Open Access Article
Open Access ArticleWearable stethoscope for lung disease diagnosis
Chundong
Qiu
ab,
Wenru
Zeng
b,
Wei
Tian
a,
Jingyi
Xu
a,
Yingnan
Tian
a,
Chao
Zhao
 *a and
Hong
Liu
*a and
Hong
Liu
 *a
*a
aState Key Laboratory of Digital Medical Engineering, School of Biological Science and Medical Engineering, Southeast University, Nanjing 210096, China. E-mail: czhao@seu.edu.cn; liuh@seu.edu.cn
bNanjing Drum Tower Hospital, Nanjing 210000, China
First published on 24th November 2023
Abstract
Lung disease is one of the most widespread types of disease, especially in the era of COVID-19. Its diagnosis is of great importance, as different types have diverse treatments and prognoses. The most popular methods are computed tomography scanning, ultrasonogram, and bioimpedance sensors, but they are not suitable for wearable applications. Here, we developed a wearable stethoscope with an accompanying algorithm for lung disease diagnosis. It was demonstrated on 18 patients in hospital with three types of lung disease. After collecting and pre-processing lung sound signals, several machine learning methods with optimized features were applied and achieved high classification metrics. The features of the low-frequency wavelets decomposed from the lung sound signals were found to be important, serving as potential biomarkers for different types of lung disease. Overall, it was proven that our wearable stethoscope could provide a more user-friendly method and find greater application scenarios for lung disease diagnosis.
Introduction
Lung diseases are one of the most common types of disease in the world. As the lung is a complex system, lung disease can be divided into many types with totally different causes and symptoms, affecting their diagnosis, treatment, and prognosis. In general, lung diseases can be classified by the affected region, such as the airways, air sacs, interstitium and others.1 For lung diseases affecting the airways, chronic obstructive pulmonary disease (COPD) is one of the most common. COPD causes obstructed air flow from the lungs, resulting in symptoms such as difficulty breathing,2 and is often triggered by long-term exposure to cigarette smoke.3 However, it is treatable by bronchodilators using inhalers after diagnosis. Among lung diseases affecting the air sacs, pneumonia is the most widespread, especially in the era of COVID-19. Pneumonia may cause the air sacs to fill with fluid or pus, and affects mostly children younger than 2 years old and people older than 65. After diagnosis, the symptoms may ease in a few days, while a feeling of tiredness can remain for a longer time. It is generally treated using antibiotics. Among lung diseases affecting the interstitium, interstitial lung disease (ILD) is prevalent, which causes progressive scarring of lung tissue due to long-term exposure to hazardous materials, such as asbestos.4 ILD is generally irreversible, so an early diagnosis is critical. Many ILD patients are initially treated with a corticosteroid and other drugs to suppress the immune system.Generally, as lung diseases occur in different parts of the respiratory system, early and accurate diagnosis is critical for different treatment strategies with various prognoses. Currently, diagnosis is performed using pulmonary function tests, which test the amount of the air inhaled and exhaled by the lungs, arterial blood gas analysis, pulse oximetry, and sputum testing. Furthermore, chest X-rays, computed tomography (CT) scans,5 and echocardiograms can be utilized to determine the severity and location of lung diseases.6,7 However, for diagnosis of relatively healthy people, portable or even wearable biomedical devices may find more application scenarios than conventional instruments in the hospital, and provide a key factor for the early diagnosis of lung diseases.8,9 Also, for people in hospital, a wearable device provides a more friendly method than disturbing the patient by moving them from their ward to the department with the relevant instruments, especially for patients in an intensive care unit (ICU).
Current wearable technology is based on sensing the physical vital signs or biochemical signals of the subject. Optical methods such as photoplethysmography (PPG) use visible or IR light to sense the signal, but penetrate only a few millimetres into the skin and thus cannot reach the lungs; it is mostly used to detect arterial blood vessels. Electrophysiological methods such as electrocardiography (ECG), electroencephalograms (EEG) or electromyography (EMG) cannot be applied to the lungs,10 because they do not generate strong enough electrical signals. Alternatively, bioimpedance devices have been studied to monitor tidal volume and respiratory rate11,12 and used to classify respiration disorders13,14 such as apnea and hypopnea.15 However, bioimpedance measurements require the injection of a weak current into the body, which can be noticeable to the subject, affecting the user experience and making this method unsuitable for long-term monitoring. Respiration causes vibrations or micromotions on the chest, which can be directly sensed by strain or inertial sensors,16 but the resulting information is not as rich as that of lung sounds.17–19 As a result, wearable lung sensing is focused on the generated sounds using ultrasonogram (USG) methods. However, the ultrasonic technique is harmful to the tissue and not recommended for usage longer than 30 min, which may result in abnormal lung sounds being missed. Additionally, its complicated piezoelectric sensor array makes it not widely affordable. Nevertheless, the stethoscope, which also detects sound signals, is the tool mostly widely used by doctors to detect lung diseases. It passively receives lung sounds, and thus it could be worn for hours or days, and experienced doctors could make an initial diagnosis of the type of lung disease directly using the stethoscope alone. In addition to the sensing method, current research is focused on different types of lung sounds, such as crackling, rhonchi, wheezing and stridor.20–27 However, the lung sound type is not directly related to the diagnosis of the type of lung disease, and one type of disease is sometimes related to several types of abnormal lung sounds.
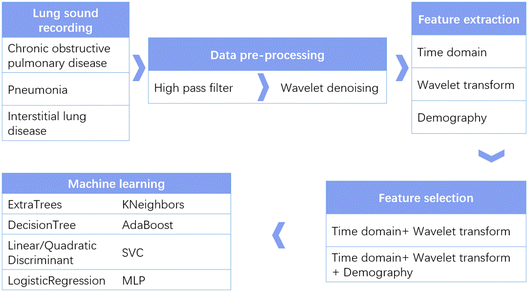
In order to take advantage of the stethoscope, make it digital, and solve the issue of lung disease diagnosis, we optimized a wearable small-scale electronic stethoscope (WSES) system previously developed by us17 and applied it to 18 patients with lung diseases in hospital. The system has a microphone IC chip with an integrated microelectromechanical system (MEMS). Its small size, low power consumption, and high signal-to-noise ratio (SNR) makes this wearable stethoscope applicable for patients in hospital. A user-friendly mobile application was developed to collect signals from the device for further diagnosis, making our device cable-free. In order to diagnose the type of lung disease, several machine learning methods were compared to obtain the best performance. Our results showed that the system could reach a diagnosis accuracy higher than 90% and could be applied in hospital to assist doctors in lung disease diagnosis. Fig. 1 shows an overall flow chart of our study design. Lung sounds from patients with different lung diseases were collected and pre-processed to remove noise and artifacts. Then, features in the time domain, from wavelet decomposition and demography, were extracted. Next, different feature sets were compared, and nine machine learning algorithms were applied to find the optimized algorithms with the highest performance metrics.
Methods
Subjects and ethical considerations
This study was conducted on 18 patients with lung disease recruited from Nanjing Drum Tower Hospital. There were nine male and nine female patients with a mean age of 74.2 years (standard deviation ± 10.4 years), with the specific demographic and pathological characteristics detailed in Table 1. All subjects had normal hearing, normal vision, and normal speech function. The subjects were conscious and able to communicate with doctors, and the gold standard for lung disease type was evaluation by the doctor before the experiment. Prior to the experiment, subjects were informed of the procedure and related precautions, and that the experiment was not harmful to humans. All participants signed an informed consent form and the study was approved by the Ethics Committee of Southeast University. Data were obtained in accordance with the guidelines of the University Ethics Committee and the ethical principles of the Declaration of Helsinki for medical research involving human beings.| No | Sex | Age | HR | SpO2 | RR | SBP | DBP |
|---|---|---|---|---|---|---|---|
| 1 | Male | 91 | 60 | 99 | 30 | 137 | 55 |
| 2 | Female | 84 | 114 | 95 | 50 | 110 | 64 |
| 3 | Male | 87 | 75 | 98 | 17 | 130 | 77 |
| 4 | Female | 88 | 114 | 97 | 18 | 128 | 79 |
| 5 | Female | 75 | 96 | 98 | 36 | 105 | 75 |
| 6 | Male | 67 | 70 | 99 | 25 | 105 | 67 |
| 7 | Female | 70 | 89 | 100 | 13 | 143 | 78 |
| 8 | Female | 72 | 84 | 99 | 26 | 101 | 51 |
| 9 | Female | 82 | 76 | 99 | 18 | 138 | 64 |
| 10 | Male | 75 | 65 | 98 | 24 | 113 | 62 |
| 11 | Female | 55 | 53 | 95 | 22 | 110 | 66 |
| 12 | Male | 79 | 70 | 99 | 14 | 111 | N/A |
| 13 | Female | 80 | 96 | 98 | 21 | 159 | 71 |
| 14 | Male | 69 | 71 | 83 | 30 | 152 | 91 |
| 15 | Male | 63 | 106 | 88 | 15 | 120 | 72 |
| 16 | Male | 55 | 62 | 96 | 17 | 98 | 64 |
| 17 | Female | 74 | 72 | 99 | 24 | 126 | 73 |
| 18 | Male | 69 | 87 | 89 | 24 | 125 | 83 |
Hardware and experimental procedure
Fig. 2 presents the design of the device and a photo of a patient wearing it. The sensing chip is an ICS-43432 (InvenSense) with a size of 3.5 mm by 3.5 mm, and the microcontroller unit (MCU) is nRF52840 (Nordic Semiconductor) with a size of 3.5 mm by 3.6 mm. The ICS-43432 senses the sound from the body and transmits the digital signal to the nRF52840, which has a Bluetooth module to send the data to a mobile phone. The device operates at a sampling rate of 2.4 kHz, which is sufficient for lung sound collection. Blue medical tape (Huaxi Sanitary Materials) was placed between the device and the patient's skin for sanitary purposes, and the battery used was 250 mA h. The device cost is tens of US dollars, and it could be afforded by the general public. The firmware was developed with uVision 5.28 based on NordicSemi nRF SDK 15.3. The algorithm was demonstrated on MATLAB R2021a and Scikit-Learn 0.24.2 in Python 3.7.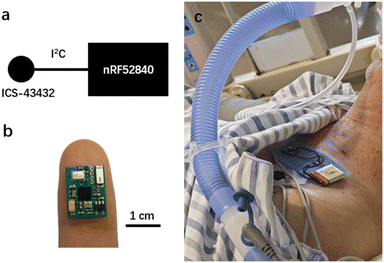 | ||
| Fig. 2 a) Schematic and b) photo of the wearable stethoscope. c) Photo of a patient wearing our device. | ||
In a quiet environment in the ward, the subjects wore our device lying flat, keeping their body posture unchanged as much as possible. We collected around three minutes of lung sounds for each patient. The signal quality was initially judged according to the frequency domain display in the app interface, which was used to decide whether to repeat the data acquisition process.
Data pre-processing and feature extraction
The app saved the lung sound data to the memory of the smartphone as a text file. The name of the file contains time information, such as “Tue Nov 08 10:09:47 GMT + 08:00 2023-282”. The first six digits of the last string are timestamps and the seventh digit is the order of collected data. The text file is read into MATLAB, and the sound data of the corresponding period were searched and intercepted according to the timestamps recorded during the experiment. The data was processed using a 100 Hz high-pass filter and a wavelet denoising algorithm.After the pre-processing, the features in the time domain, frequency domain, and nonlinear domain of the signal were extracted, as listed in Table 2. Other information listed in Table 1, such as HR, SpO2, RR, SBP and DBP, was collected but not included as features because during the collection period they were not continuously monitored. They were only utilized to confirm that the patient status was stable. Here, nine features, namely, maximum value, minimum value, maximum minus minimum value, standard deviation, mean of absolute, median of absolute, mean, kurtosis, and mean of absolute of the derivative in the time domain were extracted. Next, the mean of absolute of coefficients, mean of power of coefficients, and standard deviation of coefficients of five wavelets were extracted, resulting in 15 features. Also, the ratio between the mean of absolute of coefficients was derived as four features. Adding sex and age as features, in total there are 30 features involved.
| Time domain features | ||
|---|---|---|
| Max | Min | Max–min |
| Standard deviation | Mean of absolute | Median of absolute |
| Mean | Kurtosis | Mean of absolute of derivative |
| Wavelet decomposition features | ||
|---|---|---|
| Mean of absolute of coefficients | Mean of power of coefficients | Standard deviation of coefficients |
| Demographic features | ||
|---|---|---|
| Sex | Age | |
Machine learning models
Supervised learning is the most applicable method for lung disease classification, in which input data with labels are propagated by an algorithm, which then learns the patterns associated with each label. A supervised machine learning model was trained with extracted features as the dataset and the predetermined lung disease types as labels. This study utilizes the base classification models ExtraTreesClassifier, DecisionTreeClassifier, LinearDiscriminantAnalysis, LogisticRegression, QuadraticDiscriminantAnalysis, KNeighborsClassifier, AdaBoostClassifier, SVC, and MLPClassifier from the Python machine learning library scikit-learn. Parameter optimization was performed using RandomizedSearchCV. The performance of the above machine learning models was judged by accuracy, precision, recall, and F1 score. Accuracy, recall, precision, and F1 scores were used to evaluate our algorithms. They are related to true positives (TP), true negatives (TN), false positives (FP), and false negatives (FN). Accuracy is calculated as (TP + TN)/(TP + TN + FP + FN), recall is quantified as (TP/(TP + FN)) and precision is (TP/(TP + FP)). The F1 metric is the harmonic mean of precision and recall for overall performance evaluation. Also, a confusion matrix was studied.The features were first standardized by removing the mean and scaling to unit variance. The dataset was divided into training and test subsets at a ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The classifier was trained using the training subsets and then utilized for prediction on the test subsets. The algorithm's capability and performance to tell the difference between different lung diseases was studied. K-Fold was adopted for cross-validation. The dataset was split randomly into K parts, with one part as test subsets and the remaining as training subsets.
1. The classifier was trained using the training subsets and then utilized for prediction on the test subsets. The algorithm's capability and performance to tell the difference between different lung diseases was studied. K-Fold was adopted for cross-validation. The dataset was split randomly into K parts, with one part as test subsets and the remaining as training subsets.
Results and discussion
Feature extraction and selection
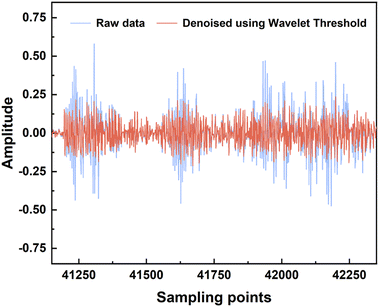
The acquired lung sound signals were filtered using a 100 Hz high-pass filter, as lung sound frequency is typically higher than 100 Hz, and then decomposed using ‘wavedec’ at level 5 with the wavelet ‘coif4’ in MATLAB. Then, ‘wthresh’ and ‘waverec’ were utilized to denoise and reconstruct the lung sound signals. Fig. 3 shows the raw data and the signals denoised using wavelet thresholds.After feature extraction, the ranking of feature importance was calculated, and is shown in Fig. 4a. Two sets of features (28 features vs. 30 features) were trialed (Table 3). Classification using 28 features in the time domain and wavelet decomposition results in an accuracy of 0.85 ± 0.04, recall of 0.84 ± 0.04, precision of 0.84 ± 0.03, and F1 of 0.83 ± 0.03, which is rather good already. Interestingly, Fig. 4b shows that after adding the demographic information, all performance metrics were boosted, with an accuracy of 0.99 ± 0.01, recall of 0.98 ± 0.02, precision of 0.99 ± 0.01, and F1 of 0.99 ± 0.01. Indeed, age and gender are the most important features, which indicates the relationships between lung disease type and demographic information. Thus, it was decided to include all 30 features for all machine learning methods. It should be noted that obviously, age and sex alone could not determine the lung disease type; all three lung disease groups included patients with a wide range of ages and of both sexes. In addition to age and sex, the features related to low-frequency wavelets ranked higher, such as coefficients from the decomposition at level 5 and level 4, which indicates that the lower-frequency lung sounds are biomarkers for diagnosis. It could also be observed that the higher the frequency of a wavelet feature, the lower its ranking in the frequency importance.
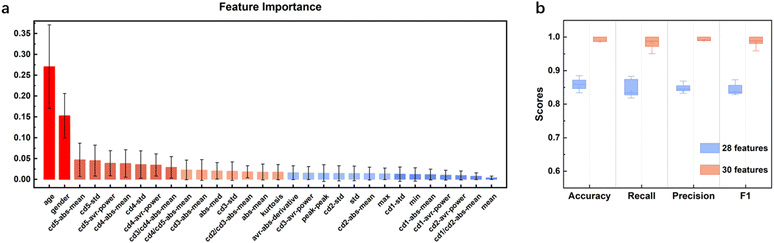 | ||
| Fig. 4 a) Feature importance of ETC and b) classification performance of ETC for two sets of features. | ||
| Accuracy | Recall | Precision | F1 | |
|---|---|---|---|---|
| 28 | 0.85 ± 0.04 | 0.84 ± 0.04 | 0.84 ± 0.03 | 0.83 ± 0.03 |
| 30 | 0.99 ± 0.01 | 0.98 ± 0.02 | 0.99 ± 0.01 | 0.99 ± 0.01 |
Classification results
After feature selection, nine machine learning methods were compared, and the model parameters were optimized. The classification results for the top four algorithms are shown in Fig. 5. K-Fold validation was utilized on accuracy, recall, precision and F1 to test the robustness of the model. The optimized hyperparameters for ExtraTreesClassifier were n_estimators = 125, min_samples_split = 3, min_samples_leaf = 1, criterion = entropy, with a mean cross-validation F1 score of 0.986 ± 0.016. The optimized hyperparameters of MLPClassifier were alpha = 0.0001, hidden_layer_sizes = (200, 200, 200), max_iter = 1000, learning_rate_init = 0.001, tol = 0.0001, beta_1 = 0.99, beta_2 = 0.99, epsilon = 1 × 10−7 with a mean cross-validation F1 score of 0.966 ± 0.015. The optimized hyperparameters of KNeighborsClassifier were algorithm = ball_tree, leaf_size = 40, n_neighbors = 30, p = 1, weights = distance with a mean cross-validation F1 score of 0.942 ± 0.022. The optimized hyperparameters of svm.SVC were C = 2, kernel = rbf, gamma = scale, tol = 0.0001 with a mean cross-validation F1 score of 0.954 ± 0.018. ETC performed the best, followed by MLPC and KNN. Fig. 5b presents the confusion matrix of ETC, with especially high metrics for PI and IPF. The only missed label was an instance of “others” wrongly classified as “PI”. Thus, it is proven that using our wearable device with the corresponding optimized diagnosis algorithm, our system could accurately diagnose lung disease and could find further applications in clinics.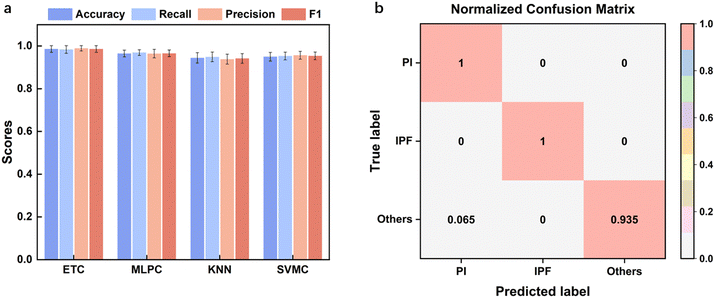 | ||
| Fig. 5 Model performance characterization. a) Performance metrics of the best four models. b) Confusion matrix of the ETC model. | ||
Nevertheless, several insights and limitations were identified. First, different biocompatible adhesives could be applied to attach our device to the patient's skin to minimize the environmental noise and match the sound impedance in order to obtain the highest SNR. Second, different types of sound could be categorized in detail to determine the relationships between the sound type and lung disease type. Third, most of the patients were more than 60 years old, and the robustness of our system in lung disease diagnosis should be proven with other ranges of ages.
Conclusions
Lung disease type classification is critical for deciding the treatment strategy and prognosis monitoring. Currently, most lung disease studies with wearable technology are focused on sound type classification, which is not clinically significant. Here, we introduced a wearable stethoscope and applied it to 18 lung disease patients with the three most widespread disease types. Machine learning methods were used to achieve high performance metrics, proving its potential for application in clinics.Author contributions
C. Z. and H. L. conceived the research idea. C. Q. and W. Z. performed experiments. W. T. analyzed the data. C. Q. and C. Z. wrote the paper. All authors discussed the results and reviewed the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Key Research and Development Program of Jiangsu Province (BE2021700), Science and Technology Development Program of Suzhou (SYG202117), the National Natural Science Foundation of China (62001104, 62271136), Natural Science Foundation of Jiangsu Province (BK20200357), Key Project and Open Research Fund of State Key Laboratory of Bioelectronics, the Fundamental Research Funds for the Central Universities and Zhishan Young Scholars of Southeast University. We thank all doctors, nurses and patients related to this paper.Notes and references
- T. K. Burki, Lancet Respir. Med., 2019, 7, 1015–1016 CrossRef PubMed.
- D. Blanco-Almazan, W. Groenendaal, M. Lozano-Garcia, L. Estrada-Petrocelli, L. Lijnen, C. Smeets, D. Ruttens, F. Catthoor and R. Jane, IEEE Trans. Biomed. Eng., 2021, 68, 298–307 Search PubMed.
- J. E. Bibault and L. Xing, Lancet Digital Health, 2020, 2, e216–e217 CrossRef PubMed.
- M. S. Wijsenbeek, C. C. Moor, K. A. Johannson, P. D. Jackson, Y. H. Khor, Y. Kondoh, S. K. Rajan, G. C. Tabaj, B. E. Varela, P. van der Wal, R. N. van Zyl-Smit, M. Kreuter and T. M. Maher, Lancet Respir. Med., 2023, 11, 97–110 CrossRef PubMed.
- Q. Dou, T. Y. So, M. Jiang, Q. Liu, V. Vardhanabhuti, G. Kaissis, Z. Li, W. Si, H. H. C. Lee, K. Yu, Z. Feng, L. Dong, E. Burian, F. Jungmann, R. Braren, M. Makowski, B. Kainz, D. Rueckert, B. Glocker, S. C. H. Yu and P. A. Heng, npj Digit. Med., 2021, 4, 60 CrossRef PubMed.
- R. Aggarwal, V. Sounderajah, G. Martin, D. S. W. Ting, A. Karthikesalingam, D. King, H. Ashrafian and A. Darzi, npj Digit. Med., 2021, 4, 65 CrossRef PubMed.
- H. Barnes, S. M. Humphries, P. M. George, D. Assayag, I. Glaspole, J. A. Mackintosh, T. J. Corte, M. Glassberg, K. A. Johannson, L. Calandriello, F. Felder, A. Wells and S. Walsh, Lancet Digital Health, 2023, 5, e41–e50 CrossRef CAS PubMed.
- S. H. Lee, Y. S. Kim, M. K. Yeo, M. Mahmood, N. Zavanelli, C. Chung, J. Y. Heo, Y. Kim, S. S. Jung and W. H. Yeo, Sci. Adv., 2022, 8, eabo5867 CrossRef PubMed.
- S. H. Lee, Y. S. Kim and W. H. Yeo, Adv. Healthcare Mater., 2021, 10, e2101400 CrossRef PubMed.
- Y. Wang, W. Tian, J. Xu, Y. Tian, C. Xu, B. Ma, Q. Hao, C. Zhao and H. Liu, IEEE Sens. J., 2023, 23, 21767–21775 Search PubMed.
- J. A. Berkebile, S. A. Mabrouk, V. G. Ganti, A. V. Srivatsa, J. A. Sanchez-Perez and O. T. Inan, IEEE Trans. Biomed. Eng., 2022, 69, 1909–1919 Search PubMed.
- A. R. Fekr, K. Radecka and Z. Zilic, IEEE J. Biomed. Health Inform., 2015, 19, 1532–1548 Search PubMed.
- A. R. Fekr, M. Janidarmian, K. Radecka and Z. Zilic, IEEE J. Biomed. Health Inform., 2016, 20, 733–747 Search PubMed.
- C. Qiu, F. Wu, W. Han and M. R. Yuce, IEEE Trans. Biomed. Eng., 2022, 69, 2970–2981 Search PubMed.
- T. Van Steenkiste, W. Groenendaal, P. Dreesen, S. Lee, S. Klerkx, R. de Francisco, D. Deschrijver and T. Dhaene, IEEE J. Biomed. Health Inform., 2020, 24, 2589–2598 Search PubMed.
- D. Jarchi, S. J. Rodgers, L. Tarassenko and D. A. Clifton, IEEE Sens. J., 2018, 18, 4981–4989 Search PubMed.
- J. Xu, W. Zeng, C. Zhao, J. Tong and H. Liu, Sens. Diagn., 2023, 2, 657–664 RSC.
- J. Xu, C. Zhao, B. Ding, X. Gu, W. Zeng, L. Qiu, H. Yu, Y. Shen and H. Liu, IEEE Sens. J., 2022, 22, 11526–11534 Search PubMed.
- C. Zhao, W. Zeng, D. Hu and H. Liu, IEEE Sens. J., 2021, 21, 15962–15971 Search PubMed.
- J. A. Dar, K. K. Srivastava and A. Mishra, Comput. Biol. Med., 2023, 164, 107311 CrossRef PubMed.
- N. S. Haider, Biomed. Signal Process. Control., 2021, 64, 102313 CrossRef.
- D.-M. Huang, J. Huang, K. Qiao, N.-S. Zhong, H.-Z. Lu and W.-J. Wang, Mil. Med. Res., 2023, 10, 44 Search PubMed.
- A. Kandaswamy, C. S. Kumar, R. P. Ramanathan, S. Jayaraman and N. Malmurugan, Comput. Biol. Med., 2004, 34, 523–537 CrossRef CAS PubMed.
- A. Mondal, P. Banerjee and H. Tang, Comput. Methods Programs Biomed., 2018, 159, 199–209 CrossRef PubMed.
- G. Shah, P. Koch and C. B. Papadias, IEEE J. Biomed. Health Inform., 2015, 19, 151–157 Search PubMed.
- K. H. Tsai, W. C. Wang, C. H. Cheng, C. Y. Tsai, J. K. Wang, T. H. Lin, S. H. Fang, L. C. Chen and Y. Tsao, IEEE J. Biomed. Health Inform., 2020, 24, 3203–3214 Search PubMed.
- W. Wang, D. Qin, S. Wang, Y. Fang and Y. Zheng, Comput. Biol. Med., 2023, 164, 107282 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |