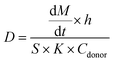Open Access Article
Open Access ArticleA gut–brain axis on-a-chip platform for drug testing challenged with donepezil†
Francesca
Fanizza
 a,
Simone
Perottoni
a,
Simone
Perottoni
 a,
Lucia
Boeri
a,
Francesca
Donnaloja
a,
Francesca
Negro
a,
Francesca
Pugli
a,
Gianluigi
Forloni
b,
Carmen
Giordano‡
a,
Lucia
Boeri
a,
Francesca
Donnaloja
a,
Francesca
Negro
a,
Francesca
Pugli
a,
Gianluigi
Forloni
b,
Carmen
Giordano‡
 *a and
Diego
Albani‡
*a and
Diego
Albani‡
 b
b
aDepartment of Chemistry, Materials and Chemical Engineering ‘Giulio Natta’, Politecnico di Milano, Milan, Italy. E-mail: carmen.giordano@polimi.it
bDepartment of Neuroscience, Istituto di Ricerche Farmacologiche Mario Negri IRCCS, Milan, Italy
First published on 12th February 2025
Abstract
Current drug development pipelines are time-consuming and prone to a significant percentage of failure, partially due to the limited availability of advanced human preclinical models able to better replicate the in vivo complexity of our body. To contribute to an advancement in this field, we developed an in vitro multi-organ-on-a-chip system, that we named PEGASO platform, which enables the dynamic culturing of human cell-based models relevant for drug testing. The PEGASO platform is composed of five independent connected units, which are based on a previously developed millifluidic organ-on-a-chip device (MINERVA 2.0), hosting human primary cells and iPSC-derived cells recapitulating key biological features of the gut, immune system, liver, blood–brain-barrier and brain that were fluidically connected and challenged to model the physiological passage of donepezil, a drug prescribed for Alzheimer's disease. The nutrient medium flow rate of the connected units was tuned to obtain suitable oxygenation and shear stress values for the cells cultured in dynamic condition. A computational model was at first developed to simulate donepezil transport within the platform and to assess the drug amount reaching the last organ-on-a-chip. Then, we demonstrated that after 24 hours of donepezil administration, the drug was actually transported though the cell-based models of the platform which in turn were found viable and functional. Donepezil efficacy was confirmed by the decreased acetylcholinesterase activity at the brain model and by the increased expression of a donepezil-relevant multi-drug transporter (P-gp). Overall, the PEGASO platform is an innovative in vitro tool for drug screening and personalized medicine applications which holds the potential to be translated to preclinical research and improve new drug development pipelines.
Introduction
The development of new drugs is a time-consuming and costly process. Despite the rigorous preclinical assessment of drug safety and efficacy, the success rate of clinical trials remains overall low, particularly in the field of neurodegenerative disorders. Preclinical trials with animal models often fail to yield the same results in human trials. Reports indicate that only a portion of drugs entering human clinical trials successfully pass phase II and phase III, with a total probability of around 9% for a drug to move from phase I to final approval.1 Additionally, a significant percentage of drug failures in the market are attributed to unanticipated toxicity in patients, underscoring the need for complementary strategies besides animal testing.2One alternative lies in the development of human cell-based microphysiological systems as organ-on-a-chip (OOC). These devices can replicate in vitro the key microarchitectures of human organs and mimic human metabolism together with resembling physiopathological features. Moreover, OOCs can be linked together in a multi-OOC platform to analyze multi-organ crosstalk, drug–organ interactions and secondary drug toxicity.3,4
However, most OOC studies have largely been based on 2D culture of non-human-derived cell lines, or featuring immortalized or primary human cells, lacking 3D complexity and having just partial physiologic relevance. To contribute to the OOC field and offer a tool to address these shortcomings, we developed the MINERVA 2.0. device:5,6 a 3D-printed millifluidic OOC consisting of two hemi-chambers, apical and basal, interfaced through a porous membrane of a commercial Transwell-like insert. The peculiar design of the millifluidic MINERVA 2.0 allows the accommodation of millimetric 3D models as hydrogel-based scaffolds and the serial connection of single devices as plug-and-play units to build human OOC platforms. Here we present a multi-organ platform that we named PEGASO (PErsonalisation of induced pluripotent stem cell-based body-on-chips for in vitro testinG of therApeutic agentS against brain disOrders). It connects five MINERVA 2.0. devices under a common circulating medium, hosting primary or iPSC-derived human cells and recapitulating key features of organs such as the gut, immune system, liver, blood–brain barrier (BBB) and brain, to allow the resembling of the physiological passage of a drug approved for treating neurological diseases. Oral administration is the most preferred drug therapy route for donepezil since it has compliance for patients.7 The PEGASO platform aims at modeling the key passages of the orally-administered donepezil from the gut to the brain (Fig. 1). In the body, donepezil is first absorbed across the intestinal barrier and then the intestinal endothelium which represents the gateway to the body's systemic distribution, reaching peak plasma levels in a few hours.8 Along this path, the monocytes from the “blood circulation” have the potential to interact with the drug leading to its modification and/or the secretion of mediators which can freely diffuse back across the intestinal barrier. Donepezil is mainly excreted unchanged in the urine, but it also undergoes hepatic metabolism8 in the liver getting biotransformed before re-entering the blood circulation. The drug next reaches the BBB, a selective permeability barrier regulating fluid extravasation and drug and metabolite diffusion, finally targeting the brain parenchyma.9
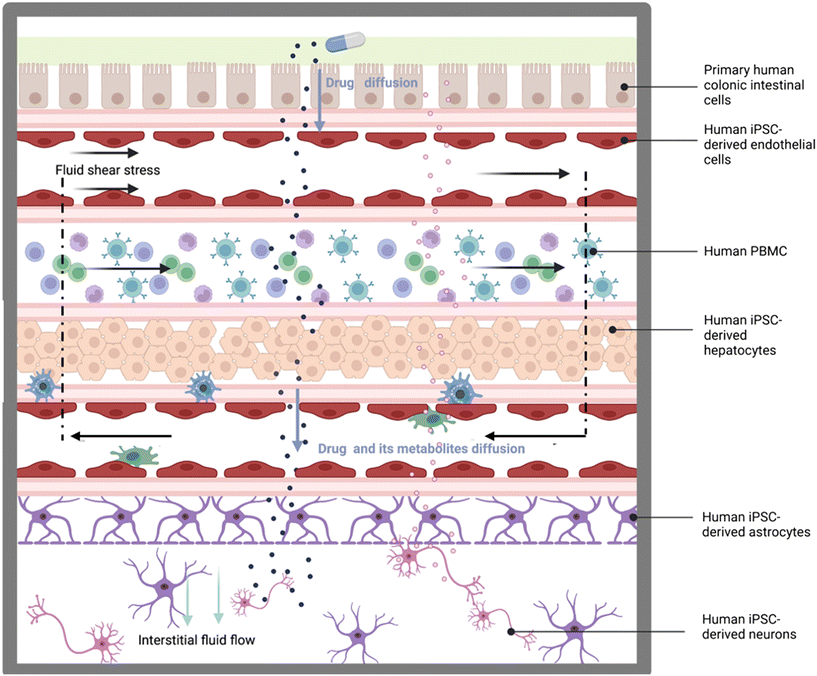 | ||
| Fig. 1 Schematic representation of the main physical and biological features that are implemented in the PEGASO multi-OOC platform (created with Biorender®). | ||
We aimed at recreating the key features of this pathway by Transwell-based models of cell barriers, the immune system and a co-culture of brain parenchymatic cells (neurons and astrocytes) connected under the action of tuned fluid flow.
The nutrient medium flow rates within the OOC were adjusted to ensure optimal oxygenation and shear stress for the cultured cells. Additionally, a computational model was developed to analyze the drug transport and predict the drug amount reaching the final OOC.
After 24 hours from donepezil administration the cells were viable and able to express physiologic functionalities indicating that the multi-OOC can be used to test basic drug pharmacokinetics and biological targeting. These results present a new in vitro tool for drug screening and potentially personalized medicine which will be of benefit to the community in improving the drug development pipeline and reducing drug failure in the field of neurodegenerative disorders.
Results
Design of the PEGASO multi-OOC platform
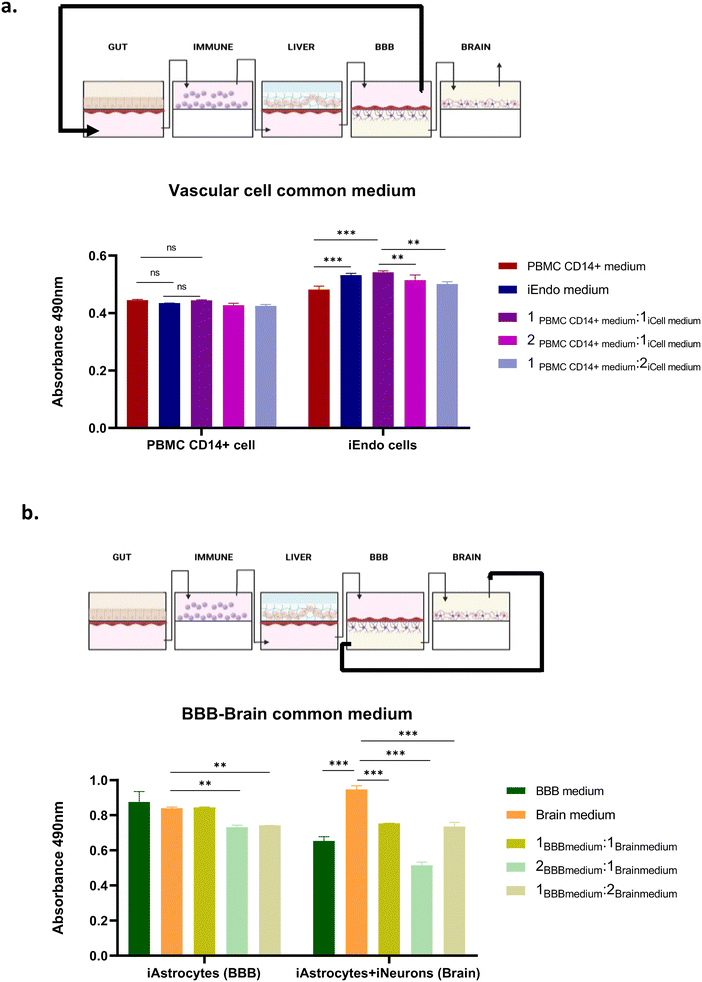
The PEGASO multi-OOC platform was designed to test the toxicity and efficacy of drugs for neurological diseases.The platform is composed of five OOCs (based on MINERVA 2.0) representing the gut, immune, liver, BBB and brain, linked together through fluidic connections (Fig. 2).
The gut OOC mimicking drug absorption was linked to the immune OOC which was coupled to the liver OOC simulating metabolism that in turn was connected to the BBB OOC, which was linked to the brain OOC, the final target of the Alzheimer's disease (AD) drug donepezil.
The peculiar design of the MINERVA 2.0 device, composed of two culture chambers, enables physical separation of two cellular constructs which communicate through the OOC porous membrane.5
The first device of the platform, namely the gut OOC, contained primary human intestinal cells facing iPSC-derived endothelial cells; the immune OOC was populated with human PBMC CD14+ cells; the liver OOC hosted a collagen–poly(ethylene)glycol (COLL–PEG) hydrogel encapsulating iPSC-derived hepatocytes interconnected with iPSC-derived endothelial cells; the BBB OOC was based on iPSC-derived endothelial cells and astrocytes while the last device, the brain OOC, contained a co-culture of iPSC-derived astrocytes and neurons.
To ensure the survival and differentiation state of human primary cells and iPSC-derived cells, a vascular common culture medium was recirculated from the basal gut compartment to the apical BBB to nourish the endothelial cells and the monocytes while the BBB–brain common culture medium recirculated between the basal BBB compartment and brain compartment to perfuse astrocytes and neurons. Differently, the intestinal cells and the hepatocytes were maintained in specific cell media recirculated in a closed loop.
The integration of recirculation of culture media allowed the elimination of waste products and medium enrichment with secreting factors beyond simulating the blood circulation.
The glass windows present at the base of each OOC allow the user to monitor daily the morphology of the cultured cells (Fig. 2b and d).
Optimization of the culturing media for the PEGASO platform
Culture conditions for the multi-OOC may differ considerably from the single-OOCs due to the growth requirements of the different cell types.To select the shared cell culture media to be circulated within the endothelial-lined vascular chambers, referred to as “vascular cell culture media” (VCCM) (Fig. 3a), and within the chambers hosting astrocytes and neurons, named “BBB–brain cell culture media” (BBCCM) (Fig. 3b), we conducted experiments with different single cell media.
In particular, we tested different media compositions onto cell-seeded inserts cultured in static condition before the insertion into the OOC.
The VCCM has to circulate through the basal chamber of the gut, the immune, the basal chamber of the liver and the apical chamber of the BBB. So, it was crucial for this medium to maintain the viability of both endothelial cells and PBMC CD14+ cells. Consequently, we cultured both cell types with different media compositions in static condition, and we found that the culture media composed of endothelial and monocyte media in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio provided optimal support for the viability of both cell types (Fig. 3a).
1 ratio provided optimal support for the viability of both cell types (Fig. 3a).
The same analysis was performed for the BBCCM that has to circulate through the basal chamber of the BBB and the brain and supports astrocytes alone and in co-culture with neurons. The viability assessment by the MTS assay onto cell-seeded inserts in static condition indicated that the brain medium ensured higher viability for both the astrocytes and neurons (Fig. 3b).
Computational model to implement the PEGASO multi-OOC platform
The flow rate for the dynamic culturing was selected by balancing the needs of each OOC. Thirty μL min−1 was able to guarantee suitable shear stress (SS) values and oxygen levels for all the cells (Table 1).11–19Fig. 4 shows the SS profiles and the oxygen concentration level for each OOC.| Model | Chamber | Numerical model | In vivo values | ||
|---|---|---|---|---|---|
| Shear stress (SS) (mPa) | Oxygen concentration (mol m−3) | Shear stress (SS) (mPa) | Oxygen concentration (mol m−3) | ||
| Gut | Apical (0.5 mm): intestinal cells | 0.745 | 0.199 | >0.67 (ref. 10) | 0.004–0.1 (ref. 11) |
| Basal (0.5 mm): endothelial cells | 0.913 | 300–9500 (ref. 12 and 13) | |||
| Immune | Apical (2 mm): PBMC CD14+ monocytes | 0.062 | 0.177 | 300–9500 (ref. 12 and 13) | ∼0.053 (ref. 14) |
| Liver | Apical (2 mm): hepatocytes | 0.022 | 0.169 | <0.2 (ref. 15 and 16) | >0.021 (ref. 17) |
| Basal (0.5 mm): endothelial cells | 0.913 | 300–9500 (ref. 12 and 13) | |||
| BBB | Apical (2 mm): endothelial cells | 0.062 | 0.151 | 0.5–2.3 (ref. 18) | ∼0.05 (ref. 19) |
| Basal (0.5 mm): astrocytes | 0.913 | ||||
| Brain | Apical (2 mm): astrocytes and neurons | 0.062 | 0.144 | 0.5–2.3 (ref. 18) | ∼0.05 (ref. 19) |
In detail, the intestinal cells experienced SS able to guarantee values higher than 0.67 mPa, which is the minimum value required to initiate epithelial cell differentiation.10
On the other hand, the PBMC CD14+ monocytes were subjected to negligible SS (average 0.062 mPa) which resembled the interstitial fluid flow conditions.
To minimize SS on hepatocytes and mimic the condition of the native liver sinusoid, a hydrogel encapsulating the hepatocytes reduced the average SS to 0.022 mPa, closer to the physiologic values,20 while for the common flow rate of 30 μL min−1, the SS reached average values in the BBB and brain of 0.913 mPa and 0.062 mPa, respectively.
However, with regard to the vascular compartment, the SS values ranging from 0.062 (in the apical chamber) to 0.913 mPa (in the basal chamber) were below the values experience by endothelial cells of the small veins.21
Fig. 4b shows the simulated oxygen concentration decaying from 0.2 mol m−3 in the gut OOC to 0.14 mol m−3 in the brain OOC. The theoretical oxygen levels in all the OOCs remained positive thus confirming that there was no oxygen deficiency.
Computational model of drug transport through the PEGASO multi-OOC platform
To demonstrate the applicability of the PEGASO multi-OOC platform as a drug screening tool, we assessed the response to donepezil, a drug for AD treatment. We modeled at first drug transport with COMSOL Multiphysics.We developed a computational model to predict donepezil diffusion though the platform to establish if an initial dose of 200 μM, injected into the gut, was sufficient to reach the final organ, namely the brain keeping a biologically active concentration.
We have considered neither drug uptake by cells or absorption by the device material nor drug metabolism by the liver OOC. The drug diffusion coefficients were measured experimentally for all the cell models and are shown in Table 3.
Fig. 5 shows the results of the computational model of donepezil diffusion through the platform.
After 24 hours from drug administration in the gut, a decay in donepezil concentration occurred from the basal chamber of the gut to the brain where it reached 141 μM. Counter-current flows for the apical and basal chambers chosen for the gut–immune–liver connection to increase mass transport allowed a slight increase in donepezil concentration.
Differently, the co-current flows of the liver–BBB–brain, chosen to reduce the tubing obstacle, determined a drop in concentration.
Moreover, a mean drug concentration of 20 μM reached the exit of the apical chamber of the liver after having crossed the basal chamber, the membrane and the hydrogel encapsulating the hepatocytes.
Experimental assessment of donepezil impact on cell viability and drug transporter expression through the platform
After having numerically assessed that an initial dose of 200 μM was sufficient to reach the brain compartment with a clinically relevant dose, we proceeded with the experimental tests.We measured through a three-way valve for media collection the donepezil concentration in the brain OOC 24 hours from drug injection (Fig. 6a). A discrepancy in drug levels between the experimental measurement and the numerical model was observed, as we found 45 μM donepezil in the brain compared to the 141 μM simulated.
To assess if donepezil induced unwanted toxic effects, we analyzed lactate dehydrogenase (LDH) levels in the cell culture media 24 hours from drug administration and by microscopic inspection which was feasible through the MINERVA 2.0 transparent bottom allowing live tissue imaging. Fig. 6b and c show the low levels of LDH (below 5%) in both untreated and treated samples indicating the absence of cytotoxicity.
The viability of all the cells in the platform after drug injection was evaluated with live/dead assay and confocal microscopy (Fig. 7). Cells were found viable 24 hours after donepezil administration as shown by the calcein green stained viable cells and no significant differences were detected.
Once the lack of evident cell toxicity was demonstrated, a biological correlation to donepezil passage through the platform was assessed by evaluating the P-glycoprotein (P-gp) expression, an ATP-dependent drug transport protein which can actively transport hydrophobic drugs such as donepezil.22
Since P-gp is predominantly found in the intestine, the liver and the BBB,23 we evaluated its expression by immunofluorescence in gut, liver and BBB OOC perfused samples treated and not-treated with donepezil (Fig. 8). An increased P-gp signal was observed in the donepezil-treated platform.
Evaluation of donepezil biologic effects on individual OOC of the platform
We then evaluated key biological features of individual OOC after donepezil exposure.The integrity of the intestinal and BBB barrier was assessed with TEER measurements. No difference in TEER values was observed for both the intestinal–endothelial barrier and the BBB, thus indicating that the drug presence did not affect the barrier functionality (Fig. 9a and d).
Hepatocyte functionality was assessed by measuring albumin and urea production (Fig. 9b and c). Albumin concentration in the cell culture media was comparable between the untreated and donepezil-treated conditions, while urea secretion showed a significant increase after drug exposure.
Finally, we evaluated the inhibitory effect of donepezil on acetylcholinesterase (AChE) activity of neurons in the brain OOC (Fig. 9d). As expected, the AChE activity greatly diminished in the treated platform.
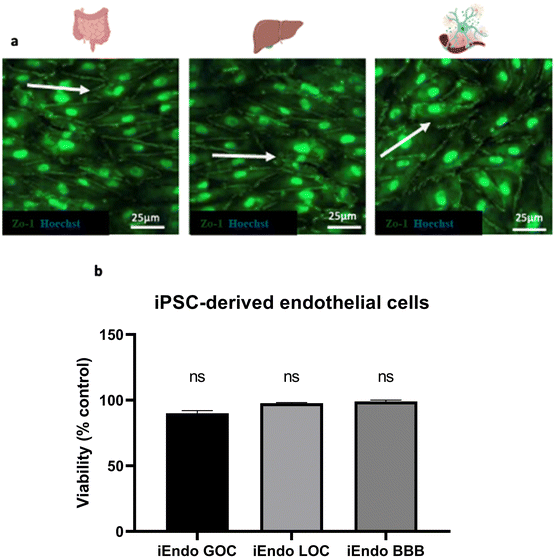
In the end, we evaluated the viability of endothelial cells present in the basal chamber of the gut and liver and in the apical chamber of the BBB after donepezil administration under continuous recirculating medium (Fig. 10). The endothelial cells in the gut, liver and BBB OOCs seemed to align along the direction of fluid flow, as evidenced by zonulin-1 (ZO-1) staining, and were viable as demonstrated by the MTS assay.
Discussion
Organ-on-a-chip (OOC) devices have been shown to recapitulate human organ-level physiology in healthy and pathological conditions as well as drug-induced toxicities.24 While these approaches highlight the value of single OOC, pharmacokinetic and pharmacodynamic analysis and multi-organ toxicity studies require multi-OOC or whole-body systems.Here we have described the design and biological characterization of the PEGASO platform, a novel human multi-OOC solution for drug testing and pharmacokinetic studies.
The PEGASO platform relies on five compartments based on millifluidic devices, hydraulically connected in series and loaded with human cell models, representing most of the key biological systems and pathways involved in pharmacokinetics of drugs for neurological disease.
Drug absorption, metabolism and distribution were introduced in the multi-OOC platform by fluidically coupling relevant OOC models of the gut,6 immune system, liver,5 BBB and brain through vascular endothelium-lined channels.
The presence of these channels enabled the multi-OOC system to be perfused with a common vascular cell culture medium (VCCM) and to mimic the systemic circulation, a desirable feature to measure drug concentration and facilitate extrapolation to human systems.
Others have already reported fluidic linking of individual OOC leading to multiple microfluidic devices, but focusing on a single type of parenchymatic organ (such as liver, lung, or kidney epithelium) and not including a separate vascular compartment.25–28
It is worth underscoring that the modular design of the platform enables the addition or removal of an OOC and isolation of organ subsystems which would be more difficult in animal models. In addition, the placing into the device of the seeded Transwell-like insert after their maturation in static condition avoids using enzymatic dissociation for cell harvesting from the cell culture support and re-plating inside the device, which could impair for example iPSC differentiation and survival, particularly for neuronal cell types.
The administration of the drug into the gut OOC takes place through a 3-way valve and can be changed to other modes of delivery, for instance, intravenous by dosing to the immune OOC or to the interconnected vascular medium.
To optimize the flow rate for the multi-OOC perfusion we set up a numerical model to evaluate the oxygen supply and shear stress (SS) profile for the cells in the OOCs serially connected.
With a flow rate of 30 μL min−1 oxygen concentrations decayed from the gut (20%) to the brain (14%) but kept positive indicating that there was no oxygen deficiency.
However, these values are above the physiological oxygen levels of human organs and tissues and were near the ambient oxygen levels. More investigation about this point is warranted, for instance by measuring the local level of oxidative stress, even if the lack of cell toxicity does not support a major issue to this respect.
Differently, the SS generated by fluidic flow was already demonstrated to be able to induce intestinal epithelial differentiation,29 mimic the physiological conditions of the human sinusoid,30 and resemble interstitial fluid flow conditions together with avoiding monocyte and astrocyte damage and detachment.31–33
With regard to the vascular compartment, the physiological SS found in cerebral capillaries is believed to be approximately 0.3–2 Pa.34 However, the range of SS endothelial cells are generally subjected to in OOCs is wide, ranging from a few mPa to values greater than 1 Pa.35–37 The values experienced by endothelial cells in our platform were below the in vivo values of the capillary beds21 due to the need of selecting a common input flowrate for multiple organ models. Nevertheless, our tests showed endothelial cells viable and functional and apparently able to elongate along the direction of the fluid flow similar to native tissue. To have a clearer picture of endothelial cell functionality more investigation should be performed, for instance by assessing the transcriptomic profile of these cells under different flow rates in our device. Moreover, future engineering of the platform would be possible in order to apply larger input flow rates in the vascular compartment, which, for example, were necessary for Harding et al.38 to reach 1.2 Pa SS on endothelial cells cultured in a Transwell-based OOC device, as our platform is.
In the view of exploiting the PEGASO platform for drug testing studies, we developed a computational model to predict the pharmacokinetic profile of the Alzheimer's disease prescribed drug donepezil through the whole multi-OOC platform.
We experimentally obtained parameters such as drug diffusion coefficients and partition coefficients for all the models, and we investigated the drug diffusion mechanism occurring though the hydrogel in the liver.
After 24 hours, starting from 200 μM donepezil concentration in the gut OOC, a decay turned out from the gut to the brain OOC model, where a value of 141 μM was predicted. To validate the numerical model, we performed experimental tests to measure the donepezil concentration in the brain OOC cell culture media. The drug concentration found was 45 μM, a biologically relevant dose but lower than the simulated numerical one.39–41 We hypothesize that the reduction of the donepezil concentration in the experimental tests was due to hepatic metabolism or other biological interactions, for instance with cell media components, not considered into the numerical model.
We recognize the importance of measuring the active metabolite concentrations in the system as they contribute to the overall therapeutic and adverse effects of a drug. Therefore, it will be relevant to quantify not only donepezil but also the levels of donepezil principal active metabolite, 6-OH donepezil, using liquid chromatography to distinguish between the biological effects caused by donepezil itself and those induced by its metabolite.42
As for the biological effect of donepezil exposure in the platform, the live/dead and LDH assays showed the absence of cytotoxicity after drug injection and a high viability of over 80% in all the OOCs of the platform.
This finding was in agreement with several studies indicating that donepezil at a dose of 200 μM is safe.43
To get a clearer picture of the donepezil effect on our platform, we evaluated hepatocyte functionality by measuring albumin and urea levels. Albumin concentration in the cell culture media was comparable between the untreated and donepezil-treated conditions, while the urea secretion increased after drug injection. A hypothesis being considered is that the presence of the drug stimulated liver activity with increased protein turnover, which could lead to an increase in protein breakdown and subsequently result in higher urea production.44
We also demonstrated that the TEER levels of gut and BBB models were unaffected by donepezil.
Moreover, a key point was to show that our platform was able to predict not only donepezil transport but also its effect on brain cells. To this respect, the enzymatic inhibitory effect of donepezil was assessed by investigating the acetylcholinesterase (AChE) activity in the brain OOC. AChE enzymatic activity greatly diminished in the multi-OOC treated platform with respect to the control untreated samples. This result demonstrated both the effective transport of donepezil or its metabolites through all the OOCs and the suitability of the platform to recapitulate donepezil efficacy as an AChE inhibitor.
The passage of donepezil through the cell barriers and the circulating system was further confirmed by the correlated expression of P-glycoprotein (P-gp), a multidrug efflux pump, in the intestinal, hepatic and BBB layers of the multi-OOC.45
To the best of our knowledge, the PEGASO platform presented in this work is the first multi-OOC platform developed to test drugs against neurodegenerative diseases.4 In fact, few studies focused on developing tools to study neurological diseases such as Alzheimer's and Parkinson's diseases but with single OOC such as the BBB or the brain and not with multi-OOC platforms.46,47 The results described in this work contribute to multi-OOC field development through the integration of multiple organ systems in mutual communication, thus being potentially suitable as a support of available animal models to investigate systemic scenarios, such as drug metabolism, PD/PK, and drug toxicity. Regarding the latter, our model represents a technological step forward starting from reported two-organ liver–brain OOCs for neurotoxicity evaluation.3 Moreover, the integration into the platform of human cells, such as primary and iPSC-derived cells, increases the physiological relevance of the model. In a scenario where only a few iPSC-based multi-OOCs have been reported to host fully differentiated and functional models48 we were able to connect five functional iPSC-based and/or primary cell-based OOCs. In this study we have implemented cell and barrier models of representative players potentially involved in orally administered drug routes. In particular, we carried out a first characterization of the immune cells hosted in the platform in the view of obtaining an immunocompetent multi-OOCs. In fact, it is believed that incorporating immune cells into OOC systems is a necessary step to get platforms capable of being efficient in detecting drug-induced toxicity, also taking into account the action of the immune system.49 This would be relevant in the light of literature evidence on donepezil modulatory effects on the immune system.50,51
We acknowledge the limitations of the multi-OOC PEGASO platform lacking other important organs, such as kidney or heart, on which donepezil or drugs in general may cause undesired effects.52–54 The inclusion of a kidney OOC would be essential for excretion modelling and evaluation of optimal drug dosages. However, the versatile platform design will allow us to implement the hosted in vitro models according to the upcoming literature and further application needs. Additionally, we have to implement our protocols for long-term drug exposure assays.
Conclusion
This study presents the PEGASO platform, a multiorgan microphysiological system hosting human cells suitable for drug screening studies. Human cells of five tissues/organs, challenged in the platform with donepezil, were viable and functional, indicating that the multi-OOC can be used to test drugs for PD/PK purposes.The developed platform is modular making it cost-affordable and versatile, especially with iPSC-derived cells. At present, our model features commercial iPSC-derived cells and human primary cells but may be improved by inserting patient-specific iPSC-derived cells to develop a “patient-on-a-chip” for testing personalized therapies. This advancement holds potential for further exploitation in the neurodegenerative disease field studded by a high drug discovery failure rate.
Materials and methods
Millifluidic device
MINERVA 2.0 is a 3D printed in nylon millifluidic OOC device compatible with commercial Transwell-like cell culture inserts.5,6Briefly, the millifluidic device consists of two nylon 3D-printed components assembled with a snap-fit closure system and a 12-well Transwell-like insert having a PET membrane with a pore diameter of 0.4 μm and density of 2 × 106 pores per cm2. Inside the device there are two hemi-chambers, apical and basal, interfaced through a porous membrane of the Transwell-like insert. The apical chamber can be 0.5 mm or 2 mm high, while the basal chamber is 0.5 mm. To implement the PEGASO multi-OOC platform using MINERVA 2.0, our device hosts Luer-lock connectors coupled to millifluidic channels with diameters of 0.5–1 mm.
MINERVA 2.0. devices were sterilized by UV rays (SafeMate cabinet) for 10 h or with hydrogen peroxide (V-PRO® 60 Low Temperature Sterilization System).
The gut OOC had a 0.5 mm high apical chamber and a basal chamber of 0.5 mm, while the immune OOC, liver OOC, BBB OOC and brain OOC had a 2 mm high apical chamber and a basal chamber of 0.5 mm.
Cell models
Endothelial cells were maintained in endothelial medium composed of VascuLife VEGF Medium (Lifeline Cell Technologies, Frederick, MD) supplemented with the complete growth factors.
Intestinal cells were maintained in Epithelial Cell Growth Medium® (iXCells Biotechnologies).
The cell-seeded inserts were maintained 7 days in static conditions before being assembled once inserted in the MINERVA 2.0.
The cell-seeded inserts were maintained for 4 hours in static conditions before being assembled once inserted in the MINERVA 2.0.
The cell-seeded inserts were maintained for 5 days in static conditions before being assembled once inserted in the MINERVA 2.0.
The membrane sides of endothelial cells and astrocytes were pre-coated with 10 μg ml−1 laminin (Sigma-Aldrich) and 30 μg ml−1 fibronectin (PromoCell), respectively. All the culture inserts were incubated at 37 °C and 5% CO2 for 7 days with medium renewal every 2 days.
Endothelial cells were maintained in endothelial medium composed of VascuLife VEGF Medium (Lifeline Cell Technologies, Frederick, MD) supplemented with the complete growth factors included in the kits. Here, only 10 mL of the glutamine solution was added per 500 mL of media and 50 mL of the CDI-provided supplement replaced the VascuLife FBS component.
Astrocytes were maintained in DMEM/F12, HEPES (ThermoFisher), FBS hyclone (GE Healthcare Life Sciences), and N2 supplement 10× (ThermoFisher).
The cell-seeded inserts were maintained for 7 days in static conditions before being assembled once inserted in the MINERVA 2.0.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) astrocytes = 4
astrocytes = 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), were seeded on the upper side of the insert membrane. Cell culture inserts were pre-coated with poly-ornithine (Sigma-Aldrich) incubated overnight at 37 °C, followed by 1 hour incubation at 37 °C with 10 μg ml−1 laminin (Roche). After seeding, all the cell culture inserts were incubated at 37 °C and 5% CO2 with medium renewal every 2 days.
1), were seeded on the upper side of the insert membrane. Cell culture inserts were pre-coated with poly-ornithine (Sigma-Aldrich) incubated overnight at 37 °C, followed by 1 hour incubation at 37 °C with 10 μg ml−1 laminin (Roche). After seeding, all the cell culture inserts were incubated at 37 °C and 5% CO2 with medium renewal every 2 days.
Astrocytes and neurons were maintained in BrainPhys Neuronal Medium (STEMCELL), iCell Neural Supplement B (CDI), iCell Nervous System Supplement (CDI), N-2 Supplement 100× (ThermoFisher), and penicillin–streptomycin 100× (ThermoFisher).
The cell-seeded inserts were maintained for 7 days in static conditions before being assembled once inserted in the MINERVA 2.0.
Organ-on-a-chip cell culture media
The cells after maturation in static condition in cell-specific culture media were transferred into the OOC, connected in series to shape the PEGASO platform and perfused with the cell culture media as reported in Table 2. In brief, the apical chamber of the gut and liver OOCs was perfused with intestinal medium and hepatocyte medium, respectively. The lower chamber of the gut OOC, the immune OOC together with the lower chamber of the liver OOC and the upper chamber of the BBB OOC were perfused with a vascular cell common medium (VCCM), while the lower chamber of the BBB OOC and the brain OOC was perfused with BBB–brain common medium (BBCM).| OOC | |||||
|---|---|---|---|---|---|
| Gut | Immune | Liver | BBB | Brain | |
| Apical chamber | Intestinal medium | Vascular common medium | Hepatocytes medium | Vascular common medium | BBB–brain common medium |
| Basal chamber | Vascular common medium | — | Vascular common medium | BBB–brain common medium | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of Blood Cell Culture Medium® (iXCells Biotechnologies) and endothelial cell medium composed of VascuLife VEGF Medium (Lifeline Cell Technologies, Frederick, MD) supplemented with the complete growth factors.
1 of Blood Cell Culture Medium® (iXCells Biotechnologies) and endothelial cell medium composed of VascuLife VEGF Medium (Lifeline Cell Technologies, Frederick, MD) supplemented with the complete growth factors.
Donepezil hydrochloride
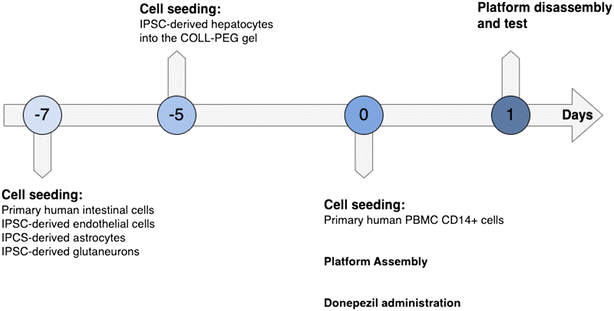
Donepezil hydrochloride (cat. #D6821, Merck) was dissolved in dimethyl sulfoxide (DMSO) to a stock concentration of 10 mM and diluted in intestinal medium to 200 μM final concentration.PEGASO platform dynamic culturing
The cell seeded inserts after 5 or 7 days of maturation were perfused in the MINERVA 2.0 for 24 hours maintaining the flow rate established for the dynamic culture of single devices (30 μl min−1) (Fig. 11).Two hours after platform assembly, donepezil hydrochloride 200 μM was dissolved into the apical compartment of the gut OOC. The PEGASO platform treated with DMSO, the vehicle of donepezil, was used as the control. After 24 hours of dynamic perfusion and donepezil administration the platform was disassembled, and biological tests were performed.
For each experiment, three platforms treated with donepezil and three with DMSO (control) were used.
Numerical evaluation of the PEGASO platform
To evaluate the shear stresses, the oxygen concentration profiles, and the amount of donepezil diffused into all the organs of the PEGASO platform connected in series, we ran Multiphysics computational simulations with the software COMSOL Multiphysics®. The geometry of the MINERVA 2.0 apical and basal chambers was extracted from the internal space of the culture chamber using Solidworks® software. The gut OOC has an apical chamber 0.5 mm high and a basal chamber 0.5 mm high, while the immune, liver, BBB and brain OOC devices have an apical chamber of 2 mm and a basal chamber of 0.5 mm. The simulations were performed with perfusion in counter-current configuration for the gut–immune–liver–BBB connections and in-current configuration for the BBB–brain connection.Fluid velocity vector u was determined using the Navier–Stokes equation in the stationary condition:
| ρ(u·∇)u = ∇·(−pI + K) + F |
| ρ ∇·u = 0 |
| K = μ(∇u + (∇u)T) |
Shear stresses were calculated as:
We estimated oxygen distribution with the equation of mass transport of diluted species:
| ∇·(−Di·∇ci) + u·∇ci = Ri |
For the 2D cell models (gut, immune, BBB and brain) we set the reaction term as null, and we assumed a homogeneous cell distribution through the membranes. The oxygen consumption rate was calculated as:  , where Vmax is the oxygen cell consumption rate, d is the cell density and Vol is the volume.
, where Vmax is the oxygen cell consumption rate, d is the cell density and Vol is the volume.
For the 3D cell model (liver), it was assumed a reaction term function of the local oxygen concentration according to the Michaelis–Menten kinetics:  , where Vmax is the maximum molar consumption rate, c is the local oxygen concentration and Km is the Michaelis–Menten constant.
, where Vmax is the maximum molar consumption rate, c is the local oxygen concentration and Km is the Michaelis–Menten constant.
We estimated donepezil distribution with the equation of mass transport of diluted species (∇·(−Di·∇ci) + u·∇ci = Ri) in which the reaction term was set as 0.
We assumed an initial AD drug donepezil equal to 0.2 mol m−3 at the inlet of the apical chamber of the gut OOC.
The drug diffusion coefficient for all the organs of the platform was estimated experimentally and calculated with the Fick's law equation for a diffusion cell:
To run the simulations, we chose the “fine” element size for mesh building.
All the characteristic parameters are summarized in Table 3.
| Parameter | Value |
|---|---|
| Inlet oxygen concentration | 0.195 mol m−3 |
| Outlet pressure | 0 Pa |
| Oxygen diffusion coefficient | 2 × 10−9 m2 s−1 |
| Condition at the wall | No slip condition |
| Density of the culture media at 37 °C | 1000 kg m−3 |
| Dynamic viscosity of the culture media | 8.1 × 10−4 Pa s |
| COLL–PEG gel permeability | 6.45 × 10−17 m2 |
| COLL–PEG gel porosity | 0.93 |
| Donepezil diffusion coefficient in the GOC | 1.11 × 10−9 m2 s−1 |
| Donepezil diffusion coefficient in the IMMUNE | 1.87 × 10−9 m2 s−1 |
| Donepezil diffusion coefficient in the LOC | 4.12 × 10−11 m2 s−1 |
| Donepezil diffusion coefficient in the BBBOC | 1.05 × 10−9 m2 s−1 |
| Donepezil diffusion coefficient in the BOC | 0.98 × 10−9 m2 s−1 |
| Intestinal cell oxygen consumption | J c = 1.14 × 10−8 mol m−2 s−1 (ref. 56) |
| Endothelial cell oxygen consumption | J c = 1.46 × 10−9 mol m−2 s−1 (ref. 57) |
| PBMC CD14+ oxygen consumption | J c = 3 × 10−8 mol m−2 s−1 (ref. 58) |
| Hepatocyte oxygen consumption | R c = 5.53 × 10−4 mol m−3 s−1 (ref. 17) |
| Astrocyte oxygen consumption | J c = 1.56 × 10−13 mol m−2 s−1 |
| Astrocyte + neuron oxygen consumption | J c = 9.37 × 10−13 mol m−2 s−1 |
Donepezil measurement
Donepezil concentration was assessed by a fluorimetric method. Medium from the gut and brain OOCs was collected at 24 h and analyzed through spectroscopy to quantify the donepezil amount. The analysis was made by means of a fluorescence microplate reader (Tecan Infinite M200) at 233 nm as the excitation wavelength and 400 nm as the emission wavelength. A standard curve with the purified drug was created and test donepezil concentration was extrapolated by the best-fitting curve analysis.Cell viability assays
To investigate cell viability a LIVE/DEAD test (Promokine) was performed. After 24 h of perfusion, devices were disassembled, and the medium was substituted with 250 μL of LIVE/DEAD solution (20 μL of ethidium bromide and 5 μL of calcein AM in 10 mL of medium without phenol red). After 1 hour of incubation, samples were observed under a confocal microscope with a 20× objective. For each sample, we collected six random acquisitions (three in the insert center and three at the periphery)The live/dead images were postprocessed in ImageJ software, and viable and dead cells were manually counted. The viability was calculated as follows:
The following equation was used to calculate the percentual toxicity of donepezil treated samples with respect to control samples:
After 24 h of static culture, 80 μL of CellTiter 96 Aqueous One Solution Reagent was pipetted into the Transwell-like insert containing the sample in 500 μL of medium.
After 2 h of incubation at 37 °C in a humidified 5% CO2 atmosphere, the amount of soluble formazan produced by cellular reduction of MTS was measured with a plate reader (490 nm absorbance).
Transepithelial electrical resistance (TEER)
After 24 h of perfusion, the gut and BBB devices were disassembled, the cell culture inserts moved into a well plate and both apical (0.5 mL) and basal (1 mL) media substituted with fresh medium. TEER was measured using EVOM (World Precision Instruments, USA) coupled with a chopstick-like electrode. Cell layer resistance (Rmeasured; Ω) was calculated by placing the shorter electrode in the apical compartment of the inserts and the longer one in contact with the plate. TEER (Ω cm2) was calculated as follows:| TEER = (Rmeasured − Rblank) × Membrane Area |
Urea and albumin production
Albumin and urea production were analyzed to assess the liver-specific functions by collecting the medium after 24 hours of culture. Albumin concentration in the medium was determined by an enzyme-linked immunosorbent assay (ELISA) kit (Bethyl Laboratories). Urea concentration was evaluated by using a urea assay kit (Sigma-Aldrich) following the manufacturer's protocol. Levels of albumin and urea secretion were quantified and normalized per hour to the sample volume and seeded cells. All collected samples of the cell supernatant were kept frozen at −80 °C prior to performing the assays. Samples were thawed to room temperature and prepared according to the manufacturer's protocol.Acetylcholinesterase activity
The activity of AChE, which is responsible for the degradation of acetylcholine in tissue samples, was determined for neurons using an acetylcholinesterase colorimetric commercial kit by Abcam (ab138871) and was expressed in U mg−1 protein. The assay was performed according to the manufacturer's instructions, and the activity was measured at 410 nm using a spectrophotometer. Levels of AChE were quantified and normalized on untreated samples.Immunofluorescence staining and data analysis
The immunofluorescence staining was performed in a 12-well plate comparing donepezil-treated and untreated perfusion cell culture conditions. Samples were rinsed three times with PBS and fixed with 4% paraformaldehyde for 20 minutes at room temperature. After washing three times with PBS, cells were permeabilized and blocked for non-specific antigens with 0.1% Triton X-100 and 4% bovine serum albumin or normal goat serum, depending on the primary antibody used. After 40 minutes, samples were incubated overnight at 4 °C with the primary antibody![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) rabbit anti-ZO1 polyclonal antibody (1
rabbit anti-ZO1 polyclonal antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, ThermoFisher). The day after samples were washed with PBS three times for 5 minutes and were incubated with anti-mouse or -rabbit Alexa Fluor 488 secondary antibody solution (Abcam) for 1 hour. After two washing steps, cell nuclei were labeled with Hoechst 33342 (1
50, ThermoFisher). The day after samples were washed with PBS three times for 5 minutes and were incubated with anti-mouse or -rabbit Alexa Fluor 488 secondary antibody solution (Abcam) for 1 hour. After two washing steps, cell nuclei were labeled with Hoechst 33342 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in PBS, Invitrogen, Thermo Fisher Scientific) for 10 minutes at room temperature. Finally, samples were washed two more times and mounted using Fluorsave reagent (EDM Millipore). Fluorescence images were acquired by Olympus Fluoview. The images were post-processed using the open-source software ImageJ (https://imagej.nih.gov/ij/index.html, USA).
000 in PBS, Invitrogen, Thermo Fisher Scientific) for 10 minutes at room temperature. Finally, samples were washed two more times and mounted using Fluorsave reagent (EDM Millipore). Fluorescence images were acquired by Olympus Fluoview. The images were post-processed using the open-source software ImageJ (https://imagej.nih.gov/ij/index.html, USA).
In more detail, the quantification of the P-gp fluorescence signal was based on the recognition of the region of interest within the acquisition area. From the segmentation of the ROI it was possible to obtain parameters like integrated density, mean gray value and area. The total fluorescence of an ROI within the acquisition area was calculated with the following formula:
| TF = integrated density − (total area × mean gray background value). |
Data availability
The raw data supporting this article are included in a Supplementary Information folder which is stored by the corresponding author of this manuscript and are available upon request.Author contributions
FF executed the experiments, developed the platform and drafted the manuscript; SP contributed to OOC and platform development; LB contributed to biological protocol implementation. FF, SP and FD developed computational analysis. FN and FP contributed to the experiment's execution. GF made the equipment at the Dept. Neuroscience, Istituto di Ricerche Farmacologiche Mario Negri IRCCS, available for the experiments. DA and CG supervised the present study and related project activities. All authors discussed the results and approved the final manuscript.Conflicts of interest
The authors have no conflicts to disclose.Acknowledgements
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: this work was funded by the Ministero dell'Istruzione, dell'Università e della Ricerca (MIUR) under the FARE 2019 program (project code R18WWPCXLY-PEGASO) and by the European Research Council (ERC) under the European Union's Horizon 2020 research and innovation program (Grant agreement No. 724734-MINERVA). The work reflects only the authors' views and the agency is not responsible for any use that may be made of the information contained.Notes and references
- M. T. Wasylewski, K. Strzebonska, M. Koperny, M. Polak, J. Kimmelman and M. Waligora, Clinical development success rates and social value of pediatric Phase 1 trials in oncology, PLoS One, 2020, 15(6), e0234911 CrossRef CAS PubMed.
- D. Sun, W. Gao, H. Hu and S. Zhou, Why 90% of clinical drug development fails and how to improve it?, Acta Pharm. Sin. B, 2022, 12(7), 3049–3062 CrossRef CAS PubMed.
- N. Picollet-D'hahan, A. Zuchowska, I. Lemeunier and S. Le Gac, Multiorgan-on-a-chip: a systemic approach to model and decipher inter-organ communication, Trends Biotechnol., 2021, 39(8), 788–810 CrossRef PubMed.
- F. Fanizza, M. Campanile, G. Forloni, C. Giordano and D. Albani, Induced pluripotent stem cell-based organ-on-a-chip as personalized drug screening tools: A focus on neurodegenerative disorders, J. Tissue Eng., 2022, 13, 20417314221095340 Search PubMed.
- F. Fanizza, L. Boeri, F. Donnaloja, S. Perottoni, G. Forloni and C. Giordano, et al., Development of an Induced Pluripotent Stem Cell-Based Liver-on-a-Chip Assessed with an Alzheimer's Disease Drug, ACS Biomater. Sci. Eng., 2023, 9(7), 4415–4430 CrossRef CAS PubMed.
- F. Donnaloja, L. Izzo, M. Campanile, S. Perottoni, L. Boeri and F. Fanizza, et al., Human gut epithelium features recapitulated in MINERVA 2.0 millifluidic organ-on-a-chip device, APL Bioeng., 2023, 7(3), 36117 CrossRef CAS PubMed.
- Z. Q. Zhao, B. Z. Chen, X. P. Zhang, H. Zheng and X. D. Guo, An Update on the Routes for the Delivery of Donepezil, Mol. Pharmaceutics, 2021, 18(7), 2482–2494 CrossRef CAS PubMed.
- B. Seltzer, Donepezil: a review, Expert Opin. Drug Metab. Toxicol., 2005, 1(3), 527–536 CrossRef CAS PubMed.
- P. Papakyriakopoulou, E. Balafas, G. Colombo, D. M. Rekkas, N. Kostomitsopoulos and G. Valsami, Nose-to-Brain delivery of donepezil hydrochloride following administration of an HPMC-Me-β-CD-PEG400 nasal film in mice, J. Drug Delivery Sci. Technol., 2023, 84, 104463 CrossRef CAS.
- H. J. Kim, D. Huh, G. Hamilton and D. E. Ingber, Human gut-on-a-chip inhabited by microbial flora that experiences intestinal peristalsis-like motions and flow, Lab Chip, 2012, 12(12), 2165–2174 RSC.
- R. Singhal and Y. M. Shah, Oxygen battle in the gut: Hypoxia and hypoxia-inducible factors in metabolic and inflammatory responses in the intestine, J. Biol. Chem., 2020, 295(30), 10493–10505 CrossRef CAS PubMed.
- A. G. Koutsiaris, S. V. Tachmitzi, N. Batis, M. G. Kotoula, C. H. Karabatsas and E. Tsironi, et al., Volume flow and wall shear stress quantification in the human conjunctival capillaries and post-capillary venules in vivo, Biorheology, 2007, 44, 375–386 Search PubMed.
- A. Villasante, M. J. Lopez-Martinez, G. Quiñonero, A. Garcia-Lizarribar, X. Peng and J. Samitier, Microfluidic model of the alternative vasculature in neuroblastoma, In Vitro Models, 2024, 3(1), 49–63 CrossRef PubMed.
- A. Carreau, B. Hafny-Rahbi El, A. Matejuk, C. Grillon and C. Kieda, Why is the partial oxygen pressure of human tissues a crucial parameter? Small molecules and hypoxia, J. Cell. Mol. Med., 2011, 15(6), 1239–1253 CrossRef CAS PubMed.
- E. L. LeCluyse, R. P. Witek, M. E. Andersen and M. J. Powers, Organotypic liver culture models: Meeting current challenges in toxicity testing, Crit. Rev. Toxicol., 2012, 42(6), 501–548 CrossRef CAS PubMed.
- H. Rashidi, S. Alhaque, D. Szkolnicka, O. Flint and D. C. Hay, Fluid shear stress modulation of hepatocyte-like cell function, Arch. Toxicol., 2016, 90(7), 1757–1761 CrossRef CAS PubMed.
- A. Ehrlich, D. Duche, G. Ouedraogo and Y. Nahmias, Challenges and Opportunities in the Design of Liver-on-Chip Microdevices, Annu. Rev. Biomed. Eng., 2019, 21, 219–239 CrossRef CAS PubMed.
- X. Wang, B. Xu, M. Xiang, X. Yang, Y. Liu and X. Liu, et al., Advances on fluid shear stress regulating blood-brain barrier, Microvasc. Res., 2020, 128, 103930 CrossRef PubMed.
- E. Ortíz-Prado, J. F. Dunn, J. E. Vásconez, D. Castillo and G. Viscor, Partial pressure of oxygen in the human body: a general review, Am. J. Blood Res., 2019, 9(1), 1–14 Search PubMed.
- H. M. M. Ahmed, S. Salerno, S. Morelli, L. Giorno and L. De Bartolo, 3D liver membrane system by co-culturing human hepatocytes, sinusoidal endothelial and stellate cells, Biofabrication, 2017, 9(2), 25022 CrossRef PubMed.
- E. Roux, P. Bougaran, P. Dufourcq and T. Couffinhal, Fluid Shear Stress Sensing by the Endothelial Layer, Front. Physiol., 2020, 11, 861 CrossRef PubMed.
- D. Spieler, C. Namendorf, T. Namendorf, M. von Cube and M. Uhr, Donepezil, a cholinesterase inhibitor used in Alzheimer's disease therapy, is actively exported out of the brain by abcb1ab p-glycoproteins in mice, J. Psychiatr. Res., 2020, 124, 29–33 CrossRef PubMed.
- J. D. Wessler, L. T. Grip, J. Mendell and R. P. Giugliano, The P-Glycoprotein Transport System and Cardiovascular Drugs, J. Am. Coll. Cardiol., 2013, 61(25), 2495–2502 CrossRef CAS PubMed.
- D. E. Ingber, Human organs-on-chips for disease modelling, drug development and personalized medicine, Nat. Rev. Genet., 2022, 23, 467–491 CrossRef CAS PubMed.
- T. Sasserath, J. W. Rumsey, C. W. McAleer, L. R. Bridges, C. J. Long and D. Elbrecht, et al., Differential monocyte actuation in a three-organ functional innate immune system-on-a-chip, Adv. Sci., 2020, 7(13), 2000323 CrossRef CAS PubMed.
- Y. S. Zhang, J. Aleman, S. R. Shin, T. Kilic, D. Kim and S. A. Mousavi Shaegh, et al., Multisensor-integrated organs-on-chips platform for automated and continual in situ monitoring of organoid behaviors, Proc. Natl. Acad. Sci. U. S. A., 2017, 114(12), E2293–E2302 CAS.
- F. Yin, X. Zhang, L. Wang, Y. Wang, Y. Zhu and Z. Li, et al., HiPSC-derived multi-organoids-on-chip system for safety assessment of antidepressant drugs, Lab Chip, 2021, 21(3), 571–581 RSC.
- C. Oleaga, A. Riu, S. Rothemund, A. Lavado, C. W. McAleer and C. J. Long, et al., Investigation of the effect of hepatic metabolism on off-target cardiotoxicity in a multi-organ human-on-a-chip system, Biomaterials, 2018, 182, 176–190 CrossRef CAS PubMed.
- V. De Gregorio, B. Corrado, S. Sbrescia, S. Sibilio, F. Urciuolo and P. A. Netti, et al., Intestine-on-chip device increases ECM remodeling inducing faster epithelial cell differentiation, Biotechnol. Bioeng., 2020, 117(2), 556–566 CrossRef CAS PubMed.
- K. Vekemans and F. Braet, Structural and functional aspects of the liver and liver sinusoidal cells in relation to colon carcinoma metastasis, World J. Gastroenterol., 2005, 11(33), 5095–5102 CrossRef CAS PubMed.
- N. Fahy, U. Menzel, M. Alini and M. J. Stoddart, Shear and Dynamic Compression Modulates the Inflammatory Phenotype of Human Monocytes in vitro, Front. Immunol., 2019, 10, 383 CrossRef CAS PubMed.
- X. Wang, B. Xu, M. Xiang, X. Yang, Y. Liu and X. Liu, et al., Advances on fluid shear stress regulating blood-brain barrier, Microvasc. Res., 2020, 128, 103930 CrossRef PubMed.
- N. M. Wakida, G. M. S. Cruz, P. Pouladian, M. W. Berns and D. Preece, Fluid Shear Stress Enhances the Phagocytic Response of Astrocytes, Front. Bioeng. Biotechnol., 2020, 8, 596577 CrossRef PubMed.
- X. Wang, B. Xu, M. Xiang, X. Yang, Y. Liu and X. Liu, et al., Advances on fluid shear stress regulating blood-brain barrier, Microvasc. Res., 2020, 128, 103930 CrossRef PubMed.
- N. Ucciferri, E. M. Collnot, B. K. Gaiser, A. Tirella, V. Stone and C. Domenici, et al., In vitro toxicological screening of nanoparticles on primary human endothelial cells and the role of flow in modulating cell response, Nanotoxicology, 2014, 8(6), 697–708 CrossRef CAS PubMed.
- R. B. Riddle, K. Jennbacken, K. M. Hansson and M. T. Harper, Endothelial inflammation and neutrophil transmigration are modulated by extracellular matrix composition in an inflammation-on-a-chip model, Sci. Rep., 2022, 12(1), 1–14 CrossRef PubMed.
- B. Elbakary and R. K. S. Badhan, A dynamic perfusion based blood-brain barrier model for cytotoxicity testing and drug permeation, Sci. Rep., 2020, 10(1), 1–12 CrossRef PubMed.
- I. C. Harding, N. R. O'Hare, M. Vigliotti, A. Caraballo, C. I. Lee and K. Millican, et al., Developing a transwell millifluidic device for studying blood–brain barrier endothelium, Lab Chip, 2022, 22(23), 4603–4620 RSC.
- N. A. Kapai, J. V. Bukanova, E. I. Solntseva and V. G. Skrebitsky, Donepezil in a narrow concentration range augments control and impaired by beta-amyloid peptide hippocampal LTP in NMDAR-independent manner, Cell. Mol. Neurobiol., 2012, 32(2), 219–226 CrossRef CAS PubMed.
- P. J. Tiseo, S. L. Rogers and L. T. Friedhoff, Pharmacokinetic and pharmacodynamic profile of donepezil HCl following evening administration, Br. J. Clin. Pharmacol., 1998, 46(Suppl 1), 13–18 CrossRef CAS PubMed.
- E. I. Solntseva, J. V. Bukanova and V. G. Skrebitsky, Donepezil in low micromolar concentrations modulates voltage-gated potassium currents in pyramidal neurons of rat hippocampus, Biochem. Biophys. Res. Commun., 2013, 430(3), 1066–1071 CrossRef CAS PubMed.
- K. Matsui, M. Mishima, Y. Nagai, T. Yuzuriha and T. Yoshimura, Absorption, Distribution, Metabolism, and Excretion of Donepezil (Aricept) after a Single Oral Administration to Rat, Drug Metab. Dispos., 1999, 27(12), 1406–1414 CrossRef CAS PubMed.
- D. Goldblum, M. Gygax, M. Böhnke and J. G. Garweg, In vitro toxicity of rivastigmine and donepezil in cells of epithelial origin, Ophthalmic Res., 2002, 34(2), 97–103 CrossRef CAS PubMed.
- F. D. Newby and S. R. Price, Determinants of protein turnover in health and disease, Miner. Electrolyte Metab., 1998, 24, 6–12 CrossRef CAS PubMed.
- J. I. Lai, Y. J. Tseng, M. H. Chen, C. Y. F. Huang and P. M. H. Chang, Clinical Perspective of FDA Approved Drugs With P-Glycoprotein Inhibition Activities for Potential Cancer Therapeutics, Front. Oncol., 2020, 10, 561936 CrossRef PubMed.
- N. Bengoa-Vergniory, E. Faggiani, P. Ramos-Gonzalez, E. Kirkiz, N. Connor-Robson and L. V. Brown, et al., CLR01 protects dopaminergic neurons in vitro and in mouse models of Parkinson's disease, Nat. Commun., 2020, 11(1), 1–14 CrossRef PubMed.
- M. Usenovic, S. Niroomand, R. E. Drolet, L. Yao, R. C. Gaspar and N. G. Hatcher, et al., Internalized tau oligomers cause neurodegeneration by inducing accumulation of pathogenic tau in human neurons derived from induced pluripotent stem cells, J. Neurosci., 2015, 35(42), 14234–14250 CrossRef CAS PubMed.
- A. P. Ramme, L. Koenig, T. Hasenberg, C. Schwenk, C. Magauer and D. Faust, et al., Autologous induced pluripotent stem cell-derived four-organ-chip, Future Sci. OA, 2019, 5(8), FSO413 CrossRef CAS PubMed.
- A. I. Morrison, M. J. Sjoerds, L. A. Vonk, S. Gibbs and J. J. Koning, In vitro immunity: an overview of immunocompetent organ-on-chip models, Front. Immunol., 2024, 15, 1373186 CrossRef CAS PubMed.
- E. Conti, L. Tremolizzo, M. E. Santarone, M. Tironi, I. Radice and C. P. Zoia, et al., Donepezil modulates the endogenous immune response: implications for Alzheimer's disease, Hum. Psychopharmacol., 2016, 31(4), 296–303 CrossRef CAS PubMed.
- M. Sochocka, E. Zaczyńska, J. Leszek, I. Siemieniec and Z. Błach-Olszewska, Effect of donepezil on innate antiviral immunity of human leukocytes, J. Neurol. Sci., 2008, 273(1–2), 75–80 CrossRef CAS PubMed.
- Z. Erbayraktar, A. Evlice, G. Yener and N. N. Ulusu, Effects of donepezil on liver and kidney functions for the treatment of Alzheimer's disease, J. Integr. Neurosci., 2017, 16(3), 335–346 Search PubMed.
- Z. Pu, W. Xu, Y. Lin, J. Shen and Y. Sun, Donepezil decreases heart rate in elderly patients with Alzheimer's disease, Int. J. Clin. Pharmacol. Ther., 2019, 57(2), 94–100 CrossRef PubMed.
- A. T. McLaren, J. Allen, A. Murray, C. G. Ballard and R. A. Kenny, Cardiovascular effects of donepezil in patients with dementia, Dementia Geriatr. Cognit. Disord., 2003, 15(4), 183–188 CrossRef CAS PubMed.
- M. Tunesi, L. Izzo, I. Raimondi, D. Albani and C. Giordano, A miniaturized hydrogel-based in vitro model for dynamic culturing of human cells overexpressing beta-amyloid precursor protein, J. Tissue Eng., 2020, 11, 2041731420945633 CrossRef PubMed.
- R. Kim, P. Attayek, Y. Wang, K. Furtado, R. Tamayo, C. Sims and N. Allbritton, An in vitro intestinal platform with a self-sustaining oxygen gradient to study the human gut/microbiome interface, Biofabrication, 2020, 12, 015006 CrossRef CAS PubMed.
- S. Krantz, Y. Kim, S. Srivastava, J. Leasure, P. Toth, G. Marsboom and J. Rehman, Mitophagy mediates metabolic reprogramming of induced pluripotent stem cells undergoing endothelial differentiation, J. Biol. Chem., 2021, 297(6), 101410 CrossRef CAS PubMed.
- J. Janssen, B. Lagerwaard, M. Porbahaie, A. Nieuwenhuizen, H. Savelkoul, R. Joost van Neerven, J. Keijer and V. de Boer, Extracellular flux analyses reveal differences in mitochondrial PBMC metabolism between high-fit and low-fit females, Am. J. Physiol., 2022, 322, E141–E153 CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4lc00273c |
| ‡ These authors are co-last authors. |
| This journal is © The Royal Society of Chemistry 2025 |