Open Access Article
Open Access ArticleCirculating microRNA detection using electrochemical biosensor for rapid diagnosis of liver disease in dogs
Appan
Roychoudhury
 a,
Federico
Diez
b,
Richard J.
Mellanby
b,
James W.
Dear
a,
Federico
Diez
b,
Richard J.
Mellanby
b,
James W.
Dear
 c and
Till T.
Bachmann
c and
Till T.
Bachmann
 *a
*a
aCentre for Inflammation Research, Institute for Regeneration and Repair, University of Edinburgh, 4-5 Little France Drive, Edinburgh EH16 4UU, UK. E-mail: till.bachmann@ed.ac.uk
bThe Royal (Dick) School of Veterinary Studies and Roslin Institute, The Hospital for Small Animals, University of Edinburgh, Easter Bush Campus, Edinburgh EH25 9RG, UK
cCentre for Cardiovascular Science, The Queen's Medical Research Institute, University of Edinburgh, 47 Little France Crescent, Edinburgh EH16 4TJ, UK
First published on 4th June 2024
Abstract
Liver disease in dogs is a major cause of morbidity and mortality. Non-invasive diagnosis of liver disease in dogs is a clinical challenge and improved tests which could done at point-of-care are highly desirable. Liver-specific circulating microRNAs have emerged as promising biomarkers for liver injury across many vertebrate species including dogs. MicroRNA-122 (miR-122), originating from the damaged hepatocytes, provides high specificity and sensitivity in detecting liver disease, compared to the traditional biomarkers. In this study, we present the development of a point-of-care compatible electrochemical biosensor for rapid, early diagnosis of liver disease in dogs by detecting miR-122 in clinical samples. Building on our prior work utilising electrochemical impedance spectroscopy (EIS) for direct and amplification-free detection of miR-122 in human drug-induced liver injury, we have used a miR-122 target-specific short probe and implemented target overhang formation during hybridisation in a flow-based sample cycling setup for enhanced detection performance and demonstrated its performance in real clinical dog samples for the first time. We determined the hybridisation performance by analysing miR-122 specificity and sensitivity achieving a limit of detection (LOD) and limit of quantification (LOQ) of 10 pM and 100 pM, respectively, and high specificity over a nearly-complementary sequence of a miR-122 precursor. Using conventional sample preparation, the developed EIS assay was used to analyse serum samples from dogs with liver disease which were identified based on an increased serum alanine aminotransferase concentration. The test successfully distinguished samples from dogs with and without liver disease in comparable performance to the gold-standard real-time polymerase chain reaction (qPCR) detection. We will further focus on developing sample-to-answer test by combining our miR-122 EIS biosensor with a compatible sample preparation to measure miR-122 from dog blood at point of care.
Introduction
Liver disease is a significant contributor to illness and death in dogs, with 1 in 8 dogs developing liver disease during their lifetime.1 The most common liver disorders observed in dogs are reactive hepatopathies and primary hepatitis.2,3 These illnesses often manifest with nonspecific clinical symptoms such as depression, lethargy, loss of appetite, vomiting, diarrhoea and weight loss. However, they also present with characteristic clinical signs like jaundice, bleeding tendencies and ascites.4 The diagnosis of liver diseases poses a challenge for veterinarians due to the non-specific symptoms or the appearance of specific signs only when there is significant hepatocellular damage. Consequently, an extensive diagnostic workup is required to make a definitive diagnosis, including clinical examination, ultrasonography, laboratory testing, and potentially cytology and/or histopathology. Initial screening for liver injury involves measuring liver enzyme activities such as alanine aminotransferase (ALT), aspartate aminotransferase (AST), alkaline phosphatase (AP), and gamma-glutamyltransferase (GGT) activity. However, these current biochemical assays have limited sensitivity and specificity in detecting histologically confirmed liver disease.5 The gold-standard non-invasive method of diagnosing liver injury involves assessing ALT activity can yield non-specific signals, resulting in false negatives and false positives, which reduces confidence in the diagnosis. Histopathological evaluation of liver biopsy samples remains necessary for most liver diseases to establish a definitive diagnosis, serving as the reference standard against which the accuracy of other tests is compared. However, liver biopsy is an invasive, expensive procedure which is associated with the risk of complications. Therefore, there is an urgent need to develop improved diagnostic tests, ideally at point of care, that enable veterinarians to diagnose liver disease in dogs earlier and more accurately, without the complications associated with biopsies.Recent research has revealed that hepatocyte-derived microRNAs can serve as more sensitive and specific biomarkers for liver injury in dogs.6,7 Circulating microRNAs possess certain properties that make them suitable biomarkers, including organ specificity, relative stability in blood circulation, and the ability to be amplified and measured using conventional polymerase chain reaction (PCR) method. MicroRNA-122 (miR-122) is distinctly and abundantly expressed in the liver, representing 70% of the total liver microRNA content.8 Our previous study demonstrated that miR-122 exhibits enhanced clinical specificity (97%) and sensitivity (77%) for diagnosing liver disease in dogs compared to standard biomarkers.7 Furthermore, miR-122 is conserved across species, including humans,9–11 cats,12 mice,13 rats14 and zebrafish,15 and can serve as an early biomarker released into blood circulation following liver injury.11,16,17
Conventional methods for microRNA detection are based on northern blotting,18 real-time reverse transcription polymerase chain reaction (RT-qPCR),19,20 microarray21 and next-generation sequencing.22 These methods have their own individual advantages and limitations, but none of them is suitable for point-of-care applications. Among these, PCR-based methods have shown high sensitivity and good detection limits, and so are considered the gold standard way of quantifying microRNAs. However, the need for a thermal cycler, multiple expensive enzymes, multiple operating steps, and a well-trained operator has led to laboratory-based methods with a higher measurement cost and limited availability in point-of-care applications.
For point-of-care detection of microRNA, we previously developed electrochemical biosensors and used them for diagnostic test development for drug-induced liver injury (DILI) in humans by detecting miR-122 in clinical samples.23,24 Our biosensor assay uses screen-printed electrodes functionalised with sequence-specific peptide nucleic acid (PNA) probes, enabling direct and amplification-free detection of the miR-122 target using electrochemical impedance spectroscopy (EIS), making it suitable for point-of-care applications. A comprehensive investigation of fundamental parameters influencing sequence-specific microRNA binding was performed by altering the probe length, orientation of probe immobilisation, target overhangs, and degrees of base overhang on the electrode surface.23 Our established model analysis and experiments showed that shorter probes target causing overhangs on the electrode surface to produce stronger EIS signals than their longer counterparts with no overhangs, and the EIS signal was proportionally higher with increasing lengths of the overhangs. This finding is particularly important for the detection of unamplified microRNA targets, enabling fast molecular diagnostics at the point of care. By using the optimised short probe and target overhangs on the electrode surface, we achieved a detection limit of 1 nM for miR-122 at no-flow and room temperature. In a subsequent publication, we optimised the biosensor assay in a continuous flow-based system using size-matched probe.24 The flow-based system indeed enhanced the assay performance by facilitating the fast transport of target molecules to the immobilised probes on the electrode surface. Following thorough characterisation of flow rate and temperature to achieve higher detection sensitivity and specificity, the optimised flow conditions showed a detection limit of 50 pM for miR-122 with the size-matched probe. We further established assay specificity using murine tissue-extracted samples and then applied our assay to human patient samples, where it demonstrated performance equivalent to qPCR (considered the gold standard) after incorporating a microRNA extraction step from blood serum samples using a commercial kit. Since the sequence of miR-122 is the same in humans and dogs, we can directly apply the knowledge gained from our human studies to the diagnosis of liver injury in dogs.
Besides our efforts in developing miR-122 biosensors using EIS, various other techniques such as resonance light scattering (RLS),25–27 colorimetric,28,29 fluorescence,30,31 dynamic chemical labelling,32,33 differential pulse voltammetry (DPV)34 and square wave voltammetry (SWV)35 have been reported in the literature. While optical biosensors (RLS, colorimetric, fluorescence, dynamic chemical labelling) offer sensitive detection of miR-122, they also present limitations, particularly in terms of instrumentation complexity, the need for labelling and susceptibility to sample interference, signal stability, limited multiplexing, cost, and accessibility, which pose challenges for their implementation in point-of-care applications.36 In contrast, electrochemical techniques, and specifically EIS, have demonstrated several advantages including simplicity, ease of miniaturization, portability, rapid and sensitive detection, cost-effectiveness and multiplexing capabilities.37,38 EIS is highly sensitive to bio-recognition events and less disruptive to measured biological interactions compared to voltammetric techniques like DPV and SWV, as measurements are conducted within a narrow range of small potentials and do not require target labelling.39,40 These advantages, coupled with the availability of portable readers, make EIS-based techniques well-suited for point-of-care applications. Furthermore, our EIS-based techniques have exhibited clinical sensitivity and achieved improved or comparable detection limits for miR-122 compared to other reported optical and electrochemical biosensors.
Assessing dogs with potential liver injury at the point of care poses significant challenges for clinicians. Therefore, the aim of this study is to develop a proof-of-concept test which could allow clinicians to quickly and accurately measure miR-122, a well-established biomarker of liver injury in dogs, in a point-of-care setting. Building on our previous research, we investigated a new combination of short probe and target overhangs during miR-122 hybridisation with a flow-based sample cycling setup, thereby significantly enhancing biosensor performance. We assessed the higher detection signal of the short probe by comparing it with a size-matched probe under flow conditions. We optimised the hybridisation temperature for the short probe in the flow condition and checked the miR-122 target specificity performance in the presence of nearly and non-complementary targets. With optimised flow and temperature, we conducted miR-122 target dose dependency studies, which showed a detection limit of 10 pM, lower than our previous finding for both the short probe in no-flow (1 nM) and the size-matched probe in flow conditions (50 pM). As miR-122 exhibits the same sequence and function across species and serves as a sensitive and specific biomarker for liver disease in dogs, we applied the new biosensor setup to liver injury diagnostics for dogs. This optimisation was validated clinically after conventional kit-based microRNA extraction, successfully distinguishing liver-injured dog serum samples from non-liver injury controls and comparing the biosensor performance with the gold-standard qPCR.
Experimental
Reagents, probes and targets
The chemical spacers, 6-mercapto-1-hexanol (MCH) and 1,6-hexanedithiol (HDT), used for electrode preparation, were purchased from ProChimia Surfaces (Gdynia, Poland). All other chemicals used in electrode preparation and electrochemical measurements, including tris(2-carboxyethyl)phosphine hydrochloride (TCEP), dimethylformamide (DMF), dimethyl sulfoxide (DMSO), monosodium phosphate (NaH2PO4), disodium phosphate (Na2HPO4), sodium chloride (NaCl), potassium ferricyanide [K3Fe(CN)6], potassium ferrocyanide [K4Fe(CN)6] and sulfuric acid (H2SO4), were purchased from Sigma-Aldrich (Gillingham, UK). The miRNeasy Serum/Plasma Advanced kit for microRNA extraction, and miRCURY LNA RT kit, miRCURY LNA SYBR Green PCR kit and miRCURY LNA miRNA PCR assay kit for reverse transcription and real-time PCR reactions, were procured from Qiagen UK (Manchester, UK). Unless otherwise stated, all chemicals used in this study were of analytical grade. Aqueous solutions and reaction buffers were prepared using deionised water (resistivity > 18 MΩ cm) from a Millipore MilliQ water purification system (Bedford, MA, USA).The size-matched and short probes specific for microRNA-122 (miR-122) target were made of peptide nucleic acid (PNA) backbone and ordered through Cambridge Research Biochemicals (Cleveland, UK), obtained from Panagene (Daejeon, South Korea). Both of PNA probes (>95% HPLC purified) were modified with a spacer comprising three ethylene glycol units (abbreviated as AEEEA) and a terminal thiol group at the N-end (equivalent to the 5′-end of DNA) for covalent binding on a clean gold substrate and for self-assembled monolayers (SAMs) formation. The size-matched (full-length) probe (P-FL22) is 22 nt in length and reverse complementary to the miR-122 target sequence. The short probe (P-S310) is 10 nt in length and made with the last 10 bases from the N-end of P-FL22 to form a 12 nt overhang target sequence on the electrode surface during hybridisation with the miR-122 target (see Table 1 for structural and sequence details of probes and targets and overhang formation). Stock solutions of PNA probes were prepared in 50% (v/v) dimethylformamide (DMF) aqueous solution and stored at −20 °C when not in use.
| Sequence name | Type | Probe sequence (C–N) | Length (nt) | PNA N-end modification |
|---|---|---|---|---|
| Target sequence (5′-3′) | ||||
| P-FL22 | PNA | ACC TCA CAC TGT TAC CAC AAA C | 22 | Thiol-C11-AEEEA |
| P-S310 | PNA | ACC TCA CAC T | 10 | Thiol-C11-AEEEA |
| miR-122 | RNA | UGG AGU GUG ACA AUG GUG UUU G | 22 | |
| miR-3591 | RNA | UUU AGU GUG AUA AUG GCG UUU GA | 23 | |
| miR-39 | RNA | UCA CCG GGU GUA AAU CAG CUU G | 22 |
The complementary (miR-122), nearly-complementary (miR-3591), and non-complementary (miR-39) RNA target sequences were purchased from Metabion (Martinsried, Germany). Stock solutions of RNA targets were prepared in nuclease-free deionised (DI) water and stored at −80 °C when not in use.
Electrode preparation and electrochemical impedance spectroscopy (EIS) measurements
Screen-printed gold electrodes (DRP-C223BT) were purchased from Metrohm DropSens (Oviedo, Spain) and functionalised with PNA probes following the protocol developed in our earlier studies.23,24,41 In short, electrodes were electrochemically cleaned using cyclic voltammetry (0–1.6 V potential range, 100 mV s−1 scan rate and 10 cycles) in 100 mM sulfuric acid aqueous solution. The cleaned gold working electrode was promptly incubated with a ternary MCH/HDT/PNA probe layer, comprising 6 μM PNA probe, 100 μM MCH, 200 μM HDT and 5 mM TCEP (reducing reagent), and kept in a humid chamber for 16 h for probe immobilisation. Subsequently, 1 mM MCH solution was used to block unspecific sites by incubating for 2 h. Lastly, the probe-functionalised electrodes were rinsed with 50% (v/v) DMSO aqueous solution followed by DI water before using for the electrochemical impedance spectroscopy (EIS) measurements.All electrochemical studies encompassing electrode cleaning and EIS measurements were conducted using a potentiostat/galvanostat electrochemical system (Metrohm Autolab, PGSTAT128N). EIS measurements (see Scheme 1A, frequency range 0.3 Hz–100 kHz, AC signal 10 mV rms at the measured open circuit potential using measurement buffer) were carried out in an EIS measurement buffer (10 mM sodium phosphate, pH 7, 20 mM sodium chloride and 0.2 mM potassium ferri/ferrocyanide redox mediator) and by using a flow-based sample cycling set-up as developed in our previous study.24 This flow-based system (Scheme 1B) was contained with sample reservoirs maintained in a heat block (Starlab, N2400-4002) to control the temperature, a multichannel dispenser peristaltic pump (ISMATEC, ISM930C) to regulate the flow rate, enclosed flow cells with functionalised electrodes placed on a hot plate for temperature-controlled hybridisation measurements, and flow cells connected with the potentiostat and display system. The total volume required to run this closed-loop sample cycling setup was 600 μL. The potentiostat operation and EIS measurements were performed using Nova 2.1.6 software. The charge transfer resistance (Rct) values were measured using a modified Randles equivalent circuit (electrolyte resistance (RS) in series with the parallel combination of the constant phase element (CPE) as a non-ideal capacitance, with Rct and the Warburg (W) element) and by fitting the recorded Nyquist plots in the faradaic EIS measurements (Scheme 1A). The EIS measurements were recorded pre and post hybridisation with the desired sample incubation time using the probe-functionalised electrodes. The increase in Rct value, obtained by dividing the post-hybridisation (sample measurement) by the pre-hybridisation (baseline measurement) Rct, was referred to as the ‘Fold change’ and utilised to plot the EIS data. All EIS measurements were performed in triplicates under identical conditions unless otherwise stated.
Collection and preparation of dog serum samples
Dog serum samples were collected from the Hospital for Small Animals (HfSA) at the Royal (Dick) School of Veterinary Studies (R(D)SVS), University of Edinburgh. Ethical clearance for the study was approved by the R(D)SVS Veterinary Ethical Review Committee (project title: Initiation of novel diagnostic assay development programme; VERC reference number: 139.19). The serum samples were surplus following the completion of a comprehensive biochemical profile of dogs attending the clinic. Briefly, blood was collected in plain tubes, and the serum separation was done by centrifugation within 4 h of collection. Alanine aminotransferase (ALT) activity was measured on the day of collection using a biochemical analyser system (Beckman Coulter AU480). A total of 30 dog serum samples were obtained from the hospital with their individual ALT values. Characteristics of the dogs including breed, sex, age, weight, clinical conditions (healthy or diagnosed with liver and non-liver diseases), and their respective ALT values are mentioned in the Table 2. The ALT values were used to categorise the samples as either liver injury (ALT > 200 U L−1) or non-liver injury (ALT 18–96 U L−1) samples. MicroRNA from the dog serum was extracted using miRNeasy Serum/Plasma Advanced kit (Qiagen) following the manufacturer's protocol. MicroRNAs were eluted in the final volume of 15 μL RNase-free water. After microRNA extraction, the gold-standard real-time polymerase chain reaction (qPCR) analysis was performed for each elute to check the miR-122 content. The extracted RNA solution was further diluted (50 times) in EIS measurement buffer prior to the EIS measurements. All the serum samples and RNA extraction elutes were stored at −80 °C when not in use.| No. | Breed | Sex (M/F) | Age (years) | Weight (kg) | Clinical diagnosis | ALT (U L−1) | miR-122 qPCR (Ct value) | miR-122 EIS (fold change) | |
|---|---|---|---|---|---|---|---|---|---|
| No. 1 to 15 diagnosed with liver diseases and 16 to 30 diagnosed with non-liver diseases. M represents male and F represents female. | |||||||||
| Dogs with liver diseases | 1 | Sheepdog-Shetland | F | 10.2 | 12.6 | Copper-associated chronic hepatitis, gallbladder mucocoele | 657 | 27.33 | 1.6 |
| 2 | Spaniel-Cocker | M | 2.6 | Acute hepatic injury, idiopathic epilepsy | 2473 | 22.92 | 2.81 | ||
| 3 | Terrier-Boston | M | 7.1 | 8.8 | Meningoencephalitis of unknown origin, suppurative and ulcerative glossitis | 551 | 27 | 1.6 | |
| 4 | Saint Bernard, short-haired | M | 4.3 | 68 | Atopic dermatitis | 275 | 28.24 | 1.5 | |
| 5 | Sheepdog-Shetland | F | 10.2 | 12.6 | Copper-associated chronic hepatitis, gallbladder mucocoele | 473 | 24.41 | 1.92 | |
| 6 | Terrier-Border | F | 12.3 | 12.35 | Peritonitis | 283 | 27.56 | 3.01 | |
| 7 | Shih-tzu | M | 2.3 | 9.2 | Hepatopathy | 1086 | 27.23 | 2.21 | |
| 8 | Collie | M | 13.3 | 25 | Liver mass | 1228 | 26.57 | 2.36 | |
| 9 | Dachshund, miniature | M | 12.2 | 5.25 | Hepatopathy | 532 | 25.64 | 2.76 | |
| 10 | Retriever-Labrador | M | 12.9 | 31.9 | Histiocytic sarcoma – disseminated | 467 | 26.75 | 3.1 | |
| 11 | Spaniel-Cocker, parti-colour | M | 5.3 | 14 | Supraventricular tachycardia | 320 | 22.99 | 2.11 | |
| 12 | Collie-Scottish Rough | M | 9.3 | 40.5 | Hepatopathy | 260 | 29.02 | 2.34 | |
| 13 | Retriever-Labrador | M | 12.1 | Hemangiosarcoma – metastatic/disseminated | 4406 | 24.85 | 3.55 | ||
| 14 | Retriever-Labrador | M | 12.4 | 35.8 | Aseptic peritonitis | 1388 | 27.31 | 2.81 | |
| 15 | Shepherd Dog-German (Alsatian) | F | 8.4 | 27.25 | Hepatopathy | 1899 | 26.55 | 1.92 | |
| Dogs with non-liver diseases | 16 | Collie-Border | F | 13.3 | 16.1 | Gastroenteritis | 36 | 32.24 | 1.45 |
| 17 | Jug (JackPug, PugRussell) | M | 4.8 | 7.4 | Gastroenteritis | 66 | 30.21 | 1.54 | |
| 18 | Terrier-Border | M | 15.3 | 11.3 | Osteoarthritis, degenerative joint disease | 23 | 29.16 | 1.85 | |
| 19 | Purebred cross | F | 6.6 | 33.7 | Chronic kidney (renal) disease – non-azotaemic | 65 | 34.99 | 2.44 | |
| 20 | Newfoundland | M | 1.7 | 60 | Ureteric stricture | 18 | 32.17 | 1.62 | |
| 21 | Spaniel-Springer | M | 9.8 | 18.95 | Anaemia – haemolytic, immune-mediated (IMHA) – primary (idiopathic) | 31 | 29.52 | 1.55 | |
| 22 | Retriever-Labrador | M | 10.2 | 38.5 | Facial abscess | 50 | 30.8 | 2.46 | |
| 23 | Crossbreed (Mongrel) | M | 12.3 | 10.7 | Cataract | 84 | 28.74 | 1.63 | |
| 24 | Spaniel-Springer | M | 2.3 | Acute myeloid leukaemia | 81 | 30.69 | 1.95 | ||
| 25 | Terrier-Jack Russell | F | 14.1 | 6.3 | Subcutaneous mast cell tumour | 96 | 30.84 | 1.77 | |
| 26 | Shepherd Dog-German (Alsatian) | M | 7.3 | 37.2 | Multicentric B-cell lymphoma | 64 | 30.18 | 1.77 | |
| 27 | Retriever-Labrador | M | 4 | 28 | Idiopathic paroxysmal dyskinesia | 21 | 29.51 | 1.68 | |
| 28 | Crossbreed (Mongrel) | F | 15.25 | Diagnosis not made | 78 | 28.86 | 1.28 | ||
| 29 | Terrier-Parson Russell | M | 10.1 | 7.7 | Anal sac abscess | 40 | 29.19 | 1.85 | |
| 30 | Retriever-Labrador | M | 10.2 | 38.5 | Facial abscess | 50 | 29.77 | 1.56 | |
Reverse transcription and real-time polymerase chain reaction (RT-qPCR)
The miRCURY LNA RT kit (Qiagen) was used to prepare cDNA following the manufacturer's instructions. In brief, 3 μL of RNA eluate was reverse transcribed into cDNA by adding it to 7 μL of the reverse transcription master mix. The reverse transcription program was run on a thermocycler (Quanta Biotech Q Cycler II) with an incubation at 42 °C for 60 min, followed by a 5 min inactivation step at 95 °C, and concluded with a 4 °C hold step. The prepared cDNA was subsequently diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30) in nuclease-free water and utilised in combination with the miRCURY LNA SYBR Green PCR kit (Qiagen) and a specific miRCURY LNA miRNA PCR assay targeting hsa-miR-122-5p (Qiagen) for the qPCR reaction. The qPCR reaction was prepared by combining 3 μL of diluted cDNA with 7 μL of PCR master mix and was conducted using a LightCycler 480 (Roche LightCycler 96) with the recommended cycling parameters (PCR initial activation step at 95 °C for 2 min; 2-step cycling, denaturation at 95 °C for 10 s, annealing/extension at 56 °C for 60 s; 45 cycles). All samples were run in duplicate, and the Ct (threshold cycles) values were collected after analysing the recorded data using the software supplied with the LightCycler instrument.
30) in nuclease-free water and utilised in combination with the miRCURY LNA SYBR Green PCR kit (Qiagen) and a specific miRCURY LNA miRNA PCR assay targeting hsa-miR-122-5p (Qiagen) for the qPCR reaction. The qPCR reaction was prepared by combining 3 μL of diluted cDNA with 7 μL of PCR master mix and was conducted using a LightCycler 480 (Roche LightCycler 96) with the recommended cycling parameters (PCR initial activation step at 95 °C for 2 min; 2-step cycling, denaturation at 95 °C for 10 s, annealing/extension at 56 °C for 60 s; 45 cycles). All samples were run in duplicate, and the Ct (threshold cycles) values were collected after analysing the recorded data using the software supplied with the LightCycler instrument.
Results and discussion
Target overhang formation during hybridisation in a flow-based sample cycling setup
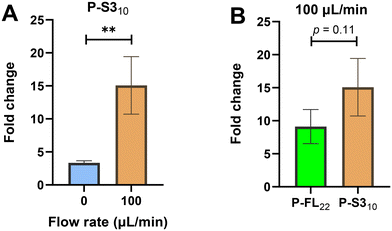
Building on our previous work, we incorporated short probe and target overhang formation during hybridisation in a closed-loop sample cycling setup we developed. In our previous assay optimisation on probe-target binding by using different probe orientation immobilisations, probe lengths and probe combinations, we found that the short probe (P-S310) producing 12 nt target overhang on the electrode surface during hybridisation with the miR-122 target, resulted significant rise in the EIS signals (enhancement in Rct) when comparing with the size-matched full length probe (P-FL22) and no target overhang in a faradaic EIS measurements at no-flow (0 μL min−1) condition.23 We verified the same observation at three different probe concentrations (1.5, 6 and 9 μM) and all these measurements showed at least one-third enhancement in EIS signal during the formation of target overhang on the electrode interface. With our systematic experiments using different lengths of the overhangs on the electrode surface, and with our experimental modelling based on Poisson–Boltzmann and Gouy–Chapman model, we found that the target overhangs close to electrode surface were limiting the access for Fe(CN)63−/4− redox couple, thus enhanced the charge transfer resistance during faradaic EIS measurements. Beside this, we developed a closed-loop sample cycling setup to facilitate the fast transport of targets to the surface immobilised probe, while maintaining a clinically relevant sample volume (total 600 μL with diluted sample in measurement buffer) requiring system.24 Using the size-matched probe (P-FL22) and no target overhang formation, we previously found a >3.5 fold rise in hybridisation signal from faradaic EIS measurements under flow conditions, as compared to no-flow, with the highest signal observed at the 100 μL min−1 flow rate. With our hybridisation experiments in flow conditions and theoretical analyses, we established that convective transport of targets to surface-immobilised probes causing faster hybridisation and higher EIS signal in the desired reaction time frame. In the present study with hybridisation experiments under flow conditions, we introduced the optimised short probe (P-S310) into our flow-based hybridisation system with 100 μL min−1 flow rate. The results show a 4.5 fold increase in the EIS signal (Rct fold change before and after target hybridisation) in the flow condition compared to no-flow (Fig. 1A). The obtained result with P-S310 reveals the similar trend that we observed before with the P-FL22 in the flow conditions.24 We compared the performance of the P-S310 and target overhang formation with the P-FL22 and no target overhang during miR-122 target hybridisation in the flow condition (100 μL min−1). Fig. 1B illustrates a nearly one-third increase in the average EIS signal (Rct fold change) for P-S310 compared to P-FL22 during miR-122 target hybridisation in the flow condition. We attribute this enhanced performance to the combined effects of convective transport of targets in the flow condition and the electrostatic barrier caused by the target overhang following hybridisation, resulting in higher EIS signals for P-S310 compared to both the no-flow condition and P-FL22 in the flow condition. Hence, in the subsequent studies for miR-122 target detection, we employed P-S310 in the sample cycling setup with a 100 μL min−1 flow rate.Optimisation of hybridisation temperature using short probe
Probes with shorter lengths than the target are shown to have lower stringency in binding and prone to bind with unrelated targets. Though, factors such as GC content in the probe and the hybridisation temperature play crucial roles in determining binding affinity and specific interaction with the desired target. Our short probe P-S310 has 50% GC content, higher than that of P-FL22 (45.5%). Although, P-S310 has a lower theoretical basic melting temperature (Tm) of 30 °C, compared to P-FL22 (53 °C) due to its shorter length during binding with the miR-122 target. We checked the effect of hybridisation temperature (21–45 °C) during the binding of miR-122 with P-S310 in the flow condition. In our EIS measurements, we observed an increase in the hybridisation signal up to 35 °C, followed by a sharp decrease in the signal continuing up to 45 °C (Fig. 2). These results closely followed the temperature profile of a typical surface melting curve, displaying a steady high signal up to the melting temperature (Tm) of the probe-target duplex, followed by a 50% signal reduction at the Tm.42 Our experimental observations, indicating an optimum hybridisation signal at 35 °C, and subsequent signal decrease up to 45 °C, potentially due to the dissociation of more than half of the targets from the probe molecules, strongly align with the calculated basic melting temperature (30 °C).Specificity for miR-122 detection at optimised assay temperature
We verified the miR-122 target specificity of P-S310 at the optimised hybridisation temperature (35 °C) under flow condition and by comparing it with hybridisation to a nearly-complementary target (miR-3591) and a non-complementary target (miR-39) through our EIS measurements. The miR-3591 is a human microRNA classified as miR-122 precursor (RF00684) in the Rfam (RNA family database), considered as a nearly-complementary target. The miR-39 is a nematode microRNA from Caenorhabditis elegans, utilised as a non-specific target (see Table 1 for sequences). Fig. 3A displays the hybridisation kinetics signals, where P-S310 shows a steady increase in the hybridisation signal with 10 nM perfectly-matched miR-122 target throughout the 60 min time window. In contrast, there is a marginal increase for miR-3591 up to 10 minutes, followed by signal saturation, and almost no increase for miR-39 with the same target concentration (10 nM each), compared to blank (no target) buffer measurements. Considering the fold change in EIS signals upon target hybridisation at 60 min (Fig. 3B), the detected EIS signal for miR-122 hybridisation demonstrates a significant rise, while there is no significant increase for miR-3591 and miR-39 hybridisation, as compared to blank measurements. These results confirm that P-S310 retained a high level of specificity for the miR-122 target at our measured hybridisation temperature (35 °C), even at half of the full target length. We envisage that optimising hybridisation temperature and consequential washing in the flow-based sample cycling setup would be beneficial in reducing unspecific binding or adsorption of interferences while studying real biological samples using the P-S310 short probe.Sensitivity of miR-122 detection
We determined the sensitivity and limit of detection (LOD) for miR-122 detection using P-S310 under optimised flow conditions (35 °C and 100 μL min−1 flow rate). With gradually decreasing miR-122 concentrations from 10 nM down to 5 pM, we observed a concentration dependent hybridisation signal in our EIS measurements. The hybridisation kinetics studies revealed a continuous enhancement in EIS signals with increasing concentrations, with no distinct signal saturation observed up to a 10 nM concentration within a 60 min reaction time (Fig. 4A). Although, our binding kinetics studies with P-S310 exhibited an overall similar behaviour to our previous observation with P-FL22 and miR-122 target hybridisation,24 and other binding kinetics studies with surface bound PNA/DNA probes,43,44 with a steadier increase and close to faster saturation at higher concentrations. At elevated concentrations, we anticipate more targets are available for binding, potentially leading to higher signals. Utilising the 60 min end-point readings at different target concentrations (5 pM–10 nM), we generated a calibration curve to calculate the analytical sensitivity, LOD, as well as limit of quantification (LOQ) for miR-122 detection (Fig. 4B). We found a linear response in between 1 and 10 nM miR-122, with analytical sensitivity of 2.09 fold change per nM for miR-122 within the linear range and a LOD of 10 pM (equivalent to 6 fmole), and LOQ of 100 pM based on the blank measurements.45 In a previous study on miR-122 detection using rat plasma samples in a paracetamol-induced liver toxicity model, it was demonstrated that miR-122 concentration can vary between 32 pM and 5.35 nM when treated with a single dose of paracetamol (1500 mg kg−1 oral).32 Our measured LOD indicates that our developed sensor with P-S310 exhibits clinical sensitivity for liver injury. We also compared the sensing parameters of our miR-122 biosensor with those reported in literature, as summarised in Table 3.| Detection method | Limit of detection (pM) | Linear range | Reaction time (min) | Ref. |
|---|---|---|---|---|
| Resonance light scattering | 98 | 200 pM–10 nM | 40 | 25 |
| Resonance light scattering | 9.4 | 0.16–4.8 nM | 40 | 26 |
| Resonance light scattering | 6.1 | 50 pM–300 nM | 60 | 27 |
| Colorimetric | 1200 | 0–100 nM | 30 | 28 |
| Colorimetric | 16 | 20 pM–1 nM | 60 | 29 |
| Dynamic chemical labelling – Simoa assay | 1.32 | 180 | 32 | |
| Dynamic chemical labelling – time-gated fluorescence | 100 | 33 | ||
| Fluorescence | 2500 | 2.5–500 nM | 40 | 30 |
| Fluorescence | 5000 | 5–1000 nM | 120 | 31 |
| Differential pulse voltammetry | 1.73 | 10 pM–10 μM | 60 | 34 |
| Square wave voltammetry | 3200 | 5–50 nM | 60 | 35 |
| Electrochemical impedance spectroscopy using short probe and target overhang formation | 1000 | 5–100 nM | 60 | 23 |
| Electrochemical impedance spectroscopy under flow condition | 50 | 1–50 nM | 60 | 24 |
| Electrochemical impedance spectroscopy after combining short probe and target overhang with the flow condition | 10 | 1–10 nM | 60 | This work |
Clinical studies by analysing dog serum samples
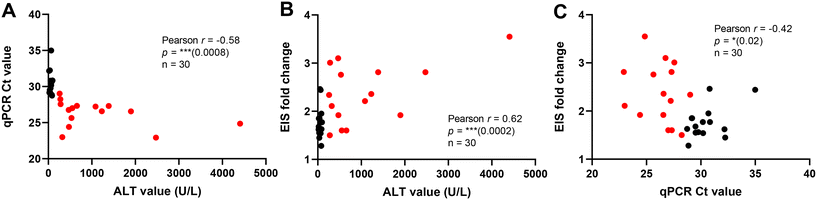
We conducted a study on dog serum samples collected from the small animal hospital to diagnose liver injury by detecting miR-122 using the assay developed with P-S310 in the flow-based sample cycling setup (see Table 2 for details on dog samples). MicroRNA sample preparation from the serum was performed using a standard bench-kit extraction, and the elutes were used in our EIS measurements. The EIS signals were significantly higher in the liver injury samples compared to the non-liver injury controls (Fig. 5A). The EIS detection of miR-122 was compared with the gold-standard real-time reverse transcription polymerase chain reaction (RT-qPCR) detection (Fig. 5C), revealing less sensitivity for EIS. This observation was made after considering the significant signal difference between liver injury and non-liver injury groups, and the area under the curve (AUC 0.79 for EIS and 0.99 for qPCR) from the receiver operator characteristics (ROC) analysis of EIS (Fig. 5B) and qPCR (Fig. 5D). While qPCR analysis exhibited better clinical sensitivity (0.93 for qPCR and 0.4 for EIS at 0.93 specificity), the EIS analysis was performed without any target amplification and labelling steps, making it more suitable for point-of-care setups and facilitating early diagnosis of liver disease in dogs. PCR methods are laboratory-based and have a relatively long time to result (TTR) of one working day, whereas EIS detection can be performed in 30 min with a low-complexity setup and at the point of care.The miR-122 detection signals obtained from EIS and qPCR studies were correlated with the alanine aminotransferase (ALT) activity present in the dog serum samples. Both the comparison between qPCR Ct value and ALT activity (Fig. 6A) and EIS fold change and ALT activity (Fig. 6B) revealed equivalent correlation parameters (Pearson r −0.58 for qPCR and 0.62 for EIS) with a moderate correlation (Pearson r −0.42) in between miR-122 signals for EIS and qPCR (Fig. 6C). The correlation between miR-122 detection signals obtained from EIS and qPCR analyses with ALT activity in dog serum samples highlights the importance of reliable biomarkers in liver health assessment. Both methods exhibited comparable correlation parameters, demonstrating consistent performance. Additionally, a moderate correlation between miR-122 signals for EIS and qPCR further supports the reliability of these detection techniques. This validation supports the role of miR-122 as a robust biomarker for liver injury, offering veterinarians valuable insights into canine liver health and potential diagnostic options for liver disease.
Conclusions
We have shown the direct detection of microRNA-122 biomarker in clinical samples from dogs without the need for target labelling and amplification. Our established assay utilising the newly-combined short probe in a closed-loop sample cycling setup demonstrated high specificity in microRNA-122 detection, while maintaining sensitivity in the clinical range. We took these findings further and did successful evaluation of dog serum samples from the liver injury, achieving performances comparable to qPCR detection following the conventional microRNA sample preparation. In future work, we will proceed towards the development of an integrated sample-to-answer test by combining our existing point-of-care compatible microRNA-122 detection platform with a compatible sample preparation step to enable direct measurement from dog whole blood. This provides the opportunity to measure microRNA-122 on-site, such as at a veterinary clinic or even at home, providing veterinarians and pet owners with an immediate option for liver injury diagnosis.Compliance with ethical standard
The study was approved by the University of Edinburgh Veterinary Ethical Review Committee (VERC reference number: 139.19) and followed the institutional guidelines. No lab animals were involved in the study, and so the relevant ethical review board for client owned animals (University of Edinburgh Veterinary Ethical Review Committee) approved the study.Author contributions
Appan Roychoudhury: conceptualization, methodology, data curation, formal analysis, investigation, validation, writing – original draft. Federico Diez: resources, validation. Richard J. Mellanby: resources, validation, writing – review & editing. James W. Dear: writing – review & editing, funding acquisition. Till T. Bachmann: conceptualization, methodology, validation, resources, writing – review & editing, funding acquisition.Conflicts of interest
The authors declare that they have no conflicts of interest except RJM, who currently works for IDEXX Laboratories Inc in which he holds shares/stock options.Acknowledgements
The authors acknowledge the Rosetrees Trust, UK for providing funding (Ref. CF1\100010) for the project ‘Early diagnosis of drug-induced liver injury using point of care detection of microRNA’. We would like to acknowledge Nila Roy Choudhury for the support with qPCR experiments.References
- P. J. Watson, A. J. A. Roulois, T. J. Scase, R. Irvine and M. E. Herrtage, J. Small Anim. Pract., 2010, 51, 566–572 CrossRef CAS PubMed
.
- J. H. Poldervaart, R. P. Favier, L. C. Penning, T. S. G. A. M. van den Ingh and J. Rothuizen, J. Vet. Intern. Med., 2009, 23, 72–80 CrossRef CAS PubMed
.
- D. Twedt, Vet. Q., 1998, 20, S46–S47 CrossRef PubMed
.
-
T. R. Tams, Handbook of small animal gastroenterology, Elsevier Health Sciences, 2nd edn, 2003 Search PubMed
.
- K. Dirksen, I. A. Burgener, J. Rothuizen, T. S. G. A. M. van den Ingh, L. C. Penning, B. Spee and H. Fieten, J. Vet. Intern. Med., 2017, 31, 1017–1027 CrossRef CAS PubMed
.
- K. Dirksen, T. Verzijl, T. S. G. A. M. van den Ingh, J. C. M. Vernooij, L. J. W. van der Laan, I. A. Burgener, B. Spee and H. Fieten, Vet. J., 2016, 211, 75–81 CrossRef CAS PubMed
.
- W. Oosthuyzen, P. W. L. Ten Berg, B. Francis, S. Campbell, V. Macklin, E. Milne, A. G. Gow, C. Fisher, R. J. Mellanby and J. W. Dear, J. Vet. Intern. Med., 2018, 32, 1637–1644 CrossRef CAS PubMed
.
- D. de Rie, I. Abugessaisa, T. Alam, E. Arner and P. Arner,
et al.
, Nat. Biotechnol., 2017, 35, 872–878 CrossRef CAS PubMed
.
- V. Bihrer, M. Friedrich-Rust, B. Kronenberger, N. Forestier, J. Haupenthal, Y. Shi, J. Peveling-Oberhag, H. H. Radeke, C. Sarrazin, E. Herrmann, S. Zeuzem, O. Waidmann and A. Piiper, Am. J. Gastroenterol., 2011, 106(9), 1663–1669 CrossRef CAS PubMed
.
- J. C. McCrae, N. Sharkey, D. J. Webb, A. D. B. Vliegenthart and J. W. Dear, Clin. Toxicol., 2016, 54, 53–55 CrossRef CAS PubMed
.
- P. J. Starkey Lewis, J. Dear, V. Platt, K. J. Simpson, D. G. N. Craig, D. J. Antoine, N. S. French, N. Dhaun, D. J. Webb, E. M. Costello, J. P. Neoptolemos, J. Moggs, C. E. Goldring and B. K. Park, Hepatology, 2011, 54, 1767–1776 CrossRef CAS PubMed
.
- S. K. Armstrong, W. Oosthuyzen, A. G. Gow, S. Salavati Schmitz, J. W. Dear and R. J. Mellanby, J. Feline Med. Surg., 2022, 24, e289–e294 CrossRef PubMed
.
- K. Wang, S. Zhang, B. Marzolf, P. Troisch, A. Brightman, Z. Hu, L. E. Hood and D. J. Galas, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4402–4407 CrossRef CAS PubMed
.
- S. Starckx, A. Batheja, G. R. Verheyen, S. D. Jonghe, K. Steemans, B. V. Dijck, M. Singer, N. Bogdan, J. Snoeys, P. Vinken, J. C. Sasaki, J. V. Gompel, P. Guzzie-Peck, A. Lampo and L. Lammens, Toxicol. Pathol., 2013, 41, 795–804 CrossRef CAS PubMed
.
- H.-S. Nam, K.-S. Hwang, Y.-M. Jeong, J.-I. Ryu, T.-Y. Choi, M.-A. Bae, W.-C. Son, K.-H. You, H.-Y. Son and C.-H. Kim, Biomed Res. Int., 2016, 1473578 Search PubMed
.
- P. J. Starkey Lewis, M. Merz, P. Couttet, O. Grenet, J. Dear, D. J. Antoine, C. Goldring, B. K. Park and J. G. Moggs, Clin. Pharmacol. Ther., 2012, 92, 291–293 CrossRef CAS PubMed
.
- A. D. B. Vliegenthart, J. M. Shaffer, J. I. Clarke, L. E. J. Peeters, A. Caporali, D. N. Bateman, D. M. Wood, P. I. Dargan, D. G. Craig, J. K. Moore, A. I. Thompson, N. C. Henderson, D. J. Webb, J. Sharkey, D. J. Antoine, B. K. Park, M. A. Bailey, E. Lader, K. J. Simpson and J. W. Dear, Sci. Rep., 2015, 5(1), 15501 CrossRef CAS PubMed
.
- A. Válóczi, C. Hornyik, N. Varga, J. Burgyán, S. Kauppinen and Z. Havelda, Nucleic Acids Res., 2004, 32, e175 CrossRef PubMed
.
- M. Lundegard, K. Nylander and K. Danielsson, Med. Oral Patol. Oral Cir. Bucal, 2015, 20, e130–e134 CrossRef PubMed
.
- C.-Y. Yu, B.-C. Yin and B.-C. Ye, Chem. Commun., 2013, 49, 8247–8249 RSC
.
- B. Wang and Y. Xi, Microarrays, 2013, 2, 34–50 CrossRef CAS PubMed
.
- S. Rahmann, M. Martin, J. H. Schulte, J. Köster, T. Marschall and A. Schramm, Methods, 2013, 59, 154–163 CrossRef CAS PubMed
.
- A. Roychoudhury, J. W. Dear and T. T. Bachmann, Biosens. Bioelectron., 2022, 212, 114404 CrossRef CAS PubMed
.
- A. Roychoudhury, J. W. Dear, M. Kersaudy-Kerhoas and T. T. Bachmann, Biosens. Bioelectron., 2023, 231, 115298 CrossRef CAS PubMed
.
- F. Chen, F. Zhang, Y. Liu and C. Cai, Talanta, 2018, 186, 473–480 CrossRef CAS PubMed
.
- S. Lv, F. Chen, C. Chen, X. Chen, H. Gong and C. Cai, Talanta, 2017, 165, 659–663 CrossRef CAS PubMed
.
- R. Zou, F. Zhang, C. Chen and C. Cai, Microchim. Acta, 2019, 186, 1–7 CrossRef CAS PubMed
.
- J. Lee, Y. K. Kim, S. Lee, S. Yoon and W. K. Kim, Sens. Actuators, B, 2019, 282, 861–867 CrossRef CAS
.
- Q. Wang, R.-D. Li, B.-C. Yin and B.-C. Ye, Analyst, 2015, 140, 6306–6312 RSC
.
- R. Liao, S. Li, H. Wang, C. Chen, X. Chen and C. Cai, Anal. Chim. Acta, 2017, 983, 181–188 CrossRef CAS PubMed
.
- R. Liao, K. He, C. Chen, X. Chen and C. Cai, Anal. Chem., 2016, 88, 4254–4258 CrossRef CAS PubMed
.
- B. López-Longarela, E. E. Morrison, J. D. Tranter, L. Chahman-Vos, J.-F. Léonard, J.-C. Gautier, S. Laurent, A. Lartigau, E. Boitier, L. Sautier, P. Carmona-Saez, J. Martorell-Marugan, R. J. Mellanby, S. Pernagallo, H. Ilyine, D. M. Rissin, D. C. Duffy, J. W. Dear and J. J. Díaz-Mochón, Anal. Chem., 2020, 92, 3388–3395 CrossRef PubMed
.
- E. Garcia-Fernandez, M. C. Gonzalez-Garcia, S. Pernagallo, M. J. Ruedas-Rama, M. A. Fara, F. J. López-Delgado, J. W. Dear, H. Ilyine, C. Ress, J. J. Díaz-Mochón and A. Orte, Chem. Commun., 2019, 55, 14958–14961 RSC
.
- S. Kasturi, Y. Eom, S. R. Torati and C. Kim, J. Ind. Eng. Chem., 2021, 93, 186–195 CrossRef CAS
.
- D. M. Mills, P. Calvo-Marzal, J. M. Pinzon, S. Armas, D. M. Kolpashchikov and K. Y. Chumbimuni-Torres, Electroanalysis, 2017, 29, 873–879 CrossRef CAS PubMed
.
- S. Rasheed, T. Kanwal, N. Ahmad, B. Fatima, M. Najam-ul-Haq and D. Hussain, TrAC, Trends Anal. Chem., 2024, 173, 117640 CrossRef CAS
.
- H. S. Magar, R. Y. A. Hassan and A. Mulchandani, Sensors, 2021, 21, 6578 CrossRef CAS PubMed
.
- N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747–1763 RSC
.
- A. Bogomolova, E. Komarova, K. Reber, T. Gerasimov, O. Yavuz, S. Bhatt and M. Aldissi, Anal. Chem., 2009, 81, 3944–3949 CrossRef CAS PubMed
.
- S. Wang, J. Zhang, O. Gharbi, V. Vivier, M. Gao and M. E. Orazem, Nat. Rev. Methods Primers, 2021, 1, 41 CrossRef CAS
.
- A. Roychoudhury, R. J. Allen, T. Curk, J. Farrell, G. McAllister, K. Templeton and T. T. Bachmann, ACS Sens., 2022, 7, 3692–3699 CrossRef CAS PubMed
.
- W. Qiao, H.-C. Chiang, H. Xie and R. Levicky, Chem. Commun., 2015, 51, 17245–17248 RSC
.
- K. K. Jensen, H. Ørum, P. E. Nielsen and B. Nordén, Biochemistry, 1997, 36, 5072–5077 CrossRef CAS PubMed
.
- A. Munir, H. Waseem, M. R. Williams, R. D. Stedtfeld, E. Gulari, J. M. Tiedje and S. A. Hashsham, Microarrays, 2017, 6, 9 CrossRef PubMed
.
-
A. D. McNaught and A. Wilkinson, Compendium of chemical terminology, Blackwell Science Oxford, 1997 Search PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |