Open Access Article
Open Access ArticleNano–bio interactions and drug delivery using soft nanoparticles: a new paradigm in pharmaceutical cargo release
Rohini
Singh
a,
Fei Rui
Long
a,
Anjali
Saini
a,
Natali
Joma
a,
Abhirup
Basu
 b,
Morteza
Mahmoudi
b,
Morteza
Mahmoudi
 c,
Hojatollah
Vali
c,
Hojatollah
Vali
 d and
Ashok
Kakkar
d and
Ashok
Kakkar
 *a
*a
aDepartment of Chemistry, McGill University, 801 Sherbrooke St West, Montréal, Quebec H3A 0B8, Canada. E-mail: ashok.kakkar@mcgill.ca
bDepartment of Chemical & Biomolecular Engineering, North Carolina State University, Raleigh, NC, USA
cDepartment of Radiology, College of Human Medicine, Michigan State University, East Lansing, MI, USA; Precision Health Program, Michigan State University, East Lansing, MI, USA
dDepartment of Anatomy & Cell Biology, McGill University, 3640 University Street, Montréal, Quebec, H3A 0C7 Canada
First published on 15th November 2024
Abstract
The bilateral relationship between nanomaterials and biological systems can play a significant role in therapeutic interventions and diagnostics. The nanomaterials may lose their synthetic identity after encountering biological fluids (e.g., serum or plasma), and it might lead to unintended outcomes in real-time applications. Despite advances in nanomedicine, clinical translation and overall patient survival using nanoformulations have largely remained elusive. The layer of biomolecules formed around nanoparticles (NPs), often referred to as protein-corona (PC), can impact their physicochemical properties, including size, surface charge/chemistry, chemical composition, solubility, etc. Recently, a few mechanistic evaluations have demonstrated that the formation of a corona layer on nanoparticles can also have a consequential effect on the release profiles of polymeric soft NPs. To evaluate their therapeutic efficacy and resolve discrepancies that exist between in vitro and in vivo results, transition of NPs from their native to the corona-coupled state and its impact on unloading of their cargo need to be understood. Here, we highlight (i) how inherent properties of polymer precursors can affect PC build-up on soft NPs and its impact on cargo-release kinetics and (ii) limitations of existing methods in analyzing PC in complex systems, with emphasis on the impact nano–bio interactions have on the soft nanoparticle-based drug delivery domain.
1. Introduction
Nanomaterials are revolutionizing medical interventions owing to their ability to target specific sites and higher safety, as measured by their therapeutic index over conventional treatments. The last four decades have seen remarkable progress in the development and application of engineered nanoparticles (NPs) in the treatment of various diseases including cancer,1–7 neurological disorders,8–11 autoimmune diseases,12–14 hepatitis,15–18 infections,19–21 muscular degeneration,22–24 and hypercholesterolemia. The scope of fine-tuning the physicochemical properties of NPs such as their size, shape, surface functionalities, and morphologies has offered a unique platform in drug delivery and diagnostics.25–30 Advantages of nanomedicine include prolonged retention times, increased solubility, improved biodistribution, reduced immunogenicity, and low systemic toxicity. NPs for biological applications have been fabricated using either hard (inorganic) or soft (organic, lipid-based, and polymeric) materials. The inorganic NPs from mesoporous silica,31,32 quantum dots,33,34 silver,35,36 and gold37–39 have been demonstrated to be highly efficient for imaging and diagnostic purposes, due to their unique magnetic, optoelectrical, and chemical properties. However, the uncertainty surrounding their cytotoxic dosage-limits has been a major hurdle in transitioning to clinical levels.40,41 Soft NPs are more suited for drug delivery due to their modular build-up, inherent biocompatible nature, ability to introduce multiple functions through targeting ligands (antibodies, antibiotics, peptides, and nucleic acids), as well as varied compositions including lipid NPs,42,43 polymeric micelles,44,45 polymersomes,46–48 dendrimers,49,50 and liposomes51–53 (Fig. 1). In addition, the ability to load multiple drugs makes them optimal candidates for combination therapy to address drug resistance, as well as for interventions targeting more than one type of pathogen or disease type, such as bacterial infections.54 Stimuli-responsive NPs offer opportunities to take advantage of internal (pH, enzymes, and small molecules) or external (magnetic field, temperature, ultrasound, electric field, and light) cues for sustained and controlled drug delivery to disease sites.55–60 | ||
| Fig. 1 The main types of nanocarriers that have been used in drug delivery, together with a list of their biophysiochemical properties. | ||
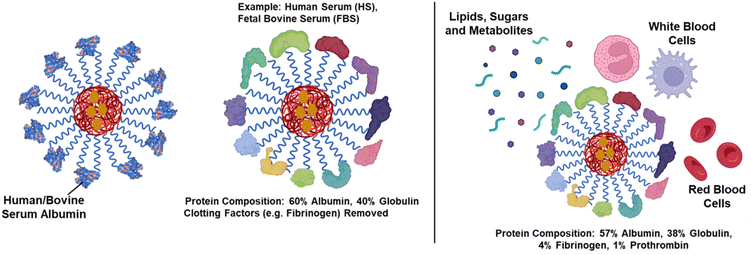
Nanocarrier-based drug delivery has been extensively investigated and its benefits in enhancing the efficacy of hydrophobic drugs through encapsulation has been well documented. However, since the effect of nano–bio interactions was not taken into consideration, many nanocarriers failed at clinical trials despite showing promising results in vitro.61,62 By re-evaluating the existing nanocarrier-based systems in more detail, it was proposed that the protein corona (PC), at least in part, is responsible for such disparities,63–66 and the biological medium alters the physicochemical properties of nanocarriers, which affects their efficacy.67,68 When nanocarriers enter the bloodstream, they interact with a complex biological environment rich in biomolecules, including proteins, lipids, and sugars. This interaction results in the adsorption of serum proteins primarily on the surface of nanoparticles, a coating often referred to as protein corona (PC).69,70 As proteins are the most abundant biomolecules in the bloodstream, they preferentially adsorb onto the nanocarrier surface, forming a protein-rich layer. This protein corona can significantly alter the physicochemical properties of nanoparticles71,72 (Fig. 2). The outer soft layer in PC is composed of molecules in an active and rapidly exchanging state with the surrounding biological environment as compared to the inner hard layer, which is closely bound to the NP surface and has a stable arrangement of biomolecules.73 It is now becoming evident that the PC can play a decisive role in immune response, retention times, targeting capability, cytotoxicity, biodistribution, clearance rates, and biodegradation of nanocarriers.74,75 It is not only the concoction of plasma proteins that affects the pharmacokinetic behavior of nanocarriers but also several other factors such as gender, demographics, and disease state that contribute to the variation in PC and eventually impact the overall in vivo efficacy of a nanocarrier.76 The underlying biological differences in the plasma compositions of animal models and humans account for the failure of clinical trials of numerous lab-scale nano-formulations successfully tested on animal models.
 | ||
| Fig. 2 A summary of factors affecting protein corona on NPs and the downstream effect of protein corona. | ||
Recently, some comprehensive reviews have summarized how the composition of plasma proteins can influence the corona build-up on NPs and eventually change their therapeutic fates.77 However, a detailed analysis of how PC can affect the release profiles of drugs from nanocarriers still remains elusive. Its understanding can change the outlook of the PC–NP relationship and its repercussions on the efficacy of nano-formulations. The build-up of PC on NPs once they enter into circulation or the targeted sites is still being explored, and the kinetics of corona formation may help in better understanding its role in the release of drugs from NPs. Only a few experimental studies are available in the literature that focus on the characterization of PC, but they are quite simplistic and limited to animal plasma such as bovine serum or single protein systems which are far different from the human biological medium.78 If PC can capture and solubilize soft NPs, it can increase the availability of drugs but its effect on the overall sustained release and efficacy is still to be identified. Hence, a detailed investigation on how PC manipulates release profiles and the drug delivery behavior of nanoformulations is imperative to understand the underlying mechanisms and to help design more efficient nanocarriers in the future.
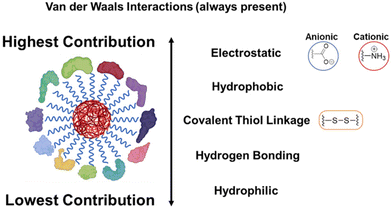
It has been shown that PC can shield the drug from release in some cases, while aiding in sustained release in others.79–82 Since the disease stage, gender, and demographics influence corona composition, it becomes evident why the same NP-based therapeutic kit produces different results when tested for a larger set of patients. It is essential to understand the evolution of PC in varied environments. Hence, to design the most effective nanocarrier for drug delivery, we need to understand the pattern and evolution of PC in humans. PC formation and its evolution have been studied in different biological systems in vitro and in humans under different environmental conditions.83–85 Such studies can provide a roadmap of the periodic evolution of PC and help design efficient NP-based therapeutic systems. It is necessary to mention that current methodologies lack the resolution to decipher the mechanism or kinetics of PC formation, its composition on NPs, and subsequent cargo release from NPs. It is unclear whether the corona build-up is a purely physical adsorption-based phenomenon or a gradual chemical process and if it could be reversed. However, some articles have reported it to be an interplay between both forces.86 Using current methodologies, we can only create a simplistic biological medium in vitro to track the temporal formation of corona. It is becoming clear that in vitro results may not represent real-life systems. It has been more than five decades since researchers proposed therapeutic NP-based formulations, but these represent less than 22% of the total market share of the pharmaceutical industry.87 It begs the question that if NP-based systems are superior to conventional drugs, as demonstrated in numerous studies, then why do most fail to make it to clinical translation. To address this, we propose the relevance of evaluating the role of PC in the release profiles of soft NPs. It is also clear that the failure at analyzing the kinetics of PC formation on NPs is becoming a limiting factor in reassessing the existing NP-based formulations. Advances in experimental investigations are required to design more effective and robust NP-based drug delivery systems and accelerate their successful clinical translation. An understanding of the structure–property relationships of surface functional groups of polymeric NPs, which play a key role in interacting with plasma proteins, is desired. Each polymeric particle is an individual platform with different composition of polymers, surface charge, functional groups, hydrophobicity, hydrophilicity, size, and shape. The intermolecular forces have a strong contribution in PC formation, with electrostatic forces as the most dominant and hydrophilic forces as the least dominant, as shown in Fig. 3.88 Without a common roadmap, it necessitates a complex examination of each one of them to understand the mechanism of their interaction with human plasma proteins.
2. Factors affecting NP–PC build-up
The human body is a highly dynamic system, and changes in serum proteins due to temporary conditions are inevitable. Human plasma is composed of more than 1500 different types of proteins,89–91 which could contribute to the formation of varied and diverse corona on the surface of NPs. This complex mixture contains antibodies, cytokines, secreted proteins, transporters, lysosomal proteins, hormones, foreign proteins due to infection, protein fragments, inter- and intracellular proteins, receptor ligands, and the most abundant (>55%) albumin protein.92 When NPs enter into circulation, interactions with serum proteins inevitably impart a new nano–bio interface to them, which may lead to changes in their targeting ability and the payload release profiles.93 A study related to PC kinetics has shown that 20 abundant proteins form the corona within less than half a minute of exposure of silica and polystyrene NPs to human serum.94 Hence, the dynamics of protein deposition and equilibrium among proteins based on their affinity and stability to stay inside the PC layer dictates the final composition of the protein layer formed on the surface of different types of NPs. Based on the abundance, serum albumin, apolipoproteins A1, and complement C3 outnumber all other proteins in participating in PC deposition. Once PC is formed, the amount of bound proteins changes, while its composition remains constant, which contradicts earlier studies. Such analyses propose that PC formation is independent of the Vroman effect, a phenomenon in protein adsorption, where the composition of proteins on the surface changes over time due to competitive binding.95 Interestingly, the molecular weight of the proteins may also have a role in corona build-up, as the most abundant proteins in the corona layer have a molecular weight >60 kDa. Bioinformatic analysis has revealed that irrespective of exposure time to serum proteins or particle surface functionalization, the protein corona exhibits a net negative charge at physiological pH, mimicking the composition of net negatively charged cells inside the human body.64,96Apart from the diverse composition of human plasma, there are multiple factors such as gender, age, demographics, disease stage, habits, and temporary conditions such as pregnancy and medication that can influence PC composition. To our knowledge, there are only two studies that carried out an extensive analysis of the impact of disease states on the composition of PC.77,97 One of these investigated the composition variation in PC in patients suffering from fauvism, hemophilia, hypofibrinogenemia, diabetes, hypercholesterolemia, and temporary conditions such as pregnancy, and in healthy individuals with lifestyle habits such as smoking, alcohol, and diet.77 The SDS-polyacrylamide gel electrophoresis (PAGE) protein analysis showed that patients suffering from the same type of disease had similar PC composition, whereas the composition varied between patients with different diseased states.77 In another independent study, clinically approved liposomes (AmBisome) were used to analyze PC formation in gastric, breast and pancreatic cancer patients.97 The results demonstrated substantial variability in PC composition among all cancer types, with liposomes in pancreatic cancer having lower negative charge than in breast or gastric cancer, indicating a higher concentration of cationic proteins in the corona. The SDS-PAGE analysis showed the presence of a band at ∼37 kDa associated with immunoglobins A (IgA), which is a marker for autoimmune response in cancer patients. Since liposomes had varied PC in all cancer types, it was anticipated that it will affect the pharmacokinetics of such nano-formulations, implicating PC as a pivotal factor if the same liposomal drug fails to generate a similar therapeutic response in another disease type. Apart from these, it is essential to analyze other factors that may affect the adsorption of proteins on the surface of NPs. Understanding how proteins interact with NPs is crucial. This includes analyzing factors that influence the formation of PC around the NP surface. In particular, the role of surface charge and functional groups in attracting or repelling proteins is important. Additionally, the hydrophobicity or hydrophilicity of the NP surface plays a critical role in determining the types of proteins that get adsorbed.
3. Dynamics and statistics of PC
It has been suggested that protein adsorption on NPs is a dynamic phenomenon. Predicting the fate of NPs in vivo requires an understanding of this behavior, as well as inclusion of the studies pertaining to the role of PC in in vitro studies. The amount of proteins adsorbed on the surface of NPs fluctuates until it reaches an equilibrium.98,99 Because of the Vroman effect, the PC changes over time as more mobile proteins attach to the surface first and are then replaced by those with higher affinity.100–104 It brings about changes in PC's physical characteristics and composition. The timeline might vary for different shapes, sizes, surface charges, and morphologies of the NPs. It has been observed that PC builds up within a few minutes, and subsequently only an exchange of low affinity proteins with higher affinity proteins occurs. In a study by S. Palchetti et al. in 2016, a remarkable difference in the number of proteins adsorbed on PEGylated liposomes was reported, when tested in static and dynamic systems.105 A total of 207 proteins were identified on liposomes in a circulating fetal bovine serum, compared to only 118 proteins that were adsorbed in a static fetal bovine system, which accounts for almost 57% variation in the results.105 These data help in elucidating discrepancies often seen in reproducibility of protein adsorption on the same particles and media but tested under relatively different flow conditions (Fig. 4). Such analysis also highlights the significance of in vivo and dynamic systems when establishing statistics of PC formation.Another comprehensive study conducted a qualitative and quantitative comparison of PC among bare liposomes, PEGylated liposomes, and monoclonal antibody IgG-functionalized liposomes.106 All three types of formulations were tested in vitro and in vivo. It was found that the number and types of proteins adsorbed had greater variation in same liposomal NPs when tested in vitro than in vivo. Among bare liposomes, 212 proteins were adsorbed in vivo, as compared to 30 in vitro exclusively, with an overlap of 241 proteins. In PEGylated liposomes, a total of 502 proteins were found in the corona layer, where 241 were exclusively in vivo and 24 in vitro, with an overlap of 237 proteins.106 In IgG targeted liposomes, which had more exposed charges, a denser layer of PC was formed with a total of 542 proteins, where 254 proteins were in vivo as compared to 31 in vitro, with an overlap of 257 proteins. These differences in the number of proteins adsorbed on the same liposomal surfaces when tested in static and dynamic systems necessitate re-investigation of existing in vitro studies in a dynamic system to establish a standard PC composition and its influence on drug release behavior and targeting capability in a more reliable manner. The study showed that in addition to the surface morphology and charge, the environment also influences corona composition and plays a pivotal role in the type and quantity of proteins adsorbed. The microscopic visualization of the PC showed a network of fibrillary structures in vitro as compared to an uneven morphology in vivo with a broken coverage of liposomal surfaces. A further investigation into the targeting capability of liposomes showed that among all three formulations tested, the PC significantly reduced the receptor binding and cellular internalization of the IgG-coated liposomes. It shows the influence of hydrophobic interactions and the propensity of higher protein adsorption on charged surfaces as compared to neutral ones.
4. PC impacting the payload release from nanocarriers
The PC not only influences the physicochemical properties of nano-formulations, but also impacts the release profiles of the NP-conjugated or encapsulated drugs. It transforms the nanocarriers into new identities, significantly changing their biodistribution and overall therapeutic efficiency. Numerous studies have been performed to study the delivery profiles of the payloads in nanocarriers, but it has become evident only in the last decade that PC may have an indispensable effect on release profiles in vivo. The significant differences between human and mouse plasma compositions contribute to the discrepancies observed between in vitro and in vivo studies. These can affect drug release kinetics and often lead to failure of promising NP-based formulations in clinical trials. Therefore, it is crucial to re-evaluate the release profiles of previously studied nanocarriers in the presence of human proteins to gain a more accurate understanding of their drug release behavior. Recently, a few studies have demonstrated that PC can strongly modulate the biotransformation and release kinetics of encapsulated drugs, and it needs further detailed investigation.4.1 Effect of PC on drug release from inorganic and soft NPs
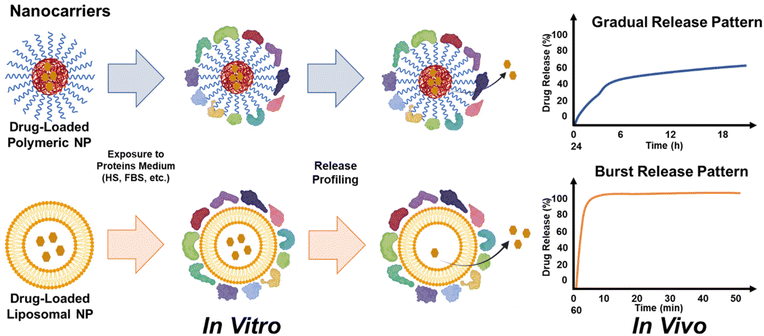
PC formation and its implications are not limited to only organic nanocarriers, but it spans over other subtypes including inorganic NPs. As more studies on metal NPs demonstrate enhancement of the targeting ability and overall efficacy as therapeutic carriers, the associated complexities such as PC formation warrant a reinvestigation with a new perspective. A study by Shahed Behzadi et al. used superparamagnetic iron oxide nanoparticles (SPIONs), polymeric nanocapsules, and albumin-bound paclitaxel (Abraxane) nanocarriers to evaluate the effect of PC on drug-release profiles.107 It was found that there is a significant difference in the thickness of corona layers surrounding these nanocarriers. The PC layer on iron oxide particles was thicker (32 nm) than nanocapsules (≈10 nm), which led to the different release profiles of the two nanocarriers. Overall, PC had a shielding effect on all the nano-formulations and particularly reduced the release of paclitaxel from albumin-bound Abraxane NPs, highlighting an important role of the nanomaterial composition in varying the propensity of NPs towards serum proteins. It was observed that the stable shell of PC around the polymeric NPs reduced the burst release of either protein-conjugated NPs or surface-loaded drug nanocarriers and the formation of a protein buffer layer made a shield onto the surface of small NPs in vivo, impeding drug release, as compared to drug-loaded liposomes that showed burst release even after the formation of a PC layer (Fig. 5).Soft and inorganic NPs, owing to their intrinsic compositional differences, will have varied NP–corona relationships.78 Several factors, such as NP material composition, surface properties, increase in particle size, corona–drug interaction, corona–NP interaction, PC composition, and modulation of the entry mechanisms into the target site, play a critical role in modulating drug release kinetics. Table 1 summarizes the role of such factors affecting corona build-up on inorganic and soft NPs. It is evident that most of the studies pertaining to corona formation have been carried out on metallic or inorganic NPs, but it is mostly soft NPs that are at clinical stages. However, very little data are available on the effect of human serum proteins on such nano-formulations, necessitating the need for more investigations. Among all the studies on payload release under the influence of PC, it has been shown that it slows down the release of drug from inorganic NPs. In contrast, in most soft NPs including liposomes, PC leads to an enhancement in the release of drug molecules. These studies emphasize that it is critical to consider PC as a driving factor in the release kinetics, while deploying lipid-based nano-formulations inside the human body.
| Types of NPs | Loaded drug/molecule | Mechanism by which PC affects release from NPs | Payload release | Ref. |
|---|---|---|---|---|
| Inorganic NPs | ||||
| (PEG) fumarate-coated superparamagnetic iron oxide | Tamoxifen citrate | Increase in particle size | Slower | 107 |
| PX albumin-bound NP formulation (Abraxane®) | Paclitaxel (PX) | Increase in particle size | Slower | 107 |
| Mesoporous silica NPs | Camptothecin | Increase in particle size | Slower | 108 |
| Poly-3-hydroxybutyrate-co-3-hydroxyhexanoate NPs | Coumarin-6 | Increase in particle size | Slower | 109 |
| Gold nanorods | Doxorubicin (DOX) | Increase in particle size | Slower | 110 |
| Solid cationic Eudragit RS | Dexamethasone | Increase in particle size and interparticle aggregation | Slower | 111 |
| NH3-dependent dissolution of silver (Ag) NPs | Silver ion | Chelation of Ag+ with the thiol and cysteine of BSA | Faster | 112 |
| Silver NPs | Silver ion | Interaction of BSA with NPs | Slower | 113 |
| Gold NPs | Poly (acrylic acid) (PAA) | Increase in size | NA | 114 |
| Gold nanorods | DNA | Depends on the protein composition (soft corona or hard corona proteins) | Faster/slower | 115 |
| Organic NPs (soft nanoparticles) | ||||
| Doxoves (a PEGylated liposomal DOX) | DOX | Interaction of PC (mainly dysopsonins) with NPs, which destabilized the liposome membrane | Faster | 116 |
| Traditional temperature-sensitive liposomes (TTSLs) | DOX | Interaction of PC with TTSL lipids, which destabilized the lipid vesicles | Faster | 117 |
| Transferrin-functionalized p(HEMA-ran-GMA) copolymer NPs | Lomerizine and YM872 | Change in pH | Slower | 118 |
| Lipid NPs | Zwitterionic lipid dioleoyl phosphatidylethanolamine (DOPE) | Increase in size | Faster | 119 |
4.2 Challenges surrounding PC formation on soft NPs
Protein adsorption on nanocarrier surfaces is influenced by structural characteristics (including surface charge, size, and shape) and environmental factors (composition, pH, and temperature).120 Moreover, PC formation is the result of intermolecular interactions (van der Waals, hydrogen bonding, coulombic and hydrophobic effects).121 Although PC is considered undesirable and could impair cellular targeting and uptake,122,123 some studies show the advantages of protein coatings. It has been shown that pre-coating NPs with serum albumin improves blood circulation time,124 while pre-coating with immunoglobulin reduces nanoparticle interaction with phagocytic cells.125 Adsorption of vitronectin glycoproteins can also enhance targeting for cancer cells,126 and AuNP conjugation with albumin and apolipoprotein E promotes organ targeting.127 Moreover, protein coating can also reduce cytotoxicity of nanoparticles, as noted for inorganic nanoparticles coated with fetal bovine serum.128 Interactions in macromolecules are responsible for the colloidal stability of polymeric NPs. Hence, soft NPs are more prone to destabilization inside the human body as electrostatic interactions with serum proteins and polymeric functional groups disturb the polymeric stability, leading to abrupt release of the cargo.116,117 Abstiens et al.129 examined the interaction of serum proteins with polymeric colloids with varying particle charge and hydrophobicity. Fluorescence resonance energy transfer (FRET) was used to quantify and measure the colloidal stability of hard PC in polymeric NPs with uncharged methoxy groups, positively charged amine groups, negatively charged carboxylic acids, or zwitterionic NPs. Zwitterionic NPs were least affected by the formation of a PC when compared to neutral methoxy groups or negatively charged carboxylate groups. This could be due to the fact that zwitterionic NPs provide fewer opportunities for serum proteins to bind using electrostatic and hydrophobic interactions. Positively charged amine-NPs, on the other hand, had strong interactions with serum proteins, resulting in a wider spectrum of proteins in the hard corona.130,131 FRET and SDS-PAGE agarose gel electrophoresis showed that the increased protein adsorption onto colloidal surfaces damaged NP integrity, and the lipophilic payload continuously leached out of the hydrophobic core. Cargo leaching was demonstrated to be directly proportional to protein concentration on SDS-PAGE. Herein, we collate all the available studies focusing on soft NPs and their interplay with PC and how these coronae affect encapsulated drugs, their release, and overall functionality of the soft NPs.PC formation on a clinically approved liposomal drug formulation (AmBisome) was examined by incubating it in blood from gastric, breast, and pancreatic cancer patients.135 It was observed that hard corona formed within few seconds of incubation and its protein quantity remained constant over time, which is commensurate with earlier reports.119 Plasma proteins did not cause any aggregation of liposomes but rather resulted in thick and heterogenous corona formation. Zeta potential measurements showed variation in surface charges after deposition of plasma proteins, which suggested that diversity in the PC of healthy and cancer patients dictated their propensity towards cationic lipids.
Another study focusing on the kinetics of protein binding on PEGylated liposomes indicated the importance of temporal evaluation of the dynamics of PC formation and its effect on drug release.136,137 Liposomes were injected into mice intravenously and extracted from circulation at specific time intervals. Extracted liposomes with PC were analyzed by dynamic light scattering (DLS), transmission electron microscopy (TEM), and mass spectrometry (MS). The findings indicated that an intricate PC was established within 10 minutes after injection. While the overall protein adsorption remained constant, the abundance of each detected protein varied over time, suggesting the occurrence of competitive exchange processes. It was observed that at different time intervals, the most abundant proteins remained the same, but their adsorbed concentration fluctuated with time. At the 10 min time interval, macroglobulin was most abundant, followed by hemoglobin and apolipoproteins at subsequent time points. This is very valuable information for designing time-based targeted nano-therapies, as it would impact the kinetics of circulation and clearance of nano-formulations. It is evident that NPs reach the targeted site at different time points, and the biological medium imparts a new identity to the NPs, which could affect their circulating times, interaction, and uptake. Despite such revelations, there are only a few studies that focus on the dynamic nature and kinetics of PC of soft NPs, and to date, true in vivo real-time analysis of NPs in humans remains elusive and demands thorough investigation.
Most of the studies have been carried out to characterize PC using in vitro static systems by end-point analyses. However, an interesting investigation by Palchetti et al. showed that there is a significant difference in the composition of PC on PEGylated liposomes when tested in vitro and in circulating systems.138 They showed that the sizes of the liposomes did not differ much, but the PC of circulating liposomes was more negative with accumulation of serum albumin, while those tested in a static system had higher accumulation of alpha-2-HS-glycoprotein. Such analysis urges that a thorough re-evaluation of the polymeric drug delivery vehicles in a dynamic circulating platform is required to establish their true efficacy and help identify disparities when tested in vitro and in vivo.105
Overall, protein adsorption was found to be independent of the chemical composition of biomacromolecules. Despite polymersomes being negatively charged, protein adsorption occurred regardless of isoelectric points and surface charges. These findings imply that electrostatic forces cannot overrule the intermolecular interactions that govern protein adsorption. This is supported by the fact that the negatively charged proteins (IgG and BSA) can bind to a surface of negatively charged vesicles. Irrespective of the chemical makeup of PC or the polymer concentration, protein-coated assemblies were less cytotoxic than uncoated NPs, demonstrating benefits of PC.
It has been shown that dendrimers bind to lipid membranes152 and soluble proteins153via non-specific interactions, including electrostatic and hydrophobic, with dendrimer generation and chemical composition playing a significant role in protein binding affinity.154 Gel electrophoretic assays revealed that G6 and G7 PAMAM dendrimers exhibited the highest protein binding. A detailed study by Åkesson et al. reported hard corona formation on cationic PAMAM dendrimers of generation G4 to G7 (with the same surface modification but different sizes), showing generation-dependent interactions with both cell membrane and cell toxicity.155 The overlap in the size and density of plasma proteins and PAMAM dendrimers makes it implausible to study PC using conventional techniques based on varying sedimentation rates of coated NPs from plasma proteins under centrifugal forces. Moreover, there is no alternative way to distinguish what is bound, from impurities in abundant proteins. To address these problems, electrophoretic mobility methods that are commonly used to separate charged small NPs that travel across the porous gel matrix, such as SDS-PAGE (Sodium Dodecyl Sulfate Polyacrylamide Gel Electrophoresis), have been utilized. The variation in plasma concentrations resulted in differences in mobilities and surface charges spanning across the generations of dendrimers. Overall, the study confirmed that larger dendrimers had a greater protein binding capacity, which was later confirmed by mass spectrometry.155
Another study156 investigated interactions between serum proteins (HSA and IgE) and PAMAM dendrimers of different sizes (G1–G4) and surface chemistry, including positively charged amine, negatively charged succinic acid (SA), and neutral hydroxyl (OH), polyethylene glycol (PEG), and phosphorylcholine. They reported that neutral PEG, and OH modifications significantly decreased protein interactions when compared to anionic or cationic surfaces. Neutral PAMAM dendrimers form sporadic interactions with proteins, whereas charged dendrimers tend to remain attached to proteins for a longer amount of time. HSA with a negative charge favored positively charged PAMAM-G1 to G4 with binding rates as high as 80%. HSA bound less frequently to negatively charged PAMAM-SA when compared to unmodified dendrimers, but substantially more than the neutral dendrimers. IgE with a net positive charge favored both positively and negatively charged PAMAM dendrimers with a similar binding frequency of 10%, which was higher than that of neutral dendrimers.156 It is noted that most studies carried out on polymersomes and dendrimers do not explore the effect of PC on payload release, which is becoming a critical factor in determining the overall efficacy of any NP-based formulation.
5. Nano–bio interactions and drug release
Interactions of plasma proteins with NPs can affect drug release kinetics in NPs, which has implications in clinical translation. Scientists have also seen a positive side of this interaction and are now using the proteins accumulated in PC as biomarkers. Studies dedicated to characterizing the PC and how it can be of diagnostic value exploiting its accumulative tendencies, are summarized below.5.1 Hurdles in the characterization of PC
To minimize misinterpretations in the analysis of PC, the complexity of the system needs to be minutely investigated. It has been shown that PC can shield NPs and can change the drug release behavior,69,82,157–159 but the exact mechanisms are still elusive, as it is very difficult to isolate individual components from drug-NPs and plasma concoction. The most common technique used to study drug release behavior is the dialysis method,160,161 but the major hurdle in quantifying the drug release in the presence of PC is the loss of drug due to adsorption on the inner surface of the tubing.162 Moreover, in the presence of serum or plasma proteins, there is the possibility of interaction between the drug and the medium. It becomes extremely challenging to isolate and identify pure forms of drugs or the components of the complex plasma (Fig. 6). Such interactions can lead to the formation of irreversible drug–protein complexes, which might result in inaccurate analyses. These possibilities are inevitable and pose a major issue in studying drug release profiles from nanocarriers. Moreover, it is extremely difficult to analyze and identify the drug inside the dialysis tubing because it might exist in a complex form with NPs or serum, significantly impacting the quantification of the released drug and ultimately the drug release profile. The advanced methods to address these issues and measure this difference are lacking and we need to revamp our methods for a better understanding of altered release profiles. As per the literature, currently the techniques are failing to establish a reliable characterization regime for PC proteins and quantify the effect on drug release.163,164 Variability in measurements of proteins and drugs can be attributed to variation in the plasma components owing to different physiological conditions such as disease state, gender, demographics, and age group. Other factors such as storage conditions, duration of storage, polydispersity of NPs, and resolution of the measurement techniques165–167 also play a major role in the differences observed in results when same samples are tested at different time points. Apart from this, the polymeric composition and surface charges attract different proteins and play a significant role in corona formation and drug release behavior, which can potentially alter the release and adsorption of drugs. Therefore, for such an analysis, we need robust characterization of the NP–PC complex, which is only possible if we have advanced and accurate technologies to identify individual components of corona, which can improve reproducibility, minimize misinterpretations, and provide an explanation for the discrepancies in results. It is imperative to emulate in vivo conditions in drug release behavior to envision a successful translation of NP-based drugs to clinical levels.5.2 PC as a biomarker for cancer detection: positive side of the protein deposition
PC could be seen as a roadblock in maintaining controlled release in NP-based targeted therapy that led to the failure in reaching clinical translation. However, it can also serve as a diagnostic platform for detection of NP-based protein biomarkers for various diseases. With the challenges faced in therapeutic interventions and diagnostic tool development during the COVID-19 pandemic, a plethora of options to detect the biomarker proteins of the corona virus have been explored, leading to an evaluation of the associated properties of PC with a particular disease type. A recent study has shown how different surface chemistries can be effectively utilized to detect Alzheimer's disease at an early stage.168 Interestingly, the disease-specific PC formed around NPs could be utilized to detect changes in the protein pattern in the plasma with >92% specificity. These results were later used to train a classifier machine learning algorithm to diagnose Alzheimer's disease at its different stages of development. Hence, the ability of NPs to pick up the disease-specific biomarker proteins at early stages can be advantageous in developing noninvasive technologies for the timely detection of various diseases with high accuracy and specificity. The relevance of personalized PC in the detection of various types of cancers has also been examined.169 As it is evident that every cancer type has specific antibodies or protein biomarkers, but in the absence of early symptoms or inability of diagnostic platforms to pick low amounts of biomarkers can lead to poor prognoses. However, NPs owing to their inherent capability of binding to plasma proteins can help pick up specific biomarkers from cancer type. Therefore, where PC may negatively impact the NP-based therapy in some cases, it certainly can prove to be a vital tool in the development of diagnostic platforms or assays for various diseases such as cancer, COVID-19, Alzheimer's, Parkinson's, cystic fibrosis, and other neurodegenerative disorders where proteins are the challenging factors.6. Conclusions
Protein corona constitutes a dynamic layer of proteins that spontaneously forms on the surface of NPs upon interaction with biological fluids. It can significantly impact the fate and bioactivity of NPs, as well as the release of their cargo. There are several knowledge gaps in understanding the effect of PC formation on NPs and its impact on drug release characteristics. A wide array of nanocarriers have been utilized in delivering potent pharmaceutics; however, accurately predicting the composition of PC in NPs in different biological environments remains a challenge. This is due to the complex interplay of factors that influence protein adsorption, such as NP surface properties, protein abundance, protein–protein interactions, and protein–drug interactions. The dynamics of PC formation and evolution, including protein exchange, conformational changes, and protein–NP dissociation, is not fully understood. This knowledge is crucial for predicting the long-term behavior of NPs in biological systems, as well as for quantification of the impact of PC formation on cargo release from NPs. Factors such as protein–cargo interactions and PC thickness can influence cargo release rates and mechanisms. Evaluating cargo release in complex biological environments, such as blood or tissue fluids, is challenging due to the presence of multiple proteins, competitive adsorption, and dynamic interactions. In addition, our ability to control PC formation to achieve specific biological outcomes, such as enhanced targeting or reduced toxicity, is limited. A detailed understanding of PC-mediated interactions is needed to rationally design NPs with tailored outcomes. To address these challenges, a multidisciplinary approach combining experimental and computational techniques is desired. Advanced analytical methods, such as proteomics and single-particle tracking, are needed to characterize PC composition, dynamics, and interactions. Computational models that incorporate knowledge of PC formation can be used to predict the behavior of NPs in biological environments, as well as to provide insights into the mechanism of PC formation and its impact on cargo release. Through such investigations, we can gain a deeper understanding of the role of PC in nanomedicine and develop more effective and safer nanotherapeutics.Understanding the influence of PC formation on nanoparticles (NPs) and its effect on the release of cargo can significantly improve the design of nanocarriers for drug delivery and other biomedical applications. Specifically, this knowledge can lead to enhanced targeting, reduced toxicity, controlled cargo release, improved stability and circulation, predictable behavior and more clinical translations. By tailoring PC composition, nanocarriers can be designed to selectively interact with specific cell types or tissues. This can improve drug delivery to the desired site of action and reduce off-target effects. Understanding how PC affects the biocompatibility of NPs can help in designing NPs that minimize toxicity and immune responses. By controlling the interactions between PC and the cargo, the release rate of drugs or other therapeutic agents can be modulated. It is known that PC can help stabilize NPs and prolong their circulation time in the bloodstream. Favorably modulating this behavior can help in building more sustained drug delivery models and improve treatment efficacy. This can enhance drug bioavailability and improve treatment outcomes. The evolving landscape of PC research not only deepens our understanding of NP biology but also propels the development of new paradigms such as using NP-based PC as a diagnostic tool in identifying protein biomarkers associated with different diseases. As we navigate the challenges posed by the dynamic PC, the collective progress in unraveling its complexities promises a future where NPs can be tailored with precision, generating next-generation NP formulations with improved functionalities and ushering in a new era of targeted and effective nanomedicine.
Author contributions
R. S., F. R. L., A. S., N. J., and A. B.: writing, review, editing, and visualization; M. M. and A. K.: conceptualization, writing, review, and editing; H. V.: conceptualization and editing.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
M.M. discloses that (i) he is a Co-founder and Director of the Academic Parity Movement (www.paritymovement.org), a non-profit organization dedicated to addressing discrimination, violence, and incivility; (ii) he is a Co-founder of and shareholder in Targets Tip Corp,; and (iii) he recieves royalities/honoraria for his published books, plenary lectures and licensed patents.Acknowledgements
AK would like to thank the Natural Sciences and Engineering Research Council (NSERC, Canada, grant RGPIN-2023-03565) and the New Frontiers in Research Fund – Exploration (NFRFE, Canada, grant number NFRFE-2022-00356) for financial assistance. Figures were created with elements from BioRender.References
- A. Basu, R. Singh and S. Gupta, Bacterial infections in cancer: A bilateral relationship, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2022, 14, e1771 Search PubMed.
- R. Singh, C. S. Kumar, M. Banerjee and S. Gupta, ACS Appl. Bio Mater., 2019, 2, 5032–5041 CrossRef CAS.
- T. Sun, Y. S. Zhang, B. Pang, D. C. Hyun, M. Yang and Y. Xia, Nanomater. Neoplasms, 2021, 31–142 Search PubMed.
- E. Pavitra, B. Dariya, G. Srivani, S. M. Kang, A. Alam, P. R. Sudhir, M. A. Kamal, G. S. R. Raju, Y. K. Han, B. V. K. S. Lakkakula, G. P. Nagaraju and Y. S. Huh, Semin. Cancer Biol., 2021, 69, 293–306 CrossRef CAS PubMed.
- D. D. Ma and W. X. Yang, Oncotarget, 2016, 7, 40882 CrossRef.
- N. Praetorius and T. Mandal, Recent Pat. Drug Delivery Formulation, 2008, 1, 37–51 Search PubMed.
- K. M. Camacho, S. Menegatti, D. R. Vogus, A. Pusuluri, Z. Fuchs, M. Jarvis, M. Zakrewsky, M. A. Evans, R. Chen and S. Mitragotri, J. Controlled Release, 2016, 229, 154–162 CAS.
- R. Poupot, D. Bergozza and S. Fruchon, Materials, 2018, 11, 270 CrossRef PubMed.
- C. Fornaguera and C. Solans, Curr. Pathobiol. Rep., 2016, 4, 189–197 Search PubMed.
- W. Zhang, A. Mehta, Z. Tong, L. Esser and N. H. Voelcker, Adv. Sci., 2021, 8, 2003937 CAS.
- D. Singh, H. Kapahi, M. Rashid, A. Prakash, A. B. A. Majeed and N. Mishra, Artif. Cells, Nanomed., Biotechnol., 2016, 44, 780–791 CAS.
- P. Serra and P. Santamaria, Eur. J. Immunol., 2018, 48, 751–756 CAS.
- P. Serra and P. Santamaria, Clin. Immunol., 2015, 160, 3–13 CrossRef CAS PubMed.
- E. Saito, S. J. Gurczynski, K. R. Kramer, C. A. Wilke, S. D. Miller, B. B. Moore and L. D. Shea, Sci. Adv., 2020, 6, 9317–9333 CrossRef PubMed.
- C. Prego, P. Paolicelli, B. Díaz, S. Vicente, A. Sánchez, Á. González-Fernández and M. J. Alonso, Vaccine, 2010, 28, 2607–2614 CrossRef CAS PubMed.
- X. Hang, H. Peng, H. Song, Z. Qi, X. Miao and W. Xu, J. Virol. Methods, 2015, 222, 150–157 CrossRef CAS PubMed.
- N. H. A. Ellah, H. M. Tawfeek, J. John and H. F. Hetta, Nanomedicine, 2019, 14, 1471–1491 CrossRef PubMed.
- J. Miao, P. Gao, Q. Li, K. He, L. Zhang, J. Wang and L. Huang, Int. J. Mol. Sci., 2021, 22, 11227 CrossRef CAS.
- B. Kischkel, S. A. Rossi, S. R. Santos, J. D. Nosanchuk, L. R. Travassos and C. P. Taborda, Front. Cell. Infect. Microbiol., 2020, 10, 463 CrossRef CAS.
- I. E. Mba and E. I. Nweze, World J. Microbiol. Biotechnol., 2020, 36, 1–20 CrossRef.
- V. Mishra, M. Singh, Y. Mishra, N. Charbe, P. Nayak, K. Sudhakar, A. A. A. Aljabali, S. H. Shahcheraghi, H. Bakshi, Á. Serrano-Aroca and M. M. Tambuwala, Appl. Sci., 2021, 11, 7119 CrossRef CAS.
- D. Huang, F. Yue, J. Qiu, M. Deng and S. Kuang, Acta Biomater., 2020, 118, 196–206 CrossRef CAS PubMed.
- M. E. Nance, C. H. Hakim, N. N. Yang and D. Duan, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2018, 10, e1472 Search PubMed.
- P. Rimessi, P. Sabatelli, M. Fabris, P. Braghetta, E. Bassi, P. Spitali, G. Vattemi, G. Tomelleri, L. Mari, D. Perrone, A. Medici, M. Neri, M. Bovolenta, E. Martoni, N. M. Maraldi, F. Gualandi, L. Merlini, M. Ballestri, L. Tondelli, K. Sparnacci, P. Bonaldo, A. Caputo, M. Laus and A. Ferlini, Mol. Ther., 2009, 17, 820–827 CrossRef CAS PubMed.
- P. Sabourian, G. Yazdani, S. S. Ashraf, M. Frounchi, S. Mashayekhan, S. Kiani and A. Kakkar, Int. J. Mol. Sci., 2020, 21, 1–20 Search PubMed.
- D. P. Otto, A. Otto and M. M. de Villiers, Expert Opin. Drug Delivery, 2015, 12, 763–777 CrossRef CAS.
- R. Zein, W. Sharrouf and K. Selting, J. Oncol., 2020, 2020, 1–16 CrossRef PubMed.
- T. M. Kiio and S. Park, J. Pharm. Invest., 2021, 51, 35–51 CrossRef CAS.
- A. Basu, L. B. Okello, N. Castellanos, S. Roh and O. D. Velev, Soft Matter, 2023, 19, 2466–2485 CAS.
- A. Basu, M. R. Clary, J. B. Tracy, C. K. Hall and O. D. Velev, ACS Nano, 2024, 18, 19814–19827 CAS.
- Y. Huang, P. Li, R. Zhao, L. Zhao, J. Liu, S. Peng, X. Fu, X. Wang, R. Luo, R. Wang and Z. Zhang, Biomed. Pharmacother., 2022, 151, 113053 CAS.
- S. Jafari, H. Derakhshankhah, L. Alaei, A. Fattahi, B. S. Varnamkhasti and A. A. Saboury, Biomed. Pharmacother., 2019, 109, 1100–1111 CrossRef CAS PubMed.
- J. Yao, P. Li, L. Li and M. Yang, Acta Biomater., 2018, 74, 36–55 CAS.
- K. J. McHugh, L. Jing, A. M. Behrens, S. Jayawardena, W. Tang, M. Gao, R. Langer and A. Jaklenec, Adv. Mater., 2018, 30, 1706356 Search PubMed.
- R. R. Miranda, I. Sampaio and V. Zucolotto, Colloids Surf., B, 2022, 210, 112254 CAS.
- P. Tan, H. S. Li, J. Wang and S. C. B. Gopinath, Biotechnol. Appl. Biochem., 2021, 68, 1236–1242 CAS.
- R. Wilson, Chem. Soc. Rev., 2008, 37, 2028–2045 RSC.
- W. Zhou, X. Gao, D. Liu and X. Chen, Chem. Rev., 2015, 115, 10575–10636 CrossRef CAS.
- E. Boisselier and D. Astruc, Chem. Soc. Rev., 2009, 38, 1759–1782 RSC.
- S. J. Soenen, P. Rivera-Gil, J. M. Montenegro, W. J. Parak, S. C. De Smedt and K. Braeckmans, Nano Today, 2011, 6, 446–465 CrossRef CAS.
- B. Fadeel and A. E. Garcia-Bennett, Adv. Drug Delivery Rev., 2010, 62, 362–374 CrossRef CAS PubMed.
- S. V. Mussi and V. P. Torchilin, J. Mater. Chem. B, 2013, 1, 5201–5209 RSC.
- M. Rajabi and S. A. Mousa, Curr. Pharm. Biotechnol., 2016, 17, 662–672 CAS.
- R. Kumar, A. Kulkarni, D. K. Nagesha and S. Sridhar, Theranostics, 2012, 2, 714 CrossRef CAS PubMed.
- H. Zhang and P. Mi, Theranostic Bionanomater., 2019, 289–302 CAS.
- A. K. Sharma, P. Prasher, A. A. Aljabali, V. Mishra, H. Gandhi, S. Kumar, S. Mutalik, D. K. Chellappan, M. M. Tambuwala, K. Dua and D. N. Kapoor, Drug Delivery Transl. Res., 2020, 10, 1171–1190 CAS.
- B. C. Paruchuri, V. Gopal, S. Sarupria and J. Larsen, Nanomedicine, 2021, 16, 2679–2693 CAS.
- T. Anajafi and S. Mallik, Ther. Delivery, 2015, 6, 521–534 CAS.
- A. M. Caminade and C. O. Turrin, J. Mater. Chem. B, 2014, 2, 4055–4066 CAS.
- H. Wang, Q. Huang, H. Chang, J. Xiao and Y. Cheng, Biomater. Sci., 2016, 4, 375–390 RSC.
- Y. B. Patil, U. S. Toti, A. Khdair, L. Ma and J. Panyam, Biomaterials, 2009, 30, 859–866 Search PubMed.
- C. J. Cheng, G. T. Tietjen, J. K. Saucier-Sawyer and W. M. Saltzman, Nat. Rev. Drug Discovery, 2015, 14(4), 239–247 CrossRef CAS PubMed.
- L. E. van Vlerken and M. M. Amiji, Expert Opin. Drug Delivery, 2006, 3, 205–216 CrossRef CAS.
- W. Yang, H. Veroniaina, X. Qi, P. Chen, F. Li and P.C. Ke, Adv. Ther., 2020, 3(1), 1900102 CrossRef PubMed.
- V. M. Martín Giménez, G. Arya, I. A. Zucchi, M. J. Galante and W. Manucha, Soft Matter, 2021, 17, 8577–8584 Search PubMed.
- W. Gao, J. M. Chan and O. C. Farokhzad, Mol. Pharm., 2010, 7, 1913–1920 CAS.
- P. K. Bolla, V. A. Rodriguez, R. S. Kalhapure, C. S. Kolli, S. Andrews and J. Renukuntla, J. Drug Delivery Sci. Technol., 2018, 46, 416–435 CAS.
- S. Jain, S. K. Cherukupalli, A. Mahmood, S. Gorantla, V. K. Rapalli, S. K. Dubey and G. Singhvi, J. Appl. Pharm. Sci., 2019, 9, 130–143 CAS.
- A. Kaushik, R. D. Jayant, V. Sagar and M. Nair, Expert Opin. Drug Delivery, 2014, 11, 1635–1646 CAS.
- R. Guduru, P. Liang, C. Runowicz, M. Nair, V. Atluri and S. Khizroev, Sci. Rep., 2013, 3, 2953 Search PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS.
- I. Kola and J. Landis, Nat. Rev. Drug Discovery, 2004, 3, 711–716 CrossRef CAS PubMed.
- N. Singh, C. Marets, J. Boudon, N. Millot, L. Saviot and L. Maurizi, Nanoscale Adv., 2021, 3, 1209–1229 RSC.
- M. H. Akhter, H. Khalilullah, M. Gupta, M. A. Alfaleh, N. A. Alhakamy, Y. Riadi and S. Md, Biomedicines, 2021, 9, 1496 CrossRef CAS.
- D. Maiolo, P. Del Pino, P. Metrangolo, W. J. Parak and F. Baldelli Bombelli, Nanomedicine, 2015, 10, 3231–3247 CrossRef CAS PubMed.
- H. Li, Y. Wang, Q. Tang, D. Yin, C. Tang, E. He, L. Zou and Q. Peng, Acta Biomater., 2021, 129, 57–72 CrossRef CAS PubMed.
- A. M. Javier, O. Kreft, A. P. Alberola, C. Kirchner, B. Zebli, A. S. Susha, E. Horn, S. Kempter, A. G. Skirtach, A. L. Rogach, J. Rädler, G. B. Sukhorukov, M. Benoit and W. J. Parak, Small, 2006, 2, 394–400 Search PubMed.
- D. Hühn, K. Kantner, C. Geidel, S. Brandholt, I. De Cock, S. J. H. Soenen, P. Riveragil, J. M. Montenegro, K. Braeckmans, K. Müllen, G. U. Nienhaus, M. Klapper and W. J. Parak, ACS Nano, 2013, 7, 3253–3263 Search PubMed.
- C. Corbo, R. Molinaro, A. Parodi, N. E. Toledano Furman, F. Salvatore and E. Tasciotti, Nanomedicine, 2016, 11, 81–100 CAS.
- Q. Peng and H. Mu, J. Controlled Release, 2016, 225, 121–132 Search PubMed.
- M. P. Monopoli, D. Walczyk, A. Campbell, G. Elia, I. Lynch, F. Baldelli Bombelli and K. A. Dawson, J. Am. Chem. Soc., 2011, 133, 2525–2534 CAS.
- W. L. Koh, P. H. Tham, H. Yu, H. L. Leo and J. C. Y. Kah, Nanomedicine, 2016, 11, 2275–2287 CAS.
- G. Berrecoso, J. Crecente-Campo and M. J. Alonso, Drug Delivery Transl. Res., 2020, 10, 730–750 CrossRef CAS.
- Q. Xiao, M. Zoulikha, M. Qiu, C. Teng, C. Lin, X. Li, M. A. Sallam, Q. Xu and W. He, Adv. Drug Delivery Rev., 2022, 186, 114356 CAS.
- S. Panico, S. Capolla, S. Bozzer, G. Toffoli, M. Dal Bo and P. Macor, Pharmaceutics, 2022, 14(12), 2605 CAS.
- C. Corbo, R. Molinaro, M. Tabatabaei, O. C. Farokhzad and M. Mahmoudi, Biomater. Sci., 2017, 5, 378–387 CAS.
- M. J. Hajipour, S. Laurent, A. Aghaie, F. Rezaee and M. Mahmoudi, Biomater. Sci., 2014, 2, 1210 CAS.
- S. Sharifi, G. Caracciolo and M. Mahmoudi, Trends Pharmacol. Sci., 2020, 41, 641–652 CAS.
- S. Bhunia, P. Saha, P. Moitra, M. A. Addicoat and S. Bhattacharya, Chem. Sci., 2022, 13, 7920–7932 RSC.
- G. Su, H. Jiang, B. Xu, Y. Yu and X. Chen, Mol. Pharm., 2018, 15, 5019–5030 CrossRef CAS.
- Z. A. Zhang, X. Xin, C. Liu, Y-H Liu, H. X. Duan, L-l Qi, Y. Y. Zhang, H-M Zhao, L. Q. Chen, M. J. Jin, Z. G. Gao and W. Huang, J. Nanobiotechnol., 2021, 19, 1–24 CrossRef CAS.
- S. Behzadi, V. Serpooshan, R. Sakhtianchi, B. Müller, K. Landfester, D. Crespy and M. Mahmoudi, Colloids Surf., B, 2014, 123, 143–149 CrossRef CAS PubMed.
- L. Zeng, J. Gao, Y. Liu, J. Gao, L. Yao, X. Yang, X. Liu, B. He, L. Hu, J. Shi, M. Song, G. Qu and G. Jiang, TrAC, Trends Anal. Chem., 2019, 118, 303–314 CrossRef CAS.
- M. Mahmoudi, Nat. Commun., 2022, 13, 1–4 Search PubMed.
- M. Mahmoudi, M. P. Landry, A. Moore and R. Coreas, Nat. Rev. Mater., 2023, 8, 422–438 CrossRef.
- S. M. Kamali Shahri, S. Sharifi and M. Mahmoudi, Expert Opin. Drug Delivery, 2021, 18, 1379–1394 CrossRef CAS.
- E. D. Namiot, A. V. Sokolov, V. N. Chubarev, V. V. Tarasov and H. B. Schiöth, Int. J. Mol. Sci., 2023, 24, 787 CAS.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CAS.
- R. Pieper, C. L. Gatlin, A. J. Makusky, P. S. Russo, C. R. Schatz, S. S. Miller, Q. Su, A. M. McGrath, M. A. Estock, P. P. Parmar, M. Zhao, S. T. Huang, J. Zhou, F. Wang, R. Esquer-Blasco, N. L. Anderson, J. Taylor and S. Steiner, Proteomics, 2003, 3, 1345–1364 CAS.
- A. A. Kliuchnikova, S. E. Novikova, E. V. Ilgisonis, O. I. Kiseleva, E. V. Poverennaya, V. G. Zgoda, S. A. Moshkovskii, V. V. Poroikov, A. V. Lisitsa, A. I. Archakov and E. A. Ponomarenko, Int. J. Mol. Sci., 2023, 24, 769 CAS.
- N. L. Anderson, N. G. Anderson, T. W. Pearson, C. H. Borchers, A. G. Paulovich, S. D. Patterson, M. Gillette, R. Aebersold and S. A. Carr, Mol. Cell. Proteomics, 2009, 8, 883–886 CAS.
- N. Kamaly, O. C. Farokhzad and C. Corbo, Nanoscale, 2022, 14, 1606–1620 RSC.
- M. Farshbaf, H. Valizadeh, Y. Panahi, Y. Fatahi, M. Chen, A. Zarebkohan and H. Gao, Int. J. Pharm., 2022, 614, 121458 CrossRef CAS PubMed.
- S. Tenzer, D. Docter, J. Kuharev, A. Musyanovych, V. Fetz, R. Hecht, F. Schlenk, D. Fischer, K. Kiouptsi, C. Reinhardt, K. Landfester, H. Schild, M. Maskos, S. K. Knauer and R. H. Stauber, Nat. Nanotechnol., 2013, 8, 772–781 CrossRef CAS.
- S. L. Hirsh, D. R. McKenzie, N. J. Nosworthy, J. A. Denman, O. U. Sezerman and M. M. M. Bilek, Colloids Surf., B, 2013, 103, 395–404 CAS.
- M. Barz, W.J. Parak and R. Zentel, Advanced Science, 2024, 11(34), 2402935 CAS.
- V. Colapicchioni, M. Tilio, L. Digiacomo, V. Gambini, S. Palchetti, C. Marchini, D. Pozzi, S. Occhipinti, A. Amici and G. Caracciolo, Int. J. Biochem. Cell Biol., 2016, 75, 180–187 CAS.
- V. P. Zhdanov, Curr. Opin. Colloid Interface Sci., 2019, 41, 95–103 CAS.
- B. Kharazian, N. L. Hadipour and M. R. Ejtehadi, Int. J. Biochem. Cell Biol., 2016, 75, 162–174 CAS.
- M. P. Monopoli, C. Åberg, A. Salvati and K. A. Dawson, Nat. Nanotechnol., 2012, 7, 779–786 CAS.
- D. Chakraborty, K. R. Ethiraj and A. Mukherjee, RSC Adv., 2020, 10, 27161–27172 RSC.
- M. A. Dobrovolskaia, B. W. Neun, S. Man, X. Ye, M. Hansen, A. K. Patri, R. M. Crist and S. E. McNeil, Nanomedicine, 2014, 10, 1453–1463 CrossRef CAS PubMed.
- F. Tavanti, A. Pedone and M. C. Menziani, New J. Chem., 2015, 39, 2474–2482 RSC.
- S. Harnisch and R. H. Müller, Eur. J. Pharm. Biopharm., 2000, 49, 41–46 CrossRef CAS.
- S. Palchetti, V. Colapicchioni, L. Digiacomo, G. Caracciolo, D. Pozzi, A. L. Capriotti, G. La Barbera and A. Laganà, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 189–196 CrossRef CAS PubMed.
- M. Hadjidemetriou, Z. Al-Ahmady, M. Mazza, R. F. Collins, K. Dawson and K. Kostarelos, ACS Nano, 2015, 9, 8142–8156 CrossRef CAS.
- M.N. Gupta and I. Roy, Mol. Pharmaceutics, 2020, 17, 725–737 CrossRef CAS.
- A. J. Paula, R. T. Araujo Júnior, D. S. T. Martinez, E. J. Paredes-Gamero, H. B. Nader, N. Durán, G. Z. Justo and O. L. Alves, ACS Appl. Mater. Interfaces, 2013, 5, 8387–8393 CrossRef CAS PubMed.
- Q. Peng, X. Q. Wei, Q. Yang, S. Zhang, T. Zhang, X. R. Shao, X. X. Cai, Z. R. Zhang and Y. F. Lin, Nanomedicine, 2015, 10, 205–214 CrossRef CAS PubMed.
- D. Chakraborty, L. Mohan, S. A. Alex, N. Chandrasekaran and A. Mukherjee, Biomater. Sci., 2019, 7, 63–75 RSC.
- K. Obst, G. Yealland, B. Balzus, E. Miceli, M. Dimde, C. Weise, M. Eravci, R. Bodmeier, R. Haag, M. Calderón, N. Charbaji and S. Hedtrich, Biomacromolecules, 2017, 18, 1762–1771 CrossRef CAS PubMed.
- A. K. Ostermeyer, C. Kostigen Mumuper, L. Semprini and T. Radniecki, Environ. Sci. Technol., 2013, 47, 14403–14410 CAS.
- M. Levak, P. Burić, M. Dutour Sikirić, D. Domazet Jurašin, N. Mikac, N. Bačić, R. Drexel, F. Meier, Ž. Jakšić and D. M. Lyons, Environ. Sci. Technol., 2017, 51, 1259–1266 CAS.
- M. P. Monopoli, F. B. Bombelli and K. A. Dawson, Nat. Nanotechnol., 2011, 6, 11–12 CAS.
- A. Cifuentes-Rius, H. De Puig, J. C. Y. Kah, S. Borros and K. Hamad-Schifferli, ACS Nano, 2013, 7, 10066–10074 CAS.
- G. Caracciolo, S. Palchetti, L. Digiacomo, R. Z. Z. Chiozzi, A. L. Capriotti, H. Amenitsch, P. M. Tentori, V. Palmieri, M. Papi, F. Cardarelli, D. Pozzi and A. Laganà, ACS Appl. Mater. Interfaces, 2018, 10, 22951–22962 CrossRef CAS.
- Z. S. Al-Ahmady, M. Hadjidemetriou, J. Gubbins and K. Kostarelos, J. Controlled Release, 2018, 276, 157–167 CrossRef CAS.
- P. S. R. Naidu, E. Denham, C. A. Bartlett, T. McGonigle, N. L. Taylor, M. Norret, N. M. Smith, S. A. Dunlop, K. S. Iyer and M. Fitzgerald, RSC Adv., 2020, 10, 2856–2869 RSC.
- A. L. Barrán-Berdón, D. Pozzi, G. Caracciolo, A. L. Capriotti, G. Caruso, C. Cavaliere, A. Riccioli, S. Palchetti and A. Laganaì, Langmuir, 2013, 29, 6485–6494 CrossRef.
- V. H. Nguyen and B. J. Lee, Int. J. Nanomed., 2017, 12, 3137–3151 CrossRef CAS PubMed.
- S. Schöttler, K. Landfester and V. Mailänder, Angew. Chem., Int. Ed., 2016, 55, 8806–8815 Search PubMed.
- S. Ritz, S. Schöttler, N. Kotman, G. Baier, A. Musyanovych, J. Kuharev, K. Landfester, H. Schild, O. Jahn, S. Tenzer and V. Mailänder, Biomacromolecules, 2015, 16, 1311–1321 CAS.
- A. Lesniak, F. Fenaroli, M. P. Monopoli, C. Åberg, K. A. Dawson and A. Salvati, ACS Nano, 2012, 6, 5845–5857 CAS.
- K. I. Ogawara, K. Furumoto, S. Nagayama, K. Minato, K. Higaki, T. Kai and T. Kimura, J. Controlled Release, 2004, 100, 451–455 CAS.
- J. Simon, L. K. Müller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailänder, Nanoscale, 2018, 10, 10731–10739 CAS.
- G. Caracciolo, F. Cardarelli, D. Pozzi, F. Salomone, G. Maccari, G. Bardi, A. L. Capriotti, C. Cavaliere, M. Papi and A. Laganà, ACS Appl. Mater. Interfaces, 2013, 5, 13171–13179 CrossRef CAS.
- M. Schäffler, F. Sousa, A. Wenk, L. Sitia, S. Hirn, C. Schleh, N. Haberl, M. Violatto, M. Canovi, P. Andreozzi, M. Salmona, P. Bigini, W. G. Kreyling and S. Krol, Biomaterials, 2014, 35, 3455–3466 CrossRef.
- A. Bhargava, A. Dev, S. J. Mohanbhai, V. Pareek, N. Jain, S. R. Choudhury, J. Panwar and S. Karmakar, Sci. Total Environ., 2021, 772, 144797 CrossRef CAS PubMed.
- K. Abstiens, S. Maslanka Figueroa, M. Gregoritza and A. M. Goepferich, Soft Matter, 2019, 15, 709–720 RSC.
- S. Pulakkat, S. A. Balaji, A. Rangarajan and A. M. Raichur, ACS Appl. Mater. Interfaces, 2016, 8, 23437–23449 CrossRef CAS.
- M. A. Qureshi, Eur. Polym. J., 2024, 208, 112865 CrossRef.
- A. I. Fernandes, A. F. Jozala, Y. Cao, X. Dong and X. Chen, Pharmaceutics, 2022, 14, 778 CrossRef.
- D. Guimarães, A. Cavaco-Paulo and E. Nogueira, Int. J. Pharm., 2021, 601, 120571 CrossRef PubMed.
- S. Palchetti, D. Caputo, L. Digiacomo, A. L. Capriotti, R. Coppola, D. Pozzi and G. Caracciolo, Pharmaceutics, 2019, 11, 31 CrossRef CAS.
- R. Cai and C. Chen, Advanced Materials, 2019, 31(45), 1805740 Search PubMed.
- M. Hadjidemetriou, Z. Al-Ahmady and K. Kostarelos, Nanoscale, 2016, 8, 6948–6957 Search PubMed.
- R. Rampado, S. Crotti, P. Caliceti, S. Pucciarelli and M. Agostini, Front. Bioeng. Biotechnol., 2020, 8, 166 CrossRef PubMed.
- S. Palchetti, V. Colapicchioni, L. Digiacomo, G. Caracciolo, D. Pozzi, A. L. Capriotti, G. La Barbera and A. Laganà, Biochim. Biophys. Acta, Biomembr., 2016, 1858, 189–196 CrossRef CAS PubMed.
- S. Balzan, C. de Almeida Quadros, R. de Cleva, B. Zilberstein and I. Cecconello, J. Gastroenterol. Hepatol., 2007, 22, 464–471 CAS.
- D. Docter, U. Distler, W. Storck, J. Kuharev, D. Wünsch, A. Hahlbrock, S. K. Knauer, S. Tenzer and R. H. Stauber, Nat. Protoc., 2014, 9, 2030–2044 Search PubMed.
- X. Zhang and P. Zhang, Curr. Nanosci., 2016, 13, 124–129 Search PubMed.
- M. J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CAS.
- P. V. Pawar, S. V. Gohil, J. P. Jain and N. Kumar, Polym. Chem., 2013, 4, 3160–3176 RSC.
- X. Hu, Y. Zhang, Z. Xie, X. Jing, A. Bellotti and Z. Gu, Biomacromolecules, 2017, 18, 649–673 CrossRef CAS PubMed.
- S. Matoori and J. C. Leroux, Mater. Horiz., 2020, 7, 1297–1309 RSC.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- F. A. de Oliveira, L. J. C. Albuquerque, C. E. Castro, K. A. Riske, I. C. Bellettini and F. C. Giacomelli, Colloids Surf., B, 2022, 213, 112387 CrossRef CAS.
- S. Svenson and D. A. Tomalia, Adv. Drug Delivery Rev., 2005, 57, 2106–2129 CAS.
- D. Chandrasekar, R. Sistla, F. J. Ahmad, R. K. Khar and P. V. Diwan, Biomaterials, 2007, 28, 504–512 CrossRef CAS.
- R. Y. Sathe and P. V. Bharatam, Adv. Colloid Interface Sci., 2022, 303, 102639 CrossRef CAS.
- J. M. Rae and B. Jachimska, Nanoscale, 2021, 13, 2703–2713 RSC.
- C. V. Kelly, M. G. Liroff, L. D. Triplett, P. R. Leroueil, D. G. Mullen, J. M. Wallace, S. Meshinchi, J. R. Baker, B. G. Orr and M. M. B. Holl, ACS Nano, 2009, 3, 1886–1896 CrossRef CAS.
- L. Giehm, C. Christensen, U. Boas, P. M. H. Heegaard and D. E. Otzen, Biopolymers, 2008, 89, 522–529 CrossRef CAS PubMed.
- J. Giri, M. S. Diallo, A. J. Simpson, Y. Liu, W. A. Goddard, R. Kumar and G. C. Woods, ACS Nano, 2011, 5, 3456–3468 CrossRef CAS PubMed.
- A. Åkesson, M. Cárdenas, G. Elia, M. P. Monopoli and K. A. Dawson, RSC Adv., 2012, 2, 11245–11248 RSC.
- B. Wang, Y. Sun, T. P. Davis, P. C. Ke, Y. Wu and F. Ding, ACS Sustainable Chem. Eng., 2018, 6, 11704 CrossRef CAS.
- N. Liu, M. Tang and J Ding, Chemosphere, 2020, 245, 125624 CrossRef CAS.
- W. Xiao and H. Gao, Int. J. Pharm., 2018, 552, 328–339 CrossRef CAS.
- A. Berardi and F. Baldelli Bombelli, Expert Opinion on Drug Delivery, 2019, 16(6), 563–566 CrossRef PubMed.
- S. Modi and B. D. Anderson, Mol. Pharm., 2013, 10, 3076–3089 CrossRef CAS.
- J. Weng, H. H. Y. Tong and S. F. Chow, Pharmaceutics, 2020, 12, 732 CrossRef CAS.
- C. Larsen, S. W. Larsen, H. Jensen, A. Yaghmur and J. Østergaard, Expert Opin. Drug Delivery, 2009, 6, 1283–1295 CrossRef CAS PubMed.
- G. Moreno-Bautista and K. C. Tam, Colloids Surf., A, 2011, 389, 299–303 CrossRef CAS.
- M. Y. Levy and S. Benita, Int. J. Pharm., 1990, 66, 29–37 CrossRef CAS.
- N. Kamaly, B. Yameen, J. Wu and O. C. Farokhzad, Chem. Rev., 2016, 116, 2602–2663 CrossRef CAS.
- S. Lappe, D. Mulac and K. Langer, Int. J. Pharm., 2017, 517, 338–347 CrossRef CAS.
- E. Izak-Nau, A. Huk, B. Reidy, H. Uggerud, M. Vadset, S. Eiden, M. Voetz, M. Himly, A. Duschl, M. Dusinska and I. Lynch, RSC Adv., 2015, 5, 84172–84185 RSC.
- C. Corbo, A. A. Li, H. Poustchi, G. Y. Lee, S. Stacks, R. Molinaro, P. Ma, T. Platt, S. Behzadi, R. Langer, V. Farias and O. C. Farokhzad, Adv. Healthc. Mater., 2021, 10, 2000948 CrossRef CAS.
- E. Quagliarini, R. Di Santo, D. Pozzi and G. Caracciolo, Sens. Int., 2020, 1, 100025 CrossRef.
| This journal is © The Royal Society of Chemistry 2025 |