Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Paper-based point of care diagnostics for cancer biomarkers
Prateek
Bhardwaj
 *a,
Bharti
Arora†
*a,
Bharti
Arora†
 a,
Survanshu
Saxena†
a,
Survanshu
Saxena†
 a,
Subhasini
Singh†
a,
Subhasini
Singh†
 a,
Pranoti
Palkar
a,
Pranoti
Palkar
 b,
Jayant Sastri
Goda
b,
Jayant Sastri
Goda
 *b and
Rinti
Banerjee
*b and
Rinti
Banerjee
 a
a
aNanomedicine Lab, Department of Biosciences and Bioengineering, Indian Institute of Technology Bombay, 400076, India. E-mail: prateekbhardwaj26@gmail.com
bDepartment of Radiation Oncology, Advanced Centre for Treatment Research and Education in Cancer, Navi Mumbai, & Homi Bhabha National Institute, Maharashtra, India. E-mail: jgoda@actrec.gov.in
First published on 23rd February 2024
Abstract
Early detection of cancers is key to a better prognosis. Advanced proteomics and genomic detection techniques offer great specificity and sensitivity, however, delayed symptomatic detection, cost, and patient-incompliant sample procurement limit routine cancer diagnosis, thus affecting treatment opportunities and patient survival. The revolutionary impact of paper-based COVID-19 antigen home test kits highlighted the importance of affordable routine diagnosis in tackling pandemics. Therefore, inexpensive, user-friendly, and sensitive paper-based biosensors can prove to be a game changer in the management of cancer. Even though the fabrication of paper-based biosensors is easy and inexpensive, their compromised sensitivity requires significant improvement for effective diagnosis. This review comprehensively and systemically focuses on highlighting the impactful advancements that occurred over the past 10 years to improve the sensitivity at different levels of paper-based detection i.e. advancements in paper chemistry, assay type, detection technique, and signal enhancement. A detailed focus has also been provided on the impact of advanced nanomaterials (classified into inorganic, organic, and amalgamation of both) in enhancing analyte detection, signal amplification, signal transmission, and signal readout to develop point-of-care systems with fast interpretation, better reliability, specificity, biocompatibility, and low detection limits for the early paper-based detection of cancer. Moreover, a specific section on the types of samples employed for cancer detection, comprehensive tabulation of validated biosensors with clinical samples, their current challenges, and future prospects can help disseminate extensive information in driving the research forward in low-cost diagnosis of cancer.
1. Introduction
Limitations in detecting minute concentrations of cancer-specific biomarkers and their unexplored presence in some malignancies account for delayed cancer detection, a major factor that indirectly affects the prognosis and mortality.1 Advancements in molecular detection with the advent of highly sensitive genomic and proteomic technologies minimized these limitations to an extent, but technique-associated cost along with the requirement of basic infrastructure and trained operators limits their affordability in resource-constrained conditions especially in developing countries.2 Therefore, paper-based diagnostic technologies that can meet WHO's ASSURED (affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-user) criteria are required.3 Such technologies majorly utilize two kinds of substrates like cellulose fibre (filter and chromatography paper) and nitrocellulose membrane (uniform pore size, 0.45 μm) for fabrication because of their porous nature with modifiable surface chemistry and optical properties.3,4 Filter papers with pore sizes 20–25 μm are generally used due to their appropriate porosity, hydrophilicity, stiffness, and lateral fluid flow rate.3,4 The utilization of inexpensive biodegradable substrates makes these point-of-care devices easily scalable and disposable. Multiple types of paper-based POCs have been developed over the years starting from the conventional dipstick and lateral flow tests (latex agglutination and radioimmunoassay) in the late 1950s to advanced microfluidic tests utilizing different detection approaches namely colorimetric, fluorometric, or electrochemical.5 To overcome the limitations associated with the sensitivity and specificity of conventional paper-based POCs, novel biomaterial-based approaches are being employed to modify the surface properties of the paper through biopolymer coatings, impregnation or surface functionalization of paper with different nanoparticles, or the use of nanoparticles as sensing molecules for signal amplification.4 Over the past 5 years, a handful of review articles highlighted the importance of low-cost paper-based diagnostics in cancer detection with individual focus on either type of paper-based diagnostics, microfluidic-based analytical devices, or the role of nanomaterials in cancer detection, however, a systemic and comprehensive review highlighting the advancements at all the fronts of paper-based cancer detection is highly desirable to drive the research in this domain.6–11This review exhaustively highlights the impactful technological advancements that occurred over the past decade at each step of biomarker detection starting from advancements in paper substrate chemistry, assay type, and detection technique, to advanced signal enhancement strategies exploited in developing extremely sensitive paper-based biosensors specifically for cancer detection. In assay type, this review comprises conventional dipstick assays to advanced 3D μPADs (microfluidic paper-based analytical devices), wherein the detection techniques comprise dot blot, advanced immunochromatography integrated with nucleic acid amplification technique, molecularly imprinted polymers, and conducting papers. Importantly, a detailed focus was imparted to the advanced nanomaterial-based detection and signal amplification strategies with a systematic description based on the use of different inorganic, organic, and hybrid nanomaterials. It also provides an exhaustive comparison of the devices used in detecting different cancer biomarkers from different clinical samples and underlines their challenges and future implications in cancer diagnosis.
2. Advancements in the substrate (paper) chemistry
Advancements in paper-based devices originate from making small alterations in the functionality of the substrate (paper) itself. Chitosan not only enhances the wet-strength of μPADs but also stabilizes the immobilized antibodies.12 Hence, chitosan-treated nitrocellulose membrane can be a good substrate for chemiluminescence ELISA (CL-ELISA) on paper.13 The poly(oligoethylene glycol methacrylate) (POEGMA) coating on paper makes it a protein repellent, thus reducing the output signal noise due to non-specific adsorption of proteins in any biological sample.14 Further, nitrocellulose sheets can be coated with a hydrophobic layer of polycaprolactone (PCL) to reduce the flow rate.15 The reduction in the fluid flow rate in paperfluidic chips increases target-probe binding, ultimately resulting in a strong signal and lower limit of detection. To automate the sequential steps employed in the lateral flow assays, a water-swellable polymer-based intermediate strip can be added to the LFA design to automatically release the secondary reagents in a timely manner.16 The synthesis of porous ‘hydrogel walls’ on the nitrocellulose paper can facilitate the capture of detection probes as they flow through the assay.17 Polymer-based biochips have been developed as superior substrates for the detection of HPV from gynecological swabs and biopsies from genital warts.18,193. Advancements in the type of paper-based assays
Paper as a substrate can be utilized in various ways to develop a variety of assays for the detection of a specific cancer biomarker in different kinds of biological samples. Significant progress from employing conventional assays like dipstick to more advanced microfluidic-based detection of cancer has been discussed below (refer to Fig. 1).3.1 Dipstick test
This rapid diagnostic method involves the dipping of an ordinary paper strip in a patient's sample to determine the type of malignancy based on color change after enzyme-catalyzed reactions, which can be correlated with the color-coded chart for a semi-quantitative determination as shown in Fig. 1. Initially, it was established in the 1950s as one of the vital point-of-care tests for the detection of glucose in the urine, however, its user-friendliness has fascinated many researchers to explore its potential for the detection of numerous diseases including cancer.20 A direct comparison of the dipstick test with an established immuno-fecal occult blood test (IFOBT) for the detection of transferrin (Tf) and fecal hemoglobin respectively in the fecal discharge of patients with colon cancer and premalignant lesions reflected its superiority over IFOBT with a specificity of 76.5% over 69% (IFOBT). The IFOBT test generally detects hemoglobin discharged in feces of colon cancer patients, but the fecal hemoglobin is more prone to lysis due to the effect of bacteria or digestive enzymes.21 Whereas, detection of transferrin discharged in feces due to bleeding by Tf dipstick test provides a more stable analysis as transferrin is resistant towards bacterial and digestive enzyme action.22 Despite easy analysis and wide applicability of test strips, the use of dipstick assay is quite limited for cancer detection due to lack of precision, limited detection range, and requirement of large sample volume. Integration of paper strips with more advanced assays like flow-based and microfluidic-based significantly minimizes these limitations and has thus gained a lot of importance in cancer diagnosis.233.2 Flow assays
Flow assays have gained significant importance in the detection of various analytes in complex samples within a short period. They have been categorized into two main types i.e. lateral and vertical flow assays depending on the directionality of the flow of the sample due to capillary mode of action.LFA is classified into two major formats; direct type and competitive type. Direct type can detect larger analytes and molecules with multiple antigenic sites in the form of the sandwich-based assay, whereas competitive type analyzes the smaller molecules with a single antigenic determinants.25 Many LFA detection kits are available in the market for the detection of various cancers (refer to Table 1).
| Company | Product name | Cancer type | Analyte | Required sample | Detection time (min) | Sensitivity | Specificity | Ref. |
|---|---|---|---|---|---|---|---|---|
| NR: not reported. | ||||||||
| CTK Biotech | On-site PSA semi-quantitative rapid test | Prostate | PSA | 60–90 ml blood/serum/plasma | 10 | 100% | 99% | 26 |
| On-site FOB rapid test | Colorectal | hHB | Human fecal specimens | 10 | NR | NR | ||
| Alere | Alere NMP22 BladderChek | Bladder | Nuclear matrix protein (NMP22) | 4 drops of urine | 30 | 99% when combined with cystoscopy | 99% NPV along with cystoscopy | 27 |
| Clearview iFOBT | Colon | Faecal occult blood | Faeces | 5 | 93.60% | 99.10% | ||
| Arbor Vita Corp. | OncoE6 cervical test | Cervical | E6 oncoproteins | Cervical swab | 150 | 84.6% | 98.5% | 28 |
| OncoE6™ oral test | Oral | E6 oncoproteins | NR | 150 | NR | NR | ||
| Quicking Biotech Co., Ltd | CA125 rapid | Ovarian | CA125 Ag | 100 μL of serum | 5–10 | 40 U ml−1 | NR | 29 |
| Innovation Biotech | AFP test | Hepato-cellular | AFP | Serum plasma | 10 | 25 ng ml−1 | NR | 30 |
| Diagnostic Automation/Cortez Diagnostics Inc. | CEA serum rapid test | Colon adenocarcinoma | CEA | Serum | 10 | 5 ng ml−1 | NR | 31 |
| LifeSign, LLC | Status BTA | Bladder | Bladder tumor associated antigen | 3 drops of urine | 5 | 67% | 70% | 32 |
| ALFA SCIENTIFIC DESIGNS | INSTANT-VIEW® | Prostate | PSA | Serum | 4–7 | 4 ng ml−1 | NR | 33 |
| AccuBioTech Co., Ltd | AFP whole blood test cassette | Hepatocellular carcinoma | AFP | Blood/serum/plasma | <10 | 99.3% | 99.0% | 34 |
| PSA whole blood test cassette | Prostate | PSA | Blood | <10 | 98.9% | 98.6% | ||
| CEA whole blood test cassette | Hepato-cellular carcinoma | CEA | Blood | <10 | 98.7% | 99.3% | ||
| FOB feces test strip | Colon | Human occult blood in feces | Blood | <10 | 94.6% | 99.3% | ||
| TÜRKLAB | CEA (carcino-embryonic antigen) test | Diagnosis of primary carcinomas | CEA | Blood/serum/plasma | <10 | 99.3% | 99.3% | 35 |
| AFP (alpha fetal protein) test | Detection of many tumors at an earlier stage/detection of fetal open neural tube defects | AFP | Blood/serum/plasma | <10 | 99.3% | 99% | ||
| FOB (fecal occult blood) test | Colorectal cancer | Hb | Human faeces | <10 | 99.9% | 97% | ||
| Ulti med | FOB cassette | Colorectal cancer | Human occult blood | Human faeces | <10 | NR | NR | 36 |
Though LFA offers advantages like easy detection, robustness, and specificity, its low reproducibility due to difficulty in controlling the sample flow rate, compromised user-friendliness owing to time-consuming sample pre-treatment, and cross-reactivity during multiplex LFA on a single strip leads to false positive errors. In addition, processing of complex samples often requires sophisticated equipment which limits the use of such LFAs in hospitals and clinics thus affecting its overall operational flexibility and self-diagnosis at home.37
3.3 Microfluidic paper-based analytical device (μPAD)
Paper-based microfluidics are point-of-care devices that consist of a set of hydrophilic cellulose fibres that direct the flow of liquid from the inlet to the outlet through permeation. The agglomerated cellulose fibres enhance the porosity of the device for the movement of the sample from the region of high concentration of analyte to the region of low concentration based on low Reynold's number i.e., the ratio of inertial force to viscous force.40 While fabricating the hydrophilic microfluidic paper, certain hydrophobic barriers are generated to restrict the flow of samples within the device to make it more sensitive and precise for detection.41 It is a modified version of the standard lateral flow assay. The principle of paper-based microfluidics is the interaction of droplets of the sample with the cellulose fibres which ensures homogenous distribution by wetting the paper surface, followed by its wicking through the capillary mode of action.42 There are three main types of microfluidic devices. Continuous microfluidic devices allow the continuous movement of samples in the presence of an external force through microporous channels by enhancing the liquid flow without any hindrance.43 The second type is the droplet-based microfluidic device which comprises the motion of 2–3 fluids simultaneously creating droplets that are separated by an immiscible liquid within the micro-carrier channel. These devices have the advantage of generating highly reactive microreactor droplets due to a high surface-to-volume ratio, thus stimulating faster reactions, and performing multiple reactions in a device.44 The third type is a new-generation digital microfluidic device that enables the movement of droplets on a series of planar electrodes. It is widely used due to its high scalability and minimum power utilization, along with a low sample requirement.43 Fabrication of the microfluidic device determines its wettability and biocompatibility. Conventionally, glass and silicone were used for fabrication purposes followed by polymer-based compounds such as polydimethylsiloxane (PDMS) and polymethylmethacrylate (PMMA) to improve the tensile strength and wettability of the device, but paper-based microfluidics has emerged as one of the most promising diagnostic tool for several decades.43 This technology was first established by Whitesides et al. in 2007 as an inexpensive and portable biosensor for the detection of glucose in urine.43 Minimal sample requirement, controlled flow, low-cost fabrication, biocompatibility, stability, and robust capillary action are a few of its advantages.45 In addition, various detection techniques (colorimetric, enzymatic, chemiluminescence, electrochemical) have also been miniaturized into a small chip for rapid analysis.43 Microfluidic devices are found in either 2D or 3D format.4. Advancements in detection techniques
Apart from using the paper in different ways for efficient cancer detection, significant improvement has occurred in functionalizing the surface of the paper for analyzing different types of cancer-specific biomarkers with enhanced sensitivity and specificity. Technologies employing the immobilization of antigen or antibodies for immune-based detection, integration of nucleic acid amplification for nucleic acid detection, and incorporation of advanced recognition elements and conducting layer onto the surface of the paper have been well described in this section (refer to Fig. 2).4.1 Quantitative dot blot assay (QBD)
The identification of cancer biomarkers on a paper-based substrate started with the dot-immunoblot assays.51 The antigen is blotted on the nitrocellulose sheet, which is further exposed to the primary antibody followed by a secondary antibody attached with an enzyme for colorimetric detection. (Fig. 2A) Alternatively, monoclonal antibodies were bound on paper discs.52 Moving from ‘enzyme-labeled’ secondary antibodies, the fluorescently labeled secondary antibodies eliminated the ‘enzymatic-reaction’ step from the assay to identify tumour-associated proteins.53 The dot-immunoblots were superseded by immuno-chromatographic assays (ICAs). A QDB assay can enhance the conventional immunoblot assay to achieve quantitative and high throughput analysis functionality. A QDB plate is engineered in a 96-well plate format with nitrocellulose paper as the binding surface on the base of each well (Fig. 2D). QDB has been used to study and quantify the profile of macrophage-capping protein (CAPG) from the prostate cancer tissue of TRAMP (transgenic adenocarcinoma of the mouse prostate) mice model.54 The speed of the quantitative dot blot assay makes it an ideal option for situations where rapid results are required. The dot blot assays do not require time-consuming steps, e.g., gel electrophoresis or several washes, compared to methods like Western blotting or ELISA. Overall, QDB assays are a valuable tool for rapid, cost-effective, and semiquantitative analysis of target molecules, but they have limitations in terms of dynamic range, specificity, and multiplexing capabilities when compared to other quantitative techniques. Even though they are called quantitative assays, dot blots have become increasingly deemed to be a semi-quantitative assay because their signal intensity is usually compared with an average curve based on known concentrations in target molecules. This may result in variability and errors of quantification, especially at extreme points on the standard curve.554.2 Immunochromatographic assays (ICAs)
In an immunochromatographic assay, the antigen moves upwards through the chromatography nitrocellulose paper strips dipped vertically in the vials and is detected by the antigen–antibody reaction in a similar scheme as the lateral flow assay (Fig. 2B). The ICA can detect binding by the measurement of fluorescence intensity using a laser FL scanner for corresponding analyte concentration evaluation. Since this technique could detect both free analyte and analyte bound to the serum protease, separation of serum from the whole blood can be eliminated.56 Many fluorescently-tagged antibody labels have been developed into lateral flow assays to detect HPV biomarkers for cervical cancer diagnosis and early identification of bile duct cancer that capture IgG antibodies from human serum.57 Detection using ICAs can be improved through precise printing of detector molecules on nitrocellulose sheets using a bio-plasmonic calligraphy system and laboratory syringe pumps.58 Apart from antibodies, nucleic acid can also be used to detect biomarkers in a colorimetric paper assay, such as for the identification of lung cancer biomarker mir21 (microRNA). The poly(vinylidene fluoride) (PVDF) paper is impregnated with a luminescent reporter – poly(3-alkoxy-4-methylthiophene) (PT) along with the complementary strand of mir21 in the form of peptide nucleic acid (PNA). In the presence of a noncomplementary (non-target) sequence, a non-fluorescent duplex due to the electrostatic interaction between non-complementary sequences and PT gets formed. This interaction leads to fluorescence quenching and thus change of colour from orange to purple.59 However, the presence of target DNA leads to the formation of fluorescing triplex (PT–PNA–mir21) of orange color. About 13 strains of HPV have been identified by multiplexed lateral flow assay using DNA-complementarity-based identification instead of antigen–antibody reaction.60 Recently, a pyrrolidinyl peptide nucleic acid (acpcPNA) probe has been engineered into a fluorogenic paper-based biosensor to detect HCV cDNA, which has become a leading cause of hepatocellular carcinoma. The overhang region of the ssDNA significantly amplifies the signal, reducing the limit of detection to as low as 5 pmol.61 Recently, the LFA platform has been extended to develop a new technique of ‘SERS-LFA’(surface-enhanced Raman spectroscopy-based lateral flow assay) where the biosensor uses aptamers as the recognition element to detect thrombin and platelet-derived growth factor-BB (PDGF-BB) from prostate cancer plasma.62 Instead of using antibody or ss-DNA, the lateral flow assay can also be modified with HPV 16 virus-like particles as probes to identify HPV antibodies from serum. Such point-of-care systems are capable of evaluating the vaccination status of a person against cervical cancer.634.3 NAAT–ICA/NAAT–LFA
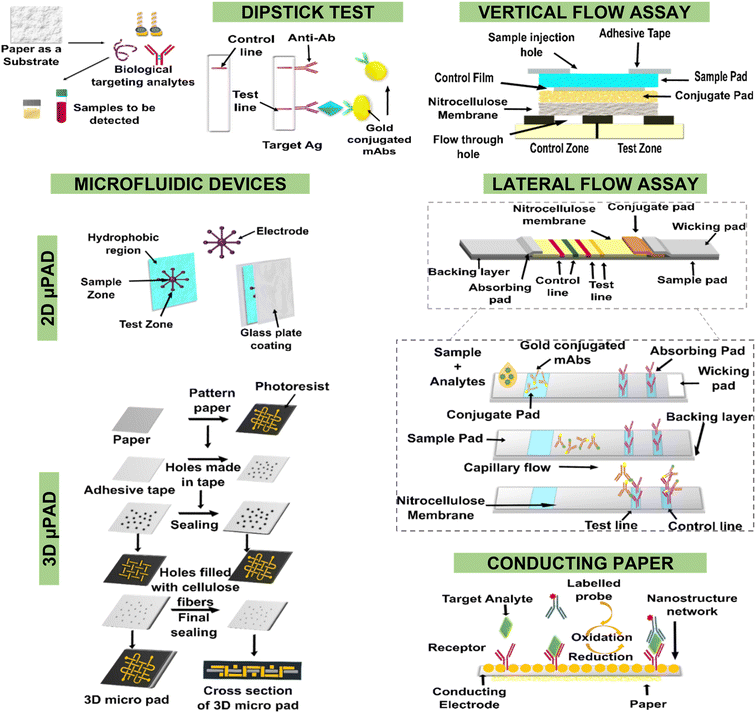
The nucleic acid amplification technique has been integrated with lateral flow assays/immunochromatographic assays to form the nucleic acid amplification test—lateral flow assay (NAAT–LFA) (Fig. 2E). Immunochromatographic tests (ICTs) are being nested with PCR (polymerase chain reaction) for the identification of HPV 16 and 18 DNA. Other amplification techniques such as loop-mediated isothermal amplification (LAMP) have also been integrated with lateral flow dipsticks (LFD) to detect cervical cancer biomarkers. In yet another approach, a combination of isothermal recombinase polymerase amplification (RPA), reverse dot-blot assay (RDB), and lateral flow dipstick (LFD) has been designed to identify various genotypes of HPV viral DNA.64–68 To reduce the time, cost, and requirement of expensive instruments to conduct PCR, interesting strategies using CRISPR–Cas systems are gaining importance. Clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 is a gene-editing technology that employs 2 components: a single guide RNA to detect the target DNA sequence and a CRISPR-associated protein (Cas) endonuclease to cleave the target sequence upon homology recognition.69 Tsou et al. developed a CRISPR–Cas12a system integrated with paper-based LFA for the point-of-care detection of circulating HPV nucleic acids in the plasma of cervical cancer patients. The system has an equal sensitivity with PCR at a relatively lesser cost and time (refer to Fig. 3).70 Such systems have also been immobilized on screen-printed electrodes (like graphene oxide, In2O3–In2S3) for the label-free detection of circulating tumour DNAs.71,72 In another approach, amplification is reached by polymerisation, where the binding of the analyte triggers a controlled free-radical polymerization which enhances the detectable signal in the paper-based device. Different cancer biomarkers such as CEA, AFP, cancer antigen 125 (CA125), and carbohydrate antigen 153 (CA153) have been identified by this method.63,73 Advanced origami paper-based devices modified with a pyrrolidinyl PNA can be used to capture prostate cancer associate (PCA3) biomarker DNA in the clinical samples. Subsequent addition of a G-quadruplex (GQD) DNAzyme reported probe binds to the exposed sequence of PNA-bound PCA3 gene and triggers a hybridization chain reaction to accumulate more GQD onto the device. Peroxidase activity of GQD-hemin oxidizes ABTS substrate and generates deep green color with a detection limit of 0.5 uM PCA3.74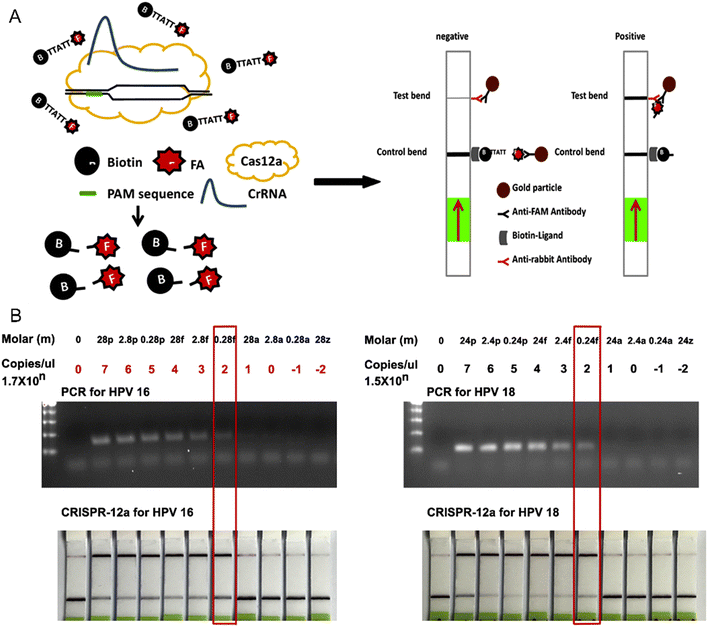 | ||
| Fig. 3 Schematic of the integrated CRISPR–Cas12a system with paper-based lateral flow assay for the detection of circulating nucleic acids (A), and comparative limit of detection of HPV 16 and 18 nucleic acids by PCR and CRISPR–Cas12a system with LFA in spiked samples (B). Figures A and B have been adapted with permission from Tsou et al., Translation Oncology, 2019.70 | ||
4.4 Molecularly imprinted polymers (MIPs)
Molecularly imprinted polymers (MIPs) have emerged as versatile synthetic probe generation tools to recognize any epitopes on the biomarker (Fig. 2F).75 Qi, J. et al. designed a movable valve microfluidic paper-based electrochemical device (Bio-MIP-ePADs), where the electropolymerization of dopamine monomers on paper substrate introduced a ‘biological’ molecularly imprinted polymer layer, which was used to detect CEA with a limit of detection in ng ml−1. The ‘artificial’ synthesis of biorecognition element (dopamine in this case), eliminates the requirement of expensive and relatively unstable antibodies.76 Fluorescent molecularly imprinting conjugated polythiophenes (FMICPs) have been used to develop MIP receptors against AFP and CEA due to their affinity for the boronate present in the synthetic receptor. This paper-based device has achieved the limit of detection in the fg ml−1 range, and, has been further validated with the detection of AFP in the clinical serum sample of liver cancer patients (refer to Fig. 4).77 Imprinting of the MIP layer on screen-printed electrodes has gained attention in the last few years. The electropolymerization of phenol has enabled the synthesis of MIP receptors directly on the paper substrate to recognize 3-nitrotyrosine from urine samples.78 The addition of poly(3,4ethylenedioxythiophene) PEDOT on the screen-printed electrodes before conjugating it with an MIP layer made of Eriochrome Black T (EBT) (as the functional monomer) can further increase the sensitivity of paper-based impedometric biosensors while detecting interleukin-β (a cancer cell proliferating biomarker whose overexpression by the inflammasomes aids in carcinogenesis).79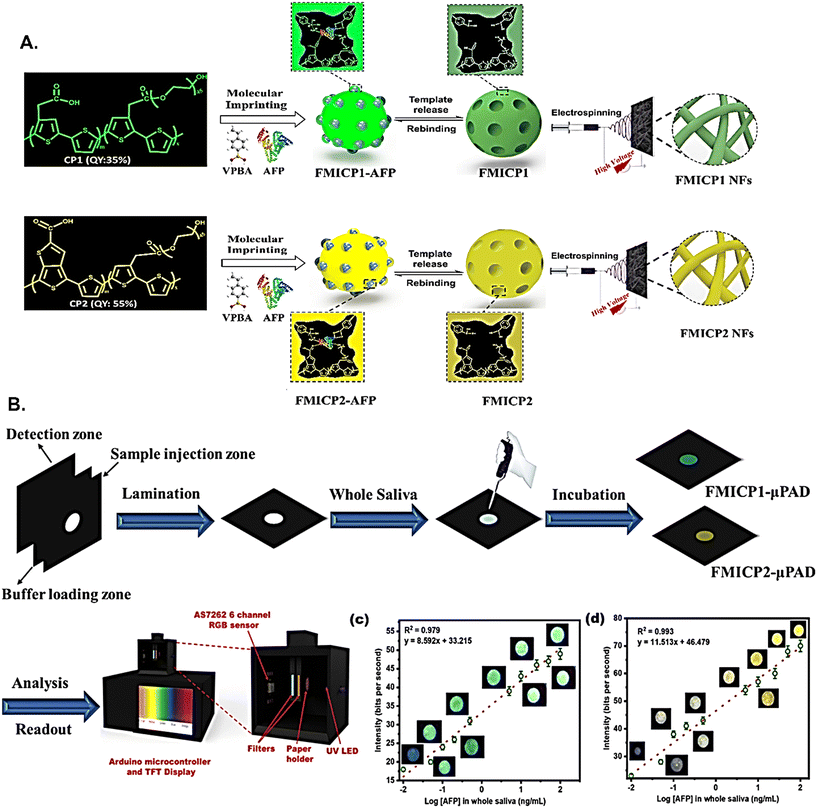 | ||
| Fig. 4 A) Synthesis of molecularly imprinted nanofibres. B) Detection of AFP and CEA cancer biomarkers using MIP-coated μPAD device. Linear correlation between AFP concentration in whole saliva and the color intensity across the photodiode on C) FMICP1-μPAD and D) FMICP2-μPAD sensors. Figures A–D have been adapted with permission from Tawfik et al., Biosensors and Bioelectronic, 2020.77 | ||
4.5 Conducting paper
Conducting paper is an ordinary paper saturated with charged nanomaterials, which increases its conductivity by generating a potential difference across the paper as shown in Fig. 2C.80 It requires the formation of a biological layer incorporated with nanomaterials for interaction with samples, which will be detected in the form of an electrochemical signal using a transducer. Development of paper electrodes requires a three-electrode system comprising a working electrode, a counter electrode, and a reference electrode. All these electrodes are deposited within the paper using conducting inks e.g., carbon inks, silver inks, or gold inks, which enhance the conductivity of the device. For manufacturing paper-based electrodes using plastics, ceramics, and other resources as matrix materials, screen printing fabrication is used.81The development of ‘conducting paper’ has led to many advancements in cancer detection. Paper electrodes are used for the detection of CEA up to 10 pg ml−1 upon immobilization of graphene/gold nanoparticles nanocomposites with high sensitivity, specificity, and stability.82 Such conducting paper strips can be developed by immersing nitrocellulose strips into poly(3,4ethylenedioxythiophene):poly(4-styrene sulfonate) (PEDOT:PSS) polymer followed by immobilization of CEA protein or CEA-aptamers. Any binding event on this conducting polymer layer translates to an electrochemically measurable signal with a high sensitivity of detection (refer to Fig. 5).83,84 Graphene ink or reduced graphene oxide is generally added to the polymer layer to increase the conductivity of paper.84,85 Detection of analyte can also be achieved using phthalocyanine-BODIPY dye to detect CYFRA 21 (a lung cancer biomarker), as it can develop a porous structure on the conducting substrate to allow a change in conductance during analyte passage.86 Recently, the ePADS (electrochemical paper-based analytical device) which uses screen-printed graphene electrodes has been automatized by incorporating dual-flow behaviors in a single paper-based platform. This eliminated the need for multi-step reagent manipulation and has been demonstrated to successfully detect HCV antigen at an LOD of 1.19 pg ml−1 from the serum of patient.87
 | ||
| Fig. 5 Surface functionalization of conductive paper-based biosensor. Figure has been adapted with permission from Yen et al., Sensors, 2020.84 | ||
5. Advancements in signal enhancement with next generation nanomaterials
Integration of a wide variety of nanomaterials to the paper-based LFAs and microfluidic devices is gaining significant importance as they can enhance the biosensor's performance by providing highly active surface, rich chemistry for easy functionalization with multiple biorecognition molecules, inherent optical properties, high electrical conductivity, and surface plasmon-related electromagnetic fields for fluorescence quenching, and color changes.88,89 These nanomaterials have been systematically classified into inorganic, organic, and hybrid and their role in amplifying the colorimetric, fluorescence, and electrochemical-based signal for cancer biomarker detection with enhanced sensitivity and limit of detection has been well explained in this section (refer to Fig. 6).5.1 Inorganic nanomaterial-based biosensors
Inorganic nanomaterials comprise quantum dots, metallic nanoparticles (gold, silver), magnetic nanoparticles, and upconversion nanoparticles. These nanoparticles are attractive for their size and shape-dependent tunable physical (optical, magnetic properties) and chemical properties (stability, inertness).90Abarghoei et al. reported a AuNCs-based Y-shaped microfluidic biosensor for the detection of prostate cancer using citrate as an early biomarker. AuNCs capped with cysteine (Cys–AuNC) possess peroxidase activity that can oxidize 3,3′,5,5′-tetramethylbenzidine (TMB) in the presence of H2O2 to blue color in the absence of citrate. However, in prostate cancer patient samples, the citrate biomarker inhibits the peroxidase activity of Cys–AuNCs and prevents the color change by blocking TMB oxidation.93 A similar approach has also been used for colorimetric aptasensing of CEA over a range of 4 to 25 ng ml−1 with 100% sensitivity and around 97% specificity. The aptasensor employs mercury ions (Hg2+) to block the TMB-reducing capability of AuNPs during CEA detection by the aptamer and in turn imparts a reduction in the blue color.94 AuNRs are also used for cancer detection due to their high surface area, improved signal-to-noise ratio, and high electrical conductivity leading to better electron transfer as compared to AuNPs.95 Electrochemical techniques such as cyclic voltammetry (CV), differential pulse voltammetry (DPV), electrochemical impedance spectroscopy (EIS), amperometry, which sense the analyte based on current change with respect to voltage, change in impedance over a range of frequencies (EIS), change in current with respect to time (amperometry) due to antigen–antibody interactions can also involve the use of AuNRs as biosensing element.96 CV measures current upon linearizing the potential of a working electrode with time. The potential is generally measured on a cyclical basis from two limits that are normally forward and backward.97 On the contrary, DPV involves the use of several possible pulses to an active electrode with low amplitude and short duration coupled onto a linear sweep of impulses.97 In comparison with traditional voltammetry techniques, the resulting current shall be recorded differentially to provide enhanced sensitivity and resolution. CV and DPV, with their distinct advantages and applications, are the most widely used electroanalytical techniques.97 While CV provides information on redox behavior and kinetics for electrochemical reactions in a range of potentials, DPV offers enhanced sensitivity and selectivity, which makes it particularly suited to trace analysis and the quantification of analytes. EIS is also a very powerful technique to analyze the electrical response of an electrochemical system, with regard to applied alternating current signal at various frequencies.98 It provides information on the electrochemical processes occurring on the surface of the electrode and on the properties of the electrolyte solution. Amperometry is used in response to chemical species undergoing oxidation or reduction on an electrode and measures the current at a specific potential. It is frequently used for the detection and quantification of analytes with high sensitivity.99 A special form of gold nanomaterial called gold nanorattles (AuNRTs), which are hollow plasmonic nanostructures, have also been used for the diagnosis of renal cancer by detecting perilipin-2 (PLIN-2) biomarkers. They consist of an AuNP core with a porous and cubic Au shell and exhibit higher refractive index sensitivity as compared to solid nanostructures (such as AuNRs) for localized surface plasmon resonance (LSPR).100
Similar to gold nanoparticles, silver nanoparticles can also be employed in paper-based electrochemical sensors owing to their high biocompatibility and electrical conductivity.91 Nanoporous silver, AuNRs, and AuNP inks can be coated onto cellulose fibres to generate paper working electrodes, that can be immobilized with cancer biomarker-specific primary antibodies for electrochemical detection of antigens like PSA, CEA, and AFP. Metallic nanoparticles and nanocomposites like porous zinc oxide spheres–silver nanoparticles (PZS–AgNPs) composite are also used to amplify biosensing signal upon immobilization with secondary antibody (refer to Fig. 7).101–103
 | ||
| Fig. 7 Schematic of the gold nanorods coated paper-based electrochemical immunosensor containing porous zinc oxide spheres–silver nanoparticles nanocomposites as detection labels (A), the effect of different detection labels on prostate-specific antigen (PSA) concentration-dependent current generation (B), and a calibration curve of PSA using electrochemical immunosensor (C). Schematic of 3-D origami multiplexed electrochemical immunodevice (D), steps involved in its generation using nanoporous silver coating as an electrode, and chitosan-coated nanoporous gold as detection labels for CEA and AFP (E), a calibration curve of CEA (F), and AFP (G). Figures A–C have been adapted with permission from Sun et al., New Journal of Chemistry, 2015.101 Figures D–G have been adapted from Li et al., Chemical Communications, 2013.102 | ||
Even though, gold and silver nanoparticles are predominant choices for electrochemical biosensors, numerous other metals like platinum, copper, nickel, palladium, and iridium also possess significant catalytic properties that can help design low-detection and high-sensitivity sensors. Even though their application has not been explored much in paper-based biosensing, they possess great potential in the detection of oligonucleotides, carbohydrate, and peptide moiety, and as electron transfer intermediates in electrochemical sensors.91,104
Graphene is a 2-dimensional flat sheet of hexagonally arranged carbon lattice. Graphene-based biosensors gained popularity in biosensing applications in the form of graphene-oxide sheets, graphene paper-electrodes, and graphene quantum dots.116 Graphene-oxide modified microfluidic analytical devices were developed for the detection of cancer cells (MCF-7, HL-60, and K562 cells). DNA-aptamer functionalized coloured quantum dots are adsorbed on the graphene paper nanosensor, where a fluorescence signal is detected based on FRET upon biomarker analyte binding.117 Graphene-oxide or reduced graphene oxide nanocomposites have also been used to modify the paper electrodes for electrochemical sensing of PSA, CEA, pancreatic cancer biomarker PEAK1, and cancer antigen 125 (refer to Fig. 8).118–121 Graphene-based nanomaterials can also be used for the development of dot immune-graphene–gold filtration assay (DIGGFA) exploiting the photothermal effect of graphene oxide nanocomposites to detect MCF-7 breast cancer cells, portable 3D-μPAD functionalized with graphene nanocomposites to detect α-fetoprotein, and graphene oxide screen printed electrodes (GPHOXE) to detect circulating tumour DNAs.71,122,123 Apart from the graphene-oxide sheets and graphene paper electrodes, graphene quantum dots are emerging as new ‘nano-biosensor’ probes for cancer detection.124 Graphene quantum dots with multi-walled carbon nanotubes (MW-CNTs) can significantly amplify the sensitivity of detection to up to 0.03 ng ml−1 for the simultaneous quantification of 2 metastatic biomarkers (IL-13Rα2 and CDH-17) from breast and colorectal cancer cell lysate samples and paraffin-embedded tumour tissues within 3 hours.125
 | ||
| Fig. 8 A and B) Paper coated graphene oxide based immunosensor for pancreatic cancer biomarker (PEAK1 tyrosine kinase) detection. Figures A and B have been adapted from Prasad et al., Sensors and Actuators B: Chemical, 2020.119 | ||
CNTs on the other hand are rolled-up large cylindrical hollow tubes of single (single-walled CNTs) or multiple sheets of graphene (multi-walled CNTs).126 Their distinctive features such as special geometrical shape, high conductivity and chemical stability, high aspect ratio, and lightweight with good mechanical strength makes it a suitable candidate for biosensing platform.127 Carbon nanotubes can offer electrical conductivity 100 times higher than copper along with mechanical strength better than that of steel and even superior thermal conductivity than diamond.128 Exemplary properties of CNTs can be imparted onto the paper-based sensors by loading antibody-modified multi-walled CNTs onto micro-porous papers. Any binding event on the surface of the antibody results in a shift of electron transfer in the sensor, leading to a change in resistance that can be recorded in impedimetric sensors.129 These conducting paper electrodes can be a great low-cost yet sensitive alternative to the advanced ionic liquid CNT-modified electrodes, advanced quantum dots attached CNTs based electrochemical sensors, or CNT field-effect transistor based biosensors.129–131 Even a primary antibody-attached multi-wall CNTs modified paper biosensor could impart a noteworthy detection limit of 1.18 ng ml−1 for PSA as compared to commercially available ELISA kits (51 pg ml−1).129 Application of CNTs has also been extended to the detection of human fetuin A (HFA) (a pancreatic and liver cancer biomarker) and miR-141 (prostate cancer biomarker) by functionalizing onto screen printed carbon electrodes (SPEs) and electroactive polymer printed electrodes respectively.132–134 These discovered ‘nano-probes’ of carbon allotropes integrated with paper-substrate can help in achieving a quantifiable and highly sensitive electrochemical/amperometric/fluorometric nano-biosensor with a strong potential for translation in clinical settings. In conclusion, superior conductivity, ease of functionalization, and tunable signal enhancement properties make inorganic nanomaterials extremely important for the development of advanced paper-based biosensors for cancer detection.
5.2 Organic nanomaterial based biosensors
In addition to inorganic nanomaterials, different kinds of organic nanoparticles (polymer dots, liposomes, dendrimers, nanofibres) have also been employed for the paper-based detection of cancer biomarkers by exploiting the unique properties of biomolecules. | ||
| Fig. 9 Schematic of FRET-created traffic light immunochromatographic strip (ICTS) for the cancer biomarker detection (A), photographic images of ICTS detecting different concentration of prostate specific antigen (PSA) biomarker upon excitation under 410 nm UV light (blue panel) and its corresponding fluorescence with a 600 nm long-pass filter (purple panel) (B), and multiplex detection of dual biomarkers (CEA and PSA) using FRET-created ICTS (C). Figures A–C have been adapted with permission from Yang et al., Analytical Chemistry, 2020.142 | ||
5.3 Multi-nanomaterial based hybrid biosensors
Each nanomaterial possesses unique properties that can be utilized to enhance antigen capture, detection, and signal amplification, however in order to enhance the overall sensitivity, specificity, and efficiency of a biosensor, a rational amalgamation of multiple nanomaterials or combination of nanomaterial with special polymers would be ideal. For example, in addition to the use of conducting polymers like PEDOT:PSS for the fabrication of conducting paper, the use of PVA-nanofibres, nanostructured iron oxide, reduced graphene oxide (rGO), and CNTs can increase the sensitivity, specificity, and robustness of a biosensor by improving its mechanical strength, electrochemical activity, and cancer biosensing properties.85,162,164,165 Similarly, a novel ink-jet printed photo paper-based biosensor can be coated with conductive silver nanoparticles (AgNPs) and rGO-based nano ink (Ag/rGO), where Ag/rGO nano-ink was used synergistically with cysteamine doped AuNPs (CysA/AuNPs) as signal amplification labels for the chronoamperometric detection of breast cancer-specific carbohydrate (CA 15-3). This increased the electron transfer efficacy and biosensor efficiency due to the synergistic use of signal amplification labels as well as hybrid nanocomposite of carbon-based (rGO) and inorganic nanomaterials (AgNPs).166–168 GQDs with Au–Pd alloy nanoparticle (NPs) probes can be used to quantify H2O2 released by tumor cells by both photoelectrochemical as well as colorimetric transduction, whereas when used with reduced-graphene quantum dots and chitosan, they can fluorescently detect alkaline phosphatase (ALP)—a bone, breast, and ovarian cancer biomarker.169,170Another interesting hybrid strategy involved the use of graphene–polyaniline modified paper electrodes (G-PANI) for the electrochemical detection of HPV nucleic acid. Anthraquinone-labeled peptide nucleic acid (ss-PNA) probe was functionalized on the G-PANI surface for capturing complementary target DNA of HPV. Upon sample addition, the PNA-target DNA construct blocks the charge transfer from anthraquinone to G-PANI conducting paper surface and reduces the current. This reduction in the current signal correlates with the presence of HPV DNA for cervical cancer diagnosis.171 Xia et al. have also reported a paper-based thin film microextraction system that can detect gaseous benzaldehyde (BA) in human exhalation as a biomarker for Lung Cancer using metal–organic framework (MOF such as NU-901) structures embedded with gold nanorods and quantum dots (GNRs-QDs@NU-901) in dual mode i.e. fluorescence detection as well as SERS signal quantification.110 Therefore, when BA encounters these structures, its aldehyde group interacts with the amine group of 4-mercaptonoaniline present on GNRs and this results in the formation of Schiff base, which in turn, causes disassembly of GNRs and QDs, resulting in fluorescence signal. On the other hand, the formed Schiff base was also used as a SERS reporter for BA detection.110
A hybrid rGO-modified paper electrode coated with branched zinc nanorods (BZR) instead of AuNRs alone can also be used for enhanced immunosensing of α-fetoprotein (a tumor biomarker) due to increased sites for antibody loading, along with the synergistic use of porous zinc oxide spheres–AuNPs (PZS@Au) nanocomposites as signal labels.103,172 A similar strategy including SiO2 nanoparticles, chitosan, and graphene was also integrated into a paper-based microfluidic device for the multiplex detection of four kinds of cancer biomarkers such as CEA, AFP, CA125, and CA153. To remove the complexity due to the presence of labels, a novel label-free microfluidic-based electrochemical (EC) immunosensor was developed for the detection of CEA by coating the working paper electrode with antibody-linked carbon-organic–inorganic (graphene–thionine–AuNPs) hybrid nanocomposites. Thionine being an electrically active substance was employed for current generation due to redox reactions, whereas amino-functionalized graphene was used for signal amplification by accelerating the electron transfer due to its excellent conductivity.82 Further advancements in the same immunosensor have been made for the simultaneous detection of dual cancer biomarkers (CEA and lung cancer-specific neuron-specific enolase (NSE)). Working electrodes were modified with graphene–thionine–AuNPs nanocomposites and Prussian blue (PB)–poly (3,4-ethylenedioxythiophene) (PEDOT)–AuNPs nanocomposites to immobilize CEA aptamer and NSE aptamers respectively and enhance electron transfer. An improved limit of detection of 2 pg ml−1 for CEA and 10 pg ml−1 for NSE was observed along with correlating efficacy with commercially available equipment for detecting biomarkers in clinical serum samples (refer to Fig. 10).116 Since EC sensors are less portable and relatively costly due to the requirement of heavy and costly instrumentations, a novel battery-triggered electrochemiluminescence (ECL-luminescence generated during electrochemical reaction) based immunosensor using dual-signal amplification strategy was developed for the detection of PSA and CEA. It is a microfluidic device with wax-patterned paper (with screen-printed electrodes) employing a hybrid of graphene oxide–chitosan–gold nanoparticle as an immunosensing platform and functionalized nanoporous silver as signal amplification labels.173
 | ||
| Fig. 10 Paper-based electrochemical aptasensor. The working electrode was modified using amino functional graphene (NG)–thionin (THI)–gold nanoparticles (AuNPs) and Prussian blue (PB)–poly (3,4-ethylenedioxythiophene) (PEDOT)–AuNPs nanocomposites for the immobilization of the CEA and the NSE biomarkers as well as to promote electron transfer. Figure has been adapted with permission from Wang et al., Biosensors and Bioelectronics, 2019.116 | ||
Immunoassay-based multi-readout biosensing of cancer biomarkers like PSA can also be achieved through a photothermal biosensing lab-on-a-chip. A PMMA paper hybrid disk consisting of a dissociative clip unit and a magazine bearer can be used to load both thermoresponsive poly(N-isopropylacrylamide)-acrylamide (NIPAM-AcAm) hydrogel and photothermal Fe3O4 nanoparticles probes. Immunocapturing of the antigen cause conversion of Fe3O4 to Prussian blue nanoparticles which in the presence of photothermal laser causes phase separation of the dye with the hydrogel and impart both colorimetric and thermal readout of the antigen sensing (refer to Fig. 11). Such sensors offer great versatility in portable paper-based analytical devices with visual quantitative signal readouts.174 Overall, most of these hybrid nanomaterial-based biosensors are preclinically validated using real samples such as serum, plasma, and urine for the detection of cancer biomarkers, which enhances their possibility for further clinical validation in the future.
 | ||
| Fig. 11 A) Development of photothermal responsive hydrogel loaded poly (methyl methacrylate)/paper hybrid disk (PT-disk) based sandwich immunosensor. B) Schematic representation of the clip and magnetic bearer assembly and tri-mode visual output of immunoassay signal from the PT-disk. Figures A and B have been adapted with permission from Fu et al., Biosensors and Bioelectronics, 2020.174 | ||
6. Different types of samples employed in paper-based cancer diagnosis
Applications of paper-based POC devices are generally studied using various biological samples such as blood, urine, sweat, tears, and saliva (refer to Table 2).| Cancer | Sample | Biomarker | Ref. |
|---|---|---|---|
| Breast | Blood | HER-2 ECD, CEA, CA15-3, LAG-3, MUC1, CA125, CA27.29, BRCA1, BRCA2, NY-BR-1, ING-1, ER/PR, miRNA-17-5p, miR-155, miRNA-222, miRNA-548b-5p, miR-532-502, 5 miRNA panel (let-7b-5p, miR-122-5p, miR-146b-5p, miR-210-3p, miR-215-5p), 3 miRNA panel (miR-21-3p, miR-21-5p, miR-99a-5p) | 194–203 |
| Lung | Blood | NY-ESO-1, CEA, CA19-9, SCC, CYFRA21-1, NSE, EGFR, PD-1, CEA, KRAS, ALK (anaplastic lymphoma kinase), miR-339-5p, miR-21, miR-141, miR-145, miR-210-3p, miRNA-31, 6 miRNA panel (miR-19b-3p, miR-21-5p, miR-221-3p, miR-409-3p, miR-425-5p, miR-584-5p), 3 miRNA panel (miR-28-5p, miR-362-5p, miR-660-5p), 3 miRNA panel (miR-16, miR-205, miR-486), 4 miRNA panel (miR-146b, miR-205, miR-29c, miR-30b), 3 miRNA panel (miR-21, miR-210, miR-486-5p) | 204–214 |
| Liver | Blood | PD-1, AFP, CEA, miR-224, miRNA-200 lncRNA-TSIX, miR-548-a-3p, SOGA1, miRNA-122a, 3 miRNA panel (miR-122, miR-148a, miR-1246) | 215–219 |
| Ovarian | Blood | CA125, CA15-3, HER2, BRCA1/2, AFP, hCG, p53, CEA, CA549, CASA, CA19-9, MCA, MOV-1, TAG72, KRAS | 220–222 |
| Gastric | Blood | CEA, HER-2 ECD, LAG-3, CA72-4, CA19-9, miR-101, miRNA-22-3p, miR-196a/b, 2 miRNA panel (miR-19b-3p, miR-106a-5p), 2 miRNA panel (miR-21, miR-222), 4 miRNA panel (miR-21, miR-93, miR-106a, miR-106b), 3 miRNA panel (miR-221, miR-376c, miR-744) | 223–232 |
| Colorectal | Blood | EGFR, CEA, KRAS, miRNA-141, miR-338-5p, miR-497, 2 miRNA panel (miR-1290, miR-320d), 7 miRNA panel (miR-103a-3p, miR-127-3p, miR-151a-5p, miR-17-5p, miR-181a-5p, miR-18a-5p, miR-18b-5p), miRNA panel (miR-144-3p, miR-425-5p, miR-1260b) | 233–240 |
| Pancreatic | Blood | PD-1, CEA, CA19-9, CA125, TIM-3, KRAS, hsa-miR-21-5p, miR-221-3p, microRNA-100, miR-21-5p, miR-182, 2 miRNA panel (miRNA-196, miRNA-200 with CA19-9), 6 miRNA panel (let-7b-5p, miR-192-5p, miR-19a-3p, miR-19b-3p, miR-223-3p, miR-25-3p), 2 miRNA panel (miR-10b, miR-30c) | 241–248 |
| Bladder | Blood | BAT, FDP, NMP22, HA-Hase, BLCA-4, CYFRA 21-1, miR-155 | 220–222, 249 |
| Urine | 2 miRNA panel (miR-93-5p, miR-516a-5p), 4 miRNA panel (let-7b-5p, miR-149-5p, miR-146a-5p, miR-423-5p) | 250, 251 | |
| Prostate | Blood | PSA, PAP, 3 miRNA panel (miR-145, miR-148, miR-185) | 221, 222, 252 |
| Esophageal | Tumor tissue | SCC | 221 |
| Blood | 6 miRNA panel (miR-106a, miR-18a, miR-20b, miR-486-5p, miR-584, miR-223-3p) | 232 | |
| Endometrial | Blood | CA 125, 6 miRNA panel (miR-143-3p, miR-195-5p, miR-20b-5p, miR-204-5p, miR-423-3p, miR-484) | 222, 253 |
| Diffuse large B-cell lymphoma (DLBCL), chronic lymphocytic leukemia (CLL) | Tumor tissue and blood | PD-L1 | 220 |
| Osteosarcoma | Tumor tissue | TIM-3, miR-663a, miR-139-5p, miR-101 | 222, 254–256 |
| Chronic myeloid leukemia | Blood and bone marrow | BCR-ABL1 | 221 |
| Non-Hodgkin's lymphoma | Blood | let-7a, miR-124 | 249 |
| Acute myeloid leukemia | Tumor tissue, blood | FLT3, IDH1/2, DNMT3A, NPM1, SRSF2, let-7a, miR-124 | 249, 257, 258 |
| Melanoma | Tumor tissue | PD-1, GP100, MAGE-A, Tyrosinase, NY-ESO-1 | 222 |
| Testicular | Blood | Alpha-fetoprotein (AFP), β-human chorionic gonadatropin, CAGE-1, ESO-1 | 221, 222 |
| Glioma | Tumor tissue, cerebrospinal fluid | IDH1, IDH2, MGMT, PPM1D, H3F3A, | 259–261 |
| Blood | miR-182, 3 miRNA panel (miR-19a-3p, miR-106a-5p, miR-181b-5p) | 262, 263 |
6.1 Blood samples
Blood has accessibility to all bodily sites, hence it can help diagnose different malignancies through abnormal tissue-specific pathological signatures. The complex nature of blood due to the presence of different kinds of cells, salts, and proteins affects the final analysis using sophisticated instruments due to volume constraints post plasma or serum separation.175 Therefore, paper-based biosensors are employed due to minimal plasma or serum sample requirements. Microfluidic devices can diagnose breast cancer by detecting human epidermal growth factor receptor (HER2) in serum samples with a 1 pM limit of detection.176 Similarly, μPADs employ gold nanoparticle-coated electrodes to detect cancer antigen-125 (CA-125) in blood samples for the diagnosis of ovarian cancer. Gold nanoparticles-based lateral flow assays are also developed for the detection of humoral hypercalcemia of malignancy (HHM) by sensing nanograms levels of parathyroid hormone-like hormone (PTHLH) secreting factor in human serum samples.177 PDGF-BB and thrombin can be detected in human serum spiked samples using LFA-aptasensors.178Since, the increased concentration of fibrinogen in serum samples is a biomarker of advanced hepatocellular carcinoma, a point-of-care paper-based biosensor to quantify plasma fibrinogen concentration has been recently developed.179 Thrombin is added to the paper strips followed by the addition of the plasma samples containing known concentrations of fibrinogen. The reaction between thrombin and fibrinogen leads to differential eluted lengths when placed in elution baths, which is inversely correlated to the fibrinogen concentration.180
6.2 Urine samples
Urine is a commonly collected biological fluid for the detection of various diseases because of the presence of various analytes (proteins, cells, hormones, drugs, bilirubin, glucose, albumin, creatinine,). It has a broad pH range of 4.5–8.0 and a viscosity of 0.6–1.2 mPa s. As compared to blood, it requires fewer sample processing steps, but in a few cases, it requires processing such as buffer dilution to remove sample turbidity and pH neutralization as urine's acidic pH might affect the reagent or antibody coated onto the paper.181 Urine contains various cancer biomarkers such as sarcosine and citrate for prostate cancer, nuclear matrix protein number 22 (NMP22) for bladder cancer, neutrophil gelatinase-associated lipocalin (NGAL), matrix metalloproteinase 9 (MMP-9), and a disintegrin and metalloprotease-12 (ADAM-12) for breast cancer, HPV (human papillomavirus) E6 oncoprotein for cervical cancer, and perilipin-2 for renal cancer.93,100,182,183 Therefore, plasmonic gold nano rattles coated and cysteine-capped gold nanoclusters coated paper-based microfluidic devices were explored for the detection of perilipin-2 and citrate, respectively.93,100 A commercial paper-based OncoE6™ cervical test is also available for the detection of HPV16-E6 oncoprotein present in urine, responsible for cervical cancer.182 Synthetic urinary biomarkers have also been reported for the paper-based detection of colorectal cancer by developing protease-sensitive peptide-coated nanoworms (NWs) with reporter probes. MMP-9 released by colorectal cancer cells cleaves the protease-sensitive linkages and facilitates the release of fluorescent probes into the urine for subsequent detection (refer to Fig. 12).184 Despite many advantages associated with the high usage of urine samples for detection purposes, sometimes due to contamination, incorrect collection as well as presence of interfering agents can result in pre-analytical errors.185 | ||
| Fig. 12 Schematic of the synthetic urinary biomarker and steps involved in its in vivo processing to excrete detectable analyte in urine (A) and its subsequent paper-based detection (B). Figures A and B have been adapted with permission from Warren et al., PNAS, 2014.184 | ||
6.3 Other samples
Salivary samples can be used due to the presence of various cancer biomarkers such as IL-6, IL-8, CYFRA-21-1, sCD44, etc.186–189 CD44 biomarker in saliva-containing oral rinse of oral cancer patients was detected using a colorimetric lateral flow assay.190 Saliva has also been used for the detection of cancer using various non-paper-based biosensors.186,191 The advantages of using saliva involve its non-invasive nature along with low sample processing costs. However, the disadvantages include the chances of interference due to various other substances present in saliva such as food, mucins, bacteria, and the presence of biomarkers in lesser quantity as compared to blood or urine.192 To detect oral and oropharyngeal cancer from oral rinses, a paper-biosensor with salivary biomarker (CD44) test strips immobilized with anti-CD44 monoclonal antibodies has been developed.190Swab samples obtained from the internal lining of the wound, respiratory tract, and cervix, consisting of mucous, cancer cells or biomarkers, and other components, can also be used for cancer detection. Multiple studies have reported the paper-based detection of cervical cancer biomarkers (HPV 6, 11, 16, and 18) in cervical swabs using lateral flow assays.67 The commercial device OncoE6™ developed for cervical cancer diagnosis through urine can also employ vaginal and cervical scrapes from the patients.182 Detection using swabs and scrapes is an inexpensive way to collect specimens for cancer testing. However, chances of infection and patient discomfort are the major concerns.
Cancer tissues can also be directly used for diagnosis after extraction and amplification of the DNA from the tissue specimen. Klapperich et al. reported a colorimetric and lateral flow paper fluidic device that detects HPV 16 DNA from cervical tissue specimens.193 In addition, various samples such as tears, sweat, and VOCs have gained major importance because of rapid analysis due to their direct usage for detection without any sample processing, but fast evaporation and low sample volume can also lead to false results. Their application for cancer detection is majorly restricted to sophisticated techniques to date and needs further advancements. In conclusion, the selection of samples for biosensing should be dependent on factors such as a higher abundance of biomarker-of-interest, non-invasive nature of the sample collection, low cost, lesser volume, and lesser sample processing.
7. Clinical validation of paper-based cancer diagnostics
In vitro diagnostics (IVDs) are generally classified as per medical device regulatory norms. Based on the level of risk involved, cancer-specific IVDs can be categorized into class III (classification system, class I–III) according to the Food and Drug Administration (FDA, USA) and class C (classification system, class A–D) as per the European Union ‘In Vitro Diagnostic Medical Devices Regulation (IVDR)’ and Central Drugs Standard Control Organization (CDSCO, India). All such devices are evaluated on their analytical performance, including biases or device inaccuracy, imprecision, analytical specificity, and sensitivity. In vitro diagnosis requires validation using clinical samples obtained directly from the patient to establish its efficacy. Multiple paper-based in vitro diagnostics are clinically approved for cancer detection.264 Transferrin detection kit and OncoE6™ test specific for colon cancer-specific transferrin and cervical cancer-specific HPV (16 and 18) E6 oncoprotein are a few such examples (refer to Table 1).182,265 In addition, numerous paper-based devices based on conventional and advanced approaches have been validated using clinical samples from cancer patients as a proof-of-concept. A comprehensive table of such devices has been provided below (refer to Table 3). Successful clinical translation of these devices requires strict adherence to the guidelines provided by the specific regulatory agencies.| Type | Cancer biomarker | Cancer type | Sample | Sensing molecule | Detection technique | Sample volume | LOD | Assay time | Ref. |
|---|---|---|---|---|---|---|---|---|---|
| Dipstick | Transferrin (Tf) | Colon cancer | Patient's faeces | Transferrin test kit | Colorimetric | NR | NR | <5 min | 265 |
| Exosomes | Hematological cancers, melanoma | Cell supernatants, plasma, urine | Gold nanoparticles | Colorimetric | 100 μl | 8.54 × 105 exosomes per μl | 15 min | 266 | |
| Lateral flow assay | HPV 16 and 18 E6 oncoprotein | Cervical cancer | Patient's urine, vaginal and cervical scrapes | OncoE6™ cervical test kits | Colorimetric | Urine (5 ml), vaginal sample (6 ml) | NR | 2 ½ hours | 182 |
| Ramos cells | B-cell, human Burkitt's lymphoma | Ramos cells spiked blood | Gold nanoparticles-aptamer conjugates | Colorimetric | 80 | 800 Ramos cells | 15 min | 267 | |
| Micro RNR's miR-210 and miR-424 | Breast cancer, gastrointestinal cancer | NR | Gold nanoparticle | Colorimetric | 50 μl | 10 pmol | NR | 268 | |
| Alpha-fetoprotein (AFP) | Liver cancer | Patient's serum | Gold nanoparticle | Colorimetric | 100 μl | 0.1 ng ml−1 | 10 min | 269 | |
| HPV-16 & HPV-18 DNA | Cervical cancer | Patient's plasma | CRISPR–Cas12a | Fluorescence | 25 μl | 0.24fM | 3 hours | 70 | |
| Parathyroid hormone-like hormone | Humoral hypercalcemia of malignancy | Patient's serum | Gold nanoparticles | Fluorescence | 150 μL | 1.42 ng ml−1 | 15–20 min | 177 | |
| Carbohydrate antigen CA19-9 | Pancreatic and colorectal cancer | Patient's plasma | Antibody conjugated gold nanoparticles | Colorimetric | 100 μL | 5 U ml−1 | 5 min | 270 | |
| Perilipin-2 (PLIN-2) | Renal cancer | Patient's urine | Anti-PLIN-2 linked gold nanorattles | Localized surface plasmon resonance | Few drops of urine | NR | 2 hours | 100 | |
| HPV DNA | Cervical cancer | Patient's cervical tissue | Antibody against amplified DNA | Colorimetric | 6 μL | 104 DNA copies | 1 hour | 193 | |
| CD44 | Oral cancer | Patient's oral rinses | Antibody against sCD44 | Colorimetric | 5 mL | NR | 10 min | 190 | |
| HPV 6, 11, 16, and 18 DNA | Cervical cancer | Patient's cervical tissue swabs | Fluorescently labelled DNA | Fluorescence | 5 μL | 10–102 plasmid DNA per μL | 30 min | 67 | |
| Volatile benzaldehyde | Lung cancer | Patient exhaled breath | Gold nanorods and quantum dots embedded metal organic framework structures (GNRs-QDs@NU-901) | Fluorescence and SERS signal | 1000 mL (∼500 mL × 2) | 1.2–0.1 ppb | 5 min | 110 | |
| Vertical flow assay | CEA, alpha-fetoprotein (AFP), Cancer antigen 199 | Multiple cancer types | Patient's serum | FITC labelled anti-CEA, anti-AFP, anti-CA199 antibodies | Fluorescence | 10 μL | CEA (0.03 ng ml−1), AFP (0.05 ng ml−1), CA199 (0.09 ng ml−1) | 220 s | 38 |
| Prostate-specific antigen (PSA), CEA, AFP | Prostate cancer | Patient's serum | Raman dyes (RDs) encoded core–shell SERS nanotags | Fluorescence | 30 μL | 0.37, 0.43, and 0.26 pg ml−1 for PSA, CEA, and AFP | 7 min | 39 | |
| μPADs | Cancer antigen (CA-125) | Ovarian cancer | Patient's blood | Gold nanoparticles | Electro-chemical | 5 μl | 3500 U ml−1 | NR | 49 |
| CA-125 | Ovarian cancer | CA-125 spiked human plasma | Silver nanoparticles/reduced graphene oxide (Ag/RGO) nano-ink and cysteamine caped gold nanoparticles (CysA/Au NPs) | Electro-chemical | 10 μl | 0.78 U ml−1 | 1 hour | 166 | |
| DNA sequences | Chronic myelogenous leukemia | Patient's blood | Cadmium selenide quantum dots with complementary DNA probes | Electro-chemical | NR | 10−15 M | NR | 271 | |
| CEA, Her-2/Neu (C-erbB-2) | Colon and breast cancer | Patient's serum and saliva | CdSe/ZnS quantum dots bio-conjugated labels | Fluorescent | NR | CEA (0.02 ng ml−1), Her2/Neu (0.27 ng ml−1) | 27 min | 272 | |
| 273 | Oral Cancer | Patient's saliva | G-rich target recognition sequence attached onto gold electrode | Electro-chemical | 5 μl | 0.02 fM | 1.5–2 hours | 273 | |
| miRNA 21 | Breast cancer | Patient's blood | Molecular beacon probes | Fluorescence | 50 μl | 0.01 μM | 30 min | 274 | |
| AFP, CEA, carbohydrate antigen 125 (CA125), CA153 | Colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Antibody coated graphene and chitosan modified screen printed paper electrode | Electro-chemical | 2.0 μL | 0.005 ng ml−1 | 5 min | 275 | |
| PSA, CEA | Prostate, colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Antibody linked graphene oxide-chitosan/AuNPs nanocomposite coated on screen printed paper electrode | Electro-chemiluminescence | 4.0 μL | 0.8 pg ml−1 | 15 min | 276 | |
| ROR1+ cancer cells | Haematological cancer (chronic lymphocytic leukemia, B-cell acute lymphoblastic leukemia) | Buffy coat blood samples | Antibody-conjugated fluorescent microparticles | Fluorescence | 10 μL | 0.1 cells per μl | <10 min | 277 | |
| Thioredoxin-1 | Breast cancer | Blood serum samples | Anti-Trx-1 antibody-conjugated with HRP | Colorimetric | 100 μL | 0–200 ng ml−1 | 14 min | 50 | |
| Conducting paper electrodes | CEA | Colorectal, lung, gastric, pancreatic, cervical cancer | Patient's serum | Anti-CEA linked graphene/thionine/gold nanoparticles (AuNPs) nocomposite coated on screen printed paper electrode | Electro-chemical | NR | 10 pg ml−1 | NR | 82 |
| CA 125 antigen | Ovarian cancer | CA 125 spiked in p plasma | Ag–DPA–GQD/CysA–Au NPs Cysteamine coped gold nanoparticles (CysA/Au NPS) | Electro-chemical | NR | 15 U ml−1 | 2 min | 278 | |
| Cancer antigen 125 (CA125) | Ovarian cancer | Patient's serum | Antibody tagged nanocomposites (reduced graphene oxide/thionine/AuNPs) | Electro-chemical | NR | 0.01 U ml−1 | 20 min | 279 | |
| Human epidermal growth factor receptor (HER2) | Breast cancer | HER2 spiked undiluted serum | Aptamers immobilized on gold micro-electrode arrays on silicon/silicon oxide wafers | Electro-chemical | 50 μl | 1 pM | 30 min | 176 | |
| CEA | Colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Anti-CEA taggedPEDOT:PSS – CNT nanocomposite | Electro-chemical | NR | 2 ng ml−1 | 15 min | 164 | |
| CEA | Colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Anti-CEA tagged PEDOT:PSS – iron oxide (Fe2O3) nanocomposite | Electro-chemical | NR | 4 ng ml−1 | 10 min | 165 | |
| CEA | Colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Anti-CEA tagged reduced graphene oxide (rGO) | Electro-chemical | 10 μl | 1 ng ml−1 | 20 s | 280 | |
| CEA | Colon, lung, ovarian, pancreatic, liver and breast cancer | Patient's serum | Anti-CEA tagged PEDOT:PSS – polyvinyl alcohol nanofibers | Electro-chemical | 10 μL | 0.2 ng ml−1 | 10 min | 162 | |
| AFP | Hepatocellular carcinoma | Patient's serum | Antibody linked branched zinc nanorods modified rGO | Electro-chemical | 20 μL | 0.08 pg ml−1 | 10 min | 172 | |
| PSA | Prostate cancer | Patient's serum | Antibody linked gold nanorods | Electro-chemical | 5.0 μL | 1.5 pg ml−1 | 1.5 hours | 101 | |
| CYFRA 21-1 | Lung Cancer | Patient's blood | Phthalocyanine-BODIPY dye | Electro-chemical | 1 mL | 0.20 pg ml−1 | 10 min | 86 |
8. Challenges and future prospects
Paper-based diagnosis has come a long way from glucose testing as a basic dipstick assay to advanced disease detection like cancer, COVID-19, etc. Advancements in the paper substrate, employing it in multiple assay formats along with the inclusion of advanced nanomaterial-based detection and signal enhancement techniques have enhanced the sensitivity and specificity of paper-based detection of cancer.Although LFA allows easy detection, the laborious sample preparation and multiplex detection limit its use. VFA conquers these limitations by allowing the vertical flow of samples for enhanced interaction and multiple detections.281 Microfluidics devices have emerged as a remarkable tool for POC detection by integrating small devices on laboratory chips with homogeneous mixing of samples.20 The progress in screen printing of paper electrodes has offered rapid analysis at the nanoscale with high sensitivity.281,282
However, a few bottlenecks such as inadequate limit-of-detection, the need for an extensive pre-processing of the sample before usage, the lesser scope for quantification of signal, fewer types of detectable samples (particularly a challenge for whole cell detection), use of a single biomarker for detection, and instability of detecting biomarkers on a paper-substrate can limit their use to only primary screening in clinical settings.37,283,284 Different approaches in terms of enhancing the device design as well as constituent detecting molecules from qualitative to quantitative detection are being explored. Upgradation of conventional qualitative assays like dot blots to quantitative formats (quantitative dot blot) and the use of newer nanomaterials (such as upconversion nanoparticles and gold nanorattles) can significantly enhance the limit-of-detection.54,100,113 Advancements in user-friendly detection strategies like colorimetric can be achieved through the impregnation of metal–organic frameworks (MOFs) like NH2-MIL-125(Ti) in the paper or by using thermochromic fibre papers by impregnating organic dye functionalized SiO2 nanoparticles for the photothermal detection of analytes in ng ml−1 range.285,286 Visual colorimetric readouts can also be coupled with electrical output using flexible piezoelectric pressure sensors of a hybrid conductive material composed of multiwalled carbon nanotubes (MWCNTs) and polyaniline urchin-like hollow spheres (PANI-UHSs) and an elastic thermoplastic polyurethane (TPU) substrate.287 Functionalizing paper electrodes with multiwalled carbon nanotube in a pressure-tight system can impart great sensitivity to pressure changes induced by analyte detection byproduct gas and impart good resistance with a detection limit of <200 pg CEA per ml.288 Introduction of conducting paper that employs the printing of carbon-based electrodes, photoelectric materials (NaNbO3 nanostructures, g-C3N4 quantum dots modified TiO2 nanospheres, Bi2O2S (BOS) nanosheets), or conducting polymers on paper can provide a wider scope of quantification through electrochemical detection in comparison to conventional colorimetric and fluorometric strategies.289–291 The amalgamation of such materials possessing unique properties can improve the limit of detection to even femtogram per ml. Nano-inks developed from inorganic-carbon-conducting polymer composites are one such example.167 Miniaturization and technological advancements in the detection process by integrating mobile cameras for easy fluorescence readout instead of bulky fluorescence equipment and the development of battery-triggered electrochemiluminescence readers can be very beneficial for robust and portable applications.276,292
Capturing cancer-derived exosomes is an emerging way of detecting malignancies through liquid biopsies like blood or urine.293,294 Gold nanoparticles tethered aptamer-based capturing of exosomes and multi-directional hybridization chain reaction using hairpin-like DNA unit 1/2 to recruit multiple signal molecules can enable electrochemical signal amplification and aptasensing of as low as 283 exosomes per μl of sample.293 Nevertheless, overcoming the bottlenecks like negating the false positives by identifying the exosomes secreted by normal cells, reduction of sample volume, and exosome stability during analysis would help unleash the true potential of these biosensors for clinical sample analysis.295 Genetic biomarker sensing would be extremely important for personalized cancer treatment, therefore, detecting cancer-specific genetic biomarkers like the BRCA1 gene from breast or ovarian cancer patients, methylguanine methyl transferase (MGMT), IDH1/2 from glioma patients, FLT3, DNMT3A from acute myeloid leukemia, etc. would be the way forward to develop next-generation paper-based diagnostics.296,297 Electrochemical biosensors can be tuned to detect the BRCA1 mutated cancer patients by capturing BRCA1 single-stranded DNA (ssDNA) using complementary DNA adhered lipid membrane-coated gold nanoparticles. Anchoring this complex onto a 2D-layer of graphene analogue i.e. MXene can several fold enhance the ssDNA detection signal and increase the limit of detection to 1zM (zeptomolar).298 Electrochemical-based biosensing of biomarker DNA up to 1.2 pM can be achieved by integrating CRISPR–Cas12a in a portable biosensor with a smartphone-based signal read for resource-limiting conditions. Semiconductors like In2O3–In2S3 can be used to modify the paper substrate for photoactivation, which can generate photocurrents based on differential G-quadruplexes cleavage by activated target DNA-bound CRIPR–Cas12a.72
To make these biosensors more user-friendly and compact, research is being driven toward making ‘all-in-one’ paper-based devices. It focuses on capturing and amplifying target nucleic acid present in the sample directly on paper and integrating it with the detection system using paper fluidics.193 Paper fluidics approach can reduce the detection time to 1 hour and also open the doors to capture more varied types of ‘biomarkers’ on paper, for instance, capturing cancerous cells directly.193,277 Since the conventional biorecognition elements such as antibodies, DNA, and PNA, are relatively less stable on paper-substrate, new bio-recognition elements such as synthetically injectable and tunable markers and MIPs are gaining importance.75,184 In addition, research focuses on modifying the paper substrate itself through the generation of nitrocellulose–polymer composites can also address the above-mentioned limitations, including the requirement of flexible substrates to capture intact cells and the printing of electrodes for enhanced detection.5,299 All these new strategies have shown promising results as a proof-of-concept, however, the efficacy of these paper-based techniques as point-of-care diagnostics will truly be validated upon testing in a large cohort of patients in clinical trials and real-world settings. Overall, paper-based sensors offer promising future applications in personalized therapy for cancer by enabling: real-time monitoring of disease biomarkers and treatment responses, facilitating immediate adjustments to therapy, augmenting precision medicine approaches through personalized molecular profiling and targeted therapy selection, and early detection of disease recurrence or treatment resistance. Integration of data analytics and machine learning to the paper-based cancer biosensing can tremendously help identify clinical biomarkers, their rapid and minimally invasive detection, and provide personalized therapy decisions in a highly cost-effective manner.300
Author contributions
Prateek Bhardwaj: conceptualization, investigation, project administration, supervision, validation, visualization, writing – original draft, writing – review & editing. Survanshu Saxena: investigation, writing – original draft. Bharti Arora: investigation, writing – original draft. Subhasini Singh: investigation, writing – original draft. Pranoti Palkar: investigation, writing – review & editing. Rinti Banerjee: supervision, resources. Jayant Sastri Gota: supervision, validation, resources, writing – review & editing.Conflicts of interest
The authors declare no conflict of interests.Acknowledgements
All the authors would like to acknowledge Prof. Rinti Banerjee (whom we lost during COVID-19 pandemic) for her guidance and scientifically grooming early career researchers like Dr. Prateek Bhardwaj, Survanshu Saxena, Bharti Arora, and Subhasini Singh.References
- N. Emrizal, Z. Mohd Zain and K. G. Heah, Asia-Pac. J. Mol. Biol. Biotechnol., 2023, 31(2), 62–70 Search PubMed.
- K. J. Land, S. Smith and R. W. Peeling, in Paper-based Diagnostics: Current Status and Future Applications, ed. K. J. Land, Springer International Publishing, Cham, 2019, pp. 1–21, DOI:10.1007/978-3-319-96870-4_1.
- M. Sher, R. Zhuang, U. Demirci and W. Asghar, Expert Rev. Mol. Diagn., 2017, 17, 351–366 CrossRef CAS PubMed.
- K. Ratajczak and M. Stobiecka, Carbohydr. Polym., 2020, 229, 115463 CrossRef CAS PubMed.
- H. Shafiee, W. Asghar, F. Inci, M. Yuksekkaya, M. Jahangir, M. H. Zhang, N. G. Durmus, U. A. Gurkan, D. R. Kuritzkes and U. Demirci, Sci. Rep., 2015, 5, 8719 CrossRef PubMed.
- B. Asci Erkocyigit, O. Ozufuklar, A. Yardim, E. Guler Celik and S. Timur, Biosensors, 2023, 13(3), 387 CrossRef CAS PubMed.
- A. Dukle, A. J. Nathanael, B. Panchapakesan and T.-H. Oh, Biosensors, 2022, 12(9), 737 CrossRef CAS PubMed.
- C. Pereira, C. Parolo, A. Idili, R. R. Gomis, L. Rodrigues, G. Sales and A. Merkoçi, Trends Chem., 2022, 4, 554–567 CrossRef CAS.
- A. A. Shalaby, C.-W. Tsao, A. Ishida, M. Maeki and M. Tokeshi, Sens. Actuators, B, 2023, 379, 133243 CrossRef CAS.
- M. Y. Azab, M. F. O. Hameed and S. S. A. Obayya, Biology, 2023, 12, 232 CrossRef CAS PubMed.
- K. Ratajczak and M. Stobiecka, Carbohydr. Polym., 2020, 229, 115463 CrossRef CAS PubMed.
- N. Yonet-Tanyeri, B. Z. Ahlmark and S. R. Little, Adv. Mater. Technol., 2021, 6(8), 2001138 CrossRef CAS PubMed.
- R. H. Tang, M. Li, L. N. Liu, S. F. Zhang, N. Alam, M. You, Y. H. Ni and Z. D. Li, Cellulose, 2020, 27, 3835–3846 CrossRef CAS.
- X. Deng, N. M. Smeets, C. Sicard, J. Wang, J. D. Brennan, C. D. Filipe and T. Hoare, J. Am. Chem. Soc., 2014, 136, 12852–12855 CrossRef CAS PubMed.
- C. T. Yew, P. Azari, J. R. Choi, F. Li and B. Pingguan-Murphy, Anal. Chim. Acta, 2018, 1009, 81–88 CrossRef CAS PubMed.
- K. Kim, H. A. Joung, G. R. Han and M. G. Kim, Biosens. Bioelectron., 2016, 85, 422–428 CrossRef CAS PubMed.
- J. A. Jenkins, L. Dube, Y. Luo, J. Chen, T. H. Fan, Y. Lei and J. Zhao, Sens. Actuators, B, 2018, 262, 493–498 CrossRef CAS.
- S. S. Acimovic, M. A. Ortega, V. Sanz, J. Berthelot, J. L. Garcia-Cordero, J. Renger, S. J. Maerkl, M. P. Kreuzer and R. Quidant, Nano Lett., 2014, 14, 2636–2641 CrossRef CAS PubMed.
- T. Brandstetter, S. Bohmer, O. Prucker, E. Bisse, A. zur Hausen, J. Alt-Morbe and J. Ruhe, J. Virol. Methods, 2010, 163, 40–48 CrossRef CAS PubMed.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- M. F. Syful Azlie, M. R. Hassan, S. Junainah and B. Rugayah, Med. J. Malays., 2015, 70, 24–30 CAS.
- S. Wang, X. Wang, S. Tang, Y. Chen, J. Jia, J. Chen, H. Shen and B. Bai, Lab. Med., 2019, 50, 130–137 CrossRef PubMed.
- A. T. Singh, D. Lantigua, A. Meka, S. Taing, M. Pandher and G. Camci-Unal, Sensors, 2018, 18(9), 2838 CrossRef PubMed.
- K. M. Koczula and A. Gallotta, Essays Biochem., 2016, 60, 111–120 CrossRef PubMed.
- B. W. Liu, D. Du, X. Hua, X. Y. Yu and Y. H. Lin, Electroanalysis, 2014, 26, 1214–1223 CrossRef CAS.
- https://ctkbiotech.com/products/?filters=product_cat[biomarkers-rapid-tests] .
- https://www.alere.com/en/home/products-services/product-lookup.html .
- https://www.arborvita.com/avproducts/ .
- https://www.quickingbio.com/ .
- https://www.bio-techne.com/diagnostics/lateral-flow-assay-development .
- https://www.rapidtest.com/index.php/Tabs/tabs/index.php?product=Cancer-Rapid-tests&cat=22 .
- https://www.lifesignmed.com/product-catalog/urology/status-bta .
- https://www.alfascientific.com/products/ .
- https://www.accubiotech.com/category?catid=2 .
- https://www.turklab.com.tr/products/product-list .
- https://www.ultimed.org/produktkategorie/rapid-tests/ .
- N. Jiang, R. Ahmed, M. Damayantharan, B. Unal, H. Butt and A. K. Yetisen, Adv. Healthcare Mater., 2019, 8, e1900244 CrossRef PubMed.
- Y. C. Jiao, C. Du, L. J. Zong, X. Y. Guo, Y. F. Han, X. P. Zhang, L. Li, C. W. Zhang, Q. Ju, J. H. Liu, H. D. Yu and W. Huang, Sens. Actuators, B, 2020, 306, 127239 CrossRef CAS.
- R. Chen, B. Liu, H. Ni, N. Chang, C. Luan, Q. Ge, J. Dong and X. Zhao, Analyst, 2019, 144, 4051–4059 RSC.
- F. A. Vicente, I. Plazl, S. P. M. Ventura and P. Znidarsic-Plazl, Green Chem., 2020, 22, 4391–4410 RSC.
- G. Luka, A. Ahmadi, H. Najjaran, E. Alocilja, M. DeRosa, K. Wolthers, A. Malki, H. Aziz, A. Althani and M. Hoorfar, Sensors, 2015, 15, 30011–30031 CrossRef CAS PubMed.
- B. Alsaeed and F. R. Mansour, Microchem. J., 2020, 155, 104664 CrossRef CAS.
- A. W. Martinez, S. T. Phillips, M. J. Butte and G. M. Whitesides, Angew. Chem., Int. Ed., 2007, 46, 1318–1320 CrossRef CAS PubMed.
- C. Özyurt, İ. Uludağ, B. İnce and M. K. Sezgintürk, J. Pharm. Biomed. Anal., 2023, 226, 115266 CrossRef PubMed.
- C. M. Pandey, S. Augustine, S. Kumar, S. Kumar, S. Nara, S. Srivastava and B. D. Malhotra, Biotechnol. J., 2018, 13, 1700047 CrossRef PubMed.
- C. Baynes and J. Y. Yoon, SLAS Technol., 2018, 23, 30–43 CrossRef CAS PubMed.
- P. Lisowski and P. K. Zarzycki, Chromatographia, 2013, 76, 1201–1214 CrossRef CAS PubMed.
- E. Lazzarini, A. Pace, I. Trozzi, M. Zangheri, M. Guardigli, D. Calabria and M. Mirasoli, Biosensors, 2022, 12, 825 CrossRef CAS PubMed.
- B. B. Nunna, D. Mandal, J. U. Lee, H. Singh, S. Zhuang, D. Misra, M. N. U. Bhuyian and E. S. Lee, Nano Convergence, 2019, 6, 3 CrossRef PubMed.
- M. J. Lee, V. Soum, S. N. Lee, J. H. Choi, J. H. Shin, K. Shin and B. K. Oh, Anal. Bioanal. Chem., 2022, 414, 3219–3230 CrossRef CAS PubMed.
- Z. Z. Shi, Y. Lu and L. Yu, in Next Generation Point-of-care Biomedical Sensors Technologies for Cancer Diagnosis, ed. P. Chandra, Y. N. Tan and S. P. Singh, Springer Singapore, Singapore, 2017, pp. 365–396, DOI:10.1007/978-981-10-4726-8_16.
- X. Zhang, G. Soori, T. J. Dobleman and G. G. Xiao, Expert Rev. Mol. Diagn., 2014, 14, 97–106 CrossRef CAS PubMed.
- Q. Q. Zhuang, H. H. Deng, S. B. He, H. P. Peng, Z. Lin, X. H. Xia and W. Chen, ACS Appl. Mater. Interfaces, 2019, 11, 31729–31734 CrossRef CAS PubMed.
- G. Tian, F. Tang, C. Yang, W. Zhang, J. Bergquist, B. Wang, J. Mi and J. Zhang, Oncotarget, 2017, 8, 58553–58562 CrossRef PubMed.
- W. Zhang, G. Yu, Y. Zhang, F. Tang, J. Lv, G. Tian, Y. Zhang, J. Liu, J. Mi and J. Zhang, Anal. Biochem., 2019, 576, 42–47 CrossRef CAS PubMed.
- X. Jiang and P. B. Lillehoj, Analyst, 2021, 146, 1084–1090 RSC.
- L. Sadaow, O. Sanpool, R. Rodpai, H. Yamasaki, W. Ittiprasert, V. H. Mann, P. J. Brindley, W. Maleewong and P. M. Intapan, Am. J. Trop. Med. Hyg., 2019, 101, 1156–1160 CrossRef CAS PubMed.
- L. M. Tian, S. Tadepalli, S. H. Park, K. K. Liu, J. J. Morrissey, E. D. Kharasch, R. R. Naik and S. Singamaneni, Biosens. Bioelectron., 2014, 59, 208–215 CrossRef CAS PubMed.
- U. H. Yildiz, P. Alagappan and B. Liedberg, Anal. Chem., 2013, 85, 820–824 CrossRef CAS PubMed.
- S. J. Chen, C. I. Rai, S. C. Wang and Y. C. Chen, Diagnostics, 2023, 13, 2255 CrossRef CAS PubMed.
- P. Teengam, N. Nisab, N. Chuaypen, P. Tangkijvanich, T. Vilaivan and O. Chailapakul, Biosens. Bioelectron., 2021, 189, 113381 CrossRef CAS PubMed.
- X. Cao, Q. Song, Y. Sun, Y. Mao, W. Lu and L. Li, Nanotechnology, 2021, 32, 445101 CrossRef CAS PubMed.
- B. D. Grant, C. A. Smith, P. E. Castle, M. E. Scheurer and R. Richards-Kortum, Vaccine, 2016, 34, 5656–5663 CrossRef PubMed.
- B. Ma, J. Fang, W. Lin, X. Yu, C. Sun and M. Zhang, Anal. Bioanal. Chem., 2019, 411, 7451–7460 CrossRef PubMed.
- R. Kumvongpin, P. Jearanaikoon, C. Wilailuckana, N. Sae-Ung, P. Prasongdee, S. Daduang, M. Wongsena, P. Boonsiri, W. Kiatpathomchai, S. S. Swangvaree, A. Sandee and J. Daduang, Mol. Med. Rep., 2017, 15, 3203–3209 CrossRef CAS PubMed.
- Y. B. Kuo, Y. S. Li and E. C. Chan, J. Virol. Methods, 2015, 212, 8–11 CrossRef CAS PubMed.
- Y. Xu, Y. Liu, Y. Wu, X. Xia, Y. Liao and Q. Li, Anal. Chem., 2014, 86, 5611–5614 CrossRef CAS PubMed.
- K. Dhama, K. Karthik, S. Chakraborty, R. Tiwari, S. Kapoor, A. Kumar and P. Thomas, Pak. J. Biol. Sci., 2014, 17, 151–166 CrossRef CAS PubMed.
- M. Redman, A. King, C. Watson and D. King, Arch. Dis. Child.: Educ. Pract. Ed., 2016, 101, 213–215 CrossRef PubMed.
- J. H. Tsou, Q. Leng and F. Jiang, Transl. Oncol., 2019, 12, 1566–1573 CrossRef PubMed.
- Z. O. Uygun, L. Yeniay and N. S. F. Gi Rgi, Anal. Chim. Acta, 2020, 1121, 35–41 CrossRef CAS PubMed.
- R. Zeng, H. Gong, Y. Li, Y. Li, W. Lin, D. Tang and D. Knopp, Anal. Chem., 2022, 94, 7442–7448 CrossRef CAS PubMed.
- Y. Wu, P. Xue, K. M. Hui and Y. Kang, Biosens. Bioelectron., 2014, 52, 180–187 CrossRef CAS PubMed.
- P. Kaewarsa, T. Vilaivan and W. Laiwattanapaisal, Anal. Chim. Acta, 2021, 1186, 339130 CrossRef CAS PubMed.
- G. Selvolini and G. Marrazza, Sensors, 2017, 17, 718 CrossRef PubMed.
- J. Qi, B. Li, N. Zhou, X. Wang, D. Deng, L. Luo and L. Chen, Biosens. Bioelectron., 2019, 142, 111533 CrossRef CAS PubMed.
- S. M. Tawfik, M. R. Elmasry, M. Sharipov, S. Azizov, C. H. Lee and Y. I. Lee, Biosens. Bioelectron., 2020, 160, 112211 CrossRef CAS PubMed.
- G. V. Martins, A. C. Marques, E. Fortunato and M. G. F. Sales, Sens. Bio-Sens. Res., 2020, 28, 100333 CrossRef.
- A. R. Cardoso, M. H. de Sa and M. G. F. Sales, Bioelectrochemistry, 2019, 130, 107287 CrossRef CAS PubMed.
- L. Syedmoradi, M. Daneshpour, M. Alvandipour, F. A. Gomez, H. Hajghassem and K. Omidfar, Biosens. Bioelectron., 2017, 87, 373–387 CrossRef CAS PubMed.
- E. Solhi, M. Hasanzadeh and P. Babaie, Anal. Methods, 2020, 12, 1398–1414 RSC.
- Y. Wang, H. Xu, J. Luo, J. Liu, L. Wang, Y. Fan, S. Yan, Y. Yang and X. Cai, Biosens. Bioelectron., 2016, 83, 319–326 CrossRef CAS PubMed.
- S. Kumar, S. Kumar, C. M. Pandey and B. D. Malhotra, J. Phys.: Conf. Ser., 2016, 704, 1900041 Search PubMed.
- Y. K. Yen, C. H. Chao and Y. S. Yeh, Sensors, 2020, 20, 1372 CrossRef CAS PubMed.
- S. Kumar, S. Kumar, S. Srivastava, B. K. Yadav, S. H. Lee, J. G. Sharma, D. C. Doval and B. D. Malhotra, Biosens. Bioelectron., 2015, 73, 114–122 CrossRef CAS PubMed.
- R. I. Stefan-van Staden, I. R. Comnea-Stancu, H. Yanik, M. Goksel, A. Alexandru and M. Durmus, Anal. Bioanal. Chem., 2017, 409, 6195–6203 CrossRef CAS PubMed.
- S. Boonkaew, A. Yakoh, N. Chuaypen, P. Tangkijvanich, S. Rengpipat, W. Siangproh and O. Chailapakul, Biosens. Bioelectron., 2021, 193, 113543 CrossRef CAS PubMed.
- S. M. Park, A. Aalipour, O. Vermesh, J. H. Yu and S. S. Gambhir, Nat. Rev. Mater., 2017, 2, 17014 CrossRef CAS PubMed.
- G. Chen, I. Roy, C. Yang and P. N. Prasad, Chem. Rev., 2016, 116, 2826–2885 CrossRef CAS PubMed.
- K. Khalid, X. Tan, H. F. Mohd Zaid, Y. Tao, C. Lye Chew, D. T. Chu, M. K. Lam, Y. C. Ho, J. W. Lim and L. Chin Wei, Bioengineered, 2020, 11, 328–355 CrossRef CAS PubMed.
- X. P. Huang, Y. F. Zhu and E. Kianfar, J. Mater. Res. Technol., 2021, 12, 1649–1672 CrossRef CAS.
- S. Medici, M. Peana, D. Coradduzza and M. A. Zoroddu, Semin. Cancer Biol., 2021, 76, 27–37 CrossRef CAS PubMed.
- S. Abarghoei, N. Fakhri, Y. S. Borghei, M. Hosseini and M. R. Ganjali, Spectrochim. Acta, Part A, 2019, 210, 251–259 CrossRef CAS PubMed.
- L. M. Yang, M. Cui, Y. P. Zhang, L. Jiang, H. Y. Liu and Z. Liu, Sens. Actuators, B, 2022, 350, 130857 CrossRef CAS.
- Q. Zhang, H. H. Yan, C. Ru, F. Zhu, H. Y. Zou, P. F. Gao, C. Z. Huang and J. Wang, Biosens. Bioelectron., 2022, 201, 113942 CrossRef CAS PubMed.
- M. Alagiri, P. Rameshkumar and A. Pandikumar, Microchim. Acta, 2017, 184, 3069–3092 CrossRef CAS.
- A. Khoshroo, A. Fattahi and L. Hosseinzadeh, J. Electroanal. Chem., 2022, 910, 116182 CrossRef CAS.
- Y. K. Yen, C. H. Chao and Y. S. Yeh, Sensors, 2020, 20, 1372 CrossRef CAS PubMed.
- M. Gutierrez-Capitan, A. Baldi and C. Fernandez-Sanchez, Sensors, 2020, 20, 967 CrossRef CAS PubMed.
- R. Hu, R. Gupta, Z. Wang, C. Wang, H. Sun, S. Singamaneni, E. D. Kharasch and J. J. Morrissey, Kidney Int., 2019, 96, 1417–1421 CrossRef CAS PubMed.
- G. Q. Sun, H. Y. Liu, Y. Zhang, J. H. Yu, M. Yan, X. R. Song and W. X. He, New J. Chem., 2015, 39, 6062–6067 RSC.
- W. P. Li, L. Li, M. Li, J. H. Yu, S. G. Ge, M. Yan and X. R. Song, Chem. Commun., 2013, 49, 9540–9542 RSC.
- M. Pavithra, S. Muruganand and C. Parthiban, Sens. Actuators, B, 2018, 257, 496–503 CrossRef CAS.
- J. Upan, N. Youngvises, A. Tuantranont, C. Karuwan, P. Banet, P. H. Aubert and J. Jakmunee, Sci. Rep., 2021, 11, 13969 CrossRef CAS PubMed.
- W.-J. Liu, L.-J. Wang and C.-Y. Zhang, Anal. Chim. Acta, 2023, 1278, 341615 CrossRef CAS PubMed.
- M. Garcia-Cortes, J. R. Encinar, J. M. Costa-Fernandez and A. Sanz-Medel, Biosens. Bioelectron., 2016, 85, 128–134 CrossRef CAS PubMed.
- S. Bock, J. An, H. M. Kim, J. Kim, H. S. Jung, X. H. Pham, W. Y. Rho and B. H. Jun, Bull. Korean Chem. Soc., 2020, 41, 989–993 CrossRef CAS.
- Z. Lin, S. Lv, K. Zhang and D. Tang, J. Mater. Chem. B, 2017, 5, 826–833 RSC.
- Z. Qiu, J. Shu and D. Tang, Anal. Chem., 2017, 89, 5152–5160 CrossRef CAS PubMed.
- Z. Xia, D. Li and W. Deng, Anal. Chem., 2021, 93, 4924–4931 CrossRef CAS PubMed.
- J. Wang, T. Sheng, X. Zhu, Q. Li, Y. Wu, J. Zhang, J. Liu and Y. Zhang, Mater. Chem. Front., 2021, 5, 1743–1770 RSC.
- K. Li, E. Hong, B. Wang, Z. Wang, L. Zhang, R. Hu and B. Wang, Photodiagn. Photodyn. Ther., 2019, 25, 177–192 CrossRef CAS PubMed.
- S. Xu, B. Dong, D. Zhou, Z. Yin, S. Cui, W. Xu, B. Chen and H. Song, Sci. Rep., 2016, 6, 23406 CrossRef CAS PubMed.
- H. He, B. Liu, S. Wen, J. Liao, G. Lin, J. Zhou and D. Jin, Anal. Chem., 2018, 90, 12356–12360 CrossRef CAS PubMed.
- D. Maiti, X. Tong, X. Mou and K. Yang, Front. Pharmacol., 2018, 9, 1401 CrossRef CAS PubMed.
- Y. Wang, J. Luo, J. Liu, S. Sun, Y. Xiong, Y. Ma, S. Yan, Y. Yang, H. Yin and X. Cai, Biosens. Bioelectron., 2019, 136, 84–90 CrossRef CAS PubMed.
- L. L. Liang, M. Su, L. Li, F. F. Lan, G. X. Yang, S. G. Ge, J. H. Yu and X. R. Song, Sens. Actuators, B, 2016, 229, 347–354 CrossRef CAS.
- Y. Fan, S. Y. Shi, J. S. Ma and Y. H. Guo, Biosens. Bioelectron., 2019, 135, 1–7 CrossRef CAS PubMed.
- K. S. Prasad, X. Y. Cao, N. Gao, Q. J. Jin, S. T. Sanjay, G. Henao-Pabon and X. J. Li, Sens. Actuators, B, 2020, 305, 127516 CrossRef CAS PubMed.
- B. Wei, K. Mao, N. Liu, M. Zhang and Z. G. Yang, Biosens. Bioelectron., 2018, 121, 41–46 CrossRef CAS PubMed.
- G. Q. Sun, L. N. Zhang, Y. Zhang, H. M. Yang, C. Ma, S. G. Ge, M. Yan, J. H. Yu and X. R. Song, Biosens. Bioelectron., 2015, 71, 30–36 CrossRef CAS PubMed.
- L. Li, L. Zhang, J. Yu, S. Ge and X. Song, Biosens. Bioelectron., 2015, 71, 108–114 CrossRef CAS PubMed.
- J. Zhou, Y. Zheng, J. Liu, X. Bing, J. Hua and H. Zhang, J. Pharm. Biomed. Anal., 2016, 117, 333–337 CrossRef CAS PubMed.
- M. Hasanzadeh and N. Shadjou, Mater. Sci. Eng., C, 2017, 71, 1313–1326 CrossRef CAS PubMed.
- V. Serafin, A. Valverde, M. Garranzo-Asensio, R. Barderas, S. Campuzano, P. Yanez-Sedeno and J. M. Pingarron, Microchim. Acta, 2019, 186, 411 CrossRef PubMed.
- Y. Koo, V. N. Shanov and Y. Yun, Micromachines, 2016, 7, 72 CrossRef PubMed.
- P. Nguyen-Tri, T. A. Nguyen, P. Carriere and C. Ngo Xuan, Int. J. Corros., 2018, 2018, 4749501 Search PubMed.
- D. C. Ferrier and K. C. Honeychurch, Biosensors, 2021, 11, 486 CrossRef CAS PubMed.
- S. Ji, M. Lee and D. Kim, Biosens. Bioelectron., 2018, 102, 345–350 CrossRef CAS PubMed.
- N. Aliakbarinodehi, G. De Micheli and S. Carrara, Anal. Chem., 2016, 88, 9347–9350 CrossRef CAS PubMed.
- F. Liu, W. Deng, Y. Zhang, S. Ge, J. Yu and X. Song, Anal. Chim. Acta, 2014, 818, 46–53 CrossRef CAS PubMed.
- E. Sanchez-Tirado, A. Gonzalez-Cortes, P. Yanez-Sedeno and J. M. Pingarron, Biosens. Bioelectron., 2018, 113, 88–94 CrossRef CAS PubMed.
- H. V. Tran, B. Piro, S. Reisberg, L. D. Tran, H. T. Duc and M. C. Pham, Biosens. Bioelectron., 2013, 49, 164–169 CrossRef CAS PubMed.
- Q. M. Feng, J. B. Pan, H. R. Zhang, J. J. Xu and H. Y. Chen, Chem. Commun., 2014, 50, 10949–10951 RSC.
- K. M. Tsoi, Q. Dai, B. A. Alman and W. C. Chan, Acc. Chem. Res., 2013, 46, 662–671 CrossRef CAS PubMed.
- S. Nikazar, V. S. Sivasankarapillai, A. Rahdar, S. Gasmi, P. S. Anumol and M. S. Shanavas, Biophys. Rev., 2020, 12, 703–718 CrossRef PubMed.
- R. Bilan, I. Nabiev and A. Sukhanova, ChemBioChem, 2016, 17, 2103–2114 CrossRef CAS PubMed.
- K. D. Wegner and N. Hildebrandt, Chem. Soc. Rev., 2015, 44, 4792–4834 RSC.
- A. Singh, M. Bezuidenhout, N. Walsh, J. Beirne, R. Felletti, S. Wang, K. T. Fitzgerald, W. M. Gallagher, P. Kiely and G. Redmond, Nanotechnology, 2016, 27, 305603 CrossRef PubMed.
- X. Zhang, J. Yu, C. Wu, Y. Jin, Y. Rong, F. Ye and D. T. Chiu, ACS Nano, 2012, 6, 5429–5439 CrossRef CAS PubMed.
- K. Pu, A. J. Shuhendler, J. V. Jokerst, J. Mei, S. S. Gambhir, Z. Bao and J. Rao, Nat. Nanotechnol., 2014, 9, 233–239 CrossRef CAS PubMed.
- Y. Q. Yang, Y. C. Yang, M. H. Liu and Y. H. Chan, Anal. Chem., 2020, 92, 1493–1501 CrossRef CAS PubMed.
- C. C. Fang, C. C. Chou, Y. Q. Yang, T. Wei-Kai, Y. T. Wang and Y. H. Chan, Anal. Chem., 2018, 90, 2134–2140 CrossRef CAS PubMed.
- H. Y. Liu, P. J. Wu, S. Y. Kuo, C. P. Chen, E. H. Chang, C. Y. Wu and Y. H. Chan, J. Am. Chem. Soc., 2015, 137, 10420–10429 CrossRef CAS PubMed.
- Y. W. More, S. D. Padghan, R. S. Bhosale, R. P. Pawar, A. L. Puyad, S. V. Bhosale and S. V. Bhosale, Sensors, 2018, 18, 3433 CrossRef PubMed.
- P. Y. You, F. C. Li, M. H. Liu and Y. H. Chan, ACS Appl. Mater. Interfaces, 2019, 11, 9841–9849 CrossRef CAS PubMed.
- T. M. Allen and P. R. Cullis, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed.
- S. Kim and H. D. Sikes, ACS Appl. Mater. Interfaces, 2019, 11, 28469–28477 CrossRef CAS PubMed.
- P. Yingchoncharoen, D. S. Kalinowski and D. R. Richardson, Pharmacol. Rev., 2016, 68, 701–787 CrossRef CAS PubMed.
- E. Frohnmeyer, N. Tuschel, T. Sitz, C. Hermann, G. T. Dahl, F. Schulz, A. J. Baeumner and M. Fischer, Analyst, 2019, 144, 1840–1849 RSC.
- J. Zheng, X. Hu, Y. Zeng, B. Zhang, Z. Sun, X. Liu, W. Zheng and Y. Chai, Anal. Chim. Acta, 2023, 1263, 341319 CrossRef CAS PubMed.
- Y. Yang, C. Li, H. Shi, T. Chen, Z. Wang and G. Li, Talanta, 2019, 192, 325–330 CrossRef CAS PubMed.
- A. Crintea, A. C. Motofelea, A. S. Sovrea, A. M. Constantin, C. B. Crivii, R. Carpa and A. G. Dutu, Pharmaceutics, 2023, 15, 1406 CrossRef CAS PubMed.
- S. Thakare, A. Shaikh, D. Bodas and V. Gajbhiye, Colloids Surf., B, 2022, 209, 112174 CrossRef CAS PubMed.
- H. Zhang, Z. Lei, R. Tian and Z. Wang, Talanta, 2018, 178, 116–121 CrossRef CAS PubMed.
- M. A. Jafri, S. A. Ansari, M. H. Alqahtani and J. W. Shay, Genome Med., 2016, 8, 69 CrossRef PubMed.
- S. Cinti, D. Moscone and F. Arduini, Nat. Protoc., 2019, 14, 2437–2451 CrossRef CAS PubMed.
- L. Rivas, M. Medina-Sanchez, A. de la Escosura-Muniz and A. Merkoci, Lab Chip, 2014, 14, 4406–4414 RSC.
- Q. Tarres, R. Aguado, M. A. Pelach, P. Mutje and M. Delgado-Aguilar, Nanomaterials, 2022, 12, 79 CrossRef CAS PubMed.
- I. Gonzalez, M. Alcala, G. Chinga-Carrasco, F. Vilaseca, S. Boufi and P. Mutje, Cellulose, 2014, 21, 2599–2609 CrossRef CAS.
- D. Quesada-Gonzalez, C. Stefani, I. Gonzalez, A. de la Escosura-Muniz, N. Domingo, P. Mutje and A. Merkoci, Biosens. Bioelectron., 2019, 141, 111407 CrossRef CAS PubMed.
- S. Kumar, P. Rai, J. G. Sharma, A. Sharma and B. D. Malhotra, Adv. Mater. Technol., 2016, 1, 1600056 CrossRef.
- C. S. Park, C. Lee and O. S. Kwon, Polymers, 2016, 8, 249 CrossRef PubMed.
- S. Kumar, M. Willander, J. G. Sharma and B. D. Malhotra, J. Mater. Chem. B, 2015, 3, 9305–9314 RSC.
- S. Kumar, M. Umar, A. Saifi, S. Kumar, S. Augustine, S. Srivastava and B. D. Malhotra, Anal. Chim. Acta, 2019, 1056, 135–145 CrossRef CAS PubMed.
- F. Bahavarnia, A. Saadati, S. Hassanpour, M. Hasanzadeh, N. Shadjou and A. Hassanzadeh, Int. J. Biol. Macromol., 2019, 138, 744–754 CrossRef CAS PubMed.
- F. Farschi, A. Saadati and M. Hasanzadeh, Heliyon, 2020, 6, e04327 CrossRef PubMed.
- F. Farshchi, A. Saadati, N. Fathi, M. Hasanzadeh and M. Samiei, Anal. Methods, 2021, 13, 1286–1294 RSC.
- L. Li, Y. Zhang, L. Zhang, S. Ge, H. Liu, N. Ren, M. Yan and J. Yu, Anal. Chem., 2016, 88, 5369–5377 CrossRef CAS PubMed.
- F. Shi, J. Li, J. Sun, H. Huang, X. Su and Z. Wang, Talanta, 2020, 207, 120341 CrossRef CAS PubMed.
- P. Teengam, W. Siangproh, A. Tuantranont, C. S. Henry, T. Vilaivan and O. Chailapakul, Anal. Chim. Acta, 2017, 952, 32–40 CrossRef CAS PubMed.
- G. Sun, H. Yang, Y. Zhang, J. Yu, S. Ge, M. Yan and X. Song, Biosens. Bioelectron., 2015, 74, 823–829 CrossRef CAS PubMed.
- W. Zhou, H. Tavakoli, L. Ma, C. Bautista and X. Li, in Multidisciplinary Microfluidic and Nanofluidic Lab-on-a-chip, ed. X. Li, C. Yang and P. C. H. Li, Elsevier, Amsterdam, 2022, pp. 325–360, DOI:10.1016/B978-0-444-59432-7.00009-1.
- G. Fu, X. Li, W. Wang and R. Hou, Biosens. Bioelectron., 2020, 170, 112646 CrossRef CAS PubMed.
- H. Torul, H. Ciftci, D. Cetin, Z. Suludere, I. H. Boyaci and U. Tamer, Anal. Bioanal. Chem., 2015, 407, 8243–8251 CrossRef CAS PubMed.
- S. K. Arya, P. Zhurauski, P. Jolly, M. R. Batistuti, M. Mulato and P. Estrela, Biosens. Bioelectron., 2018, 102, 106–112 CrossRef CAS PubMed.
- A. Chamorro-Garcia, A. de la Escosura-Muniz, M. Espinosa-Castaneda, C. J. Rodriguez-Hernandez, C. de Torres and A. Merkoci, Nanomedicine, 2016, 12, 53–61 CrossRef CAS PubMed.
- G. Liu, A. S. Gurung and W. Qiu, Molecules, 2019, 24, 756 CrossRef PubMed.
- X. Zhang and Q. Long, Medicine, 2017, 96, e6694 CrossRef CAS PubMed.
- M. Bialkower, H. McLiesh, C. A. Manderson, R. F. Tabor and G. Garnier, Analyst, 2019, 144, 4848–4857 RSC.
- D. C. Christodouleas, B. Kaur and P. Chorti, ACS Cent. Sci., 2018, 4, 1600–1616 CrossRef CAS PubMed.
- C. M. Oliveira, L. W. Musselwhite, N. de Paula Pantano, F. L. Vazquez, J. S. Smith, J. Schweizer, M. Belmares, J. C. Possati-Resende, M. A. Vieira, A. Longatto-Filho and J. Fregnani, PLoS One, 2020, 15, e0232105 CrossRef CAS PubMed.
- C. Bax, B. J. Lotesoriere, S. Sironi and L. Capelli, Cancers, 2019, 11, 1244 CrossRef CAS PubMed.
- A. D. Warren, G. A. Kwong, D. K. Wood, K. Y. Lin and S. N. Bhatia, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 3671–3676 CrossRef CAS PubMed.
- J. Delanghe and M. Speeckaert, Biochem. Med., 2014, 24, 89–104 CrossRef CAS PubMed.
- S. Kumar, S. Kumar, S. Tiwari, S. Srivastava, M. Srivastava, B. K. Yadav, S. Kumar, T. T. Tran, A. K. Dewan, A. Mulchandani, J. G. Sharma, S. Maji and B. D. Malhotra, Adv. Sci., 2015, 2, 1500048 CrossRef PubMed.
- S. Aziz, S. S. Ahmed, A. Ali, F. A. Khan, G. Zulfiqar, J. Iqbal, A. A. Khan and M. Shoaib, Cancer Invest., 2015, 33, 318–328 CrossRef CAS PubMed.
- P. K. Mankapure, S. R. Barpande, J. D. Bhavthankar and M. Mandale, J. Appl. Oral Sci., 2015, 23, 491–496 CrossRef PubMed.
- Q. Wang, P. Gao, X. Wang and Y. Duan, Clin. Chim. Acta, 2014, 427, 79–85 CrossRef CAS PubMed.
- E. J. Franzmann and M. J. Donovan, Expert Rev. Mol. Diagn., 2018, 18, 837–844 CrossRef CAS PubMed.
- S. Kumar, J. G. Sharma, S. Maji and B. D. Malhotra, RSC Adv., 2016, 6, 77037–77046 RSC.
- S. Prasad, A. K. Tyagi and B. B. Aggarwal, Exp. Biol. Med., 2016, 241, 783–799 CrossRef CAS PubMed.
- N. M. Rodriguez, W. S. Wong, L. Liu, R. Dewar and C. M. Klapperich, Lab Chip, 2016, 16, 753–763 RSC.
- X. Gu, J. Q. Xue, S. J. Han, S. Y. Qian and W. H. Zhang, Cancer Biomarkers, 2016, 16, 395–403 CAS.
- R. Hamam, A. M. Ali, K. A. Alsaleh, M. Kassem, M. Alfayez, A. Aldahmash and N. M. Alajez, Sci. Rep., 2016, 6, 25997 CrossRef CAS PubMed.
- D. Huo, W. M. Clayton, T. F. Yoshimatsu, J. Chen and O. I. Olopade, Oncotarget, 2016, 7, 55231–55248 CrossRef PubMed.
- M. Li, X. Zou, T. Xia, T. Wang, P. Liu, X. Zhou, S. Wang and W. Zhu, Cancer Med., 2019, 8, 7006–7017 CrossRef CAS PubMed.
- N. Satomi-Tsushita, A. Shimomura, J. Matsuzaki, Y. Yamamoto, J. Kawauchi, S. Takizawa, Y. Aoki, H. Sakamoto, K. Kato, C. Shimizu, T. Ochiya and K. Tamura, PLoS One, 2019, 14, e0222024 CrossRef CAS PubMed.
- M. Swellam, R. F. K. Zahran, H. Abo El-Sadat Taha, N. El-Khazragy and C. Abdel-Malak, Arch. Physiol. Biochem., 2019, 125, 456–464 CrossRef CAS PubMed.
- C. Yan, J. Hu, Y. Yang, H. Hu, D. Zhou, M. Ma and N. Xu, Mol. Med. Rep., 2019, 20, 3991–4002 CAS.
- X. Yu, J. Liang, J. Xu, X. Li, S. Xing, H. Li, W. Liu, D. Liu, J. Xu, L. Huang and H. Du, J. Breast Cancer, 2018, 21, 363–370 CrossRef PubMed.
- Z. Zeng, X. Chen, D. Zhu, Z. Luo and M. Yang, Yonsei Med. J., 2017, 58, 697–702 CrossRef CAS PubMed.
- X. Zou, M. Li, Z. Huang, X. Zhou, Q. Liu, T. Xia and W. Zhu, Gene, 2020, 722, 144104 CrossRef CAS PubMed.
- T. Aiso, K. Ohtsuka, M. Ueda, S. Karita, T. Yokoyama, S. Takata, N. Matsuki, H. Kondo, H. Takizawa, A. A. Okada, T. Watanabe and H. Ohnishi, Oncol. Lett., 2018, 16, 6643–6651 CAS.
- L. L. Li, L. L. Qu, H. J. Fu, X. F. Zheng, C. H. Tang, X. Y. Li, J. Chen, W. X. Wang, S. X. Yang, L. Wang, G. H. Zhao, P. P. Lv, M. Zhang, Y. Y. Lei, H. F. Qin, H. Wang, H. J. Gao and X. Q. Liu, Oncotarget, 2017, 8, 45399–45414 CrossRef PubMed.
- M. Sromek, M. Glogowski, M. Chechlinska, M. Kulinczak, L. Szafron, K. Zakrzewska, J. Owczarek, P. Wisniewski, R. Wlodarczyk, L. Talarek, M. Turski and J. K. Siwicki, Cell. Oncol., 2017, 40, 529–536 CrossRef CAS PubMed.
- Y. Sun, H. Mei, C. Xu, H. Tang and W. Wei, Pathol., Res. Pract., 2018, 214, 119–125 CrossRef CAS PubMed.
- W. Z. Switlik, M. S. Karbownik, M. Suwalski, J. Kozak and J. Szemraj, Genet. Test. Mol. Biomarkers, 2019, 23, 353–358 CrossRef CAS PubMed.
- L. Wu, B. Hu, B. Zhao, Y. Liu, Y. Yang, L. Zhang and J. Chen, Oncotarget, 2017, 8, 42173–42188 CrossRef PubMed.
- H. J. Yan, J. Y. Ma, L. Wang and W. Gu, Med. Sci. Monit., 2015, 21, 722–726 CrossRef CAS PubMed.
- X. Yang, Q. Zhang, M. Zhang, W. Su, Z. Wang, Y. Li, J. Zhang, D. G. Beer, S. Yang and G. Chen, Int. J. Biol. Sci., 2019, 15, 1712–1722 CrossRef CAS PubMed.
- Y. Yang, K. Chen, Y. Zhou, Z. Hu, S. Chen and Y. Huang, OncoTargets Ther., 2018, 11, 587–597 CrossRef PubMed.
- Y. Zhao, Int. J. Clin. Exp. Pathol., 2018, 11, 2597–2604 Search PubMed.
- X. Zhou, W. Wen, X. Shan, W. Zhu, J. Xu, R. Guo, W. Cheng, F. Wang, L. W. Qi, Y. Chen, Z. Huang, T. Wang, D. Zhu, P. Liu and Y. Shu, Oncotarget, 2017, 8, 6513–6525 CrossRef PubMed.
- Y. Wang, C. Zhang, P. Zhang, G. Guo, T. Jiang, X. Zhao, J. Jiang, X. Huang, H. Tong and Y. Tian, Cancer Med., 2018, 7, 1670–1679 CrossRef CAS PubMed.
- A. Weis, L. Marquart, D. A. Calvopina, B. Genz, G. A. Ramm and R. Skoien, Int. J. Mol. Sci., 2019, 20, 864 CrossRef CAS PubMed.
- A. Habieb, M. Matboli, H. El-Tayeb and F. El-Asmar, Mol. Biol. Rep., 2019, 46, 4581–4590 CrossRef PubMed.
- E. G. E. El-Ahwany, L. Mourad, M. M. K. Zoheiry, H. Abu-Taleb, M. Hassan, R. Atta, M. Hassanien and S. Zada, Arch. Med. Sci., 2019, 15, 1454–1461 CrossRef CAS PubMed.
- S. A. Dhayat, A. Husing, N. Senninger, H. H. Schmidt, J. Haier, H. Wolters and I. Kabar, PLoS One, 2015, 10, e0140066 CrossRef PubMed.
- S. Mummareddy, S. Pradhan, A. K. Narasimhan and A. Natarajan, Biosensors, 2021, 11, 500 CrossRef CAS PubMed.
- I. E. Tothill, Semin. Cell Dev. Biol., 2009, 20, 55–62 CrossRef CAS PubMed.
- H. Khan, M. R. Shah, J. Barek and M. I. Malik, TrAC, Trends Anal. Chem., 2023, 158, 116813 CrossRef CAS.
- T. H. Chen, C. T. Chiu, C. Lee, Y. Y. Chu, H. T. Cheng, J. T. Hsu, R. C. Wu, T. S. Yeh and K. H. Lin, Dig. Dis. Sci., 2018, 63, 2301–2308 CrossRef CAS PubMed.
- S. S. Emami, R. Nekouian, A. Akbari, A. Faraji, V. Abbasi and S. Agah, J. Cancer Res. Ther., 2019, 15, 115–119 CrossRef CAS PubMed.
- T. Imamura, S. Komatsu, D. Ichikawa, M. Miyamae, W. Okajima, T. Ohashi, J. Kiuchi, K. Nishibeppu, T. Kosuga, H. Konishi, A. Shiozaki, K. Okamoto, H. Fujiwara and E. Otsuji, Oncotarget, 2017, 8, 106538–106550 CrossRef PubMed.
- H. Imaoka, Y. Toiyama, M. Okigami, H. Yasuda, S. Saigusa, M. Ohi, K. Tanaka, Y. Inoue, Y. Mohri and M. Kusunoki, Gastric Cancer, 2016, 19, 744–753 CrossRef CAS PubMed.
- X. Jiang, W. Wang, Y. Yang, L. Du, X. Yang, L. Wang, G. Zheng, W. Duan, R. Wang, X. Zhang, L. Wang, X. Chen and C. Wang, Oncotarget, 2017, 8, 65132–65142 CrossRef PubMed.
- M. Y. Song, K. F. Pan, H. J. Su, L. Zhang, J. L. Ma, J. Y. Li, Y. Yuasa, D. Kang, Y. S. Kim and W. C. You, PLoS One, 2012, 7, e33608 CrossRef CAS PubMed.
- M. M. Tsai, C. S. Wang, C. Y. Tsai, C. G. Huang, K. F. Lee, H. W. Huang, Y. H. Lin, H. C. Chi, L. M. Kuo, P. H. Lu and K. H. Lin, Eur. J. Cancer, 2016, 64, 137–148 CrossRef CAS PubMed.
- N. Wang, L. Wang, Y. Yang, L. Gong, B. Xiao and X. Liu, Biochem. Biophys. Res. Commun., 2017, 493, 1322–1328 CrossRef CAS PubMed.
- G. Zhao, T. Jiang, Y. Liu, G. Huai, C. Lan, G. Li, G. Jia, K. Wang and M. Yang, BMC Cancer, 2018, 18, 676 CrossRef PubMed.
- X. Zhou, W. Wen, J. Zhu, Z. Huang, L. Zhang, H. Zhang, L. W. Qi, X. Shan, T. Wang, W. Cheng, D. Zhu, Y. Yin, Y. Chen, W. Zhu, Y. Shu and P. Liu, Oncotarget, 2017, 8, 34468–34480 CrossRef PubMed.
- E. Bilegsaikhan, H. N. Liu, X. Z. Shen and T. T. Liu, J. Dig. Dis., 2018, 19, 404–410 CrossRef CAS PubMed.
- H. Cheng, L. Zhang, D. E. Cogdell, H. Zheng, A. J. Schetter, M. Nykter, C. C. Harris, K. Chen, S. R. Hamilton and W. Zhang, PLoS One, 2011, 6, e17745 CrossRef CAS PubMed.
- X. Liu, X. Xu, B. Pan, B. He, X. Chen, K. Zeng, M. Xu, Y. Pan, H. Sun, T. Xu, X. Hu and S. Wang, J. Cancer, 2019, 10, 43–50 CrossRef PubMed.
- L. Min, S. Zhu, L. Chen, X. Liu, R. Wei, L. Zhao, Y. Yang, Z. Zhang, G. Kong, P. Li and S. Zhang, J. Extracell. Vesicles, 2019, 8, 1643670 CrossRef CAS PubMed.
- Y. Tan, J. J. Lin, X. Yang, D. M. Gou, L. Fu, F. R. Li and X. F. Yu, Cancer Epidemiol., 2019, 60, 67–76 CrossRef PubMed.
- M. L. Wikberg, R. Myte, R. Palmqvist, B. van Guelpen and I. Ljuslinder, Cancer Med., 2018, 7, 1697–1705 CrossRef CAS PubMed.
- H. Zhang, M. Zhu, X. Shan, X. Zhou, T. Wang, J. Zhang, J. Tao, W. Cheng, G. Chen, J. Li, P. Liu, Q. Wang and W. Zhu, Gene, 2019, 687, 246–254 CrossRef CAS PubMed.
- G. Zou, R. Wang and M. Wang, Cancer Biomarkers, 2019, 25, 11–18 CAS.
- Q. Chen, L. Yang, Y. Xiao, J. Zhu and Z. Li, Med. Oncol., 2014, 31, 225 CrossRef PubMed.
- P. Karasek, N. Gablo, J. Hlavsa, I. Kiss, P. Vychytilova-Faltejskova, M. Hermanova, Z. Kala, O. Slaby and V. Prochazka, Cancer Genomics Proteomics, 2018, 15, 321–327 CrossRef CAS PubMed.
- X. Lai, M. Wang, S. D. McElyea, S. Sherman, M. House and M. Korc, Cancer Lett., 2017, 393, 86–93 CrossRef CAS PubMed.
- F. Li, J. W. Xu, L. Wang, H. Liu, Y. Yan and S. Y. Hu, Artif. Cells, Nanomed., Biotechnol., 2018, 46, 482–487 CrossRef CAS PubMed.
- K. Qu, X. Zhang, T. Lin, T. Liu, Z. Wang, S. Liu, L. Zhou, J. Wei, H. Chang, K. Li, Z. Wang, C. Liu and Z. Wu, Sci. Rep., 2017, 7, 1692 CrossRef PubMed.
- P. Skrha, A. Horinek, E. Pazourkova, J. Hajer, P. Fric, J. Skrha and M. Andel, Pancreatology, 2016, 16, 839–843 CrossRef CAS PubMed.
- A. J. Stroese, H. Ullerich, G. Koehler, V. Raetzel, N. Senninger and S. A. Dhayat, J. Cancer Res. Clin. Oncol., 2018, 144, 2377–2390 CrossRef CAS PubMed.
- X. Zou, J. Wei, Z. Huang, X. Zhou, Z. Lu, W. Zhu and Y. Miao, Cancer Med., 2019, 8, 2810–2822 CrossRef CAS PubMed.
- T. Kim and C. M. Croce, Exp. Mol. Med., 2023, 55, 1314–1321 CrossRef CAS PubMed.
- H. Lin, X. Shi, H. Li, J. Hui, R. Liu, Z. Chen, Y. Lu and W. Tan, BMC Cancer, 2021, 21, 1293 CrossRef CAS PubMed.
- J. T. Lin and K. W. Tsai, Int. J. Mol. Sci., 2021, 22, 4278 CrossRef CAS PubMed.
- D. Coradduzza, T. Solinas, F. Balzano, N. Culeddu, N. Rossi, S. Cruciani, E. Azara, M. Maioli, A. Zinellu, M. R. De Miglio, M. Madonia, M. Falchi and C. Carru, J. Mol. Diagn., 2022, 24, 1171–1180 CrossRef CAS PubMed.
- X. Fan, X. Zou, C. Liu, W. Cheng, S. Zhang, X. Geng and W. Zhu, Biosci. Rep., 2021, 41, BSR20210111 CrossRef CAS PubMed.
- C. Huang, Y. Sun, S. Ma, A. S. Vadamootoo, L. Wang and C. Jin, Pathol., Res. Pract., 2019, 215, 152411 CrossRef CAS PubMed.
- L. Zhou, X. Ma, J. Yue, T. Chen, X. Y. Wang, Z. W. Wang, J. Pan and Y. Lin, Cancer Biomarkers, 2018, 23, 561–567 CAS.
- Z. S. Yao, C. Li, D. Liang, X. B. Jiang, J. J. Tang, L. Q. Ye, K. Yuan, H. Ren, Z. D. Yang, D. X. Jin, S. C. Zhang, J. Y. Ding, Y. C. Tang, J. X. Xu, K. Chen, W. X. Xie, D. Q. Guo and J. C. Cui, Cancer Biomarkers, 2018, 22, 127–133 CAS.
- D. Ouyang, N. Ye, Y. Jiang, Y. Wang, L. Hu, S. Chao, M. Yarmush, M. Tuner, Y. Li and B. Tang, Biomed. Microdevices, 2023, 26, 3 CrossRef PubMed.
- J. Prada-Arismendy, J. C. Arroyave and S. Röthlisberger, Blood Rev., 2017, 31, 63–76 CrossRef CAS PubMed.
- J. Zhao and Y. Lin, Front. Oncol., 2023, 13, 1132628 CrossRef PubMed.
- H. Sareen, Y. Ma, T. M. Becker, T. L. Roberts, P. de Souza and B. Powter, Int. J. Mol. Sci., 2022, 23, 8835 CrossRef CAS PubMed.
- S. Tamai, T. Ichinose and M. Nakada, Brain Tumor Pathol., 2023, 40, 66–77 CrossRef PubMed.
- Y. Xiao, L. Zhang, Z. Song, C. Guo, J. Zhu, Z. Li and S. Zhu, Med. Sci. Monit., 2016, 22, 855–862 CrossRef CAS PubMed.
- F. Zhi, N. Shao, R. Wang, D. Deng, L. Xue, Q. Wang, Y. Zhang, Y. Shi, X. Xia, S. Wang, Q. Lan and Y. Yang, Neuro-Oncology, 2015, 17, 383–391 CrossRef CAS PubMed.
- T. Zhang, F. Ding, Y. Yang, G. Zhao, C. Zhang, R. Wang and X. Huang, Biosensors, 2022, 12, 485 CrossRef CAS PubMed.
- J. Q. Sheng, S. R. Li, Z. T. Wu, C. H. Xia, X. Wu, J. Chen and J. Rao, Cancer Epidemiol., Biomarkers Prev., 2009, 18, 2182–2185 CrossRef CAS PubMed.
- M. Oliveira-Rodriguez, S. Lopez-Cobo, H. T. Reyburn, A. Costa-Garcia, S. Lopez-Martin, M. Yanez-Mo, E. Cernuda-Morollon, A. Paschen, M. Vales-Gomez and M. C. Blanco-Lopez, J. Extracell. Vesicles, 2016, 5, 31803 CrossRef PubMed.
- G. Liu, X. Mao, J. A. Phillips, H. Xu, W. Tan and L. Zeng, Anal. Chem., 2009, 81, 10013–10018 CrossRef CAS PubMed.
- A. Javani, F. Javadi-Zarnaghi and M. J. Rasaee, Anal. Biochem., 2017, 537, 99–105 CrossRef CAS PubMed.
- P. Preechakasedkit, W. Siangproh, N. Khongchareonporn, N. Ngamrojanavanich and O. Chailapakul, Biosens. Bioelectron., 2018, 102, 27–32 CrossRef CAS PubMed.
- K. Baryeh, S. Takalkar, M. Lund and G. D. Liu, J. Pharm. Biomed. Anal., 2017, 146, 285–291 CrossRef CAS PubMed.
- A. S. Ghrera, C. M. Pandey, M. A. Ali and B. D. Malhotra, Appl. Phys. Lett., 2015, 106, 193703 CrossRef.
- J. V. Jokerst, A. Raamanathan, N. Christodoulides, P. N. Floriano, A. A. Pollard, G. W. Simmons, J. Wong, C. Gage, W. B. Furmaga, S. W. Redding and J. T. McDevitt, Biosens. Bioelectron., 2009, 24, 3622–3629 CrossRef CAS PubMed.
- D. Y. Zhang, Y. L. Wang, X. Y. Jin, Q. Xiao and S. Huang, Analyst, 2022, 147, 2412–2424 RSC.
- B. Salim, M. V. Athira, A. Kandaswamy, M. Vijayakumar, T. Saravanan and T. Sairam, Biomed. Microdevices, 2017, 19, 89 CrossRef PubMed.
- Y. Wu, P. Xue, Y. Kang and K. M. Hui, Anal. Chem., 2013, 85, 8661–8668 CrossRef CAS PubMed.
- W. Li, M. Li, S. Ge, M. Yan, J. Huang and J. Yu, Anal. Chim. Acta, 2013, 767, 66–74 CrossRef CAS PubMed.
- T. H. Ulep, R. Zenhausern, A. Gonzales, D. S. Knoff, P. A. Lengerke Diaz, J. E. Castro and J. Y. Yoon, Biosens. Bioelectron., 2020, 153, 112042 CrossRef CAS PubMed.
- A. Saadati, S. Hassanpour, F. Bahavarnia and M. Hasanzadeh, Anal. Methods, 2020, 12, 1639–1649 RSC.
- Y. Fan, S. Shi, J. Ma and Y. Guo, Biosens. Bioelectron., 2019, 135, 1–7 CrossRef CAS PubMed.
- S. Kumar, S. Kumar, S. Srivastava, B. K. Yadav, S. H. Lee, J. G. Sharma, D. C. Doval and B. D. Malhotra, Biosens. Bioelectron., 2015, 73, 114–122 CrossRef CAS PubMed.
- A. T. Singh, D. Lantigua, A. Meka, S. Taing, M. Pandher and G. Camci-Unal, Sensors, 2018, 18, 2838 CrossRef PubMed.
- C. Desmet, C. A. Marquette, L. J. Blum and B. Doumeche, Biosens. Bioelectron., 2016, 76, 145–163 CrossRef CAS PubMed.
- Y. Q. Wu, Z. W. Sun, F. C. Gao, Y. D. Wang and Y. Y. Jiang, Adv. Mater. Technol., 2023, 8, 2300357 CrossRef CAS.
- M. H. Son, S. W. Park, H. Y. Sagong and Y. K. Jung, BioChip J., 2023, 17, 44–67 CrossRef CAS.
- Z. Yu, H. Gong, F. Xue, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2022, 94, 13233–13242 CrossRef CAS PubMed.
- S. Lv, Y. Tang, K. Zhang and D. Tang, Anal. Chem., 2018, 90, 14121–14125 CrossRef CAS PubMed.
- L. Huang, R. Zeng, J. Xu and D. Tang, Anal. Chem., 2022, 94, 13278–13286 CrossRef CAS PubMed.
- Z. Yu, Y. Tang, G. Cai, R. Ren and D. Tang, Anal. Chem., 2019, 91, 1222–1226 CrossRef CAS PubMed.
- Z. Yu, H. Gong, Y. Li, J. Xu, J. Zhang, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2021, 93, 13389–13397 CrossRef CAS PubMed.
- Z. Yu, H. Gong, J. Xu, Y. Li, Y. Zeng, X. Liu and D. Tang, Anal. Chem., 2022, 94, 3418–3426 CrossRef CAS PubMed.
- Z. Yu, Q. Lin, H. Gong, M. Li and D. Tang, Biosens. Bioelectron., 2023, 223, 115028 CrossRef CAS PubMed.
- M. Magiati, A. Sevastou and D. P. Kalogianni, Microchim. Acta, 2018, 185, 314 CrossRef PubMed.
- L. L. Wang, L. P. Zeng, Y. R. Wang, T. T. Chen, W. Q. Chen, G. Y. Chen, C. Y. Li and J. H. Chen, Sens. Actuators, B, 2021, 332, 129471 CrossRef CAS.
- S. H. Luo, Y. Wu, W. L. Pan, G. Z. Zhong, B. Situ, B. Li, X. Y. Ye, X. J. Jiang, W. B. Li, Y. Zhang, L. Zheng and Q. Wang, Sens. Actuators, B, 2023, 374, 132792 CrossRef CAS.
- M. Mahmoodpour, B. A. Kiasari, M. Karimi, A. Abroshan, D. Shamshirian, H. Hosseinalizadeh, A. Delavari and H. Mirzei, Front. Oncol., 2023, 13, 1131435 CrossRef CAS PubMed.
- M. Butler, L. Pongor, Y. T. Su, L. Xi, M. Raffeld, M. Quezado, J. Trepel, K. Aldape, Y. Pommier and J. Wu, Trends Cancer, 2020, 6, 380–391 CrossRef CAS PubMed.
- J. Prada-Arismendy, J. C. Arroyave and S. Rothlisberger, Blood Rev., 2017, 31, 63–76 CrossRef CAS PubMed.
- K. P. Divya, S. Keerthana, C. Viswanathan and N. Ponpandian, Microchim. Acta, 2023, 190, 116 CrossRef CAS PubMed.
- M. S. Draz, M. Moazeni, M. Venkataramani, H. Lakshminarayanan, E. Saygili, N. K. Lakshminaraasimulu, K. M. Kochehbyoki, M. K. Kanakasabapathy, S. Shabahang, A. Vasan, M. A. Bijarchi, A. Memic and H. Shafiee, Adv. Funct. Mater., 2018, 28, 1707161 CrossRef PubMed.
- E. R. Kim, C. Joe, R. J. Mitchell and M. B. Gu, Trends Biotechnol., 2023, 41, 374–395 CrossRef CAS PubMed.
Footnote |
| † All three authors have contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2024 |