 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A stress-responsive p38 signaling axis in choanoflagellates†
Florentine U.
Rutaganira
 *ab,
Maxwell C.
Coyle‡
*ab,
Maxwell C.
Coyle‡
 d,
Maria H. T.
Nguyen
d,
Maria H. T.
Nguyen
 c,
Iliana
Hernandez
a,
Alex P.
Scopton
e,
Arvin C.
Dar§
c,
Iliana
Hernandez
a,
Alex P.
Scopton
e,
Arvin C.
Dar§
 e and
Nicole
King
e and
Nicole
King
 d
d
aDepartment of Biochemistry, Stanford University School of Medicine, Stanford, CA 94305, USA. E-mail: funr@stanford.edu
bDepartment of Developmental Biology, Stanford University School of Medicine, Stanford, CA 94305, USA
cDepartment of Biology, Stanford University, Stanford, CA 94305, USA
dHoward Hughes Medical Institute and the Department of Molecular and Cell Biology, University of California, Berkeley, CA 94720, USA
eDepartment of Oncological Sciences, Tisch Cancer Institute, Icahn School of Medicine at Mount Sinai, New York, NY 10029, USA
First published on 1st April 2025
Abstract
Animal kinases regulate cellular responses to environmental stimuli, including cell differentiation, migration, survival, and response to stress, but the ancestry of these functions is poorly understood. Choanoflagellates, the closest living relatives of animals, encode homologs of diverse animal kinases and have emerged as model organisms for reconstructing animal origins. However, efforts to identify key kinase regulators in choanoflagellates have been constrained by the limitations of currently available genetic tools. Here, we report on a framework that combines small molecule-driven kinase discovery with targeted genetics to reveal kinase function in choanoflagellates. To study the physiological roles of choanoflagellate kinases, we established two high-throughput platforms to screen the model choanoflagellate Salpingoeca rosetta with a curated library of human kinase inhibitors. We identified 95 diverse kinase inhibitors that disrupt S. rosetta cell proliferation. By focusing on one inhibitor, sorafenib, we identified a p38 kinase as a regulator of the heat shock response in S. rosetta. This finding reveals a conserved p38 function between choanoflagellates, animals, and fungi. Moreover, this study demonstrates that existing kinase inhibitors can serve as powerful tools to examine the ancestral roles of kinases that regulate modern animal development.
Introduction
Phenotypic screens with libraries of small molecules have revolutionized cell biology by providing chemical tools to study protein function.1–4 Because aberrant kinase activity can lead to human disease,5,6 many tools have been developed to inhibit kinase activity and detect protein phosphorylation. Small molecules that target the kinase active site coupled with assays of kinase inhibition have resulted in effective therapeutic strategies to counter the functions of misregulated kinases, including inhibition of aberrant cell growth and proliferation caused by oncogenic kinases.5,7We sought to test whether kinase-regulated physiology in choanoflagellates, the closest living relatives of animals, could be revealed by kinase inhibitors. Choanoflagellates possess homologs of diverse animal kinases (Fig. S1, ESI†)8–10 and, due to their phylogenetic placement, are well-suited for studies of the ancestral functions of animal cell signaling proteins.11,12 Indeed, a previous study showed that two broad-spectrum kinase inhibitors disrupt cell proliferation in the choanoflagellate Monosiga brevicollis.13 However, this study did not demonstrate whether kinases were directly targeted or identify specific pathways regulated by kinase signaling.
Using a library of well-characterized kinase inhibitors that vary in their human kinase inhibition profile, we treated cultures of Salpingoeca rosetta, a model choanoflagellate, in a multiwell format. We found that treatment of S. rosetta cultures with a set of kinase inhibitors disrupted cell proliferation and led to global inhibition of S. rosetta phosphotyrosine signaling. Using one of these inhibitors, sorafenib, followed by reverse genetics, we found that an S. rosetta p38 kinase homolog is activated by environmental stressors and signals downstream of sorafenib-inhibited kinases.
Results
Screening of a human kinase inhibitor library reveals small molecules that inhibit S. rosetta kinase signaling and cell proliferation
To investigate whether kinase activity regulates S. rosetta cell proliferation, we treated S. rosetta cultures with characterized human kinase inhibitors. Because the highest conservation between choanoflagellate and human kinases occurs in the kinase domain,8–10 we focused on kinase inhibitors that bind in the kinase active site. As a proof-of-concept, we first assayed staurosporine, a well-characterized broad-spectrum kinase inhibitor and inducer of cell death in diverse organisms.14,15 Our initial screen used flow cytometry to measure the density of S. rosetta cells individually treated with staurosporine in multiwell plates. Staurosporine significantly and reproducibly reduced S. rosetta cell density (Fig. S2A–C, ESI†) and tyrosine phosphorylation (Fig. S2D, ESI†) in a dose-dependent manner.After validating the flow cytometry pipeline with staurosporine, we expanded our study to screen 1255 inhibitors of diverse human kinases (Fig. S1 and Table S1, ESI†). We screened S. rosetta cultures with each of the molecules in the library at 10 μM and measured cell density at a 24-hour endpoint (Fig. 1A, B and Table S1, ESI†). This screen revealed 44 compounds (3.5% of the library; Fig. 1A and Fig. S3, ESI†) that significantly decreased S. rosetta cell counts compared to DMSO controls.
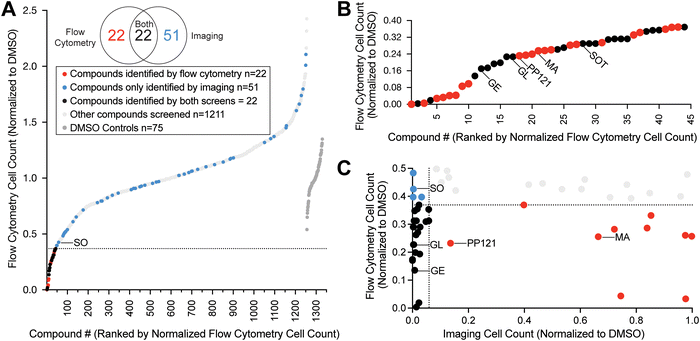 | ||
| Fig. 1 High-throughput screening of a small molecule library revealed inhibitors of S. rosetta cell proliferation. (A) Treatment of S. rosetta cultures with 1255 different small molecules (see Table S1, ESI†) resulted in a distribution of cell counts, assessed by flow cytometry, at the 24-hour endpoint. S. rosetta cell counts were normalized to the average of DMSO controls within the same plate (dark grey). Compounds determined to significantly inhibit S. rosetta cell proliferation by flow cytometry (based on two-tailed p-value <0.05 calculated from z-score), fall below the dotted line and are indicated in red. Compounds that were not detected as significant inhibitors by flow cytometry but were identified by imaging (based on two-tailed p-value <0.05 calculated from z-score) are in blue. Compounds that were not significant inhibitors for either screen are indicated in light grey. Sorafenib (SO), a focus of this study, is labeled. (B) The range of normalized cell counts measured by flow cytometry for compounds that significantly inhibited S. rosetta cell proliferation. Compounds that were the focus of further study – genistein (GE), glesatinib (GL), PP121, masitinib (MA), sotrastaurin (SOT) – are labeled. (C) Comparison of normalized values of compounds that inhibited S. rosetta cell proliferation, assessed by flow cytometry and the corresponding normalized values determined by imaging. Compounds determined to significantly inhibit S. rosetta cell proliferation (based on two-tailed p-value <0.05 calculated from z-score) by flow cytometry fall below the dotted line on the y-axis and by imaging, to the left of the dotted line on the x-axis. | ||
As a complementary assay, we pursued an imaging-based workflow to measure cell density. Although S. rosetta cell density could be measured by flow cytometry without staining reagents, our co-culture system (S. rosetta cultured with prey bacterium Echinicola pacifica) and treatment paradigm presented two potential sources of inaccuracy: compound aggregation due to low solubility in choanoflagellate media and choanoflagellate-sized clumps of bacterial biofilm. Therefore, we developed an imaging pipeline to enumerate S. rosetta cells by segmenting fixed-cell immunofluorescence micrographs at a 48-hour endpoint. Our imaging pipeline distinguished wells with staurosporine-treated cells from DMSO controls (Fig. S2E and F, ESI†) with comparable z′ standard statistics to flow cytometry (Fig. S2B and E, ESI†).16 This orthogonal approach identified 22 compounds (1.8% of the library) that overlapped with the flow cytometry screen and 51 additional molecules (4.4% of the library) that inhibited S. rosetta cell proliferation but were not identified in our flow cytometry screen (Fig. 1A, C and Fig. S2G, S3, ESI†). In total, 95 ATP-competitive inhibitors of human protein and lipid kinases (Fig. S4A–C, ESI†) that ranged in selectivity (Fig. S4D, ESI†) were identified as potential inhibitors of S. rosetta cell proliferation.
Choanoflagellates are predicted to express kinases that regulate animal cell growth, including mitotic kinases,17 the serine–threonine kinase Akt,18 and a diverse set of tyrosine kinases.8 We identified GSK461364 and Volasertib, inhibitors of polo-like kinase 1 (PLK1), a mitotic kinase, in both screens (Fig. S3, ESI†). In addition, both screens identified diverse inhibitors of human Akt and tyrosine kinases (Fig. S3, ESI†). We also identified inhibitors of S. rosetta cell proliferation that disrupt human kinase signaling indirectly, i.e. without binding to a kinase (Fig. S3, ESI†). For example, genistein, a natural product that indirectly alters kinase activity,19,20 inhibited S. rosetta cell proliferation and was previously shown to inhibit the growth of M. brevicollis.13
Some inhibitors of S. rosetta cell proliferation also disrupt S. rosetta phosphotyrosine signaling
After identifying human tyrosine kinase inhibitors that inhibited S. rosetta cell proliferation, we used biochemical methods to determine if the phenotype observed was correlated with inhibition of S. rosetta kinase activity. Widely available phospho-specific antibodies that distinguish phosphorylated amino acids from their unphosphorylated cognates can reveal kinase activity in diverse organisms13,21–24 and have been used to detect tyrosine phosphorylation of peptides or animal proteins by heterologously expressed choanoflagellate kinases.25–28 We triaged our set of 95 identified proliferation inhibitors to focus on those compounds that inhibit human tyrosine kinases (Fig. S3, ESI†), as opposed to those that inhibit serine or threonine phosphorylation, in part because of the relative lack of specificity of commercially-available phosphoserine and phosphothreonine antibodies.22–24In particular, we focused on four inhibitors – sorafenib, glesatinib, masitinib and PP121 – that were identified by either or both screening paradigms (Table S1 and Fig. S3, ESI†) and have a narrower range of kinase targets than staurosporine.29–32 In humans, sorafenib, glesatinib, masitinib and PP121 inhibit select receptor tyrosine kinases (RTKs), among other targets. Sorafenib also inhibits some non-receptor tyrosine kinases, tyrosine kinase-like serine–threonine kinases, and p38 stress-responsive kinases.30 Masitinib and PP121 also inhibit Src kinase and PP121 additionally inhibits PI3K.29–31 Specific residues shared in the kinase domains of human and choanoflagellate tyrosine kinases suggested that these inhibitors might effectively inhibit choanoflagellate tyrosine kinases. For example, choanoflagellate tyrosine kinases have residues within the predicted active site that are necessary for kinase activity in other organisms (e.g. “K” in VAIK, “D” in HRD and DFG) (Fig. S5A, ESI†)8,33–35 and additional residues that confer sensitivity towards tyrosine kinase inhibitors (Fig. S5B–D, ESI†).36,37
Of the four inhibitors tested, S. rosetta cultures were most sensitive to sorafenib and glesatinib. Although 1 μM masitinib and PP121 were sufficient to reduce S. rosetta cell proliferation over the first 40 hours of treatment (Fig. 2A), masitinib- and PP121-treated cultures recovered within 85 hours. Masitinib and PP121 are effective in human culture media for 48–72 hours,31,38 so we have no reason to believe this recovery is due to reduced inhibitor stability, although we can’t rule it out. Of note, both compounds were found to be ineffective inhibitors of cell proliferation in the imaging pipeline, which had an endpoint of 48 hours (Fig. 1C).
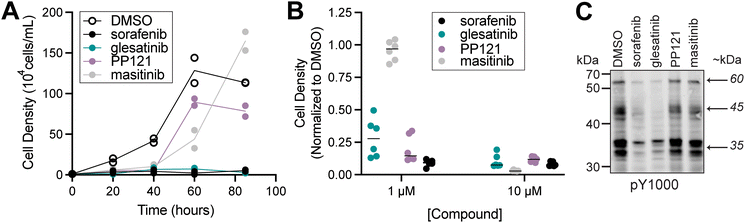 | ||
| Fig. 2 Glesatinib and sorafenib, two multi-target tyrosine kinase inhibitors, disrupt S. rosetta cell proliferation and tyrosine phosphosignaling. (A) Treatment of S. rosetta cultures with 1 μM sorafenib and glesatinib led to a complete block of cell proliferation, while treatment with 1 μM masitinib or PP121 led to a partial reduction in cell proliferation relative to DMSO-treated cultures. Two biological replicates were conducted per treatment, and each point represents the mean of three measurements from each biological replicate. For timepoints at 40, 60, and 85 hours, cell densities of inhibitor-treated cultures were significantly different from vehicle (DMSO) (p-value <0.01). Significance was determined by a two-way ANOVA multiple comparisons test. (B) S. rosetta cultures treated with 1 μM or 10 μM sorafenib, glesatinib, or PP121 for 24 hours had reduced normalized cell density, whereas masitinib only had reduced normalized cell density at 10 μM. Normalized cell densities were determined to be reduced if differences between treatments and vehicle (DMSO) were significant (p-value <0.01) Significance was determined by determined by a two-way ANOVA multiple comparisons test. Movies show S. rosetta cells treated with 10 μM glesatinib that undergo cell lysis (Movie S1, ESI†) and sorafenib, that have cell body deformation (Movies S2 and S3, ESI†), in comparison to DMSO control (Movie S4, ESI†). (C) Western blot analysis of S. rosetta cultures treated with 1 μM sorafenib and glesatinib for 1 hour showed a decrease in tyrosine phosphorylation of proteins at ∼60 kDa, ∼45 kDa, and ∼35 kDa (indicated by arrows and detected with pY1000 anti-phosphotyrosine antibody) compared to vehicle (DMSO) control. Masitinib and PP121 did not reduce the phosphotyrosine signal. | ||
In contrast, sorafenib and glesatinib inhibited cell proliferation throughout the 85-hour growth experiment at 1 μM (Fig. 2A) and decreased cell density at 1 μM and 10 μM (Fig. 2B). Glesatinib treatment induced cell lysis (Movie S1, ESI†), whereas sorafenib induced cell body elongation (Movies S2 and S3, ESI†), in comparison to the DMSO control (Movie S4, ESI†). Importantly, treatment of S. rosetta cultures with 1 μM sorafenib or glesatinib led to a global decrease in phosphotyrosine signal, while treatment with 1 μM masitinib or PP121 did not decrease phosphotyrosine levels as detected by western blot (Fig. 2C).
These findings showed that some kinase inhibitors, including sorafenib and glesatinib, could disrupt both S. rosetta cell proliferation and tyrosine kinase signaling. To further investigate patterns among human kinase inhibitors that showed this effect and identify kinases that might be relevant for the observed inhibition, we tested a panel of 17 human TK inhibitors (Fig. S6, ESI†) that share overlapping kinase targets with sorafenib and glesatinib. Of the 17 compounds tested, treatment with four additional small molecules (regorafenib, AD80, milciclib, and vemurafenib) led to a global decrease in phosphotyrosine staining (Fig. S6A, “*”, ESI†) but not phosphoserine and phosphothreonine staining (Fig. S7, ESI†) as detected by western blot. These four inhibitors mildly reduced the rate of cell proliferation (Fig. S6B and S8, ESI†). Other tyrosine kinase inhibitors impaired S. rosetta cell proliferation but did not disrupt phosphotyrosine signaling, including PP2 (Fig. S6A, C and S8, ESI†), consistent with prior findings in M. brevicollis.13
Sorafenib binds to S. rosetta p38 kinase
To identify specific S. rosetta kinases whose activity might regulate S. rosetta cell physiology, we focused on sorafenib, an inhibitor that binds TKs and serine–threonine kinases30 with well-characterized structure–activity relationships.39,40 We started by using ActivX ATP and ADP probes41,42 to covalently enrich for kinases and high-affinity kinase interactors present within S. rosetta lysates after pretreatment with vehicle (DMSO) or sorafenib. While sorafenib is predicted to bind to the active site of a select subset of kinases, ActivX probes target ATP and ADP binding proteins more broadly. Therefore, pretreatment of S. rosetta lysates with sorafenib was predicted to competitively inhibit binding of ActivX probes to sorafenib targets. By identifying kinases from S. rosetta lysates that were more often bound to ActivX probes after DMSO pretreatment of S. rosetta lysate compared with those that were recovered by ActivX probes after sorafenib pretreatment, we aimed to identify likely targets of sorafenib. Through this strategy, we identified a predicted S. rosetta p38 kinase (Fig. S9A, ESI†) that bound 10-fold less well to ActivX probe in the presence of sorafenib (Fig. 3 and Table S2, ESI†). Hereafter, we refer to this serine–threonine kinase as Sr-p38.43 | ||
| Fig. 3 S. rosetta p38 binds to sorafenib. The ActivX ATP probe was used to pull down kinases from S. rosetta lysates that were pretreated with either DMSO or the ATP-competitive inhibitor sorafenib. We found that pretreatment with sorafenib reduced the level of p38 recovered using the ActivX ATP probe, indicating that sorafenib and p38 interact and outcompete ActivX ATP probe binding. Kinases plotted are only those that were identified in both vehicle and sorafenib pre-treatments. For full kinase enrichment list, see Table S2 (ESI†), and for alignment of S. rosetta p38 with those from animals and fungi, see Fig. S9A (ESI†). | ||
Kinase enrichment with ActivX probes and other approaches have previously identified human p38 kinases as primary targets of sorafenib.39,44 In animals, sorafenib preferentially binds kinases with threonine gatekeeper residues,39 including tyrosine kinases and two of four vertebrate p38 kinase paralogs with threonine gatekeepers39 (Fig. S9A, ESI†). Sr-p38, contains a threonine gatekeeper (Fig. S9A, ESI†) and lysine residues necessary for ActivX probe binding. Because sorafenib blocked ActivX probe binding to Sr-p38, we infer that sorafenib binds to Sr-p38 directly. Although sorafenib treatment reduces S. rosetta cell proliferation and sorafenib binds to Sr-p38, these two findings did not directly implicate Sr-p38 in the regulation of cell proliferation. Therefore, we next sought to understand how Sr-p38 regulates S. rosetta cell physiology.
S. rosetta p38 is a heat-responsive kinase
Environmental stressors activate p38 kinases in animals and fungi43,45–51 and p38 kinase is present in diverse choanoflagellates (Fig. S9B, ESI†). However, the roles of p38 kinase in choanoflagellate biology are unknown. Although a previous study identified a nutrient-sensitive protein in M. brevicollis with a molecular weight similar to p38 kinases,13 the identity and function of this protein were not studied directly.We wondered if stressors relevant to choanoflagellates would activate Sr-p38 signaling. Some choanoflagellates, including S. rosetta, live in sun-lit water zones that undergo daily and yearly fluctuations in temperature and nutrients.52–55 Antibodies specific to phospho-p38 are commercially available, allowing us to detect p38 phosphorylation under different conditions. We found two – p38 MAPK (pThr180/pTyr182) from Biorad and anti-ACTIVE® p38 from Promega – that recognized a heat stress-induced protein with minimal background. By generating knockouts of Sr-p38 with a recently established protocol58 (Methods), we confirmed that the heat stress-induced protein detected by these antibodies was Sr-p38 (Fig. 4A–C). (Unfortunately, the anti-ACTIVE® p38 antibody from Promega is no longer commercially available, but we provide these results as additional evidence for the connection of Sr-p38 to S. rosetta cell physiology.)
 | ||
| Fig. 4 S. rosetta p38 phosphorylation is induced by environmental stressors. (A) Strategy for generating Sr-p38 and Sr-JNK knockout cell lines. The Sr-p38 and Sr-JNK loci were targeted by a guide RNA complexed with Cas9 that anneals before the kinase domain and directs Cas9 to introduce a double-strand break downstream of codon 15 (codon 15 is serine in Sr-p38 and alanine in Sr-JNK), indicated by (*). The Cas9-guide RNA complex was coupled with a double-stranded homology-directed repair to introduce a palindromic premature termination stop sequence and a puromycin resistance cassette. The resulting truncated proteins, Sr-p381–15 and Sr-JNK,1–15 lack the kinase domain and phosphorylation sites (indicted by the extended circle) recognized by both phospho-p38 antibodies used in this study. Protein diagrams were created with IBS 2.0.56 (B) Heat shock induces Sr-p38 phosphorylation in wild-type cells and the phospho-p38 signal is recognized by the anti-ACTIVE® p38 antibody (Promega #V1211). This phospho-p38 signal is decreased in Sr-p381–15 knockout cell lines but not Sr-JNK1–15 knockout lines, indicating that the Anti-ACTIVE® p38 antibody (Promega #V1211) detects Sr-p38 and that Sr-p38, but not Sr-JNK, responds to heat shock. Three biological replicates of wild-type cells, ten clones of Sr-p381–15 and five clones of Sr-JNK1–15 strains were incubated at 37 °C for one hour. Lysates from the treated cultures were analyzed by western blot with the Anti-ACTIVE® p38 antibody and quantified by densitometry to identify if any changes in Sr-p38 phosphorylation occurred. Significance was determined by a one-way ANOVA multiple comparisons test between wild-type cells and Sr-p381–15 or Sr-JNK1–15. (C) Similar to (B), the phospho-p38 signal recognized by the p38 MAPK pThr180/pTyr182 (Biorad #AHP905) antibody in heat shocked wild-type cells is decreased in Sr-p381–15 knockout cells but not Sr-JNK1–15 knockout cells. (D) S. rosetta cells, normally cultured at 22 °C were incubated at 37 °C to induce heat shock. Lysates from the treated cultures were analyzed by western blot with the Anti-ACTIVE® p38 antibody (Promega #V1211) to identify if any changes in Sr-p38 phosphorylation occurred. 30 minutes of heat shock was sufficient to induce Sr-p38 phosphorylation. (E) S. rosetta cells were treated with hydrogen peroxide, a form of oxidative stress for 10 min and 30 min 10 min of treatment with 0.5 M H2O2 at 22 °C was sufficient to induce Sr-p38 phosphorylation detected by the anti-ACTIVE® p38 antibody (Promega #V1211) (F) Sr-p381–15 and Sr-JNK1–15 strains grow similarly to wild-type. Four wild-type cultures and four randomly selected Sr-p381–15 and Sr-JNK1–15 clones were grown in 24-well plates over a 96-hour growth course and showed similar growth. Significance was determined by a two-way ANOVA multiple comparisons test. (G) The induction of Sr-p38 phosphorylation by heat shock was kinase-dependent. S. rosetta cultures pretreated with 10 μM or 1 μM sorafenib for 30 minutes followed by 30 minutes of heat shock at 37 °C and probed with the Anti-ACTIVE® p38 antibody (Promega #V1211) had decreased Sr-p38 phosphorylation. APS6-46 treated cultures were not different from vehicle (DMSO) control. In (D), (E) and (G), 4–12% bis-Tris SDS-PAGE gels were used to resolve the bands observed. | ||
To investigate if Sr-p38 is activated in response to environmental stressors, we exposed S. rosetta cultures to heat shock and oxidative stress. When S. rosetta cells cultured at ambient temperature were subjected to heat and oxidative stress, we observed an increase in phosphorylation (Fig. 4B–E and Fig. S10A, B, ESI†). Within 30 minutes of heat shock at 37 °C, the phospho-p38 signal increased relative to pre-treatment (Fig. 4D and Fig. S10A, ESI†). We also observed an increase in phosphorylation of an ∼45 kDa protein in cell lysates treated with 0.5 M hydrogen peroxide (Fig. 4E and Fig. S10B, ESI†).
In animals, three stress-activated kinases mediate responses to heat shock and oxidative stress: p38, c-Jun N-terminal kinase (JNK), and extracellular signal-related kinase 5 (ERK5). Upstream dual-specificity kinases (MAP2Ks) activate these kinases through dual phosphorylation of threonine and tyrosine in a short motif: “TGY,” “TPY,” and “TEY” for p38, JNK, and ERK5, respectively49,57 (Fig. S9A, ESI†). Although neither of the phospho-specific p38 antibodies used in this study recognize phosphorylated human JNK or ERK5, the size of the band recognized by these antibodies (∼45 kDa) is closer to the predicted size for S. rosetta JNK (39 kDa) than for S. rosetta p38 (60 kDa). To test whether Sr-p38 and Sr-JNK were responsive to heat shock and oxidative stress, we also generated Sr-JNK knockout cell lines (Fig. 4A).
The phospho-p38 signal was reduced in heat-shocked Sr-p381–15 lines but not in Sr-JNK1–15 lines (Fig. 4B, C and Fig. S12A, B, ESI†), demonstrating that Sr-p38 is the stress-responsive protein detected in our assays. In contrast, the hydrogen peroxide-induced protein phosphorylation signal was preserved in both Sr-p381–15 lines and Sr-JNK1–15 lines (Fig. S12C and D, ESI†). Therefore, heat shock induces phosphorylation of Sr-p38, but not Sr-JNK. These findings reveal that the connection between heat shock stress and the activation of p38 kinase is conserved between yeast, animals, and choanoflagellates.43,59,60
Sorafenib inhibits S. rosetta cell proliferation separately from the stress-responsive p38 kinase signaling axis
Surprisingly, we found that the growth rates of Sr-p381–15 and Sr-JNK1–15 clones were indistinguishable from those of wild-type cells, indicating that Sr-p38 and Sr-JNK are dispensable for the regulation of cell proliferation (Fig. 4F). Moreover, mutants with reduced sorafenib binding39 maintained sensitivity to sorafenib (Fig. S13, ESI†). Together, these two findings suggest that kinases other than Sr-p38 were more relevant to sorafenib's effect on proliferation.Previous studies have linked phosphotyrosine signaling to p38 kinase activation in animals in response to multiple stimuli (e.g. heat shock, oxidative stress, growth factors, ultraviolet light).49,61 Sorafenib blocks this signaling axis by inhibiting p38 and upstream kinases (e.g. tyrosine kinases, dual-specificity kinases).39,40 Therefore, we set out to test whether the signaling axis between upstream kinases and p38 in animals is conserved in S. rosetta. To this end, we tested whether sorafenib could reduce the observed phosphorylation of Sr-p38 in S. rosetta cultures subjected to heat shock. As a control, we used APS6-46, a sorafenib analog that shares sorafenib's core structure but has modifications that make it too large to bind to most sorafenib kinase targets.40 Under standard growth conditions, APS6-46 did not inhibit S. rosetta tyrosine phosphorylation or cell division (Fig. S14, ESI†), and a previous study found that similar sorafenib analogs could control for off-target inhibition of non-kinase proteins by sorafenib.62 In heat-shocked cells, Sr-p38 signaling was not activated in S. rosetta cultures pretreated with sorafenib, whereas cultures pretreated with APS6-46 (Fig. 4G) or selective p38 kinase inhibitors (Fig. S15, ESI†) retained Sr-p38 phosphorylation comparable to the DMSO control. Because sorafenib treatment blocked Sr-p38 activation and inhibited phosphotyrosine signaling, we infer that sorafenib blocks Sr-p38 signal transduction through the inhibition of upstream kinases that transduce the heat stress response in S. rosetta (Fig. 5). Future studies will be required to identify additional binding targets of sorafenib that regulate S. rosetta cell proliferation and heat-responsive activation of Sr-p38 (Fig. 5).
Discussion
To investigate the relevance of kinase signaling in choanoflagellate cell physiology, we have established two high-throughput phenotypic screens of cells treated with a small molecule library. By treating S. rosetta cultures with validated human kinase inhibitors, we uncovered molecules that revealed the physiological relevance of kinases as regulators of S. rosetta cell proliferation. Moreover, we identified a biologically relevant environmental stressor that activates p38 kinase signaling in S. rosetta.Until recently, the functions of stress-responsive kinases have only been characterized in animals (p38, JNK, and ERK5)49,57 and fungi (Hog1, a homolog of p38).43,59,60 Our discovery that S. rosetta phosphorylates p38 and other proteins in response to heat and oxidative shock (Fig. 4B–E) demonstrates that choanoflagellates undergo stress-responsive signaling. Because the phosphorylation of Sr-p38 can be inhibited by sorafenib (Fig. 4G), we infer that Sr-p38 functions within a signaling axis downstream of sorafenib-targeted kinases. Further exploration of this heat-responsive pathway in S. rosetta will be necessary to uncover regulators upstream of Sr-p38 and if those regulators mirror function in animals or fungi.
Our approach of using sorafenib to uncover the role of Sr-p38 and other sorafenib-targeted kinases in S. rosetta allowed us to assess Sr-p38's role before undertaking targeted genetics. Vertebrates express four p38 paralogs; the p38α knockout is embryonic lethal, whereas knockouts for the other p38 paralogs are viable.49S. rosetta is predicted to encode a family of stress-responsive kinases, including Sr-p38, Sr-JNK, and a homolog of ERK5 (EGD76774) (Fig. S9A, ESI†). We observed an increase in phosphorylation of a ∼45 kDa protein that was recognized by two independent phospho-p38 antibodies in S. rosetta cultures that were subjected to heat shock (Fig. 4A, B and Fig. S10A, B, ESI†). The signal was lost in Sr-p381–15 knockout strains (Fig. 4B–D and Fig. S12A, B, ESI†), implicating Sr-p38, in the heat shock response. The viability of Sr-p381–15 cells and the sensitivity of Sr-p38 mutants with reduced sorafenib binding (Fig. 4F and Fig. S13C, ESI†) suggest that sorafenib's impact on cell proliferation is mediated by binding to other kinases.
Choanoflagellates have dynamic life histories and express diverse kinase families, including p38 kinases, that are found in animals.8–10,17,43,63 How kinases regulate additional aspects of choanoflagellate physiology, including life history transitions, remains to be investigated. We infer that fast-acting inhibition of kinase activity will be a powerful approach to study the roles of kinases that regulate cell state transitions in choanoflagellates. Because kinase inhibitors allow the enzymatic activity of kinases to be disrupted while preserving kinase localization and scaffolding functions, using small molecules as tools can distinguish whether kinase catalytic activity or other kinase functions are necessary for choanoflagellate development.9,64,65 Insight into the roles of individual kinases during the emergence of new cell–cell signaling networks (e.g. receptor tyrosine kinase signaling in the last common ancestor of animals, choanoflagellates, and their closest relatives8–10,13,63,66,67) will be fundamental to understanding the contributions of kinase signaling to the origin of animal multicellularity.
Materials and methods
Co-culturing of S. rosetta with the prey bacterium Echinicola pacifica
Choanoflagellates are bacterivores and require prey bacteria that are co-cultured in choanoflagellate media.68Echinicola pacifica, a Bacteriodetes bacterium, grown in seawater-based media enriched with glycerol, yeast extract, and peptone sustains S. rosetta growth.69 This co-culture of S. rosetta and E. pacifica, publicly available from the American Type Culture Collection (ATCC) as ATCC PRA-390 and also known as “SrEpac”,69 was used in this study.High-throughput chemical screening in S. rosetta
To quantify changes in S. rosetta cell proliferation after small molecule treatment, we established a high-throughput screening pipeline.We first assembled a library of 1255 compounds (Table S1, ESI†) from commercial (Selleckchem Kinase Inhibitor Library Catalog #L1200 Lot: Z1316458) and academic (Kevan M. Shokat, University of California, San Francisco) sources. 98% of molecules in the library were characterized as human kinase inhibitors and 2% were compounds that are cytotoxic to other protists. Most of the kinase inhibitors in the library modulate human kinase activity by binding to the kinase active site and are ATP-competitive. Because we did not know if inhibitors designed to bind to human kinases would bind to choanoflagellate kinase homologs with the same potency or selectivity, we chose inhibitors with a range of selectivity: 75% of the human kinome is targeted by at least one inhibitor in the library, and the library includes inhibitors of all classified human kinase groups (Fig. S1, ESI†). Compounds dissolved at 10 mM in dimethyl sulfoxide (DMSO, Sigma #D8418) were placed into individual wells of 384-well deep well master plates (Corning #3342). We generated deep well stock plates with solutions that could be directly transferred to assay plates. Liquid handling (Agilent V11 Bravo) was used to dilute compound master plates (containing 10 mM compound in 100% DMSO) into deep well stock plates (containing 450 μM compound in 4.5% DMSO).
In a primary screen, S. rosetta cell counts were determined by analysis of acquired flow cytometry events after a 24-hour incubation. Assay plates were generated by plating 2 μL of the deep well stock plates into 384-well assay plates (Thermo Scientific #142761 using the Agilent V11 Bravo). 88 μL of SrEpac cultured in high-nutrient media (5% Sea Water Complete)70 at exponential phase (∼9 × 105 cells per mL) was diluted to 2 × 104 cells per mL in high-nutrient media and dispensed into the assay plate (ThermoFisher Multidrop™ Combi with long standard dispensing tube cassette #24072677) to treat the SrEpac culture at 10 μM compound and 0.1% DMSO. After a 24-hour incubation, assay plates were individually placed into an autosampler (BD Biosciences High Throughput Sampler, HTS) running in high-throughput mode. 40 μL of each well was mixed twice (at 180 μL second−1), and 10 μL of cells from each well were loaded onto a flow cytometer (BD Biosciences LSR II) at 1 μL second−1. In between each well, the autosampler needle was washed with 400 μL of sheath fluid (1× phosphate-buffered-saline pH 7.4). Loaded cell samples were acquired on the cytometer with forward scatter (FSC) and side scatter (SSC) parameter voltages set to 538 and 308, respectively. A polygon gate from a DMSO well within a plate (D23 for Plate 1 and C23 for Plates 2–4 due to a shift in the distribution of observed events) was used to analyze events and quantify cell counts for all wells in an individual assay plate using FlowJo v10.8™ (BD Biosciences) (Fig. S2A, ESI†).
In a secondary screen, S. rosetta cell counts were determined by enumerating segmented cell objects from immunofluorescence images taken on an Opera Phenix high-content imager after a 48-hour incubation. Assay plates were generated by adding 4 μL of compound in the deep well stock plates into 96-well assay plates containing 176 μL of SrEpac in low-nutrient medium (1% cereal grass medium and 1% sea water complete71) using an electronic multichannel pipetter (Rainin E4 Multi Pipette E8-20XLS+). Cells were initially expanded in high-nutrient media containing 4% cereal grass medium and 4% sea water complete71 and at exponential phase (∼1 × 106 cells per mL), diluted to 2 × 104 cells per mL with AK-seawater. After a 48-hour incubation, cells were mixed in a thermomixer for two minutes at 800 rpm at room temperature to dislodge any cells attached to biofilm at the bottom of the 96-well plate. 100 μL of cells were transferred to poly-D-lysine (Sigma # P6407) coated 384-well imaging plates (PerkinElmer Cell Carrier Ultra Plates #6057302) using an electronic multichannel pipetter (Rainin E4 Multi Pipette Multi E12-200XLS+). To optimize the screen, after 40 minutes of adherence, 1 μL of FM 1–43X mixture (1 μL of 500 μg mL−1 mixture of FM 1–43X dye made by dissolving tube in 200 μL of methanol) was added to the cells and incubated for 15 minutes. 50 μL of the cell dye-mixture was removed followed by fixation and washing as described next for the full screen. For the full screen, after 40 minutes of adherence, 50 μL of cells were removed, and the remaining 50 μL were washed once with 50 μL 4× PBS. After incubating in the 4× PBS for 5 minutes, 50 μL was removed. Cells were fixed for 20 minutes at room temperature by adding 50 μL of 4% formaldehyde in PEM buffer (100 mM PIPES pH 7, 1 mM EGTA, 1 mM MgSO4). After fixation, cells were washed by removing 50 μL of solution from the plate and adding 50 μL of PEM buffer three times. After the final wash, 75 μL of solution was removed. At this point, cells for the optimization screen were imaged on a PerkinElmer Opera Phenix with the following imaging specifications for the fluorescein (FITC) channel: 20× water objective (NA 1.0, working distance 1.7 mm, 646 μm2 field of view), 60 ms and a three plane z stack at −8 μm, −6 μm and −4 μm. For the full screen, after washing the fixative, cells were blocked by adding 75 μL of 2% BSA and 0.6% Triton-X100 in PEM for 30 minutes at room temperature. After blocking 25 μL was removed and 25 μL of primary antibody solution in 1% BSA and 0.3% Triton-X100 in PEM was added to stain the cell body and flagella overnight at 4 °C (anti-tubulin, Abcam #ab6161, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 dilution). Due to the amount of time required for imaging, plates were staggered and processed one plate each day. On each imaging day, a plate was brought to room temperature and the primary antibody was washed three times by adding 50 μL of 1% BSA and 0.3% Triton-X100 in PEM and removing 50 μL of solution from the plate three times. Secondary antibody (Goat anti-rat Alexa Fluor 488, Invitrogen #A-11006, 1
1000 dilution). Due to the amount of time required for imaging, plates were staggered and processed one plate each day. On each imaging day, a plate was brought to room temperature and the primary antibody was washed three times by adding 50 μL of 1% BSA and 0.3% Triton-X100 in PEM and removing 50 μL of solution from the plate three times. Secondary antibody (Goat anti-rat Alexa Fluor 488, Invitrogen #A-11006, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 dilution) and nuclear stain (DRAQ5, Thermo Scientific #62251, 1
300 dilution) and nuclear stain (DRAQ5, Thermo Scientific #62251, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 dilution) were added in 25 μL of 1% BSA and 0.3% Triton-X100 in PEM and incubated for 2 hours at room temperature. After incubation, the secondary antibody was washed three times by adding 50 μL of PEM and removing 50 μL of solution from the plate three times. To stain the cell collar and obtain a second cell body marker, rhodamine phalloidin (Invitrogen #R415, 1
500 dilution) were added in 25 μL of 1% BSA and 0.3% Triton-X100 in PEM and incubated for 2 hours at room temperature. After incubation, the secondary antibody was washed three times by adding 50 μL of PEM and removing 50 μL of solution from the plate three times. To stain the cell collar and obtain a second cell body marker, rhodamine phalloidin (Invitrogen #R415, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300 dilution) and Cell Tracker cmDIL (Invitrogen # C7000, 250 nM) were added for 25 minutes. To preserve the staining, 50 μL of solution was removed from the plate and 50 μL of 50% glycerol in PEM was added. 22 fields of view in 4 planes (each plane separated by 1 μm) of each well in the 384-well plate were imaged with a 40× water immersion objective (NA 1.1, working distance 0.62 mm, 323 μm2 field of view) on a PerkinElmer Opera Phenix with optimized imaging specifications for each channel (Alexa 488 – tubulin – 20 ms; TRITC – Cell Tracker cmDIL/rhodamine phalloidin – 100 ms; brightfield – 100 ms; Alexa Fluor 647 – DRAQ5/Nuclei – 1 s). Due to restrictions on the available image area on the Opera Phenix, columns 1–23 of each 384-well plate were imaged first followed by a second scan with column 24 alone.
300 dilution) and Cell Tracker cmDIL (Invitrogen # C7000, 250 nM) were added for 25 minutes. To preserve the staining, 50 μL of solution was removed from the plate and 50 μL of 50% glycerol in PEM was added. 22 fields of view in 4 planes (each plane separated by 1 μm) of each well in the 384-well plate were imaged with a 40× water immersion objective (NA 1.1, working distance 0.62 mm, 323 μm2 field of view) on a PerkinElmer Opera Phenix with optimized imaging specifications for each channel (Alexa 488 – tubulin – 20 ms; TRITC – Cell Tracker cmDIL/rhodamine phalloidin – 100 ms; brightfield – 100 ms; Alexa Fluor 647 – DRAQ5/Nuclei – 1 s). Due to restrictions on the available image area on the Opera Phenix, columns 1–23 of each 384-well plate were imaged first followed by a second scan with column 24 alone.
For both assays, the quantified cell counts were normalized to the average cell count of all DMSO wells within an individual plate to account for any plate-to-plate variation. Compounds were determined to significantly inhibit S. rosetta cell proliferation if the resulting normalized cell count had a p-value <0.05 (based on two-tailed p-value calculated from z-score of all treated samples). The resulting normalized cell count data were plotted using GraphPad Prism 9.3.1™ (GraphPad San Diego, California, USA) (Fig. 1 and Fig. S2B, E, G, ESI†).
Follow-up S. rosetta cell proliferation assays in response to compound treatment
Compound treatments post-library screening were conducted with commercially available inhibitors; sorafenib analogs previously synthesized;40 AD80 previously synthesized;72 and imatinib, PP1, and sunitinib provided by K. Shokat (UC San Francisco). Sorafenib (#S1040), regorafenib (#S5077), dasatinib (#S7782), PP2 (#S7008), and milciclib (#S2751) were purchased from Selleckchem. Glesatinib (#HY-19642A), masitinib (#HY-10209), lapatinib (#HY-50898), PP121 (#HY-10372), gliteritinib (#HY-12432), brigatinib (#HY-12857), RAF265 (#HY-10248), vemurafenib (#HY12057), skepinone-L (#HY-15300), BIRB 796 (#HY-10320), were purchased from MedChem Express. SU6656 (#13338) was purchased from Cayman Chemical. R406 (#A5880) was purchased from ApexBio Technlology.SrEpac cultured in high-nutrient media (4% sea water complete with 4% cereal grass)71 in exponential phase (∼5 × 105–9 × 105 cells per mL) was diluted to lower density (1 × 104 cells per mL), and 1 mL or 100 μL of cells were plated into 24-well or 96-well multiwell plates (Corning #3526, Thermo Scientific #260251), respectively. Cells were treated with compound by adding 1 μL of a 1000× compound stock in DMSO to a well in the 24-well plate or adding 1 μL of a 100× compound stock in DMSO. Equal volumes of DMSO were added to vehicle control wells so (v/v%) DMSO in controls was the same as compound treated wells. At set timepoints, cells were harvested. For cell assays in 24-well plates, cells in the well were pipetted up and down to resuspend the well and the well contents were transferred to a 1.5 mL Eppendorf tube. Cells were fixed by adding 40 μL of 37% formaldehyde (methanol-stabilized, Sigma-Aldrich #252549). For 96-well plates, 1 μL of 37% formaldehyde was added to each well by using a multichannel pipette, and a pierceable aluminum plate seal (USA Scientific TempPlate® Sealing Foil) was added to cover the plate. The plate was vortexed at 2000 rpm (24-well plates) or 3000 rpm (96-well plates) in a plate vortexer (Eppendorf ThermoMixer C) to ensure equal fixation of cells in the well. Fixed cells were immediately counted or placed at 4 °C for up to 2 weeks before counting. The cell density of sample timepoints along the full growth course was determined by analysis of micrographs taken on a Widefield microscope (Carl Zeiss AG Axio Observer.Z1/7, Oberkochen, Germany),71 or brightfield imaging using a cell counter (Logos Biosystems LUNA-FL™). The resulting normalized cell density data at each timepoint was plotted using GraphPad Prism 9.3.1™ (GraphPad San Diego, California, USA) (Fig. 2A, 4F and Fig. S6B–E, S13B, S14C, ESI†). For comparisons between growth curves and phosphotyrosine signal (see assessment of S. rosetta kinase signaling by western blotting) the area under the growth curve (AUC) was analyzed using GraphPad Prism 9.3.1™ (GraphPad San Diego, California, USA) with baseline at Y = 0 and minimum peak height >10% above the baseline to maximum Y value (Fig. S8 and S14B, ESI†).
For dose–response assays, SrEpac cultured in high-nutrient media (5% sea water complete70 or 4% sea water complete with 4% cereal grass71) in exponential phase (∼5 × 105–9 × 105 cells per mL) was diluted to lower density. Starting density used varied based on treatment length: for 24-hour treatments and less, cells were plated at 2 × 105 cells per mL; for 24–48 hour treatments, cells were plated at 1 × 105 cells per mL; and for 48+ hour treatments, cells were plated at 5 × 104 cells per mL. Cell density was determined at the treatment endpoint using the same approach as the treatment growth curves described in the preceding paragraph. The resulting normalized cell density data at each dose was plotted using GraphPad Prism 9.3.1™ (GraphPad San Diego, California, USA), (Fig. 2B and Fig. S2C, S13C, S14D, ESI†).
Live imaging of treated S. rosetta cultures
Cells were imaged by differential interference contrast (DIC) using a 100× (oil immersion, Plan-Apochromat, 1.4 NA) Zeiss objective mounted on a Zeiss Observer Z.1 with a Hamamatsu Orca Flash 4.0 V2 CMOS camera (C11440-22CU). Movies were annotated using the Annotate_movie73 plugin on Fiji (v 2.3.0/1.53q)74 (Movies S1–S4, ESI†).Assessment of S. rosetta kinase signaling by western blotting
After compound treatment, the SrEpac culture was harvested and lysed to quantify protein abundance and immunoblotting by western blot. S. rosetta cells were harvested by centrifugation at 6000g for five minutes in Falcon tubes in a swinging bucket centrifuge and transferred into 1.5 mL Eppendorf tubes and washed two times with 4× phosphate buffered saline (PBS, 6.2 mM potassium phosphate monobasic, 621 mM sodium chloride, 10.8 mM sodium phosphate dibasic) with centrifugation at 6000g for 5 minutes in a fixed angle centrifuge at room temperature in between each wash. Cells were resuspended and lysed in digitonin lysis buffer (20 mM Tris pH 8, 150 mM potassium chloride, 5 mM magnesium chloride, 250 mM sucrose, 1 mM Pefabloc® SC serine protease inhibitor (Sigma-Aldrich Cat# 76307), 8 mM digitonin, 1 mM dithiothreitol, 0.06 U μL−1 benzonase nuclease, 1× Roche PhosSTOP phosphatase inhibitor cocktail, 1× Roche cOmplete protease inhibitor cocktail) for 30 minutes. Lysed cells were spun at 18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes at 4 °C and supernatants were isolated. Protein concentration in supernatants was determined by Bradford assay. Samples were boiled in loading dye (LiCOR #928-40004) and equal protein amounts for each sample were loaded onto NuPAGE™ 4–12% bis-Tris SDS-PAGE gels (Invitrogen Cat#s WG1402BOX, WG1403BOX, NP0335BOX, NP0336BOX). To resolve bands recognized by the phospho-p38 antibody (Promega #V1211), a NuPAGE™ 12% bis-Tris SDS-PAGE gel (Invitrogen Fisher NP0349BOX) was used. PageRuler™ or PageRuler Plus™ prestained protein ladder (Thermo Scientific #26614 and #26620), EGF-stimulated A431 cell lysate control (Sigma Aldrich #12-302), and E. pacifica control lysate (lysed as for S. rosetta cells) were added to wells and gels were run in Novex NuPAGE MES buffer (Invitrogen Cat# NP000202). Gels were transferred to 0.45 μm nitrocellulose (Bio-Rad) in Tris-glycine buffer (Bio-Rad #1610734) with 10% methanol. After transfer, blots were stained with LI-COR Revert™ 700 and imaged for total protein. After total protein was stained, blots were blocked with 5% bovine serum albumin in Tris buffered saline (TBS, 25 mM Tris, 150 mM sodium chloride) for 1 hour. Primary antibodies (see next paragraph) were added in TBS with Tween (0.1%) and left overnight. Blots were washed with TBS-Tween four times. LI-COR secondary IRDyes® were added in TBS-Tween at 1
000g for 15 minutes at 4 °C and supernatants were isolated. Protein concentration in supernatants was determined by Bradford assay. Samples were boiled in loading dye (LiCOR #928-40004) and equal protein amounts for each sample were loaded onto NuPAGE™ 4–12% bis-Tris SDS-PAGE gels (Invitrogen Cat#s WG1402BOX, WG1403BOX, NP0335BOX, NP0336BOX). To resolve bands recognized by the phospho-p38 antibody (Promega #V1211), a NuPAGE™ 12% bis-Tris SDS-PAGE gel (Invitrogen Fisher NP0349BOX) was used. PageRuler™ or PageRuler Plus™ prestained protein ladder (Thermo Scientific #26614 and #26620), EGF-stimulated A431 cell lysate control (Sigma Aldrich #12-302), and E. pacifica control lysate (lysed as for S. rosetta cells) were added to wells and gels were run in Novex NuPAGE MES buffer (Invitrogen Cat# NP000202). Gels were transferred to 0.45 μm nitrocellulose (Bio-Rad) in Tris-glycine buffer (Bio-Rad #1610734) with 10% methanol. After transfer, blots were stained with LI-COR Revert™ 700 and imaged for total protein. After total protein was stained, blots were blocked with 5% bovine serum albumin in Tris buffered saline (TBS, 25 mM Tris, 150 mM sodium chloride) for 1 hour. Primary antibodies (see next paragraph) were added in TBS with Tween (0.1%) and left overnight. Blots were washed with TBS-Tween four times. LI-COR secondary IRDyes® were added in TBS-Tween at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 dilution and incubated for 1 hour at room temperature. Blots were washed with TBS-Tween four times followed by TBS one time and imaged with LI-COR Odyssey® imager (Fig. 2C, 4D, E, G and Fig. S2D, S6A, S7, S10, S12, S14E, S15, ESI†). Staining intensities were quantified with Image Studio Lite 5.2.5 (LI-COR, 2014) (Fig. 4B, C and Fig. S2D, S10A, S12A, B, S14B, E, ESI†) and the resulting normalized cell density data at each dose was plotted using GraphPad Prism 9.3.1™ (Fig. 4B and C).
000 dilution and incubated for 1 hour at room temperature. Blots were washed with TBS-Tween four times followed by TBS one time and imaged with LI-COR Odyssey® imager (Fig. 2C, 4D, E, G and Fig. S2D, S6A, S7, S10, S12, S14E, S15, ESI†). Staining intensities were quantified with Image Studio Lite 5.2.5 (LI-COR, 2014) (Fig. 4B, C and Fig. S2D, S10A, S12A, B, S14B, E, ESI†) and the resulting normalized cell density data at each dose was plotted using GraphPad Prism 9.3.1™ (Fig. 4B and C).
Primary antibodies for pY1000 (#8954), anti-phospho-Erk (anti-phospho p44/42 MAPK #4370), anti-phospho (Ser/Thr)-Phe (#9631), anti-phosphotheronine (#9381) and anti-phosphothreonine-proline (#9391) were purchased from Cell Signaling Technology. Anti-phospho p38 MAPK (anti-ACTIVE® p38, #V1211) was purchased from Promega or Biorad (p38 MAPK pThr180/pTyr182, #AHP905). Anti-phosphoserine (Rb X, #AB1603) was purchased from Millipore Sigma. Anti-alpha-tubulin (YOL 1/34, #ab6161) was purchased from Abcam.
ActivX mass spectrometry workflow to identify S. rosetta proteins that bind sorafenib
For all mass spectrometry experiments, SrEpac cultures were grown to a high density in Pyrex baffled flasks without shaking. In tall necked flasks, cultures were grown in the maximum volume of culture media, and an aquarium pump was used to bubble air into the foam-plugged PYREX® Delong flask pierced with a serological pipette at a bubbling rate of approximately one bubble per second.75 For wide 2.8 L PYREX® Fernbach flasks, bubbling was not needed. S. rosetta cells were harvested by spinning in 200 mL Nalgene conicals at 2000g for 10 minutes in a swinging bucket centrifuge at room temperature to obtain cell pellets. Cells were first washed by resuspending cell pellets from four conicals in approximately 45 mL of 4× PBS, transferring cells to 50 mL Falcon tubes, and spinning at 2000g for 10 minutes in a swinging bucket centrifuge at room temperature. Cells were washed a second time by resuspending cells in ∼15 mL of 4× PBS per 50 mL Falcon tube, transferring cells to 15 mL Falcon tubes, and spinning at 2000g for 10 minutes in a swinging bucket centrifuge at room temperature. Cells were then ready for the ActivX probe workflow.For ActivX probe enrichment, cells were lysed in digitonin lysis buffer (see assessment of S. rosetta kinase signaling by western blotting) and 500 μL of 5 mg mL−1 supernatants were obtained as previously described.42 20 mM of manganese chloride cofactor was added to the lysate and incubated for 5 minutes followed by the addition of 100 μM sorafenib or 1% DMSO (vehicle control) and incubation for 10 minutes. 20 μM ActivX probe was added for kinase capture. Biotinylated proteins were captured using streptavidin beads in the presence of 6 M urea/immunoprecipitation (IP) lysis buffer, and samples were washed with 6 M urea/IP lysis buffer. Protein samples were provided to the University of California, Davis mass spectrometry facility (https://cmsf.ucdavis.edu) on beads and underwent standard tryptic digestion with Promega ProteaseMAX™ Surfactant, Trypsin Enhancer. Briefly, samples were first reduced at 56 °C for 45 minutes in 5.5 mM DTT followed by alkylation for one hour in the dark with iodoacetamide added to a final concentration of 10 mM. Trypsin was added at a final enzyme![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) substrate mass ratio of 1
substrate mass ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 and digestion carried out overnight at 37 °C. The reaction was quenched by flash freezing in liquid nitrogen and the digest was lyophilized. Digest was reconstituted in 0.1% TFA with 10% acetonitrile prior to injection. For quantification of peptides within each sample, 500 fmol of Hi3 E. coli standard reagent ClpB (Waters, Milford, MA) was placed into each sample and injected with 1.0 μg of total digest. Each sample was run in triplicate.
50 and digestion carried out overnight at 37 °C. The reaction was quenched by flash freezing in liquid nitrogen and the digest was lyophilized. Digest was reconstituted in 0.1% TFA with 10% acetonitrile prior to injection. For quantification of peptides within each sample, 500 fmol of Hi3 E. coli standard reagent ClpB (Waters, Milford, MA) was placed into each sample and injected with 1.0 μg of total digest. Each sample was run in triplicate.
The mass spectrometry instrument used to analyze the samples was a Xevo G2 QTof coupled to a nanoAcquity UPLC system (Waters, Milford, MA). Samples were loaded onto a C18 Waters Trizaic nanotile of 85 μm ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ×
× ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 mm; 1.7 μm (Waters, Milford, MA). The column temperature was set to 45 °C with a flow rate of 0.45 mL min−1. The mobile phase consisted of A (water containing 0.1% formic acid) and B (acetonitrile containing 0.1% formic acid). A linear gradient elution program was used: 0–40 min, 3–40% (B); 40–42 min, 40–85% (B); 42–46 min, 85% (B); 46–48 min, 85–3% (B); 48–60 min, 3% (B). Mass spectrometry data were recorded for 60 minutes for each run and controlled by MassLynx 4.2 SCN990 (Waters, Milford, MA). Acquisition mode was set to positive polarity under resolution mode. Mass range was set form 50–2000 Da. Capillary voltage was 3.5 kV, sampling cone at 25 V, and extraction cone at 2.5 V. Source temperature was held at 110 °C. Cone gas was set to 25 L h−1, nano flow gas at 0.10 bar, and desolvation gas at 1200 L h−1. Leucine–enkephalin at 720 pmol μL−1 (Waters, Milford, MA) was used as the lock mass ion at m/z 556.2771 and introduced at 1 μL min−1 at 45 second intervals with a 3 scan average and mass window of ±0.5 Da. The MSe data were acquired using two scan functions corresponding to low energy for function 1 and high energy for function 2. Function 1 had collision energy at 6 V and function 2 had a collision energy ramp of 18–42 V.
100 mm; 1.7 μm (Waters, Milford, MA). The column temperature was set to 45 °C with a flow rate of 0.45 mL min−1. The mobile phase consisted of A (water containing 0.1% formic acid) and B (acetonitrile containing 0.1% formic acid). A linear gradient elution program was used: 0–40 min, 3–40% (B); 40–42 min, 40–85% (B); 42–46 min, 85% (B); 46–48 min, 85–3% (B); 48–60 min, 3% (B). Mass spectrometry data were recorded for 60 minutes for each run and controlled by MassLynx 4.2 SCN990 (Waters, Milford, MA). Acquisition mode was set to positive polarity under resolution mode. Mass range was set form 50–2000 Da. Capillary voltage was 3.5 kV, sampling cone at 25 V, and extraction cone at 2.5 V. Source temperature was held at 110 °C. Cone gas was set to 25 L h−1, nano flow gas at 0.10 bar, and desolvation gas at 1200 L h−1. Leucine–enkephalin at 720 pmol μL−1 (Waters, Milford, MA) was used as the lock mass ion at m/z 556.2771 and introduced at 1 μL min−1 at 45 second intervals with a 3 scan average and mass window of ±0.5 Da. The MSe data were acquired using two scan functions corresponding to low energy for function 1 and high energy for function 2. Function 1 had collision energy at 6 V and function 2 had a collision energy ramp of 18–42 V.
RAW MSe files were processed using Protein Lynx Global Server (PLGS) version 3.0.3 (Waters, Milford, MA). Processing parameters consisted of a low energy threshold set at 200.0 counts, an elevated energy threshold set at 25.0 counts, and an intensity threshold set at 1500 counts. Each sample was searched against the Salpingoeca rosetta genome hosted on Ensembl Genomes.17,76 Each databank was randomized within PLGS and included the protein sequence for ClpB. Possible structure modifications included for consideration were methionine oxidation, asparagine deamidation, glutamine deamidation, serine dehydration, threonine dehydration, and carbamidomethylation of cysteine. For viewing, PLGS search results were exported in Scaffold v4.4.6 (Proteome Software Inc., Portland, OR). We quantified absolute protein abundance77 and focused our attention on kinases that were present in both DMSO and sorafenib pretreated samples but were enriched in multiple DMSO replicates, represented by higher PLGS scores.77 The resulting data was plotted with GraphPad Prism 9.3.1™ (Fig. 3).
Generation of Sr-p381–15 and Sr-JNK1–15 strains by genome editing
Candidate guide RNA sequences that targeted early in the Sr-p38 and Sr-JNK open reading frame were identified using the EuPaGDT tool (https://grna.ctegd.uga.edu/) and the S. rosetta genome17 hosted on Ensembl Protists (Ensembl 108).78 Guide RNA length was set at 15 and an YRNGRSGGH PAM sequence was used. Guide RNA candidates were filtered for guides with one on-target hit (including making sure the guides do not span exon–exon boundaries), zero off-target hits (including against the genome of the co-cultured bacterium E. pacifica), lowest strength of the predicted secondary structure (assessed using the RNAfold web server: https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), and annealing near codon 15 of Sr-p38 and Sr-JNK. A crRNA with the guide sequence TGCAAGTCTGTGTAGCACGA for Sr-p38 and TTGTCGATGTGTGGAAGCAG for Sr-JNK, as well as universal tracrRNAs, were ordered from IDT (Integrated DNA Technologies, Coralville, IA). Repair templates were produced by PCR using a previously published plasmid encoding the pEFL-pac-5′Act (Addgene ID pMS18, catalog number #225681) cassette as a template. The premature termination sequence (5′-TTTATTTAATTAAATAAA-3′) was included in custom primers with 50 bp homology arms to the Sr-p38 or Sr-JNK locus. PCR reactions were set up in 50 μL reactions using Q5 high fidelity DNA polymerase with 50 ng plasmid as template, 200 μM dNTPs, 0.25 μM of each primer and 0.02 U μL−1 Q5 polymerase. The following PCR program was run on an Applied Biosystems Veriti 96-well Thermal Cycler: 30′′ 98 °C; 40× (10′′ 98 °C; 30′′ 68 °C; 1′ 72 °C); 2′ 72 °C. The PCR product size (expected 2 kb) was visually checked by running 2 μL of the reaction on a 1% (w/v) agarose gel containing ethidium bromide at 1 μg mL−1 run in TAE buffer and visualized with the Alpha Innotech 2000 Photo Imaging System Ultraviolet Transilluminator (Fig. S11A and B, ESI†). PCR products were purified using a PCR purification kit (NEB Monarch Cat #T1130L and SydLabs Tiniprep columns Cat# MB0110L) and the final product was eluted with pre-warmed 20 μL milliQ water which was left to incubate on the column for 10 minutes before eluting. 1.5 μL was used to measure DNA concentration using a NanoDrop spectrophotometer (ThermoFisherScientific NanoDrop 2000). 6 μg of the remaining DNA was concentrated in 2 μL by evaporation for 2 hours at 55 °C or lyophilized and resuspended into 2 μL warm milliQ water and then used as a repair template for nucleofection. Genome editing proceeded as described previously,58 but with 3H Buffer (75 mM Tris–HCl, 20 mM HEPES, 90 mM NaCl, 15 mM MgCl2, 5 mM KCl, 10 mM glucose, 0.4 mM calcium nitrate) replacing Lonza SF buffer. Puromycin (80 μg mL−1) was added after 24 hours. At 72 hours post addition of puromycin, the wild-type cells had died, and puromycin-resistant cells appeared for Sr-JNK1–15 nucleofections. Puromycin-resistant cells for Sr-p381–15 appeared at 96 hours. Genotyping primers used to confirm insertion of the premature termination sequence and puromycin resistance cassettes58 were AACAGGAGGCACAGTTACGA (forward) and GAACAAGCAACACACCACCA (reverse) for Sr-p38; CGTTAATCGACGACGCCAA (forward) and ATGAGCTGGATGTGGGGGA (reverse) for Sr-JNK. Premium PCR Sequencing was performed by Plasmidsaurus using Oxford Nanopore Technology with custom analysis and annotation to genotype clones. Base calls in the insertion region for Sr-p381–15 are in Fig. S11C (ESI†) and Sr-JNK1–15 in Fig. S11D (ESI†).Generation of Sr-p38T110M strains by genome editing
Candidate guide RNA sequences were obtained for Sr-p38 using the EuPaGDT tool (https://grna.ctegd.uga.edu/) and the S. rosetta genome17 hosted on Ensembl Protists (Ensembl 108)78 as previously published.71 Guide RNA length was set at 15, and an NGG PAM sequence was used. Guide RNA candidates were filtered for guides with one on-target hit (including making sure the guides do not span exon–exon boundaries), zero off-target hits (including against the genome of the co-cultured bacterium E. pacifica), lowest strength of the predicted secondary structure (assessed using the RNAfold web server: https://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi), and annealing near codon 110 of Sr-p38. A crRNA with the guide sequence CTACATCATCACAGAGAAGA, as well as universal tracrRNAs, were ordered from IDT (Integrated DNA Technologies, Coralville, IA). Repair templates were designed as single-stranded DNA oligos, in the same sense strand as the guide RNA, with 50 base pairs of genomic sequence on either side of the DSB cut site. The repair oligo GGTTGACCTGTACATCTCCGACGCGCGTGACATCTACATCATCATGGAGAAGATGGTGTGTATCTTTGACGCGGGTTGACTGGCCGTGATGGCGCGTGTT was ordered from IDT as an Ultramer. Genotyping primers were AGATTCGTTCCAGCGGAATACA (forward) and GGGAAGAAGTGCGGAGTGAA (reverse). Genome editing proceeded as described previously.71 Clones were Sanger sequenced at the UC Berkeley DNA Sequencing Facility.Bioinformatic analysis of the S. rosetta kinome
The S. rosetta kinome was annotated based on previously predicted kinases17 and orthoDB ortholog annotation79 of S. rosetta and human protein sequences in Uniprot80 (Fig. S1, ESI†). For prediction of choanoflagellate p38 kinases, a HMMER profile was generated from human and previously predicted S. rosetta p38 kinases17,43 and searched against available genomes and transcriptomes of choanoflagellates9,12,81,82 (Fig. S6A and B, ESI†). Protein targets of individual human kinase inhibitors were manually annotated and plotted using CORAL83 (Fig. S1, ESI†). References for kinase inhibitory data for each compound is available in Table S1 (ESI†). To analyze conservation within the kinase domain, kinase sequence alignments of predicted kinases were generated with Clustal Omega84 (Fig. S5B, C and S9B, ESI†) and amino acid logos were generated with WebLogo85 (Fig. S5A, ESI†).Author contributions
F. U. R., M. C., A. C. D., and N. K. designed research; F. U. R. performed screening and choanoflagellate research, F. U. R., M. C., M. H. T. N., and I. H. generated choanoflagellate strains, and A. P. S. synthesized sorafenib analogs; F. U. R., A. C. D., and N. K. analyzed data; and F. U. R., and N. K. wrote the paper.Data availability
Data supporting this article are included in the ESI.† Raw data for this article, including flow cytometry fcs files, genotyping, image segmentation output files, sequence alignments, raw counts and cell densities, mass spectrometry peptides, raw western blot images, western blot cropping and western blot quantitation, are available at Figshare at https://doi.org/10.6084/m9.figshare.20669730.v3.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank D. Booth, M. Carver, A. Garcia De Las Bayonas and J. Reyes-Rivera for critical reading of the manuscript; K. Shokat (University of California, San Francisco) for the contribution of kinase inhibitors in the screened library and Shokat lab members for discussions; M. West and P. He of the High-Throughput Screening Facility (QB3 HTSF) at UC Berkeley who provided access to the Agilent V11 Bravo liquid handling system for screening plate generation and Opera Phenix for plate imaging through funding from the National Institutes of Health (S10OD021828); H. Nolla, A. Valeros and the CRL-FACS facility and staff at UC Berkeley for their support and providing access to the BD LSRII flow cytometer and BD HTS sample loading robot; A. Andaya of the mass spectrometry facility at UC Davis. The A. C. D. laboratory receives support from the NIH (R01s CA227636, CA258736, CA256480, and R56 AG066712), the Mark Foundation for Cancer Research (20-030-ASP, 21-039-ASP) and Alex's Lemonade Stand Foundation for Childhood Cancer. N. K. and F. U. R.'s work was supported by an Investigator award (N. K.) and the Hanna H. Gray Fellows Program (F. U. R.), both from the Howard Hughes Medical Institute.References
- P. Avasthi, A. Marley, H. Lin, E. Gregori-Puigjane, B. K. Shoichet, M. von Zastrow and W. F. Marshall, A chemical screen identifies class a G-protein coupled receptors as regulators of cilia, ACS Chem. Biol., 2012, 7, 911–919 CrossRef CAS PubMed.
- T. U. Mayer, T. M. Kapoor, S. J. Haggarty, R. W. King, S. L. Schreiber and T. J. Mitchison, Small molecule inhibitor of mitotic spindle bipolarity identified in a phenotype-based screen, Science, 1999, 286, 971–974 CrossRef CAS PubMed.
- J. R. Peterson and T. J. Mitchison, Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton, Chem. Biol., 2002, 9, 1275–1285 CrossRef CAS PubMed.
- D. P. Walsh and Y.-T. Chang, Chemical genetics, Chem. Rev., 2006, 106, 2476–2530 CrossRef CAS PubMed.
- P. Blume-Jensen and T. Hunter, Oncogenic kinase signalling, Nature, 2001, 411, 355–365 CrossRef CAS PubMed.
- M. A. Lemmon and J. Schlessinger, Cell signaling by receptor tyrosine kinases, Cell, 2010, 141, 1117–1134 CrossRef CAS PubMed.
- R. Roskoski Jr, Properties of FDA-approved small molecule protein kinase inhibitors: a 2024 update, Pharmacol. Res., 2024, 200, 107059 CrossRef PubMed.
- G. Manning, S. L. Young, W. T. Miller and Y. Zhai, The protist, Monosiga brevicollis, has a tyrosine kinase signaling network more elaborate and diverse than found in any known metazoan, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 9674–9679 CrossRef CAS PubMed.
- N. King, M. J. Westbrook, S. L. Young, A. Kuo, M. Abedin, J. Chapman, S. Fairclough, U. Hellsten, Y. Isogai, I. Letunic, M. Marr, D. Pincus, N. Putnam, A. Rokas, K. J. Wright, R. Zuzow, W. Dirks, M. Good, D. Goodstein, D. Lemons, W. Li, J. B. Lyons, A. Morris, S. Nichols, D. J. Richter, A. Salamov, J. G. I. Sequencing, P. Bork, W. A. Lim, G. Manning, W. T. Miller, W. McGinnis, H. Shapiro, R. Tjian, I. V. Grigoriev and D. Rokhsar, The genome of the choanoflagellate Monosiga brevicollis and the origin of metazoans, Nature, 2008, 451, 783–788 CrossRef CAS PubMed.
- W. Yeung, A. Kwon, R. Taujale, C. Bunn, A. Venkat and N. Kannan, Evolution of Functional Diversity in the Holozoan Tyrosine Kinome, Mol. Biol. Evol., 2021, 38, 5625–5639 CrossRef CAS PubMed.
- N. Ros-Rocher, A. Pérez-Posada, M. M. Leger and I. Ruiz-Trillo, The origin of animals: an ancestral reconstruction of the unicellular-to-multicellular transition, Open Biol., 2021, 11, 200359 CrossRef CAS PubMed.
- D. J. Richter, P. Fozouni, M. B. Eisen and N. King, Gene family innovation, conservation and loss on the animal stem lineage, eLife, 2018, 7, e34226 CrossRef PubMed.
- N. King, C. T. Hittinger and S. B. Carroll, Evolution of key cell signaling and adhesion protein families predates animal origins, Science, 2003, 301, 361–363 CrossRef PubMed.
- R. Bertrand, E. Solary, P. O’Connor, K. W. Kohn and Y. Pommier, Induction of a common pathway of apoptosis by staurosporine, Exp. Cell Res., 1994, 211, 314–321 CrossRef PubMed.
- M. Deponte, Programmed cell death in protists, Biochim. Biophys. Acta, 2008, 1783, 1396–1405 CrossRef PubMed.
- J. H. Zhang, T. D. Chung and K. R. Oldenburg, A simple statistical parameter for use in evaluation and validation of high throughput screening assays, J. Biomol. Screen., 1999, 4, 67–73 CrossRef PubMed.
- S. R. Fairclough, Z. Chen, E. Kramer, Q. Zeng, S. Young, H. M. Robertson, E. Begovic, D. J. Richter, C. Russ, M. J. Westbrook, G. Manning, B. F. Lang, B. Haas, C. Nusbaum and N. King, Premetazoan genome evolution and the regulation of cell differentiation in the choanoflagellate Salpingoeca rosetta, Genome Biol., 2013, 14, R15 CrossRef PubMed.
- T. J. P. van Dam, F. J. T. Zwartkruis, J. L. Bos and B. Snel, Evolution of the TOR pathway, J. Mol. Evol., 2011, 73, 209–220 CrossRef PubMed.
- M. Belenky, J. Prasain, H. Kim and S. Barnes, DING, a genistein target in human breast cancer: a protein without a gene, J. Nutr., 2003, 133, 2497S–2501S CrossRef PubMed.
- Z.-C. Dang, V. Audinot, S. E. Papapoulos, J. A. Boutin and C. W. G. M. Löwik, Peroxisome proliferator-activated receptor gamma (PPARgamma) as a molecular target for the soy phytoestrogen genistein, J. Biol. Chem., 2003, 278, 962–967 CrossRef PubMed.
- J. B. Müller, P. E. Geyer, A. R. Colaço, P. V. Treit, M. T. Strauss, M. Oroshi, S. Doll, S. Virreira Winter, J. M. Bader, N. Köhler, F. Theis, A. Santos and M. Mann, The proteome landscape of the kingdoms of life, Nature, 2020, 582, 592–596 CrossRef PubMed.
- J. Reinders and A. Sickmann, State-of-the-art in phosphoproteomics, Proteomics, 2005, 5, 4052–4061 CrossRef PubMed.
- M. Mann, S. E. Ong, M. Grønborg, H. Steen, O. N. Jensen and A. Pandey, Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome, Trends Biotechnol., 2002, 20, 261–268 CrossRef PubMed.
- B. M. Sefton and S. Shenolikar, Overview of protein phosphorylation, Curr. Protoc. Protein Sci., 2001, ch. 13, unit13.1 Search PubMed.
- Y. Segawa, H. Suga, N. Iwabe, C. Oneyama, T. Akagi, T. Miyata and M. Okada, Functional development of Src tyrosine kinases during evolution from a unicellular ancestor to multicellular animals, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 12021–12026 CrossRef PubMed.
- W. Li, S. L. Young, N. King and W. T. Miller, Signaling properties of a non-metazoan Src kinase and the evolutionary history of Src negative regulation, J. Biol. Chem., 2008, 283, 15491–15501 CrossRef CAS PubMed.
- K. P. Schultheiss, B. P. Craddock, M. Tong, M. Seeliger and W. T. Miller, Metazoan-like signaling in a unicellular receptor tyrosine kinase, BMC Biochem., 2013, 14, 4 CrossRef CAS PubMed.
- S. U. Aleem, B. P. Craddock and W. T. Miller, Constitutive Activity in an Ancestral Form of Abl Tyrosine Kinase, PLoS One, 2015, 10, e0131062 CrossRef PubMed.
- S. H. Myers, V. G. Brunton and A. Unciti-Broceta, AXL inhibitors in cancer: a medicinal chemistry perspective, J. Med. Chem., 2016, 59, 3593–3608 CrossRef CAS PubMed.
- M. I. Davis, J. P. Hunt, S. Herrgard, P. Ciceri, L. M. Wodicka, G. Pallares, M. Hocker, D. K. Treiber and P. P. Zarrinkar, Comprehensive analysis of kinase inhibitor selectivity, Nat. Biotechnol., 2011, 29, 1046–1051 CrossRef CAS PubMed.
- B. Apsel, J. A. Blair, B. Gonzalez, T. M. Nazif, M. E. Feldman, B. Aizenstein, R. Hoffman, R. L. Williams, K. M. Shokat and Z. A. Knight, Targeted polypharmacology: discovery of dual inhibitors of tyrosine and phosphoinositide kinases, Nat. Chem. Biol., 2008, 4, 691–699 CrossRef CAS PubMed.
- M. S. Lopez, J. W. Choy, U. Peters, M. L. Sos, D. O. Morgan and K. M. Shokat, Staurosporine-derived inhibitors broaden the scope of analog-sensitive kinase technology, J. Am. Chem. Soc., 2013, 135, 18153–18159 CrossRef CAS PubMed.
- S. S. Taylor, M. M. Keshwani, J. M. Steichen and A. P. Kornev, Evolution of the eukaryotic protein kinases as dynamic molecular switches, Philos. Trans. R. Soc., B, 2012, 367, 2517–2528 CrossRef CAS PubMed.
- J. A. Adams, Kinetic and catalytic mechanisms of protein kinases, Chem. Rev., 2001, 101, 2271–2290 CrossRef CAS PubMed.
- G. Manning, D. B. Whyte, R. Martinez, T. Hunter and S. Sudarsanam, The protein kinase complement of the human genome, Science, 2002, 298, 1912–1934 CrossRef CAS PubMed.
- Y. Liu, A. Bishop, L. Witucki, B. Kraybill, E. Shimizu, J. Tsien, J. Ubersax, J. Blethrow, D. O. Morgan and K. M. Shokat, Structural basis for selective inhibition of Src family kinases by PP1, Chem. Biol., 1999, 6, 671–678 CrossRef CAS PubMed.
- C. Zhang, M. S. Lopez, A. C. Dar, E. Ladow, S. Finkbeiner, C.-H. Yun, M. J. Eck and K. M. Shokat, Structure-guided inhibitor design expands the scope of analog-sensitive kinase technology, ACS Chem. Biol., 2013, 8, 1931–1938 CrossRef CAS PubMed.
- P. Dubreuil, S. Letard, M. Ciufolini, L. Gros, M. Humbert, N. Castéran, L. Borge, B. Hajem, A. Lermet, W. Sippl, E. Voisset, M. Arock, C. Auclair, P. S. Leventhal, C. D. Mansfield, A. Moussy and O. Hermine, Masitinib (AB1010), a potent and selective tyrosine kinase inhibitor targeting KIT, PLoS One, 2009, 4, e7258 CrossRef PubMed.
- J. X. Yu, A. J. Craig, M. E. Duffy, C. Villacorta-Martin, V. Miguela, M. Ruiz de Galarreta, A. P. Scopton, L. Silber, A. Y. Maldonado, A. Rialdi, E. Guccione, A. Lujambio, A. Villanueva and A. C. Dar, Phenotype-Based Screens with Conformation-Specific Inhibitors Reveal p38 Gamma and Delta as Targets for HCC Polypharmacology, Mol. Cancer Ther., 2019, 18, 1506–1519 CrossRef CAS PubMed.
- M. Sonoshita, A. P. Scopton, P. M. U. Ung, M. A. Murray, L. Silber, A. Y. Maldonado, A. Real, A. Schlessinger, R. L. Cagan and A. C. Dar, A whole-animal platform to advance a clinical kinase inhibitor into new disease space, Nat. Chem. Biol., 2018, 14, 291–298 CrossRef CAS PubMed.
- J. G. Villamor, F. Kaschani, T. Colby, J. Oeljeklaus, D. Zhao, M. Kaiser, M. P. Patricelli and R. A. L. van der Hoorn, Profiling protein kinases and other ATP binding proteins in Arabidopsis using Acyl-ATP probes, Mol. Cell. Proteomics, 2013, 12, 2481–2496 CrossRef CAS PubMed.
- M. P. Patricelli, A. K. Szardenings, M. Liyanage, T. K. Nomanbhoy, M. Wu, H. Weissig, A. Aban, D. Chun, S. Tanner and J. W. Kozarich, Functional interrogation of the kinome using nucleotide acyl phosphates, Biochemistry, 2007, 46, 350–358 CrossRef CAS PubMed.
- V. Shabardina, P. R. Charria, G. B. Saborido, E. Diaz-Mora, A. Cuenda, I. Ruiz-Trillo and J. J. Sanz-Ezquerro, Evolutionary analysis of p38 stress-activated kinases in unicellular relatives of animals suggests an ancestral function in osmotic stress, Open Biol., 2023, 13, 220314 CrossRef CAS PubMed.
- R. Rudalska, D. Dauch, T. Longerich, K. McJunkin, T. Wuestefeld, T.-W. Kang, A. Hohmeyer, M. Pesic, J. Leibold, A. von Thun, P. Schirmacher, J. Zuber, K.-H. Weiss, S. Powers, N. P. Malek, M. Eilers, B. Sipos, S. W. Lowe, R. Geffers, S. Laufer and L. Zender, In vivo RNAi screening identifies a mechanism of sorafenib resistance in liver cancer, Nat. Med., 2014, 20, 1138–1146 CrossRef CAS PubMed.
- J. L. Brewster, T. de Valoir, N. D. Dwyer, E. Winter and M. C. Gustin, An osmosensing signal transduction pathway in yeast, Science, 1993, 259, 1760–1763 CrossRef CAS PubMed.
- J. L. Brewster and M. C. Gustin, Hog1: 20 years of discovery and impact, Sci. Signal., 2014, 7, re7 Search PubMed.
- H. C. Causton, B. Ren, S. S. Koh, C. T. Harbison, E. Kanin, E. G. Jennings, T. I. Lee, H. L. True, E. S. Lander and R. A. Young, Remodeling of yeast genome expression in response to environmental changes, Mol. Biol. Cell, 2001, 12, 323–337 CrossRef CAS PubMed.
- C. Schüller, J. L. Brewster, M. R. Alexander, M. C. Gustin and H. Ruis, The HOG pathway controls osmotic regulation of transcription via the stress response element (STRE) of the Saccharomyces cerevisiae CTT1 gene, EMBO J., 1994, 13, 4382–4389 CrossRef PubMed.
- A. Cuadrado and A. R. Nebreda, Mechanisms and functions of p38 MAPK signalling, Biochem. J., 2010, 429, 403–417 CrossRef CAS PubMed.
- S. Dorion, H. Lambert and J. Landry, Activation of the p38 signaling pathway by heat shock involves the dissociation of glutathione S-transferase Mu from Ask1, J. Biol. Chem., 2002, 277, 30792–30797 CrossRef CAS PubMed.
- J. Rouse, P. Cohen, S. Trigon, M. Morange, A. Alonso-Llamazares, D. Zamanillo, T. Hunt and A. R. Nebreda, A novel kinase cascade triggered by stress and heat shock that stimulates MAPKAP kinase-2 and phosphorylation of the small heat shock proteins, Cell, 1994, 78, 1027–1037 CrossRef CAS PubMed.
- M. Carr, B. S. C. Leadbeater, R. Hassan, M. Nelson and S. L. Baldauf, Molecular phylogeny of choanoflagellates, the sister group to Metazoa, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 16641–16646 CrossRef CAS PubMed.
- B. S. C. Leadbeater, The Choanoflagellates, Cambridge University Press, 2015 Search PubMed.
- L. Tomanek, Variation in the heat shock response and its implication for predicting the effect of global climate change on species’ biogeographical distribution ranges and metabolic costs, J. Exp. Biol., 2010, 213, 971–979 CrossRef CAS PubMed.
- N. Ros-Rocher, J. Reyes-Rivera, U. Horo, Y. Foroughijabbari, C. Combredet, B. T. Larson, M. C. Coyle, E. A. T. Houtepen, M. J. A. Vermeij, J. L. Steenwyk and T. Brunet, bioRxiv, 2024, preprint DOI:10.1101/2024.03.25.586565.
- Y. Xie, H. Li, X. Luo, H. Li, Q. Gao, L. Zhang, Y. Teng, Q. Zhao, Z. Zuo and J. Ren, IBS 2.0: an upgraded illustrator for the visualization of biological sequences, Nucleic Acids Res., 2022, 50, W420–W426 CrossRef CAS PubMed.
- Y. Suzaki, M. Yoshizumi, S. Kagami, A. H. Koyama, Y. Taketani, H. Houchi, K. Tsuchiya, E. Takeda and T. Tamaki, Hydrogen peroxide stimulates c-Src-mediated big mitogen-activated protein kinase 1 (BMK1) and the MEF2C signaling pathway in PC12 cells: potential role in cell survival following oxidative insults, J. Biol. Chem., 2002, 277, 9614–9621 CrossRef CAS PubMed.
- C. Combredet and T. Brunet, bioRxiv, 2024, preprint, bioRxiv:2024.07.13.603360 DOI:10.1101/2024.07.13.603360.
- A. Winkler, C. Arkind, C. P. Mattison, A. Burkholder, K. Knoche and I. Ota, Heat stress activates the yeast high-osmolarity glycerol mitogen-activated protein kinase pathway, and protein tyrosine phosphatases are essential under heat stress, Eukaryot. Cell, 2002, 1, 163–173 CrossRef CAS PubMed.
- M. C. Gustin, J. Albertyn, M. Alexander and K. Davenport, MAP kinase pathways in the yeast Saccharomyces cerevisiae, Microbiol. Mol. Biol. Rev., 1998, 62, 1264–1300 CrossRef PubMed.
- Y. Devary, R. A. Gottlieb, T. Smeal and M. Karin, The mammalian ultraviolet response is triggered by activation of Src tyrosine kinases, Cell, 1992, 71, 1081–1091 CrossRef CAS PubMed.
- S. J. Dixon, D. N. Patel, M. Welsch, R. Skouta, E. D. Lee, M. Hayano, A. G. Thomas, C. E. Gleason, N. P. Tatonetti, B. S. Slusher and B. R. Stockwell, Pharmacological inhibition of cystine-glutamate exchange induces endoplasmic reticulum stress and ferroptosis, eLife, 2014, 3, e02523 CrossRef PubMed.
- H. Suga, M. Dacre, A. de Mendoza, K. Shalchian-Tabrizi, G. Manning and I. Ruiz-Trillo, Genomic Survey of Premetazoans Shows Deep Conservation of Cytoplasmic Tyrosine Kinases and Multiple Radiations of Receptor Tyrosine Kinases, Sci. Signaling, 2012, 5, ra35 Search PubMed.
- M. A. Shogren-Knaak, P. J. Alaimo and K. M. Shokat, Recent advances in chemical approaches to the study of biological systems, Annu. Rev. Cell Dev. Biol., 2001, 17, 405–433 CrossRef CAS PubMed.
- W. A. Weiss, S. S. Taylor and K. M. Shokat, Recognizing and exploiting differences between RNAi and small-molecule inhibitors, Nat. Chem. Biol., 2007, 3, 739–744 CrossRef PubMed.
- A. Sebé-Pedrós, M. I. Peña, S. Capella-Gutiérrez, M. Antó, T. Gabaldón, I. Ruiz-Trillo and E. Sabidó, High-Throughput Proteomics Reveals the Unicellular Roots of Animal Phosphosignaling and Cell Differentiation, Dev. Cell, 2016, 39, 186–197 CrossRef PubMed.
- H. Suga and W. T. Miller, Src signaling in a low-complexity unicellular kinome, Sci. Rep., 2018, 8, 5362 CrossRef PubMed.
- N. King, S. L. Young, M. Abedin, M. Carr and B. S. C. Leadbeater, Starting and maintaining Monosiga brevicollis cultures, Cold Spring Harb. Protoc., 2009, 2009, db.prot5148 CrossRef PubMed.
- T. C. Levin and N. King, Evidence for sex and recombination in the choanoflagellate Salpingoeca rosetta, Curr. Biol., 2013, 23, 2176–2180 Search PubMed.
- A. Woznica, A. M. Cantley, C. Beemelmanns, E. Freinkman, J. Clardy and N. King, Bacterial lipids activate, synergize, and inhibit a developmental switch in choanoflagellates, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 7894–7899 CrossRef CAS PubMed.
- D. S. Booth and N. King, Genome editing enables reverse genetics of multicellular development in the choanoflagellate Salpingoeca rosetta, eLife, 2020, 9, e56193 CrossRef PubMed.
- A. C. Dar, T. K. Das, K. M. Shokat and R. L. Cagan, Chemical genetic discovery of targets and anti-targets for cancer polypharmacology, Nature, 2012, 486, 80–84 Search PubMed.
- S. Daetwyler, C. D. Modes and R. Fiolka, Fiji plugin for annotating movies with custom arrows, Biol. Open, 2020, 9(11), bio056200 Search PubMed.
- J. Schindelin, I. Arganda-Carreras, E. Frise, V. Kaynig, M. Longair, T. Pietzsch, S. Preibisch, C. Rueden, S. Saalfeld, B. Schmid, J.-Y. Tinevez, D. J. White, V. Hartenstein, K. Eliceiri, P. Tomancak and A. Cardona, Fiji: an open-source platform for biological-image analysis, Nat. Methods, 2012, 9, 676–682 CrossRef CAS PubMed.
- M. A. Sigg, T. Menchen, C. Lee, J. Johnson, M. K. Jungnickel, S. P. Choksi, G. Garcia 3rd, H. Busengdal, G. W. Dougherty, P. Pennekamp, C. Werner, F. Rentzsch, H. M. Florman, N. Krogan, J. B. Wallingford, H. Omran and J. F. Reiter, Evolutionary Proteomics Uncovers Ancient Associations of Cilia with Signaling Pathways, Dev. Cell, 2017, 43, 744–762.e11 CrossRef CAS PubMed.
- A. D. Yates, J. Allen, R. M. Amode, A. G. Azov, M. Barba, A. Becerra, J. Bhai, L. I. Campbell, M. Carbajo Martinez, M. Chakiachvili, K. Chougule, M. Christensen, B. Contreras-Moreira, A. Cuzick, L. Da Rin Fioretto, P. Davis, N. H. De Silva, S. Diamantakis, S. Dyer, J. Elser, C. V. Filippi, A. Gall, D. Grigoriadis, C. Guijarro-Clarke, P. Gupta, K. E. Hammond-Kosack, K. L. Howe, P. Jaiswal, V. Kaikala, V. Kumar, S. Kumari, N. Langridge, T. Le, M. Luypaert, G. L. Maslen, T. Maurel, B. Moore, M. Muffato, A. Mushtaq, G. Naamati, S. Naithani, A. Olson, A. Parker, M. Paulini, H. Pedro, E. Perry, J. Preece, M. Quinton-Tulloch, F. Rodgers, M. Rosello, M. Ruffier, J. Seager, V. Sitnik, M. Szpak, J. Tate, M. K. Tello-Ruiz, S. J. Trevanion, M. Urban, D. Ware, S. Wei, G. Williams, A. Winterbottom, M. Zarowiecki, R. D. Finn and P. Flicek, Ensembl Genomes 2022: an expanding genome resource for non-vertebrates, Nucleic Acids Res., 2022, 50, D996–D1003 Search PubMed.
- J. C. Silva, M. V. Gorenstein, G.-Z. Li, J. P. C. Vissers and S. J. Geromanos, Absolute Quantification of Proteins by LCMSE, Mol. Cell. Proteomics, 2006, 5, 144–156 Search PubMed.
- F. Cunningham, J. E. Allen, J. Allen, J. Alvarez-Jarreta, M. R. Amode, I. M. Armean, O. Austine-Orimoloye, A. G. Azov, I. Barnes, R. Bennett, A. Berry, J. Bhai, A. Bignell, K. Billis, S. Boddu, L. Brooks, M. Charkhchi, C. Cummins, L. Da Rin Fioretto, C. Davidson, K. Dodiya, S. Donaldson, B. El Houdaigui, T. El Naboulsi, R. Fatima, C. G. Giron, T. Genez, J. G. Martinez, C. Guijarro-Clarke, A. Gymer, M. Hardy, Z. Hollis, T. Hourlier, T. Hunt, T. Juettemann, V. Kaikala, M. Kay, I. Lavidas, T. Le, D. Lemos, J. C. Marugán, S. Mohanan, A. Mushtaq, M. Naven, D. N. Ogeh, A. Parker, A. Parton, M. Perry, I. Piližota, I. Prosovetskaia, M. P. Sakthivel, A. I. A. Salam, B. M. Schmitt, H. Schuilenburg, D. Sheppard, J. G. Pérez-Silva, W. Stark, E. Steed, K. Sutinen, R. Sukumaran, D. Sumathipala, M.-M. Suner, M. Szpak, A. Thormann, F. F. Tricomi, D. Urbina-Gómez, A. Veidenberg, T. A. Walsh, B. Walts, N. Willhoft, A. Winterbottom, E. Wass, M. Chakiachvili, B. Flint, A. Frankish, S. Giorgetti, L. Haggerty, S. E. Hunt, G. R. IIsley, J. E. Loveland, F. J. Martin, B. Moore, J. M. Mudge, M. Muffato, E. Perry, M. Ruffier, J. Tate, D. Thybert, S. J. Trevanion, S. Dyer, P. W. Harrison, K. L. Howe, A. D. Yates, D. R. Zerbino and P. Flicek, Ensembl 2022, Nucleic Acids Res., 2022, 50, D988–D995 Search PubMed.
- E. M. Zdobnov, D. Kuznetsov, F. Tegenfeldt, M. Manni, M. Berkeley and E. V. Kriventseva, OrthoDB in 2020: evolutionary and functional annotations of orthologs, Nucleic Acids Res., 2021, 49, D389–D393 CrossRef CAS PubMed.
- The UniProt Consortium, UniProt: the universal protein knowledgebase in 2021, Nucleic Acids Res., 2021, 49, D480–D489 CrossRef PubMed.
- T. Brunet, B. T. Larson, T. A. Linden, M. J. A. Vermeij, K. McDonald and N. King, Light-regulated collective contractility in a multicellular choanoflagellate, Science, 2019, 366, 326–334 Search PubMed.
- D. M. Needham, S. Yoshizawa, T. Hosaka, C. Poirier, C. J. Choi, E. Hehenberger, N. A. T. Irwin, S. Wilken, C.-M. Yung, C. Bachy, R. Kurihara, Y. Nakajima, K. Kojima, T. Kimura-Someya, G. Leonard, R. R. Malmstrom, D. R. Mende, D. K. Olson, Y. Sudo, S. Sudek, T. A. Richards, E. F. DeLong, P. J. Keeling, A. E. Santoro, M. Shirouzu, W. Iwasaki and A. Z. Worden, A distinct lineage of giant viruses brings a rhodopsin photosystem to unicellular marine predators, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20574–20583 Search PubMed.
- K. S. Metz, E. M. Deoudes, M. E. Berginski, I. Jimenez-Ruiz, B. A. Aksoy, J. Hammerbacher, S. M. Gomez and D. H. Phanstiel, Coral: Clear and Customizable Visualization of Human Kinome Data, Cell Syst, 2018, 7, 347–350.e1 CrossRef CAS PubMed.
- F. Madeira, M. Pearce, A. R. N. Tivey, P. Basutkar, J. Lee, O. Edbali, N. Madhusoodanan, A. Kolesnikov and R. Lopez, Search and sequence analysis tools services from EMBL-EBI in 2022, Nucleic Acids Res., 2022, 50(W1), W276–W279 CrossRef CAS PubMed.
- G. E. Crooks, G. Hon, J.-M. Chandonia and S. E. Brenner, WebLogo: a sequence logo generator, Genome Res., 2004, 14, 1188–1190 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00122b |
| ‡ Current address: Department of Molecular and Cellular Biology, Harvard University, Cambridge, MA, 02138, USA. |
| § Current address: Chemical Biology Program, Sloan Kettering Institute, Memorial Sloan Kettering Cancer Center, New York, NY 10065, USA. |
| This journal is © The Royal Society of Chemistry 2025 |

