 Open Access Article
Open Access ArticleDemocratizing nucleic acid-based molecular diagnostic tests for infectious diseases at resource-limited settings – from point of care to extreme point of care
Suman
Chakraborty

Mechanical Engineering Department, Indian Institute of Technology Kharagpur, Kharagpur 721302, India. E-mail: suman@mech.iitkgp.ac.in
First published on 28th February 2024
Abstract
The recurring instances of infectious disease outbreaks, coupled with complications such as comorbidity challenges and antibiotic resistance, consistently underscore the limitations arising from the absence of diagnostic tests that are both accurate and affordable. There is a pressing need for scalable, accessible, and user-friendly diagnostic solutions that can be deployed effectively in resource-constrained settings. The development of such tests is crucial to address the challenges posed by infectious diseases, providing timely and reliable information to mitigate the impact of outbreaks and enhance public health response strategies. The gold-standard nucleic acid amplification tests (NAATs) are technologically robust but challenged by the requirements of expensive equipment, high-end infrastructure, stringent process-control and expert technicians, all of which are scarce at remote locations. This review provides a treatise on the developments and recent advancements of simplified variants of these tests that carry their promises of being deployable at the grass-root level. Various aspects of technology disruptions, ranging from sample-management, test-protocol and device-design innovations including disease-tracking wearables to the infusion of data-sciences are discussed, and their current restrictions are emphasized. The aspects of covering massive geographical areas and large populations all at once, inclusive aspects accommodating humans and their environment in a connected manner are also brought into perspective. Emphasis is laid on transformational considerations such as innovations to take care of the scarcity of power supply, storage and environmental control, expert personnel, materials supply chain and other inhibitors compounding to strong barriers in accessibility and affordability of the diagnostic test. Finally, it is highlighted that the core technological considerations take care of only one aspect of the intervention, whereas more holistic aspects such as measures of overcoming social barriers, ensuring due protection of individual patient's data (security and privacy) and adhering to ethical norms for clinical trials and validation, connectivity with the livelihood of challenged communities including underprivileged women, interlacing with sustainability issues and employment creation may often turn out to be even more imperative considerations for lab-to-field adaptation. It is envisaged that this inclusive paradigm appears to be the future of infectious disease management, catering the underserved, with no differential treatment of the rich and poor.
1. Introduction
Infectious diseases that emerged and spread via several outbreaks in the past centuries, decades and years, including the most recent COVID-19 pandemic, have been continuing to disrupt human lives and livelihoods time and again. Because of their lethal spread via different modes of transmission as well as the dynamic evolution and mutation of the disease spreading pathogens, the spotlight for their management inevitably is focused on community-centric perspectives rather than considering the affected individuals in isolation. This has brought in an imperative need to address the underlying challenges holistically, accounting for multi-faceted aspects that may be interconnected in a rather complex manner. For instance, climate change and global warming, exemplified by the loss of Arctic sea ice, as well as contamination of the environment,1 pose severe threats to biodiversity and human and animal health. Recent viral outbreaks, possibly of zoonotic origin, underscore the risks. The interface between humans and animals amplifies antimicrobial resistance. Proactive measures are crucial to safeguard against infectious diseases and their implications.While vaccination is a proven means of arresting several types of infections at the first place, its limited outreach and loss of efficacy at instances due to genetic mutations of the pathogens, challenged cold-chain and storage and transportation issues render it rather impractical to deploy the same as a stand-alone measure against the deadly infections. From practical considerations, therefore, significant emphasis needs to be laid on consolidating other common public health measures such as effective sanitation, physical distancing and various other infection prevention means including arresting water stagnation, accumulation of garbage and sanitary sewage induced contamination. Boosted by the phenomenal advancements in artificial intelligence (AI), machine learning (ML), big data analytics, and cyber-physical technologies, the possibilities of remote-monitoring of all these factors with high efficacy also seem to be ever-increasing, although its current state of the art in terms of ground level implementation remains to be somewhat rudimentary and often scientifically non-specific in terms of providing red-signals on particular diseased conditions, particularly considering that several different types of infections may have common symptomatic expressions and similar epidemiological features.2 For example, a plethora of upper respiratory tract infections such as those due to COVID 19, different variants of influenza, and adenovirus may have some common symptoms such as cough, cold, fever and severe respiratory distress. This has continued to prompt the primary healthcare providers either misdiagnosing or over-treating diseases using non-specific and empirical therapies. The consequent non-judicious use of broad-spectrum antibiotics has now been established to culminate into a hugely damaging influence on otherwise treatable infectious such TB, HIV/AIDS, malaria, and pneumonia. This establishes an imperative need for decentralized diagnostic testing facilities for disease-specific conditions, albeit without the loss of accuracy as compared to the established laboratory gold standards. This challenge is more compelling for the molecular diagnostic tests for infectious disease detection, which entail examining the DNA or RNA, the distinctive genetic code present in the cells. These tests are inherently prone to contamination when executed outside stringent process-controlled facilities, as compared to several other types of routine pathological procedures. Moreover, the reagents for infectious disease detection are commonly ultra-sensitive to the transport and storage conditions. Furthermore, any befitting compact instrumentation that can run the molecular diagnostic test protocol via adequate process control (for example, temperature control) as a substitute of the usual high-end apparatus used in the labs for detecting infectious diseases (such as automated analyzers, PCR, and ELISA kits) may not be available at the first place. The use of compact albeit complex instrumentation as a substitute of the bulky laboratory equipment may be a viable technological remedy to this challenge but may otherwise bring in adverse cost penalties that may not be conducive for deployment at the underserved settings. Not only that, other preferred attributes of point-of-care (POC) tests such as the execution with minimal user involvement, amenability to analyze unprocessed human samples (for example, direct testing of body fluids without requiring specialized kits for nucleic acid extraction from the same) and simplicity in the overall test flow put in additional constraints. Several of these aspects are addressed the best if the process-flow for the diagnostic test is largely automated, but the same is likely to conflict with the essential requirements of simplicity and affordability. As a result, striking a balance between the POC-centric demands and accuracy of the test by itself may be a practical hindrance. Consequently, many of the POC test outcomes turn out to be unacceptable in the benchmark of established clinical reference standards despite their technological merits and inventiveness. This requires a paradigm shift in infusing some of the field-deployment-oriented perspectives in the laboratory-innovation stage itself rather than exploring the same after the first protype is developed. Such requirements stem from the need of developing diagnostic solutions that not can only function at the POC settings but also continue to be effective if the testing conditions are severely challenged by extreme harsh weather conditions, alternatively termed as the extreme POC diagnostics paradigm. This proposition, however, is more easily stated than implemented for molecular pathology since this process involves analysing all the way down to the genetic sequence levels to identify indicators that may implicate the possible onset of a particular disease in a pin-pointed manner.3
Despite the above-mentioned challenge, one particular aspect that turns out to be favourable towards advancing different molecular diagnostic tests is the fact that a broad class of such procedures, such as the nucleic acid-based tests, can be applied to detect the distinctive infections due to different pathogens in a highly selective manner. These tests, therefore, are increasingly being put forward as the preferred option for infectious disease detection against a possible combination of more non-specific tests as well as otherwise sensitive but cumbersome culture-based procedures. The nucleic acid amplification tests (NAAT), which analyse the RNA or DNA from appropriate body-fluid samples to detect the genetic sequences specific to the invading pathogen, represent the most preferred molecular diagnostic procedures from the scientific and technological robustness considerations, where the nucleic acid is amplified in terms of the number of its copies (just as multiplex photocopies in a photo-copying machine) as the test progresses, enabling the detection of the target gene sequence at some level of amplification that can resolve the specific region of interest of the concerned gene.4,5
The real-time polymerase chain reaction (PCR) is a scientifically established gold standard for the above kinds of tests where thermal cycling is used to amplify the nucleic acids to amplify them in large numbers. However, these tests are hindered by the need of sophisticated laboratory infrastructure and technicians, their restricted availability for only a limited panel of tests in established diagnostic outlets, and their limitations in offering insights on antimicrobial susceptibility unless specially adapted, leaving apart the prohibitive cost burden. The advent of portable PCR platforms such as Cepheid GeneXpert for tuberculosis6 as well as some panels for Blood Culture Identification could offer early promises to combine pathogen detection along with its possible antibiotic resistance assessment. However, the treatment with antibiotics may by itself be questioned if the infection is not of bacterial origin. This brings in an emphatic need of demarcating bacterial versus non-bacterial (such as viral) infection at the first place. Common means of assessing the same implicitly by ascertaining the concentrations of different biomarkers in blood such as neutrophil, lymphocyte, C-reactive protein (CRP) and procalcitonin are usual. However, since many of these parameters are generic indicators of inflammation or infection, relying on them alone to pinpoint any particular infection appears unfeasible. More specific tests unveiling the particular patterns of the host RNA expression in blood hold the capability of resolving such paradox unambiguously but are not easily available in even the most advanced diagnostic lab settings. While this is merely an isolated example, it evidences the concerning divergence of the advances in laboratory-centric molecular diagnostic procedures and the developments of tests that effectively work in practice at the grass root level where intensive laboratory-dependent technologies are more likely to fail than succeed.
The innovation of a diagnostic test for infectious disease detection that may withstand all-weather conditions, therefore, is indeed of great importance because of the reasons mentioned as above, but remains challenging because of the extreme boundary conditions that the test has to satisfy. On the one hand, it should meet the most stringent scientific requirements to attain the high level of quality benchmarks established for molecular diagnostics, and on the other hand, it should function satisfactorily even amidst extreme deviations from high-end laboratory facilities such as quality power supply, highly controlled environment, reliable supply chain of the materials and reagents, and availability of highly skilled technicians. In addition, the test needs to remain affordable if it is to be deployed at the grassroot level without any subsidy for long-term sustenance. These constraints call for disruptive innovations in molecular diagnostic technologies with pointers that are not commonly emphasised in traditional life science research; for summarized depiction, see Table 1. The scope of this article is premised on the above considerations and is aimed at shedding light on the developments, advancements and perspectives of the lab-to-field adaptation of emerging NAATs for infectious disease detection and management in resource poor settings, in particular, addressing the common challenges of accessibility and affordability as the two major concerns inhibiting their benefit to public health.
| Parameter | Challenge | Conventional | Innovation |
|---|---|---|---|
| Power | Erratic/non-availability, poor quality, massive surges | Depends on continuous supply of ‘good quality’ power | Both battery and power-operated, solar, hand-operated |
| Environment | Heat, dirt, dust, humidity | Sensitive to these conditions | Robust to ignore these |
| Reagent | Fridge/air-conditioners do not work except at centralized location; degradation of reagents may occur due to uncontrolled handling and transportation | Reagent storage requires strict temperature/humidity-controlled environment | All weather reagents |
| Supply-chain | Does not exist | Long periods of non-performance as supply takes long time | Easy availability |
| Skill sets | Trained technicians not available | Require highly skilled personnel | Easy to train & simple to operate with minimal training |
| Cost | Price-sensitivity | Highly expensive | Needs to be inexpensive |
2. Democratized diagnostics for infectious diseases – the key factors and inclusive considerations
2.1. Metrics of accuracy
Any innovations in NAATs diagnostic test, be it POC or lab-based, needs to be reasonably accurate. The metrices of accuracy7 are commonly assessed in terms of diagnostic sensitivity (an index of how likely the test is to correctly identify those individuals having the disease) and specificity (an index of how likely the test is to correctly identify those individuals without the disease). The use of additional parameters such as the positive predictive value (PPV), which is a screening test's probability of correctly identifying from among a set of samples those that are actually disease-positive and not categorizing the disease-negative ones from the same sample as disease-positive at the same time; and the negative predictive value (NPV), which is a screening test's probability of correctly identifying from among a set of samples those that are actually disease-negative and not categorizing the disease-positive ones from the same sample as disease-negative at the same time as the more preferred quantifiers of accuracy continue to be debated. A pragmatic recommendation to that end may be to alter the acceptable limits of clinical sensitivity and specificity for particular diseases in tune with the statistical variations in the PPV and NPV. However, common regulatory considerations tend to apply the same uniform benchmarks as for the laboratory-intensive test procedures, disregarding such scopes of modifying the bars based on the relative prevalence of disease positive or disease negative cases in the target population cohort.2.2. Satisfying the ‘ASSURED’ and ‘REASSURED’ criteria of WHO
Another important aspect of democratizing infectious disease detection is to make it simplified for common use. Rapid antigen or antibody tests would in-principle fit well with such proposition, but their inherent scientific and technological limits would commonly disallow realizing the envisaged high benchmarks of sensitivity and specificity as realized by the NAATs. This consideration led to an upsurge in advancing the POC variants of the NAATs to their further simplified field-adaptable formats. A guiding principle to assess the indices of efficacy of such tests was originally laid down by the World Health Organization (WHO) in 2003 by the ASSURED criteria,8 which is an acronym for the following desirable attributes of an effective POC test: affordable, sensitive, specific, user-friendly, rapid and robust, equipment-free, and deliverable to end-users. It has not been an easy proposition for the NAATs to satisfy these criteria in totality, attributable to some of the stringent controls that cannot be easily relaxed or compromised for the highly contamination-prone molecular pathology protocols. This brings in obvious difficulties in the field adaptation of many of the promising laboratory advancements of POC-based NAATs. Synchronizing with the emerging needs, WHO subsequently appended a couple of more desirable features to their original ASSURED criteria to make it ‘REASSURED’, emphasizing two additional pointers, namely, real-time connectivity (R) and ease of specimen collection (E), thereby setting up more stringent albeit preferred attributes for the POC-based NAATS in meeting up the expected standards.2.3. The use of wearable detectors
With the recent boom in smartwatches, smart clothes and bands, the proposition of continuous monitoring of the body vitals including diagnosing infections appears to be progressively more realistic than ever before, offering promises of bringing in the realm of infectious disease screening to routine lifestyle practice.9 Smartwatches capable of monitoring the heart rate, exercises, sleeping pattern, diet, etc., on a daily basis are now available in the market in a variety of commercial brands and price structures. Even around a decade back, Google Glass was presented as a rapid diagnostic test (RDT) reader platform capable of both qualitative and quantitative assessments of diverse lateral flow immunochromatographic assays and other biomedical diagnostic tests. The advent of the skin-attached flexible sensors further facilitated the development of adhesively bonded patches that can in-principle be used for the automated collection and analysis of samples as delicate as human blood.10–13 Advancements on skin-interfaced electronics, using items as frugal as paper and pencil, may also be combined judiciously for real-time monitoring of several body vital parameters that fall beyond the scope of the standard smartwatches,14 providing not only a wider variety of data for establishing the disease correlations but also arriving at inferences on other comorbidity factors (such as hypertension, diabetes, cardiac ailment, and kidney diseases) that may interplay intensely towards dictating the severity of any infection. Furthermore, if more specialized data on accelerometery and photoplethysmography waveforms may be acquired from the wearable, the same may be used to ascertain specific comorbidity indicators of cardiovascular origin.15,16 Thus, the wearable sensors hold the capabilities of performing a wide variety of diagnostic tests that have strong clinical bearings with infectious disease management, be it in a non-invasive or minimally-invasive manner.It was reported17 during the recent pandemic that even the non-specific and pre-symptomatic data from consumer smartwatches can be used for detecting the onset of a respiratory infectious disease such as COVID-19 by analysing the physiological and activity data with particular reference to alterations in their heart rates, number of daily steps and sleeping hours using AI. ML may also be used for automated feature extractions from a plethora of such correlated data on multiple time-varying physiological parameters including the specific diagnostic test outcome.18 However, since the body vital parameters, even the physiologically referenced ones such as the respiration rate, skin temperature, blood oxygen saturation and electrocardiogram readings are too generic to indicate different possible physical conditions, they may at the best be used as the first level of screening indicators and need not be confused with any presumptive diagnosis indicators unless co-opted with disease-specific diagnostic test outcome. Thus, the promises of health condition tracking by wearables come with the caution of not getting over-awed by the digital technology itself as the same needs to be authenticated with established gold standards with utmost care. For example, in a wearable health tracking mode of infectious disease detection, it may be ideal to include a benchmarked POC variant of the particular NAAT being miniaturized via the wearable device, and efforts must be directed towards achieving a pragmatic trade-off among the access, cost, rapidity of the test execution via the wearable versus its accuracy towards maximizing the benefit to the stakeholders for effective disease management.19
2.4. Diagnostics for ‘one-health’ towards infection-tracing
Another relatively emerging perspective of infectious disease detection and management pertains to ‘one health’, connecting agriculture, live-stock, water quality and human health in a common thread rather than addressing them in isolation. One particular well-known catastrophe is the transmission of zoonotic diseases from animals to humans, as has been experienced by the human being multiple times during the different outbreaks of emerging coronaviruses (creating severe acute respiratory syndrome-SARS and Middle East respiratory syndrome-MERS).20 Fortunately, the diagnostic testing of soil, plants, water, food, animals and humans for pathogens is conceptually similar; therefore, the same considerations of decentralizing the lab-centric tests as relevant for humans do apply univocally, including the POC-based NAATs. For instance, while the testing of foods for pathogens may be centralized in-principle, one single lab may not be specialized in testing all the critical contaminants. Further, for the testing, the food samples often need to be shipped to central labs with stringent temperature and humidity control so as to prevent any alterations in the food constitution before the testing. The centralized testing of food also leads to an unwarranted delay in food supply chain and an eventual cost penalty that has to be borne by the consumers at the end. Therefore, the possibilities of testing the food at the packaging centres, manufacturing units, markets and stores and at-home conditions may be immensely beneficial. Similar considerations apply for the water- and soilborne pathogens, and of course, for all the animals that share the same food-chain with the humans.3. Different variants of NAATs for infectious disease detection
3.1. The polymerase chain reaction (PCR) test – a gold standard
PCR21,22 is essentially an enzymatic process in which a specific region of the DNA is replicated in cycles to yield many copies of a particular sequence. The PCR works on the extracted (released) target nucleic acid that is added to the reaction mix (master mix) containing all the necessary reactants (primers, nucleotides, covalent ions, buffer, and enzyme) and placed into a thermal cycler to undergo amplification. While the scientific foundation and technological robustness of the PCR tests are beyond question, the most critical concern for their implementation in resource-limited settings is the prohibitively expensive infrastructure and its supportive maintenance. The real time PCR machine (RT-PCR) is very expensive, but that is not just a matter of one-time investment. It requires high-end laboratory facilities and specialized technicians, cumulatively leading to operation and maintenance concerns if the recurring resources for such purposes are not available. In the COVID-19 pandemic, pro-active measures were taken by various authorities to result in dramatic reductions in the cost of RT-PCR test kits. But it remains underemphasized that even zero-cost kits would be not of much help if the supportive infrastructure for sustaining the RT-PCR test facility is by itself a challenge. Since such infrastructure at very remote locations cannot be established overnight and maintained perpetually, exploring simplified variants of NAATs that may be disseminated with much lower level of resources emerging as the central theme of several innovations in the POC diagnostics space. While portable PCR machines have off late been developed to overcome these barriers largely, their complexities and prohibitive costs continue to keep them away from the affordability of most of the small to medium scale diagnostic labs and other similar units established at under-resourced settings.3.2. Exploring beyond PCR – the isothermal NAATs
The motivation of developing POC versions of the NAATs triggered many interesting developments in the recent past. Much effort towards these developments is emphasized on substituting the PCR with much simplified thermal protocols. While the class of such alternative test protocols is by itself quite broad, one common thread for the same is the use of isothermal reactions as against thermal-cycle mediated processes. These isothermal tests include the loop-mediated isothermal amplification,23 nicking endonuclease amplification reaction,24 transcription mediated amplification,25 and the helicase-mediated chain reactions,26 as briefly discussed subsequently.Recombinase polymerase amplification (RPA). Recombinase polymerase amplification (RPA) is one variant of the isothermal NAAT that utilizes recombinase, single-stranded DNA binding proteins (SSBPs) and strand-displacing DNA polymerase for catalysing the strand invasion of a primer into double stranded (ds) DNA.27,28 One key benefit of the RPA assay is its relatively less sensitive dependence on precise temperature control due to the enzyme properties, leading to the possibility of utilizing the body heat for a designed thermal processing at 37–40 °C.29 Further, a simple visual depiction of the positive or negative test result can be realized by integrating the protocol with a lateral flow assay (LFA).30 Since the deployment of strand-displacing DNA polymerase eliminates the need of DNA template denaturation, this test is inherently very sensitive. However, one scientific challenge is the primer-dimer formation on account of the relatively low temperature of the designed amplification reaction.31
Loop-mediated isothermal amplification (LAMP). The above challenge may be circumvented using the loop-mediated isothermal amplification (LAMP)-based assays,32 which appear to be the backbones of the most extensively developed isothermal NAATs for infectious disease detection as per the present state of the art.33 The LAMP assay commonly needs a primer-set (pairs of inner and outer primers, 4 or 6 primers), six or eight gene regions, and Bacillus stearothermophilus (Bst) DNA polymerase as essential ingredients of the test-reagent master mix, along with additional probes and enzymes for test-specific adaptation (for example, reverse transcriptase for converting RNA to DNA before the latter can be amplified). 4–6 primers recognize 6–8 distinct regions of the target DNA. A strand-displacing DNA polymerase initiates synthesis and 2 of the primers form loop structures to facilitate subsequent rounds of amplification. Bst DNA polymerase is known to be inhibitor-resistant, eliminating the need of a high level of process control for the RNA extraction. This implicates that it may even be possible to substitute the formal RNA extraction procedure with an equivalent sample purification step that can be achieved by simple heat activation so that the entire sample-to-answer workflow may be executed in a complete POC framework without any lab-centric control, although the derived advantage of elegance in execution may be offset to some extent by a compromise in the test sensitivity. Recently, there have been endeavours to combine the RPA and the LAMP assay in a hybrid format in a single sealed tube to harness their respective benefits.34,35 However, the extensive clinical validation of such assays is yet to be reported.
CRISPR-based diagnostics. The quest towards progressively more independence from complex and expensive laboratory equipment and sensitive reagents for rapid NAATs with no compromise in the sensitivity and specificity led to the emergence of CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats)/cas (CRISPR associated proteins)-based diagnostics.47 CRISPRs represent regions in the bacterial genome that help defend against invading pathogens. These regions are composed of short DNA repeats and spacers. When a previously unseen virus, for instance, infects a bacterium, a new spacer derived from the virus is incorporated amongst existing spacers. The CRISPR sequence is transcribed and processed to generate short CRISPR RNA molecules. The CRISPR RNA associates with and guides the bacterial molecular machinery to a matching target sequence in the invading virus. The molecular machinery cuts up and destroys the invading viral genome. Comparative genomics analyses of closely related Streptococcus thermophilus strains have previously shown that genetic polymorphism predominantly occurs at hypervariable loci, such as the eps and rps operons, along with two clustered regularly interspaced short palindromic repeats (CRISPR) loci. CRISPR loci typically comprise multiple noncontiguous direct repeats separated by variable sequences known as spacers and are frequently found in proximity to Cas genes (CRISPR-associated). Various hypotheses have been proposed, suggesting roles for CRISPR and Cas genes, including providing immunity against foreign genetic elements through a mechanism based on RNA interference.48 While the CRISPR/Cas9 system is a known gene-editing technology for over a decade,49 the finding that Cas nucleases such as Cas12 and Cas13 can cleave collateral ssDNAs/ssRNAs that do not complement the guide RNA sequence is relatively more recent. This triggered the development of new diagnostic assays for COVID-19 such as DETECTR (DNA endonuclease-targeted CRISPR trans reporter) and SHERLOCK (specific high-sensitivity enzymatic reporter unlocking).50 The AIOD-CRISPR assay, consisting of consists of two Cas12a-crRNA complexes, was subsequently developed to detect both SARS-CoV-2 and human immunodeficiency virus type 1 (HIV-1), with a single incubation step at 37 °C.51 In response to the COVID-19 pandemic crisis, different CRISPR/Cas-based assays received emergency use authorization for POC based testing, in conjunction with loop-mediated or recombinase-aided amplification.52,53 These CRISPR/Cas combinations are known to feature prokaryotic adaptive immune system enabling the memorization of previous infections by interlacing short sequences of invading genomes-termed spacers-into the CRISPR locus, but their innovative adaptations to novel diagnostic technologies effectively opened up a new perspective of enhancing the accuracy of the NAATs, making up for some shortcomings that may originate due to obvious deviations from the resourced lab tests commonly built upon the PCR-based framework.54–59
Despite the scientific merits, the complexity of the CRISPR/Cas-based protocols created the need of further simplifying the assay towards making it amenable for POC-based applications. Deploying more types of Cas enzymes was proposed to be a possibility, though the requirements of custom-built equipment and nucleic acid extraction could not be obviated via this measure.60,61 Converting multiple enzymatic steps into individual reactions could also simplify the assay to an extent.62,63 Another possible approach of the assay simplification could be through a three-step process inclusive of calibration, incubation and rinsing that may be organized in a portable digital reader aided by a reaction buffer, as opposed to more common multi-step protocols that require significantly more reagent components, thereby opening up the possibilities of performing full-genome analysis on-chip.64 One more simplified variant of the CRISPR/Cas family of tests concerned the development of a disposable microfluidic cartridge deployed in a portable instrument for automated testing in about an hour for SARS-CoV2 detection.65 This test used an extraction-free direct lysis of saliva using shelf-stable and low-cost reagents, multiplexed isothermal RNA amplification, subsequent T7 transcription, followed by Cas13-aided cleavage of a quenched fluorophore. Patchsung et al. reported the clinical validation of the specific high-sensitivity enzymatic reporter unlocking (SHERLOCK) assay, deploying the Cas13a enzyme from Leptotrichia wadei for detecting SARS-CoV2 in a multiplex lateral-flow strip having an internal control for safeguarding against ribonuclease contamination.66 The evaluation of a cost-effective CRISPR-based automated detection method for the rapid detection of SARS-CoV2 was reported by Xu et al., elucidating its high efficacy.67
Like several other NAATs, the field deployment of CRISPR-based diagnostics at resource-limited settings brings in some critical challenges that cannot be offset trivially. First, the sample processing involves lengthy protocols and often relies on instrumentation. Separating the amplification and detection steps into a two-step procedure would enhance the risk of contamination amidst the rapid production of the amplicons, which cannot be easily safeguarded in a POC setting. Secondly, the reaction components must be stored and transported at ultralow temperatures. Table 2 provides an at-a-glance comparison of the PCR, LAMP, RPA and the CRISPR assays, with an emphasis on their relative advantages and shortcomings for infectious disease detection. Many articles were put forward in the recent years on the relative quantitative performances of these techniques, particularly for COVID-19 detection, but their outcome often appears to be conflicting and confusing as their assessment boundary constraints were different for different studies and cannot be universally normalized. Chan et al.68 investigated the impact of comparative stochastic performance on retesting outcomes in six commercial SARS-CoV-2 NAATs. This study aimed to evaluate how variations in stochastic performance among different NAATs influence the results of retesting procedures. Using a serially diluted panel of viral material, covering a range of 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 copies per mL, the performance of six commercial NAATs was assessed. The analysis revealed three distinct stochasticity profiles, indicating differences in sensitivity and accuracy beyond the limit of detection (LOD). Incorporating a subset of results with high cycle thresholds, a rapid PCR assay was employed for retesting to simulate common retesting protocols. The study found that the ability of the rapid PCR assay to accurately reproduce a true weak positive specimen was limited by its own stochastic performance at the corresponding pathogen concentration. These findings underscored the importance of considering the stochastic behaviour of NAATs, particularly at low pathogenic loads, as it can significantly impact retesting outcomes. Their results thus revealed that relying solely on retesting to distinguish false positives may lead to missed true positives, highlighting the need for a nuanced approach in confirming the test results, even when employing more sensitive assays for confirmatory testing, possibly opening up a never-ending debate: is the gold-standard for NAATs a true performance benchmark?
000 copies per mL, the performance of six commercial NAATs was assessed. The analysis revealed three distinct stochasticity profiles, indicating differences in sensitivity and accuracy beyond the limit of detection (LOD). Incorporating a subset of results with high cycle thresholds, a rapid PCR assay was employed for retesting to simulate common retesting protocols. The study found that the ability of the rapid PCR assay to accurately reproduce a true weak positive specimen was limited by its own stochastic performance at the corresponding pathogen concentration. These findings underscored the importance of considering the stochastic behaviour of NAATs, particularly at low pathogenic loads, as it can significantly impact retesting outcomes. Their results thus revealed that relying solely on retesting to distinguish false positives may lead to missed true positives, highlighting the need for a nuanced approach in confirming the test results, even when employing more sensitive assays for confirmatory testing, possibly opening up a never-ending debate: is the gold-standard for NAATs a true performance benchmark?
| PCR | LAMP | RPA | CRISPR | |
|---|---|---|---|---|
| Key assay steps | (1) Denaturation of the double-stranded DNA templates to separate single strands(2) Annealing, featuring the binding of short DNA molecules called primers to the flanking regions of the target DNA; and (3) Extension of the 3′ end of each primer along the template strands Steps (1)–(3) are cycled 25–35 times to exponentially amplify the copies of the target DNA. DNA polymerases (https://www.thermofisher.com/search/browse/category/us/en/90226286) are used for synthesizing the new complementary strands from the single-stranded DNA templates. All DNA polymerases possess 5′ → 3′ polymerase activity, featuring the incorporation of nucleotides to extend primers at their 3′ ends in the 5′ to 3′ direction |
Deploys a DNA polymerase with strand-displacement activity, along with two inner primers (forward and backward inner primers FIP and BIP) and two outer primers (F3 and B3) that recognize six separate regions within a target DNA sequence(1) 2F2 region of FIP hybridizes to the F2c region of the target DNA and initiates complementary strand synthesis (2) Outer primer F3 hybridizes to the F3c region of the target DNA and extends, displacing the FIP-linked complementary strand (3) This displaced strand forms a loop at the 5′ end. This single stranded DNA with a loop at the 5′ end serves as a template for BIP. B2 hybridizes to the B2c region of the template DNA. DNA synthesis is then initiated, resulting in the formation of a complementary strand and opening of the 5′ end loop (4) The outer primer B3 hybridizes to the B3c region of the target DNA and extends, displacing the BIP-linked complementary strand, resulting in a dumbbell-shaped DNA (5) The nucleotides are added to the 3′ end of F1 by DNA polymerase, which extends and opens up the loop at the 5′ end. The dumbbell shaped-DNA then gets converted to a stem loop structure. This structure serves as an initiator for LAMP cycling, which is the second stage of the LAMP reaction (6) To initiate LAMP cycling, the FIP hybridizes to the loop of the stem-loop DNA structure. Strand synthesis is thus initiated. As the FIP hybridizes to the loop, the F1 strand is displaced and forms a new loop at the 3′ end (7) Nucleotides are added to the 3′ end of B1. The extension takes place displacing the FIP strand. This displaced strand again forms a dumbbell-shaped DNA. Following self-primed strand displacement DNA, the synthesis results in one complementary structure of the original stem loop DNA and one gap repaired stem loop DNA (8) Both these products serve as template for a BIP-primed strand displacement reaction in the next cycles with no change in the temperature required. A LAMP target sequence is amplified 13 times every half cycle |
DNA is exponentially amplified by proteins involved in cellular DNA recombination, repair and synthesis. The corresponding reagents include recombinase, single-stranded binding proteins (SSB) and a strand displacing Bsu polymerase, a crowding agent, ATP and an additional single-stranded binding protein support recombinase loading and disassembly for exponential amplification(1) Recombinase proteins form complexes with each primer that scans the DNA for homologous sequences (2) The primers are subsequently inserted at the cognate site by the strand-displacement activity of the recombinase (3) Single stranded binding proteins stabilise the displaced DNA chain (4) The recombinase subsequently disassembles, leaving the 3′-end of the primers accessible to a strand displacing DNA polymerase (5) The polymerase elongates the primer (6) Exponential amplification occurs by repetition of these steps in cycles |
Method #1: (1) RNA targets are amplified through nucleic acid sequence-based amplification (NASBA), which amplifies RNA from either an RNA or DNA target and utilizes avian myeloblastosis virus reverse transcriptase, RNase H and T7 RNA polymerase. The process starts with reverse transcription (RT) to complementary DNA using a sequence-specific primer, which appends a trigger sequence (magenta) for the toehold sensor(2) The RNA from the RNA/DNA hybrid is destroyed by RNase H, enabling a primer that contains a T7 promoter to bind and create a complementary second DNA strand (3) T7 transcription of the double stranded DNA template creates the target RNA sequence, which can be used as starting material for a new NASBA cycle or detected by the toehold sensor (4) If a protospacer-adjacent motif (PAM) sequence is present in the double stranded DNA amplicon, Cas protein-mediated cleavage leads to a truncated template for T7 transcription. This generates a shorter target RNA that cannot activate the toehold sensor (5) In the absence of the PAM sequence, a full-length target RNA containing the trigger is transcribed, which produces a visible colorimetric signal Method #2 (1) DNA or RNA are amplified by RPA or RT-RPA, respectively (2) For RNA-targeting CRISPR enzymes (including Cas13a), the amplified RPA product is T7-transcribed into RNA (3) Binding of the crRNA to the complementary target sequence activates the Cas enzyme and triggers collateral cleavage of the quenched fluorescent reporters. Accordingly, Cas13a (used in SHERLOCK for COVID-19 detection) or Cas12a (used in DETECTR for COVID-19 detection) indicate the presence of RNA or DNA target sequences, respectively |
| Advantage | (1) Gold standard, currently the most common detection method(2) Higher sensitivity compared to culture and staining-based methods (3) Generic test format and can be applied for detecting uncommon pathogens, for which no established diagnostic methods exist (4) Can be utilized for antimicrobial resistance assessment, genomic analysis etc. |
(1) Rapid test(2) Reasonably sensitive and specific. Highly specific amplification is facilitated by designing 4 primers to recognize 6 distinct regions on the target gene (3) Does not need any thermal cycler, which means that a simplified instrument for maintaining isothermal condition would work (4) Thermally denaturing double-stranded DNA is not compulsorily required (5) Less sensitive to PCR inhibitors (6) Can be used in resource limited settings |
(1) Near body-temperature amplification gives the possibility of using body heat for the amplification without needing specialized instrumentation(2) Well-resistant to inhibitors (3) Tolerates more mismatches |
(1) Relies on a relatively simple and highly effective genome editing technology(2) Highly specific tests (3) Short turnaround time of the diagnostic test due to the exclusion of RNA isolation and amplification |
| Challenge | (1) DNA polymerase can lead to multiple mutations in the generated fragment, resulting in potential non-specific binding of a primer to other simpler sequences of the target DNAThis may result in a compromise in the specificity although the sensitivity can be high69 (2) Need for narrow list of the pathogens resulting in the infection to use specific primers (3) Need for sophisticated equipment and/or high-end infrastructure as well as highly trained personnel to run the test |
(1) Rapid amplification of the DNA produces large number of amplicons that are prone to carryover contamination if exposed to uncontrolled ambience(2) Unlike the PCR machine, instrumentation for LAMP-based tests is yet to be standardized (3) Performance criteria are not straight forward to establish because of lack of established benchmarks on rapid nucleic acid-based testing (4) Primer–primer interactions may affect specificity (5) Multiplexing disease detection in a single step is yet to be standardized (6) Quantification of the test outcome is not trivial |
(1) Challenged primer design (no standard software) and reagent supply chain(2) Prone to non-specific amplification | (1) Although some Cas proteins have been identified to recognize and cut double stranded DNA in a PAM-dependent manner and thereby target specific gene sequences, some highly specific sequences are not yet available in the gene editing tool base. This makes it necessary to strengthen the CRISPR tool for target selection(2) The vast existence of RNA enzymes results in RNA instability, thereby affecting the diagnostic accuracy when implemented outside highly controlled lab settings (3) A high level of thermal activation of the CRISPR-Cas-based methods may conflict with their compatibility with other enzymes used for the test |
| Open questions | (1) Can the PCR instrument be simplified to an extent that it is portable and affordable at the same time?(2) Can the PCR test protocol be implemented without high-end process control and operational skill? (3) Can the alternative forms of PCR (such as droplet PCR) replace the traditional PCR? |
(1) Can LAMP replace traditional culture-based methods?(2) How is LAMP sensitivity affected by the type of sample as well dissemination at the field as against controlled lab settings? (3) Can LAMP-based tests be standardized like their PCR counterparts? (4) Can the benefits in innovations on LAMP-based tests be brought out of research labs for commercial sustainability? (5) Can LAMP be used as a rapid POC test for assessing anti-microbial resistance by identifying drug-resistant strains quickly? (6) Can droplet-based digital LAMP emerge to be highly effective for large field-based adaptation? |
(1) Can RPA be implemented and standardized as a field-deployable POC test?(2) Can the body-heat be harnessed for the DNA amplification in a standardized manner that remains to be reasonably accurate and does not give rise to physical discomfort and logistic issues for the patient at the same time? | (1) Can the different workflows of CRISPR-based diagnostics be systematically integrated into user-friendly devices, going beyond the realm of mere streamlining of individual assay steps?(2) Can the extensive use of lyophilized reagents ensure the applicability of CRISPR diagnostics in low-resource settings irrespective of unfavourable transportation and storage conditions? (3) Can standardized and validated CRISPR data analysis procedures be framed to enable uniform data reporting and comparison of the outcome with different calibrated assays? |
3.3. Beyond the considerations of routine diagnostic assays
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths annually may be attributed to this treatment-disaster, with the rate of annual growth in the numbers of such patients ramping up to 10 million if not more.73 Therefore, an a-priori estimation of possible antibiotic resistance before starting a treatment with antibiotic may render to be much more effective than commonly perceived. This requirement leads to the need of developing a multiplex diagnostic assay that can not only can probe multiple disease-causing pathogens, typically, the ones known to have similar symptomatic manifestations, but also predict the potential antibiotic resistance profiles on treatment. This specific insight can not only minimize the adversities due to empirical or heuristic antibiotic drug prescriptions but also arrest the possibilities of newly emerging mechanisms of further antibiotic resistance.74–77
000 deaths annually may be attributed to this treatment-disaster, with the rate of annual growth in the numbers of such patients ramping up to 10 million if not more.73 Therefore, an a-priori estimation of possible antibiotic resistance before starting a treatment with antibiotic may render to be much more effective than commonly perceived. This requirement leads to the need of developing a multiplex diagnostic assay that can not only can probe multiple disease-causing pathogens, typically, the ones known to have similar symptomatic manifestations, but also predict the potential antibiotic resistance profiles on treatment. This specific insight can not only minimize the adversities due to empirical or heuristic antibiotic drug prescriptions but also arrest the possibilities of newly emerging mechanisms of further antibiotic resistance.74–77
While different methods of assessing antimicrobial drug resistances do exist, these are mostly culture-based, often requiring upto 72 hours for providing a conclusive evidence,78–80 whereas many antibiotic treatments are time-critical and cannot be delayed to that extent. Moreover, the sensitivity to antibiotics as determined from the cell culture may also be attributed to a possible lack of gene expression or potentially poor growth, leaving ample room for false negative or inconclusive results.81–84 This leads to a shift in the gold-standard itself from the culture-based tests to the NAATs that can potentially detect antibiotic resistant genes.78 However, like the basic detection of the presence or absence of the infection itself, this aspect of diagnostics should also be ideally simple enough for adaptability in resource-limited settings.85
The advent of microfluidics and nanomaterials ushered promises to overcome these constraints, but the resulting assays often exhibited insensitive clinical performance or inadequacy in determining more than one antibiotic resistance strains.86–92 Ayfan et al. reported the rapid and compact molecular detection assay of Neisseria gonorrhoeae and ciprofloxacin susceptibility cases by combining an RPA assay with LFA-mediated detection.93 Abdou Mohamed et al. were among the early researchers94 to develop a multiplex nucleic acid enzyme–gold nanoparticle-based diagnostic platform for identifying the target pathogens alongside the respective antibiotic resistant genes within a span of 2 hours. Their test deployed an enzyme comprising two nucleic acid strands instead of amino acids, having the capability of selective DNA cleaving. The specific target domain of the DNA has a particular sequence that does not bind and becomes active only if the two strands of the enzyme come sufficiently close to each other.95 Their assay could be adapted in a standard isothermal NAAT framework, by executing RPA reactions at 37–40 °C. Post amplification, the products were denatured using NaOH for obtaining single-stranded sequences. The denatured products were mixed with short blocking oligonucleotide strands. The neutralization of the resulting solution with HCl facilitated the hybridization of the blocking strands with the specific target gene. The subsequent mixing of the blocked amplicons with the enzyme solution enabled the hybridization of the blocked amplicons with the enzyme sensor arms. Without target-specific binding, the inactive enzyme retained the double stranded DNA integrity where the nanoparticles were cross-linked by the linker DNA to produce a dark purple colour in the solution. On target-specific binding to both of the DNA strands alongside the enzyme, the catalytic activity of the enzyme could be triggered so that it could bind with the linker DNA and cleave the same. In this way, the sequential cleavage of multiple linker strands could be realized, resulting in an amplification of the detection signal. Further, this could release the nanoparticles and make them monodispersed, displaying discernible red colour in the solution. This technology could emerge to be a valuable tool for the rapid assessment of life-threatening conditions due to infection, for example, evaluating a potential life-threatening organ dysfunction resulting from a dysregulated immune response to infection, also known as sepsis.96 The heterogeneity of such life-threatening complication demands personalized monitoring and treatment based on the pathogen information and its host-response speculation (commonly derived from the quantitative measures on the plasma proteins and cell-surface proteins, via data analytics and classification tools into different endotypes),97–99 sequencing data, genetic predisposition and epigenetic alterations as the sepsis progresses.100 While there could be several other life-threatening instances where similar strategies for critical care could be envisioned, the particular example of sepsis management clearly emphasizes the need of a rapid and highly accurate NAAT-tool for an inclusive life-saving intervention that could potentially be provided even in primary hospital care settings.
4. Field adaptation challenges – technology confronts practicality
4.1. The major technology barriers
With an accounting of a phenomenal array of technological breakthroughs in developing and advancing various isothermal NAATs, it may appear to be rather surprising but indeed a ground reality that their successful field, and market adaptations have been rare. One technology-specific remedy to this inadequacy is the development of a standardized portable, scalable albeit affordable instrument for executing the isothermal reactions, akin to the PCR machine that can perform the more sophisticated variants of these tests. However, this aspect gives only a partial view of the scenario since the requirements of stringent process controls for the NAATs are hard to satisfy outside controlled lab settings. For the isothermal NAATs, the resulting challenge is particularly glaring as the rapid amplification of the nucleic acid leads to large numbers of amplicons being produced almost all suddenly, which render the products to be highly contamination prone if exposed to uncontrolled ambience. The reagent mixture production recipe and its supply chain cum dispensing also need tight quality control. Further, the qualitative readout obtained from the POC test (commonly colorimetric), if observed and interpreted manually, may result in ambiguities that could render the detection error-prone, although the use of digital technologies has proven to improve the accuracy of the detection considerably.101 In addition, there are challenges associated with sample collection and dispensing accurately enough in a setting where the same is not performed by a specialized technician to make the test outcome reliable. This brings in challenges in executing the test reliably enough. While process automation can be an obvious remedy, the complexity and hence the resulting cost penalty may render it prohibitive for consideration when targeted for the grassroot level use. Summarily, while technological simplification from the PCR paradigm is emphatically warranted, safeguards based on further technological innovations to assure that the same does not result in unacceptably inaccurate test results are extremely critical, albeit without additional cost penalty.4.2. Towards simplified and user-friendly dissemination of NAATs
For blood sample-based NAATs, a microneedle patch may seamlessly interface as a sample collection layer in an integrated wearable, whereas simplifying this technology for extensive and routine use on a daily basis would leave much scopes of design innovation. Another commonly ignored aspect stems from the different methods of blood sample collection such as by finger-prick, a dedicated collection device, and microneedle patches, which may effectively concern fluids of different inherent constitutions, may be difficult to normalize for diagnostic test interpretation purposes unless spelt out in the standard operating procedure. The blood-based tests may further require plasma separation, for which in situ arrangement without much complexity in the system design should be ideally sought for. Otherwise, separate arrangements of centrifugation or equivalent may render the method inappropriate for resource-limited settings, unless inventive interventions are adapted. One notable development in this regard has been the introduction of a lightweight (2 g), human-powered paper centrifuge (‘paperfuge’), drawing inspirations from the historic whirligig (or buzzer) toys, which can rotate at 125000 r.p.m. to separate plasma from whole blood in about 75 s,108 though integrating the same with the other process flows of a NAAT protocol would not remain trivial. Another aspect of sample handling concerns its enrichment towards amplifying the test signal. Yang et al. developed a sample enrichment platform integrating four microfluidic channels where the output signals from RT-LAMP reaction were transduced to portable commercial pregnancy test strips by the means of highly specific human chorionic gonadotropin (hCG)-conjugated toehold-mediated strand exchange probes.109 Summarily, sample collection and processing issues should be accounted for in the test readout normalization and calibration readjustments to interpret the test readout correctly, although not much systematic work on the same has been reported thus far.
For a schematic depiction of the concerned sample-to-answer workflow, see Fig. 1. The device eliminates the need for additional interventions like fluidic systems, offering a time-synchronized workflow for essential nucleic acid testing components, including sample purification, reverse transcription, nucleic acid amplification, and specific nucleic acid-based probing steps. The PINAT test protocol can be executed by unskilled operators in a single-step swab-to-result format, enabling rapid onsite diagnosis of COVID-19 with minimal user intervention outside a controlled laboratory environment. In a laboratory-based in vitro validation experiment using 200 clinical samples collected during the first wave of the pandemic in India (August 2020), the PINAT system demonstrated sensitivity and specificity comparable to the gold-standard RT-PCR test for detecting SARS-CoV-2 infection. In a subsequent field trial during the second wave of the pandemic (June 2021), the PINAT successfully tested 170 patient samples at the point of collection in a single-step swab-to-result format, exhibiting satisfactory levels of sensitivity and specificity compared to other POC tests commonly used in community healthcare settings. Additionally, the PINAT system demonstrated efficacy in detecting Influenza A virus RNA directly from virion particles, showcasing a high level of detection accuracy. The specific innovations in their diagnostic procedure include single-dispensing-step with single-microchamber integration of all possible process elements for nucleic acid-based testing including sample extraction/lysis, reverse transcription, amplification, and specific hybridization probing (with provision for multiple nucleic acid probing) in a thermo-kinetically controlled, automated, time-synchronized manner without any external field-mediated operative intervention. This innovation led to seamless sample-to-result integration in an inexpensive, scalable, pre-programmable and customizable portable device, with mobile application-integrated interpretation and analytics involving minimal manually-operative procedures that can be undertaken by unskilled personnel outside the controlled lab environment in a resource-constrained setting.
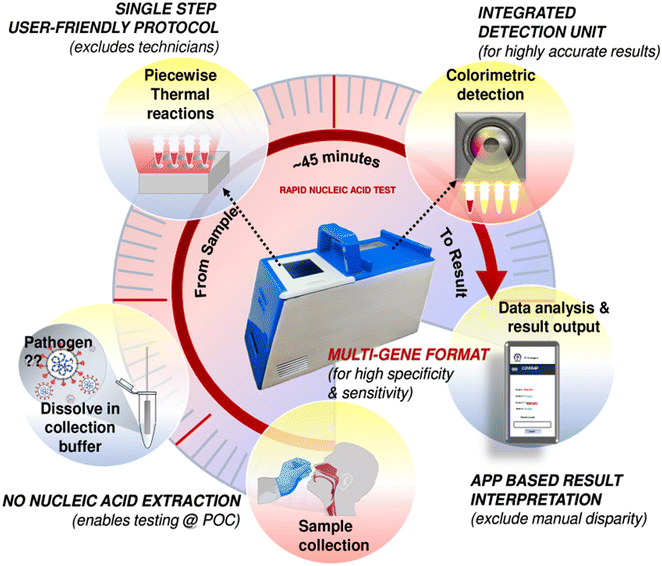 | ||
| Fig. 1 Process flow from sample collection to the test result readout as per the PINAT technology, all disseminated via the integrated low-cost device, as illustrated for COVID-19 detection. The specific steps are as follows: (1) the sample collected from the patient briefly heated isothermally in the device; no RNA extraction is needed; (2) the heat lysed sample is then mixed with the reagents in dedicated reaction-chambers; (3) the PINAT reactions are executed synchronously in the portable device in a pre-programed manner without intermediate manual intervention; (4) the products are detected by smartphone-interfaced colorimetric analysis, with the entire sample-to-result integration occurring within 45 minutes. Retaining the reaction chambers were completely sealed till the end of the test ensures no unwarranted contamination on account of exposure to uncontrolled ambience. Figure developed in-house of the author, following the work of his research group, as reported in Banerjee et al.133 | ||
Electricity-free heating realized by the two-prong intervention of phase change materials along with designed exothermic reactions to achieve the desired temperature for the thermal processing steps of the NAAT also appeared to be quite promising.131 For maintaining the temperature of the reaction chambers during isothermal heating, these thermal units were kept in contact with a synthetic fat-based compound having a high specific heat capacity and melting point around the reaction temperature. A common food-storing insulated housing with two chambers could be customized for this purpose so that the bottom chamber could house the exothermic reaction, whereas the top chamber contained the phase change material and the reaction wells. An aluminium “honeycomb” material was deployed in the upper chamber before introducing the phase change material so as to ensure effective heat transfer to the reaction wells that were embedded within the same. Yang et al. developed a light-weight (∼300 gram) and reusable device for RT-Recombinase Aided Amplification (RAA) reaction (executed at 37–44 °C) powered by solar photothermal energy conversion, embodying a convex lens for focusing the sunlight, sunflower-like light tracking system for efficient light utilization, alongside a simple visual detection scheme using ultraviolet flashlight.132 A solar panel with stored energy could provide adequate power for the light-sensitive platform. Seok et al. developed a two-stage single-pot isothermal NAAT for POC-based applications.133 The first stage executed RPA reactions before the LAMP reactions occurred in the second stage. Their specially designed buffer could maximize the volume of the products of the RPA reaction, which could subsequently be added to the LAMP master mix minus the inhibition, forward and backward primers. The reaction pot was compartmentalised with a barrier. In the second stage, the LAMP reaction temperature could melt away the barrier, thereby facilitating the mixing of the products of the first stage with the reactants of the second stage for triggering the amplification. The test could be applied for detecting hepatitis B and C viruses, with the viral load calibrated based on the signal threshold time.
In the digital technology space, stand-alone sensors and smartphones may need to be further augmented to develop a broader integrated Digital Molecular Diagnostic system that can simultaneously measure multiple analytes and process the resulting quantitative raw data in real time, including quantitative data generated from molecular diagnostics.138,139 As a consequence, it may be possible to establish panels of target pathogen genes and biomarkers to distinguish between different classes of pathogens,140 and the key biomarkers may be combined in a single test.141,142 This may enable detecting the pathogen and the molecular markers of its resistance to common drugs, relating the invasion of a pathogen with its illness-creating potential143 using risk scores from the gene expression levels,144 prognosis,145 and the extent of transmissibility.146,147
Another aspect that may substantially advance with the digital technologies is a quantification of the infection severity, an aspect that is not commonly in the scope of the existing NAATs that commonly gives a yes/no type of answer on infection positivity or otherwise. Despite the phenomenal advancements in data sciences, the aspect of quantifying pathogenic loads in low-cost POC devices is at its infancy. One particular scientific challenge inhibiting much progress on this aspect lies in the low signal-to-noise ratios of the isothermal NAATs. The digital NAAT, apparently ideal for such purposes, may not be effective for technology translation in low-resource settings.148,149 Therefore, further research may be directed to innovate the means of reducing the noises from the test signal, for example, using established stabilizing chemicals such as hydroxynaphthol blue in the LAMP master mix to dampen out the noises in the measurement.150 Recently, Biswas et al. deployed an ML-based approach to extract quantitative readout from colorimetric LAMP tests, opening up the possibilities of utilizing the established image analysis principles for arriving at the pathogenic load (CT-value equivalent of a PCR) with the aid of a simple smartphone or device-integrated camera.151
4.3. Diagnostic inclusiveness and holistic surveillance
The diagnostics of infectious disease by testing on humans is one of the most imperative measures for early detection, but spotting every individual in the community and bringing them into the ambit of such testing may have ground level implementation challenges. Considering this constraint, disease-specific wastewater-based epidemiology (WBE) has progressively been attracting attention over the past couple of years for early monitoring and red-flagging of community-specific infectious disease outbreaks.152 This approach can focus its spotlight on a collection of populations based on demography, habitat, etc., as against isolated individuals. It is attractive for its cost-effectiveness and rapid coverage of a relatively vast region at once. Post onset of the recent COVID-19 pandemic, it was evidenced that the traces of SARS-CoV-2 RNA get shed in feces, saliva, and sputum, while these body fluids often get mingled with waste water. Therefore, an endeavour of detecting these traces in the wastewater by the same molecular diagnostic procedures as for the human samples may be a pragmatic approach towards understanding such epidemiological aspects. The efficacy of this approach, however, may be best realized if highly accurate POC-based NAAT sensors are interconnected by the Internet of Things (IoT) for real time analytics and dissemination in support with other digital technologies in a coherent manner. In its most simplified albeit user-friendly form, the smartphone, which in the present era is no less powerful than a miniaturized computer, can by itself act as a standalone surveillance tool for this purpose thanks to the outstanding recent advancements in 4G and 5G internet connectivity, easy access to cloud-based services, near-field communication, and wireless and Bluetooth connection.The first layer of metadata for infection surveillance may include the outcome of the NAAT-sensors, along with essential geographical and environmental data. With a red flag from this layer of screening, the additional layer of clinical and symptom-based indicators collected from sample populations in the target region may be integrated to correlate with the findings and recommendation. Krivoňáková et al. outlined one such development by mathematically correlating the outcome of the RT-qPCR tests of SARS-CoV-2 (viral particles) in wastewater samples and death cases or positive PCR-tests in Bratislava-Slovakia.153 They emphasized the criticality of the location-specific attributes of waste water (temperature, pH, (bio)chemical constituents, etc.) for making the outcome effective and meaningful. Another aspect to consider here is the fact that the action of numerous sensors in a network would require harnessing ubiquitous energy from ambient environment for empowering them, considering the fact that while individual sensors may be minimally power-hungry, their collective integration would demand significant electrical power that may not be available from the external grid at remote locations.
A multi-layer disease screening strategy with progressively more specific diagnostic inputs embedded in the deeper layers may lay a robust conceptual foundation of disease diagnosis decision support as against over-relying on standalone diagnostic tests. This may be achieved via multi-layer screening, where vulnerable patients are screened out at different levels and then subjected to progressively more inclusive diagnostic tests, whereas unnecessary diagnostic tests are avoided for the personnel who are clearly out of risk. The first layer of screening need not require any instruments and may be completely based on the available patient metadata as well as past medical history. Red flag cases from this screening are to be subsequently tested using some essential body signal acquisition in a non-invasive manner with no over-burdening of the resources. The cases that are predicted to be vulnerable from this screening by a medical algorithm-driven decision-making software are then to be tested by POC technologies for approaching from presumptive to confirmatory diagnosis. In this way, the valuable resources for POC testing are not used as an ‘overkill’ but used only judiciously when the clinical decision support cannot be made with high level of confidence without the same. The data set acquired from each patient in this manner may be subjected to patient-specific disease modelling, risk categorization, identification of infectious zone hot-spots and policy decision support.
4.4. Seamless field adaptation and establishment of efficacy
One major utility of the different variants of the isothermal NAAT-based tests is its potential direct adaptation on a mass-scale for arresting infectious disease progression, including self-testing using saline gargle solution (instead of swab). Such tests could be mechanized largely by pseudonymized sample handling, automated nucleic acid extraction, and the amplification cum detection using a semiquantitative multiplex colorimetric RT-LAMP. Lou et al. performed more than 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 such tests, establishing its capability to run amidst challenged supply chains.154 Kundrod et al. developed a quantitative sample-to-answer RT-LAMP test for detecting SARS-CoV-2 from saliva, nasal, or nasopharyngeal swabs, using an extraction-free simple lysis and sample deactivation protocol.155 The authors piloted a University-wide surveillance testing program covering upto 400 human subjects per day. Gibani et al. assessed the clinical accuracy of a POC-based NAAT for SARS-CoV-2.22 Karlikow et al. field-validated a cell-free expression system based on NAAT and toehold-switch reactions, with computer vision-empowered image analysis, reporting accuracies at par with real time q-PCR for detecting Zika and chikungunya viruses.156 Morris et al. assessed the performance of a POC-based PCR test on-field for sexual health assessment, deploying a compact, transportable, single-use instrument.157
000 such tests, establishing its capability to run amidst challenged supply chains.154 Kundrod et al. developed a quantitative sample-to-answer RT-LAMP test for detecting SARS-CoV-2 from saliva, nasal, or nasopharyngeal swabs, using an extraction-free simple lysis and sample deactivation protocol.155 The authors piloted a University-wide surveillance testing program covering upto 400 human subjects per day. Gibani et al. assessed the clinical accuracy of a POC-based NAAT for SARS-CoV-2.22 Karlikow et al. field-validated a cell-free expression system based on NAAT and toehold-switch reactions, with computer vision-empowered image analysis, reporting accuracies at par with real time q-PCR for detecting Zika and chikungunya viruses.156 Morris et al. assessed the performance of a POC-based PCR test on-field for sexual health assessment, deploying a compact, transportable, single-use instrument.157
4.5. Pool testing
One aspect that may potentially revolutionize molecular diagnostics via the fusion of digital technologies with body fluid-based physical testing concerns an extensive upscaling of the numbers of tests by pool testing, with thousands of samples being tested in a single run. In a pooled test, all the collected samples are mixed to perform the initial run. If the outcome is infection-negative, then all the samples are estimated as negative. On the other hand, if the outcome is positive, then each sample is to be retested. Recent research evidenced that the next-generation sequencing of molecular barcode-tagged pooled samples can upscale the parallel testing of samples massively (thousands of samples in a single run).158 Using a synthetic RNA standard as reference, they could establish an extraction-free format of the NAAT for the end-point quantification so that the test can automatically terminate when all the primers in the reaction master mix are consumed without needing to pre-set any cycle threshold for the PCR. Further, post-PCR multiplexing and library generation can enable a single operator handling thousands of samples in parallel. With the aid of an automated decapper, a single liquid handler and a NAAT instrument, a daily testing load in the tune of 3000 can be covered, thereby institutionalizing routine testing in different medium-sized organizations quite successfully. Rapid and frequently repeated testing (mostly on a daily basis), contact tracing and isolating the potentially infected individuals in this way can successfully arrest infection outbreaks, even if the testing device comprises of the analytical sensitivity in preference to the other benefits derived.159 While robotic handling and automation may appear to be a practical deterrent in achieving this idealized paradigm, a simplification of the same could be realized utilizing swarms of electromagnetically actuated magnets for pooling the liquid samples using LAMP reactions, where the ferrofluid droplets carry the test master mixes.160 The magnets by themselves act as ‘ferrobots’, which, in conjunction with automated structures, and low-voltage electric fields may produce electromagnetic effects for sample processing and mixing.5. Technology gap areas – challenges, opportunities and remedies
5.1. Lab research perspectives versus field adaptation
With such phenomenal advancements in NAAT-based diagnostic tests for infectious diseases that can potentially be deployed at the POC, it may appear to be somewhat ironical that their on-field adaptations in extreme resource-poor settings have remained to be somewhat elusive so far, although the corresponding lab protypes had indeed proven to be technologically disruptive from several fronts. This deficit, though apparently surprising, may be grossly rationalized by appreciating the fact that while scientific innovations appear to be the prime movers for lab-research, lab-to-field translation of these technologies enforce several stringent boundary conditions that by themselves demand radical modifications in the protype to make it amenable to a product of real-life utilities. For example, many of the lab innovations stress on developing different non-integrated stand-alone assays with particular capabilities such as plasma separation from the whole blood, nucleic acid extraction, nucleic acid amplification and test signal readout. Integrating these elements into a complete workflow, in the current practice, emerges more commonly as an afterthought rather than a compact pre-design, keeping some inevitable loopholes that tend to get exposed on implementation without specialized infrastructure and process control.5.2. Sample management-related issues
The lack of standardization in sample collection and processing is a matter of concern, as mentioned earlier, and the normalization of the same to draw an acceptable clinical reference is commonly elusive. For instance, the outcome of testing based on nasal, nasopharyngeal, oropharyngeal swabs and saliva samples, and even the same type of sample collected by different personnel may be grossly contrasting albeit with no change in the technology platform used for their analysis. There can be significant disparities in the analytical outcome based on finger-pricked capillary blood and the traditional venepuncture blood samples. Furthermore, any sample collected reflects the conditions of the same at the instant of collection that gets frozen in time, whereas the disease progression is often very dynamic and rapid. The precise metering and dispensing of samples in extreme POC settings is another matter of concern. Pipetting or automated dispensing is known to address such issues quite routinely in more sophisticated settings, but it may be ideal, in terms of implementation by minimally trained personnel, by the much simpler dispensing by some simple-to-use droppers or direct absorption onto the test kit without precise in situ metering. The design for manufacturing of such kits, for on-field-issues, therefore may preferably accommodate such lack of control in sample acquisition and dispensing and offset the same through innovations that ensure an assured level of precision of the sample management by the internal fluidic design of the testing platform. Further, since the POC-based tests may be conducted in a wide variety of locations ranging from centralized labs, hospital emergency rooms, clinician's office, community centres and the patient's home, particular aspects of process control in each of these environments may be normalized by infusing adequate ‘training’ data in a data-driven analytic framework so that any bias in the testing environment may be suitably adjusted.5.3. Manufacturing aspects
The integration of different work elements of the test protocol may also compromise with the scalability of device manufacturing. Notably, in molecular diagnostics, minimizing the numbers of manual steps and avoiding exposure of the reagents to uncontrolled ambience are two key aspects that need to be in-built with the protype design stage itself so as to safeguard from carryover contamination that may spoil the test altogether. The inclusion of complex embodiments including external field-mediated effects (such as electrical and magnetic) in the diagnostic device technology may eliminate these issues but would otherwise make the device complex, non-scalable (due to the dependence on lab-based microfabrication technologies) and expensive. Most of the lab-centric manufacturing technologies are only suitable for proof-of-concept demonstration and prototyping but not efficient for large-scale manufacturing because of their low throughput and non-standardized quality control with an industrially-acceptable benchmark. The fabrication of paper-based devices, performed by different printing technologies, is inherently more scalable than traditional polymer-microfluidics fabrication paradigms,161 although the large-scale manufacturing of different types of paper architectures commonly used for NAATs (such as folded paper kits) remains to be standardized on a large-scale, unlike the 2D LFA strips. The sample-to-answer NAAT devices in the format of a portable box appear to be more amenable to manufacturing upscaling, considering the established industrial standards of injection moulding. The holistic purview of design-integrated manufacturing including such favourable scalability aspects, materials selection and digital convergence (such as Industry 4.0) may lead to an ecosystem-based new model of manufacturing and supply chain of low-cost diagnostic devices as an alternative to established medical device production approaches. The infusion of extreme POC diagnostic tests in the commercialization space demands a convergence of the aspects of new (emerging) materials, design and automation, manufacturing in the modern digital twin (cyber-physical) framework, fostering innovation-driven start-ups and a complete end-to-end solution including supply-chain ecosystem for affordable and accessible diagnostic tests of infectious diseases executed at resource-limited settings.5.4. Validation
Clinical validation at different levels before the test authentication is also a strong hindrance. The enormous heterogeneity in the test samples, attributable to extremely personalized patient-centric features, cannot be sufficiently generalized in organized clinical validation. Often, the validation is done with the help of skilled personnel in reasonably controlled settings whereas the on-field resource-limited adaptations of the same test are expected to perform in more testing circumstances, emphasizing the shift of focus from POC to extreme POC.162 Currently, the methods of validation as per regulatory guidelines are over-structured, leaving little room for getting credits due to assay simplification and instrumentation inventiveness, whereas establishing the sensitivity and specificity limits as per classical NAAT-based norms remains the central considerations. Ironically, many regulatory authorities establish the acceptability of all types of NAATs by approaching the PCR test standards, whereas the rapid antigen or antibody tests have much less stringent acceptance criteria. This leaves little scope of promoting POC-based NAATs that may be effectively formatted like a rapid test in a sample-to-answer framework. Such examples emphasize setting up the validation framework as per the envisaged merits of the test and its utilitarian importance rather than a straight-jacketed guideline that applies univocally for all the NAATs.5.5. Reagent supply chain
The supply chain, transportation and storage of the reagents for the test may emerge to be a serious bottleneck for remote continuum healthcare. More stable reagents at elevated temperatures (as close to the ambient as possible) with enhanced shelf life are the most desirable and may be best realized using their lyophilized versions that may be diluted in any standard buffer solution while running the test. Lyophilized reagents,163 commonly used in molecular diagnostics, undergo a specific freeze-drying process to enhance their stability and facilitate long-term storage. In this process, the lyophilized reagent is initially cooled to a low temperature, typically 85 °C or lower, within a dedicated freeze-drying apparatus. The primary drying phase involves reducing the pressure in the surrounding environment below the triple point of water. Applying a modest amount of heat then activates the free ice crystals to undergo sublimation, transitioning directly from a solid to a gaseous phase. This phase removes approximately 95 percent of the water, transforming the product or sample from a glassy frozen state into a predominantly dry powder. Subsequently, the secondary drying phase eliminates residual water molecules adsorbed to the product or sample, resulting in a lyophilized powder with a water content of one to two percent. This lyophilized powder is significantly lighter and smaller in volume than the original product or sample. To prepare the lyophilized powder for use, a process called reconstitution is employed. This entails adding sterile, distilled water or an appropriate buffer to form a solution. Lyophilization, by avoiding the need for dehydration through heating, provides a convenient and safe method for the extended storage of molecular reagents and assay products while maintaining their activity. After reconstitution, all reagents become saturated with water in solution. Lyophilized molecular reagents can be stored at ambient temperature for up to two years, reducing costs, packaging requirements, and environmental impact. The use of pre-formulated lyophilized reagents containing all necessary components for NAATs simplifies processes, minimizes hands-on time, and is crucial for enhancing laboratory throughput and obtaining time-sensitive results. For applications in democratized diagnostics, where infrastructure limitations and point-of-use considerations may hinder low-temperature shipping, the stability of room temperature-stable lyophilized reagents becomes crucial. Despite the special resources required for their preparation, the benefits of lyophilization facilities can be realized globally, contributing to improved accessibility and efficiency in diagnostic procedures across diverse locations.5.6. Detection readout
Simple visual readouts such as discernible colours to indicate the presence or absence of infection are greatly advantageous for low-resource settings but may result in ambiguities in manual interpretation when the colour changes are subtler because of off-design conditions during uncontrolled operation. This kind of subjectivity may be largely offset by the automated analysis of the test readouts using the digital technologies and AI tools, often mediated by a smartphone app. This intervention also opens up the possibilities of quantitative analytics and interpretation on the pathogenic load, instead of giving a mere yes/no type answer in the presence of the infection or otherwise. However, a suitable normalization of the camera settings, background illumination, etc., needs to be exercised so that inaccuracies do not creep in due to lack of expertise or control in acquiring the image and processing the optical readout.6. Summary and outlook: beyond the technology boundaries
Isothermal NAATs have covered a wide ground quite rapidly to emerge as a very promising alternative to the resource-intensive PCR tests for infectious disease detection for resource-poor settings. However, a trend in many such developments, which is simultaneously heartening and alarming, is the over-emphasis of achieving higher scientific pursuits to an extent that often beats the central goal of extreme POC diagnostics, its simplicity from all considerations. Perhaps, the common perception of equivalencing the efficacy of a technology with its scientific complexity here acts as the primary deterrent. A pioneer of the High Renaissance, the famous Leonardo Da Vinci, was the proponent of the motto “simplicity is the ultimate sophistication”. A simple and affordable means of achieving sample-to-answer integration in a POC-based NAAT for infectious diseases detection is by no means an ordinary proposition, and in effect is elegant at its best, which is much more preferred than a more complex design that may yield marginally more accurate solutions but cannot be implemented by any means in uncontrolled settings. The design of any emerging extreme POC-based NAAT, therefore, needs to address multiple challenges with robust yet simple approach rather than aiming for innovations that are very much unlikely to have success without supporting sophistication. While the aspect of simplicity in the design by itself is somewhat subjective, it can be envisioned by organizing open-source hardware (such as Arduino) and software to the extent feasible and minimize the use of disposables per test. The use of “green” materials and forms of energy for sustainable, biodegradable and affordable dissemination of the otherwise delicate NAATs, miniaturization with minimal component embodiment and maximining the exploitation of the versatilities of the microcontrollers, LED lamps, PCBs as well as smartphones, use of functional nanomaterials for test signal amplification, IoT integration and use of established image processing, feature extraction, pattern recognition and denoising algorithms and data analytics via AI/ML, nucleic acid extraction-free test protocols, minimizing the ambient exposure and pipetting of the sample and inclusion of a control sensor to ensure that the patient sample has adequate quantity and quality of the nucleic acid that may safeguard against erroneous test results despite extreme lack of control in sample collection and processing may all conform to user-friendly steps needing no specialized knowledge in molecular biology. Such interventions can make the test simultaneously accessible and affordable, and to an extent transformative, by democratizing its reach to the poorest segments of the community. For an example of early initiation of such efforts, one may refer to the DIY Diagnostics focus of the Freshman Research Initiative, University of Texas at Austin (https://fri.cns.utexas.edu/research-streams/diy-diagnostics). The aim of this stream is developing straightforward diagnostic tools accessible to individuals with minimal or no medical or scientific training. Taking inspiration from the user-friendly home pregnancy test, they adopted an open approach, not restricted to specific technology platforms, by integrating molecular biology, chemistry, electronics, and computer programming in diverse combinations. By exploring various avenues of technology, such endeavours strive to create diagnostics that are simple, user-friendly, and can be easily utilized by individuals without extensive scientific expertise. Quality assured benchmarking and compliance are likely to miss in the early stages of these innovations but may be ensured strategically as such outcomes claim up the ladders of progressively higher technology readiness levels (TRLs).In a similar spirit, using open source software for primer design and adapting home-made recipes for making probes and enzymes may enable a cottage-industry like framework for the test reagents, which simply need quality-controlled environment and processes to be fit enough for the intended test. This spirit of converting creative pursuit to a diagnostic technology with minimal user steps in between can greatly help in facilitating the sensing of traces of pathogens in our bodies, food and environment. This knowledge base may improve the efficacy of any clinical decision dramatically by considering the diagnostic data as against the traditional intuition or experience-based approaches. As per the famous quote of William Thomson (Lord Kelvin), “if you can't measure it, you can't improve it”. Therefore, democratization in access to diagnostic data may be transformational in making the world a better place for living and safeguard us against the adversities of future epidemics and pandemics.
During the COVID-19 pandemic, several POC adaptations of the NAAT-based tests emerged, some of which got emergency authorization for on-field adaptation, whereas some others struggled to get approvals simply because any rational framework for POC NAATs simply did not exist and the existing RT-PCR test norms were mostly referred to instead, creating a situation as if apples are compared with oranges. For example, while rapid antigen tests with much lower sensitivity got approved quite easily because of satisfying the concerned benchmark, rapid NAATs of much higher efficacy but equivalent user-friendliness, simplicity and cost structure, which struggled hard to get cleared because of undue benchmarking with the RT-PCR tests that demand the kinds of infrastructure that are unimaginable in the underserved settings. Several noteworthy efforts in bringing out novel molecular diagnostic technologies for the purpose of COVID-19 testing got buried because of inconsistent regulatory policies where, instead of the criticalities associated with satisfactory performance in extreme POC settings, the aspects of clinical sensitivity and specificity were given the prime consideration. Another commonly ignored aspect in benchmarking the NAATs is to assess its efficacy in a clinical setting whereas off-design conditions on the field may destroy the performance dramatically. The regulatory guidelines of the respective National and territorial regulatory bodies such as the Food and Drug Administration (FDA) of the United States of America, Conformité Européenne (CE) of the European Union use their own classifications of the medical diagnostic tests based on their risk to the users and intended use. These classifications, however, are widely varying in specifications and have enough rooms of ambiguous interpretation based on the nature of the test kit, test method or the testing instrument. While these are indeed helping guidelines for certain checks and balances, their rigid interpretation is likely to deter the timely market entry of disruptive new simple technologies only because no established regulatory guidelines truly cover the scope of their performance. Having said that, adherence to universally accepted manufacturing standards as evidenced by ISO13485 certification from the prototyping level itself may greatly benefit the lab-to-market journey of the diagnostic test and in fact may enable expediting the same by virtue of quality assurance in all the stages of product development, encompassing fabrication, packaging, labelling, storage, transportation, deployment and troubleshooting.
Fortunately, with the boom in AI and ML, many of the common limitations of the POC-based diagnostic procedures mentioned as above appear to be overcome in a more pragmatic way than ever before. However, this also brings in the danger of infusing an ‘over-dose’ of AI into the diagnostic technology framework and undermine many of the gold standard physical principles that can by no means be ignored. A possible balancing measure here is perhaps not to attempt using AI as a stand-alone technology for the diagnostic purposes but amalgamate the same judiciously with the first principles of physics, chemistry, physiology and device technology so as to capture even the faintest of the disease-specific signals from the test sample with high efficacy. This balancing act may, in principle, offset several inevitable on-field constraints such as practical limitations in the skills of the frontline health workers or in some cases the patients themselves working in a self-testing mode, who are by no means nearly as experts as specialized and trained technicians in executing the test. If interlaced carefully, such cocktail of AI and physical systems (often recognized as cyber-physical systems) may emerge to be effective in carrying the burden of reasonable accurate detection by efficient feature extraction from the raw sensing data and denoising the same. Moreover, the algorithmic aspect of the detection may not only facilitate capturing the onset of the disease precisely at its first place but also for assessing the epidemiology of the infection towards the management of its outbreak. On the flip side, over-emphasis on the data-science centric aspects may hold the possibility of masking the due central focus on the core diagnostic technology itself (for example, over-claiming the efficacies of complete non-invasive tests as against the standard invasive procedures), giving rise to unrealistic claims of efficacy and unwarranted potential distractions brought in by other peripheral features (for example, attractively smart mobile application interface and unnecessary fancy electronic embodiments in response to consumerism).
Beyond the data-science infused diagnostic tests, the exploitation of digital transformation in revolutionizing the dissemination of NAATs at the bottom of the community pyramid is indeed promising but needs to be exercised with utmost care with due consideration of the end objective, which is not to showcase the muscle power of the state-of-the-art information and data sciences but to result in a perspective that brings in benefits to all the concerned stake holders. In a physical examination of a patient, a doctor aims to interlace different holistic aspects of clinical practice and hardly depends on the outcome of any stand-alone diagnostic test as the sole guiding factor, at least on the first place. This natural algorithmic approach emulating a doctor's wisdom is something that needs to complement the outcome of the POC-based NAATs even in the modern digital era. Otherwise, the digital approach of managing the infection would merely turn the same into some number-crunching exercise as against addressing the concerns of human patients in a personalized manner. The POC-adapted NAATs, therefore, must be sensitive to the target product profile (TPP) for the intended use in harmony with the overall patient management ecosystem.164 This should ideally include the particular measures to ensure data privacy and security2,165 and to ensure widespread and sustainable implementation.166 Mobile-based Unified Payment Interface (UPI) can be leveraged for connecting the receipts and payments made by the relevant stakeholders, be it patient, healthcare provider or insurance provider or pharmacy personnel.167
While outlining the technical aspects of concern, it also needs to be emphasized that the success or lack of the same on any POC diagnostic technology for the resource-limited and underserved settings delves critically into several other aspects and concerns (socio-economic, cultural, legal, ethical, regulatory and other policy-driven factors) that cumulatively may hold the decisive key towards its functional success, often in preference to the technological innovations alone. For instance, the fear of stigmatization of any patient being detected as positive, as evidenced from being singled-out in their communities, work places, or even families, is a known deterrent against going for diagnostic testing even if the test is done for free.168,169 Further, any field trial without informed consent of the participating human subject is ethically incorrect. Compromising with the privacy and security of the medical data may often be hard to eliminate completely because of the direct interfacing of the patient with frontline personnel who may work with smartphone-based interfaces for sharing the patient's diagnostic outcome with other stake holders (for example, remote doctors), which may also raise concern for the protection of the private medical data. There is progressively more use of emerging technologies such as blockchain to ensure the security and privacy of such data at different layers as well as adequate encryption and decryption of the same as per prescribed norms. Infectious disease testing policies of any significant impact in global health indeed need to be sensitive to all such factors and should take adequate measures to deal with them acceptably rather than ignoring it summarily. For example, there should be a sensitive assessment and mitigation of any potential adverse social response, for instance, on conducting HIV screening at dedicated centres that can stigmatize and single-out individuals inadvertently, probing criminal familial association from NAATs, or potential fear of loss of privacy due to the accessibility of data for some socially-sensitive infections to others who may have undue bias or prejudice developed from some social taboo. Overall, in the field testing as well as community centric adaptation stage, these aspects need to be managed both strategically and ethically, ensuring no misalignment with the expectations from the concerned communities.
Ensuring an alignment of all the stakeholders, preferably via an ecosystem approach, appears to be central in introducing the new diagnostic tools to address any concerns of the common people. Safety assurance, compliance to regulations and evidenced efficacy must always precede in a transparent manner before any ground level implementation rather than hurriedly infusing new technologies without adequate checks and balances. Measures of surveillance need to ensure community security, albeit without harming individual's rights to decision-making and freedom at the same time and not imposing any diagnostic intervention as a forceful measure. Field adaptations that are imposed top-down in a rigid manner by influential or powerful authorities from well-to-do to emerging communities are unethical unless exercised with due care to ensure no harm of the participants. One major gap in this regard is the lack of established universal standards unlike the traditional industrial frameworks. Making a structured effort to formulate robust standards for NAATs for POC-based applications may indeed ensure compliance to regulations, standards and ethics and at the same time benefit all the stakeholders. Programmed awareness campaigns for the target individuals and potential beneficiaries may be organized to ensure no hiccups during grassroot level implementation. An all-inclusive social engagement, encompassing capacity building or workforce development and absorbing them into the diagnostic framework as a financially benefitting stakeholder (for example, engaging them in device fabrication, test kit supply-chain and sample collection and testing with some assured margin from the revenue generated) may not only address the problems of healthcare but in isolation also foster self-employment opportunities for the underprivileged youth and women, as per the direct experience of the author in working with diagnostic technologies in resource-limited settings.
Finally, the most important aspect to keep in purview for the best outcome for any singled out diagnostic test is that it forms the basis of only one aspect of the disease diagnosis and management but does not alone hold the capability of providing any deterministic clinical decision unless the test is itself a laboratory-benchmarked gold standard, which is not the scenario of the simplified POC-based tests. Keeping this in view, the diagnosis accuracy (not diagnostic accuracy) of simplified molecular tests may be enhanced by interlacing the diagnostic test outcome with other supporting evidences such as the patient metadata and demographic information, body vitals, medical history and co-morbidity, travel history, and symptoms presented. This approach of amalgamating the diagnostic test results with other supporting factors akin to what a doctor does in a clinical setting and translating the same with a logic-based algorithm that aims to mimic a ‘doctor's mind’ albeit without having any inevitable distraction or fatigue of a physical doctor holds the future of arriving at a judicious decision on any presumptive or suspected infection and its prompt management in a low-resource setting. On the one hand, training the diagnostic technology with datasets on high-frequency measurements of validated biomarkers under different conditions covering the instances of potential cross-interferences with other biomarkers using AI-based tools may improve the test accuracy despite all simplifications in the underlying technology. On the other side, coding the other supportive evidences into a scoring system framework that applies complementary analytic models may de-convolute inevitable heterogeneity in different forms of evidence and algorithmically extract patient-specific insights on the probability of being infected. Combinedly, this approach could enable arriving at a data-substantiated and evidence-based clinical decision support with personalized treatment strategies towards the holistic management of infectious diseases. In the modern digital era, this might be a true weapon to the physical doctor to arrive at the diagnosis and therapeutic judgment quickly and accurately. The key here is to balance the AI and the physical technology judiciously and not being overawed by the capabilities of AI alone. With such a delicate amalgamation of physiological reference and data science without prejudice, the extremely difficult task of infectious disease detection and management at the bottom of the pyramid may become a reality not by eliminating the role of a specialist doctor but by combining holistic insights on the potential disease into a logical decision framework that turns out to be a friendly companion not only for the aid of the doctors and the patient but also for the benefit of the socio-economic system at large by enhancing healthcare and underserved livelihood at the same time.
Conflicts of interest
The author declares no conflict of interest.Acknowledgements
The author acknowledges the help of Dr. Sudip Nag for thorough proofreading and further inputs on this topic. The author acknowledges Dr. Arindam Mondal, Dr. Aditya Bandopadhyay and Dr. Satadal Saha for the stimulating discussions on some of the contents of this work. The author further acknowledges gratefully SERB, Department of Science and Technology, Government of India, for his J. C. Bose National Fellowship and DSIR, Government of India, for their support under the Common Research and Technology Development Hub (CRTDH) program.References
- J. A. Soller, T. Bartrand, N. J. Ashbolt, J. Ravenscroft and T. J. Wade, Water Res., 2010, 44, 4736–4747 CrossRef CAS PubMed.
- Geneva: World Health Organization, 2021. Licence: CC BY-NC-SA 3.0 IGO.
- D. C. Christodouleas, B. Kaur and P. Chorti, ACS Cent. Sci., 2018, 4, 1600–1616 CrossRef CAS PubMed.
- Y. Zhu, M. Zhang, Z. Jie and S. Tao, Front. Microbiol., 2022, 13, 1074289 CrossRef PubMed.
- P. Maffert, S. Reverchon, W. Nasser, C. Rozand and H. Abaibou, Eur. J. Clin. Microbiol. Infect. Dis., 2017, 36, 1717–1731 CrossRef CAS PubMed.
- W. Muttamba, W. Ssengooba, R. Sekibira, B. Kirenga, A. Katamba and M. Joloba, PLoS One, 2018, 13(3), e0194741 CrossRef PubMed.
- A. K. Akobeng, Acta Paediatr., 2006, 96, 338–341 CrossRef PubMed.
- H. Kettler, K. White and S. Hawkes, J & UNDP/World Bank/WHO Special Programme for Research and Training in Tropical Diseases, 2004.
- B. Yang, J. Kong and X. Fang, Talanta, 2019, 204, 685–692 CrossRef CAS PubMed.
- C. Pang, C. Lee and K. Y. Suh, J. Appl. Polym. Sci., 2013, 130, 1429–1441 CrossRef CAS.
- D. Son, J. Lee, S. Qiao, R. Ghaffari, J. Kim, J. E. Lee, C. Song, S. J. Kim, D. J. Lee, S. W. Jun, S. Yang, M. Park, J. Shin, K. Do, M. Lee, K. Kang, C. S. Hwang, N. Lu, T. Hyeon and D. H. Kim, Nat. Nanotechnol., 2014, 9, 397–404 CrossRef CAS PubMed.
- S. Wang, T. Chinnasamy, M. A. Lifson, F. Inci and U. Demirci, Trends Biotechnol., 2016, 34, 909–921 CrossRef CAS PubMed.
- Kenry, J. C. Yeo and C. T. Lim, Microsyst. Nanoeng., 2016, 2, 16043 CrossRef CAS PubMed.
- Y. Xu, G. Zhao, L. Zhu, Q. Fei, Z. Zhang, Z. Chen, F. An, Y. Chen, Y. Ling, P. Guo, S. Ding, G. Huang, P. Y. Chen, Q. Cao and Z. Yan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 18292–18301 CrossRef CAS PubMed.
- D. Jarchi, P. Charlton, M. Pimentel, A. Casson, L. Tarassenko and D. A. Clifton, Healthc. Technol. Lett., 2019, 6, 19–26 CrossRef PubMed.
- P. H. Charlton, D. A. Birrenkott, T. Bonnici, M. A. F. Pimentel, A. E. W. Johnson, J. Alastruey, L. Tarassenko, P. J. Watkinson, R. Beale and D. A. Clifton, IEEE Rev. Biomed. Eng., 2018, 11, 2–20 Search PubMed.
- T. Mishra, M. Wang, A. A. Metwally, G. K. Bogu, A. W. Brooks, A. Bahmani, A. Alavi, A. Celli, E. Higgs, O. Dagan-Rosenfeld, B. Fay, S. Kirkpatrick, R. Kellogg, M. Gibson, T. Wang, E. M. Hunting, P. Mamic, A. B. Ganz, B. Rolnik, X. Li and M. P. Snyder, Nat. Biomed. Eng., 2020, 4, 1208–1220 CrossRef CAS PubMed.
- F. E. Shamout, T. Zhu, P. Sharma, P. J. Watkinson and D. A. Clifton, IEEE J. Biomed. Health. Inform., 2020, 24, 437–446 Search PubMed.
- Y. Baik, L. Cilloni, E. Kendall, D. Dowdy and N. Arinaminpathy, Epidemics, 2022, 41, 100631 CrossRef CAS PubMed.
- Z. Zhu, X. Lian, X. Su, W. Wu, G. A. Marraro and Y. Zeng, Respir. Res., 2020, 21, 224 CrossRef CAS PubMed.
- L. Garibyan and N. Avashia, J. Invest. Dermatol., 2013, 133, e6 CrossRef CAS PubMed.
- M. M. Gibani, C. Toumazou, M. Sohbati, R. Sahoo, M. Karvela, T. K. Hon, S. D. Mateo, A. Burdett, K. Y. F. Leung, J. Barnett, A. Orbeladze, S. Luan, S. Pournias, J. Sun, B. Flower, J. Bedzo-Nutakor, M. Amran, R. Quinlan, K. Skolimowska, C. Herrera, A. Rowan, A. Badhan, R. Klaber, G. Davies, D. Muir, P. Randell, D. Crook, G. P. Taylor, W. Barclay, N. Mughal, L. S. P. Moore, K. Jeffery and G. S. Cooke, Lancet Microbe, 2020, 1, e300–e307 CrossRef CAS PubMed.
- V. Lalande, L. Barrault, S. Wade, C. Eckert, J. C. Petit and F. Barbut, J. Clin. Microbiol., 2011, 49, 2714–2716 CrossRef PubMed.
- J. Bell, A. Bonner, D. M. Cohen, R. Birkhahn, R. Yogev, W. Triner, J. Cohen, E. Palavecino and R. Selvarangan, J. Clin. Virol., 2014, 61, 81–86 CrossRef CAS PubMed.
- A. J. Gorzalski, H. Tian, C. Laverdure, S. Morzunov, S. C. Verma, S. VanHooser and M. W. Pandori, J. Clin. Virol., 2020, 129, 104501 CrossRef CAS PubMed.
- J. W. P. Teo, D. Chiang, R. Jureen and R. T. P. Lin, Diagn. Microbiol. Infect. Dis., 2015, 83, 261–262 CrossRef CAS PubMed.
- E. K. Rames and J. Macdonald, Appl. Microbiol. Biotechnol., 2019, 103, 8115–8125 CrossRef CAS PubMed.
- I. M. Lobato and C. K. O'Sullivan, Trends Analyt. Chem., 2018, 98, 19e35 Search PubMed.
- P. Wang, C. Ma, X. Zhang, L. Chen, L. Yi, X. Liu, Q. Lu, Y. Cao and S. Gao, Front. Cell. Infect. Microbiol., 2021, 11, 680728 CrossRef CAS PubMed.
- I. Torjesen, BMJ, 2021, 372, n287 CrossRef PubMed.
- H. Wu, P. Zhao, X. Yang, J. Li, J. Zhang, X. Zhang, Z. Zeng, J. Dong, S. Gao and C. Lu, Front. Microbiol., 2020, 11, 1015 CrossRef PubMed.
- V. L. Dao Thi, K. Herbst, K. Boerner, M. Meurer, L. P. Kremer, D. Kirrmaier, A. Freistaedter, D. Papagiannidis, C. Galmozzi, M. L. Stanifer, S. Boulant, S. Klein, P. Chlanda, D. Khalid, I. Barreto Miranda, P. Schnitzler, H. G. Kräusslich, M. Knop and S. Anders, Sci. Transl. Med., 2020, 12, eabc7075 CrossRef CAS PubMed.
- T. Notomi, Y. Mori, N. Tomita and H. Kanda, J. Microbiol., 2015, 53, 1–5 CrossRef CAS PubMed.
- J. Song, C. Liu, M. G. Mauk, S. C. Rankin, J. B. Lok, R. M. Greenberg and H. H. Bau, Clin. Chem., 2017, 63, 714–722 CrossRef CAS PubMed.
- M. El-Tholoth, E. Anis and H. H. Bau, Analyst, 2021, 146, 1311 RSC.
- Y. Mori, T. Hirano and T. Notomi, BMC Biotechnol., 2006, 6, 3 CrossRef PubMed.
- E. A. Phillips, T. J. Moehling, S. Bhadra, A. D. Ellington and J. C. Linnes, Anal. Chem., 2018, 90, 6580–6586 CrossRef CAS PubMed.
- M. P. De Baar, E. C. Timmermans, M. Bakker, E. de Rooij, B. Van Gemen and J. Goudsmit, J. Clin. Microbiol., 2001, 39, 1895–1902 CrossRef CAS PubMed.
- G. D. Suarez, D. A. Suarez, Y. Y. K. Tang, J. X. Zhang, J. Li, S. Nagl and P. P. H. Cheung, Anal. Methods, 2022, 14, 378 RSC.
- C. H. Woo, S. Jang, G. Shin, G. Y. Jung and J. W. Lee, Nat. Biomed. Eng., 2020, 12, 1168–1179 CrossRef PubMed.
- J. G. Carter, L. O. Iturbe, J. H. A. Duprey, I. R. Carter, C. D. Southern, M. Rana, C. M. Whalley, A. Bosworth, A. D. Beggs, M. R. Hicks, J. H. R. Tucker and T. R. Dafforn, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100347118 CrossRef CAS PubMed.
- M. Gavrilov, J. Y. C. Yang, R. S. Zou, W. Ma, C. Y. Lee, S. Mohapatra, J. Kang, T. W. Liao, S. Myong and T. Ha, Nat. Commun., 2022, 13, 6312 CrossRef CAS PubMed.
- J. M. Rosenbohm, C. M. Klapperich and M. Cabodi, Anal. Chem., 2022, 94, 3956–3962 CrossRef CAS PubMed.
- F. Bošković, J. Zhu, R. Tivony, A. Ohmann, K. Chen, M. F. Alawami, M. Đorđević, N. Ermann, J. Pereira-Dias, M. Fairhead, M. Howarth, S. Baker and U. F. Keyser, Nat. Nanotechnol., 2023, 18, 290–298 CrossRef PubMed.
- P. Trémeaux, S. Lhomme, F. Abravanel, S. Raymond, C. Mengelle, J. M. Mansuy and J. Izopet, J. Clin. Virol., 2020, 129, 104541 CrossRef PubMed.
- M. Leelawong, S. L. Mitchell, R. C. Fowler, E. Gonzalez, S. Hughes, M. P. Griffith, J. W. Marsh, L. H. Harrison and J. L. Rakeman, Diagn. Microbiol. Infect. Dis., 2021, 101, 115468 CrossRef CAS PubMed.
- R. Barrangou and L. A. Marraffini, Mol. Cell, 2014, 54, 234–244 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval, S. Moineau, D. A. Romero and P. Horvath, Science, 2007, 315, 1709–1712 CrossRef CAS PubMed.
- E. V. Koonin and K. S. Makarova, RNA Biol., 2013, 10, 679–686 CrossRef CAS PubMed.
- T. Hou, W. Zeng, M. Yang, W. Chen, L. Ren, J. Ai, J. Wu, Y. Liao, X. Gou, Y. Li, X. Wang, H. Su, B. Gu, J. Wang and T. Xu, PLoS Pathog., 2020, 16, e1008705 CrossRef CAS PubMed.
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, Nat. Microbiol., 2020, 5, 536–544 CrossRef PubMed.
- M. M. Kaminski, O. O. Abudayyeh, J. S. Gootenberg, F. Zhang and J. J. Collins, Nat. Biomed. Eng., 2021, 5, 643–656 CrossRef CAS PubMed.
- R. Gupta, T. A. Kazi, D. Dey, A. Ghosh, V. Ravichandiran, S. Swarnakar, S. Roy, S. R. Biswas and D. Ghosh, Appl. Microbiol. Biotechnol., 2021, 105, 7593–7605 CrossRef CAS PubMed.
- W. Feng, H. Peng, J. Xu, Y. Liu, K. Pabbaraju, G. Tipples, M. A. Joyce, H. A. Saffran, D. L. Tyrrell, S. Babiuk, H. Zhang and X. C. Le, Anal. Chem., 2021, 93, 12808–12816 CrossRef CAS PubMed.
- C. S. Talwar, K. H. Park, W. C. Ahn, Y. S. Kim, O. S. Kwon, D. Yong, T. Kang and E. Woo, Biosensors, 2021, 11, 301 CrossRef CAS PubMed.
- J. Gong, G. Zhang, W. Wang, L. Liang, Q. Li, M. Liu, L. Xue and G. Tang, Sci. Rep., 2021, 11, 12800 CrossRef CAS PubMed.
- J. Zhao, C. Ao, Z. Wan, E. E. Dzakah, Y. Liang, H. Lin, H. Wang and S. Tang, Virus Res., 2021, 303, 198505 CrossRef CAS PubMed.
- Y. You, P. Zhang, G. Wu, Y. Tan, Y. Zhao, S. Cao, Y. Song, R. Yang and Z. Du, Front. Microbiol., 2021, 12, 700016 CrossRef PubMed.
- L. Jiang, W. Zeng, W. Wu, Y. Deng, F. He, W. Liang, M. Huang, H. Huang, Y. Li, X. Wang, H. Su, S. Pan and T. Xu, Emerging Infect. Dis., 2021, 27, 2379–2388 CrossRef CAS PubMed.
- P. Fozouni, S. Son, M. D. de León Derby, G. J. Knott, C. N. Gray, M. V. D'Ambrosio, C. Zhao, N. A. Switz, G. R. Kumar, S. I. Stephens, D. Boehm, C. L. Tsou, J. Shu, A. Bhuiya, M. Armstrong, A. R. Harris, P. Y. Chen, J. M. Osterloh, A. Meyer-Franke, B. Joehnk, K. Walcott, A. Sil, C. Langelier, K. S. Pollard, E. D. Crawford, A. S. Puschnik, A. Phelps, M. Kistler, J. L. DeRisi, J. A. Doudna, D. A. Fletcher and M. Ott, Cell, 2021, 184, 323–333 CrossRef CAS PubMed.
- T. Y. Liu, G. J. Knott, D. C. J. Smock, J. J. Desmarais, S. Son, A. Bhuiya, S. Jakhanwal, N. Prywes, S. Agrawal, M. D. de León Derby, N. A. Switz, M. Armstrong, A. R. Harris, E. J. Charles, B. W. Thornton, P. Fozouni, J. Shu, S. I. Stephens, G. R. Kumar, C. Zhao, A. Mok, A. T. Iavarone, A. M. Escajeda, R. McIntosh, S. Kim, E. J. Dugan, IGI Testing Consortium, K. S. Pollard, M. X. Tan, M. Ott, D. A. Fletcher, L. F. Lareau, P. D. Hsu, D. F. Savage and J. A. Doudna, Nat. Chem. Biol., 2021, 17, 982–988 CrossRef CAS PubMed.
- J. Joung, A. Ladha, M. Saito, N. G. Kim, A. E. Woolley, M. Segel, R. P. J. Barretto, A. Ranu, R. K. Macrae, G. Faure, E. I. Ioannidi, R. N. Krajeski, R. Bruneau, M. W. Huang, X. G. Yu, J. Z. Li, B. D. Walker, D. T. Hung, A. L. Greninger, K. R. Jerome, J. S. Gootenberg, O. O. Abudayyeh and F. Zhang, N. Engl. J. Med., 2020, 383, 1492–1494 CrossRef CAS PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm, S. Siddiqui, B. M. Shaw, G. Adams, T. F. Kosoko-Thoroddsen, M. E. Kemball, J. N. Uwanibe, F. V. Ajogbasile, P. E. Eromon, R. Gross, L. Wronka, K. Caviness, L. E. Hensley, N. H. Bergman, B. L. MacInnis, C. T. Happi, J. E. Lemieux, P. C. Sabeti and C. Myhrvold, Nat. Commun., 2020, 11, 5921 CrossRef CAS PubMed.
- R. Hajian, S. Balderston, T. Tran, T. deBoer, J. Etienne, M. Sandhu, N. A. Wauford, J. Y. Chung, J. Nokes, M. Athaiya, J. Paredes, R. Peytavi, B. Goldsmith, N. Murthy, I. M. Conboy and K. Aran, Nat. Biomed. Eng., 2019, 3, 427–437 CrossRef CAS PubMed.
- J. G. Carter, L. Orueta Iturbe, J. H. A. Duprey, I. R. Carter, C. D. Southern, M. Rana, C. M. Whalley, A. Bosworth, A. D. Beggs, M. R. Hicks, J. H. R. Tucker and T. R. Dafforn, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100347118 CrossRef CAS PubMed.
- M. Patchsung, K. Jantarug, A. Pattama, K. Aphicho, S. Suraritdechachai, P. Meesawat, K. Sappakhaw, N. Leelahakorn, T. Ruenkam, T. Wongsatit, N. Athipanyasilp, B. Eiamthong, B. Lakkanasirorat, T. Phoodokmai, N. Niljianskul, D. Pakotiprapha, S. Chanarat, A. Homchan, R. Tinikul, P. Kamutira, K. Phiwkaow, S. Soithongcharoen, C. Kantiwiriyawanitch, V. Pongsupasa, D. Trisrivirat, J. Jaroensuk, T. Wongnate, S. Maenpuen, P. Chaiyen, S. Kamnerdnakta, J. Swangsri, S. Chuthapisith, Y. Sirivatanauksorn, C. Chaimayo, R. Sutthent, W. Kantakamalakul, J. Joung, A. Ladha, X. Jin, J. S. Gootenberg, O. O. Abudayyeh, F. Zhang, N. Horthongkham and C. Uttamapinant, Nat. Biomed. Eng., 2020, 4, 1140–1149 CrossRef CAS PubMed.
- J. Xu, Y. Ma, Z. Song, W. Sun, Y. Liu, C. Shu, H. Hua, M. Yang and Q. Liang, Heliyon, 2023, 9, e13190 CrossRef CAS PubMed.
- H. T. Chan, M. H. T. Keung, I. Nguyen, E. L. O. Ip, S. M. Chew, D. Siler, M. Saville and D. Hawkes, J. Clin. Virol. Plus, 2022, 2, 100079 CrossRef PubMed.
- L. Garibyan and N. Avashia, J. Invest. Dermatol., 2013, 133, 1–4 CrossRef PubMed.
- CDC, U.S. Department of Health and Human Services, CDC, 2019.
- A. C. Singer, H. Shaw, V. Rhodes and A. Hart, Front. Microbiol., 2016, 7, 1728 Search PubMed.
- F. Prestinaci, P. Pezzotti and A. Pantosti, Pathog. Global Health, 2015, 109, 309–318 CrossRef PubMed.
- World Health Organization, Report to the Secretary-General of the United Nations, 2019.
- J. H. Jorgensen and M. J. Ferraro, Clin. Infect. Dis., 2009, 49, 1749–1755 CrossRef CAS PubMed.
- E. A. Mothershed and A. M. Whitney, Clin. Chim. Acta, 2006, 363, 206–220 CrossRef CAS PubMed.
- A. Deshpande, J. Gans, S. W. Graves, L. Green, L. Taylor, H. B. Kim, Y. A. Kunde, P. M. Leonard, P. E. Li, J. Mark, J. Song, M. Vuyisich and P. S. White, J. Microbiol. Methods, 2010, 80, 155–163 CrossRef CAS PubMed.
- A. Deshpande and P. S. White, Expert Rev. Mol. Diagn., 2012, 12, 645–659 CrossRef CAS PubMed.
- P. A. Wayne, 30th ed CLSI supplement M100, Clinical and Laboratory Standards Institute, 2020.
- E. M. Marlowe and M. J. Bankowski, J. Clin. Microbiol., 2011, 49, S53–S56 CrossRef PubMed.
- S. Leekha, C. L. Terrell and R. S. Edson, Mayo Clin. Proc., 2011, 86, 156–167 CrossRef PubMed.
- H. Wong, L. Louie, R. Y. Lo and A. E. Simor, J. Clin. Microbiol., 2010, 48, 4546–4551 CrossRef PubMed.
- J. Xu, C. Shi, M. Song, X. Xu, P. Yang, G. Paoli and X. Shi, J. Food Sci., 2014, 79, M635–M642 CAS.
- M. Haveri, S. Suominen, L. Rantala, T. Honkanen-Buzalski and S. Pyörälä, Vet. Microbiol., 2005, 106, 97–102 CrossRef CAS PubMed.
- M. N. Thaker, L. Kalan, N. Waglechner, A. Eshaghi, S. N. Patel, S. Poutanen, B. Willey, B. Coburn, A. McGeer, D. E. Low and G. D. Wright, Antimicrob. Agents Chemother., 2015, 59, 1405–1410 CrossRef PubMed.
- K. J. Land, D. I. Boeras, X. S. Chen, A. R. Ramsay and R. W. Peeling, Nat. Microbiol., 2019, 4, 46–54 CrossRef CAS PubMed.
- W. S. Chan, B. S. Tang, M. V. Boost, C. Chow and P. H. Leung, Biosens. Bioelectron., 2014, 53, 105–111 CrossRef CAS PubMed.
- J. J. Storhoff, A. D. Lucas, V. Garimella, Y. P. Bao and U. R. Müller, Nat. Biotechnol., 2004, 22, 883–887 CrossRef CAS PubMed.
- O. Piepenburg, C. H. Williams, D. L. Stemple and N. A. Armes, PLoS Biol., 2006, 4, e204 CrossRef PubMed.
- B. Veigas, D. Machado, J. Perdigão, I. Portugal, I. Couto, M. Viveiros and P. V. Baptista, Nanotechnology, 2010, 21, 415101 CrossRef PubMed.
- R. Ramakrishnan, W. Buckingham, M. Domanus, L. Gieser, K. Klein, G. Kunkel, A. Prokhorova and P. V. Riccelli, Clin. Chem., 2004, 50, 1949–1952 CrossRef CAS PubMed.
- M. Valiadi, S. Kalsi, I. G. Jones, C. Turner, J. M. Sutton and H. Morgan, Biomed. Microdevices, 2016, 18, 18 CrossRef PubMed.
- M. C. Keays, M. O'Brien, A. Hussain, P. A. Kiely and T. Dalton, Bioengineered, 2016, 7, 79–87 CrossRef CAS PubMed.
- A. K. S. Ayfan, J. Macdonald, A. D. Irwin, H. M. Zowawi, B. M. Forde, D. L. Paterson, M. M. Lahra and D. M. Whiley, J. Antimicrob. Chemother., 2022, 77, 2933–2936 CrossRef CAS PubMed.
- M. A. Abdou Mohamed, H. N. Kozlowski, J. Kim, K. Zagorovsky, M. Kantor, J. J. Feld, S. Mubareka, T. Mazzulli and W. C. W. Chan, ACS Nano, 2021, 15, 9379–9390 CrossRef CAS PubMed.
- E. Mokany, S. M. Bone, P. E. Young, T. B. Doan and A. V. Todd, J. Am. Chem. Soc., 2010, 132, 1051–1059 CrossRef CAS PubMed.
- B. J. R. Reddy, U. Hassan, C. Seymour, D. C. Angus, T. S. Isbell, K. White, W. Weir, L. Yeh, A. Vincent and R. Bashir, Nat. Biomed. Eng., 2018, 2, 640–648 CrossRef PubMed.
- B. W. Haak and W. J. Wiersinga, Lancet Gastroenterol. Hepatol., 2017, 2, 135–143 CrossRef PubMed.
- R. P. Dickson, B. H. Singer, M. W. Newstead, N. R. Falkowski, J. R. Erb-Downward, T. J. Standiford and G. B. Huffnagle, Nat. Microbiol., 2016, 1, 16113 CrossRef CAS PubMed.
- K. F. Budden, S. L. Gellatly, D. L. Wood, M. A. Cooper, M. Morrison, P. Hugenholtz and P. M. Hansbro, Nat. Rev. Microbiol., 2017, 15, 55–63 CrossRef CAS PubMed.
- K. Bomsztyk, D. Mar, D. An, R. Sharifian, M. Mikula, S. A. Gharib, W. A. Altemeier, W. C. Liles and O. Denisenko, Crit. Care, 2015, 19, 225 CrossRef PubMed.
- J. Rodriguez-Manzano, K. Malpartida-Cardenas, N. Moser, I. Pennisi, M. Cavuto, L. Miglietta, A. Moniri, R. Penn, G. Satta, P. Randell, F. Davies, F. Bolt, W. Barclay, A. Holmes and P. Georgiou, ACS Cent. Sci., 2021, 7, 307–317 CrossRef CAS PubMed.
- M. Kong, Z. Li, J. Wu, J. Hu, Y. Sheng, D. Wu, Y. Lin, M. Li, X. Wang and S. Wang, Talanta, 2019, 205, 120155 CrossRef CAS PubMed.
- S. Khaliliazar, I. Öberg Månsson, A. Piper, L. Ouyang, P. Réu and M. M. Hamedi, Adv. Healthcare Mater., 2021, 10, 2100034 CrossRef CAS PubMed.
- J. Tu, R. M. Torrente-Rodríguez, M. Wang and W. Gao, Adv. Funct. Mater., 2020, 30, 1906713 CrossRef CAS.
- F. Wen, T. He, H. Liu, H. Chen, T. Zhang and C. Lee, Nano Energy, 2020, 78, 105155 CrossRef CAS.
- D. Li, H. Zhou, X. Hui, X. He, H. Huang, J. Zhang, X. Mu, C. Lee and Y. Yang, Adv. Sci., 2021, 8, 2101879 CrossRef CAS PubMed.
- P. Q. Nguyen, L. R. Soenksen, N. M. Donghia, N. M. Angenent-Mari, H. de Puig, A. Huang, R. Lee, S. Slomovic, T. Galbersanini, G. Lansberry, H. M. Sallum, E. M. Zhao, J. B. Niemi and J. J. Collins, Nat. Biotechnol., 2021, 39, 1366–1374 CrossRef CAS PubMed.
- M. Bhamla, B. Benson, C. Chai, G. Katsikis, A. Johri and M. Prakash, Nat. Biomed. Eng., 2017, 1, 0009 CrossRef CAS.
- M. Yang, Y. Tang, L. Qi, S. Zhang, Y. Liu, B. Lu, J. Yu, K. Zhu, B. Li and Y. Du, Anal. Chem., 2021, 93, 11956–11964 CrossRef CAS PubMed.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije, M. E. Grunberg, T. F. Kosoko-Thoroddsen, N. L. Welch, P. P. Pillai, S. Mantena, G. Kim, J. N. Uwanibe, O. G. John, P. E. Eromon, G. Kocher, R. Gross, J. S. Lee, L. E. Hensley, B. L. MacInnis, J. Johnson, M. Springer, C. T. Happi, P. C. Sabeti and C. Myhrvold, Nat. Biomed. Eng., 2022, 6, 932–943 CrossRef CAS PubMed.
- Z. A. Crannell, B. Rohrman and R. Richards-Kortum, PLoS One, 2014, 9, e112146 CrossRef PubMed.
- D. Nam, S. Kim, J. H. Kim, S. Lee, D. Kim, J. Son, D. Kim, B. S. Cha, E. S. Lee and K. S. Park, Biosensors, 2023, 13, 367 CrossRef CAS PubMed.
- B. S. Alladin-Mustan, C. J. Mitran and J. M. Gibbs-Davis, Chem. Commun., 2015, 51, 9101–9104 RSC.
- B. A. Rabe and C. Cepko, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 24450–24458 CrossRef CAS PubMed.
- P. Q. M. Nguyen, M. Wang, N. A. Maria, A. Y. Li, H. Y. Tan, G. M. Xiong, M.-K. M. Tan, A. A. S. Bhagat, C. W. M. Ong and C. T. Lim, Microsyst. Nanoeng., 2022, 8, 82 CrossRef CAS PubMed.
- N. Odiwuor, J. Xiong, F. Ogolla, W. Hong, X. Li, F. M. Khan, N. Wang, J. Yu and H. Wei, Anal. Chim. Acta, 2022, 1200, 339590 CrossRef CAS PubMed.
- N. Kaur and B. J. Toley, Analyst, 2018, 143, 2213–2234 RSC.
- A. V. Govindarajan, S. Ramachandran, G. D. Vigil, P. Yagera and K. F. Böhringer, Lab Chip, 2012, 12, 174–181 RSC.
- N. M. Rodriguez, W. S. Wong, L. Liu, R. Dewarbc and C. M. Klapperich, Lab Chip, 2016, 16, 753–763 RSC.
- L. K. Lafleur, J. D. Bishop, E. K. Heiniger, R. P. Gallagher, M. D. Wheeler, P. Kauffman, X. Zhang, E. C. Kline, J. R. Buser, S. Kumar, S. A. Byrnes, N. M. J. Vermeulen, N. K. Scarr, Y. Belousov, W. Mahoney, B. J. Toley, P. D. Ladd, B. R. Lutza and P. Yager, Lab Chip, 2016, 16, 3777–3787 RSC.
- K. T. L. Trinh, T. N. D. Trinh and N. Y. Lee, Biosens. Bioelectron., 2019, 135, 120–128 CrossRef CAS PubMed.
- J. Reboud, G. Xu, A. Garrett, M. Adriko, Z. Yang, E. M. Tukahebwa, C. Rowell and J. M. Cooper, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 4834–4842 CrossRef CAS PubMed.
- W. W. McConnell, C. Davis, S. R. Sabir, A. Garrett, A. Bradley-Stewart, P. Jajesniak, J. Reboud, G. Xu, Z. Yang, R. Gunson, E. C. Thomson and J. M. Cooper, Nat. Commun., 2021, 12, 6994 CrossRef PubMed.
- X. Guo, M. A. Khalid, I. Domingos, A. L. Michala, M. Adriko, C. Rowel, D. Ajambo, A. Garrett, S. Kar, X. Yan, J. Reboud, E. M. Tukahebwa and J. M. Cooper, Nat. Electron., 2021, 4, 615–624 CrossRef CAS.
- T. Zhang, R. Deng, Y. Wang, C. Wu, K. Zhang, C. Wang, N. Gong, R. Ledesma-Amaro, X. Teng, C. Yang, T. Xue, Y. Zhang, Y. Hu, Q. He, W. Li and J. Li, Nat. Biomed. Eng., 2022, 6, 957–967 CrossRef CAS PubMed.
- F. Mazur, A. D. Tjandra, Y. Zhou, Y. Gao and R. Chandrawati, Nat. Rev. Bioeng., 2023, 1, 180–192 CrossRef PubMed.
- S. Banerjee, S. K. Biswas, N. Kedia, R. Sarkar, A. De, S. Mitra, S. Roy, R. Chowdhury, S. Samaddar, A. Bandopadhyay, I. Banerjee, S. Jana, R. Goswami, S. Dutta, M. Chawla-Sarkar, S. Chakraborty and A. Mondal, ACS Sens., 2021, 6, 3753–3764 CrossRef CAS PubMed.
- G. Xun, S. T. Lane, V. A. Petrov, B. E. Pepa and H. Zhao, Nat. Commun., 2021, 12, 2905 CrossRef CAS PubMed.
- R. Snodgrass, A. Gardner, A. Semeere, V. L. Kopparthy, J. Duru, T. Maurer, J. Martin, E. Cesarman and D. Erickson, Nat. Biomed. Eng., 2018, 2, 657–665 CrossRef CAS PubMed.
- E. E. Lin, U. A. Razzaque, S. A. Burrows and S. K. Smoukov, PLoS One, 2022, 17, e0259886 CrossRef CAS PubMed.
- P. LaBarre, K. R. Hawkins, J. Gerlach, J. Wilmoth, A. Beddoe, J. Singleton, D. Boyle and B. Weigl, PLoS One, 2011, 6, e19738 CrossRef CAS.
- X. Yang, C. Guo, Q. Zhang, Y. Chen, Y. Liu and X. Zhang, Talanta, 2023, 258, 124422 CrossRef CAS PubMed.
- Y. Seok, Q. Yin, H. Bai and H. H. Bau, Anal. Chem., 2022, 94, 1778–1786 CrossRef CAS PubMed.
- A. W. Martinez, S. T. Phillips, E. Carrilho, S. W. Thomas 3rd, H. Sindi and G. M. Whitesides, Anal. Chem., 2008, 80, 3699–3707 CrossRef CAS PubMed.
- L. Shen, J. A. Hagen and I. Papautsky, Lab Chip, 2012, 12, 4240–4243 RSC.
- N. K. Thom, G. G. Lewis, K. Yeung and S. T. Phillips, RSC Adv., 2014, 4, 1334–1340 RSC.
- V. Turbé, C. Herbst, T. Mngomezulu, S. Meshkinfamfard, N. Dlamini, T. Mhlongo, T. Smit, V. Cherepanova, K. Shimada, J. Budd, N. Arsenov, S. Gray, D. Pillay, K. Herbst, M. Shahmanesh and R. A. McKendry, Nat. Med., 2021, 27, 1165–1170 CrossRef PubMed.
- S. F. Berlanda, M. Breitfeld, C. L. Dietsche and P. S. Dittrich, Anal. Chem., 2021, 93, 311–331 CrossRef CAS PubMed.
- J. Rodriguez-Manzano, A. Moniri, K. Malpartida-Cardenas, J. Dronavalli, F. Davies, A. Holmes and P. Georgiou, Anal. Chem., 2019, 91, 2013–2020 CrossRef CAS.
- J. A. Herberg, M. Kaforou, V. J. Wright, H. Shailes, H. Eleftherohorinou, C. J. Hoggart, M. Cebey-López, M. J. Carter, V. A. Janes, S. Gormley, C. Shimizu, A. H. Tremoulet, A. M. Barendregt, A. Salas, J. Kanegaye, A. J. Pollard, S. N. Faust, S. Patel, T. Kuijpers, F. Martinón-Torres, J. C. Burns, L. J. Coin and M. Levin, JAMA, 2016, 316, 835–845 CrossRef PubMed.
- H. H. Dolin, T. J. Papadimos, S. Stepkowski, X. Chen and Z. K. Pan, Shock, 2018, 49, 364–370 CrossRef CAS PubMed.
- R. L. Zemans, S. Jacobson, J. Keene, K. Kechris, B. E. Miller, R. Tal-Singer and R. P. Bowler, Respir. Res., 2017, 18, 117 CrossRef PubMed.
- M. F. Abasıyanık, K. Wolfe, H. Van Phan, J. Lin, B. Laxman, S. R. White, P. A. Verhoef, G. M. Mutlu, B. Patel and S. Tay, Nat. Commun., 2020, 11, 2607 CrossRef PubMed.
- S. T. Anderson, M. Kaforou, A. J. Brent, V. J. Wright, C. M. Banwell, G. Chagaluka, A. C. Crampin, H. M. Dockrell, N. French, M. S. Hamilton, M. L. Hibberd, F. Kern, P. R. Langford, L. Ling, R. Mlotha, T. H. M. Ottenhoff, S. Pienaar, V. Pillay, J. A. G. Scott, H. Twahir, R. J. Wilkinson, L. J. Coin, R. S. Heyderman, M. Levin and B. Eley, N. Engl. J. Med., 2014, 370, 1712–1723 CrossRef CAS PubMed.
- K. E. Poti, D. J. Sullivan, A. M. Dondorp and C. J. Woodrow, Trends Parasitol., 2020, 36, 112–126 CrossRef.
- J. Budd, B. S. Miller, E. M. Manning, V. Lampos, M. Zhuang, M. Edelstein, G. Rees, V. C. Emery, M. M. Stevens, N. Keegan, M. J. Short, D. Pillay, E. Manley, I. J. Cox, D. Heymann, A. M. Johnson and R. A. McKendry, Nat. Med., 2020, 26, 1183–1192 CrossRef CAS PubMed.
- C. S. Wood, M. R. Thomas, J. Budd, T. P. Mashamba-Thompson, K. Herbst, D. Pillay, R. W. Peeling, A. M. Johnson, R. A. McKendry and M. M. Stevens, Nature, 2019, 566, 467–474 CrossRef CAS PubMed.
- G. Nixon, J. A. Garson, P. Grant, E. Nastouli, C. A. Foy and J. F. Huggett, Anal. Chem., 2014, 86, 4387–4394 CrossRef CAS PubMed.
- J. Kuypers and K. R. Jerome, J. Clin. Microbiol., 2017, 55, 1621–1628 CrossRef CAS PubMed.
- J. E. Kong, Q. Wei, D. Tseng, J. Zhang, E. Pan, M. Lewinski, O. B. Garner, A. Ozcan and D. Di Carlo, ACS Nano, 2017, 11, 2934–2943 CrossRef CAS PubMed.
- S. K. Biswas, A. Bairagi, S. Nag, A. Bandopadhyay, I. Banerjee, A. Mondal and S. Chakraborty, Int. J. Biol. Macromol., 2023, 253, 127137 CrossRef CAS PubMed.
- T. Mahmoudi, T. Naghdi, E. Morales-Narváez and H. Golmohammadi, Trends Analyt. Chem., 2022, 153, 116635 CrossRef CAS PubMed.
- N. Krivoňáková, A. Šoltýsová, M. Tamáš, Z. Takáč, J. Krahulec, A. Ficek, M. Gál, M. Gall, M. Fehér, A. Krivjanská, I. Horáková, N. Belišová, P. Bímová, A. B. Škulcová and T. Mackuľak, Sci. Rep., 2021, 11, 19456 CrossRef PubMed.
- D. Lou, M. Meurer, S. Ovchinnikova, R. Burk, A. Denzler, K. Herbst, I. A. Papaioannou, Y. Duan, M. L. Jacobs, V. Witte, D. Ürge, D. Kirrmaier, M. Krogemann, K. Gubicza, K. Boerner, C. Bundschuh, N. M. Weidner, U. Merle, B. Knorr, A. Welker, C. M. Denkinger, P. Schnitzler, H. G. Kräusslich, V. L. Dao Thi, M. De Allegri, H. T. Nguyen, A. Deckert, S. Anders and M. Knop, EMBO Rep., 2023, 24, e57162 CrossRef CAS.
- K. A. Kundrod, M. E. Natoli, M. M. Chang, C. A. Smith, S. Paul, D. Ogoe, C. Goh, A. Santhanaraj, A. Price, K. W. Eldin, K. P. Patel, E. Baker, K. M. Schmeler and R. Richards-Kortum, PLoS One, 2022, 17, e0264130 CrossRef CAS.
- M. Karlikow, S. J. R. da Silva, Y. Guo, S. Cicek, L. Krokovsky, P. Homme, Y. Xiong, T. Xu, M. A. Calderón-Peláez, S. Camacho-Ortega, D. Ma, J. J. F. de Magalhães, B. N. R. F. Souza, D. G. de Albuquerque Cabral, K. Jaenes, P. Sutyrina, T. Ferrante, A. D. Benitez, V. Nipaz, P. Ponce, D. G. Rackus, J. J. Collins, M. Paiva, J. E. Castellanos, V. Cevallos, A. A. Green, C. Ayres, L. Pena and K. Pardee, Nat. Biomed. Eng., 2022, 6, 246–256 CrossRef CAS PubMed.
- S. R. Morris, C. C. Bristow, M. R. Wierzbicki, M. Sarno, L. Asbel, A. French, C. A. Gaydos, L. Hazan, L. Mena, P. Madhivanan, S. Philip, S. Schwartz, C. Brown, D. Styers, T. Waymer and J. D. Klausner, Lancet Infect. Dis., 2021, 21, 668–676 CrossRef CAS PubMed.
- J. S. Bloom, L. Sathe, C. Munugala, E. M. Jones, M. Gasperini, N. B. Lubock, F. Yarza, E. M. Thompson, K. M. Kovary, J. Park, D. Marquette, S. Kay, M. Lucas, T. Love, A. S. Booeshaghi, O. F. Brandenberg, L. Guo, J. Boocock, M. Hochman, S. W. Simpkins, I. Lin, N. LaPierre, D. Hong, Y. Zhang, G. Oland, B. J. Choe, S. Chandrasekaran, E. E. Hilt, M. J. Butte, R. Damoiseaux, C. Kravit, A. R. Cooper, Y. Yin, L. Pachter, O. B. Garner, J. Flint, E. Eskin, C. Luo, S. Kosuri, L. Kruglyak and V. A. Arboleda, Nat. Biomed. Eng., 2021, 5, 657–665 CrossRef CAS PubMed.
- L. Mutesa, P. Ndishimye, Y. Butera, J. Souopgui, A. Uwineza, R. Rutayisire, E. L. Ndoricimpaye, E. Musoni, N. Rujeni, T. Nyatanyi, E. Ntagwabira, M. Semakula, C. Musanabaganwa, D. Nyamwasa, M. Ndashimye, E. Ujeneza, I. E. Mwikarago, C. M. Muvunyi, J. B. Mazarati, S. Nsanzimana, N. Turok and W. Ndifon, Nature, 2021, 589, 276–280 CrossRef CAS PubMed.
- H. Lin, W. Yu, K. A. Sabet, M. Bogumil, Y. Zhao, J. Hambalek, S. Lin, S. Chandrasekaran, O. Garner, D. Di Carlo and S. Emaminejad, Nature, 2022, 611, 570–577 CrossRef CAS PubMed.
- D. M. Cate, J. A. Adkins, J. Mettakoonpitak and C. S. Henry, Anal. Chem., 2015, 87, 19–41 CrossRef CAS PubMed.
- M. M. Gong, B. D. MacDonald, T. V. Nguyen, K. Van Nguyen and D. Sinton, Lab Chip, 2014, 14, 957–963 RSC.
- S. Bhadra, A. Pothukuchy, R. Shroff, A. W. Cole, M. Byrom, J. W. Ellefson, J. D. Gollihar and A. D. Ellington, PLoS One, 2018, 13, e0201681 CrossRef PubMed.
- P. Cocco, A. Ayaz-Shah, M. P. Messenger, R. M. West and B. Shinkins, BMC Med., 2020, 18, 119 CrossRef.
- T. Jandoo, Digit. Health, 2020, 6, 2055207619898984 Search PubMed.
- D. Stibbe, S. Reid and J. Gilbert, The Partnering Initiative and UN DESA, 2018.
- S. Smith, R. Koech, D. Nzorubara, M. Otieno, L. Wong, G. Bhat, E. van den Bogaart, M. Thuranira, D. Onchonga and T. F. Rinke de Wit, BMC Med. Inform. Decis. Mak., 2019, 19, 1–12 CrossRef PubMed.
- B. Person, F. Sy, K. Holton, B. Govert and A. Liang, Emerging Infect. Dis., 2004, 10, 358–363 CrossRef PubMed.
- E. Lieber, L. Li, Z. Wu, M. J. Rotheram-Borus and J. Guan, Aids Behav., 2006, 10, 463–471 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |

