial peptides, etc., and enhancing antibacterial effects, reducing bacterial resistance risks, and improving biosafety.Despite ongoing debates about the mechanisms underlying the antimicrobial activity of carbon nanomaterials, their potential applications in the antimicrobial field remain highly promising. As our understanding of the key functional parameters required for effective bacterial targeting at the molecular level deepens, continued efforts to optimize the design of carbon nanomaterials are expected. Moreover, advancing the exploration of their antimicrobial mechanisms will contribute to significant breakthroughs in both basic research and technological applications. In conclusion, this rapidly developing field has already achieved notable progress. We are optimistic that further exploration of the antimicrobial properties of carbon nanomaterials will accelerate the development of new agents against Gram-negative and resistant bacteria. In the near future, carbon nanomaterials are poised to play a key role in the clinical treatment of infections caused by Gram-negative and resistant bacteria.
Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Author contributions
Chengwei Zhang, Ying Zheng, and Xiaoyan Lin: data curation, writing – original draft, writing – review & editing. Shaohuang Weng: conceptualization, supervision, project administration, writing – review &editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank the financial support from the Joint Funds for the Innovation of Science and Technology, Fujian Province (2021Y9007), and the National Science Foundation of Fujian Province (2022J01493).Notes and references
- C. A. Bradley, Nat. Rev. Drug Discovery, 2018, 17, 394 CrossRef CAS PubMed.
- Z. Wang, H. Zhang, J. Han, H. Xing, M. C. Wu and T. Yang, Infect. Control Hosp. Epidemiol., 2017, 38, 758–759 CrossRef PubMed.
- G. V. Asokan and A. Vanitha, Perspect. Public Health, 2018, 138, 87–88 CrossRef PubMed.
- A. Upadhayay, J. Ling, D. Pal, Y. Xie, F. F. Ping and A. Kumar, Drug Resistance Updates, 2023, 66, 100890 CrossRef CAS PubMed.
- Z. Breijyeh, B. Jubeh and R. Karaman, Molecules, 2020, 25, 1340 CrossRef CAS PubMed.
- G. M. Carlone, M. J. Valadez and M. J. Pickett, J. Clin. Microbiol., 1982, 16, 1157–1159 CrossRef CAS PubMed.
- Y. Fujiwara, K. Ohnishi, H. Horlad, Y. Saito, D. Shiraishi, H. Takeya, D. Yoshii, S. Kaieda, T. Hoshino and Y. Komohara, Clin. Transl. Immunol., 2020, 9, e1162 CrossRef CAS PubMed.
- G. Mamou, F. Corona, R. Cohen-Khait, N. G. Housden, V. Yeung, D. Sun, P. Sridhar, M. Pazos, T. J. Knowles and C. Kleanthous, Nature, 2022, 606, 953–959 CrossRef CAS PubMed.
- P. R. Murray, K. S. Rosenthal and M. A. Pfaller, Medical Microbiology, Elsevier Health Sci., 2020 Search PubMed.
- M. C. Sousa, Nature, 2019, 576, 389–390 CrossRef CAS PubMed.
- S. I. Miller, mBio, 2016, 7 DOI:10.1128/mbio.01541-16.
- V. Gupta and P. Datta, Indian J. Med. Res., 2019, 149, 97–106 CrossRef CAS PubMed.
- M. Exner, S. Bhattacharya, B. Christiansen, J. Gebel, P. Goroncy-Bermes, P. Hartemann, P. Heeg, C. Ilschner, A. Kramer and E. Larson, GMS Hyg Infect. Control, 2017, 12, Doc05 Search PubMed.
- M. Bassetti, A. Vena, C. Sepulcri, D. R. Giacobbe and M. Peghin, Antibiotics, 2020, 9, 632 CrossRef CAS PubMed.
- E. Massa, E. Michailidou, D. Agapakis, S. Papadopoulos, T. Tholioti, I. Aleuroudis, T. Bargiota, M. Passakiotou, M. Daoudaki and N. Antoniadis, Transplant. Proc., 2019, 51, 454–456 CrossRef CAS PubMed.
- J. Chen, S. M. Andler, J. M. Goddard, S. R. Nugen and V. M. Rotello, Chem. Soc. Rev., 2017, 46, 1272–1283 RSC.
- M. H. Kollef and K. D. Betthauser, Curr. Opin. Infect. Dis., 2019, 32, 169–175 CrossRef PubMed.
- Y. Liu, W. Wang, H. Yan, D. Wang, M. Zhang and S. Sun, Future Microbiol., 2019, 14, 899–915 CrossRef CAS PubMed.
- S. Ghosh, S. Sen, M. Jash, S. Ghosh, A. Jana, R. Roy, N. Mukherjee, D. Mukherjee, J. Sarkar and S. Ghosh, ACS Infect. Dis., 2024, 10, 1267–1285 CrossRef CAS PubMed.
- H. Douafer, V. Andrieu, O. Phanstiel IV and J. M. Brunel, J. Med. Chem., 2019, 62, 8665–8681 CrossRef CAS PubMed.
- X. Gao, J. Ding, C. Liao, J. Xu, X. Liu and W. Lu, Adv. Drug Delivery Rev., 2021, 179, 114008 CrossRef CAS PubMed.
- A. Adak, S. Ghosh, V. Gupta and S. Ghosh, Biomacromolecules, 2019, 20, 1889–1898 CrossRef CAS PubMed.
- A. Adak, V. Castelletto, B. Mendes, G. Barrett, J. Seitsonen and I. W. Hamley, ACS Appl. Bio Mater., 2024, 7, 5553–5565 CrossRef CAS PubMed.
- A. Adak, V. Castelletto, A. de Sousa, K. A. Karatzas, C. Wilkinson, N. Khunti, J. Seitsonen and I. W. Hamley, Biomacromolecules, 2024, 25, 1205–1213 CrossRef CAS PubMed.
- R. Samat, S. Sen, M. Jash, S. Ghosh, S. Garg, J. Sarkar and S. Ghosh, ACS Infect. Dis., 2024, 10, 3098–3125 CrossRef CAS PubMed.
- S. Sen, R. Samat, M. Jash, S. Ghosh, R. Roy, N. Mukherjee, S. Ghosh, J. Sarkar and S. Ghosh, J. Med. Chem., 2023, 66, 11555–11572 CrossRef CAS PubMed.
- M. Hamad, F. Al-Marzooq, G. Orive and T. H. Al-Tel, Drug Discovery Today, 2019, 24, 2225–2228 CrossRef PubMed.
- Q. Xin, Q. Liu, L. Geng, Q. Fang and J. R. Gong, Adv. Healthcare Mater., 2017, 6, 1601011 CrossRef PubMed.
- R. Van Noorden, Nature, 2011, 469, 14 CrossRef CAS PubMed.
- D. Nagarajan, D. Gangadharan and S. Venkatanarasimhan, Carbon Dots in Analytical Chemistry, Elsevier, 2023, pp. 1–13 Search PubMed.
- J. Xu, B. Huang, C. M. Lai, Y. S. Lu and J. W. Shao, J. Photochem. Photobiol., B, 2024, 255, 112920 CrossRef CAS PubMed.
- J. Kong, Y. Wei, F. Zhou, L. Shi, S. Zhao, M. Wan and X. Zhang, Molecules, 2024, 29, 2002 CrossRef CAS PubMed.
- S. Paveethra, H. Manisekaran and S. Sasidharan, Asian Pac. J. Cancer Prev., 2024, 25, 3393 CrossRef CAS PubMed.
- Y. P. Sun, B. Zhou, Y. Lin, W. Wang, K. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang and H. Wang, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- S. Santiago, T. N. Lin, C. H. Chang, Y. A. Wong, C. A. J. Lin, C. T. Yuan and J. Shen, Phys. Chem. Chem. Phys., 2017, 19, 22395–22400 RSC.
- V. Nguyen, L. Yan, J. Si and X. Hou, J. Appl. Phys., 2015, 117, 084304 CrossRef.
- L. Cui, X. Ren, J. Wang and M. Sun, Mater. Today Nano, 2020, 12, 100091 CrossRef.
- V. Nguyen, N. Zhao, L. Yan, P. Zhong and P. H. Le, Mater. Res. Express, 2020, 7, 015606 CrossRef CAS.
- A. Menazea and M. Ahmed, Nano-Struct. Nano-Objects, 2020, 22, 100464 CrossRef CAS.
- K. S. Khashan, G. M. Sulaiman and R. Mahdi, Artif. Cells, Nanomed., Biotechnol., 2017, 45, 1699–1709 CrossRef CAS PubMed.
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- F. Chao-Mujica, L. Garcia-Hernández, S. Camacho-López, M. Camacho-López, M. Camacho-López, D. Reyes Contreras, A. Pérez-Rodríguez, J. Peña-Caravaca, A. Páez-Rodríguez and J. Darias-Gonzalez, J. Appl. Phys., 2021, 129, 163301 CrossRef CAS.
- Y. Lin, M. Zhou, X. Tai, H. Li, X. Han and J. Yu, Matter, 2021, 4, 2309–2339 CrossRef CAS.
- J. Ge, M. Lan, B. Zhou, W. Liu, L. Guo, H. Wang, Q. Jia, G. Niu, X. Huang and H. Zhou, Nat. Commun., 2014, 5, 4596 CrossRef CAS PubMed.
- P. G. Luo, F. Yang, S. T. Yang, S. K. Sonkar, L. Yang, J. J. Broglie, Y. Liu and Y. P. Sun, RSC Adv., 2014, 4, 10791–10807 RSC.
- H. Li, J. Huang, Y. Song, M. Zhang, H. Wang, F. Lu, H. Huang, Y. Liu, X. Dai and Z. Gu, ACS Appl. Mater. Interfaces, 2018, 10, 26936–26946 CrossRef CAS PubMed.
- X. Liu, L. Yu, F. Liu, L. Sheng, K. An, H. Chen and X. Zhao, J. Mater. Sci., 2012, 47, 6086–6094 CrossRef CAS.
- B. D. Mansuriya and Z. Altintas, Nanomaterials, 2021, 11, 2525 CrossRef CAS PubMed.
- L. Cui, X. Ren, M. Sun, H. Liu and L. Xia, Nanomaterials, 2021, 11, 3419 CrossRef CAS PubMed.
- S. Zhuo, M. Shao and S. T. Lee, ACS Nano, 2012, 6, 1059–1064 CrossRef CAS PubMed.
- H. Huang, Y. Cui, M. Liu, J. Chen, Q. Wan, Y. Wen, F. Deng, N. Zhou, X. Zhang and Y. Wei, J. Colloid Interface Sci., 2018, 532, 767–773 CrossRef CAS PubMed.
- Y. Wu, Y. Liu, J. Yin, H. Li and J. Huang, Talanta, 2019, 205, 120121 CrossRef CAS PubMed.
- S. Bi, C. Hang, J. Qi, W. Zhang and X. Jiang, Adv. Biol., 2023, 7, 2200297 CrossRef CAS PubMed.
- M. Al-Salih, S. Samsudin and S. S. Arshad, J. Genet. Eng. Biotechnol., 2021, 19, 76 CrossRef PubMed.
- A. Barhoum, A. Meftahi, M. S. Kashef Sabery, M. E. Momeni Heravi and F. Alem, J. Mater. Sci., 2023, 58, 13531–13579 CrossRef CAS.
- A. M. Khalil, M. A. Gharieb, S. M. Abdelaty and A. M. El-Khatib, Mater. Res. Express, 2024, 11, 075006 CrossRef CAS.
- F. Fenniche, A. Henni, Y. Khane, D. Aouf, N. Harfouche, S. Bensalem, D. Zerrouki and H. Belkhalfa, J. Inorg. Organomet. Polym. Mater., 2022, 32, 1011–1025 CrossRef CAS.
- F. Karkeh-Abadi, F. Soofivand, H. Safardoust-Hojaghan, Q. A. Yousif and M. Salavati-Niasari, J. Mater. Res. Technol., 2022, 18, 4413–4426 CrossRef CAS.
- N. H. Hussen, A. H. Hasan, Y. M. FaqiKhedr, A. Bogoyavlenskiy, A. R. Bhat and J. Jamalis, ACS Omega, 2024, 9, 9849–9864 CrossRef CAS PubMed.
- S. Ross, R. S. Wu, S. C. Wei, G. M. Ross and H. T. Chang, J. Food Drug Anal., 2020, 28, 677 CAS.
- P. D. Modi, V. N. Mehta, V. S. Prajapati, S. Patel and J. V. Rohit, Carbon Dots in Analytical Chemistry, Elsevier, 2023, pp. 15–29 Search PubMed.
- S. Pandiyan, L. Arumugam, S. P. Srirengan, R. Pitchan, P. Sevugan, K. Kannan, G. Pitchan, T. A. Hegde and V. Gandhirajan, ACS Omega, 2020, 5, 30363–30372 CrossRef CAS PubMed.
- Z. Liang, L. Zeng, X. Cao, Q. Wang, X. Wang and R. Sun, J. Mater. Chem. C, 2014, 2, 9760–9766 RSC.
- A. M. Alam, B. Y. Park, Z. K. Ghouri, M. Park and H. Y. Kim, Green Chem., 2015, 17, 3791–3797 RSC.
- J. Briscoe, A. Marinovic, M. Sevilla, S. Dunn and M. Titirici, Angew. Chem., Int. Ed., 2015, 54, 4463–4468 CrossRef CAS PubMed.
- S. Min, P. Ezati, K. S. Yoon and J. W. Rhim, Int. J. Biol. Macromol., 2023, 231, 123493 CrossRef CAS PubMed.
- S. Nangan, K. Kanagaraj, G. Kaarthikeyan, A. Kumar, M. Ubaidullah, B. Pandit, R. Govindasamy and T. Natesan, Environ. Res., 2023, 237, 116990 CrossRef CAS PubMed.
- G. Arora, N. S. Sabran, C. Y. Ng, F. W. Low and H. Jun, Heliyon, 2024, 10, e35543 CrossRef CAS PubMed.
- P. C. Hsu and H. T. Chang, Chem. Commun., 2012, 48, 3984–3986 RSC.
- S. Liu, J. Tian, L. Wang, Y. Zhang, X. Qin, Y. Luo, A. M. Asiri, A. O. Al-Youbi and X. Sun, Adv. Mater., 2012, 24, 2037 CrossRef CAS PubMed.
- S. Sahana, A. Gautam, R. Singh and S. Chandel, Nat. Prod. Bioprospect., 2023, 13, 51 CrossRef PubMed.
- D. S. Ghataty, R. I. Amer, M. A. Amer, M. F. Abdel Rahman and R. N. Shamma, Pharmaceutics, 2023, 15, 234 CrossRef CAS PubMed.
- S. Wei, X. Shi, C. Wang, H. Zhang, C. Jiang, G. Sun and C. Jiang, Spectrochim. Acta, Part A, 2023, 287, 122039 CrossRef CAS PubMed.
- G. Ma, R. Wang, M. Zhang, Z. Dong, A. Zhang, M. Qu, L. Gao, Y. Wei and J. Wei, Spectrochim. Acta, Part A, 2023, 289, 122178 CrossRef CAS PubMed.
- J. Peng, W. Gao, B. K. Gupta, Z. Liu, R. Romero-Aburto, L. Ge, L. Song, L. B. Alemany, X. Zhan and G. Gao, Nano Lett., 2012, 12, 844–849 CrossRef CAS PubMed.
- G. Magdy, H. Elmansi, M. M. Samaha, E. Said and N. El-Enany, Sustainable Chem. Pharm., 2024, 37, 101424 CrossRef CAS.
- S. J. Lee, Y. Y. Zheng, W. M. Chen and Y. H. Hsueh, ACS Omega, 2024, 9, 36453–36463 CAS.
- M. K. Kumawat, M. Thakur, R. B. Gurung and R. Srivastava, ACS Sustainable Chem. Eng., 2017, 5, 1382–1391 CrossRef CAS.
- P. M. Olmos-Moya, S. Velazquez-Martinez, C. Pineda-Arellano, J. R. Rangel-Mendez and L. F. Chazaro-Ruiz, Carbon, 2022, 187, 216–229 CrossRef CAS.
- S. Yalshetti, B. Thokchom, S. M. Bhavi, S. R. Singh, S. R. Patil, B. Harini, M. Sillanpää, J. Manjunatha, B. Srinath and R. B. Yarajarla, Sci. Rep., 2024, 14, 9915 CrossRef CAS PubMed.
- M. M. Osman, R. El-Shaheny and F. A. Ibrahim, Anal. Chim. Acta, 2024, 1319, 342946 CrossRef CAS PubMed.
- G. Ge, L. Li, D. Wang, M. Chen, Z. Zeng, W. Xiong, X. Wu and C. Guo, J. Mater. Chem. B, 2021, 9, 6553–6575 RSC.
- R. Liu, D. Wu, S. Liu, K. Koynov, W. Knoll and Q. Li, Angew. Chem., Int. Ed., 2009, 48, 4598–4601 CrossRef CAS PubMed.
- J. Zong, Y. Zhu, X. Yang, J. Shen and C. Li, Chem. Commun., 2011, 47, 764–766 RSC.
- J. Lv, H. Tian, L. Pan, Z. Chen, M. Li, R. A. Ghiladi, Z. Qin and X. Yin, Chem. Eng. Sci., 2024, 295, 120084 CrossRef CAS.
- Y. Zhao, X. Wu, S. Sun, L. Ma, L. Zhang and H. Lin, Carbon, 2017, 124, 342–347 CrossRef CAS.
- C. Shuai, X. Shi, K. Wang, Y. Gu, F. Yang and P. Feng, Bio-Des. Manuf., 2024, 7, 105–120 CrossRef CAS.
- B. Khan, J. Zhang, S. Durrani, H. Wang, A. Nawaz, F. Durrani, Y. Ye, F. G. Wu and F. Lin, ACS Appl. Mater. Interfaces, 2024, 16, 47257–47269 CrossRef CAS PubMed.
- T. J. Antony and K. Jagannathan, Ceram. Int., 2024, 50, 16343–16351 CrossRef CAS.
- M. Zhang, H. Yang, K. Yang, Q. Yang, W. Liu and X. Yang, J. Chromatogr. A, 2024, 1725, 464926 CrossRef CAS PubMed.
- Y. Liu, C. Huang, W. Yue, X. Wang, Y. Sun, W. Bi, L. Wang and Y. Xu, Carbon, 2024, 224, 119085 CrossRef CAS.
- P. Kodithuwakku, D. Jayasundara, I. Munaweera, R. Jayasinghe, T. Thoradeniya, A. Bogahawatta, K. J. Manuda, M. Weerasekera and N. Kottegoda, ACS Omega, 2024, 9, 26568–26581 CrossRef CAS PubMed.
- Z. Li, M. Zhou, L. Zhao, W. Hu, X. Wang, H. Wei, X. Tong, C. Yao, X. Li and Y. Zhang, Chem. Eng. J., 2024, 498, 155081 CrossRef CAS.
- B. Thangaraj, N. Wongyao, P. R. Solomon, V. Gupta, A. Abdullah, S. Abedrabbo and J. Hassan, J. Environ. Chem. Eng., 2024, 12, 113474 CrossRef CAS.
- S. He, J. Huang, Q. Zhang, W. Zhao, Z. Xu and W. Zhang, Adv. Funct. Mater., 2021, 31, 2105198 CrossRef CAS.
- Y. Bai, C. Shi, X. Ma, J. Li, S. Chen, N. Guo, X. Yu, C. Yang and Z. Zhang, Chem. Eng. J., 2022, 447, 137545 CrossRef CAS.
- R. Elizabeth Roy, N. KS, S. Salim, S. Sugathan and A. John, Chem. Biodiversity, 2025, 22, e202401350 CrossRef CAS PubMed.
- R. Venkatesan, P. Sivaprakash, I. Kim, G. E. Eldesoky and S. C. Kim, J. Environ. Chem. Eng., 2023, 11, 110194 CrossRef CAS.
- J. Luan, W. L. Duan, Y. X. Li, F. B. Meng, Y. Liu, X. S. Zhang, J. Zhou, W. Z. Li and Y. Fu, ACS Sustain. Chem. Eng., 2023, 11, 12423–12434 CrossRef CAS.
- B. Yang, L. Gao, M. Xue, H. Wang, Y. Hou, Y. Luo, H. Xiao, H. Hu, C. Cui and H. Wang, Materials, 2021, 14, 7356 CrossRef CAS PubMed.
- S. Kim, Y. Song and M. J. Heller, Adv. Mater., 2017, 29, 1701845 CrossRef PubMed.
- S. N. Khan, B. M. Weight, B. J. Gifford, S. Tretiak and A. Bishop, J. Phys. Chem. Lett., 2022, 13, 5801–5807 CrossRef CAS PubMed.
- E. Haque, J. Kim, V. Malgras, K. R. Reddy, A. C. Ward, J. You, Y. Bando, M. S. A. Hossain and Y. Yamauchi, Small Methods, 2018, 2, 1800050 CrossRef.
- A. Ghaffarkhah, E. Hosseini, M. Kamkar, A. A. Sehat, S. Dordanihaghighi, A. Allahbakhsh, C. van der Kuur and M. Arjmand, Small, 2022, 18, 2102683 CrossRef CAS PubMed.
- C. Zhao, X. Song, Y. Liu, Y. Fu, L. Ye, N. Wang, F. Wang, L. Li, M. Mohammadniaei and M. Zhang, J. Nanobiotechnol., 2020, 18, 1–32 CrossRef PubMed.
- Z. Luo, D. Yang, C. Yang, X. Wu, Y. Hu, Y. Zhang, L. Yuwen, E. K. L. Yeow, L. Weng and W. Huang, Appl. Surf. Sci., 2018, 434, 155–162 CrossRef CAS.
- H. H. Huang, A. Anand, C. J. Lin, H. J. Lin, Y. W. Lin, S. G. Harroun and C. C. Huang, Carbon, 2021, 174, 710–722 CrossRef CAS.
- Y. Yu, L. Mei, Y. Shi, X. Zhang, K. Cheng, F. Cao, L. Zhang, J. Xu, X. Li and Z. Xu, J. Mater. Chem. B, 2020, 8, 1371–1382 RSC.
- I. X. Yin, J. Y. Niu, M. L. Mei, J. Tang, W. K. K. Wu and C. H. Chu, Int. J. Nanomed., 2024, 19, 11195–11212 CrossRef PubMed.
- G. Yang, L. Wang, C. Zhang, P. Li, H. Du, Y. Mao, M. Qiu, Q. Li, D. Hao and Q. Wang, Sep. Purif. Technol., 2023, 312, 123433 CrossRef CAS.
- X. Tian, A. Zeng, Z. Liu, C. Zheng, Y. Wei, P. Yang, M. Zhang, F. Yang and F. Xie, Int. J. Nanomed., 2020, 15, 6519–6529 CrossRef CAS PubMed.
- W. Q. Li, Z. Wang, S. Hao, L. Sun, M. Nisic, G. Cheng, C. Zhu, Y. Wan, L. Ha and S. Y. Zheng, Nanoscale, 2018, 10, 3744–3752 RSC.
- W. Su, R. Guo, F. Yuan, Y. Li, X. Li, Y. Zhang, S. Zhou and L. Fan, J. Phys. Chem. Lett., 2020, 11, 1357–1363 CrossRef CAS PubMed.
- M. Hassan, V. G. Gomes, A. Dehghani and S. M. Ardekani, Nano Res., 2018, 11, 1–41 CrossRef CAS.
- P. Devi, S. Saini and K.-H. Kim, Biosens. Bioelectron., 2019, 141, 111158 CrossRef CAS PubMed.
- Y. Bai, Y. Wang, L. Cao, Y. Jiang, Y. Li, H. Zou, L. Zhan and C. Huang, Anal. Chem., 2021, 93, 16466–16473 CrossRef CAS PubMed.
- A. S. Krishna, C. Radhakumary, M. Antony and K. Sreenivasan, J. Mater. Chem. B, 2014, 2, 8626–8632 RSC.
- K. Wang, Q. Ji, H. Li, F. Guan, D. Zhang, H. Feng and H. Fan, J. Inorg. Biochem., 2017, 166, 64–67 CrossRef CAS PubMed.
- Y. Lu, L. Li, M. Li, Z. Lin, L. Wang, Y. Zhang, Q. Yin, H. Xia and G. Han, Bioconjugate Chem., 2018, 29, 2982–2993 CrossRef CAS PubMed.
- P. Li, S. Liu, G. Zhang, X. Yang, W. Cao, X. Gong and X. Xing, ACS Appl. Bio Mater., 2020, 3, 1105–1115 CrossRef CAS PubMed.
- J. Liang, W. Li, J. Chen, X. Huang, Y. Liu, X. Zhang, W. Shu, B. Lei and H. Zhang, J. Mater. Chem. A, 2022, 10, 23384–23394 RSC.
- Y. Liu, Y. Zhao, S. Guo, D. Qin, J. Yan, H. Cheng, J. Zhou, J. Ren, L. Sun and H. Peng, Carbohydr. Polym., 2024, 346, 122656 CrossRef CAS PubMed.
- M. J. Meziani, X. Dong, L. Zhu, L. P. Jones, G. E. LeCroy, F. Yang, S. Wang, P. Wang, Y. Zhao and L. Yang, ACS Appl. Mater. Interfaces, 2016, 8, 10761–10766 CrossRef CAS PubMed.
- A. Pandey, A. Devkota, Z. Yadegari, K. Dumenyo and A. Taheri, Nanomaterials, 2021, 11, 2012 CrossRef CAS PubMed.
- L. N. Wu, Y. J. Yang, L. X. Huang, Y. Zhong, Y. Chen, Y. R. Gao, L. Q. Lin, Y. Lei and A. L. Liu, Carbon, 2022, 186, 452–464 CrossRef CAS.
- Q. Xin, H. Shah, A. Nawaz, W. Xie, M. Z. Akram, A. Batool, L. Tian, S. U. Jan, R. Boddula and B. Guo, Adv. Mater., 2019, 31, 1804838 CrossRef CAS PubMed.
- V. TK, A. K. Patel and V. Muthuvijayan, J. Biomater. Sci., Polym. Ed., 2023, 34, 419–434 CrossRef PubMed.
- S. Shen, H. Qi, T. Yi, T. Jing, J. Li, Y. Gao, Q. Zeng and H. Zhao, J. Mol. Struct., 2025, 1322, 140620 CrossRef CAS.
- Y. Fu, F. Wang, H. Sheng, M. Xu, Y. Liang, Y. Bian, S. A. Hashsham, X. Jiang and J. M. Tiedje, Carbon, 2020, 163, 360–369 CrossRef CAS.
- M. K. Ibrahim, A. Haria, N. V. Mehta and M. S. Degani, Future Med. Chem., 2023, 15, 2113–2141 CrossRef CAS PubMed.
- S. H. Kim, D. Semenya and D. Castagnolo, Eur. J. Med. Chem., 2021, 216, 113293 CrossRef CAS PubMed.
- A. R. Gomes, C. L. Varela, A. S. Pires, E. J. Tavares-da-Silva and F. M. Roleira, Bioorg. Chem., 2023, 138, 106600 CrossRef CAS PubMed.
- A. Dong, Y. J. Wang, Y. Gao, T. Gao and G. Gao, Chem. Rev., 2017, 117, 4806–4862 CrossRef CAS PubMed.
- R. Liu, X. C. Qing, Z. B. Feng, X. M. Zhang, Y. Zhang, M. Y. Zhang, B. Y. Zhu, Y. Li and L. Xie, Macromol. Rapid Commun., 2023, 36, 564–573 Search PubMed.
- R. Eldawud, A. Wagner, C. Dong, T. A. Stueckle, Y. Rojanasakul and C. Z. Dinu, NanoImpact, 2018, 9, 72–84 CrossRef PubMed.
- A. S. Bati, L. Yu, M. Batmunkh and J. G. Shapter, Nanoscale, 2018, 10, 22087–22139 RSC.
- C. J. Barnett, C. E. Gowenlock, K. Welsby, A. Orbaek White and A. R. Barron, Nano Lett., 2018, 18, 695–700 CrossRef CAS PubMed.
- S. Khan, P. Jawlikar, S. Lahoti, O. Bhusnure, S. Chitlange and J. Sangshetti, Curr. Pharm. Des., 2021, 27, 2454–2467 CrossRef CAS PubMed.
- J. Ackermann, J. T. Metternich, S. Herbertz and S. Kruss, Angew. Chem., Int. Ed., 2022, 61, e202112372 CrossRef CAS PubMed.
- M. R. M. Radzi, N. A. Johari, W. F. A. W. M. Zawawi, N. A. Zawawi, N. A. Latiff, N. A. N. N. Malek, A. A. Wahab, M. I. Salim and K. Jemon, Biomater. Adv., 2022, 134, 112586 CrossRef CAS PubMed.
- T. Mocan, C. T. Matea, T. Pop, O. Mosteanu, A. D. Buzoianu, S. Suciu, C. Puia, C. Zdrehus, C. Iancu and L. Mocan, Cell. Mol. Life Sci., 2017, 74, 3467–3479 CrossRef CAS PubMed.
- S. Kang, M. Pinault, L. D. Pfefferle and M. Elimelech, Langmuir, 2007, 23, 8670–8673 CrossRef CAS PubMed.
- S. Kang, M. Herzberg, D. F. Rodrigues and M. Elimelech, Langmuir, 2008, 24, 6409–6413 CrossRef CAS PubMed.
- L. M. Pasquini, S. M. Hashmi, T. J. Sommer, M. Elimelech and J. B. Zimmerman, Environ. Sci. Technol., 2012, 46, 6297–6305 CrossRef CAS PubMed.
- M. Assis, J. R. Santos, M. H. Cipriano, R. Y. Reis, L. K. Ribeiro, L. H. Mascaro, E. Longo and J. Andrés, Surf. Interfaces, 2024, 53, 105074 CrossRef CAS.
- K. Yan, Y. Qin, J. Xue, J. Wang, G. Wang, N. Wang and Y. Chu, Chem. Eng. J., 2024, 484, 149363 CrossRef CAS.
- J. Wang, X. Mu and M. Sun, Nanomaterials, 2019, 9, 218 CrossRef CAS PubMed.
- M. Sang, J. Shin, K. Kim and K. J. Yu, Nanomaterials, 2019, 9, 374 CrossRef CAS PubMed.
- D. Kireev, S. K. Ameri, A. Nederveld, J. Kampfe, H. Jang, N. Lu and D. Akinwande, Nat. Protoc., 2021, 16, 2395–2417 CrossRef CAS PubMed.
- M. Qin, Y. Xu, H. Gao, G. Han, R. Cao, P. Guo, W. Feng and L. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 35255–35263 CrossRef CAS PubMed.
- J. Han, I. Johnson and M. Chen, Adv. Mater., 2022, 34, 2108750 CrossRef CAS PubMed.
- Z. Li, W. Zhang and F. Xing, Int. J. Mol. Sci., 2019, 20, 2461 CrossRef PubMed.
- L. Sun, Y. Zhang, Y. Wang, Y. Yang, C. Zhang, X. Weng, S. Zhu and X. Yuan, Nanoscale, 2018, 10, 1759–1765 RSC.
- H. Gu, H. Tang, P. Xiong and Z. Zhou, Nanomaterials, 2019, 9, 130 CrossRef PubMed.
- Y. Esmaeili, E. Bidram, A. Zarrabi, A. Amini and C. Cheng, Sci. Rep., 2020, 10, 18052 CrossRef CAS PubMed.
- N. Karki, H. Tiwari, C. Tewari, A. Rana, N. Pandey, S. Basak and N. G. Sahoo, J. Mater. Chem. B, 2020, 8, 8116–8148 RSC.
- A. M. Oliveira, M. Machado, G. A. Silva, D. B. Bitoque, J. Tavares Ferreira, L. A. Pinto and Q. Ferreira, Nanomaterials, 2022, 12, 1149 CrossRef CAS PubMed.
- K. Joshi, B. Mazumder, P. Chattopadhyay, N. S. Bora, D. Goyary and S. Karmakar, Curr. Drug Delivery, 2019, 16, 195–214 CrossRef CAS PubMed.
- D. de Melo-Diogo, R. Lima-Sousa, C. G. Alves and I. J. Correia, Biomater. Sci., 2019, 7, 3534–3551 RSC.
- W. Hu, C. Peng, W. Luo, M. Lv, X. Li, D. Li, Q. Huang and C. Fan, ACS Nano, 2010, 4, 4317–4323 CrossRef CAS PubMed.
- Y. Liu, J. Wen, Y. Gao, T. Li, H. Wang, H. Yan, B. Niu and R. Guo, Appl. Surf. Sci., 2018, 436, 624–630 CrossRef CAS.
- R. K. Matharu, T. A. Tabish, T. Trakoolwilaiwan, J. Mansfield, J. Moger, T. Wu, C. Lourenço, B. Chen, L. Ciric and I. P. Parkin, J. Colloid Interface Sci., 2020, 571, 239–252 CrossRef CAS PubMed.
- X. Guo, X. Zhang, M. Yu, Z. Cheng, Y. Feng and B. Chen, J. Colloid Interface Sci., 2024, 661, 802–814 CrossRef CAS PubMed.
- A. Chatterjee, E. Perevedentseva, M. Jani, C. Y. Cheng, Y. S. Ye, P. H. Chung and C. L. Cheng, J. Biomed. Opt., 2015, 20, 051014 CrossRef PubMed.
- H. Wang, M. Zhang, Y. Ma, B. Wang, M. Shao, H. Huang, Y. Liu and Z. Kang, J. Mater. Chem. B, 2020, 8, 2666–2672 RSC.
- J. Wang, Y. Wei, X. Shi and H. Gao, RSC Adv., 2013, 3, 15776–15782 RSC.
- V. T. Pham, V. K. Truong, M. D. Quinn, S. M. Notley, Y. Guo, V. A. Baulin, M. Al Kobaisi, R. J. Crawford and E. P. Ivanova, ACS Nano, 2015, 9, 8458–8467 CrossRef CAS PubMed.
- J. Shangguan, Z. Wu, C. Qiao, Y. Zhang, L. Li, Q. Li, Y. Gao, H. Yan and W. Liu, ACS Omega, 2024, 9, 7034–7042 CrossRef CAS PubMed.
- Z. Ruan, Z. Xu, T. Liu, L. Chen, X. Liu, K. Chen and C. Zhao, Heliyon, 2024, 10, e38177 CrossRef CAS PubMed.
- D. Zhao, H. Liu, M. Xu, C. Yin, X. Xiao and K. Dai, Spectrochim. Acta, Part A, 2024, 314, 124195 CrossRef CAS PubMed.
- J. Wang, Y. Wang, H. Zhang, W. Zhu and L. Liu, Exp. Biol. Med., 2023, 248, 2227–2236 CAS.
- H. Li, J. Huang, Y. Song, M. Zhang, H. Wang, F. Lu, H. Huang, Y. Liu, X. Dai, Z. Gu, Z. Yang, R. Zhou and Z. Kang, ACS Appl. Mater. Interfaces, 2018, 10, 26936–26946 CrossRef CAS PubMed.
- M. Mortimer, Y. Wang and P. A. Holden, Front. Bioeng. Biotechnol., 2021, 9, 683520 CrossRef PubMed.
- G. E. Villalpando-Rodriguez and S. B. Gibson, Oxid. Med. Cell. Longevity, 2021, 2021, 9912436 CrossRef PubMed.
- S. Gurunathan, J. W. Han, A. A. Dayem, V. Eppakayala and J. H. Kim, Int. J. Nanomed., 2012, 7, 5901–5914 CrossRef CAS PubMed.
- C. D. Vecitis, K. R. Zodrow, S. Kang and M. Elimelech, ACS Nano, 2010, 4, 5471–5479 CrossRef CAS PubMed.
- W. Bing, H. Sun, Z. Yan, J. Ren and X. Qu, Small, 2016, 12, 4713–4718 CrossRef CAS PubMed.
- T. Cui, Y. Fan, Y. Liu, X. Fan, Y. Sun, G. Cheng and J. Cheng, Foods, 2023, 13, 67 CrossRef PubMed.
- R. Rosato, G. Santarelli, A. Augello, G. Perini, M. De Spirito, M. Sanguinetti, M. Papi and F. De Maio, Int. J. Mol. Sci., 2024, 25, 8033 CrossRef CAS PubMed.
- L. Sun, Y. Zhao, H. Peng, J. Zhou, Q. Zhang, J. Yan, Y. Liu, S. Guo, X. Wu and B. Li, J. Nanobiotechnol., 2024, 22, 210 CrossRef PubMed.
- W. Xia, Z. Wu, B. Hou, Z. Cheng, D. Bi, L. Chen, W. Chen, H. Yuan, L. Koole and L. Qi, Mater. Today Bio, 2025, 30, 101428 CrossRef CAS PubMed.
- J. W. Kim, E. V. Shashkov, E. I. Galanzha, N. Kotagiri and V. P. Zharov, Lasers Surg. Med., 2007, 39, 622–634 CrossRef PubMed.
- S. Tan, X. Wu, Y. Xing, S. Lilak, M. Wu and J. X. Zhao, Colloids Surf., B, 2020, 185, 110616 CrossRef CAS PubMed.
- R. Sun, H. Chen, L. Sutrisno, N. Kawazoe and G. Chen, Sci. Technol. Adv. Mater., 2021, 22, 404–428 CrossRef CAS PubMed.
- B. Z. Ristic, M. M. Milenkovic, I. R. Dakic, B. M. Todorovic-Markovic, M. S. Milosavljevic, M. D. Budimir, V. G. Paunovic, M. D. Dramicanin, Z. M. Markovic and V. S. Trajkovic, Biomaterials, 2014, 35, 4428–4435 CrossRef CAS PubMed.
- W. S. Kuo, H. H. Chen, S. Y. Chen, C. Y. Chang, P. C. Chen, Y. I. Hou, Y. T. Shao, H. F. Kao, C. L. L. Hsu and Y. C. Chen, Biomaterials, 2017, 120, 185–194 CrossRef CAS PubMed.
- W. S. Kuo, Y. T. Shao, K. S. Huang, T. M. Chou and C. H. Yang, ACS Appl. Mater. Interfaces, 2018, 10, 14438–14446 CrossRef CAS PubMed.
- M. Zühlke, T. T. Meiling, P. Roder, D. Riebe, T. Beitz, I. Bald, H.-G. Löhmannsröben, T. Janßen, M. Erhard and A. Repp, ACS Omega, 2021, 6, 23742–23749 CrossRef PubMed.
- P. Kadyan, P. Thillai Arasu and S. K. Kataria, Int. J. Biomater., 2024, 2024, 2626006 Search PubMed.
- M. P. Mingeot-Leclercq and J. L. Décout, MedChemComm, 2016, 7, 586–611 RSC.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan and H. Fang, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- X. Liu and K. L. Chen, Langmuir, 2015, 31, 12076–12086 CrossRef CAS PubMed.
- P. Yi and K. L. Chen, Environ. Sci. Technol., 2013, 47, 5711–5719 CrossRef CAS PubMed.
- W. Zhu, A. von dem Bussche, X. Yi, Y. Qiu, Z. Wang, P. Weston, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12374–12379 CrossRef CAS PubMed.
- T. Mashino, N. Usui, K. Okuda, T. Hirota and M. Mochizuki, Bioorg. Med. Chem., 2003, 11, 1433–1438 CrossRef CAS PubMed.
- M. Wu, R. Kempaiah, P. J. J. Huang, V. Maheshwari and J. Liu, Langmuir, 2011, 27, 2731–2738 CrossRef CAS PubMed.
- J. Liu, S. Fu, B. Yuan, Y. Li and Z. Deng, J. Am. Chem. Soc., 2010, 132, 7279–7281 CrossRef CAS PubMed.
- Y. J. Li, S. G. Harroun, Y. C. Su, C. F. Huang, B. Unnikrishnan, H. J. Lin, C. H. Lin and C. C. Huang, Adv. Healthcare Mater., 2016, 5, 2545–2554 CrossRef CAS PubMed.
- Y. Yao, T. Zhang and M. Tang, Environ. Pollut., 2023, 317, 120676 CrossRef CAS PubMed.
- I. E. M. Carpio, C. M. Santos, X. Wei and D. F. Rodrigues, Nanoscale, 2012, 4, 4746–4756 RSC.
- J. Chen, H. Peng, X. Wang, F. Shao, Z. Yuan and H. Han, Nanoscale, 2014, 6, 1879–1889 RSC.
- M. Dallavalle, M. Calvaresi, A. Bottoni, M. Melle-Franco and F. Zerbetto, ACS Appl. Mater. Interfaces, 2015, 7, 4406–4414 CrossRef CAS PubMed.
- H. Chen, B. Wang, D. Gao, M. Guan, L. Zheng, H. Ouyang, Z. Chai, Y. Zhao and W. Feng, Small, 2013, 9, 2735–2746 CrossRef CAS PubMed.
- J. Qi, T. Zhang, R. Zhang, J. Liu, M. Zong, Q. Zhang, Y. Ping, Y. Gong, B. Zhang and X. Liu, Sustainable Mater. Technol., 2024, 41, e01087 CrossRef CAS.
- Y. Wang, T. Li, L. Lin, D. Wang and L. Feng, RSC Adv., 2024, 14, 27873–27882 RSC.
- S. Zhang, L. Wang, T. Xu and X. Zhang, ACS Appl. Mater. Interfaces, 2023, 15, 9110–9119 CrossRef CAS PubMed.
- J. Liu, S. Lu, Q. Tang, K. Zhang, W. Yu, H. Sun and B. Yang, Nanoscale, 2017, 9, 7135–7142 RSC.
- G. Liang, H. Shi, Y. Qi, J. Li, A. Jing, Q. Liu, W. Feng, G. Li and S. Gao, Int. J. Nanomed., 2020, 15, 5473–5489 CrossRef CAS PubMed.
- T. J. Simmons, S. H. Lee, T. J. Park, D. P. Hashim, P. M. Ajayan and R. J. Linhardt, Carbon, 2009, 47, 1561–1564 CrossRef CAS.
- H. Z. Zardini, A. Amiri, M. Shanbedi, M. Maghrebi and M. Baniadam, Colloids Surf., B, 2012, 92, 196–202 CrossRef CAS PubMed.
- X. Dong, E. McCoy, M. Zhang and L. Yang, J. Environ. Sci., 2014, 26, 2526–2534 CrossRef PubMed.
- S. Aslan, M. Deneufchatel, S. Hashmi, N. Li, L. D. Pfefferle, M. Elimelech, E. Pauthe and P. R. Van Tassel, J. Colloid Interface Sci., 2012, 388, 268–273 CrossRef CAS PubMed.
- M. Wang, Y. Su, Y. Liu, Y. Liang, S. Wu, N. Zhou and J. Shen, J. Colloid Interface Sci., 2022, 608, 973–983 CrossRef CAS PubMed.
- X. Bai, X. Zhang, J. Xiao, X. Lin, R. Lin, R. Zhang, X. Deng, M. Zhang, W. Wei, B. Lan, S. Weng and M. Chen, Adv. Healthcare Mater., 2024, 13, 2302873 CrossRef CAS PubMed.
- M. Mazaheri, O. Akhavan and A. Simchi, Appl. Surf. Sci., 2014, 301, 456–462 CrossRef CAS.
- S. Faghihi, M. Gheysour, A. Karimi and R. Salarian, J. Appl. Phys., 2014, 115, 244307 CrossRef.
- W. Z. Weng, Study on the treatment of infectious bone defects and wound scars after open fractures with carbon nanomaterials, PhD thesis, Second Military Medical University of the Chinese People's Liberation Army, Shanghai, 2017.
- Y. Ma, D. Bai, X. Hu, N. Ren, W. Gao, S. Chen, H. Chen, Y. Lu, J. Li and Y. Bai, ACS Appl. Mater. Interfaces, 2018, 10, 3002–3010 CrossRef CAS PubMed.
- N. K. de Moura, G. F. d. M. Morgado, E. F. Martins, C. A. Escanio, E. Esposito, J. Marini and F. R. Passador, Macromol. Symp., 2022, 406, 2200020 CrossRef CAS.
- R. Gayathri, K. ArulJothi, K. H. Raj and S. Gnanavel, Results Surf. Interfaces, 2024, 17, 100287 CrossRef.
- X. Zhang, X. Bai, X. Deng, K. Peng, Z. Zheng, J. Xiao, R. Zhang, Z. Huang, J. Huang, M. Chen and S. Weng, Carbon, 2023, 213, 118229 CrossRef CAS.
- Á. Serrano-Aroca, K. Takayama, A. Tuñón-Molina, M. Seyran, S. S. Hassan, P. Pal Choudhury, V. N. Uversky, K. Lundstrom, P. Adadi and G. Palù, ACS Nano, 2021, 15, 8069–8086 CrossRef PubMed.
- Y. Wu, C. Li, H. C. van der Mei, H. J. Busscher and Y. Ren, Antibiotics, 2021, 10, 623 CrossRef CAS PubMed.
- C. Zhang, L. Zhang, X. Liu, L. Chen, Y. Yang and S. Yu, Mater. Rev., 2020, 34, 53–57 Search PubMed.
- R. Teixeira-Santos, L. C. Gomes, R. Vieira, F. Sousa-Cardoso, O. S. Soares and F. J. Mergulhão, Nanomaterials, 2023, 13, 2604 CrossRef CAS PubMed.
- Y. Li, X. Liu and T. Wu, China Environ. Sci., 2021, 41, 4402–4414 CAS.
- I. S. Raja, S.-J. Song, M. S. Kang, Y. B. Lee, B. Kim, S. W. Hong, S. J. Jeong, J.-C. Lee and D.-W. Han, Nanomaterials, 2019, 9, 1214 CrossRef CAS PubMed.
- M. Kim, D. Goerzen, P. V. Jena, E. Zeng, M. Pasquali, R. A. Meidl and D. A. Heller, Nat. Rev. Mater., 2024, 9, 63–81 CrossRef CAS.
- Z. Peng, X. Liu, W. Zhang, Z. Zeng, Z. Liu, C. Zhang, Y. Liu, B. Shao, Q. Liang, W. Tang and X. Yuan, Environ. Int., 2020, 134, 105298 CrossRef CAS PubMed.
- S. Chen, Y. Jiang, Z. Zhu, Q. Zhang, C. Zhang, Q. Zhang, W. Qian, S. Zhang and F. Wei, Adv. Sci., 2024, 11, 2306355 CrossRef CAS PubMed.
- N. Nasirzadeh, Y. Mohammadian, Y. Rasoulzadeh, M. Rezazadeh Azari and F. Khodagholi, Tanaffos, 2022, 21, 391–400 Search PubMed.
- Y. Chiou, C. Lin, S. Harroun, Y. Chen, L. Chang, A. Wu, F. Chang, Y. Lin, H. Lin, A. Anand, B. Unnikrishnan, A. Naing and C. Huang, Nanoscale, 2022, 14, 11719–11730 RSC.
- M. Zhang, Y. Wang, C. Miao, S. Lin, Y. Zheng, X. Lin, Y. Wang, X. Lin, X. Zhu and S. Weng, Acta Biomater., 2025, 200, 591–609 CrossRef PubMed.
Footnote |
| † Equally contributed. |
| This journal is © The Royal Society of Chemistry 2025 |

