Open Access Article
Open Access ArticleUltrasensitive detection of flap endonuclease 1 using a chemiluminescence optical fiber biosensor with hybridization chain reaction†
Jinlan
Yang
a,
Yile
Qin
a,
Dingsong
Li
a,
Yaxue
Jia
a,
Jiahui
Fu
*b and
Li
Yang
 *a
*a
aDepartment of Chemistry, Northeast Normal University, 5268 Renmin Street, Changchun, Jilin Province 130024, China. E-mail: yangl330@nenu.edu.cn
bThe First Hospital of Jilin University, Changchun, Jilin Province, China. E-mail: 18343160896@163.com
First published on 22nd November 2023
Abstract
Flap endonuclease 1 (FEN1) is an important biomarker involved in tumor progression and can catalyze the cleavage of DNA with a bifurcated structure at the 5′ end, while its expression is so low in cancer cells that accurate and sensitive quantification is quite challenging. In this study, we propose a novel sensing method for sensitively detecting FEN1 in HeLa cells by combining chemiluminescent optical fiber sensors (COFSs) with an enzyme-free hybridization chain reaction (HCR) amplification strategy. The free 5′-flap produced by FEN1-specific cleavage of the bifurcated structure double-stranded DNA is captured by the COFS modified with a hairpin capture probe to trigger HCR amplification. Signal conversion is achieved by introducing the streptavidin-modified horseradish peroxidase on the fiber surface to catalyze chemiluminescent substrates based on the specific binding between biotin and streptavidin. The chemiluminescence emission is transmitted by the same optical fiber and recorded as the sensing response. With this approach, excellent analytical performance for FEN1 detection is achieved, with a wide linear range of 10 fM to 75 pM, a limit-of-detection as low as 3.4 fM and good specificity. The method is successfully applied for the determination of FEN1 in HeLa cell lysate samples with satisfactory accuracy, the results of which are consistent with those from the ELISA assays. Our study indicates that with the essential advantages of high sensitivity, excellent integration and portability, the COFS combined with a suitable efficient amplification strategy can perform ultrasensitive determination of biomarkers and will have valuable potential applications in various fields of point-of-care testing.
Introduction
Flap endonuclease 1 (FEN1) is a structure-specific nuclease and plays a key role in DNA replication and repair by catalyzing the cleavage of DNA with a bifurcated structure at the 5′ end.1 Since FEN1 is involved in the maintenance process of cell proliferation, the upregulated level of FEN1 can be found in cells with a high replication rate, such as cancer cells.2,3 Studies have shown that persistent overexpression of FEN1 is associated with poor prognosis of lung cancer, metastasis of triple-negative breast cancer, and increased risk of colorectal cancer.4–6 On the other hand, the expression level and activity of FEN1 are very low, especially in the early stages of cancer, thus, it is urgent to develop highly sensitive assays for the detection of FEN1 in the field of clinical diagnosis.7–9Traditional testing methods, including western blotting and enzyme-linked immunoassay, have been developed for FEN1 assay due to its clinical significance.10,11 These two methods directly read the molecule number of FEN1 rather than the enzyme activity, which are performed without any signal conversion and amplification, resulting in a limited detection sensitivity with only qualitative or semi-quantitative results.12,13 In recent years, novel sensing methods based on a signal amplification strategy have been proposed for more sensitive and rapid detection of FEN1. For example, Yang et al. reported a dual-signal amplification method by combining Bst DNA polymerase and nicking enzyme Nt.BstNBI.14 Li et al. designed a flapped dumbbell DNA probe by using T4 DNA ligase-based rolling circle amplification.15 Chen's group developed a detection system based on the trans-cleavage of CRISPR/Cas12a.16 Li's group developed two kinds of fluorescent biosensors based on the exponential amplification reaction and CRISPR/Cas13a system to realize the rapid and sensitive detection of FEN1.17,18 Very recently, a hybridization chain reaction (HCR)-based strategy has been developed for FEN1 detection, showing the great potential of enzyme-free amplification in improving the detection sensitivity for FEN1 assay.19
Owing to its several essential advantages for biomarker assays, chemiluminescent optical fiber sensors (COFSs) have now emerged as a very promising approach in various analytical fields.20 With the well-known silicon chemistry, it is very simple to modify specific recognition molecules on the surface of an optical fiber, making it a suitable support for developing a biosensor.21 In addition, the same optical fiber is also employed to transmit chemiluminescence signals directly to the detector without any external light source, avoiding the background interference and enhancing the signal-to-noise ratio.22,23 Moreover, the feature of COFSs makes them beneficial for designing portable and automatic sensing devices which would have great potential applications in the field of point-of-care testing (POCT).24
Herein, we extend the application of COFSs for sensitive determination of FEN1 in biological samples. To improve the detection sensitivity, we combine an enzyme-free HCR signal amplification strategy with COFSs by modifying a hairpin capture probe (CP) on the fiber surface. As FEN1 can specifically recognize and cleave the bifurcated structure double-stranded DNA, the produced short-strand DNA, i.e., the dissociative 5′-flap, is captured by the CP modified on the fiber based on the principle of complementary base pairing, which then triggers the HCR amplification. The opened hairpin CP acts as an initiator to trigger the cascade reaction of biotin-modified H1 and H2, producing a long DNA structure containing a large number of biotin-binding sites on the surface of the fiber. Based on the specific binding between biotin and streptavidin, the streptavidin-modified horseradish peroxidase (SA-HRP) is introduced into the sensing system to achieve signal amplification by catalyzing the chemiluminescent substrate. We show that our strategy can achieve highly sensitive and specific determination of FEN1 in HeLa cell lysate samples.
Experimental section
Reagents and apparatus
The sequences of DNA used in the present study are shown in Table S1.† All oligonucleotides, endonucleases (HindIII, ECoRI, and Dral), exonucleases (Exo I and Exo III) and HeLa cell lysate were obtained from Sangon Biotech Co., Ltd (Changchun, China). FEN1 was bought from Beijing Solarbio Science & Technology Co., Ltd. (Beijing, China). 3-Aminopropyl diethoxymethylsilane (3-ADMS), carbodiimide hydrochloride (EDC), N-hydroxy-succinimide (NHS), oxidized glutathione (GSSG), bovine serum albumin (BSA) and streptavidin-modified horseradish peroxidase (SA-HRP) were obtained from Sigma Aldrich Co. Ltd (MO, USA). Streptavidin-labeled magnetic beads (MBs, 1 μM) and the chemiluminescent substrate were bought from Solarbio Co., Ltd (Beijing, China). A human FEN1 ELISA kit was purchased from Fenon Biotechnology Co., Ltd. (Wuhan, China). Hydrofluoric acid (48%, w/w), NaOH, Na2HPO4, NaH2PO4, KCl, Tris-HCl, (NH4)2SO4 and MgSO4 were purchased from Beijing Chemical Works (Beijing, China). Agarose gel, DNA marker and 5× TBE buffer solution were purchased from Tiangen Biochemical Co., Ltd (Beijing, China). Ultrapure water provided with a Milli-Q Water System (MA, USA) is used in the experiments. All solutions were filtered using a 0.22 μm micromembrane before use.The chemiluminescence signal was detected and recorded using a photon-counting detector (PCD, CH326 module, Beijing Hamamatsu Photon Technique INC). Data acquisition was performed using a CH297-01 counting unit and its software package (Beijing Hamamatsu Photon Technique Inc., Beijing). FT-IR (Nicolet 6700 FT-IR spectrometer, Thermo Fisher, USA) and UV-vis spectroscopy (Micro-spectrophotometer, KAIAO k5600, China) were used to characterize the fiber sensor. ELISA kit assay was performed using a microplate reader (Varioskan LUX, Thermo Fisher, USA).
Construction of HCR-COFS for FEN1 assay
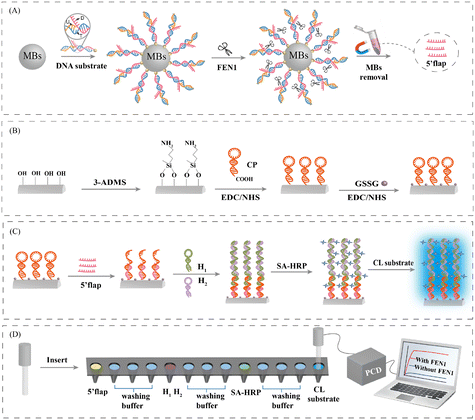
A bare optical fiber (SFS400/440/700 T, numerical aperture 0.22. Wyoptics Technology Co., Ltd., Shanghai, China) with a length of 4 cm was used to construct the HCR-COFS. The optical fiber was firstly pretreated to remove the cladding (details are shown in the ESI†). The HCR-COFS was then prepared by immobilizing the carboxy-modified CP on the fiber surface, according to the following procedures:1) Annealing treatment of the CP. The CP solution was heated at 95 °C for 5 min and then slowly cooled to room temperature. 2) Introduction of the amino group to the fiber surface. The pre-treated optical fiber was placed in 1% (v/v) 3-ADMS ethanol solution for 1 h to introduce the amino group to the surface. 3) Immobilization of the CP. After washing with ethanol three times, the optical fiber was placed in PBS (0.01 M, pH 7.4) solution containing 0.015 M EDC, 0.030 M NHS, and 0.5 μM CP for 1 h to achieve the immobilization of the CP. 4) Removing non-specific adsorption. After washing with PBS (0.01 M, pH 7.4) three times, the fiber was incubated in 0.015 M EDC, 0.030 M NHS and 10 mg mL−1 GSSG solution for 2 h to close the residual active sites to reduce non-specific adsorption. Finally, the prepared HCR-COFS was thoroughly washed with PBS three times and stored at 4 °C for further use.
Preparation of DNA substrate-loaded MBs
Double-stranded DNA substrates were prepared by hybridization of three complementary sequences including the upstream strand (U), downstream strand (D), and template strands (T) in PBS buffer (0.01 M, pH 7.4) at 95 °C for 5 min, followed by slow cooling to room temperature. To prepare the DNA substrate-loaded MBs, 200 μL DNA substrates (0.1 μM) were incubated with 200 μL MBs (1 mg mL−1) at room temperature for 45 min. To remove the excess DNA substrates, the DNA substrate-loaded MBs were washed with the reaction buffer (15 mM KCl, 30 mM Tris-HCl, 15 mM (NH4)2SO4, 3 mM MgSO4, pH 8.8) three times and then redispersed in 200 μL reaction buffer for further use.FEN1 concentration detection assay and sample analysis
200 μL DNA substrate-loaded MBs (1 mg mL−1) were incubated with FEN1 (concentration in the range of 10 fM to 75 pM) in the reaction buffer at 37 °C for 2 h. The DNA substrate-loaded MBs were subsequently discarded by magnetic separation to obtain the dissociative 5′-flap ssDNA. The sensing region of the HCR-COFS was inserted into the solution containing dissociative 5′-flap ssDNA for 2 h to complete the hybridization reaction. After washing with PBS buffer three times, the sensing region was immersed into 100 μL PBS buffer (0.01 M, pH 7.4) containing 1 μM H1 and H2 at 37 °C for 1 h. The optical fiber was finally rinsed with PBS and incubated with the freshly-prepared SA-HRP solution for 30 min at 37 °C. After washing with PBS buffer three times, the sensing region was inserted into the freshly prepared chemiluminescent substrate solution, and the other end of the fiber was connected to the detection window of the PCD. The response of the HCR-COFS biosensor was recorded by chemiluminescence detection in the dark.We use recovery experiments to analyze the FEN1 concentration in actual samples. All experiments were performed in accordance with the Guidelines of Northeast Normal University and approved by the Ethics Committee of Northeast Normal University. For the recovery experiments, FEN1 with different concentrations (0.5, 1.0, and 1.5 pM) were respectively added to the HeLa cell lysate. Real samples were measured to determine FEN1 using the HCR-COFS approach.
ELISA assay for FEN1
The standard FEN1 samples (100 μL), HeLa cell lysate samples (100 μL), and biotin-conjugated antibody (100 μL) were successively added to the 96-well plate, and the unbound components were washed with PBS solution. Then the 100 μL HRP–streptavidin was added into the 96-well plate. After removing the HRP–streptavidin with PBS, a 90 μL TMB substrate solution was added to each hole for color rendering. The enzyme–substrate reaction was terminated by the addition of sulfuric acid solution (100 μL), and the absorbance value was measured by using a microplate reader at a wavelength of 450 nm.Results and discussion
Working principle of the HCR-COFS-based method for FEN1 assay
The working principle of the HCR-COFS for FEN1 assay is shown in Fig. 1. FEN1 in a sample specifically recognizes and cleaves the bifurcated structure at the 5′ end of the DNA substrates, which are loaded on the surface of MBs. The dissociative 5′-flap ssDNA can be easily separated from the DNA substrate-loaded MBs by magnetic separation (Fig. 1(A)). The prepared HCR-COFS containing the hairpin CP (Fig. 1(B)) captures the dissociative 5′-flap ssDNA based on the principle of complementary base pairing, which opens up the hairpin structure of the CP. The unoccupied part of the CP then acts as an initiator of the HCR to hybridize with the toehold of H1, which in turn experiences a chain reaction with H2, producing a long DNA structure containing a large number of biotin-binding sites on the fiber surface (Fig. 1(C)). Based on the affinity interaction between biotin and streptavidin, SA-HRP is connected with the fiber sensor, which is used to catalyze the chemiluminescent substrate for signal amplification. The emission is measured using a PCD, the intensity of which quantitatively represents the response of sensing FEN1. The analytical process is schematically shown in Fig. 1(D). The sensing region of the HCR-COFS was inserted into the solution containing the 5′-flap to complete target capture. After washing with PBS buffer three times, the sensing region was immersed into 100 μL PBS buffer (0.01 M, pH 7.4) containing 1 μM H1 and H2 for HCR amplification. The optical fiber was finally rinsed with PBS and incubated with the freshly-prepared SA-HRP solution for signal molecule capture. After washing with PBS buffer three times, the sensing region was inserted into the freshly prepared chemiluminescent substrate solution, and the other end of the fiber was connected to the detection window of the PCD. The response of the HCR-COFS biosensor was recorded by chemiluminescence detection in the dark. A picture of the sensor system in Fig. 1(D) is shown in Fig. S1.†Feasibility of the HCR-COFS-based method for FEN1 assay
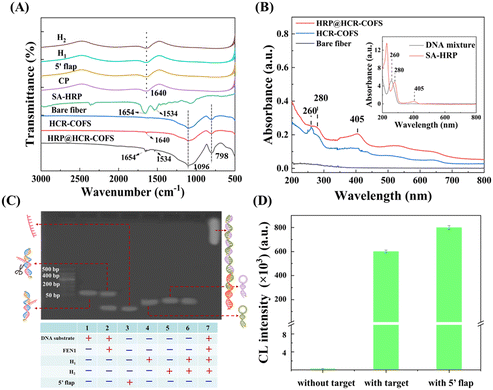
FT-IR and UV-vis spectroscopy are employed to characterize the fabrication of the HCR-COFS. As shown in Fig. 2A, the FT-IR spectra of H1, H2, the 5′ flap, and the CP show the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N characteristic stretching at 1640−1 cm in aromatic rings from DNA, which can be also observed in the FT-IR spectrum of the HCR-COFS. Consistent with the results of FT-IR, the characteristic absorption of DNA at 260 nm can also be observed in the UV-vis spectrum of the HCR-COFS. These results prove the effective loading of the CP on the fiber surface. In addition to the antisymmetric stretching vibration peak of Si–O–Si (1096 cm−1) and symmetric stretching vibration of Si–O (798 cm−1) from the bare fiber, the FT-IR spectrum of HRP@HCR-COFS shows two peaks at 1654 cm−1 and 1534 cm−1, which are the characteristic vibration bands of HRP, corresponding to the C
N characteristic stretching at 1640−1 cm in aromatic rings from DNA, which can be also observed in the FT-IR spectrum of the HCR-COFS. Consistent with the results of FT-IR, the characteristic absorption of DNA at 260 nm can also be observed in the UV-vis spectrum of the HCR-COFS. These results prove the effective loading of the CP on the fiber surface. In addition to the antisymmetric stretching vibration peak of Si–O–Si (1096 cm−1) and symmetric stretching vibration of Si–O (798 cm−1) from the bare fiber, the FT-IR spectrum of HRP@HCR-COFS shows two peaks at 1654 cm−1 and 1534 cm−1, which are the characteristic vibration bands of HRP, corresponding to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of primary amide and N–H bending vibration of secondary amide, respectively.25,26 The characteristic absorption peak at 280 nm and 405 nm from SA-HRP can be also observed in the UV-vis spectrum of HRP@HCR-COFS (Fig. 2B). The FT-IR and UV-vis spectra of HRP@HCR-COFS confirm that SA-HRP is successfully modified on the fiber surface after HCR amplification.
O stretching vibration of primary amide and N–H bending vibration of secondary amide, respectively.25,26 The characteristic absorption peak at 280 nm and 405 nm from SA-HRP can be also observed in the UV-vis spectrum of HRP@HCR-COFS (Fig. 2B). The FT-IR and UV-vis spectra of HRP@HCR-COFS confirm that SA-HRP is successfully modified on the fiber surface after HCR amplification.
Agarose gel electrophoresis is used to show the feasibility for FEN1 assays (Fig. 2C). Lane 2 displays the two DNA bands from the product of co-incubation of FEN1 and the DNA substrate, corresponding to the DNA substrate (lane 1) and the 5′-flap (lane 3). This result indicates that FEN1 specifically recognizes and cleaves the bifurcated structure at the 5′ end of the double-stranded DNA substrate. While no DNA in lanes 1–6 is larger than 150 bp, the number of DNA substrate + FEN1 + H1 + H2 in lane 7 is far beyond 500 bp, indicating that HCR amplification can be carried out efficiently in this sensing system.
The chemiluminescence signals detected using the HCR-COFS in different compositions are recorded to further confirm the feasibility of the proposed method for FEN1 assays. As shown in Fig. 2D, when there is no target in the system, amplification is not triggered and only a weak signal can be detected. After the target or 5′-flap is directly added to the sensing system, a significantly enhanced chemiluminescence signal is observed, since the 5′-flap can trigger the HCR by opening up the CP on the fiber. The amplified signal intensity triggered by the target is approximately 2400 times that without the target. All of the above results demonstrate the feasibility and reliability of the developed HCR-COFS for ultra-sensitive detection of FEN1 with effective signal amplification.
Analytical performance of the HCR-COFS
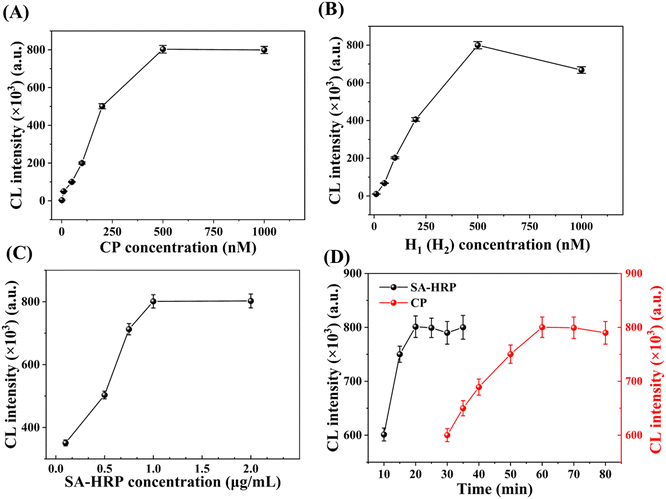
We first investigate and optimize several experimental conditions that could affect the analytical performance of the HCR-COFS, including the concentration of each HCR reactant (the CP, H1, and H2), the concentration of signal molecule SA-HRP, the CP immobilization time and the SA-HRP incubation time. As shown in Fig. 3A and B, the chemiluminescence intensity gradually increases and reaches the maximum value at 500 nM as the concentration of the CP and H1 (or H2) gradually increases. While the increase of the concentration of the HCR reactant can produce more double-stranded DNA on the optical fiber surface for binding signal molecules, the HCR will reach saturation due to the steric hindrance effect generated by the DNA double strand. In Fig. 3C, we show the chemiluminescence signal intensity as a function of the amount of SA-HRP under the optimal HCR conditions. The signal molecule SA-HRP is introduced to the sensor for catalyzing the generation of chemiluminescence by connecting with the amplified DNA sequence, thus the chemiluminescence signal intensity is enhanced by increasing the amount of SA-HRP and becomes stable when the SA-HRP concentration is larger than 1 μg mL−1. The effects of immobilization time of the CP and the incubation time of SA-HRP on the sensor response are also evaluated and optimized, as shown in Fig. 3D. The results show that the CP immobilization based on EDC/NHS crosslinking can be completed in 60 min, and the incubation time required for specific binding between streptavidin and biotin is around 20 min. Therefore, 60 min for the CP immobilization time and 20 min for the SA-HRP incubation time are used in the following assays.Under the optimal experimental conditions, we examine the analytical performance of the HCR-COFS for sensing FEN1. The response of the sensor to various concentrations of FEN1 (10 fM to 75 pM) is recorded as shown in Fig. 4A. A good linear correlation between the signal intensity and the logarithm of FEN1 concentration (Y = 1.63 × 105X + 2.28 × 106, R2 = 0.9910) is observed in a wide range of 10 fM–75 pM. The limit of detection (LOD) is determined according to the formula, 3.3δ/LOD = signal intensity (A)/concentration (A), where δ is the standard deviation of the signal values of 11 blank signals, and A is the lowest concentration point.27 The LOD for FEN1 assays using the present HCR-COFS is calculated to be 3.4 fM. In view of the fact that our proposed HCR-COFS can specifically identify the short-strand DNA 5′-flap, we employ the 5′-flap as the model target to evaluate the performance of the proposed method for detection of short-chain nucleic acids. The results are shown in Fig. 4B. A wide linear range of 1 fM-1 nM with a LOD as low as 0.27 fM can be achieved for the detection of the 5′-flap, indicating the capability of the HCR-COFS for sensitive sensing of short-chain nucleic acid biomarkers.
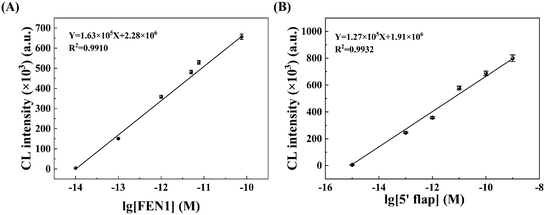 | ||
| Fig. 4 Linear response of the proposed HCR-COFS to (A) FEN1 concentrations and (B) 5′-flap concentrations. | ||
We further evaluated the specificity of the HCR-COFS for detecting FEN1, by selecting BSA, GSSG, HRP, HindIII, EcoRI, Dral, Exo I and Exo III as the interfering substances. Among these interfering substances, BSA, GSSG, and HRP are foreign proteins widely used to process biological samples; HindIII, EcoRI, Dral, Exo I, and Exo III are the common endonucleases or exonucleases with nucleic acid cleavage ability. As shown in Fig. 5, only FEN1 produces an apparent chemiluminescence signal response, which is over 3000 times higher than any other interfering substance. This is attributed to the fact that the interfering substances cannot trigger the HCR amplification by catalyzing the cleavage of DNA with a bifurcated structure at the 5′ end. The results show that the proposed HCR-COFS presents excellent selectivity for FEN1 determination, which is the result of the rational design of the DNA substrate based on the catalytic properties of FEN1.
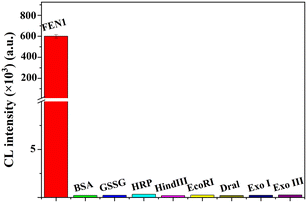 | ||
| Fig. 5 Selectivity of the proposed HCR-COFS for FEN1 assays (FEN1: 75 pM; BSA, GSSG, and HRP: 10 mg mL−1; HindIII, EcoRI, Dral, Exo I, and Exo III: 10 U μL−1). | ||
There are several merits of the proposed method that ensure excellent analytical performance for sensing FEN1 as we discussed above. The efficient HCR amplification on the fiber surface and the lossless light conductivity of the fiber allow a high signal-to-noise ratio of the sensor, which can significantly improve the detection sensitivity. In addition, no auxiliary DNA polymerase is used in the assays, which eliminates the interference of possible false positives. It is also worth mentioning that the proposed method requires no expensive and bulky instruments, making it possible to design a portable device that is suitable for POC diagnosis.
Analysis of real samples based on HCR-COFS sensors
To evaluate the potential application of the HCR-COFS for real sample analysis, we employ the sensor to detect FEN1 in HeLa cell lysate. As shown in Table 1, the sensor can directly detect 1.98 pM FEN1 in the cell lysate without dilution, showing sufficient sensitivity for analysis in biological matrices. A standard addition method is used to further prove the practical application of the HCR-COFS. The recoveries for the spiked samples are calculated as 98.7–102.3% with a RSD of 2.1–3.6% (n = 3), indicating good accuracy and reproducibility of the HCR-COFS for FEN1 detection. In Fig. S2,† we compare the results of the proposed method to those of the standard ELISA method for the analysis of both the spiked and unspiked samples. A good correlation between the ELISA method and HCR-COFS for the determination of HeLa cell lysate with a concentration range of 2.0–50 pM is achieved with a correlation efficiency of 0.999 within the 95% confidence interval. The results prove that the present HCR-COFS is suitable for the detection of FEN1 in complex biological matrices, which is expected to be a valuable diagnostic tool for clinical applications.| Sample | Spiked (pM) | Detected (pM) | Recovery (%) | RSD (n = 3, %) | ELISA (pM) | RSD (n = 3, %) |
|---|---|---|---|---|---|---|
| HeLa cells | 0 | 1.98 ± 0.04 | — | 2.1 | 1.96 ± 0.08 | 4.2 |
| 0.5 | 2.51 ± 0.06 | 101.2 ± 2.4 | 2.2 | 2.48 ± 0.13 | 5.0 | |
| 1.0 | 2.97 ± 0.06 | 98.7 ± 3.0 | 3.6 | 2.92 ± 0.18 | 5.9 | |
| 1.5 | 3.56 ± 0.08 | 102.3 ± 2.3 | 2.2 | 3.54 ± 0.11 | 3.0 |
Conclusions
In summary, we have proposed a novel FEN1 sensing approach by combining COFSs and a HCR amplification strategy. This approach is simply accomplished by capturing the 5′-flap sequences produced by FEN1-specific cleavage of DNA to trigger the HCR amplification reaction on the surface of the COFS. Chemiluminescence emission is recorded as the sensing response, which is achieved by SA-HRP connected with the fiber sensor via the affinity interaction between biotin and streptavidin to catalyze the substrates for signal amplification. Benefitting from the effective signal amplification and lossless signal output, the developed HCR-COFS exhibits excellent specificity and high sensitivity, with a LOD as low as 3.4 fM for FEN1 detection. The method is successfully applied for the determination of FEN1 in HeLa cell lysate samples, the results of which are consistent with those from the ELISA assay. Our study will pave the way to utilize COFSs as a powerful tool for real-time and on-site detection of disease biomarkers.Author contributions
Jinlan Yang: data curation and writing – original draft. Dingsong Li: data curation and supervision. Yaxue Jia: validation and investigation. Jiahui Fu: visualization. Li Yang: methodology, writing – review & editing, investigation and project administration.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (22074014), the Fundamental Research Funds for the Central Universities (CGZH202204; 2412022QD007), and the Science and Technology Project of Jilin Provincial Department of Education (JJKH20231300KJ). L. Yang would also like to thank the Jilin Provincial Department of Education and the Jilin Provincial Key Laboratory of Micro-Nano Functional Materials (Northeast Normal University) for the support.References
- S. E. Tsutakawa, S. Classen, B. R. Chapados, A. S. Arvai, L. D. Finger, G. Guenther, C. G. Tomlinson, P. Thompson, A. H. Sarker, B. Shen, P. K. Cooper, J. A. Grasby and J. A. Tainer, Cell, 2011, 145, 198–211 CrossRef CAS.
- L. Balakrishnan and R. A. Bambara, Annu. Rev. Biochem., 2013, 82, 119–138 CrossRef CAS PubMed.
- R. S. Williams and T. A. Kunkel, Cell, 2011, 145, 171–172 CrossRef CAS PubMed.
- K. Zhang, S. Keymeulen, R. Nelson, T. R. Tong, Y. C. Yuan, X. Yun, Z. Liu, J. Lopez, D. J. Raz and J. Y. Kim, Am. J. Pathol., 2018, 188, 242–251 CrossRef CAS PubMed.
- L. Xu, J. L. Qu, N. Song, L. Y. Zhang, X. Zeng, X. F. Che, K. Z. Hou, S. Shi, Z. Y. Feng, X. J. Qu, Y. P. Liu and Y. E. Teng, Oncol. Rep., 2020, 44, 2443–2454 CrossRef CAS PubMed.
- L. Liu, C. Zhou, L. Zhou, L. Peng, D. Li, X. Zhang, M. Zhou, P. Kuang, Q. Yuan, X. Song and M. Yang, Carcinogenesis, 2012, 33, 119–123 CrossRef CAS PubMed.
- H. Zhang, S. Ba, D. Mahajan, J. Y. Lee, R. Ye, F. Shao, L. Lu and T. Li, Nano Lett., 2018, 18, 7383–7388 CrossRef CAS PubMed.
- Y. Wang, S. Li, L. Zhu, J. Zou, X. Jiang, M. Chen and B. Chen, Clin. Transl. Oncol., 2019, 21, 1026–1033 CrossRef CAS.
- Y. Zhang, X. Liu, L. Liu, J. Chen, Q. Hu, S. Shen, Y. Zhou, S. Chen, C. Xue, G. Cui and Z. Yu, Dis. Markers, 2020, 2020, 2514090 Search PubMed.
- L. He, Y. Zhang, H. Sun, F. Jiang, H. Yang, H. Wu, T. Zhou, S. Hu, C. S. Kathera, X. Wang, H. Chen, H. Li, B. Shen, Y. Zhu and Z. Guo, EBioMedicine, 2016, 14, 32–43 CrossRef CAS.
- E. Zhao, C. Zhou and S. Chen, Clin. Res. Hepatol. Gastroenterol., 2021, 45, 101455 CrossRef CAS PubMed.
- X. Liu, Y. Zhao and F. Li, Biosens. Bioelectron., 2021, 173, 112832 CrossRef CAS PubMed.
- Y. Zhu, J. Zhu, Y. Gao, J. Shi and P. Miao, ChemElectroChem, 2023, 10, e202300020 CrossRef CAS.
- H. Yang, C. Wang, E. Xu, W. Wei, Y. Liu and S. Liu, Anal. Chem., 2021, 93, 6567–6572 CrossRef CAS PubMed.
- B. Li, A. Xia, S. Xie, L. Lin, Z. Ji, T. Suo, X. Zhang and H. Huang, Anal. Chem., 2021, 93, 3287–3294 CrossRef CAS PubMed.
- C. Cui, C. H. Lau, L. T. Chu, H. K. Kwong, C. Tin and T. H. Chen, Biosens. Bioelectron., 2023, 220, 114859 CrossRef CAS PubMed.
- B. Zhou, L. Lin and B. Li, Sens. Actuators, B, 2021, 346, 130457 CrossRef CAS.
- Q. Han, X. Bian, Y. Chen and B. Li, Sens. Actuators, B, 2023, 393, 134265 CrossRef CAS.
- L. Zhang, X. Liu, N. Zhang, X. Liu and W. Jiang, Sens. Actuators, B, 2022, 353, 131147 CrossRef CAS.
- O. Salama, S. Herrmann, A. Tziknovsky, B. Piura, M. Meirovich, I. Trakht, B. Reed, L. I. Lobel and R. S. Marks, Biosens. Bioelectron., 2007, 22, 1508–1516 CrossRef CAS.
- S. A. Pidenko, N. A. Burmistrova, A. A. Shuvalov, A. A. Chibrova, Y. S. Skibina and I. Y. Goryacheva, Anal. Chim. Acta, 2018, 1019, 14–24 CrossRef CAS.
- X.-D. Wang and O. S. Wolfbeis, Anal. Chem., 2019, 92, 397–430 CrossRef PubMed.
- M. Li, R. Singh, Y. Wang, C. Marques, B. Zhang and S. Kumar, Biosensors, 2022, 12, 843 CrossRef CAS PubMed.
- E. Torelli, M. Manzano and R. S. Marksd, Sens. Actuators, B, 2017, 247, 868–874 CrossRef CAS.
- H. Wang, J. Han, Z. Li and Z. Wang, J. Hazard. Mater., 2023, 447, 130797 CrossRef CAS.
- J. Han, H. Wang, Z. Li and Z. Wang, J. Chromatogr. A, 2021, 1652, 462373 CrossRef.
- D. Kong, J. Zhao, S. Tang, W. Shen and H. K. Lee, Anal. Chem., 2021, 93, 12156–12161 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sd00230f |
| This journal is © The Royal Society of Chemistry 2024 |