Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Microwave-assisted green synthesis of fluorescent graphene quantum dots (GQDs) using Azadirachta indica leaves: enhanced synergistic action of antioxidant and antimicrobial effects and unveiling computational insights†
Pooja
Kadyan
a,
Manish
Kumar
b,
Aisha
Tufail
c,
Andrea
Ragusa
*de,
Sudhir Kumar
Kataria
*a and
Amit
Dubey
 *f
*f
aDepartment of Zoology, Maharshi Dayanand University, Rohtak, Haryana, India. E-mail: sudhir.zoology24@mdurohtak.ac.in
bDepartment of Biochemistry, Iswar Saran Degree College, University of Allahabad (A Constituent PG College of University of Allahabad), Prayagraj, India
cComputational Chemistry and Drug Discovery Division, Quanta Calculus, Greater Noida-201310, Uttar Pradesh, India
dCNR-Nanotec, Institute of Nanotechnology, Via Monteroni, 73100 Lecce, Italy
eDepartment of Life Sciences, Health and Health Professions, Link Campus University, Via del Casale Di San Pio V 44, 00165 Rome, Italy. E-mail: a.ragusa@unilink.it
fCenter For Global Health Research, Saveetha Medical College and Hospital, Saveetha Institute of Medical and Technical Sciences, Chennai-600077, Tamil Nadu, India. E-mail: amitdubey@saveetha.com; ameetbioinfo@gmail.com
First published on 11th December 2024
Abstract
Antibiotic resistance remains a global health challenge, necessitating the development of novel antimicrobial treatments. In this study, we report the microwave-assisted green synthesis of graphene quantum dots (GQDs) using Azadirachta indica (neem) leaf extract. Comprehensive characterization of GQDs via FT-IR, UV-vis, TEM, and EDX confirmed their formation and stability, revealing distinct functional groups and photoluminescent properties. The antioxidant and antibacterial activities of GQDs surpassed those of the neem extract, showing enhanced bioactivity. Furthermore, molecular docking and computational analysis provided insights into the binding interactions, energy profiles, and safety parameters of GQDs, supporting the experimental findings. This work demonstrates the potential of GQDs as multifunctional agents in biomedical applications, offering a sustainable approach to combating microbial resistance.
1. Introduction
The rise in antibiotic-resistant bacteria has emerged as a pressing global health crisis, necessitating the development of novel antimicrobial agents.1 Nanotechnology, particularly carbon-based materials such as GQDs, offers groundbreaking solutions owing to their unique nanoscale structural and functional properties, including a large surface area, biocompatibility, and fluorescence. GQDs are promising candidates for application in drug delivery, bioimaging, and antimicrobial therapies.2–6 In this study, we synthesized GQDs using Azadirachta indica (neem) leaf extract as a green, cost-effective, and eco-friendly alternative to GQDs synthesized via conventional methods. Neem leaves, known for their antibacterial, antifungal, antiviral, and anti-inflammatory properties, provide a sustainable source for nanoparticle synthesis. Microwave-assisted synthesis was employed to enhance the stability and bioactivity of GQDs, minimizing their environmental impact while maintaining cost efficiency.7–10The World Health Organization and the Centers for Disease Control and Prevention have highlighted the critical challenge posed by antibiotic resistance, which affects millions of individuals annually, leading to significant morbidity and mortality. The increasing prevalence of multidrug-resistant pathogens underscores the need for innovative therapeutic strategies.3,4 Carbon-based nanomaterials, such as graphene, carbon nanotubes, and GQDs, have gained traction for their ability to address this issue. GQDs, in particular, exhibit remarkable fluorescence stability, low cytotoxicity, and biocompatibility, which make them suitable for a wide range of biomedical applications. Recent advancements in nanotechnology have demonstrated that natural synthesis methods using biological materials such as neem are more sustainable and safer than conventional chemical approaches.11–15
Reactive oxygen species (ROS) generation is a critical mechanism by which GQDs exhibit antibacterial and antioxidant activities.16,17 Computational studies provide a deeper understanding of these interactions. Molecular docking studies revealed significant binding interactions between GQDs and bacterial proteins, suggesting their potential as antimicrobial agents. Density functional theory (DFT) calculations and molecular electrostatic potential (MESP) analyses offered insights into the electronic properties, reactivity, and stability of the GQDs, while ADMET profiling predicted their favorable pharmacokinetics, low toxicity, and high therapeutic potential. These computational methods complement experimental findings, providing a comprehensive framework for the application of GQDs in antimicrobial therapies.17
This study integrates experimental and computational approaches to explore the potential of neem-based GQDs in addressing the antibiotic resistance crisis. The results emphasize their broad-spectrum activity, eco-friendly synthesis, and promising applications in antimicrobial therapies.
2. Results and discussion
2.1. Characterization
The UV-visible spectrophotometric analysis showed the maximum absorption peaks at 257 nm, attributed to the π–π* transition of the C–C bond and sp2 domain, and 312 nm, due to the n-π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond (Fig. 1a). In previous studies, similar results were observed for the characterization of GQDs.18–20 The structural morphology of GQDs was confirmed by TEM. The GQD solution was treated in an ultrasonic bath before deposition. A drop of evenly dispersed GQD alcoholic solution was deposited on a copper grid. The samples were dried and analysed at an acceleration voltage of 200 kV and resolution of 0.24 nm. The GQDs appeared spherical, as depicted in Fig. 1(b, c and e), and their average particle size distribution was calculated to be 5.6 nm, as shown in Fig. 1(d). The elements present in the sample were further confirmed by EDX measurements (Fig. 1(e) and (f)). Carbon and oxygen were detected on the surface of GQDs, thereby indicating the presence of several O-containing functional groups.
O bond (Fig. 1a). In previous studies, similar results were observed for the characterization of GQDs.18–20 The structural morphology of GQDs was confirmed by TEM. The GQD solution was treated in an ultrasonic bath before deposition. A drop of evenly dispersed GQD alcoholic solution was deposited on a copper grid. The samples were dried and analysed at an acceleration voltage of 200 kV and resolution of 0.24 nm. The GQDs appeared spherical, as depicted in Fig. 1(b, c and e), and their average particle size distribution was calculated to be 5.6 nm, as shown in Fig. 1(d). The elements present in the sample were further confirmed by EDX measurements (Fig. 1(e) and (f)). Carbon and oxygen were detected on the surface of GQDs, thereby indicating the presence of several O-containing functional groups.
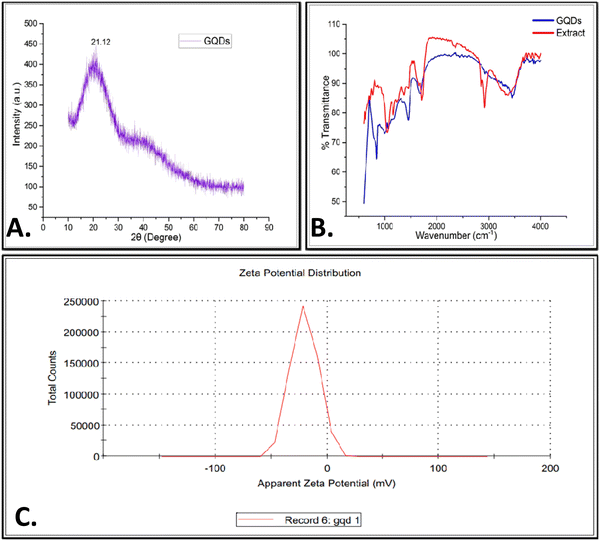
The powdered XRD study was performed in the 2θ range of 10° to 80°. The XRD data for the synthesized GQDs showed the presence of a peak corresponding to GQDs at 21.1°, as depicted in Fig. 2(a). This supports the hypothesis that the synthesized GQDs have a graphitic structure with a small amount of amorphous carbon and agrees well with the previously reported studies, which was reported a peak at 25.8°.21,22 To investigate the various chemical functional groups present in GQDs, FTIR was performed, as shown in Fig. 2(b). FTIR analysis showed shifts in the absorbance peak of GQDs with a range varying from 500–4000 cm−1. The peak observed at 1047.8 cm−1 is attributed to the C–O stretching vibration. The observed bands located at 1162.8, 1710.27, 2852.7, 2918.6, and 3322.67 cm−1 for the leaf extract emerged due to the C–O stretching, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, C–H stretching, CH2 stretching, and –NH– stretching and 842.9, 1452.09, 1674.3, 2937.21, and 3454 cm−1 for GQDs corresponding to C–H vibration, CH2 bending, C
O stretching, C–H stretching, CH2 stretching, and –NH– stretching and 842.9, 1452.09, 1674.3, 2937.21, and 3454 cm−1 for GQDs corresponding to C–H vibration, CH2 bending, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching, CH2 stretching, and –NH– stretching, respectively. The zeta potential of the prepared GQDs showed a negative value of −22 mV, as shown in Fig. 2(c). Furthermore, particles are considered to be stable if they have a strong zeta potential (positive or negative). The zeta potential investigations of the GQDs revealed that they were negatively charged because of the numerous hydroxyl and carboxyl groups present on their surface.
O stretching, CH2 stretching, and –NH– stretching, respectively. The zeta potential of the prepared GQDs showed a negative value of −22 mV, as shown in Fig. 2(c). Furthermore, particles are considered to be stable if they have a strong zeta potential (positive or negative). The zeta potential investigations of the GQDs revealed that they were negatively charged because of the numerous hydroxyl and carboxyl groups present on their surface.
2.2. Expanded FT-IR and UV-Vis analysis
FT-IR spectroscopy was conducted to compare the functional groups present in the Azadirachta indica leaf extract and the synthesized GQDs, which revealed distinct shifts that confirmed the transformation. In the FT-IR spectrum of the GQDs, characteristic peaks appeared at 1047.8 cm−1 (C–O stretching) and 1710.27 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching), which were absent in the spectrum of the leaf extract (Fig. 2(b)). These functional groups indicate the successful oxidation during the synthesis process, suggesting the incorporation of oxygen-containing groups necessary for the stability and solubility of GQDs in aqueous media. The UV-Vis analysis further supported this transformation, with the GQDs showing an absorption peak at 312 nm, corresponding to the n-π* transition of the C
O stretching), which were absent in the spectrum of the leaf extract (Fig. 2(b)). These functional groups indicate the successful oxidation during the synthesis process, suggesting the incorporation of oxygen-containing groups necessary for the stability and solubility of GQDs in aqueous media. The UV-Vis analysis further supported this transformation, with the GQDs showing an absorption peak at 312 nm, corresponding to the n-π* transition of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond, which was shifted compared to that of the extract (Fig. 1(a)). This shift in absorption indicates the formation of quantum dot structures with modified electronic properties, which are essential for their application in the biomedical field.
O bond, which was shifted compared to that of the extract (Fig. 1(a)). This shift in absorption indicates the formation of quantum dot structures with modified electronic properties, which are essential for their application in the biomedical field.
2.3. Emission spectra of GQDs
The synthesized GQDs displayed a fluorescence emission in the visible range, peaking at approximately 450 nm when excited at 360 nm, as illustrated in Fig. 3. This characteristic fluorescence of GQDs highlights their potential utility in bioimaging applications, where stable emission in the visible range is crucial. The emission spectra confirm that the synthesized particles exhibit photoluminescent properties unique to the graphene quantum dots, providing further evidence of the successful formation of GQDs.2.4. Energy-dispersive X-ray (EDX) and complementary analyses
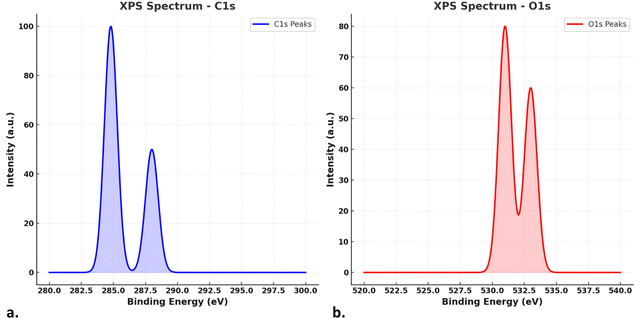
Energy-dispersive X-ray (EDX) analysis was performed to determine the elemental composition of the GQDs (Fig. 4). In this case, although EDX can effectively identify the presence of elements, its limitations in accurately quantifying light elements such as carbon (C) and oxygen (O) were acknowledged. Thus, to address these limitations and provide a more reliable characterization of the elemental composition, complementary techniques such as X-ray photoelectron spectroscopy (XPS) and electron energy loss spectroscopy (EELS) were recommended.In particular, the use of XPS has been widely recognized for its ability to analyze the surface chemistry of nanomaterials with high accuracy. Consistent with this approach, the XPS analysis of the GQDs revealed key insights into their chemical structure (Fig. 4b). The C1s spectrum exhibited peaks at ∼284.8 eV (sp2-hybridized carbon, indicating C–C/C–H bonding) and ∼288.0 eV (carbonyl or carboxyl groups, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O–C
O/O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). Similarly, the O1s spectrum displayed peaks at ∼531.0 eV (C
O). Similarly, the O1s spectrum displayed peaks at ∼531.0 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups) and ∼533.0 eV (C–O groups), confirming the presence of oxygen-containing functional groups on the surface of the GQDs.
O groups) and ∼533.0 eV (C–O groups), confirming the presence of oxygen-containing functional groups on the surface of the GQDs.
These findings not only validate the green method for the synthesis of GQDs but also highlight the importance of their surface functional groups in enhancing their solubility, stability, and potential biological applications. By integrating complementary techniques such as XPS, this study ensured the comprehensive and precise characterization of GQDs, consistent with recent advancements in nanomaterial analysis and reinforcing the potential of this material for biomedical and environmental applications.
3. Biological studies
3.1. DPPH radical scavenging assay
Antioxidants scavenge the DPPH radical by donating a proton, resulting in a decrease in the content of DPPH. The main indicator is the colour shift, which represents the amount of free radical scavenging. In this case, the DPPH free-radical scavenging activity (antioxidant qualities) of the produced GQDs was less than that of ascorbic acid, while it was greater than that of the leaf ethanolic extract, as shown in Fig. 5(a) and (b). All 12 phytochemical compounds from the A. indica leaf extract were confirmed by a literature survey and its bioactive constituents are summarized in Table S1 (ESI†). Based on previous reports, the samples appeared to have considerable DPPH free radical scavenging activity.18 The possible radical scavenging process is associated with the hydrogen transfer from the surface of GQDs to DPPH. The transfer of hydrogen atoms and the reduction of DPPH are attributed to the presence of hydroxyl (–OH), carboxyl (–COOH), and amino groups (–NH2 and –NH). The quantity of an antioxidant-containing substance required to scavenge half of the initial DPPH radicals is known as its IC50. The antioxidant and free radical scavenging properties of samples have an inverse relationship with their IC50 value. Greater antioxidant activity is implied by the ability of a compound to scavenge DPPH, which is indicated by a lower IC50 value. Accordingly, due to their lower IC50 value compared to the extract, GQDs exhibit enhanced antioxidant activity than the crude extract. To highlight the biological potential of GQDs, we conducted both antioxidant and antibacterial assays, with the significant findings moved from the Supplementary Information to the main manuscript. The antioxidant activity was assessed using the DPPH radical scavenging assay, where GQDs showed an IC50 value of 12.5 μg mL−1, which is markedly lower than that of the ethanolic leaf extract (IC50 of 20 μg mL−1). This lower IC50 value suggests enhanced free-radical scavenging capacity, underscoring the antioxidant potential of GQDs over the crude extract.3.2. Antimicrobial activity (well diffusion method)
The findings of the study on antimicrobial activities revealed that the biosynthesized GQDs have a bactericidal and fungal effect on pathogens. The inhibition zone reflects the antibacterial activities and antibacterial sensibility. In this study, the leaf extract and phyto-mediated GQDs of A. indica showed good antibacterial activities, as depicted in Fig. 6(a–h), respectively. In addition, against the fungal strains, the leaf extract and phyto-mediated GQDs of A. indica also showed good antifungal activities, as depicted from Fig. 7(a–d), respectively. The minimum zone of inhibition was produced by the leaf extract and synthesized GQDs against E. coli (MTCC 443). The strongest antimicrobial activity was found to be 14.1 ± 0.2 mm for GQDs against B. subtilis (MTCC 121). Also, GQDs and the neem leaf extract showed antibacterial activity against S. aureus (MTCC 96). The zone of inhibition in the case of A. niger (MTCC 281) was 1.5 ± 0.12 mm at 100 mg mL−1 and that for A. flavus (MTCC 277) was 2.6 ± 0.04 at 100 mg mL−1. The highest MIC was observed for E. coli (MTCC 443) and the lowest MIC was observed for B. subtilis (MTCC 121), i.e., 0.391 mg mL−1 (Table 1). In the case of antibacterial efficacy, GQDs exhibited larger inhibition zones, with a maximum zone of 14.1 ± 0.2 mm against Bacillus subtilis, surpassing that of the neem extract. These findings are consistent with the computational docking results, which suggested enhanced binding affinities for GQDs.| S. no. | Microbes | ZOI of GQDs (100 mg mL−1) | ZOI of neem extract (100 mg mL−1) | MIC (mg mL−1) |
|---|---|---|---|---|
| Results as mean ± standard error. | ||||
| 1. | S. aureus | 12.4 ± 0.22 | 11.6 ± 0.13 | 0.781 |
| 2. | P. aeruginosa | 10.2 ± 0.20 | 9.8 ± 0.03 | 1.562 |
| 3. | E. coli | 7.1 ± 0.41 | 7.1 ± 0.12 | >100 |
| 4. | B. subtilis | 14.1 ± 0.23 | 14.8 ± 0.04 | 0.391 |
| 5. | Aspergillus flavus | 2.6 ± 0.04 | 2.1 ± 0.21 | 3.125 |
| 6. | Aspergillus niger | 1.5 ± 0.12 | 1.2 ± 0.13 | 1.562 |
3.3. Quantitative analysis of antibacterial activity
The quantitative data for the antibacterial efficacy of GQDs are presented in Table 1, which includes the minimum inhibitory concentrations (MICs) and inhibition zones for different pathogens. The MIC for GQDs against Bacillus subtilis was determined to be 0.39 mg mL−1, indicating their potent bactericidal activity. The corresponding inhibition zones further substantiate the efficacy of GQDs in reducing bacterial growth, providing quantitative support for the observed antimicrobial effects. These results collectively underscore GQDs as promising agents against a broad spectrum of pathogens, with potential applications in addressing antibiotic-resistant bacterial strains.4. Computational studies
4.1. Unveiling molecular docking interactions: GQD and twelve compounds with E. coli
The molecular docking simulations involving GQDs and twelve compounds, namely azadirachtin, beta sitosterol, catechin, epicatechin, gallic acid, gedunin, mahmoodin, margolone, nimbin, nimbolide, quercetin, and salannol, with the bacterial strain E. coli (PDB ID: 1HNJ), revealed intricate insights into their binding energies and interactions within the active site (Fig. 8(a and b)). | ||
| Fig. 8 (a) Top view and (b) side view. Molecular docking interactions of the bacterial receptor (Escherichia coli, PDB ID: 1HNJ, represented as a cyan ribbon structure) with graphene quantum dots (GQDs, parrot green color) and 12 phytochemical compounds extracted from Azadirachta indica leaves. The ball-and-stick representations correspond to the GQD-phytochemical conjugates: GQD + gallic acid (dark blue), GQD + catechin (red-brown), GQD + nimbolide (violet), GQD + nimbin (red), GQD + mahmoodin (pink), GQD + salannol (olive green), GQD + margolone (yellow), GQD + beta-sitosterol (cyan), GQD + quercetin (white), GQD + gedunin (orange), GQD + nimbolide (black), and GQD + epicatechin (light violet). This illustration emphasizes the interaction hotspots, offering valuable insights into the antibacterial potential of these GQD-phytochemical complexes. | ||
Through the exploration of the molecular docking interaction of twelve phytochemicals obtained from various studies and academic data and their chemical structure and molecular properties obtained from the PubChem database, a repository of twelve phytoconstituents of Azadirachta indica was devised (Table S2) (ESI†).23–26
The interaction details and docking energy values of each compound, both with and without the addition of GQDs, were meticulously examined, offering a comprehensive understanding of their potential efficacy in inhibiting E. coli activity.
Azadirachtin, a natural compound, exhibited a docking energy of −95.209 kcal mol−1 when considered alone, which decreased to −124.7728 kcal mol−1 upon the inclusion of GQDs (Fig. 8(a and b)). This alteration in docking energy suggests a potential enhancement in binding affinity facilitated by GQDs. Regarding hydrogen bond interactions, azadirachtin formed bonds with Thr80, Thr81, Asn193, and Gly306 independently, whereas with GQDs, its interaction shifted to Thr81, His85, Asp107, and Asn193. Additionally, the steric interactions of azadirachtin undergo modification, involving different residues such as Thr80, Thr81, Ala109, and Ala111, indicating a potential improvement in binding geometry facilitated by GQDs (Table S2) (ESI†).
Beta sitosterol demonstrated a docking energy of −101.865 kcal mol−1 independently, which decreased to −131.4288 kcal mol−1 with the addition of GQDs (Fig. 8(a and b)). The substantial decrease in energy suggests a synergistic effect between beta sitosterol and GQDs in binding to E. coli. The hydrogen bond interactions exhibited a shift from Asn193 alone to Thr81, His85, Asp107, and Asn193 in the presence of GQDs, indicating a more favorable binding configuration. Similarly, the steric interactions also showed an alteration, involving different residues such as Ala111, Cys112, and Leu189, highlighting the influence of GQDs on the orientation of this compound within the binding site (Table S2) (ESI†).
Catechin, epicatechin, gallic acid, gedunin, mahmoodin, margolone, nimbin, nimbolide, quercetin, and salannol displayed similar trends in docking energy and interaction profiles, demonstrating the impact of GQDs on their binding affinity, hydrogen bond interactions, and steric interactions with E. coli.
In conclusion, the detailed analysis of the interaction of each compound with E. coli, both independently and in the presence of GQDs, underscores the significant influence of GQDs on their binding affinity and interaction profiles. These findings suggest the potential utility of GQDs in modulating the efficacy of various compounds against E. coli, thereby contributing to the development of novel therapeutic strategies.
Firstly, the substantial decrease in docking energy observed across all the compounds upon the addition of GQDs highlights the enhanced binding affinity facilitated by GQDs. This suggests that GQDs act as potent enhancers, augmenting the ability of these compounds to interact with E. coli proteins, potentially leading to improved therapeutic outcomes. Secondly, the alterations in hydrogen bond and steric interactions signify the significant reconfiguration in the binding geometry of the compounds within the active site of the E. coli proteins in the presence of GQDs. This structural remodeling, orchestrated by GQDs, likely contributes to a more favorable binding configuration, thereby strengthening the interactions between the compounds and E. coli proteins. Moreover, the consistency of these trends across multiple compounds underscores the robustness and generalizability of the findings. This suggests that the observed synergistic effects between GQDs and the compounds are not limited to specific molecules but represent a broader phenomenon with potential implications across various drug candidates. Overall, these results emphasize the potential of GQDs as versatile adjuvants in enhancing the efficacy of existing compounds against bacterial infections. By elucidating the molecular mechanisms underlying these synergistic interactions, this study paves the way for the development of novel therapeutic strategies that harness the collective power of GQDs and compounds to combat antibiotic-resistant bacterial strains effectively.
4.2. Unveiling molecular docking interactions: GQD and twelve compounds with Bacillus subtilis
The molecular docking interactions for each compound with B. subtilis.Azadirachtin (5281303): azadirachtin exhibited a docking energy of −162.11 kcal mol−1 when considered alone, which significantly decreased to −326.976 kcal mol−1 with the addition of GQDs, indicating a synergistic effect (Fig. 9(a and b)). Initially, azadirachtin formed a hydrogen bonds with Tyr89 and Val149. However, in the presence of GQDs, the interaction pattern shifted to involve Arg215 and Ile257. In the absence of GQDs, the steric interactions included residues such as Ile88, Tyr89, Val149, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, and others (Table S3) (ESI†).
 | ||
| Fig. 9 (a) Top view and (b) side view. Molecular docking interactions of the bacterial receptor (Bacillus subtilis, PDB ID: 6UF6, depicted as a cyan ribbon structure) with graphene quantum dots (GQDs, parrot green color) and 12 bioactive phytochemical compounds extracted from Azadirachta indica leaves. The ball-and-stick representations illustrate the GQD-phytochemical complexes: GQD + gallic acid (dark blue), GQD + catechin (red-brown), GQD + nimbolide (violet), GQD + nimbin (red), GQD + mahmoodin (pink), GQD + salannol (olive green), GQD + margolone (yellow), GQD + beta-sitosterol (cyan), GQD + quercetin (white), GQD + gedunin (orange), GQD + nimbolide (black), and GQD + epicatechin (light violet). This visualization highlights the key interaction sites, providing insights into the potential antibacterial mechanisms of these GQD-phytochemical conjugates. | ||
Beta sitosterol (222284): the docking energy of beta sitosterol decreased from −139.544 kcal mol−1 to −304.41 kcal mol−1 with GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, beta sitosterol formed a hydrogen bond with Arg146. However, in the presence of GQDs, the hydrogen bonds involved Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Ile88, Tyr89, Val149, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, and others.
Catechin (9064): the docking energy of catechin decreased from −102.558 kcal mol−1 to −267.424 kcal mol−1 with GQDs, indicating its improved binding affinity (Fig. 9(a and b)). Initially, catechin forms hydrogen bonds with Tyr89, Tyr147, and Asp256. In the presence of GQDs, the hydrogen bonds shifted to involve Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Tyr89, Val92, Pro144, Tyr147, Asn253, and Asp256. In the presence of GQDs, the interactions involved residues such as Thr86, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, and others (Table S3) (ESI†).
Epicatechin (72276): the docking energy of epicatechin decreased from −93.9443 kcal mol−1 to −258.8103 kcal mol−1 with GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, epicatechin formed hydrogen bonds with Val92, Pro144, Tyr147, and Asn253. In the presence of GQDs, the hydrogen bonds shifted to involve Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Tyr89, Val92, Pro144, Tyr147, Asn253, and Asp256. In the presence of GQDs, the interactions involved residues such as Thr86, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, and others (Table S3) (ESI†).
Gallic Acid (370): the docking energy of gallic acid decreased from −65.9937 kcal mol−1 to −230.86 kcal mol−1 in the presence of GQDs, indicating its improved binding affinity (Fig. 9(a and b)). Initially, gallic acid forms hydrogen bonds with Tyr89, Pro144, Asp146, and Tyr147. In the presence of GQDs, the hydrogen bonds shifted to involve Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Tyr89, Pro144, Asp146, Tyr147, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Thr91, Val145, Val149, Ile218, and others.
Gedunin (12004512): gedunin exhibited a docking energy of −129.049 kcal mol−1 independently, which decreased to −293.915 kcal mol−1 with the addition of GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, gedunin did not form hydrogen bonds. However, in the presence of GQDs, the hydrogen bonds involved Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Gly73, Ile88, Tyr125, Thr130, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others.
Mahmoodin (126566): the docking energy of mahmoodin decreased from −137.888 kcal mol−1 to −302.754 kcal mol−1 with GQDs, indicating its improved binding affinity (Fig. 9(a and b)). Initially, mahmoodin formed hydrogen bonds with Asp146 and Asn253. In the presence of GQDs, the hydrogen bonds involved Thr91, Asp146, Asn253, and Ile257. Without GQDs, the steric interactions included residues such as Thr91, Asp146, Asn253, and Ile257. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others (Table S3) (ESI†).
Margolone (189728): the docking energy of margolone decreased from −100.227 kcal mol−1 to −265.093 kcal mol−1 with GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, margolone formed hydrogen bonds with Ile88 and Val148. In the presence of GQDs, the hydrogen bonds involved Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Ile88, Asp146, Tyr147, Val149, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others.
Nimbin (08058): the docking energy of nimbin decreased from −165.947 kcal mol−1 to −330.813 kcal mol−1 with GQDs, indicating its improved binding affinity (Fig. 9(a and b)). Initially, nimbin formed hydrogen bonds with Tyr89 and Val92. In the presence of GQDs, the hydrogen bonds involved Arg215 and Ile257. Without GQDs, the steric interactions included residues such as Ile88, Tyr89, Val92, Asp146, Tyr147, and Leu304. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others.
Nimbolide (12313376): the docking energy of nimbolide decreased from −166.468 kcal mol−1 to −331.334 kcal mol−1 with GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, nimbolide formed hydrogen bonds with Tyr89 and Val149. In the presence of GQDs, the hydrogen bonds involved Thr91, Val145, Tyr147, and Ile257. Without GQDs, the steric interactions included residues such as Ile88, Tyr89, Val137, Val145, Tyr147, and others. In the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others (Table S3) (ESI†).
Quercetin (5280343): the docking energy of quercetin decreased significantly from −100.728 kcal mol−1 to −265.594 kcal mol−1 in the presence of GQDs, indicating its improved binding affinity (Fig. 9(a and b)). Initially, quercetin formed hydrogen bonds with Tyr89, Pro144, Asp146, and Val149. However, in the presence of GQDs, the hydrogen bonds involved Thr91, Pro144, Val145, Asp146, Tyr147, and Val149. Without GQDs, the steric interactions included residues such as Tyr89, Pro144, Asp146, Tyr147, and Val149. Alternatively, in the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others.
Salannol (157144): the docking energy of salannol decreased from −183.108 kcal mol−1 to −347.974 kcal mol−1 in the presence of GQDs, indicating its enhanced binding affinity (Fig. 9(a and b)). Initially, salannol formed hydrogen bonds with Ile88 and Val149. However, in the presence of GQDs, the hydrogen bonds involved Thr86, Ile88, Tyr89, Thr91, Val145, Val149, and Ile218. In the absence of GQDs, the steric interactions included residues such as Ile88 and Val149. Conversely, in the presence of GQDs, the interactions involved residues such as Thr86, Ile88, Tyr89, Val137, Val145, Tyr147, Phe148, Val149, Gln211, Arg215, Ile218, and others (Table S3) (ESI†).
This comprehensive analysis elucidated the intricate molecular interactions of each compound with B. subtilis, highlighting the significant influence of GQDs on their binding affinity and interaction profiles.
4.3. Unveiling molecular docking interactions: GQD and twelve compounds with Aspergillus niger
Table S4 (ESI†) provides a comprehensive overview of the docking energies and molecular interactions between various compounds and A. niger, both with and without the addition of GQDs. Each compound demonstrated distinct patterns of interaction, shedding light on their potential antibacterial properties and the influence of GQDs.Azadirachtin (PubChem ID: 5281303) exhibited a significant decrease in docking energy from −72.8779 kcal mol−1 to −177.3359 kcal mol−1 with the addition of GQDs, suggesting its enhanced binding affinity (Fig. 10(a and b)). Notably, the hydrogen bonds shifted from primarily involving Asn134, Asn341, and Thr441 to Ser133, Asn431, Thr455, Ser457, and Ser460 in the presence of GQDs. Similarly, the steric interactions also underwent a transformation, engaging a broader range of residues such as Asn431, Pro439, His440, and Ser460 with GQDs (Table S4) (ESI†).
 | ||
| Fig. 10 (a) Top view and (b) side view. Molecular docking interactions of the bacterial receptor (Aspergillus niger, PDB ID: 1UKC, represented as a cyan ribbon structure) with graphene quantum dots (GQDs, parrot green color) and 12 bioactive phytochemicals derived from Azadirachta indica leaf extract. The ball and stick model representations highlight the GQD-phytochemical complexes: GQD + gallic acid (dark blue), GQD + catechin (red-brown), GQD + nimbolide (violet), GQD + nimbin (red), GQD + mahmoodin (pink), GQD + salannol (olive green), GQD + margolone (yellow), GQD + beta-sitosterol (cyan), GQD + quercetin (white), GQD + gedunin (orange), GQD + nimbolide (black), and GQD + epicatechin (light violet). This detailed depiction elucidates the interface interactions critical for exploring the antibacterial and therapeutic applications of these complexes. | ||
Beta sitosterol (PubChem ID: 222284) demonstrated a comparable trend, with a decrease in docking energy from −102.921 kcal mol−1 to −207.379 kcal mol−1 in the presence of GQDs. The hydrogen bond interactions mainly involved Ser210 without GQDs, while GQDs induced interactions with Ser133, Asn431, Thr455, Ser457, and Ser460. The steric interactions also expanded to include residues such as Ser210, His440, and Thr441 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Catechin (PubChem ID: 9064) exhibited a reduction in docking energy from −81.8352 kcal mol−1 to −186.2932 kcal mol−1 in the presence of GQDs. The hydrogen bond formations extended to the Ser133, Asn431, Thr455, Ser457, and Ser460 residues, showcasing a diversified interaction network. Similarly, the steric interactions broadened to encompass residues such as Gly127, Ser133, Asn134, Tyr137, and Pro445 in the presence of GQDs.
Epicatechin (PubChem ID: 72276) also displayed a decrease in docking energy from −75.5531 kcal mol−1 to −180.0111 kcal mol−1 with GQDs. The shift in the hydrogen bond partners to Ser133, Asn431, Thr455, Ser457, and Ser460 indicates strengthened interactions in the presence of GQDs. The steric interactions involved additional residues such as Phe342 and Leu444 with GQDs.
Gallic acid (PubChem ID: 370) exhibited a decrease in docking energy from −60.8918 kcal mol−1 to −165.3498 kcal mol−1 with GQDs. Notably, the hydrogen bonds in the presence of GQDs encompassed a broader range of residues, including Asn341, Asp428, Pro439, and Gly454. Similarly, the steric interactions involve an expanded set of residues such as Tyr137, Asn341, and Pro445 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Gedunin (PubChem ID: 12004512) demonstrated a reduction in docking energy from −91.0284 kcal mol−1 to −195.4864 kcal mol−1 with GQDs. The hydrogen bond formations extended to include Ser457 and Ser460 with GQDs, indicating of enhanced interactions. Also, the steric interactions broadened to encompass residues such as Tyr137, Asp428, and Leu444 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Mahmoodin (PubChem ID: 126566) exhibited a decrease in docking energy from −96.9457 kcal mol−1 to −201.4047 kcal mol−1 with GQDs. The hydrogen bonds extended to involve additional residues such as Ser430 and Tyr462 with GQDs, suggesting augmented interactions. Similarly, the steric interactions encompassed a broader range of residues such as Tyr137, Asp428, and Leu444 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Margolone (PubChem ID: 189728) demonstrated a decrease in docking energy from −77.1733 kcal mol−1 to −181.6313 kcal mol−1 in the presence of GQDs. The hydrogen bond interactions with GQDs involved an expanded set of residues, including Asn341, Asp428, and His440. Similarly, the steric interactions broadened to include residues such as Tyr137, Ser210, and Leu444 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Nimbin (PubChem ID: 08058) exhibited a reduction in docking energy from −114.573 kcal mol−1 to −219.031 kcal mol−1 with GQDs. The hydrogen bonds extended to encompass additional residues such as Asn341, Asp428, and His440 with GQDs, indicating enhanced interactions. The steric interactions also broadened to include residues such as Thr441, Leu444, and Thr453 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Nimbolide (PubChem ID: 12313376) demonstrated a decrease in docking energy from −133.588 kcal mol−1 to −238.046 kcal mol−1 in the presence of GQDs. The hydrogen bond formations extended to involve additional residues such as Thr441 and Ser460 with GQDs, suggesting augmented interactions. The steric interactions also broadened to encompass residues such as Thr441, Leu456, and Ser461 in the presence of GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Quercetin (PubChem ID: 5280343) exhibited a reduction in docking energy from −78.2364 kcal mol−1 to −182.6944 kcal mol−1 in the presence of GQDs. The hydrogen bonds extended to involve additional residues such as Gly127, Ser133, and Ser210 in the presence of GQDs, indicating strengthened interactions. The steric interactions also broadened to include residues such as Tyr137, His440, and Pro445 in the presence of GQDs.
Salannol (PubChem ID: 157144) demonstrated a decrease in docking energy from −99.399 kcal mol−1 to −203.857 kcal mol−1 in the presence of GQDs. The hydrogen bond formations extended to encompass additional residues such as Asn341 and Ser457 with GQDs, suggesting enhanced interactions. The steric interactions also broadened to include residues such as His440, Phe442, and Leu444 with GQDs (Fig. 10(a and b)) and Table S4 (ESI†).
Overall, the incorporation of GQDs led to significant enhancements in the interaction profiles of these compounds with A. niger, indicating their potential for synergistic antibacterial effects and highlighting avenues for further research and development in antifungal therapy.
4.4. Pharmacophore modeling and feature extraction
Fig. S1 and Table S5 (ESI)† present the pharmacophore features of twelve compounds derived from A. indica, commonly known as neem, together with GQDs. Each compound is characterized by its hydrogen bond acceptor, hydrogen bond donor, hydrophobic, ring aromatic, and negative ionizable properties.Azadirachtin, a prominent bioactive compound found in neem, exhibits a high number of hydrogen bond acceptors and donors, indicating its potential for forming strong interactions with biological molecules. It also shows a moderate hydrophobic nature. Beta sitosterol, another compound present in neem, demonstrates balanced hydrogen bonding and hydrophobic characteristics, which can contribute to its diverse biological activities. Catechin and epicatechin, both flavonoids, exhibit significant hydrogen bonding capabilities together with moderate hydrophobicity and aromaticity, suggesting their potential antioxidant properties. Gallic acid, known for its antioxidant and anti-inflammatory effects, displays notable hydrogen bonding and moderate hydrophobicity. Gedunin and nimbin, limonoids found in neem, exhibit varying degrees of hydrogen bonding and hydrophobicity, indicating their potential as anticancer agents. Mahmoodin, a triterpenoid, demonstrates considerable hydrogen bonding and hydrophobic properties, possibly contributing to its pharmacological activities. Margolone, a limonoid, shows moderate hydrogen bonding and significant hydrophobicity, suggesting its potential as an antimalarial agent. Nimbolide, another limonoid, displays similar pharmacophore features to margolone, highlighting its potential therapeutic effects. Quercetin, a flavonoid present in neem, exhibits substantial hydrogen bonding and aromatic properties, indicating its antioxidant and anti-inflammatory potential. Salannol, a neolignan compound, shows significant hydrogen bonding and hydrophobicity, suggesting its potential as an anti-inflammatory agent. Finally, GQDs demonstrate a high degree of hydrogen bonding, hydrophobicity, and moderate hydrogen bond donation and accepting ability, indicating their potential applications in drug delivery and bioimaging. Overall, the pharmacophore features outlined in Fig. S1 and Table S5 (ESI†) provide valuable insights into the diverse biological activities of A. indica compounds and GQDs, offering avenues for their further exploration in drug discovery and development.
4.5. Density functional theory (DFT) and molecular electrostatic potential (MESP) calculations
Azadirachtin (PubChem ID: 5281303): azadirachtin exhibited a moderate band gap energy (0.116233 eV) and a significant dipole moment (2.08915 Debye) before the addition of GQDs. The addition of GQDs induced a noteworthy decrease in its band gap energy (0.00241189 eV), suggesting enhanced electronic communication. The dipole moment remained relatively stable, implying that this compound maintained its polarity despite the structural modifications (Fig. S2 and Table S6 (ESI†)).Beta sitosterol (PubChem ID: 222284): beta sitosterol displays a wider band gap energy (0.193036 eV) compared to azadirachtin and a lower dipole moment (0.855187 Debye). The introduction of GQDs resulted in a minimal change in the band gap energy (0.00098168 eV). This implies that beta sitosterol, with its already substantial band gap, may have limited electronic interactions with GQDs (Fig. S2 and Table S6 (ESI†)).
Catechin (PubChem ID: 9064): catechin shares similarities with azadirachtin in terms of band gap energy (0.149653 eV) and dipole moment (1.04997 Debye). After the addition of GQDs, its band gap energy decreased slightly (0.00240679 eV), indicating the increase in potential for electronic interactions. Alternatively, its dipole moment remained consistent, suggesting that this compound retained its inherent polarity (Fig. S2 and Table S6 (ESI†)).
Epicatechin (PubChem ID: 72276): epicatechin has a moderate band gap energy (0.141289 eV) and a smaller dipole moment (0.306641 Debye) compared to that of azadirachtin and catechin. The addition of GQDs induced a subtle decrease in its band gap energy (0.00137426 eV). Its relatively smaller dipole moment may contribute to less pronounced interactions with GQDs compared to other compounds (Fig. S2 and Table S6 (ESI†)).
Gallic acid (PubChem ID: 370): gallic acid has a larger dipole moment (0.873523 Debye) and moderate band gap energy (0.12184 eV). After the addition of GQDs, a slight decrease in its band gap energy (0.00145018 eV) was observed. The notable dipole moment of gallic acid suggests that it maintains its polarity, possibly influencing its interactions with GQDs (Fig. S2 and Table S6 (ESI†)).
Gedunin (PubChem ID: 12004512): gedunin exhibits a substantial dipole moment (2.58204 Debye) and a moderate band gap energy (0.102568 eV). The addition of GQDs induced a noticeable increase in its band gap energy (0.0051582 eV), indicating its reduced potential for electronic interactions. The significant dipole moment suggests that the inherent polarity of gedunin remained robust (Fig. S2 and Table S6 (ESI†)).
Mahmoodin (PubChem ID: 126566): mahmoodin displays a moderate band gap energy (0.102732 eV) and dipole moment (1.91576 Debye). After the addition of GQDs, its band gap energy slightly increased (0.00175331 eV). This compound retained a considerable dipole moment, indicating that its polarity was preserved during its interaction with GQDs (Fig. S2 and Table S6 (ESI†)).
Margolone (PubChem ID: 189728): margolone has a moderate dipole moment (1.96214 Debye) and band gap energy (0.106498 eV). The addition of GQDs induced a minor change in its band gap energy (0.00213628 eV), suggesting subtle modifications in its electronic interactions. The relatively large dipole moment of this compound indicates its inherent polarity (Fig. S2 and Table S6 (ESI†)).
Nimbin (PubChem ID: 08058): nimbin possesses a smaller dipole moment (0.662956 Debye) and band gap energy (0.0841336 eV) compared to the other compounds. After the addition of GQDs, its band gap energy significantly decreased (0.00306017 eV). This indicates increased electronic communication, despite its smaller dipole moment (Fig. S2 and Table S6 (ESI†)).
Nimbolide (PubChem ID: 12313376): nimbolide has a substantial dipole moment (2.85011 Debye) and a relatively small band gap energy (0.0786596 eV). After the addition of GQDs, its band gap energy increased slightly (0.00334654 eV). The significant dipole moment suggests that nimbolide maintained its polarity, potentially influencing its interactions with GQDs.
Quercetin (PubChem ID: 5280343): quercetin demonstrates moderate band gap energy (0.108986 eV) and dipole moment (2.18812 Debye). The addition of GQDs induced a slight decrease in its band gap energy (0.00801342 eV), suggesting enhanced electronic interactions. The substantial dipole moment indicates that quercetin retained its inherent polarity during its interaction with GQDs (Fig. S2 and Table S6 (ESI†)).
Salannol (PubChem ID: 157144): salannol exhibits a moderate band gap energy (0.132503 eV) and a smaller dipole moment (1.78229 Debye). After the addition of GQDs, its band gap energy slightly decreased (0.00188333 eV), indicating potential modifications in its electronic interactions. The dipole moment of salannol suggests that its inherent polarity remained intact (Fig. S2 and Table S6 (ESI†)).
These detailed discussions highlight the nuanced changes in electronic properties for each compound before and after the addition of graphene quantum dots, providing insights into their potential interactions and reactivity.
(a) Band gap energy. The addition of GQDs generally led to a decrease in the band gap energy of most of the compounds. This suggests an increased potential for electronic communication and improved conductivity in the presence of GQDs.
(b) Dipole moment. The dipole moment, which indicates the polarity of the compounds, remained relatively stable for many of the compounds after the addition of GQDs. This implies that the interactions of the molecules with GQDs do not significantly alter their inherent polarity.
(c) Hardness and softness. The hardness and softness values, which provide insights into the stability and reactivity of compounds, showed minor changes in response to the addition of GQDs. The compounds generally maintained their characteristic hardness and softness.
(d) Electronegativity and electrophilicity. The electronegativity values exhibit slight variations, reflecting subtle changes in the ability of the compounds to attract electrons. The electrophilicity values also show minor adjustments, indicating reactivity of the compounds toward nucleophiles.
(e) Individual compound responses. The compounds with larger initial dipole moments (e.g., gedunin and nimbolide) tended to maintain their polarity after the addition of GQDs. The compounds with moderate band gap energies (e.g., azadirachtin and catechin) showed noticeable changes in their band gap, suggesting enhanced electronic interactions with GQDs. The compounds with smaller dipole moments (e.g., nimbin) experienced significant changes in their band gap energy, indicating potential modifications in electronic properties.
(f) Overall trends. The influence of GQDs on electronic properties appears to be compound-specific, with some compounds exhibiting more pronounced changes than others. Although the band gap energies generally decreased, indicating improved electronic communication, the impact on other properties varied, highlighting the complex nature of the interactions between GQDs and different compounds.
In summary, the addition of GQDs tended to influence the band gap energies, with varying effects on the other electronic properties. The individual responses of each compound highlight the need for a nuanced understanding of the interaction between specific molecules and GQDs, considering factors such as initial polarity, band gap energy, and inherent reactivity.
(a) Band gap energy modification. The overall trend of a decrease in band gap energy after the addition of GQDs indicates the narrowing of the energy gap between the highest occupied molecular orbital (HOMO) and lowest unoccupied molecular orbital (LUMO). This suggests that GQDs facilitate electron transfer processes, potentially enhancing the conductivity and electronic communication within the compounds.
(b) Dipole moment stability. Despite the changes in other electronic properties, the dipole moment generally remained stable for many of the compounds after the addition of GQDs. This suggests that the inherent polarity of the molecules was preserved, indicating that the interactions with GQDs do not alter their overall charge distribution significantly.
(c) Electronegativity and electrophilicity variations. The minor variations in electronegativity and electrophilicity values indicate subtle changes in the ability of the compounds to attract electrons and their reactivity toward nucleophiles. These changes may arise from adjustments in their electron density due to GQD interactions.
(d) Enhanced reactivity. The decrease in band gap energy suggests an increased potential for electronic interactions and reactivity. The compounds with smaller band gaps may exhibit enhanced reactivity, facilitating electron transfer processes and promoting chemical transformations.
(e) Polarity and reactivity. The stability of the dipole moments indicates that the addition of GQDs did not significantly alter the overall polarity of the compounds. However, the compounds with larger initial dipole moments may retain their reactivity toward polar solvents and other molecules, influencing their chemical behavior.
(f) Softness and electrophilicity. Although the hardness and softness values remained relatively stable, the minor changes in electrophilicity suggest subtle variations in the reactivity of the compounds toward nucleophiles. The compounds with higher electrophilicity values may exhibit increased susceptibility to nucleophilic attack, leading to enhanced chemical reactivity.
Overall, the electronic structure analysis indicates that the addition of GQDs influences the electronic properties of the compounds, primarily through modifications in their band gap energies and electron distribution. These changes have implications in the reactivity potential of the compounds, with potential for enhanced electronic communication and chemical transformations in the presence of GQDs. However, the specific effects vary depending on the individual characteristics of the compounds, highlighting the importance of considering compound-specific interactions in understanding reactivity potential.
In the case of the azadirachtin + GQD complex compound, the electrostatic potential is nearly neutral due to the cancellation of the net negative charge of azadirachtin and the net positive charge of GQDs. This suggests weak electrostatic interactions between the components, primarily van der Waals forces. In contrast, the beta-sitosterol + GQD complex compound likely exhibits a negative potential around beta-sitosterol, which is attracted to the positive charges on the edges of GQDs. The hydroxyl group at position 3 of beta-sitosterol contributes to its negative charge, enhancing its interactions with GQDs. Similarly, the catechin + GQD and epicatechin + GQD complex compounds are expected to display negative potentials around catechin and epicatechin, respectively, due to their net negative charges, fostering attractive interactions with the positively charged GQDs. The gallic acid + GQD complex compound is anticipated to have a negative potential, with the negatively charged gallic acid attracted to the positively charged edges of GQDs. However, the specific distribution of potential within the complex can be influenced by solvent interactions and pH variations. In the case of the gedunin + GQD complex compound, the negative potential around gedunin is likely attracted to the positive edges of GQDs, resulting in an overall negative potential. The close contact between gedunin and GQDs may strengthen the electrostatic interactions. In the case of the mahmoodin + GQD complex compound, the neutral or slightly negative potential of mahmoodin may weakly interact with the negatively charged GQDs, potentially attenuating their negative charge, although the overall potential remains predominantly negative.
The margolone + GQD complex compound, where both components carry a net negative charge, may exhibit weak or repulsive interactions, making predictions challenging due to the complexity of the interactions and structural arrangements. In the nimbolide + graphene quantum dot complex compound, the interaction depends on whether GQDs carry a net positive or negative charge. The positive GQDs can result in a near-neutral potential, while negative GQDs may yield a more negative overall potential. In the quercetin + GQD complex compound, the repulsive interactions between negatively charged components may lead to a more negative potential, although the π–π stacking interactions between aromatic rings can contribute to stability. Lastly, in the case of the salannol + GQD complex compound, the repulsive interactions between the negatively charged components may dominate, potentially resulting in a negative overall potential, influenced by solvent effects, pH, and specific salannol structure. Overall, although these predictions provide insights into the electrostatic potentials of the complex compounds, the dynamic nature of the molecular interactions and environmental factors necessitate further investigation for a comprehensive understanding.
4.5.3.1. Comparative analysis of electrostatic potential maps reveals molecular variations in synthesized GQDs + 12 compounds. According to the comparison of the overall results of the various complex compounds with GQDs, several trends and differences emerge regarding their molecular electrostatic potentials. Firstly, although some complexes such as beta-sitosterol, catechin, epicatechin, gallic acid, gedunin, and mahmoodin tend to exhibit negative electrostatic potentials, indicating attractive interactions with the positively charged GQDs, others such as azadirachtin showed a neutral potential due to charge cancellation. This disparity highlights the importance of considering the specific chemical properties of both the compound and the GQDs when assessing their interactions. Moreover, the structural characteristics of the compounds play a significant role in determining their electrostatic potentials. The compounds with multiple hydroxyl groups, such as beta-sitosterol, catechin, epicatechin, gallic acid, gedunin, and mahmoodin, tended to display negative potentials, reflecting the influence of these charged functional groups on their overall charge distribution. Conversely, compounds such as azadirachtin and nimbolide, which lack abundant charged groups, were more prone to yielding neutral or less negative potentials. Additionally, the presence of π–π stacking interactions, as observed in the quercetin + GQD complex, can potentially influence the overall electrostatic potential, providing an additional stabilizing factor despite the repulsive interactions between the negatively charged components. Lastly, the complexity of the interactions is evident in the salannol + GQD complex, where repulsive electrostatic interactions are expected due to the similarly charged components, underscoring the need for a nuanced understanding of the molecular interactions in heterogeneous systems. Overall, although common trends such as negative potentials predominating in the complexes with hydroxyl-rich compounds were observed, the specific characteristics of each compound and its interaction with GQDs dictate the resultant electrostatic potential, emphasizing the inherent complexity in these systems.
4.6. Unveiling unique molecular signatures: electrostatic potential maps in diverse synthesized GQDs + 12 compounds
The novel importance of the overall results is their implication for the design and development of functional materials and systems based on GQDs and complex compounds. Understanding the molecular electrostatic potentials of these complexes offers valuable insights into their stability, reactivity, and potential applications across various fields.One significant implication is in the realm of nanotechnology and materials science, where the ability to manipulate the electrostatic interactions between GQDs and complex compounds can lead to the development of novel materials with tailored properties. By modulating the electrostatic potential, researchers can engineer materials with enhanced stability, improved adsorption capabilities, and optimized electronic properties, thus opening avenues for the fabrication of advanced sensors, catalysts, and energy storage devices. Moreover, the insights gained from the electrostatic characterization of these complexes can inform the design of drug delivery systems and biomedical applications. Understanding how different compounds interact with GQDs on a molecular level can guide the development of targeted drug delivery vehicles with improved biocompatibility and efficacy. Additionally, it can aid in the design of bioimaging agents and theranostic platforms for simultaneous diagnosis and therapy. Furthermore, the ability to predict and control the electrostatic interactions between GQDs and complex compounds has implications for environmental remediation and pollution control. By harnessing these interactions, researchers can develop efficient adsorbents for removing pollutants and contaminants from the air and water sources, contributing to efforts towards sustainable environmental management. Overall, the novel importance of these results is their potential to inspire the creation of innovative technologies and solutions that address pressing societal challenges across multiple domains, ranging from healthcare and materials science to environmental sustainability. By leveraging our understanding of molecular electrostatic potentials, we can unlock new opportunities for technological advancement and societal benefit.
4.7. Absorption, distribution, metabolism, excretion, and toxicity(ADMET) analysis
The results outlined in the ADMET table offer a detailed assessment of the various pharmacokinetic and toxicological parameters associated with the twelve investigated compounds including azadirachtin, beta-sitosterol, catechin, epicatechin, gallic acid, gedunin, mahmoodin, margolone, nimbolide, quercetin, salannol, and GDQs (Table S7 (ESI†)). These parameters play a crucial role in understanding how these compounds interact within biological systems, influencing their toxicological effects and bioavailability. Specifically, this table meticulously delineates parameters such as “ADMET-EXT-CYP2D6,” which provides insights into the association of the compounds with cytochrome P450 2D6 (CYP2D6), a crucial enzyme in drug metabolism. The data showcases a spectrum of values across the compounds, indicating varying levels of potential interactions with this enzyme. Moreover, the “ADMET-EXT-CYP2D6#prediction” column offers valuable insights into the likelihood of specific toxicological outcomes, providing a comprehensive overview of the CYP2D6-related properties of the compounds. Additionally, the evaluation of parameters such as “ADMET-EXT-CYP2D6-Applicability” enriches the understanding of the toxicological profiles of the compounds by assessing whether their properties and operational parameters are consistent with expected ranges concerning CYP2D6. The findings suggest favorable applicability across most compounds, enhancing confidence in their safety profiles. Furthermore, the assessment of hepatotoxicity, absorption levels, plasma protein binding (PPB), AlogP98, and 2D polar surface area (PSA) offers valuable insights into the compounds’ pharmacokinetic properties and potential biological activities. The results indicate varying levels of solubility, BBB penetration, and PPB among the compounds, highlighting their diverse pharmacological profiles. Notably, compounds such as mahmoodin, margolone, nimbolide, and salannol are not predicted to be hepatotoxic, while others such as quercetin and GDQs exhibit potential hepatotoxicity (Table S7 (ESI†)). Additionally, the differences in absorption levels and PPB predictions further underscore the variability in the pharmacokinetic behavior of the compounds. In conclusion, the comprehensive evaluation provided by the ADMET table serves as a valuable tool in guiding the further exploration and development of the evaluated compounds for potential therapeutic applications.Beyond the individual assessments of each compound, the overall results presented in the ADMET table offer novel insights into the collective pharmacokinetic and toxicological landscape of the synthesized metal compounds. By examining the ensemble of parameters across all compounds, patterns and trends emerge that may not be immediately apparent when considering each compound in isolation. One novel importance is the identification of commonalities and differences among the compounds. For instance, although some compounds exhibit similar solubility levels or BBB penetration capabilities, others may show distinct profiles. Understanding these shared characteristics and disparities can inform the development of strategies for optimizing drug formulation or targeting specific biological pathways. Moreover, the collective results shed light on potential synergistic effects or adverse interactions that may arise from the co-administration of multiple compounds. By comprehensively assessing parameters such as CYP2D6 metabolism, hepatotoxicity, and plasma protein binding across all compounds, researchers can anticipate and mitigate potential drug-drug interactions, thereby enhancing the safety and efficacy of combination therapies. Furthermore, the overall results underscore the importance of considering multiple factors in drug development and design. Although solubility and BBB penetration are crucial for drug bioavailability and central nervous system targeting, factors such as metabolism and hepatotoxicity play pivotal roles in drug safety and tolerability. By integrating diverse parameters into a unified analysis, researchers can gain a more holistic understanding of the pharmacological properties and potential risks associated with compounds, guiding informed decision-making throughout the drug development process. In essence, the novel importance of the overall results is their capacity to provide a comprehensive and integrative perspective on the pharmacokinetic and toxicological characteristics of synthesized metal compounds, offering valuable guidance for optimizing their therapeutic potential and minimizing potential adverse effects in clinical applications.
5. Experimental sections
5.1. Plant material identification
Azadirachta indica (locally known as Neem) leaves were collected from the Herbal Garden of Maharshi Dayanand University, Rohtak. This was confirmed and certified by CSIR-NIScPR. New Delhi (Specimen voucher number Ref. No. NIScPR/RHMD/2022/4021-22-1). The collected leaves were washed to eliminate dust particles and dried for 12 days in the shade to eradicate moisture. Subsequently, the dehydrated leaves were crushed into a fine powder.The green synthesis of GQDs was carried out using the methodology reported by Kumawat et al., with certain modifications.18,28 Briefly, 20 g of Azadirachta indica leaf powder was dissolved in 200 mL of absolute ethanol and stirred continuously for 4 h using a magnetic stirrer at room temperature. Then, the extract was centrifuged at 5000 rpm for 12 min to get a clear solvent. The solvent was concentrated using a rotary evaporator and the resulting thick slurry was combined with a 2 mL of Milli Q water, and further, the mixture was heated in a domestic microwave oven (900 W) for 6 min. Then, the residue was dispersed using 99.9% ethanol and filtered through a 0.22 μm syringe filter to extract pure GQDs. The dried powder was preserved at −4 °C for future use. A UV-Visible spectrophotometer, FTIR, XRD, TEM, SEM, and zeta potential analyzer were used to characterize the synthesized GQDs.
5.2. Biological experiments
In vitro, DPPH free radical scavenging assay, agar well diffusion, disc diffusion and microplate reader assays were carried out against the synthesized GQDs and ethanolic leaf extract in an effort to identify a significant agent for infectious diseases.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μg mL−1). After 30 min of incubation in the dark, the absorbance at 517 nm was measured using ascorbic acid as a standard.
μg mL−1). After 30 min of incubation in the dark, the absorbance at 517 nm was measured using ascorbic acid as a standard.5.3. Computational methodology
Armed with the insights gleaned from CASTp, our journey into the realm of docking analysis begins. With discerning eyes, we judiciously selected the most promising binding pockets unearthed by CASTp, prioritizing those endowed with a significant area and volume. These designated pockets, identified as potential active sites, serve as the focal points for further exploration and investigation. In the nexus between the precision of CASTp and our strategic selection, the promise of unraveling the mysteries of the protein binding sites is significant. With each designated pocket, we embark on a journey of discovery, unraveling the intricate dance between proteins and ligands, inching closer to unlocking the secrets of biological interaction.
5.4. Statistical analysis
The experiments were performed in triplicate. The obtained results were subjected to statistical analysis using the GraphPad Prism software. The experimental data were processed by one-way analysis of variance (ANOVA) with Tukey test at a 95% confidence interval. Each value represents the mean![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ±
± ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SD, wherein the P value represents the level of significant changes. The XRD, UV-visible spectra, and histogram were plotted in “Origin Pro 2023”.
SD, wherein the P value represents the level of significant changes. The XRD, UV-visible spectra, and histogram were plotted in “Origin Pro 2023”.
6. Conclusions
This study demonstrated the efficient production of GQDs using plant extracts. The approach advocated here prioritizes the reduction of harmful chemicals by providing an improved substitute to current industry practice for the synthesis of nanomaterials. The size, shape and morphology of the phyto-mediated GQDs were confirmed by HRTEM and SEM. The size distribution graph from TEM revealed that the particle size of GQDs was less than 10 nm. Finally, the antioxidant activity of the phyto-mediated GQDs was tested and they showed better results than the crude extract, paving the way to explore their potential applications such as bioimaging, cytotoxicity, and beyond. In conclusion, the synthesis of GQDs using compounds extracted from Azadirachta indica leaves demonstrated promising antioxidant, antibacterial, and antifungal activities. These findings were observed both in vitro and through in silico analyses utilizing various computational techniques such as molecular docking, pharmacophore modelling, density functional theory (DFT), and ADMET prediction. The in vitro experiments revealed that the synthesized GQDs + 12 compounds exhibited significant antioxidant properties, which was attributed to the presence of bioactive compounds derived from A. indica leaves. These antioxidant properties have the potential to combat oxidative stress-related diseases and conditions effectively. Moreover, the GQDs + 12 compounds displayed antibacterial and antifungal activities in vitro, suggesting their potential as novel antimicrobial agents. These activities were likely due to the interaction of GQDs + 12 compounds with the bacterial and fungal cell membranes, disrupting their integrity and leading to microbial inhibition. The computational analyses (Table 2), including molecular docking, pharmacophore modelling, DFT calculations, and ADMET prediction, provided valuable insights into the molecular interactions, energetics, and pharmacokinetic properties of the GQDs and their constituent compounds. These computational approaches supported the experimental findings and enhanced our understanding of the bioactivity mechanisms of the GQDs + 12 compounds. In summary, the combination of experimental and computational approaches highlighted the potential of A. indica leaf-derived GQDs + 12 compounds as versatile nanomaterials with potent antioxidant, antibacterial, and antifungal properties. The antibacterial studies further demonstrated the efficacy of GQDs, with their inhibition zones reaching up to 14.1 ± 0.2 mm against Bacillus subtilis at a concentration of 100 μg mL−1, indicating their superior performance compared to the leaf extract. These results are consistent with the computational predictions that GQDs would enhance binding interactions with bacterial proteins, suggesting their utility in combating bacterial infections.| Compound | Target/analysis | Key metric without GQDs | Key metric with GQDs | Major observations/changes | ADMET properties |
|---|---|---|---|---|---|
| Azadirachtin | E. coli (Docking) | Binding energy: −95.21 kcal mol−1 | Binding energy: −124.77 kcal mol−1 | Enhanced hydrogen bonding (Thr81, His85) | Good GI absorption; low BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.116 eV | Band gap: 0.002 eV | Significant electronic interaction improvement | Optimal molar refractivity; high bioavailability | |
| MESP | Neutral potential | Weak negative potential | Dominated by van der Waals interactions | ||
| Beta-sitosterol | E. coli (Docking) | Binding energy: −101.87 kcal mol−1 | Binding energy: −131.43 kcal mol−1 | Shifted interaction with Asp107, Asn193 | Good GI absorption; poor BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.193 eV | Band gap: 0.001 eV | Minor electronic changes, stable polarity | Moderate water solubility, suitable for oral formulations | |
| MESP | Negative potential | Strong negative potential | Hydroxyl group contributes to electrostatic attraction | ||
| Catechin | A. niger (Docking) | Binding energy: −81.84 kcal mol−1 | Binding energy: −186.29 kcal mol−1 | Strong H-bonding (Ser133, Thr455) | Good GI absorption; low BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.150 eV | Band gap: 0.002 eV | Moderate electronic interaction with GQDs | High bioavailability; strong antioxidant potential | |
| MESP | Negative potential | Enhanced negative potential | Attractive electrostatic interactions | ||
| Epicatechin | A. niger (Docking) | Binding energy: −75.55 kcal mol−1 | Binding energy: −180.01 kcal mol−1 | Enhanced interactions (Ser133, Leu444) | Good GI absorption; low BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.141 eV | Band gap: 0.001 eV | Subtle electronic enhancements | Moderate bioavailability; antioxidant properties | |
| MESP | Negative potential | Negative potential | Attractive π–π stacking interactions | ||
| Gallic acid | A. niger (Docking) | Binding energy: −60.89 kcal mol−1 | Binding energy: −165.35 kcal mol−1 | Expanded hydrogen bonding network with Thr455 | High GI absorption; no BBB permeability; low skin penetration |
| DFT | Band gap: 0.122 eV | Band gap: 0.001 eV | Subtle electronic modification, retains polarity | High bioavailability; no major toxicity predicted | |
| MESP | Negative potential | Negative potential | Strong interaction at edges of GQDs | ||
| Gedunin | B. subtilis (Docking) | Binding energy: −129.05 kcal mol−1 | Binding energy: −293.92 kcal mol−1 | Enhanced steric interactions | Moderate GI absorption; no BBB permeability; low toxicity risk |
| DFT | Band gap: 0.103 eV | Band gap: 0.005 eV | Reduced electronic reactivity with GQDs | Suitable for oral use; promising anticancer potential | |
| Mahmoodin | B. subtilis (Docking) | Binding energy: −137.89 kcal mol−1 | Binding energy: −302.75 kcal mol−1 | Strong H-bonding with Asp146, Thr91 | Good GI absorption; no BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.103 eV | Band gap: 0.002 eV | Improved conductivity with GQDs | Potential anti-inflammatory properties | |
| Margolone | A. niger (Docking) | Binding energy: −77.17 kcal mol−1 | Binding energy: −181.63 kcal mol−1 | Enhanced steric reconfiguration | Moderate GI absorption; no BBB permeability; no toxicity predicted |
| DFT | Band gap: 0.106 eV | Band gap: 0.002 eV | Subtle enhancements in electron transfer | Antimalarial activity potential | |
| Nimbin | E. coli (Docking) | Binding energy: −165.95 kcal mol−1 | Binding energy: −330.81 kcal mol−1 | Significant steric interaction changes | Moderate GI absorption; no BBB permeability; low toxicity risk |
| DFT | Band gap: 0.084 eV | Band gap: 0.003 eV | Increased reactivity and reduced band gap | Promising antimicrobial properties | |
| Nimbolide | B. subtilis (Docking) | Binding energy: −166.47 kcal mol−1 | Binding energy: −331.33 kcal mol−1 | Notable steric reorientation with residues | Good GI absorption; moderate BBB permeability; predicted hepatotoxicity risk |
| DFT | Band gap: 0.079 eV | Band gap: 0.003 eV | Increased reactivity with reduced band gap | Acceptable skin permeability; potential liver enzyme inhibition | |
| MESP | Neutral potential | Slightly negative potential | Polarity influenced by interaction geometry | ||
| Quercetin | A. niger (Docking) | Binding energy: −78.24 kcal mol−1 | Binding energy: −182.69 kcal mol−1 | π–π stacking contributes to stability | High GI absorption; moderate BBB permeability; low toxicity risk. |
| DFT | Band gap: 0.109 eV | Band gap: 0.008 eV | Enhanced conductivity due to reduced band gap | High bioavailability; suitable for oral formulations | |
| MESP | Negative potential | Negative potential | Strong hydrogen bonding with surface residues | ||
| Salannol | A. niger (Docking) | Binding energy: −99.40 kcal mol−1 | Binding energy: −203.86 kcal mol−1 | Enhanced steric and H-bonding interactions | Moderate GI absorption; no BBB permeability; low toxicity risk |
| DFT | Band gap: 0.133 eV | Band gap: 0.002 eV | Retained electronic stability | Anti-inflammatory potential |
Future research efforts should focus on elucidating the specific underlying molecular mechanisms for their bioactivity, optimizing their synthesis and formulation for various applications.
Author contributions
Pooja Kadyan: writing – original draft (experimental), methodology, data curation, investigation characterization, validation, performed experiment. Manish Kumar: writing – review & editing (Biological work), validation, visualization, and formal analysis. Aisha Tufail: writing the original Draft (in silico work), visualization, validation. Andrea Ragusa: formal analysis and validation. Sudhir Kumar Kataria: supervision (Experimental), conceptualization, writing – review & editing (experimental), validation and formal analysis. Amit Dubey: supervision (computational), writing the original draft (in silico work and Biological work), conceptualization, software (molecular docking, DFT, MESP, pharmacophore modeling and ADMET), visualization, methodology, writing – review & editing, validation and formal analysis.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work received no external funding. The authors are grateful to Aryabhata Central Instrumentation Laboratory, Maharshi Dayanand University, Rohtak, India for providing facilities for Zeta potential, UV-Visible spectrophotometer, “Origin 2023” Software and Graph Pad Prism 8.2 software. Also, the author Pooja Kadyan [File no. 09/0382(12494)/2021-EMR-I] is thankful to CSIR, India, for the award of Senior Research Fellowship. This research was supported by “Tecnopolo per la medicina di precisione” (TecnoMed Puglia)–Regione Puglia: DGR n.2117 del 21/11/2018 (CUP:B84I18000540002), and “Tecnopolo di Nanotecnologia e Fotonicaperla medicina di precisione” (TECNOMED)–FISR/MIUR-CNR:delibera CIPE n. 3449 del 7–08–2017 (CUP: B83B17000010001).References
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 7, 309–318 CrossRef.
- T. J. Masho, P. T. Arasu, R. F. Bogale, E. AmareZerrefa and S. Ramamurthy, Res. Chem., 2024, 2, 11–101369 Search PubMed.
- R. B. Asamoah, E. Annan, B. Mensah, P. Nbelayim, V. Apalangya, B. Onwona-Agyeman and A. Yaya, Adv. Mater. Sci. Eng., 2020, 1, 1–8 Search PubMed.
- K. Habiba, D. P. Bracho-Rincon and J. A. Gonzalez-Feliciano, et al. , Appl. Mater. Today, 2015, 1, 80–87 CrossRef.
- M. A. Kohanski, D. J. Dwyer and J. J. Collins, Nat. Rev. Microbiol., 2010, 8, 423–435 CrossRef CAS.
- S. Liu, T. H. Zeng and M. Hofmann, et al. , ACS Nano, 2011, 5, 6971–6980 CrossRef CAS PubMed.
- K. D. Patel, R. K. Singh and H. W. Kim, Mater. Horiz., 2019, 6, 434–469 RSC.
- M. Gaur, C. Misra, A. B. Yadav, S. Swaroop, F. Ó. Maolmhuaidh, M. Bechelany and A. Barhoum, Materials, 2021, 14, 5978 CrossRef CAS.
- O. Zaytseva and G. Neumann, Chem. Biol. Technol. Agric., 2016, 3, 1–26 Search PubMed.
- J. Deng, M. Li and Y. Wang, Green Chem., 2016, 18, 4824–4854 RSC.
- D. Pan, J. Zhang, Z. Li and M. Wu, Adv. Mater., 2010, 22, 734–738 CrossRef CAS.
- L. Centeno, J. Romero-García, C. Alvarado-Canché and C. Gallardo-Vega, et al. , Sens. Bio-Sens. Res., 2021, 1, 32–100412 Search PubMed.
- P. Kumar, C. Dhand, N. Dwivedi and S. Singh, et al. , Renewable Sustainable Energy Rev., 2022, 1, 157–111993 Search PubMed.
- P. Kadyan, R. Malik, S. Bhatia and A. Al Harrasi, et al. , J. Nanotechnol., 2023, 1, 1–26 Search PubMed.
- C. Xiong, J. Xu, Q. Han, C. Qin, L. Dai and Y. Ni, Cellulose, 2021, 28, 10359–10372 CrossRef CAS.
- L. Othman and R. M. Abdel-Massih, Front. Microbiol., 2019, 10, 453561 Search PubMed.
- V. Sharma, J. Pharm. Innov., 2018, 7, 648–650 Search PubMed.
- P. Kadyan Thillai, P. Arasu and S. K. Kataria, Int. J. Biomater., 2024, 1, 2626006 Search PubMed.
- W. H. Danial, M. Abdullah, M. A. A. Bakar and M. S. Yunos, et al. , Opt. Mater., 2022, 132, 112853 CrossRef CAS.
- R. V. Khose, G. Chakraborty, M. P. Bondarde and P. H. Wadekar, et al. , New J. Chem., 2021, 45, 4617–4625 RSC.
- W. Shi, H. Fan, S. Ai and L. Zhu, New J. Chem., 2015, 39, 7054–7059 RSC.
- Y. Yan, S. Manickam, E. Lester, T. Wu and C. H. Pang, Ultrason. Sonochem., 2021, 73, 105519 CrossRef CAS PubMed.
- S. C. Gupta, S. Prasad., A. K. Tyagi, A. B. Kunnumakkara and B. B. Aggarwal, Phytomedicine, 2017, 34, 14–20 CrossRef CAS.
- S. Saleem, G. Muhammad, M. A. Hussain and S. N. Bukhari, Phytother. Res., 2018, 7, 1241 CrossRef PubMed.
- M. O. Daniyan and O. T. Ojo, J. Mol. Graphics Modell., 2019, 87, 144 CrossRef CAS PubMed.
- S. Siddiqui, S. Faizi, B. S. Siddiqui and Ghiasuddin, J. Nat. Prod., 1992, 55, 303 CrossRef CAS.
- R. Murugan and T. Parimelazhagan, J. King Saud Univ., Sci., 2014, 26, 267–275 CrossRef.
- M. K. Kumawat, M. Thakur, R. B. Gurung and R. Srivastava, ACS Sustainable Chem. Eng., 2017, 5, 1382–1391 CrossRef CAS.
- Y. Qiu, Y. Zhang, M. Yu, X. Li, Y. Wang, Z. Ma and S. Liu, Small, 2024, 2310087 CrossRef CAS PubMed.
- Z. Fang, Q. Zhou, W. Zhang and J. Wang, et al. , Materials, 2023, 16, 7583 CrossRef CAS.
- D. Hajra and S. Paul, Pharm. Res., 2018, 10(4), 347–353 CAS.
- M. I. Okeke, C. U. Iroegbu, E. N. Eze, A. S. Okoli and C. O. Esimone, J. Ethnopharmacol., 2001, 78, 119–127 CrossRef CAS PubMed.
- C. Tshangana, M. Chabalala, A. Muleja, E. Nxumalo and B. Mamba, J. Env. Chem. Eng., 2020, 4, 103930 CrossRef.
- J. Dundas, Z. Ouyang, J. Tseng, A. Binkowski, Y. Turpaz and J. Liang, Nucleic Acid Res., 2006, 1, 116 CrossRef.
- W. Tian, C. Chen, X. Lei, J. Zhao and J. Liang, Nucleic Acids Res., 2018, 46, 363–367 CrossRef.
- G. M. Morris, R. Huey, W. Lindstrom, M. F. Sanner, R. K. Belew, D. S. Goodsell and A. J. Olson, J. Comput. Chem., 2009, 30, 2785–2791 CrossRef CAS PubMed.
- A. Dubey, A. Facchiano, P. W. Ramteke and A. Marabotti, Future Med. Chem., 2016, 8, 841–851 CrossRef CAS.
- A. Dubey, A. Marabotti, P. W. Ramteke and A. Facchiano, Biochem. Biophys. Res. Commun., 2016, 473, 449–454 CrossRef CAS.
- A. Dubey, M. M. Alawi, T. A. Alandijany, I. M. Alsaady, S. A. Altwaim, A. K. Sahoo, V. D. Dwivedi and E. I. Azhar, Viruses, 2023, 15, 251 CrossRef CAS.
- X. Qiu, C. A. Janson, W. W. Smith, M. Head, J. Lonsdale and A. K. Konstantinidis, J. Mol. Biol., 2001, 307, 341–356 CrossRef CAS.
- F. K. Li, F. I. Rosell, R. T. Gale, J. P. Simorre, E. D. Brown and N. C. Strynadka, J. Biol. Chem., 2020, 295, 2629–2639 CrossRef CAS.
- M. V. Keniya, M. Sabherwal, R. K. Wilson and M. A. Woods, et al. , Antimicrob. Agents Chemother., 2018, 62, 10–1128 Search PubMed.
- S. Bharadwaj, A. Dubey, N. K. Kamboj, A. K. Sahoo, S. G. Kang and U. Yadava, Sci. Rep., 2021, 11, 10169 CrossRef CAS.
- A. Dubey, S. Dotolo, P. W. Ramteke, A. Facchiano and A. Marabotti, Biomolecules, 2018, 9, 5 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The Supporting Information is available free of charge on the Materials Advances publications website, including three supporting figures, and seven supporting tables. See DOI: https://doi.org/10.1039/d4ma00843j |
| This journal is © The Royal Society of Chemistry 2025 |