 Open Access Article
Open Access ArticleDeploying nucleic acids-loaded plant-derived exosomes as green nano gadget in cancer gene therapy
Marola Paula
Fawzy
a,
Hatem A. F. M.
Hassan
 b,
Muhammad Umair
Amin
c,
Eduard
Preis
c,
Udo
Bakowsky
b,
Muhammad Umair
Amin
c,
Eduard
Preis
c,
Udo
Bakowsky
 *c and
Sherif Ashraf
Fahmy
*c and
Sherif Ashraf
Fahmy
 *c
*c
aDepartment of Chemistry, School of Life and Medical Sciences, University of Hertfordshire Hosted by Global Academic Foundation, R5 New Garden City, New Capital, Cairo 11835, Egypt
bMedway School of Pharmacy, University of Kent, Chatham Maritime, Kent ME4 4TB, UK
cDepartment of Pharmaceutics and Biopharmaceutics, University of Marburg, Robert-Koch-Str. 4, 35037 Mar-burg, Germany. E-mail: ubakowsky@aol.com; sheriffahmy@aucegypt.edu
First published on 13th January 2025
Abstract
The pursuit of effective drug delivery systems is critical in advancing cancer therapies, particularly in the realms of chemotherapy, radiotherapy and immunotherapy. This review focuses on plant-based extracellular vesicles (PBEs) as innovative nanoplatforms for encapsulating and delivering genetic materials, including microRNAs (miRNAs), small interfering RNAs (siRNAs), and mitochondrial DNA (mtDNA). We explore the unique properties of PBEs that enhance the stability and bioavailability of these therapeutic molecules, particularly their resistance to degradation by ribonucleases (RNases) and their ability to withstand gastrointestinal digestion. By improving the stability and facilitating cellular uptake of these genetic particles, PBEs offer a significant enhancement of their therapeutic efficacy through nuclear gene modulation. This review highlights the transformative potential of PBEs in developing novel drug delivery systems for cancer treatment, paving the way for future research and clinical advancements in RNA-based therapies and beyond.
Introduction
Cancer is characterized by unregulated cell proliferation that spreads from its origin to distant sites within the body, frequently resulting in death, hence marked as a major global health issue and a leading cause of death worldwide. Consequently, early detection and treatment are crucial for preventing disease progression, elevating overall survival rates, and reducing mortality rates.1Henceforth, nanotechnology is being increasingly applied in cancer research, with significant advancements in areas such as gene therapy, molecular imaging, drug delivery, and cancer detection and diagnosis. These applications have demonstrated considerable promise, offering new avenues for more effective and targeted treatment strategies with minimized side effects.2,3
This is because nanomaterials, being biological in nature, can easily traverse cellular barriers while exhibiting active and passive targeting capabilities. Despite the availability of various medications for cancer, their sensitivity often leads to suboptimal results, along with numerous side effects and damage to healthy cells.4 Consequently, the shift in interest in cancer research has been focused on exploring different types of nanomaterials, such as plant-based exosomes (PBEs), indicating that nanoparticles can effectively balance the enhancement of therapeutic efficacy with the reduction of drug toxicity in treating cancer.2,5
Building on this, nanomaterials have opened new avenues for delivering biofunctional agents across various biological applications, including diagnostics, imaging, and therapy. For instance, exosome-based targeted delivery systems have emerged following the successful use of liposomes in both passive and active cargo delivery. As naturally occurring nanocarriers, exosomes facilitate various physiological and pathological processes by encapsulating diverse bioactive molecules within a phospholipid bilayer membrane. It is noteworthy that the intercellular transfer of proteins via exosomes may also play a role in cancer development.6,7
Furthermore, recent evidence suggests that microRNAs (miRNAs) function as immune modulators within tumors. Acting as either tumor suppressors or oncogenes, these miRNAs can affect anti-tumor immunity and facilitate interactions between tumor cells and nearby immune cells. Given their specific regulatory capabilities, miRNAs are emerging as potential prognostic biomarkers, as well as therapeutic targets in immunotherapy.8,9 Therefore, to the best of our knowledge, in this review, we are the first to highlight the successful encapsulation and delivery of genetic materials to cancerous cells using plant-derived exosomes (PBEs). These exosomes have been evaluated in various cancer cell lines to assess their potential to enhance therapeutic efficacy, reduce side effects, and minimize the toxicity of immune molecules. By exploring the delivery of genetic cargo within PBEs, this study aims to advance the development of more targeted and effective cancer therapies.
PBEs a potential drug delivery system
Drug delivery systems (DDS) address various challenges associated with drug administration, such as enhancing circulation, reducing degradation, minimizing side effects, and achieving specific therapeutic objectives, all of which contribute to improved efficacy of active pharmaceutical ingredients (APIs). The demand for safe and effective drug delivery methods has driven the scientific community to explore various alternatives. Recently, mammalian-derived exosomes have garnered interest due to their characteristics as natural carriers with inherent specialized targeting capabilities. However, concerns regarding their immunogenicity and cytotoxicity have been raised, which may pose challenges for clinical translation. The need for a sufficient and cost-effective source of nanoparticles has led to the exploration of plant-derived nanoparticles, which can overcome these limitations.10,11 These naturally occurring nanoparticles can be engineered to enhance their therapeutic properties, offering a novel approach to precision medicine in cancer treatment.12 Their inherent biocompatibility and low toxicity make them ideal candidates for drug delivery systems. Recent developments have concentrated on modifying these exosomes to improve their targeting of cancer cells, thereby increasing their potential as tools for precision medicine.13Numerous constituents present in consumable plants, including antioxidants, anti-inflammatory agents, and anti-cancer compounds, demonstrate biological efficacy. Phytochemicals, also referred to as secondary metabolites, are widely acknowledged to possess these properties. Furthermore, research has shown that PBEs closely mimic the structures of mammalian exosomes and are adept at transporting plant-derived components, such as short RNAs, into human cells, offering various therapeutic advantages and facilitating communication between different species. Furthermore, PBEs containing miRNAs can regulate gene expression post-transcriptionally in both plant and animal cells, traversing biological boundaries and inhibiting detrimental interactions in plants.14,15 Additionally, PBEs can be modified by altering their surface characteristics and cargo to enhance targeting and therapeutic efficacy. By manipulating surface proteins, researchers can improve the ability of exosomes to recognize and bind to specific signals from cancer cells.16 For instance, exosomes derived from ginger and grapes can be engineered to carry ligands that target receptors overexpressed in certain cancer types. This targeted approach ensures that the therapeutic agents within the exosomes are delivered directly to cancer cells, minimizing off-target effects and enhancing treatment specificity. Additionally, the cargo of these exosomes can include chemotherapeutic drugs, RNA molecules, or other therapeutic agents, thereby boosting their anti-cancer effects.17
Overview of plant-based exosomes (PBEs)
Extracellular vesicles (EVs) comprise a heterogeneous group of lipid bilayer-enclosed particles that are actively generated and released by numerous cell types into the extracellular space. This is a universal process found across all life forms, including prokaryotes and eukaryotes, and takes place under a broad spectrum of conditions, from normal physiological processes to disease-related states.18 EVs comprise a variety of subtypes, classified based on their mechanisms of synthesis and release, such as exosomes, apoptotic blebs, and other distinct EV groups. These vesicles can also be categorized according to their cell of origin (e.g., platelet-derived or endothelial cell-derived) or the physiological state of the cells, such as “oncosomes” released from cancer cells or “prostasomes” derived from the prostate. The primary types of EVs include microvesicles, exosomes, and apoptotic bodies (Fig. 1),19 though recent research has identified additional forms, including large oncosomes, migrasomes, ectosomes, exomeres, supermeres, and membrane particles (Table 1). EVs are widely distributed across human bodily fluids, such as breast milk, cerebrospinal fluid, urine, saliva, and blood, and can be detected in both healthy and disease states. Notably, the characteristics of these fluids—along with the associated diseases and underlying conditions—are closely linked to variations in the quantity, tissue origin, molecular composition, and functional properties of EVs.18 | ||
| Fig. 1 A visual representation of the diversity and sources of extracellular vesicles (EVs) illustrates their broad range of bioactive contents, including proteins, nucleic acids, and lipids. These components not only contribute to the structural integrity of EVs but also carry distinct cellular markers. Cells from a variety of tissue types utilize EVs as mediators of intercellular communication, secreting them into surrounding body fluids. In humans, a significant portion of EVs are derived from stem cells. Furthermore, EVs are not exclusive to humans; organisms across different kingdoms, from plants to bacteria, also actively produce and release EVs into their respective environments. This figure is reproduced from ref. 18 with permission from Nature, copyright (2024). | ||
| EV subtypes | Origin | Size (nm) | Mechanism of biogenesis | Ref. |
|---|---|---|---|---|
| Exosomes | Multivesicle body | 50–150 | Endosomes mature into late endosomes, forming multivesicular bodies (MVBs) with intraluminal vesicles that fuse with the plasma membrane for release (dependent or independent of ESCRT) | 20 |
| Microvesicles | Plasma membrane | 100–1000 | Calcium influx and cortical cytoskeleton remodeling cause direct plasma membrane budding and cleavage. | 21 |
| Apoptotic bodies | Plasma membrane | 100–5000 | Cytoplasmic fragmentation during programmed cell death | 22 |
| Exomeres | Secreting from cells | ≤50 | Cleavage of large cytoplasmic extensions from cell body | 23 |
| Migrasomes | Retraction fibers | 500–3000 | Because of cell migration and actin polarization/migrasomes are formed at the tip or by bifurcation of the retraction fibers during migration | 24 |
| Oncosomes | The shedding of non-apoptotic plasma membrane blebbing | 1000–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Released by cancer cells with amoeboid movement | 25 |
| Supermeres | Unknown | ∼35 (<50) | Unknown | 26 |
Specifically, exosomes are membrane-bound vesicles secreted into the extracellular space in mammalian or plant cells, primarily through the multivesicular body (MVB) sorting pathway, resulting in mammalian-based or plant-based exosomes (PBEs), respectively. These vesicles range in size from 30 to 150 nm and consist of a lipid bilayer enclosing biologically active components. Initially, exosomes were considered merely artifacts or mechanisms for cellular waste disposal.27 However, current research has established their significant role in cell-to-cell communication, including the transfer of biologically active substances and regulation of gene expression. Plant-derived exosomes gained attention when their essential role in plant immunity against pathogen attacks was identified.28,29 The biological components of PBEs exosomes, including proteins, miRNAs, RNA, and different metabolites, are similar to those found in mammals.30 While mammalian exosomes are widely recognized for their potential as disease biomarkers, there is growing interest in exclusively studying PBEs due to their inherent therapeutic activities that can be exploited for human health. Unlike mammalian-based exosomes, PBEs demonstrate several advantages, such as a more resourceful, economical large-scale production, non-toxic and non-immunogenic properties, and the ability to encapsulate various therapeutic and genetic molecules. As a result, substantial research efforts focus on identifying and developing nanosystems based on plant exosomes.31 Thus, herein, we exclusively spotlight the recent advances reported on the biosynthesis, composition, isolation, and purification of PBEs from various plant origins. In addition, their innate anticancer activities and their exploitation as potential nanocarriers for gene delivery will be discussed.
Biogenesis of PBEs
The biogenesis of exosomes (MBEs or PBEs) is a highly regulated process, as summarized in Fig. 2 and 3. Generally, exosomes can originate from the plasma membrane via three distinct pathways: (i) individual endosomes are transformed into MVBs, and the resulting exosomes are released upon fusion with the plasma membrane, (ii) exosomes can be expelled directly by vesicles that emerge from the plasma membrane, or (iii) they can be formed directly from dysregulated compartments associated with the internal plasma membrane (IPMC).32 MBEs are produced via the endocytic route, where, throughout late endocytosis, intraluminal vesicles (ILVs) are formed by membrane invagination inside multivesicular bodies (MVBs). The ILVs are produced when early endosomes are incorporated with the adjoining membrane layer. These intraluminal vesicles (ILVs) develop into exosomes, which are primarily released into the extracellular space with the help of the endosomal sorting complex required for transport (ESCRT).33 ESCRT plays a vital role in the biogenesis of exosomes, and it is categorized into four complexes (ESCRT-0, -I, -II, -III) comprising different proteins (Table 2).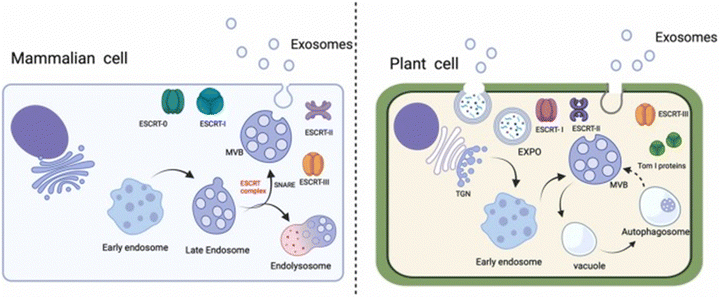 | ||
| Fig. 2 Illustrates the biogenesis process of exosomes in mammalian and plant cells. This figure is reproduced from ref. 34 with permission from ScienceDirect, copyright (2023). | ||
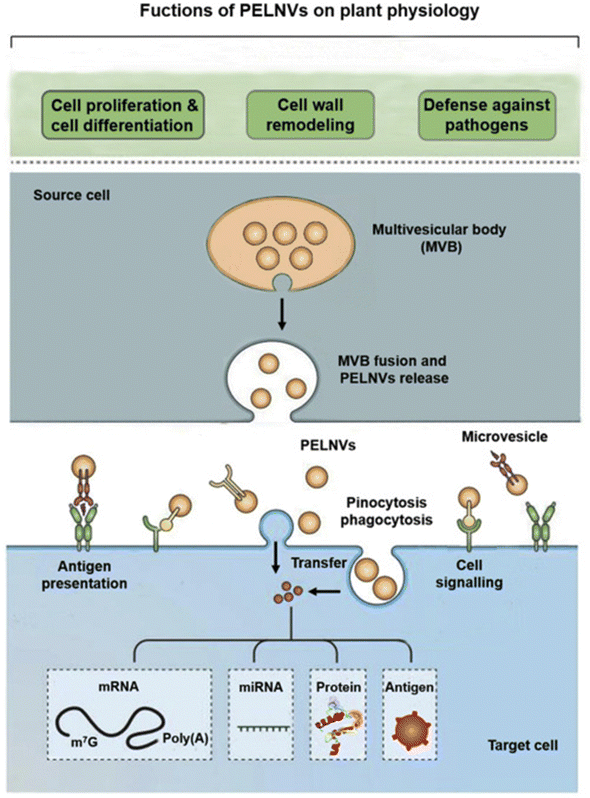 | ||
| Fig. 3 The formation of plant extracellular lipid nanovesicles and their roles in plant physiological processes. This figure is reproduced from ref. 35 with permission from Molecular Therapy, copyright (2021). | ||
| ESCRT type | Composition | Role |
|---|---|---|
| ESCRT-0 | It comprised two subunits: hepatocyte growth-factor regulated tyrosine kinase substrate (Hrs) and signal transducing adaptor molecule (STAM) | It also interacts with the TSG101 protein of ESCRT-1 and delivers the complex to the site. |
| ESCRT-I | Is a heterotetrametric complex | It is responsible for placing the cargo into the MVBs and initiates a concentration of the cargo proteins through interaction with the ESCRT-II complex. |
| ESCRT-II | is a hetero-tetramer comprised of EAP45, EAP30, and EAP 20 proteins. | It is responsible for the concentration of the cargo proteins. |
| ESCRT-III | It is an oligomer of charged multivesicular body protein (CHMP). | It is the most important functional component in triggering the release of exosomes: it initiates the establishment of membrane-binding spirals with the aid of VpS4p/SKD1 ATPase (source of energy). This results in the release of the vesicle buds from the membrane. Then, the vesicles interact with N-ethylmaleimide sensitive factor attachment protein receptors (SNARE), facilitating its release. |
Exosome secretion occurs via well-established pathways, including ESCRT and tetraspanin routes.36 ESCRT proteins, such as Hrs, STAM1, and TSG101, play essential roles in exosome formation and depend on ubiquitin-binding for their functionality.37 Cells contain various ubiquitinated soluble molecules, such as MHC-II, which facilitate the enrichment of ILVs. However, non-ubiquitinated MHC-II has also been identified in exosomes that are released into the extracellular space.32,38 Rab31 and GTPase have been identified as regulators of lipid-mediated exosome formation through mechanisms that function independently of the ESCRT and tetraspanin pathways.39 Exosome secretion mediated by the ESCRT pathway, which comprises a cytoplasmic multi-subunit system, is essential for membrane remodeling. This system facilitates vesicle budding and the segregation of contents into MVBs, relying on four key ESCRT complexes: ESCRT-0, ESCRT-I, ESCRT-II, and ESCRT-III, along with related proteins such as ALIX, VPS4, and TSG10.40 The ESCRT system is pivotal in exosome biogenesis, particularly in forming ILVs. Aggregates of PLD2-phosphatidic acid promote the fusion of endosomes with the ESCRT-I complex, thereby enhancing the secretion of exosomes from the relevant cells.41 The interaction between the MVB12B-MABP complex and PLD2-phosphatidic acid promotes exosome secretion while compromising the integrity of the MM (MVB12B-MABP)-PPA pathway. This disruption adversely affects late endosome budding, resulting in a marked reduction in the quantity of exosomes released.41 In contrast to the standard ESCRT pathway, when internal MVB transmembrane proteins and late endosomes are transported in vitro, ALIX facilitates the aggregation of ESCRT-III within late endosomes.42 This process enhances the extracellular secretion of transmembrane proteins through the cell membrane. Furthermore, in this mechanism, ESCRT-III directly interacts with lysobisphosphatidic acid (LBPA), circumventing the more complex ESCRT pathway.43 On the other hand, in the case of PBEs, the proteins are internalized by introversion, swelling, and the formation of transportation vesicles at the plasma membrane. Then, they are transferred to the trans Golgi network (TGN), a subunit that develops into MVBs. Like MBEs, ESCRT protein complexes are remarkably preserved in plants and execute akin roles in the ordering and maturation of MVBs.29 In addition, unlike animals, plants have a distinctive route to produce exosomes employing double membranous structures similar to autophagosomes called exocyst positive organelles (EXPOs). EXPOs adopt an endocytic pathway and unite with the plasma membrane to release uni-vesicles into the cell wall (exosomes produced by EXPO).29
The molecular composition of PBEs
PBEs share structural similarities with mammalian exosomes. The main components of PBEs consist of proteins, lipids, and nucleic acids (Fig. 4), with their specificity determined mainly by their cellular origin.44 These three components are crucial, as each plays a distinct role. Generally, the protein profiles of PBEs influence the uptake mechanism, with lipids being vital for efficient cellular absorption and nucleic acids functioning within recipient cells.45 | ||
| Fig. 4 The schematic representation illustrates the structural composition of exosomes derived from plants. This figure is reproduced from ref. 10 with permission from Drug Delivery and Translational Research, copyright (2024). | ||
Their protein content largely determines the functionality of PBEs. PBEs can act as signal transducers through protein kinases and G proteins, serve as intracellular transporters via annexins, facilitate exosome biogenesis through proteins such as ubiquitin, clathrin, and ALIX, and promote cell adhesion via integrin-like proteins such as lactadherin.46 While both animals and PBEs are rich in proteins, their roles and mechanisms of action differ significantly. In humans, CD63 is recognized as the most common protein biomarker for exosomes and is widely used as a key marker in cancer research.47 In contrast, SYP121 is a plant-specific syntaxin protein that provides resistance against fungal infections. When plant leaves are infected with pathogens, the production of PBEs increases, facilitating cellular uptake. The quantity of digested glycoprotein-derived vesicles (GDVs) is significantly lower than that of undigested GDVs, and the digestion of GDVs is closely linked to their interaction with type II lectin. Consequently, inhibiting the binding of GDVs to type II lectin would reduce GDV internalization in cells. During this process, the protein sites on the surface of GDVs can interact with CD98 and type II lectin. By binding to CD98, GDVs can decrease their affinity for type II lectin, thereby enhancing their capacity for cellular uptake.48
Their lipid composition primarily influences the uptake of exosomes. PBEs are rich in phospholipids, including phosphatidic acids (PA), phosphatidylethanolamines (PE), and phosphatidylcholine (PC). Among these, PA is the most abundant lipid and plays a crucial role in exosome delivery; for instance, PA has been shown to modulate pathogenicity by affecting the interactions between exosomes and hemopexin in periodontal pathogenic bacteria. Additionally, research has indicated that PC-enriched PBEs are preferentially absorbed by Ruminococcus, suggesting that the specific lipid composition is significant in facilitating targeted uptake of exosomes by cells.31 The importance of lipid composition in absorption was further evidenced by the successful uptake of modified nanoparticles by intestinal cells in mice. Imaging studies indicated that the reduced size and higher concentration of PA in the edible-labeled PBEs contributed to the formation of nanoparticles within mouse enterocytes. Consequently, the lipid composition of PBEs can be modified and organized into nanocarriers with distinctive architectures, enhancing their potential for use as drug delivery systems.32,49
PBEs frequently contain a substantial amount of non-coding RNAs that regulate gene expression in recipient cells, thereby influencing processes such as cell proliferation, apoptosis, metabolism, and immune responses.50 Exogenous PBEs have been shown to be absorbed by rat intestinal epithelial cells (IEC6), taking up miR-156a, miR-168a, and miR-166a, which subsequently modulates TNF-α production in mammalian adipocytes.51 However, these findings have yet to be validated in humans.52 This suggests that PBEs could serve as effective drug delivery carriers in gene therapy applications. Eukaryotic argonaute (AGO) proteins can selectively bind to siRNAs and guide the complex to specific gene targets by leveraging base pairing between mRNA and siRNA. Subsequent studies have demonstrated that AGO proteins can specifically associate with small RNAs derived from PBEs in Arabidopsis.53 There are ten different types of AGO proteins, each exhibiting distinct capabilities for binding siRNAs. Notably, PBEs and their associated siRNAs were found to bind exclusively to the AGO1 protein,54 suggesting that the siRNAs influence the protein metabolism of PBEs. Furthermore, siRNAs within PBEs have been shown to enhance the secretion of IL-23 and TNF-α from macrophages infected with Helicobacter pylori, thereby inducing an inflammatory response in target cells. This was achieved by reducing the expression of specific antigenic genes while maintaining cellular homeostasis.55
Isolation and purification methodologies of PBEs
Exosomes can be purified through various methods, which depend on factors such as their size, flotation density, shape, and the presence of specific molecular markers on their surface. Standard techniques for isolating exosomes from cell culture media or homogenates include ultracentrifugation, differential centrifugation, size exclusion chromatography, and the use of commercial kits. Several innovative technologies are also being developed that leverage exosomes’ physicochemical and hydrodynamic properties. The choice of purification method is influenced by the source material from which the exosomes are derived, as well as the intended downstream applications of the final product.34 Some of the conventional and new isolation methods are presented in Fig. 5. Exosomes are gaining prominence in research for their isolation and purification. Nonetheless, it is important to note that there is currently no standardized method for exosome extraction, as various techniques offer unique advantages and drawbacks; the choice of separation method primarily depends on the characteristics of the samples and the intended applications.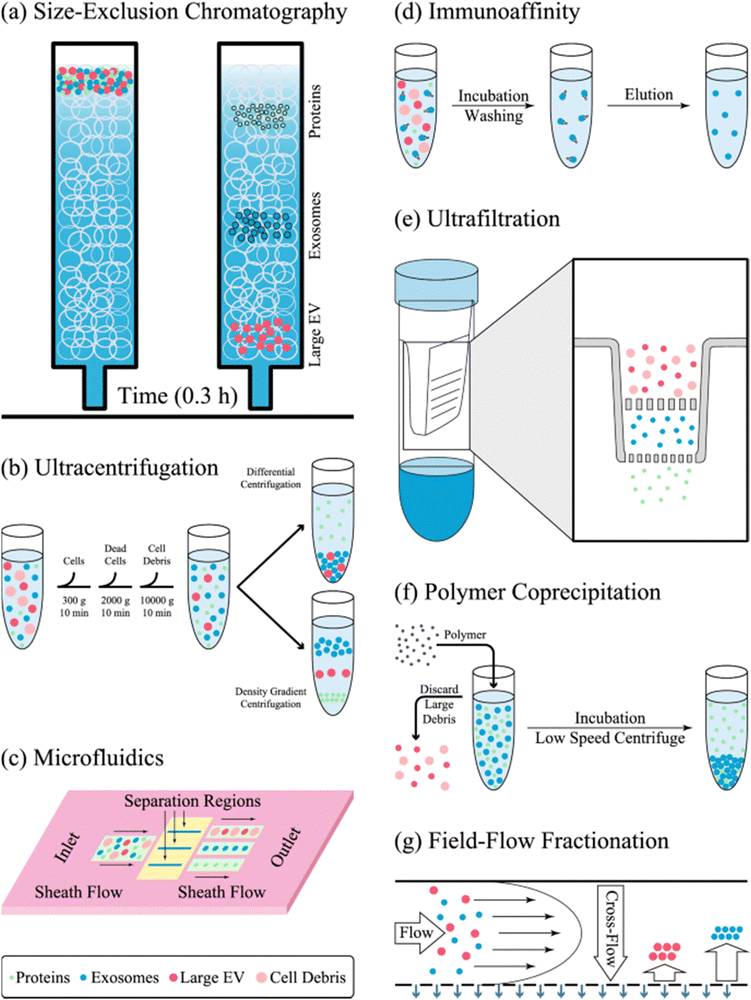 | ||
| Fig. 5 A schematic representation of the exosome separation techniques. (a) Size exclusion chromatography separates particles based on size differences. (b) Ultracentrifugation is frequently employed to extract exosomes according to their size, sedimentation characteristics, or density in sucrose gradients. (c) Microfluidics-based methods utilize both physical properties, such as size and density, and chemical properties, including binding to exosome surface antigens. (d) Immunoaffinity techniques capture exosomes based on their specific interactions with antibodies or magnetic nanoparticles. (e) Ultrafiltration involves passing particles through a filter to separate them according to the pore size of the filter. (f) The polymer co-precipitation method, which relies on steric exclusion, aggregates particles into clumps that can be easily precipitated through low-speed centrifugation. (g) In field flow fractionation, particles separate at different positions on a membrane according to their size. This figure is reproduced from ref. 56 with permission from the Journal of Translational Medicine, copyright (2022). | ||
Traditional methods for PBEs isolation
Ultracentrifugation is the primary method for isolating exosomes from plant sources, relying on the differences in size and density between exosomes and the surrounding materials (Fig. 6). The process involves subjecting plant homogenates or juice to sequential centrifugation at increasing speeds. Pellets from centrifugation below 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g, which do not contain exosomes, are discarded. The remaining pellet is washed, centrifuged, and collected. While effective, this method is labor-intensive and requires expensive equipment. Ultracentrifugation can lead to protein aggregates, which can be minimized using a density gradient. Exosomes float in a 30% sucrose gradient, while most contaminants do not. Increasing centrifugation speed to 135
000 × g, which do not contain exosomes, are discarded. The remaining pellet is washed, centrifuged, and collected. While effective, this method is labor-intensive and requires expensive equipment. Ultracentrifugation can lead to protein aggregates, which can be minimized using a density gradient. Exosomes float in a 30% sucrose gradient, while most contaminants do not. Increasing centrifugation speed to 135![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g with sucrose fractionation yields purer exosome preparations, though it reduces overall yield.34,35,57
000×g with sucrose fractionation yields purer exosome preparations, though it reduces overall yield.34,35,57
 | ||
| Fig. 6 The ultracentrifugation technique is utilized for the isolation of exosomes from plant sources. This method involves subjecting the sample to high-speed centrifugation, which separates the exosomes based on their size and density, allowing for their effective extraction and purification. This figure is reproduced from ref. 35 with permission from Molecular Therapy, copyright (2021). | ||
A schematic illustration of the immunoaffinity-based exosome isolation process is presented in Fig. 7. The immunoaffinity method for isolating exosomes involves labeling specific abundant membrane proteins, such as CD9, CD63, CD81, ALIX, EPCAM, RAB5, and ANNEXIN. This method can be categorized into three types based on antibody substrates: chromatographic stationary phase separation, magnetic bead immune separation, and enzyme-linked immunosorbent separation. The enzyme-linked immunosorbent assay (ELISA) is commonly used for isolating and quantifying exosomes via immunoaffinity chromatography (IAC), utilizing surface biomarkers to create specific binding sites for corresponding antibodies. Advances in this technology include incorporating noncovalent interactions to improve the release of exosomes from magnetic beads, enhancing purity.58
 | ||
| Fig. 7 (A) The foundational concept of the immunoaffinity-based approach for the extraction of exosomes. This figure is reproduced from ref. 58 with permission from Biosensors, copyright (2023). | ||
Immunoaffinity-based exosome isolation typically results in ultrapure preparations, making it ideal for evaluating exosome immunogenicity and therapeutic potential. However, a major limitation of this approach is the need for specific markers, which are often poorly characterized in certain systems, especially in plants. Moreover, this technique is not suitable for large-scale exosome extraction due to the additional processing required to remove the attached antibodies.34,59
Ultrafiltration (UF) isolates particles within a specific size range using a membrane with a defined pore diameter or molecular weight cut-off (MWCO), making it a size-based separation technique. Two main types of UF devices are tandem-configured and sequential ultrafiltration. The choice of filter is crucial for exosome recovery, with research indicating that a 10 kDa cellulose membrane provides optimal efficiency. UF is advantageous in reducing processing time and does not require specialized equipment, though membrane fouling, which decreases efficiency and shortens membrane lifespan, remains a challenge. Tangential flow filtration (TFF), a type of cross-flow filtration, effectively addresses this issue by preventing clogging and improving exosome yield and bioactivity. TFF also ensures greater consistency across batches. However, UF faces limitations due to pore occlusion and the presence of nanoparticles similar in size to exosomes, which can be mitigated by combining UF with other methods.34,60,61 Different types of exosomes UF methods are presented in Fig. 8.
 | ||
| Fig. 8 Schematic representation of exosome isolation via ultrafiltration. (A) Tandem ultrafiltration: in this configuration, biofluids are processed through two membranes. The larger vesicles, such as cell debris, apoptotic bodies, and whole cells, are captured by the 200-nm membrane, whereas vesicles ranging from 20 to 200 nm in diameter are retained by the lower 20-nm filter. (B) Sequential ultrafiltration: initially, larger particles (e.g., cells or cell debris) are eliminated using a 1000-nm filter. The resultant filtrate is then processed through a secondary filter with a 500-kD cutoff to remove free proteins. Ultimately, exosomes with diameters between 50 and 200 nm are collected from the filtrate using a 200-nm filter. (C) Tangential flow filtration. This figure is reproduced from ref. 61 with permission from Analytical and Bioanalytical Chemistry, copyright (2022). | ||
In 1955, Grant H. L. and Colin R. R. introduced size-exclusion chromatography (SEC) to isolate solutes based on molecular weight by passing an aqueous solution through a column of starch and water, as presented in Fig. 9. During this process, molecules with different hydrodynamic radii exhibit distinct behaviors. Smaller molecules enter the stationary phase's pores and are delayed, while larger molecules, unable to enter the pores, bypass them and elute more quickly.62 SEC effectively separates molecules by size, and unlike ultrafiltration, it preserves the structural integrity of exosomes through passive gravity flow. This method retains exosome biological functionality and requires small sample volumes. However, it may result in contamination by lipoproteins and protein aggregates.63
 | ||
| Fig. 9 Principle of size-exclusion chromatography for exosome isolation. Size-exclusion chromatography relies on the passage of a solution through a stationary phase composed of porous resin particles, allowing for the separation of molecules based on size (A). Molecules with hydrodynamic radii smaller than the pore size of the stationary phase enter the pores and travel a longer distance within them, while larger particles, unable to penetrate the pores, move around the resin (B). This differential movement results in varying retention times for particles of different sizes, thereby enabling size-based separation (C). This figure is reproduced from ref. 60. | ||
Exosome separation can be performed cost-effectively using polymer-induced precipitation (Fig. 10), where highly hydrophilic polymers like polyethylene glycol (PEG) create a hydrophobic microenvironment that induces exosome precipitation. PEG, with a molecular weight between 6000 and 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, is ideal for this process due to its non-toxic and inert nature. While this method offers high yield, it can compromise preparation quality, as PEG may also precipitate hydrophilic extracellular impurities.56,60
000, is ideal for this process due to its non-toxic and inert nature. While this method offers high yield, it can compromise preparation quality, as PEG may also precipitate hydrophilic extracellular impurities.56,60
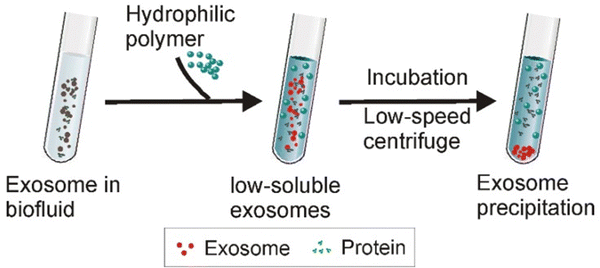 | ||
| Fig. 10 Schematic of the polymer precipitation strategy: when highly hydrophilic polymers are introduced to a solution containing exosomes, these polymers bind to the surrounding water molecules, which decreases the solubility of the exosomes and prompts their precipitation. This allows for the easy collection of exosomes through low-speed centrifugation. This figure is reproduced from ref. 60 with permission from Theranostics, copyright (2020). | ||
Novel methods for PBEs isolation
Affinity capture can be used to isolate and enrich exosomes by employing specific antibodies that target surface proteins and markers. When exosome-containing materials are incubated with solid matrices linked to these antibodies, exosomes are enriched and can be washed away. This method can be enhanced by using technologies like micro-bead-assisted flow cytometry. However, antibodies have drawbacks, including high costs, limited stability, and the need for specialized personnel. To overcome these limitations, aptamers and peptides provide a more stable and cost-effective alternative. Recent innovations include thermophoretic aptasensors with CD63 aptamers on microbeads and CD63 aptamer-based graphene composites for exosome capture. Additionally, methods utilizing phosphatidylserine, highly expressed in exosomes, are being explored, such as using titanium dioxide-modified magnetic beads or Tim4-coated magnetic beads for exosome isolation.64–66Molecularly imprinted polymers (MIPs) provide nanocavities that replicate the shape and charge-based binding characteristics of exosomes (Fig. 11). Purification of exosomes can be achieved through electrostatic binding and conformational recognition using biomimetic materials, which offer high specificity and stability as alternatives to antibodies. Hybrid structures, such as MIP-aptamers, have been used to enhance specificity further. Molecularly imprinted polymers (MIPs) have successfully isolated exosomes from tear samples, enabling purification without needing specific marker identification. However, a limitation of this method is that macromolecules in the sample can block the exosome binding sites. Despite this, MIP development is still in its early stages, and ongoing research may lead to improved strategies for exosome separation.34,67–69
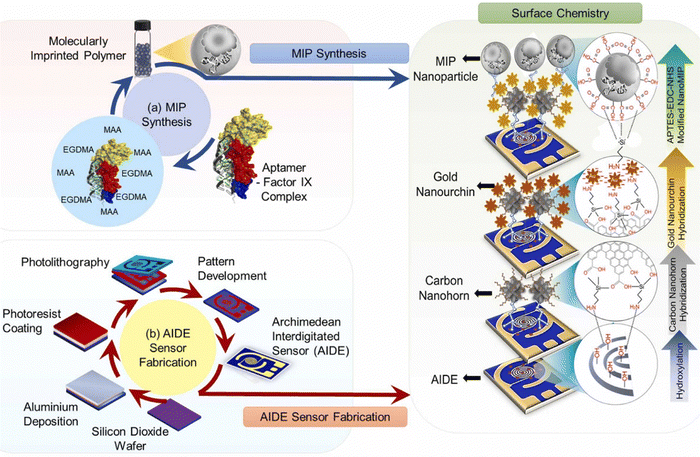 | ||
| Fig. 11 Schematic overview of the entire investigation: this includes the synthesis of molecularly imprinted polymers (MIPs) (a) and the fabrication of the AIDE sensor through photolithography (b). This figure is reproduced from ref. 69 with permission from Elsevier, copyright (2022). | ||
Microfluidics, which involves controlling small volumes of fluids within micrometer-sized channels, has been used to develop exosome purification methods. Immuno-microfluidics-based separation employs antibodies immobilized on chips to target specific molecular markers on exosome surfaces (Fig. 12). Microfluidic chips are modified with antibodies, aptamers, or peptides to improve selectivity in exosome separation. Various microchannel designs have been developed, including nanowires, Y-shaped micropillars, and trapping microchannels. Key advantages of microfluidics for exosome isolation include reduced sample volumes and the elimination of labels, as separation depends on the physical properties of the exosomes. This approach is gaining attention for its potential to make exosome separation more cost-effective and reliable.34,70–73
 | ||
| Fig. 12 Diagram illustrating the operating principle of the microfluidic device: this innovative microfluidic system employs a dual filtration method. A new sample is introduced via the sample input and initially filtered through a 2 μm filter. After this step, the sample enters the bottom chamber and flows through a channel to the 15 nm/30 nm filter for additional filtration. Exosomes are then collected from the collection channel inlet using a pipette. This figure is reproduced from ref. 73 with permission from Sensors, copyright (2023). | ||
Asymmetrical flow field-flow fractionation (AF4) is an analytical technique that separates nanoparticles from larger molecules based on size. This separation occurs due to differences in mobility within a flow field created by liquid moving over a membrane and through a channel, primarily influenced by the intrinsic diffusion coefficients of the particles, as presented in Fig. 13. AF4 has effectively isolated extracellular vesicles from plasma.74 Notably, this method allows for the gentle separation of exosomes without relying on specific markers while still achieving high-size resolution. However, a limitation of this technique is the potential contamination of the preparation by particles with the same hydrodynamic size as exosomes.73,75
 | ||
| Fig. 13 Asymmetric field-flow fractionation (AF4): prior to separation, samples are concentrated on the membrane. Particles diffuse off the membrane based on their size, aggregating in regions where the forces of diffusion and cross flow are balanced. Concurrently, laminar flow directs the particles toward the detector located at the end of the flow channel. This figure is reproduced from ref. 73 with permission from Sensors, copyright (2023). | ||
The liposome fusion technique, which shares structural similarities with exosomes, particularly in their bilayer membranes, allows for flexibility during fusion, as shown in Fig. 14. Recent advancements include antibody-linked lipid patch microarrays developed for isolating extracellular vesicles from cancer samples. This technology optimizes exosome surface properties to reduce immunogenicity, enhance stability, and extend their half-life in circulation.76 However, the hydrophobic nature of lipids complicates their direct loading into exosomes. Genetically modifying exosome lipid membranes is also challenging due to complex lipid production processes and unclear mechanisms of lipid sorting. To overcome these challenges, a novel technique involves creating engineered hybrid exosomes by fusing exosome and liposome membranes through a freeze–thaw method, allowing surface modification with peptides or antibodies.77
 | ||
| Fig. 14 Schematic of the procedure for creating exosome–liposome hybrids. This figure is reproduced from ref. 77 with permission from Acta Pharmacologica Sinica, copyright (2022). | ||
Loading methods of nucleic acids in PBEs
Nucleic acids are characterized by high hydrophilicity and negative charges. However, these particles face significant challenges in cellular uptake due to susceptibility to nucleases and clearance by the liver and kidneys, leading to rapid degradation. The inherent instability and degradability of nucleic acids substantially hinder their effective delivery, making them highly dependent on protective drug delivery systems to ensure their stability and facilitate cellular delivery.78 As a result, the use of exosomes for loading and delivering nucleic acid drugs has gained attention. Compared to lipid nanoparticles, exosomes are less cytotoxic and offer superior cargo expression, making them more efficient carriers that protect nucleic acid drugs from degradation and enhance their stability.79 Exosomes naturally facilitate the delivery of miRNAs and siRNAs, which bind to target mRNAs in recipient cells, leading to mRNA degradation or the inhibition of protein synthesis. This mechanism, known as RNA interference, enables the silencing of specific genes. Consequently, researchers have increasingly focused on developing small RNA-based therapies leveraging this process.80It is worth mentioning that exosomes loaded with therapeutic molecules face several challenges, including competition from endogenous exosomes, internalization and clearance by the mononuclear phagocyte system, and difficulties with precise targeting. To address these issues, specific modifications to the exosome surface are required. These modifications can be chemical or biological (Fig. 15). Chemical modifications typically involve the biological binding of targeted ligands to surface proteins, although this approach may lead to surface protein inactivation or exosome aggregation. On the other hand, biological modifications involve displaying functional ligands on the exosome membrane, but this strategy requires plasmid construction and the overexpression of proteins in donor cells. Despite their limitations, both approaches have successfully improved some therapeutic potential.81
 | ||
| Fig. 15 Chemical and biological modifications of exosome surfaces are crucial strategies for enhancing their therapeutic potential and addressing challenges related to targeting, stability, and cargo delivery. This figure is retrieved from ref. 81 with permission from Nature, copyright (2022). | ||
Currently, drug loading methods are commonly categorized as passive and active loading approaches. Passive loading involves co-incubating plant extracellular lipid nanoparticles with drug molecules at specified temperatures, relying on diffusion and lipophilic interactions between the drug and the vesicle membrane for binding. While this method is relatively simple, it tends to lower encapsulation efficiency. In contrast, active loading techniques, including ultrasound-assisted loading, electroporation, extrusion, and freeze–thaw cycles, have been reported to substantially enhance the loading capacity of PELNs, with increases exceeding 11-fold.82
Several loading methods are available for PBEs. However, due to the previously discussed challenges, only specific loading techniques are suitable for incorporating nucleic acids. These methods include co-incubation, electroporation, sonication, and transfection, as shown in Fig. 16.
 | ||
| Fig. 16 Common techniques for loading nucleic acids into plant-derived extracellular vesicles (EVs) include several approaches designed to efficiently encapsulate therapeutic agents while preserving the integrity and functionality of the EVs. These approaches include: Incubation (A), Transfection (B), Sonication (C), Extrusion (D) and Electroporation (E). This figure is reproduced from ref. 10 with permission from Link Springer, copyright (2024). | ||
Incubation method
The incubation method involves co-incubating exosomes with nucleic acid molecules for a defined period at room temperature, facilitating the loading of these molecules into the exosomes. This approach is simple to implement and preserves the structural integrity of the exosomes. The loading efficiency is determined mainly by the polarity of the small molecule. Lipophilic small molecules with low to medium molecular weight, such as catalase, are more readily incorporated into exosomes through this process.83 The aqueous core and bilayer lipid membrane of exosomes facilitate the loading of both hydrophilic and hydrophobic drugs through co-incubation, enhancing drug encapsulation efficiency.84Electroporation method
Electroporation is a technique that utilized to load nucleic acid molecules into exosomes by applying an external electric field near the exosome membrane, using brief high-voltage pulses to disrupt the membrane. Under optimal conditions, this causes a rapid rupture of the lipid bilayer, inducing a transient increase in membrane permeability that allows nucleic acid molecules to diffuse into the exosomes. After electroporation, exosomes are typically incubated for a period to allow the restoration of membrane integrity.85 This method is increasingly recognized as a safer alternative for loading therapeutic molecules into exosomes. This method minimizes the risks associated with more invasive techniques, offering a more controlled and effective approach to drug encapsulation while maintaining the integrity of the exosomes.81Sonication method
Nucleic acid molecules can be loaded into exosomes using sonication, a process in which an ultrasound probe generates mechanical shear forces to deform the exosome membrane, facilitating the entry of drugs into the exosomes. This method has demonstrated promising outcomes, with several studies reporting high drug loading efficiency and improved antitumor effects. The mechanical disruption caused by sonication enables a more effective encapsulation of drugs, enhancing their therapeutic potential, particularly in cancer treatment.86Extrusion method
Extrusion-based methods are widely utilized to load mainly small-molecule drugs and protein-based drugs into isolated exosomes. In this technique, a combination of the drug and exosomes is pushed through a lipid extruder with membranes with pores ranging from 100 to 400 nm, all while maintaining specific temperature conditions. This procedure disrupts the exosome membrane, promoting effective drug loading. It can also be applied to the hybridization of exosome membranes.84Transfection method
The process of transferring specific plasmids into exosome donor cells for the expression of a desired nucleic acid drug, which is then encapsulated and secreted by exosomes, typically involves the use of transfection reagents. These reagents facilitate the uptake of plasmid DNA by the donor cells, enabling the subsequent production and packaging of the nucleic acid drug into exosomes for delivery.87,88 This method has been shown to offer higher loading efficiency and improved protein stability; however, they are less desirable due to the potential toxicity and side effects associated with transfection reagents, which can interfere with regular cellular gene expression. These adverse effects can limit the applicability of transfection-based approaches, particularly in therapeutic contexts where minimizing cellular disruption is critical.81Genetic materials delivery by PBEs from various origins
As previously discussed, genetic materials and PBEs have demonstrated promising therapeutic effects on cancer cells, including inhibiting cell proliferation, induction of apoptosis, and reducing associated toxicity. Given these benefits, the combinatorial strategy of encapsulating genetic molecules within PBEs represents a promising approach for advancing cancer treatment. Despite its potential, only a limited number of studies have explored the loading of genetic components into PBEs. In the following section, we will review and discuss these key studies, highlighting their contributions to this emerging area of cancer therapy.Ginger based genetic vehicle
Ginger is a complex spice that consists of a natural composition of water, proteins, lipids, fibers, carbohydrates, and volatile essential oils. Chemical analyses of the essential oil, utilizing advanced mass spectrometry techniques, have revealed that the root contains hundreds of distinct compounds. The primary chemical families present in ginger include gingerols, shogaols, zigiberenes, and zingerone. Additionally, some ginger varieties contain phytosterols, vitamins, minerals, and terpene compounds such as bisabolene, farnesene, sesquiphellandrene, and curcumene, albeit in smaller quantities. Consequently, the intricate array of bioactive compounds in ginger, particularly the prominent phenolic groups like gingerols, shogaols, zingiberene, paradol, and zingerone (Fig. 17), underpins its nutraceutical importance.89–92 | ||
| Fig. 17 Presenting the molecular geometry of the bioactive components of ginger. This figure is reproduced from ref. 90 with permission from Seminars in Cancer Biology, copyright (2021). | ||
The use of ginger as a cancer remedy is not a novel concept; it has a long history in traditional medicine worldwide and has demonstrated efficacy in various field studies. In India, both the juice and decoction of the rhizome are administered orally. In Singapore, cooked rhizome is utilized as a preventive measure against cancer. Palestinians prepare an infusion of the rhizome to help prevent breast cancer and treat liver and stomach cancers. While in Ghana, ginger is traditionally applied as a paste, with the crushed rhizome or root either consumed or used topically for treating brain and stomach cancer.91–93 Therefore, recent pharmacological research has investigated the validity of the claims of the efficacy and safety of ginger and its bioactive components possessing anticancer properties.90
As a result, a study aimed to explore ginger as an effective vehicle for the oral delivery of drugs to treat colorectal cancer (CRC). It provided an overview of the structure and function of CRISPR–Cas9 (Fig. 18), along with current challenges in its delivery technology. Furthermore, it presented evidence supporting PBEs as promising nanocarriers for orally administered CRISPR/Cas9-based therapies targeting the lower digestive tract. Consequently, proposing the oral administration of PBEs loaded with CRISPR/Cas9 plasmid DNA targeting specifically long non-coding RNAs (lncRNAs), such as LINC01296, to inhibit the growth of colon cancer cells, specifically the SW480 and SW620 cell lines.94 LINC01296 was specifically chosen in this study since cell proliferation is influenced by long non-coding RNAs (lncRNAs), with dysregulation of these molecules contributing to the proliferation of colon cancer cells. For instance, the overexpression of LINC01082 is associated with the downregulation of p21-activated kinases 2 (PAK2) in malignant colon tissues, inhibiting SW480 colon adenocarcinoma cell growth. Additionally, reducing the expression of LINC01296 in SW480 and SW620 colon cancer cells has decreased cell proliferation through gene expression suppression.95 The successful purification of exosomes derived from ginger exhibited a negative transmembrane potential on their surface, ranging from −12 mV to −17.1 mV and a size of less than 300 in neutral pH. Moreover, CRISPR-based gene therapies can be introduced into the gastrointestinal (GI) and large intestine through the use of PBEs, as studies have proved its efficient stability and direct delivery (Fig. 19). As well, these NPs exhibited lower immunogenicity, effective cellular uptake, reduced toxicity, and favorable biocompatibility.94
 | ||
| Fig. 18 The illustration depicts the structural characteristics of the CRISPR–Cas9 protein, highlighting its complex with the single-guide RNA (sgRNA) that is attached to the target gene. Additionally, the figure illustrates the mechanisms of non-homologous end joining (NHEJ) repair and homologous directed repair (HDR). This figure is retrieved from ref. 94 with permission from Genes, copyright (2023). | ||
 | ||
| Fig. 19 The diagram demonstrates the application of plasmid-extracellular vesicles (PEVs) loaded with CRISPR/Cas9 for the treatment of colon cancer cells. It outlines the oral administration of these PEVs to target long non-coding RNAs (lncRNAs) associated with colon cancer. (A) The illustration shows the breakdown of CRISPR/Cas9-loaded PEVs within the colon. (B) It depicts the uptake of these PEVs by intestinal epithelial cells through the process of endocytosis. (C) The figure concludes with the elimination of malignant colonocytes following the treatment of lncRNAs in colon cancer cells with Cas9. This figure is reproduced from ref. 94 with permission from Frontiers in Oncology, copyright (2023). | ||
Another study aimed to isolate ginger-derived exosome-like nanoparticles (GELNs) using a novel approach that combines differential centrifugation with an extraction kit. Subsequently, researchers constructed and compared short RNA libraries from both ginger and GELNs. Through target gene prediction and functional analysis, the most highly expressed miRNAs in GELNs were identified, which were bdi-miR5179, csi-miR396e-5p, and ptc-miR396g-5p which were afterwards tested for their efficacy on colorectal adenocarcinoma (Caco-2) cell lines. The TEM images of the GELNs displayed spherical particles with characteristic cup-shaped membrane structures with appropriate particle sizes of 156 ± 36 nm, a negative surface charge of −26.6 ± 5 mV, and a protein concentration of 2391.43 ± 750 ng μL−1. Also, GELNs have been shown to be effectively internalized in Caco-2 cell lines and resided near the nucleus through caveolin-mediated endocytosis mechanism LPS treatment resulted in and diminished the expression of NF-κB and other inflammatory cytokines, proposing successive tumor regression.96
Broccoli based genetic vehicle
The role of cruciferous vegetables in cancer prevention has garnered considerable attention in recent years. In 1997, Fahey et al. were among the first to document the anticarcinogenic properties of broccoli sprouts (BSp).97 A substantial body of research indicates that the consumption of broccoli and other cruciferous vegetables (which belong to the Brassicaceae or Cruciferae family) may lower the overall risk of various cancers, particularly breast, prostate, colorectal, lung, and bladder cancer. Due to its health benefits, broccoli (Brassica oleracea var. italica) is a prominent green vegetable frequently incorporated into the Mediterranean diet. One of its most recognized bioactive effects is its potential anticancer properties. Broccoli is rich in various active biochemical compounds, including carotenoids, vitamin C, and glucosinolates as presented in Fig. 20. Glucosinolates are the most essential compounds in broccoli due to their anticancer properties. Notably, approximately 200 different glucosinolates have been identified. Among these, broccoli is particularly rich in the sulfur-containing compound sulforaphane (1-isothiocyanato-4-methylsulfinylbutane), which is part of the isothiocyanate family of phytochemicals. The –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S functional group is considered the most significant active component. On the other hand, sulforaphane has been thoroughly investigated for its chemopreventive properties by inhibiting histone deacetylases (HDAC), which play a critical role in tumor gene expression.98–100
S functional group is considered the most significant active component. On the other hand, sulforaphane has been thoroughly investigated for its chemopreventive properties by inhibiting histone deacetylases (HDAC), which play a critical role in tumor gene expression.98–100
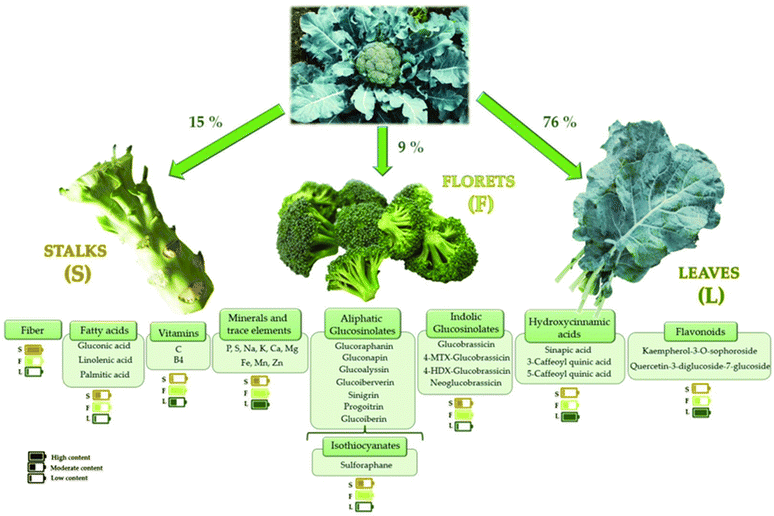 | ||
| Fig. 20 Introducing the bioactive compounds of broccoli. This figure is reproduced from ref. 100 with permission from MDPI, copyright (2023). | ||
In light of the above, miRNAs from broccoli may play a role in the health benefits associated with consuming cruciferous vegetables. Consequently, broccoli-derived EVs were evaluated in vitro as effective carriers for miRNA transport and delivery, demonstrating the potential to produce biological effects on host cells. The ultracentrifugation cycle (1UC) yielded EVs with particle sizes ranging from 75 to 400 nm, averaging 146.7 ± 7.2 nm. The nanoparticle tracking analysis (NTA) of the pooled F8–F10 samples indicated that the EVs varied in size from 35 to 300 nm, with a mean size of 174.3 ± 5.5 nm. Electron microscopy confirmed the presence of spherical EVs of various sizes, each enclosed by a plasma membrane. Moreover, the most abundant miRNAs in both the tissues and exosomes were ath-miR159a, ath-miR166b-3p, ath-miR319a and ath-miR403-3p (Fig. 21), hence were selected for further testing on colorectal adenocarcinoma (Caco-2) cell lines. In addition, results showed that the transportation of miRNAs through broccoli-derived exosomes was more successful and efficient due to the safeguarding of RNA cargo against degradation by RNase A, which resulted in higher expression of miRNAs in Caco-2 cells. Exogenous miRNA-loaded broccoli exosomes demonstrated a detrimental effect on Caco-2 cells, decreasing their viability by approximately 30% when using a volume of 5% EVs, suggesting their ability to inhibit cancer cell proliferation.101
 | ||
| Fig. 21 Secondary structure predictions for plant miRNAs were conducted, selecting the structures with the lowest estimated free energy of formation (ΔG) as the most probable configurations. This figure was retrieved from ref. 101 with permission from ScienceDirect, copyright (2022). | ||
A recent study, investigated the isolation and characterization of broccoli-derived extracellular vesicles (BDEVs) from selenium-enriched broccoli (Se-BDEVs) and conventional broccoli (cBDEVs), assessing their miRNA expression through miRNA sequencing (miRNA-seq). There was an effective purification of broccoli exosomes with a primary particle size peak. Se-BDEVs were found to be 127.1 nm, whereas cBDEVs were 117.4 nm. Additionally, transmission electron microscopy (TEM) analysis of the EV pellets revealed spherical, membrane-bound structures approximately 100 nm in diameter. Bioinformatic analyses, along with target gene (TG) prediction and functional assessments, indicated that BDEVs might play a significant role in pancreatic cancer; therefore, further testing was performed on PANC-1 cell lines. Notably, Se-BDEVs exhibited greater anti-pancreatic adenocarcinoma (PAAD) activity than cBDEVs, attributed to the increased expression of miR167a. Additionally, the findings of this study indicated that both Se-BDEVs and cBDEVs possess PAAD activity, with Se-BDEVs demonstrating greater efficacy. The data suggest that the elevated concentration of miR167a in Se-BDEVs contributes to promoting apoptosis, thereby inhibiting the growth of pancreatic cancer cells by targeting insulin receptor substrate – 1 (IRS1) as presented in Fig. 22.102
 | ||
| Fig. 22 Graphical representation of a study which tested the isolation of broccoli derived exosomes encapsulating miR167a inducing apoptosis in pancreatic cancer cells lines through insulin receptor substrate – 1 (IRS1). (1) and (2) Illustrating the isolation and purification of exosomes derived from broccoli annotated as Se-BDEVs that could be internalized in human cells. (3) and (4) Se-BDEVs encapsulating miR167a induced apoptosis in PANC-1 cell lines through IRS1. This figure is retrieved from ref. 102 with permission from International Journal of Nanomedicine, copyright (2023). | ||
Apples based genetic vehicle
Apples (Malus domestica), the most widely consumed fruit, contain a variety of bioactive compounds, including vitamins, pectins, dietary fibers, triterpenic acids, oligosaccharides, and phenolic compounds such as dihydrochalcones, flavonols, hydroxybenzoic acids, and anthocyanidins. They are particularly noted for their high antioxidant capacity, attributed to a substantial concentration of these phenolic compounds, exceeding 20 mmol TE per kg. Given their rich content of dietary fiber and bioactive compounds, apples and apple by-products are increasingly utilized to enhance food products. These items, which are rich in natural antioxidants, present a viable alternative to synthetic additives.103Accordingly, a recent study aimed to investigate whether apple-derived exosomes could establish active and direct communication with mammalian cells. In this context, researchers utilized ADNVs extracted from the golden delicious variety of Malus domestica encapsulating miR-146 with the goal of inducing activation of THP-1 (monocyte cell extracted from acute monocytic leukemia patient) cells into type I macrophages to assess their safety and biological activity. The physical characteristics of ADNVs were consistent, exhibiting a circular shape and varying in size from 80 to 250 nm with a mean radius of 157 nm. Moreover, the primary function of miR-146a and miR-146b is to inhibit the transcription of IRAK1 (interleukin 1 receptor-associated kinase 1) and TRAF6 (tumor necrosis factor receptor-associated factor 6) mRNA during the early stages of inflammation, thereby impacting the canonical NF-κB (nuclear factor kappa-light-chain-enhancer) pathway, which is also activated by TNFα. Differential analysis concluded that following exposure to ADNVs, activated macrophages exhibited inhibition of the NF-κB pathway, as evidenced by the overexpression of miR-146a and the downregulation of IL-1β. Furthermore, treatment with ADNVs in fibroblast and THP-1 cells was shown to reduce levels of tumor necrosis factor-alpha (TNFα), leading to a subsequent decrease in interleukin 8 (IL-8) and interleukin 1β (IL-1β), which collectively contribute to diminished inflammation and cell proliferation.104
Moreover, given that plant mRNAs are frequently found in food-derived nanoparticles and typically influence gene expression by promoting the degradation of the corresponding mRNA, a study specifically investigated the potential role of mRNAs derived from apples. This study utilized colorectal adenocarcinoma (Caco-2) cells as a model of gastrointestinal epithelial cells to examine the effects of apple-derived nanoparticles (ADNVs) on the expression and function of intestinal transporters. The resultant NPs had a size ranging from 100–200 nm with a mean diameter of approximately 170 nm and good cell internalization properties within ∼6 hours. The effect of ADNVs on mRNA transporters expression concluded that the expression of breast cancer resistance protein (BCRP) mRNA increased and there was a significant reduction in the expression of levels of sodium bile acid transporter (ASBT), organic anion transporting polypeptide 2B1 (OATP2B1), and organic cation transporter 2 (OCTN2) mRNAs. Overall, the results of this study indicate that ADNVs can modulate the expression of intestinal transporter mRNAs specifically, decreased OATP2B1 expression in Caco-2 cells, affecting transport activity, protein levels, and mRNA expression. Gene truncation tests and luciferase assays suggested that this activity involves large molecules, which are likely miRNAs. This finding challenges the prevailing notion that food-derived large molecules do not interact directly with intestinal tissues due to their instability and limited membrane permeability, distinguishing it from the known effects of small molecules.105
Artemisia annua based genetic vehicle
Artemisia annua, commonly referred to as “annual absinthe”, is an annual herbaceous plant, which is reflected in its name “annua.” This plant is cultivated across various regions, including Australia, India, Central and Eastern Europe, Asia, temperate areas of America, Africa, and tropical climates. In the temperate regions of Asia, particularly in China and Korea, it is widely employed as a dietary spice, herbal tea, and medicinal herb. Due to its abundant phytochemical composition, Artemisia annua possesses a wide range of additional biological properties, including anti-asthmatic activities, antioxidant, hepatoprotective, anti-inflammatory, antifungal, and antitumor.106Subsequently, a group of researchers isolated and purified exosomes from Artemisia annua. These newly identified vesicles, termed ADNVs (artemisia-derived nanovesicles), reprogrammed tumor-associated macrophages (TAMs) from a pro-tumor phenotype to a pro-inflammatory one, exhibiting significant anti-tumor effects. Through the cGAS-STING pathway illustrated in Fig. 23, artemisia-derived mitochondrial DNA (mtDNA) was taken up by TAMs via the vesicles, leading to an enhanced cytotoxic T cell response that contributed to tumor regression. Additionally, our findings indicate that ADNVs improved the efficacy of αPD-L1-mediated immunotherapy by activating the STING pathway. The characterization of ADNVs revealed that the vesicles exhibited a spherical morphology and a bilayer membrane structure. The measured average hydrodynamic diameter was 106.8 nm, with a concentration of approximately 1.25 × 1011 vesicles per mL in solution. The ADNVs displayed a negative zeta potential of −22.5 mV. Furthermore, the analysis composition of the pure ADNVs showed significant amounts of nucleic acids and proteins encapsulated within the vesicles. In order to test the safety and efficacy of these NPs, Lewis lung carcinoma (LLC) cells were inoculated into mice. The results demonstrated that ADNVs therapy effectively inhibited tumor progression, reducing malignant characteristics such as cellular proliferation, invasion, and resistance to apoptosis in tumors while not exhibiting noticeable toxic effects in vivo and in vitro.107
 | ||
| Fig. 23 Presenting the mechanism of action of ADNV which activates cGAS-STING pathway that promotes tumor regression. This figure is retrieved from ref. 107 with permission from Journal of Nanobiotechnology, copyright (2023). | ||
Brucea javanica based genetic vehicle
Brucea javanica (BJ) is a prominent Chinese medicinal plant from the Simaroubaceae family, characterized as an evergreen shrub that is widely distributed in Southeast Asia and northern Australia. The fruit of BJ, commonly referred to as Yadanzi, is utilized for medicinal purposes. In Southeast Asian countries such as Indonesia and Cambodia, BJ is commonly employed as an antimalarial, antitrypanosomal, and hypoglycemic agent. In Australia, it is utilized for its analgesic properties, while in China, it is used to treat diarrhea, malaria, and skin conditions. However, recent research has highlighted the extensive pharmacological effects of BJ metabolites (Fig. 24), including anti-cancer, anti-inflammatory, and antiviral activities. Much of the focus on BJ metabolites has centered on quassinoids, which have been demonstrated to exhibit antitumor effects, while alkaloids and acidic metabolites showed toxic effects, and terpenoids were found to possess anti-inflammatory properties.108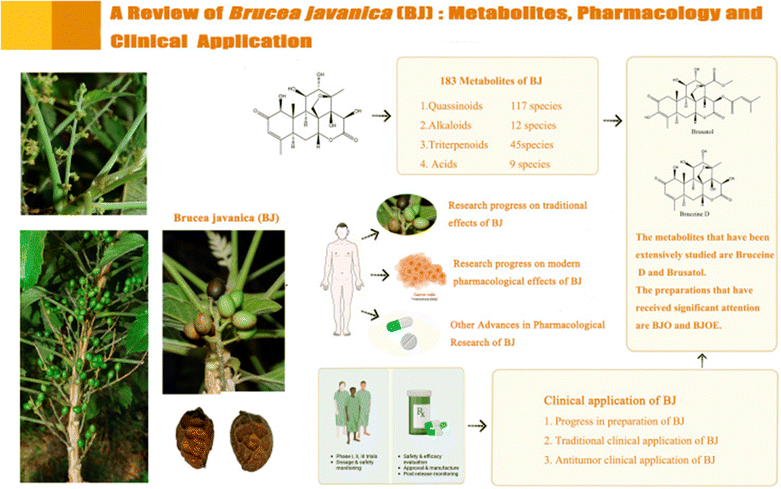 | ||
| Fig. 24 Illustrating the metabolites, pharmacology and clinical applications of BJ. This figure is reproduced from ref. 108 with permission from Frontiers in Pharmacology, copyright (2024). | ||
Hence, a study was conducted for the first time to isolate and characterize exosome-like nanoparticles (ELNs) derived from the fruits of BJ (BF-Exos). The study also evaluated the potential of BF-Exos as a nanoplatform for delivering functional miRNAs from BJ, as well as their impact on molecular interference in 4T1 breast cancer cells and the surrounding tumor microenvironment. The isolated exosomes exhibited a cup-like morphology and were distributed uniformly with an average particle size of 104.6 ± 29.4 nm, and a zeta potential of −9.2 ± 0.1 mV. In addition, cellular uptake findings indicate that BF-Exos possesses the capability to deliver 10 functional miRNA cargos (miR-9993a, miR-6240, miR-22, let-7a, miR-125b, miR-100, let-7f, miR-92a, miR-99b, and miR-29a), positioning them as promising biotherapeutics for targeting 4T1 breast cancer cell lines. The results indicated that BF-Exo treatment led to the down-regulation of the AMPK pathway, suggesting that AMPK signaling may serve as the primary pathway through which BF-Exos exerts their anti-tumor effects in 4T1 cells. In addition, the expression levels of critical phosphorylated proteins (p-PI3K, p-Akt, and p-mTOR) were significantly reduced, indicating inhibition of the PAM pathway. Overall, BF-Exos has been shown to inhibit angiogenesis, tumor growth, invasion, metastasis and modulation of the tumor microenvironment as presented in Fig. 25.109
 | ||
| Fig. 25 A schematic illustration of the molecular mechanism by which BF-Exos function as biotherapeutics in tumor progression and angiogenesis in the context of 4T1 breast cancer cell lines. This figure is reproduced from ref. 109 with permission from ScienceDirect, copyright (2024). | ||
Acerola based genetic vehicle
Malpighia fruits (Malpighiae emarginata DC/Malpighia glabra L.), commonly known as acerola, Guarani cherry, Barbados cherry, or wild pancake myrtle, are produced by a shrub native to the Yucatán Peninsula. Acerola exhibits several intriguing and beneficial health-promoting properties. It is increasingly recognized in the market for exotic vegetable raw materials, particularly for its high vitamin C (ascorbic acid) content, as well as polyphenolic compounds (such as anthocyanins) and carotenoids. Previous studies have highlighted acerola fruit (including pomace, seeds, and juice) and its leaves as valuable sources of carotenoids (such as lutein and β-carotene), polyphenolic compounds (including hydroxycinnamic acids and flavonoids), pheophytin, and chlorophyll derivatives, which may serve as ingredients for potential natural nutraceuticals.110 Acerola is an excellent source of ascorbic acid, and its consumption may significantly enhance immune function by increasing both the quantity and activity of immune cells, such as lymphocytes.111 Additionally, acerola pomace has the highest concentration of naringenin, which is associated with anti-atherosclerotic effects and estrogen-like activity.112 Moreover, acerola fruit and leaves are believed to possess anticancer properties due to the presence of tetranorditerpenes known as acerolanin A, B, and C (tetranorditerpenoids featuring a rare 2H-benz[e]inden-2-one substructure), which have been shown to reduce the viability of breast cancer cells (MCF-7 cell line) and colon cancer cells (HCT-116 cell line). Consequently, acerola leaf extracts may play a significant role in nutritional prophylaxis for individuals at high risk of developing breast cancer, including those with known BRCA1 and BRCA2 gene mutations or a family history.113Thus, due to the previously reported benefits of acerola, a study assessed the feasibility of developing a drug delivery system (DDS) for the oral administration of nucleic acids specifically, has-miR-340, hsa-miR-146a, utilizing acerola-derived exosome-like nanoparticles (AELNs). The purification of acerola was done using ultracentrifugation, exoEasy midi kit, and ExoQuick methods. The ultracentrifugation method resulted in non-uniform particles with an average diameter of 352 ± 180 nm. In contrast, exoEasy midi kit particles were uniform with an average size of 245 ± 132 nm, and ExoQuick particles were not uniform with a diameter of 340 ± 172 nm. Luckily, the concentration of has-miR-340 present in the recovered AELNs reached a maximum of 3.9 × 1022 particles per mL. These nucleic acids presented with degradation inhibition from RNase when delivered via AELNs, showing their remarkable stability. Furthermore, AELNs were tested on human cervical squamous cell carcinoma cell line (SiHa cells), showing increased expression level of matrix metalloproteinase-2 (MMP2), upregulation of hsa-miR-146a and downregulation of nuclear factor kB (NF-kB) suggesting the induction of apoptosis, suppression of cell proliferation and limiting tumor neovascularization.114
Tangerine based genetic vehicle
Flavonoids are a class of natural compounds characterized by their polyphenolic structures. These phytochemicals are commonly present in a variety of sources, including fruits, vegetables, tea, cocoa, and certain beverages. To date, nearly 9000 distinct flavonoids have been identified across diverse plant species. Flavonoids are categorized into six main types: flavones, flavonols, flavanones, isoflavonoids, anthocyanins, and chalcones. They possess a fifteen-carbon skeleton composed of two benzene rings (designated A and B) connected by a heterocyclic ring (C). Flavonoids can be found in forms such as glycosides, aglycones, and methylated derivatives.115 Various flavonoids demonstrate anti-cancer activity by targeting multiple molecular pathways. Among them, tangeretin (Fig. 26), a polymethoxylated flavone abundant in citrus fruits, has been extensively studied. Research indicates that tangeretin induces cytotoxicity in a range of cancer cell lines, arrests the cell cycle, triggers apoptosis, inhibits angiogenesis, and prevents metastasis. Additionally, it acts as a free radical scavenger, neutralizing DPPH, hydroxyl, hydrogen peroxide, and superoxide anion radicals by modulating various antioxidant enzymes. Co-treatment with tangeretin has also been shown to enhance drug sensitivity in cancers of the colon, urinary bladder, lymphoblastoid leukemia, and ovaries. Furthermore, it increases radiation sensitivity, inhibits metastasis, and influences cellular metabolic energy in animal models.116 | ||
| Fig. 26 Presenting the structure of tangeretin. This figure is reproduced from ref. 99 with permission from ScienceDirect, copyright (2020). | ||
A study tested the extraction and characterization of nanovesicles from tangerine juice (TNVs) and developed a method for loading exogenous siRNAs targeting DDHD1gene, which is associated with increased proliferation rates in colon cancer cells due to its overexpression. TNVs were isolated from the fruit juice using differential centrifugation in which TEM results showed that the vesicles exhibited a characteristic cup-like or nearly circular shape, with diameters ranging from 40 to 150 nm, a negative z-potential of −5.94 mV and a polydispersity index of 0.26. In addition, results demonstrate that DDHD1-siRNA were loaded successfully into TNVs via electroporation with Cy3-siRNA loading efficiency of 13%. Also, TNV encapsulating siRNA concluded the successful internalization of siRNA in SW480 cells (human colorectal cancer cells) and the downregulation of DDHD1 gene, which reduced cell viability by up to 23%.117
Four food-derived exosomes based genetic vehicle
Another study employed size-exclusion chromatography (SEC) and ultracentrifugation to isolate four different food-derived EVs (broccoli, pomegranates, apples, and oranges), which were subsequently analyzed for their potential as carriers of enhanced synthetic miRNA delivery. To investigate the biological effects of miRNA-loaded food-derived EVs, they performed transcriptomic analysis on a human intestinal cell line (Caco-2). Additionally, they examined the capacity of food-derived EVs to transport dietary polyphenols and other secondary metabolites, including glucosinolates from broccoli, in an endogenous context. The results of this research showed that the particle size distribution of the EVs varied between 50 and 500 nm, with mean sizes of 213 ± 15 nm, 268 ± 14 nm, 221 ± 7.2 nm, and 191 ± 9.9 nm for broccoli, pomegranate, apple and orange respectively. In addition, the zeta potential of the exosomes was measured, with values ranging from −21.23 ± 1.45 mV for broccoli, −26.06 ± 3.39 mV for pomegranate, −20.31 ± 0.60 mV for orange, and −27.11 ± 1.01 mV for apple. Regarding the encapsulation of genetic particles, four miRNA families—miR159, miR162, miR166, and miR396—were found to be common to both total tissue and isolated exosomes across all four foods examined. Subsequently, four specific miRNAs (ath-miR159a, ath-miR162a-3p, ath-miR166b-3p, and ath-miR396-5p), representing these common miRNA families, were selected for further analysis based on their abundance. The fruit exosomes showed that the RNA cargos were shielded from degradation by RNase A. Also, broccoli-derived exosomes reduced cell viability by 50%, while pomegranate and apple EVs decreased survival to approximately 60% when tested on Caco-2 cells. In contrast, HepG2 cell viability was unaffected by apple-derived EVs, although pomegranate EVs led to a reduction of around 25% in cell viability. Broccoli-derived EVs induced approximately 50% apoptosis in Caco-2 cells, primarily observed at the sub-G1 stage. In addition, all PBEs exerted gene modulation.118The reviewed research papers discussing the delivery of genetic molecules as cargo in PBEs have been summarized in Table 3.
| PBE | Encapsulated genetic material | Physicochemical properties | Target cancer cell | Advantage mechanism | Ref. | |
|---|---|---|---|---|---|---|
| Size (nm) | Zeta potential (mV) | |||||
| Ginger | LINC01296 | <300 | −12 mV to −17.1 | Colorectal cancer | Efficient GIT stability, direct delivery, lower immunogenicity, effective cellular uptake, reduced toxicity, and favorable biocompatibility | 94 |
| Ginger | bdi-miR5179, csi-miR396e-5p, and ptc-miR396g-5p | 156 ± 36 | −26.6 ± 5 | Colorectal adenocarcinoma (Caco-2) | Diminished expression of NF-κB and other inflammatory cytokines hence, successive tumor regression | 96 |
| Broccoli | ath-miR159a, ath-miR166b-3p, ath-miR319a and ath-miR403-3p | 146.7 ± 7.2 nm | — | Colorectal adenocarcinoma (Caco-2 cells) | Avoided RNase degradation, higher expression of miRNAs, decreasing the viability and proliferation of cancer cells | 101 |
| Broccoli | miR167a | – Se-BDEVs: 127.1 – cBDEVs: 117.4 | — | Pancreatic adenocarcinoma (PANC-1) | Greater efficacy, promotion of apoptosis | 102 |
| Apple | mRNA transporters | ∼170 | — | Colorectal adenocarcinoma (Caco-2) | Inhibition of NF-κB pathway, reduce levels of TNFα, decrease in IL-8 and IL-1β, diminished inflammation and cell proliferation | 105 |
| Apple | miR-146 | 80–250 | — | Fibroblast and THP-1 cells | Decreased OATP2B1 affecting transport activity, protein levels, and mRNA expression | 104 |
| Artemisia annua | mtDNA | 106.8 | −22.5 | Lewis lung carcinoma (LLC) | Reduced cellular proliferation, invasion, and resistance to apoptosis, no toxic effects | 107 |
| Brucea javanica | miR-9993a, miR-6240, miR-22, let-7a, miR-125b, miR-100, let-7f, miR-92a, miR-99b, and miR-29a | 104.6 ± 29.4 | −9.2 ± 0.1 | Breast cancer | Inhibit angiogenesis, tumor growth, invasion, metastasis and modulation of the tumor microenvironment | 109 |
| Acerola | has-miR-340, hsa-miR-146a | – Ultracentrifugation: 352 ± 180 – exoEasy midi kit: 245 ± 132 – ExoQuick: 340 ± 172 nm | — | SiHa cells | Increased expression of MMP2, hsa-miR-146a and downregulation of NF-kB, induction of apoptosis, suppression of cell proliferation and limited tumor neovascularization | 114 |
| Tangerine | siRNA | 40–150 | −5.94 | SW480 cells (human colorectal cancer cells) | Successful internalization of siRNA, downregulation of DDHD1 gene, reduction of cell viability | 117 |
| Broccoli, pomegranates, apples, and oranges | ath-miR159a, ath-miR162a-3p, ath-miR166b-3p, and ath-miR396-5p | – Broccoli: 213 ± 15 – pomegranate: 268 ± 14 – apple: 221 ± 7.2 – orange: 191 ± 9.9 | Broccoli: −21.23 ± 1.45, pomegranate: −26.06 ± 3.39, orange −20.31 ± 0.60, apple −27.11 ± 1.01 | Colorectal adenocarcinoma (Caco-2), and hepatocellular carcinoma (HepG2) | Protection from RNAse degradation, enhanced uptake by cancer cells, induced apoptosis, and gene modulation | 118 |
Clinical trials
Exosomes used in clinical trials can be classified into two categories: derived from plants and human specimens. To date, clinical trials involving exosomes from human specimens have yielded complete results, whereas plant-derived exosomes are still in the early stages, with patient recruitment yet to begin. Due to their vesicular structure, exosomes are also being explored as drug carriers in clinical trials. Three plant-derived exosome sources (grape, ginger, and aloe) have been registered in clinical trials by the University of Louisville, but they are not currently recruiting. Grape-derived exosomes are being studied to treat radiation- and chemotherapy-induced oral mucositis, with preclinical research suggesting the benefits of tissue regeneration. Ginger- and aloe-derived exosomes are being tested for polycystic ovary syndrome, targeting insulin resistance and inflammation. Additionally, plant-derived exosomes are being explored as drug carriers, such as for curcumin in intestinal diseases. While plant-derived exosomes are animal-free and linked to traditional herbal medicine, they face challenges due to slow life cycles and limited production and characterization data. In contrast to mammalian exosomes, which are harvested from culture media, plant exosomes are extracted from apoplastic vesicles in leaves, roots, and fruit juices. Despite limited information on markers, existing techniques for animal exosome characterization can be applied to plant-derived exosomes, with GMP production improvements needed.119Conclusions and future perspectives
EVs hold great potential as drug delivery systems but face significant challenges in clinical translation due to size variability, heterogeneity, and batch-to-batch differences, which pose higher risks compared to synthetic nanocarriers. EVs can be categorized into three types: natural, hybrid (drug-modified), and EV-inspired liposomes, and are classified as biological medicines. These biologically complex therapeutics are susceptible to production variability influenced by cell culture conditions. Unlike cell therapies, EVs do not adapt after harvesting, making quality control more difficult. A “quality-by-design” approach has been suggested, but EVs' size and complexity complicate product characterization and quality assurance. Despite these challenges, EV-based systems offer significant advantages over traditional drugs, particularly in their biodistribution properties.120The bioengineering of EVs adds extra steps, including selecting robust donor cells and genetic modifications, complicating production and purification, especially when dealing with unwanted substances. Traditional isolation methods such as ultracentrifugation and tangential flow filtration (TFF) face issues with purity and biological activity, and bioengineered EVs require more stringent quality control. Shelf life can be improved through cryopreservation or lyophilization, though stability concerns persist. Characterization of EVs involves techniques like flow cytometry and mass spectrometry, but bioengineered EVs require stricter monitoring to ensure consistency and potency. The regulatory landscape for EVs is complex, with bioengineered versions falling into different categories depending on modifications, underscoring the need for more precise guidelines to ensure safety and address regulatory challenges.121
Overall, PBEs have emerged as promising candidates for future pharmacological applications. Their unique properties position them as a potential answer to many of the challenges currently facing drug delivery systems. However, before definitive conclusions regarding the manufacturing challenges, therapeutic efficacy and safety of PBEs can be drawn. This review highlights the substantial potential of PBEs to enhance the stability and targeted delivery of a wide variety of extracellular molecules, with particularly promising implications for the development of genetic-based therapies and drug delivery systems more broadly. Despite these promising prospects, only a limited number of studies have investigated the encapsulation of genetic materials within PBEs specifically for targeting cancerous cell lines. Therefore, we advocate for further research in this area, as it holds significant promise for advancing cancer therapies and optimizing drug delivery mechanisms.
Data availability
This manuscript does not involve any experimental work.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Dr. Sherif Ashraf Fahmy acknowledges the financial support and sponsorship received from the Alexander von Humboldt Foundation, Germany. Open Access funding was provided by the Open Access Publishing Fund of Philipps-Universität Marburg to Prof. Udo Bakowsky.References
- M. P. Fawzy, H. A. F. M. Hassan, N. K. Sedky, M. S. Nafie, R. A. Youness and S. A. Fahmy, Revolutionizing cancer therapy: nanoformulation of miRNA-34 – enhancing delivery and efficacy for various cancer immunotherapies: a review, Nanoscale Adv., 2024, 6, 5220–5257, 10.1039/D4NA00488D.
- M. Y. Ali, I. Tariq, M. F. Sohail, M. U. Amin, S. Ali, S. R. Pinnapireddy, A. Ali, J. Schäfer and U. Bakowsky, Selective anti-ErbB3 aptamer modified sorafenib microparticles: in vitro and in vivo toxicity assessment, Eur. J. Pharm. Biopharm., 2019, 145, 42–53 CrossRef PubMed.
- N. K. Sedky, N. M. Abdel-Kader, M. Y. Issa, M. M. M. Abdelhady, S. N. Shamma, U. Bakowsky and S. A. Fahmy, Co-Delivery of Ylang Ylang Oil of Cananga odorata and Oxaliplatin Using Intelligent pH-Sensitive Lipid-Based Nanovesicles for the Effective Treatment of Triple-Negative Breast Cancer, Int. J. Mol. Sci., 2023, 24, 8392, DOI:10.3390/ijms24098392.
- V. K. Chaturvedi, A. Singh, V. K. Singh and M. P. Singh, Cancer Nanotechnology: A New Revolution for Cancer Diagnosis and Therapy, Curr. Drug Metab., 2018, 20, 416–429, DOI:10.2174/1389200219666180918111528.
- S. A. Fahmy, N. K. Sedky, A. Ramzy, M. M. M. Abdelhady, O. A. A. Alabrahim and S. N. Shamma, et al., Green extraction of essential oils from Pistacia lentiscus resins: Encapsulation into Niosomes showed improved preferential cytotoxic and apoptotic effects against breast and ovarian cancer cells, J. Drug Delivery Sci. Technol., 2023, 87, 104820, DOI:10.1016/J.JDDST.2023.104820.
- K. Stefańska, M. Józkowiak, A. Angelova Volponi, J. A. Shibli, A. Golkar-Narenji and P. Antosik, et al., The Role of Exosomes in Human Carcinogenesis and Cancer Therapy—Recent Findings from Molecular and Clinical Research, Cells, 2023, 12, DOI:10.3390/CELLS12030356.
- H. M. E. S. Azzazy, A. M. Sawy, A. Abdelnaser, M. R. Meselhy, T. Shoeib and S. A. Fahmy, Peganum harmala Alkaloids and Tannic Acid Encapsulated in PAMAM Dendrimers: Improved Anticancer Activities as Compared to Doxorubicin, ACS Appl. Polym. Mater., 2022, 4, 7228–7239, DOI:10.1021/ACSAPM.2C01093/ASSET/IMAGES/LARGE/AP2C01093_0008.JPEG.
- T. Abaza, M. K. A. El-Aziz, K. A. Daniel, P. Karousi, M. Papatsirou, S. A. Fahmy, N. M. Hamdy, C. K. Kontos and R. A. Youness, Emerging Role of Circular RNAs in Hepatocellular Carcinoma Immunotherapy, Int. J. Mol. Sci., 2023, 24, 16484, DOI:10.3390/ijms242216484.
- R. A. Youness, A. H. Mohamed, E. K. Efthimiadou, R. Y. Mekky, M. Braoudaki and S. A. Fahmy, A snapshot of photoresponsive liposomes in cancer chemotherapy and immunotherapy: opportunities and challenges, ACS Omega, 2023, 8(47), 44424–44436 CrossRef PubMed.
- M. D. Langellotto, G. Rassu, C. Serri, S. Demartis, P. Giunchedi and E. Gavini, Plant-derived extracellular vesicles: a synergetic combination of a drug delivery system and a source of natural bioactive compounds, Drug Delivery Transl. Res., 2024, 1–15, DOI:10.1007/S13346-024-01698-4/TABLES/2.
- S. A. Fahmy, N. K. Sedky, H. A. F. M. Hassan, N. M. Abdel-Kader, N. K. Mahdy, M. U. Amin, E. Preis and U. Bakowsky, Synergistic Enhancement of Carboplatin Efficacy through pH-Sensitive Nanoparticles Formulated Using Naturally Derived Boswellia Extract for Colorectal Cancer Therapy, Pharmaceutics, 2024, 16, 1282, DOI:10.3390/pharmaceutics16101282.
- S. A. Fahmy and W. Mamdouh, Garlic oil–loaded PLGA nanoparticles with controllable size and shape and enhanced antibacterial activities, J. Appl. Polym. Sci., 2018, 135(16), 46133 CrossRef.
- M. A. Di Bella, Overview and Update on Extracellular Vesicles: Considerations on Exosomes and Their Application in Modern Medicine, Biology, 2022, 11, 804, DOI:10.3390/BIOLOGY11060804.
- Y. Akao, Y. Kuranaga, K. Heishima, N. Sugito, K. Morikawa and Y. Ito, et al., Plant hvu-MIR168-3p enhances expression of glucose transporter 1 (SLC2A1) in human cells by silencing genes related to mitochondrial electron transport chain complex I, J. Nutr. Biochem., 2022, 101, 108922, DOI:10.1016/J.JNUTBIO.2021.108922.
- J. Schulze, J. Lehmann, S. Agel, M. U. Amin, J. Schaefer and U. Bakowsky, In Ovo Testing Method for Inhalants on a Chorio-Allantoic Membrane, ACS Appl. Bio Mater., 2021, 4(11), 7764–7768 CrossRef.
- Z. Jin, J. Na, X. Lin, R. Jiao, X. Liu and Y. Huang, Plant-derived exosome-like nanovesicles: A novel nanotool for disease therapy, Heliyon, 2024, 10, DOI:10.1016/J.HELIYON.2024.E30630/ASSET/53E05DC1-0CE2-462B-8486-F29D98F6B39A/MAIN.ASSETS/GR2.JPG.
- R. Dhar, N. Mukerjee, D. Mukherjee, A. Devi, S. K. Jha and S. Gorai, Plant-derived exosomes: A new dimension in cancer therapy, Phytother. Res., 2024, 38, 1721–1723, DOI:10.1002/PTR.7828.
- M. A. Kumar, S. K. Baba, H. Q. Sadida, S. A. Marzooqi, J. Jerobin and F. H. Altemani, et al., Extracellular vesicles as tools and targets in therapy for diseases, Signal Transduction Targeted Ther., 2024, 9, 1–41, DOI:10.1038/s41392-024-01735-1.
- R. Ono, Y. Yoshioka, Y. Furukawa, M. Naruse, M. Kuwagata and T. Ochiya, et al., Novel hepatotoxicity biomarkers of extracellular vesicle (EV)-associated miRNAs induced by CCl4, Toxicol. Rep., 2020, 7, 685–692, DOI:10.1016/J.TOXREP.2020.05.002.
- M. Xu, J. Ji, D. Jin, Y. Wu, T. Wu and R. Lin, et al., The biogenesis and secretion of exosomes and multivesicular bodies (MVBs): Intercellular shuttles and implications in human diseases, Genes Dis., 2022, 10, 1894, DOI:10.1016/J.GENDIS.2022.03.021.
- J. W. Clancy, M. Schmidtmann and C. D’Souza-Schorey, The ins and outs of microvesicles, FASEB BioAdv., 2021, 3, 399, DOI:10.1096/FBA.2020-00127.
- M. Battistelli and E. Falcieri, Apoptotic Bodies: Particular Extracellular Vesicles Involved in Intercellular Communication, Biology, 2020, 9, 21, DOI:10.3390/BIOLOGY9010021.
- S. Anand, M. Samuel and S. Mathivanan, Exomeres: A New Member of Extracellular Vesicles Family, Subcell. Biochem., 2021, 97, 89–97, DOI:10.1007/978-3-030-67171-6_5.
- X. Zhang, L. Yao, Y. Meng, B. Li, Y. Yang and F. Gao, Migrasome: a new functional extracellular vesicle, Cell Death Discovery, 2023, 9, 1–7, DOI:10.1038/s41420-023-01673-x.
- C. Ciardiello, R. Migliorino, A. Leone and A. Budillon, Large extracellular vesicles: Size matters in tumor progression, Cytokine Growth Factor Rev., 2020, 51, 69–74, DOI:10.1016/J.CYTOGFR.2019.12.007.
- Q. Zhang, D. K. Jeppesen, J. N. Higginbotham, R. Graves-Deal, V. Q. Trinh and M. A. Ramirez, et al., Supermeres are functional extracellular nanoparticles replete with disease biomarkers and therapeutic targets, Nat. Cell Biol., 2021, 23, 1240–1254, DOI:10.1038/s41556-021-00805-8.
- X. Lu, Q. Han, J. Chen, T. Wu, Y. Cheng and F. Li, et al., Celery (Apium graveolens L.) Exosome-like Nanovesicles as a New-Generation Chemotherapy Drug Delivery Platform against Tumor Proliferation, J. Agric. Food Chem., 2023, 71, 8413–8424, DOI:10.1021/ACS.JAFC.2C07760/SUPPL_FILE/JF2C07760_SI_001.PDF.
- J. Dai, Y. Su, S. Zhong, L. Cong, B. Liu and J. Yang, et al., Exosomes: key players in cancer and potential therapeutic strategy, Signal Transduction Targeted Ther., 2020, 5, 1–10, DOI:10.1038/s41392-020-00261-0.
- D. Subha, K. Harshnii, K. G. Madhikiruba, M. Nandhini and K. S. Tamilselvi, Plant derived exosome- like Nanovesicles: an updated overview, Plant Nano Biol., 2023, 3, 100022, DOI:10.1016/J.PLANA.2022.100022.
- M. Nemati, B. Singh, R. A. Mir, M. Nemati, A. Babaei and M. Ahmadi, et al., Plant-derived extracellular vesicles: a novel nanomedicine approach with advantages and challenges, Cell Commun. Signaling, 2022, 20, 1–16, DOI:10.1186/S12964-022-00889-1.
- T. Karamanidou and A. Tsouknidas, Plant-Derived Extracellular Vesicles as Therapeutic Nanocarriers, Int. J. Mol. Sci., 2021, 23, 191, DOI:10.3390/IJMS23010191.
- X. Wei, X. Li, Y. Zhang, J. Wang and S. Shen, Advances in the Therapeutic Applications of Plant-Derived Exosomes in the Treatment of Inflammatory Diseases, Biomedicines, 2023, 11, 1554, DOI:10.3390/BIOMEDICINES11061554.
- Q. F. Han, W. J. Li, K. S. Hu, J. Gao, W. L. Zhai and J. H. Yang, et al., Exosome biogenesis: machinery, regulation, and therapeutic implications in cancer, Mol. Cancer, 2022, 21, 1–26, DOI:10.1186/S12943-022-01671-0.
- D. Subha, K. Harshnii, K. G. Madhikiruba, M. Nandhini and K. S. Tamilselvi, Plant derived exosome- like Nanovesicles: an updated overview, Plant Nano Biol., 2023, 3, 100022, DOI:10.1016/J.PLANA.2022.100022.
- H. A. Dad, T. W. Gu, A. Q. Zhu, L. Q. Huang and L. H. Peng, Plant Exosome-like Nanovesicles: Emerging Therapeutics and Drug Delivery Nanoplatforms, Mol. Ther., 2021, 29, 13–31, DOI:10.1016/J.YMTHE.2020.11.030/ASSET/C270D6FD-1E15-4271-B739-DEAABA0A6127/MAIN.ASSETS/GR3.JPG.
- A. Kumar and G. Deep, Hypoxia in tumor microenvironment regulates exosome biogenesis: Molecular mechanisms and translational opportunities, Cancer Lett., 2020, 479, 23–30, DOI:10.1016/J.CANLET.2020.03.017.
- S. M. Migliano, E. M. Wenzel and H. Stenmark, Biophysical and molecular mechanisms of ESCRT functions, and their implications for disease, Curr. Opin. Cell Biol., 2022, 75, 102062, DOI:10.1016/J.CEB.2022.01.007.
- K.-J. Cho, S. Ishido, L. C. Eisenlohr and P. A. Roche, Activation of Dendritic Cells Alters the Mechanism of MHC Class II Antigen Presentation to CD4 T Cells, J. Immunol., 2020, 204, 1621–1629, DOI:10.4049/JIMMUNOL.1901234.
- D. Wei, W. Zhan, Y. Gao, L. Huang, R. Gong and W. Wang, et al., RAB31 marks and controls an ESCRT-independent exosome pathway, Cell Res., 2020, 31, 157–177, DOI:10.1038/s41422-020-00409-1.
- P. Fernandez, A. P. López and Y. Olmos, The ESCRT Machinery: Remodeling, Repairing, and Sealing Membranes, Membranes, 2022, 12, 633, DOI:10.3390/MEMBRANES12060633.
- A. L. Egea-Jimenez, S. Audebert, M. Castro-Cruz, J.-P. Borg, G. David, L. Camoin, et al.PLD2-phosphatidic acid recruit ESCRT-I to late endosomes for exosome biogenesis, bioRxiv, 2020, preprint, BioRxiv:2020.11.25.398396. DOI:10.1101/2020.11.25.398396.
- S. Hu, B. Li, F. Wu, D. Zhu, J. Zouhar and C. Gao, et al., Plant ESCRT protein ALIX coordinates with retromer complex in regulating receptor-mediated sorting of soluble vacuolar proteins, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2200492119, DOI:10.1073/PNAS.2200492119/SUPPL_FILE/PNAS.2200492119.SAPP.PDF.
- J. Larios, V. Mercier, A. Roux and J. Gruenberg, ALIX- And ESCRT-III-dependent sorting of tetraspanins to exosomes, J. Cell Biol., 2020, 219, DOI:10.1083/JCB.201904113/133723/ALIX-AND-ESCRT-III-DEPENDENT-SORTING-OF.
- X. R. Guo, Y. Ma, Z. M. Ma, T. S. Dai, S. H. Wei and Y. K. Chu, et al., Exosomes: The role in mammalian reproductive regulation and pregnancy-related diseases, Front. Physiol., 2023, 14, 1056905, DOI:10.3389/FPHYS.2023.1056905/BIBTEX.
- S. Suharta, A. Barlian, A. C. Hidajah, H. B. Notobroto, I. D. Ana and S. Indariani, et al., Plant-derived exosome-like nanoparticles: A concise review on its extraction methods, content, bioactivities, and potential as functional food ingredient, J. Food Sci., 2021, 86, 2838–2850, DOI:10.1111/1750-3841.15787.
- J. Kim, S. Li, S. Zhang and J. Wang, Plant-derived exosome-like nanoparticles and their therapeutic activities, Asian J. Pharm. Sci., 2022, 17, 53–69, DOI:10.1016/J.AJPS.2021.05.006.
- Y. An, T. Jin, Y. Zhu, F. Zhang and P. He, An ultrasensitive electrochemical aptasensor for the determination of tumor exosomes based on click chemistry, Biosens. Bioelectron., 2019, 142, 111503, DOI:10.1016/J.BIOS.2019.111503.
- H. Song, B. S. B. Canup, V. L. Ngo, T. L. Denning, P. Garg and H. Laroui, Internalization of garlic-derived nanovesicles on liver cells is triggered by interaction with CD98, ACS Omega, 2020, 5, 23118–23128, DOI:10.1021/ACSOMEGA.0C02893/ASSET/IMAGES/LARGE/AO0C02893_0008.JPEG.
- Y. Wang, Y. Wei, H. Liao, H. Fu, X. Yang and Q. Xiang, et al., Plant Exosome-like Nanoparticles as Biological Shuttles for Transdermal Drug Delivery, Bioengineering, 2023, 10, 104, DOI:10.3390/BIOENGINEERING10010104.
- J. H. Cho, Y. D. Hong, D. Kim, S. J. Park, J. S. Kim and H. M. Kim, et al., Confirmation of plant-derived exosomes as bioactive substances for skin application through comparative analysis of keratinocyte transcriptome, Appl. Biol. Chem., 2022, 65, 1–9, DOI:10.1186/S13765-022-00676-Z/FIGURES/4.
- K. Aquilano, V. Ceci, A. Gismondi, S. De Stefano, F. Iacovelli and R. Faraonio, et al., Adipocyte metabolism is improved by TNF receptor-targeting small RNAs identified from dried nuts, Commun. Biol., 2019, 2, 1–13, DOI:10.1038/s42003-019-0563-7.
- Y. Ito, K. Taniguchi, Y. Kuranaga, N. Eid, Y. Inomata and S. W. Lee, et al., Uptake of MicroRNAs from Exosome-Like Nanovesicles of Edible Plant Juice by Rat Enterocytes, Int. J. Mol. Sci., 2021, 22, 3749, DOI:10.3390/IJMS22073749.
- D. B. Krishnatreya, P. M. Baruah, B. Dowarah, S. Chowrasia, T. K. Mondal and N. Agarwala, Genome-wide identification, evolutionary relationship and expression analysis of AGO, DCL and RDR family genes in tea, Sci. Rep., 2021, 11, 1–18, DOI:10.1038/s41598-021-87991-5.
- B. He, Q. Cai, L. Qiao, C. Y. Huang, S. Wang and W. Miao, et al., RNA-binding proteins contribute to small RNA loading in plant extracellular vesicles, Nat. Plants, 2021, 7, 342–352, DOI:10.1038/s41477-021-00863-8.
- Q. Zhou, K. Ma, H. Hu, X. Xing, X. Huang and H. Gao, Extracellular vesicles: Their functions in plant–pathogen interactions, Mol. Plant Pathol., 2022, 23, 760–771, DOI:10.1111/MPP.13170.
- A. Amiri, R. Bagherifar, E. Ansari Dezfouli, S. H. Kiaie, R. Jafari and R. Ramezani, Exosomes as bio-inspired nanocarriers for RNA delivery: preparation and applications, J. Transl. Med., 2022, 20, 1–16, DOI:10.1186/S12967-022-03325-7/TABLES/3.
- C. Coughlan, K. D. Bruce, O. Burgy, T. D. Boyd, C. R. Michel and J. E. Garcia-Perez, et al., Exosome Isolation by Ultracentrifugation and Precipitation: A Comparison of Techniques for Downstream Analyses, Curr. Protoc. Cell Biol., 2020, 88, e110, DOI:10.1002/CPCB.110.
- S. Sonbhadra, Mehak and L. M. Pandey, Biogenesis, Isolation, and Detection of Exosomes and Their Potential in Therapeutics and Diagnostics, Biosensors, 2023, 13, 802, DOI:10.3390/BIOS13080802.
- Y. Huang, S. Wang, Q. Cai and H. Jin, Effective methods for isolation and purification of extracellular vesicles from plants, J. Integr. Plant Biol., 2021, 63, 2020, DOI:10.1111/JIPB.13181.
- D. Yang, W. Zhang, H. Zhang, F. Zhang, L. Chen and L. Ma, et al., Progress, opportunity, and perspective on exosome isolation – efforts for efficient exosome-based theranostics, Theranostics, 2020, 10, 3684–3707, DOI:10.7150/THNO.41580.
- W. Z. Liu, Z. J. Ma and X. W. Kang, Current status and outlook of advances in exosome isolation, Anal. Bioanal. Chem., 2022, 414, 7123–7141, DOI:10.1007/S00216-022-04253-7/FIGURES/6.
- D. Yang, W. Zhang, H. Zhang, F. Zhang, L. Chen and L. Ma, et al., Progress, opportunity, and perspective on exosome isolation – efforts for efficient exosome-based theranostics, Theranostics, 2020, 10, 3684, DOI:10.7150/THNO.41580.
- K. Sidhom, P. O. Obi and A. Saleem, A Review of Exosomal Isolation Methods: Is Size Exclusion Chromatography the Best Option?, Int. J. Mol. Sci., 2020, 21, 1–19, DOI:10.3390/IJMS21186466.
- Y. Pang, J. Shi, X. Yang, C. Wang, Z. Sun and R. Xiao, Personalized detection of circling exosomal PD-L1 based on Fe3O4@TiO2 isolation and SERS immunoassay, Biosens. Bioelectron., 2020, 148, 111800, DOI:10.1016/J.BIOS.2019.111800.
- M. Chang, Q. Wang, W. Qin, X. Shi and G. Xu, Rational Synthesis of Aptamer-Functionalized Polyethylenimine-Modified Magnetic Graphene Oxide Composites for Highly Efficient Enrichment and Comprehensive Metabolomics Analysis of Exosomes, Anal. Chem., 2020, 92, 15497–15505, DOI:10.1021/ACS.ANALCHEM.0C03374/SUPPL_FILE/AC0C03374_SI_001.PDF.
- Y. Li, J. Deng, Z. Han, C. Liu, F. Tian and R. Xu, et al., Molecular Identification of Tumor-Derived Extracellular Vesicles Using Thermophoresis-Mediated DNA Computation, J. Am. Chem. Soc., 2021, 143, 1290–1295, DOI:10.1021/JACS.0C12016/SUPPL_FILE/JA0C12016_SI_001.PDF.
- Q. Zhou, Z. Xu and Z. Liu, Molecularly Imprinting–Aptamer Techniques and Their Applications in Molecular Recognition, Biosensors, 2022, 12, 576, DOI:10.3390/BIOS12080576.
- L. Liu, J. Liu, W. Zhou, Z. Sui, J. Liu and K. Yang, et al., An artificial antibody for exosome capture by dull template imprinting technology, J. Mater. Chem. B, 2022, 10, 6655–6663, 10.1039/D2TB00494A.
- H. Krishnan, S. C. B. Gopinath, M. K. M. Arshad, H. I. Zulhaimi, P. Anbu and S. Subramaniam, Molecularly imprinted polymer enhances affinity and stability over conventional aptasensor for blood clotting biomarker detection on regimented carbon nanohorn and gold nanourchin hybrid layers, Sens. Actuators, B, 2022, 363, 131842, DOI:10.1016/J.SNB.2022.131842.
- M. Tayebi, Y. Zhou, P. Tripathi, R. Chandramohanadas and Y. Ai, Exosome Purification and Analysis Using a Facile Microfluidic Hydrodynamic Trapping Device, Anal. Chem., 2020, 92, 10733–10742, DOI:10.1021/ACS.ANALCHEM.0C02006/SUPPL_FILE/AC0C02006_SI_004.MP4.
- N. Sun, Y. Te Lee, R. Y. Zhang, R. Kao, P. C. Teng and Y. Yang, et al., Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring, Nat. Commun., 2020, 11, 1–12, DOI:10.1038/s41467-020-18311-0.
- Z. Yu, S. Lin, F. Xia, Y. Liu, D. Zhang and F. Wang, et al., ExoSD chips for high-purity immunomagnetic separation and high-sensitivity detection of gastric cancer cell-derived exosomes, Biosens. Bioelectron., 2021, 194, 113594, DOI:10.1016/J.BIOS.2021.113594.
- N. Ramnauth, E. Neubarth, A. Makler-Disatham, M. Sher, S. Soini and V. Merk, et al., Development of a Microfluidic Device for Exosome Isolation in Point-of-Care Settings, Sensors, 2023, 23, 8292, DOI:10.3390/S23198292.
- B. Wu, X. Chen, J. Wang, X. Qing, Z. Wang and X. Ding, et al., Separation and characterization of extracellular vesicles from human plasma by asymmetrical flow field-flow fractionation, Anal. Chim. Acta, 2020, 1127, 234–245, DOI:10.1016/J.ACA.2020.06.071.
- M. Marioli and W. T. Kok, Continuous asymmetrical flow field-flow fractionation for the purification of proteins and nanoparticles, Sep. Purif. Technol., 2020, 242, 116744, DOI:10.1016/J.SEPPUR.2020.116744.
- H. Y. Liu, R. Kumar, C. Zhong, S. Gorji, L. Paniushkina and R. Masood, et al., Rapid Capture of Cancer Extracellular Vesicles by Lipid Patch Microarrays, Adv. Mater., 2021, 33, DOI:10.1002/ADMA.202008493.
- A. Mukherjee, B. Bisht, S. Dutta and M. K. Paul, Current advances in the use of exosomes, liposomes, and bioengineered hybrid nanovesicles in cancer detection and therapy, Acta Pharmacol. Sin., 2022, 43(11), 2759–2776, DOI:10.1038/s41401-022-00902-w.
- D. E. Murphy, O. G. de Jong, M. J. W. Evers, M. Nurazizah, R. M. Schiffelers and P. Vader, Natural or synthetic RNA delivery: A stoichiometric comparison of extracellular vesicles and synthetic nanoparticles, Nano Lett., 2021, 21, 1888–1895, DOI:10.1021/acs.nanolett.1c00094.
- Y. Y. Lee, H. Kim and V. N. Kim, Sequence determinant of small RNA production by DICER, Nature, 2023, 615(7951), 323–330, DOI:10.1038/s41586-023-05722-4.
- I. K. Herrmann, M. J. A. Wood and G. Fuhrmann, Extracellular vesicles as a next-generation drug delivery platform, Nat. Nanotechnol., 2021, 16, 748–759, DOI:10.1038/s41565-021-00931-2.
- Y. Zhang, Q. Liu, X. Zhang, H. Huang, S. Tang and Y. Chai, et al., Recent advances in exosome-mediated nucleic acid delivery for cancer therapy, J. Nanobiotechnol., 2022, 20, 1–29, DOI:10.1186/S12951-022-01472-Z.
- N. Mu, J. Li, L. Zeng, J. You, R. Li and A. Qin, et al., Plant-Derived Exosome-Like Nanovesicles: Current Progress and Prospects, Int. J. Nanomed., 2023, 18, 4987–5009, DOI:10.2147/IJN.S420748.
- H. Zeng, S. Guo, X. Ren, Z. Wu, S. Liu and X. Yao, Current Strategies for Exosome Cargo Loading and Targeting Delivery, Cells, 2023, 12, DOI:10.3390/cells12101416.
- S. I. Ohno, M. Takanashi, K. Sudo, S. Ueda, A. Ishikawa and N. Matsuyama, et al., Systemically injected exosomes targeted to EGFR deliver antitumor microRNA to breast cancer cells, Mol. Ther., 2013, 21, 185–191, DOI:10.1038/MT.2012.180.
- G. Wu, J. Zhang, Q. Zhao, W. Zhuang, J. Ding and C. Zhang, et al., Molecularly Engineered Macrophage-Derived Exosomes with Inflammation Tropism and Intrinsic Heme Biosynthesis for Atherosclerosis Treatment, Angew. Chem., Int. Ed., 2020, 59, 4068–4074, DOI:10.1002/anie.201913700.
- S. Le Saux, H. Aarrass, J. Lai-Kee-Him, P. Bron, J. Armengaud and G. Miotello, et al., Post-production modifications of murine mesenchymal stem cell (mMSC) derived extracellular vesicles (EVs) and impact on their cellular interaction, Biomaterials, 2020, 231, 119675, DOI:10.1016/J.BIOMATERIALS.2019.119675.
- L. J. Born, K. H. Chang, P. Shoureshi, F. Lay, S. Bengali and A. T. W. Hsu, et al., HOTAIR-Loaded Mesenchymal Stem/Stromal Cell Extracellular Vesicles Enhance Angiogenesis and Wound Healing, Adv. Healthcare Mater., 2022, 11, 2002070, DOI:10.1002/ADHM.202002070.
- Y. Ma, L. Sun, J. Zhang, C. L. Chiang, J. Pan and X. Wang, et al., Exosomal mRNAs for Angiogenic–Osteogenic Coupled Bone Repair, Adv. Sci., 2023, 10, 2302622, DOI:10.1002/ADVS.202302622.
- D. Pan, C. Zeng, W. Zhang, T. Li, Z. Qin and X. Yao, et al., Non-volatile pungent compounds isolated from Zingiber officinale and their mechanisms of action, Food Funct., 2019, 10, 1203–1211, 10.1039/C8FO02019A.
- M. F. Mahomoodally, M. Z. Aumeeruddy, K. R. R. Rengasamy, S. Roshan, S. Hammad and J. Pandohee, et al., Ginger and its active compounds in cancer therapy: From folk uses to nano-therapeutic applications, Semin. Cancer Biol., 2021, 69, 140–149, DOI:10.1016/J.SEMCANCER.2019.08.009.
- S. M. Nachvak, D. Soleimani, M. Rahimi, A. Azizi, M. Moradinazar and M. H. Rouhani, et al., Ginger as an anticolorectal cancer spice: A systematic review of in vitro to clinical evidence, Food Sci. Nutr., 2023, 11, 651–660, DOI:10.1002/FSN3.3153.
- M. Zadorozhna and D. Mangieri, Mechanisms of Chemopreventive and Therapeutic Proprieties of Ginger Extracts in Cancer, Int. J. Mol. Sci., 2021, 22, 6599, DOI:10.3390/IJMS22126599.
- K. R. Shanmugam, B. Shanmugam, G. Venkatasubbaiah, S. Ravi and K. S. ReddyRecent Updates on the Bioactive Compounds of Ginger (Zingiber officinale) on Cancer: A Study with Special Emphasis of Gingerol and Its Anticancer Potential: Effect of Ginger and Its Compounds in Cancer Subjects, Handbook of Oxidative Stress in Cancer: Therapeutic Aspects, 2022, vol. 1, pp. 489–506 DOI:10.1007/978-981-16-5422-0_188/FIGURES/5.
- T. Hillman, The use of plant-derived exosome-like nanoparticles as a delivery system of CRISPR/Cas9-based therapeutics for editing long non-coding RNAs in cancer colon cells, Front. Oncol., 2023, 13, 1194350, DOI:10.3389/FONC.2023.1194350/BIBTEX.
- S. Chen and X. Shen, Long noncoding RNAs: functions and mechanisms in colon cancer, Mol. Cancer, 2020, 19, 1–13, DOI:10.1186/S12943-020-01287-2/FIGURES/2.
- L. Yin, L. Yan, Q. Yu, J. Wang, C. Liu and L. Wang, et al., Characterization of the MicroRNA Profile of Ginger Exosome-like Nanoparticles and Their Anti-Inflammatory Effects in Intestinal Caco-2 Cells, J. Agric. Food Chem., 2022, 70, 4725–4734, DOI:10.1021/ACS.JAFC.1C07306/ASSET/IMAGES/LARGE/JF1C07306_0012.JPEG.
- J. W. Fahey, Y. Zhang and P. Talalay, Broccoli sprouts: An exceptionally rich source of inducers of enzymes that protect against chemical carcinogens, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 10367–10372, DOI:10.1073/PNAS.94.19.10367/ASSET/A702873D-8055-446E-8376-B3626625816B/ASSETS/GRAPHIC/PQ1872098005.JPEG.
- A. Gasmi, A. Gasmi Benahmed, M. Shanaida, S. Chirumbolo, A. Menzel and W. Anzar, et al., Anticancer activity of broccoli, its organosulfur and polyphenolic compounds, Crit. Rev. Food Sci. Nutr., 2024, 64, 8054–8072, DOI:10.1080/10408398.2023.2195493.
- D. B. Nandini, R. S. Rao, B. S. Deepak and P. B. Reddy, Sulforaphane in broccoli: The green chemoprevention!! Role in cancer prevention and therapy, J. Oral Maxillofac. Pathol., 2020, 24, 405, DOI:10.4103/JOMFP.JOMFP_126_19.
- F. Artés-Hernández, L. Martínez-Zamora, M. Cano-Lamadrid, S. Hashemi and N. Castillejo, Genus Brassica By-Products Revalorization with Green Technologies to Fortify Innovative Foods: A Scoping Review, Foods, 2023, 12, 561, DOI:10.3390/FOODS12030561.
- L. del Pozo-Acebo, M. C. López de las Hazas, J. Tomé-Carneiro, A. del Saz-Lara, J. Gil-Zamorano and L. Balaguer, et al., Therapeutic potential of broccoli-derived extracellular vesicles as nanocarriers of exogenous miRNAs, Pharmacol. Res., 2022, 185, 106472, DOI:10.1016/J.PHRS.2022.106472.
- X. Wang, B. Wu, G. Sun, W. He, J. Gao and T. Huang, et al., Selenium Biofortification Enhanced miR167a Expression in Broccoli Extracellular Vesicles Inducing Apoptosis in Human Pancreatic Cancer Cells by Targeting IRS1, Int. J. Nanomed., 2023, 18, 2431, DOI:10.2147/IJN.S394133.
- U. Asma, K. Morozova, G. Ferrentino and M. Scampicchio, Apples and Apple By-Products: Antioxidant Properties and Food Applications, Antioxidants, 2023, 12, 1456, DOI:10.3390/ANTIOX12071456.
- M. Trentini, F. Zanotti, E. Tiengo, F. Camponogara, M. Degasperi and D. Licastro, et al., An Apple a Day Keeps the Doctor Away: Potential Role of miRNA 146 on Macrophages Treated with Exosomes Derived from Apples, Biomedicines, 2022, 10, 415, DOI:10.3390/BIOMEDICINES10020415.
- D. Fujita, T. Arai, H. Komori, Y. Shirasaki, T. Wakayama and T. Nakanishi, et al., Apple-Derived Nanoparticles Modulate Expression of Organic-Anion-Transporting Polypeptide (OATP) 2B1 in Caco-2 Cells, Mol. Pharmaceutics, 2018, 15, 5772–5780, DOI:10.1021/ACS.MOLPHARMACEUT.8B00921/ASSET/IMAGES/LARGE/MP-2018-009212_0007.JPEG.
- A. Septembre-Malaterre, M. L. Rakoto, C. Marodon, Y. Bedoui, J. Nakab and E. Simon, et al., Artemisia annua, a Traditional Plant Brought to Light, Int. J. Mol. Sci., 2020, 21, 4986, DOI:10.3390/IJMS21144986.
- J. Liu, J. Xiang, C. Jin, L. Ye, L. Wang and Y. Gao, et al., Medicinal plant-derived mtDNA via nanovesicles induces the cGAS-STING pathway to remold tumor-associated macrophages for tumor regression, J. Nanobiotechnol., 2023, 21, 1–19, DOI:10.1186/S12951-023-01835-0/FIGURES/7.
- J. Chen, D. Yu, X. Li, Q. Deng, H. Yang and L. Chen, et al., A review of Brucea javanica: metabolites, pharmacology and clinical application, Front. Pharmacol., 2024, 14, 1317620, DOI:10.3389/FPHAR.2023.1317620.
- G. Yan, Q. Xiao, J. Zhao, H. Chen, Y. Xu and M. Tan, et al., Brucea javanica derived exosome-like nanovesicles deliver miRNAs for cancer therapy, J. Controlled Release, 2024, 367, 425–440, DOI:10.1016/J.JCONREL.2024.01.060.
- P. Poletto, G. Álvarez-Rivera, G. D. López, O. M. A. Borges, J. A. Mendiola and E. Ibáñez, et al., Recovery of ascorbic acid, phenolic compounds and carotenoids from acerola by-products: An opportunity for their valorization, LWT, 2021, 146, 111654, DOI:10.1016/J.LWT.2021.111654.
- I. Carneiro Ferreira, V. Pereira da Silva, J. C. Vilvert, F. de França Souza, S. T. de Freitas and M. dos Santos Lima, Brazilian varieties of acerola (Malpighia emarginata DC.) produced under tropical semi-arid conditions: Bioactive phenolic compounds, sugars, organic acids, and antioxidant capacity, J. Food Biochem., 2021, 45, e13829, DOI:10.1111/JFBC.13829.
- N. Carvalho Gualberto, C. Santos de Oliveira, J. Pedreira Nogueira, M. Silva de Jesus, H. Caroline Santos Araujo and M. Rajan, et al., Bioactive compounds and antioxidant activities in the agro-industrial residues of acerola (Malpighia emarginata L.), guava (Psidium guajava L.), genipap (Genipa americana L.) and umbu (Spondias tuberosa L.) fruits assisted by ultrasonic or shaker extraction, Food Res. Int., 2021, 147, 110538, DOI:10.1016/J.FOODRES.2021.110538.
- R. Olędzki and J. Harasym, Acerola (Malpighia emarginata) Anti-Inflammatory Activity—A Review, Int. J. Mol. Sci., 2024, 25, 2089, DOI:10.3390/IJMS25042089.
- T. Umezu, M. Takanashi, Y. Murakami, S. I. Ohno, K. Kanekura and K. Sudo, et al., Acerola exosome-like nanovesicles to systemically deliver nucleic acid medicine via oral administration, Mol. Ther.–Methods Clin. Dev., 2021, 21, 199–208, DOI:10.1016/j.omtm.2021.03.006.
- M. Ashrafizadeh, Z. Ahmadi, R. Mohammadinejad and E. Ghasemipour Afshar, Tangeretin: a mechanistic review of its pharmacological and therapeutic effects, J. Basic Clin. Physiol. Pharmacol., 2020, 31, DOI:10.1515/JBCPP-2019-0191/MACHINEREADABLECITATION/RIS.
- W. Raza, S. Luqman and A. Meena, Prospects of tangeretin as a modulator of cancer targets/pathways, Pharmacol. Res., 2020, 161, 105202, DOI:10.1016/J.PHRS.2020.105202.
- N. Rabienezhad Ganji, O. Urzì, V. Tinnirello, E. Costanzo, G. Polito and A. Palumbo Piccionello, et al., Proof-of-Concept Study on the Use of Tangerine-Derived Nanovesicles as siRNA Delivery Vehicles toward Colorectal Cancer Cell Line SW480, Int. J. Mol. Sci., 2024, 25, 546, DOI:10.3390/IJMS25010546/S1.
- M. C. López de las Hazas, J. Tomé-Carneiro, L. del Pozo-Acebo, A. del Saz-Lara, L. A. Chapado and L. Balaguer, et al., Therapeutic potential of plant-derived extracellular vesicles as nanocarriers for exogenous miRNAs, Pharmacol. Res., 2023, 198, 106999, DOI:10.1016/J.PHRS.2023.106999.
- Y. S. Chen, E. Y. Lin, T. W. Chiou and H. J. Harn, Exosomes in clinical trial and their production in compliance with good manufacturing practice, Tzu Chi Med. J., 2019, 32, 113, DOI:10.4103/TCMJ.TCMJ_182_19.
- I. K. Herrmann, M. J. A. Wood and G. Fuhrmann, Extracellular vesicles as a next-generation drug delivery platform, Nat. Nanotechnol., 2021, 16(7), 748–759, DOI:10.1038/s41565-021-00931-2.
- Y. Ma, S. Dong, A. J. Grippin, L. Teng, A. S. Lee and B. Y. S. Kim, et al., Engineering therapeutical extracellular vesicles for clinical translation, Trends Biotechnol., 2024, 43(1), 61–82, DOI:10.1016/J.TIBTECH.2024.08.007.
| This journal is © The Royal Society of Chemistry 2025 |
