Open Access Article
Open Access ArticlePhotoaffinity enabled transcriptome-wide identification of splice modulating small molecule–RNA binding events in native cells†
Raven
Shah
 ,
Wanlin
Yan
,
Joyce
Rigal
,
Steve
Mullin
,
Lin
Fan
,
Lynn
McGregor
,
Andrew
Krueger
,
Nicole
Renaud
,
Andrea
Byrnes
* and
Jason R.
Thomas
*
,
Wanlin
Yan
,
Joyce
Rigal
,
Steve
Mullin
,
Lin
Fan
,
Lynn
McGregor
,
Andrew
Krueger
,
Nicole
Renaud
,
Andrea
Byrnes
* and
Jason R.
Thomas
*
Novartis Biomedical Research, Discovery Sciences, Cambridge, MA, USA. E-mail: andrea.byrnes@novartis.com; jason.thomas@novartis.com
First published on 3rd April 2025
Abstract
Splice modulating small molecules have been developed to promote the U1 snRNP to engage with pre-mRNAs with strong and altered sequence preference. Transcriptomic profiling of bulk RNA from compound treated cells enables detection of RNAs impacted; however, it is difficult to delineate whether transcriptional changes are a consequence of direct compound treatment or trans-acting effects. To identify RNA targets that bind directly with splice modulating compounds, we deployed a photoaffinity labeling (PAL)-based Chem-CLIP approach. Through this workflow, we identify the telomerase lncRNA (TERC) as a previously unknown target of this class of clinically relevant small molecules. Using cellular ΔSHAPE-MaP, we orthogonally validate and further define the compound binding site as likely to be the conserved CR4/5 domain. Additionally, a thorough analysis of the PAL-based Chem-CLIP data reveals that considering competed RNAs, irrespective of magnitude of enrichment, adds a rich dimension of hit calling.
Introduction
Splicing modulation via RNA-targeting small molecules is an established effective therapeutic modality in the treatment of neurodegenerative diseases such as spinal muscular atrophy (SMA) and Huntington's disease (HD).1–7 These splicing-modulating compounds interact directly with pre-mRNA/U1snRNP complexes to induce therapeutically favorable changes in splicing to alter disease progression. In the case of SMA, exclusion of exon 7 in the SMN2 gene prevents canonical RNA transcript expression, leading to an aberrant isoform that is quickly degraded through nonsense mediated decay pathways.1,2,4 Treatment with branaplam and risdiplam (Evrysdi®) can promote exon 7 inclusion by enhancing the engagement of U1snRNP with the nonconical GGA 3′ exon ending motif.1,2,4Branaplam and structurally related analogs show therapeutic efficacy in the treatment of HD, which is caused by CAG trinucleotide expansion repeats in exon 1 of the HTT gene. CAG repeat expansion leads to mutant HTT protein aggregation and downstream progressive neurodegeneration.2,7 Splicing modulation can deplete mutant HTT mRNA via inclusion of a “poison” cryptic exon that leads to frameshift-induced rapid transcript degradation.2,7 Other recent expression profiling studies with branaplam and risdiplam treatment have identified additional compound-induced cryptic exons in disease-relevant mRNAs, where inclusion of these exons led to significant reduction in transcript levels. These findings highlight the potential to use RNA-interacting small molecules to regulate target RNA steady-state levels to treat a broad spectrum of genetic disorders.8–10
Branaplam and risdiplam function as “molecular glues” bringing together the splicing machinery to nGA 3′ exonic ends in an U1 snRNP-dependent manner.1,2 Within the context of SMN2, proposed mechanisms suggest that branaplam and risdiplam act at an unpaired adenosine bulge at the −1 position (−1A) of the pre-mRNA splice site.3,11 Recent single-molecule imaging studies not only validate the −1A bulge hypothesis but also suggest that branaplam alters U1 snRNP splice site recognition. This occurs through a sequential binding mechanism where compounds bind to the U1 snRNP/U1-C complex after the initial engagement and stabilization of the −1A bulged residue at the splice junction.12 Time-dependent transcriptomic profiling of branaplam and risdiplam suggest these compounds have diverse target profiles that fluctuate across treatment duration. Though, it remains to be understood whether the extent to which these transcriptional fluctuations are a consequence of direct compound engagement or of splicing events that have already occurred.2,11,13
Capturing direct engagement between small molecule and their RNA targets can provide improved insight into compound selectivity, RNA/RNP ligandability, and functional nodes of compound regulation. The applications of electrophilic or PAL moieties, widely used in the areas of covalent drug discovery and chemoproteomics,14–17 have been adapted to enable covalent enrichment of small molecule bound RNAs coupled with next-generation sequencing (NGS).18–22 To identify the RNA interactors for the branaplam series of splicing modulators, we adopted a PAL-based Chem-CLIP workflow, as this strategy has been successfully applied to capture small molecule–RNA engagement in a PreQ1 riboswitch system20 and RNA interactors of a dihydropyrimidine 1 photoaffinity probe.19 Our aim was to establish a workflow for profiling transcriptome-wide compound-RNA binding events in native cellular environments using mechanistically well-understood tool compounds.
Results and discussion
Bioactivity characterization of diazirine-based PAL chemical probe
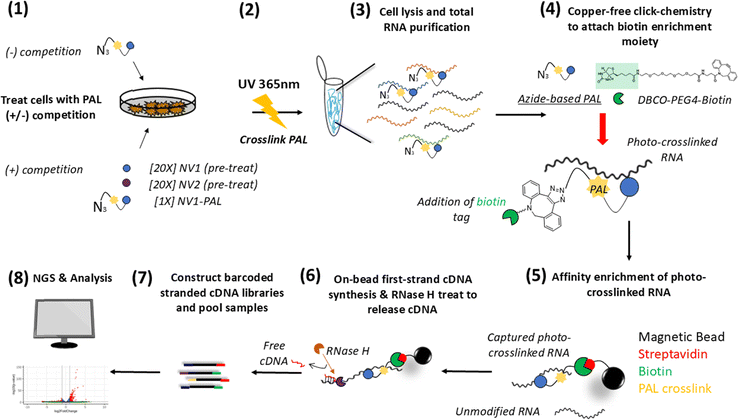
A photoaffinity probe based on NV1 was designed to incorporate a photocrosslinkable diazirine moiety and a terminal azide to enable crosslinking and subsequent enrichment (Fig. 1(a)). Compound bioactivity and cytotoxicity were assessed using a stably integrated SMN2 luciferase mini-gene reporter SH-SY5Y cell line. The reporter construct design and readout were as previously reported1. In this cell model, corrective splicing induction of exon7 within the SMN2 cassette is monitored via luciferase activity. The NV1-based PAL probe, NV1-PAL, retains comparable splicing activity as NV1 (Fig. 1(b)). Additionally, NV2 exhibited no increase in luciferase activity, consistent with previously data demonstrating that this structurally related compound lacked the ability correct SMN2 splicing.1 Also, no statistically significant change in cell viability was observed with up to 10 μM NV1-PAL treatment for 24 h (Fig. 1(c)). Splicing activity at the SMN2 genetic locus was comparable for NV1 and NV1-PAL at 1 μM treatment at the 3 h mark (Fig. 1(d)). Additionally, we confirmed that NV1-PAL retained the ability to crosslink with target RNA in cells (Fig. 1(e)). Confident that NV1-PAL maintained splicing modulating activity and is capable of photocrosslinking to RNA targets, we next set out to apply a streamlined workflow for unbiased profiling of small molecule RNA interactions in cells.Applying PAL-based Chem-CLIP in a native cellular environment to identify putative RNA targets
To enable transcriptome-wide profiling of endogenous RNA-small molecule interactions, we applied a PAL-based Chem-CLIP workflow as represented schematically in Fig. 2. Consistent with other studies,20,21 two criteria were selected for identifying RNAs likely to be direct interactors of NV1: (1) a target RNA should be enriched by NV1-PAL and (2) the enrichment of that RNA should be diminished by pre-treatment with excess NV1; i.e., competition. We multiplexed PAL-based Chem-CLIP samples with identically and contemporaneously treated samples for bulk transcriptomic profiling. We anticipated that this additional layer of multiplexing would enable correlations of enrichment for NV1-PAL-labeled RNAs with changes in expression or splicing. Also, this additional layer of multiplexing would enable direct assessment of PAL enriched transcripts with their abundance prior to enrichment across all treatment conditions.Native, wildtype SH-SY5Y cells were initially treated with DMSO, 20 μM NV1, or 20 μM NV2 for 1 h. This pre-treatment step allows the compounds to occupy binding sites on target RNAs and thus, prevent subsequent PAL enrichment. After pre-treatment, cells are then treated with 1 μM NV1-PAL for 3 h. To distinguish enrichment of RNAs driven by NV1 scaffold from enrichment driven by the PAL moiety, cells were also treated with 1 μM B1-PAL probe. Upon UV irradiation, the diazirine is converted into a carbene intermediate which crosslinks with probe-bound RNAs or is quenched by bulk solvent. Upon total RNA isolation, RNA can undergo enrichment for Chem-CLIP or simultaneously processed for bulk RNAseq.
Identification of putative RNA targets for NV1 considering enrichment and competition
High-quality cDNA libraries were generated (Fig. S1(a) and (b), ESI†) and pooled together for short-read sequencing (∼30–70 million reads/library). The majority of reads for all libraries were enriched in exonic regions of the transcriptome (Fig. S1(c), ESI†). In the analysis of the PAL-based Chem-CLIP samples, RNAs were defined as enriched if the relative abundance of the RNA following NV1-PAL treatment was at least two-fold greater than B1-PAL (p-value ≤ 0.05); for NV1-PAL 391 RNAs were enriched above B1-PAL (Fig. 3(a)). RNAs were defined as competed if the relative abundance was decreased by at least two-fold in the presence of 20 μM NV1 or NV2 (p-value ≤ 0.05); 641 RNAs were competed by pre-treatment with NV1 (Fig. 3(b)). | ||
| Fig. 3 Unbiased cellular PAL-based chem-CLIP profiling of NV1 reveals novel potential RNA targets (A) volcano plot of the relative transcript abundance comparing enrichment with NV1-PALvs.B1-PAL. (B) Volcano plot of relative transcript abundance comparing enrichment with or without 20 μM NV1. Transcripts with differential enrichment were deemed hits if the fold-change ≥2 and p-value ≤ 0.05, as calculated using DESeq2.23 (C) Scatterplot of differential enrichment (NV1-PALvs.B1-PAL) vs. competitive probe labeling (20 μM vs. 0 μM NV1). Each circle denotes an individual gene (total transcripts/gene) with p-value ≤0.05. Gray points = fold change ≤2, dark yellow = NV1-PAL enriched above B1-PAL, blue = NV1 competed RNAs, and red = enriched and competed RNAs. Dotted lines denote fold-change cut-offs (FC ≥ 2). R2 coefficient = 0.59. (D) Read density of TERC across all conditions. Greater read density is observed for the NV1-PAL treatment alone. Genome track was visualized using integrated genomics viewer (IGV).24 | ||
Based on established design criteria, RNAs statistically enriched beyond B1-PAL and whose probe labeling was competed by NV1 were defined as hits; a total 161 RNAs were enriched and competed (Fig. 3(c) and Table S1, ESI†). Many of the top RNA hits were protein-coding mRNAs not previously shown to be modulated by NV1. We also observed a diverse set of putative RNA targets (Fig. S2(a), ESI†) with a broad range of respective transcript abundances (Fig. S2(b), ESI†). The strongest putative mRNA targets for NV1 include ARL3, MICOS10, COPS6, and ATPSPF. An additional putative RNA target of NV1 is the noncoding RNA TERC (telomerase lncRNA), the RNA component of the telomerase RNP complex which is localized to Cajal bodies and indispensable for telomere maintenance.25,26 Read density from PAL-based Chem-CLIP experiments for TERC is shown in Fig. 3(d). In the NV1-PAL + NV1 samples, TERC expression was found to increase relative to NV1-PAL alone, NV1-PAL + NV2, and B1-PAL treatments (not statistically significant); however, it is unclear whether this change in transcript abundance is a result of NV1-PAL engagement (Fig. S3, ESI†). For further reference the read density for GAPDH, a transcript that does not show enrichment or competition, is shown in Fig. S4 (ESI†). For TERC, similar degrees of decreased read density resulting from competition of probe labeling with NV1 and NV2 are observed. Competitive binding in the presence of NV2 suggests this putative binding interaction is unlikely to be functionally relevant in the context of splicing modulation. These data suggest a ligandable RNA/RNP binding pocket within TERC which has also been shown previously to bind a structurally unrelated riboswitch targeting metabolite, PreQ1, via PAL-based Chem-CLIP20. With engagement observed with PreQ1 and NV1, we next wanted to ensure that TERC engagement was not an artifact of diazirine-based PAL labeling but rather NV1 binding to a highly evolutionarily conserved ncRNA with important potential therapeutic relevance.
Cellular ΔSHAPE-MaP profiling defines the CR4/5 domain of TERC as a likely binding site for NV1
For a PAL-independent cellular engagement assay, we turned to differential 2′-hydroxyl acylation reactivity coupled to mutational profiling (ΔSHAPE-MaP)27–29 as a method to provide orthogonal evidence of binding by NV1 in the same native cellular context as the PAL-based Chem-CLIP experiments. ΔSHAPE-MaP enables characterization of the small molecule binding site at single nucleotide resolution and provides insight into conformational changes in RNA secondary structure induced by compound engagement.28,30,31 Coupled to mutational profiling (MaP), chemically acylated nucleotides are miscoded during reverse transcription, and these mutational mismatches can be detected and read-out via NGS and read alignment.To improve sequence coverage of TERC lncRNA, target TERC RNA was further enriched post cDNA generation via capture oligonucleotide probes. Sufficient sequencing depth was achieved to ensure 300× coverage of each residue of TERC (Fig. S5(a) and (b), ESI†). Significantly increased mutation frequencies were observed for only NAI probe treated samples (Fig. S5(c), ESI†). For the DMSO-treated cells, TERC exhibits distinct regions of high and low chemical probe reactivity indicating a highly structured RNA in cells (Fig. S6(a), ESI†). SHAPE-MaP performed in the presence of NV1 produces a similar overall pattern of regions of differential high and low reactivity, suggesting that NV1 does not globally alter the secondary structure of TERC RNA (Fig. 4(a)).
 | ||
| Fig. 4 Cellular differential SHAPE-MaP of TERC lncRNA without and with NV1. (A) Single nucleotide resolution of differential reactivity for DMSO vs. 10 μM NV1 treated samples. Blue dots indicate regions of significant differences in reactivity (z-score ≥ 2). Positive values indicate an increase in reactivity in the presence of NV1, while negative values indicate a decrease in reactivity. Noteworthy changes in reactivity are indicated by the boxed region within the conserved CR4/5 domain of TERC lncRNA. (B) Secondary structure within the CR4/5 domain of TERC lncRNA. Boxed region indicates site of significant clustered changes in chemical reactivity in the presence of compound. The bulged residue 249C shows the most significant reduction in reactivity and is likely important for NV1 binding. Secondary structures with SHAPE reactivity constraints were visualized using Forna.32 Nucleotides are colored based on structure (stems = green, multiloops/junctions = red, interior loops = yellow, hairpin loops = blue, 5′ and 3′ unpaired region = orange). All SHAPE-MaP experiments were performed with biological replicates (N = 2) and include no NAI probe controls. Differential reactivities plotted 5′ → 3′ orientation of TERC lncRNA. | ||
Applying ΔSHAPE, which compares chemical probe reactivity measurements between DMSO and NV1-treated samples, statistically significant changes in SHAPE reactivity were identified. NV1-treatment yielded both increases and decreases in SHAPE reactivity, indicating secondary structure rearrangements or differences in protein binding at the RNA binding interface upon NV1 engagement. TERC RNA contains 5 distinct structural regions: core pseudoknot, CR7, the H/ACA binding region, the hypervariable region, and the C4/5 domain.26,33 Of these domains, the highly conserved C4/5 domain contained the greatest number and the strongest cluster of residues with decreased chemical probe reactivity in the presence of NV1 (Fig. 4(a)). We note that within the CR4/5 domain, C249 shows the strongest decrease in probe reactivity in the presence of NV1 and may hint at this residue's importance for NV1 binding to TERC RNA (Fig. 4(b) and Fig. S6(b), ESI†). The observed changes in SHAPE-MAP reactivity provides the intended orthogonal validation that NV1 likely binds directly with TERC RNA. Given, the number and magnitude of significant changes in SHAPE reactivity localized within the CR4/5 domain we propose that this domain is the likely site of compound engagement.
SAR-dependent competition as a hit calling criteria enables identification of on-mechanism mRNAs
While further characterization of the NV1-TERC interaction via SHAPE reactivity profiling validates the PAL experiment itself, there were only a few mRNA expected targets1,13 of NV1 that were also enriched and competed. While 161 RNAs were both enriched over B1-PAL and competed by NV1, many of these RNAs were ncRNAs and not expected on-mechanism mRNA splice targets (3′ nGA exon ending). In reviewing the two criteria for hit calling, we noticed that while known targets were able to meet the at least 2-fold competition criteria, they consistently failed to show greater than 2-fold enrichment over B1-PAL. Attempts to normalize or use other criteria to assess enrichment (differential peak calling, normalizing chem-CLIP read counts with bulk RNAseq, and transcript-dependent factor normalizations) repeatedly failed to show known targets as enriched (Fig. S7, ESI†) and thus we could not find a method of analysis where the known targets would meet the designed two criteria for hit calling (enrichment and competition).One of the advantages of multiplexing PAL-based Chem-CLIP samples with identically treated bulk RNASeq samples is that it allows for a direct comparison of enrichment through binding affinity versus transcript abundance. Comparing the relative transcript abundance following PAL-based Chem-CLIP with bulk RNASeq samples shows no covariation (Fig. 5(a)). This suggests that PAL-based affinity enrichment has an altered rank order of transcript abundance. In contrast, the relationship between transcript abundance in the competition samples for PAL-based Chem-CLIP versus bulk RNASeq shows a higher degree of positive covariation (R2 = 0.49) (Fig. 5(b)). This implies the competition samples (20 μM NV1 + 1 μM NV1-PAL) are more like bulk RNASeq samples despite the presence of NV1-PAL. We interpret this to indicate that presence of competitor decreases PAL labeling, thus returning the relative abundance back towards their nonenriched states. Taken together, these observations indicate that competition, even without significant enrichment, may contain more relevant target engagement information than previously appreciated.
A potential cause for concern in using competition by NV1, irrespective of the magnitude of enrichment, as a hit calling criteria is that decreases in transcript abundance following NV1 treatment may bias the data. It is well established that splicing modulation is often associated with a decrease in respective transcript levels.8,10,13 Indeed, we observe that RNAs competed by NV1 have significantly lower transcript abundances relative to NV1-PAL treatment alone (Fig. 5(c)). However, statistically significant fold changes between competed transcripts (PAL-based Chem-CLIP) vs. transcripts with decreased abundance upon compound treatment (RNASeq) were distinctly different (Fig. 5(d) and Fig S8, ESI†).
We note that while compound-competed transcripts were differentiated from transcripts with decrease abundance, there was some degree of overlap between these groups. To identify on-target splicing related hits, we filtered for transcripts whose enrichment was decreased by at least 2-fold in the presence of NV1 and unaffected by NV2 (inactive) (Fig. 5(e)). Recall that 20 μM NV1 competed the enrichment of 641 transcripts. Of these, 202 RNAs were competed by both NV1 and NV2, while 439 RNAs were selectively competed by NV1 and not NV2. Interestingly, among the 202 RNAs competed by NV1 and NV2, many of these RNAs were also successfully enriched above B1-PAL. These include ARL3, MICOS10, COPS6, ATPSPF, and TERC (Fig. S9, ESI†).
In terms of selective competition, 439 RNAs were exclusively competed by NV1 and of these 235 were found to be differentially spliced in their corresponding bulk RNASeq samples (Fig. 5(f) and Table S2, ESI†). In contrast, 207 RNAs were competed by NV2 alone and none of these were found to be differentially spliced in corresponding bulk RNASeq samples. The observation that only NV1 competed RNAs were differentially spliced underscores that the known splicing specific mechanisms can be captured in the competition data despite any apparent sign of enrichment.
SAR-dependent competition and not just competition alone is critical for hit calling
An important disclaimer about the competitor treated samples: we do not believe that most differentially spliced transcripts in the competition samples are the result of the U1snRNP molecular glue mechanism. Upon 1 μM NV1-PAL treatment, we observe 6116 unique differentially spliced genes relative to B1-PAL (background) using DEXseq.34 In contrast, the NV1 competition samples (20 μM NV1 + 1 μM NV1-PAL) showed 9161 differentially spliced transcripts. This large difference in number of differentially spliced genes highlights a principal weakness of our study, namely using too large of an excess of competitor ligand. Hence, our focus on only RNAs selectively competed by NV1 but not NV2 as potential NV1 targets.Novel putative target identified from unbiased profiling
One of the additional value propositions of unbiased profiling is the access to novel target engagement data. While assessing expected targets of NV1, we noticed that GATA3 was competed by NV1 and differentially spliced (Fig. 6(a)), suggesting possible target engagement. GATA3 is a transcription factor implicated in breast cancer prognosis35. GATA3 has a 3′ AGA exon ending containing transcript isoform. Interestingly, NV1 treatment was associated with a decrease of exon inclusion of the 3′ AGA exon within GATA3, as well as a decrease in inclusion of other exons within the transcript. These splicing changes were also associated with a significant reduction in total GATA3 transcript abundance in the (20 μM NV1 + 1 μM NV1-PAL)-treated samples (Fig. 6(b)). Collectively, these data suggest NV1 engagement inhibits exon inclusion and ultimately leads to decrease in GATA3 transcript abundance. Of the 235 RNAs competed and differentially spliced in the presence of NV1, 10 RNAs contain the expected 3′ nGA exon motif (Fig. 6(c)). It is important to recall that these experiments were performed with a 4 h treatment duration and likely fail to capture the total 3′ nGA exons that were likely compound-induced in other reported experiments with longer treatment times (typically > 24).Conclusion
Previous RNA target identification studies using covalent chemical probes18–20,22,36 have demonstrated feasibility of covalent probe-mediated RNA enrichment coupled to NGS. These pioneering studies leveraged exogenous expression of high-affinity aptamers19,20 or were performed in cellular lysates, establishing critical proof-of-concept and forming foundational guidelines for transcriptome-wide profiling of RNA-small molecule interactions. Building on these prior studies, our aim was to profile the RNA targeting landscape of a class of clinically relevant splicing modulators under endogenous cellular conditions.Using conventional hit calling criteria, we identified the lncRNA TERC as a previously unknown target of NV1 and NV2. Further characterizing the NV1-TERC interaction with ΔSHAPE-MaP highlighted a defined region within the CR4/5 domain of TERC showing decreased chemical reactivity. Further work is needed to better define the binding of NV1 with TERC at a molecular level. While we propose initially focusing such efforts on the CR4/5 domain, we remain open to the possibility that NV1-TERC engagement is protein-mediated or via a binary RNA-interacting mechanism analogous to BIBR1532, a previously described TERC inhibitor.37
To enable the advancement of translating PAL-based Chem-CLIP into an endogenous cellular system, we leveraged knowledge starting from a well-characterized splicing tool compound to determine experimental parameters and target hit calling criteria. The extensive characterization of U1snRNP-dependent molecular glues enabled us to pinpoint that lack of enrichment as the failing criteria in our study. Also, we leveraged this information to focus on competition, even in absence of enrichment, as a meaningful way to interpret the data. It is difficult to discern why anticipated targets consistently failed to demonstrate enrichment. In the design of the PAL probe we focused on identifying a PAL probe with comparable potency as NV1. Perhaps a deeper SAR exploration of the linker chemistry and reactive group would furnish a second-generation probe with potent cellular activity and selective SMN2 enrichment/competition.
While we demonstrate that early binding events for NV1 are consistent with expected on-mechanism mRNA hit calling from expression profiling studies, we also capture engagement with diverse RNA biotypes that may be a product of compound cellular distribution or suggest engagement with structured RNA/RNP active sites. The latter would indicate potential druggable RNAs that could be developed into functional modulators through traditional medicinal chemistry or by imparting functionality into the compounds; for example, RIBOTACs or covalent RNA binders.38–40
Methods
General details for photoaffinity probe synthesis and characterization
Detailed chemical synthesis schemes with corresponding characterization of chemical photoaffinity probe can be found in the ESI.† General operating procedures of LC/MS, high resolution mass spectral analysis, and HNMR can be found below.Cell culture conditions
The adherent human neuroblastoma bone marrow biopsy-derived cell line, SH-SY5Y cells (Sigma catalog: 86012802), were cultured in DMEM/F-12 (Gibco, cat: 11320033) supplemented with 10% FBS and 1% penicillin/streptomycin. All cells were cultured at 37 °C in 5% CO2 in humidified incubators and were free from mycoplasma (MycoAlert Detection Kit, Lonza).Bioactivity characterization of diazirine-based PAL chemical probes
Bioactivity of chemical probes relative to parent compound was tested using the SMN2 mini-gene luciferase reporter (SH-SY5Y cells). 1.2 × 104 cells were seeded into individual wells of two separate 384-well plates for activity and cytotoxicity assays and allowed to recover at 37 °C with 5% CO2 overnight. The following day, serial dilution of chemical probe or parent compound was added, then following 24 h incubation, the luciferase and cytotoxicity assays readings were performed. Luciferase expression and cytotoxicity was assessed using the Promega Bright-Glo luciferase assay and CellTiter-Glo luminescent cell viability assay, respectively. All assays were performed triplicate.Cellular treatment for PAL-based Chem-CLIP workflow
For the PAL-based Chem-CLIP workflow, all treatment conditions were performed in triplicate. 1.2 × 107 SH-SY5Y cells were seeded into individual 10 cm2 cell culture plates using FluoroBrite DMEM media (Gibco, cat: A1896701) supplemented with 1% FBS and allowed to incubate at 37 °C with 5% CO2 overnight. Cells were then treated with either DMSO or 20 μM competitor compound for 1 h at 37 °C with 5% CO2. Next, cells were treated with either DMSO or 1 μM PAL probe for 3 h at 37 °C with 5% CO2. Prior to photocrosslinking, cells were washed once with DPBS (Gibco, cat: 14190144), after which 5 mL of cold DPBS was added to each plate. Photocrosslinking was performed for 20 min at 4 °C with 365 nm wavelength light (UVP Blak-Ray XX-40BLB UV bench lamp, 40 Watt). Cells are then maintained at 4 °C and harvested via scraping. The detached cells and media were pelleted (300 × g for 5 minutes at 4 °C). The supernatant was aspirated and the cell pellets flash frozen and stored at −80 °C until further processing.RNA extraction for PAL-based Chem-CLIP workflow
Cells are lysed and RNA is extracted following the Qiagen RNeasy Plus (cat: 74134) following manufacturer's protocol. Cell pellets were resuspended in 600 μL of RLT lysis buffer and homogenized using the Qiagen QIAshredder (cat: 79656). 600 μL of 70% ethanol was added to the flow-through of the QIAshredder column and transferred to a gDNA column provided by the RNeasy kit. The remainder of the RNeasy protocol is followed for total RNA isolation including on-column DNase digestion using the Qiagen DNase set (cat: 79254). Quality and quantification of purified RNA is evaluated on a Nanodrop prior to further processing.Copper-free click chemistry for PAL-based Chem-CLIP workflow
Copper-free click reactions were performed in nuclease-free water with 10 μg of total RNA, 10 μM final concentration of DBCO-PEG4-desthiobiotin (Sigma Aldrich cat: 760749) with 1 unit per μL final concentration of SUPERase·In™ RNase Inhibitor (Invitrogen cat: AM2694). The click-reaction was allowed to proceed at room temperature for 2 h with gentle agitation. Following completion of the copper-free click chemistry reaction, total RNA was purified from other reaction components using a Zymo RNA spin column (Zymo Research, cat = C1003-50). RNA was eluted from the columns with 22 μL of nuclease-free water. Biotinylated-RNA was stored at −20 °C until ready for processing.Enrichment of biotinylated RNAs for PAL-based Chem-CLIP workflow
Magnetic streptavidin beads (Invitrogen Dynabeads MyOneStreptavidin C1 (cat# 65002)) were blocked in batch at room temperature in a blocking buffer (0.03% BSA, 10 μM yeast tRNA (Invitrogen, cat # AM7120G)) with end-over-end agitation. Ensure sufficient beads to allow for 20 μL of beads/sample during enrichment step. Following blocking, beads are washed three times with wash buffer (50 mM Tris–HCl (pH 7.4), 100 mM NaCl, 10 mM EDTA (pH 8.0), 0.2% Tween-20, 1 unit per μL SUPERase·In™ RNase Inhibitor), then resuspended to make a slurry and aliquoted to individual tubes to provide 20 μL of beads/sample. When ready to proceed to final enrichment step, the beads were separated from the wash buffer using a magnetic stand, then spun at 200 × g for 1 min and any residual wash buffer was removed by aspiration.For enrichment with streptavidin, 20 μL of Zymo purified RNA from the click reaction was added to individual aliquots of beads. Incubation of streptavidin beads with RNA proceeded for 2 h at room temperature with end-over-end agitation. Following this incubation, the supernatant was removed and beads were washed three times with 200 μL wash buffer, then with 200 μL of nuclease-free water to remove residual wash buffer which contains EDTA and may impair reverse transcription efficiency.
On-bead reverse transcription and cDNA generation for PAL-based Chem-CLIP workflow
Following streptavidin-mediated enrichment of photocrosslinked RNAs, reverse transcription is performed directly on beads. The SMARTer Stranded Total RNA-seq kit v3 – Pico input (Takara, cat: 634486) was used with some modification from the manufacturer's protocol. Magnetic streptavidin beads with bound biotinylated target RNAs were resuspended in 10 μL of nuclease-free water.To fragment probe-labeled RNAs, 8 μL of resuspended RNA-bead mixture was added to individual wells of a PCR plate containing 1 μLSMART Pico Oligos Mix v3, 4 μL 5× First-Stranded Buffer and incubated at 94C for 3.5 min, then immediately placed on ice to halt further fragmentation. The manufacturer's protocol for reverse transcription reaction was followed. It is worth noting to take extra precautions to homogenize the reverse transcription reactions as the solution is viscous.
Following completion of the first strand synthesis reaction, 20 μL of each reaction mixture (beads and solution) was transferred into individual wells of a PCR plate. RNase H cleavage reactions were initiated with the addition of 5 μL 10× RNase H reaction buffer, 24 μL nuclease-free water, and 1 μL RNase H (New England Biosciences, cat #M0523). RNase H cleavage reaction proceeded at 37 °C for 20 minutes with gentle agitation. RNase H cleavage reaction was quenched with addition 1 μL0.5 M EDTA (pH 8.0).
Magnetic streptavidin beads were removed from the reaction solution with aid of magnetic stand. First strand synthesized cDNAs were further purified using RNAclean XP beads (Beckman Coulter, cat# A63987) with the addition of 100 μL of RNAclean XP beads to each reaction mixture, incubated at room temp for 5 min. Supernatant was removed with aid of magnetic stand. RNAclean XP beads were washed twice with 100 μL70% ethanol. Carefully pipette residual ethanol from samples and allow beads to air dry for 10 minutes. Single-stranded cDNA was eluted from beads with 22 μL nuclease-free water and proceed to first PCR amplification step in library generation per manufacturer protocol including ribodepletion until final PCR amplification step. The number of final PCR cycles may require optimization depending on crosslinking, enrichment efficiency, and RNA quality. Our final libraries were generated with 14 PCR cycles for PAL-based Chem-CLIP cDNA libraries. Final library quality was assessed using a D1000 TapeStation system (Agilent, cat = 5067–5582).
Reverse transcription and cDNA generation for bulk RNAseq
Wildtype SH-SY5Y cells were treated identically and in parallel with cellular treatment for PAL-based Chem-CLIP workflow. RNA was harvested identically and in parallel with RNA extraction for PAL-based Chem-CLIP workflow. Reverse transcription and cDNA generation were performed as described for On-bead reverse transcription and cDNA generation for PAL-based Chem-CLIP workflow, except that 10 ng of total RNA input was used in place of Magnetic streptavidin beads bound to biotinylated target RNAs. Also, during the final PCR amplification step, 10 PCR cycles were performed.NGS sequencing parameters for PAL-based chem-CLIP and bulk RNAseq
Libraries were diluted to 5 nM in TE buffer (pH 8.0), pooled, denatured, and loaded on the Illumina NovaSeq 6000 instrument (Part number 20012850) for sequencing following the manufacturer's instructions. The PAL-based Chem-CLIP libraries were loaded at a final concentration of 300 pM with a 6% PhiX spike in to increase sequence diversity. The bulk RNAseq libraries were loaded at a final concentration of 100 pM with a 1% PhiX spike, per manufacturer's recommendation. Each library set was independently loaded and sequenced using the NovaSeq 6000 SP Reagent Kit v1.5 (Part number 20027466) per the manufacturer's instructions. The PAL-based Chem-CLIP libraries were sequenced 53 base pair paired-end with 8 base dual indexes and trimmed to 50 base pairs. The bulk RNAseq libraries were sequenced 51 base pair (paired-end) with 10 base dual indexes and trimmed to 50 base pairs. The sequence intensity files were generated using the Illumina Real Time Analysis software (RTA3). An internal data processing pipeline was used to demultiplex, create, extract and trim the FASTQ files using Illumina's bcl2fastq software (v2.20). An average of 30 million reads and 40 million reads per sample were obtained for the PAL-based Chem-CLIP and bulk RNAseq libraries, respectively, with a minimum of 23 million reads per sample.Analysis of PAL-based Chem-CLIP and RNAseq datasets
A previously reported exon quantification pipeline (version 2.5)41 with STAR (version 2.7.3a)42 was used to align the reads against the human genome reference from the latest build (GRCh38). Differential gene expression and differential PAL-based Chem-CLIP enrichment were determined using DESeq2 (v1.38.3).23 RNAs were defined to be differentially expressed or enriched, respectively, if |log2FC| ≥ 2 and p-value ≤ 0.05 relative to B1-PAL. Alternative strategies for normalization of PAL-based Chem-CLIP read counts apply transcript-dependent normalization, where enriched read counts are matched with a bulk RNAseq (transcript abundance) read count matrix. Normalization matrices are then set to a row-wise geometric mean ∼1. Sample code can be found in ESI,† Fig S7.Splicing analysis of bulk RNAseq samples was performed using the DEXseq splicing analysis pipeline, an algorithm for detecting differentially spliced exons by surveying junction reads.34 Changes in splicing and gene expression were correlated to PAL-based Chem-CLIP to understand if compound engagement leads to a functional response.
RT-qPCR to assess splicing efficiency of PAL probes & qPCR read-out of PAL-enriched cDNA libraries
SMN2 mini-gene expressing SH-SY5Y cells were plated into individual wells of 6-well plate (1 × 106 cells per well) and allowed to recover at 37 °C with 5% CO2 overnight. Cells were treated 1 μM for the indicated time points. Cells are harvested, and total RNA is isolated as described in RNA extraction for PAL-based Chem-CLIP workflow. Reverse transcription is performed following the manufacturer's protocol for SuperScript II (Invitrogen, cat# 18064-014) and qPCR was performed using the 2X SYBR Green qPCR master mix (Invitrogen, cat = A46109) using standard cycling conditions (50 °C for 2 minutes (min), 95 °C for 2 min followed by 40 cycles of 95 °C for 15 seconds (s) and 60 °C for 60 seconds). Primer sequences used to assess splicing kinetics was 5′-GAAGGAAGGTGCTCACATTCCT-3′ (SMN2 exon7, Forward) and 5′-GGCGCCGGGCCTTTCTTTATG-3′ (Luciferase, Reverse).PAL-enriched cDNA libraries can also be generated using the SuperScript II protocol, performing the RT reaction directly on the magnetic streptavidin beads. Following first-strand cDNA synthesis, a subsequent RNase H reaction and cDNA clean-up is performed as detailed in the chem-CLIP protocol to release single-stranded cDNA from the photocrosslinked RNA templates. The final cDNA can then be used as input for qPCR using the 2X SYBR Green qPCR master mix. This procedure is how we tested photocrosslinking to target RNAs in cells in Fig. 1(e).
Cellular SHAPE-MaP cDNA library generation:
SH-SY5Y cells were plated in 10 cm2 cell culture plates with DMEM/F-12 antibiotic-free media supplemented with 10% FBS. The following day, cells were treated with DMSO or 10 μM NV1 for four hours. Following treatment duration, cells were harvested (cell scraper), spun down (300 × g), resuspended in 950 μL of room temperature PBS. 50 μL of DMSO (background mutation control) or 50 μL of 2M NAI (EMD Millipore; cat# 03-310) were added to the resuspended cell mixture for 15 minutes at 37 °C with gentle agitation. DMSO samples were processed initially to minimize over acylation. Following NAI treatment, samples were spun down in 4C centrifuge (1000 × g) for 5 minutes, and supernatant was aspirated without disturbing the cellular pellet. Pellets were washed and resuspended in chilled PBS. Samples were spun down again, and PBS was aspirated without disturbing pellets. Cellular pellets were flash frozen and stored at −80 °C. Total RNA was isolated as described in RNA extraction for PAL-based Chem-CLIP workflow, except that the RNA was eluted in 50 μL nuclease-free water. Final sample quality was assessed on an RNA TapeStation and concentration/purity was assessed with the Nanodrop.10 μg of total RNA isolated from each SHAPE-treated sample was subjected to rRNA depletion, and 100 ng of rRNA-depleted RNA samples were fragmented and subsequently phosphorylated in preparation for ligation. After phosphorylation, barcoded RNA adapters were ligated to the 3′ end of the fragmented RNA. RNA was reverse transcribed in the presence of MnCl2 and betaine as previously described27 to induce mutations in the cDNA followed by ssDNA adapter ligation, quantitative PCR, and PCR amplification to generate sequencing libraries. 500 ng of each final library was pooled, and probe hybridization was performed followed by biotin-based enrichment of target sequences. 5′- biotinylated enrichment probes were designed to tile the target sequence using the FASTA sequence for ENSG00000270141, and a custom panel was created by mixing equimolar concentrations of each probe. Custom probes were designed and purchased from Integrated DNA Technologies. Following hybridization to target TERC lncRNA, biotinylated probes hybridized to the target complex were enriched via magnetic streptavidin beads. Beads were washed using the wash buffer (50 mM Tris–HCl (pH 7.4), 100 mM NaCl, 10 mM EDTA (pH 8.0), 0.2% Tween-20, 1 unit per μL SUPERase·In™ RNase Inhibitor). Post-enrichment amplification was performed on the purified target sequences and quantified on an Agilent 4200 TapeStation. The enriched cDNA pool was sequenced on a NextSeq 1000/2000.
Cellular SHAPE-MaP analysis
Reads were barcode trimmed with cutadapt 1.14 and mapped to a custom reference genome generated using the primary sequence of TERC lncRNA. There was incomplete coverage using the sequence for ENSG00000270141, thus the primary sequence for telomerase-vert (RF00024) was used (ENSG00000277925) to generate the reference genome. Star aligner version 2.4.0i was used for reference alignment, and duplicate reads were filtered using umItools 0.5.0. Downstream analysis was performed as previously described27 using the ShapeMapper 2.0 pipeline.27,43,44The output of SHAPE reactivity scores were then used as constraints for secondary structure prediction using RNAstructure.45 The secondary structure generated using SHAPE reactivities was exported to the forna visualization software32 to compare secondary structures of DMSO with NV1 treated samples.
Curation of differentially expressed exons with 3′ nGA exon endings for quantification of 3′ nGA exon ending enrichment in NV1 probe target profiles
Starting with the splicing output from DEXseq34 to compile splicing junctions of NV1 treated cells from the bulk RNAseq analysis, bed files were created using the exon boundaries. The primary sequence of these respective boundaries from the GRCh38 reference genome build were extracted using bedtools.46 Exons with 3′ nGA endings were then further extracted and cross-referenced back to the NV1-PAL RNA target profiles to identify putative RNA interactors with possible 3′ nGA ending cryptic exons.Data availability
PAL-based Chem-CLIP, bulk RNAseq, and SHAPE-MaP raw sequencing fastq files have been deposited on SRA with the associated BioProject (PRJNA1187381). Custom analysis scripts for data processing and quality control can be provided upon request. Data supporting the findings of this study not found within the main text of the paper can be found in the ESI† files.Conflicts of interest
All authors on this paper are associated with Novartis Biomedical Research.Acknowledgements
The authors would like to thank Razvan Nutiu for his support in earliest phases of these studies and Steve Canham for engaging discussions and review of the manuscript. We also thank Eclipse Bioinnovations for their help with SHAPE-MaP sample preparation and analysis. R. S. was supported by the Novartis Biomedical Research Postdoctoral Office.References
- J. Palacino, S. E. Swalley, C. Song, A. K. Cheung, L. Shu and X. Zhang, et al., SMN2 splice modulators enhance U1-pre-mRNA association and rescue SMA mice, Nat. Chem. Biol., 2015, 11(7), 511–517 CrossRef CAS PubMed.
- C. G. Keller, Y. Shin, A. M. Monteys, N. Renaud, M. Beibel and N. Teider, et al., An orally available, brain penetrant, small molecule lowers huntingtin levels by enhancing pseudoexon inclusion, Nat. Commun., 2022, 13(1), 1150 CrossRef CAS PubMed.
- F. Malard, A. C. Wolter, J. Marquevielle, E. Morvan, A. Ecoutin and S. H. Rüdisser, et al., The diversity of splicing modifiers acting on A-1 bulged 5′-splice sites reveals rules for rational drug design, Nucleic Acids Res., 2024, 52(8), 4124–4136 CrossRef CAS PubMed.
- H. Ratni, M. Ebeling, J. Baird, S. Bendels, J. Bylund and K. S. Chen, et al., Discovery of Risdiplam, a Selective Survival of Motor Neuron-2 (SMN2) Gene Splicing Modifier for the Treatment of Spinal Muscular Atrophy (SMA), J. Med. Chem., 2018, 61(15), 6501–6517 CrossRef CAS PubMed.
- M. Sivaramakrishnan, K. D. McCarthy, S. Campagne, S. Huber, S. Meier and A. Augustin, et al., Binding to SMN2 pre-mRNA–protein complex elicits specificity for small molecule splicing modifiers, Nat. Commun., 2017, 8(1), 1476 CrossRef PubMed.
- A. Garcia-Lopez, F. Tessaro, H. R. A. Jonker, A. Wacker, C. Richter and A. Comte, et al., Targeting RNA structure in SMN2 reverses spinal muscular atrophy molecular phenotypes, Nat. Commun., 2018, 9(1), 2032 CrossRef PubMed.
- F. Krach, J. Stemick, T. Boerstler, A. Weiss, I. Lingos and S. Reischl, et al., An alternative splicing modulator decreases mutant HTT and improves the molecular fingerprint in Huntington's disease patient neurons, Nat. Commun., 2022, 13(1), 6797 Search PubMed.
- Z. L. McLean, D. Gao, K. Correia, J. C. L. Roy, S. Shibata and I. N. Farnum, et al., Splice modulators target PMS1 to reduce somatic expansion of the Huntington's disease-associated CAG repeat, Nat. Commun., 2024, 15(1), 3182 Search PubMed.
- E. W. Ottesen, N. N. Singh, D. Luo and R. N. Singh, High-affinity RNA targets of the Survival Motor Neuron protein reveal diverse preferences for sequence and structural motifs, Nucleic Acids Res., 2018, 46(20), 10983–11001 CAS.
- B. Fair, C. B. A. Najar, J. Zhao, S. Lozano, A. Reilly and G. Mossian, et al. Global impact of aberrant splicing on human gene expression levels, bioRxiv, 2023.
- Y. Ishigami, M. S. Wong, C. Martí-Gómez, A. Ayaz, M. Kooshkbaghi and S. M. Hanson, et al., Specificity, synergy, and mechanisms of splice-modifying drugs, Nat. Commun., 2024, 15(1), 1880 CrossRef CAS PubMed.
- D. S. White, B. M. Dunyak, F. H. Vaillancourt and A. A. Hoskins A Sequential Binding Mechanism for 5′ Splice Site Recognition and Modulation for the Human U1 snRNP, bioRxiv, 2024.
- E. W. Ottesen, N. N. Singh, D. Luo, B. Kaas, B. J. Gillette and J. Seo, et al., Diverse targets of SMN2-directed splicing-modulating small molecule therapeutics for spinal muscular atrophy, Nucleic Acids Res., 2023, 51(12), 5948–5980 CrossRef CAS PubMed.
- F. Offensperger, G. Tin, M. Duran-Frigola, E. Hahn, S. Dobner and C. W. A. Ende, et al., Large-scale chemoproteomics expedites ligand discovery and predicts ligand behavior in cells, Science, 2024, 384(6694), eadk5864 CrossRef CAS PubMed.
- M. L. Matthews, L. He, B. D. Horning, E. J. Olson, B. E. Correia and J. R. Yates 3rd, et al., Chemoproteomic profiling and discovery of protein electrophiles in human cells, Nat. Chem., 2017, 9(3), 234–243 CrossRef CAS PubMed.
- C. Wang, E. Weerapana, M. M. Blewett and B. F. Cravatt, A chemoproteomic platform to quantitatively map targets of lipid-derived electrophiles, Nat. Methods, 2014, 11(1), 79–85 CrossRef CAS PubMed.
- J. R. Thomas, S. M. Brittain, J. Lipps, L. Llamas, R. K. Jain and M. Schirle, A Photoaffinity Labeling-Based Chemoproteomics Strategy for Unbiased Target Deconvolution of Small Molecule Drug Candidates, Methods Mol. Biol., 2017, 1647, 1–18 CrossRef PubMed.
- A. N. Sexton, L. E. Vandivier, J. C. Petter, H. Mukherjee and J. C. Blain, Determination of RNA-ligand interactions with the photoaffinity platform PEARL-seq, Methods, 2022, 205, 83–88 CrossRef CAS PubMed.
- H. Mukherjee, J. C. Blain, L. E. Vandivier, D. N. Chin, J. E. Friedman and F. Liu, et al., PEARL-seq: A Photoaffinity Platform for the Analysis of Small Molecule–RNA Interactions, ACS Chem. Biol., 2020, 15(9), 2374–2381 CrossRef CAS PubMed.
- S. Balaratnam, C. Rhodes, D. D. Bume, C. Connelly, C. C. Lai and J. A. Kelley, et al., A chemical probe based on the PreQ(1) metabolite enables transcriptome-wide mapping of binding sites, Nat. Commun., 2021, 12(1), 5856 CrossRef CAS PubMed.
- B. M. Suresh, W. Li, P. Zhang, K. W. Wang, I. Yildirim and C. G. Parker, et al., A general fragment-based approach to identify and optimize bioactive ligands targeting RNA, Proc. Natl. Acad. Sci. U. S. A., 2020, 117(52), 33197–33203 CrossRef CAS PubMed.
- L. Fang, W. A. Velema, Y. Lee, L. Xiao, M. G. Mohsen and A. M. Kietrys, et al., Pervasive transcriptome interactions of protein-targeted drugs, Nat. Chem., 2023, 15(10), 1374–1383 CrossRef CAS PubMed.
- M. I. Love, W. Huber and S. Anders, Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2, Genome Biol., 2014, 15(12), 550 CrossRef PubMed.
- J. T. Robinson, H. Thorvaldsdóttir, W. Winckler, M. Guttman, E. S. Lander and G. Getz, et al., Integrative genomics viewer, Nat. Biotechnol., 2011, 29(1), 24–26 CrossRef CAS PubMed.
- H. Laprade, E. Querido, M. J. Smith, D. Guérit, H. Crimmins and D. Conomos, et al., Single-Molecule Imaging of Telomerase RNA Reveals a Recruitment-Retention Model for Telomere Elongation, Mol. Cell, 2020, 79(1), 115–126 CrossRef CAS PubMed.
- Q. Zhang, N. K. Kim and J. Feigon, Architecture of human telomerase RNA, Proc. Natl. Acad. Sci. U. S. A., 2011, 108(51), 20325–20332 CrossRef CAS PubMed.
- M. J. Smola and K. M. Weeks, In-cell RNA structure probing with SHAPE-MaP, Nat. Protoc., 2018, 13(6), 1181–1195 CrossRef CAS PubMed.
- S. Martin, C. Blankenship, J. W. Rausch and J. Sztuba-Solinska, Using SHAPE-MaP to probe small molecule–RNA interactions, Methods, 2019, 167, 105–116 CrossRef CAS PubMed.
- A. M. Mustoe, S. Busan, G. M. Rice, C. E. Hajdin, B. K. Peterson and V. M. Ruda, et al., Pervasive Regulatory Functions of mRNA Structure Revealed by High-Resolution SHAPE Probing, Cell, 2018, 173(1), 181–195 CrossRef CAS PubMed.
- M. J. Zeller, O. Favorov, K. Li, A. Nuthanakanti, D. Hussein and A. Michaud, et al., SHAPE-enabled fragment-based ligand discovery for RNA, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(20), e2122660119 CrossRef CAS PubMed.
- Y. Wang, S. Parmar, J. S. Schneekloth and P. Tiwary, Interrogating RNA-Small Molecule Interactions with Structure Probing and Artificial Intelligence-Augmented Molecular Simulations, ACS Cent. Sci., 2022, 8(6), 741–748 CrossRef CAS PubMed.
- P. Kerpedjiev, S. Hammer and I. L. Hofacker, Forna (force-directed RNA): Simple and effective online RNA secondary structure diagrams, Bioinformatics, 2015, 31(20), 3377–3379 CrossRef CAS PubMed.
- L. Chen, C. M. Roake, A. Freund, P. J. Batista, S. Tian and Y. A. Yin, et al., An Activity Switch in Human Telomerase Based on RNA Conformation and Shaped by TCAB1, Cell, 2018, 174(1), 218–230 CrossRef CAS PubMed.
- S. Anders, A. Reyes and W. Huber, Detecting differential usage of exons from RNA-seq data, Genome Res., 2012, 22(10), 2008–2017 CrossRef CAS PubMed.
- F. Afzaljavan, A. S. Sadr, S. Savas and A. Pasdar, GATA3 somatic mutations are associated with clinicopathological features and expression profile in TCGA breast cancer patients, Sci. Rep., 2021, 11(1), 1679 CrossRef CAS PubMed.
- Y. Tong, P. R. A. Zanon, X. Yang, X. Su, J. L. Childs-Disney and M. D. Disney Transcriptome-wide mapping of small-molecule RNA-binding sites in live cells, bioRxiv, 2024.
- E. Pascolo, C. Wenz, J. Lingner, N. Hauel, H. Priepke and I. Kauffmann, et al., Mechanism of human telomerase inhibition by BIBR1532, a synthetic, non-nucleosidic drug candidate, J. Biol. Chem., 2002, 277(18), 15566–15572 CrossRef CAS PubMed.
- Y. Tong, Y. Lee, X. Liu, J. L. Childs-Disney, B. M. Suresh and R. I. Benhamou, et al., Programming inactive RNA-binding small molecules into bioactive degraders, Nature, 2023, 618(7963), 169–179 CrossRef CAS PubMed.
- Y. Tong, P. Zhang, X. Yang, X. Liu, J. Zhang and M. Grudniewska, et al., Decreasing the intrinsically disordered protein α-synuclein levels by targeting its structured mRNA with a ribonuclease-targeting chimera, Proc. Natl. Acad. Sci. U. S. A., 2024, 121(2), e2306682120 CrossRef CAS PubMed.
- Y. Song, J. Cui, J. Zhu, B. Kim, M. L. Kuo and P. R. Potts, RNATACs: Multispecific small molecules targeting RNA by induced proximity, Cell Chem. Biol., 2024, 31(6), 1101–1117 CrossRef CAS PubMed.
- S. Schuierer and G. Roma, The exon quantification pipeline (EQP): a comprehensive approach to the quantification of gene, exon and junction expression from RNA-seq data, Nucleic Acids Res., 2016, 44(16), e132 CrossRef PubMed.
- A. Dobin, C. A. Davis, F. Schlesinger, J. Drenkow, C. Zaleski and S. Jha, et al., STAR: ultrafast universal RNA-seq aligner, Bioinformatics, 2013, 29(1), 15–21 CrossRef CAS PubMed.
- M. J. Smola, G. M. Rice, S. Busan, N. A. Siegfried and K. M. Weeks, Selective 2′-hydroxyl acylation analyzed by primer extension and mutational profiling (SHAPE-MaP) for direct, versatile and accurate RNA structure analysis, Nat. Protoc., 2015, 10(11), 1643–1669 CrossRef CAS PubMed.
- S. Busan and K. M. Weeks, Accurate detection of chemical modifications in RNA by mutational profiling (MaP) with ShapeMapper 2, RNA, 2018, 24(2), 143–148 CrossRef CAS PubMed.
- S. Bellaousov, M. Kayedkhordeh, R. J. Peterson and D. H. Mathews, Accelerated RNA secondary structure design using preselected sequences for helices and loops, RNA, 2018, 24(11), 1555–1567 CrossRef CAS PubMed.
- A. R. Quinlan and I. M. Hall, BEDTools: a flexible suite of utilities for comparing genomic features, Bioinformatics, 2010, 26(6), 841–842 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00266k |
| This journal is © The Royal Society of Chemistry 2025 |