Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Monitoring the kinetic evolution of mesenchymal stem cell differentiation using Raman microspectroscopy†
F.
Ravera
 *a,
E.
Efeoglu
*a,
E.
Efeoglu
 b and
H. J.
Byrne
b and
H. J.
Byrne
 a
a
aFOCAS Research Institute, Technological University Dublin, City Campus, Dublin 8, Ireland. E-mail: D16126527@mytudublin.ie
bSchool of Food Science and Environmental Health, Technological University Dublin, Dublin, Ireland
First published on 8th May 2025
Abstract
Raman microspectroscopy (RMS) offers a powerful, non-destructive approach for in situ monitoring of dynamic biochemical processes within cells. However, the ability to reliably data-mine the spectroscopic signatures and their evolution and extract meaningful information can be challenging. Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS) regression analysis is a powerful chemometric technique that can potentially address this challenge by deconvoluting the spectra into individual component spectra, each representing a specific biochemical species, and regressing the solutions against kinetic constraints. In this study, MCR-ALS analysis was performed on spectral data of differentiation process of mesenchymal stem cells into chondrocytes, carried out on two different substrates, collagen 3-dimensional hydrogel and the conventional 2-dimensional culture model at time intervals of 1–7, 14, 21 days. The kinetic evolution of the chondrogenesis was modelled according to a phenomenological rate equation model, in an attempt to describe the biochemical evolution of the cell composition within the process. Moreover, the ability of the algorithm to faithfully extract the correct reaction rates and spectral profiles has been explored. The results indicated that the differentiation process originates in the nucleolar regions, subsequently extending to the nuclear and cytoplasmic compartments and corroborated a more rapid differentiation rate in cell cultures grown on 3D collagen hydrogels compared to 2D substrates. The combination of Raman microspectroscopy and MCR-ALS offers a powerful approach for elucidating the complex mechanisms underlying chondrogenesis and developing innovative strategies for regenerative medicine.
1. Introduction
Cell therapy is a rapidly emerging field of regenerative medicine that involves the injection or implantation of living cells into a patient to treat a variety of diseases and injuries.1 By leveraging the body's inherent healing mechanisms, cell therapies aim to restore damaged tissues and organs. The development of cell-based therapies, particularly those employing stem cells, holds significant promise for regenerative medicine. However, to realise the full potential of these therapies, a comprehensive understanding of cellular behaviour and function is essential. This necessitates the development of robust monitoring methods that can track cell-based therapies throughout their entire lifecycle, from initial cell isolation to clinical application. Such monitoring can enable researchers and clinicians to optimise culture conditions, ensure product quality, and assess safety and efficacy.1Within the context of stem cell-based therapies, a particular focus lies on chondrogenesis, the process of cartilage formation from mesenchymal stem cells (MSCs).2 Chondrocytes play a pivotal role in the maintenance of cartilage through the secretion of growth factors and enzymes that regulate its synthesis and extracellular matrix (ECM).3 Chondrogenesis is regulated by multiple complex signalling paths, and, during the first phase of intense growth, known as pre-cartilaginous condensation, MSCs cluster together and become a flock of mesenchymal progenitor cells and nodules.3–5 This pre-cartilaginous condensation is an extremely important intermediate phase that provides the fundamental scaffold for the skeletal elements, and it is provoked by a reduction of intercellular distances and consequent increase of cell to cell contacts.3 Within these densely packed cellular aggregates, the MSCs further differentiate into chondrocytes, which proliferate and continue the deposition of extracellular matrix.3,5
In order to translate cellular metabolism and biochemical insights into clinical applications, it is imperative to address the inherent complexity of cells, particularly when assessing their safety and efficacy for therapeutic purposes. Substantial efforts have been devoted to optimising cell growth conditions,6–10 necessitating precise generation of the desired cell product while mitigating contamination risks. However, identifying an effective monitoring technique remains a significant challenge.
The expression of pivotal chondrogenic genes, including SOX9, collagen type I, type II, aggrecan, and fibronectin, has been previously assessed using real-time polymerase chain reaction (RT-PCR).11–16 Histological staining with Alcian Blue, Blyscan, and immunofluorescence techniques have confirmed the presence of proteoglycans and collagen II, further substantiating MSC differentiation into chondrocytes.3,11,12,15,17–19 Immunohistochemical analysis facilitated the identification and quantification of collagen II accumulation, thereby evaluating the organisation of the extracellular matrix (ECM) and the expression of chondrogenic markers, such as collagen II and aggrecan, throughout differentiation.6,10,12,20,21 Additionally, the quantification of glycosaminoglycan (GAG) content, normalised to DNA, has demonstrated a significant increase in extracellular matrix (ECM) deposition on softer matrices, suggesting that mechanical properties influence chondrogenic commitment.11,15,18 Despite the advancements provided by these techniques, a major challenge remains in achieving in situ, real-time monitoring of chondrogenic differentiation. The conventional bioassays used to analyse differentiation stages are often destructive, time-consuming, and require large sample quantities,10,22–30 limiting their clinical and translational applications.10,22–30
Vibrational spectroscopy can overcome numerous limitations and has been extensively explored over the last decades as a diagnostic and prognostic tool for biomedical applications.31,32 Raman microspectroscopy (RMS), has previously been used to provide an in situ and real time assessment of stem cell quality and differentiation potential.29,33,34
Expanding upon the work of Ravera et al. (2024) on the benefits of 3D hydrogels compared to conventional 2D cell culture for in vitro chondrogenesis, this study aims to investigate and quantitatively compare the kinetic evolution and different rates of differentiation of MSCs on different substrates. In this work, it is proposed that the different stages of chondrocyte development, from undifferentiated MSCs to pre-chondrogenic condensed cells, and early differentiated chondrocytes, respectively, can be represented by distinct spectral profiles identified in the Raman spectral profile of the cell population. The kinetics of the evolution process can thus be modelled according to a phenomenological rate equation model, in which a series of ordinary differential equations are used to describe the biochemical evolution of the cell composition from their undifferentiated state, through pre-chondrogenic condensation to differentiated chondrocytes. The kinetic model will then be used as a set of constraints applied to the multivariate curve resolution-alternating least squares (MCR-ALS) algorithm to extract the independent spectral profiles (Components) from the measured spectra as a function of time, as well as kinetic rate constants to characterise the differentiation process.35
MCR-ALS regression analysis is an unmixing method which can provide an accurate molecular decomposition of the spectroscopic information contained in a data set.35,36 The algorithm operates under the assumption that any given complex spectrum within the analysis can be represented as a linear combination of pure component spectra. These components are weighted based on their relative abundance at each point in the analysis. Within the context of MCR-ALS image analysis, a “pure component” can represent a singular chemical compound or a distinct stage within a process that exhibits a consistent spectral signature.35–37 MCR-ALS is rapidly emerging as a powerful tool for spectral analysis, offering valuable insights. Perez-Guaita et al. previously used MCR-ALS combined with pharmacokinetic modelling to analyse the uptake of drugs by cells in vitro. This approach allowed them to extract the spectral signatures and concentration profiles of the drug and its metabolites over time.38 Moreover, MCR-ALS has been applied for innovative analysis of skin tissues, providing in vivo spectral profiles corresponding to the contribution of the optical system and skin components.39 By utilising MCR-ALS to analyse the kinetic evolution of the differentiation process, according to a model of A → B, B → C, with rate constants of k1 and k2, respectively, this study aims to extract the under- lying spectral components and their corresponding concentration profiles from the Raman spectra of MSCs cultured on both 2D and 3D hydrogel substrates and moreover quantify the process in terms of the rate constants of the progression. This approach will enable a detailed analysis of the progressive evolution of the spectral features associated with the different phases of chondrogenesis, including the initial condensation of MSCs, the subsequent differentiation into chondrocytes, and the maturation of the extracellular matrix. Furthermore, this study will focus on the subcellular localisation of spectral changes, examining the evolution of the organelles quantitatively comparing the kinetic profiles of these subcellular regions in MSCs differentiated on 2D and 3D substrates, quantifying their rates of change, providing valuable insights into the influence of the microenvironment on the chondrogenic process.
Overall, this study leverages the power of MCR-ALS to unravel the complex spectral signatures of MSCs undergoing chondrogenesis, offering a deeper understanding of the molecular and cellular mechanisms involved in this critical developmental process, and demonstrating the potential of Raman microspectroscopy as a tool for monitoring the development of cell-based therapies.
2. Materials and methods
2.1. Summary
The manuscript presents a further analysis of the spectral data previously presented in Ravera et al.33,40 For completeness, the Materials and Methods of that study are summarised here.Briefly, rat mesenchymal stem cells (rMSCs) from GIBCO® (Thermo Fisher Scientific) were cultured in Dulbecco's Modified Eagle's medium (DMEM) supplemented with 10% foetal bovine serum (FBS) and 5 μg mL−1 gentamicin. Sub-culturing was performed every 3–4 days at 60–80% confluency to maintain a healthy and expanding cell population. As a well studied model for the differentiation process, a commercially available rodent mesenchymal stem cell line was chosen. Chondrogenic differentiation has previously been demonstrated by positive proteoglycan Alcian blue staining after 18 days of culturing,12,41 and by monitoring the content of sulphated glycosaminoglycans after 14, 21 and 28 days.18,42,43 To induce chondrogenesis, a commercially available StemPro™ Chondrogenesis Differentiation Kit (Thermo Fisher Scientific) was employed. The comparison of two distinct culture environments was carried out, 3D and 2D, for 3 weeks. 3D cell cultures were prepared according to previously described protocol40 with collagen hydrogel-coated dishes (Collagen I, rat tail (3 mg mL−1) GIBCO®) and seeded with rMSCs at a density of 5 × 106 cells per mL.
2D cell cultures were seeded onto polystyrene plates at a density of 8 × 106 cells per mL and then seeded onto CaF2 disks (Crystran, UK) at a specific density of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per disk for Raman analysis. All samples were fixed with 10% formalin solution, rinsed with and stored in distilled water (dH2O) for Raman measurements.
000 cells per disk for Raman analysis. All samples were fixed with 10% formalin solution, rinsed with and stored in distilled water (dH2O) for Raman measurements.
2.2. Raman spectral data
For Raman analysis of rMSCs, control and differentiated cultures, samples were prepared by directly fixing the samples with formaldehyde solution. Cells were washed three times with Dulbecco's Phosphate-Buffered Saline (D-PBS, containing no calcium, magnesium, or phenol red), then fixed with 10% formalin for 15 minutes. The formalin solution was removed after 15 minutes of incubation at room temperature, cells were rinsed with dH2O three times and kept in 2 mL of dH2O for Raman measurements.A Horiba Jobin–Yvon LabRAM HR800 spectrometer, equipped with a 785 nm diode laser as source, was used throughout the study. All measurements were acquired in water to reduce scatter and minimise the risk of photodamage,44,45 by using a ×100 water immersion objective (LUMPlanF1, Olympus, N.A. 1), of spot size ∼1 μm. The spectrometer was calibrated to the 520.7 cm−1 line of silicon prior to spectral acquisition. Although the high resolution HR800 instrument can achieve a dispersion of ∼0.25 cm−1 per pixel with higher density gratings, 300 lines per mm grating was chosen to capture the whole fingerprint region of the spectrum in a single window, with a spectral dispersion of approximately 1.5 cm−1 per pixel. A 100 μm confocal pinhole was used for all measurements. The spectra were dispersed onto a 16-bit dynamic range Peltier cooled CCD detector and the spectral range from 400 to 1800 cm−1, the so-called fingerprint region, was chosen. Point spectra from the cytoplasm, nucleus, and nucleolus (see Fig. S1†) of 20 to 40 cells were acquired for each differentiation time point, and spectra were acquired for 3 × 30 seconds at each spot.
2.3. Spectral data processing
Prior to analysis, raw spectral data underwent pre-processing in Matlab (Mathworks) to remove unwanted background signal and noise. A Savitzky–Golay filter (3rd order, 9 points) was applied to smooth the spectra. The background signal, primarily originating from water, due to the immersion objective geometry,45,46 was removed using the adapted Extended Multiplicative Signal Correction (EMSC) algorithm.47 EMSC was applied to the smoothed spectra using a reference spectrum of cells on collagen hydrogel and a 7th-order polynomial baseline. Finally, the data was vector normalised to minimise instrumental variation. Each dataset contained 20–40 spectra (observations), representing points in an n-dimensional space defined by the wavenumbers (400–1800 cm−1).2.4. Multivariate curve resolution-alternating least squares modelling
Multivariate Curve Resolution-Alternating Least Squares (MCR-ALS) was performed to resolve the multiple component responses within the measured spectral responses, as explained in detail by de Juan and Tauler.37The MCR-ALS Graphical User Interface (GUI) 2.0 was employed for all analyses presented in this study.37,48 It is freely available from the Multivariate Curve Resolution Homepage (https://mcrals.wordpress.com/). The GUI was run in Matlab 2023b (Mathworks). Singular value decomposition was used to initially specify the number of components as 3. Initial estimates of the components were then performed using Evolving Factor Analysis. The fitting procedure was subjected to non-negativity (NNLS) constraints in both the concentration and spectral domains, and no further normalisation was applied. Kinetic constraints of A → B → C were applied to the concentration domain, 100 iterations were performed, and the convergence criterion was set at 0.01.
Following the definition of the kinetic model across the specified time frame (t1 to tn), the program performs an optimisation routine, resulting in the generation of the best-fit kinetic profiles for component concentrations (copt) and their corresponding spectral profiles (sopt). Notably, the optimised kinetic parameters are also recorded for each model variation, such as those arising from adjustments to the time window or time step.
3. Results and discussion
3.1. Tracking subcellular biochemical changes in MSC differentiation with Raman microspectroscopy and MCR-ALS
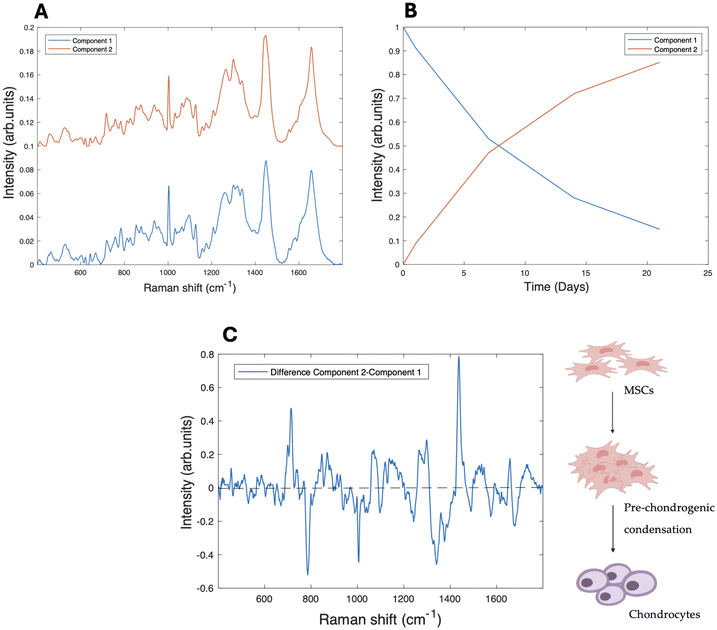
Raman microspectroscopic analysis was performed on the subcellular regions of nucleolus, nucleus and cytoplasm of rMSC cultures in both 2D and 3D environments, over a period of 1–21 days.33,40MCR-ALS was initially used to examine the spectral information gathered from Raman microspectroscopy of subcellular regions of differentiating MSCs towards chondrocytes cultured and differentiated on collagen hydrogel substrates. Singular value decomposition (SVD) was used to first establish the number of components to be used in the model, which was specified as 3, imposing a three-stage model of initial differentiation from rMSC (A) to the condensation stage (B), followed by the last differentiation stage (C), on the evolution of the Raman spectra of the nucleolus over the 21 days of differentiation. The resultant spectral components can be seen in Fig. 1A, for the case of the nucleoli. The kinetic evolution of the components, constrained to an evolution model of A → B, B → C, with rate constants of k1 and k2 (per day), respectively, can be seen in Fig. 1B. The fit parameters are tabulated in Table 1. To better visualise the spectroscopic changes occurring between the stages, the difference spectra (Component 2–Component 1) and (Component 3–Component 2), are shown in Fig. 1C.
| 3D | 2D | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| k1 | k2 | Sig 1 | Sig 2 | k1 | k2 | Sig 1 | Sig 2 | ||
| A → B | Nucleolus | 0.47 | 0.04 | 0.05 | 0.003 | 0.25 | 0.04 | 0.01 | 0.002 |
| B → C | Nucleus | 0.22 | 0.025 | 0.03 | 0.003 | 0.12 | 0.052 | 0.01 | 0.006 |
| Cytoplasm | 1.19 | 0.230 | 0.16 | 0.03 | 0.15 | 0.049 | 0.01 | 0.005 | |
| B → C | 0.16 | 0.02 | |||||||
A difference spectrum is a visual representation of the calculated difference between two spectra, obtained by subtracting one spectrum from the other, point by point. Difference spectra are particularly useful for highlighting subtle changes that might be difficult to discern when comparing the original spectra directly. They can also help identify changes in peaks and visualise the level of differences between spectra. Additionally, difference spectra can reveal the main peaks of the spectra being compared, as these peaks will often appear as prominent features in the difference spectrum.
Component 1 is seen to decrease dramatically after one day of differentiation, while component 2 increases concomitantly and continues to dominate throughout 21 days of differentiation. Component 3 shows a steady increase throughout the differentiation process, although it only accounts for ∼40% of the spectral profile after 21 days. As expected, the main spectral changes observed in the nucleolus (Fig. 1A) from Components 1 to 3 are associated with nucleic acid structures, including an increased intensity of the phosphate stretching vibration of RNA/DNA content, such as the band at ∼782–86 cm−1, manifest as a sharp peak, compared to the other components.49
Fig. 1C illustrates the difference spectra between the three components and allows a clearer identification of the spectral changes occurring between the stages of the differentiation process. The peak at ∼1093 cm−1, corresponding to the stretching vibration of RNA/DNA, is present in the spectral representation of both difference spectra,50 and there are further changes in the DNA/RNA related peak at ∼1336 cm−1.51 However, further significant changes are also observed in the intensities of protein and lipid related peaks, for example the peak at ∼1004 cm−1, assigned to phenylalanine,52 and the range of 1303–1343 cm−1, in which the most relevant peaks (1303 cm−1, 1322 cm−1 and 1343 cm−1) correspond to the contribution of CH2CH3 bending in collagen.53 Changes are also observed at the peaks at ∼1123 cm−1 (C–C stretching mode of lipids and proteins), ∼1453 cm−1, and ∼1468 cm−1 (CH bending mode of structural proteins and lipids).54
An intense nucleolar activity in the early phases of differentiation, or pre-chondrogenic condensation, has previously been observed with Raman microspectroscopy33,55 and indicates that the nucleolus is the primary site of biochemical activity in the initial stages of differentiation.56,57 This decrease is closely linked to a reduction in transcriptional activity and cell proliferation, as stem cells transition from a highly proliferative state to a more specialised, functionally distinct phenotype.29,33,55
This intense activity is reflected in the changes occurring in Component 2 in the first days of the differentiation of the pure components analysis with MCR-ALS. Interestingly, many of the spectral changes evident in the difference spectrum representing the first phase of differentiation appear to be reversed in the second phase, as evidenced by the almost mirror image nature of the second difference spectrum.
During chondrogenic differentiation a decrease in sphingomyelins and phosphatidylcholines has previously been detected.58 Lipid changes are crucial for differentiation progression, and the identified markers could aid in monitoring chondrogenesis and improving cartilage regeneration. The findings emphasise the role of lipids in cell growth, differentiation, and cartilage matrix production.
To further explore the effectiveness of MCR-ALS analysis, a comparative study of the spectroscopic results of rMSCs chondrogenic differentiation analysis performed on cells grown on a 2D CaF2 was performed. The equivalent plot for the analysis of the datasets for the nucleolus, nucleus and cytoplasm in a 2D environment are shown in the ESI Fig. S2–S4.† The results showed interesting similarities and differences when compared with the equivalent data obtained on different growth substrate but otherwise, the same differentiation conditions.
Interestingly, the rate of differentiation from undifferentiated rMSCs towards the pre-chondrogenic phase (k1 = 0.47 ± 0.05 per day) appears higher on a 3D hydrogel substrate than the equivalent observed organelle grown onto a 2D substrate (k1 = 0.25 ± 0.01 per day) (Table 1). However, the rate of differentiation from the second to the third component k2 is significantly lower (k2 = 0.040 ± 0.003 per day) on the 3D substrate, and a similar value is observed for the equivalent process on 2D (k2 = 0.04 ± 0.002 per day).
A similar analysis was performed on the nucleus, and the spectral profiles and their kinetic evolution are shown in Fig. 2A & B and the difference spectra in Fig. 2C. In terms of the evolution kinetics, Component 1 is again seen to have completely decayed by day 7, while Component 2 still represents ∼50% of the spectral profile after 21 days (Fig. 2B). Similarly to the nucleolus, as differentiation progresses, the increase of Component 2 (from day 1 onwards) represents the development of the pre-chondrogenic condensation phase, in which intense transcriptional activity is known to occur.4,59 This is further supported by the observed increase in peaks associated with RNA/DNA content (786 cm−1, 1093 cm−1) within component B.49 However, the difference spectra between components of the nucleus present distinctive features compared to the equivalent nucleolar spectra. Firstly, it appears that the main molecular changes take place between the second and third component, and therefore in the second phase of the differentiation. This is manifested in the difference spectrum Fig. 2C (red), in which more prominent peaks associated with relevant spectral changes are observable, while the spectrum generated by the difference between the first and the second components (blue) shows fewer remarkable peaks.
The main changes are observed in the range ∼1300–1340 cm−1 which shows differences of intensity similar to those observed for the nucleolus. Other peaks are attributable to the contribution of nucleic acids of DNA/RNA (1263 cm−1, ∼1342 cm−1), CH2CH3 cm−1 of collagen (1300–1304 cm−1), and 1318–37 cm−1 (DNA/RNA and Amide III).60
Interestingly, peaks at ∼1004 cm−1 (assigned to phenylalanine), 1064 cm−1–1088 cm−1 C–O, C–C stretching (carbohydrates) show a prominent increase, potentially reflecting changes in protein and GAGs synthesis associated with the formation of the chondrogenic extracellular matrix.61,62
As is the case for the nucleolus, the comparison with the nuclear spectral data obtained onto 2D substrate reveals a higher kinetic constant k1 of the cell culture on 3D than on 2D (3D = 0.22 ± 0.02 per day and 2D = 0.12 ± 0.01 per day). In this case, however, the rate of the second phase, k2, appears to be higher on the 2D substrate (3D = 0.025 ± 0.003 per day and 2D = 0.052 ± 0.006 per day).
Fig. 3 illustrates the kinetic profile of the third observed subcellular region, the cytoplasm. The observation of the cytoplasmic features, alongside those from the nucleolus, provides a comprehensive picture of the dynamic biochemical changes occurring during chondrocyte differentiation. As was the case for the other cellular regions, singular value decomposition (SVD) of the data allowed for the identification of three spectral components. In the kinetic analysis, Component 1 is seen to rapidly decay at a rate of k1 = 1.2 ± 0.2 per day, while, in contrast to the other cellular regions, Component 2 also decays rapidly, such that Component 3 completely dominates after 21 days. In fact, examination of the spectra of Component 1 and 2 indicates a high degree of similarity, and the difference spectrum of Fig. 3C is relatively weak. It could therefore be considered that the evolution of the cytoplasmic region, in the cells grown on the 3D hydrogel matrix, is better described by a two stage process A → B, at a single rate k1.
Fig. 4 illustrates the results of the analysis of the spectroscopic data of the cytoplasm on 3D substrates according to a two stages kinetic model A → B. Component 1 decays almost completely over the 21 day period, while Component 2 increases concomitantly to a value of ∼0.85. Fig. 4C shows the main observed peaks, associated with amide II (1257 cm−1), and lipids (1300 cm−1), ∼1314–21 cm−1 CH3CH2 twisting (collagen assignment) and suggests that the initiation of extracellular matrix formation is observed in the transition from the first to the second component (Fig. 4B).50,63 More prominent peaks in the spectrum include those at ∼1290–1400 cm−1 and ∼840–860 cm−1, corresponding to CH bending and the C–O–C group of polysaccharides, ∼1266 cm−1, ∼1314–21 cm−1, 1335 cm−1, 1342 cm−1 corresponding to CH3CH2 contributions of collagen, amide III and glycosaminoglycans (GAGs) respectively, the essential building blocks of the chondrogenic matrix.49,52,53,64
In such a 2 stage model, the rate of change, k1, is determined to be 0.16 ± 0.02 per day. Analysis of the data for differentiation on a 2D substrate fit well to a 3 Component model, with k1 = 0.15 ± 0.01 per day, k2 = 0.049 ± 0.005 per day.
3.2. Discussion
The approach explored in this work enables the observation of molecular evolution without perturbing the system, making it invaluable for studying dynamic cellular processes. While biochemical based assays can establish the presence of specific biomarkers of the progression of the differentiation process at fixed points in time, RMS analysis monitors the progression in a more holistic fashion, shedding more light on the process subcellularly, as well as within the cell population. By offering non-destructive, label-free insights at both cellular and extracellular levels, RMS provides a comprehensive view of biochemical transitions occurring during chondrogenesis and cartilage formation.65–67 Importantly, RMS has further demonstrated its capacity to distinguish between different stages of chondrogenesis by detecting key biochemical changes, including the synthesis and organisation of crucial ECM components.33,34,68,69 A key advantage of RMS resides in its ultimate capacity for real-time, monitoring of biochemical alterations during chondrogenesis, thereby providing a dynamic and continuous assessment that contrasts with the fixed time-point data obtained from conventional biochemical assays. Additionally, it facilitates the detection of a broad range of extracellular matrix (ECM) constituents, encompassing both sulphated and non-sulphated glycosaminoglycans (GAGs). This capability distinguishes it from dye-based assays, such as Blyscan and Alcian Blue, which exhibit selectivity towards sulphated GAGs, by affording a more comprehensive molecular characterisation of the differentiating tissue.19,64 The combination of RMS with multivariate curve resolution-alternating least squares (MCR-ALS) represents a significant advancement in extracting kinetic data from complex biological systems.36 By quantifying dynamic cellular composition changes over time, this technique offers valuable insights into the biochemical signatures of chondrogenesis at the single-cell level. While similar techniques have been used before, this study presents a novel application by integrating these methods for real-time monitoring of subcellular progression during chondrogenesis, offering a more nuanced and comprehensive analysis compared to traditional methods. Kinetic studies have previously described the biphasic nature of chondrogenesis, with distinct ECM production phases.70,71Sorrell et al. (2018) investigated the kinetics of chondrogenesis in hMSC-derived aggregates at defined time points and used microarray analysis to monitor time-dependent mRNA expression profiles of ECM-related genes, including early markers like PAPSS2, PRG4, and fibronectin, and later cartilage-specific genes such as COL2A1 and ACAN.71 These data revealed the biphasic pattern in which early matrix production involved fibrous proteins and small proteoglycans, followed by later accumulation of cartilage-specific ECM components.71
This dynamic pattern of ECM regulation aligns with the findings presented, providing a valuable framework for understanding the temporal regulation of ECM synthesis during cartilage differentiation. In addition, a recent study applied RMS combined with MCR-ALS to monitor biochemical changes in chondrocytes during osteoarthritis progression, focusing on apoptotic processes at the single-cell level. The study highlighted how MCR-ALS can deconvolve complex Raman spectra to isolate molecular components like lipids, DNA, and proteins, assessing their relative changes in response to OA-induced degeneration.72 By applying this approach to both two-dimensional (2D) and three-dimensional (3D) culture systems,40 the investigation reveals notable differences in the pace and pattern of differentiation. While the initial study indicated that the cytoplasmic regions displayed a distinct behaviour compared to the nuclear regions, the comparative study of the differentiation process in a 3D culture environment compared to the 2D culture, further indicated that the differentiation process overall was more rapid in the 3D environment.40 The application of MCR-ALS enables a more quantitative approach to the analysis of the progression of the differentiation process, as well as a quantitative comparison of the progression in different cell culture environments, in terms of the rate constants of the different kinetic stages.38 The optical resolution of Raman microspectroscopy furthermore enables a quantitative comparison of the progression rates within the different subcellular regions, in this study, the nucleolus, nucleus and cytoplasmic regions, which may lay the foundation for an improved understanding of the differentiation and condensation process.
The analysis indicates that the nucleolus and the nucleus are the primary sites of biochemical activity during chondrogenesis, significant spectral changes being observed early in the differentiation process, while the cytoplasm is a direct manifestation of those changes, and region in which the extracellular matrix is generated. In both 3D and 2D culture environments, the initial rate of change of the nucleolar region is highest, followed by the surrounding nuclear regions. In the 2D culture, the rate of subsequent evolution of the cytoplasmic region appears to be similar to that of the nuclear region, while in the 3D culture environment, the condensation stage associated with the growth of the extracellular matrix appears to begin from the outset. Notably, a similar difference was observed in the evolution of the condensation pellet in 2D and 3D,40 whereby a progressive evolution of the pellet from the edges to the centre was observed in 2D, whereas a much more spatially uniform evolution was observed in the 3D environment. This is consistent with the earlier deposition of the extracellular matrix in the 3D environment, potentially prompted by the biocompatible nature of the collagen hydrogel.
The MCR-ALS analysis is thus a promising analytic tool to quantify the kinetics of the evolution of the differentiation process, and also to identify the specific biochemical signatures at the different stages. It is noted, however, that the dataset should be considered sparse, in that the measurement intervals are relatively long compared to the differentiation stages, and the process has not been completed by the stage of the last measurement, at 21 days. With the exception of Component A at t = 0, there is considerable overlap of all components at all other measurement times, and thus the analysis is subject to ambiguity and essentially has difficulty in completely unmixing the components.73 This is potentially the reason for the near mirror image nature of the difference spectra in Fig. 1B & 3B, which could be improved by more regular, ultimately real-time sampling over the full time course. Nevertheless, the study demonstrates the potential benefits of kinetic monitoring of the label free spectroscopic signatures of the differentiation process at a subcellular and cell population level.
Growth conditions represent a critical determinant in directing mesenchymal stem cell (MSC) chondrogenesis, and therefore the differentiation rates. The complex interplay between cell confluence, three-dimensional architecture, and substrate porosity orchestrates both the biochemical and mechanical cues necessary for differentiation.3,4,74 The significance of microenvironmental modifications accompanying the differentiation of MSCs into chondrocytes has been extensively investigated.75 Changes in the extracellular matrix (ECM), shaped by the synthesis and remodelling of its components, are crucial for successful chondrogenesis, as evidenced by a marked upregulation of collagen type II and ACAN, alterations in collagen type I, and increased synthesis of proteoglycans and glycoproteins during chondrogenic differentiation. The density at which MSCs are cultured can influence gene expression, proliferation behaviour, and overall morphology.76 High confluence not only promotes a rounded cell morphology, an essential feature of chondrocyte-like behaviour, but also significantly enhances the synthesis of cartilage-specific ECM proteins, such as collagen type II and aggrecan, through increased cell-to-cell interactions and mechanical cues.77 Moreover, the role of optimised high-density, three-dimensional (3D) culture conditions in promoting chondrogenesis has been widely explored.10,15,78–82 Three-dimensional culture systems exhibit elevated expression of chondrogenic markers such as COL2A1, SOX9, and ACAN, which are minimally expressed in two-dimensional (2D) monolayers. In contrast, hypertrophic markers, including COL10A1, RUNX2, and ALP, are significantly upregulated in 2D cultures but suppressed in 3D environments.78,82–84 Glycosaminoglycan (GAG) content has been reported to be significantly higher in 3D cultures, whereas cell proliferation is predominantly observed in 2D systems, suggesting divergent pathways that favour matrix production versus cellular expansion, respectively.
4. Conclusions
This study demonstrates the efficacy of Raman microspectroscopy (RMS) as a powerful tool for in situ monitoring and potentially quality process control of cell-based therapies. Using the model example of chondrogenesis from mesenchymal stem cells, in vitro, the sequential biochemical changes in the nucleolar, nuclear and cytoplasmic regions can be identified. MCR-ALS analysis of Raman spectra furthermore provides detailed insights into the biochemical changes underlying this process, allowing for the identification of specific molecular markers and the quantification of their relative contributions and evolution rates.The study seeks to address the limitations of conventional fixed-timepoint assays, such as immunostaining, PCR, or histological analysis, which require destructive sample preparation and provide only a static view of cellular processes at a single time point. This inherently restricts their capacity to track dynamic changes and transient biochemical states throughout differentiation. In contrast, Raman microspectroscopy allows non-invasive, real-time monitoring of the same sample, thereby enabling continuous observation of molecular evolution and offering a detailed kinetic profile of chondrogenesis over time.
The analysis confirms that the differentiation process is initiated in the nucleolar regions, before progressing in the nuclear and cytoplasmic regions. The analysis also proves that the process is more rapid in cell cultures grown on the 3D collagen hydrogel environment, than those on the 2D substrate. This non-destructive and sensitive technique, therefore, offers a valuable approach for studying cellular differentiation and optimising culture conditions for tissue engineering applications. By combining Raman microspectroscopy and MCR-ALS, researchers can gain a deeper understanding of the complex mechanisms involved in chondrogenesis and develop more effective strategies for regenerative medicine.
Data availability
The data that support the findings of this study are available from the corresponding author Francesca Ravera, (D16126527@mytudublin.ie) upon request.Conflicts of interest
There are no conflicts to declare.Acknowledgements
FR acknowledges financial support from the TU Dublin Fiosraigh Postgraduate Scholarship programme.References
- S. Rangan, et al., Applications of Raman spectroscopy in the development of cell therapies: state of the art and future perspectives, Analyst, 2020, 145(6), 2070–2105 RSC.
- S. M. Richardson, et al., Mesenchymal stem cells in regenerative medicine: Focus on articular cartilage and intervertebral disc regeneration, Methods, 2016, 99, 69–80 CrossRef CAS PubMed.
- C. Tacchetti, et al., Cell condensation in chondrogenic differentiation, Exp. Cell Res., 1992, 200(1), 26–33 CrossRef CAS PubMed.
- B. K. Hall and T. Miyake, Divide, accumulate, differentiate: cell condensation in skeletal development revisited, Int. J. Dev. Biol., 1995, 39(6), 881–893 CAS.
- B. K. Hall and T. Miyake, The membranous skeleton: the role of cell condensations in vertebrate skeletogenesis, Anat. Embryol., 1992, 186(2), 107–124 CrossRef CAS PubMed.
- H. A. Owida, et al., Vibrational spectroscopic monitoring and biochemical analysis of pericellular matrix formation and maturation in a 3-dimensional chondrocyte culture model, Analyst, 2018, 143(24), 5979–5986 RSC.
- A. D. Doyle, Generation of 3D Collagen Gels with Controlled Diverse Architectures, Curr. Protoc. Cell Biol., 2016, 72, 10.20.1–10.20.16 Search PubMed.
- V. Moulisová, et al., Hybrid Protein-Glycosaminoglycan Hydrogels Promote Chondrogenic Stem Cell Differentiation, ACS Omega, 2017,(2470–1343), 7609–7620 CrossRef PubMed.
- J. Yang, et al., The negatively charged microenvironment of collagen hydrogels regulates the chondrogenic differentiation of bone marrow mesenchymal stem cells in vitro and in vivo, J. Mater. Chem. B, 2020, 8(2050–7518), 4680–4693 RSC.
- U. Nöth, et al., Chondrogenic differentiation of human mesenchymal stem cells in collagen type I hydrogels, J. Biomed. Mater. Res., Part A, 2007, 83(3), 626–635 CrossRef PubMed.
- S. Ghosh, M. Laha, S. Mondal, S. Sengupta and D. L. Kaplan, In vitro model of mesenchymal condensation during chondrogenic development, Biomaterials, 2009, 30, 6530–6540 CrossRef CAS PubMed.
- S. Grässel, S. Stöckl and Z. Jenei-Lanzl, Isolation, culture, and osteogenic/chondrogenic differentiation of bone marrow-derived mesenchymal stem cells, in Methods Mol Biol, 2012, pp. 203–267 Search PubMed.
- V. Lefebvre, et al., SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1(II) collagen gene, Mol. Cell. Biol., 1997, 17(4), 2336–2346 CrossRef CAS PubMed.
- H. Akiyama, et al., The transcription factor Sox9 has essential roles in successive steps of the chondrocyte differentiation pathway and is required for expression of Sox5 and Sox6, Genes Dev., 2002, 16(0890–9369), 2813–2828 CrossRef CAS PubMed.
- Z. Miao, et al., Collagen, agarose, alginate, and Matrigel hydrogels as cell substrates for culture of chondrocytes in vitro: A comparative study, J. Cell. Biochem., 2018, 119(10), 7924–7933 CrossRef CAS PubMed.
- T. Schwarzl, et al., Transcriptional profiling of early differentiation of primary human mesenchymal stem cells into chondrocytes, Sci. Data, 2023, 10, 758 CrossRef CAS PubMed.
- M. Solursh, T. F. Linsenmayer and K. L. Jensen, Chondrogenesis from single limb mesenchyme cells, Dev. Biol., 1982, 94(1), 259–264 CrossRef CAS PubMed.
- L. A. Solchaga, K. J. Penick and J. F. Welter, Chondrogenic differentiation of bone marrow-derived mesenchymal stem cells: tips and tricks, in Methods in molecular biology, Clifton, N.J., 2011, vol. 698, pp. 253–278 Search PubMed.
- H. T. Mohamed, et al., Implementation of infrared and Raman modalities for glycosaminoglycan characterization in complex systems, Glycoconjugate J., 2017, 34(3), 309–323 CrossRef CAS PubMed.
- S. Grässel, et al., Gene and protein expression profile of naive and osteo-chondrogenically differentiated rat bone marrow-derived mesenchymal progenitor cells, Int. J. Mol. Med., 2009, 23(6), 745–755 Search PubMed.
- N. Ahmed, et al., CD45-positive cells of haematopoietic origin enhance chondrogenic marker gene expression in rat marrow stromal cells, Int. J. Mol. Med., 2006, 18(2), 233–240 CAS.
- Y. Xia, E. M. Darling and W. Herzog, Functional properties of chondrocytes and articular cartilage using optical imaging to scanning probe microscopy, J. Orthop. Res., 2018, 36(2), 620–631 CrossRef CAS PubMed.
- Á. Fernández-Iglesias, et al., A simple method based on confocal microscopy and thick sections recognizes seven subphases in growth plate chondrocytes, Sci. Rep., 2020, 10(1), 6935 CrossRef PubMed.
- J. Aigner, et al., Distribution and viability of cultured human chondrocytes in a three-dimensional matrix as assessed by confocal laser scan microscopy, In Vitro Cell. Dev. Biol.: Anim., 1997, 33(6), 407–409 CrossRef CAS PubMed.
- I. Walter, et al., Confocal laser scanning microscopy of chondrocytes in vitro: cytoskeletal changes after quinolone treatment, Scanning, 1998, 20(7), 511–515 CrossRef CAS PubMed.
- M. Zayed, et al., Donor-Matched Comparison of Chondrogenic Potential of Equine Bone Marrow- and Synovial Fluid-Derived Mesenchymal Stem Cells: Implications for Cartilage Tissue Regeneration, Front. Vet. Sci., 2016, 3, 121 Search PubMed.
- Y. Lin, et al., Chemical analysis of single cells, Anal. Chem., 2011, 83(12), 4369–4392 CrossRef CAS PubMed.
- C. Bour-Dill, et al., Determination of intracellular organelles implicated in daunorubicin cytoplasmic sequestration in multidrug-resistant MCF-7 cells using fluorescence microscopy image analysis, Cytometry, 2000, 39(1), 16–25 CrossRef CAS PubMed.
- A. Ghita, et al., Applications of Raman micro-spectroscopy to stem cell technology: label-free molecular discrimination and monitoring cell differentiation, EPJ Tech. Instrum., 2015, 2(1), 6 CrossRef PubMed.
- A. D. Murdoch, et al., Chondrogenic differentiation of human bone marrow stem cells in transwell cultures: generation of scaffold-free cartilage, Stem Cells, 2007, 25(11), 2786–2796 CrossRef CAS PubMed.
- H. J. Byrne, et al., Spectropathology for the next generation: quo vadis?, Analyst, 2015, 140(7), 2066–2073 RSC.
- M. J. Baker, et al., Clinical applications of infrared and Raman spectroscopy: state of play and future challenges, Analyst, 2018, 143(8), 1735–1757 RSC.
- F. Ravera, E. Efeoglu and H. J. Byrne, Monitoring stem cell differentiation using Raman microspectroscopy: chondrogenic differentiation, towards cartilage formation, Analyst, 2021,(1364–5528), 322–337 RSC.
- J. J. Lazarevic, et al., Probing primary mesenchymal stem cells differentiation status by micro-Raman spectroscopy, Spectrochim. Acta, Part A, 2019, 213, 384–390 CrossRef CAS PubMed.
- A. de Juan and R. Tauler, Chemometrics applied to unravel multicomponent processes and mixtures: Revisiting latest trends in multivariate resolution, Anal. Chim. Acta, 2003, 500(1–2), 195–210 CrossRef CAS.
- J. Felten, et al., Vibrational spectroscopic image analysis of biological material using multivariate curve resolution-alternating least squares (MCR-ALS), Nat. Protoc., 2015, 10(1750–2799), 217–240 CrossRef CAS PubMed.
- A. de Juan, J. Jaumot and R. Tauler, Multivariate Curve Resolution (MCR). Solving the mixture analysis problem, Anal. Methods, 2014, 6, 4964–4976 RSC.
- D. Pérez-Guaita, et al., Combining Pharmacokinetics and Vibrational Spectroscopy:MCR-ALS Hard-and-Soft Modelling of Drug Uptake In Vitro Using Tailored Kinetic Constraints, Cells, 2022, 11, 1555 CrossRef PubMed.
- I. Matveeva, et al., Multivariate Curve Resolution Alternating Least Squares Analysis of In Vivo Skin Raman Spectra, Sensors, 2022, 22, 9588 CrossRef CAS PubMed.
- F. Ravera, E. Efeoglu and H. J. Byrne, A comparative analysis of stem cell differentiation on 2D and 3D substrates using Raman microspectroscopy, Analyst, 2024, 149(15), 4041–4053 RSC.
- P. Sangeetha, et al., Mesenchymal stem cells derived from rat bone marrow (rbm msc): techniques for isolation, expansion and differentiation, J. Stem Cell Res. Ther., 2017, 3(3), 272–277 Search PubMed.
- A. Lotfy, et al., Comparative study of biological characteristics of mesenchymal stem cells isolated from mouse bone marrow and peripheral blood, Biomed. Rep., 2019, 11(4), 165–170 CAS.
- H. Zhang, et al., The ability to form cartilage of NPMSC and BMSC in SD rats, Int. J. Clin. Exp. Med., 2015, 8(4), 4989–4996 Search PubMed.
- F. Bonnier, et al., In vitro analysis of immersed human tissues by Raman microspectroscopy, J. Raman Spectrosc., 2011, 42(5), 888–896 CrossRef CAS.
- F. Bonnier, et al., Analysis of human skin tissue by Raman microspectroscopy: Dealing with the background, Vib. Spectrosc., 2012, 61, 124–132 CrossRef CAS.
- R. Boyack and E. C. Le Ru, Investigation of particle shape and size effects in SERS using T-matrix calculations, Phys. Chem. Chem. Phys., 2009, 11(34), 7398–7405 RSC.
- L. T. Kerr and B. M. Hennelly, A multivariate statistical investigation of background subtraction algorithms for Raman spectra of cytology samples recorded on glass slides, Chemom. Intell. Lab. Syst., 2016, 158, 61–68 CrossRef CAS.
- J. Jaumot, et al., A graphical user-friendly interface for MCR-ALS: a new tool for multivariate curve resolution in MATLAB, Chemom. Intell. Lab. Syst., 2005, 76(1), 101–110 CrossRef CAS.
- Z. Liu, et al., Circulation and long-term fate of functionalized, biocompatible single-walled carbon nanotubes in mice probed by Raman spectroscopy, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 1410–1415 CrossRef CAS PubMed.
- J. W. Chan, et al., Micro-Raman spectroscopy detects individual neoplastic and normal hematopoietic cells, Biophys. J., 2006, 90(2), 648–656 CrossRef CAS PubMed.
- N. Stone, et al., Raman spectroscopy for identification of epithelial cancers, Faraday Discuss., 2004, 126, 141–157 RSC . discussion 169–83.
- A. C. S. Talari, et al., Raman Spectroscopy of Biological Tissues, Appl. Spectrosc. Rev., 2014, 50(1), 46–111 CrossRef.
- F. Bonnier and H. J. Byrne, Understanding the molecular information contained in principal component analysis of vibrational spectra of biological systems, Analyst, 2012, 137(2), 322–332 RSC.
- Z. Huang, et al., Near-infrared Raman spectroscopy for optical diagnosis of lung cancer, Proc. Natl. Acad. Sci. U. S. A., 2003, 107(0020–7136), 1047–1052 CAS.
- I. Notingher, I. Bisson, A. E. Randle, W. L. Polak, J. M. P. Hench and L. Larry, In situ spectroscopic study of nucleic acids in differentiating embryonic stem cells, Vib. Spectrosc., 2004, 35(1), 199–203 CrossRef CAS.
- M. J. Seibel, S. P. Robins and J. P. Bilezikian, Dynamics of Bone and Cartilage Metabolism, in Principles and Clinical Applications, Elsevier, 2nd edn, 2006 Search PubMed.
- H. Chen, et al., Runx2 regulates endochondral ossification through control of chondrocyte proliferation and differentiation, J. Bone Miner. Res., 2014,(1523–4681) Search PubMed.
- B. Rocha, et al., Characterization of lipidic markers of chondrogenic differentiation using mass spectrometry imaging, Proteomics, 2015, 15(4), 702–713 CrossRef CAS PubMed.
- B. K. Hall and T. Miyake, All for one and one for all: condensations and the initiation of skeletal development, Bioessays, 2000, 22(2), 138–147 CrossRef CAS PubMed.
- R. J. Lakshmi, et al., Tissue Raman spectroscopy for the study of radiation damage: brain irradiation of mice, Radiat. Res., 2002, 157, 175–182 CrossRef CAS.
- N. Mainreck, et al., Rapid characterization of glycosaminoglycans using a combined approach by infrared and Raman microspectroscopies, J. Pharm. Sci., 2011, 100(2), 441–450 CrossRef CAS PubMed.
- R. Ellis, E. Green and C. P. Winlove, Structural Analysis of Glycosaminoglycans and Proteoglycans by Means of Raman Microspectrometry, Connect. Tissue Res., 2009, 50(1), 29–36 CrossRef PubMed.
- R. K. Dukor, Vibrational Spectroscopy in the Detection of Cancer, in in Handbook of Vibrational Spectroscopy, ed. J. M. Chalmers, P. R. Griffiths, J. M. Chalmers and P. R. Griffiths, Handbook of Vibrational Spectroscopy, 2006 Search PubMed.
- S. Brézillon, et al., Probing glycosaminoglycan spectral signatures in live cells and their conditioned media by Raman microspectroscopy, Analyst, 2017, 142, 1333–1341 RSC.
- M. S. Bergholt, A. Serio and M. B. Albro, Raman Spectroscopy: Guiding Light for the Extracellular Matrix, Front. Bioeng. Biotechnol., 2019, 7, 303 CrossRef PubMed.
- M. S. Bergholt, et al., Raman Spectroscopy Reveals New Insights into the Zonal Organization of Native and Tissue-Engineered Articular Cartilage, ACS Cent. Sci., 2016, 2(12), 885–895 CrossRef CAS PubMed.
- M. B. Albro, et al., Raman spectroscopic imaging for quantification of depth-dependent and local heterogeneities in native and engineered cartilage, npj Regener. Med., 2018, 3, 3 CrossRef CAS PubMed.
- A. Bonifacio, et al., Chemical imaging of articular cartilage sections with Raman mapping, employing uni- and multi-variate methods for data analysis, Analyst, 2010, 135(12), 3193–3204 RSC.
- C. C. Moura, et al., Raman spectroscopy and coherent anti-Stokes Raman scattering imaging: prospective tools for monitoring skeletal cells and skeletal regeneration, J. R. Soc. Interface, 2016, 13(118), 20160182 CrossRef PubMed.
- I. Sekiya, et al., In vitro cartilage formation by human adult stem cells from bone marrow stroma defines the sequence of cellular and molecular events during chondrogenesis, Proc. Natl. Acad. Sci. U. S. A., 2002, 99(7), 4397–4402 CrossRef CAS PubMed.
- J. M. Sorrell, R. A. Somoza and A. I. Caplan, Human mesenchymal stem cells induced to differentiate as chondrocytes follow a biphasic pattern of extracellular matrix production, Inc. J. Orthop. Res., 2018, 36(6), 1757–1766 CrossRef CAS PubMed.
- G. Uppal, T. Goyal, A Kumar and R Kumar, Unravelling changes in single-cell osteoarthritic chondrocytes through coupling of Raman spectroscopy and multivariate curve resolution-alternating least square (MCR-ALS) algorithm, Microchem. J., 2025, 212, 113264 CrossRef CAS.
- A. M. de Juan, M. Martínez, M. Tauler and R. Kumar, Combining hard- and soft-modelling to solve kinetic problems, Chemom. Intell. Lab. Syst., 2000, 54(2), 123–141 CrossRef CAS.
- Y. S. Lee and C. M. Chuong, Adhesion molecules in skeletogenesis: I. Transient expression of neural cell adhesion molecules (NCAM) in osteoblasts during endochondral and intramembranous ossification, J. Bone Miner. Res., 1992, 7(12), 1435–1446 CrossRef CAS PubMed.
- F. Djouad, et al., Microenvironmental changes during differentiation of mesenchymal stem cells towards chondrocytes, Arthritis Res. Ther., 2007, 9(2), R33 CrossRef PubMed.
- A. C. Bean and R. Tuan, Fiber diameter and seeding density influence chondrogenic differentiation of mesenchymal stem cells seeded on electrospun poly(ε-caprolactone) scaffolds, Biomed. Mater., 2015, 10, 015018 CrossRef PubMed.
- P. Su, C. Xu, J. Yang, W. Yu and D. Huang, Chondrogenic differentiation of human mesenchymal stem cells: a comparison between micromass and pellet culture systems, Biotechnol. Lett., 2010, 32(9), 1339 CrossRef PubMed.
- M. M. J. Caron, et al., Redifferentiation of dedifferentiated human articular chondrocytes: comparison of 2D and 3D cultures, Osteoarthr. Cartil., 2012, 20(10), 1170–1178 CrossRef CAS PubMed.
- T. Wang and F. Yang, A comparative study of chondroitin sulfate and heparan sulfate for directing three-dimensional chondrogenesis of mesenchymal stem cells, Stem Cell Res. Ther., 2017, 8(1757–6512), 284 CrossRef PubMed.
- H. Naito, et al., The advantages of three-dimensional culture in a collagen hydrogel for stem cell differentiation., J. Biomed. Mater. Res., Part A, 2013, 10(1552–4965), 2838–2845 CrossRef PubMed.
- G. Z. Jin and H. W. Kim, Effects of Type I Collagen Concentration in Hydrogel on the Growth and Phenotypic Expression of Rat Chondrocytes, Tissue Eng. Regener. Med., 2017, 14(4), 383–391 CrossRef CAS PubMed.
- D. Bosnakovski, et al., Chondrogenic differentiation of bovine bone marrow mesenchymal stem cells (MSCs) in different hydrogels: influence of collagen type II extracellular matrix on MSC chondrogenesis, Biotechnol. Bioeng., 2006,(0006–3592), 1152–1163 CrossRef CAS PubMed.
- N. Bhardwaj and C. K. Subhas, Chondrogenic differentiation of rat MSCs on porous scaffolds of silk fibroin/chitosan blends, Biomaterials, 2012, 33(10), 2848–2857 CrossRef CAS PubMed.
- S. Takeda, et al., Continuous expression of Cbfa1 in nonhypertrophic chondrocytes uncovers its ability to induce hypertrophic chondrocyte differentiation and partially rescues Cbfa1-deficient mice, Genes Dev., 2001, 15(4), 467–481 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4an01509f |
| This journal is © The Royal Society of Chemistry 2025 |