 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Machine learning-guided optimization for ionic liquid-based polyethylene terephthalate waste recycling†
Ji
Gao‡
a,
Wenbo
Peng‡
a,
Andres
Galindo
a,
Ethan
Slaton
b,
Jose
Perez Martinez
a,
Guanghui
Lan
c and
Zhaohui
Tong
 *a
*a
aSchool of Chemical & Biomolecular Engineering, Georgia Institute of Technology, 311 Ferst Dr, Atlanta, 30318, GA, USA. E-mail: zhaohui.tong@chbe.gatech.edu
bDepartment of Chemical Engineering, University of Florida, Gainesville, Florida 32603, USA
cH. Milton Stewart School of Industrial and Systems Engineering, Georgia Institute of Technology, 755 Ferst Drive, Atlanta, 30332, GA, USA
First published on 20th May 2025
Abstract
Ionic liquid (IL)-catalyzed polyethylene terephthalate (PET) glycolysis has emerged as a promising method for recycling valuable monomers for high-quality polymer production. However, traditional approaches rely heavily on trial-and-error and time-consuming experiments to explore the large search space with multiple design factors. Here, we introduce a novel multi-objective optimization framework that integrates a graph neural network with process simulation for simultaneous IL design and reaction optimization towards unified economic and environmental metrics. We identified seven ILs unseen in the literature. Experimental validation demonstrates that approximately 47% of the optimized IL and reaction condition combinations outperform the best-reported literature values. This results in an average cost reduction of 29% and CO2 emissions reduction of 2.6% compared to the literature results. This work demonstrates the potential of machine learning to guide reaction optimization towards cost-effective and low-carbon targets for the PET recycling process.
Green foundation1. Our research focuses on enhancing consumer PET plastic recycling through ionic liquid-based glycolysis. We employed data-driven methodologies that guide catalyst selection and reaction optimization, minimizing the need for trial-and-error experimentation.2. Compared to the data collected from the literature, our screened ionic liquid under optimized reaction conditions demonstrates improvements. On average, it achieves a 43% reduction in cost and a 20% reduction in CO2 emissions compared to the top 60 literature values. It also achieves a 29% cost reduction and a 2% CO2 emission reduction compared to the average of the top 10 literature values. 3. Future research should focus on specific types of metal-free ionic liquids with reduced toxicity, and processes requiring less energy and fewer steps in their synthesis. This would minimize energy consumption and costs while also reducing pollution from the production process. |
Introduction
Over eighty-two million tons of single-use polyethylene terephthalate (PET) are annually produced worldwide1 and its extensive use has led to a significant accumulation of plastic waste.2 Increasing recycled content in products has become an important sustainability goal to mitigate plastic pollution and reduce dependence on petroleum-based resources.3–6 Life-cycle analysis demonstrates that solvolysis processes, including glycolysis, have a low CO2 footprint, making them environmentally favorable.7 Glycolysis stands out for its technical feasibility, economic viability, and reduced environmental impact, making it a promising method for efficient PET waste management, which has already been applied industrially.8–10 However, traditional salt-based catalysts for PET glycolysis present challenges, such as long reaction times, high reaction temperatures, and difficulties in separating the product.11,12 Ionic liquid (IL)-based glycolysis has emerged as a promising alternative. ILs offer several advantages, including improved reaction times, temperatures, and yields, while their easy separation from the solid product further enhances the overall efficiency of the process.13,14Substantial progress has been made in utilizing ILs as effective catalysts for PET glycolysis since 2006.8,13,15 Exploring novel IL combinations has shown promise in increasing yields under less vigorous reaction conditions. Efforts have also been put into seeking ILs with reduced toxicity and lower environmental impact to mitigate environmental concerns.15–17 Despite the progress in developing green processes for PET glycolysis, significant challenges still exist. First, identifying optimal ILs from the vast pool of possible combinations of cations and anions remains a major hurdle.18 Currently, the catalyst selection process relies heavily on time-consuming, trial-and-error-based experiments, limiting its efficiency. Second, optimization of reaction conditions requires time-consuming and expensive factorial design of experiments, which hinders the exploration of diverse ILs, especially under various reaction parameters. These parameters, often multi-dimensional and inter-correlated (e.g., catalyst loading, solvent ratio, reaction time, temperature, etc.), pose a significant challenge for optimization due to the non-linear and potentially non-convex nature of the problem. Third, the lack of a comprehensive evaluation framework beyond yield and conversion assessment limits our understanding of the entire process. For instance, while high yield is crucial for process efficiency, it alone cannot capture the full picture, neglecting environmental impact, energy efficiency, and costs associated with reaction conditions. Thus, a comprehensive solution capable of tackling all these challenges has yet to be achieved, shaping the outlook since 2012.19 Therefore, it is essential to consider these interconnected factors from a more holistic perspective. This necessitates a comprehensive approach that seamlessly combines IL catalyst design and reaction parameter optimization with integrated performance measures to achieve economically feasible and environmentally friendly processes.1
Computational approaches, such as the conductor-like screening model for realistic solvents (COSMO-RS), molecular dynamics, and quantum chemical computations, are common screening techniques for IL applications.20–22 However, their use in reactive systems such as IL-catalyzed PET glycolysis may face limitations. COSMO-RS is efficient for solubility prediction tasks, but may struggle with dynamic polymer–IL interactions and catalytic modeling due to their parameterization dependence. Molecular dynamics simulations, while suited for non-reactive dissolution, are faced with computational limitations and force field inaccuracies when used in large, reactive systems. While quantum chemistry can handle reactivity, it requires accurate mechanistic insights and becomes computationally expensive for complex polymer systems. One common drawback of these physics-based methods is that they rely on simplified models (like polymer fragments) and mechanistic databases absent in this context.23 Data-driven approaches have advantages in this case: they avoid the need for force field estimation, reaction mechanisms, or physical parameters by learning patterns directly from experimental data, making them well-suited for addressing this challenge.
Machine learning (ML) has become a powerful tool in molecular and reaction science.24,25 Researchers have predicted reaction yield under different reaction conditions for PET recycling using ML.26,27 Existing studies in the field of ILs primarily utilize ML for predicting physical properties like density and viscosity.28,29 However, the application of ML to IL-based reactions lags, specifically for PET glycolysis reactions when a large pool of various ILs exists. The adoption of graph neural networks (GNNs)30 offers a promising tool to bridge this gap. A GNN is a type of neural network that is useful when working with data structured as graphs. In chemistry, molecules are often represented as graphs: atoms as the nodes (or vertices) of the graph, with features such as their atomic number, charge, and bonds between atoms as the edges that connect the nodes. GNNs are designed to work on these graphs by learning how each atom's environment affects its properties by considering both the atom's features and its neighbors. GNNs excel at efficiently representing molecules, making them effective for tasks like reaction prediction and catalyst design in various fields.31,32 Introducing GNNs in IL-catalyst molecular design presents a significant opportunity to push the boundaries and explore their potential for complex tasks, such as simultaneous catalyst design and optimization of the IL-catalyzed PET glycolysis reaction.
In this study (Fig. 1), we develop a GNN model that predicts reaction yield according to not only different reaction parameters but also IL molecular structures based on approximately four hundred data points from the literature. This information is then incorporated into a process model to estimate the cost and carbon emissions associated with the entire PET glycolysis process. We identify the ideal combinations of IL catalysts and reaction conditions for a more efficient, economic, and eco-friendly glycolysis process, guided by the combined economic and environmental performance indicators. The experimental validation results show that these new ILs and reaction conditions demonstrate significantly better PET glycolysis performance compared with the literature values in terms of cost and CO2 emission. Our GNN-based ML model allows for simultaneous catalyst design and reaction condition optimization that eliminates the need for individual adjustment of each reaction parameter. Through optimization towards tailored performance indicators, our integrated model enables a smooth transition from laboratory to process for an industrial-oriented design.
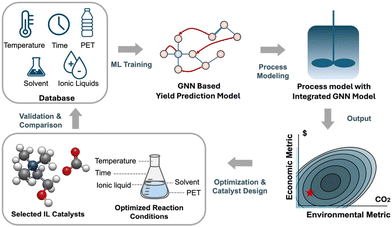 | ||
| Fig. 1 Schematic representation for simultaneous IL catalyst design and reaction optimization towards integrated economic and environmental metrics. | ||
Results and discussion
Development of a GNN ML model for PET glycolysis yield prediction
To initiate our data-driven prediction and catalyst design, we developed a GNN-based model for yield prediction of BHET (bis(2-hydroxyethyl) terephthalate) in the PET glycolysis reaction, which is schematically illustrated in Fig. 2A. The GNN model harnesses the rich structural information about the IL, encoding it into a compact vector to featurize its molecular information. These vectors were concatenated with other PET degradation reaction conditions and fed into a fully connected layer of the regression problem for predicting the yield of BHET, the main product of PET glycolysis. To train our model effectively, we compiled a dataset of approximately four hundred experimental data points from diverse literature sources (Data S1†), focused on the IL-catalyzed glycolysis of PET. Each data point captures the essential reaction parameters, including the quantity of IL, solvent (EG), and PET, the source and size of PET, and the reaction temperature and time. To gauge the performance of each reaction, we chose to use the yield of BHET as a key indicator. This metric (eqn (1)) encompasses both selectivity, which measures the preferred formation of the desired product (BHET) over undesired byproducts, and conversion, which reflects the overall extent of PET consumed in the reaction. To make this equation valid, we make two assumptions. First, we assume that the major depolymerization product is BHET, with minimal intermediate oligomers in the final mixture. This is supported by NMR analysis (Fig. 5A), which shows clear peaks corresponding to high-purity BHET. Second, we assume that the reaction is irreversible under the selected conditions. The re-polymerization of BHET into oligomers is thermodynamically possible, especially at high BHET concentrations and lower temperatures. However, this process generally requires much higher temperatures (e.g., 280 °C) than those used here.33 The resulting dataset comprises 68 distinct types of ILs, featuring 14 cations and 37 anions, which form the basis for learning an informative model from existing research. | (1) |
We implemented a three-stage validation strategy to evaluate the effectiveness of our GNN model. First, we employed cross-validation to assess the model's performance (Fig. 2D). Next, we utilized model explanation techniques on ILs (Fig. 2B) and reaction conditions (Fig. 2C) to gain deeper insight into the model's decision-making process. By comparing the ML results with existing literature results, we confirmed the alignment with established findings. Finally, we validated the ML model's predictions against actual experimental outcomes, which serve as a holdout testing set (in the Experimental validation and performance section), providing a critical test of its real-world applicability.
To gauge the predictive power of our GNN model, we focus on three key performance metrics: R2, which captures the proportion of variance explained; root mean squared error (RMSE), which measures the residual prediction error with stronger weighting on large errors; and mean absolute error (MAE), which measures the residual prediction error with equal weighting on all data. We carried out hyperparameter optimization with Optuna34 for nine parameters including batch size, number of GNN layers, GNN output size (size of vector to embed one IL), fully connected layer dimensions, GNN dropout rate, fully connected layer dropout rate, optimizer learning rate, optimizer weight decay, and whether to use a scheduler, with details shown in the ESI.† The tuned model demonstrated an average validation R2 of 0.64, an MAE of 0.12, and an RMSE of 0.16 on a stratified 10-fold cross-validation. The stratification criterion was based on reaction yield, dividing the data according to yield intervals between 0, 20, 40, 60, 80, and 100 to ensure balanced representation across different yield ranges. We carried out a bias/variance analysis on the training and validation loss curves to analyze the potential model overfitting with limited training data (ESI†). We found that through a combination of strong regularization (a large dropout35 value of 70% applied to the GNN part of the model, 5% dropout applied to each fully connected layer, and optimizer regularization) and early stopping, the overfitting was effectively controlled. The aggregated results of the cross-validation testing of datasets are shown in Fig. 2D. It can be seen that there exist regions of overestimation and underestimation. This is represented by the larger RMSE compared to MAE. While the R2 and RMSE values are not perfect, they align with the anticipated challenges associated with the literature data, characterized by a scarcity of training data, heterogeneity in experimental conditions, and the complex interplay of molecular and reaction variables. Considering these challenges, we can conclude that the attained metrics are acceptable and indicative of the model's practical utility for guiding experimental efforts. Finally, we used the tuned parameters to train the model on the entire dataset for the prediction and testing of unseen cases.
To gain further insights into the molecular structural significance and contribution of the ILs, we utilized GNNExplainer,36 which allowed us to compare the atomic-level contribution with established reaction mechanisms documented in the literature, serving as a valuable validation step. The GNNExplainer aggregates the node and edge importance scores onto each atom. In this way, we were able to pinpoint the comparative contribution of each atom to the final yield prediction. We observed that anions provide a much higher contribution, which aligns with experimental findings.37 But this makes the relatively weaker contribution of the cation atoms less visible. To improve visualization, we normalized the contributions of cation and anion atoms separately and enhanced clarity by using different colors for cations (red) and anions (blue). Fig. 2B showcases an example involving choline formate, where the model recognizes the roles of the negatively charged oxygen atoms and the positively charged nitrogen atom in the cleavage of the ester bonds between PET monomers. Our GNN model's explanation graphs, when compared to the established reaction mechanisms,38 showed a certain degree of alignment (Fig. S1†). However, we must note that the explanation graph only illustrates the comparative importance of different atoms to yield prediction, providing hints regarding potential mechanistic insights rather than direct mechanistic implications. This alignment suggests that the GNN model may be a potential tool for comparing different ILs in glycolysis reactions. To further explain the model, the feature representations learned by the GNN are analyzed. We applied t-distributed stochastic neighbor embedding (t-SNE) to the GNN-featurized IL outputs (the GNN output before the fully connected layer), followed by a cluster analysis with density-based spatial clustering of applications (DBSCAN)39 to study the clusters and outliers (ESI†). We observed that specific groups of ILs form distinct clusters (Fig. S24 and S26†), reinforcing the model's ability to capture meaningful molecular features.
The multilayer perceptron part of the GNN model (prediction layer) offers valuable insights into understanding the reaction parameters. Specifically, we employed the Shapley additive explanation (SHAP)40 analysis to decouple the intercorrelated reaction conditions, revealing their underlying effects on the prediction results, as shown in Fig. 2C. Among these features, temperature emerges as the most important factor because temperature has the greatest effect on the reaction rate. Other factors, such as the reaction time, catalyst loading, and the solvation effect of EG, were also found to significantly affect the predicted yield. PET source (bottle, powder, or pellet) is another important factor due to the distinct properties exhibited by different types of PET, potentially leading to different reaction rates.
Finally, the performance of our GNN model was assessed through multiple sets of testing experiments that varied reaction conditions and catalyst types. These experiments yielded a compelling result, with the model achieving an MAE of 0.08 and RMSE of 0.12, which is aligned with the k-fold cross-validation results. This confirms the effectiveness of our approach in guiding the molecular design of IL catalysts and the optimization of PET glycolysis reactions.
ML-based simulation for the PET glycolysis process
To facilitate the design of sustainable glycolysis processes, we developed an integrated model that combines the GNN with process simulation using BioSteam41 (Fig. 3). Our focus was on the reactor section, as the preprocessing and finishing stages are similar across different catalysts and reaction conditions. The process model includes a heat exchanger, a solvent recovery evaporator (with a 90% recycling rate), pumps, and a glycolysis reactor that accounts for heat loss to the environment. A stoichiometric reactor simulator was employed, allowing us to focus solely on mass and energy balances without the need for dynamic reaction simulation. The reactor model is governed by yield, with yield predictions inferred from the GNN model based on the quantities of reactants, temperature, and reaction time. The feed stream and process operating time are set to achieve an annual BHET production of one thousand tons for comparison. BioSteam is used to solve mass and energy balances while estimating key process metrics, such as profit, raw material and utility costs, and carbon emissions. This setup enables a comparison of trade-offs in parameters like reactant quantities, reaction temperature, and yield.For the economic evaluation, we gathered pricing data for ILs and utilities from futures markets and wholesale vendors (ESI†), along with carbon emission factors sourced from the EPA42 and EU legislation43 (ESI†). During the development of our model, we adopted a simplified approach by assuming uniform carbon emission values for all ILs. This approximation stems from the scarcity of carbon emission data for the ILs, making it challenging to integrate precise values. Our process model remains a valuable resource, offering insights for informed decision-making. This integrated approach empowers us to move beyond traditional yield or conversion optimization. Instead, it prioritizes both economic and environmental considerations for the development of eco-friendly and commercially viable glycolysis processes.
Process and reaction optimization
The effectiveness or optimized performance of a chemical reaction is determined by many factors in practice. However, the current literature is predominantly focused on yield (or conversion and selectivity) as the sole performance metric. These approaches overlook a crucial aspect of the chemical reaction process: a high yield achieved with an expensive catalyst and under harsh reaction conditions may impair the economic viability of the process and increase carbon emissions. This can be attributed to the lack of methods integrating the process and its economic and environmental impact indicators. Thus, in contrast, our goal is to optimize production while simultaneously optimizing reaction temperature, time, catalyst, and solvent usage. These criteria are ultimately simplified to two key factors: production cost and environmental impact. In this work, equipped with the ability to estimate both cost and carbon emissions, we formulate an optimization problem using a weighted sum method, as stated in eqn (2). By varying reaction conditions (p), we aim to minimize a weighted sum of cost (c) and carbon emissions (e) for solvent, catalyst, and utilities (i). These costs and emissions can be efficiently calculated using our ML-based process model (f). The objective function assigns weights to the cost (wc) and emission (we) components, reflecting their relative importance in the process subject to customizable needs: | (2) |
In this study, we assumed no specific preferences and assigned equal weights to the costs and emissions. We conducted around 3.8 × 105 simulations with data enumerated from various ILs and reaction conditions (ESI†), yielding a ranked list of candidate combinations (with a full distribution plot shown in Fig. S2†). We first carried out the analysis by visualizing the objective function for the top 105 performance cases, as shown in Fig. 4A. Clearly, an optimization target is located on the lower left corner of the plot, where both cost and carbon emissions are minimized. We observed an overall correlated relationship between cost and carbon emissions, suggesting that good combinations of catalyst and reaction conditions could help reduce the performance indicators synergistically. Near the optimum at the bottom left corner of the plot, a convex Pareto front can be observed, showing the trade-offs between costs and carbon emissions.
We acknowledge the inherent uncertainty associated with the yield predictions from the GNN-based ML model and performance estimation by the process simulation model. These uncertainties indicate that the absolute ranking within the entire dataset may not be definitive, and solving the optimization problem directly is not applicable. Therefore, we recommend focusing on the top-ranked combination cases of ILs and reaction conditions, where the most promising candidates reside. We narrowed down the observation to the top 104 (around the top 2.6%) cases (shown in Data S1†), which have been calculated from the best combinations of the production cost and environmental impact, including the factors considered in eqn (2). Differing from the experimental screening methods targeting only an optimal yield, our model considers a balance between a moderately high yield (approximately 60%–70%) coupled with a shortened reaction time and elevated temperature for optimized economic and environmental performance (Fig. 4B). Thus, the cost and carbon emission incurred by the reaction (temperature, time, catalyst, solvent, and yield), and process (reactor, solvent recycling, energy, and utilities) can be considered synergistically.
From the cost breakdown analysis shown in Fig. 4C, we identify that even with a recycling unit in place, solvent remains the major expense, which is consistent with the analysis presented in the literature.44 This is because a high amount of EG is needed to immerse and dissolve the original PET, and more solvent directly translates into increased energy demand for heating and catalyst addition. The breakdown of carbon emissions is not shown here since more than 99% of the total emissions are from utilities. This result supports that our simplification and assumption of using constant catalyst carbon emissions for catalysts is reasonable.
Experimental validation and performance
We successfully identified optimal reaction conditions and screened new IL catalysts for PET glycolysis by combining our process simulation model with the GNN reaction model. To validate the accuracy and effectiveness of the integrated GNN-simulation model in predicting reaction performance, we conducted PET glycolysis reactions using seven combinations of newly screened ILs and reaction conditions from the top rankings with representative cations (imidazolium, tetraalkylammonium, guanidinium, and choline) and anions (metal-based chlorozincate). These combinations were chosen based on their predicted economic and emission benefits and have not been previously reported in the literature for PET glycolysis. The selected ILs include tetramethylamine trichlorozincate ([N1111][ZnCl3]), tetramethylamine tetrachlorozincate ([N1111]2[ZnCl4]), 1,1,3,3-tetra-methylguanidinium tetrachlorozincate ([TMG]2[ZnCl4]), choline trichlorozincate ([Ch][ZnCl3]), choline tetrachlorozincate ([Ch]2[ZnCl4]), 1-ethyl-3-methylimidazolium trichlorozincate ([Emim][ZnCl3]), and 1-ethyl-3-methylimidazolium tetrachlorozincate ([Emim]2[ZnCl4]). These ILs are synthesized in our laboratory, and the details are shown in the Experimental section (synthesis of new ILs). Their chemical structures were characterized and confirmed by both NMR and UV-Vis spectra, as shown in Fig. S5–S23†.We then selected the reaction conditions according to the model prediction results. Our model suggests a 90 minute reaction time, and an IL loading (the mass ratio of IL to PET) of 2% to achieve the top performance while simultaneously considering the trade-off between BHET yield and cost.45 In the validation experiments, the solvent-to-PET ratio was set at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, which ensures sufficient immersion of the shredded PET substrate while maintaining minimum production costs using a lower solvent volume. This moderate solvent amount also ensures mixture fluidity and facilitates mixing.46Fig. 5B compares these model-selected reaction conditions with the optimized conditions reported in the literature. The maximum temperature was constrained to 190 °C, which is sufficiently high to promote glycolysis but still below the boiling point of EG (196 °C).47 Our model emphasizes reducing solvent usage, reaction time, and catalyst loading. However, it offsets this by using higher temperatures to achieve better overall performance in terms of cost and CO2 emissions. By utilizing these model-predicted reaction conditions, we compared the experimental results with the predictions from our GNN model (Fig. 5C). Using shredded consumer plastic bottles with a particle size of around 5 mm for PET glycolysis experiments, we observed an average deviation of only 4% between the predicted and experimental values. All data points fell within overlapping error bars. This agreement validates the GNN model's ability to predict reaction yield. The NMR characterization of the BHET product from PET glycolysis proves the high purity of the BHET monomer from the PET glycolysis using the screened ILs and under the selected reaction conditions (Fig. 5A and Fig. S14†). The peak at δ 8.19 ppm signifies the presence of the four aromatic protons in the benzene ring. The multiple peaks range from δ 4.12 to 3.92 ppm and from δ 3.59 to 3.43 ppm, corresponding to the methylene protons of COO–CH2 and CH2–OH, respectively.
1, which ensures sufficient immersion of the shredded PET substrate while maintaining minimum production costs using a lower solvent volume. This moderate solvent amount also ensures mixture fluidity and facilitates mixing.46Fig. 5B compares these model-selected reaction conditions with the optimized conditions reported in the literature. The maximum temperature was constrained to 190 °C, which is sufficiently high to promote glycolysis but still below the boiling point of EG (196 °C).47 Our model emphasizes reducing solvent usage, reaction time, and catalyst loading. However, it offsets this by using higher temperatures to achieve better overall performance in terms of cost and CO2 emissions. By utilizing these model-predicted reaction conditions, we compared the experimental results with the predictions from our GNN model (Fig. 5C). Using shredded consumer plastic bottles with a particle size of around 5 mm for PET glycolysis experiments, we observed an average deviation of only 4% between the predicted and experimental values. All data points fell within overlapping error bars. This agreement validates the GNN model's ability to predict reaction yield. The NMR characterization of the BHET product from PET glycolysis proves the high purity of the BHET monomer from the PET glycolysis using the screened ILs and under the selected reaction conditions (Fig. 5A and Fig. S14†). The peak at δ 8.19 ppm signifies the presence of the four aromatic protons in the benzene ring. The multiple peaks range from δ 4.12 to 3.92 ppm and from δ 3.59 to 3.43 ppm, corresponding to the methylene protons of COO–CH2 and CH2–OH, respectively.
 | ||
| Fig. 5 Experimental results. (A) Photographs of raw materials and the product of the PET glycolysis reaction, and the NMR spectrum of BHET. (B) Comparison of optimized reaction conditions and the literature data. The shaded regions indicate the distribution of the literature-reported reaction conditions. The vertical orange lines indicate the reaction conditions selected in our study. (C and D) Comparison between experimental and prediction results of BHET yield for PET bottle and powder, respectively. Error bars for experimental results are the standard deviations of repeated experiments. Error bars for predicted values are model MAE. A scatter plot showing the same comparison is shown in Fig. S3.† (E) Performance comparison between the top 60 literature-reported reaction conditions and ILs versus experimental results of optimized reaction conditions and ILs. The data used with detailed information are shown in Data S1.† | ||
Additionally, we consider the validation of a crucial but often overlooked factor, the original PET material type, and its impact on PET glycolysis performance. Our collected dataset indicates that the size and physical properties of the starting PET material significantly influence PET glycolysis performance. Fig. 2C demonstrates that the PET type is a significant factor besides temperature, which may affect the accuracy of model prediction. Therefore, instead of shredded PET bottles, we conducted glycolysis experiments using PET powders with a much finer average particle size of 0.075 mm (Fig. 5D) for validation. The model predictions deviated slightly more from the experimental results, with an average difference of 12%. The experimental yield was higher for shredded PET bottles than for the PET powder. One plausible explanation for this phenomenon could be attributed to the differences in their crystallinity. Higher crystallinity leads to lower BHET yield due to the decreased free volume in PET flakes, which impedes the diffusion of solvent and IL to the ester's active sites, thereby affecting the efficiency of the depolymerization process.48 With relatively higher crystallinity, the conversion and BHET yield values from PET powder with almost all ILs were 18% lower on average than those of shredded PET bottles, confirming our speculation. In addition, it was observed that PET powder tended to aggregate at the bottom of the flask, even under stirring conditions. This aggregation impeded efficient heat transfer and restricted the interaction between the reactants and the inner PET molecules. Furthermore, it was noted that PET bottle slices typically took around 30 minutes to melt completely under the provided reaction conditions. In contrast, PET powder required approximately 50–60 minutes for full melting. Therefore, despite the larger surface area of PET powder compared to PET slices, the glycolysis process was conducted at a slower rate for the former under identical experimental conditions.
The experimental validation data also confirm that the combinations of new ILs and reaction conditions identified by the model significantly enhance both economic and environmental performance compared to literature benchmarks. As depicted in Fig. 5E, employing the screened ILs under optimized conditions results in an average decrease of 43% in cost and 20% in CO2 emissions compared to the top 60 literature values. Notably, our optimized PET glycolysis processes achieved a 29% cost reduction and 2% CO2 emissions reduction compared to the mean of the top 10 literature values. Moreover, the performance of approximately 47% (19 out of 40 experiments) of the selected combinations surpasses that of the best-reported literature values in both cost and emission reduction. These findings underscore the outstanding prediction accuracy of our model and its significant potential for optimizing industrial PET glycolysis processes, thereby delivering substantial economic and environmental benefits.
Conclusions
In this study, we integrated a graph neural network (GNN) prediction model with a process simulation model to identify cost-effective and low-carbon catalysts for optimized IL-based PET glycolysis. We combined catalyst selection, reaction conditions, conversion, and yield into industry-aware performance indicators (cost and CO2 emissions), which facilitate industrial-oriented design beyond laboratory-scale experiments. The GNN model demonstrates robust predictive capabilities on PET glycolysis yield, leading to the discovery of seven promising new ILs for PET glycolysis, considering both economic and environmental impacts.However, limitations exist mainly due to the quantity and quality of data. Unlike direct property predictions for ILs such as CO2 solubility,49 which can utilize a relatively large database with more than ten thousand data points, the IL-based glycolysis reaction data are limited (on the scale of hundreds). This prevents us from exploring broader systems and conditions. This study only considered a single ionic liquid (IL) system without including other factors like co-solvents, supports, or external stimuli such as microwave heating, which are used in the current state-of-the-art study. The small quantity of non-uniform data from various literature studies and laboratories also introduces more uncertainty in prediction, and can propagate through our framework to downstream estimations.
The limitation in data availability also limits the potential for GNN applications and generalizability. While GNNs can be applied to ILs with new cations and anions, in this study, we restricted our analysis to combinations of existing cations and anions from the collected data. Constructing sufficient and diverse experimental datasets would enable the GNN to be leveraged for the prediction of completely new ILs. Another way to address the data limitation is through incorporating mechanistic or computational data augmentation, such as density functional theory, kinetics study, and fluid mechanics simulation via hybrid modeling and training approaches.50,51 Integrating such computational approaches in future studies could bridge the gap between empirical data and mechanistic insights, resulting in more generalizable models, and improving their predictive capabilities.
We implemented a simplified process model to simulate real-world production scenarios for cost and emissions estimation. Although this simplified model offers valuable insights, it may still diverge significantly from actual industrial processes. Hence, it is only appropriate for the use of process-informed guidance for laboratory-scale discovery of ILs and glycolysis reaction optimization. Process scale-up or application on the real process is complicated and process-specific. It requires detailed process design and techno-economic analysis, which is not a direct objective of our study. Nevertheless, the importance of this work is that our new framework is highly customizable, allowing users to integrate more detailed process simulation models and data to optimize catalyst design and reaction conditions for PET glycolysis.
Experimental
GNN model
Multi-head attention-based graph convolution52,53 is applied to the GNN model using the PyTorch Geometric54 package. The output for each head at each GNN layer is computed as follows:Here,  denotes the d dimensional hidden information for nodes (N) within the graphs;
denotes the d dimensional hidden information for nodes (N) within the graphs;  denotes the de dimensional edge features between the nodes;
denotes the de dimensional edge features between the nodes; 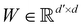 is the shared weighting to be trained; and α denotes the normalized attention coefficients. The normalized attention coefficients between nodes are calculated as follows:
is the shared weighting to be trained; and α denotes the normalized attention coefficients. The normalized attention coefficients between nodes are calculated as follows:
Function σ is the leaky rectified linear unit activation function:
| σ(x) = max(0.2x,x). |
The global mean pool is used to construct graph-level outputs by computing the average of node features. To avoid overfitting with the limited amount of data, we applied a large dropout of 70% at the penultimate graph neural network layer, and 5% for each intermediate layer of the fully connected network. The implementation details of the featurization layer (GNN) and the prediction layer (fully connected network) can be found in the ESI†. The training and testing metrics for the cross-validation process are shown in Fig. S4.†
Materials
The PET powder was purchased from Guangyuan Inc. in China. PET beverage bottles were purchased from a local supermarket (Publix). The bottles were washed, dried, and cut into approximately 0.5 cm by 0.5 cm slices for degradation experiments. All the chemicals for validation experiments were purchased from Fisher Scientific International, Inc.Experimental procedures
The synthesis of the ILs closely followed the previous works on the synthesis of ILs, which contained either the cation or the anion of the screened ILs.55–59 The experimental procedure adhered to the guidelines outlined in previous studies.57,60 NMR and UV-Vis spectrometry were utilized to characterize the synthesized ILs, confirming the presence of the specified cations and anions as reported in prior studies.60 The yield of BHET was calculated by determining the molar ratio of BHET to PET monomers. A comprehensive analysis of the reaction outcomes was achieved through these methods. Additionally, BHET was further characterized by NMR to verify its high purity. This experimental validation process enabled us to assess the accuracy and reliability of ML models in predicting reaction yields, thereby validating their potential applicability and performance in future predictive tasks.All NMR analysis was performed with a Bruker AVIII-400 5 mm broadband probe. Chemical shifts for 1H NMR were reported in ppm on a δ scale. All UV-Vis characterization studies were conducted with a Cary 5000 UV-Vis-NIR spectrophotometer. A deuterium light source was used in all experiments. Measurements were carried out under ambient conditions using a quartz UV cuvette with a 1 cm path length.
Synthesis of new ILs
To synthesize the desired ionic liquids, a general procedure was followed. A specific amount of the corresponding halide salt ([N1111][Cl], [TMG][Cl], [Ch][Cl], or [Emim][Cl]) was dissolved in a minimal amount of water, and then a stoichiometric amount of ZnCl2 dissolved in water with a few drops of concentrated HCl was added slowly. The mixture was then heated to 60 °C–70 °C under vigorous stirring for 4 hours to ensure complete reaction. The resulting solution was dried in a vacuum oven overnight to remove any residual solvent and obtain the desired ionic liquid product. The detailed synthesis procedure for each IL in the study can be found in the ESI.†Glycolysis of PET powder
In a 150 mL Erlenmeyer flask, about 4 g of PET powder, 14.4 mL (∼16 g) of EG, and ∼0.08 g of catalyst were mixed and heated to 190 °C for 90 min. After that, the mixture was cooled to about 70 °C and filtered, and the solid was washed with about 100 mL of DI water. The precipitate was collected, dried in a vacuum oven, and weighed to obtain the mass of unreacted PET. The filtrate was then concentrated to around 30 mL by heating and placed in the fridge overnight. After that, the product, BHET, was precipitated and collected by filtration. It was then dried and weighed to obtain the dry mass.Glycolysis of shredded PET bottles
In a 150 mL Erlenmeyer flask, about 2 g of PET slices from a disposable water bottle (with dimensions of around 0.5 cm by 0.5 cm), 7.2 mL (∼8 g) of EG, and ∼0.04 g of catalyst were mixed and heated to 190 °C for 90 min. The subsequent steps were the same as those for the PET powder.Author contributions
W.P., A.G., J.P.M., and J.G. conducted the experiments. E.S. and J.G. conducted data collection. J.G. performed computations and simulations. J.G. and W.P. drafted the manuscript. Z.T. and G.L. contributed to the conceptual design, supervised the project, and edited the paper.Data availability
The database generated from the literature, experimental results, prediction results, and screening results of this study is available within Data S1.† The price and simulation data are available in the ESI.† The source code of models is available on GitHub at https://github.com/TongSustainabilityGroup/gnn_ionic_liquid.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the support from the NSF AI Institute for Advances in Optimization (2112533) and NSF-Cyber-Physical Systems (CPS)-NIFA (2024-67021-43862).References
- National Renewable Energy Laboratory, Researchers Engineer Microorganisms to Tackle PET Plastic Pollution, https://www.nrel.gov/news/program/2021/researchers-engineer-microorganisms-to-tackle-pet-plastic-pollution.html, (accessed 17 April 2024).
- OECD, Global Plastics Outlook, OECD, 2022 Search PubMed.
- European Commission , Single-use Plastics, https://environment.ec.europa.eu/topics/plastics/single-use-plastics_en, (accessed 17 April 2024).
- L. Smith, Plastic Waste, 2024 Search PubMed.
- M. McCabe , Whitehouse, Doggett Reintroduce Bicameral Bill Act to Tackle Plastic Pollution Crisis and Hold Polluters Accountable, https://www.whitehouse.senate.gov/news/release/whitehouse-doggett-reintroduce-bicameral-bill-act-to-tackle-plastic-pollution-crisis-and-hold-polluters-accountable/, (accessed 17 April 2024).
- S. Hadley , Recycled PET Market – Supply & Demand Market Issues, https://www.plasticcollective.co/recycled-pet-market-supply-demand-market-issues/, (accessed 17 April 2024).
- I. Vollmer, M. J. F. Jenks, M. C. P. Roelands, R. J. White, T. van Harmelen, P. de Wild, G. P. van der Laan, F. Meirer, J. T. F. Keurentjes and B. M. Weckhuysen, Angew. Chem., Int. Ed., 2020, 59, 15402–15423 CrossRef CAS PubMed.
- M. Zunita, H. P. Winoto, M. F. K. Fauzan and R. Haikal, Polym. Degrad. Stab., 2023, 211, 110320 CrossRef CAS.
- C. Shuangjun, S. Weihe, C. Haidong, Z. Hao, Z. Zhenwei and F. Chaonan, J. Therm. Anal. Calorim., 2021, 143, 3489–3497 CrossRef.
- Z. Ju, W. Xiao, X. Lu, X. Liu, X. Yao, X. Zhang and S. Zhang, RSC Adv., 2018, 8, 8209–8219 RSC.
- A. M. Al-Sabagh, F. Z. Yehia, A.-M. M. F. Eissa, M. E. Moustafa, G. Eshaq, A.-R. M. Rabie and A. E. ElMetwally, Ind. Eng. Chem. Res., 2014, 53, 18443–18451 CrossRef CAS.
- E. Barnard, J. J. Rubio Arias and W. Thielemans, Green Chem., 2021, 23, 3765–3789 RSC.
- J. Xin, Q. Zhang, J. Huang, R. Huang, Q. Z. Jaffery, D. Yan, Q. Zhou, J. Xu and X. Lu, J. Environ. Manage., 2021, 296, 113267 CrossRef CAS PubMed.
- K. Ghosal and C. Nayak, Mater. Adv., 2022, 3, 1974–1992 RSC.
- T. Christoff-Tempesta and T. H. Epps, ACS Macro Lett., 2023, 12, 1058–1070 CrossRef CAS PubMed.
- A. M. Al-Sabagh, F. Z. Yehia, G. Eshaq, A. M. Rabie and A. E. ElMetwally, Egypt. J. Pet., 2016, 25, 53–64 CrossRef.
- Y. Liu, X. Yao, H. Yao, Q. Zhou, J. Xin, X. Lu and S. Zhang, Green Chem., 2020, 22, 3122–3131 RSC.
- S. K. Singh and A. W. Savoy, J. Mol. Liq., 2020, 297, 112038 CrossRef CAS.
- L. Bartolome, M. Imran, B. G. Cho, W. A. Al-Masry and D. H. Kim, in Material Recycling - Trends and Perspectives, ed. D.Achilias, InTech, 2012 Search PubMed.
- X. Liu, K. E. O'Harra, J. E. Bara and C. H. Turner, J. Phys. Chem. B, 2021, 125, 3653–3664 CrossRef CAS PubMed.
- X. Zhang, Z. Liu and W. Wang, AIChE J., 2008, 54, 2717–2728 CrossRef CAS.
- P. Zhou, J. Yu, K. L. Sánchez-Rivera, G. W. Huber and R. C. Van Lehn, Green Chem., 2023, 25, 4402–4414 RSC.
- Y. Liu, X. Yao, H. Yao, Q. Zhou, J. Xin, X. Lu and S. Zhang, Green Chem., 2020, 22, 3122–3131 RSC.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Nature, 2018, 559, 547–555 CrossRef CAS PubMed.
- M. Meuwly, Chem. Rev., 2021, 121, 10218–10239 CrossRef CAS PubMed.
- G.-S. Ha, M. A. M. Rashid, D. H. Oh, J.-M. Ha, C.-J. Yoo, B.-H. Jeon, B. Koo, K. Jeong and K. H. Kim, Waste Manage., 2024, 174, 411–419 CrossRef CAS PubMed.
- Y. Wang, S. Li, K. Meng, Y. Zhang, Z. Fang and S. Sun, ACS Sustainable Chem. Eng., 2024, 12, 5415–5426 CrossRef CAS.
- K. Baran and A. Kloskowski, J. Phys. Chem. B, 2023, 127, 10542–10555 CrossRef CAS PubMed.
- S. Koutsoukos, F. Philippi, F. Malaret and T. Welton, Chem. Sci., 2021, 12, 6820–6843 RSC.
- J. Zhou, G. Cui, S. Hu, Z. Zhang, C. Yang, Z. Liu, L. Wang, C. Li and M. Sun, AI Open, 2020, 1, 57–81 CrossRef.
- Z. Wu, J. Wang, H. Du, D. Jiang, Y. Kang, D. Li, P. Pan, Y. Deng, D. Cao, C.-Y. Hsieh and T. Hou, Nat. Commun., 2023, 14, 2585 CrossRef CAS PubMed.
- P. Reiser, M. Neubert, A. Eberhard, L. Torresi, C. Zhou, C. Shao, H. Metni, C. van Hoesel, H. Schopmans, T. Sommer and P. Friederich, Commun. Mater., 2022, 3, 93 CrossRef CAS PubMed.
- K. Tomita, Polymer, 1973, 14, 50–54 CrossRef CAS.
- T. Akiba, S. Sano, T. Yanase, T. Ohta and M. Koyama, arXiv, 2019, preprint, arXiv:1907.10902 Search PubMed.
- N. Srivastava, G. Hinton, A. Krizhevsky, I. Sutskever and R. Salakhutdinov, J. Mach. Learn. Res., 2014, 15, 1929–1958 Search PubMed.
- Z. Ying, D. Bourgeois, J. You, M. Zitnik and J. Leskovec, in Advances in Neural Information Processing Systems, ed. H. Wallach, H. Larochelle, A. Beygelzimer, F. d Alché-Buc, E. Fox and R. Garnett, Curran Associates, Inc., 2019, vol. 32 Search PubMed.
- L. Liu, H. Yao, Q. Zhou, D. Yan, J. Xu and X. Lu, ACS Eng. Au, 2022, 2, 350–359 CrossRef CAS.
- Y. Liu, X. Yao, H. Yao, Q. Zhou, J. Xin, X. Lu and S. Zhang, Green Chem., 2020, 22, 3122–3131 RSC.
- M. Ester, H.-P. Kriegel, J. Sander and X. Xu, in Proceedings of the Second International Conference on Knowledge Discovery and Data Mining, AAAI Press, 1996, pp. 226–231.
- S. M. Lundberg and S.-I. Lee, in Advances in Neural Information Processing Systems 30, ed. I. Guyon, U. V. Luxburg, S. Bengio, H. Wallach, R. Fergus, S. Vishwanathan and R. Garnett, Curran Associates, Inc., 2017, pp. 4765–4774 Search PubMed.
- Y. Cortes-Peña, D. Kumar, V. Singh and J. S. Guest, ACS Sustainable Chem. Eng., 2020, 8, 3302–3310 CrossRef.
- US EPA, 2015, preprint, https://www.epa.gov/climateleadership/ghg-emission-factors-hub.
- 2018, preprint, https://data.europa.eu/eli/reg_impl/2018/2066/oj/eng.
- T. Uekert, A. Singh, J. S. DesVeaux, T. Ghosh, A. Bhatt, G. Yadav, S. Afzal, J. Walzberg, K. M. Knauer, S. R. Nicholson, G. T. Beckham and A. C. Carpenter, ACS Sustainable Chem. Eng., 2023, 11, 965–978 CrossRef CAS.
- A. McNeeley and Y. A. Liu, Ind. Eng. Chem. Res., 2024, 63(8), 3400–3424 CrossRef CAS.
- S. Marullo, C. Rizzo, N. T. Dintcheva and F. D'Anna, ACS Sustainable Chem. Eng., 2021, 9, 15157–15165 CrossRef CAS.
- A. Asueta, S. Arnaiz, R. Miguel-Fernández, J. Leivar, I. Amundarain, B. Aramburu, J. I. Gutiérrez-Ortiz and R. López-Fonseca, Polymers, 2023, 15, 4196 CrossRef CAS PubMed.
- H. Lu, D. J. Diaz, N. J. Czarnecki, C. Zhu, W. Kim, R. Shroff, D. J. Acosta, B. R. Alexander, H. O. Cole, Y. Zhang, N. A. Lynd, A. D. Ellington and H. S. Alper, Nature, 2022, 604, 662–667 CrossRef CAS PubMed.
- Z. Song, H. Shi, X. Zhang and T. Zhou, Chem. Eng. Sci., 2020, 223, 115752 CrossRef CAS.
- L. von Rueden, S. Mayer, R. Sifa, C. Bauckhage and J. Garcke, in Advances in Intelligent Data Analysis XVIII 2020, vol. 12080, pp. 548–560 Search PubMed.
- J. Willard, X. Jia, S. Xu, M. Steinbach and V. Kumar, ACM Comput. Surv., 2023, 55, 1–37 CrossRef.
- S. Brody, U. Alon and E. Yahav, arXiv, 2021, preprint, arXiv:2105.14491.
- P. Veličković, G. Cucurull, A. Casanova, A. Romero, P. Liò and Y. Bengio, arXiv, 2017, preprint, arXiv:1710.10903.
- M. Fey and J. E. Lenssen, arXiv, 2019, preprint, arXiv:1903.02428.
- L. Liu, H. Yao, Q. Zhou, D. Yan, J. Xu and X. Lu, ACS Eng. Au, 2022, 2, 350–359 CrossRef CAS.
- G. Kumar, K. Kumar and A. Bharti, Ind. Eng. Chem. Res., 2024, 63, 6024–6046 CrossRef CAS.
- Y. Liu, X. Yao, H. Yao, Q. Zhou, J. Xin, X. Lu and S. Zhang, Green Chem., 2020, 22, 3122–3131 RSC.
- Q. Wang, X. Yao, S. Tang, X. Lu, X. Zhang and S. Zhang, Green Chem., 2012, 14, 2559–2566 RSC.
- L. Wang, G. A. Nelson, J. Toland and J. D. Holbrey, ACS Sustainable Chem. Eng., 2020, 8, 13362–13368 CrossRef CAS.
- Q. Wang, Y. Geng, X. Lu and S. Zhang, ACS Sustainable Chem. Eng., 2015, 3, 340–348 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5gc01998b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |





