Open Access Article
Open Access ArticleStochastic generalization models learn to comprehensively detect volatile organic compounds associated with foodborne pathogens via Raman spectroscopy†
Bohong Zhang *a,
Anand K. Nambisana,
Abhishek Prakash Hungunda,
Xavier Jonesb,
Qingbo Yang*b and
Jie Huang
*a,
Anand K. Nambisana,
Abhishek Prakash Hungunda,
Xavier Jonesb,
Qingbo Yang*b and
Jie Huang *a
*a
aDepartment of Electrical and Computer Engineering, Missouri University of Science and Technology, Rolla, Missouri 65409, USA. E-mail: bzdtx@mst.edu; jieh@mst.edu
bCooperative Research, College of Agriculture, Environmental and Human Sciences, Lincoln University of Missouri, Jefferson City, MO 65101, USA. E-mail: YangQ@lincolnu.edu
First published on 13th February 2025
Abstract
Ensuring food safety requires continuous innovation, especially in the detection of foodborne pathogens and chemical contaminants. In this study, we present a system that combines Raman spectroscopy with machine learning (ML) algorithms for the precise detection and analysis of VOCs linked to foodborne pathogens in complex liquid mixtures. A remote fiber-optic Raman probe was developed to collect spectral data from 42 distinct VOC mixtures, representing contamination scenarios with dilution levels ranging from undiluted to highly diluted states. A dataset comprising 1445 Raman spectra was analyzed using classification and regression ML models, including multi-layer perceptron (MLP), random forest, and extreme gradient boosting decision trees (XGBDT). The optimized ML models achieved over 90% classification accuracy for pure VOCs and demonstrated robust performance in identifying mixtures containing up to six VOCs at concentrations as low as 0.25% (400-fold dilution). Additionally, regression analysis effectively predicted VOC concentrations at levels as low as 1% (100-fold dilution), with the best model achieving an R2 value exceeding 0.82. This approach demonstrates the potential for rapid and real-time food safety monitoring, effectively overcoming the limitations of traditional methods such as culture-based or qPCR techniques, while its ability to reliably classify complex VOC mixtures makes it a valuable tool for on-site food safety assessments and quality control applications across various industries.
Introduction
The global demand for fresh, organic, and minimally processed foods has steadily increased in recent years, driven by advancements in agricultural technology and consumer preference for healthier, preservative-free options. While this trend offers significant health benefits, it also introduces greater risks of contamination by foodborne pathogens due to the reduced use of chemical preservatives and the complex logistics involved in the production, transportation, and retailing of these products. Organic and fresh foods, particularly those that rely on minimal processing, are highly susceptible to contamination by pathogens such as Listeria monocytogenes, Salmonella spp., and Escherichia coli during various stages of the supply chain. The complexity of food production, coupled with compromised cold-chain management and the inherent vulnerability of fresh products, has resulted in increased incidents of foodborne disease outbreaks which underscore the urgent need for more efficient and reliable detection methods in the food industry.Traditional detection methods, such as agar plate cultures, quantitative polymerase chain reaction (qPCR), and enzyme-linked immunosorbent assay (ELISA) kits, remain the gold standard for pathogen identification in food. These methods are widely used by regulatory agencies such as the USDA and FDA to ensure food safety compliance. However, these techniques present several challenges when applied to large-scale food production. For example, agar plate cultures require extended incubation times, often taking days to produce results, making them unsuitable for real-time monitoring in fast-paced food production environments.1 Similarly, qPCR and ELISA-based methods, while more rapid, still require specialized laboratory equipment, skilled technicians, and substantial sample preparation, limiting their feasibility for on-site detection and their scalability across large sample volumes.2,3 Moreover, these conventional methods struggle with detecting low levels of pathogens, particularly in cases where the pathogens enter a viable but non-culturable (VBNC) state due to environmental stressors.4 In such conditions, bacteria can evade detection by traditional culturing methods despite remaining viability and pathogenic potential, thus posing an undetected threat to public health.5 Additionally, the complexity of modern food matrices where multiple types of foods and contaminants co-exist further complicating the detection process. Current methods are often limited in their ability to analyze complex, mixed samples, which may contain multiple VOCs and various interfering substances that mask the presence of pathogens.6 Recent technological advancements, such as electronic nose (E-nose) devices, offer new possibilities for pathogen detection in food.7 These devices measure the conductivity or permittivity changes in the air surrounding food products to detect spoilage or contamination by monitoring VOCs. However, while E-nose technology shows promise for providing general information on food quality, it lacks the ability needed to identify specific compounds in a complex VOC mixture. E-nose devices also struggle with detecting pathogens in low concentrations and have yet to achieve the sensitivity required for widespread use in the food industry. Early-stage spoiled food emits VOCs at 5–100 ppm, with levels increasing exponentially as spoilage progresses. Under refrigeration, VOC release is inhibited, even in highly contaminated samples. For instance, indole—a VOC linked to E. coli—can reach 20–40 ppm in refrigerated seafood but often requires heating to 100 °C for GC-MS detection.8
Given these limitations, there is a critical need for the development of new detection systems that are faster, more sensitive, and capable of processing complex food samples in real-time. One promising solution is the application of Raman spectroscopy. Raman spectroscopy is a non-destructive optical technique that detects molecular vibrations, providing detailed information about a sample's chemical composition.9–11 It has shown significant potential for identifying pathogen-specific VOCs in food, offering high sensitivity and selectivity at a relatively low cost.12–14 However, while Raman spectroscopy has been applied successfully in simple sample matrices, its use in complex food systems has been limited by challenges in interpreting the resulting spectra. To overcome these challenges, integrating Raman spectroscopy with ML algorithms offers a powerful approach. ML algorithms, including classification and regression models, can analyze the vast amounts of spectral data generated by Raman systems and detect subtle differences in VOC profiles, even within complex mixtures. By automating the analysis process, ML can enhance the precision and speed of VOC detection, allowing for the real-time identification of foodborne pathogens in diverse food matrices.13,15 However, the benchtop Raman system is limited by the need for complex sample preparation and its inability to support remote or on-site monitoring. To overcome these challenges, this study introduces a system that integrates Raman spectroscopy with advanced ML algorithms, utilizing a fiber-optic-based Raman probe for real-time, high-throughput detection and classification of pathogen-specific VOC signatures in food samples. Building on our previous study,14 this work advances the methodology by optimizing ML algorithms to improve both accuracy and efficiency for much more complex and mixture samples with low concentration levels. The big number of datasets and the introduction of regression analysis as an additional layer of data analysis, enabling more precise quantification of pathogen concentrations. By leveraging the power of machine learning, the system efficiently extracts molecular information from Raman spectra and accurately identifies VOCs at varying concentrations. Compared to traditional detection systems, this approach offers significant advantages, including portability, rapid on-site analysis, and the ability to process complex samples without extensive preparation. These enhancements hold potential for transforming food safety monitoring, providing a reliable, cost-effective, and scalable solution for real-time pathogen detection in the food industry.
Materials & methods
Chemicals & sample preparation
In this study, we utilized 42 different VOC mixtures as target analytes, comprising combinations of compounds such as 2-nonanone, 2-undecanone, 1-dodecanol, 3-methyl-butanoic acid, acetoin, octanol, methyl-trisulfide, and 3-hydroxy-2-butanone. The VOCs selected—such as indole, benzothiazole, and 3-methylbutanal—are well-documented as biochemical markers associated with foodborne pathogens. For example, indole and benzothiazole are characteristic of E. coli,16,17 while Listeria monocytogenes produces 3-methylbutanal and dodecanal.18 These compounds act as fingerprints, enabling the identification and prediction of food spoilage.19,20 To ensure the accuracy and reliability of the VOC mixtures, a stringent preparation protocol was followed. Pure samples of each selected VOC, with a purity greater than 99.99%, were initially prepared in individual 2 mL volumes. These pure VOCs were then combined in predetermined ratios to create two-, three-, and four-component mixtures. The mixtures were organized into distinct classes, with each receiving a class label reflecting the unique chemical combinations involved, as outlined in Fig. 1. Each VOC mixture was diluted using acetonitrile (ACN) solvent to create a series of concentrations, ranging from undiluted (0×, 100%) to progressively lower levels at 5× (20%), 10× (10%), 20× (5%), and 400× (0.25%). These dilutions, which extended down to parts-per-million (ppm) levels, allowed for a comprehensive assessment of the system's sensitivity across a wide range of concentrations. The corresponding VOC concentrations at each dilution factor, expressed in units per milliliter (unit/mL), percentage (%), and parts per million (ppm), are detailed in Table S2 of the ESI.† The wide range of chemical combinations was designed to reflect the complexity of real-world samples, such as those found in spoiled food products or pathogen-contaminated environments. To further expand the dataset for ML algorithms, five Raman spectra were collected for each dilution within each class label. This generated a comprehensive dataset of 42 distinct classes, resulting in 289 data sets and 1445 Raman spectra, ensuring that the model could robustly learn and predict VOC patterns across different concentrations and combinations.Raman spectra analysis of VOC samples
To simulate real-world scenarios of foodborne pathogen detection, we created a broad dataset by preparing complex VOC mixtures at varying dilution levels. Firstly, understanding the Raman spectra of both pure and mixed VOC samples is essential for accurate analysis. Fig. 2(a–c) exemplified the Raman spectra of pure VOCs – 2-nonanone, 2-undecanone where distinct peaks are clearly observed and linked to specific molecular vibrations. For instance, the symmetric C–H stretching bond at 2888 cm−1 and asymmetric C–H stretching bond at 2921 cm−1 in 2-nonanone;21 the CH2–CH3 stretching bond at 2879 cm−1 and C–H stretching at 2913 cm−1 and 2966 cm−1 in 3-methyl-1-butanethiol;22 and the CH3 stretching bond at 2946 cm−1 in acetoin, sharp peaks.23 These pure spectra provide a baseline for understanding the behavior of individual VOCs in Raman spectroscopy. As the VOCs are mixed and their Raman spectra recorded at 5× (20%), 20× (10%), and 100× (1%) dilution levels in Fig. 2(d–f), notable differences in peak intensity and spectral clarity emerge. At the 5× (20%) dilution, characteristic Raman peaks of the pure VOCs are still identifiable, although some overlap occurs due to the combination of compounds. The distinct chemical bond features, such as the symmetric C–H stretching bond, are still visible, albeit at reduced intensity compared to the undiluted samples. As the dilution increases to 20× (5%), the VOC-specific peaks become noticeably weaker in the overlap region. By the time the dilution reaches 100× (1%), the Raman peaks corresponding to the VOCs are significantly diminished or almost absent. Instead, the spectra are increasingly dominated by the peaks of the acetonitrile (ACN) solvent, particularly the C–H and C![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) N bonds, which become the most prominent features. This shift presents a significant challenge for detecting and identifying VOCs at higher dilutions, as the spectral signatures of the VOCs are increasingly masked by the solvent's peaks. In particular, the Raman spectra at the 100× dilution (1%), make it difficult to pinpoint specific VOCs because the solvent's peaks overwhelm the already faint signals from the VOCs. To overcome this challenge, advanced ML techniques are applied to analyze and deconvolute the complex and overlapping spectra. These optimized models are designed to detect subtle variations in the Raman signals that are not easily discernible through conventional methods, allowing for more accurate detection and classification of VOCs even at extreme dilutions. Fig. 3 illustrates the workflow for Raman spectroscopy-based detection and analysis of VOCs, covering experimental setup, data acquisition, and computational analysis. The setup includes a 3D optical stage for precise alignment of a fiber-optic Raman probe, connected to a laser source and a spectrometer for signal acquisition. Raman spectra were collected using a 532 nm excitation beam with a laser power of 100 mW, a focusing lens with a working distance of 0.8 cm, and a spot size of approximately 100 micrometers. Each spectrum was acquired over 10 seconds. The spectrometer measured wavenumbers from 0 to 4574 cm−1 with a resolution of about 6 cm−1, while the machine learning analysis focused on the 2600–3200 cm−1 range, which encompasses the dominant Raman peaks of the selected VOCs. This ensures detailed spectral analysis and captures subtle features influencing model performance. The acquired spectra were preprocessed with baseline correction using polynomial fitting, smoothing via a 3rd-order Savitzky–Golay filter, and normalization to the most prominent peak. These processed spectra were then analyzed using machine learning models for classification and regression to predict VOC concentrations and identify specific compounds in mixtures.
N bonds, which become the most prominent features. This shift presents a significant challenge for detecting and identifying VOCs at higher dilutions, as the spectral signatures of the VOCs are increasingly masked by the solvent's peaks. In particular, the Raman spectra at the 100× dilution (1%), make it difficult to pinpoint specific VOCs because the solvent's peaks overwhelm the already faint signals from the VOCs. To overcome this challenge, advanced ML techniques are applied to analyze and deconvolute the complex and overlapping spectra. These optimized models are designed to detect subtle variations in the Raman signals that are not easily discernible through conventional methods, allowing for more accurate detection and classification of VOCs even at extreme dilutions. Fig. 3 illustrates the workflow for Raman spectroscopy-based detection and analysis of VOCs, covering experimental setup, data acquisition, and computational analysis. The setup includes a 3D optical stage for precise alignment of a fiber-optic Raman probe, connected to a laser source and a spectrometer for signal acquisition. Raman spectra were collected using a 532 nm excitation beam with a laser power of 100 mW, a focusing lens with a working distance of 0.8 cm, and a spot size of approximately 100 micrometers. Each spectrum was acquired over 10 seconds. The spectrometer measured wavenumbers from 0 to 4574 cm−1 with a resolution of about 6 cm−1, while the machine learning analysis focused on the 2600–3200 cm−1 range, which encompasses the dominant Raman peaks of the selected VOCs. This ensures detailed spectral analysis and captures subtle features influencing model performance. The acquired spectra were preprocessed with baseline correction using polynomial fitting, smoothing via a 3rd-order Savitzky–Golay filter, and normalization to the most prominent peak. These processed spectra were then analyzed using machine learning models for classification and regression to predict VOC concentrations and identify specific compounds in mixtures.
Data processing & ML algorithms
Data preprocessing & feature extraction
The data preprocessing step involves splitting all the Raman VOC spectral data into train set, test set, and validation set. The train and hold out test sets are split first with split ratio 0.2, and then the remaining train set is split into train and validation sets again with 0.2 split ratio. Therefore, the validation set size is calculated based on the train set size after the test set is split. Before the data is used for training any ML model, key features need to be extracted from the Raman spectra and pre-processed before the model training. The features extracted from the Raman spectra of each VOC type, shown in Fig. 4 are tabulated in a data frame. A feature extraction algorithm is developed to extract wavenumber and frequency domain features from each Raman spectrum and appended to the data frame. Each wavenumber and frequency domain features attempts to capture different characteristics of the Raman spectrum. A total of 96 features were extracted from each spectrum. The features most influential in classification and prediction during training and testing are highlighted in the feature importance plots shown in Fig. 4. The selection of features in this study was inspired by a previous study.24 Feature importance metrics were calculated using Shapley Additive Explanations (SHAP) values, computed through the Kernel Explainer module in Python. SHAP values here indicate how much each feature in the dataset contributed either positively or negatively to the predicted output and it is computed by using a weighted linear regression method. The feature importance plots for the other two models can be found in Fig. S1 in the ESI.† | ||
| Fig. 4 Feature importance plots for 3 prominent pipeline classifiers. FI plot for (a) rank-1 MLP classifier, (b) rank-3 random forest classifier, and (c) rank-2 XGBDT classifier. The plots are tabulated according to the pure VOCs' class labels S1 to S8. When these VOCs are mixed to obtain a mixture VOC, the feature importance (or mean of SHAP values) of that mixture will be a derivative of the mean SHAP value of the pure VOCs used to obtain the mixture. The feature aliases in the x-axis of the bar plots are defined in the Table S1 in the ESI.† These plots give detailed insight into the impact of certain features on the model pipeline performance and are plotted right to left with the most important feature (i.e., the feature with largest mean SHAP value) starting at the right. Higher mean SHAP values suggest a greater impact on the model's output. Features with larger bars are more influential in the model's predictions (P. S. more detailed and clear information can be found in Fig. S4 of the ESI†). | ||
Features in this context encompass statistical metrics (e.g., mean, standard deviation, skewness), spectral peaks (e.g., intensity, position, width) derived from the wavenumber (Raman shift) domain, as well as frequency-domain characteristics (e.g., Fourier transforms, power spectral density). Some features exhibit potential correlations between the wavenumber and frequency domains. For instance, in the wavenumber (Raman shift) domain, statistical features such as signal minima, maxima, mean, and median collectively summarize the central tendency and range of the Raman signal, providing insights into its overall distribution and variation. In frequency domain, features such as max frequency power and max frequency magnitude can be correlated. Even the histogram features can have correlations depending on the distribution of the Raman spectra. Having correlations in data is not necessarily an issue with the exception that between similar Raman signals, it might lead to multi-collinearity causing inaccuracies in regression models. This has been overcome by the implementation of feature selection or dimensionality reduction techniques such as principal component analysis (PCA) to remove the redundant features in the feature extraction algorithm. The PCA resulted in a drop in the performance, which was rectified by dropping the most highly correlated features.
Model training & performance metrics
The improved cross-validation (CV) algorithm shown in Fig. 5, first initializes the stratified CV and arrays to store the evaluation metrics for each fold or train/test split. It then loops over each split provided by the stratified CV and prepares the training and validation data. The data for the classifier is kept separate from the data for the regressor because they are predicting different targets. It then constructs a pipeline for the classifier, which includes scaling the features and the classifier itself. Then, grid search is performed on this pipeline, passing the training data and the classification targets. It uses the best classifier found by the grid search to predict the training and validation data and adds these predictions as new features to the regression inputs. It constructs a similar pipeline for the regressor, which includes the scaler, the classifier's predictions as features, and the regressor itself. It performs grid search on this pipeline as well, passing the regression inputs and targets. It uses the best regressor found by the grid search to predict the validation data. It calculates various performance metrics (f1, precision, recall, mean squared error, and R2) on the validation set, where the f1 score is given by:
 | (1) |
 | (2) |
 | (3) |
 | (4) |
 | (5) |
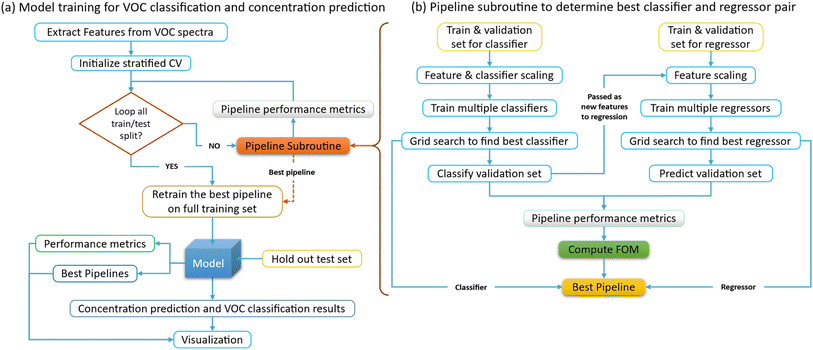 | ||
| Fig. 5 Improved cross-validation algorithm for stacked generalization model training to classify VOC and predict its concentration. (a) Model training for VOC classification and concentration prediction. (b) Pipeline subroutine to determine best classifier and regressor pair. The pipeline method for stochastic generalization models in ML boosts efficiency by optimizing each stage individually and ensures consistent, reliable results by maintaining uniform processes. It is scalable to accommodate large feature datasets that can be trained by complex models, while enhancing reproducibility across different environments. It also provides a structured workflow, making it easier to iterate on models and experiments. The figure of merit shown in eqn (5) is used to determine the best model pipeline, which depends on both the f1 score of the classifier and R2 value of the regressor. | ||
After all folds have been processed, it retrains the best pipeline on the full training set and evaluates this final model on the holdout test set, calculates the same performance metrics as before, and returns these along with the CV metrics and the best pipeline. The stacked generalization scheme involves model training for both classification and regression, where the VOC needs to be classified, and its concentration needs to be predicted. To optimally decide the combination of a regressor and classifier, a 5-fold cross-validation algorithm is implemented for this multi-label classification and regression. Grid search inside each cross-validation (CV) fold is performed to ensure that the hyperparameters of the model are optimized separately for each fold. The primary purpose of CV is to estimate how well the trained model generalizes to unseen data.
To make a robust estimate, the model is trained and tested on different subsets of VOC data multiple times. Grid search CV is then used to find the best hyperparameters for the model on a particular fold. This is done by training multiple versions of a model on the same data but with different hyperparameters, then selecting the hyperparameters that produce the best performance according to f1 score metric. This approach provides a more robust estimate of the model's performance because it minimizes data leakage—i.e., information from the validation set influencing the model training process. If the hyperparameters were optimized on the full data before performing CV, information from the validation set would indirectly influence the training process, leading to an overly optimistic performance estimate. This approach requires more computational resources since it needs to perform grid search CV (which is already computationally intensive) multiple times—i.e., once for each CV fold. Further, a pipeline system is defined for both the regression and classification models. An improved version of CV function is implemented to keep track of the best pipeline and not just select the best individual classifier and regression models. The flow of the implemented CV and training algorithm is shown in Fig. 5. This process ensures that the algorithm not only finds the best combination of parameters for each model but also the best combination of models (classifier and regressor) for this specific task. This is achieved by evaluating each pipeline on a validation set that is kept separate during the grid search. The combination of stratified CV and grid search allows the algorithm to evaluate each pipeline fairly and comprehensively. The choice of using the average of f1 and R2 scores as the selection criterion for FOM was made since both tasks, classification, and regression, are equally important for the overall performance of our model. The f1 score is a commonly used metric for classification problems, especially in cases where the data may be unbalanced. It combines precision and recall giving a single measure of the quality of the classifier. On the other hand, the R2 score (coefficient of determination) is a statistical measure that represents the proportion of the variance for the dependent variable that's explained by the independent variables in a regression model. If one of the tasks (classification or regression) is more important depending on the application, a weighted average can be used that can give more prominence to the metric corresponding to that task (either f1 or R2). Alternatively, other factors such as training time, complexity of the model, and interpretability, could be considered and added to the model selection criterion. Both f1 and R2 scores are single-value summaries of the performance of the trained model.
Results & discussion
VOC classification
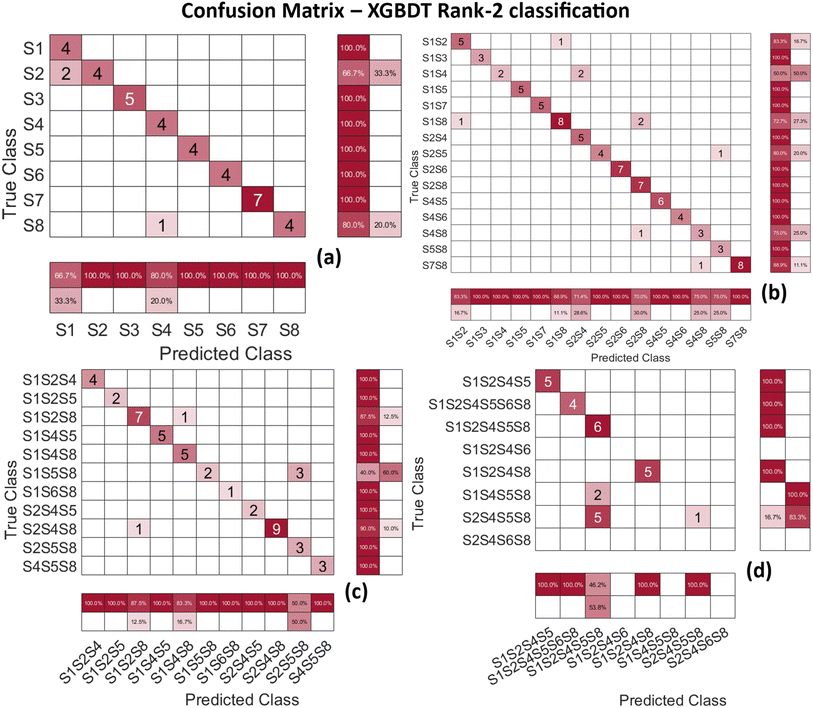
The MLP rank-1 classification results are shown in Fig. 6. Classes S1, S2 and S3, correspond to the pure VOCs 2-nonanone, 2-undecanone, and 1-dodecanol respectively. The chemical structure of these VOCs shows striking similarity between each other. All three of them have a zig-zag carbon–hydrogen chain with an oxygen atom at the end of the chain. The only difference is the position of the oxygen atom in the chain and the number of C–H links in the chain. This results in large similarity with very few minor differences in Raman spectra of these 3 VOCs. When features are extracted, they end up having high correlation leading to misclassification. These misclassifications only increase with the mixtures due to the increase in correlation between features. The detailed explanation can be found in Fig. S2 in the ESI.† The use of recurrent neural networks (RNN) can alleviate this issue when dealing with multi-collinear data. This avenue is being pursued as part of future work.Next, Random Forest rank-3 classifier is tested, and its classification results are shown in the confusion matrix in Fig. 7 below. When compared to the MLP rank-1 classifier in the previous Fig. 6, there is a drastic improvement in the classification performance of mixture VOCs classification when the rank is increased. The S3 class label corresponding to 1-dodecanol VOC particularly shows a high degree of similarity with S1 class label corresponding to 2-nonanone VOC due to their chemical composition being near identical. The blank spaces along the diagonal of the confusion matrix are due to the lack of samples for those mixtures as shown (right side) in Fig. 7(b) and (d). Particularly observing the misclassified samples in Fig. 7(b), for example, class label S2S4 which corresponds to a mixture of 2-undecanone and 3-methyl-butanoic acid, is misclassified as class label S1S4 which corresponds to a mixture of 2-nonanone and 3-methyl-butanoic acid. This is notable because it tells the misclassification of the mixture VOC is due to the pure VOC samples themselves being misclassified as shown in Fig. 7(a).
In Fig. 8(a), the pure VOCs are classified by the XGBDT R2 classifier presents the best classification amongst all the classifiers with a weight mean f1 score of 0.90. When compared to the performance of the previous classifiers in Fig. 6 and 7, the XGBDT rank-2 classifier performs much better as the number of misclassified samples is drastically reduced. Obtaining high classification accuracy poses a challenge due to the dataset being imbalanced i.e., the number of samples in each class vary greatly or there is no equal amount of data for each VOC. This imbalance arises due to dropping of highly correlated features. Hence, f1 score presents a better measure of the classification problem in this case. It is defined as the harmonic mean of the precision and recall as shown in eqn (1). The performance metrics for all the classifiers trained in the model pipeline are shown in Table 1. Both macro and weighted averages are computed to be flexible with the data. Initially macro average is calculated to determine the classifier performance by giving equal importance to each class regardless of the class distribution. Finally, the weighted average is calculated, where the metrics for each label are determined independently, and weight is given to those labels based on the number of samples in the dataset.
| Classifier | Macro avg f1-score | Macro avg precision | Macro avg recall | Weighted avg f1-score | Weighted avg precision | Weighted avg recall |
|---|---|---|---|---|---|---|
| XGBDT rank-1 | 0.92 | 0.92 | 0.93 | 0.90 | 0.88 | 0.93 |
| XGBDT rank-2 | 0.92 | 0.92 | 0.93 | 0.90 | 0.88 | 0.93 |
| MLP | 0.80 | 0.82 | 0.80 | 0.82 | 0.82 | 0.82 |
| RF rank-2 | 0.79 | 0.94 | 0.75 | 0.88 | 0.91 | 0.87 |
| RF rank-3 | 0.79 | 0.94 | 0.75 | 0.88 | 0.91 | 0.87 |
Regression for dilution prediction
Knowing the dilution levels of VOCs is crucial when detecting foodborne pathogens because it helps to ensure the accuracy and sensitivity of detection method employed. VOCs are often used as indicators of microbial contamination, and their concentration can affect the reliability of the results. Proper dilution ensures that the detection methods are not overwhelmed by high concentrations of VOCs, which could lead to false positives or negatives. If the dilution levels are too high, the concentration of VOCs may fall below the detection threshold, leading to inaccurate or missed detections. This precision is essential for early detection and control of foodborne pathogens, ultimately enhancing food safety and public health. The max dilution level for effective detection depends on several factors, like the sensitivity of detection method, the specific VOCs being measured, and the type of foodborne pathogen.Regression analysis was conducted using five different algorithms, with the three that produced the best results shown in Fig. 9: Random Forest rank-1 regression, Random Forest rank-3 regression, and XGBDT rank-2 decision tree regression. The remaining two algorithms are included in Fig. S3 in the ESI.† In each graph, the shaded red area around the regression line represents the interquartile range (IQR), where a smaller IQR indicates more consistent predictions. As the dilution level increases, the IQR broadens, reflecting greater inconsistency in the predictions. The mean serves as a measure of central tendency, while the standard deviation indicates the extent of variation or dispersion in the data. A lower standard deviation suggests that the values are clustered more closely around the mean. The R2 value, also called the coefficient of determination, represents the proportion of variance in the dependent variable that can be explained by the independent variables. Higher R2 values indicate a better model fit, making R2 a suitable performance metric for evaluating the regressor. Among the models, the rank-2 XGBDT regression has the highest R2 value, making it the most effective regressor in the pipeline. The XGBoost rank-2 regression model stands out as the best overall model, with the lowest errors across most metrics, making it highly reliable for predicting VOC dilution levels. In contrast, the Random Forest models exhibit slightly higher errors, suggesting greater sensitivity to data variations or outliers. Both XGBDT rank-1 and rank-2 models show lower Mean Squared Error (MSE), higher R2 values, and greater explained variance, indicating they provide more accurate predictions compared to the Random Forest models. Additionally, the Random Forest rank-1, rank-2, and rank-3 models display progressively higher MSE, reflecting less accurate predictions. The XGBDT models also have the lowest mean and median absolute errors, further emphasizing their superior performance. Moreover, all error metrics are influenced by increasing dilution levels, which affect the overall accuracy of predictions (Table 2).
| Regression model | Mean squared error (MSE) | Coefficient of determination (R2) | Explained variance | Maximum error | Mean absolute error | Median absolute error |
|---|---|---|---|---|---|---|
| XGBDT rank-1 | 200.36 | 0.80 | 0.81 | 47.67 | 10.68 | 7.61 |
| XGBDT rank-2 | 191.76 | 0.81 | 0.82 | 48.14 | 10.52 | 8.03 |
| RF rank-1 | 252.40 | 0.75 | 0.75 | 71.57 | 11.80 | 9.35 |
| RF rank-2 | 268.37 | 0.73 | 0.74 | 61.75 | 12.23 | 9.4 |
| RF rank-3 | 274 | 0.73 | 0.73 | 63.45 | 12.30 | 9.57 |
Advancements, limitations, and future directions in Raman spectroscopy-based foodborne pathogens detection
This study presents a comprehensive integration of Raman spectroscopy with advanced ML techniques to enable the detection and quantification of VOCs associated with foodborne pathogens. Our approach is built on a robust methodological foundation, inspired by prior work that explored the synergistic use of spectroscopy and ML for chemical detection. Recent advancements in Raman spectroscopy, when combined with machine learning (ML) and artificial intelligence (AI), have shown significant potential in the rapid detection and classification of foodborne pathogens. For instance, researchers have successfully utilized Raman spectroscopy and decision tree algorithms to analyze single-cell spectra, achieving serotype-level discrimination of bacterial strains with accuracy rates of up to 95.8%.25 However, the cumbersome culture process and large sample requirements often hinder practical applications. To address this, generative adversarial networks (GANs) and support vector machines (SVMs) have been employed, demonstrating improved classification accuracy and reduced sample requirements by generating synthetic training data.26 Similarly, deep learning and machine learning approaches, such as one-dimensional convolutional neural networks (1D-CNNs) and random forest, have been integrated with portable Raman devices, enabling detection of pathogens in contaminated food samples with high accuracies above 95%.27 Surface-enhanced Raman scattering (SERS) platforms have further enhanced sensitivity through nanostructures like hydrophobic Si substrates, which improve local enrichment and, when paired with ML, achieve 100% classification accuracy for multiple bacterial species.28 These advancements are complemented by innovative photonics-based sensing systems, which use ML to process real-time optical signals, achieving detection accuracies of up to 95% in fresh produce samples.29 Additionally, convolutional neural networks (CNNs) and SERS have been leveraged to address challenges such as weak signals, complex spectra, and limited datasets, ensuring applicability across diverse environments and food safety scenarios.30 Drawing from these advancements, our study employs a stacked generalization pipeline to address the challenges of spectral overlap, data complexity, and high-dimensionality inherent in Raman spectroscopy. The proposed system achieves figures of merit, including a classification accuracy of 90% for pure VOCs, robust regression results with R2 values exceeding 0.82, and successful detection of analytes at concentrations as low as 0.1% for regression tasks and 0.25% for classification. These results place our approach ahead of traditional methods such as agar plate cultures,31 qPCR,32 and ELISA,33 which, despite their sensitivity, require extensive laboratory infrastructure, significant sample preparation, and extended processing times, making them unsuitable for real-time, on-site applications. Similarly, our system outperforms emerging technologies such as electronic noses (E-nose),34 which lack the specificity to resolve individual VOCs in complex mixtures. Our integration of Raman spectroscopy with ML enables rapid, high-throughput, and non-invasive analysis with minimal sample preparation, underscoring its potential as a transformative solution for food safety monitoring.Despite these strengths, the proposed system faces limitations. The inherently low Raman cross-section of certain analytes, particularly alkanes, reduces sensitivity at extreme dilutions. Challenges such as spectral overlap in multiplex mixtures and dataset imbalances further complicate both classification and regression tasks. To mitigate these issues, we incorporated advanced feature extraction, preprocessing techniques like Savitzky–Golay smoothing and baseline correction, and weighted performance metrics such as f1 scores to improve model accuracy and generalizability. The VOCs selected for this study—such as 2-nonanone, acetoin, and methyl-trisulfide—were chosen based on their documented association with pathogens like Listeria monocytogenes, Escherichia coli, and Salmonella spp., and their relevance to contamination scenarios in food supply chains. Importantly, our system's modular design ensures adaptability; changes in the analyte set would only require the collection of new spectral data and retraining of the ML models, demonstrating the flexibility and scalability of the approach.
Future efforts will focus on expanding the library of VOCs to enhance the system's coverage and relevance across diverse applications. Additionally, real-world validation under operational conditions will be conducted to evaluate the system's performance in dynamic environments. Enhancing the portability of the hardware will further facilitate its deployment in on-site settings. By addressing these challenges and building on its foundational strengths, our future approach is to establish a scalable, cost-effective, and innovative framework for advancing pathogen detection and monitoring in food safety and environmental applications.
Conclusions
In conclusion, this study highlights the successful integration of Raman spectroscopy with advanced ML algorithms for the detection and quantification of VOCs associated with foodborne pathogens in complex liquid mixtures. Leveraging a remote fiber-optic Raman probe, spectral data were collected and analyzed from 42 distinct VOC mixtures. The classification model achieved 90% accuracy in identifying pure VOCs and demonstrated robust performance in mixtures containing up to six VOCs, at concentrations as low as 0.25% (400-fold dilution). Furthermore, regression models effectively predicted VOC concentrations down to 1% (100-fold dilution), achieving an R2 value of 0.82. Specific VOCs, such as 2-nonanone (Listeria monocytogenes), acetoin (Escherichia coli), and methyl-trisulfide (Salmonella spp.), were effectively identified, demonstrating the system's ability to detect pathogen-specific compounds in complex food matrices. The proposed system addresses the challenges of traditional methods like culture-based assays and qPCR, offering faster, non-invasive, and high-throughput detection without extensive sample preparation. Its integration of stochastic generalization models enhanced prediction accuracy by resolving spectral overlaps and accommodating complex VOC interactions. The methodology demonstrated scalability in analyzing diverse mixtures, making it a valuable tool for real-time, on-site food safety monitoring. Future work will aim to expand the VOC library, enhance model robustness for unseen mixtures, and validate the system in real-world scenarios. By integrating the proposed fiber-optic Raman spectroscopy system with ML, this approach provides a practical, portable, and cost-effective solution to foodborne pathogen detection. These advancements have significant implications for improving food safety, ensuring supply chain compliance, and enabling broader applications in environmental monitoring and quality control across various industries.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Bohong Zhang: supervision, investigation, validation, visualization, writing – original draft. Anand K. Nambisan: validation, visualization, writing – original draft. Abhishek Prakash Hungund: validation, visualization, writing – original draft, Xavier Jones: data acquisition, Qingbo Yang: supervision, investigation, writing – review and editing, Jie Huang: supervision, investigation, writing – review and editing.Conflicts of interest
The authors state that there is no conflict of interest in this paper.Acknowledgements
This work is financially supported by the USDA NIFA Evans–Allen Program (project number MOLU2021YANGQ), the USDA-NIFA-NEXTGEN project (grant number: 2023-70440-40147), and the Ewing Marion Kauffman Foundation (Prime Award No. RG-202101-9858).References
- B. D. Jett, K. L. Hatter, M. M. Huycke and M. S. Gilmore, Simplified agar plate method for quantifying viable bacteria, Biotechniques, 1997, 23(4), 648–650 CrossRef CAS PubMed.
- A. T. Perestam, K. K. Fujisaki, O. Nava and R. S. Hellberg, Comparison of real-time PCR and ELISA-based methods for the detection of beef and pork in processed meat products, Food Control, 2017, 71, 346–352 CrossRef CAS.
- H. Z. Senyuva, I. B. Jones, M. Sykes and S. Baumgartner, A critical review of the specifications and performance of antibody and DNA-based methods for detection and quantification of allergens in foods, Food Addit. Contam.,: Part A, 2019, 36(4), 507–547 CrossRef CAS PubMed.
- X. Zhao, J. Zhong, C. Wei, C.-W. Lin and T. Ding, Current perspectives on viable but non-culturable state in foodborne pathogens, Front. Microbiol., 2017, 8, 580 Search PubMed.
- M. Aladhadh, A review of modern methods for the detection of foodborne pathogens, Microorganisms, 2023, 11(5), 1111 CrossRef CAS PubMed.
- K. M. Tripathi, T. Y. Kim, D. Losic and T. T. Tung, Recent advances in engineered graphene and composites for detection of volatile organic compounds (VOCs) and non-invasive diseases diagnosis, Carbon, 2016, 110, 97–129 CrossRef CAS.
- T. Kuchmenko, U. Ruslan and L. Larisa, E-nose for the monitoring of plastics catalytic degradation through the released Volatile Organic Compounds (VOCs) detection, Sens. Actuators, B, 2020, 322, 128585 CrossRef CAS.
- S. L. Snellings, N. E. Takenaka, Y. Kim-Hayes and D. W. Miller, Rapid colorimetric method to detect indole in shrimp with gas chromatography mass spectrometry confirmation, J. Food Sci., 2003, 68(4), 1548–1553 CrossRef CAS.
- B. Zhang, H. Tekle, R. J. O'Malley, T. Sander, J. D. Smith, R. E. Gerald and J. Huang, In situ and real-time mold flux analysis using a high-temperature fiber-optic Raman sensor for steel manufacturing applications, J. Lightwave Technol., 2023, 41(13), 4419–4429 CAS.
- B. Zhang, W. Liao, H. Ma and J. Huang, In situ monitoring of the hydration of calcium silicate minerals in cement with a remote fiber-optic Raman probe, Cem. Concr. Compos., 2023, 142, 105214 CrossRef CAS.
- B. Zhang, H. Tekle, R. J. O'Malley, J. D. Smith, R. E. Gerald and J. Huang, In situ high-temperature Raman spectroscopy via a remote fiber-optic Raman probe, IEEE Trans. Instrum. Meas., 2023, 72, 1–8 Search PubMed.
- C. L. Wong, U. S. Dinish, M. S. Schmidt and M. Olivo, Non-labeling multiplex surface enhanced Raman scattering (SERS) detection of volatile organic compounds (VOCs), Anal. Chim. Acta, 2014, 844, 54–60 CrossRef CAS PubMed.
- J. Wang, Q. Chen, T. Belwal, X. Lin and Z. Luo, Insights into chemometric algorithms for quality attributes and hazards detection in foodstuffs using Raman/surface enhanced Raman spectroscopy, Compr. Rev. Food Sci. Food Saf., 2021, 20(3), 2476–2507 CrossRef PubMed.
- B. Zhang, Md A. Rahman, J. Liu, J. Huang and Q. Yang, Real-time detection and analysis of foodborne pathogens via machine learning based fiber-optic Raman sensor, Measurement, 2023, 217, 113121 CrossRef.
- Y. Feng, Y. Wang, B. Beykal, M. Qiao, Z. Xiao and Y. Luo, A mechanistic review on machine learning-supported detection and analysis of volatile organic compounds for food quality and safety, Trends Food Sci. Technol., 2023, 104297 Search PubMed.
- C. S. DeJong, D. I. Wang, A. Polyakov, A. Rogacs, S. J. Simske and S. Viktor, Bacterial detection and differentiation via direct volatile organic compound sensing with surface enhanced Raman spectroscopy, ChemistrySelect, 2017, 2(27), 8431–8435 CrossRef CAS.
- W. J. Thrift, A. Cabuslay, A. Benjamin Laird, S. Ranjbar, A. I. Hochbaum and R. Ragan, Surface-enhanced Raman scattering-based odor compass: Locating multiple chemical sources and pathogens, ACS Sens., 2019, 4(9), 2311–2319 CrossRef CAS PubMed.
- M. C. Lemfack, J. Nickel, M. Dunkel, R. Preissner and B. Piechulla, mVOC: a database of microbial volatiles, Nucleic Acids Res., 2014, 42, D744–D748 CrossRef CAS PubMed.
- E. Tait, J. D. Perry, S. P. Stanforth and J. R. Dean, Use of volatile compounds as a diagnostic tool for the detection of pathogenic bacteria, TrAC, Trends Anal. Chem., 2014, 53, 117–125 CrossRef CAS.
- Y. Wang, Y. Li, J. Yang, R. Jia and S. Chengjun, Microbial volatile organic compounds and their application in microorganism identification in foodstuff, TrAC, Trends Anal. Chem., 2016, 78, 1–16 CrossRef CAS.
- H. Okabayashi and T. Kitagawa, Assignments of the CH stretching Raman lines of hydrocarbon chains. Raman spectra of normal Cn-1H2n-1COOK (n= 3, 4, 6, 8, 10, 12, 14, 16, and 18) and their specifically deuterated derivatives, J. Phys. Chem., 1978, 82(16), 1830–1836 CrossRef CAS.
- H. Vašková, and M. Tomeček, Rapid spectroscopic measurement of methanol in water-ethanol-methanol mixtures, in MATEC Web of Conferences, EDP Sciences, 2018 Search PubMed.
- S. Duraipandian, W. Zheng, J. Ng, J. J. H. Low, A. Ilancheran and Z. Huang, Simultaneous fingerprint and high-wavenumber confocal Raman spectroscopy enhances early detection of cervical precancer in vivo, Anal. Chem., 2012, 84(14), 5913–5919 CrossRef CAS PubMed.
- S. Rozov, Machine Learning and Deep Learning methods for predictive modelling from Raman spectra in bioprocessing, arXiv, 2020, preprint, arXiv:2005.02935, DOI:10.48550/arXiv.2005.02935.
- S. Yan, S. Wang, J. Qiu, M. Li, D. Li, D. Xu, D. Li and Q. Liu, Raman spectroscopy combined with machine learning for rapid detection of food-borne pathogens at the single-cell level, Talanta, 2021, 226, 122195 CrossRef CAS PubMed.
- Y. Du, D. Han, S. Liu, X. Sun, B. Ning, T. Han, J. Wang and Z. Gao, Raman spectroscopy-based adversarial network combined with SVM for detection of foodborne pathogenic bacteria, Talanta, 2022, 237, 122901 CrossRef CAS PubMed.
- S. Sharma and T. Lokesh, Optical sensing for real-time detection of food-borne pathogens in fresh produce using machine learning, Sci. Prog., 2024, 107(2), 00368504231223029 CrossRef CAS PubMed.
- M. H.-U. Rahman, R. Sikder, M. Tripathi, M. Zahan, T. Ye, G. Z. Etienne, B. K. Jasthi, A. B. Dalton and G. Venkataramana, Machine learning-assisted raman spectroscopy and SERS for bacterial pathogen detection: clinical, food safety, and environmental applications, Chemosensors, 2024, 12(7), 140 CrossRef CAS.
- H. Kang, J. Lee, J. Moon, T. Lee, J. Kim, Y. Jeong and E. -K. Lim, et al., Multiplex Detection of Foodborne Pathogens using 3D Nanostructure Swab and Deep Learning-Based Classification of Raman Spectra, Small, 2024, 2308317 CrossRef CAS PubMed.
- Y. Jeon, S. Lee, Yu-J. Jeon, D. Kim, J.-H. Ham, D.-H. Jung, H.-Y. Kim and J. You, Rapid identification of pathogenic bacteria using data preprocessing and machine learning-augmented label-free surface-enhanced Raman scattering, Sens. Actuators, B, 2025, 425, 136963 CrossRef CAS.
- B. Swaminathan and P. Feng, Rapid detection of food-borne pathogenic bacteria, Annu. Rev. Microbiol., 1994, 48, 401–427 CrossRef CAS PubMed.
- S. Ishii, T. Segawa and S. Okabe, Simultaneous quantification of multiple food-and waterborne pathogens by use of microfluidic quantitative PCR, Appl. Environ. Microbiol., 2013, 79(9), 2891–2898 CrossRef CAS PubMed.
- Il-H. Cho and J. Irudayaraj, In-situ immuno-gold nanoparticle network ELISA biosensors for pathogen detection, Int. J. Food Microbiol., 2013, 164(1), 70–75 CrossRef CAS PubMed.
- E. Bonah, X. Huang, J. Harrington Aheto and R. Osae, Application of electronic nose as a non-invasive technique for odor fingerprinting and detection of bacterial foodborne pathogens: A review, J. Food Sci. Technol., 2020, 57, 1977–1990 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra08316d |
| This journal is © The Royal Society of Chemistry 2025 |