 Open Access Article
Open Access ArticleA dual-functional substrate for quantitation of substrate levels and GCase activity in living cells†
Ben
Tiet
a,
Sha
Zhu
a,
Xi
Chen
a,
Nadia
Anastasi
c,
Nicholas W.
See
a,
Matthew C.
Deen
b,
Eva
Harde
c and
David J.
Vocadlo
 *ad
*ad
aDepartment of Chemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada. E-mail: dvocadlo@sfu.ca
bLaboratory of Organic Chemistry, ETH Zurich, D-CHAB, Zurich 8093, Switzerland
cRoche Pharma Research and Early Development, Neuroscience and Rare Diseases Discovery and Translational Area, Roche Innovation Center Basel, F. Hoffmann-La Roche Ltd, Basel, Switzerland
dDepartment of Molecular Biology and Biochemistry, Simon Fraser University, Burnaby, BC V5A 1S6, Canada
First published on 3rd July 2025
Abstract
Loss of function mutations in the gene GBA1, which encodes the lysosomal glycoside hydrolase β-glucocerebrosidase (GCase) cause Gaucher's disease (GD). Moreover, one mutant allele of GBA1 is the most common genetic risk factor for the development of Parkinson's disease (PD). To gain a better understanding how these mutations drive development of PD and how GCase is regulated within cells, the field needs chemical reporters of GCase activity that can be used within living cells. Fluorogenic substrates are one method that can be used to quantify enzyme activities within cells yet existing substrates for GCase have limitations. In particular, the inability to monitor cellular uptake of substrate limits the ability to disentangle impairments in uptake of substrate from impairments in lysosomal GCase activity. Here we report on the preparation and biological characterisation of LysoRF-GBA – a new chemical tool which can be used to quantitatively measure both the cellular levels of intact substrate and lysosomal GCase activity within lysosomes. We demonstrate that, by using LysoRF-GBA, endogenous GCase activity can be measured within live neuroblastoma cells. The selectivity of this substrate for GCase, relative to other cellular enzymes, was validated by genetic and pharmacological perturbation of GCase. By using LysoRF-GBA and concomitantly monitoring levels of both cleaved product and intact substrate, we were able to measure GCase engagement with a known pharmacological chaperone and discriminate between pharmacological agents that affect GCase activity from those that impair endocytosis. Further, the ability to monitor intracellular levels of intact LysoRF-GBA also enabled us to measure its time dependent accumulation within cells, providing insight into when steady state levels of this substrate are reached. LysoRF-GBA therefore shows high potential to be exploited as a tool for the discovery of compounds that could beneficially modulate its activity for benefit in diseases including PD.
Introduction
Lysosomal glycoside hydrolase, β-glucocerebrosidase (GCase; CAZy family GH30, EC 3.2.1.45), catalyzes the hydrolysis of glucosylceramide into glucose and ceramide.1–3 Deficiency in GCase activity due to biallelic mutations in GBA1 causes Gaucher's disease (GD), a lysosomal storage disorder that can lead to severe neurodegeneration.3,4 Parkinson's disease (PD) is the second most common neurodegenerative disorder, affecting over 3% of individuals aged 65 and older. Depending on the population studied, approximately 5–30% of Parkinson's disease (PD) patients carry mutations in GBA1, making it the most common genetic risk factor for PD.5–8 Indeed, the risk of developing PD is increased by up to 30-fold by these mutations depending on the age, ethnicity, and mutations involved in the analysis.3–10 Although significant efforts have been made to study the biological processes underlying the relationship between GCase activity and PD, the molecular mechanisms involved are not well understood.1,4 The ability to measure GCase activity in its native cellular environment has the potential to grant insight into these mechanisms, yet limitations in the methods for measuring cellular GCase activity have hindered the development of GCase-directed therapies.One approach to addressing this need for GCase assays is the development of fluorogenic assays for GCase. These assays provide a convenient and accurate means of measuring the catalytic activity of this enzyme. The most widely used substrate, 4-methylumbelliferyl β-glucopyranoside (4MU-Glc), has seen widespread use in basic research1,11–14 as well as in clinical settings to quantify the amount of GCase activity in blood samples from new-borns.15 Yet, using such simple fluorogenic substrates to monitor GCase activity in buffered enzyme assays leads to an inability to detect cellular factors that might affect the activity of this enzyme such as endogenous regulatory factors that influence GCase activity and maturation.1,16,17 For example, studies show that GCase activity is stimulated by anionic lipids, and inhibited by cationic lipids, yet this has only been demonstrated in vitro.16 Thus, while existing tools can provide some use in the search for modulators of GCase activity, they are unable to capture factors that can influence GCase within the cellular environment.
Indeed, assays using tissue lysates where the cells and lysosomes have been broken up disrupt the natural environment of the lysosomes and result in a significant dilution of lysosomal proteins and possible modulators of GCase through the addition of reaction buffer, leading to an incomplete understanding of GCase activity within cells.1,18 Furthermore, lysate assays measure the total cellular activity of GCase instead of the functionally important lysosomal pool of GCase. Accordingly, to overcome these limitations, there is interest in the creation of new ways to accurately measure lysosomal GCase activity within living cells.
Recently, new fluorogenic substrates that allow quantitative measurement of GCase activity directly within cells have been developed.17–21 These fluorescence-quenched substrates have shown much improved properties relative to the earliest approach that involved the simple arylglucoside, 5-(pentafluorobenzylamino) fluorescein di-β-glucopyranoside (PFB-FDG),11,22–25 which has to be used at very high concentration to alkylate cell surface proteins, displays problematic pH-dependent fluorescence in the physiological range, and is rapidly cleared from cells.1 The development of fluorescence-quenched GCase substrates LysoFQ-GBA18 and LysoFix-GBA21 are dark-to-light substrates that are switched on by enzymatic cleavage of the glycosidic bond to liberate a quencher, which in turn allows detection of a bright pH-insensitive fluorophore.17–21
Although powerful tools for some applications, these dark-to-light fluorescent substrates still have some limitations. As the intact substrate is not fluorescent, one cannot determine the intracellular levels of substrate at any given time point. As such, a clear limitation is that one cannot be certain whether a measured decrease in apparent activity is due to a decrease in enzymatic turnover of the substrate or simply decreased uptake of the substrate. We envisioned that generating a GCase substrate capable of circumventing this and other limitations associated with these previous GCase substrates by allowing concomitant relative quantification of intact intracellular substrate as well as its enzymatically produced fluorescent product.
Ratiometric substrates have demonstrated advantages for imaging enzyme activity. Their dual emission wavelengths provide greater precision and reliability compared to substrates whose turnover is measured at a single wavelength.26,27 Various ratiometric substrates have been generated for detecting the enzymatic activity of proteases and glycosidases in biological systems, though fewer have been applied to quantifying enzymatic activity in a rigorous manner. To our knowledge, none have been used to examine the kinetics of substrate uptake or quantifying relative substrate levels within cells.28–40 In light of these considerations, we set out to create a dual-functional substrate for GCase that is capable of ratiometric imaging to enable improved understanding regarding the factors that influence reliable measurements of enzymatic activity using fluorogenic cell imaging substrates. Furthermore, such a substrate would enable us to quantify the levels of substrate within cells as well as the fraction of substrate turned over at any given time.
Towards this goal, we were inspired by this previous work,18,36,37 to create a selective dual emissive substrate (LysoRF-GBA) that can engage in Förster resonance energy transfer (FRET) and be used to quantify the activity of endogenous lysosomal GCase (Fig. 1).17–20 This substrate is designed such that the intact substrate produces an emission signal which differs from that of the enzymatically cleaved substrate. We show that this substrate can be used in a ratiometric manner or a dual emissive mode that allows for tracking of time-dependent accumulation of intracellular substrate, the study of lysosomal GCase activity, monitoring of the effects of exogenous compounds on GCase catalysis, and disentanglement of the effects of disrupting GCase activity from those influencing endocytic uptake of substrate.
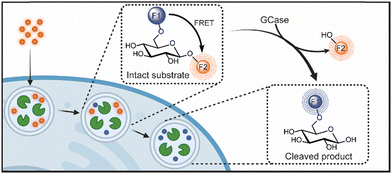 | ||
| Fig. 1 A conceptual image for the uptake and turnover of LysoRF-GBA by active GCase in living cells. Created in BioRender. Tiet, B. (2025) https://BioRender.com/1l1tmh3. | ||
Results and discussion
Synthesis of LysoRF-GBA
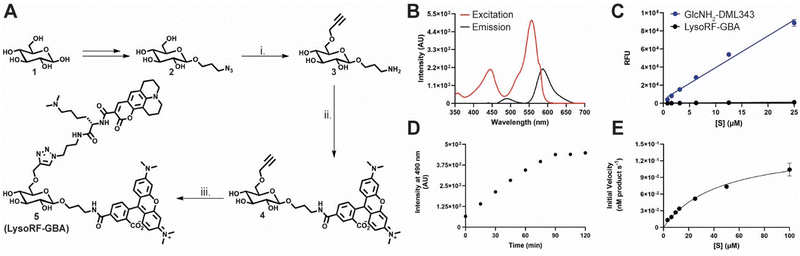
To create a FRET-based substrate for GCase that can report on both enzyme activity and intact substrate, we leveraged the design of a previously described substrate, LysoFQ-GBA,18,21 in which the O-6 position was modified to achieve high selectivity for GCase. Notably, this substrate exhibited very efficient fluorescence quenching, which we anticipated arose from the close proximity of its fluorophore-quencher pair leading either to FRET quenching or contact quenching. On the assumption, that the quenching observed for LysoFQ-GBA arises predominantly from FRET, we aimed to replace the fluorophore-quencher pair with two fluorophores that could interact through FRET, though recognizing the potential deleterious contact quenching effects arising from possible collision of these groups. After consideration, we selected TAMRA and coumarin343 as our FRET pair for the design of LysoRF-GBA. Schultz and coworkers have demonstrated the success of this fluorophore pair for generation of ratiometric reporters.36,37 When irradiated in the excitation band of coumarin343 (438 nm; donor species), we proposed that efficient FRET should occur to result in emission from TAMRA (590 nm; acceptor species). This emission would only be detected if the substrate is structurally intact (Fig. 1). However, upon GCase-catalyzed hydrolysis of the glycosidic bond of LysoRF-GBA, the TAMRA moiety would be liberated and diffuse away from the coumarin343 fluorophore. This would terminate the FRET process such that irradiation at 438 nm would lead to emission being detected at 483 nm, which would arise from emission from coumarin343. By monitoring both these wavelengths simultaneously, we envisioned that we should be able to use LysoRF-GBA to monitor both enzyme activity and the cellular levels of substrate within living cells (Fig. 1). Finally, we considered that the inclusion of an N,N-dimethyl-L-lysine (DML) residue as a lysosomotropic moiety would improve retention of the cleaved product and enhance substrate solubility.18,21The preparation of 5 (LysoRF-GBA) was commenced from D-glucose 1 which was synthetically elaborated to intermediate azide 2 using previously established methods.20 Azide 2 was then regioselectively alkylated at the primary hydroxyl with 3-chloro-1-propyne and sodium hydride in N,N-dimethylformamide. Without further purification, the azide was reduced with trimethyl phosphine under Staudinger conditions to yield amine 3. Coupling with the commercially available NHS-ester of 5-TAMRA under basic conditions provided TAMRA-alkyne 4 in moderate yield (45%). Finally, to produce our desired fluorescent substrate (LysoRF-GBA), we used copper-catalyzed azide–alkyne cycloaddition (CuAAC) chemistry to couple a coumarin343–DML conjugate, which was prepared in a parallel synthetic sequence. LysoRF-GBA was fully characterised by nuclear magnetic resonance (NMR) spectroscopy and high-resolution mass spectrometry (MS).
Upon completing synthesis of LysoRF-GBA (Fig. 2(A)), we assessed its in vitro characteristics starting with its basic photophysical properties. First, we acquired the excitation and emission spectra of LysoRF-GBA (Fig. 2(B)). LysoRF-GBA exhibited excitation maxima at approximately 440 and 550 nm corresponding to the major absorption bands associated with coumarin343 and TAMRA, respectively. Additionally, emission maxima were observed at 490 and 590 nm, which are consistent with the emission bands seen for coumarin343 and TAMRA, respectively. Next, we analyzed the FRET efficiency of LysoRF-GBA in comparison to an analogue which did not contain the FRET acceptor functionality (GlcNH2-DML343 13, synthetic details are provided in the ESI†). LysoRF-GBA showed an excellent FRET efficiency of 98.7%, which compares favourably with other FRET-based pairs in the literature (Fig. 2(C)).17,18,20,21 We next examined GCase catalyzed turnover of LysoRF-GBA. We first treated LysoRF-GBA with a high concentration of GCase and collected spectral scans at specified intervals over a 120-minute period (Fig. S1, ESI†). The emission intensity at 490 nm over the time course was then measured and showed that the fluorescence intensity had plateaued after 90 minutes. This is consistent with complete turnover of the substrate (Fig. 2(D)). Next, using a concentration of GCase well below that of substrate, we acquired a series initial rate measurements (Fig. 2(E)). Using this approach, we were able to observe Michaelis–Menten kinetics. Using a non-linear regression, an estimated Michaelis constant (KM) of 45 ± 5 μM and a first order rate constant (kcat) of 0.00585 ± 0.0003 s−1 were obtained. We also determined a second order rate constant for GCase catalyzed turnover of LysoRF-GBA (kcat/KM = 130 ± 20 M−1 s−1) that showed a surprising 100-fold increase over the fluorescence quenched substrate LysoFQ-GBA.18,20,21 Finally, incubation with the functionally related non-lysosomal glucocerebrosidase (GBA2),41,42 which also cleaves β-glucosidases, showed this enzyme was unable to cleave LysoRF-GBA (Fig. S2, ESI†). These findings show that LysoRF-GBA is an effective dual emissive fluorescent substrate for selective monitoring GCase activity over other cellular β-glucosidases.
Live cell imaging of GCase activity in human neuroblastoma cells
Having established suitability of LysoRF-GBA as a selective substrate for GCase, we next assessed its turnover within live cells. We used a human neuroblastoma cell line (SK-N-SH) as these cells express are known to express GCase. We first investigated if LysoRF-GBA could report on endogenous GCase activity selectively and quantitatively. To ensure that our product signal was derived strictly from GCase activity, we measured the time-dependent change in fluorescence of coumarin343 (ex = 438/24 nm, em = 483/32 nm) that is associated with cleavage of LysoRF-GBA in the presence and absence of AT3375, a highly selective GCase inhibitor.18,20,43 Over a four-hour incubation period, in the absence of inhibitor, we observed significant coumarin343 fluorescence derived from the cleaved product in a manner that depended on the concentration of substrate. In contrast, the conditions containing the inhibitor showed no significant turnover of the substrate (Fig. 3(A)), in accordance with the preliminary in vitro data which showed LysoRF-GBA is selective for lysosomal GCase. Furthermore, these data also show that there is no adventitious cleavage of any other amide bonds within the molecule that could confound the interpretation of the data. To examine the feasibility of using LysoRF-GBA as a substrate to quantify turnover by GCase within cells, we examined the time dependent turnover of substrate at different concentrations. This was performed by incubating the neuroblastoma cells with varying concentrations of LysoRF-GBA and allowing cleavage by GCase for specified intervals prior to quenching the enzymatic reactions with inhibitor and imaging. We found that the coumarin343 fluorescence associated with turnover of LysoRF-GBA by GCase exhibited a linear response with respect to time at all substrate concentrations examined (Fig. 3(B)) and exhibited a punctate pattern consistent with lysosomal distribution (Fig. 3(C)). Colocalization studies showed this signal overlapped well with lysotracker DND-26 (PCC = 0.61, Fig. S3, ESI†). An alternate analysis, using the ratio of the fluorescence intensities of our product relative to our substrate signal, also showed a linear response that was superior at 10 μM of LysoRF-GBA over the four-hour period (Fig. 3(D)). These analyses indicated that 10 μM of LysoRF-GBA was an optimal concentration for measuring GCase activity in live cells. Next, we determined whether the fluorescent product was retained within cells, which is important to enable reliable quantitation of cellular enzymatic activity. Using a stopped time-course experiment, we incubated LysoRF-GBA (10 μM) with SK-N-SH cells for 2 hours before removing excess substrate by washing the cells and blocking any further GCase activity using a high concentration of AT3375. We observed excellent retention of the fluorescent product (ex = 438/24 nm, em = 483/32 nm) over the 2.5-hour assay period (Fig. 3(E)). Therefore, the fluorescent cleaved product accumulates and remains trapped within lysosomes. These data collectively show that LysoRF-GBA serves as a selective GCase substrate that provides precise quantification of relative GCase activity within live cells.We then assessed GCase activity in cell lines that had been perturbed either chemically, using AT3375, or genetically, by knocking out (KO) GBA1. A distinct difference was observed in the GCase activity detected when using 10 μM LysoRF-GBA in wild-type (WT) cells as compared to the cell lines that were genetically or chemically perturbed (Fig. S4, ESI†). As LysoRF-GBA turnover was not observed in KO cell lines, we evaluated if lysosomal activity could be rescued by treatment with a GCase replacement therapeutic (Cerezyme). We observed that addition of 100 or 1000 nM of Cerezyme was able to rescue lysosomal GCase activity. However, addition of Cerezyme at lower concentrations (10 nM) was insufficient to recover GCase activity based on the fluorescence detected (ex = 488 nm, em = 525/25 nm) for the cleaved product (Fig. 4). A similar trend was observed when treated with 20 μM of LysoRF-GBA (Fig. S5, ESI†). These results further support LysoRF-GBA being selective for lysosomal GCase.
Distinguishing between endocytosis and GCase inhibition
The principle benefit of using substrates to quantify cellular enzyme activity is the ability to assess pharmacological agents or cellular factors that have either direct effects on the enzyme activity or act indirectly to alter the cellular environment in a manner that influences the activity of the enzyme of interest. In this regard, one limitation of many substrates, is that they are actively endocytosed. For this reason, the inhibition of endocytosis results in decreased substrate turnover that cannot be readily distinguished from inhibition of GCase itself. Assuming that the TAMRA-containing product can diffuse out of the cells, intact LysoRF-GBA within cells can be quantified by directly measuring TAMRA fluorescence (ex = 543/36, em = 590/36 nm). The cleaved product can be detected by monitoring coumarin343 fluorescence (ex = 438/24 nm, em = 483/32 nm), we recognized that LysoRF-GBA could be used to distinguish between the pharmacological effects of compounds acting directly on GCase from those affecting its uptake. As an initial step to assess this idea, we used LysoRF-GBA by monitoring product formation through coumarin343 (ex = 438/24 nm, em = 483/32 nm) to quantify GCase engagement with AT3375 and found a cellular IC50 value of 11 ± 2 nM with a Hill slope of −1.2 that further supports AT3375 acting directly as a GCase inhibitor within cells (Fig. 5(A)). Interestingly, because we could also detect the levels of intact substrate within cells based on TAMRA fluorescence (ex = 543/36 nm, em = 590/36 nm), we were able to calculate an IC50 value (15 ± 2 nM) based on remaining amount of substrate present within cells that showed a Hill slope of 1.0 (Fig. 5(B)). Notably, the close concordance of these IC50 values, being within error, supports the idea that the TAMRA-containing product diffuses out of cells. Therefore, LysoRF-GBA can be used to measure target engagement by GCase inhibitors and provides quantitation of the inhibition of GCase activity in live cells. Furthermore, as the intact substrate can be monitored using TAMRA fluorescence, LysoRF-GBA offers insight into the remaining quantities of substrate that remain.We next tested the effects of pharmacological inhibitors of endocytosis on cellular levels of substrate. By monitoring fluorescence of both coumarin343 (GCase activity) and TAMRA (intact substrate levels) we were able to assess the effects of these compounds relative to our positive and negative controls which consisted of DMSO and AT3375, respectively. Here we used casin, dyngo-4a, pitstop-2, and wortmannin to antagonize different endocytosis pathways.44–47 For all endocytosis inhibitors, except for pitstop-2, we observed a decrease in turnover of substrate (Fig. 5(C)). When analyzing the levels of intact substrate by measuring TAMRA FRET signal (ex = 438/24, em = 590/36 nm, Fig. 5(D)) and TAMRA fluorescence (Fig. 5(E)), we found that these endocytosis inhibitors all yielded considerably lower levels of substrate compared to both DMSO and AT3375 treated cells. Thus, most of these endocytosis inhibitors cause a reduction in apparent GCase activity measured when one measures a decrease in the extent of product formation, but this can be distinguished from true inhibition of activity by monitoring the fluorescence associated with intact substrate that occurs from blocking the uptake of substrate.
Measuring substrate accumulation and defining the steady state levels
Traditional approaches relying on dark-to-light fluorescence-quenched substrates do not offer the possibility of detecting the substrate. This is relevant to the study of enzymes within living cells because scenarios are known to arise where substrate diffusion or uptake into the cells becomes rate limiting, causing the rate of the enzymatic turnover of the substrate to be masked.1,39,48,49 Despite this recognized problem, no studies, to our knowledge, have examined the uptake and accumulation of synthetic substrates within cells. We were therefore curious if we could use LysoRF-GBA to uncover how this substrate accumulates within living cells as a function of time (Fig. 6(A)). We therefore set out to develop a general strategy that could be used to quantify changes in the relative levels of intact substrate when cells are incubated with varying levels of substrate. Such experiments would also help define when a steady state pseudo-equilibrium might be achieved wherein the rate of substrate uptake by cells becomes equivalent to the rate of substrate release. We therefore designed an experiment in which we first treated SK-N-SH cells with 10 μM of AT3375 to completely inhibit GCase and prevent any enzymatic turnover of substrate.In our preliminary experiment, we added varying concentrations of substrate and measured the intact substrate levels inside the cells at various time points extending to four hours. In this preliminary experiment performed both in the presence and absence of inhibitor, we confirm our expectation that blockade of enzymatic turnover led to more substrate being present (Fig. 6(B)). More detailed time-dependent measurements made for FRET-derived fluorescence of TAMRA (ex = 438/24, em = 590/36 nm), corresponding to the intact substrate, showed a linear increase over four hours for the 5 and 10 μM concentrations, as well as a linear response up to three hours for the 20 μM treatment with LysoRF-GBA (Fig. 6(C)). Importantly, no significant increase in coumarin343 fluorescence (ex = 438/24 nm, em = 483/32 nm) was observed under any conditions over the entire four-hour assay period (Fig. S6, ESI†) indicating that GCase activity was completely blocked under these conditions. We next investigated the extent of substrate accumulation over longer incubation periods to determine if a steady state level of substrate was eventually reached. We therefore incubated cells with LysoRF-GBA for up to 72 hours and monitored FRET-based TAMRA fluorescence (ex = 438/24, em = 590/36 nm), which corresponds to the intact substrate. We found that the substrate signal plateaus after 24 hours, suggesting that an equilibrium for uptake of the substrate is reached by this time. Notably, we found that using concentrations higher than 10 μM led to diminishing increases in the steady state accumulation of the substrate, suggesting saturation of the uptake mechanisms. Also notable is that at lower concentrations, such as 3 μM, substrate accumulation is linear over the entire 72-hour time course. Notably, some lysosomotropic agents that are rapidly taken up have in some cases been shown to modestly increase the pH of the lysosome even at μM concentrations.50–52 Accordingly, we note that in cases where such alterations may be important, pH should be examined when using LysoRF-GBA, particularly in cell lines that exhibit fast uptake of this substrate. Regardless, these data collectively support 10 μM as a useful and effective operating concentration for LysoFQ-GBA with these cells (Fig. 6(D)).
To our knowledge, this is the first example where the time-dependent accumulation of an exogenous substrate has been measured inside cells. Our findings using LysoRF-GBA show that cellular uptake of substrate occurs in a linear manner over an incubation of up to 4 hours before a steady state is attained at 24 hours. Furthermore, such accumulation shows concentration dependence and approaches a maximum level, which we attribute to saturation in cellular uptake.
Conclusions
In summary, we have developed LysoRF-GBA as a new chemical tool that can be used to measure lysosomal GCase activity in living cells. This substrate is actively taken up by living human neuroblastoma cells where it is selectively turned over by lysosomal GCase. The coumarin fluorescence of the cleaved coumarin343-containing product of LysoRF-GBA provides an accurate representation of lysosomal GCase activity because it does not readily diffuse out of cells. LysoRF-GBA was shown in this way to be useful in measuring endogenous GCase activity in a time-and dose-dependent manner when monitoring product formation based on coumarin343 fluorescence. Under conditions where GCase turnover was blocked by inclusion of AT3775 or genetically perturbed by knockout of the GBA1 gene, no increase in coumarin343 fluorescence was observed, thus confirming its selectivity. Because the TAMRA-containing product diffuses out of cells, one can also quantify intact substrate levels by directly measuring TAMRA fluorescence. However, it is difficult to be certain that all TAMRA product has diffused out of the cell at any given time point, therefore, a more direct method to quantify intact substrate within cells is to measure FRET-based TAMRA fluorescence, which relies on close proximity of the coumarin343 fluorophore. We therefore used the FRET-based TAMRA fluorescence of intact LysoRF-GBA to investigate the point at which steady state levels of intact substrate were reached within cells, which we expect are defined by the rates of intact substrate influx and efflux. Indeed, in this way, we showed that LysoRF-GBA uptake occurs in a largely linear manner over an incubation period of up to 4 hours before steady state levels are reached at 24 hours. Such accumulation was also shown to be concentration dependent. The effects of pharmacological inhibitors of endocytosis were also evaluated using LysoRF-GBA and compared to conditions in the presence and absence of AT3375. These experiments showed that LysoRF-GBA could distinguish between the effects of GCase and endocytosis inhibitors by monitoring both intracellular GCase activity and intact substrate levels. This is advantageous as it provides insight into how exogenous substrates are taken up by cells and how such mechanisms can affect their subsequent turnover. The collective features of LysoRF-GBA support this fluorogenic substrate as being a valuable quantitative live cell imaging tool that can discriminate the effects of different pharmacological agents. We anticipate that it will be of great use to practitioners in the field as LysoRF-GBA offers a promising new tool to study GCase regulation and discover potential therapeutic compounds for GCase related diseases. Moreover, we expect that the general approaches described herein should also aid in the advance of precision substrates that can be used in a more quantitative manner to assay various cellular enzyme activities.Conflicts of interest
N. A. and E. H. were employees of Roche. The materials described in this manuscript are subject of a patent application assigned to Simon Fraser University.Data availability
The experimental section, figures and tables; NMR spectra; and other details are provided within the ESI.†Acknowledgements
The authors are grateful for support from the Natural Sciences and Engineering Research Council (NSERC) of Canada (RGPIN-05426), the Canadian Glycomics Network (RG-1), and the Michael J. Fox Foundation (MJFF) for Parkinson Research (16536 and 14249). D. J. V. thanks the Canada Research Chairs program for support as a Tier I Canada Research Chair in Chemical Biology. M. C. D. was supported by a NSERC PGS-D scholarship. The authors also thank the Centre for High-Throughput Chemical Biology (HTCB) for access to core facilities. We also thank Gideon Davies for providing the sample of GBA2 enzyme. NWS is grateful to Michael Smith Health Research BC, Pacific Parkinson's Research Institute and Parkinson Society British Columbia for support with a postdoctoral fellowship. Scientific illustrations (Fig. 1 and Fig. 6a) were created using Biorender.Notes and references
- D. Ysselstein, T. J. Young, M. Nguyen, S. Padmanabhan, W. D. Hirst, N. Dzamko and D. Krainc, Mov. Disord, 2021, 36, 2719–2730 CrossRef CAS PubMed.
- J. Do, C. McKinney, P. Sharma and E. Sidransky, Mol. Neurodegener., 2019, 14, 36 CrossRef PubMed.
- M. L. Furderer, E. Hertz, G. J. Lopez and E. Sidransky, Int. J. Mol. Sci., 2022, 23, 5842 CrossRef CAS PubMed.
- E.-J. Bae, N. Y. Yang, C. Lee, H.-J. Lee, S. Kim, S. P. Sardi and S.-J. Lee, Exp. Mol. Med., 2015, 47, e153 CrossRef CAS PubMed.
- L. Smith and A. H. V. Schapira, Cells, 2022, 11, 1261 CrossRef CAS PubMed.
- S. R. L. Vieira and A. H. V. Schapira, J. Neural Transm., 2022, 129, 1105–1117 CrossRef PubMed.
- K. A. Senkevich, A. E. Kopytova, T. S. Usenko, A. K. Emelyanov and S. N. Pchelina, Acta Nat., 2021, 13, 70–78 CrossRef CAS PubMed.
- A. Migdalska-Richards and A. H. V. Schapira, J. Neurochem., 2016, 139, 77–90 CrossRef CAS PubMed.
- E. Sidransky, M. A. Nalls, J. O. Aasly, J. Aharon-Peretz, G. Annesi, E. R. Barbosa, A. Bar-Shira, D. Berg, J. Bras, A. Brice, C.-M. Chen, L. N. Clark, C. Condroyer, E. V. De Marco, A. Dürr, M. J. Eblan, S. Fahn, M. J. Farrer, H.-C. Fung, Z. Gan-Or, T. Gasser, R. Gershoni-Baruch, N. Giladi, A. Griffith, T. Gurevich, C. Januario, P. Kropp, A. E. Lang, G.-J. Lee-Chen, S. Lesage, K. Marder, I. F. Mata, A. Mirelman, J. Mitsui, I. Mizuta, G. Nicoletti, C. Oliveira, R. Ottman, A. Orr-Urtreger, L. V. Pereira, A. Quattrone, E. Rogaeva, A. Rolfs, H. Rosenbaum, R. Rozenberg, A. Samii, T. Samaddar, C. Schulte, M. Sharma, A. Singleton, M. Spitz, E.-K. Tan, N. Tayebi, T. Toda, A. R. Troiano, S. Tsuji, M. Wittstock, T. G. Wolfsberg, Y.-R. Wu, C. P. Zabetian, Y. Zhao and S. G. Ziegler, N. Engl. J. Med., 2009, 361, 1651–1661 CrossRef CAS PubMed.
- L. Oftedal, J. Maple-Grødem, M. G. G. Førland, G. Alves and J. Lange, Sci. Rep., 2020, 10, 22098 CrossRef CAS PubMed.
- E. Aflaki, D. K. Borger, N. Moaven, B. K. Stubblefield, S. A. Rogers, S. Patnaik, F. J. Schoenen, W. Westbroek, W. Zheng, P. Sullivan, H. Fujiwara, R. Sidhu, Z. M. Khaliq, G. J. Lopez, D. S. Goldstein, D. S. Ory, J. Marugan and E. Sidransky, J. Neurosci., 2016, 36, 7441–7452 CrossRef CAS PubMed.
- C. E. G. Leyns, A. Prigent, B. Beezhold, L. Yao, N. G. Hatcher, P. Tao, J. Kang, E. Suh, V. M. Van Deerlin, J. Q. Trojanowski, V. M. Y. Lee, M. E. Kennedy, M. J. Fell and M. X. Henderson, NPJ Parkinsons Dis., 2023, 9, 74 CrossRef CAS PubMed.
- J. R. Mazzulli, F. Zunke, T. Tsunemi, N. J. Toker, S. Jeon, L. F. Burbulla, S. Patnaik, E. Sidransky, J. J. Marugan, C. M. Sue and D. Krainc, J. Neurosci., 2016, 36, 7693–7706 CrossRef CAS PubMed.
- S. Bannink, K. O. Bila, J. Van Weperen, N. A. M. Ligthart, M. J. Ferraz, R. G. Boot, D. Van Der Vliet, D. E. C. Boer, H. S. Overkleeft, M. Artola and J. M. F. G. Aerts, J. Lipid Res., 2024, 65, 100670 CrossRef CAS PubMed.
- T. Sawada, J. Kido, K. Sugawara, S. Yoshida, S. Matsumoto, T. Shimazu, Y. Matsushita, T. Inoue, S. Hirose, F. Endo and K. Nakamura, Mol. Genet. Metab. Rep., 2022, 31, 100850 CAS.
- M. Abdul-Hammed, B. Breiden, G. Schwarzmann and K. Sandhoff, J. Lipid Res., 2017, 58, 563–577 CrossRef CAS PubMed.
- S. Cecioni, R. A. Ashmus, P.-A. Gilormini, S. Zhu, X. Chen, X. Shan, C. Gros, M. C. Deen, Y. Wang, R. Britton and D. J. Vocadlo, Nat. Chem. Biol., 2022, 18, 332–341 CrossRef CAS PubMed.
- M. C. Deen, Y. Zhu, C. Gros, N. Na, P.-A. Gilormini, D. L. Shen, S. Bhosale, N. Anastasi, R. Wang, X. Shan, E. Harde, R. Jagasia, F. C. Lynn and D. J. Vocadlo, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2200553119 CrossRef CAS PubMed.
- S. Cecioni and D. J. Vocadlo, J. Am. Chem. Soc., 2017, 139, 8392–8395 CrossRef CAS PubMed.
- A. K. Yadav, D. L. Shen, X. Shan, X. He, A. R. Kermode and D. J. Vocadlo, J. Am. Chem. Soc., 2015, 137, 1181–1189 CrossRef CAS PubMed.
- S. Zhu, M. C. Deen, Y. Zhu, P.-A. Gilormini, X. Chen, O. B. Davis, M. Y. Chin, A. G. Henry and D. J. Vocadlo, Angew. Chem., Int. Ed., 2023, 62, e202309306 CrossRef CAS PubMed.
- P. Gaspar, W. W. Kallemeijn, A. Strijland, S. Scheij, M. Van Eijk, J. Aten, H. S. Overkleeft, A. Balreira, F. Zunke, M. Schwake, C. Sá Miranda and J. M. F. G. Aerts, J. Lipid Res., 2014, 55, 138–145 CrossRef CAS PubMed.
- A. Labrador-Garrido, S. Zhong, L. Hughes, S. Keshiya, W. S. Kim, G. M. Halliday and N. Dzamko, Front. Cell. Neurosci., 2023, 17, 1229213 CrossRef CAS PubMed.
- J. R. Mazzulli, F. Zunke, O. Isacson, L. Studer and D. Krainc, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 1931–1936 CrossRef CAS PubMed.
- D. Ysselstein, M. Nguyen, T. J. Young, A. Severino, M. Schwake, K. Merchant and D. Krainc, Nat. Commun., 2019, 10, 5570 CrossRef CAS PubMed.
- K. Kikuchi, Chem. Soc. Rev., 2010, 39, 2048 RSC.
- M. H. Lee, J. S. Kim and J. L. Sessler, Chem. Soc. Rev., 2015, 44, 4185–4191 RSC.
- K. Gu, Y. Xu, H. Li, Z. Guo, S. Zhu, S. Zhu, P. Shi, T. D. James, H. Tian and W.-H. Zhu, J. Am. Chem. Soc., 2016, 138, 5334–5340 CrossRef CAS PubMed.
- X. Hou, J. Peng, F. Zeng, C. Yu and S. Wu, Mater. Chem. Front., 2017, 1, 660–667 RSC.
- T. Komatsu, K. Kikuchi, H. Takakusa, K. Hanaoka, T. Ueno, M. Kamiya, Y. Urano and T. Nagano, J. Am. Chem. Soc., 2006, 128, 15946–15947 CrossRef CAS PubMed.
- H. W. Lee, V. Juvekar, D. J. Lee, S. M. Kim and H. M. Kim, Anal. Chem., 2021, 93, 14778–14783 CrossRef CAS PubMed.
- Y. Li, H. Wang, J. Li, J. Zheng, X. Xu and R. Yang, Anal. Chem., 2011, 83, 1268–1274 CrossRef CAS PubMed.
- L. Shi, C. Yan, Y. Ma, T. Wang, Z. Guo and W.-H. Zhu, Chem. Commun., 2019, 55, 12308–12311 RSC.
- G.-Y. Yang, C. Li, M. Fischer, C. W. Cairo, Y. Feng and S. G. Withers, Angew. Chem., Int. Ed., 2015, 54, 5389–5393 CrossRef CAS PubMed.
- G.-Q. Zhang, W. Feng, Z. Gao, G.-L. Zhang, X. Wu, Y. Xiao, X. Li, L. Zheng, D. Ding, J. Guo and B. Situ, Aggregate, 2023, 4, e286 CrossRef CAS.
- A. Cobos-Correa, J. B. Trojanek, S. Diemer, M. A. Mall and C. Schultz, Nat. Chem. Biol., 2009, 5, 628–630 CrossRef CAS PubMed.
- M. Guerra, V. S. Halls, J. Schatterny, M. Hagner, M. A. Mall and C. Schultz, J. Am. Chem. Soc., 2020, 142, 20299–20305 CrossRef CAS PubMed.
- Y. Cai, J. Zhan, H. Shen, D. Mao, S. Ji, R. Liu, B. Yang, D. Kong, L. Wang and Z. Yang, Anal. Chem., 2016, 88, 740–745 CrossRef CAS PubMed.
- Z. Huang, R. An, S. Wei, J. Wang and D. Ye, Analyst, 2021, 146, 1865–1871 RSC.
- Y. Jin, K. Xu, Y. Huang, H. Zhong and R. Zhao, Anal. Chem., 2021, 93, 2045–2052 CrossRef CAS PubMed.
- R. G. Boot, M. Verhoek, W. Donker-Koopman, A. Strijland, J. Van Marle, H. S. Overkleeft, T. Wennekes and J. M. F. G. Aerts, J. Biol. Chem., 2007, 282, 1305–1312 CrossRef CAS PubMed.
- H. G. Körschen, Y. Yildiz, D. N. Raju, S. Schonauer, W. Bönigk, V. Jansen, E. Kremmer, U. B. Kaupp and D. Wachten, J. Biol. Chem., 2013, 288, 3381–3393 CrossRef PubMed.
- A. Gehrlein, V. Udayar, N. Anastasi, M. L. Morella, I. Ruf, D. Brugger, S. Von Der Mark, R. Thoma, A. Rufer, D. Heer, N. Pfahler, A. Jochner, J. Niewoehner, L. Wolf, M. Fueth, M. Ebeling, R. Villaseñor, Y. Zhu, M. C. Deen, X. Shan, Z. Ehsaei, V. Taylor, E. Sidransky, D. J. Vocadlo, P.-O. Freskgård and R. Jagasia, Nat. Commun., 2023, 14, 2057 CrossRef CAS PubMed.
- D. Vercauteren, R. E. Vandenbroucke, A. T. Jones, J. Rejman, J. Demeester, S. C. De Smedt, N. N. Sanders and K. Braeckmans, Mol. Ther., 2010, 18, 561–569 CrossRef CAS PubMed.
- A. McCluskey, J. A. Daniel, G. Hadzic, N. Chau, E. L. Clayton, A. Mariana, A. Whiting, N. N. Gorgani, J. Lloyd, A. Quan, L. Moshkanbaryans, S. Krishnan, S. Perera, M. Chircop, L. Von Kleist, A. B. McGeachie, M. T. Howes, R. G. Parton, M. Campbell, J. A. Sakoff, X. Wang, J. Sun, M. J. Robertson, F. M. Deane, T. H. Nguyen, F. A. Meunier, M. A. Cousin and P. J. Robinson, Traffic, 2013, 14, 1272–1289 CrossRef CAS PubMed.
- D. J. Spiro, W. Boll, T. Kirchhausen and M. Wessling-Resnick, MBoC, 1996, 7, 355–367 CrossRef CAS PubMed.
- L. von Kleist, W. Stahlschmidt, H. Bulut, K. Gromova, D. Puchkov, M. J. Robertson, K. A. MacGregor, N. Tomilin, A. Pechstein, N. Chau, M. Chircop, J. Sakoff, J. P. von Kries, W. Saenger, H.-G. Kräusslich, O. Shupliakov, P. J. Robinson, A. McCluskey and V. Haucke, Cell, 2011, 146, 471–484 CrossRef CAS PubMed.
- M. C. Deen, P.-A. Gilormini and D. J. Vocadlo, Curr. Opin. Chem. Biol., 2023, 77, 102403 CrossRef CAS PubMed.
- A. Zotter, F. Bäuerle, D. Dey, V. Kiss and G. Schreiber, J. Biol. Chem., 2017, 292, 15838–15848 CrossRef CAS PubMed.
- F. Marceau, M.-T. Bawolak, R. Lodge, J. Bouthillier, A. Gagné-Henley, R. C. Gaudreault and G. Morissette, Toxicol. Appl. Pharmacol., 2012, 259, 1–12 CrossRef CAS PubMed.
- S. Lu, T. Sung, N. Lin, R. T. Abraham and B. A. Jessen, PLoS One, 2017, 12, e0173771 CrossRef PubMed.
- M. Hollemans, R. O. Elferink, G. Philip, D. Groot, A. Strijland and J. M. Tager, Biochim. Biophys. Acta, Biomembr., 1981, 643, 140–151 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d5cb00045a |
| This journal is © The Royal Society of Chemistry 2025 |

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:


