Open Access Article
Open Access ArticleAptamer-functionalized nanopipettes: a promising approach for viral fragment detection via ion current rectification†
Shekemi
Denuga
 a,
Dominik
Duleba
a,
Dominik
Duleba
 a,
Pallavi
Dutta
a,
Guerrino
Macori
a,
Pallavi
Dutta
a,
Guerrino
Macori
 cd,
Damion K.
Corrigan
cd,
Damion K.
Corrigan
 b,
Séamus
Fanning
b,
Séamus
Fanning
 ce and
Robert P.
Johnson
ce and
Robert P.
Johnson
 *ac
*ac
aSchool of Chemistry, University College Dublin, Belfield, Dublin 4, Ireland. E-mail: robert.johnson@ucd.ie
bCentre for Advanced Measurement Science and Health Translation, Department of Pure and Applied Chemistry, University of Strathclyde, G1 1XL, UK
cUCD-Centre for Food Safety, University College Dublin, Belfield, Dublin 4, Ireland
dSchool of Biology and Environmental Science, University College Dublin, Belfield, Dublin 4, Ireland
eSchool of Public Health, Physiotherapy & Sports Science, University College Dublin, Belfield, Dublin 4, Ireland
First published on 10th May 2024
Abstract
In this report, ion current rectification, an electrochemical phenomenon observed in asymmetric nanopipettes, is used for the label-free detection of SARS-CoV-2 viral fragments in nasopharyngeal samples. Quartz nanopipettes are functionalized with aptamers targeting the spike protein S1 domain, wherein changes to the surface charge magnitude, distribution, and ion transport behavior modulate the current–voltage response upon binding. The aptamer-modified nanopipette provides a selective and sensitive method for detecting SARS-CoV-2, with a limit of detection in the laboratory of 0.05 pg mL−1. The effectiveness of this low-cost platform was demonstrated by sensing SARS-CoV-2 in nasopharyngeal samples, successfully discriminating between positive and negative cases with minimal template preparation, highlighting the platform's potential as a versatile sensing strategy for infectious disease detection in clinical diagnosis.
Introduction
Nanopore-based electrochemical sensing platforms have emerged as promising technologies for detecting various analytes due to their simplicity, versatility, and potential for integration as low-cost, label-free point-of-care devices.1–3 The vast majority of these nanopore systems utilize resistive pulse sensing, a technique involving the detection of molecules translocating across a pore via a momentary drop in current relative to that of the open, unblocked pore.4 Since its discovery in 1996, this methodology has been extensively utilized for the characterization and detection of single molecules and DNA, as well as for protein sequencing applications.5–9 An alternative paradigm involves using current–voltage (I–V) measurements in nanopores that display ion current rectification (ICR).10,11 ICR refers to the non-ohmic I–V response exhibited by asymmetric nanopores, where the current magnitudes measured at equal but opposite potentials differ.12,13 This non-linearity arises in pores with a charged surface primarily due to the interactions between the charged interface and the electrolyte species, resulting in a diffuse layer of counter ions along the nanopore wall, thus forming an electric double layer (EDL).14 For geometrically asymmetric pores such as conical-shaped glass nanopipettes, the EDL overlaps at the pore mouth, giving rise to ion flux perm-selectivity. Consequently, depending on the applied potential, there is a local accumulation and depletion of ions, leading to a high and low conductivity state within the nanopore, resulting in a rectified I–V response.12,15–17 Given that the EDL arises to compensate for the interfacial charge at the nanopore surface, nanopores exhibiting ICR can, therefore, be used for sensing by leveraging this surface charge dependence. When the immobilization of an analyte onto the nanopore walls via surface-bound biorecognition elements induces changes in the surface charge, analyte binding is then transduced by the modulation in the magnitude of the rectification observed.18,19 Ion current-rectifying nanopores have been widely reported for detecting metal ions,20 small organic molecules,21 proteins,22 and nucleic acids18,23 utilizing the inherent sensitivity and selectivity between biorecognition elements and their target analyte. Thus, to achieve ICR-based sensing, the internal surface of the nanopore must first be functionalized with biorecognition elements that are highly selective towards the analyte of interest.24,25Aptamers are synthetic, single-stranded nucleic acid biorecognition elements designed to bind with analytes of interest with high affinity and selectivity.26 They are widely used in biosensor development due to their exceptional affinity and specificity to target molecules, increased stability, reproducibility, and low production costs.27,28 Aptamers are generally designed and selected to bind to the analyte of interest through a series of in vitro screening iterations using the Systematic Evolution of Ligands by Exponential Enrichment (SELEX) technique, mimicking natural selection.29,30 In recent years, researchers have integrated aptamers as biorecognition elements within nanopipettes to enable ultra-sensitive and label-free detection of various analytes of interest.31–33 Notably, Actis et al. developed an aptamer-functionalized nanopore sensor capable of detecting thrombin spiked in human serum in phosphate buffer silane (PBS) (up to 20%), demonstrating that nanopore sensors can be utilized for sensing in complex media.34 Similarly, Nakatsuka et al. also utilized aptamer-modified nanopipettes to detect neurotransmitters such as dopamine and serotonin in human serum based on changes in ICR.25,35 More recently, Ma et al. reported the detection of the nucleocapsid-protein of the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) using an aptamer-functionalized nanopipette, achieving a detection limit of 73 pg mL−1 in spiked serum samples.36 Despite these advancements, to the best of our knowledge, aptamer-functionalized nanopipettes are yet to be utilized to detect biomarkers such as viral fragments in real clinical samples.
Extensive research has been carried out to develop rapid, sensitive, and selective sensors aimed at detecting and limiting the dissemination of SARS-CoV-2, the causative agent of the COVID-19 pandemic.37–41 Given the global impact and depth of knowledge on SARS-CoV-2, as well as the wide availability of clinical samples, it serves as an attractive model system for ICR-based sensing. We selected the spike protein (S), S1 subunit (Fig. S1†), as our target of interest due to its surface localization and involvement in host-cell receptor recognition.42 Further, an aptamer with a high affinity towards the S1 domain has been well-studied by previous publications.43 In this report, the internal surface of a quartz nanopipette was functionalized with this S1 selective aptamer. Subsequently, these aptamer-functionalized nanopipettes were employed for COVID-19 detection and quantification with synthetic spike protein and patient nasal swab samples. Our results are complemented with finite element simulations using COMSOL®, which support our supposition that the observed current–voltage response modulation upon target binding is driven by changes in the local surface charge when a fraction of the aptamer sites capture the spike-protein or spike-protein-containing virus. Our results show that the nanopipette sensing platform for SARS-CoV-2 is highly sensitive and selective, demonstrating the potential of aptamer-modified nanopipettes as a universal sensing strategy for detecting infectious diseases in clinically relevant samples.
Experimental methods
Materials and reagents
All reagents were purchased from Merck Chemicals and used without further purification unless otherwise stated. Ag/AgCl electrodes were prepared by dipping 0.25 mm diameter silver wire into a potassium hypochlorite solution overnight before thoroughly rinsing. All solutions were prepared using Milli-Q-water from an Elga Purelab DV 35 water purification system. A 33-mer S1-binding aptamer (modified with a thiol group at the 5′ terminus) and buffer system were purchased from Aptamer Group (York, UK). Further details on the aptamer system are provided in a previous publication and the ESI.†43 The S1 glycoprotein and IL-6 protein were obtained from Abcam.Nanopipette fabrication
Nanopipettes with a mean pore radius of 109 ± 20 nm were fabricated from filamented quartz capillaries (#Q100-70-7.5, outer diameter: 1.0 mm, inner diameter: 0.7 mm) using a laser-based pipette puller (P-2000, Sutter Instruments) with the following instrument-specific parameters: heat: 580, filament: 3, velocity: 55, delay: 128, and pull: 110.44 The size of the nanopipettes was characterized using scanning electron microscopy (SEM), and conductivity measurements, as described in the ESI.†Functionalization procedure
A three-step procedure was used for functionalizing the internal walls of quartz nanopipettes with an S1 subunit selective aptamer. A 5% (v/v) solution of (3-aminopropyl)triethoxysilane (APTES) in ethanol was backfilled into the quartz pipette using a microfil syringe, and the nanopipettes were left to incubate at room temperature for 1 h. The nanopipettes were then thoroughly rinsed with ethanol to remove unreacted APTES and baked at 70 °C for 1 h. Next, the APTES-functionalized nanopipettes were filled with a solution of 1 mM 3-(maleimido)propionic acid N-hydroxysuccinimide ester dissolved in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 9 (v/v) DMSO
9 (v/v) DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1× PBS overnight at room temperature, and then rinsed with 1× PBS to remove any unreacted MPS present in the nanopipette. The aptamer solution was then diluted to 1 μM in the Aptamer Group buffer system. To prepare the aptamer for functionalization, it was first denatured at 95 °C for 5 minutes and cooled to room temperature. Then, the nanopipettes were backfilled and incubated in the aptamer solution for 4 h at room temperature. The nanopipettes were rinsed thoroughly with deionized water. Cyclic voltammetry was used to confirm successful modification, as described in the results section.
1× PBS overnight at room temperature, and then rinsed with 1× PBS to remove any unreacted MPS present in the nanopipette. The aptamer solution was then diluted to 1 μM in the Aptamer Group buffer system. To prepare the aptamer for functionalization, it was first denatured at 95 °C for 5 minutes and cooled to room temperature. Then, the nanopipettes were backfilled and incubated in the aptamer solution for 4 h at room temperature. The nanopipettes were rinsed thoroughly with deionized water. Cyclic voltammetry was used to confirm successful modification, as described in the results section.
Sample testing
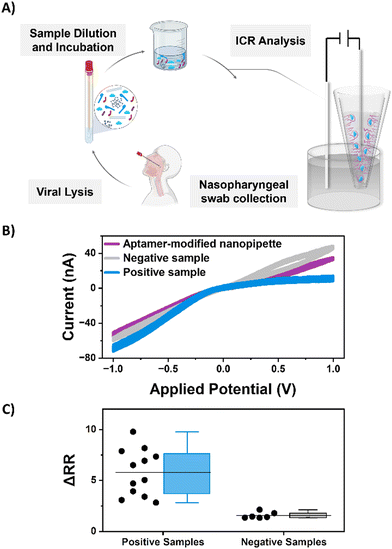
For testing of the S1 subunit, the spike protein was diluted in 1× PBS to prepare the desired target concentration for detection. The aptamer-functionalized nanopipettes were incubated in the target solution overnight at room temperature. For real sample testing, SARS-CoV-2 specimens were collected from positive nasal rapid antigen detection tests (RADTs) from voluntary participants working in meat processing plants in Ireland. For this study, sample extraction was performed in containment biosafety laboratory category 2 (BSL-2) facilities using standard BSL-2 work practices following a procedure described by Macori et al.45 The SARS-CoV-2 positive RADTs were inoculated on-site and were provided in 1.5 mL tubes. A volume of 700 μL of viral lysis buffer (AVL) was added to each sample. Following an incubation of 10 min, the RADT was transferred into a sterile 50 mL sample tube, sealed, and maintained at 4 °C before being delivered to the laboratory. Subsequently, the lysed suspension was diluted in 1× PBS (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) for ICR analysis. Twelve samples were randomly selected from a larger dataset of positive RADTs, including Abbott Panbio™ (Abbott Laboratories Ltd., USA) and Clinitest® Rapid COVID-19 Antigen Test (Siemens Healthineers, Germany). The aptamer-functionalized nanopipettes were then dipped in this solution overnight. All experiments were performed in compliance with the Guidelines for Ethical Conduct in Human Research. The team received ethical approval from the UCD Human Research Ethics Committee (no.: LS-E-20-196-Mulcahy), including informed consents from human participants of this study.
1) for ICR analysis. Twelve samples were randomly selected from a larger dataset of positive RADTs, including Abbott Panbio™ (Abbott Laboratories Ltd., USA) and Clinitest® Rapid COVID-19 Antigen Test (Siemens Healthineers, Germany). The aptamer-functionalized nanopipettes were then dipped in this solution overnight. All experiments were performed in compliance with the Guidelines for Ethical Conduct in Human Research. The team received ethical approval from the UCD Human Research Ethics Committee (no.: LS-E-20-196-Mulcahy), including informed consents from human participants of this study.
Electrochemical measurements and analysis
All I–V measurements were performed using a 0.01 M KCl electrolyte. Nanopipettes were backfilled with the electrolyte, and an Ag/AgCl wire working electrode was placed inside the pipette. The nanopipette was placed in a bulk electrolyte solution containing an Ag/AgCl wire reference electrode. The current was measured using a Biologic SP-200 potentiostat, with the applied voltage swept from −1 V to +1 V relative to the reference electrode at a scan rate of 0.1 V s−1. Measurements were performed with a filter bandwidth of 50 kHz. Noise was further filtered numerically using Biologic's EC-Lab software by applying a moving average filter with a window size of 11 points. Data are reported as mean ± standard error, calculated from a minimum of five data points.Results and discussion
Nanopipette functionalization with an aptamer specific for the SARS-CoV-2 S1 subunit
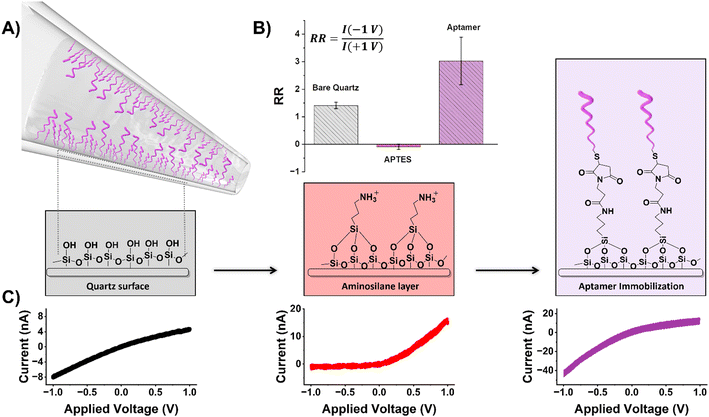
A thiolated aptamer with a high binding affinity (KD = 10.7 ± 0.07 nM) (Fig. S4†) for the spike protein S1 domain of SARS-COV2 was covalently immobilized onto the inner surface of a glass nanopipette (Fig. 1) following a three-step process. As the response of the ion-current rectifying nanopore sensor is strongly dependent on the surface charge, the success of each modification step was monitored via the extent of rectification, quantified by the rectification ratio (eqn (1)).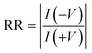 | (1) |
| SiOH ↔ SiO− + H+ | (2) |
Aptamer-functionalized nanopipettes exhibit high sensitivity
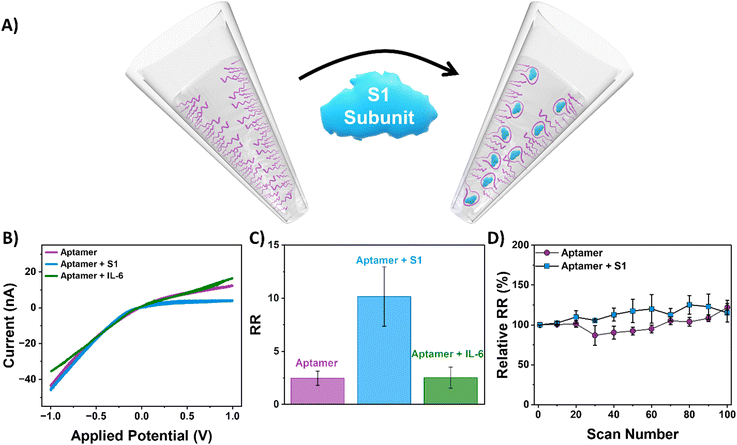
The 33-mer S1 binding aptamer immobilized to the surface of the nanopipette was used to target the S protein of SARS-CoV-2 by incubating it in an aqueous solution containing 50 ng mL−1 S1 subunit of the spike protein overnight. Following exposure of the aptamer-functionalized nanopipettes to the S1 protein, a pronounced increase in rectification from 2.4 ± 0.7 to 10.1 ± 2.8 was observed (Fig. 2). We propose the mechanism of sensing in our device is therefore as follows. When an aptamer probe is present on the internal walls of the conical nanopore, the electrical double-layer region, which encompasses all of the pore orifice, selectively excludes the transport of anions through the pore. Anion exclusion results in the depletion and accumulation of ions at positive and negative voltages, respectively, which, in turn, manifests as low and high current states at equal but opposite voltages (as measured by the RR). The S1 subunit, with a pI between 5.2 and 6.2, is negatively charged at physiological pH.50,51 The capture of these proteins at the internal nanopipette walls, therefore, increases the rectification ratio through an increase in local negative charge density at the pore walls. Thus, upon binding of the spike protein, the perm-selectivity at the pore orifice increases, the accumulation and depletion states become more pronounced, and the RR increases. Our observations are supported by a finite element simulation model constructed with COMSOL®, which predicts the same change in the direction of the rectification ratio when the charge at the pipette walls becomes increasingly negative, with the magnitude of the change dependent on the number of proteins units captured (Fig. S7†). To validate the specificity of the ICR response observed, the aptamer functionalized nanopipettes were also immersed in a solution of interleukin 6 (IL-6), and the current–voltage response was recorded. IL-6 was chosen because SARS-CoV-2 is associated with elevated levels of pro-inflammatory cytokine production, with IL-6 consequently a likely interferent in patients' samples. When exposed to IL-6, the RR remained at 2.5 ± 0.9 (Fig. 2C). The sensor response was also found to be stable with continuous interrogation (Fig. 2D and S8†).To determine the analytical range of the nanopipette sensing platform and noting the significant reported variability in viral load of SARS-COV-2 infected patients,52 we investigated the platform response to different concentrations of S1 subunit across a broad range, 100 ng mL−1 to 5 × 10−9 ng mL−1. When the concentration of spike protein is plotted as a function of the change in rectification ratio (ΔRR, i.e., the difference between the rectification ratio of the target and the aptamer), we observed that higher concentrations of the spike protein gave rise to higher changes in rectification (Fig. 3), with spike protein concentrations as low as 5 × 10−9 ng mL−1 detected. The developed sensor exhibits responsiveness across a wide range of spike protein concentrations, with a limit of detection (LOD) of 0.05 pg mL−1.53
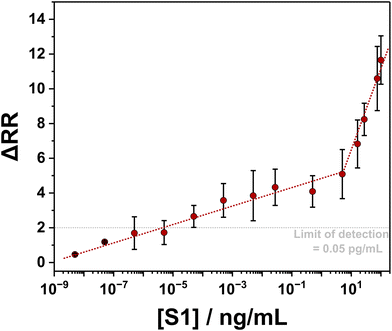 | ||
| Fig. 3 Quantification of S1 concentration using the aptamer-functionalized nanopipette. Two lines of best fit are drawn (red dotted lines) to highlight the relationship between the change in RR and S1 concentration. The limit of detection (indicated by the grey dotted line) is calculated as the signal at zero analyte concentration with 1.645 times its standard deviation plus 1.645 times the standard deviation of a low-concentration sample.53 Error bars represent the standard error from a minimum of six measurements, each with a different functionalized nanopipette. | ||
Aptamer functionalized nanopipettes can be used directly with patient nasal samples
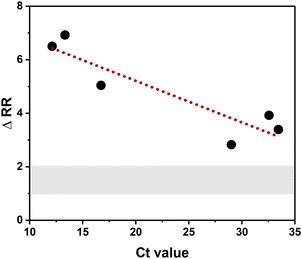
To evaluate the performance of the aptamer-functionalized nanopipettes for real-sample sensing, twelve SARS-CoV-2 samples extracted from positive nasal RADTs were analyzed by ICR, as shown in Fig. 4. Nanopipettes exposed to a positive SARS-CoV-2 sample exhibited increased rectification, with a mean change of 5.8 ± 0.6. As discussed earlier, this increase in RR arises due to the heightened anionic charge density at the nanopore surface upon aptamer-S1 binding. For comparison, nanopipettes incubated in a negative COVID-19 sample showed minimal change in rectification, with a mean change in rectification ratio of 1.6 ± 0.2. This small change in RR is likely attributed to the non-specific interactions between the negative charge of the aptamer and other interferents in biological matrixes. Interestingly, sensors exposed to different positive clinical samples were prone to considerable variation in RR response. We speculated that this variation was due to the differing viral loads of individual samples, with a concentration-dependent relationship between RR and analyte concentration. In comparison, a tighter distribution was observed for pipettes exposed to the same negative SARS-CoV-2 sample. Despite the variability in RR observed for the SARS-CoV-2 positive samples, the developed nanopipette sensor could clearly detect the S1 subunit in all twelve of the clinical nasal samples tested and distinguish them from a negative SARS-CoV-2 patient sample baseline.To further evaluate the diagnostic potential of our nanopore sensor on clinical samples, a sub-set of six of the positive samples was selected for qualitative reverse transcription-based polymerase chain reaction (RT-qPCR) analysis, and the cycle threshold (Ct) value, which is indicative of the minimum number of amplification cycles required for a positive PCR result, recorded. Higher viral loading necessitates a lower number of PCR cycles, and thus, RT-qPCR on such samples exhibit lower Ct values, while lower viral loadings require a high number of cycles and exhibit higher Ct values. Fig. 5 compares the results obtained by the RT-qPCR to the change in rectification response from the aptamer-functionalized nanopipette, where a clear correlation between viral load (measured from the Ct value) and sensor response (measured from the ΔRR) is observed. The rectification response increases with decreasing Ct values, demonstrating that the variation in ΔRR corresponds to the variation in viral loading in samples. The LOD obtained with the nanopore response corresponds to a Ct value of 33.45, which meets the POC test requirements set by the WHO.54 These results show that the aptamer-functionalized nanopipette presents a promising detection platform for biomarkers indicative of infectious disease.
Conclusion
Herein, we have developed an ion-current rectifying nanopore sensor for detecting COVID-19 using an aptamer-functionalized nanopipette with high affinity for the S1 domain of SARS-CoV-2. Quantitative determination of the S1 protein analyte is achieved through a mechanism in which binding of the analyte to the selective aptamer immobilized on the nanopore internal walls increases the anionic surface charge, enhancing the observed ion-current rectification of the nanopore. These findings are supported by the Finite Element simulation performed using COMSOL® Multiphysics. The biosensor exhibited a large dynamic range from 100 ng mL−1 to 5 × 10−6 pg mL−1 with a detection limit of 0.05 pg mL−1. Further, the sensor successfully discriminates between positive and negative patient samples in nasal swabs with minimal sample preparation. Our study showcases the viability of ion-rectifying nanopipettes for clinical applications, where the ability to work with minimal sample preparation, high sensitivity and selectivity, low fabrication costs, and large dynamic range are all significant advantages of the developed methodology. Notwithstanding these advantages, it should be noted that the sensor's sensitivity and selectivity hinge on the binding affinity between the bioreceptor immobilized on the nanopore surface and the analyte of interest. Furthermore, challenges related to the nanopipette's reusability remain to be addressed. ICR-based sensing presents a relatively new and promising sensing platform with ongoing technical challenges, such as improving the robustness of the nanopipette, as well as conducting a more rigorous study of the limit of detection and limit of quantification for this technology, being imperative to maximizing its potential for effective detection of infectious diseases, particularly in a clinical setting. Nevertheless, aptamer-functionalized nanopipettes are an attractive electrochemical sensing platform with great potential for sensing in clinically relevant samples.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge funding from Science Foundation Ireland under the Frontiers for the Future Programme (Project No. 20/FFP-P/8728). SD acknowledges funding from the National University of Ireland for a Travelling Doctoral Scholarship. DD acknowledges Postgraduate scholarship funding from the Irish Research Council (Project No. GOIPG/2022/1648). We gratefully acknowledge Emer Farrell for her support with illustrations in this manuscript.References
- J. Stanley and N. Pourmand, Nanopipettes—The past and the present, APL Mater., 2020, 8(10), 100902, DOI:10.1063/5.0020011
.
- R. E. Özel, S. Kahnemouyi, H. Fan, W. H. Mak, A. Lohith, A. Seger, M. Teodorescu and N. Pourmand, Smartphone Operated Signal Transduction by Ion Nanogating (STING) Amplifier for Nanopore Sensors: Design and Analytical Application, ACS Sens., 2016, 1(3), 265–271, DOI:10.1021/acssensors.5b00289
.
- W. Shi, A. K. Friedman and L. A. Baker, Nanopore Sensing, Anal. Chem., 2017, 89(1), 157–188, DOI:10.1021/acs.analchem.6b04260
.
- S. Denuga, D. E. Whelan, S. P. O'Neill and R. P. Johnson, Capture and analysis of double-stranded DNA with the α-hemolysin nanopore: Fundamentals and applications, Electrochem. Sci. Adv., 2022, 2(5), e2200001, DOI:10.1002/elsa.202200001
.
- J. J. Kasianowicz, E. Brandin, D. Branton and D. W. Deamer, Characterization of individual polynucleotide molecules using a membrane channel, Proc. Natl. Acad. Sci. U. S. A., 1996, 93(24), 13770–13773, DOI:10.1073/pnas.93.24.13770
.
- J. Clarke, H.-C. Wu, L. Jayasinghe, A. Patel, S. Reid and H. Bayley, Continuous base identification for single-molecule nanopore DNA sequencing, Nat. Nanotechnol., 2009, 4(4), 265–270, DOI:10.1038/nnano.2009.12
.
- I. Nir, D. Huttner and A. Meller, Direct Sensing and Discrimination among Ubiquitin and Ubiquitin Chains Using Solid-State Nanopores, Biophys. J., 2015, 108(9), 2340–2349, DOI:10.1016/j.bpj.2015.03.025
.
- S. Cai, R. Ren, J. He, X. Wang, Z. Zhang, Z. Luo, W. Tan, Y. Korchev, J. B. Edel and A. P. Ivanov, Selective Single-Molecule Nanopore Detection of mpox A29 Protein Directly in Biofluids, Nano Lett., 2023, 23(24), 11438–11446, DOI:10.1021/acs.nanolett.3c02709
.
- M. Wanunu, T. Dadosh, V. Ray, J. Jin, L. McReynolds and M. Drndić, Rapid electronic detection of probe-specific microRNAs using thin nanopore sensors, Nat. Nanotechnol., 2010, 5(11), 807–814, DOI:10.1038/nnano.2010.202
.
- C. Wei, A. J. Bard and S. W. Feldberg, Current Rectification at Quartz Nanopipet Electrodes, Anal. Chem., 1997, 69(22), 4627–4633, DOI:10.1021/ac970551g
.
- D. Duleba and R. P. Johnson, Sensing with ion current rectifying solid-state nanopores, Curr. Opin. Electrochem., 2022, 34, 100989, DOI:10.1016/j.coelec.2022.100989
.
- D. Woermann, Analysis of non-ohmic electrical current–voltage characteristic of membranes carrying a single track-etched conical pore, Nucl. Instrum. Methods Phys. Res., Sect. B, 2002, 194(4), 458–462, DOI:10.1016/S0168-583X(02)00956-4
.
- Z. S. Siwy, Ion-Current Rectification in Nanopores and Nanotubes with Broken Symmetry, Adv. Funct. Mater., 2006, 16(6), 735–746, DOI:10.1002/adfm.200500471
.
- Z. Siwy, E. Heins, C. C. Harrell, P. Kohli and C. R. Martin, Conical-Nanotube Ion-Current Rectifiers: The Role of Surface Charge, J. Am. Chem. Soc., 2004, 126(35), 10850–10851, DOI:10.1021/ja047675c
.
- C.-Y. Lin, L.-H. Yeh and Z. S. Siwy, Voltage-Induced Modulation of Ionic Concentrations and Ion Current Rectification in Mesopores with Highly Charged Pore Walls, J. Phys. Chem. Lett., 2018, 9(2), 393–398, DOI:10.1021/acs.jpclett.7b03099
.
- D. Woermann, Electrochemical transport properties of a cone-shaped nanopore: high and low electrical conductivity states depending on the sign of an applied electrical potential difference, Phys. Chem. Chem. Phys., 2003, 5(9), 1853–1858, 10.1039/B301021J. 10.1039/B301021J
.
- E. B. Farrell, D. Duleba and R. P. Johnson, Aprotic Solvent Accumulation Amplifies Ion Current Rectification in Conical Nanopores, J. Phys. Chem. B, 2022, 126(30), 5689–5694, DOI:10.1021/acs.jpcb.2c03172
.
- Y. Fu, H. Tokuhisa and L. A. Baker, Nanopore DNA sensors based on dendrimer-modified nanopipettes, Chem. Commun., 2009, 4877–4879, 10.1039/B910511E
.
- I. Vlassiouk, T. R. Kozel and Z. S. Siwy, Biosensing with Nanofluidic Diodes, J. Am. Chem. Soc., 2009, 131(23), 8211–8220, DOI:10.1021/ja901120f
.
- R. Gao, Y.-L. Ying, X. Wu, B.-Y. Yan, P. Iqbal and J. A. Preece, Ultrasensitive determination of mercury(II) using glass nanopores functionalized with macrocyclic dioxotetraamines, Microchim. Acta, 2016, 183(1), 491–495, DOI:10.1007/s00604-015-1634-1
.
- S. Zhang, A. Bao, T. Sun, E. Wang and J. Wang, PEI/Zr4+−coated nanopore for selective and sensitive detection of ATP in combination with single-walled carbon nanotubes, Biosens. Bioelectron., 2015, 63, 287–293, DOI:10.1016/j.bios.2014.07.062
.
- M. Ali, B. Yameen, R. Neumann, W. Ensinger, W. Knoll and O. Azzaroni, Biosensing and Supramolecular Bioconjugation in Single Conical Polymer Nanochannels. Facile Incorporation of Biorecognition Elements into Nanoconfined Geometries, J. Am. Chem. Soc., 2008, 130(48), 16351–16357, DOI:10.1021/ja8071258
.
- M. Ali, R. Neumann and W. Ensinger, Sequence-Specific Recognition of DNA Oligomer Using Peptide Nucleic Acid (PNA)-Modified Synthetic Ion Channels: PNA/DNA Hybridization in Nanoconfined Environment, ACS Nano, 2010, 4(12), 7267–7274, DOI:10.1021/nn102119q
.
- S. Zhang, W. Chen, L. Song, X. Wang, W. Sun, P. Song, G. Ashraf, B. Liu and Y.-D. Zhao, Recent advances in ionic current rectification based nanopore sensing: a mini-review, Sens. Actuators Rep., 2021, 3, 100042, DOI:10.1016/j.snr.2021.100042
.
- N. Nakatsuka, A. Faillétaz, D. Eggemann, C. Forró, J. Vörös and D. Momotenko, Aptamer Conformational Change Enables Serotonin Biosensing with Nanopipettes, Anal. Chem., 2021, 93(8), 4033–4041, DOI:10.1021/acs.analchem.0c05038
.
- H. Y. Kong and J. Byun, Nucleic Acid aptamers: new methods for selection, stabilization, and application in biomedical science, Biomol. Ther., 2013, 21(6), 423–434, DOI:10.4062/biomolther.2013.085
.
- S. Song, L. Wang, J. Li, C. Fan and J. Zhao, Aptamer-based biosensors, TrAC, Trends Anal. Chem., 2008, 27(2), 108–117, DOI:10.1016/j.trac.2007.12.004
.
- S. Tian, L. Huang, Y. Gao, Z. Yu and D. Tang, A nucleic acid-based magnetic potentiometric aptasensing platform for indirect detection of prostate-specific antigen with catalytic hairpin assembly, Sens. Diagn., 2023, 2(3), 707–713, 10.1039/D3SD00059A. 10.1039/D3SD00059A
.
- J. Xu, J. Zhang, R. Zeng, L. Li, M. Li and D. Tang, Target-induced photocurrent-polarity-switching photoelectrochemical aptasensor with gold nanoparticle-ZnIn2S4 nanohybrids for the quantification of 8-hydroxy-2′-deoxyguanosine, Sens. Actuators, B, 2022, 368, 132141, DOI:10.1016/j.snb.2022.132141
.
- S. Lv, K. Zhang, Q. Zhou and D. Tang, Plasmonic enhanced photoelectrochemical aptasensor with D-A F8BT/g-C3N4 heterojunction and AuNPs on a 3D-printed device, Sens. Actuators, B, 2020, 310, 127874, DOI:10.1016/j.snb.2020.127874
.
- L. Reynaud, A. Bouchet-Spinelli, C. Raillon and A. Buhot, Sensing with Nanopores and Aptamers: A Way Forward, Sensors, 2020, 20(16), 4495, DOI:10.3390/s20164495
.
- S.-L. Cai, S.-H. Cao, Y.-B. Zheng, S. Zhao, J.-L. Yang and Y.-Q. Li, Surface charge modulated aptasensor in a single glass conical nanopore, Biosens. Bioelectron., 2015, 71, 37–43, DOI:10.1016/j.bios.2015.04.002
.
- S. Zhang, H. Chai, K. Cheng, L. Song, W. Chen, L. Yu, Z. Lu, B. Liu and Y.-D. Zhao, Ultrasensitive and regenerable nanopore sensing based on target induced aptamer dissociation, Biosens. Bioelectron., 2020, 152, 112011, DOI:10.1016/j.bios.2020.112011
.
- P. Actis, A. Rogers, J. Nivala, B. Vilozny, R. A. Seger, O. Jejelowo and N. Pourmand, Reversible thrombin detection by aptamer functionalized STING sensors, Biosens. Bioelectron., 2011, 26(11), 4503–4507, DOI:10.1016/j.bios.2011.05.010
.
- A. Stuber, A. Douaki, J. Hengsteler, D. Buckingham, D. Momotenko, D. Garoli and N. Nakatsuka, Aptamer Conformational Dynamics Modulate Neurotransmitter Sensing in Nanopores, ACS Nano, 2023, 17(19), 19168–19179, DOI:10.1021/acsnano.3c05377
.
- W. Ma, W. Xie, R. Tian, X. Zeng, L. Liang, C. Hou, D. Huo and D. Wang, An ultrasensitive aptasensor of SARS-CoV-2 N protein based on ion current rectification with nanopipettes, Sens. Actuators, B, 2023, 377, 133075, DOI:10.1016/j.snb.2022.133075
.
- P. Zhou, X.-L. Yang, X.-G. Wang, B. Hu, L. Zhang, W. Zhang, H.-R. Si, Y. Zhu, B. Li and C.-L. Huang,
et al., A pneumonia outbreak associated with a new coronavirus of probable bat origin, Nature, 2020, 579(7798), 270–273, DOI:10.1038/s41586-020-2012-7
.
- M. Ciotti, M. Ciccozzi, A. Terrinoni, W.-C. Jiang, C.-B. Wang and S. Bernardini, The COVID-19 pandemic, Crit. Rev. Clin. Lab. Sci., 2020, 57(6), 365–388, DOI:10.1080/10408363.2020.1783198
.
- T. Chaibun, J. Puenpa, T. Ngamdee, N. Boonapatcharoen, P. Athamanolap, A. P. O'Mullane, S. Vongpunsawad, Y. Poovorawan, S. Y. Lee and B. Lertanantawong, Rapid electrochemical detection of coronavirus SARS-CoV-2, Nat. Commun., 2021, 12(1), 802, DOI:10.1038/s41467-021-21121-7
.
- S. S. Timilsina, N. Durr, P. Jolly and D. E. Ingber, Rapid quantitation of SARS-CoV-2 antibodies in clinical samples with an electrochemical sensor, Biosens. Bioelectron., 2023, 223, 115037, DOI:10.1016/j.bios.2022.115037
.
- H. Qi, Z. Hu, Z. Yang, J. Zhang, J. J. Wu, C. Cheng, C. Wang and L. Zheng, Capacitive Aptasensor Coupled with Microfluidic Enrichment for Real-Time Detection of Trace SARS-CoV-2 Nucleocapsid Protein, Anal. Chem., 2022, 94(6), 2812–2819, DOI:10.1021/acs.analchem.1c04296
.
- B. C. Dhar, Diagnostic assay and technology advancement for detecting SARS-CoV-2 infections causing the COVID-19 pandemic, Anal. Bioanal. Chem., 2022, 414(9), 2903–2934, DOI:10.1007/s00216-022-03918-7
.
- P. Lasserre, B. Balansethupathy, V. J. Vezza, A. Butterworth, A. Macdonald, E. O. Blair, L. McAteer, S. Hannah, A. C. Ward and P. A. Hoskisson,
et al., SARS-CoV-2 Aptasensors Based on Electrochemical Impedance Spectroscopy and Low-Cost Gold Electrode Substrates, Anal. Chem., 2022, 94(4), 2126–2133, DOI:10.1021/acs.analchem.1c04456
.
- D. Duleba, P. Dutta, S. Denuga and R. P. Johnson, Effect of Electrolyte Concentration and Pore Size on Ion Current Rectification Inversion, ACS Meas. Sci. Au, 2022, 2(3), 271–277, DOI:10.1021/acsmeasuresciau.1c00062
.
- G. Macori, T. Russell, G. Barry, S. C. McCarthy, L. Koolman, P. Wall, D. Sammin, G. Mulcahy and S. Fanning, Inactivation and Recovery of High Quality RNA From Positive SARS-CoV-2 Rapid Antigen Tests Suitable for Whole Virus Genome Sequencing, Front. Public Health, 2022, 10, 863862, DOI:10.3389/fpubh.2022.863862
.
- G.-C. Liu, M.-J. Gao, W. Chen, X.-Y. Hu, L.-B. Song, B. Liu and Y.-D. Zhao, pH-modulated ion-current rectification in a cysteine-functionalized glass nanopipette, Electrochem. Commun., 2018, 97, 6–10, DOI:10.1016/j.elecom.2018.09.017
.
- G.-C. Liu, W. Chen, M.-J. Gao, L.-B. Song, X.-Y. Hu and Y.-D. Zhao, Ion-current-rectification-based customizable pH response in glass nanopipettes via silanization, Electrochem. Commun., 2018, 93, 95–99, DOI:10.1016/j.elecom.2018.06.015
.
- C. Pick, C. Argento, G. Drazer and J. Frechette, Micropatterned Charge Heterogeneities via Vapor Deposition of Aminosilanes, Langmuir, 2015, 31(39), 10725–10733, DOI:10.1021/acs.langmuir.5b02771
.
- L. Martínez-Jothar, S. Doulkeridou, R. M. Schiffelers, J. Sastre Torano, S. Oliveira, C. F. van Nostrum and W. E. Hennink, Insights into maleimide-thiol conjugation chemistry: Conditions for efficient surface functionalization of nanoparticles for receptor targeting, J. Controlled Release, 2018, 282, 101–109, DOI:10.1016/j.jconrel.2018.03.002
.
- O. Areo, P. U. Joshi, M. Obrenovich, M. Tayahi and C. L. Heldt, Single-Particle Characterization of SARS-CoV-2 Isoelectric Point and Comparison to Variants of Interest, Microorganisms, 2021, 9(8), 1606, DOI:10.3390/microorganisms9081606
.
- P. Piotr, Additional Positive Electric Residues in the Crucial Spike Glycoprotein S Regions of the New SARS-CoV-2 Variants, Infect. Drug Resist., 2021, 14, 5099–5105, DOI:10.2147/IDR.S342068
.
- Q. Yang, T. K. Saldi, P. K. Gonzales, E. Lasda, C. J. Decker, K. L. Tat, M. R. Fink, C. R. Hager, J. C. Davis and C. D. Ozeroff,
et al., Just 2% of SARS-CoV-2−positive individuals carry 90% of the virus circulating in communities, Proc. Natl. Acad. Sci. U. S. A., 2021, 118(21), e2104547118, DOI:10.1073/pnas.2104547118
.
- D. A. Armbruster and T. Pry, Limit of blank, limit of detection and limit of quantitation, Clin. Biochem. Rev., 2008, 29(Suppl 1), S49–S52 Search PubMed
.
- W. H. Organization, Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.10, 2020 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Additional experimental details on nanopipette characterization and sensor stability, aptamer affinity data, and Comsol® modelling of the nanopipette sensor response are available in supporting information. See DOI: https://doi.org/10.1039/d4sd00097h |
| This journal is © The Royal Society of Chemistry 2024 |