 Open Access Article
Open Access ArticleA pilot study of metaproteomics and DNA metabarcoding as tools to assess dietary intake in humans†
Brianna L.
Petrone
 ab,
Alexandria
Bartlett
ac,
Sharon
Jiang
a,
Abigail
Korenek
c,
Simina
Vintila
c,
Christine
Tenekjian
d,
William S.
Yancy
Jr.
de,
Lawrence A.
David
*a and
Manuel
Kleiner
ab,
Alexandria
Bartlett
ac,
Sharon
Jiang
a,
Abigail
Korenek
c,
Simina
Vintila
c,
Christine
Tenekjian
d,
William S.
Yancy
Jr.
de,
Lawrence A.
David
*a and
Manuel
Kleiner
 *c
*c
aDepartment of Molecular Genetics and Microbiology, Duke University School of Medicine, Durham, NC, USA. E-mail: lawrence.david@duke.edu
bMedical Scientist Training Program, Duke University School of Medicine, Durham, NC, USA
cDepartment of Plant and Microbial Biology, North Carolina State University, Raleigh, NC, USA. E-mail: manuel_kleiner@ncsu.edu
dDuke Lifestyle and Weight Management Center, Durham, NC, USA
eDepartment of Medicine, Duke University School of Medicine, Durham, NC, USA
First published on 12th December 2024
Abstract
Objective biomarkers of food intake are a sought-after goal in nutrition research. Most biomarker development to date has focused on metabolites detected in blood, urine, skin, or hair, but detection of consumed foods in stool has also been shown to be possible via DNA sequencing. An additional food macromolecule in stool that harbors sequence information is protein. However, the use of protein as an intake biomarker has only been explored to a very limited extent. Here, we evaluate and compare measurement of residual food-derived DNA and protein in stool as potential biomarkers of intake. We performed a pilot study of DNA sequencing-based metabarcoding and mass spectrometry-based metaproteomics in five individuals’ stool sampled in short, longitudinal bursts accompanied by detailed diet records (n = 27 total samples). Dietary data provided by stool DNA, stool protein, and written diet record independently identified a strong within-person dietary signature, identified similar food taxa, and had significantly similar global structure in two of the three pairwise comparisons between measurement techniques (DNA-to-protein and DNA-to-diet record). Metaproteomics identified proteins including myosin, ovalbumin, and beta-lactoglobulin that differentiated food tissue types like beef from dairy and chicken from egg, distinctions that were not possible by DNA alone. Overall, our results lay the groundwork for development of targeted metaproteomic assays for dietary assessment and demonstrate that diverse molecular components of food can be leveraged to study food intake using stool samples.
Introduction
Biomarkers of food intake are a promising tool for measuring human nutrition, which is the leading preventable risk factor for mortality worldwide.1 Food-derived compounds measured in biological specimens can provide objective and accurate data on what is being eaten. Objective measurement strategies can overcome limitations of self-reported dietary data, which are collected by asking individuals to describe what they eat via written record, standardized surveys, or a trained interviewer. While self-reports have provided key insights into the relationship between diet and health, they are at best considered a semi-quantitative strategy with well-characterized sources of error,2 including participant memory, cognitive ability, language and cultural suitability of the collection tool, and social desirability bias (e.g. a tendency to underreport food intake3). Most self-report tools are either administered only once to assess habitual diet for up to the last year (“food frequency questionnaires”), or a handful of times—though there are notable exceptions4—to collect repeated data on short-term intake (“24 hours recalls”). Repeated self-reporting increases the likelihood of reactivity bias, in which individuals alter their diet to make it simpler to report: most existing dietary assessments take between 30 minutes to upwards of an hour to complete, placing both time and cognitive burdens on participants, and depending on the method, research staff. Depending on the specimen type and underlying physiology, biomarkers can facilitate denser sampling strategies to provide high-resolution data on daily variation in diet. Development and validation of dietary biomarkers therefore has the potential to generate more accurate and comprehensive dietary data from large numbers of individuals and to enable retrospective investigations in studies where dietary data were not initially collected.5Prior work on biomarkers of food intake has largely focused on metabolites present in blood, urine, skin, or hair. Stool has been used to a much lesser extent although it is both widely collected in studies and produced directly from consumed foods. Driven by increased research on gut microbiota, large studies now routinely sample stool from hundreds to thousands of individuals.6–9 Stool contains molecular information aggregated from host, microbial, dietary, clinical, and environmental factors, and is increasingly leveraged at the population scale with wastewater epidemiology to monitor infectious disease, illicit drug use, or whole-community microbiome composition. After microbial biomass, the largest portion of the dry weight of organic solids in feces is derived from unabsorbed dietary carbohydrate (∼25%), protein (2–25%), and fat (2–15%), with exact proportions varying with the specific foods consumed.10 However, much of the biomarker development in stool to date has relied on measuring proxies for residual food material, rather than the food itself. For example, recent efforts have measured fecal metabolite shifts in response to supplementation11 or substitution of the source12 of dietary protein, or used gut microbiota to predict which of six food items was included in the diet as a controlled intervention.13
Direct assessment of food tissue, however, is feasible even after degradation in the gastrointestinal tract. Here, we focus on two macromolecules, DNA and protein, which can provide information on consumed food. Both can be measured with omic-scale tools (“metabarcoding” or “FoodSeq” for DNA, and “metaproteomics” for protein) that quantitatively survey residual food DNA or proteins in stool samples, analogous to qualitative surveys that ask individuals to report their diet. Dietary DNA was first used to study foods consumed by wild animals before being applied to free-living human populations.14 In its current form, FoodSeq uses high-throughput sequencing technology to amplify and identify marker DNA regions from consumed food genomes. Early animal dietary studies also relied on detection of protein from consumed prey tissues and demonstrated that protein epitopes from prey could resist digestion for up to several days.15 Metaproteomics, which is the large-scale identification and quantification of proteins from microbiomes using high-resolution mass spectrometry,16 has since been applied to query diverse measures in microbial communities17 and used to study nutrient flows in biological systems.18 Recent work has developed high-throughput metaproteomic methods for stool19 that were applied to a human cohort undergoing a dietary intervention.20 Plant proteins were observed in the dataset but not systematically identified and analyzed due to lack of a food proteome reference database. This is representative of broader metaproteomics studies of the gut microbiome, which exclude the dietary proteins contained in the mass spectrometry data by not including them in protein references or removing them as “contaminants”.
To evaluate and compare molecular assessment of dietary proteins and DNA in stool, we applied both metaproteomics and FoodSeq to a pilot set of samples collected in short longitudinal bursts (most in runs of three to five days) from five individuals with detailed accompanying diet records. We began by developing the infrastructure for dietary detection with metaproteomics, which to our knowledge has not yet been applied in humans. To this end, we created the first protein sequence database curated for the identification of dietary peptides and evaluated strategies for analyzing the resulting data. We evaluated broad correspondence between DNA, protein, and diet record measurements, then specifically compared molecular dietary measures to conventional diet records as a reference technique. Finally, we used the high detail of metaproteomics to provide examples of candidate food tissue-specific biomarkers that could be further developed for intake of exact food items.
Materials and methods
Human study design, sample collection, and diet composition
Samples were drawn from two human studies based at Duke University in Durham, NC: a behavioral intervention that returned daily gut microbiome data to participants by text message to incentivize fiber intake (NCT04037306, here “Intervention”) and a protocol for stool collection from healthy donors (with approval of the Duke Health Institutional Review Board [IRB] Pro00049498, “Habitual Diet”). All participants provided written informed consent and authorized future use of their de-identified stool samples for research, which was performed in compliance with Duke Health IRB guidelines. Application of FoodSeq and metaproteomics was a secondary analysis and determined exempt by the Duke Health IRB (Pro00100567).The data collected by each study are compared side-by-side in Fig. S1.† In brief, participants in both studies collected daily stool samples when permitted by their natural bowel habits. All participants first collected their entire stool in a plastic collection tub suspended above the toilet bowl. Intervention participants subsampled 3–5 g amounts with scoop-cap tubes in triplicate. The Habitual Diet participant returned the entire stool sample, which was aliquoted into a similar 3–5 g amount. All samples were stored at −20 °C immediately after collection.
Participants in the “Intervention” study were clients of a residential-style, medically supervised weight loss center, with all their weekday meals prepared in the center's cafeteria and consumed on site. Diets were low-calorie (ranging from ∼1200 to 1700 kcal day−1, depending on participant body size) but were nutrient dense, included many unique foods, and varied day-to-day. A digital menu system used by clients to order meals recorded the exact amount of food served, except for beverages, a daily fruit offering, and a salad bar, which could be freely chosen. Participants were encouraged to dine out or prepare their own meals on weekends and submitted food diaries recording Saturday and Sunday intake. The “Habitual Diet” participant was a healthy donor with normal BMI who ate a freely selected diet with no restriction on caloric intake. Foods consumed included commercially prepared and home-cooked meals and were recorded in a food diary without information on amount, calorie, or nutrient content.
Diet detection with metaproteomics
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 minutes to pellet debris. The supernatant was transferred to fresh tubes and centrifuged 3 minutes at 21
000g for 5 minutes to pellet debris. The supernatant was transferred to fresh tubes and centrifuged 3 minutes at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to pellet any remaining debris. We then prepared tryptic digests (13.5 hours digestion) using the filter-aided sample preparation (FASP) protocol21 with all centrifugations performed at 14
000g to pellet any remaining debris. We then prepared tryptic digests (13.5 hours digestion) using the filter-aided sample preparation (FASP) protocol21 with all centrifugations performed at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. Briefly, we combined 60 μL of the lysate supernatant with 400 μL UA solution (8 M urea in 0.1 M Tris/HCl pH 8.5) in a 10 kDa MWCO 500 μl centrifugal filter unit (VWR International) and centrifuged for 40 minutes. Filters were washed with 200 μL of UA solution and centrifuged for 40 minutes. 100 μL IAA (0.05 M iodoacetamide in UA solution) was added to the filters, incubated 20 minutes, and centrifuged for 20 minutes. Filters were then washed with 100 μL of UA and centrifuged three times, and the buffer changed to 50 mM ammonium bicarbonate by adding 100 μl and centrifuging three times. For digestion, we added 0.95 μg of MS grade trypsin (Thermo Scientific Pierce, Rockford, IL, USA) in 40 μl of 50 mM ammonium bicarbonate to the filters and incubated 13.5 hours in a wet chamber at 37 °C. Following digestion, filters were centrifuged for 20 minutes, washed with 50 μL of 0.5 M NaCl, and centrifuged for an additional 20 minutes. Peptide concentrations were measured with the Pierce Micro BCA assay (Thermo Scientific Pierce).
000g. Briefly, we combined 60 μL of the lysate supernatant with 400 μL UA solution (8 M urea in 0.1 M Tris/HCl pH 8.5) in a 10 kDa MWCO 500 μl centrifugal filter unit (VWR International) and centrifuged for 40 minutes. Filters were washed with 200 μL of UA solution and centrifuged for 40 minutes. 100 μL IAA (0.05 M iodoacetamide in UA solution) was added to the filters, incubated 20 minutes, and centrifuged for 20 minutes. Filters were then washed with 100 μL of UA and centrifuged three times, and the buffer changed to 50 mM ammonium bicarbonate by adding 100 μl and centrifuging three times. For digestion, we added 0.95 μg of MS grade trypsin (Thermo Scientific Pierce, Rockford, IL, USA) in 40 μl of 50 mM ammonium bicarbonate to the filters and incubated 13.5 hours in a wet chamber at 37 °C. Following digestion, filters were centrifuged for 20 minutes, washed with 50 μL of 0.5 M NaCl, and centrifuged for an additional 20 minutes. Peptide concentrations were measured with the Pierce Micro BCA assay (Thermo Scientific Pierce).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and maximum injection time of 200 ms. We performed data-dependent MS2 for the 15 most abundant ions at resolution of 15
000 and maximum injection time of 200 ms. We performed data-dependent MS2 for the 15 most abundant ions at resolution of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and maximum injection time of 100 ms (Intervention) or 200 ms (Habitual Diet). The instrument parameters were as follows: 445.12003 lock mass, normalized collision energy equal to 24 and exclusion of ions with +1 charge state. We used a 25 s dynamic exclusion for Intervention samples and a 15 s dynamic exclusion for Habitual Diet samples. Method differences between the two sample sets are the result of LC-MS/MS method optimization.
000 and maximum injection time of 100 ms (Intervention) or 200 ms (Habitual Diet). The instrument parameters were as follows: 445.12003 lock mass, normalized collision energy equal to 24 and exclusion of ions with +1 charge state. We used a 25 s dynamic exclusion for Intervention samples and a 15 s dynamic exclusion for Habitual Diet samples. Method differences between the two sample sets are the result of LC-MS/MS method optimization.
A preliminary search before clustering against the most abundant human proteins revealed that some host digestive proteins (e.g. Homo sapiens alpha-amylase) were misidentified as dietary proteins (e.g. cattle, or Bos Taurus, alpha-amylase). To address these misidentifications, we concatenated the individual dietary proteomes and used cd-hit-2d to remove dietary proteins with an identity threshold of at least 50% to the 15 most abundant human proteins identified in the samples (Table S2†). This additional clustering step allowed us to be more confident that the dietary proteins we did identify were true dietary proteins and not cross-species identifications of host proteins. The final database contained 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 942
942![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 188 protein sequences.
188 protein sequences.
Proteinaceous biomass was calculated as previously described25 by considering proteins with ≥2 protein unique peptides to provide high confidence that the protein originated from a specific taxon in addition to the above 5% FDR condition. Total PSMs from the remaining proteins were then summed within human, microbial taxa, and dietary taxa.
We only considered identified dietary proteins that were the master proteins of their protein group and that had at least 5 cumulative PSMs in the dataset. After summing PSMs from proteins identified to the same food taxon, we also only considered a taxon “detected” if it had ≥5 PSMs.
Because the filter we applied to the protein reference database to address misidentification of host or microbial proteins as dietary was likely incomplete, we applied an additional filter to animal-derived proteins in the dataset. We reasoned that due to a closer evolutionary relationship, animal proteins were more likely to be misidentified as human than those derived from fungi or plants. We therefore manually categorized each of these proteins with >5 PSMs as “muscle”, “egg”, “dairy” or “other” and then used regular expressions to automatically label the remainder based on the names identified in the manual pass. The regular expressions for muscle-specific proteins included matches to terms like “actin”, “titin”, “sarco-”, and “myo-”; for egg to “ovo-” and “vitello-”; and for dairy to “casein” and “butyrophilin”. In downstream analyses, we excluded the “other” category to remove potential misidentifications, though we note that this strategy also excluded uncharacterized proteins.
Diet detection with FoodSeq
For both trnL and 12SV5, each PCR batch included a positive and negative control, and samples were only advanced to the secondary PCR if controls performed as expected (otherwise, the entire batch was repeated). Secondary PCR amplification to add Illumina adapters and dual 8 bp indices for sample multiplexing was performed in a 50 μl volume containing 5 μl of 2.5 μM forward and reverse indexing primers, 10 μl of 5X KAPA HiFi buffer, 1.5 μl of 10 mM dNTPs, 0.5 μl of 100X SYBR Green I, 0.5 μl KAPA HiFi polymerase, 22.5 μl nuclease free water, and 5 μl of primary PCR product diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in nuclease-free water.
100 in nuclease-free water.
Sequence data were screened for contamination on a per-PCR batch basis using decontam v1.8.036 using DNA quantitation data from the library pooling step, and suspected contaminants were removed. ASV count tables, taxonomic assignments, and metadata were organized using phyloseq v1.32.0. As with metaproteomic data, we considered a taxon “detected” if it had ≥5 sequence reads.
Dietary data collection and processing
Statistical analysis
To assess taxon overlap between the datasets, area-proportional Euler diagrams were visualized using the euler function of package eulerR v7.0.0.40 Exact tests of multi-set interaction were done with function MSET of package SuperExactTest v.1.1.0.41 The number of taxa detected within each sample was compared with repeated-measures ANOVA using the lm function of stats v4.2.2 and Anova of car v3.1.2.42
We compared DNA- or protein-based presence or absence to the presence or absence of the same food taxon in the menu record from 1 to 2 days prior to account for the mean (28 h) and typical variation of measured gastrointestinal transit times in humans (28, 29). Responses were coded as true positives (TP, food present by both molecular detection and menu), true negatives (TN, absent by both molecular detection and menu), false positives (FP, present by molecular detection, not by menu), and false negatives (FN, absent by molecular detection, but present in menu). Recall was calculated as TP/(TP + FN). Precision was calculated as TP/(TP + FP). Two-tailed Spearman correlations between precision and recall were performed using the cor.test function from R stats v4.1.3.
Results
DNA- and protein-based detection of consumed foods
We conducted paired assessment of food-derived proteins and DNA in stool collected by five individuals: four volunteers consuming interventional diets at a medically supervised weight loss center, and one donor consuming their typical diet (“Intervention” and “Habitual Diet”, respectively, n = 27 total samples). The samples shared two features that enabled the joint evaluation of metaproteomics and metabarcoding (FoodSeq): (1) they were accompanied by detailed, longitudinal dietary records for external validation (Fig. 1a), and (2) the majority were collected in runs of several consecutive days to investigate the kinetics of molecular dietary signals (Fig. S1†). Measurement of peptide spectra by metaproteomics, amplification of the trnL-P6 chloroplast marker region (“plant FoodSeq”), and amplification of the V5 region of the mitochondrial 12S rRNA gene (“animal FoodSeq”) was successful for 26, 23, and 27 of the samples, respectively. In successful samples, we detected a median of 1985 metaproteomic peptide-spectrum matches (range 794–17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 472), 68
472), 68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 256 trnL reads (range 9744–95
256 trnL reads (range 9744–95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 249), and 7647 12SV5 reads (range 404–22
249), and 7647 12SV5 reads (range 404–22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 705; Fig. S2).† To identify foods by their peptide spectra or DNA sequences, we generated reference databases of protein or marker gene sequences from a manually curated list of 246 foods known to be consumed in human diets. The protein reference database included proteomes from 180 plants, 56 animals, and 8 fungi (Table S1†) and was refined to exclude animal proteins similar to the human proteins that are most abundant in human fecal material (Table S2†). The DNA reference database contained 909 sequences representing 591 plants and 31 animals. Collectively, peptides from 8273 unique food-derived proteins and 93 DNA amplicon sequence variants (ASVs; 82 [88%] from trnL, and 11 [12%] from 12SV5) were detected in the sample set.
705; Fig. S2).† To identify foods by their peptide spectra or DNA sequences, we generated reference databases of protein or marker gene sequences from a manually curated list of 246 foods known to be consumed in human diets. The protein reference database included proteomes from 180 plants, 56 animals, and 8 fungi (Table S1†) and was refined to exclude animal proteins similar to the human proteins that are most abundant in human fecal material (Table S2†). The DNA reference database contained 909 sequences representing 591 plants and 31 animals. Collectively, peptides from 8273 unique food-derived proteins and 93 DNA amplicon sequence variants (ASVs; 82 [88%] from trnL, and 11 [12%] from 12SV5) were detected in the sample set.
 | ||
| Fig. 1 Dietary landscapes of study participants by written records and stool-based measurements. (a) Participant diets included food items derived from 27 food groups, shown as a heatmap of presence (gray) or absence (white) of each food group (x-axis) in the recorded diet on the day prior to stool sampling (y-axis). The x-axis dendrogram reflects food group relationships as structured by the Interagency Food Safety Analytics Collaboration (IFSAC)44 and the y-axis dendrogram the clustering of recorded diets by relatedness of the food groups they contain. Although food groups are displayed for clarity, dietary records provided data resolved to the level of individual food species (e.g. the column “vegetables, vegetable row crops” summarizes data on 13 unique food items, shown in inset). (b) Principal coordinate analysis (PCoA) and principal component analysis (PCA) ordinations of samples in dietary space derived from either recorded diet (presence-absence of food), metaproteomic detection of food proteins in stool (number of PSMs per food), or FoodSeq detection of DNA in stool (number of sequence reads per food). Points represent individual stool samples, or for menu data, the day of eating prior to sample collection. Samples are colored by participant and by diet type, either habitual diet (HD) or interventional diet (ID1 to ID4). Metaproteomic data are filtered with the final criteria described in the text (5% FDR, ≥1 UP and >5 PSMs for the food taxon). (c) Results of Mantel tests comparing distance matrices of points in (b) between datasets, interpretable as correlation coefficients (1, perfect positive correlation; 0, uncorrelated; −1, perfect negative correlation). | ||
In the metaproteomic dataset, proteinaceous biomass was dominated by microbiota-derived proteins (74% of all peptide-spectrum matches), with smaller contributions from host (14%) and dietary (11%) proteins (Fig. S3†). To tune the sensitivity and stringency of the metaproteomic analysis for dietary proteins, we tested two filtering methods: (1) selecting dietary proteins identified with a 5% false-discovery rate (FDR) cutoff and at least one protein unique peptide and (2) selecting dietary proteins identified with an FDR of 5% and at least one protein group unique peptide. The first analysis restricted proteins to those definitively identified by one or more peptides matching to a single protein sequence, whereas the second included proteins that did not have a unique peptide match, but for which there was a unique peptide match to the protein group composed of very similar homologous sequences. Because we observed only slight variations in the overall results between the protein-unique peptide (PUP) strategy and the unique peptide (UP) analysis and the UP analysis included 39% more PSMs, 2616 additional proteins, and 18 additional food taxa (Fig. S4†), we selected the UP analysis for all downstream steps.
Despite the steps taken to remove dietary proteins similar to human proteins from the metaproteomic reference database, we noted detection of a high number of food species (n = 18) in every sample, including staple foods like wheat, corn, and soy, but also less common items like goat and salmon. We also observed persistent detection of digestive tract and intestinal epithelial proteins (e.g. progastricin, enteropeptidase, glycoprotein 2). We therefore categorized every protein name from an animal species with ≥5 cumulative PSMs in the full dataset as “muscle”, “egg”, “dairy”, or “other” (n = 280 manual categorizations) and used regular expressions to automatically label the remainder (animal-derived proteins with <5 cumulative PSMs, n = 1170) with the same dietary categories (see Table S3†). In downstream analyses of animal taxa, we excluded the “other” category, which included likely host and microbial cross-identifications as well as uncharacterized proteins. When considering food taxa identified by metaproteomics generally, we filtered to only those taxa with five or more PSMs (46 taxa removed); we performed no additional filtering when analyzing data at the level of individual proteins. Across samples, 105 foods were detected by metaproteomics with the most abundant ones being rice (Oryza sativa), oats (Avena sativa), and chicken (Gallus gallus); out of 90 foods detected by FoodSeq, the most prevalent items were carrot family (a DNA sequence variant shared by carrots, celery, parsley, and parsnips), peppers, avocado, cattle, chicken, and turkey (Table S4†).
Overall dietary assessment by record, DNA, and protein
Because there is no gold-standard method for dietary assessment, we first evaluated the consistency between FoodSeq, metaproteomics, and conventional diet records as tools to capture inter- and intra-individual dietary differences. Participant identity was significantly associated with the overall composition of the diet provided by all three techniques (Fig. 1b). Roughly 22% of the variation in dietary composition captured in menu records could be attributed to differences between the participants (PERMANOVA on Jaccard dissimilarity, R2 = 0.22, p = 0.001). This individual-specific dietary signature was also present in both food DNA and protein composition in stool, and was stronger for DNA-based than protein-based data (PERMANOVA on Aitchison distance R2 = 0.45 for FoodSeq, and 0.28 for metaproteomics, both with p = 0.001; Fig. 1b).We tested if sample-to-sample differences were similar between the three measures of diet composition using the Mantel test, which evaluates correlation between distance matrices. When we used distance metrics that incorporated abundance data (Aitchison distance on the number of metaproteomic peptide-spectrum matches or DNA sequencing read counts), inter-sample distances were significantly and positively correlated for DNA and protein (Mantel r = 0.265, p = 0.001), DNA and menu (r = 0.307, p = 0.002), but not for menu and protein (r = −0.0453, p = 0.659; all visualized in Fig. 1c). Interestingly, only the relationship between menu- and DNA-based diet composition was preserved when we used a presence/absence-based distance, indicating potentially significant quantitative information present in the molecular datasets (Jaccard dissimilarity, Fig. S5†).
Direct comparison of molecular detection to recorded diet
We next evaluated FoodSeq and metaproteomic data in relationship to recorded dietary data. In doing so, we note that neither cohort's dietary data represented a “ground truth” comparator. The manually recorded diet of the healthy donor lacked information on amount of food and exact ingredients of prepared dishes that were present in the detailed digital menus from the Intervention center; the Intervention menus reported only food served (rather than consumed) and did not track items that could be freely chosen (e.g. fresh fruit and salad bar offerings that varied daily and potential off-menu eating that occurred when clients were not at the center). However, exact menu records or diet diaries are established methods of dietary assessment, so we evaluated molecular measures against them.All food items were coded to a plant, animal, or fungal source taxon. We manually synchronized taxon names across the three datasets to reconcile naming differences that arose by data type (see Methods, Table S5†). To align with estimates of 24–48 hours for gastrointestinal transit time,45,46 detected taxa were compared to foods recorded in the diet records from two days prior to sampling. There was significant overlap between taxa recorded or identified by the diet records, DNA, and protein, with the intersection size of foods observed by all three measures unlikely to be detected by chance (multi-set intersection test p = 0.03 for plant and p = 0.07 for animal taxa, Fig. 2a). 68% of plant taxa and 41% of animal taxa were detected by at least two measures. Comparing the number of food taxa identified within single samples, there was no difference between the number of plants or animals recorded in diet records from two days prior to sampling and detected by metaproteomics (repeated-measures ANOVA, Benjamini–Hochberg adjusted p = 0.72 and 0.69, respectively). FoodSeq identified significantly fewer plant and animal taxa in comparison to diet records (repeated-measures ANOVA, Benjamini–Hochberg adjusted p = 10−10 and 10−7 for plants and animals, respectively) and metaproteomics identified significantly more fungi within individual samples (paired t-test p = 0.002; Fig. 2b).
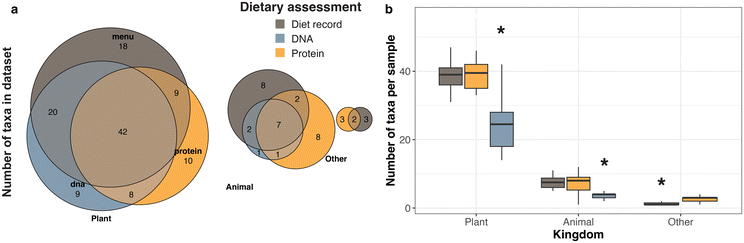 | ||
| Fig. 2 Diet records and stool measurements include similar food taxa. (a) Overlap between food taxa detected by diet record, stool DNA, or stool protein, separated by kingdom. “Detection” is any record of food intake, or any sequence read count or PSM count ≥5 in each sample. Note that the FoodSeq assay does not include a marker for fungi, seaweed, or bacteria-derived foods (i.e. xanthan gum), which are shown in the “Other” category. Menu data included from records that occurred 1 or 2 days prior to any collected sample. Molecular data is only included from samples with successful molecular detection by both protein- and DNA-based methods, resulting in a total number of taxa that differs slightly from that in all samples reported in Table S4.† (b) On a per-sample basis, dietary richness (the number of unique food items recorded or detected) was significantly lower by FoodSeq than for the two other measures. | ||
We next calculated FoodSeq and metaproteomic performance in comparison to the prior two days of recorded diet at the level of food taxon. Cumulatively, performance was higher for FoodSeq than metaproteomics across taxonomic ranks (Fig. 3a; two-way ANOVA p < 10−5 for performance by dataset, p = 0.4 for performance by taxonomic level) and, within each measure, significantly higher for animal compared to plant taxa (unpaired Mann–Whitney test, p < 10−5 and p = 0.005 for DNA and protein, respectively; Fig. 3b). Per-taxon performance varied widely, with some taxa in near-perfect agreement with menu data and others dominated by false positives, false negatives, or a mixture (Fig. 3c).
 | ||
| Fig. 3 Performance of DNA- and protein-based dietary assessments in comparison to recorded diet. (a) Summary of molecular detection consistency with menu data across taxonomic levels of analysis. All data are treated as presence-absence, described in Methods. “All” levels indicates no aggregation of taxa, preserving each individual taxon at the level to which it can most accurately be specified by the three methods (a mix of family, genus, and species designations). The “predictive performance” measure used here is the F-measure, which is the harmonic mean of precision and recall and ranges from 0 (completely inaccurate detection) to 1 (perfect precision and recall). Black bars indicate the median. (b) Comparison of performance by DNA- and protein-based assessment in comparison to recorded diet between food taxa of animal and plant origin. (c) Protein and DNA detections in comparison to the recorded diet from the two days prior to sample collection. For ease of visualization, data are presented at the family level; see Fig. S6† for a visualization of the full dataset. Detections were coded as true positives (TP, food present by both molecular measure and menu), true negatives (TN, absent by both molecular measure and menu), false positives (FP, present by molecular measure, not by menu), and false negatives (FN, absent by molecular measure, but present in menu). A gray bar indicates that the food was never detected by the molecular measure in any sample in the dataset; therefore, we cannot confirm that detection is possible and do not interpret the absence of detection as a true or false negative. Taxa are aggregated to the family level and ranked by their F measure statistic, which is the harmonic mean of their sensitivity and positive predictive value. | ||
Because of the limitations of the menu data noted above, we also directly compared FoodSeq and metaproteomic performance to determine if divergences from menu data were shared or distinct. For each measure, we calculated recall (or true positive rate, the proportion of recorded foods that were detected by molecular signal) and precision (or positive predictive value, the proportion of positive molecular signals that were confirmed by a menu entry). On a per-taxon basis, FoodSeq and metaproteomic recall were moderately correlated with one another (Spearman ρ = 0.02–0.52, p ∼ 10−1–10−4 across varying taxonomic rank), but for precision this correlation was markedly stronger (Spearman ρ = 0.80–0.88, p ∼ 10−11–10−14). This finding was consistent across taxonomic levels (Fig. 4). Recall and precision differ by only one term in their denominator: the number of false negatives (recall) or the number of false positives (precision). The increased correlation observed for precision indicated that the false positive structure between the two molecular measures was stronger than the false negative structure. In other words, a false positive by either measure was more likely to be shared by the alternate approach, and could be reflective of a real divergence from the recorded menu (a food or beverage not captured by diet records like tea [Theaceae], one commonly eaten but not reported like chocolate [Malvaceae], or an error in assuming a constant two day lag in our analysis). However, failures to detect recorded items were less correlated with one another, and therefore likely to be due to method-specific limitations or biases. From this comparison, we also noted extremes of detection: some foods were reliably detected by both measures (chicken, peppers, and carrots), better detected by FoodSeq (tilapia, peas), better detected by metaproteomics (citrus), and poorly suited for detection by either method (olives or their oil, sugarcane).
 | ||
| Fig. 4 Per-food detections by metaproteomics and FoodSeq are more correlated for precision than recall. Each point represents an individual food taxon, summarized to the taxonomic level as labeled at the top of each facet. “All” includes every taxon in the dataset without any aggregation; these can differ in their taxonomic level due to a variable degree of resolution in the FoodSeq data. As in Fig. 3, precision and recall are calculated based on presence-absence data for both recorded intake and molecular detection. | ||
Metaproteomics enables distinction of food type from the same source species
Finally, we selected a subset of foods for analysis to highlight the tissue-specificity of metaproteomics and support its use as a complementary measure to FoodSeq. Unlike DNA signatures, which are common across all tissues, metaproteomics provides data on individual protein constituents that reflect tissue of origin and thus food type. We began by specifically investigating Gallus gallus (chicken) and Bos Taurus (cattle), which are food species with prominent tissue-specific consumption of meat versus egg or dairy. In the metaproteomic dataset, we labeled 28, 10, and 4 proteins as meat-specific, egg-specific, and dairy-specific, respectively (Table S3†) and used them to categorize intake from stool. The key proteins identified for meat were myosin, actin, and titin; for egg, avidin, ovotransferrin, ovalbumin (abundant in egg white), and vitellogenins (abundant in yolks); and for dairy, caseins, butyrophilin (the major protein associated with fat droplets), and beta-lactoglobulin (the major whey protein in cow's milk). For all tissue types but dairy, detections by tissue-specific proteins had no significant difference from presence or absence of the same food tissue in diet records from the day prior to sampling (McNemar test p = 1.0 for egg, p = 0.7 for chicken meat, p = 0.7 for cattle meat, and p = 0.0008 for dairy; Fig. 5). Metaproteomics detected dairy intake less frequently than menu records (dairy detected in 50% of samples compared to 100% of the diet records one or two days prior to sampling), potentially reflecting the impact of processing, digestion, or lower protein content per gram of tissue.We next tested if we could identify a set of candidate protein biomarkers for specific foods in the metaproteomic dataset that would lend themselves to future development of targeted proteomic assays for high throughput detection at reduced cost. Considering foods detected in ≥5 samples (n = 85), we identified individual proteins that were present in more than half the cases an individual food taxon was detected by metaproteomics. Of 3392 candidate proteins for the initial 85 foods, we identified 67 candidates with this strategy that could serve as potential standalone indicators of intake, covering food species including corn, oats, chocolate, Brussels sprouts, almond, grape, and chicken (Table S6†). In addition, we also noted interesting cases that did not meet these criteria (potentially due to mixing of true signal with spurious positive detections from food proteins that share similarity to human proteins). Two specific additional proteins we want to highlight as examples are ananain and bromelain, which are both proteases that are highly specific to pineapple. Although daily fruit offerings in Intervention participants were not always recorded to the species level, three intake events for bromelain came shortly after recorded meals with pineapple as an ingredient in this cohort and ananain was detected in the Habitual Diet participant only in the sample collected the day after written pineapple consumption.
Discussion
We have shown that residual dietary DNA and protein from consumed foods are present in human stool, readily detected with existing techniques, and can be identified with reference databases curated from publicly available genomes of food species. Our findings demonstrate that both FoodSeq and metaproteomics detect a broad range of foods and have a significantly similar global structure. This detection is possible despite forces that may degrade the molecular components of food prior to detection, including thermal and chemical degradation by cooking and enzymatic degradation in the gastrointestinal tract by host proteases and nucleases. In our data, dietary sources still accounted for a substantial proportion of total proteinaceous biomass of stool (∼11% on average). Comparable data on the prevalence of dietary DNA in the overall pool of extracted stool DNA is sparse but supportive of a lesser contribution: in a wild primate model, 0.004–0.008% of metagenomic reads were dietary in origin,47 which agrees with anecdotal communications we have heard from researchers working in humans (unidentified metagenomic sequences assumed to be dietary and <1% of total reads). An increased prevalence of diet-derived stool protein in comparison to DNA also agrees with prior work in invertebrate animals showing that protein epitopes, especially those internal to the protein, survive in the gut for longer than DNA fragments.15Despite likely lower input, we found that on average, DNA-based assessment had higher detection performance than protein when we considered individual foods in direct comparison to the diet record (Fig. 3a). However, some foods were preferentially detected by protein, likely due to nuances of their composition and digestion (points falling above the unit line in Fig. 4). FoodSeq protocols include an amplification step that metaproteomics does not, potentially allowing DNA to more reliably report food items present in lower initial abundance like herbs and spices. We hypothesized this would lead to predominantly false negative detections by protein (protein failing to detect a recorded item). However, we found that false positive detections were the most common discrepancy between protein and menu data. 23 food taxa were detected in >90% of samples by metaproteomics: these included several species that were supported by diet records and could plausibly be consumed daily by the US-based participants in this study (corn, chicken, soy) but others that were not (dates, Napa cabbage, cassava). We were able to reduce putative false positive signals from prevalent animal detections (goat, duck) by considering only animal-derived proteins that we could definitively identify as originating from skeletal muscle, egg, or milk. We suspect that remaining animal false positives may be due to continued misidentification of host-derived proteins as dietary ones by our necessarily large reference database.
We did not perform a similar filtering strategy for plant foods due to the higher variation in tissue types consumed (e.g. roots, tubers, leaves, fruits, seeds, nuts, grains, etc.). For plant taxa, possibly erroneous false positives are likely attributable to (1) fruit or salad items consumed by Intervention participants that were not recorded on the menu (e.g. pineapple or Bromeliaceae), (2) foods consumed but not reported by participants (e.g. chocolate or Malvaceae), and (3) the incompleteness of some reference food genomes in public databases, which can lead to mis-identification of close relatives. To this final point, if sufficient similarity between protein sequences is present, proteins from a species with an incomplete proteome may instead be identified as one or more related taxa. For example, very few protein sequences are currently available for broccoli (Brassica oleracea var. italica). Therefore, if a participant consumed high quantities of broccoli and thus many mass spectra of broccoli peptides are acquired from stool samples, these spectra will likely match to proteins from other members of the cabbage family (Brassicaceae) of which broccoli is a member. Future sequencing efforts of food species genomes or genes encoding dominant food proteins should improve database quality and resolve associated taxonomic resolution issues. The false positives caused by database incompleteness may also have contributed to the overall weaker correspondence we observed between proteomic data globally and dietary records by Mantel test. We therefore expect that future work can improve proteomic performance compared to known intake.
Nevertheless, proteomic data can provide more dietary information than DNA alone. DNA cannot distinguish foods with an identical sequence at the marker region used: this is the case not only for foods derived from different parts of the same organism but often for foods derived from closely related species. For instance, a single trnL sequence variant is shared by dill, carrot, cumin, parsley, fennel, and parsnip, but these foods are readily distinguished by protein constituents in our data. Tissue type conveys important nutritional information (e.g. the fat content of chicken breast versus egg) that is unmeasurable by FoodSeq but readily assessed by metaproteomics. In our data, we identified multiple cases where metaproteomics had higher resolution than FoodSeq. Protein signals differentiated durum wheat (most commonly used in pasta) from bread wheat and identified distinct tissues from the same food species (e.g. chicken and egg, beef and dairy). For future biomarker development, food-derived proteins provide a large number of candidate targets, some of which are very abundant in specific tissues. Additionally, the detection limit of protein-based biomarkers could be increased by developing targeted mass spectrometry approaches that are much more sensitive than the discovery-focused untargeted approach that we used.48 Specific food groups of interest could also be targeted with DNA barcodes designed to delineate their members (e.g. grains, carrot family members, cruciferous vegetables) that cannot be distinguished at the trnL region.
Features of the consumed diet and its record limited our analyses, and we can therefore make specific recommendations for the design of future dietary studies and follow-on validation. To assess the performance of FoodSeq and metaproteomics in real-world diets and across many foods, we included participants with diets that included a range of items and varied day-to-day. However, in some cases, this limited our analysis: for example, most participants consumed both beef and dairy or both chicken and egg in the days prior to sampling, which precluded a clear connection between detected proteins and a specific episode of prior intake. Designing diet interventions that include non-repeating food items or introduction of a specific item into the baseline diet may help to adjust for this effect by more accurately linking molecular and menu data. Even though the dietary records of both Intervention and Habitual Diet participants were detailed, they had notable limitations: some foods were not tracked by the digital menu system, and others could be eaten off-menu or left on the plate (intervention); quantity was not recorded, and complex meals were not always separated into their component ingredients (Habitual Diet). Future studies could include paired sampling of food inputs and stool outputs to reduce errors from ingredients inferred during the process of coding written records to food items.
The data shared here can be re-analyzed for food-specific candidate biomarkers that would lend themselves to development in targeted proteomic assays that would allow for high throughput and reduced cost. Clear frameworks for establishing biomarker validity from nutritional epidemiology49 can then guide in vitro and in vivo testing. With additional development, metaproteomics may be helpful not only for detecting what is present in stool, but to determine what is absent from foods known to be consumed, and thus being degraded farther upstream and absorbed by the host or fermented by microbiota. Future work may also develop the ability of molecular measures to measure not only identity but amount of food consumed, which has previously been shown to be possible for a subset of foods with trnL FoodSeq.14 Quantitative assays could be similarly developed for metaproteomics, with the caveat that protein absorption (and thus residual amount in stool) varies with source, matrix, and processing,50 and may need to be evaluated on a per-food basis. A central theme in dietary assessment has been developing tools with different sources of error that can be used to validate one another. By expanding the range of tools available for dietary assessment, our work provides independent measures of food and tissue constituents in the diet that can be used in future studies.
Author contributions
LAD, MK, BLP, and AB conceptualized the study. Funding acquistion was by BLP, MK and LAD. BLP, AB, and SJ carried out investigation and SJ curated the data. AB, AK, SV, WSY, and CT provided resources. Formal analysis was performed by BLP. BLP, AB, LAD, and MK wrote, revised, and edited the paper. The work was supervised by LAD and MK.Data availability
The metaproteomics data and protein sequence database have been deposited to the PRIDE repository43 with the dataset identifiers PXD029334 https://www.ebi.ac.uk/pride/login for the participant on the habitual diet and PXD030004 for the diet intervention participants. FoodSeq data, reference databases, and code used to generate the figures in the article are available at https://github.com/bpetrone/diet-detection.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank our study volunteers for their participation, Verónica Palacios for human study support, and Tonya Snipes, Lisa Alston-Latta, and Margaret Huggins for keeping our lab spaces and glassware clean. We thank Angie Mordant and Clara Tang for assistance in metaproteomics data generation. This work was supported by the National Institute Of General Medical Sciences of the National Institutes of Health under Award Numbers R35GM138362 (MK) and R01DK116187 (LAD), the Foundation for Food and Agriculture Research Grant ID: 593607 (MK), the Duke Microbiome Center (LAD), the Triangle Center for Evolutionary Medicine (BLP), the Integrative Bioinformatics for Investigating and Engineering Microbiomes Graduate Student Fellowship (BLP), and the Ruth L. Kirschstein National Research Service Award to the Duke Medical Scientist Training Program. This work used a high-performance computing facility partially supported by grants 2016-IDG-1013 (“HARDAC+: Reproducible HPC for Next-generation Genomics”) and 2020-IIG-2109 (“HARDAC-M: Enabling memory-intensive computation for genomics”) from the North Carolina Biotechnology Center. We made all LC-MS/MS measurements in the Molecular Education, Technology, and Research Innovation Center (METRIC) at North Carolina State University.References
- GBD 2015 Risk Factors Collaborators, Global, regional, and national comparative risk assessment of 79 behavioural, environmental and occupational, and metabolic risks or clusters of risks in 188 countries, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet, 2015, 386, 2287–2323 CrossRef PubMed.
- V. Kipnis, D. Midthune, L. Freedman, S. Bingham, N. E. Day, E. Riboli, P. Ferrari and R. J. Carroll, Bias in dietary-report instruments and its implications for nutritional epidemiology, Public Health Nutr., 2002, 5, 915–923 CrossRef PubMed.
- A. F. Subar, Using intake biomarkers to evaluate the extent of dietary misreporting in a large sample of adults: The OPEN study, Am. J. Epidemiol., 2003, 158, 1–13 CrossRef.
- A. J. Johnson, P. Vangay, G. A. Al-Ghalith, B. M. Hillmann, T. L. Ward, R. R. Shields-Cutler, A. D. Kim, A. K. Shmagel, A. N. Syed, J. Walter, R. Menon, K. Koecher and D. Knights, Daily sampling reveals personalized diet-microbiome associations in humans, Cell Host Microbe, 2019, 25, 789–802 CrossRef CAS.
- P. Maruvada, J. W. Lampe, D. S. Wishart, D. Barupal, D. N. Chester, D. Dodd, Y. Djoumbou-Feunang, P. C. Dorrestein, L. O. Dragsted, J. Draper, L. C. Duffy, J. T. Dwyer, N. J. Emenaker, O. Fiehn, R. E. Gerszten, F. B. Hu, R. W. Karp, D. M. Klurfeld, M. R. Laughlin, A. R. Little, C. J. Lynch, S. C. Moore, H. L. Nicastro, D. M. O'Brien, J. M. Ordovás, S. K. Osganian, M. Playdon, R. Prentice, D. Raftery, N. Reisdorph, H. M. Roche, S. A. Ross, S. Sang, A. Scalbert, P. R. Srinivas and S. H. Zeisel, Perspective: Dietary biomarkers of intake and exposure—exploration with omics approaches, Adv. Nutr., 2020, 11, 200–215 CrossRef PubMed.
- The Human Microbiome Project Consortium, Structure, function and diversity of the healthy human microbiome, Nature, 2012, 486, 207–214 CrossRef PubMed.
- D. Zeevi, T. Korem, N. Zmora, D. Israeli, D. Rothschild, A. Weinberger, O. Ben-Yacov, D. Lador, T. Avnit-Sagi, M. Lotan-Pompan, J. Suez, J. A. Mahdi, E. Matot, G. Malka, N. Kosower, M. Rein, G. Zilberman-Schapira, L. Dohnalová, M. Pevsner-Fischer, R. Bikovsky, Z. Halpern, E. Elinav and E. Segal, Personalized nutrition by prediction of glycemic responses, Cell, 2015, 163, 1079–1094 CrossRef CAS.
- D. McDonald, E. Hyde, J. W. Debelius, J. T. Morton, A. Gonzalez, G. Ackermann, A. A. Aksenov, B. Behsaz, C. Brennan, Y. Chen, L. DeRight Goldasich, P. C. Dorrestein, R. R. Dunn, A. K. Fahimipour, J. Gaffney, J. A. Gilbert, G. Gogul, J. L. Green, P. Hugenholtz, G. Humphrey, C. Huttenhower, M. A. Jackson, S. Janssen, D. V. Jeste, L. Jiang, S. T. Kelley, D. Knights, T. Kosciolek, J. Ladau, J. Leach, C. Marotz, D. Meleshko, A. V. Melnik, J. L. Metcalf, H. Mohimani, E. Montassier, J. Navas-Molina, T. T. Nguyen, S. Peddada, P. Pevzner, K. S. Pollard, G. Rahnavard, A. Robbins-Pianka, N. Sangwan, J. Shorenstein, L. Smarr, S. J. Song, T. Spector, A. D. Swafford, V. G. Thackray, L. R. Thompson, A. Tripathi, Y. Vázquez-Baeza, A. Vrbanac, P. Wischmeyer, E. Wolfe, Q. Zhu, The American Gut Consortium and R. Knight, American Gut: An open platform for citizen science microbiome research, mSystems, 2018, 3, e00031–e00018 CrossRef CAS.
- F. Asnicar, S. E. Berry, A. M. Valdes, L. H. Nguyen, G. Piccinno, D. A. Drew, E. Leeming, R. Gibson, C. Le Roy, H. A. Khatib, L. Francis, M. Mazidi, O. Mompeo, M. Valles-Colomer, A. Tett, F. Beghini, L. Dubois, D. Bazzani, A. M. Thomas, C. Mirzayi, A. Khleborodova, S. Oh, R. Hine, C. Bonnett, J. Capdevila, S. Danzanvilliers, F. Giordano, L. Geistlinger, L. Waldron, R. Davies, G. Hadjigeorgiou, J. Wolf, J. M. Ordovás, C. Gardner, P. W. Franks, A. T. Chan, C. Huttenhower, T. D. Spector and N. Segata, Microbiome connections with host metabolism and habitual diet from 1,098 deeply phenotyped individuals, Nat. Med., 2021, 27, 321–332 CrossRef CAS.
- C. Rose, A. Parker, B. Jefferson and E. Cartmell, The characterization of feces and urine: A review of the literature to inform advanced treatment technology, Crit. Rev. Environ. Sci. Technol., 2015, 45, 1827–1879 CrossRef CAS.
- M. Beaumont, K. J. Portune, N. Steuer, A. Lan, V. Cerrudo, M. Audebert, F. Dumont, G. Mancano, N. Khodorova, M. Andriamihaja, G. Airinei, D. Tomé, R. Benamouzig, A.-M. Davila, S. P. Claus, Y. Sanz and F. Blachier, Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: a randomized, parallel, double-blind trial in overweight humans, Am. J. Clin. Nutr., 2017, 106, 1005–1019 CrossRef CAS.
- Y. Zhu, X. Shi, X. Lin, K. Ye, X. Xu, C. Li and G. Zhou, Beef, chicken, and soy proteins in diets induce different gut microbiota and metabolites in rats, Front. Microbiol., 2017, 8, 1395 CrossRef PubMed.
- L. M. Shinn, Y. Li, A. Mansharamani, L. S. Auvil, M. E. Welge, C. Bushell, N. A. Khan, C. S. Charron, J. A. Novotny, D. J. Baer, R. Zhu and H. D. Holscher, Fecal bacteria as biomarkers for predicting food intake in healthy adults, J. Nutr., 2021, 151, 423–433 CrossRef PubMed.
- B. L. Petrone, A. Aqeel, S. Jiang, H. K. Durand, E. P. Dallow, J. R. McCann, H. K. Dressman, Z. Hu, C. B. Tenekjian, W. S. Yancy, P.-H. Lin, J. J. Scialla, P. C. Seed, J. F. Rawls, S. C. Armstrong, J. Stevens and L. A. David, Diversity of plant DNA in stool is linked to dietary quality, age, and household income, Proc. Natl. Acad. Sci. U. S. A., 2023, 120, e2304441120 CrossRef CAS.
- W. O. C. Symondson, Molecular identification of prey in predator diets, Mol. Ecol., 2002, 11, 627–641 CrossRef CAS.
- F. Salvato, R. L. Hettich and M. Kleiner, Five key aspects of metaproteomics as a tool to understand functional interactions in host-associated microbiomes, PLoS Pathog., 2021, 17, e1009245 CrossRef CAS.
- M. Kleiner, Metaproteomics: Much more than measuring gene expression in microbial communities, mSystems, 2019, 4, e00115–e00119 CrossRef CAS PubMed.
- M. Kleiner, X. Dong, T. Hinzke, J. Wippler, E. Thorson, B. Mayer and M. Strous, Metaproteomics method to determine carbon sources and assimilation pathways of species in microbial communities, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(24), e5576–e5584 CrossRef CAS.
- C. G. Gonzalez, H. C. Wastyk, M. Topf, C. D. Gardner, J. L. Sonnenburg and J. E. Elias, High-throughput stool metaproteomics: Method and application to human specimens, mSystems, 2020, 5, e00200–e00220 CrossRef CAS PubMed.
- H. C. Wastyk, G. K. Fragiadakis, D. Perelman, D. Dahan, B. D. Merrill, F. B. Yu, M. Topf, C. G. Gonzalez, W. Van Treuren, S. Han, J. L. Robinson, J. E. Elias, E. D. Sonnenburg, C. D. Gardner and J. L. Sonnenburg, Gut-microbiota-targeted diets modulate human immune status, Cell, 2021, 184, 4137–4153 CrossRef CAS PubMed.
- J. R. Wiśniewski, A. Zougman, N. Nagaraj and M. Mann, Universal sample preparation method for proteome analysis, Nat. Methods, 2009, 6, 359–362 CrossRef.
- A. Mordant and M. Kleiner, Evaluation of sample preservation and storage methods for metaproteomics analysis of intestinal microbiomes, Microbiol. Spectrum, 2021, 9, e01877–e01821 CAS.
- W. Li and A. Godzik, Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences, Bioinformatics, 2006, 22, 1658–1659 CrossRef CAS.
- P. J. McMurdie and S. Holmes, phyloseq: An R Package for Reproducible Interactive Analysis and Graphics of Microbiome Census Data, PLoS One, 2013, 8, e61217 CrossRef CAS PubMed.
- M. Kleiner, E. Thorson, C. E. Sharp, X. Dong, D. Liu, C. Li and M. Strous, Assessing species biomass contributions in microbial communities via metaproteomics, Nat. Commun., 2017, 8, 1558 CrossRef PubMed.
- W. Shehzad, T. Riaz, M. A. Nawaz, C. Miquel, C. Poillot, S. A. Shah, F. Pompanon, E. Coissac and P. Taberlet, Carnivore diet analysis based on next-generation sequencing: application to the leopard cat (Prionailurus bengalensis) in Pakistan: LEOPARD CAT DIET, Mol. Ecol., 2012, 21, 1951–1965 CrossRef CAS PubMed.
- M. De Barba, C. Miquel, F. Boyer, C. Mercier, D. Rioux, E. Coissac and P. Taberlet, DNA metabarcoding multiplexing and validation of data accuracy for diet assessment: application to omnivorous diet, Mol. Ecol. Resour., 2014, 14, 306–323 CrossRef CAS PubMed.
- Economic Research Service (ERS) of the U.S. Department of Agriculture (USDA), USDA ERS - Food Availability (Per Capita) Data System, https://www.ers.usda.gov/data-products/food-availability-per-capita-data-system/, (accessed 9 March 2023).
- C. Lachat, J. E. Raneri, K. W. Smith, P. Kolsteren, P. Van Damme, K. Verzelen, D. Penafiel, W. Vanhove, G. Kennedy, D. Hunter, F. O. Odhiambo, G. Ntandou-Bouzitou, B. De Baets, D. Ratnasekera, H. T. Ky, R. Remans and C. Termote, Dietary species richness as a measure of food biodiversity and nutritional quality of diets, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 127–132 CrossRef CAS.
- B.-E. Van Wyk, Food plants of the world: an illustrated guide, Timber Press, Portland, Or, 1st edn, 2005 Search PubMed.
- D. J. Winter, rentrez: An R package for the NCBI eUtils API, R J., 2017, 9, 520 CrossRef.
- bcl2fastq Conversion Software, https://emea.support.illumina.com/sequencing/sequencing_software/bcl2fastq-conversion-software.html, (accessed 4 April 2024).
- B. Bushnell , BBMap, https://sourceforge.net/projects/bbmap/, (accessed 4 April 2024).
- M. Martin, Cutadapt removes adapter sequences from high-throughput sequencing reads, EMBnet j., 2011, 17, 10 CrossRef.
- B. J. Callahan, P. J. McMurdie, M. J. Rosen, A. W. Han, A. J. A. Johnson and S. P. Holmes, DADA2: High-resolution sample inference from Illumina amplicon data, Nat. Methods, 2016, 13, 581–583 CrossRef CAS PubMed.
- N. M. Davis, D. M. Proctor, S. P. Holmes, D. A. Relman and B. J. Callahan, Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data, Microbiome, 2018, 6, 226 CrossRef.
- J. Oksanen, vegan: Community Ecology Package Search PubMed.
- L. Lahti and S. Shetty, microbiome R package Search PubMed.
- R Core Team, 2022.
- J. Larsson and P. Gustafsson, A case study in fitting area-proportional Euler diagrams with ellipses using eulerr, Proceedings of International Workshop on Set Visualization and Reasoning, 2018, 2116, 84–91 Search PubMed.
- M. Wang and Y. Zhao, SuperExactTest: Exact test and visualization of multi-set intersections.
- J. Fox and S. Weisberg, An R Companion to Applied Regression, Sage, Thousand Oaks, CA, 2019.
- Y. Perez-Riverol, J. Bai, C. Bandla, D. García-Seisdedos, S. Hewapathirana, S. Kamatchinathan, D. J. Kundu, A. Prakash, A. Frericks-Zipper, M. Eisenacher, M. Walzer, S. Wang, A. Brazma and J. A. Vizcaíno, The PRIDE database resources in 2022: a hub for mass spectrometry-based proteomics evidences, Nucleic Acids Res., 2022, 50, D543–D552 CrossRef CAS PubMed.
- L. C. Richardson, M. C. Bazaco, C. C. Parker, D. Dewey-Mattia, N. Golden, K. Jones, K. Klontz, C. Travis, J. Z. Kufel and D. Cole, An updated scheme for categorizing foods implicated in foodborne disease outbreaks: A tri-agency collaboration, Foodborne Pathog. Dis., 2017, 14, 701–710 CrossRef.
- F. Asnicar, E. R. Leeming, E. Dimidi, M. Mazidi, P. W. Franks, H. Al Khatib, A. M. Valdes, R. Davies, E. Bakker, L. Francis, A. Chan, R. Gibson, G. Hadjigeorgiou, J. Wolf, T. D. Spector, N. Segata and S. E. Berry, Blue poo: impact of gut transit time on the gut microbiome using a novel marker, Gut, 2021, 70, 1665–1674 CrossRef CAS.
- G. K. Nandhra, E. B. Mark, G. L. Di Tanna, A. Haase, J. Poulsen, S. Christodoulides, V. Kung, M. W. Klinge, K. Knudsen, P. Borghammer, K. O. Andersen, L. Fynne, N. Sutter, V. Schlageter, K. Krogh, A. M. Drewes, M. Birch and S. M. Scott, Normative values for region–specific colonic and gastrointestinal transit times in 111 healthy volunteers using the 3D–Transit electromagnet tracking system: Influence of age, gender, and body mass index, Neurogastroenterology & Motility, 2020, 32, e13734 Search PubMed.
- A. Srivathsan, A. Ang, A. P. Vogler and R. Meier, Fecal metagenomics for the simultaneous assessment of diet, parasites, and population genetics of an understudied primate, Front. Zool., 2016, 13, 17 CrossRef.
- A. Doerr, Targeted proteomics, Nat. Methods, 2011, 8, 43–43 CrossRef CAS.
- L. O. Dragsted, Q. Gao, A. Scalbert, G. Vergères, M. Kolehmainen, C. Manach, L. Brennan, L. A. Afman, D. S. Wishart, C. Andres Lacueva, M. Garcia-Aloy, H. Verhagen, E. J. M. Feskens and G. Praticò, Validation of biomarkers of food intake—critical assessment of candidate biomarkers, Genes Nutr., 2018, 13, 14 CrossRef CAS.
- A. Bartlett and M. Kleiner, Dietary protein and the intestinal microbiota: An understudied relationship, iScience, 2022, 25, 105313 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4fo02656j |
| This journal is © The Royal Society of Chemistry 2025 |

