Open Access Article
Open Access ArticleSetting new benchmarks in AI-driven infrared structure elucidation†
Marvin
Alberts
 *abc,
Federico
Zipoli
ab and
Teodoro
Laino
*abc,
Federico
Zipoli
ab and
Teodoro
Laino
 ab
ab
aIBM Research Europe, Säumerstrasse 4, 8803 Rüschlikon, Switzerland. E-mail: marvin.alberts@ibm.com
bNCCR Catalysis, Switzerland
cUniversity of Zurich, Department of Chemistry, Winterthurerstrasse 190, 8057 Zurich, Switzerland
First published on 25th June 2025
Abstract
Automated structure elucidation from infrared (IR) spectra represents a significant breakthrough in analytical chemistry, having recently gained momentum through the application of Transformer-based language models. In this work, we improve our original Transformer architecture, refine spectral data representations, and implement novel augmentation and decoding strategies to significantly increase performance. We report a Top-1 accuracy of 63.79% and a Top-10 accuracy of 83.95% compared to the current performance of state-of-the-art models of 53.56% and 80.36%, respectively. Our findings not only set a new performance benchmark but also strengthen confidence in the promising future of AI-driven IR spectroscopy as a practical and powerful tool for structure elucidation. To facilitate broad adoption among chemical laboratories and domain experts, we openly share our models and code.
1 Introduction
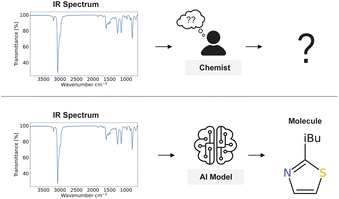
Infrared (IR) spectroscopy is a valuable analytical technique widely utilised across chemistry, pharmaceuticals, environmental science, and forensic investigations due to its rapid, non-destructive, and cost-effective characterisation of molecular structures and functional groups.1–4 Although nuclear magnetic resonance (NMR) spectroscopy and tandem mass spectrometry (MS/MS) have gained prominence in structure elucidation,5,6 IR spectroscopy provides distinct advantages, including minimal sample preparation, low operational costs, rapid measurement times, and direct observation of vibrational modes that correspond to specific functional groups. The characteristic absorption bands in IR spectra enable rapid identification of molecular features that may be challenging or time-consuming to discern through other analytical methods.7However, despite its widespread use, determining the complete molecule structure from an IR spectrum remains notoriously challenging. Interpretation of the spectra is often limited to the manual identification of a few functional groups or relies on the use of spectral databases and reference tables for comparison.8–10 The complexity of overlapping bands and coupled vibrations in the fingerprint region (500–1500 cm−1) further complicates the interpretation of the spectra.11 This often limits the amount of information that can be reliably extracted to a few select functional groups (Fig. 1).
The emergence of computational chemistry provided new a framework for understanding vibrational spectroscopy, with various computational approaches enabling the simulation of IR spectra. These techniques aided in the interpretation of IR spectra based on the molecular structure and shed new insights on the relation between vibrations in the molecular structures and the peaks observed in the spectra.12–14 However, the inverse problem, i.e. predicting molecular structures or functional groups directly from experimental IR spectra, has remained largely unsolved through traditional computational approaches.
Recently, artificial intelligence (AI) has developed into a transformative tool across chemistry. Machine learning approaches have shown remarkable success in interpreting NMR spectra,15–21 analysing MS/MS spectra,22–24 and the prediction of functional groups from IR spectra.25–29 These developments have demonstrated the potential for AI-driven methods to overcome traditional limitations in the analysis of spectroscopic data.
Building on this foundation, it has been demonstrated that artificial intelligence can directly predict the molecular structure from IR spectra.30–32 This opened new avenues in analytical chemistry and inspired subsequent perspectives33 and developments.34,35 Wu et al.34 advanced the methodology further, achieving notable accuracy improvements, Kanakala et al.35 developed a contrastive retrieval system to match molecules with spectra and Priessner et al.36 developed a multimodal approach combining IR spectra with other spectroscopic modalities. In this work, we push the boundaries even further by addressing previous architectural limitations, adopting a patch-based spectral representation method, and refining augmentation and decoding strategies. These modifications substantially improve our model's performance, raising the Top-1 accuracy from 44.39% to 63.79% and Top-10 accuracy from 69.79% to 83.95%. The model presented in this work exceeds the previous best-in-class by approximately 9%, effectively becoming the new state-of-the-art. Our findings redefine what is possible, showing that the full potential of IR spectra for structure elucidation is now within reach through specifically tailored architecture and data engineering strategies.
2 Results and discussion
2.1 Model architecture ablation
Our model directly predicts chemical structures as SMILES37 using only a compound's chemical formula and its infrared (IR) spectrum. Previously, we represented IR spectra as discretized text, limiting each spectrum to 400 data points with absorbance values quantized into 100 bins.32 Although effective, this discretization significantly reduced spectral resolution, resulting in a considerable loss of information.Recently, Wu et al.34 addressed this limitation by introducing a patch-based Transformer model for IR spectral analysis, inspired by Vision Transformers (ViT) originally developed for image data.38 This approach segments the IR spectrum into smaller fixed-size segments or “patches,” effectively preserving richer, fine-grained spectral details. Patch-based Transformers have proven successful across multiple data modalities beyond images, including audio and time-series data, due to their enhanced representational capabilities.39,40 Based on these insights, we implemented a patch-based representation of IR spectra, resulting in substantial improvements in performance.
However, the patch-based representation is not the only recent advancement in Transformer architectures. Xiong et al.41 introduced post-layer normalization, replacing the original pre-layer normalization approach of the vanilla Transformer. This modification optimizes gradient flow during training, leading to more effective and efficient model convergence. Similarly, Gated Linear Units (GLUs), introduced by Shazeer,42 represent an improvement over traditional activation functions such as the Rectified Linear Unit (ReLU) and the Gaussian Error Linear Unit (GeLU). GLUs allow for enhanced model parametrization without additional depth, thus improving model expressivity.43 In this study, we also replaced the standard sinusoidal positional encodings with learned positional embeddings,44 enabling the model to develop more adaptive sequence representations throughout training.
We conducted comprehensive ablation studies evaluating the impact of each of these architectural changes, summarized in Table 1. During pretraining, we incorporated both simulated data from our original study and additional spectra introduced in our recent multimodal dataset,45 substantially increasing our training samples from 634![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 585 to 1
585 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 399
399![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 806 spectra. For each architectural configuration (as detailed in the table rows), we pretrained a model on simulated spectra, followed by fine-tuning on 3453 experimental spectra from the NIST database—the same dataset utilized in our previous work, obtained in full compliance with NIST's data usage policies.46 To ensure robust evaluation, we implemented 5-fold cross-validation during fine-tuning. Comprehensive results, including Top-5 accuracies, are provided in the ESI, Section 1.†
806 spectra. For each architectural configuration (as detailed in the table rows), we pretrained a model on simulated spectra, followed by fine-tuning on 3453 experimental spectra from the NIST database—the same dataset utilized in our previous work, obtained in full compliance with NIST's data usage policies.46 To ensure robust evaluation, we implemented 5-fold cross-validation during fine-tuning. Comprehensive results, including Top-5 accuracies, are provided in the ESI, Section 1.†
| Layer normalisation | Pos. encoding | GLUs | Patch size | Simulated | Experimental | ||
|---|---|---|---|---|---|---|---|
| Top-1 ↑ | Top-10 ↑ | Top-1 ↑ | Top-10 ↑ | ||||
| Pre- | Sinusoidal | ✗ | 125 | 20.84 | 47.29 | 42.59 ± 2.64 | 78.04 ± 2.81 |
| Post- | Sinusoidal | ✗ | 125 | 39.86 | 66.52 | 48.36 ± 3.14 | 81.58 ± 2.08 |
| Post- | Learned | ✗ | 125 | 39.78 | 67.19 | 49.55 ± 1.77 | 82.39 ± 0.83 |
| Post- | Learned | ✓ | 125 | 42.94 | 69.47 | 50.01 ± 1.53 | 83.09 ± 1.83 |
In Table 1, we demonstrate that each newly introduced architectural component contributes incrementally to improved performance. Throughout these experiments, we maintained a fixed patch size of 125 data points, corresponding to 15 patches per spectrum. Based on these findings, all subsequent experiments employed models incorporating post-layer normalization, learned positional embeddings, and Gated Linear Units (GLUs).
Next, we evaluated the optimal patch size by training models with patch sizes ranging from 25 to 150 data points (Table 2). Performance on experimental data steadily improved with increasing patch sizes, reaching a maximum at a patch size of 75 before subsequently declining. Interestingly, this trend contrasted with the performance observed on simulated data, where smaller patches consistently yielded better results. This discrepancy suggests that, while smaller patches may enhance the model's ability to capture detailed spectral features, they could also promote overfitting during fine-tuning. Supporting this interpretation, the training metrics showed that models using a patch size of 25 had a higher average validation loss than those using a patch size of 75. Complete validation loss curves are provided in the ESI, Section 2.† Based on these results, we selected a patch size of 75 for all subsequent experiments.
| Layer normalisation | Pos. encoding | GLUs | Patch size | Simulated | Experimental | ||
|---|---|---|---|---|---|---|---|
| Top-1 ↑ | Top-10 ↑ | Top-1 ↑ | Top-10 ↑ | ||||
| Post- | Learned | ✓ | 25 | 45.73 | 71.30 | 49.81 ± 3.49 | 81.26 ± 1.71 |
| Post- | Learned | ✓ | 50 | 44.48 | 70.89 | 51.03 ± 2.82 | 82.35 ± 2.83 |
| Post- | Learned | ✓ | 75 | 44.23 | 70.68 | 52.25 ± 2.71 | 83.00 ± 2.14 |
| Post- | Learned | ✓ | 100 | 43.49 | 69.72 | 51.72 ± 3.08 | 82.62 ± 2.19 |
| Post- | Learned | ✓ | 125 | 42.97 | 69.40 | 50.57 ± 2.59 | 83.57 ± 1.67 |
| Post- | Learned | ✓ | 150 | 41.52 | 68.93 | 48.36 ± 3.11 | 82.07 ± 2.13 |
2.2 Augmentations
In our previous work,32 data augmentation proved to be one of the most effective strategies to increase model performance. Among the augmentation techniques evaluated, horizontal shifting provided the greatest benefit, followed by Gaussian smoothing of spectra. In the current study, we further extend our augmentation strategy by introducing two additional methods: SMILES augmentation and pseudo-experimental spectra generation.SMILES augmentation, originally proposed by Bjerrum,47 involves enriching the training dataset by including non-canonical SMILES representations. This approach has successfully improved generalization in various molecular prediction tasks, ranging from retrosynthesis to structure elucidation.48–50 By presenting the model with alternative yet chemically equivalent SMILES representations, we encourage better generalization and robustness.
The primary challenge in our pretraining–fine-tuning approach is the significant sim-to-real gap between simulated and experimental IR spectra. This gap leads to a considerable domain shift that the model must bridge during fine-tuning. To address this issue, we introduce a novel augmentation method called pseudo-experimental spectra, defined as simulated spectra transformed to closely mimic experimental spectra. We achieve this transformation using a transfer function implemented as a multilayer perceptron (MLP) with a bottleneck layer. Given the limited availability of experimental IR spectra, we trained the transfer function on 2000 pairs of simulated and experimental spectra. Additionally, molecular fingerprints were included as auxiliary inputs to further improve transformation accuracy. A visual overview of this methodology is provided in Fig. 2, and further details on the architecture, hyperparameter optimization, and performance evaluation are available in the Methods section and ESI, Section 3.†
During pretraining, we expanded our dataset by adding 700![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 pseudo-experimental spectra. In the fine-tuning phase, we incorporated an additional, smaller subset of 3000 pseudo-experimental spectra matching the distribution of our experimental dataset. For consistency, two additional augmented spectra per sample were generated using all other augmentation techniques. As shown in Table 3, we observed substantial performance gains when augmenting the dataset with non-canonical SMILES and pseudo-experimental spectra, with all four augmentation techniques demonstrating a synergistic effect when combined. Interestingly, different augmentations contributed distinctly to model performance: pseudo-experimental spectra primarily improved Top-5 and Top-10 accuracies (see ESI, Section 1†), likely due to increased molecular diversity in the training data. In contrast, non-canonical SMILES augmentation significantly boosted Top-1 accuracy but slightly reduced Top-5 and Top-10 accuracies, possibly due to the model encountering multiple equivalent SMILES representations—either facilitating precise prediction or increasing ambiguity. Consequently, we employed all four augmentation strategies for all subsequent experiments.
000 pseudo-experimental spectra. In the fine-tuning phase, we incorporated an additional, smaller subset of 3000 pseudo-experimental spectra matching the distribution of our experimental dataset. For consistency, two additional augmented spectra per sample were generated using all other augmentation techniques. As shown in Table 3, we observed substantial performance gains when augmenting the dataset with non-canonical SMILES and pseudo-experimental spectra, with all four augmentation techniques demonstrating a synergistic effect when combined. Interestingly, different augmentations contributed distinctly to model performance: pseudo-experimental spectra primarily improved Top-5 and Top-10 accuracies (see ESI, Section 1†), likely due to increased molecular diversity in the training data. In contrast, non-canonical SMILES augmentation significantly boosted Top-1 accuracy but slightly reduced Top-5 and Top-10 accuracies, possibly due to the model encountering multiple equivalent SMILES representations—either facilitating precise prediction or increasing ambiguity. Consequently, we employed all four augmentation strategies for all subsequent experiments.
| Pretraining augmentation | Fine tuning augmentation | Simulated | Experimental | ||
|---|---|---|---|---|---|
| Top-1 ↑ | Top-10 ↑ | Top-1 ↑ | Top-10 ↑ | ||
| a Horizontal shifting as implemented in Alberts et al.32 b Gaussian smoothing as implemented in Alberts et al.32 | |||||
| Hori.a | None | 43.18 | 68.95 | 50.33 ± 2.37 | 83.15 ± 1.19 |
| Smoothingb | None | 42.94 | 67.84 | 48.60 ± 1.74 | 81.90 ± 0.56 |
| Pseudo | None | 45.26 | 73.97 | 50.45 ± 1.13 | 83.64 ± 0.93 |
| SMILES | None | 50.86 | 70.51 | 54.62 ± 3.06 | 82.65 ± 1.51 |
| Hori.a + smoothingb SMILES + pseudo | None | 50.62 | 72.39 | 55.58 ± 1.75 | 84.19 ± 1.78 |
| Hori.a + smoothingb SMILES + pseudo | Hori.a | 50.62 | 72.39 | 57.49 ± 1.86 | 84.25 ± 1.46 |
| Hori.a + smoothingb SMILES + pseudo | Smoothingb | 50.62 | 72.39 | 56.04 ± 1.85 | 85.06 ± 2.05 |
| Hori.a + smoothingb SMILES + pseudo | Pseudo | 50.62 | 72.39 | 55.10 ± 3.00 | 85.19 ± 1.99 |
| Hori.a + smoothingb SMILES + pseudo | SMILES | 50.62 | 72.39 | 59.80 ± 1.64 | 80.99 ± 1.33 |
| Hori.a + smoothingb SMILES + pseudo | Hori.a + smoothingb SMILES + pseudo | 50.62 | 72.39 | 60.75 ± 1.54 | 81.92 ± 1.74 |
2.3 Formula constrained generation
During training, we provide the model with both the chemical formula and the corresponding IR spectrum. However, the generated molecular structures do not always strictly conform to the provided chemical formula. To address this issue, we introduce a formula-constrained generation method, inspired by constrained decoding techniques developed within natural language processing.51–55 Our approach enforces constraints during model inference, ensuring that the generated molecules exactly match the desired chemical formula.Specifically, we implement three constraint conditions: (1) preventing sequence termination if the partially generated molecule's chemical formula remains incomplete (e.g., missing atoms); (2) enforcing immediate termination when the target chemical formula is satisfied and the SMILES string is valid; and (3) prohibiting token selections that would cause the generated molecule to exceed the atom counts defined by the target formula. This procedure is illustrated in Fig. 3. Applying these constraints on both the filtered NIST dataset (containing molecules with 6–13 heavy atoms) and the complete NIST dataset model performance, achieving an approximately 2% accuracy increase, as detailed in Table 4.
| Dataset | N–Molecules | Constrained generation | Experimental | ||
|---|---|---|---|---|---|
| Top-1 ↑ | Top-5 ↑ | Top-10 ↑ | |||
| NIST (6-13 heavy atoms) | 3455 | ✗ | 60.75 ± 1.54 | 77.12 ± 1.43 | 81.92 ± 1.74 |
| NIST (6-13 heavy atoms) | 3455 | ✓ | 63.25 ± 1.95 | 79.15 ± 1.09 | 83.56 ± 1.91 |
| NIST (5-35 heavy atoms) | 5024 | ✗ | 56.71 ± 1.40 | 71.64 ± 1.68 | 75.62 ± 1.88 |
| NIST (5-35 heavy atoms) | 5024 | ✓ | 59.94 ± 1.18 | 74.96 ± 0.86 | 78.46 ± 0.25 |
2.4 Comparison to baselines
In this section, we provide a comprehensive comparison between our enhanced model (this work), our original model (Alberts et al.32), and the recently published model by Wu et al.34 The evaluation spans multiple aspects of the model's performance, including molecular accuracy, scaffold accuracy, and it's capability to accurately predict the presence of functional groups within target molecules. To ensure an unbiased and rigorous comparison, we fine-tuned the model proposed by Wu et al.34 on the NIST dataset using five-fold cross-validation.Table 5 summarizes the performance of all three models across the evaluation metrics. The results demonstrate that our enhanced model consistently outperforms both our previous model and the model by Wu et al.34 across all primary metrics. Notably, for molecular prediction accuracy, our model improves Top-1 accuracy by approximately 19 percentage points over our original model, and by around 10 percentage points compared to the model proposed by Wu et al.34
| Structure | Scaffold | |||||
|---|---|---|---|---|---|---|
| Top-1 ↑ | Top-5 ↑ | Top-10 ↑ | Top-1 ↑ | Top-5 ↑ | Top-10 ↑ | |
| Alberts et al.32 | 44.39 ± 5.31 | 66.85 ± 3.08 | 69.79 ± 2.48 | 83.23 ± 1.91 | 91.92 ± 0.88 | 93.11 ± 0.60 |
| Wu et al.34 | 53.56 ± 1.13 | 74.18 ± 0.79 | 80.36 ± 0.70 | 89.24 ± 1.10 | 93.48 ± 0.53 | 94.60 ± 0.61 |
| Ours | 63.25 ± 1.95 | 79.15 ± 1.09 | 83.56 ± 1.91 | 91.02 ± 1.71 | 94.45 ± 0.81 | 95.36 ± 0.78 |
Additionally, to evaluate the accuracy of functional group prediction, we employed three metrics: mean F1-score, weighted average F1-score, and molecular perfection rate, calculated across 16 functional groups as defined by Fine et al.56 These metrics provide complementary insights into the predictive capabilities of the models. Specifically, the molecular perfection rate measures the model's ability to identify all functional groups in a molecule without error. As shown in Table 6, our enhanced model achieves superior performance across all metrics, further underscoring its advantage over existing alternatives.
To further characterise the performance of the three, we analysed their performance with regards to the heavy atom count, functional group composition, and Tanimoto similarity between predicted and ground truth molecules. We observed a decrease in model accuracy with an increase in the heavy atom count across all three models. When examining performance on subsets containing specific functional groups, our model demonstrates superior Top-1 accuracy compared to Wu et al. across all functional groups, while achieving better Top-5 and Top-10 performance for 11 of 16 functional groups. These findings validate our approach, with further details on the analysis provided in ESI Section 4.†
2.5 Limitations
The primary limitation of our work stems from the availability of experimental data. As in our previous work,32 we utilised the NIST EPA gas-phase database and consequently fine tuned our models exclusively on gas-phase IR spectra. When applied beyond this domain, such as to IR spectra obtained from other instrument types (e.g., ATR-IR), degraded performance can be expected. Similarly, we expect reduced performance for out-of-distribution molecules, particularly those with significantly higher heavy atom counts. These limitations can be largely addressed through the incorporation of additional experimental datasets spanning different types of instruments and larger molecular diversity.3 Conclusion
In this paper, we demonstrate significant performance improvements in our transformer-based approach for automated prediction of chemical structures from IR spectra. By adopting a patch-based spectral representation, developing a novel constrained decoding strategy, and substantially enhancing our data augmentation methods, we achieve substantial improvements in predictive accuracy. Specifically, our enhanced model achieves a 19 percentage point increase in Top-1 accuracy and a 14 percentage point increase in Top-5 accuracy compared to our original implementation.32 Furthermore, it outperforms the current state-of-the-art by 10 percentage points in Top-1 accuracy and 5 percentage points in Top-5 accuracy, respectively.34Rather than replacing human expertise, we envision these models to be used within a collaborative workflow where AI models provide rapid initial predictions from spectroscopic data, enabling chemists to concentrate their expertise on verification, refinement, and interpretation of results. Within such a framework, performance improvements directly enhance system reliability and usability. Our 10% accuracy improvement reduces false suggestions requiring investigation, strengthens initial hypotheses, and increases overall efficiency. These results provide robust evidence that we are at the cusp of a new era in analytical chemistry. Advanced language model architectures hold the potential to revitalise analytical methods previously overlooked due to their modest human interpretability, unlocking unprecedented opportunities for rapid, precise molecular identification using low-cost instrumentation.
4 Methods
4.1 Data processing
Our workflow can be divided into two stages: Pretraining and fine tuning. For pretraining we use the 634![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 585 IR spectra published with our original article32 and add to this 794
585 IR spectra published with our original article32 and add to this 794![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 403 IR spectra sourced from Alberts et al.45 Spectra sourced from both datasets were simulated using molecular dynamics with the PCFF and GAFF forcefield respectively. From each spectrum we sampled 1625 datapoints with the range of 650 to 3900 cm−1 and a resolution of 2 cm−1. No further preprocessing of the spectra was performed. The heavy atom count of all molecules in this combined dataset falls within the range of 5 to 35 and the elements are limited to carbon, hydrogen, oxygen, nitrogen, sulphur, phosphorus, silicon, boron and the halogens.
403 IR spectra sourced from Alberts et al.45 Spectra sourced from both datasets were simulated using molecular dynamics with the PCFF and GAFF forcefield respectively. From each spectrum we sampled 1625 datapoints with the range of 650 to 3900 cm−1 and a resolution of 2 cm−1. No further preprocessing of the spectra was performed. The heavy atom count of all molecules in this combined dataset falls within the range of 5 to 35 and the elements are limited to carbon, hydrogen, oxygen, nitrogen, sulphur, phosphorus, silicon, boron and the halogens.
For fine tuning we use the NIST EPA Gas phase library consisting of 5228 molecules.46 Two sets were selected from this dataset: One matching the set used in our original paper, consisting of molecules with a heavy atom count ranging from 6 to 13 and elements only including carbon, oxygen, nitrogen, sulphur, phosphorus and the halogens. This reduced the number of samples from 5228 to 3455. We selected a second set matching the heavy atom count and element distribution in our pretraining set consisting of 5024 molecules. We sampled 1625 datapoints from the experimental spectra with the range and resolution matching the spectra in the pretraining set.
4.2 Tokenisation
Chemical formulae were tokenised by splitting them into their constituent elements and numbers. IR spectra were first split into patches based on a given patch size before being projected into the embedding dimension with an MLP. SMILES were tokenised using the following regular expression following Schwaller et al.:48(\[[^\]]+]|Br?|Cl?|N|O|S|P|F|I|b|c|n|o|s|p|\
(|\)|\.|=|#|-|\+|\\\\|\/|:|∼|@|\?|>|\|*\$|\%
[0-9]{2}|[0-9])
4.3 Model training
Our model adopts, an encoder-decoder architecture based on the vanilla transformer. In addition, we investigate the effects of post-layer normalisation, learned positional embeddings and gated linear units. For each change to the original transformer architecture, we follow the implementation outlined by Xiong et al.,41 Gehring et al.44 and Shazeer42 respectively. Each model is trained for 60 epochs before the best checkpoint is evaluated. All further hyperparameters are listed below:Layers: 6.
Heads: 8.
Embedding dimension: 512.
Feedforward dimension: 2048.
Optimiser: AdamW.
Learning rate: 0.001.
Dropout: 0.1.
Warmup steps: 8000.
Adam beta_1: 0.9.
Adam beta_2: 0.999.
Batch size: 128.
4.4 Augmentation
We used four different augmentation methods while training our model. For both horizontal shifting and smoothing we used the same implementation as described in Alberts et al.32 For the SMILES augmentation, we use RDKit to generate non-canonical SMILES strings. The last augmentation, pseudo experimental spectra, requires a model to be trained to model a transfer function from simulated to experimental spectra. The hyperparameters for the best model are shown below. More information on the transfer function can be found in ESI† ??Loss function: SID.
Activation function: Sigmoid.
Learning rate: 0.001.
Layer: 4.
Bottleneck dimension: 258.
4.5 Evaluation
During inference, ten ranked SMILES strings per sample are generated using beam search or formula-constrained generation. Each generated SMILES string is canonicalised and compared to the ground truth. The Top–N accuracy is defined as the percentage of generated molecules exactly matching the ground truth based on N. As an example, the Top-5 accuracy measures whether the ground truth is present among the Top-5 generated molecules. Similarly, the Top–N scaffold accuracy measures the occurrence of the ground truth scaffold among the Top–N generated scaffolds. For this metric we used the Murcko scaffold. [cite]To evaluate the model's ability to predict the correct functional groups, three metrics were used: Mean and average weighted F1-score as well as the molecular perfection rate. The metrics were calculated based on the Top-1 generated molecule for each sample. For each of the 16 functional groups defined by Fine et al.56 the F1-score was calculated and based on the mean as well as average weighted F1-score across the 16 was measured. The molecular perfection rate, as defined by Fine et al.56 was measured by comparing the functional groups present in the ground truth and those in the Top-1 generated molecule.
Code availability
The code supporting the findings of this work is available at: https://github.com/rxn4chemistry/MultimodalAnalytical. Specific instructions to reproduce the results can be found at https://github.com/rxn4chemistry/MultimodalAnalytical/tree/main/paper_replication/ir. A persistent record of the codebase as of 18.06.2025 is available on Zenodo (DOI: 10.5281/zenodo.15692637, https://zenodo.org/records/15692638).Data availability
All data used in this study were either published with the original article (https://zenodo.org/records/7928396, DOI: 10.5281/zenodo.7928395) or as part of our multimodal dataset (https://zenodo.org/records/14770232, DOI: 10.5281/zenodo.11611177). The NIST database was used for fine tuning (https://www.nist.gov/srd/nist-standard-reference-database-35).Author contributions
M. A. conceptualised the project, developed the methods, optimised the machine learning model and analysed the data. F. Z. and T. L conceptualised the pseudo-experimental spectra with F. Z. implementing them. The manuscript was written by M. A. and T. L.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This publication was created as part of NCCR Catalysis (grant numbers 180544 and 225147), a National Centre of Competence in Research funded by the Swiss National Science Foundation.Notes and references
- B. H. Stuart, Infrared Spectroscopy: Fundamentals and Applications, John Wiley & Sons, Ltd, 2004 Search PubMed.
- T. Visser, Encyclopedia of Analytical Chemistry, John Wiley & Sons, Ltd, 2006 Search PubMed.
- M. Avram and G. D. Mateescu, Infrared Spectroscopy: Applications in Organic Chemistry, R. E. Krieger Publishing Company, 1978 Search PubMed.
- J. E. Chalmers, H. G. M. Edwards and M. D. Hargreaves, Infrared and Raman Spectroscopy in Forensic Science, John Wiley & Sons, Ltd, 2012 Search PubMed.
- P. Hore, Nuclear Magnetic Resonance - Paperback - Peter Hore - Oxford University Press, Oxford University Press, Oxford, 2015 Search PubMed.
- W. M. A. Niessen and R. A. C. Correa, Interpretation of MS-MS Mass Spectra of Drugs and Pesticides, John Wiley & Sons, Ltd, 2017 Search PubMed.
- P. R. Griffiths and J. A. de Haseth, Fourier Transform Infrared Spectrometry, John Wiley & Sons, Ltd, 2007 Search PubMed.
- KnowItAll IR Spectral Database Collection, https://sciencesolutions.wiley.com/solutions/technique/ir/knowitall-ir-collection/.
- D. Lin-Vien, N. B. Colthup, W. G. Fateley and J. G. Grasselli, The Handbook of Infrared and Raman Characteristic Frequencies of Organic Molecules, Academic Press, 1991 Search PubMed.
- W. M. Niessen and M. Honing, Structure Elucidation in Organic Chemistry, John Wiley & Sons, Ltd, 2015, pp. 105–144 Search PubMed.
- J. Coates, Encyclopedia of Analytical Chemistry, John Wiley & Sons, Ltd, 2006 Search PubMed.
- V. Barone, M. Biczysko, J. Bloino, M. Borkowska-Panek, I. Carnimeo and P. Panek, Int. J. Quantum Chem., 2012, 112, 2185–2200 CrossRef CAS.
- M. A. Palafox, Phys. Sci. Rev, 2018, 3(6), 20170184 Search PubMed.
- V. Barone, S. Alessandrini, M. Biczysko, J. R. Cheeseman, D. C. Clary, A. B. McCoy, R. J. DiRisio, F. Neese, M. Melosso and C. Puzzarini, Nat Rev Methods Primers, 2021, 1, 1–27 CrossRef.
- E. Jonas, NeurIPS, 2019, vol. 32 Search PubMed.
- B. Sridharan, S. Mehta, Y. Pathak and U. D. Priyakumar, J. Phys. Chem. Lett., 2022, 13, 4924–4933 CrossRef CAS PubMed.
- M. Alberts, F. Zipoli and A. Vaucher, Learning the Language of NMR: Structure Elucidation from NMR spectra using Transformer Models, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-8wxcz.
- O. Schilter, M. Alberts, F. Zipoli, A. C. Vaucher, P. Schwaller and T. Laino, NeurIPS 2023, AI4Science Workshop, 2023 Search PubMed.
- F. Hu, M. S. Chen, G. M. Rotskoff, M. W. Kanan and T. E. Markland, ACS Cent. Sci., 2024, 10, 2162–2170 CrossRef CAS.
- S. Devata, B. Sridharan, S. Mehta, Y. Pathak, S. Laghuvarapu, G. Varma and U. D. Priyakumar, Digital Discovery, 2024, 3, 818–829 RSC.
- M. Alberts, N. Hartrampf and T. Laino, From Spectra to Structure: AI-Powered 31P-NMR Interpretation, ChemRxiv, 2025, preprint, DOI:10.26434/chemrxiv-2025-5bd0b.
- K. Dührkop, L.-F. Nothias, M. Fleischauer, R. Reher, M. Ludwig, M. A. Hoffmann, D. Petras, W. H. Gerwick, J. Rousu, P. C. Dorrestein and S. Böcker, Nat. Biotechnol., 2021, 39, 462–471 CrossRef PubMed.
- F. Huber, S. van der Burg, J. J. J. van der Hooft and L. Ridder, J. Cheminf., 2021, 13, 84 Search PubMed.
- M. A. Stravs, K. Dührkop, S. Böcker and N. Zamboni, Nat. Methods, 2022, 19, 865–870 CrossRef CAS.
- J. A. Fine, A. A. Rajasekar, K. P. Jethava and G. Chopra, Chem. Sci., 2020, 11, 4618–4630 RSC.
- A. A. Enders, N. M. North, C. M. Fensore, J. Velez-Alvarez and H. C. Allen, Anal. Chem., 2021, 93(28), 9711–9718 CrossRef CAS PubMed.
- G. Jung, S. G. Jung and J. M. Cole, Chem. Sci., 2023, 14, 3600–3609 RSC.
- G. Chandan Kanakala, B. Sridharan and U. Deva Priyakumar, Digital Discovery, 2024, 3, 2417–2423 RSC.
- G. Lee, H. Shim, J. Cho and S.-I. Choi, ACS Omega, 2025, 10, 12717–12723 CrossRef CAS.
- M. C. Hemmer and J. Gasteiger, Anal. Chim. Acta, 2000, 420, 145–154 CrossRef CAS.
- J. D. Ellis, R. Iqbal and K. Yoshimatsu, IEEE Trans. Artif. Intell, 2024, 5, 634–646 Search PubMed.
- M. Alberts, T. Laino and A. C. Vaucher, Commun. Chem., 2024, 7, 1–11 CrossRef PubMed.
- K. Guo, Y. Shen, G. A. Gonzalez-Montiel, Y. Huang, Y. Zhou, M. Surve, Z. Guo, P. Das, N. V. Chawla, O. Wiest and X. Zhang, arXiv preprint arXiv:2502.09897, 2025 Search PubMed.
- W. Wu, A. Leonardis, J. Jiao, J. Jiang and L. Chen, The Journal of Physical Chemistry, 2025, 129, 2077–2085 CrossRef CAS PubMed.
- G. C. Kanakala, B. Sridharan and U. D. Priyakumar, Digital Discovery, 2024, 3, 2417–2423 RSC.
- M. Priessner, R. Lewis, J. P. Janet, I. Lemurell, M. Johansson, J. Goodman and A. Tomberg, Enhancing Molecular Structure Elucidation: MultiModalTransformer for both simulated and experimental spectra, ChemRxiv, 2024, preprint, DOI:10.26434/chemrxiv-2024-zmmnw.
- D. Weininger, J. Chem. Inf. Model., 1988, 28(1), 31–36 CrossRef CAS.
- A. Dosovitskiy, L. Beyer, A. Kolesnikov, D. Weissenborn, X. Zhai, T. Unterthiner, M. Dehghani, M. Minderer, G. Heigold, S. Gelly, J. Uszkoreit and N. Houlsby, An Image is Worth 16x16 Words: Transformers for Image Recognition at Scale, 2021, arXiv:2010.11929 Search PubMed.
- Y. Nie, N. H. Nguyen, P. Sinthong and J. Kalagnanam, A Time Series is Worth 64 Words: Long-term Forecasting with Transformers, 2023, arXiv:2211.14730 Search PubMed.
- Y. Gong, Y.-A. Chung and J. Glass, AST: Audio Spectrogram Transformer, 2021, arXiv:2104.01778 Search PubMed.
- R. Xiong, Y. Yang, D. He, K. Zheng, S. Zheng, C. Xing, H. Zhang, Y. Lan, L. Wang and T.-Y. Liu, On Layer Normalization in the Transformer Architecture, 2020, arXiv:2002.04745 Search PubMed.
- N. Shazeer, GLU Variants Improve Transformer, 2020, arXiv:2002.05202 Search PubMed.
- E. Almazrouei, H. Alobeidli, A. Alshamsi, A. Cappelli, R. Cojocaru, M. Debbah, E. Goffinet, D. Hesslow, J. Launay, Q. Malartic, D. Mazzotta, B. Noune, B. Pannier and G. Penedo, The Falcon Series of Open Language Models, 2023, arXiv:2311.16867 Search PubMed.
- J. Gehring, M. Auli, D. Grangier, D. Yarats and Y. N. Dauphin, Convolutional Sequence to Sequence Learning, 2017, arXiv:1705.03122 Search PubMed.
- M. Alberts, O. Schilter, F. Zipoli, N. Hartrampf and T. Laino, NeurIPS, 2024, 37, 125780–125808 Search PubMed.
- S. E. Stein, NIST Standard Reference Database 35: NIST/EPA Gas-Phase Infrared Database - JCAMP Format, 2008, https://www.nist.gov/srd/nist-standard-reference-database-35, Accessed: 2025-03-28 Search PubMed.
- E. J. Bjerrum, SMILES Enumeration as Data Augmentation for Neural Network Modeling of Molecules, 2017, arXiv:1703.07076 Search PubMed.
- P. Schwaller, T. Laino, T. Gaudin, P. Bolgar, C. A. Hunter, C. Bekas and A. A. Lee, ACS Cent. Sci., 2019, 5, 1572–1583 CrossRef CAS PubMed.
- I. V. Tetko, P. Karpov, E. Bruno, T. B. Kimber and G. Godin, Artificial Neural Networks and Machine Learning – ICANN 2019: Workshop and Special Sessions, 2019, pp. 831–835 Search PubMed.
- J. Arús-Pous, S. V. Johansson, O. Prykhodko, E. J. Bjerrum, C. Tyrchan, J.-L. Reymond, H. Chen and O. Engkvist, J. Cheminf., 2019, 11, 71 Search PubMed.
- J. E. Hu, H. Khayrallah, R. Culkin, P. Xia, T. Chen, M. Post and B. Van Durme, Proceedings of the 2019 Conference of the North American Chapter of theAssociation for Computational Linguistics, 2019, pp. 839–850 Search PubMed.
- M. Post and D. Vilar, Fast Lexically Constrained Decoding with Dynamic Beam Allocation for Neural Machine Translation, 2018, arXiv:1804.06609 Search PubMed.
- Z. Li, X. Ding, T. Liu, J. E. Hu and B. V. Durme, Proceedings of the Twenty-Ninth International Joint Conference on Artificial Intelligence, 2020, pp. 3629–3636 Search PubMed.
- P. Anderson, B. Fernando, M. Johnson and S. Gould, Guided Open Vocabulary Image Captioning with Constrained Beam Search, 2017, arXiv:1612.00576 Search PubMed.
- L. Beurer-Kellner, M. Fischer and M. Vechev, Guiding LLMs The Right Way: Fast, Non-Invasive Constrained Generation, 2024, arXiv:2403.06988 Search PubMed.
- J. Fine, A. Rajasekar, K. Jethava and G. Chopra, Chem. Sci., 2020, 11, 4618–4630 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5dd00131e |
| This journal is © The Royal Society of Chemistry 2025 |