Open Access Article
Open Access ArticleGenistein alleviates rheumatoid arthritis by inhibiting fibroblast-like synovial exosome secretion regulated by the Rab27/nSMase2/Mfge8 pathway
JiaJia
Liu†
a,
Jinyang
Shi†
a,
Sijia
Niu†
a,
Ziyan
Liu
a,
XinHua
Cui
a,
Yuli
Song
b,
Xudong
Tang
c,
Junwen
Fan
d,
Hongyue
Xu
a,
Wanlu
Yu
a,
Mingmei
Zhu
a,
Baochun
Lu
a,
Ning
Liao
a,
Danping
Peng
a,
Yang
Wang
*a and
Lu
Yu
 *a
*a
aState Key Laboratory for Diagnosis and Treatment of Severe Zoonotic Infectious Diseases, Key Laboratory for Zoonosis Research, Ministry of Education, Institute of Zoonosis, College of Veterinary Medicine Jilin University, Center of Infectious Diseases and Pathogen Biology, Department of Infectious Diseases, First Hospital of Jilin University, Changchun 130000, China. E-mail: wyang@jlu.edu.cn; yu_lu@jlu.edu.cn
bShenzhen Liyunde Biotechnology Co., Ltd, Shenzhen 518057, China
cKey Lab for New Drug Research of TCM, Research Institute of Tsinghua University in Shenzhen, Shenzhen 518057, China
dBeijing Center for Animal Disease Control and Prevention, Beijing 102629, China
First published on 3rd February 2025
Abstract
Genistein (GEN), the predominant soy isoflavone in legumes, exhibits potential anti-rheumatoid arthritis (RA) effects. This study aims to explore the role of GEN in alleviating RA by regulating exosome secretion and the inflammatory microenvironment through the Rab27/nSMase2/Mfge8 pathway in collagen-induced arthritis (CIA) mice and in vitro. In vivo studies revealed that GEN treatment significantly reduced paw swelling in CIA mice and protected the integrity of knee and ankle joints in CIA mice. GEN supplementation caused a significant decrease in the levels of MMP-9 and pro-inflammatory factors TNF-α, IL-1β and IL-6. GEN also significantly diminished the expressions of β-catenin and exosomal Dvl3 and miR-221-3p in fibroblast-like synoviocytes (FLS). Molecular docking results showed that GEN had strong binding energy with Rab27a, nSMase2 and Mfge8, the key regulators of exosome secretion, respectively, which was confirmed by CETSA and DARTS detection. In vitro mechanism analysis demonstrated that GEN treatment simultaneously downregulated the expression of Rab27a, nSMase2 and Mfge8, and phenotypic analysis verified that GEN prevented the secretion of Alix+Hsp70+CD63+ exosomes induced by type II collagen (CII) from FLS. Further analysis showed that GEN inhibited the expression of the Wnt signaling pathway protein β-catenin and exosomal Dvl3 in FLS. Additionally, GEN inhibited CII-induced secretion of MMP-9 and TNF-α, IL-1β and IL-6 in FLS, and GEN also significantly inhibited CII-induced FLS migration. Notably, GEN inhibited the expression of miR-221-3p in FLS exosomes and enhanced osteoblast differentiation and mineralization in vitro. Collectively, this study clarified that GEN alleviates RA by inhibiting the secretion of FLS exosomes regulated by the Rab27/nSMase2/Mfge8 pathway and by inhibiting Dvl3/β-catenin and miR-221-3p.
Introduction
Rheumatoid Arthritis (RA) is a chronic, systemic autoimmune disorder characterized by sustained synovial inflammation, leading to damage of the articular cartilage and underlying bone.1 In RA, the endothelial layer of the synovium exhibits visible alterations, becoming inflamed and hyperplastic, which facilitates the infiltration of inflammatory factors into the adjacent cartilage and bone.2 Consequently, this inflammatory process often results in swollen and painful joints, with stiffness that is typically more pronounced in the morning or following prolonged periods of inactivity.3 Synovial fibroblasts (FLS), particularly those derived from RA patients (RA-FLS), play a pivotal role in the pathogenesis of RA.4,5 Activated FLS are central to perpetuating chronic inflammation, contributing to joint destruction, and facilitating the pathology to adjacent, unaffected joints.6,7 These fibroblasts regulate tissue homeostasis and orchestrate the inflammatory response by producing an array of cytokines, chemokines, and matrix metalloproteinases (MMPs) that amplify the recruitment and activation of inflammatory and immune cells.8 Furthermore, FLS actively attract monocytes and possess the capacity to autonomously mediate cartilage degradation, under their critical involvement in the progression of RA.9In terms of mechanisms, exosomes are small extracellular vesicles that mediate intercellular communication by transferring biomolecules. RA-FLS-secreted exosomes can degrade bone and cartilage through direct metalloproteinase activity or autocrine induction of MMPs released from FLS. RA-FLS exosomes contain a membrane-bound tumor necrosis factor (TNF), which has an autocrine effect and subsequently stimulates the transcription factor-κB (NF-κB) signaling pathway.10 In addition, microRNAs are also thought to play an important role in RA pathogenesis.11–13 MicroRNAs (miRNAs or miRs) are small non-coding RNAs that interact with complementary sequences in target miRNAs’ 3′ untranslated region (3′-UTR), leading to translational repression or degradation of these mRNAs.14 Dysregulation of miRNAs in FLS, osteoblasts, and T lymphocytes contributes to joint destruction by promoting inflammation, degrading the extracellular matrix, and modulating the invasive behavior of cells, thereby facilitating disease progression.15 Exosomes have gained recognition as crucial vehicles for miRNA delivery. The expression levels of miRNA such as miR-221-3p have been shown to correlate with the disease stage and activity in conditions such as RA.11 In particular, miR-221-3p was reported to be induced in a TNF-driven model of arthritis and FLS from RA patients,16 and over-expression of miR-221-3p suppresses calvarial osteoblast differentiation and mineralization in vitro.13
Existing studies indicate that Rab27a, Mfge8 and nSMase2 are critical exosome secretion and function mediators significantly influencing disease pathogenesis by regulating intercellular signaling and immune responses.17–20 Inhibition of nSMase significantly diminishes exosome secretion; however, it does not completely abrogate this process.20 Targeting Rab27a further reduces the vesicular secretion of certain established exosome markers, including CD63, Tsg101, Alix and Hsc70, while leaving the secretion of others, such as Mfge8, unaffected.18,21 Therefore, the simultaneous targeting of nSMase2, Rab27a and Mfge8 is likely to substantially inhibit exosome secretion and attenuate their functional impacts. Dvl3 (dishevelled-3), a protein that plays a key role in the Wnt signaling pathway, is involved in promoting the phenotypic transformation of RA-FLS.22 Exosomal Dvl3 has been shown to exacerbate cartilage destruction and apoptosis in collagen-induced arthritis (CIA), additionally influencing downstream activities of β-catenin and RhoA.22 Thus, targeting nSMase2, Rab27a and Mfge8-regulated FLS exosome secretions and subsequently inhibiting exosomal Dvl3 and FLS β-catenin as well as exosomal miR-221-3p may yield novel insights and therapeutic strategies for managing RA.
While several medications are available for the treatment of RA, their adverse side effects and high costs have prompted an increasing number of patients to seek natural remedies.5,22 Genistein (4′,5,7-trihydroxy isoflavone) (GEN), a soy isoflavone predominantly found in legumes including lupin, broad beans, and garbanzo beans (chickpeas), has shown promise in alleviating RA symptoms in preclinical trials.23 Mechanisms of GEN include the downregulation of vascular endothelial growth factor (VEGF) and MMPs (MMP-1, MMP-2 and MMP-9) in CIA models.22 Additionally, GEN modulates reactive oxygen species (ROS)/Akt/nuclear factor-κB and adenosine monophosphate-activated protein kinase (AMPK) pathways in human arthritic synovioblast MH7A cells.24 GEN further inhibits TNFα-induced inflammatory responses and angiogenesis through the IL-6/JAK2/STAT3/VEGF pathway.23 In addition, herbal plants, such as Withania somnifera, hemp, Emblica officinalis, Terminalia bellerica, etc., have been shown to have anti-inflammatory and antioxidant activities.5 Natural products exhibiting these properties may be utilized alone or in conjunction with established anti-arthritis therapies to enhance the treatment of RA.5 However, the precise molecular mechanisms underlying anti-RA effects of bioactive ingredients, especially GEN, remain to be elucidated.
Given these findings, we aimed to investigate the protective effect of GEN on CIA mice and elucidate the mechanism by which GEN alleviates the development of RA by inhibiting exosomal secretion and by inhibiting the Dvl3/β-catenin pathway and miR-221-3p.
Materials and methods
Animal model of CIA
All experimental procedures followed the guidelines approved by the Institutional Animal Care and Use Committee of Jilin University. Male DBA/1 mice (8 weeks old) were obtained from GemPharmatech and the animals were housed in a standard artificial environment with a constant temperature of 24–26 °C, 50–60% humidity, well-ventilated conditions, standard food and water supply, and artificial lighting (6.00–18.00). Mice were randomly assigned to five groups (n = 8 per group): CK (control group), CIA (bovine CII-induced RA modelling) group, GEN-L (CIA + Gen 10 mg kg−1) group, GEN-M (CIA + Gen 20 mg kg−1) group, and GEN-H (CIA + Gen 40 mg kg−1) group. The mice were weighed and administered GEN every other day.The model for RA was established following previously reported methods.25 Briefly, 150 μg of bovine CII (Chondrex, Redmond, WA) was dissolved in 0.01 M acetic acid overnight at 4 °C and mixed with an equal volume of complete Freund's adjuvant (CFA, Chondrex). A total of 0.1 mL of this emulsion was injected intradermally into the tails of the mice. Three weeks later, 150 μg of bovine CII was reconstituted and injected alongside incomplete Freund's adjuvant (IFA, Difco Laboratories) in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio at the same site. Arthritis scores were recorded and photographed every other day starting from day 28 of primary immunization, using a scale ranging from 0 to 3 (0 = normal; 1 = slight swelling and/or erythema; 2 = marked swelling; 3 = ankylosis). Blood was collected from the submandibular venous plexus of mice.
1 ratio at the same site. Arthritis scores were recorded and photographed every other day starting from day 28 of primary immunization, using a scale ranging from 0 to 3 (0 = normal; 1 = slight swelling and/or erythema; 2 = marked swelling; 3 = ankylosis). Blood was collected from the submandibular venous plexus of mice.
Histological examination and immunohistochemistry (IHC)
Knee and ankle joints from the model animals were excised and subjected to histological analysis using hematoxylin and eosin (H&E) and Safranin O staining, followed by visualization at ×200 and ×400 magnification.For immunohistochemical staining, synovial tissue slices were subjected to antigen retrieval using citric acid, followed by blocking with 5% BSA and visualization with DAB after incubation with primary and secondary antibodies. Histological and immunohistochemical images were acquired using the 3D HISTECH Digital Pathology System.
Computational docking
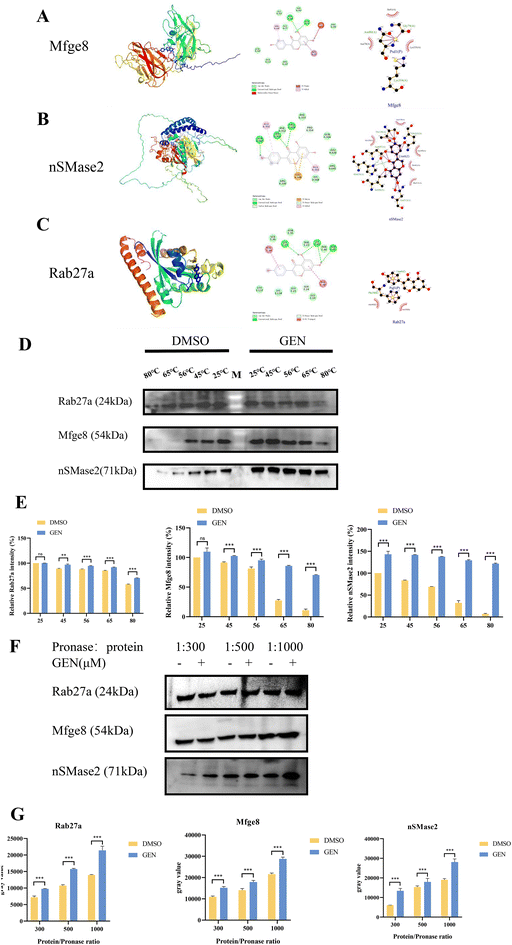
AutoDock Vina was used to predict the binding affinity of GEN to the active sites of exosome secretion-associated proteins Rab27a, nSMase2, and Mfge8. AutoDock Vina scores indicate the binding activity of GEN to the proteins, with higher negative values representing stronger binding affinity. Discovery Studio and LigPlus software were used to visualize GEN with Rab27a, nSMase2, and Mfge8 proteins.Cellular thermal shift assay (CETSA)
The binding efficiency of GEN to target proteins in FLS cells was determined by the CETSA method. FLS cells were cultured in 100 × 20 mm dishes (5 × 105 cells per mL). Cells were harvested and lysed using RIPA lysis buffer containing a protease inhibitor cocktail, and the supernatant was collected. The liquid portion of the sample was divided into three equal parts and treated with DMSO and 20 μM GEN for 1 h at ambient temperature. The proteins in the cells were then transferred to PCR tubes and denatured by heating in a PCR instrument for 3 min at different temperatures (25, 45, 56, 65 and 80 °C). After heating, the lysate is centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 min at 4 °C to separate it from the soluble fraction. The precipitate and soluble fraction were then separated in an ice bath. We transferred the supernatant and analysed it by SDS-PAGE and western blotting.
000 rpm for 20 min at 4 °C to separate it from the soluble fraction. The precipitate and soluble fraction were then separated in an ice bath. We transferred the supernatant and analysed it by SDS-PAGE and western blotting.
Drug affinity responsive target stability (DARTS)
FLS cells were treated in M-PER buffer (Thermo Fisher Scientific Inc., Indianapolis, IN, USA, 10165921001) supplemented with 1 mmol L−1 NaF, 1 × protease inhibitor mixture, and 1 mmol L−1 Na3VO4. Supernatants were treated with GEN or DMSO for 30 min, followed by centrifugation at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min. After GEN incubation, 20 μg of protein was digested in various concentrations of pronase (Roche Diagnostics, Indianapolis, IN, USA, 10165921001) for 30 min, and then the enzymatic digestion was stopped with 4 × sample buffer solution and heated for 5 min at 95 °C. The samples were analyzed by western blotting analysis.
000 rpm for 15 min. After GEN incubation, 20 μg of protein was digested in various concentrations of pronase (Roche Diagnostics, Indianapolis, IN, USA, 10165921001) for 30 min, and then the enzymatic digestion was stopped with 4 × sample buffer solution and heated for 5 min at 95 °C. The samples were analyzed by western blotting analysis.
Cell isolation and culture
Purified FLS was isolated from mouse synovial tissue by enzymatic digestion.26 Briefly, mixed synovial cells were obtained from small pieces of synovial tissue and then digested with trypsin and collagenase.27 These cells were then filtered through nylon mesh and collected by centrifugation at 800g for 5 min. The cells were cultured in exosome-free DMEM (Yobibio, China) supplemented with 100 U mL−1 penicillin, 100 μg mL−1 streptomycin, and 10% fetal bovine serum (Gibco, Grand Island, USA), and incubated at 37 °C under a humidified atmosphere with 5% CO2. We conducted experiments after 3–6 passages.We isolated cranial osteoblasts from newborn mice (postnatal day 0 to postnatal day 2) using collagenase digestion and maintained them under standard conditions.13,28 We co-cultured primary osteoblasts and FLS, and then harvested them for alkaline phosphatase (ALP) (Beyotime Biotechnology, Jiangsu, China), Von Kossa staining (Solarbio, China) and DKK2 protein expression.
Cell treatment
FLS were pretreated with GEN at the concentration required for the experiment for 2 h, and then the cells were stimulated with collagen type II (Sigma-Aldrich, St Louis, MO, USA) (40 μg ml−1) for 8 h.Exosome extraction
RA-FLS seeds were placed in a 10 cm dish and incubated with an exosome-free serum DMEM medium. The cell supernatant was collected by centrifugation at 300 g for 10 min at 4 °C. The supernatant was absorbed carefully, taking care to avoid inhalation of cells or cell fragments, and then, centrifuged again for 3000g × 20 min at 4 °C and the supernatant was collected. The supernatant was filtered through a 0.22 μm filter. Subsequently, the collected supernatant was isolated and purified using an exosome isolation and purification kit (EX100C, Shanghai Gefan Biotechnology Co., Ltd, China). Exosome separation and purification solutions were added overnight or for >14 h, centrifuged at 4 °C for 1500g × 30 min, and then centrifuged at 4 °C for 1500g × 5 min, and the residual solution was removed and resuspended with 100 μl PBS.Transmission electron microscopy (TEM)
Exosomes precipitated in PBS were dropped into a carbon-coated copper grid, and stained with 2% phosphotungstic acid for 1 minute. A small portion of exosome suspension was taken and allocated with a pipette to a copper mesh containing a thin film (such as a carbon film copper mesh). It was air dried for about 10–20 minutes to ensure that the sample is evenly spread and not excessive. The prepared mesh was placed into the sample compartment of a transmission electron microscope to prepare for observation. Observations were then made using the TEM (Hitachi H-7650, Japan).Immunofluorescence and confocal image acquisition
Immunofluorescence microscopy was employed to confirm the specificity of FLS. A confocal laser scanning microscope (CLSM) was used to identify the subcellular localization of the exosome marker protein CD63, along with the target proteins Rab27a (Affinity Biosciences, USA), nSMase2 (Affinity Biosciences, USA), Mfge8 (Bioworld Technology, USA), and Dvl3 (Bioworld Technology, USA) in FLS. Cells were grown on confocal dishes. Upon harvesting, cells were fixed with 4% paraformaldehyde and permeabilized with 0.01% Triton X-100 for 10 min. Subsequently, cells were probed with primary antibodies against vimentin (Bioworld Technology, USA), Rab27, nSMase2, Mfge8, and Dvl3, followed by incubation with fluorescently labeled secondary antibodies (488 nm and 594 nm). All samples were counterstained with Hoechst 342 to visualize nuclei. Confocal imaging was conducted using a Nikon confocal microscope.ELISA assay
TNF-α, IL-1β, IL-6 and MMP9 levels were measured using ELISA kits (Byabscience, Nanjing, China). The absorbance value at 450 nm was measured and the sample concentration was calculated from the standard concentration.Real-time quantitative RT-PCR (qRT-PCR)
To assess whether GEN can inhibit the production of microRNA-211-3p by exosomes, total RNA was extracted from the cell supernatant (ZScience Biotechnology, USA). Subsequently, reverse transcription followed by qRT-PCR was conducted using the SYBR Green qPCR reagent kit (GenePharma, Suzhou, China). Gene and miRNA expression was normalized to cel-mir-39 or GAPDH expression. The cel-mir-39 primer sequences were obtained from miRbase.29 The GAPDH forward primer sequence was ACAACTTTGGTATCGTGGAAGG; the reverse primer sequence was GCCATCACGCCACAGTTTC.22 qRT-PCR was performed under the following conditions: 95 °C for 3 min, 40 cycles of 95 °C for 12 s, and 60 °C for 40 s. The 2−ΔΔct values were calculated to represent the mRNA expression levels of the genes.Western blot analysis
Cell or tissue samples were lysed in RIPA buffer (Beyotime Biotechnology, Jiangsu, China) supplemented with a protease inhibitor (Pierce), and protein concentrations were determined using the BCA method (Beyotime Biotechnology, Jiangsu, China). Equal amounts of protein (40 μg) were separated by 10% SDS-PAGE and subsequently transferred to PVDF membranes. The membranes were blocked for 1 h at room temperature. Following this, membranes were incubated overnight at 4 °C with primary antibodies, and then treated with HRP-conjugated secondary antibodies for 1 h at room temperature. Protein bands were visualized using ECL fluorescence imaging.Migration and invasion assays
The Transwell migration assay was conducted in 24-well plates using Transwell invasion chambers (Corning, VA), following established protocols.22 FLS were seeded, and after treatment with established GEN and collagen type II, their number was counted in 3–5 randomly selected areas per well to assess their invasive capacity. After the matrix gel was embedded, the invasion chamber was placed in a cell culture incubator until the gel (Medchemexpress, USA) had fully solidified. Subsequent steps were carried out following the standard procedures for the Transwell migration assay.Alkaline phosphatase assay and Von Kossa staining
Primary osteoblasts and FLS from CIA mice were extracted and cellular alkaline phosphatase (ALP) levels were measured using an ALP assay kit (Beyotime Biotechnology, Jiangsu, China). Absorbance was measured at 405 nm after 3, 7, and 14 days of cell culture, and enzyme activity units were calculated. Definition of the ALP activity unit: in diethanolamine (DEA) buffer at pH 9.8, at 37 °C, every minute of hydrolysis of the para-nitrophenyl phosphate chromogenic substrate to yield 1 μmol of p-nitrophenol per minute at 37 °C was defined as one unit of enzyme activity.After 14 and 21 days of culture of primary osteoblasts and FLS, Von Kossa staining assays were performed per procedure.13 For quantitative analysis, the stains were extracted with 10% cetylpyridium chloride (Sigma-Aldrich, St Louis, MO, USA) in 10 mM sodium phosphate (pH 7.0) for 15 min. Von Kossa staining was then quantified by measuring absorbance at a wavelength of 561 nm, using a microplate reader (BioTek Instruments Inc., Winooski, VT, USA).
Statistical analysis
Data are presented as mean ± standard error (SE). T-test and ANOVA tests were employed to analyse differences among different groups. For ANOVA, Tukey's HSD post hoc test method was used to determine which groups had significant differences. Statistical analyses were performed using GraphPad Prism software. A *P-value of <0.05 was considered statistically significant, **P < 0.01 means the difference is highly significant, and ***P < 0.001 means the difference is extremely significant. Each experiment was repeated at least three times.Results
GEN alleviated arthritis and inflammation in CIA mice in vivo
To examine the effects of GEN on RA, a CIA mouse model was established (Fig. 1A). CIA mice treated with three concentrations of GEN all exhibited significant symptomatic improvements, particularly in the hind paws, where paw thickness in the GEN-M/GEN-H group was markedly decreased; especially, the arthritis scores were significantly lower in the medium- and high-dose groups compared to that in the control group after 15 days of booster immunization (P < 0.05) (Fig. 1B). To assess the effects of GEN on inflammatory factors in CIA mice, ELISA was performed on mouse serum.Subsequently, we analyzed the ankle and knee joints of CIA mice using hematoxylin and eosin (HE) staining and Safranine O-Fast Green Staining for pathological and morphological evaluations. HE staining of the ankle joint (Fig. 1D) revealed complete destruction of the articular cartilage in the CIA group, with chondrocytes absent. In contrast, the three concentrations of GEN treatment all preserved the cartilage structure, showing an increase in chondrocyte numbers, and the hyperplastic synovial membranes covered the cartilage surface, forming areas of vascularization. The control group exhibited no lesions. HE staining of the knee joint (Fig. 1D) revealed similar findings, with severe degradation of the articular cartilage in the CIA group characterized by a reduction in chondrocyte numbers and increased neutrophil infiltration. In the three GEN treatment groups, cartilage structure was all notably protected, with a higher density of chondrocytes and synovial membrane hyperplasia evident on the cartilage surface; the control group also showed no lesions.
Furthermore, Safranine O-Fast Green Staining of the knee and ankle joints (Fig. 1E) indicated that the three concentrations of GEN groups all significantly preserved the integrity of the articular cartilage, evidenced by an increase in chondrocytes and diminished loss of cartilage matrix compared to the CIA group. Histological analysis of the knee joints in the control group revealed smooth and flat cartilage surfaces, well-organized chondrocytes, and a uniformly stained cartilage matrix, with no apparent loss of cartilage integrity. In summary, the results presented herein illustrate the beneficial effects of GEN treatment, including a reduction in clinical signs and serum inflammatory markers in CIA mice.
GEN decreased the expression of β-catenin, exosomal Dvl3 and exosomal miR-221-3p in FLS from CIA mice
As mentioned above, exosomal Dvl3 has been shown to exacerbate cartilage destruction in CIA and influence downstream activities of β-catenin,22 and exosomal miR-221-3p is related to the disease stage of RA11 as well as osteoblast differentiation and mineralization.13 We assessed the effect of GEN on the mRNA levels of β-catenin, exosomes carrying Dvl3 and miR-221-3p in FLS from CIA mice using qRT-PCR and the IHC assay. The qRT-PCR results showed that the expression of β-catenin, exosomal Dvl3 and exosomal miR-221-3p in FLS from CIA mice were significantly higher than that in the control group (P < 0.001, P < 0.001, and P < 0.001), while three concentrations of GEN treatments significantly decreased these increase (P < 0.05, P < 0.01, and P < 0.001) (Fig. 2A–C). Subsequently, we employed immunohistochemistry to assess the expression of Dvl3 and β-catenin proteins in synovial tissues (Fig. 2D and E). Notably, no positive staining was observed in either control group. In contrast, cells within the CIA group exhibited pronounced dark brown coloration, indicative of elevated protein expression. Remarkably, following GEN treatment, there was a marked attenuation of these positive signals. Moreover, IHC scores showed significantly elevated Dvl3 and β-catenin expression in the synovium of the CIA group than that of the control group (P < 0.001), while three concentrations of GEN treatments significantly decreased these elevations (P < 0.05, P < 0.001, and P < 0.001). In conclusion, these data demonstrated that GEN treatment can inhibit the in vivo expressions of β-catenin, exosomal Dvl3 and exosomal miR-221-3p in FLS from CIA mice.GEN interacted with Mfge8, nSMase2 and Rab27a by molecular docking in silico
AutoDock analysis Vina revealed that the binding affinity of GEN to three exosome formation-related proteins (Rab27a, nSMase2 and Mfge8) was below 0.0 kcal mol−1. The calculated binding energies for these interactions were −8.3 kcal mol−1 for Rab27a, −7.5 kcal mol−1 for nSMase2, and −6.3 kcal mol−1 for Mfge8, indicating that Rab27a exhibited the strongest affinity for GEN, while Mfge8 demonstrated a comparatively weaker binding interaction. Additionally, visualization analyses conducted with Discovery Studio and LigPlus illustrated the nature of the interactions between GEN and these proteins, highlighting critical hydrogen bonds and H–π interactions (Fig. 3A–C).The CESTA WB results showed that the intensity of Rab27a, Mfge8, nSMase2 protein in the GEN-treated group was significantly higher than that in the DMSO group with increasing temperature at 25–85 °C, suggesting that the Rab27a, Mfge8 and nSMase2 proteins in the GEN-treated group had higher thermal stability (Fig. 3D). Based on the grayscale analysis of the CESTA western blot results, it is clear that the expression of the GEN treatment group proteins increased significantly with increasing temperature (P < 0.001; Fig. 3E). Subsequently, we performed DARTS experiments to identify the proteins that bind to GEN. Through DARTS western blotting and gray value analyses, we found that an increase in stability after GEN treatment was evident in the treated group (P < 0.001, P < 0.001, and P < 0.001; Fig. 3F and G). Generally, these results suggested that GEN interacted with exosome secretion-related proteins Mfge8, nSMase2 and Rab27a.
GEN inhibited the secretion of CII-induced Alix+Hsp70+CD63+ FLS-derived exosomes through downregulating Mfge8/nSMase2/Rab27a expression in vitro
Based on the protective effect of GEN on CIA mice, we then explored the mechanism of RA-FLS-derived exosomes and exosome carriers related to the occurrence of this effect. To assay the effect of GEN on the secretion of CII-induced FLS-derived exosomes, we extracted and purified FLS from mouse synovial tissue, and immunofluorescence analysis with vimentin, a specific marker for FLS, confirmed the presence of FLS (Fig. 4A). Subsequently, TEM and particle size analysis confirmed the presence of CII-induced FLS-derived exosomes with spherical three-dimensional structures and an average particle size of 100.26 ± 0.28 nm (Fig. 4B and C). Further western blot analysis showed that the exosome markers Alix and Hsp70 were present in FLS-derived exosomes in both the CII induction group and GEN treatment group; however, no bands were observed for GAPDH or the negative control protein calnexin (Fig. 4D and E). Notably, the GEN treatment (20 μM) significantly reduced the expression levels of the CII-inducing proteins Alix and Hsp70 (P < 0.05) (Fig. 4D and E). Then, western blot analysis revealed that GEN resulted in significantly lower levels of key regulators of exosome secretion such as Mfge8, nSMase2, and Rab27a proteins compared to those of the CII-induced group (P < 0.05; P < 0.05; and P < 0.05; Fig. 4F and G). Finally, CLSM assays were used to investigate the expression and co-localization between the exosome secretion regulators Mfge8, nSMase2 and Rab27a and the exosome marker CD63. Immunofluorescence staining showed that the Mfge8, nSMase2, Rab27a and CD63 expression and colocalization between Mfge8, nSMase2, Rab27a and CD63 were significantly increased (P < 0.01) in the CII induction group compared with the control group (P < 0.05), and there was a significant decrease in the GEN treatment group compared with the CII induction group (Fig. 4H). Altogether, these findings suggest that GEN inhibited the exosome secretion by reducing the expression of Mfge8, nSMase2, and Rab27a proteins in FLS in vitro, which may contribute to the protective effect of GEN on cartilage destruction and inflammation in CIA mice.GEN suppressed the expression of β-catenin and exosomal Dvl3 in CII-induced FLS in vitro
To further investigate the impact of GEN on the Wnt signaling pathway in FLS, we employed western blotting, qRT-PCR and CLSM to assess the expression of key proteins involved in this cascade. Western blot analysis revealed that the protein level of exosomal Dvl3 was significantly upregulated in CII-induced FLS compared with that of the control group (P < 0.01). However, treatment with GEN resulted in a significant reduction of exosomal Dvl3 expression (P < 0.01) (Fig. 5A). The qRT-PCR results demonstrated similar findings in mRNA levels (Fig. 5B). CLSM further demonstrated that Dvl3 co-localized with the exosome marker CD63, with fluorescence intensity significantly lower in the GEN treatment group than that in the CII induction group (Fig. 5E). Similarly, the qRT-PCR assay indicated there was a significant increase in the mRNA level of β-catenin expression in FLS in both the CII induction group and GEN treatment group compared to the control (P < 0.01, Fig. 5D). However, the mRNA level of β-catenin in FLS was significantly decreased in the GEN-treated group relative to that in the CII-induced group (P < 0.01), and western blot analysis of the protein level confirmed the qRT-PCR results (Fig. 5C and D). Collectively, these results suggest that GEN inhibits the expression of the exosomal protein Dvl3 and reduces downstream β-catenin levels in FLS in the Wnt signaling pathway, which may contribute to the protective effect of GEN on cartilage destruction in CIA mice.GEN inhibited the migration, invasion, and inflammatory response of CII-induced FLS
To determine the effect of GEN treatment on the migratory and invasive abilities of CII-induced FLS, transwell migration assays and western blotting were performed. Transwell migration assays indicated that the addition of CII significantly increased FLS migration and invasiveness through the matrix gel (P < 0.001, and P < 0.001), whereas GEN treatment significantly reduced the increase (P < 0.001, and P < 0.001) (Fig. 6A and B). Meanwhile, we found that there was no difference between the GEN group and the CK group in migration and invasion experiments. Western blot analysis revealed that CII significantly induced the expression of MMP-9 (P < 0.01), while GEN treatment significantly inhibited the expression (P < 0.05) (Fig. 6C). Consistently, ELISA results demonstrated a substantial decrease in MMP-9 levels upon GEN administration (P < 0.01) (Fig. 6D).Next, we assessed the impact of GEN on inflammatory factors from CII-induced FLS. The results of ELISA analysis showed that CII significantly induced the levels of pro-inflammatory cytokines TNF-α, IL-1β, and IL-6 (P < 0.01, P < 0.01, and P < 0.01). In contrast, GEN supplementation significantly inhibited the induction of CII on these pro-inflammatory factors (P < 0.01, P < 0.01, and P < 0.01) (Fig. 6E). Collectively, these results indicate that GEN inhibits migration, invasion, and inflammatory responses in CII-induced FLS, contributing to the suppression of disease progression.
GEN promoted the differentiation and mineralization of primary osteoblasts through reducing miR-221-3p expression in FLS-derived exosomes and subsequently inducing DKK2 expression in osteoblasts and FLS
Osteoblasts play a critical role in bone formation through the processes of proliferation, differentiation, and mineralization. And miR-221-3p was found to probably suppress the expression of Dkk2 in primary osteoblasts, and subsequent osteoblast maturation, contributing to the inhibition of bone formation in RA erosions.13 Thus, miR-221-3p expression in exosomes from various groups of FLS was assessed using qRT-PCR. The results revealed that the expression levels of miR-211-3p in exosomes were significantly elevated following CII induction (P < 0.01), whereas treatment with the GEN resulted in a significant decrease in its expression (Fig. 7A; P < 0.01).Then, primary osteoblasts were co-cultured with FLS treated with or without CII, and the expression of DKK2 was determined by western blot analysis. After induction of CII, the expression of DKK2 in cells was significantly decreased (Fig. 7B; P < 0.05). However, treatment with GEN resulted in a substantial increase in DKK2 expression in CII-treated cells (P < 0.05). Furthermore, alkaline phosphatase (ALP) assays and Von Kossa staining were performed to evaluate osteoblasts’ differentiation and mineralization capabilities, and ALP assays demonstrated that the activity of ALP in primary osteoblasts from GEN-treated cells was markedly facilitated, with a more pronounced promotion observed over time (Fig. 7C; P < 0.05). Von Kossa staining showed that after 14 days of treatment with GEN, calcified nodules formed, which exhibited a distinctly brown coloration compared to both control and CII-induced cells (Fig. 7D). Notably, at 21 days, the staining intensity increased further, with cells showing a more pronounced dark brown coloration relative to the 14 day group. At this time point, calcium deposits in the GEN group were significantly more abundant than those in both control and CII groups, indicating that GEN effectively promotes the differentiation and mineralization of primary osteoblasts. Collectively, these findings provide robust evidence that GEN serves as a potent inducer of osteoblast differentiation and mineralization.
Discussion
RA is a multifaceted autoimmune disorder characterized by intricate interactions among diverse cell types and molecular pathways.30 Despite extensive research, its pathogenesis remains incompletely understood. FLS play a pivotal role in RA, particularly through the release of cellular exosomes intimately linked to pathological immune interactions.31 This study elucidates the protective role of GEN, the predominant soy isoflavone, highlighting the inhibitory effect of GEN on FLS-derived exosome secretion in CIA mouse models and in vitro. This finding provides a new potential mechanism for the treatment of RA.In terms of the in vivo experiments, our findings demonstrate that GEN effectively mitigates inflammation and joint damage, as evidenced by alleviated paw swelling and preserved structural integrity of the knee and ankle joints in CIA mice. These results support the therapeutic promise of GEN in RA management, which is characterized by persistent inflammation and degradation of joint tissues. Moreover, GEN supplementation leads to a marked reduction in the levels of MMP-9 as well as pro-inflammatory cytokines, including TNF-α, IL-1β, and IL-6, key mediators in the pathological progression of RA. By downregulating these inflammatory factors, GEN addresses one of the core aspects of RA pathology: the promotion of a locally inflammatory microenvironment that exacerbates joint damage. Moreover, we observed that GEN significantly diminished the expression of β-catenin and exosomal Dvl3 in FLS, indicating its inhibitory effects on the Wnt signaling pathway, a critical regulator of inflammation, cellular proliferation, and migration in synovial fibroblasts.32,33 These findings show striking similarities to previous studies,34,35 but here we observed that GEN weakening RA correlates with exosome secretion.
The interaction of GEN with key regulators of exosome secretion – Rab27a, nSMase2, and Mfge8 – illustrates a novel mechanism through which GEN may modulate extracellular vesicle (EV) release and thereby influence intercellular communication within the synovial environment.19–21,36 The validation of these interactions through CETSA and DARTS underscores the direct impact of GEN on these molecular targets.37
Further in vitro mechanistic studies revealed that GEN not only downregulates the expression of Rab27a, nSMase2, and Mfge8, but also inhibits the secretion of Alix+Hsp70+CD63+ exosomes, a marker profile indicative of exosomes derived from FLS.10,38 This finding is particularly significant, given the emerging role of exosomal signaling in the pathogenesis of RA, where exosomes can propagate inflammatory signals and contribute to joint destruction.
Additionally, GEN's inhibition of CII-induced cell migration and its impact on MMP-9 in FLS and miR-221-3p expression in FLS exosomes suggest a multifaceted strategy for mitigating RA pathophysiology. MiR-221-3p, known to regulate various signaling pathways associated with inflammation and cell differentiation, may play an integral role in the modulation of RA-related processes.13 MMP-9, an important matrix metalloproteinase, plays a key role in the remodeling and degradation of the extracellular matrix. By degrading the extracellular matrix, MMP-9 plays a facilitating role in cell migration, which is important for cell migration and infiltration during inflammation.39–41 Importantly, our investigation also demonstrated that GEN enhances osteoblast differentiation and mineralization in vitro, a critical aspect of promoting bone health in RA patients who often exhibit bone loss and erosive changes due to persistent inflammation.12,42 The dual action of GEN to inhibit inflammatory mediators while promoting bone-forming cells highlights its therapeutic potential in addressing RA inflammation and bone pathology.
The potential clinical application of GEN in RA management is particularly noteworthy when comparing it with existing therapies, such as non-steroidal anti-inflammatory drugs (NSAIDs),43 disease-modifying anti-rheumatic drugs (DMARDs),44,45 and biological agents. While NSAIDs and DMARDs primarily focus on reducing symptoms and inhibiting disease progression, they may be limited by side effects and may not fully address the underlying pathological mechanisms, such as exosome-mediated inflammation. In contrast, GEN's mechanism of action, particularly its ability to inhibit exosome secretion and modulate inflammatory pathways, may complement these existing therapies by targeting one of the root causes of RA pathology: aberrant immune interactions facilitated by exosomes. Moreover, due to its natural origin and relatively favorable safety profile, GEN could potentially enhance patient compliance compared to synthetic therapies, making it an attractive adjunct treatment. In addition, GEN could work synergistically with current therapies,46 such as methotrexate or biologics, by providing a multi-targeted approach for RA management. Such a combination could optimize therapeutic outcomes, while minimizing the adverse effects associated with higher doses of conventional medications.
Despite these promising findings, this study has limitations that must be addressed in future research. The long-term effects of GEN on joint health and its potential impact when combined with existing RA therapies require further investigation in clinical settings. Additionally, while our in vitro and in vivo studies provide valuable insights, more comprehensive clinical trials are needed to confirm the efficacy and safety of GEN in RA patients. Future studies should also explore the specific mechanisms underlying the synergy between GEN and conventional RA therapies, as well as the potential for individualized treatment approaches based on patient-specific factors, such as genetic markers and disease severity. Further elucidation of the full spectrum of GEN's effects is essential, including its pharmacokinetics and dynamics in RA patients.
In summary, GEN holds significant potential as a complementary therapy in RA, addressing unmet needs in current therapeutic strategies. Continued research into its mechanisms, clinical applications, and potential synergy with existing treatments will be critical in advancing RA management.
Author contributions
J. L., J. S., and S. N. contributed equally. J. L., J. S., and S. N. executed experiments, gathered and analysed data, interpreted results, and contributed to manuscript preparation. X. C., Z. L., and Y. S. conducted the experiments. X. T., J. F., and H. X. conducted the in vivo experiments and were responsible for data analyses. W. Y., M. Z., B. L., N. L., and D. P. conducted the in vivo experiments and were responsible for data analyses. Y. W. and L. Y. designed experiments and acquired financial support and sponsorship. All authors reviewed and approved the final version of the manuscript revision.Ethical statement
DBA/1 mice were purchased from GemPharmatech (Nanjing, China). All animal procedures were performed in accordance with the Guidelines for Care and Use of Laboratory Animals of Jilin University and approved by the Animal Ethics Committee of Jilin University (number of permit: SY202407301).Data availability
All primary data were collected by the research team itself. Data can be obtained by contacting the authors by request.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the National Nature Science Foundation of China (32473029), the National Key Research and Development Program of China (2021YFC2600200), the Science and Technology Research Project of the Jilin Provincial Department of Education (JKH20211179KJ; 2016444), and the Jilin Provincial Nature Science Foundation of Jilin Provincial Department of Science and Technology (20210101341JC). This work was also supported by the National Natural Science Foundation of China (No. 81801972). We would like to thank the imaging platform in the core facility of the First Hospital of Jilin University, for their essential support and services which significantly contributed to our research.References
- D. L. Scott, F. Wolfe and T. W. Huizinga, Rheumatoid Arthritis, Lancet, 2010, 376(9746), 1094–1108 CrossRef PubMed.
- J. Falconer, A. N. Murphy, S. P. Young, A. R. Clark, S. Tiziani, M. Guma and C. D. Buckley, Review: Synovial Cell Metabolism and Chronic Inflammation in Rheumatoid Arthritis, Arthritis Rheumatol., 2018, 70(7), 984–999 CrossRef CAS PubMed.
- P. P. Katz and L. A. Criswell, Differences in Symptom Reports between Men and Women with Rheumatoid Arthritis, Arthritis Rheum., 1996, 9(6), 441–448 CrossRef CAS PubMed.
- L. C. Huber, O. Distler, I. Tarner, R. E. Gay, S. Gay and T. Pap, Synovial Fibroblasts: Key Players in Rheumatoid Arthritis, Rheumatology, 2006, 45 (6), 669–675 CrossRef PubMed.
- A. Sharma and A. Goel, Pathogenesis of Rheumatoid Arthritis and Its Treatment with Anti-Inflammatory Natural Products, Mol. Biol. Rep., 2023, 50(5), 4687–4706 CrossRef CAS PubMed.
- T. Pap, New Insights into Integrin Signalling: Implications for Rheumatoid Arthritis Synovial Fibroblasts, Arthritis Res. Ther., 2003, 5(3), 154–155 CrossRef CAS PubMed.
- S.-H. Kok, L.-D. Lin, K.-L. Hou, C.-Y. Hong, C.-C. Chang, M. Hsiao, J.-H. Wang, E. H.-H. Lai and S.-K. Lin, Simvastatin Inhibits Cysteine-Rich Protein 61 Expression in Rheumatoid Arthritis Synovial Fibroblasts through the Regulation of Sirtuin-1/FoxO3a Signaling, Arthritis Rheum., 2013, 65(3), 639–649 CrossRef CAS PubMed.
- J. H. W. Distler, A. Jüngel, L. C. Huber, C. A. Seemayer, C. F. Reich, R. E. Gay, B. A. Michel, A. Fontana, S. Gay, D. S. Pisetsky and O. Distler, The Induction of Matrix Metalloproteinase and Cytokine Expression in Synovial Fibroblasts Stimulated with Immune Cell Microparticles, Proc. Natl. Acad. Sci. U. S. A., 2005, 102(8), 2892–2897 CrossRef CAS PubMed.
- Y. Zou, S. Xu, Y. Xiao, Q. Qiu, M. Shi, J. Wang, L. Liang, Z. Zhan, X. Yang, N. Olsen, S. G. Zheng and H. Xu, Long Noncoding RNA LERFS Negatively Regulates Rheumatoid Synovial Aggression and Proliferation, J. Clin. Invest., 2018, 128(10), 4510–4524 CrossRef PubMed.
- F. Tavasolian, A. S. Moghaddam, F. Rohani, E. Abdollahi, E. Janzamin, A. A. Momtazi-Borojeni, S. A. Moallem, T. Jamialahmadi and A. Sahebkar, Exosomes: Effectual Players in Rheumatoid Arthritis, Autoimmun. Rev., 2020, 19(6), 102511 CrossRef CAS PubMed.
- G. Evangelatos, G. E. Fragoulis, V. Koulouri and G. I. Lambrou, MicroRNAs in Rheumatoid Arthritis: From Pathogenesis to Clinical Impact, Autoimmun. Rev., 2019, 18(11), 102391 CrossRef CAS PubMed.
- C. Ospelt, S. Gay and K. Klein, Epigenetics in the Pathogenesis of RA, Semin. Immunopathol., 2017, 39(4), 409–419 CrossRef PubMed.
- Y. Maeda, N. H. Farina, M. M. Matzelle, P. J. Fanning, J. B. Lian and E. M. Gravallese, Synovium–Derived MicroRNAs Regulate Bone Pathways in Rheumatoid Arthritis, J. Bone Miner. Res., 2017, 32(3), 461–472 CrossRef CAS PubMed.
- D. P. Bartel, MicroRNAs: Genomics, Biogenesis, Mechanism, and Function, Cell, 2004, 116(2), 281–297 CrossRef CAS PubMed.
- Z. Chen, H. Wang, Y. Xia, F. Yan and Y. Lu, Therapeutic Potential of Mesenchymal Cell–Derived miRNA-150-5p–Expressing Exosomes in Rheumatoid Arthritis Mediated by the Modulation of MMP14 and VEGF, J. Immunol., 2018, 201(8), 2472–2482 CrossRef CAS PubMed.
- I. Pandis, C. Ospelt, N. Karagianni, M. C. Denis, M. Reczko, C. Camps, A. G. Hatzigeorgiou, J. Ragoussis, S. Gay and G. Kollias, Identification of microRNA-221/222 and microRNA-323-3p Association with Rheumatoid Arthritis via Predictions Using the Human Tumour Necrosis Factor Transgenic Mouse Model, Ann. Rheum. Dis., 2012, 71(10), 1716–1723 CrossRef CAS PubMed.
- W. Li, D. Mu, F. Tian, Y. Hu, T. Jiang, Y. Han, J. Chen, G. Han and X. Li, Exosomes Derived from Rab27a-overexpressing Tumor Cells Elicit Efficient Induction of Antitumor Immunity, Mol. Med. Rep., 2013, 8(6), 1876–1882 CrossRef CAS PubMed.
- Y. Feng, X. Zhong, T.-T. Tang, C. Wang, L.-T. Wang, Z.-L. Li, H.-F. Ni, B. Wang, M. Wu, D. Liu, H. Liu, R.-N. Tang, B.-C. Liu and L.-L. Lv, Rab27a Dependent Exosome Releasing Participated in Albumin Handling as a Coordinated Approach to Lysosome in Kidney Disease, Cell Death Dis., 2020, 11(7), 1–17 Search PubMed.
- Y. Lu, L. Liu, J. Pan, B. Luo, H. Zeng, Y. Shao, H. Zhang, H. Guan, D. Guo, C. Zeng, R. Zhang, X. Bai, H. Zhang and D. Cai, MFG-E8 Regulated by miR-99b-5p Protects against Osteoarthritis by Targeting Chondrocyte Senescence and Macrophage Reprogramming via the NF-κB Pathway, Cell Death Dis., 2021, 12(6), 1–15 Search PubMed.
- N. Kosaka, H. Iguchi, K. Hagiwara, Y. Yoshioka, F. Takeshita and T. Ochiya, Neutral Sphingomyelinase 2 (nSMase2)-Dependent Exosomal Transfer of Angiogenic MicroRNAs Regulate Cancer Cell Metastasis*, J. Biol. Chem., 2013, 288(15), 10849–10859 CrossRef CAS PubMed.
- M. Ostrowski, N. B. Carmo, S. Krumeich, I. Fanget, G. Raposo, A. Savina, C. F. Moita, K. Schauer, A. N. Hume, R. P. Freitas, B. Goud, P. Benaroch, N. Hacohen, M. Fukuda, C. Desnos, M. C. Seabra, F. Darchen, S. Amigorena, L. F. Moita and C. Thery, Rab27a and Rab27b Control Different Steps of the Exosome Secretion Pathway, Nat. Cell Biol., 2010, 12(1), 19–30 CrossRef CAS PubMed.
- W. Tan, N. Chen, Y. Qiu, X. Feng, S. Li, Y. Zhang, H. Li, J. Gao and D. Zhao, Exosomal Dvl3 Promoted the Aggressive Phenotypic Transformation of RA-FLS via Wnt Pathway, Autoimmunity, 2022, 55(5), 285–293 CrossRef CAS PubMed.
- W.-X. Cheng, H. Huang, J.-H. Chen, T.-T. Zhang, G.-Y. Zhu, Z.-T. Zheng, J.-T. Lin, Y.-P. Hu, Y. Zhang, X.-L. Bai, Y. Wang, Z.-W. Xu, B. Song, Y.-Y. Mao, F. Yang and P. Zhang, Genistein Inhibits Angiogenesis Developed during Rheumatoid Arthritis through the IL-6/JAK2/STAT3/VEGF Signalling Pathway, J. Orthop. Transl., 2020, 22, 92–100 Search PubMed.
- Y. H. Gao and M. Yamaguchi, Inhibitory Effect of Genistein on Osteoclast-like Cell Formation in Mouse Marrow Cultures, Biochem. Pharmacol., 1999, 58(5), 767–772 CrossRef CAS PubMed.
- T. Nakasa, H. Shibuya, Y. Nagata, T. Niimoto and M. Ochi, The Inhibitory Effect of microRNA-146a Expression on Bone Destruction in Collagen-Induced Arthritis, Arthritis Rheum., 2011, 63(6), 1582–1590 CrossRef CAS PubMed.
- T. Zimmermann, E. Kunisch, R. Pfeiffer, A. Hirth, H. D. Stahl, U. Sack, A. Laube, E. Liesaus, A. Roth, E. Palombo-Kinne, F. Emmrich and R. W. Kinne, Isolation and Characterization of Rheumatoid Arthritis Synovial Fibroblasts from Primary Culture–Primary Culture Cells Markedly Differ from Fourth-Passage Cells, Arthritis Res., 2001, 3(1), 72–76 CrossRef CAS PubMed.
- J. Li, J. Li, Y. Yue, Y. Hu, W. Cheng, R. Liu, X. Pan and P. Zhang, Genistein Suppresses Tumor Necrosis Factor α-Induced Inflammation via Modulating Reactive Oxygen Species/Akt/Nuclear Factor κB and Adenosine Monophosphate-Activated Protein Kinase Signal Pathways in Human Synoviocyte MH7A Cells, Drug Des., Dev. Ther., 2014, 8, 315–323 CrossRef PubMed.
- J. Pratap, M. Galindo, S. K. Zaidi, D. Vradii, B. M. Bhat, J. A. Robinson, J.-Y. Choi, T. Komori, J. L. Stein, J. B. Lian, G. S. Stein and A. J. van Wijnen, Cell Growth Regulatory Role of Runx2 during Proliferative Expansion of Preosteoblasts, Cancer Res., 2003, 63(17), 5357–5362 CAS.
- S. Griffiths-Jones, H. K. Saini, S. van Dongen and A. J. Enright, miRBase: Tools for microRNA Genomics, Nucleic Acids Res., 2008, 36(Database issue), D154–D158 CAS.
- A. Di Matteo, J. M. Bathon and P. Emery, Rheumatoid Arthritis, Lancet, 2023, 402(10416), 2019–2033 CrossRef PubMed.
- Y. Zhang, J. Dong, P. He, W. Li, Q. Zhang, N. Li and T. Sun, Genistein Inhibit Cytokines or Growth Factor-Induced Proliferation and Transformation Phenotype in Fibroblast-Like Synoviocytes of Rheumatoid Arthritis, Inflammation, 2012, 35(1), 377–387 CrossRef CAS PubMed.
- M. M. Matzelle, M. A. Gallant, K. W. Condon, N. C. Walsh, C. A. Manning, G. S. Stein, J. B. Lian, D. B. Burr and E. M. Gravallese, Resolution of Inflammation Induces Osteoblast Function and Regulates the Wnt Signaling Pathway, Arthritis Rheum., 2012, 64(5), 1540–1550 CrossRef CAS PubMed.
- R. Baron and M. Kneissel, WNT Signaling in Bone Homeostasis and Disease: From Human Mutations to Treatments, Nat. Med., 2013, 19(2), 179–192 CrossRef CAS PubMed.
- A. Sharma, A. Goel and Z. Lin, Analysis of Anti-Rheumatic Activity of Nyctanthes Arbor-Tristis via in Vivo and Pharmacovigilance Approaches, Front. Pharmacol., 2023, 14, 1307799 CrossRef CAS PubMed.
- A. Sharma and A. Goel, Inflammatory Cytokines in Rheumatoid Arthritis: Diagnostic Challenges, Pathogenic Mechanisms and Their Role in Depression and Management, Curr. Top. Med. Chem., 2023, 23(27), 2535–2551 CrossRef CAS PubMed.
- E. Albus, K. Sinningen, M. Winzer, S. Thiele, U. Baschant, A. Hannemann, J. Fantana, A. Tausche, H. Wallaschofski, M. Nauck, H. Völzke, S. Grossklaus, T. Chavakis, M. C. Udey, L. C. Hofbauer and M. Rauner, Milk Fat Globule–Epidermal Growth Factor 8 (MFG–E8) Is a Novel Anti–inflammatory Factor in Rheumatoid Arthritis in Mice and Humans, J. Bone Miner. Res., 2016, 31(3), 596–605 CrossRef CAS PubMed.
- F. Sun, Y. Ma, F. Li, C. Li and Z. Yang, Exosomes Derived from Berberine-Treated Bone Marrow Mesenchymal Stem Cells Ameliorate Inflammatory Arthritis in Rats with Collagen-Induced Rheumatoid Arthritis, Food Agric. Immunol., 2023, 34(1), 2220566 CrossRef.
- Y. Liu, P. Jiang, Y. Qu, C. Liu, D. Zhang, B. Xu and Q. Zhang, Exosomes and Exosomal miRNAs: A New Avenue for the Future Treatment of Rheumatoid Arthritis, Heliyon, 2024, 10(6), e28127 CrossRef PubMed.
- K. Nabeshima, T. Inoue, Y. Shimao and T. Sameshima, Matrix Metalloproteinases in Tumor Invasion: Role for Cell Migration, Pathol. Int., 2002, 52(4), 255–264 CrossRef CAS PubMed.
- C. Colnot, Z. Thompson, T. Miclau, Z. Werb and J. A. Helms, Altered Fracture Repair in the Absence of MMP9, Development, 2003, 130(17), 4123–4133 CrossRef CAS PubMed.
- C. Legrand, C. Gilles, J.-M. Zahm, M. Polette, A.-C. Buisson, H. Kaplan, P. Birembaut and J.-M. Tournier, Airway Epithelial Cell Migration Dynamics: MMP-9 Role in Cell–Extracellular Matrix Remodeling, J. Cell Biol., 1999, 146(2), 517–529 CrossRef CAS PubMed.
- G. S. Firestein, Evolving Concepts of Rheumatoid Arthritis, Nature, 2003, 423(6937), 356–361 CrossRef CAS PubMed.
- B. Möller, M. Pruijm, S. Adler, A. Scherer, P. M. Villiger and A. Finckh, Swiss Clinical Quality Management in Rheumatic Diseases (SCQM) Foundation, CH-8048 Zurich, Switzerland. Chronic NSAID Use and Long-Term Decline of Renal Function in a Prospective Rheumatoid Arthritis Cohort Study, Ann. Rheum. Dis., 2015, 74(4), 718–723 CrossRef PubMed.
- A. Kerschbaumer, A. Sepriano, S. A. Bergstra, J. S. Smolen, D. Heijde, R. van der Caporali, C. J. Edwards, P. Verschueren, S. Souza, J. E. de Pope, T. Takeuchi, K. L. Hyrich, K. L. Winthrop, D. Aletaha, T. A. Stamm, J. W. Schoones and R. B. M. Landewé, Efficacy of Synthetic and Biological DMARDs: A Systematic Literature Review Informing the 2022 Update of the EULAR Recommendations for the Management of Rheumatoid Arthritis, Ann. Rheum. Dis., 2023, 82(1), 95–106 CrossRef CAS PubMed.
- S. Ramiro, C. Gaujoux-Viala, J. L. Nam, J. S. Smolen, M. Buch, L. Gossec, D. Heijde, K. van der Winthrop and R. Landewé, Safety of Synthetic and Biological DMARDs: A Systematic Literature Review Informing the 2013 Update of the EULAR Recommendations for Management of Rheumatoid Arthritis, Ann. Rheum. Dis., 2014, 73(3), 529–535 CrossRef CAS PubMed.
- P. M. Brown, A. G. Pratt and J. D. Isaacs, Mechanism of Action of Methotrexate in Rheumatoid Arthritis, and the Search for Biomarkers, Nat. Rev. Rheumatol., 2016, 12(12), 731–742 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2025 |