Open Access Article
Open Access ArticleRecent advances in sensor arrays aided by machine learning for pathogen identification
Xin
Wang
,
Ting
Yang
 * and
Jian-Hua
Wang
* and
Jian-Hua
Wang

Research Center for Analytical Sciences, Department of Chemistry, College of Sciences, Northeastern University, Shenyang 110819, China. E-mail: yangting@mail.neu.edu.cn
First published on 10th September 2024
Abstract
The development of rapid and accurate pathogen detection methods is of paramount importance for slowing the evolution of antibiotic resistance in bacteria. However, the high similarity between different pathogens, especially between antibiotic-sensitive and antibiotic-resistant strains of the same species, presents great challenges for the precise discrimination of pathogens. In recent years, chemical nose strategies, i.e. sensor arrays, have achieved certain success in pathogen discrimination. Currently, chemical nose strategies for identifying pathogens are mainly designed from two perspectives: the disparity in extrinsic properties (biomolecules, charge, and hydrophobicity of the bacterial surface) and intrinsic properties (processes and products mediated by bacterial enzymes) among different pathogens. Biosensing probes capable of responding to these properties are introduced for pathogen detection. The output signals are then processed and analyzed by machine learning algorithms to visualize the multidimensional detection results and achieve pathogen discrimination. This paper introduces the latest developments in sensor arrays for pathogen identification based on the extrinsic and intrinsic nature of bacteria, highlights the recognition mechanism of probes for bacteria, and outlines the current challenges and prospects of sensor arrays for pathogen discrimination.
1. Introduction
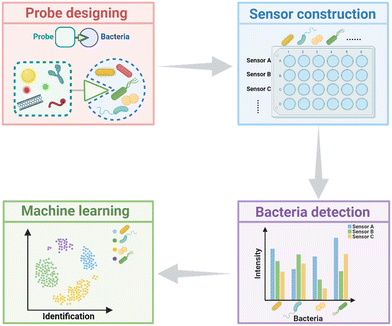
Pathogens, which can be transported through air, water, and food, are intimately linked to daily life and pose significant threats to human health.1,2 Antibiotics inhibit bacteria growth or kill bacteria through various mechanisms such as inhibiting bacterial cell wall synthesis, disrupting protein synthesis, etc. While antibiotics have saved countless lives, their overuse and misuse have led to a rise in antimicrobial resistance.3–5 Therefore, the differentiation and identification of pathogens, especially antibiotic-resistant strains, are crucial for the selection of appropriate antibiotics for the treatment of bacterial infections and slowing down the evolution of antimicrobial resistance. Conventional methods, including bacterial culture, microscopic observation, and gene sequencing techniques, are accurate but time-consuming, which may delay the optimal time for treatment, lead to deterioration of the disease, and endanger patients' lives.6–9 Therefore, the development of rapid and accurate bacterial identification and differentiation methods is of great importance in clinical diagnosis. Emerging bacterial detection techniques, such as polymerase chain reaction (PCR),10 surface-enhanced Raman scattering (SERS),11 and mass spectrometry (MS),12,13 although faster than bacterial culture, require expensive and sophisticated instruments and specialized technicians, making them difficult to implement widely.7,14 Therefore, a variety of simple and convenient biosensors for detecting bacteria have been developed based on ligand–receptor-specific recognition. These methods exhibit high specificity, as each probe can only detect a particular bacterial strain, but they are limited in detecting unknown bacterial samples in real-life situations such as food samples and clinical samples.To address these issues, chemical nose strategies designed based on simulated animal olfaction have been widely proposed.15 The chemical nose strategy, also known as the sensor array approach, usually adopts multiple recognition probes capable of recognizing targets, and sensitively recognizing the subtle differences among them.15–17 These differences are then converted into optical or electrical signals through the probes, processed with statistical methods, and then aided by appropriate machine learning algorithms to realize the classification of different targets (Scheme 1).18,19 To be brief, array-based biosensors can spontaneously identify multiple targets, with differences in their physical and chemical properties that can be recognized by the sensor array.
 | ||
| Scheme 1 Schematic illustrating the workflow of sensor arrays for differentiating bacteria. Scheme 1 was created with https://BioRender.com. | ||
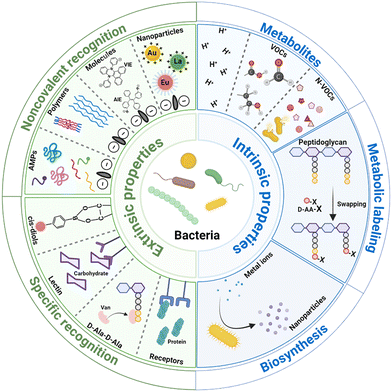
Based on the discrimination principles, the sensor arrays are mainly divided into two types (Scheme 2).16,19 (1) Pathogen discrimination through differences in the surface properties. The bacterial surface contains a variety of biomolecules, including functional groups, proteins, polysaccharides, etc., which can be recognized by probes for identifying bacteria; additionally, these biomolecules co-determine the bacterial surface charge and hydrophobicity. With probes binding to bacteria through electrostatic and hydrophobic interactions, the disparity in the surface charge and hydrophobicity of pathogens can be detected and pathogen discrimination can be achieved. (2) Pathogen discrimination through differences in the intrinsic characteristics of bacteria. The intrinsic characteristics mainly refer to the biochemical processes mediated by enzymes produced by bacteria. As bacteria possess various enzyme systems, their biochemical processes are diverse. Various metabolites are produced by bacterial metabolism, primarily including H+, volatile organic compounds, and non-volatile organic compounds; bacteria incorporate exogenous D-amino acids (D-AAs) into their peptidoglycan through the mediation of transpeptidases to label the peptidoglycan; bacteria reduce metal ions into nanoparticles with various shapes, sizes, and crystalline structures through some reductases. Based on these differences among bacteria, each bacterium possesses a unique fingerprint response spectrum. To discriminate bacteria more quickly and accurately, appropriate algorithms are selected to preprocess and analyze the signal output by the sensors. In this review, recent developments of sensor arrays for bacterial recognition and differentiation are discussed in terms of the intrinsic and extrinsic properties of bacteria. The working principles, analysis performance, advantages, and disadvantages of the reported chemical nose strategies for pathogen identification in recent years are compared and discussed. Finally, the challenges and perspectives for array-based biosensors in bacterial identification are summarized.
2. Identification mediated by bacterial surface properties
Bacteria are generally categorized into Gram-positive and Gram-negative based on the differences in their cell walls. The surface of Gram-positive bacteria is mainly composed of peptidoglycan (which accounts for 50% to 80% of the total dry weight) and teichoic-acid, and Gram-negative bacterial cell walls mainly consist of lipopolysaccharide, peptidoglycan, phospholipid, and lipoprotein. These composites are mainly responsible for the overall surface charge of bacteria.20–22 Due to the genomic difference, the composition of the surface varies among different genera/species/strains, resulting in differences in the overall surface charge or hydrophobicity. For the same strains, culture environments such as evolutionary pressures also lead to differences in surface compositions.22–24For sensor arrays based on the difference in bacterial surface properties to achieve bacteria identification, they can be roughly divided into two categories: those based on the overall difference in bacterial surface charge or hydrophobicity through noncovalent recognition between bacteria and probes, and those employing probes that specifically recognize a certain type of composite on the bacteria surface.
2.1. Noncovalent recognition – electrostatic and hydrophobic interaction
The principle to distinguish bacteria via the disparity of the surface charge and hydrophobicity is that, either the difference in bacterial gene or culture environment would lead to the compositional difference in the bacteria surface, and thereby affect the overall surface charge and hydrophobicity of a certain type of bacterium at a specular situation.25–28 By sensing the differences in bacterial surface potential and hydrophobicity with a series of probes, rapid distinguishment of bacteria can be achieved. These proposed approaches do not require complex instrumentation or specialized technicians, and the detection coverage is broad. In this section, we will discuss sensor arrays constructed by using various sensing materials (nanomaterials, small molecules, and conjugated polymers) that are capable of identifying bacteria through electrostatic and hydrophobic interactions.Gold nanomaterials exhibit low biological toxicity, large surface area, ease of functionalization, and tunable optical property. These advantages facilitate fruitful applications of gold nanomaterials for bacteria detection and identification. For instance, a novel three-dimensional optical sensing array based on gold nanoparticle/gold nanocluster (AuNP/AuNC) composites was developed by our group to discriminate various Gram-negative bacteria (Fig. 1A).33 The nanocomposites consist of cetyltrimethylammonium bromide-coated gold nanoparticles (CTAB-AuNPs, positively charged) and vancomycin templated gold nanoclusters (Van-AuNCs, negatively charged). CTAB imparts positive charges and hydrophobicity to AuNPs. Bacteria with negative charges compete with Van-AuNPs to bind with CTAB-AuNPs, leading to the dissociation of the nanocomposite, and resulting in changes in three types of optical signals. With the assistance of linear discriminant analysis (LDA), the sensor array can selectively identify 10 strains of Gram-negative bacteria (including 3 antibiotic-resistant strains), and is able to analyze bacterial samples with low concentration (OD600 = 0.015).
 | ||
| Fig. 1 Bacterial recognition through noncovalent interactions between bacteria and nanoparticles. A) A 3D sensing array constructed by using AuNP/AuNC nanocomposites for discriminating Gram-negative bacteria. Reproduced with permission.33 Copyright 2019, Royal Society of Chemistry. B) Identification of antibiotic resistance in ESKAPE pathogens by plasmonic nanosensors through electrostatic and hydrophobic interactions. Reproduced with permission.34 Copyright 2023, American Chemical Society. C) Identification of bacterial biofilm based on lanthanide nanoparticles. Reproduced with permission.35 Copyright 2022, American Chemical Society. | ||
Furthermore, Xianyu's group developed plasmonic nanosensors to determine the antibiotic-sensitive and antibiotic-resistant phenotypes among ESKAPE pathogens based on peptide-functionalized AuNPs (P-AuNPs) differing in surface plasmon resonance (SPR) wavelength, diameter, hydrophobicity, and surface charge (Fig. 1B).34 The six types of P-AuNPs interact with ESKAPE through hydrophobic and electrostatic interactions, which, with different binding forces, result in the attachment of different amounts of P-AuNPs on the bacterial surface. The SPR spectra of the P-AuNPs display a blue shift after incubating with bacteria, resulting in distinct SPR fingerprint profiles among different bacteria. Processing SPR spectra with machine learning algorithms can differentiate antibiotic-sensitive and resistant phenotypes of ESKAPE pathogens within 20 min, with an overall accuracy of 89.74%.
Lanthanide nanoparticles possess excellent photostability, tunable multicolor emission, weak autofluorescence background, and strong resistance to photobleaching, greatly improving the sensitivity and reliability of detection.36 Wang et al. reported a multi-emission sensor array based on lanthanide encoding, where lanthanide nanoparticles with different surface charges and hydrophobicity were prepared as recognition probes to identify biofilms (Fig. 1C).35 Three lanthanide nanoparticles, which emit red, yellow, and green fluorescence, respectively, are bound to the biofilm through hydrophobic and electrostatic interactions. Different biofilms differ in physicochemical properties, leading to variable binding abilities to lanthanide nanoparticles and generating characteristic luminescence signal patterns. Then, when the random forest algorithm was used to train the output signals, biofilm identification could be achieved within a few minutes, with an accuracy exceeding 80%.
 | ||
| Fig. 2 Bacterial recognition through noncovalent interaction between bacteria and small luminescent molecules. A) Fluorescent array based on AIEgens for bacterial identification. Reproduced with permission.37 Copyright 2014, Wiley-VCH. B) Identification microbial lysate through competitive interactions by AIEgens and GO.38 Copyright 2018, American Chemical Society. C) Ratiometric sensor array for phenotype identification of methicillin-resistant Staphylococcus aureus based on VIEgens. Reproduced with permission.39 Copyright 2022, American Chemical Society. | ||
Tang's group proposed a competitive binding strategy based on AIEgen/graphene oxide (GO) complexes for the identification of different microbial lysates (Fig. 2B).38 The sensor array consists of one negatively charged, five positively charged, and one neutral AIEgen combined with GO. The aggregation induced emission properties of the AIEgens together with the fluorescence quenching properties of GO reduced background fluorescence, thereby greatly enhancing the sensitivity of the sensor.
Another promising luminescent probe, VIEgens, was first reported by Tian and co-workers in 2015, who discovered this interesting phenomenon while studying the dynamic luminescence properties of dihydrodibenzo[a,c]phenazine derivatives.40,41 VIEgens emit blue fluorescence in the “saddle”-shaped conformation and emit red fluorescence in the planar conformation. Their conformation is sensitive to the surrounding microenvironment, making VIEgens promising probes for biosensing. Hu et al. proposed a sensor array constructed from eight VIE probes for phenotypic identification of methicillin-resistant Staphylococcus aureus (MRSA) (Fig. 2C).39 Around the VIEgen core, these probes carry two quaternary ammonium salts across different substitution sites. The substituents determine the structure of VIEgens, leading to differences in interactions with negatively charged bacterial cell walls, which in turn determine the molecular conformation of the VIEgens. When VIEgens interact with bacteria, they change into a “saddle” conformation with an enhanced blue fluorescence. After the blue-to-red fluorescence intensity ratio of VIEgens generated by different bacteria was processed by the principal component analysis (PCA) algorithm, this sensor array was able to identify six phenotypes of Staphylococcus aureus, with analysis results that were consistent with those obtained from polymerase chain reaction analysis.
Wang et al. developed a single-component multichannel array based on composites of graphene oxide (GO) and polyethyleneimine derivatives (PEI, Fig. 3A).23 These PEI derivatives altered the surface physicochemical properties of GO, including charge, polarity, and hydrophobicity, resulting in diverse binding strengths with bacteria. Through electrostatic and hydrophobic interactions with bacteria, the sensor array successfully identified 10 types of strains within minutes. Moreover, the sensor array could effectively recognize bacteria in complex biological samples (such as urine) (OD600 = 0.25), achieving an accuracy of 94%.
 | ||
| Fig. 3 Bacterial recognition through noncovalent interaction between bacteria and polymers. A) One-component multichannel sensor array for rapid identification of bacteria based on polymers.23 Copyright 2022, American Chemical Society. B) A polymer-based sensor for differentiating the resistant phenotypes of antibiotic-resistant bacteria. Reproduced with permission.44 Copyright 2022, American Chemical Society. C) Fluorescent polymer-based chemical-nose systems for recognizing gut microbiota. Reproduced with permission.45 Copyright 2022, Royal Society of Chemistry. D) A dual fluorescence turn-on sensor array formed by polymer and AIEgens for recognizing bacteria. Reproduced with permission.46 Copyright 2024, Wiley-VCH. | ||
Roy et al. have developed a sensor array capable of differentiating antibiotic-resistant bacteria and grouping bacteria with the same resistance phenotype, based on three cationic benzyl-functionalized polymers and three different fluorescent transducers to build four ratio channels (Fig. 3B).44 By interacting with bacteria through electrostatic interactions, the sensor array sensitively recognized subtle differences on the bacterial surface induced by antibiotics within 30 min, generating unique responses and providing useful information for bacteria resistance.
Gut microbiota is closely associated with the development of diseases. Tomita et al. developed a sensor array capable of identifying gut microbiota (Fig. 3C). This sensor array was composed of twelve polyethylene glycol-block-poly-L-lysine (PEG-b-PLL) derivatives bearing AIE fluorophores.45 These probes bind through various interactions with the bacterial surface, generating distinctive fluorescent fingerprint responses. By further processing the generated fluorescent responses with machine learning algorithms, this array sensor could categorize gut bacteria into strains, species, and phylums and distinguish the gut microbiota of obese and healthy individuals by detecting artificially simulated fecal samples.
In order to improve the sensitivity of bacterial recognition, Han's group developed a dual-channel fluorescence “turn-on” sensor array consisting of six electrostatic complexes formed by six negatively charged poly(para-aryleneethynylene) polymers (PPE) and six positively charged AIE fluorophores (Fig. 3D).46 AIE quenches the fluorescence of PPE upon complexing with PPE, whereas bacteria could compete with PPE for binding with AIE and recover the fluorescence of PPE. Meanwhile, as AIE fluorophores aggregated on the bacterial surface, their emission also enhanced. This unique dual “turn-on” response, based on a six-element sensor array, assisted by various machine learning algorithms, enabled the identification of 20 bacteria at low concentrations (OD600 = 0.001), achieving 96.7% accuracy in distinguishing urinary tract infection patients from healthy individuals.
Similarly, Zhang's group recognized and identified bacteria based on the differences in recognition and disintegration by four short antimicrobial peptides (SAMPs) against different bacterial strains.54 Processing of the collected unique fluorescence spectra of bacteria by LDA enabled the identification and differentiation of six types of bacteria within 30 min, with a classification accuracy of 100%.
2.2. Specific recognition – recognize biomarkers on bacterial surfaces
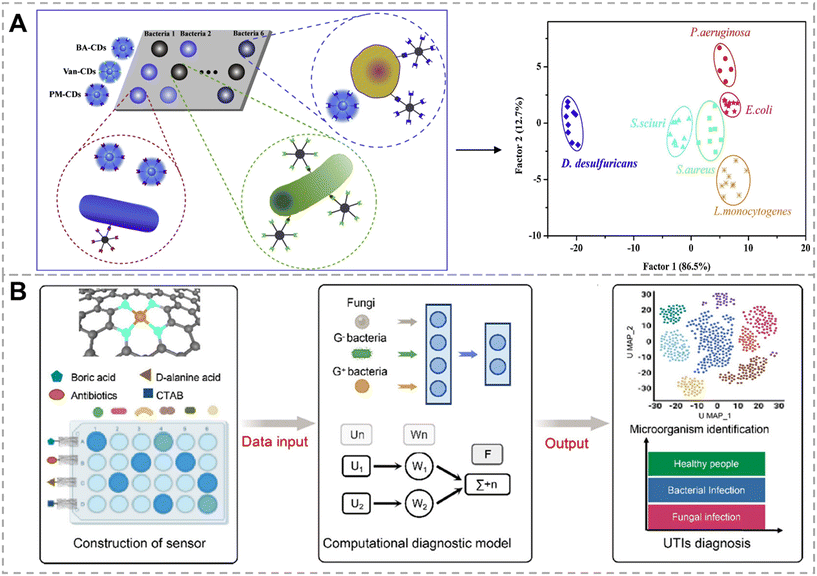
Bacterial surfaces contain a variety of biomarkers, such as carbohydrates, proteins, short peptides, etc. These biomarkers can serve as target molecules interacting specifically with recognition probes that are functionalized with receptor proteins, carbohydrates, or antibiotics, providing insights for the development of bacterial detection approaches. In this section, we summarize and discuss the identification approaches for bacteria based on various biomarker recognition mechanisms.Yan et al. proposed a colorimetric sensor array based on Wulff-type 4-mercaptophenylboronic acid (MPBA)-mercaptoethylamine (MA) co-functionalized AgNPs (MPBA-MA@AgNPs) for bacterial detection and identification (Fig. 4A).60 Wulff-type boronic acid is a unique adjacent amine boronic acid that can form cyclic ester bonds with cis-diol through intramolecular B–N bonds, allowing boronic acid and cis-diol to form cyclic esters under neutral conditions. Under neutral and alkaline conditions, MPBA binds to cis-diol on the bacterial surface, leading to the dispersion of AgNPs. Due to the presence of different carbohydrates on the bacterial surface, MPBA exhibits different affinities for the cis-diol in different carbohydrates, resulting in the color change of AgNPs. Under acidic conditions, the B–N bond breaks, and the positively charged MA binds to the negatively charged bacteria via electrostatic interactions, causing AgNPs to aggregate on the bacterial surface, and the color of AgNPs varies due to differences in bacterial surface potentials. Therefore, the MPBA-MA@AgNPs show variable binding ability to bacteria at different pH buffer solutions, and generate distinct color responses. By using LDA for data processing, 17 strains were successfully differentiated. This colorimetric method, based on specific affinity and electrostatic interactions, holds promise for the identification of bacteria in clinical urine and serum specimens; however, the color changes of various bacteria in this method are slight, which suggests that relying on colorimetry alone to differentiate bacteria is not reliable. Additionally, our group proposed a sensor array fabricated by using boronic acid modified g-C3N4 (BA-g-CN) nanosheets.61 Under acidic conditions, the positively charged BA-g-CN nanosheets interact with the negatively charged bacteria through electrostatic interaction. Under neutral or alkaline conditions, BA-g-CN nanosheets bond to the bacteria by forming covalent bonds through boronic acid and carbohydrates on the bacterial cell wall. These interactions lead to bacterial sedimentation and fluorescence reduction in the supernatant. By using acidic, neutral, and alkaline conditions as three different sensing channels, the identification of nine strains can be achieved. This sensing strategy focuses on distinguishing Gram-negative bacteria, particularly four strains of Escherichia coli, ensuring timely and targeted treatment of Gram-negative bacteria. Recently, Wang et al. constructed a dual-mode bacterial sensor array based on colorimetric and photothermal detection.62 This sensor uses boronic acid-functionalized Au–Fe3O4 nanoparticles (BA-GMNPs) as probes, which not only exhibited localized surface plasmon resonance properties but also demonstrated superparamagnetism. BA-GMNPs formed B–N bonds with cis-diol on the bacterial surface, dispersing on the bacterial surface and weakening the aggregation of BA-GMNPs induced by NaCl. The content of cis-diol on the surface of different bacteria varied, resulting in different quantities of BA-GMNPs bound to bacteria and different amounts of BA-GMNPs aggregated in the supernatant. After magnetic separation, the RGB values and UV-vis spectra of the aggregated BA-GMNPs in the supernatant were measured; the precipitate was irradiated with an 808 nm laser, and temperature changes were monitored using a thermal imager. With the assistance of LDA, six bacteria were successfully distinguished, and even bacteria in real samples, such as milk, were successfully identified, with a recognition accuracy of 93.3%.
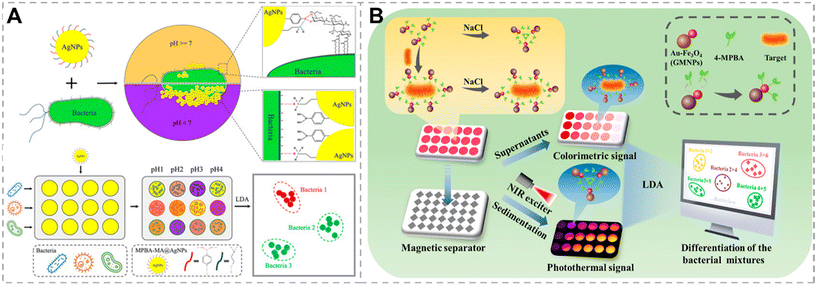 | ||
| Fig. 4 Bacterial recognition through the interaction between boronic acid and the cis-diol group. A) A colorimetric sensor array based on Wulff-type boronate functionalized AgNPs for bacterial identification. Reproduced with permission.60 Copyright 2019, American Chemical Society. B) A dual-mode sensor array based on colorimetric and photothermal detection for bacterial identification. Reproduced with permission.62 Copyright 2023, American Chemical Society. | ||
 | ||
| Fig. 5 Bacterial recognition through the interaction between lectins and glycan. A) Discrimination of bacteria by ratiometric analysis of their carbohydrate binding profile. Reproduced with permission.68 Copyright 2016, Royal Society of Chemistry. B) A sensor array consisting of multivalent glycosylated Cu:CdS quantum dots for rapid bacterial discrimination. Reproduced with permission.69 Copyright 2017, Elsevier. | ||
 | ||
| Fig. 6 Bacterial recognition through the interaction between vancomycin and D-ala-D-ala. A) Identification of bacteria by a fluorescent sensor array based on boronic acid, vancomycin, and polymyxin functionalized carbon dots. Reproduced with permission.75 Copyright 2019, Elsevier. B) A colorimetric sensor array for rapid diagnosis of urinary tract infection by a multi-recognition mechanism, which includes the recognition between vancomycin and D-ala-D-ala. Reproduced with permission.76 Copyright 2024, American Chemical Society. | ||
 | ||
| Fig. 7 Bacterial recognition through the interaction between proteins and specific receptors on bacterial surfaces. A) Protein-AuNC-based fluorescence sensor array for discrimination of bacteria. Reproduced with permission.78 Copyright 2018, Wiley-VCH. B) Multichannel sensor array based on muti-functionalized AuNCs for gut microbiota detection. Reproduced with permission.83 Copyright 2023, American Chemical Society. | ||
The composition and activity of the gut microbiota are crucial for health management and disease treatment. Liu et al. developed a rapid sensor array for distinguishing gut bacteria (Fig. 7B).83 Gold nanoclusters (AuNCs) were modified with Van, bacitracin, and lysozyme as signal-reporting probes that specifically identified D-ala-D-ala moieties, pyrophosphate groups, and peptidoglycans, respectively. Ti3C2 transition-metal carbides (MXenes) were used as fluorescence quenchers. When three recognition probes were mixed with Ti3C2 MXenes, they were tightly bound through hydrogen bonding, leading to energy transfer and quenching the fluorescence of AuNCs. Both bacteria and three functionalized AuNCs could bind with Ti3C2 MXenes but the binding strengths vary. After adding bacteria to Ti3C2 MXenes-AuNCs composites, they either restore the fluorescence of AuNCs by competing with AuNCs for binding with Ti3C2 MXenes or lead to further fluorescence quenching. Due to the association between gut bacteria and colorectal disease, this strategy coupled with pattern recognition algorithms accurately differentiated colorectal cancer patients from healthy individuals within 30 min, providing a robust and simple platform for the clinical diagnosis of colorectal cancer.
3. Identification mediated by bacterial intrinsic properties
Bacterial metabolism includes catabolism and anabolism, which are the most fundamental processes in bacterial life activity. Enzymatic catalysis is required for bacterial metabolic processes, with different bacteria possessing distinct enzyme systems, leading to diverse metabolic behaviors among different bacteria.84–883.1. Bacteria identification by metabolites
Variations exist among bacteria in the enzymes involved in the decomposition of nutrients such as sugars and proteins, resulting in diverse decomposition capabilities, pathways, and products. Methods for the identification of bacteria have been widely proposed based on metabolites among different bacteria.87,88 In this section, we will discuss the sensor arrays constructed based on the recognition of various metabolites of bacteria.Liu's group developed a colorimetric sensor array for microbial discrimination and antimicrobial susceptibility testing, using three pH indicators, including methyl red (MER), bromothymol blue (BTB), and bromocresol green (BCG), as H+-sensing probes (Fig. 8A).89 The bacterial metabolism of glucose generates acids, including acetates, lactates, and other acidic byproducts, causing changes in the acidity of the bacterial culture microenvironment. Due to variations in the ability of different bacteria to metabolize glucose, the acids produced by different bacteria also vary in terms of both types and concentrations, resulting in significant differences in the color change of the three pH indicators. Combined with the LDA algorithm, this sensor array facilitated the identification of 11 types of bacteria.
 | ||
| Fig. 8 Bacterial recognition through metabolites produced by bacterial metabolism. A) The glucose-metabolism-triggered colorimetric sensor array based on MER, BTB, and BCG for differentiation and antibiotic susceptibility testing of bacteria. Reproduced with permission.89 Copyright 2023, Elsevier. B) Agar gel-based colorimetric sensor array for identification of bacteria in household refrigerators. Reproduced with permission.90 Copyright 2023, Royal Society of Chemistry. MER: methyl red. BTB: bromothymol blue. BCG: bromocresol green. | ||
Real-time monitoring and identification of bacteria are highly elusive goals in the field of bacterial detection. Shaulof et al. introduced a capacitive artificial nose for the VOC-identification-based bacteria discrimination method using interdigitated electrodes (IDEs) modified with carbon dots (C-dots) exhibiting different polarities (C-dot-IDE).98 VOCs metabolized by bacteria induce rapid changes in the capacitance of the C-dot-IDE, which is intimately related to the polarity of gas molecules. With the application of machine learning algorithms, the C-dot-IDE gas sensor exhibits excellent recognition performance, enabling continuous monitoring of bacteria proliferation and differentiation.
 | ||
| Fig. 9 Bacterial recognition through nonvolatile compounds produced by bacterial metabolism. A) Gold nanoparticle chemiresistor capable of identifying bacteria based on metabolites released by bacteria. Reproduced with permission.100 Copyright 2015, Elsevier. B) A fluorescent nanosensor based on SWCNTs for clinically essential bacteria discrimination. Reproduced with permission.101 Copyright 2020, Nature. SWCNTs: single-walled carbon nanotubes. | ||
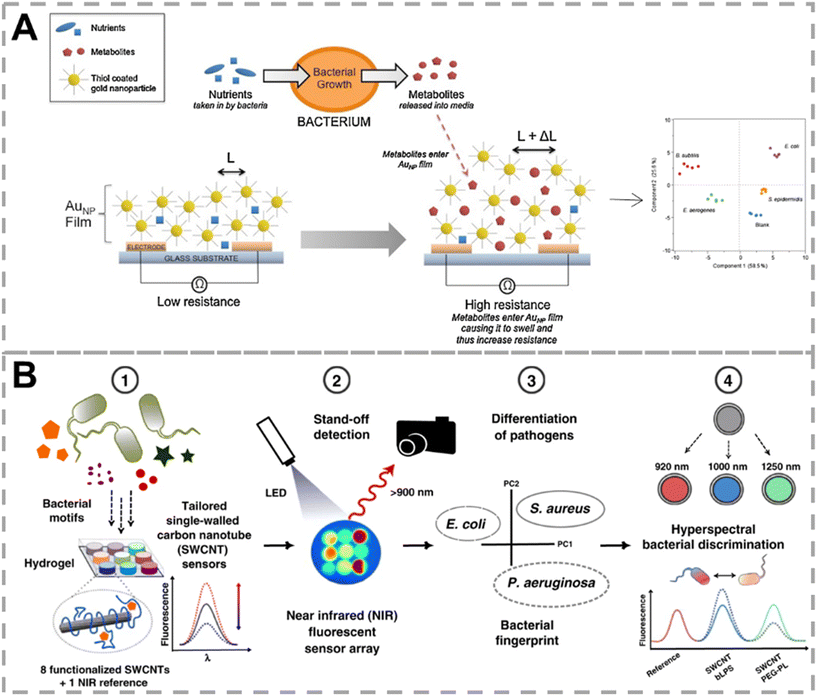
Nonvolatile metabolites are diverse among bacteria. For instance, endotoxin is a lipopolysaccharide released upon the lysis of Gram-negative bacteria, which is a unique metabolite of Gram-negative bacteria. Siderophores are a class of low molecular weight compounds synthesized by microorganisms under conditions of low iron concentration. They exhibit a specific high affinity for Fe3+ and are a common pathway for bacteria to acquire iron from the environment. Nißler et al. developed a set of near-infrared fluorescent nanosensors for remote identification of clinical bacteria (Fig. 9B).101 Nine sensing units, consisting of functionalized single-walled carbon nanotubes (SWCNTs) specific for lipopolysaccharide, siderophores, deoxyribonucleases, and proteases, along with four generic low-selective SWCNTs and one stable near-infrared fluorescent reference, were integrated into a biocompatible hydrogel. This system successfully differentiated 6 common bacteria in the clinic aided by PCA. Multiplexing was achieved by spectral encoding (900 nm, 1000 nm, 1250 nm), which was capable of differentiating Pseudomonas aeruginosa and Staphylococcus aureus. The near-infrared fluorescence also endowed this approach with the ability of tissue penetration (>5 mm).
3.2. Identification of bacteria by metabolic labeling
Metabolic labeling is an effective method to study the metabolic processes of living systems, allowing efficient and non-destructive incorporation of functionalized metabolites into target biomolecules through biosynthetic mechanisms.102,103 Peptidoglycan, which is widely present in bacterial cell walls, is a multilayered network structure composed of glycans and short peptides. Functionalized monosaccharides and amino acids can be incorporated into peptidoglycan by metabolic labeling.104,105 A sensing strategy combining metabolic labeling with fluorophores was proposed by Hong et al., with bacteria metabolizing three different fluorescent metabolic derivatives to identify different bacterial species or microbiota types (Fig. 10A).106 Due to disparities in metabolism ability, unique colorimetric fingerprints for different bacterial species and microbiota after metabolic labeling were obtained. By utilizing a portable spectrometer combined with a smartphone for direct reading of fluorescent RGB values, discrimination of different bacterial species and microbiota types was achieved.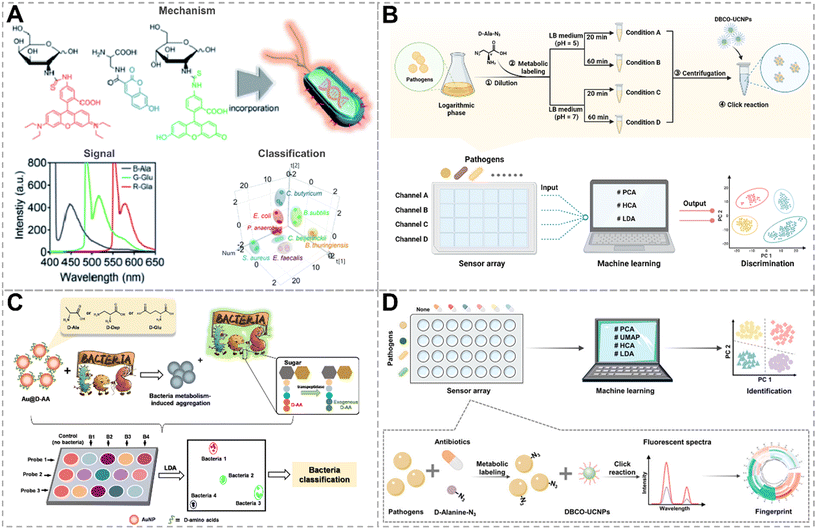 | ||
| Fig. 10 Bacterial recognition through metabolic labeling. A) The RGB-emitting probes for the detection of bacteria by labeling peptidoglycan. Reproduced with permission.106 Copyright 2020, Royal Society of Chemistry. B) A fluorescent sensor array for identifying bacteria based on metabolic labeling at various acidities and times. Reproduced with permission.107 Copyright 2022, Elsevier. C) Metabolism-triggered colorimetric sensor array constructed from three types of D-AA functionalized AuNPs for bacterial identification. Reproduced with permission.108 Copyright 2022, American Chemical Society. D) Identification of antibiotic-resistant bacteria based on metabolic labeling and click reaction. Reproduced with permission.109 Copyright 2023, American Chemical Society. | ||
D-AAs are persistent almost exclusively in bacterial cell walls, endowing them with unique advantages in bacteria labeling. Unnatural D-AAs can be incorporated into bacterial cell walls by swapping amino acids at the 5th or 4th position of the pentapeptide, mediated by D,D-transpeptidase or L,D-transpeptidase, respectively. Recently, our group proposed a chemical nose approach based on metabolic labeling and click reactions (Fig. 10B).107 Bacteria first metabolized clickable handle tagged D-AAs at two acidities (pH = 5 and 7) for 20 min and 60 min, respectively, followed by clicking with up-conversion nanoparticles (UCNPs), generating four-dimensional signal outputs. Multidimensional fluorescent response analysis by machine learning successfully classified eight types of bacteria in the training set on strain, genus, and Gram phenotype. The differences in signal response under four incubation conditions reflected variations in the quantity and activity of enzymes involved in metabolic labeling, endowing this strategy with good anti-interference capabilities. This approach can accurately classify unknown samples from the validation set into clusters independent of the training set and can accurately identify their Gram properties.
Culture-based bacterial detection and antibiotic susceptibility testing (AST) are the gold standard methods for bacterial detection, but they are time-consuming. In such cases, clinicians often resort to broad-spectrum antibiotics for treatment at an early stage, which worsens the issue of antimicrobial resistance. Therefore, the development of accurate and rapid methods for bacterial differentiation and AST is urgently needed. Liu's group constructed a colorimetric sensor array for bacterial identification using D-ala, D-2,3-diaminopropionic acid, and D-glutamate-functionalized gold nanoparticles (AuNPs) as probes (Fig. 10C).108 Bacteria were competitively metabolized with exogenous D-AAs on the surface of AuNPs through the mediation of D,D-transpeptidase, causing the AuNPs to become unstable and aggregate. AuNPs exhibit distinct aggregation phenomena due to the different metabolic capabilities of bacteria towards various D-AAs. By analyzing the unique responses by LDA, eight bacterial species, including an antibiotic-resistant strain, were successfully differentiated. Furthermore, by monitoring the bacterial metabolic activity of D-AAs under different antibiotic treatments, a colorimetric-based rapid AST method was developed, and the whole procedure was completed in 24 h, promoting the clinical practicability of this approach.
For applying array sensor approaches in the clinic, it should be sensitive enough to detect low-concentration bacteria in real clinical samples. Our group constructed a sensor array based on metabolic labeling and the “antibiotic responsive spectrum” for distinguishing antibiotic-sensitive and resistant strains at bacteria concentrations as low as 105 CFU mL−1 (Fig. 10D).109 This strategy incorporates D-ala with azido groups into bacterial cell walls through bacterial metabolism, followed by clicking with dibenzocyclooctyne-functionalized UCNPs (DBCO-UCNPs). By proceeding with metabolic labeling in the presence of six antibiotics, an “antibiotic responsive spectrum” was obtained as a result of a unique fluorescent fingerprint corresponding to a certain type of bacterium. By combining with various machine learning algorithms, eight bacterial strains were effectively clustered based on species, Gram phenotype, and antibiotic resistance. This metabolic labeling strategy exhibits excellent anti-interference capabilities, accurately identifying bacteria (concentration: 105 CFU mL−1) in artificial urinary tract infection samples and determining their resistance.
3.3. Identification of bacteria by biosynthesis
Bacteria, as efficient biofactories, can utilize various enzymes, proteins, peptides, and electron transfer pathways in vivo to regulate inorganic ions and reduce them into inorganic nanoparticles of different shapes and sizes. Up till now, researchers have employed bacteria to synthesize over 100 types of inorganic nanoparticles.110,111 Enzymes are the primary biomolecules responsible for the reduction of inorganic ions. Different microorganisms produce various reductases, including nitrate and nitrite reductases, nicotinamide adenine dinucleotide phosphate-dependent reductases, and sulfate (SO42−) and sulfite (SO32−) reductases, which co-regulate microbial biosynthesis.112–115 Xianyu's group constructed a novel sensor array based on differences in the properties of microbially biosynthesized AuNPs, such as particle size, surface plasmon resonance (SPR) spectra, and surface potential (Fig. 11).116 By employing PCA, LDA, and random forest (RF) algorithms to explore the relationships between features of microbial-synthesized AuNPs and microbial types, classification models were established. Microorganisms were classified at the levels of kingdom, order, genus, and species. This microbial discrimination result was in accordance with the results obtained by traditional classification approaches.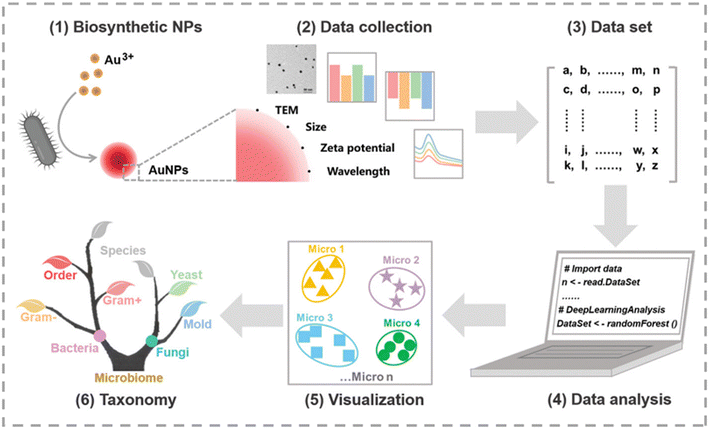 | ||
| Fig. 11 Bacterial recognition through biosynthetic nanoparticles from bacteria. Reproduced with permission.116 Copyright 2022, Wiley-VCH. | ||
4. Statistical analysis and machine learning
Machine learning algorithms can rank the significance of input variables, identify trends in the data, and classify the data, enhancing its visualization and readability.117 Since chemical nose strategies are typically constructed using multiple sensor units, resulting in large volumes of high-dimensional data with poor readability, combining machine learning with sensor arrays can extract more valuable information.118,119 In this section, we categorize the machine learning algorithms used for statistical analysis into two types: clustering methods and classification methods.4.1. Clustering methods
Clustering algorithms, including HCA, PCA, and t-distributed stochastic neighbor embedding clustering (t-SNE), are unsupervised algorithms capable of clustering data. In high-dimensional space, HCA typically uses Ward's minimum variance as the clustering criterion and Euclidean distance as the distance metric to cluster data.120,121 The HCA dendrogram intuitively expresses the similarity between samples and can effectively convey relationships between similar species. However, since the HCA dendrogram has only one quantitative axis (distance) and the orientation of the dendrogram does not affect data clustering, it may have misinterpretation issues due to changes in the relative position of the linkage axes.PCA is a commonly used dimensionality reduction algorithm in sensor arrays.76 It transforms linear combinations of the initial variables into a set of orthogonal combinations, projecting data from sensor arrays into 2D or 3D space to represent the feature relationships of data.120–122 PCA projects the original data onto a new coordinate system, where the first coordinate axis (the first component) is determined by finding the maximum variance of the dataset along this vector. Subsequent components are orthogonal to previous ones and follow the principle of maximum variance. The score plots after dimensional reduction by PCA are more visualizable than the original data. However, biological data tends to be nonlinear, and PCA, being a linear dimensionality reduction method, fails to capture these nonlinear relationships, potentially leading to information loss. t-SNE, a nonlinear dimension reduction algorithm, is more suitable for analyzing high-dimensional biological information.123 t-SNE creates a probability distribution for each data point by calculating Euclidean distances between data points, with closer points representing higher similarity. It then creates a similarity matrix based on data similarities and can preserve both local and global structures of the data, which is beneficial for discovering more relationships within biological information. All the above algorithms are unsupervised and cannot predict unknown samples. When new data is added to the dataset, the entire clustering must be recalculated.76
4.2. Classification methods
The classification method, which usually belongs to supervised algorithms, enables the prediction of unknown samples. In sensor arrays for microbial identification, the classification method utilizes a dataset labeled with microbial species as the training set. It discovers the relationship between microbial species and corresponding data through the training set and establishes a mathematical model.15,76 By inputting the data of unknown samples into the trained mathematical model, an accurate classification of unknown samples can be obtained. Common classification algorithms in sensor arrays include LDA, support vector machines (SVM), and convolutional neural networks (CNNs).In recent years, LDA has been widely applied in sensor arrays. It performs orthogonal transformations on the potentially linear dependence original variables to nonlinear dependence variables, similar to the PCA algorithm. Unlike PCA, LDA is a supervised algorithm that can predict unknown samples by establishing a mathematical model.124 It follows the classification principle of minimizing intraclass variance and maximizing interclass variance, meaning that it aims to make projections of data with the same label as close as possible while keeping projections of data with different labels as far apart as possible, in an attempt to find the dimension with the best separation. The LDA score plot can intuitively evaluate the effectiveness of classification and discrimination. However, LDA is not suitable for classification with high-dimensional data but few classes. SVM is an optimization-based algorithm aimed at finding the optimal hyperplane for classifying data. When dealing with linearly separable models, SVM achieves optimal classification by maximizing the margin between classes. For linearly inseparable models, it maps data to a high-dimensional space to achieve linear separation.125,126 SVM performs exceptionally well with data of high-dimensional and small sample populations; however, as sample populations increase, its processing efficiency decreases. The evolution of deep learning algorithms is promoted by the rapid development of hardware and software technology of computers. CNN is a type of supervised deep learning algorithm that extracts features from data through convolution and pooling, mapping the input data into a high-dimensional space, and then performing classification or regression of the features through dense layers.127 CNNs are characterized by strong feature extraction capability and high robustness, and they have been widely used in the field of biosensing.
5. Outlook and perspectives
Machine learning-assisted sensor arrays are an effective strategy for identifying bacteria, enabling rapid qualitative analysis of various bacteria, which plays an important role in detecting bacteria and inhibiting their spread in real life. We compared the reported sensor arrays in terms of recognition principle, types of interactions between bacteria and the sensor, probe materials, types of pathogens that can be identified, channel numbers employed, reaction time, minimum bacteria concentrations that can be detected (not the limit of detection), and signal readout of the sensor arrays, with details listed in Table 1.| Recognition mechanism or substance | Type of identification | Probes | Types of pathogens identified | Number of channels | Reaction time | Bacteria concentration | Signal readout | Ref. |
|---|---|---|---|---|---|---|---|---|
| Note: Ref. represents reference. | ||||||||
| Electrostatic and hydrophobic interactions | Noncovalent interaction | AuNP/AuNCs | 10 (3 antibiotic-resistant strains) | 3 | 10 min | OD600 = 0.015 | Fluorescence, UV-vis absorption and light-scattering | 33 |
| Electrostatic and hydrophobic interactions | Noncovalent interaction | AuNPs | 12 (6 antibiotic-resistant strains) | 6 | 20 min | Surface plasmon resonance spectra | 34 | |
| Electrostatic and hydrophobic interactions | Noncovalent interaction | Lanthanide NPs | 5 | 3 | A few minutes | Fluorescence | 35 | |
| Electrostatic interaction | Noncovalent interaction | AIEgens | 8 (2 antibiotic-resistant strains) | 5 | 30 min | OD600 = 0.002 | Fluorescence | 37 |
| Electrostatic interaction | Noncovalent interaction | AIEgen/GO | 6 | 7 | 2 h | OD600 = 1 | Fluorescence | 38 |
| Electrostatic interaction | Noncovalent interaction | VIEgens | 10 (2 antibiotic-resistant strains) | 8 | 60 min | 104 CFU mL−1 | Fluorescence | 39 |
| Electrostatic and hydrophobic interactions | Noncovalent interaction | Conjugated polymers and GO | 10 | 3 | Within minutes | OD600 = 0.25 | Fluorescence | 23 |
| Electrostatic interaction | Noncovalent interaction | Conjugated polymers and fluorescent transducers | 4 | 4 | 30 min | OD600 = 0.25 | Fluorescence | 44 |
| Electrostatic, hydrophobic, π–π, and hydrogen bonding interactions | Noncovalent interaction | Conjugated polymers | 16 | 12 × 4 | <30 min | OD600 = 0.04 | Fluorescence | 45 |
| Electrostatic interaction | Noncovalent interaction | Conjugated polymers and AIE | 20 | 6 | 30 s | OD600 = 0.001 | Fluorescence | 46 |
| Electrostatic and hydrophobic interactions (according to reference) | Noncovalent interaction | AMPs and conjugated polymers | 14 | 4 | 30 min | OD600 = 0.01 | Fluorescence | 53 |
| AMPs – lipid and membrane (according to reference) | Specific recognition | Short antimicrobial peptides | 6 | 4 | 30 min | 1 × 108 CFU mL−1 | Fluorescence | 54 |
| Mercaptophenylboronic acid–cis-diol interactions and electrostatic interactions | Covalent and noncovalent interactions | AgNPs | 17 | 8 | 15 min | OD600 = 0.05 | UV-vis absorption | 60 |
| BA–cis-diol interactions | Covalent interaction | Au–Fe3O4 | 6 | 5 | 20 min | OD600 = 0.1 | UV-vis absorption and temperature | 62 |
| Glycan–lectin interactions | Specific recognition | Well plate | 9 | 5 | 1.5 h | OD600 = 1 | Fluorescence | 68 |
| Glycan–lectin interactions | Specific recognition | Cu:CdS quantum dots | 3 | 6 | 30 min | OD600 = 0.3 | Fluorescence | 69 |
| Van–D-ala-D-ala, polymyxin–phospholipid and BA–cis-diol interactions | Covalent interaction and specific recognition | Carbon dots | 6 | 3 | 60 min | OD600 = 1 | Fluorescence | 75 |
| Van–D-ala-D-ala, BA–cis-diol and electrostatic interaction, and metabolic labeling | Covalent and noncovalent interactions | Fe single-atom nanozymes | 10 | 4 | 30 min | 104 CFU mL−1 | UV-vis absorption | 76 |
| HSA–cell walls, Lf–lactoferrin receptors, Lys–peptidoglycans, and Van–D-ala- D-ala interactions | Specific recognition | AuNCs | 6 (2 antibiotic-resistant strains) | 4 | 60 min | OD600 = 1 | Fluorescence | 78 |
| Van–D-ala-D-ala, bacitracin–pyrophosphate groups, and Lys – peptidoglycans | Covalent interaction and specific recognition | AuNCs and Ti3C2 MXenes | 5 | 3 | 30 min | OD600 = 0.2 | Fluorescence | 83 |
| Metabolites (H+) | Chemical reaction | Acid–base indicators | 11 (2 antibiotic-resistant strains) | 3 | 6 h | 107 CFU mL−1 | UV–vis absorption | 89 |
| Metabolites (VOCs) | Chemical reaction | — | 6 | 12 | 3 h | OD600 = 1 | PTR-MS and FGC-PTR-MS | 97 |
| Metabolites (VOCs) | Chemical reaction | Dye-containing agar gel | 4 | 16 × 3 | 12 h | 103 CFU mL−1 | — | 90 |
| Metabolites (VOCs) | Chemical reaction | C-Dot-IDE | 4 | 3 | 1.3 s, continuous monitoring | OD600 = 0.5 | Capacitance | 98 |
| LPS-binding peptide – LPS, hemin [Fe3+]–siderophores, etc. | Specific and nonspecific recognitions | SWCNTs | 6 | 9 | Incubation after 1 h, acquired in 30 s intervals | — | Fluorescence | 101 |
| Metabolic labeling | Covalent interaction | Small molecules | 3 | 3 | 12 h | OD600 = 0.4 | Fluorescence | 106 |
| Metabolic labeling and click reaction | Covalent interaction | UCNPs | 8 (2 antibiotic-resistant strains) | 4 | 100 min | 106 CFU mL−1 | Fluorescence | 107 |
| Metabolic labeling and hydrophobic interactions | Covalent interaction (metabolism) and noncovalent interaction (AuNPs aggregation) | AuNPs | 8 (2 antibiotic-resistant strains) | 3 | 6 h | OD600 = 0.1 | UV-vis absorption | 108 |
| Metabolic labeling and click reaction | Covalent interaction | UCNPs | 8 (3 antibiotic-resistant strains) | 7 | 3 h | 105 CFU mL−1 | Fluorescence | 109 |
| Biosynthesis | AuNPs | 17 (1 antibiotic-resistant strains) | 3 | 12 h | OD600 = 0.5 | UV–vis absorption, zeta potential, and particle size | 116 | |
Array-based sensing strategies have achieved certain success in bacterial discrimination, but these methods are still in their infancy and need to be further improved to meet the requirements of practical applications. The sample matrices in real bacterial-containing samples are complex, unknown, and with low concentrations of bacteria. However, current sensing strategies are mostly based on known bacterial strains cultivated in the laboratory, which may fail in practical applications due to the following limitations. First of all, as the surface charge and hydrophobicity vary among bacteria due to environmental changes, noncovalent interactions are easily affected by environmental disturbances, leading to low interference tolerance; the expression of proteins, sugars, and other biomolecules on the bacterial surface may differ as bacteria growth environments vary. Therefore, it is difficult to detect complex real samples solely by recognizing the external properties of bacteria. The sensor arrays based on the detection of the products of bacterial enzymatic catalysis processes typically require sample pretreatment to minimize interference from the sample matrices. Therefore, it is necessary to develop strategies for detecting bacteria based on multi-recognition. Secondly, sensor arrays are constructed based on a library of laboratory strains and can only identify bacteria within the library, which may pose limitations in detecting unknown bacteria in real samples. Therefore, it is necessary to develop a sensing strategy to detect a large number of clinical samples to construct a training set based on actual samples, making it possible to apply it to clinical samples. On the other hand, it is necessary to develop sensor arrays that are capable of clustering bacteria by genus, species, Gram properties, and antibiotic sensitivity, enabling rapid pre-classification of bacteria outside the database for targeted detection in the next step. Thirdly, currently developed sensor arrays are greatly influenced by bacteria concentration and struggle to achieve fingerprint recognition. For sensors that differentiate bacteria populations in samples, signal intensity is also related to bacterial concentration. Therefore, appropriate preprocessing of the signal to eliminate interference from concentration is necessary. Additionally, bacterial heterogeneity, especially the heterogeneity of antibiotic-resistant bacteria caused during antibiotic treatment, is a key focus for future monitoring. Fourthly, current data analysis of chemical nose strategies for bacterial identification primarily relies on linear dimensionality reduction methods, such as PCA, which struggle to capture the complex structure of high-dimensional data. However, in deep learning, CNN can better process data distribution in high-dimensional spaces through nonlinear dimensionality reduction. Therefore, it is essential to continuously optimize mathematical classification models and develop more robust machine learning algorithms to achieve more reliable identification results.
In summary, to enable the clinical translation of chemical nose strategies, sensor arrays capable of discriminating bacteria at the single bacterium level are helpful to avoid the concentration-dependence issue of the current sensor arrays. Secondly, sufficient strains isolated from clinical samples should be incorporated into the training set to increase the accuracy of the responses to real clinical samples. Finally, machine learning algorithms suitable for biological samples should be selected and optimized for eliminating signal fluctuations caused by possible interferences and discovering more correlations among strains, thus assisting in the qualitative analysis of bacteria outside the training set. In addition to these three aspects, to achieve the widespread application of chemical nose strategies, efforts should also focus on simplifying the preprocessing procedures and constructing portable, precise, and low-noise instrumentation.
Data availability
No data, models, or code were generated or used in this review paper.Author contributions
Xin Wang: investigation, writing – original draft, review & editing. Ting Yang: writing – review & editing, funding acquisition, resources, project administration, supervision. Jian-Hua Wang: funding acquisition, resources, supervision.Conflicts of interest
The authors have no conflicts of interest to declare.Acknowledgements
Financial support from the National Natural Science Foundation of China (22174011, 22074011, 22374014, 22334003), the Fundamental Research Funds for the Central Universities (N2305018) and the Liaoning Revitalization Talents Program (XLYC2007102) is highly appreciated.References
- F. C. Cui, Y. L. Ye, J. F. Ping and X. L. Sun, Carbon dots: Current advances in pathogenic bacteria monitoring and prospect applications, Biosens. Bioelectron., 2020, 156, 112085 CrossRef CAS PubMed.
- R. Laxminarayan, A. Duse, C. Wattal, A. K. M. Zaidi, H. F. L. Wertheim, N. Sumpradit, E. Vlieghe, G. L. Hara, I. M. Gould, H. Goossens, C. Greko, A. D. So, M. Bigdeli, G. Tomson, W. Woodhouse, E. Ombaka, A. Q. Peralta, F. N. Qamar, F. Mir, S. Kariuki, Z. A. Bhutta, A. Coates, R. Bergstrom, G. D. Wright, E. D. Brown and O. Cars, Antibiotic resistance-the need for global solutions, Lancet Infect. Dis., 2013, 13, 1057–1098 CrossRef PubMed.
- S. B. Levy and B. Marshall, Antibacterial resistance worldwide: causes, challenges and responses, Nat. Med., 2004, 10, S122–S129 CrossRef CAS PubMed.
- C. J. L. Murray, K. S. Ikuta, F. Sharara, L. Swetschinski, G. R. Aguilar, A. Gray, C. Han, C. Bisignano, P. Rao, E. Wool, S. C. Johnson, A. J. Browne and M. G. Chipeta, Global burden of bacterial antimicrobial resistance in 2019: a systematic analysis, Lancet, 2022, 399, 629–655 CrossRef CAS PubMed.
- K. E. Boehle, J. Gilliand, C. R. Wheeldon, A. Holder, J. A. Adkins, B. J. Geiss, E. P. Ryan and C. S. Henry, Utilizing Paper-Based Devices for Antimicrobial-Resistant Bacteria Detection, Angew. Chem., Int. Ed., 2017, 56, 6886–6890 CrossRef CAS PubMed.
- X. Didelot, R. Bowden, D. J. Wilson, T. E. A. Peto and D. W. Crook, Transforming clinical microbiology with bacterial genome sequencing, Nat. Rev. Genet., 2012, 13, 601–612 CrossRef CAS PubMed.
- L. Váradi, J. L. Luo, D. E. Hibbs, J. D. Perry, R. J. Anderson, S. Orenga and P. W. Groundwater, Methods for the detection and identification of pathogenic bacteria: past, present, and future, Chem. Soc. Rev., 2017, 46, 4818–4832 RSC.
- M. Gwinn, D. MacCannell and G. L. Armstrong, Next-Generation Sequencing of Infectious Pathogens, JAMA, J. Am. Med. Assoc., 2019, 321, 893–894 CrossRef PubMed.
- P. Tissari, A. Zumla, E. Tarkka, S. Mero, L. Savolainen, M. Vaara, A. Aittakorpi, S. Laakso, M. Lindfors, H. Piiparinen, M. Mäki, C. Carder, J. Huggett and V. Gant, Accurate and rapid identification of bacterial species from positive blood cultures with a DNA-based microarray platform: an observational study, Lancet, 2010, 375, 224–230 CrossRef CAS PubMed.
- S. H. Park and S. C. Ricke, Development of multiplex PCR assay for simultaneous detection of Salmonella genus, Salmonella subspecies I, Salm. Enteritidis, Salm. Heidelberg and Salm. Typhimurium, J. Appl. Microbiol., 2015, 118, 152–160 CrossRef CAS PubMed.
- W. Liu, L. B. Wei, D. M. Wang, C. Y. Zhu, Y. T. Huang, Z. J. Gong, C. Y. Tang and M. K. Fan, Phenotyping Bacteria through a Black-Box Approach: Amplifying Surface-Enhanced Raman Spectroscopy Spectral Differences among Bacteria by Inputting Appropriate Environmental Stress, Anal. Chem., 2022, 94, 6791–6798 CrossRef CAS PubMed.
- N. Li, W. F. Zhang, J. Lin, G. W. Xing, H. F. Li and J. M. Lin, A Specific Mass-Tag Approach for Detection of Foodborne Pathogens Using MALDI-TOF Mass Spectrometry, Anal. Chem., 2022, 94, 3963–3969 CrossRef CAS PubMed.
- Y. C. Dai, C. Y. Li, J. Yi, Q. Qin, B. H. Liu and L. Qiao, Plasmonic Colloidosome-Coupled MALDI-TOF MS for Bacterial Heteroresistance Study at Single-Cell Level, Anal. Chem., 2020, 92, 8051–8057 CrossRef CAS PubMed.
- O. Lazcka, F. J. Del Campo and F. X. Muñoz, Pathogen detection:: A perspective of traditional methods and biosensors, Biosens. Bioelectron., 2007, 22, 1205–1217 CrossRef CAS PubMed.
- Z. Li, J. R. Askim and K. S. Suslick, The Optoelectronic Nose: Colorimetric and Fluorometric Sensor Arrays, Chem. Rev., 2019, 119, 231–292 CrossRef CAS PubMed.
- T. Yu and Y. L. Xianyu, Array-Based Biosensors for Bacteria Detection: From the Perspective of Recognition, Small, 2021, 17, 2006230 CrossRef CAS PubMed.
- T. Li, X. Y. Zhu, X. Hai, S. Bi and X. J. Zhang, Recent Progress in Sensor Arrays: From Construction Principles of Sensing Elements to Applications, ACS Sens., 2023, 8, 994–1016 CrossRef CAS PubMed.
- J. Janata, Environmental chemical sensors - New challenge and opportunity, Crit. Rev. Anal. Chem., 1998, 28, 27–34 CrossRef CAS.
- J. Y. Yang, S. S. Lu, B. Chen, F. X. Hu, C. M. Li and C. X. Guo, Machine learning-assisted optical nano-sensor arrays in microorganism analysis, TrAC, Trends Anal. Chem., 2023, 159, 116945 CrossRef CAS.
- S. Brown, J. P. S. Maria and S. Walker, Wall Teichoic Acids of Gram-Positive Bacteria, Annu. Rev. Microbiol., 2013, 67, 313–336 CrossRef CAS PubMed.
- J. E. Hudak, D. Alvarez, A. Skelly, U. H. von Andrian and D. L. Kasper, Illuminating vital surface molecules of symbionts in health and disease, Nat. Microbiol., 2017, 2, 17099 CrossRef CAS PubMed.
- S. Lebeer, J. Vanderleyden and S. C. J. De Keersmaecker, Host interactions of probiotic bacterial surface molecules: comparison with commensals and pathogens, Nat. Rev. Microbiol., 2010, 8, 171–184 CrossRef CAS PubMed.
- H. Wang, L. J. Zhou, J. J. Qin, J. H. Chen, C. Stewart, Y. M. Sun, H. Huang, L. Xu, L. X. Li, J. S. Han and F. Li, One-Component Multichannel Sensor Array for Rapid Identification of Bacteria, Anal. Chem., 2022, 94, 10291–10298 CrossRef CAS PubMed.
- T. Yu, Y. Fu, J. He, J. Zhang and Y. Xianyu, Identification of Antibiotic Resistance in ESKAPE Pathogens through Plasmonic Nanosensors and Machine Learning, ACS Nano, 2023, 17, 4551–4563 CrossRef CAS PubMed.
- M. Z. Zhang, J. Hou, J. Xia, J. Wu, L. Z. Miao and D. L. Ji, Antibiotics can alter the bacterial extracellular polymeric substances and surface properties affecting the cotransport of bacteria and antibiotics in porous media, J. Hazard. Mater., 2024, 461, 132569 CrossRef CAS PubMed.
- Y. Yuan, M. P. Hays, P. R. Hardwidge and J. Kim, Surface characteristics influencing bacterial adhesion to polymeric substrates, RSC Adv., 2017, 7, 14254–14261 RSC.
- M. Fernández-Grajera, M. A. Pacha-Olivenza, A. M. Gallardo-Moreno, M. L. González-Martín, C. Pérez-Giraldo and M. C. Fernández-Calderón, Modification of physico-chemical surface properties and growth of Staphylococcus aureus under hyperglycemia and ketoacidosis conditions, Colloids Surf., B, 2022, 209, 112137 CrossRef PubMed.
- X. X. Li, R. Wan, Y. Y. Zha, Y. G. Chen, X. Zheng and Y. L. Su, Identification of CO2 induces oxidative stress to change bacterial surface properties, Chemosphere, 2021, 277, 130336 CrossRef CAS PubMed.
- A. T. Krasley, E. G. E. Li, J. M. Galeana, C. Bulumulla, A. G. Beyene and G. S. Demirer, Carbon Nanomaterial Fluorescent Probes and Their Biological Applications, Chem. Rev., 2024, 124, 3085–3185 CrossRef CAS PubMed.
- J. Y. Yang, X. Wang, Y. Y. Sun, B. Chen, F. X. Hu, C. X. Guo and T. Yang, Recent Advances in Colorimetric Sensors Based on Gold Nanoparticles for Pathogen Detection, Biosens. J., 2023, 13, 29 CrossRef CAS PubMed.
- M. Y. Yin, C. Jing, H. J. Li, Q. L. Deng and S. Wang, Surface chemistry modified upconversion nanoparticles as fluorescent sensor array for discrimination of foodborne pathogenic bacteria, J. Nanobiotechnol., 2020, 18, 41 CrossRef CAS PubMed.
- B. Duncan, N. D. B. Le, C. Alexander, A. Gupta, G. Y. Tonga, M. Yazdani, R. F. Landis, L. S. Wang, B. Yan, S. Burmaoglu, X. N. Li and V. M. Rotello, Sensing by Smell: Nanoparticle-Enzyme Sensors for Rapid and Sensitive Detection of Bacteria with Olfactory Output, ACS Nano, 2017, 11, 5339–5343 CrossRef CAS PubMed.
- J. Y. Yang, X. D. Jia, X. Y. Wang, M. X. Liu, M. L. Chen, T. Yang and J. H. Wang, Discrimination of antibiotic-resistant Gram-negative bacteria with a novel 3D nano sensing array, Chem. Commun., 2020, 56, 1717–1720 RSC.
- T. Yu, Y. Fu, J. T. He, J. Zhang and Y. L. Xianyu, Identification of Antibiotic Resistance in ESKAPE Pathogens through Plasmonic Nanosensors and Machine Learning, ACS Nano, 2023, 17, 4551–4563 CrossRef CAS PubMed.
- J. Wang, Z. R. Jiang, Y. R. Wei, W. J. Wang, F. B. Wang, Y. B. Yang, H. Song and Q. Yuan, Multiplexed Identification of Bacterial Biofilm Infections Based on Machine-Learning-Aided Lanthanide Encoding, ACS Nano, 2022, 16, 3300–3310 CrossRef CAS PubMed.
- A. Nsubuga, K. Zarschler, M. Sgarzi, B. Graham, H. Stephan and T. Joshi, Towards Utilising Photocrosslinking of Polydiacetylenes for the Preparation of “Stealth” Upconverting Nanoparticles, Angew. Chem., Int. Ed., 2018, 57, 16036–16040 CrossRef CAS PubMed.
- W. W. Chen, Q. Z. Li, W. S. Zheng, F. Hu, G. X. Zhang, Z. Wang, D. Q. Zhang and X. Y. Jiang, Identification of Bacteria in Water by a Fluorescent Array, Angew. Chem., Int. Ed., 2014, 53, 13734–13739 CrossRef CAS PubMed.
- J. L. Shen, R. Hu, T. T. Zhou, Z. M. Wang, Y. R. Zhang, S. W. Li, C. Gui, M. J. Jiang, A. J. Qin and B. Z. Tang, Fluorescent Sensor Array for Highly Efficient Microbial Lysate Identification through Competitive Interactions, ACS Sens., 2018, 3, 2218–2222 CrossRef CAS PubMed.
- X. L. Hu, H. Q. Gan, Z. Y. Qin, Q. Liu, M. Li, D. J. Chen, J. L. Sessler, H. Tian and X. P. He, Phenotyping of Methicillin-Resistant Staphylococcus aureus Using a Ratiometric Sensor Array, J. Am. Chem. Soc., 2023, 145, 8917–8926 CrossRef CAS PubMed.
- W. Huang, L. Sun, Z. W. Zheng, J. H. Su and H. Tian, Colour-tunable fluorescence of single molecules based on the vibration induced emission of phenazine, Chem. Commun., 2015, 51, 4462–4464 RSC.
- Z. Y. Zhang, Y. S. Wu, K. C. Tang, C. L. Chen, J. W. Ho, J. H. Su, H. Tian and P. T. Chou, Excited-State Conformational/Electronic Responses of Saddle-ShapedN,N′-Disubstituted-Dihydrodibenzo[a,c]phenazines: Wide-Tuning Emission from Red to Deep Blue and White Light Combination, J. Am. Chem. Soc., 2015, 137, 8509–8520 CrossRef CAS PubMed.
- M. A. Reppy and B. A. Pindzola, Biosensing with polydiacetylene materials: structures, optical properties and applications, Chem. Commun., 2007, 4317–4338 RSC.
- H. T. Bai, H. Lu, X. C. Fu, E. D. Zhang, F. T. Lv, L. B. Liu and S. Wang, Supramolecular Strategy Based on Conjugated Polymers for Discrimination of Virus and Pathogens, Biomacromolecules, 2018, 19, 2117–2122 CrossRef CAS PubMed.
- S. B. Roy, A. Nabawy, A. N. Chattopadhyay, Y. Y. Geng, J. M. Makabenta, A. Gupta and V. M. Rotello, A Polymer-Based Multichannel Sensor for Rapid Cell-Based Screening of Antibiotic Mechanisms and Resistance Development, ACS Appl. Mater. Interfaces, 2022, 14, 27515–27522 CrossRef PubMed.
- S. Tomita, H. Kusada, N. Kojima, S. Ishihara, K. Miyazaki, H. Tamaki and R. Kurita, Polymer-based chemical-nose systems for optical-pattern recognition of gut microbiota, Chem. Sci., 2022, 13, 5830–5837 RSC.
- Y. Yu, W. Ni, Q. Hu, H. Li, Y. Zhang, X. Gao, L. Zhou, S. Zhang, S. Ma, Y. Zhang, H. Huang, F. Li and J. Han, A Dual Fluorescence Turn-On Sensor Array Formed by Poly(para-aryleneethynylene) and Aggregation-Induced Emission Fluorophores for Sensitive Multiplexed Bacterial Recognition, Angew. Chem., Int. Ed., 2024, 63, e202318483 CrossRef CAS PubMed.
- V. Lázár, A. Martins, R. Spohn, L. Daruka, G. Grézal, G. Fekete, M. Számel, P. K. Jangir, B. Kintses, B. Csörgo, A. Nyerges, A. Györkei, A. Kincses, A. Dér, F. R. Walter, M. A. Deli, E. Urbán, Z. Hegedus, G. Olajos, O. Méhi, B. Bálint, I. Nagy, T. A. Martinek, B. Papp and C. Pál, Antibiotic-resistant bacteria show widespread collateral sensitivity to antimicrobial peptides, Nat. Microbiol., 2018, 3, 718–731 CrossRef PubMed.
- J. S. Khara, S. Obuobi, Y. Wang, M. S. Hamilton, B. D. Robertson, S. M. Newton, Y. Y. Yang, P. R. Langford and P. L. R. Ee, Disruption of drug-resistant biofilms using designed short α-helical antimicrobial peptides with idealized facial amphiphilicity, Acta Biomater., 2017, 57, 103–114 CrossRef CAS PubMed.
- B. Bechinger and S. U. Gorr, Antimicrobial Peptides: Mechanisms of Action and Resistance, J. Dent. Res., 2017, 96, 254–260 CrossRef CAS PubMed.
- M. Zasloff, Antimicrobial peptides of multicellular organisms, Nature, 2002, 415, 389–395 CrossRef CAS PubMed.
- J. D. Hale and R. E. Hancock, Alternative mechanisms of action of cationic antimicrobial peptides on bacteria, Expert Rev. Anti-Infect. Ther., 2007, 5, 951–959 CrossRef CAS PubMed.
- H. X. Yuan, Z. Liu, L. B. Liu, F. T. Lv, Y. L. Wang and S. Wang, Cationic Conjugated Polymers for Discrimination of Microbial Pathogens, Adv. Mater., 2014, 26, 4333–4338 CrossRef CAS PubMed.
- J. Han, H. Cheng, B. Wang, M. S. Braun, X. Fan, M. Bender, W. Huang, C. Domhan, W. Mier, T. Lindner, K. Seehafer, M. Wink and U. H. F. Bunz, A Polymer/Peptide Complex-Based Sensor Array That Discriminates Bacteria in Urine, Angew. Chem., Int. Ed., 2017, 56, 15246–15251 CrossRef CAS PubMed.
- P. Qi, Y. Wang, D. Zhang, Y. Sun and L. Zheng, Multichannel bacterial discrimination based on recognition and disintegration disparity of short antimicrobial peptides, Anal. Biochem., 2020, 600, 113764 CrossRef CAS PubMed.
- Y. Y. Aung, A. N. Kristanti, H. V. Lee and M. Z. Fahmi, Boronic-Acid-Modified Nanomaterials for Biomedical Applications, ACS Omega, 2021, 6, 17750–17765 CrossRef CAS PubMed.
- L. Liu, X. H. Ma, Y. Chang, H. Guo and W. Q. Wang, Biosensors with Boronic Acid-Based Materials as the Recognition Elements and Signal Labels, Biosens. J., 2023, 13, 785 CrossRef CAS PubMed.
- H. Y. Li, H. He and Z. Liu, Recent progress and application of boronate affinity materials in bioanalysis, TrAC, Trends Anal. Chem., 2021, 140, 116271 CrossRef CAS.
- W. L. Zhai, X. L. Sun, T. D. James and J. S. Fossey, Boronic Acid-Based Carbohydrate Sensing, Chem. – Asian J., 2015, 10, 1836–1848 CrossRef CAS PubMed.
- J. Yan, G. Springsteen, S. Deeter and B. H. Wang, The relationship among pKa, pH, and binding constants in the interactions between boronic acids and diols—it is not as simple as it appears, Tetrahedron, 2004, 60, 11205–11209 CrossRef CAS.
- P. Yan, Z. Ding, X. Z. Li, Y. H. Dong, T. Fu and Y. Y. Wu, Colorimetric Sensor Array Based on Wulff-Type Boronate Functionalized AgNPs at Various pH for Bacteria Identification, Anal. Chem., 2019, 91, 12134–12137 CrossRef CAS PubMed.
- J.-Y. Yang, X.-D. Jia, R.-X. Gao, M.-L. Chen, T. Yang and J.-H. Wang, Discrimination of pathogenic bacteria with boronic acid modified protonated g-C3N4 nanosheets at various pHs, Sens. Actuators, B, 2021, 340, 129951 CrossRef CAS.
- Y. Wang, J. Li, H. Liu, X. Du, L. Yang and J. Zeng, Single-Probe-Based Colorimetric and Photothermal Dual-Mode Identification of Multiple Bacteria, Anal. Chem., 2023, 95, 3037–3044 CrossRef CAS PubMed.
- T. J. Foster, J. A. Geoghegan, V. K. Ganesh and M. Höök, Adhesion, invasion and evasion: the many functions of the surface proteins of, Nat. Rev. Microbiol., 2014, 12, 49–62 CrossRef CAS PubMed.
- L. H. Liu, H. Dietsch, P. Schurtenberger and M. D. Yan, Photoinitiated Coupling of Unmodified Monosaccharides to Iron Oxide Nanoparticles for Sensing Proteins and Bacteria, Bioconjugate Chem., 2009, 20, 1349–1355 CrossRef CAS PubMed.
- S. H. Cho, J. Y. Park and C. H. Kim, Systemic Lectin-Glycan Interaction of Pathogenic Enteric Bacteria in the Gastrointestinal Tract, Int. J. Mol. Sci., 2022, 23, 1451 CrossRef CAS PubMed.
- L. Zhao, Y. F. Chen, J. Yuan, M. H. Chen, H. Zhang and X. H. Li, Electrospun Fibrous Mats with Conjugated Tetraphenylethylene and Mannose for Sensitive Turn-On Fluorescent Sensing of, ACS Appl. Mater. Interfaces, 2015, 7, 5177–5186 CrossRef CAS PubMed.
- C. Xue, S. Velayudham, S. Johnson, R. Saha, A. Smith, W. Brewer, P. Murthy, S. T. Bagley and H. Liu, Highly Water-Soluble, Fluorescent, Conjugated Fluorene-Based Glycopolymers with Poly(ethylene glycol)-Tethered Spacers for Sensitive Detection of, Chem. – Eur. J., 2009, 15, 2289–2295 CrossRef CAS PubMed.
- L. Otten, E. Fullam and M. I. Gibson, Discrimination between bacterial species by ratiometric analysis of their carbohydrate binding profile, Mol. BioSyst., 2016, 12, 341–344 RSC.
- P. Qi, X. Chen, Y. Sun and D. Zhang, Multivalent glycosylated Cu:CdS quantum dots as a platform for rapid bacterial discrimination and detection, Sens. Actuators, B, 2018, 254, 431–436 CrossRef CAS.
- S. Gardete and A. Tomasz, Mechanisms of vancomycin resistance in, J. Clin. Invest., 2014, 124, 2836–2840 CrossRef PubMed.
- S. E. Pidgeon and M. M. Pires, Vancomycin-Dependent Response in Live Drug-Resistant Bacteria by Metabolic Labeling, Angew. Chem., Int. Ed., 2017, 56, 8839–8843 CrossRef CAS PubMed.
- B. W. Simpson and M. S. Trent, Pushing the envelope: LPS modifications and their consequences, Nat. Rev. Microbiol., 2019, 17, 403–416 CrossRef CAS PubMed.
- M. Wu, G. Qi, X. Liu, Y. Duan, J. Liu and B. Liu, Bio-Orthogonal AIEgen for Specific Discrimination and Elimination of Bacterial Pathogens via Metabolic Engineering, Chem. Mater., 2019, 32, 858–865 CrossRef.
- E. Kuru, H. V. Hughes, P. J. Brown, E. Hall, S. Tekkam, F. Cava, M. A. de Pedro, Y. V. Brun and M. S. VanNieuwenhze, In Situ Probing of Newly Synthesized Peptidoglycan in Live Bacteria with Fluorescent D-Amino Acids, Angew. Chem., Int. Ed., 2012, 51, 12519–12523 CrossRef CAS PubMed.
- L. B. Zheng, P. Qi and D. Zhang, Identification of bacteria by a fluorescence sensor array based on three kinds of receptors functionalized carbon dots, Sens. Actuators, B, 2019, 286, 206–213 CrossRef CAS.
- J. Y. Yang, G. Li, S. H. Chen, X. Z. Su, D. Xu, Y. M. Zhai, Y. H. Liu, G. X. Hu, C. X. Guo, H. B. Yang, L. G. Occhipinti and F. X. Hu, Machine Learning-Assistant Colorimetric Sensor Arrays for Intelligent and Rapid Diagnosis of Urinary Tract Infection, ACS Sens., 2024, 1945–1956 CrossRef CAS PubMed.
- J. A. Geoghegan and T. J. Foster, Cell Wall-Anchored Surface Proteins of Staphylococcus aureus: Many Proteins, Multiple Functions, Curr. Top. Microbiol., 2017, 409, 95–120 CAS.
- H. W. Ji, L. Wu, F. Pu, J. S. Ren and X. G. Qu, Point-of-Care Identification of Bacteria Using Protein-Encapsulated Gold Nanoclusters, Adv. Healthcare Mater., 2018, 7, 1701370 CrossRef PubMed.
- Y. Y. Wu, B. Wang, K. Wang and P. Yan, Identification of proteins and bacteria based on a metal ion-gold nanocluster sensor array, Anal. Methods, 2018, 10, 3939–3944 RSC.
- A. J. Beddek and A. B. Schryvers, The lactoferrin receptor complex in gram negative bacteria, BioMetals, 2010, 23, 377–386 CrossRef CAS PubMed.
- S. D. GrayOwen and A. B. Schryvers, Bacterial transferrin and lactoferrin receptors, Trends Microbiol., 1996, 4, 185–191 CrossRef CAS PubMed.
- T. T. Wu, Q. Q. Jiang, D. Wu, Y. Q. Hu, S. G. Chen, T. Ding, X. Q. Ye, D. H. Liu and J. C. Chen, What is new in lysozyme research and its application in food industry? A review, Food Chem., 2019, 274, 698–709 CrossRef CAS PubMed.
- Z. Liu, X. Zhu, Q. Lu, M. Liu, H. Li, Y. Zhang, Y. Liu and S. Yao, Recognition Engineering-Mediated Multichannel Sensor Array for Gut Microbiota Sensing, Anal. Chem., 2023, 95, 5911–5919 CrossRef CAS PubMed.
- J. M. Rand, T. Pisithkul, R. L. Clark, J. M. Thiede, C. R. Mehrer, D. E. Agnew, C. E. Campbell, A. L. Markley, M. N. Price, J. Ray, K. M. Wetmore, Y. Suh, A. P. Arkin, A. M. Deutschbauer, D. Amador-Noguez and B. F. Pfleger, A metabolic pathway for catabolizing levulinic acid in bacteria, Nat. Microbiol., 2017, 2, 1624–1634 CrossRef CAS PubMed.
- Y. Ardö, Flavour formation by amino acid catabolism, Biotechnol. Adv., 2006, 24, 238–242 CrossRef PubMed.
- A. R. J. Curson, J. D. Todd, M. J. Sullivan and A. W. B. Johnston, Catabolism of dimethylsulphoniopropionate: microorganisms, enzymes and genes, Nat. Rev. Microbiol., 2011, 9, 849–859 CrossRef CAS PubMed.
- M. Gralka, S. Pollak and O. X. Cordero, Genome content predicts the carbon catabolic preferences of heterotrophic bacteria, Nat. Microbiol., 2023, 8, 1799–1808 CrossRef CAS PubMed.
- M. H. Saier, S. Chauvaux, J. Deutscher, J. Reizer and J. J. Ye, Protein-Phosphorylation and Regulation of Carbon Metabolism in Gram-Negative Versus Gram-Positive Bacteria, Trends Biochem. Sci., 1995, 20, 267–271 CrossRef CAS PubMed.
- X. Zhou, H. T. Wu, X. Y. Chen, W. R. Li, J. J. Zhang, M. Q. Wang, J. Zhang, S. Wang and Y. Q. Liu, Glucose-metabolism-triggered colorimetric sensor array for point-of-care differentiation and antibiotic susceptibility testing of bacteria, Food Chem., 2024, 438, 137983 CrossRef CAS PubMed.
- Y. Zhang, G. X. Qi, Y. L. Yu, M. X. Liu and S. Chen, A machine learning-based colorimetric sensor array for high-precision pathogen identification in household refrigerators, Chem. Commun., 2023, 59, 7603–7606 RSC.
- H. D. Bean, J. M. D. Dimandja and J. E. Hill, Bacterial volatile discovery using solid phase microextraction and comprehensive two-dimensional gas chromatography-time-of-flight mass spectrometry, J. Chromatogr., B, 2012, 901, 41–46 CrossRef CAS PubMed.
- T. Dan, D. Wang, S. M. Wu, R. L. Jin, W. Y. Ren and T. S. Sun, Profiles of Volatile Flavor Compounds in Milk Fermented with Different Proportional Combinations of Lactobacillus delbrueckii subsp. bulgaricus and Streptococcus thermophilus, Molecules, 2017, 22, 1633 CrossRef PubMed.
- B. Audrain, M. A. Farag, C. M. Ryu and J. M. Ghigo, Role of bacterial volatile compounds in bacterial biology, FEMS Microbiol. Rev., 2015, 39, 222–233 CrossRef CAS PubMed.
- F. Monedeiro, M. Milanowski, I. A. Ratiu, H. Zmyslowski, T. Ligor and B. Buszewski, VOC Profiles of Saliva in Assessment of Halitosis and Submandibular Abscesses Using HS-SPME-GC/MS Technique, Molecules, 2019, 24, 2977 CrossRef CAS PubMed.
- C. K. Koo, S. L. Wang, R. L. Gaur, F. Samain, N. Banaei and E. T. Kool, Fluorescent DNA chemosensors: identification of bacterial species by their volatile metabolites, Chem. Commun., 2011, 47, 11435–11437 RSC.
- J. Chen, J. N. Tang, H. Shi, C. Tang and R. Zhang, Characteristics of volatile organic compounds produced from five pathogenic bacteria by headspace-solid phase micro-extraction/gas chromatography-mass spectrometry, J. Basic Microbiol., 2017, 57, 228–237 CrossRef CAS PubMed.
- W. Xu, X. Zou, Y. T. Ding, Q. Zhang, Y. L. Song, J. Zhang, M. Yang, Z. Liu, Q. Zhou, D. L. Ge, Q. L. Zhang, W. C. Song, C. Q. Huang, C. Y. Shen and Y. N. Chu, Qualitative and quantitative rapid detection of VOCs differentially released by VAP-associated bacteria using PTR-MS and FGC-PTR-MS, Analyst, 2024, 149, 1447–1454 RSC.
- N. Shauloff, A. Morag, K. Yaniv, S. Singh, R. Malishev, O. Paz-Tal, L. Rokach and R. Jelinek, Sniffing Bacteria with a Carbon-Dot Artificial Nose, Nano-Micro Lett., 2021, 13, 112 CrossRef CAS PubMed.
- A. W. Boots, J. J. B. N. van Berkel, J. W. Dallinga, A. Smolinska, E. F. Wouters and F. J. van Schooten, The versatile use of exhaled volatile organic compounds in human health and disease, J. Breath Res., 2012, 6, 027108 CrossRef PubMed.
- M. S. Webster, J. S. Cooper, E. Chow, L. J. Hubble, A. Sosa-Pintos, L. Wieczorek and B. Raguse, Detection of bacterial metabolites for the discrimination of bacteria utilizing gold nanoparticle chemiresistor sensors, Sens. Actuators, B, 2015, 220, 895–902 CrossRef CAS.
- R. Nißler, O. Bader, M. Dohmen, S. G. Walter, C. Noll, G. Selvaggio, U. Gross and S. Kruss, Remote near infrared identification of pathogens with multiplexed nanosensors, Nat. Commun., 2020, 11, 5995 CrossRef PubMed.
- J. Rong, J. Han, L. Dong, Y. H. Tan, H. Q. Yang, L. S. Feng, Q. W. Wang, R. Meng, J. Zhao, S. Q. Wang and X. Chen, Glycan Imaging in Intact Rat Hearts and Glycoproteomic Analysis Reveal the Upregulation of Sialylation during Cardiac Hypertrophy, J. Am. Chem. Soc., 2014, 136, 17468–17476 CrossRef CAS PubMed.
- M. Kufleitner, L. M. Haiber and V. Wittmann, Metabolic glycoengineering - exploring glycosylation with bioorthogonal chemistry, Chem. Soc. Rev., 2023, 52, 510–535 RSC.
- A. J. F. Egan, J. Errington and W. Vollmer, Regulation of peptidoglycan synthesis and remodelling, Nat. Rev. Microbiol., 2020, 18, 446–460 CrossRef CAS PubMed.
- Y. P. Hsu, E. Hall, G. Booher, B. Murphy, A. D. Radkov, J. Yablonowski, C. Mulcahey, L. Alvarez, F. Cava, Y. V. Brun, E. Kuru and M. S. VanNieuwenhze, Fluorogenic D-amino acids enable real-time monitoring of peptidoglycan biosynthesis and high-throughput transpeptidation assays, Nat. Chem., 2019, 11, 335–341 CrossRef CAS PubMed.
- S. Hong, D. W. Zheng, Q. L. Zhang, W. W. Deng, W. F. Song, S. X. Cheng, Z. S. Zhi and X. Z. Zhang, An RGB-emitting molecular cocktail for the detection of bacterial fingerprints, Chem. Sci., 2020, 11, 4403–4409 RSC.
- X. Wang, H. Li, C. Wu, J. Yang, J. Wang and T. Yang, Metabolism-triggered sensor array aided by machine learning for rapid identification of pathogens, Biosens. Bioelectron., 2024, 255, 116264 CrossRef CAS PubMed.
- X. Gao, M. M. Li, M. Y. Zhao, X. K. Wang, S. Wang and Y. Q. Liu, Metabolism-Triggered Colorimetric Sensor Array for Fingerprinting and Antibiotic Susceptibility Testing of Bacteria, Anal. Chem., 2022, 94, 6957–6966 CrossRef CAS PubMed.
- X. Wang, H. Li, J. Yang, C. Wu, M. Chen, J. Wang and T. Yang, Chemical Nose Strategy with Metabolic Labeling and “Antibiotic-Responsive Spectrum” Enables Accurate and Rapid Pathogen Identification, Anal. Chem., 2023, 427–436 Search PubMed.
- Y. Choi and S. Y. Lee, Biosynthesis of inorganic nanomaterials using microbial cells and bacteriophages, Nat. Rev. Chem., 2020, 4, 638–656 CrossRef CAS PubMed.
- S. Iravani and R. S. Varma, Bacteria in Heavy Metal Remediation and Nanoparticle Biosynthesis, ACS Sustainable Chem. Eng., 2020, 8, 5395–5409 CrossRef CAS.
- S. S. Yao, B. A. Jin, Z. M. Liu, C. Y. Shao, R. B. Zhao, X. Y. Wang and R. K. Tang, Biomineralization: From Material Tactics to Biological Strategy, Adv. Mater., 2017, 29, 1605903 CrossRef PubMed.
- V. Bansal, D. Rautaray, A. Ahmad and M. Sastry, Biosynthesis of zirconia nanoparticles using the fungus, J. Mater. Chem., 2004, 14, 3303–3305 RSC.
- D. Rautaray, A. Sanyal, S. D. Adyanthaya, A. Ahmad and M. Sastry, Biological synthesis of strontium carbonate crystals using the fungus, Langmuir, 2004, 20, 6827–6833 CrossRef CAS PubMed.
- S. Mirzadeh, E. Darezereshki, F. Bakhtiari, M. H. Fazaelipoor and M. R. Hosseini, Characterization of zinc sulfide (ZnS) nanoparticles Biosynthesized by, Mater. Sci. Semicond. Process., 2013, 16, 374–378 CrossRef CAS.
- T. Yu, S. X. Su, J. Hu, J. Zhang and Y. L. Xianyu, A New Strategy for Microbial Taxonomic Identification through Micro-Biosynthetic Gold Nanoparticles and Machine Learning, Adv. Mater., 2022, 34, 2109365 CrossRef CAS PubMed.
- P. M. M. Zhang and C. Y. Tan, Cross-Reactive Fluorescent Sensor Array for Discrimination of Amyloid Beta Aggregates, Anal. Chem., 2022, 94, 5469–5473 CrossRef CAS PubMed.
- J. R. Askim, M. Mahmoudi and K. S. Suslick, Optical sensor arrays for chemical sensing: the optoelectronic nose, Chem. Soc. Rev., 2013, 42, 8649–8682 RSC.
- Z. Li and K. S. Suslick, The Optoelectronic Nose, Acc. Chem. Res., 2021, 54, 950–960 CrossRef CAS PubMed.
- J. F. Hair, B. Black, B. Babin, R. E. Anderson and R. L. TathamMultivariate data analysis, Pearson Prentice Hall, Upper Saddle River, N.J., 6th edn, 2006 Search PubMed.
- D. L. Massart and L. Kaufman, The interpretation of analytical chemical data by the use of cluster analysis, Wiley, New York, 1983 Search PubMed.
- M. Ringnér, What is principal component analysis?, Nat. Biotechnol., 2008, 26, 303–304 CrossRef PubMed.
- D. Kobak and P. Berens, The art of using t-SNE for single-cell transcriptomics, Nat. Commun., 2019, 10, 5416 CrossRef PubMed.
- S. Pandit, T. Banerjee, I. Srivastava, S. M. Nie and D. P. J. Pan, Machine Learning-Assisted Array-Based Biomolecular Sensing Using Surface-Functionalized Carbon Dots, ACS Sens., 2019, 4, 2730–2737 CrossRef CAS PubMed.
- J. R. Askim, Z. Li, M. K. LaGasse, J. M. Rankin and K. S. Suslick, An optoelectronic nose for identification of explosives, Chem. Sci., 2016, 7, 199–206 RSC.
- C. C. Chang and C. J. Lin, LIBSVM: A Library for Support Vector Machines, ACM T. Intel. Syst. Tec., 2011, 2, 1961189–1961199 Search PubMed.
- S. Min, B. Lee and S. Yoon, Deep learning in bioinformatics, Briefings Bioinf., 2017, 18, 851–869 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |