Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Data-driven analysis of text-mined seed-mediated syntheses of gold nanoparticles†
Sanghoon
Lee
 ab,
Kevin
Cruse
ab,
Kevin
Cruse
 bc,
Samuel P.
Gleason
cd,
A. Paul
Alivisatos
bcde,
Gerbrand
Ceder
bc,
Samuel P.
Gleason
cd,
A. Paul
Alivisatos
bcde,
Gerbrand
Ceder
 bc and
Anubhav
Jain
*a
bc and
Anubhav
Jain
*a
aEnergy Technologies Area, Lawrence Berkeley National Laboratory, Berkeley, CA, USA. E-mail: AJain@lbl.gov
bDepartment of Materials Science and Engineering, University of California, Berkeley, CA, USA
cMaterials Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA, USA
dDepartment of Chemistry, University of California, Berkeley, CA, USA
eKavli Energy NanoScience Institute, University of California Berkeley, CA, USA
First published on 22nd November 2024
Abstract
Gold nanoparticles (AuNPs) are widely used functional nanomaterials that exhibit adjustable properties depending on their shapes and sizes. Creating a comprehensive dataset of AuNP syntheses is useful for understanding how to control their morphology and size. Here, we employed search-based algorithms and fine-tuned the Llama-2 large language model to extract 492 multi-sourced seed-mediated AuNP synthesis recipes from the literature. With this dataset which we share online, we verified that the type of seed capping agent such as CTAB or citrate plays a crucial role in determining the morphology of the AuNPs, aligning with established findings in the field. We also observe a weak correlation between the final AuNR aspect ratio and silver concentration, although a large variance reduces the significance of this relationship. Overall, our work demonstrates the value of literature-based datasets in advancing knowledge in the field of nanomaterial synthesis for further exploration and better reproducibility.
Introduction
Gold nanoparticles (AuNPs) have been widely used for diverse applications such as semiconductors,1 catalysis,2 and biomedicine.3,4 Notably, AuNPs offer the advantage of tunable physical and chemical properties depending on their shape and size. For instance, gold nanostars exhibit distinct properties compared to other shapes of AuNPs closely related to their design, with amplified electromagnetic fields at the tips and enhanced hydrophilic environments at the indentations, making them useful in localized chemical manipulation and imaging applications.5 As another example, a specific size (28 × 8 nm) of gold nanorods has been found to be the most effective size for plasmonic photothermal therapy in cancer treatment.6Control of nanoparticle morphology7–10 has been established for some recipes and limited ranges of synthesis conditions, while the theory is still developing not only for various other conditions of gold nanoparticles11–14 but also for other metals15,16 and semiconductors.17,18 To advance the understanding of the underlying mechanisms that play a role in each synthesis protocol, computational approaches including Density Functional Theory (DFT) have been employed.19,20 However, it is impractical to simulate a sufficiently large system in a solution environment due to the high computational cost. Furthermore, it is important to note that achieving reproducibility in nanoparticle synthesis can be challenging, given the sensitivity of the process to numerous experimental variables including human factors,7 reagent impurities21 and stock solution age, which are often missing or poorly described in the recipes.
Among various AuNP synthesis protocols, seed-mediated growth of gold nanoparticles has been a popular method to produce AuNPs with anisotropic shapes.12,22 This method separates the nucleation and growth processes in separate flasks leading to more controlled growth stages. Briefly, spherical gold seeds are formed through the reduction of Au(III) to Au using a strong reducing agent such as sodium borohydride. This results in a solution of Au seed particles, which are typically 1–4 nm in size. Then, an aliquot of this seed solution is mixed with a growth solution containing partially reduced Au(I), made by mixing an Au(III) source, a weak reducing agent, and other precursors.23
According to Personick and Mirkin, the seed-mediated growth process for gold nanoparticles can be understood through two primary pathways: kinetic growth and selective surface passivation.12 In the kinetically controlled pathway, both the effective concentration of Au(I) ion and the reduction potential of Au(I) are key factors influencing growth. In this context, adding ascorbic acid would increase the Au(I) reduction rate, thereby promoting morphologies with higher energy surfaces (e.g., trisoctahedra > cube > octahedra). On the other hand, in the surface passivation pathway, growth can also be modulated by the deposition of a small amount of Ag onto the Au surface, referred to as Ag underpotential deposition (UPD). As the Ag monolayer coverage on the Au surface increases, shapes with more open facets (e.g., concave cubes > bipyramids) are favored.
Burrows et al. conducted a series of experiments, systematically varying multiple factors to understand the seed-mediated growth of gold nanorods (AuNRs) in particular.7 Their work revealed insightful correlations between 8 synthesis factors such as silver amount and seed aging time, and 5 different AuNR outcome responses including shape yield and AuNR size. By using fractional factorial design of experiments, Burrows et al. effectively explored many synthesis factors within a fixed protocol, conducting a total of 42 experiments. Despite the study's scope, which primarily involved two levels (e.g., NaBH4 amounts of either 0.0378 g or 0.0450 g) for each synthesis factor, their approach enabled efficient sampling of the synthesis space. Furthermore, Burrows et al. highlighted a notable challenge concerning the limited reproducibility in human-controlled nanoparticle syntheses. Specifically, despite employing the same recipe and the same chemical stock, noticeable variations in synthesis outcomes were observed among different experimenters within a single research group. Recently, Park et al.24 conducted an in-depth investigation into the correlations between similar synthesis factors for multiple growth stages. Notably, they expanded on Burrows et al.'s work by exploring a broader range of levels for each factor.
These studies motivate gathering a comprehensive, multi-sourced dataset as a foundation for data-driven studies, rather than one-variable-at-a-time (OVAT) or design of experiments (DoE) approaches.25 Efforts in the field of solid-state synthesis have demonstrated the value of this approach, with researchers constructing large, structured datasets through text-mining of the existing literature on solid-state synthesis,26–28 and training machine learning (ML) models for predicting the target stoichiometry,29 recommending precursor(s)30 and other synthesis conditions.31–34
Cruse et al. published an AuNP synthesis dataset from the literature.35 They utilized a series of machine-learned Named Entity Recognition (NER) to extract entities such as precursor materials, AuNP morphologies, and sizes. Notably, named entities are extracted but they are not connected through relation extraction (RE). To illustrate, if NER detected morphology entities such as “nanospheres” and “nanorods” from a paragraph along with precursor material entities and their amounts, relation extraction would identify which precursors were used for which morphology. Such linking requires further processing, and would ultimately lead to a more complete dataset of structured recipes.
Recently, Large Language Models (LLMs) including GPT-3/3.5/4 (ref. 36–38) and Llama-2 (ref. 39) have introduced exciting opportunities in materials science for structured information extraction and direct prediction. LLMs can be fine-tuned to output a sequence in the desired format,40,41 or they also can be used as-is (zero-shot) or with some context or examples provided42–44 (few-shot). Walker et al.41 published an AuNR recipe dataset using fine-tuned GPT-3 (ref. 36) and Llama-2.39,45 The structured recipes were extracted directly via joint Named Entity Recognition and Relation Extraction.46 This work showed that fine-tuning GPT-3 for the NERRE task achieved 76% accuracy with 240 paper annotations, and comparable performance was achieved with Llama-2-13B. Building on this prior work, we expanded our scope to include all shapes of AuNPs using seed-mediated synthesis. In addition, we manually validated all the recipes, which significantly elevated the quality of the dataset, facilitating further analyses in synthesis science.
In this work, we used a hybrid approach to exploit the advantages of both search-based parsers and LLMs. Briefly, we developed a highly precise search-based parser to construct a seed-mediated AuNP dataset from more straightforward recipes and used it to fine-tune an LLM for the NERRE task that extracts structured recipes from more challenging examples. We then manually validated the resulting datasets for further analysis to obtain 492 seed-mediated recipes for AuNPs. We used ML models to generate data-driven hypotheses and verify several known statements on the correlation between synthesis conditions and the morphology and size of synthesized AuNPs using a multi-sourced, text-mined dataset from the literature. To our knowledge, this compilation represents the largest manually validated structured database available for AuNPs using seed-mediated growth with a high level of depth and completeness.
Methods
Synthesis recipe and outcome extraction
The dataset in this study includes seed-mediated gold nanoparticle syntheses using chemical reduction. Specifically, the seed solution and growth solution should start from an Au(III) source along with other precursors for nucleation (of spherical seeds) and growth, respectively. Then, an aliquot of seed solution is mixed into the growth solution to produce the final product. Syntheses involving purchased seeds, multiple growths, or substrates are not included in the scope of this dataset. The solution that was used for diluting seed solution after its aging is regarded as part of the growth solution.We started from an initial database of nearly 5 million articles. Details for the initial database acquisition and filtration have been described by Kononova et al.26 Briefly, publications are sourced from materials science-related journals from Springer, Wiley, Elsevier, the Royal Society of Chemistry, the Electrochemical Society, and the American Chemical Society. Then, we collected 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 108
108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 803 nanomaterial papers by performing a keyword search for papers containing “nano*”, where “*” could be any contiguous set of characters.
803 nanomaterial papers by performing a keyword search for papers containing “nano*”, where “*” could be any contiguous set of characters.
Fig. 1 depicts the schematics of the synthesis extraction process. Before describing this process in detail, we provide a brief overview of the procedure. First, a “clean” dataset of 328 recipes was obtained mainly via search-based parser components. Then, a subset of this dataset was used to fine-tune Llama-2, which was then deployed to process “hard” and “partial” cases to obtain 91 and 73 validated recipes, respectively. “Hard” cases were those where the search-based parser failed to parse a recipe, and “partial” cases were those where it extracted a partially complete recipe. Additionally, we extracted precursor-only recipes, disregarding the amounts, which we referred to as the ‘categorical’ dataset, in contrast to the dataset containing complete amounts. The categorical dataset comprised 139 and 124 recipes from hard and partial cases, respectively. Overall, we obtained a dataset consisting of 492 recipes with complete amounts and 591 in the categorical format.
Next, we describe each step in detail. Note that the source code of the procedure is available (see Data and code availability). Fig. 1 illustrates the process where a paper undergoes AuNP acronym conversion and AuNP morphology detection to obtain AuNP-related papers. Using regular expressions (regex), acronyms used in each paper were searched and converted back to their full forms, and then morphology-related keywords in the paper were identified. Although acronyms for well-defined morphologies are already included in the keywords for search-based morphology extraction, this step was necessary due to confusing or overlapping acronyms. As an example, “AuNS” is used as an acronym for “gold nanosphere” in some cases and for “gold nanostar” in others, while “AuNR” is almost always identified and classified as “gold nanorod”. To resolve this, text strings that initially defined an acronym, such as “AuNSs (gold nanospheres)”, were searched for throughout the paper using regex, and then all strings of “AuNS” in the paper were replaced with “Gold nanosphere”. Then, the morphology detection algorithm seeks mentions of a pre-defined set of nanoparticle shapes (e.g., “nanosphere”, “nanorod”, and “NR”). Many miscellaneous morphologies that are rare (e.g., nanopopcorn and nanoberries) are still included in this set of patterns but are categorized as ‘other’ shapes. While non-Au NPs are filtered out, NP mentions with unspecified materials are still detected as potential AuNP morphologies. Instances involving unspecified shapes (referred to as ‘NP’, encompassing cases like ‘nanocrystals' and ‘nanoparticles') are considered low priority and are classified as such when no other specific shapes are explicitly mentioned in the paper. Then, we filtered out publications that have no AuNP morphologies. In cases that have only two morphologies including nanosphere, the sphere is ignored because spheres are often mentioned either as the gold seed shape or as the byproduct. In this process, 60 papers mentioning more than one anisotropic gold morphology were identified as having multiple syntheses and were filtered out, as handling these cases would require additional steps for recipe separation and linking, which were beyond the scope of this study.
Following morphology detection, seed-mediated recipes were screened by matching keywords including “seed-mediated” and “growth solution”. Publications that had two or more methods including seed-mediated growth and other non-seed-mediated methods (with keywords ‘one-pot’ or ‘seedless') were regarded as having multiple recipes and were therefore filtered out.
The next step in the workflow is recipe paragraph classification. The goal of this step is to identify which of the paragraphs in the full text contain information on AuNP synthesis recipes (as opposed to introduction, conclusion, etc.). Candidate recipe paragraphs were identified using regex to screen for paragraphs containing AuNP or keywords like “synthesize” and “prepared”. After this filter, the recipe paragraphs were identified using a trained classifier for enhanced precision of the recipe filter. Briefly, we trained a Support Vector Classifier (SVC), using MatBERT embeddings28 of each paragraph. Recipe paragraphs and non-recipe paragraphs were sampled and annotated to construct a dataset of 920 paragraphs, which was split 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 into training and test sets. Test set accuracy was 0.89 for this SVC classifier. Using MatBERT embeddings (784 dimension) as features showed classification performance comparable to training using GPT-3 embeddings (1536 dimension, test set accuracy 0.90).
1 into training and test sets. Test set accuracy was 0.89 for this SVC classifier. Using MatBERT embeddings (784 dimension) as features showed classification performance comparable to training using GPT-3 embeddings (1536 dimension, test set accuracy 0.90).
Seed and growth solution recipes (subrecipes) were identified from the recipe paragraph(s). We devised a keyword-based algorithm to separate seed solution, growth solution recipes and the mixing process. Briefly, given a recipe text, this algorithm searches for keywords for each subrecipe (e.g., “seeds” for seed solution, “grow” or “separate flask” for growth solution and “aliquot of seeds” for the mixing process). Once it identifies the first subrecipe keyword it reads through the text while searching for the other subrecipe to switch. As this algorithm covers clearer cases separable by distinct keywords, a significant portion of papers were filtered out and collected as the “hard” cases to be processed by the fine-tuned LLM. However, this screening process also filters out cases where a subrecipe is indeed missing, for instance, directly purchased or when authors choose to reference a recipe from a previous publication instead of explicitly detailing the procedure.
Next, precursors were extracted using regular expressions of 41 precursors of interest (full list provided in the ESI†) for each subrecipe. This specific set of precursors was identified by the most frequent precursors within the NER-detected dataset,35 with a small number of precursors manually added. These include the gold source (AuCl4−), capping agents (citrate and CTAB), reducing agents (ascorbic acid and NaBH4), and other additives (AgNO3 and HCl). This predefined set of precursors is first categorized into one of 54 precursor formulae and then normalized to one of 41 precursors using domain knowledge. For instance, ‘NaAuCl4’, ‘gold chloride’, ‘HAuCl4’, and its various hydrates are normalized as ‘AuCl4−’.
The amount of each precursor is parsed based on regular expression, via strict proximity-and-pattern-based linking using each precursor match position. Then each subrecipe is represented as a maximum 41-dimension vector, where each component of this vector corresponds to the precursor's molar concentration in the resulting solution. However, there existed several publications with recipes for which the amounts parsing returned errors (e.g., precursor typos, or the seed solution recipe being entirely missing). These cases are regarded as “hard” cases to be re-extracted by the fine-tuned LLM.
Following the amounts parsing, these recipes were post-processed to calculate the final concentration of each precursor in its solution. Here, partially complete recipes were identified, for instance, those missing the gold source, missing amounts (e.g., of HCl), or mentioning only the concentration but not the volume. These recipes were regarded as “partial” cases to be processed by fine-tuned LLM.
All extracted recipes underwent manual validation to confirm that the entire recipe had been correctly extracted. This is a strict metric, as a recipe is marked correct when all entities of the parser (precursors and amounts) are correctly parsed. If applicable, the incorrect recipes were further corrected to be included in the final dataset.
The search-based parser extracted 350 complete seed-mediated recipes with the morphological outcomes, all of which were manually corrected to give 331 recipes. A sample of 224 recipes from this along with 22 recipes sampled from the “hard” dataset were used as training data to fine-tune Llama-2 (13B, 8-bit quantized)39 so that, from a given seed-mediated recipe text, the model can extract a structured recipe in the JSON format. The annotations of these recipes were carefully curated to use only the text as presented verbatim in the input text, thereby attempting to avoid hallucination artifacts. As LLMs are sequence-to-sequence models, output order could cause variations in the output, so annotations were made to reflect the order in which the precursors were presented in the input text. The keys of the output JSON were ‘seed solution’, ‘growth solution’, and ‘mixed seed’.
We deployed this fine-tuned LLM to extract recipes from hard and partial cases. After running the fine-tuned LLM, the outputs were further post-processed to screen for complete recipes, using the same post-processing algorithm as the clean cases. Also, we applied a hallucination filter to check that every output token is present in the input text, which is consistent with the training data annotation scheme. Then, these datasets were manually validated to check that the target morphology and recipe (precursors and amounts) were correctly extracted, also categorizing the incorrect extractions as “not in scope”, “missing info” or “parser fault”. In validating hard and partial recipes, bipyramid, cube, and star syntheses were corrected if possible (in cases where the recipe was still in scope but the parser output was wrong), and the other morphology recipes were validated but not necessarily corrected. In hard cases, we identified 91 recipes with complete precursor amounts, while for 139 recipes, the emphasis was placed on accurate precursor categories regardless of amounts (referred to as ‘categorical’ recipes). As for partial cases, we observed 73 recipes with complete amounts and 124 categorical recipes. The categorical recipes correspond to both cases where the amounts were indeed missing in the original synthesis description and cases where the extracted amounts were incorrect.
For a subset of nanorod recipes using the most common precursor set, a search-based size detection algorithm was used for further analysis, correlating the recipe and size. This algorithm searches size indicators (e.g., “diameter”, “length” and “aspect ratio”) and size measurements (e.g., “between 30 and 40 nm” and “3 ± 0.5”). Then it attempts to link the indicator–measurement pair in a single sentence, and outputs a blank list if the link is not straightforward. The extraction and linking of the size to the synthesis target of interest were challenging because the size characterization section was often distant from the synthesis recipes in the document. Furthermore, many papers present the size information of the ‘final’ synthesized AuNRs after modifications (e.g., another seeded growth using NR seeds) and it was not always clear, without the figures, what the presented size information referred to. To ensure the quality of the relation extraction, we manually validated the entire dataset for further analysis.
Data-driven hypotheses from machine learning
To learn the factors controlling morphology for seed-mediated growth recipes, we trained classifier models that predict the resulting shape for a given precursor vector from a synthesis recipe. Morphology classes with 17 data points or more (bipyramids, stars, cubes, and rods) were included in the analysis.The concentration features were transformed in two different ways for comparison. The first was the final molar concentrations of the corresponding (seed or growth) solution before the reaction. The second was log(107[X] + 1), where [X] is the molar concentration.
This log transformation puts the concentration features in a form that is more typical of reaction rates, as explained below. Briefly, logistic regression is based on linear predictors, which are linear combinations of the features. Using original features, this linear combination would be in the form of  where w is the model weight vector to be trained. When using feature transformation proposed in this work, this predictor would take the form of
where w is the model weight vector to be trained. When using feature transformation proposed in this work, this predictor would take the form of  which involves an initial-reaction-rate-like form. A detailed discussion of the feature processing is in the ESI.†
which involves an initial-reaction-rate-like form. A detailed discussion of the feature processing is in the ESI.†
To train ML models with the dataset, it was important to address the asymmetric size of each morphology class with the majority being AuNR syntheses (379 recipes) and the nanostar syntheses (21 recipes) being the next most common. We first separated the dataset into AuNR recipes and non-rod recipes. AuNR and non-rod recipes were then split with a 98![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 6
2 and 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 train/test ratio, respectively. This would give a test set with a rod class size similar to the star class size, both with around 7 data points. After the split, training data for non-rod recipes were copied and duplicated by 30. This number was chosen to make each of the non-rod class size similar to the rod class size in the training set. We tested with 8 different seed values for the splits, and the accuracy scores were averaged across 8 seeds. For this classification task, we trained various models including logistic regression, nearest neighbor, decision tree, and multi-layer perceptron using scikit-learn.47 For deployment, we used all the data for training while repeating data points from non-rod classes 20 times (again to make non-rod class size similar to rod class size). We trained decision tree with depth 3 and logistic regression (with regularization parameter C = 0.5) to obtain interpretable models.
4 train/test ratio, respectively. This would give a test set with a rod class size similar to the star class size, both with around 7 data points. After the split, training data for non-rod recipes were copied and duplicated by 30. This number was chosen to make each of the non-rod class size similar to the rod class size in the training set. We tested with 8 different seed values for the splits, and the accuracy scores were averaged across 8 seeds. For this classification task, we trained various models including logistic regression, nearest neighbor, decision tree, and multi-layer perceptron using scikit-learn.47 For deployment, we used all the data for training while repeating data points from non-rod classes 20 times (again to make non-rod class size similar to rod class size). We trained decision tree with depth 3 and logistic regression (with regularization parameter C = 0.5) to obtain interpretable models.
Results
Evaluation of parser accuracy
We evaluated the performance of our data extraction parsers during the manual validation process. It should be noted that the comparison of parsers is not straightforward, given that the inputs are different (clean cases for the search-based parser and partial and hard cases for the fine-tuned LLM), and parser outputs underwent additional filtration before manual validation to ensure recipe completeness and eliminate hallucinations.The precision is defined as the number of correct outputs divided by the number of post-processed parser outputs. The overall precision of the search-based parser and LLM is 0.78 and 0.42, respectively (ESI, Table S1†). We comment on the merits and drawbacks of both methods in the Discussion section of this work.
Dataset
We categorize each recipe based on the precursors used in seed solution and growth solution along with the target morphology. As shown in Table 1, the majority of the morphologies are non-spherical. The primary composition of the dataset is AuNR synthesis using AuCl4− (or its hydrates), BH4− (as a reducing agent), CTAB (as a capping agent) in seed solution and AuCl4−, AgNO3, CTAB, and ascorbic acid (as a weak reducing agent) with or without a strong acid in growth solution. A synthesis protocol using these seven precursors (established by Nikoobakht and El-Sayed23) is commonly observed in our dataset to produce AuNRs of various dimensions.| Shape | Count | Most common precursor set |
|---|---|---|
| Rod | 377 | AuCl4−, CTAB, BH4−; AuCl4−, CTAB, AA, AgNO3 (261) + HCl (65)/H2SO4 (18)/HCl + OlA (6)/5-BrSA (6) |
| Star | 21 | AuCl4−, citrate; AuCl4−, AA, AgNO3 (2) + HCl (6) |
| AuCl4−, citrate, BH4−; AuCl4−, AA, AgNO3 + CTAB (2)/BSA (2) | ||
| AuCl4−, TritonX-100, BH4−; AuCl4−, TritonX-100, AA, AgNO3 (3) | ||
| Cube | 19 | AuCl4−, CTAB, BH4−; AuCl4−, CTAB, AA (17) |
| AuCl4−, CTAC, BH4−; AuCl4−, CTAC, AA, AgNO3, HCl (2) | ||
| Bipyramid | 17 | AuCl4−, citrate, BH4−; AuCl4−, CTAB, AA, AgNO3, HCl (10) |
| AuCl4−, CTAC, citrate, BH4−; AuCl4−, CTAB, AA, AgNO3, HCl (3) | ||
| Sphere | 15 | AuCl4−, CTAB, BH4−; AuCl4−, CTAB, AA (3) + AgNO3 (4) |
| NP | 32 | AuCl4−, CTAB, BH4−; AuCl4−, CTAB, AA (8) + AgNO3 (4) |
| Other | 11 | — |
| Total | 492 | — |
For other morphologies, we observe that the size of each dataset is not as large as that of the rods. Still, it is worth pointing out that some target morphologies are associated with certain precursor sets more commonly than others. As for gold nanocube syntheses, 2 recipes using AgNO3 and HCl in growth solution create concave nanocubes.48 Most of the recipes use AuCl4−, BH4−, and CTAB in seed solution and AuCl4−, CTAB, and ascorbic acid in growth solution.9,49 It has been discussed that nanocubes are favored over other morphologies with lower CTAB and higher AA concentration.9,10 To synthesize nanobipyramids (BiPym), the majority of recipes use the precursor set used by Liu and Guyot-Sionnest,50 and all the recipes use AuCl4−, citrate and BH4− (3 with the addition of CTAC) in seed solution, which is known to promote penta-twinned seeds. This precursor set is also used in 6 out of 21 nanostar recipes, while 10 recipes use the Turkevich method51 (AuCl4− and citrate under boiling conditions) for synthesizing seeds.
Data-driven morphology classification from machine learning
Table 2 shows the mean test set accuracies from morphology classifiers trained using different featurization methods. Overall, most classifiers show high performance (>0.8) for determining one of the four classes of morphologies. We note a high score of 0.94 from multinomial logistic regression using log-transformed concentrations, which is comparable with neural network performance. Performance of models using log-based featurization is comparable to those using linear features, with slight preference for using log-based features. We rationalize this preference for log-based features as reflecting the initial reaction rates ([AuCl4−]a[BH4−]b…), as discussed in depth in the ESI.†| Feature for precursor X | Accuracy [X] | log(107[X] + 1) |
|---|---|---|
| Multinomial logistic regression | 0.91 | 0.89 |
| Multinomial logistic regression (C = 0.2) | 0.58 | 0.94 |
| Multinomial logistic regression (C = 0.5) | 0.78 | 0.94 |
| 1-Nearest neighbor | 0.82 | 0.89 |
| 3-Nearest neighbor | 0.86 | 0.88 |
| 5-Nearest neighbor | 0.88 | 0.90 |
| Decision tree (depth = 3) | 0.84 | 0.85 |
| Decision tree (depth = 4) | 0.85 | 0.85 |
| Neural net (MLP, hidden_layer_sizes = 100) | 0.92 | 0.95 |
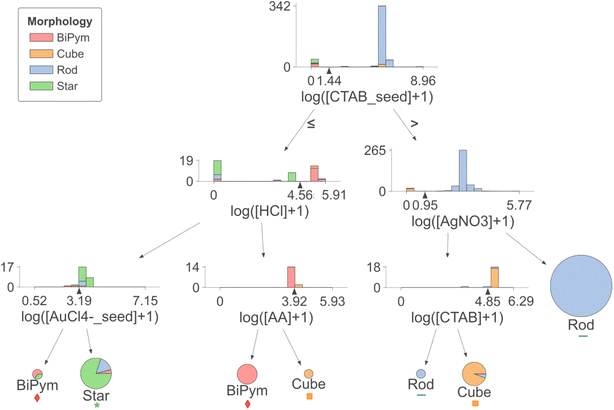
The data-driven decision boundaries obtained from the decision tree are illustrated in Fig. 2. Fig. 2 depicts a decision tree of depth 3 trained with the whole dataset while Fig. S2† shows a 2-dimensional precursor space (with CTAB in seed solution and HCl in growth solution), obtained from the first two decision boundaries.
Prior research has found that citrate-capped gold seeds have a penta-twinned structure,10 while CTAB-capped seeds are single crystalline; hence the former lead to bipyramids and the latter lead to rods.22,50 The first decision boundary in the decision tree divides the dataset into recipes that use CTAB as the seed capping agent leading to rods and cubes, and recipes that use other capping agents (including citrate + BH4−) leading to bipyramids and stars, indicating that the model has learned trends backed up by expert observations.
It is interesting that the second and third criteria (bottom left of Fig. 2) distinguish star and bipyramid syntheses by HCl concentration in growth solution (higher HCl favoring bipyramids over stars) and AuCl4− in seed solution (higher AuCl4− favors stars over bipyramids). This is consistent with the prior study that adding HCl in growth solution slows the reaction rate allowing growth of more anisotropic shapes.52 The third decision regards the concentration of AuCl4− in seed solution. Although this decision is made with very little support for bipyramids and we could not find a direct statement related to this decision, Langille et al. reviewed that the growth of gold nanostars is promoted by rapid kinetic growth.9,11,53 Higher pH in growth solution and higher concentration of AuCl4− in seed solution could be in line with the fast growth rate.
For recipes using CTAB in seed solution, two competing morphologies are nanocubes and nanorods. Most cube recipes in the dataset utilize a consistent set of precursors, specifically without AgNO3. Our decision tree analysis identified the concentration of AgNO3 as a fundamental factor (second decision point in Fig. 2) separating rods and cubes. For the latter case without AgNO3, we found the opposite trend (third decision point) to the one reported by Sau and Murphy, although we note that this was based on very few nanorod data points in this region. Sau and Murphy reported that a lower CTAB concentration resulted in the formation of nanocubes, as it promotes Au deposition on {111} faces and formation of {100} faces.9,10,54 Following this, Meena and Sulpizi have shown via molecular dynamics simulation that CTAB has higher packing density on Au(100) surfaces than on (111).55 This indicates the need for further exploration in the nanorod/nanocube formation in the absence of AgNO3.
A synthesis (precursor) space with clearer decision boundaries from regularized logistic regression is shown in Fig. 3, showing a 2D precursor space with multiple precursors. Instead of using popular dimensionality reduction methods like PCA or t-SNE, which ignore class labels, we utilized a learned weight matrix from multinomial logistic regression to better highlight decision boundaries between each class of morphology. To demonstrate a low-dimensional space, we selected two columns from this 38-feature by 4-morphology-class matrix, corresponding to bipyramids and rods. This matrix (dimension of 38 by 2) was then multiplied with the design matrix with dimensions (434 samples, 38 features), revealing one of the low-dimensional spaces using the learned weight matrix from the multinomial logistic regression model. Since these weights act as linear coefficients of the features after the log transformation, they are represented as the exponents of each precursor in the axes of Fig. 3, as discussed in the Methods section.
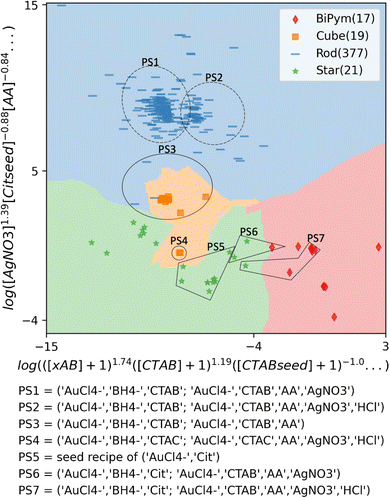 | ||
| Fig. 3 2D precursor space with morphology classification obtained from logistic regression. 3-Nearest neighbor boundaries in this 2D space are also shown in lighter colors. All the precursor concentrations are preprocessed and transformed with 107[X] + 1 where [X] is molar concentration of precursor X. ‘xAB’ represents ammonium bromide precursors excluding CTAB such as cetyltripropylammonium bromide (CTPAB), or didodecyldimethylammonium bromide (DDAB). Solid line circles or polygons indicate that all data points in them have the corresponding precursor set, while dashed line circles indicate that most of the data points in the circle have the corresponding precursor set. The full description of the axes is provided in the ESI.† | ||
Compared to Fig. S2,† clearer boundaries distinguishing stars, cubes, and rods are observed. For each dimension, the order of precursors is sorted by the absolute values of the linear coefficients (shown as exponents), and the full description of the axes and grouping of precursor sets are provided in the ESI.† We observe that various precursor sets used in our dataset can be mapped to form clusters in this synthesis space after dimensionality reduction. Specifically, the two most common precursor sets in nanorod synthesis, shown in Table 1, are observed as two somewhat distinct clusters (labeled as PS1 without acid and PS2 with HCl in Fig. 3). The most common precursor set for nanocube is observed as PS3 in Fig. 3, which also includes three nanorod recipes that appear in the bottom right histogram in Fig. 2. PS4 represents concave cube syntheses and is distinguished from the rest of the nanocube syntheses due to the use of AgNO3 and HCl. For star and bipyramid syntheses, two precursor sets (PS6 and PS7 in Fig. 3) are observed in both syntheses, but these are separated by multiple factors including HCl in growth solution and AuCl4− in seed solution, consistent with decision criteria in Fig. 2.
Correlation between the AuNR aspect ratio and AgNO3
Several past studies reported the use of silver nitrate to control the size and aspect ratio of AuNRs7,22,23 when using the most common precursor set for AuNR in the first row of Table 1. To investigate the correlation between precursor amounts and the aspect ratio (AR), a subset of the dataset (169 AuNR recipes with AR) using seven precursors (AuCl4−, BH4−, and CTAB in seed solution and AuCl4−, AgNO3, CTAB, and ascorbic acid in growth solution) without acid (133 recipes) or with HCl (36 recipes) were obtained from the search-based size detection algorithm and manual validation. In some cases, only the aspect ratio is reported without the corresponding lengths and widths, so our analysis focuses on the AR to incorporate as much data as possible. Additionally, a plot showing the correlation between AgNO3 and AuNR length and width is provided in the ESI (Fig. S3†).Previous studies that span this set of seven precursors indicate achievable rod aspect ratios between 1.5 and 5.0,56 which is also reflected in the dataset (blue scatter in Fig. 4) (two recipes57,58 reported aspect ratios ∼5.2). Aspect ratios of the nanorods are often controlled by the Ag+ amount.23 Specifically, as the silver nitrate concentration was increased in the controlled experiments of Nikoobakht and El-Sayed, the aspect ratio increased until [AgNO3] = 0.00012 M (or log[AgNO3] = −3.92) after which the aspect ratio decreased. They attributed this decrease to an increased interaction between silver and gold with the surfactant (CTAB) counterions. This trend is qualitatively shown in our dataset, as the maximum aspect ratio for a given silver concentration increases and then decreases around the region of log[AgNO3] = −3.92 (mean-squares quadratic trendline in Fig. 4). However, in our case this result is obtained by extracting a variety of synthesis recipes from the literature in contrast to the controlled experimental setting of the rest of the precursors' amounts and other synthesis factors.
 | ||
| Fig. 4 Aspect ratios of the synthesized AuNRs using AuCl4−, CTAB, and BH4− in seed solution and growth solution with AuCl4−, CTAB, AgNO3, and ascorbic acid with (scatter in orange, 36 recipes) and without HCl (scatter in blue, 133 recipes). The scatter plot with error bars shows range (or variance) of AR indicated in each publication. Dark orange circles are ‘reference’ recipes that were not included in the original database and were manually extracted from Nikoobakht and El-Sayed.23 Blue circles within a single column of [AgNO3] show duplicate recipes reproduced in different publications that had identical precursor final concentrations but reported nanorods with different (>20%) aspect ratios. Each scatter point is from a different publication except for Nikoobakht and El-Sayed. All data points are manually validated. | ||
It is known that lowering the pH by adding strong acids such as HCl leads to longer rods.59 Conversely, as shown in Fig. 4, comparing syntheses from the seven-precursor set and seven-precursor plus acid did not show a significantly different distribution of aspect ratios, although the concentrations of acid were mostly considerable ([HCl] = 0.001–0.08 M). Specifically, we observed a comparable distribution of ARs in the region of log[AgNO3] between −4.05 and −4.0 for recipes with HCl (ranging from a 2.0 to 5.2, with an average of 3.71) and those without HCl (from 2.0 to 5.0 and an average of 3.58).
Apparently identical AuNR recipes with different aspect ratios
We note that in our dataset, similar or apparently identical recipes result in different reported aspect ratios. We identified five groups of recipes each with identical concentrations of all seven precursors and a relative seed solution amount to the growth solution, but with significantly different ARs (by more than 20%) and these are highlighted in blue circles in Fig. 4 at log[AgNO3] = −4.224, −4.222, −4.11, −4.01, −3.52. Here, we note that all scatters circled in blue within a vertical column (i.e. with the same AgNO3 amount) are apparently identical recipes in terms of precursor concentration. The difference in ARs was up to 86% (5 and 2.69) which implies that there might be other highly significant factors beyond precursor amounts that impact the synthesis outcome. This large variance in the aspect ratio may also arise from how the aspect ratio is measured, specifically the sampling of the nanorods. While some studies mention sampling details (e.g., that the authors sampled >50 rods in the TEM image) this information is not always provided.Table 3 presents an overview of these identical recipes examined in depth, with detailed information available in the ESI (Table S2†). Notably, in recipe group A, different aging durations yielded distinct outcomes. Specifically, recipe index 3 resulted in shorter rods following overnight aging, contrasting with longer rods observed in recipe index 1 with shorter aging time. This finding aligns with a previous observation by Park et al., who noted that the length increases faster than the width in the earlier stages of growth.13
| Index | Group | log[AgNO3] | Seed aging | Mixture aging | AR | Ref. |
|---|---|---|---|---|---|---|
| 1 | A | −4.01 | 25 °C at least for 2 h | 27–30 °C for 20 min | 3.3 | 60 |
| 2 | A | −4.01 | — | — | 3.61 | 61 |
| 3 | A | −4.01 | 27 °C, 2–3 h | Overnight at 27 °C | 2.375 | 62 |
| 4 | B | −4.224 | 27 °C for at least 2 h | 12 h at 27 °C | 3–4 | 63 |
| 5 | B | −4.224 | 6 h | Overnight | 5 | 64 |
| 6 | B | −4.224 | 2 h at 25 °C | 3 h at 30 °C | 2.69 | 65 |
| 7 | B | −4.224 | 30 °C for 2 h | 30 °C for at least 3 h | 3.15 | 66 |
| 8 | B | −4.224 | 28 °C for at least 2 h | At least 3 h | 3.2 | 67 |
| 9 | C | −4.11 | 25 °C | 30 °C | 2.6 ± 0.3 | 68 |
| 10 | C | −4.11 | 25 °C | 27–30 °C | 4.0 | 69 |
| 11 | C | −4.11 | — | 30 °C, 30 min | 4 | 70 |
| 12 | D | −4.222 | 25 °C, 2 h | At least 3 h | 2–3 | 71 |
| 13 | D | −4.222 | — | 3 h | 3.21 | 72 |
| 14 | D | −4.222 | 25 °C, more than 2 h | Over 3 h | 3.6 ± 0.5 | 73 |
However, the inference of a missing factor is not always clear. For instance, in recipe group C, no discernible synthesis factors led to significant differences in aspect ratios, despite referencing the same work by Nikoobakht and El-Sayed. Similarly, although all mixtures in recipe group D were aged for at least 3 hours, they exhibited over a 20% difference in resulting aspect ratios. Intriguingly, recipe group B showed a wide range of ARs from 2.69 to 5. Among these, recipes 6, 7, and 8 showed no significant differences in aging conditions yet displayed an 18% variation in the AR (2.69 to 3.2). Moreover, although recipe index 4 features extended mixture aging and recipe index 5 has longer seed aging, the investigation for the effect of these extended (>6 hours) aging times requires further data points.
Discussion
Recipe extraction: hybrid parser using a search-based algorithm and LLM
Overcoming data scarcity and sparsity32 is critical for data-driven predictions in materials synthesis. In this paper, we presented a hybrid approach for synthesis extraction, mixing conventional rule-based search methods with a fine-tuned LLM. By adding the LLM we were able to obtain about 50% more data compared to when only using the search-based method. This LLM-based approach showed flexibility in separating the seed and growth subrecipes within a seed-mediated growth recipe. In contrast, there were cases where a straightforward separation algorithm utilizing the transition keyword (e.g., “For growth solution”) failed, resulting in the loss of a considerable number of relevant recipes. Also, the LLM showed flexibility in extracting precursor amounts. Specifically, the LLM could extend the predefined set of precursors of interest (such as 5-BrSA, Triton X-100, SDS, LSB and NH2OH which were not included in the original precursor set for regex), while considering the multitude of organic reagents, and including all of the possible precursors to a list of regular expressions can easily be an arduous process. Furthermore, the LLM could also easily adapt to out-of-distribution presentations of the precursor amounts. This was particularly interesting as we initially expected the patterns of the amounts of a chemical to be very limited, and regarded the others as the edge cases. For example, the LLM could also correctly parse amounts from the phrase “a mixture of (1 mL, 2 mM) HAuCl4 and (3 mL, 4 mM) CTAB” which was not included in our search-based detection and linking for the precursors.However, there exist limitations in this approach because the LLM tended to hallucinate the missing information when presented with partially complete recipes. Although we carefully constructed the training data to ensure the LLM generates tokens used in the input text, we still encountered “weak hallucinations” where the model generated information not found in the input text. For instance, there were cases where a seed solution recipe was completely missing in the source text. Indeed, we confirmed multiple cases where (i) seeds were purchased and used as-is, (ii) the recipe was described without details (e.g., “seeds were synthesized using borohydride reduction”) or (iii) the recipe was completely omitted. In such cases, we found that the model often confused the growth solution precursors and amounts with seed solution parameters, and these data were extracted as the seed solution recipe. Therefore, we had to manually filter cases and verify whether the information was indeed missing from the input text or not.
Recipe representation and missing data
For recipe representation, a synthesis procedure would be ‘completely’ described by a sequence of actions or graphs, especially considering the sensitivity of the nanoparticle syntheses. In this study, we represent a recipe as its precursor vector. While this representation effectively trains a morphology classifier, it falls short in tasks such as AuNR AR regression. In such cases, some data points in Table 3 indicate that additional features including aging temperature and time are necessary.There are challenges in achieving this complete description of the recipe. First, the information presented in the literature is often missing or varying in the level of detail. In addition to cases where the entire recipe or parts of the precursor amounts were missing (discussed in the above subsection), information such as shape yield, seed characterization, reaction/aging temperature and time is often simply missing in most cases. Furthermore, the order of mixing is not always explicitly presented. While some papers write that “Precursor A was injected into Precursor B and then Precursor C was mixed with that solution”, other papers write “A solution mixture of Precursor A, B, and C was mixed with another solution consisting of D and E”. We note that in the case of solid-state synthesis, Malik et al.29 found that adding the action sequence for product prediction did not show a significant increase in prediction power, which would imply that in a low-dataset-size regime, it is better to select some features of recipe representation rather than adding as many features as possible. Therefore, it is unclear whether more precise extraction of the action sequences would result in better model accuracy in this instance, but for larger data set sizes or for scientific understanding such sequence descriptions may prove to be useful.
Hypotheses generation from the dataset
Using a multi-source dataset extracted from the literature, we successfully identified and verified correlations between experimental conditions and morphological outcomes for seed-mediated gold nanoparticle synthesis. Specifically, we showed that the seed capping agent is crucial in shape determination of seed-mediated growth syntheses using purely statistical learning.For AuNR aspect ratio determination, we investigated the dataset using a fixed synthesis protocol with the most reproduced set of precursors. This revealed a very weak trend (Fig. 4), which may not be immediately exploitable. However, this general trend obtained from literature data is more robust against human factors and also encompasses a wider range of factor levels for each precursor, compared to existing OVAT studies. We also identified the challenges of this large data analysis by consolidating data from multiple sources in the literature. First is a bias in popularity. As shown in Table 1, most of the extracted recipes focused on nanorod synthesis, with the majority of them using a precursor set from Nikoobakht and El-Sayed.23 The second challenge is the lack of negative data, i.e., synthesis with lower shape selectivity. As shown in Fig. 3 and S2,† there are regions yet to be filled with data points for further exploration or to confirm a low shape yield. Overall, we believe that expanding on the Design of Experiments (DoE) studies including those by Burrows et al.,7 can highlight potential opportunities in further exploration and exploitation within the synthesis space. However, in the absence of controlled studies, this dataset can serve as a current map for known procedures.
Conclusion
In this study, we utilized a hybrid approach combining search-based algorithms and LLM to collect 492 seed-mediated gold nanoparticle (AuNP) synthesis protocols from various literature sources available online. Notably, while our dataset aligns with some established understandings in the field (e.g., the seed capping agent largely determines the AuNP morphology), we also encountered large variability (e.g., in AuNR aspect ratios from identical recipes) and even discrepancies. These inconsistencies could point to over-generalizations in human-centric approaches, underscoring the value of our data-driven methodology in revealing new insights and challenging existing assumptions.Looking ahead, this data-driven approach offers exciting opportunities for broader applications. Beyond AuNP synthesis, similar methodologies could be applied easily to other metal nanoparticles (e.g., AgNPs, CuNPs and PtNPs), or semiconductor nanoparticles for applications ranging from biomedicine to catalysis. Expanding this framework could inspire future developments in data-driven materials science, enabling more efficient and automated exploration of complex synthesis spaces.
Data and code availability
The dataset and code used are available at https://github.com/slee-lab/AuNP-seedmed-recipes.74Author contributions
S. L.: conceptualization, methodology, software, validation, formal analysis, investigation, writing – original draft, visualization. K. C.: writing – reviewing and editing, data curation. S. G.: writing – reviewing and editing. A. P. A.: resources, writing – reviewing and editing, supervision, project administration, funding acquisition. G. C.: resources, writing – reviewing and editing, supervision, project administration, funding acquisition. A. J.: conceptualization, methodology, resources, writing – reviewing and editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was intellectually led by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, and Materials Sciences and Engineering Division under Contract no. DE-AC02-05CH11231 (D2S2 program KCD2S2). This research used resources of the National Energy Research Scientific Computing Center (NERSC), a Department of Energy Office of Science User Facility supported by the Office of Science of the U.S. Department of Energy under Contract no. DE-AC02-05CH11231 using NERSC award BES-ERCAP0028631.References
- K. Sandeep, B. Manoj and K. G. Thomas, Gold nanoparticle on semiconductor quantum dot: do surface ligands influence Fermi level equilibration, J. Chem. Phys., 2020, 152, 044710 CrossRef CAS PubMed.
- D. T. Thompson, Using gold nanoparticles for catalysis, Nano Today, 2007, 2, 40–43 CrossRef.
- E. C. Dreaden, A. M. Alkilany, X. Huang, C. J. Murphy and M. A. El-Sayed, The golden age: gold nanoparticles for biomedicine, Chem. Soc. Rev., 2012, 41, 2740–2779 RSC.
- N. Elahi, M. Kamali and M. H. Baghersad, Recent biomedical applications of gold nanoparticles: a review, Talanta, 2018, 184, 537–556 CrossRef CAS PubMed.
- Z. Xi, R. Zhang, F. Kiessling, T. Lammers and R. M. Pallares, Role of Surface Curvature in Gold Nanostar Properties and Applications, ACS Biomater. Sci. Eng., 2024, 10, 38–50 CrossRef CAS.
- M. A. Mackey, M. R. K. Ali, L. A. Austin, R. D. Near and M. A. El-Sayed, The most effective gold nanorod size for plasmonic photothermal therapy: theory and in vitro experiments, J. Phys. Chem. B, 2014, 118, 1319–1326 CrossRef CAS PubMed.
- N. D. Burrows, S. Harvey, F. A. Idesis and C. J. Murphy, Understanding the Seed-Mediated Growth of Gold Nanorods through a Fractional Factorial Design of Experiments, Langmuir, 2017, 33, 1891–1907 CrossRef CAS PubMed.
- Y. Jiang, D. Salley, A. Sharma, G. Keenan, M. Mullin and L. Cronin, An artificial intelligence enabled chemical synthesis robot for exploration and optimization of nanomaterials, Sci. Adv., 2022, 8, eabo2626 CrossRef CAS PubMed.
- T. K. Sau and C. J. Murphy, Room temperature, high-yield synthesis of multiple shapes of gold nanoparticles in aqueous solution, J. Am. Chem. Soc., 2004, 126, 8648–8649 CrossRef CAS.
- C. J. Johnson, E. Dujardin, S. A. Davis, C. J. Murphy and S. Mann, Growth and form of gold nanorods prepared by seed-mediated, surfactant-directed synthesis, J. Mater. Chem., 2002, 12, 1765–1770 RSC.
- M. R. Langille, M. L. Personick, J. Zhang and C. A. Mirkin, Defining rules for the shape evolution of gold nanoparticles, J. Am. Chem. Soc., 2012, 134, 14542–14554 CrossRef CAS.
- M. L. Personick and C. A. Mirkin, Making Sense of the Mayhem behind Shape Control in the Synthesis of Gold Nanoparticles, J. Am. Chem. Soc., 2013, 135, 18238–18247 CrossRef CAS.
- K. Park, L. F. Drummy, R. C. Wadams, H. Koerner, D. Nepal, L. Fabris and R. A. Vaia, Growth Mechanism of Gold Nanorods, Chem. Mater., 2013, 25, 555–563 CrossRef CAS.
- M. J. Walsh, W. Tong, H. Katz-Boon, P. Mulvaney, J. Etheridge and A. M. Funston, A Mechanism for Symmetry Breaking and Shape Control in Single-Crystal Gold Nanorods, Acc. Chem. Res., 2017, 50, 2925–2935 CrossRef CAS.
- Y. Xia, Y. Xiong, B. Lim and S. Skrabalak, Shape-Controlled Synthesis of Metal Nanocrystals: Simple Chemistry Meets Complex Physics?, Angew. Chem., Int. Ed., 2009, 48, 60–103 CrossRef CAS PubMed.
- Y. Xia, X. Xia and H.-C. Peng, Shape-Controlled Synthesis of Colloidal Metal Nanocrystals: Thermodynamic versus Kinetic Products, J. Am. Chem. Soc., 2015, 137, 7947–7966 CrossRef CAS.
- D. V. Talapin, A. L. Rogach, M. Haase and H. Weller, Evolution of an ensemble of nanoparticles in a colloidal solution: Theoretical study, J. Phys. Chem. B, 2001, 105, 12278–12285 CrossRef CAS.
- H. Yang, L. S. Hamachi, I. Rreza, W. Wang and E. M. Chan, Design Rules for One-Step Seeded Growth of Nanocrystals: Threading the Needle between Secondary Nucleation and Ripening, Chem. Mater., 2019, 31, 4173–4183 CrossRef CAS.
- C. A. McCandler, J. C. Dahl and K. A. Persson, Phosphine-Stabilized Hidden Ground States in Gold Clusters Investigated via a Aun(PH3)m Database, ACS Nano, 2023, 17, 1012–1021 CrossRef CAS.
- I. Chakraborty and T. Pradeep, Atomically Precise Clusters of Noble Metals: Emerging Link between Atoms and Nanoparticles, Chem. Rev., 2017, 117, 8208–8271 CrossRef CAS.
- L. M. Liz-Marzán, C. R. Kagan and J. E. Millstone, Reproducibility in Nanocrystal Synthesis? Watch out for Impurities!, ACS Nano, 2020, 14(6), 6359–6361 CrossRef.
- M. Grzelczak, J. Pérez-Juste, P. Mulvaney and L. M. Liz-Marzán, Shape control in gold nanoparticle synthesis, Chem. Soc. Rev., 2008, 37, 1783–1791 RSC.
- B. Nikoobakht and M. A. El-Sayed, Preparation and growth mechanism of gold nanorods (NRs) using seed-mediated growth method, Chem. Mater., 2003, 15, 1957–1962 CrossRef CAS.
- K. Park, M. Ouweleen and R. A. Vaia, Product Metrics for the Manufacturability of Single-Crystal Gold Nanorods via Reaction Engineering, ACS Appl. Mater. Interfaces, 2023, 15, 52827–52842 CAS.
- E. M. Williamson and R. L. Brutchey, Using Data-Driven Learning to Predict and Control the Outcomes of Inorganic Materials Synthesis, Inorg. Chem., 2023, 62, 16251–16262 CrossRef CAS.
- O. Kononova, H. Huo, T. He, Z. Rong, T. Botari, W. Sun, V. Tshitoyan and G. Ceder, Text-mined dataset of inorganic materials synthesis recipes, Sci. Data, 2019, 6(1), 203 CrossRef.
- L. Weston, V. Tshitoyan, J. Dagdelen, O. Kononova, A. Trewartha, K. A. Persson, G. Ceder and A. Jain, Named Entity Recognition and Normalization Applied to Large-Scale Information Extraction from the Materials Science Literature, J. Chem. Inf. Model., 2019, 59, 3692–3702 CrossRef CAS PubMed.
- A. Trewartha, N. Walker, H. Huo, S. Lee, K. Cruse, J. Dagdelen, A. Dunn, K. A. Persson, G. Ceder and A. Jain, Quantifying the advantage of domain-specific pre-training on named entity recognition tasks in materials science, Patterns, 2022, 3(4), 100488 CrossRef PubMed.
- S. A. Malik, R. E. Goodall and A. A. Lee, Predicting the Outcomes of Material Syntheses with Deep Learning, Chem. Mater., 2021, 33, 616–624 CrossRef CAS.
- T. He, W. Sun, H. Huo, O. Kononova, Z. Rong, V. Tshitoyan, T. Botari and G. Ceder, Similarity of Precursors in Solid-State Synthesis as Text-Mined from Scientific Literature, Chem. Mater., 2020, 32, 7861–7873 CrossRef CAS.
- H. Huo, C. J. Bartel, T. He, A. Trewartha, A. Dunn, B. Ouyang, A. Jain and G. Ceder, Machine-Learning Rationalization and Prediction of Solid-State Synthesis Conditions, Chem. Mater., 2022, 34, 7323–7336 CrossRef CAS.
- E. Kim, K. Huang, S. Jegelka and E. Olivetti, Virtual screening of inorganic materials synthesis parameters with deep learning, npj Comput. Mater., 2017, 3(1), 53 CrossRef.
- E. Kim, K. Huang, A. Tomala, S. Matthews, E. Strubell, A. Saunders, A. McCallum and E. Olivetti, Machine-learned and codified synthesis parameters of oxide materials, Sci. Data, 2017, 4, 170127 CrossRef CAS PubMed.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials Synthesis Insights from Scientific Literature via Text Extraction and Machine Learning, Chem. Mater., 2017, 29, 9436–9444 CrossRef CAS.
- K. Cruse, A. Trewartha, S. Lee, Z. Wang, H. Huo, T. He, O. Kononova, A. Jain and G. Ceder, Text-mined dataset of gold nanoparticle synthesis procedures, morphologies, and size entities, Sci. Data, 2022, 9(1), 234 CrossRef.
- T. Brown, et al., Language Models are Few-Shot Learners, Adv. Neural Inf. Process. Syst., 2020, 1877–1901 Search PubMed.
- L. Ouyang and et al., Training Language Models to Follow Instructions With Human Feedback, 2022 Search PubMed.
- OpenAI, GPT-4 Technical Report, 2023 Search PubMed.
- H. Touvron et al. , Llama 2: Open Foundation and Fine-Tuned Chat Models, 2023, https://www.arxiv.org/abs/2307.09288 Search PubMed.
- J. Dagdelen, A. Dunn, S. Lee, N. Walker, A. S. Rosen, G. Ceder, K. Persson and A. Jain, Structured information extraction from scientific text with large language models, Nat. Commun., 2024, 15, 1418 CrossRef CAS PubMed.
- N. Walker, S. Lee, J. Dagdelen, K. Cruse, S. Gleason, A. Dunn, G. Ceder, A. P. Alivisatos, K. A. Persson and A. Jain, Extracting structured seed-mediated gold nanorod growth procedures from scientific text with LLMs, Digital Discovery, 2023, 2(6), 1768–1782 RSC.
- S. J. Yang, S. Li, S. Venugopalan, V. Tshitoyan, M. Aykol, A. Merchant, E. D. Cubuk and G. Cheon, Accurate Prediction of Experimental Band Gaps from Large Language Model-Based Data Extraction, 2023 Search PubMed.
- T. Guo, K. Guo, B. Nan, Z. Liang, Z. Guo, N. V. Chawla, O. Wiest and X. Zhang, What Can Large Language Models Do In Chemistry? A Comprehensive Benchmark On Eight Tasks, 2023 Search PubMed.
- M. Thway, A. K. Y. Low, S. Khetan, H. Dai, J. Recatala-Gomez, A. P. Chen and K. Hippalgaonkar, Harnessing GPT-3.5 for text parsing in solid-state synthesis – case study of ternary chalcogenides, Digital Discovery, 2024, 3, 328–336 RSC.
- E. J. Hu, Y. Shen, P. Wallis, Z. Allen-Zhu, Y. Li, S. Wang, L. Wang and W. Chen, LoRA: Low-Rank Adaptation of Large Language Models, 2021, https://www.arxiv.org/abs/2106.09685 Search PubMed.
- B. Townsend, E. Ito-Fisher, L. Zhang and M. May, Doc2Dict: Information Extraction as Text Generation, 2021 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, Others Scikit-learn: Machine learning in Python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
- J. Zhang, M. R. Langille, M. L. Personick, K. Zhang, S. Li and C. A. Mirkin, Concave Cubic Gold Nanocrystals with High-Index Facets, J. Am. Chem. Soc., 2010, 132, 14012–14014 CrossRef CAS.
- P. N. Sisco and C. J. Murphy, Surface-Coverage Dependence of Surface-Enhanced Raman Scattering from Gold Nanocubes on Self-Assembled Monolayers of Analyte, J. Phys. Chem. A, 2009, 113, 3973–3978 CrossRef CAS.
- M. Liu and P. Guyot-Sionnest, Mechanism of silver(I)-assisted growth of gold nanorods and bipyramids, J. Phys. Chem. B, 2005, 109, 22192–22200 CrossRef CAS PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, A study of the nucleation and growth processes in the synthesis of colloidal gold, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- L. Roach, P. L. Coletta, K. Critchley and S. D. Evans, Controlling the Optical Properties of Gold Nanorods in One-Pot Syntheses, J. Phys. Chem. C, 2022, 126, 3235–3243 CrossRef CAS.
- P. S. Kumar, I. Pastoriza-Santos, B. Rodríguez-González, F. J. G. de Abajo and L. M. Liz-Marzán, High-yield synthesis and optical response of gold nanostars, Nanotechnology, 2007, 19, 015606 CrossRef.
- N. R. Jana, L. Gearheart and C. J. Murphy, Wet chemical synthesis of high aspect ratio cylindrical gold nanorods, J. Phys. Chem. B, 2001, 105(19), 4065–4067 CrossRef CAS.
- S. K. Meena and M. Sulpizi, Understanding the Microscopic Origin of Gold Nanoparticle Anisotropic Growth from Molecular Dynamics Simulations, Langmuir, 2013, 29, 14954–14961 CrossRef CAS.
- S. E. Lohse and C. J. Murphy, The quest for shape control: a history of gold nanorod synthesis, Chem. Mater., 2013, 25(8), 1250–1261 CrossRef CAS.
- S. Zhao, X. Zhu, C. Cao, J. Sun and J. Liu, Transferrin modified ruthenium nanoparticles with good biocompatibility for photothermal tumor therapy, J. Colloid Interface Sci., 2018, 511, 325–334 CrossRef CAS.
- R.-D. Jean, W.-D. Cheng, M.-H. Hsiao, F.-H. Chou, J.-S. Bow and D.-M. Liu, Highly electrostatically-induced detection selectivity and sensitivity for a colloidal biosensor made of chitosan nanoparticle decorated with a few bare-surfaced gold nanorods, Biosens. Bioelectron., 2014, 52, 111–117 CrossRef CAS.
- L. Scarabelli, A. Sánchez-Iglesias, J. Pérez-Juste and L. M. Liz-Marzán, A “Tips and Tricks” Practical Guide to the Synthesis of Gold Nanorods, J. Phys. Chem. Lett., 2015, 6, 4270–4279 CrossRef CAS PubMed.
- S. Fateixa, M. R. Correia and T. Trindade, Resizing of Colloidal Gold Nanorods and Morphological Probing by SERS, J. Phys. Chem. C, 2013, 117, 20343–20350 CrossRef CAS.
- Z. Yang, Y.-W. Lin, W.-L. Tseng and H.-T. Chang, Impacts that pH and metal ion concentration have on the synthesis of bimetallic and trimetallic nanorods from gold seeds, J. Mater. Chem., 2005, 15, 2450–2454 RSC.
- D. Fernando, T. A. Nigro, I. Dyer, S. M. Alia, B. S. Pivovar and Y. Vasquez, Synthesis and catalytic activity of the metastable phase of gold phosphide, J. Solid State Chem., 2016, 242, 182–192 CrossRef CAS.
- C. Zhang and J. Y. Lee, Synthesis of Au Nanorod@Amine-Modified Silica@Rare-Earth Fluoride Nanodisk Core–Shell–Shell Heteronanostructures, J. Phys. Chem. C, 2013, 117, 15253–15259 CrossRef CAS.
- K. Kang, H. Jang and Y.-K. Kim, The influence of polydopamine coating on gold nanorods for laser desorption/ionization time-of-flight mass spectrometric analysis, Analyst, 2017, 142, 2372–2377 RSC.
- M. Ukaegbu, N. Enwerem, O. Bakare, V. Sam, W. Southerland, A. Vivoni and C. Hosten, Probing the adsorption and orientation of 2,3-dichloro-5,8-dimethoxy-1,4-naphthoquinone on gold nano-rods: a SERS and XPS study, J. Mol. Struct., 2016, 1114, 197–205 CrossRef CAS.
- J. Zhang, Y. Feng, J. Mi, Y. Shen, Z. Tu and L. Liu, Photothermal lysis of pathogenic bacteria by platinum nanodots decorated gold nanorods under near infrared irradiation, J. Hazard. Mater., 2018, 342, 121–130 CrossRef CAS.
- Q. Shou, M. Ebara, J. Wang, Q. Wang, X. Liang, H. Liu and T. Aoyagi, Preparation of phase diagram of gold nanorods in mixture solvent of DMSO and water and its application for efficient surface-modification, Appl. Surf. Sci., 2018, 457, 264–270 CrossRef CAS.
- G. Chandrasekar, K. Mougin, H. Haidara, L. Vidal and E. Gnecco, Shape and size transformation of gold nanorods (GNRs) via oxidation process: a reverse growth mechanism, Appl. Surf. Sci., 2011, 257, 4175–4179 CrossRef CAS.
- H.-Q. Chen, F. Yuan, S.-Z. Wang, J. Xu, Y.-Y. Zhang and L. Wang, Near-infrared to near-infrared upconverting NaYF4: Yb3+, Tm3+ nanoparticles-aptamer-Au nanorods light resonance energy transfer system for the detection of mercuric(ii) ions in solution, Analyst, 2013, 138, 2392–2397 RSC.
- K. Khaletskaya, J. Reboul, M. Meilikhov, M. Nakahama, S. Diring, M. Tsujimoto, S. Isoda, F. Kim, K.-i. Kamei, R. A. Fischer, S. Kitagawa and S. Furukawa, Integration of Porous Coordination Polymers and Gold Nanorods into Core–Shell Mesoscopic Composites toward Light-Induced Molecular Release, J. Am. Chem. Soc., 2013, 135, 10998–11005 CrossRef CAS PubMed.
- H. J. Parab, H. M. Chen, T.-C. Lai, J. H. Huang, P. H. Chen, R.-S. Liu, M. Hsiao, C.-H. Chen, D.-P. Tsai and Y.-K. Hwu, Biosensing, Cytotoxicity, and Cellular Uptake Studies of Surface-Modified Gold Nanorods, J. Phys. Chem. C, 2009, 113, 7574–7578 CrossRef CAS.
- J.-H. Park, J.-Y. Byun, W.-B. Shim, S. U. Kim and M.-G. Kim, High-sensitivity detection of ATP using a localized surface plasmon resonance (LSPR) sensor and split aptamers, Biosens. Bioelectron., 2015, 73, 26–31 CrossRef CAS.
- S. Chung, H. Lee, H.-S. Kim, M.-G. Kim, L. P. Lee and J. Y. Lee, Transdermal thiol–acrylate polyethylene glycol hydrogel synthesis using near infrared light, Nanoscale, 2016, 8, 14213–14221 RSC.
- S. Lee, https://www.github.com/slee-lab/AuNP-seedmed-recipes, 2024, DOI:10.5281/zenodo.13947755.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00158c |
| This journal is © The Royal Society of Chemistry 2025 |