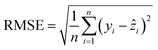Insights into ionic liquid-enhanced membrane protein stability through machine learning and molecular simulations
Ju
Liu
a,
Guiming
Zhang
bc,
Cheng
Song
d,
Yanlei
Wang
 *b,
Jing
Ren
*e and
Hongyan
He
*b,
Jing
Ren
*e and
Hongyan
He
 a
a
aCenter of Ionic Liquids and Green Energy, Beijing Key Laboratory of Solid State Battery and Energy Storage Process, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China
bSchool of Chemistry and Life Resources, Renmin University of China, Beijing, 100872, China. E-mail: ylwang17@ruc.edu.cn
cCollege of Chemistry and Life Science, Beijing University of Technology, 100124, China
dSchool of Population and Health, Renmin University of China, Beijing, 100872, China
eDepartment of Plastic and Reconstructive Surgery, the First Medical Center, Chinese PLA General Hospital, Beijing 100853, China. E-mail: eggy119@163.com
First published on 7th October 2025
Abstract
Protein stability plays a critical role in structural elucidation, enzyme activity, and the storage of protein drugs, where ionic liquids (ILs) have emerged as promising protein stabilizers due to their exceptional biocompatibility and superior solubility. However, the underlying mechanisms by which ILs modulate protein stability, particularly through the regulation of hydrogen bonding and interfacial structures, remain inadequately understood. Herein, a machine learning-based framework, integrating molecular docking, unsupervised learning, molecular dynamics simulations and correlation analysis, is applied to clarify the mechanism of ILs enhancing membrane protein stability. It is found that ILs form clusters that are adsorbed on the protein surface, with ILs entering the hydration layer of the protein and forming intermolecular hydrogen bonds with the protein surface, thereby improving stability, consistent with experiments. Furthermore, a predictive model for protein stability is established by supervised learning and verification of the mechanism through interpretability analysis. Our framework quantitatively reveals the influence of hydrogen bonds and interface structures on membrane protein stability. Overall, these quantitative results not only deepen our understanding of the interactions between ILs and protein but also shed light on the rational design of protein stabilizers.
1. Introduction
The structural analysis of membrane proteins remains a pivotal challenge in the life sciences.1–3 Although advanced prediction tools like AlphaFold have achieved remarkable accuracy in determining the three-dimensional structure of proteins,4,5 experimental validation is still needed to establish the structure–function relationships.6 However, a major obstacle is that membrane proteins tend to lose their native conformation during extraction and purification,7 which necessitates optimized methods to preserve their stability. Ionic liquids (ILs) have emerged as effective agents in the extraction and purification of membrane proteins,8–11 owing to their excellent solubility, biocompatibility, and thermal stability.12,13 ILs not only enhance extraction efficiency and increase protein yields but also act as stabilizers.12,14 For instance, Brogan et al. demonstrated via circular dichroism spectroscopy that the half-denaturation temperature of myoglobin increased by 55 °C in pyrrole-based ILs, even surpassing water's boiling point.15 Similarly, Banerjee et al. found that insulin maintained its secondary structure in the presence of choline and ILs at 4 °C for up to 4 months.16 These results highlight the significant role of ILs in the field of protein stabilization.Understanding the stabilization mechanism of proteins by ILs involves investigating two major factors: hydrogen bonding and protein–IL interface structure (Fig. 1). Some studies, such as those by Phillips et al., demonstrated using wide-angle X-ray scattering that imidazolium ILs primarily disrupt internal hydrogen bonds within the protein, thereby reducing the structural stability of proteins.17 In contrast, Ghanta et al. showed through molecular dynamics (MD) simulations that ILs predominantly break the hydrogen bonds between the protein and water, while forming new hydrogen bonds with the protein surface to enhance its stability.18 Moreover, while certain studies using UV-visible absorption spectroscopy and far-UV circular dichroism suggested that ILs mainly affect the hydration layer of proteins by replacing water molecules, thereby enhancing protein stability,19–22 other investigations proposed that the aggregation of cations into ionic clusters encapsulates the protein surface, thereby reducing structural fluctuations.8,9,23,24 The interplay between the internal and interfacial hydrogen bonds, along with the various protein–IL interfacial interactions, critically determines overall protein stability. As those interactions can enhance or disrupt structural integrity, developing a unified quantitative framework to explain IL-mediated stabilization remains a significant challenge.
In this study, the membrane was not included in the MD simulations, as our focus was on the storage process of the protein after extraction from the membrane. Nevertheless, our previous studies demonstrated that ionic liquids preferentially insert at the protein–lipid interface while preserving protein structural stability.9,22 Building on these results, we develop a machine learning (ML)-based framework to quantitatively elucidate how ILs enhance membrane protein stability (Fig. 1). Our approach encompasses five major steps: (i) dataset generation: we compile a dataset of 317 different anions paired with imidazolium cations and perform molecular docking simulations with aquaporin-2 (AQP2) and sodium channel protein (Nav) to calculate binding energies between anions and protein (EB). (ii) Classification: an unsupervised learning model is used to classify the anions into nine clusters, integrating their structural features with the EB data to construct a ML model. (iii) Screening of IL candidates: we sample and evaluate ILs using more rigorous MD simulations based on EB and clusters. In contrast to every sample investigated, we screenedby focusing on anions with the highest EB and representative candidates from each cluster. (iv) Mechanistic analysis: key descriptors related to hydrogen bonding and interface structure are extracted from the trajectories of MD simulations, and correlation analysis is performed to identify the key factors influencing protein stability. (v) Model verification: finally, we verify the proposed mechanism by constructing a supervised learning model utilizing an extra trees regressor, with interpretability provided by the Shapley additive explanation (SHAP) analysis.
2. Results and discussion
2.1. Dataset for ionic liquids and binding energies with protein
Initially, we compiled a dataset of 317 anions, including their SMILES codes and structural files (Fig. S1), obtained from the ionic liquid database.25 We then selected aquaporin 2 (AQP2) and sodium channel protein (Nav) as our model proteins because they play crucial roles in cellular water and ion transport (Fig. S2), making their stability essential for both physiological functions and biomedical research. Subsequently, we performed molecular docking studies of these anions with the target proteins AQP2 and Nav to obtain binding energies (EB) and identify the corresponding binding sites,26 thereby establishing a comprehensive dataset (Fig. 2a and Fig. S3). This dataset includes the SMILES codes of the anions, their respective PDB structural files, EB, and the corresponding binding sites. Anions that exhibit stronger binding energies to AQP2 also display stronger binding energies to Nav. Accordingly, we selected AQP2 for further analysis. Given that the EB is closely related to the specific binding site,27 an analysis of these sites (Fig. 2b) revealed four distinct regions: the outside cavity, channel center, inside cavity, and hydrophobic surface. Notably, the outside and inside cavities of AQP2 are characterized by predominantly carrying negative and positive charges, respectively.28 Among these, the inside cavity accounts for 73.7% of all binding sites (Fig. 2c), which can be attributed to the fact that under electrostatic interactions, anions tend to bind to the positively charged inside cavity of AQP2.We next performed hierarchical clustering analysis on the 317 anions using the Ward algorithm, resulting in 9 distinct clusters containing 19, 19, 34, 21, 38, 13, 68, 18, and 87 anions, respectively (Fig. S1). Notably, anions grouped within the same cluster exhibit similar structural motifs, indicating a rational basis for this classification. When correlating these structural features with their corresponding EB, we found that anions within the same cluster exhibit comparable EB values, indicating that we have established a robust structure–activity model that validates the rationality of our classification.
To further refine the structure–activity model of the anions, we analyzed both the functional groups of the anions and the distribution of their EB values (Fig. 2d). Anions in clusters I to III all contain carboxyl groups but differ in their overall composition.
Specifically, cluster I anions are primarily monocarboxylic compounds, exhibiting EB values ranging from −3 to −5 kcal mol−1. In contrast, cluster III anions not only contain carboxyl groups but also incorporate other functional groups such as ether, amine, and hydroxyl groups. Particularly, cluster VII anions generally exhibit higher EB, ranging from −4 to −10 kcal mol−1. This cluster mainly comprises fluorinated anions with sulfonic acid groups, which can form hydrogen bonds with the protein (Fig. S4), suggesting the significant role of hydrogen bonding in these interactions.7 Additionally, cluster IX anions, characterized by a wide structural diversity that includes sulfonate groups, imidazole rings, azides, halogens, etc., exhibit a broad distribution of EB values spanning from −1 to −8 kcal mol−1.
2.2. High-throughput screening through molecular dynamics simulations
To investigate the effects of ILs on protein stability through MD simulations, we selected imidazolium cations with varying alkyl chain lengths [Cnmim]+ (n = 4–14)13,29 and chloride as the counter ion due to its high water solubility.8 As shown in Fig. S5, with the increase in alkyl chain length, the adsorption state of ILs on the protein surface evolves from disordered adsorption to membrane-like adsorption, while the clustering structures in solution gradually transition from free ions to large clusters. Protein stability was characterized by root-mean-square deviation (RMSD) and α-helix content; as shown in Fig. 3a and b, the two-dimensional plot of α-helices and RMSD shifts from the center to the bottom-left corner, before returning to the center as the side chain of the cation elongates. This trend suggests an initial enhancement in stability, followed by a decline, peaking at [C6mim]+. Consequently, [C6mim]+ was chosen for further investigation.Subsequently, representative ILs were selected for MD simulations based on EB and classification. The criteria involved selecting the anion with the highest EB across all structures, followed by gradient selection of anions with varying EB within the same cluster, and choosing anions with identical EB from different clusters. Firstly, we chose the top 7 anions with the highest EB (Fig. 3, named T1 to T7), all exceeding −6.9 kcal mol−1. These anions typically have large molecular masses and contain fluorine and oxygen groups, and were paired with [C6mim]+. As shown in Fig. 3c, the ILs with the highest EB all enhanced protein stability, with RMSD values of 1.53, 1.95, and 2.15 Å for T4, T3, and control groups, respectively. Simulation snapshots reveal that these ILs form large clusters and adsorb on the protein (Fig. S6). Additionally, due to the hydrophilic sulfonic acid groups and hydrophobic fluorinated alkyl chains present in T2, T4, and T7 anions (Fig. 3d), they resemble phospholipid structures,30 promoting the formation of an ordered membrane-like structure around the protein and further stabilizing it.
To assess the influence of anions from the same cluster with different EB, 7 anions with different EB from cluster IX (E1 to E7) were selected, as shown in Fig. S7. The EB values for E1, E5, and E7 were −2.7, −5.3, and −7.4 kcal mol−1, respectively, with the corresponding protein RMSD values of 1.71, 1.59, and 1.58 Å (Fig. S8), indicating that the effect of anions from the same cluster on protein stability is not linearly correlated with the change of EB. In systems containing E1 to E3, ILs limited adsorption on the protein surface (Fig. S9), resulting in minimal stabilization effects. However, ILs in systems with E4 to E7 form clusters around the protein, leading to marked improvements in stability. Therefore, anions with higher EB within the same cluster significantly enhance protein stability.
Since E5 with a binding energy of 5.3 kcal mol−1 demonstrated the most significant enhancement in protein stability, we further selected anions with EB values around 5.3 kcal mol−1 from different clusters (CI to CIX), as shown in Fig. 3d. Among these, anions from clusters CVIII, CV, and CIV exhibited the lowest RMSD values of 1.48, 1.48, and 1.52 Å, respectively. These ILs form clusters adsorbed on the protein surface (Fig. S10), contributing to enhanced stability. To further evaluate their performance, the five screened ILs with the lowest RMSD were compared to three conventional protein stabilizers: polysorbate 20 (Tw20),31 polyethylene glycol (PEG),32 and Nonidet P-40 (NP-40).12 As shown in Fig. 3c and Fig. S11 and S12, the screened ILs outperformed commercial stabilizers, reducing RMSD by 12–18%. We further evaluated structural stability using root mean square fluctuation (RMSF) and solvent accessible surface area (SASA) of the hydrophobic core. As shown in Fig. S13a, the CVIII system shows a markedly reduced RMSF relative to the control, with fluctuations ranging from 10 to 40 Å for the control and 0 to 10 Å for CVIII. Consistently, the hydrophobic core is less exposed in CVIII, with a SASA of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 953 Å2versus 24
953 Å2versus 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 005 Å2 for the control (Fig. S13b). These metrics corroborate that the candidate ILs enhance the structural stability of the membrane protein. Additionally, these ILs exhibited low synthetic accessibility score33 (SAscore) (Fig. S14), with the Cv anion scoring 2.19, indicating favorable synthetic feasibility. Consequently, these ILs are potential protein stabilizers.
005 Å2 for the control (Fig. S13b). These metrics corroborate that the candidate ILs enhance the structural stability of the membrane protein. Additionally, these ILs exhibited low synthetic accessibility score33 (SAscore) (Fig. S14), with the Cv anion scoring 2.19, indicating favorable synthetic feasibility. Consequently, these ILs are potential protein stabilizers.
2.3. Machine learning revealing the mechanism of ionic liquids stabilizing membrane proteins
To elucidate the mechanism by which ILs enhance protein stability, two categories of descriptors were extracted from the simulation results: hydrogen bond and interface structure. As shown in Fig. 4a, hydrogen bond descriptors include the number of internal hydrogen bonds within the protein (HBpro), the number of hydrogen bonds between the protein surface and ILs (HBpro-IL), and the number of hydrogen bonds between the protein and water (HBpro-wat). Interface structure descriptors include the number of water molecules in the first and second hydration layers (CI-wat, CII-wat), anions (CI-an, CII-an), cations (CI-ca, CII-ca) near the protein surface, as well as the overall radius of gyration of the ILs (RIL). Through correlation analysis, the Pearson correlation coefficients (r) between these descriptors were calculated to quantify their influence on protein stability (Fig. 4b). The sign of r indicates the direction of correlation: a positive sign for a positive correlation and a negative sign for an inverse correlation.As shown in Fig. 4b and c, HBpro-IL exhibits a moderate negative correlation with RMSD (r = −0.39), indicating that an increased hydrogen bonds between the protein surface and ILs is associated with enhanced structural stability. This aligns with previous experiments.34,35 However, HBpro has a negligible correlation (r = −0.05), indicating that the ILs have limited influence on the internal hydrogen bonds of the protein and mainly affects surface interactions, consistent with reports that the structure of membrane proteins is stable and that ILs have difficulty disrupting their internal interactions.22 On the other hand, for the interface structure, the CI-wat, CII-wat, CI-an, and CII-an have moderate correlations with RMSD, with all |r| values exceeding 0.36, indicating the importance of the hydration layer in protein stability. Notably, CII-an is strongly negatively correlated with CII-wat (r = −0.94), suggesting that IL anions can stabilize proteins by replacing water in the hydration layer. Moreover, the correlation between CII-ca and RMSD (|r| = 0.27) is weaker than that of CII-an (|r| = 0.39), indicating that anions play a more significant role in stabilizing proteins, consistent with previous studies.36 Additionally, RIL shows a moderate positive correlation with RMSD (Fig. 4a) (r = 0.31), implying that more compact IL structures are associated with greater protein stability.24,25
In addition, to comprehensively analyze the impact of hydrogen bonds and interface structures on protein stability, canonical correlation analysis was performed. The strength of correlations between the two descriptor sets was evaluated using the first canonical correlation coefficient (rc) in Fig. 4d, which ranges from 0 (no linear relationship) to 1 (perfect linear relationship). The hydrogen bond set (HBcom) and the interface structure set (Scom) exhibit moderate correlations with RMSD, with rc = 0.44 and 0.41, respectively, indicating that both contribute to protein stability, aligning with previous findings.7 Additionally, a strong correlation was observed between HBcom and Scom, with rc = 0.88, further emphasizing the close relationship between hydrogen bonds and interface structures. Based on the above analysis, the results suggest that ILs enhance protein stability by forming ionic clusters adsorbed onto the protein surface, penetrating the hydration layer, and establishing intermolecular hydrogen bonds with the surface. Moreover, the role of anions is more pronounced than that of cations.
To validate the proposed mechanism, supervised learning was employed to predict the RMSD of protein using the extra trees regressor. Descriptors selected from MD simulations and correlation analysis were used as input features. The model was optimized to minimize the root-mean-squared error (RMSE) between predicted and actual RMSD values. A Pearson correlation coefficient of 0.913 was achieved on the test set (Fig. 4e and Fig. S15), indicating high predictive accuracy and supporting the relevance of the selected descriptors to protein stability. SHAP analysis indicated that the top four most influential features are RIL, CII-wat, HBpro-IL and CII-an (Fig. 4f). Particularly, decreased RIL and CII-wat, along with increased HBpro-IL and CII-an, were associated with enhanced predicted stability. These results provide strong support for the proposed mechanism.
This study systematically elucidates the mechanisms by which ILs influence protein stability. A notable strength lies in the broad selection of ILs with diverse chemical properties, enhancing the generalizability of the findings. Building on prior work, we integrated multiple stability-related factors and applied statistical analyses to quantitatively evaluate their contributions. The proposed mechanisms are consistent with experimental observations. However, as the analysis focused on membrane proteins with inherently strong internal interactions, future studies should extend to other protein classes, such as globular and fibrous proteins, to establish a more comprehensive understanding of IL-induced stabilization effects.
3. Conclusions
In conclusion, we developed a machine learning-integrated framework combining molecular docking, unsupervised learning, MD simulations, and correlation analysis to elucidate how ILs enhance membrane protein stability. This framework was successfully applied to elucidate the mechanisms by which ILs enhance membrane protein stability, focusing on hydrogen bonds and interface structures. Initially, we employed molecular docking and unsupervised learning to develop a structure–activity model that correlates IL structures with EB. The screened ILs showed a 12–18% improvement in performance compared to conventional stabilizers. Next, correlation analysis reveals the mechanism that ILs form ionic clusters that are adsorbed on the protein surface, with ILs penetrating into the protein hydration layer. These ILs form intermolecular hydrogen bonds with the protein surface, thereby stabilizing the protein. Finally, the model predicts the RMSD of the protein using an extra trees regressor, and SHAP values were utilized to identify the key explanatory variables influencing RMSD, which verified the mechanism. This work reveals the mechanism by which ILs improve protein stability, which is of great significance for understanding the interaction between ILs and proteins and developing new membrane protein extraction and storage systems.4. Models and methods
4.1. Molecular docking
The molecular docking between aquaporin-2 (AQP2), sodium channel protein (Nav) and ILs was conducted using the AutoDock Vina program.37 The crystal structure of AQP2 (PDB: 4nef28) and Nav (PDB: 5vb8) were utilized, and the protein and ILs’ input files were prepared by merging non-polar hydrogen atoms and adding charges. The target protein remained fixed. The docking results were visualized using PyMOL, while the protein–ligand interaction was calculated using the protein–ligand interaction profiler web service.384.2. Unsupervised learning
The dendrogram function in the SciPy package is used to implement agglomerative hierarchical clustering (AHC).39 In this method, each sample begins as its own individual cluster and progressively merges with others based on similarity as it moves up the hierarchy. The result of AHC is a bottom-up hierarchical tree diagram. In this diagram, the Euclidean distance between branches serves as the measure of similarity, while the Ward algorithm is employed to calculate the dissimilarity between nodes.404.3. Molecular dynamics simulations
The IL–AQP2 system was constructed as shown in Fig. 3c, [Cnmim]+ (n = 4–14) cations were orderly arranged around AQP2,41 and anions were randomly placed using Packmol.42 Water molecules surrounded the protein and ILs. The simulation box dimensions were set to 13.6 × 13.6 × 13.6 nm3. Periodic boundary conditions were applied in all three directions. The MD simulations were performed using Amber22 software.43 The time step was 2 fs. The protein was described using the protein.ff14SB force field, while the second-generation General AMBER Force Field (GAFF2) was employed to describe the ILs,44,45 which has been shown to reliably reproduce their structural and dynamical behaviors.8,46–48 The TIP3P force field was used for water molecules.46 Non-bonded interactions included electrostatic and van der Waals terms. The former was computed using the particle-mesh Ewald algorithm,47,48 while the latter was described using the Lennard-Jones potential with a cutoff distance of 1.0 nm. The SHAKE algorithm was employed to constrain high-frequency vibrations of hydrogen atoms.49 Temperature and pressure were controlled using the Berendsen thermostat50 and the Berendsen barostat,51 respectively, with a coupling constant of 1.0 ps. The system was first equilibrated for 10 ns under the NPT ensemble at 310 K and 1 bar, followed by 300 ns of equilibration under the NVT ensemble at 310 K. The last 50 ns trajectory was used to analyze the impact of ILs on protein stability.4.4. Correlation analysis and the supervised learning model
Correlation analysis was performed using the Statistical Package for the Social Sciences (SPSS)52 to correlate the number of internal hydrogen bonds within the protein (HBpro), the number of hydrogen bonds between the protein surface and ILs (HBpro-IL), the number of hydrogen bonds between the protein and water (HBpro-wat), and the number of water molecules in the first and second hydration layers of the protein surface (CI-wat and CII-wat), the number of anions in the first and second hydration layers of the protein surface (CI-an and CII-an), the number of cations in the first and second hydration layers of the protein surface (CI-ca and CII-ca), and the overall radius of gyration of the ILs (RIL) for the root mean square deviation (RMSD) of protein. The above descriptors are extracted from the last 50 ns trajectory of each MD to build a dataset.where di is the Euclidean distance between atom i in the two structures and N is the total number of atoms.
The model training was conducted using scikit-learn.53 The dataset was randomly divided into two groups: 80% for model training and 20% for model testing. The extra trees regressor was used, the maximum depth of the trees is unrestricted, and each leaf node must contain at least one sample. The ensemble model consists of 100 trees, and its performance is measured using the root mean square error (RMSE) calculated by:
Author contributions
Yanlei Wang and Jing Ren conceived and led the project. Ju Liu conducted the simulations and drafted the manuscript. Ju Liu and Guiming Zhang conducted the research. Cheng Song, Yanlei Wang, and Hongyan He contributed to this work by analyzing the data and revising the manuscript. Yanlei Wang and Hongyan He supervised the project. All authors discussed the results and commented on the manuscript.Conflicts of interest
The authors declare no competing financial interests.Data availability
The data supporting this article have been included as part of the supplementary information (SI). Supplementary information: the relevant data on the enhancement of protein stability by ILs. See DOI: https://doi.org/10.1039/d5cp02130h.Acknowledgements
We are thankful to the Beijing Natural Science Foundation (2252011), the National Natural Science Foundation of China (22522817 and 22508386), U35 (New Teacher Start up Fund) Project of Renmin University of China (25XNKJ13), the Beijing Nova Program (20230484478) and the China Postdoctoral Science Foundation (GZC20250782 and 2025M77150) for financial support. This research was supported by Public Computing Cloud and Big Data and Responsible Artificial Intelligence for National Governance, Renmin University of China. The authors sincerely appreciate Prof. Suojiang Zhang (IPE, CAS) for his careful academic guidance and great support.References
- Y. Lu, C.-X. Yue, L. Zhang, D. Yao, Y. Xia, Q. Zhang, X. Zhang, S. Li, Y. Shen and M. Cao, et al., Structural basis for inositol pyrophosphate gating of the phosphate channel XPR1, Science, 2024, 386(6723), eadp3252 CrossRef CAS PubMed.
- C. Shen, S. Chang, Q. Luo, K. C. Chan, Z. Zhang, B. Luo, T. Xie, G. Lu, X. Zhu and X. Wei, et al., Structural basis of BAM-mediated outer membrane β-barrel protein assembly, Nature, 2023, 617(7959), 185–193 CrossRef CAS PubMed.
- R. Sexton, M. Fazel, M. Schweiger, S. Pressé and O. Beckstein, Bayesian nonparametric analysis of residence times for protein–lipid interactions in molecular dynamics simulations, J. Chem. Theory Comput., 2025, 21(8), 4203–4220 CrossRef CAS PubMed.
- J. Abramson, J. Adler, J. Dunger, R. Evans, T. Green, A. Pritzel, O. Ronneberger, L. Willmore, A. J. Ballard and J. Bambrick, et al., Accurate structure prediction of biomolecular interactions with AlphaFold 3, Nature, 2024, 630(8016), 493–500 CrossRef CAS PubMed.
- D. Coskun, M. Lihan, J. P. G. L. M. Rodrigues, M. Vass, D. Robinson, R. A. Friesner and E. B. Miller, Using AlphaFold and experimental structures for the prediction of the structure and binding affinities of gpcr complexes via induced fit docking and free energy perturbation, J. Chem. Theory Comput., 2024, 20(1), 477–489 CrossRef CAS PubMed.
- H. Liu, P. Yan, Z. Zhang, H. Han, Q. Zhou, J. Zheng, J. Zhang, F. Xu and W. Shui, Structural mass spectrometry captures residue-resolved comprehensive conformational rearrangements of a G protein-coupled receptor, J. Am. Chem. Soc., 2024, 146(29), 20045–20058 CrossRef CAS PubMed.
- P. Bharmoria, A. A. Tietze, D. Mondal, T. S. Kang, A. Kumar and M. G. Freire, Do ionic liquids exhibit the required characteristics to dissolve, extract, stabilize, and purify proteins? Past-present-future assessment, Chem. Rev., 2024, 124(6), 3037–3084 CrossRef CAS PubMed.
- J. Liu, Y. Wang, C. Wang, J. Gao, W. Cui, B. Zhao, L. Zhang, H. He and S. Zhang, Thermodynamical origin of nonmonotonic inserting behavior of imidazole ionic liquids into the lipid bilayer, J. Phys. Chem. Lett., 2021, 12(40), 9926–9932 CrossRef CAS PubMed.
- J. Liu, J. Ren, S. Li, H. He and Y. Wang, Protein interface regulating the inserting process of imidazole ionic liquids into the cell membrane, J. Phys. Chem. B, 2024, 128(18), 4456–4463 CrossRef CAS PubMed.
- Q. Zhao, H. Chu, B. Zhao, Z. Liang, L. Zhang and Y. Zhang, Advances of ionic liquids-based methods for protein analysis, Trac-Trends Anal. Chem., 2018, 108, 239–246 CrossRef CAS.
- E. Suarez Garcia, C. F. Miranda, M. T. Cesario, R. H. Wijffels, C. van den Berg and M. H. M. Eppink, Ionic liquid-assisted selective extraction and partitioning of biomolecules from macroalgae, ACS Sustainable Chem. Eng., 2023, 11(5), 1752–1762 CrossRef CAS PubMed.
- H. Chu, Q. Zhao, J. Liu, K. Yang, Y. Wang, J. Liu, K. Zhang, B. Zhao, H. He and Y. Zheng, et al., Ionic liquid-based extraction system for in-depth analysis of membrane protein complexes, Anal. Chem., 2022, 94(2), 758–767 CrossRef CAS PubMed.
- Q. Zhao, F. Fang, Y. Liang, H. Yuan, K. Yang, Q. Wu, Z. Liang, L. Zhang and Y. Zhang, 1-dodecyl-3-methylimidazolium chloride-assisted sample preparation method for efficient integral membrane proteome analysis, Anal. Chem., 2014, 86(15), 7544–7550 CrossRef CAS PubMed.
- M. Nuruzzaman, B. M. Colella, C. P. Uzoewulu, A. E. Meo, E. J. Gross, S. Ishizawa, S. Sana, H. Zhang, M. E. Hoff, B. T. W. Medlock, E. C. Joyner, S. Sato, E. A. Ison, Z. Li and J. Ohata, Hexafluoroisopropanol as a bioconjugation medium of ultrafast, tryptophan-selective catalysis, J. Am. Chem. Soc., 2024, 146(10), 6773–6783 CrossRef CAS PubMed.
- A. P. Brogan and J. P. Hallett, Solubilizing and stabilizing proteins in anhydrous ionic liquids through formation of protein-polymer surfactant nanoconstructs, J. Am. Chem. Soc., 2016, 138(13), 4494–4501 CrossRef CAS PubMed.
- A. Banerjee, K. Ibsen, T. Brown, R. Chen, C. Agatemor and S. Mitragotri, Ionic liquids for oral insulin delivery, Proc. Natl. Acad. Sci. U. S. A., 2018, 115(28), 7296–7301 CrossRef CAS PubMed.
- D. M. Phillips, L. F. Drummy, D. G. Conrady, D. M. Fox, R. R. Naik, M. O. Stone, P. C. Trulove, H. C. De Long and R. A. Mantz, Dissolution and regeneration of bombyx mori silk fibroin using ionic liquids, J. Am. Chem. Soc., 2004, 126(44), 14350–14351 CrossRef CAS PubMed.
- K. P. Ghanta, T. Pal, S. Mondal and S. Bandyopadhyay, Microscopic understanding of the effect of ionic liquid on protein from molecular simulation studies, J. Phys. Chem. B, 2020, 124(19), 3909–3921 CrossRef CAS PubMed.
- A. Sanchez-Fernandez, M. Basic, J. Xiang, S. Prevost, A. J. Jackson and C. Dicko, Hydration in deep eutectic solvents induces non-monotonic changes in the conformation and stability of proteins, J. Am. Chem. Soc., 2022, 144(51), 23657–23667 CrossRef CAS PubMed.
- H. Cui, L. Zhang, C. B. Yildiz, L. Eltoukhy, L. Cheng, K.-E. Jaeger, U. Schwaneberg and M. D. Davari, Enzyme hydration: How to retain resistance in ionic liquids, ACS Sustainable Chem. Eng., 2022, 10(46), 15104–15114 CrossRef CAS.
- S. Sahoo, T. Pal, S. Mondal, K. P. Ghanta and S. Bandyopadhyay, Conformational properties of Aβ peptide oligomers in aqueous ionic liquid solution: Insights from molecular simulation studies, J. Phys. Chem. B, 2023, 127(51), 10960–10973 CrossRef CAS PubMed.
- J. Liu, Y. Wang, B. Gao, K. Zhang, H. Li, J. Ren, F. Huo, B. Zhao, L. Zhang and S. Zhang, et al., Ionic liquid gating induces anomalous permeation through membrane channel proteins, J. Am. Chem. Soc., 2024, 146(19), 13588–13597 CrossRef CAS PubMed.
- V. Piccoli and L. Martínez, Competitive effects of anions on protein solvation by aqueous ionic liquids, J. Phys. Chem. B, 2024, 128(32), 7792–7802 CrossRef CAS PubMed.
- Y. Wang, J. Liu, X. Yuan, M. Wang, Y. Nie, S. Zhang and H. He, Ionic liquid-based aggregation towards the mesoscale mechanism and their applications, Chem. Eng. J., 2023, 455, 140370 CrossRef CAS.
- Z. Q. Zhang, X. M. Lu, X. X. Wang and C. H. Lu, IPE ionic liquid database, Institute of process engineering, Chinese academy sciences, Beijing, 100190. https://www.ildatabase.com/ Search PubMed.
- Y. Li, X. Cui, Z. Xiong, B. Liu, B.-Y. Wang, R. Shu, N. Qiao and M.-H. Yung, Quantum molecular docking with a quantum-inspired algorithm, J. Chem. Theory Comput., 2024, 20(15), 6687–6694 CrossRef CAS PubMed.
- B. Ni, H. Wang, H. K. S. Khalaf, V. Blay and D. R. Houston, Autodock-ss: Autodock for multiconformational ligand-based virtual screening, J. Chem. Inf. Model., 2024, 64(9), 3779–3789 CrossRef CAS PubMed.
- A. Frick, U. K. Eriksson, F. de Mattia, F. Oberg, K. Hedfalk, R. Neutze, W. J. de Grip, P. M. Deen and S. Tornroth-Horsefield, X-ray structure of human aquaporin 2 and its implications for nephrogenic diabetes insipidus and trafficking, Proc. Natl. Acad. Sci. U. S. A., 2014, 111(17), 6305–6310 CrossRef CAS PubMed.
- K. S. Egorova, E. G. Gordeev and V. P. Ananikov, Biological activity of ionic liquids and their application in pharmaceutics and medicine, Chem. Rev., 2017, 117(10), 7132–7189 CrossRef CAS PubMed.
- S. J. Marrink, V. Corradi, P. C. T. Souza, H. I. Ingolfsson, D. P. Tieleman and M. S. P. Sansom, Computational modeling of realistic cell membranes, Chem. Rev., 2019, 119(9), 6184–6226 CrossRef CAS PubMed.
- N. A. Erkamp, M. Oeller, T. Sneideris, H. Ausserwoger, A. Levin, T. J. Welsh, R. Qi, D. Qian, N. Lorenzen and H. Zhu, et al., Multidimensional protein solubility optimization with an ultrahigh-throughput microfluidic platform, Anal. Chem., 2023, 95(12), 5362–5368 CrossRef CAS PubMed.
- S. Foser, A. Schacher, K. A. Weyer, D. Brugger, E. Dietel, S. Marti and T. Schreitmüller, Isolation, structural characterization, and antiviral activity of positional isomers of monopegylated interferon α-2a (PEGASYS), Protein Expr. Purif., 2003, 30(1), 78–87 CrossRef CAS PubMed.
- P. Ertl and A. Schuffenhauer, Estimation of synthetic accessibility score of drug-like molecules based on molecular complexity and fragment contributions, J. Cheminform., 2009, 1(1), 8 CrossRef PubMed.
- L. Bui-Le, C. J. Clarke, A. Bröhl, A. P. S. Brogan, J. A. J. Arpino, K. M. Polizzi and J. P. Hallett, Revealing the complexity of ionic liquid–protein interactions through a multi-technique investigation, Comm. Chem., 2020, 3(1), 55 CrossRef CAS PubMed.
- S. K. Thayallath, S. M. Shet, M. Bisht, P. Bharadwaj, M. M. Pereira, G. Franklin, S. K. Nataraj and D. Mondal, Designing protein nano-construct in ionic liquid: A boost in efficacy of cytochrome C under stresses, Chem. Commun., 2023, 59(39), 5894–5897 RSC.
- L. Bui-Le, C. J. Clarke, A. Bröhl, A. P. S. Brogan, J. A. J. Arpino, K. M. Polizzi and J. P. Hallett, Revealing the complexity of ionic liquid–protein interactions through a multi-technique investigation, Comm. Chem., 2020, 3, 1 CrossRef PubMed.
- O. Trott and A. J. Olson, Autodock vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31(2), 455–461 CrossRef CAS PubMed.
- S. Salentin, S. Schreiber, V. J. Haupt, M. F. Adasme and M. Schroeder, Plip: Fully automated protein-ligand interaction profiler, Nucleic Acids Res., 2015, 43(1), 443–447 CrossRef PubMed.
- P. Virtanen, R. Gommers, T. E. Oliphant, M. Haberland, T. Reddy, D. Cournapeau, E. Burovski, P. Peterson, W. Weckesser and J. Bright, et al., Scipy 1.0: Fundamental algorithms for scientific computing in python, Nat. Methods, 2020, 17(3), 261–272 CrossRef CAS PubMed.
- F. Murtagh and P. Legendre, Ward's hierarchical agglomerative clustering method: Which algorithms implement ward's criterion?, J. Classif., 2014, 31(3), 274–295 CrossRef.
- S. Lee, A. Mao, S. Bhattacharya, N. Robertson, R. Grisshammer, C. G. Tate and N. Vaidehi, How do short chain nonionic detergents destabilize G-protein-coupled receptors?, J. Am. Chem. Soc., 2016, 138(47), 15425–15433 CrossRef CAS PubMed.
- L. Martinez, R. Andrade, E. G. Birgin and J. M. Martinez, Packmol: A package for building initial configurations for molecular dynamics simulations, J. Comput. Chem., 2009, 30(13), 2157–2164 CrossRef CAS PubMed.
- D. A. Pearlman, D. A. Case, J. W. Caldwell, W. S. Ross, T. E. Cheatham, S. DeBolt, D. Ferguson, G. Seibel and P. Kollman, Amber, a package of computer programs for applying molecular mechanics, normal mode analysis, molecular dynamics and free energy calculations to simulate the structural and energetic properties of molecules, Comput. Phys. Commun., 1995, 91(1), 1–41 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, Development and testing of a general amber force field, J. Comput. Chem., 2004, 25(9), 1157–1174 CrossRef CAS PubMed.
- D. Vassetti, M. Pagliai and P. Procacci, Assessment of GAFF2 and OPLS-AA general force fields in combination with the water models TIP3P, SPCE, and OPC3 for the solvation free energy of druglike organic molecules, J. Chem. Theory Comput., 2019, 15(3), 1983–1995 CrossRef CAS PubMed.
- W. L. Jorgensen, J. Chandrasekhar, J. D. Madura, R. W. Impey and M. L. Klein, Comparison of simple potential functions for simulating liquid water, J. Chem. Phys., 1983, 79(2), 926–935 CrossRef CAS.
- T. Darden, D. York and L. Pedersen, Particle mesh ewald: An N.log(N) method for Ewald sums in large systems, J. Chem. Phys., 1993, 98(12), 10089–10092 CrossRef CAS.
- R. Salomon-Ferrer, A. W. Götz, D. Poole, S. Le Grand and R. C. Walker, Routine microsecond molecular dynamics simulations with AMBER on GPUs. 2. explicit solvent particle mesh Ewald, J. Chem. Theory Comput., 2013, 9(9), 3878–3888 CrossRef CAS PubMed.
- H. C. Andersen, Rattle: A “velocity” version of the shake algorithm for molecular dynamics calculations, J. Comput. Phys., 1983, 52(1), 24–34 CrossRef CAS.
- H. J. C. Berendsen, J. P. M. Postma, W. F. V. Gunsteren, A. DiNola and J. R. Haak, Molecular dynamics with coupling to an external bath, J. Chem. Phys., 1984, 81(8), 3684–3690 CrossRef CAS.
- Y. Zhang, S. E. Feller, B. R. Brooks and R. W. Pastor, Computer simulation of liquid/liquid interfaces. I. Theory and application to octane/water, J. Chem. Phys., 1995, 103(23), 10252–10266 CrossRef CAS.
- IBM SPSS statistics for windows, version 25.0*, IBM Corp., Armonk, NY, 2017 Search PubMed.
- F. Pedregosa, G. Varoquaux, A. Gramfort, V. Michel, B. Thirion, O. Grisel, M. Blondel, P. Prettenhofer, R. Weiss and V. Dubourg, et al., Scikit-learn: Machine learning in python, J. Mach. Learn. Res., 2011, 12, 2825–2830 Search PubMed.
| This journal is © the Owner Societies 2025 |