Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Diagnosis of infectious diseases: complexity to convenience
Sahil
Syed†
 a,
Alisha
Rahaman†
a,
Abhijit
Mondal
a,
Shraddha
Shaligram
a,
Alisha
Rahaman†
a,
Abhijit
Mondal
a,
Shraddha
Shaligram
 *a and
Shrikant P.
Pawar
*b
*a and
Shrikant P.
Pawar
*b
aHematology and Biochemistry Division, Department of Serology and Microbiome, Mylab Discovery Solutions Pvt. Ltd., Global Innovation Center, 5th Floor, Amar Paradigm, Opposite Croma Showroom, Baner-411045, Maharashtra, India. E-mail: shraddha.shaligram@mylabglobal.com
bDepartment of Serology and Microbiome, Mylab Discovery Solutions Pvt. Ltd., Global Innovation Center, 5th Floor, Amar Paradigm, Opposite Croma Showroom, Baner-411045, Maharashtra, India. E-mail: shrikant.p@mylabglobal.com
First published on 19th January 2024
Abstract
Infectious diseases are one of the leading causes of mortality around the world. Among the various infectious diseases, malaria, HIV, HCV, HBV, tuberculosis and influenza are amongst the most prevalent. Recently in 2019, the world had to suffer through the COVID-19 pandemic which has led to many deaths across the globe. Such infectious diseases can be prevented and contained and their transmission can be limited by their early detection and screening with the help of various diagnostic techniques. The development of systems that provide faster results with similar or better sensitivity and specificity is of utmost importance as they can play a critical role in the epidemiology and progression of a disease. Due to this, there is a need for the development of highly sensitive, specific and accurate techniques for infection control and prevention. In this paper, we include and discuss the recent advancements in diagnostic techniques that include improvements in molecular diagnostics, biosensor-based disease diagnosis and lateral flow assays and their capability to be used as point-of-care tests. The techniques have been compared with respect to their sensitivity, specificity and limit of detection. The goal of this review is to summarize the recent developments in the field of disease diagnostics that provide us with numerous new platforms. Discussion of such techniques which have future potential for point-of-care application could thus be used as a tool for directing research related to development or modification of techniques for disease diagnostics.
Introduction
Diseases caused by live pathogens such as bacteria, viruses and parasites which have the capability of fast transmission and infection among human or animal vectors by inoculation and airborne or waterborne transmission, are known as infectious diseases.1–5 They are the main cause of morbidity and mortality globally6 and have been identified by the World Health Organization (WHO) as the second major contributor to the death toll in humans.7,8 The latest addition to the list of infectious diseases was severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) which caused a pneumonia outbreak which began in December 2019 and caused an extensive amount of risk to global health.9–11 Until 21 June 2023, it led to 768![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 187
187![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 confirmed cases of coronavirus disease 2019 (COVID-19), including 6
096 confirmed cases of coronavirus disease 2019 (COVID-19), including 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 945
945![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 714 deaths, reported to the WHO.12 The International Monetary Fund estimated that the world economy decreased by 4.4% in the year since the pandemic began.13 Similar losses have been experienced by humans throughout history, for example, the influenza pandemic of 1918,14,15 the SARS pandemic in 2003,16–18 the middle east respiratory syndrome (MERS) pandemic in 2012 (ref. 19 and 20) and the Ebola outbreak in West Africa during 2013 to 2016.21,22 The appearance of these infectious diseases and their transmission considerably impacted the economy, health and daily life of humans all around the globe.23–25
714 deaths, reported to the WHO.12 The International Monetary Fund estimated that the world economy decreased by 4.4% in the year since the pandemic began.13 Similar losses have been experienced by humans throughout history, for example, the influenza pandemic of 1918,14,15 the SARS pandemic in 2003,16–18 the middle east respiratory syndrome (MERS) pandemic in 2012 (ref. 19 and 20) and the Ebola outbreak in West Africa during 2013 to 2016.21,22 The appearance of these infectious diseases and their transmission considerably impacted the economy, health and daily life of humans all around the globe.23–25
For control of these diseases, three strategies are proposed which include 1) restraining the source of infection, 2) stopping the means of communication of disease, and 3) securing the people who are at risk of infection.26–30 As a method for preventing further infection and protecting the susceptible population, identifying and controlling the source of infection is of utmost importance.31–33 Towards this end, development of specific, sensitive and fast detection systems is an important step and a major lesson from previous experiences.26 The traditional methods of disease diagnosis involved detection of pathogens by using microscopy or by culturing the pathogens and observing the culture characteristics. However, these techniques are not reliable as they often do not yield results or are wrongfully interpreted and have longer turnaround time.33–36 Diagnostic methods have undergone developments and techniques such as enzyme-linked immunosorbent assay (ELISA), nucleic acid testing (NAT) and clustered regularly interspaced short palindromic repeat (CRISPR) and CRISPR-associated protein (Cas)-based diagnostics have emerged which can detect specific antigens, antibodies or nucleic acids of the target organism. But these techniques have their drawbacks such as the requirement of well-equipped laboratories and skilled personnel, and due to their sensitive nature, they are prone to contamination which has a major impact on the results. Also, the cost of associated reagents is very high.37–39 These shortcomings make them unsuitable for use as point-of-care (POC) solutions, especially in areas where the availability of resources is limited and the chances of transmission and the need for diagnosis are high.
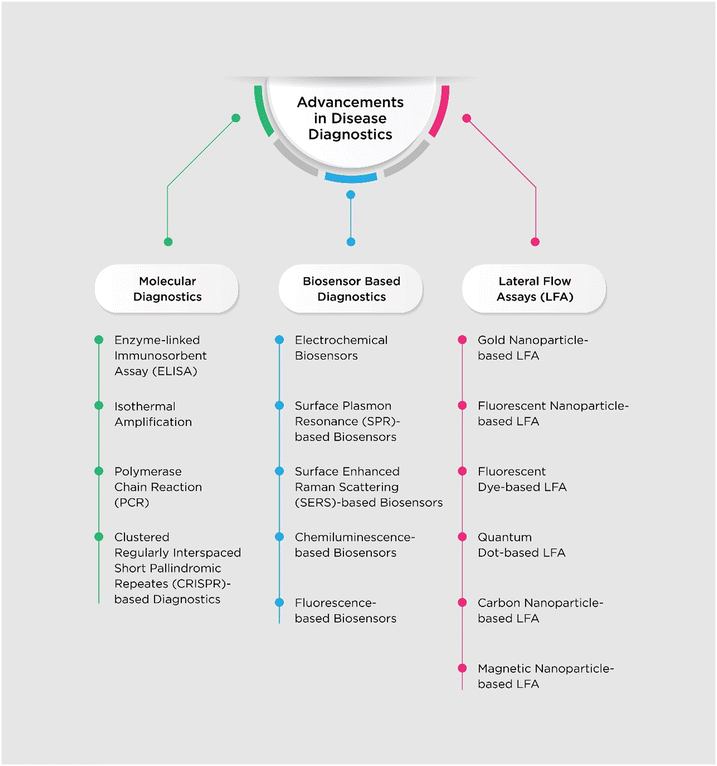
To overcome the known shortcomings, many efforts have been made and novel platforms that utilize surface enhanced Raman scattering (SERS), fluorescence, chemiluminescence, surface plasmon resonance (SPR), quantum dots (QDs), electrochemical (ECL) biosensors, chemiluminescent biosensors, colorimetric nanoparticles, fluorescent nanoparticles and carbon nanoparticles (CNPs) have been developed.40–50 Due to the backdrop of the COVID-19 pandemic, the necessity for and importance of accurate, on-site, sensitive and rapid detection systems was highlighted and research in this regard was boosted. In this review, we have tried to summarize the developments that have been made in the field of diagnostics of various infectious diseases. Also, focus has been given to new technologies which aim to provide sensitive, low-cost and accurate POC testing systems. Fig. 1 shows an overview of this study.
Molecular diagnostics
Molecular diagnostics is a collection of techniques used to analyze biological markers in the genome and proteome. The basic principles on which molecular diagnostics relies are detection of proteins and nucleic acids.36 Molecular diagnostic techniques are preferred because of their high sensitivity and specificity51,52 which lead to a precise conclusion of results. In medicine, these techniques are used to diagnose and monitor disease, detect risk, and decide which therapies will work best for individual patients.Enzyme-linked immunosorbent assay (ELISA)
ELISA is a widely used laboratory technique for detecting and quantifying the presence of an antigen or antibody in a sample.53 It is a versatile technique that can be used to detect a wide variety of molecules, including proteins, viruses, and bacteria.54 Towards improvement of this conventional technique, many efforts have been made. Attempts have been made to incorporate nanoparticles with ELISA to increase sensitivity and lower the limit of detection (LOD). Song et al., 2016 developed an invader assisted ELISA, wherein oligonucleotide probes and gold nanoparticles (AuNPs) were coupled with the assay. The group was able to detect antigens specific to hepatitis B. The LOD achieved was as low as 24 pg mL−1 hepatitis B surface antigen (HBsAg) by both the naked eye and a spectrophotometer. When compared to the conventional ELISA, it was found to be 102 times more sensitive.55 Owing to their high extinction coefficient and strong plasmon resonance properties, silver nanoparticles (AgNPs) are considered to be an optimal choice for colorimetric indicators.56 By using a combination of gold and silver nanoparticles, Khoris et al., 2019 were able to detect norovirus. They found the LOD to be 10.8 pg mL−1, which was 102 and 103 times higher in sensitivity than the traditionally used ELISA and gold-immunoassay, respectively.57 Fluorometry has the ability to detect concentrations that are manifold lower than the detection limits of colorimetric techniques. The improvement in the sensitivity of AuNP-based ELISA for detection of the Opisthorchis viverrini antigen (OvAg) was achieved by developing fluorescence AuNPs-LISA. The new assay showed dynamic linear detection of OvAg concentration in the range of 34.18 ng mL−1 to 273.44 ng mL−1 with a LOD of 36.97 ng mL−1. A 1200-fold enhanced detection sensitivity was observed when compared with colorimetric AuNPs-LISA.58 To increase the reach of this platform to places where infection with Opisthorchis viverrini is common, a smartphone-based portable fluorometer was designed to be used as a POC device for diagnosis of OvAg in urine samples.59 Wu et al., 2017 were able to develop an enhanced fluorescence ELISA system which utilized human alpha-thrombin triggering fluorescence turn-on signals. This system was able to detect HBsAg at levels of 5 × 10−4 IU mL−1; this level was 104 and 106 times lower than those of the conventional fluorescence assays and conventional ELISAs, respectively.60 To further enhance the detection of specific proteins, a system was developed which amplified the signals produced by the antigen–antibody reaction during ELISA. For achieving this, researchers combined sandwich ELISA with enzyme cycling and created an ultrasensitive ELISA.61 A 3α-hydroxysteroid dehydrogenase (3α-HSD) reaction system was used for this purpose, in which the 3α-HSD acts as a catalyst for substrate cycling between 3α-hydroxysteroid and 3-ketosteroid while excess amounts of NADH and thio-NAD as the cofactors are present. Thio-NAD cycling refers to the series of reactions that occur during the aforementioned substrate cycling.56,62 This ultrasensitive ELISA with thio-NAD cycling has been employed to detect two characteristic proteins of SARS-CoV2. Spike protein and nucleocapsid protein were detectable with LODs of 2.62 × 10−19 moles per assay and 2.95 × 10−17 moles per assay.63,64 Using a similar platform, live Mycobacterium tuberculosis specific protein-MPT64 was detectable with a turnaround time of 5 hours and LODs of 0.15 pg mL−1 and 0.10 pg mL−1 by Sakashita et al., 2020 and Wang et al., 2020, respectively.65,66Mycobacterium tuberculosis was also detected by employing traditional ELISA but using extracellular vesicles (EVs) as a biomarker.67 EVs are a new biomarker candidate for the detection of a multitude of diseases.68,69 Glycolipid lipoarabinomannan (LAM) and membrane protein LprG which are responsible for the virulence of Mycobacterium tuberculosis were detected using EVs isolated from Mycobacterium tuberculosis-infected macrophage cultures in EV-ELISA. The same group of researchers developed a portable and smartphone operable nanoparticle enhanced EV immunoassay (NEI) read by dark field microscopy and reported that their system could detect LAM and LprG biomarkers which could be used to detect Mycobacterium tuberculosis infection as well as to differentiate between latent and pulmonary tuberculosis.67Polymerase chain reaction (PCR)
PCR is a type of NAT, which amplifies specific nucleic acid sequences from the target. During the entire process of PCR, it undergoes a series of temperature cycles in the presence of a polymerase enzyme and free nucleotides that yield an exponentially amplified target sequence.70 PCR is highly sensitive and specific as it is a nucleic acid-based technique and hence it allows for accurate detection of a wide range of bacteria, viruses and parasites. Since the discovery of this technique, owing to its vast potential it has undergone modifications and development. Multiplex PCR was introduced when researchers multiplexed six polymorphic minisatellite loci in one PCR tube.71 Multiplexing PCR uses multiple primers that target different DNA or RNA sequences in a single reaction. This allows for the simultaneous detection of multiple pathogens or for the analysis of gene expression.72 PCR techniques became quantitative when the method to monitor PCR kinetics in real time was developed.73 Real-time PCR uses a fluorescent dye or probe that binds to the amplified DNA or RNA. As the DNA or RNA is amplified, the amount of fluorescence increases, which can be monitored in real time. This allows for the detection of even very small amounts of DNA or RNA, and it can also be used to quantify the amount of DNA or RNA that is present.74 The involvement of reverse transcriptase in PCR for the detection of RNA75 paved the way for the development of reverse transcriptase PCR (RT-PCR) and quantitative RT-PCR (qRT-PCR) as we know them today. Another offshoot from the PCR technology is the digital PCR (dPCR) in which a sample is split into millions of subsamples to digitize the overall content of DNA.76 Droplet-based systems have also evolved, which have cells in their sample that is loaded onto a microfluidic chip composed of different regions for cell lysis, DNA extraction and its purification followed by PCR and then detection of the fluorescence signal.77,78 Since this system is used for single-cell based studies, it is also known as single-cell PCR.79 An amalgamation of dPCR and droplet-based PCR gave rise to droplet-based digital PCR (ddPCR). The ddPCR is advantageous over qPCR in exhibiting higher precision and a lower coefficient of variation.80 A droplet magnetofluidic-based PCR system was presented by Shin et al., 2018 with potential for POC application. The platform was designed as a single step assay where only the sample was to be loaded into the cartridge with magnetic particles and the cartridge was to be loaded into the instrument. They demonstrated the usability of this platform by quantifying hepatitis C virus (HCV) RNA viral load with a LOD of 45 IU per 10 μL of sample.81 The most recent developments in PCR involve ultrafast photonic PCR. The researchers who developed it modified the usual PCR process and fabricated a thin Au film-based light-to-heat converter to heat the PCR solution over 150 °C by harnessing gold plasmon-assisted high optical absorption. Their system could complete 30 cycles within 5 minutes.82 Cho et al., 2019 utilized photonic PCR in developing a light-driven integrated cell lysis and PCR on a chip with gravity-driven cell enrichment health technology (LIGHT) platform for diagnosis of pathogens. They were able to detect 103 CFU mL−1 of E. coli within 20 minutes.83 Similarly, Nabuti et al., 2023 used Au nanofilms on a photonic PCR system and reported that with a sample volume of 20 μL, 30 cycles could be completed in 7.5 minutes with better uniformity and reliability in temperature distribution with no noticeable deviations.84 Using the well-established qRT-PCR, Bazié et al., 2023 studied the impact of viral replication and antiretroviral therapy on large and small EVs' mitochondrial DNA content in the case of human immunodeficiency virus (HIV).85 Exosomes which are a subpopulation of EVs contain miRNA. A set of four exosomal miRNAs which could be used as a biomarker for pneumonia in adenovirus-infected children were identified using qRT-PCR.86Isothermal amplification
The requirement of thermal cycling for PCR is a major blockade while developing a new platform using PCR. To counter this, researchers have discovered isothermal amplification. It overcomes the need for thermal cycling as the entire amplification process takes place at a constant temperature.70 Based on isothermal amplification, techniques such as loop mediated isothermal amplification (LAMP), rolling circle amplification (RCA), nucleic acid sequence-based amplification (NASBA) and recombinase polymerase amplification (RPA) have been derived.87–90 Among the above-mentioned techniques, two or more primers are used for amplification by NASBA, LAMP and RPA, whereas RCA utilizes one functional template to amplify target nucleic acids as primers. Apart from RCA and LAMP, other techniques rely not only on DNA polymerase but also on other additional enzymes and proteins.91 Parida et al., 2004 developed real time accelerated reverse transcriptase LAMP (RT-LAMP) for detection of West Nile (WN) virus. They reported that RT-LAMP had 10-fold higher sensitivity as compared to RT-PCR with a detection limit of 0.1 PFU of virus.92 LAMP mediated detection of Newcastle disease virus (NDV) was achieved by Pham et al., 2005 with a LOD of 0.5 pg of plasmid per reaction.93 Similarly, Leishmania donovani detection with the help of LAMP was demonstrated by Puri et al., 2021. Their research reported the development of a portable LAMP device with a reaction time of 1 hour and a LOD of 100 fg.94 NASBA-based detection of dengue viral RNA was reported by Usawattanakul et al., 2002. The NASBA assay could detect as low as 1 PFU mL−1 of all four dengue viral serotypes being tested.95 NASBA-based detection was also done for La Crosse (LAC) virus which showed a LOD of 0.0175 PFU.96 Another infectious disease that is a major threat to health is tuberculosis. Thus, RPA-based detection of Mycobacterium tuberculosis complex (MTC) DNA was developed by Boyle et al., 2014. MTC specific targets IS6110 and IS1081 were targeted and the LOD was found to be 6.25 fg and 20 fg respectively. They reported the completion of this assay in less than 20 minutes.97 For the detection of HIV-1 proviral DNA for early infant diagnosis, a RPA-based assay which could detect 3 copies of proviral DNA within 20 minutes was designed.98 RPA was also used for the detection of Plasmodium falciparum. Lateral flow RPA was developed and it demonstrated a LOD of 100 fg of genomic P. falciparum DNA within 15 minutes.99 As an aid to sensitive and fast diagnosis of SARS-CoV-2, real time RT-LAMP was developed which could detect 1 copy per reaction of viral RNA in 30 minutes with 100% sensitivity and specificity.100 Due to the high demand for rapid detection systems for SARS-CoV-2, similar RT-LAMP-based systems were developed by other researchers as well.101–103 Another detection system for SARS-CoV-2 was developed using NASBA, which had a LOD of 200 copies per mL for nucleocapsid and RNA-dependent RNA polymerase genes.104 A suitcase lab for detection of SARS-CoV-2 based on RPA was developed. The assay was deployed on-field in three laboratories in Africa and helped in the rapid diagnosis of infection, especially in resource poor settings. It could give results within 15 minutes and was able to detect 2, 15 and 15 RNA molecules of molecular standard/reaction of RNA-dependent RNA polymerase, envelope protein and nucleocapsid protein genes.105Clustered regularly interspaced short palindromic repeats (CRISPR)
The CRISPR–Cas system is a defense mechanism evolved by bacteria. CRISPR sequences are acquired by bacteria during infections from bacteriophages, and upon subsequent infection, they produce Cas protein that helps to cleave the viral nucleic acid.106–108 CRISPR–Cas9 is a widely known gene-editing tool, but it is just one of the many such systems that exist. The CRISPR–Cas systems are classified into two groups, class 1 containing a multi-effector cascade and class 2 containing single effector proteins.109 These two classes are further classified into distinct subtypes, class 1 contains types I, III, and IV and class 2 consists of types II, V and VI.110,111 After their discovery, the systems have been widely used for gene editing owing to their high specificity, but recently, developments have been made in regard to their use in diagnostics.Chen et al., 2018 created a method viz. DNA endonuclease targeted CRISPR trans reporter (DETECTR) by merging Cas12a ssDNase activation with isothermal amplification. They reported that DETECTR was able to specifically detect human papilloma virus (HPV) 16 and HPV 18 from human samples with attomolar sensitivity.112 Detection of HPV 16 and HPV 18 was also demonstrated in research undertaken by Wang et al., 2018, wherein they developed a method for detection and typing of target DNA based on Cas9 nuclease. It was abbreviated as ctPCR representing CRISPR-typing PCR. They reported that their method could detect DNA molecules at a concentration of 400 copies per μL. Also, they reported a time of 3 to 4 hours for the entire process of ctPCR.113 Similarly, Zhang et al., 2018 developed a technique named Cas9/sgRNA-associated reverse PCR (CARP) for the detection of HPV 16 and HPV 18. The authors reported that their system had a LOD of 0.002 ng of genomic DNA (cervical carcinoma cells) and 0.02 ng DNA (clinical samples) with a time requirement of 3 hours.114
Gootenberg et al., 2017 developed a detection platform named specific high-sensitivity enzymatic reporter unlocking (SHERLOCK).115 The SHERLOCK system employed Cas13 combined with isothermal amplification. It was able to specifically detect Zika virus and dengue virus at a concentration of 2 aM. Following this, SHERLOCK version 2 (SHERLOCKv2) was introduced which had advancements such as better sensitivity, multiplexity and portable nucleic acid detection. SHERLOCKv2 used Cas12 and Csm6 along with Cas13 which resulted in better multiplexing capability and amplified signal detection, respectively. The researchers reported that their system could detect samples at zeptomolar concentrations.116 To further enhance this technique and facilitate instrument-free detection directly from bodily fluids, Myhrvold et al., 2018 developed a protocol – heating unextracted diagnostic samples to obliterate nucleases (HUDSON) – that could pair with SHERLOCK. They reported detection of Zika virus and dengue virus from body fluids in less than 2 hours and less than 1 hour, respectively, with a LOD of 1 copy per mL.117 Using the same platform as employed by Myhrvold et al., 2018, Barnes et al., 2020 developed a diagnostic platform for Ebola virus and Lassa virus. Along with it, they also developed a mobile application for result interpretation and reporting. They reported a LOD of 10 copies per μL for Ebola virus, 10 copies per μL for Lassa virus clade II and 100 copies per μL for Lassa virus clade IV.118
Pardee et al., 2016 developed a cell-free, paper-based sensor for detection of Zika virus. They demonstrated the use of Cas9 protein to cleave dsDNA to detect the genotype of Zika virus after isothermal amplification of viral RNA. They reported that their system could detect viral strains which have a variation of even a single base.119 Another group of researchers developed a Cas13a-based microfluidic device which could fluorometrically detect Ebola virus. It had a detection limit of 20 pfu mL−1 and a detection time of 5 minutes.120 Due to its small size, specificity, sensitivity, short detection time and amplification-free nature, it showed substantial potential to be used as a POC diagnostic platform. Combinatorial arrayed reactions for multiplexed evaluation of nucleic acids (CARMEN) was developed by Ackerman et al., 2020 which facilitated detection of multiple targets using a single microfluidic chip. By combining CARMEN with Cas13 detection, they were able to assay a panel of 169 human associated viruses simultaneously. They also displayed the flexibility and robustness of their system by detection of Zika virus, detection and subtyping of influenza A virus and detection of drug resistance mutations in HIV.121
A PCR-based CRISPR–Cas13a (PCR-CRISPR) system was developed by combining conventional PCR amplification and specificity provided by CRISPR. They demonstrated its sensitivity and specificity by detecting hepatitis B virus (HBV) and its drug resistant variants. They found that PCR-CRISPR could detect HBV DNA as low as 1 copy per test in 15 minutes post PCR-amplification step.122 Eliminating the need for nucleic acid extraction which is usually required for CRISPR-based diagnostics, a detection system was developed that could detect SARS-CoV-2 RNA directly from clinical samples with a detection limit of 15 fM in less than 20 minutes.123 To overcome the performance drawbacks presented by RT-qPCR when using plasma samples for detection of SARS-CoV-2, Ning et al., 2021 developed a one-step CRISPR-enhanced reverse transcriptase (RT)–RPA fluorescence detection system (CRISPR-FDS). In this assay, SARS-CoV-2 RNA present in EVs was detected directly from plasma using a specific antibody and then fused with liposomes containing reagents for RT-RPA and CRISPR–Cas12a reactions using a workflow resembling ELISA.124
Biosensors
Biosensors convert the signals from a biological reaction into electrical or optical signals.125 Biosensors are being studied widely because of their ability to perform tests in a short period of time with high sensitivity.50 These involve electrochemical biosensors, SPR-based biosensors, SERS-based biosensors, fluorescent biosensors, colorimetric biosensors, and chemiluminescent biosensors. Their user-friendliness makes them compatible to be used in POC tests and can be used as a self-testing device.126 The working principle of a biosensor is depicted in Fig. 2.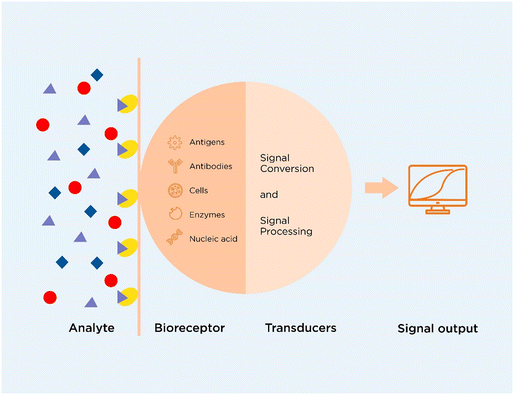 | ||
| Fig. 2 Schematic diagram of a biosensor (adapted from ref. 125 with permission from Springer Nature, copyright 2017). | ||
Electrochemical (ECL) biosensors
These biosensors transduce biological reactions into electrical signals such as in the form of current (amperometric), potential difference (voltammetric), charge (potentiometric), and impedance (impedimetric). Studies have been carried out to detect infectious diseases caused by SARS-CoV2, Zika virus, HIV, Ebola virus, etc.126–128Fig. 3 depicts a general scheme of the analytical principle of ECL biosensors.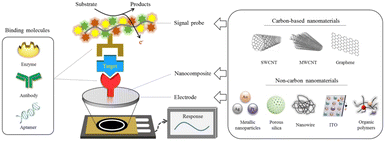 | ||
| Fig. 3 Scheme of the analytical principle for electrochemical biosensors based on carbon and non-carbon nanomaterials (reproduced from ref. 127 with permission from Springer Nature, copyright 2020). | ||
During the COVID-19 pandemic, the development and use of rapid tests have been demanded to prevent the transmission of the virus. Scientists have studied the development of electrochemical biosensors for rapid and highly sensitive detection of the virus in a short period of time. Mahari et al., 2020 developed a carbon electrode-based electrochemical biosensor that measures electrical conductivity. It utilised immobilized nCOVID-19 antibodies to detect the virus spike antigen within a time of 30 seconds. The LOD was reported to be 90 fM.45 Similarly, Hai et al., 2018 studied the development of a biosensor for the detection of human influenza A. It consists of an organic electrochemical transistor grafted with a trisaccharide against the hemagglutinin present in the spike protein of the virus. The change in the current is measured by the device when the virus particles adsorb and bind to the trisaccharide. The LOD when compared to a commercial lateral flow assay was found to be lower.129 Islam et al., 2019 also developed a sensor to detect HIV-p24 along with cTNI and citrullinated peptide for rheumatoid arthritis. The device uses anti-HIV-p24 antibodies conjugated on amine-functionalized graphene that measures the resistance change during the antigen and antibody interaction. The study reported a LOD of 100 fg mL−1 for HIV-p24 and 10 fg mL−1 for cTNI and citrullinated peptide.130
Surface plasmon resonance (SPR)-based biosensors
SPR-based biosensors are primarily used for label-free detection of molecules based on the surface plasmon resonance properties of metallic nanoparticles. Such biosensors measure the change in the refractive index due to the binding interaction and kinetics of biomolecules on the functionalized surface.50,131,132 The principle of a SPR-based biosensor is explained in Fig. 4.133 These sensors have been employed in various fields such as food studies, environmental studies, enzymology, pharmacokinetics, etc.132 Diao et al., 2018 developed a SPR-based biosensor to detect HIV DNA. The sensor is based on the use of entropy-driven strand displacement reactions (ESDRs) with double-layer DNA tetrahedrons (DDTs). The sample DNA becomes amplified to form double stranded DNA products which then bind to DDTs which are measured in 60 minutes. The device has a LOD of 48 fM with high accuracy and reproducibility.134 A similar biosensor was developed by Kim et al., 2018 for detection of HBsAg which works on the principle of the antigen–antibody reaction. The study reported the lowest detection limit of 100 fg mL−1 within a time of 10–15 minutes.135 Sun et al., 2017 studied the detection of Mycobacterium tuberculosis by detecting the antibodies present in the body against fusion protein antigen CFP10-ESAT6. The biosensor detects the antibody by measuring the change in the localized surface plasmon resonance (LSPR) properties of gold nanorods (AuNRs) achieving a sensitivity and specificity of 79% and 92%, respectively.44 Adegoke et al., 2017 developed a LSPR-mediated fluorescent biosensor for detection of Zika virus RNA. Four nanoparticles were functionalized with 3-mercaptopropionic acid (MPA) to form MPA-AgNPs, MPA-AuNPs, core/shell (CS) Au/AgNPs, and alloyed AuAgNPs which were conjugated to a Zika virus DNA probe and L-glutathione-capped CdSeS alloyed quantum dots (QDs). When viral RNA binds to the DNA probe, an enhanced fluorescence signal is observed. The study shows that a better LOD of 1.7 copies per mL was achieved when alloyed AuAgNPs conjugated to Qdot molecular beacons were used for detection.136 Similarly, a highly sensitive dengue detection device was developed by Omar et al., 2020. They built a SPR sensor consisting of a functionalized substrate that captures and detects DENV 1 E-protein in a range of 0.08–0.5 pM.137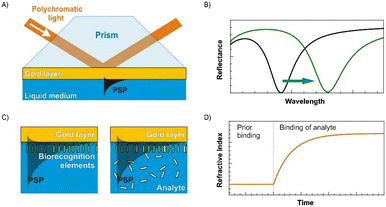 | ||
| Fig. 4 Concept of a SPR biosensor. (A) Excitation of propagating surface plasmons by polychromatic light in the Kretschmann geometry of the ATR method. (B) Spectrum of reflected light with a characteristic SPR dip for two different values of the refractive index at the gold layer. (C) SPR sensor surface with immobilized probes (left) and binding of the target molecules to the probes (right). (D) Temporal evolution of the refractive index caused by the molecular interaction (reproduced from ref. 133 with permission from Elsevier, copyright 2013). | ||
Surface enhanced Raman scattering (SERS)-based biosensors
SERS is based on the phenomenon of Raman scattering. Raman scattering is a phenomenon where the incident light when falling on a molecule leads to inelastic scattering. This scattering pattern is unique for every molecule and generates a fingerprint spectrum.48 But Raman scattering is a very weak process and requires a highly concentrated sample or longer time to generate a spectrum. Thus, the nanoparticles employed in SERS enhance the weak signals obtained after Raman scattering due to the phenomenon of LSPR. SERS signal enhancement is observed due to electromagnetic and chemical enhancement with contributions of electromagnetic enhancement.50,138,139 SERS-based biosensors are used for diagnosis based on two methods – label-free direct and label-dependent indirect methods. In the direct method, the analyte binds directly to the SERS-substrate and the detection is based on the analyte's unique Raman fingerprint. Meanwhile, the indirect method employs tags or reporter molecules that capture the analyte, and the detection occurs by measuring the change in the Raman signal of the reporter molecule bound to the substrate.140 The fabrication and working principle of a SERS-based biosensor are illustrated in Fig. 5.141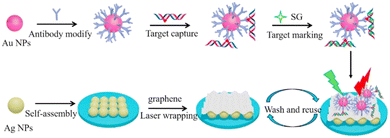 | ||
| Fig. 5 Fabrication and working principle of a carbon-based 2D reusable laser-wrapped graphene–Ag array SERS biosensor for trace detection of genomic DNA methylation (reproduced from ref. 141 with permission from Elsevier, copyright 2016). | ||
Gribanyov et al., 2021 studied the use of substrate-free SERS-based sensors that employ aptamers for specific detection of influenza A virus. The aptamer is functionalized on AgNPs which are then aggregated to produce SERS signals. The aptamer that specifically captures the virus via the hemagglutinin present on its surface leads to an increase in SERS-signals indicating a quantitative measurement of the viral load. The technique is simple due to its substrate-free nature and the preparation of AgNPs is easy, leading to a reduced detection time of below 15 minutes and achieving a detection range of 2 × 105 to 2 × 106 VP mL−1 and a LOD of 2 × 105 VP mL−1.40 A similar biosensor was developed by Zavyalova et al., 2021 for detection of SARS-CoV2 based on a phenomenon described by Gribanyov et al., 2021. The biosensor achieved a LOD of 5.5 × 104 TCID50 mL−1 and detected the virus within a time of 7 minutes with high specificity.40,142 Yang et al., 2021 described a biosensor that they developed to detect SARS-CoV2 from a water sample within a time of 5 minutes. The sensor utilises gold nanostructures that are functionalized with human angiotensin-converting-enzyme 2 (ACE2). The ACE2 captures the virus by binding to the S-protein on the virus by achieving a low detection limit of 80 copies per mL. A 109-fold SERS enhancement is observed and a 106-fold virus enhancement is achieved by the biosensor.143
Fluorescence-based biosensors
Fluorescence-based biosensors are primarily based on the principle of fluorescence, which is emitted by fluorophores, fluorescent nanoparticles, QDs, etc. The fluorophores are excited with a laser of shorter wavelength that leads to emission of light energy at a longer wavelength that is detected by a detector. Such fluorescent particles can be conjugated with biomolecules such as antibodies, aptamers, and nucleic acids and can be used as reporter molecules for detection of analytes. Fig. 6 presents a diagram explaining the principle of a fluorescence-based biosensor.144 The Stokes shift of a fluorophore plays an important role while choosing a fluorescent molecule for its application in in vitro diagnostics. The significance will be discussed in detail in later sections.145,146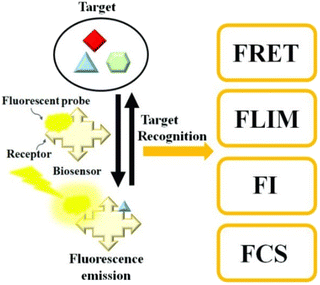 | ||
| Fig. 6 Schematic diagram of a fluorescence-based biosensor. The target analyte can be determined by Förster resonance energy transfer (FRET), fluorescence lifetime imaging (FLIM), changes in fluorescence intensity (FI), or fluorescence correlation spectroscopy (FCS) (reproduced from ref. 144 with permission from MDPI, copyright 2020). | ||
Teengam et al., 2021 described a paper-based biosensor for the detection of HCV. The detection is based on capturing the viral nucleic acid with a highly specific pyrrolidinyl peptide nucleic acid (acpcPNA) probe that is functionalized on partially oxidized cellulose paper. In the presence of the viral nucleic acid in the sample, it binds to the acpcPNA probe which, when treated with the ssDNA specific fluorescent dye, emits fluorescence which is detected on a smartphone-based detector. The sensor has an LOD of 5 pmol per spot and can be used as a POC device for HCV detection.41 Similarly, Iwanaga, 2021 designed a biosensor based on the principle of nucleic acid hybridization for detection of SARS-CoV-2. The biosensor consists of Cys-streptavidin as binding molecules functionalized on a silicon-on-insulator nanorod substrate. To simulate SARS-CoV-2 RNA, the authors used oligo-ssDNA which, when bound to the binding molecule, emits fluorescence. The authors reported a LOD of 100 fM.147 Mok et al., 2021 developed a fluorescence-based aptasensor for dengue NS1 detection. During their study with the dengue virus-derived NS1-binding aptamer (DBA), they found that the structure of the DBA becomes destroyed upon NS1 binding. This property was exploited to develop the sensor for the virus detection. For the sensor, the DBA was conjugated with 6-carboxyfluorescein (FAM) at the 5′-end in which, upon binding with NS1, the fluorescence becomes quenched and is then quantitatively measured while achieving an LOD of 2.51 nM and 8.13 nM for buffer and spiked serum conditions, respectively.148
Chemiluminescence-based biosensors
Chemiluminescence-based biosensors are based on two methods – direct and indirect methods. In the direct method, two molecules undergo a chemical reaction to produce a molecule of an electronically excited state which leads to the production of a chemiluminescence signal. On the other hand, in the indirect method, energy transfer takes place between excited molecules and a fluorophore which leads to the generation of a chemiluminescence signal.149 Such sensors have been widely used such as for food quality monitoring,150 studying antigen–antibody interactions,151 detection of hormones or biomolecules,152–154 monitoring of drug delivery and tumour growth155 and so on. Here, we have discussed studies which have included the application of chemiluminescence-based biosensors for detection of infectious diseases.In a study by Acharya et al., 2016, they developed a biosensor for the detection of Zika virus from human biological fluids by using anti-ZIKV monoclonal antibodies functionalized with polystyrene beads with electrogenerated chemiluminescent (ECL) labels. The study achieved an LOD of 1 PFU in a cost-effective way.43 Fan et al., 2022 developed a biosensor for detection of the RNA-dependent RNA-polymerase gene of SARS-CoV-2 based on the entropy-driven and DNA walker amplification strategy. In this study, they described that the binding of the gene initiates the first cycle leading to bandage formation which, when interacting with two single strands S1 and S2, causes the formation of a DNA bipedal walker. The walker then removes the hairpin structures from the top of the DNA tetrahedrons thereby allowing it to combine with the surface of Au–g-C3N4 with the help of PEI-Ru@Ti3C2@AuNPs-S7 probes. This binding and interaction lead to a change in the signal which is measured due to electrochemiluminescence resonance energy transfer (ECL-RET) attaining a LOD of 7.8 aM with high sensitivity and specificity.156 Similarly, Gutiérrez-Gálvez et al., 2022 designed and developed an electro-chemiluminescent biosensor for detection of SARS-CoV-2. The detection is based on the hybridization of the viral DNA with AuSPE/AuNMs/ProbeORF-SH electrodes which is detected by detecting the change in the electrochemiluminescence signal that is generated due to a reaction between carbon dots and [Ru(bpy)3]2+ while attaining a detection limit of 514 aM.157
Lateral flow assays (LFAs)
LFAs are primarily based on the principle of antigen–antibody interaction on a paper-strip. LFAs are widely used as POC test devices in low resource settings due to their ability to provide highly sensitive and specific detection results, low cost, rapid results and portability. LFAs have two types – lateral flow immunoassays (LFIAs) which allow detection of antigens or antibodies and nucleic acid lateral flow assays which detect nucleic acids.158 Typically, lateral flow assays have been used for qualitative and quantitative detection of biomolecules such as antigens, antibodies, nucleic acids, surface biomarkers, proteins, bacteria, or viruses. Because these tests are simple to run and do not need any chemical requirements or laboratory setup, they may be used in virtually every field of diagnosis even for cancer detection. Two types of LFIAs are majorly studied, which are as follows:159Direct assay: the target analyte binds to the monoclonal antibodies present on the test line indicating a positive test. This technique involves sandwich assays in which the analyte is captured between two antibodies – a capture antibody and a detection antibody. The detection antibody is coupled to a label that provides a visual signal when it binds to the target analyte and is immobilized on the test line coated with the capture antibody.
Competitive assay: in this type of LFIA, the tagged detection antibody competes with the target analyte in the sample for binding to the immobilized capture antibody in competitive assays. The quantity of the labelled detection antibody that binds to the capture antibody is inversely proportional to the concentration of the target analyte in the specimen. Hence, no test line indicates a positive test and vice versa.
Working principle of LFIAs: when the sample is added onto the sample well, it flows from the sample pad to the conjugate pad and then towards the nitrocellulose (NC) membrane. At the conjugate pad, the analyte interacts with the labelled antibody which, when flowing through the NC membrane, binds to the monoclonal antibody present on the test line. The remaining unbound labelled-antibodies specific for the antibody present on the control line bind to the antibodies present on the control line. When a test line is observed, it indicates a positive/negative test depending on the assay type and a control line when observed indicates proper liquid flow through the strip and test validity.158,160,161 A diagrammatic representation of this is shown in Fig. 7.
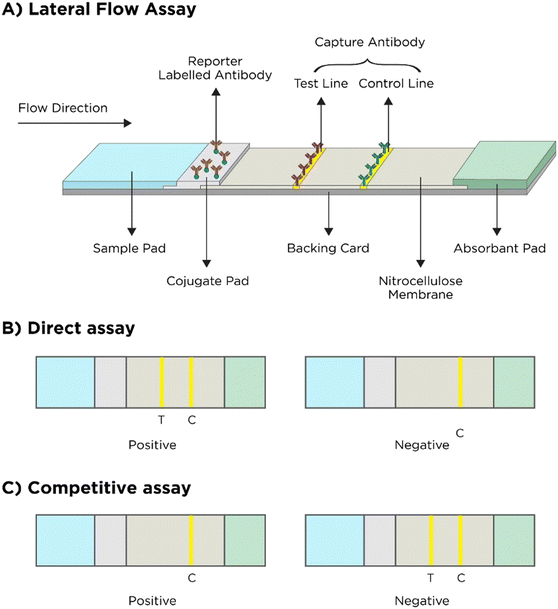 | ||
| Fig. 7 Working principle of LFA. A) General principle, B) result interpretation of the direct LFA and C) result interpretation of the competitive LFA (adapted from ref. 161 with permission from Ivyspring International Publisher, copyright 2021). | ||
Significance of detection probes and labels
The nucleic acid-based lateral flow test (NALFT), also known as a nucleic acid lateral flow immunoassay (NALFIA), is a diagnostic test used to identify and analyse certain nucleic acid sequences, such as DNA or RNA, in a sample with the help of nucleic acids or primers as detection molecules. Wang et al., 2020 in their study described a LFIA for detection of SARS-CoV-2 RNA based on DNA–RNA hybridization between a DNA probe (against ORF1ab, E-protein and N-protein of the virus) and viral RNA that is first detected by fluorescence labelled-antibodies and then is captured by S9.6 antibodies on the test line.42 Yrad et al., 2019 described a similar LFA based on nucleic acid sandwich-type hybridization to detect dengue virus. The LFA utilises an AuNP-labelled DNA reporter probe as a detection molecule to bind the dengue-1 RNA which is captured by a dengue-1 specific DNA probe as a capture molecule. The authors reported a LOD of 1.2 × 104 pfu mL−1 with a 20 minute rapid LFA test.164
In an aptamer-based lateral flow assay, aptamers are used as detection elements which comprise single-stranded DNA or RNA and peptide molecules that have high affinity and specificity for binding to certain target molecules, such as DNA, RNA, or protein.
Le et al., 2017 developed an aptamer-based LFA named DRELFA that employs both aptamers and antibodies to overcome the issue related to the cross-reactivity in the case of antibodies and the slow binding kinetics of aptamers. The authors described that in the presence of a sample, biotinylated aptamers and gold-oval conjugated antibodies (GNPs) both bind to the virus allowing its capture by the streptavidin molecules that are coated on the test line indicating a positive test due to the colour of the GNPs. The remaining unbound GNP-antibodies will bind to the control line. This method was used to specifically detect multiple strains of H3N2 influenza such as H3N2/Panama, H3N2/Udorn and H3N2/Aichi while attaining a detection limit of 2 × 106 virus particles.165 Tasbasi et al., 2019 developed a LFA for detection of Listeria monocytogenes using aptamer-gated molecules and TMB loaded mesoporous silica nanoparticles. In this LFA, in the presence of the bacteria, the aptamers bind to the bacteria and the TMB is released which then interacts with the horse-radish peroxidase (HRP) and produces a blue colour. The authors reported an LOD of 53 cells using the strategy and described that similar strategies can be used to detect any other microorganisms.166
Gold nanoparticles (AuNPs)
Colloidal AuNPs that are used in LFAs are screened by the size, quality, and power of the colour of the particle. AuNPs of size 15–20 nm are generally employed as labels for different qualitative LFAs. To the naked eye, AuNPs appear red in colour and thus are used for conjugation with the detection antibody, which helps in visual observation of test and control lines. Other than this, different morphologies of AuNPs have also been employed in SERS-based LFAs as they exhibit plasmonic absorption peaks in the visible area, which provide a red shift thereby allowing quantitative determination of the analyte concentration.167 In a study by Prakash et al., 2021, the authors used colloidal gold nanoparticles to construct a LFA for the detection of Brucella spp., reporting a detection limit of 107 CFU mL−1.47 A highly sensitive multiplex LFA was developed by Sánchez-Purrà et al., 2017 which employs gold nanostars as SERS-tags for SERS-based detection and distinguishing Zika NS1 and dengue NS1 antigens and reports an LOD of 0.7 ng mL−1 and 7.67 ng mL−1 for Zika virus and dengue virus, respectively.168 Another multiplex LFA was developed and was targeted towards EVs. It used AuNPs conjugated with an anti-CD63 antibody as a detector probe and anti-CD81 and anti-CD9 antibodies as capture antibodies. The device exhibited a LOD of 3.4 × 106 EVs per μL for CD81 and CD9 and required 15 minutes for completion of the test.169Fluorescent nanoparticles
Due to the low stability and sensitivity of AuNPs, fluorescent nanoparticles are widely used to provide a signal that can be detected and measured using a fluorescence reader. A comparative analysis of the gold-based LFA and the fluorescence-based LFA/fluorescence immunoassay is shown in Table 1. An appropriate fluorophore is selected based on the Stokes shift. The Stokes shift is the difference in the maxima of the absorption and emission bands. A larger Stokes shift means a faster excitation of the fluorescent molecule leading to a faster emission of fluorescence. It also diminishes self-absorption preventing self-quenching, thereby leading to higher photostability of the fluorescent molecule.48,170 Liang et al., 2017 developed a competitive fluorescent LFA for the quantitative detection of anti-HBc (hepatitis B core antigen) using europium microparticles as reporter molecules. In the assay, in the presence of the desired analyte, a less intense test-line is observed and in the absence of the analyte, the T-line intensity is increased. The authors reported a detection range of 0.63–640 IU mL−1 and a LOD of 0.31 IU mL−1.171 Similarly, a fluorescent europium chelate was employed by Ryu et al., 2018 for detection of HBsAg and HCV antigens and antibodies against HBsAg and HCV. The fluorescence signal generated due to the antigen–antibody interaction was measured using an automated fluorescence immunoassay system while achieving a sensitivity and specificity of 99.8% and 99.3% for the HBV antigen test, 100.0% and 100.0% for the anti-HBs test, and 98.8% and 99.1% for the anti-HCV test, respectively.172| Title/parameter | Fluorescence-based LFA | Gold-nanoparticle-based LFA | Ref. |
|---|---|---|---|
| Detector molecule | Fluorescent microspheres | Gold nanoparticles | 162, 173 |
| Sensitivity | Highly sensitive | Less sensitive comparatively | 174 |
| Limit of detection | Low LOD | Comparatively higher LOD | 159 |
| Qualitative/quantitative | Qualitative and huge range for quantitative results | Qualitative but very low quantitative dynamic range | 175 |
| Multiplexity | High multiplexicity capability | Low multiplexicity | 175 |
| Surface chemistries | Variety of different surface chemistries are available | No such availability unless different functional groups are conjugated as per need | 175 |
| Stability | Comparatively stable at changing pH and temperatures | Sensitive to changes in pH, temperature, and organics | 175 |
| Linear range | A wide linear range | Not a very wide linear range comparatively | 176 |
Fluorescent dyes
Due to their great sensitivity and compatibility with various detection systems, fluorescent dyes such as fluorescein isothiocyanate (FITC), rhodamine, cyanine dyes (e.g., Cy3 and Cy5), and Alexa Fluor dyes are widely utilized in LFAs. Upon excitation by their respective excitation wavelength, a higher wavelength is emitted, which is recorded concurrently by the fluorescence reader. Bamrungsap et al., 2014 doped Cy5 in silica nanoparticles, providing improved stability and sensitivity for influenza detection.177Quantum dots (QDs)
QDs are semiconductor nanocrystals with specific fluorescence properties, making them compatible for use in a wide range of diagnostic applications, including LFAs. In terms of sensitivity, quantum dots are commonly used in LFAs due to their strong emission and low absorption spectra. They address the shortcomings of fluorescent tags because of their strong emission peaks, photostability, brightness, low backgrounds and long-term stability. QDs can also be employed in place of fluorophores when encapsulated in nanocarriers. QDs increase chemical and photostability, signal intensity, hydrophilicity, and dispersion, make conjugation easier, and have fewer environmental consequences.50,178–180 Because various QDs have varied quantum yields for improved brightness, QDs have increased sensitivity. Photobleaching occurs under prolonged fluorescence excitation but QDs do not denature or deteriorate under such conditions, hence signal detection is simple and dependable over long periods. Rong et al., 2019 used quantum dots conjugated to the Zika NS1 antibody which in the presence of an antigen forms an antigen–antibody complex. The complex flows through the NC membrane and binds to the monoclonal antibodies present on the test line indicating a positive test and reporting a LOD of 0.045 and 0.15 ng mL−1 in buffer and serum, respectively.46Li et al., 2021 in a similar study developed a LFA to detect SARS-CoV-2 antibodies. In the assay, polystyrene-based fluorescent QDs were used and were conjugated to spike RBD protein. Upon binding of the antibody to the QD conjugated protein, a complex is formed and binds to the test line containing RBD protein, thereby indicating a positive test.181 In another study by Deng et al., 2018, a LFA was constructed based on the principle of strand displacement amplification that employs two DNA hairpin structures H1 and CdTe (quantum dot) labelled H2 for the detection of HIV DNA. In the presence of the viral DNA, H1 interacts with the viral DNA, followed by its despiralization and interaction with CdTe–H2 allowing it to then bind to the t-DNA on the test line present on the NC membrane. With this technique, the authors were able to achieve a detection range of 1 pM to 10 nM and a lowest detection limit of 0.76 pM.182
Carbon nanoparticles (CNPs)
CNPs, also known as carbon nanodots or carbon QDs, are nanoscale carbon-based compounds with exceptional optical characteristics that can replace standard fluorophores conventionally used in LFAs. Carbon nanotubes,183 carbon nanoparticles49 and carbon nanostrings184,185 are the most used and favoured materials for conjugation with biomolecules such as antibodies or nucleic acids. They showed several advantages over other fluorophores, such as carbon nanoparticles can be easily functionalized or changed with diverse compounds, have a large surface area and strong optical and electrical properties, enable customisation dependent on assay needs, are nontoxic, exhibit low cytotoxicity, controllable fluorescence, high photostability, and minimal background noise, and lastly can be used for cost-effective diagnostic assays.186 Wiriyachaiporn et al., 2017 utilised carbon nanostrings conjugated with antibodies to detect influenza A virus such as H1N1 and H3N2 strains based on a sandwich immunoassay. In the assay, in the presence of the virus, an antibody–antigen–carbon nanostring antibody complex is formed that allows the observation of the test to be visualized, thereby indicating a positive test and achieving a LOD of 350 TCID50 mL−1.187 In another study by Xu et al., 2019, the detection of severe fever with thrombocytopenia syndrome virus (SFTSV) as low as 10 pg mL−1 was reported when fluorescent carbon dots/SiO2 nanospheres (CSNs) were employed as reporter molecules conjugated with specific antibodies.188Magnetic nanoparticles (MNPs)
MNPs are nanoparticles (10–400 nm) with both colour and superparamagnetic properties, capable of producing dark lines and magnetic signals. In terms of magnetic detection, coil magnetometers and magnetic sensors that gather signals from test lines are the most often used detectors. In a fundamental LFA concept, MNPs coupled with antibodies captured at the test line produce magnetic signals that are recorded with a magnetometer, helping in the improvement of quantitative analysis. Multiplex detection allows a combination of targets to be separated by magnetic separation and caught by dispensed detecting areas, which works similarly to chromatography.50,189,190 Because antibody-based LFAs are challenging to screen, Bu et al., 2020 developed an antibiotic affinity-based LFA using ampicillin (Amp) coated with MNPs. Because bacteria have a high affinity for antibiotics, the Amp-MNP combination detects Salmonella enteritidis (S. enteritidis) by exceptional binding, and consequently, magnetic separation can be done for sample complexes.191 Similarly, Liu et al., 2023 in their study described a SERS-LFA in which they employed dual-layer DTNB-modified Fe3O4@Au MNPs conjugated with anti-H1N1/SARS-CoV-2/RSV antibodies to aid in simultaneous detection of influenza A virus (H1N1), SARS-CoV2 and respiratory syncytial virus (RSV) with a reported LOD of 85 copies per mL, 8 pg mL−1, and 8 pg mL−1 for H1N1, SARS-CoV-2 and RSV, respectively.192CRISPR-based LFA
Combining the techniques of CRISPR and LFAs, researchers have created a platform which could utilise the POC ability of LFAs along with sensitivity and specificity provided by CRISPR. Due to its high efficiency, CRISPR-based LFA diagnostics was thoroughly investigated during and after the COVID-19 pandemic. A CRISPR–Cas12-based DETECTR LFA for detection of SARS-CoV-2 from respiratory swab RNA extracts was developed by Broughton et al., 2020. Their assay employed isothermal amplification of viral RNA followed by a lateral flow readout of the amplified sample. They reported that this system could achieve a LOD of 10 copies per μL and results could be read in <40 minutes.193 Another group of researchers developed a CRISPR/Cas9-mediated LFA for simultaneous detection of the envelope (E) and open reading frame 1ab (Orf1ab) genes of SARS-CoV-2. They combined multiplex RT-RPA with a LFA and reported that their assay had a turnaround time of less than 1 hour. The LOD achieved for the E and Orf1ab genes was 10 copies per reaction (25 μL).194 Similarly, Ali et al., 2022 developed a biotin-coupled specific CRISPR-based assay for nucleic acid detection (Bio-SCAN) for detection of SARS-CoV-2. It could give colorimetric results on lateral flow strips. The reported sample to result time was less than 1 hour and the LOD was 4 copies per μL.195 A diagnostic tool named streamlined highlighting of infections to navigate epidemics (SHINE) was developed by Arizti-Sanz et al., 2020 and was further modified to develop SHINE version 2 (SHINEv2).196,197 This RNA-extraction-free technology enabled the direct use of samples. SHINEv2 could detect SARS-CoV-2 in nasopharyngeal samples in less than 90 minutes, using lateral-flow technology and incubation in a heat block at 37 °C. Also, it was demonstrated that body heat could be used to run the test without hampering the performance. SHINEv2 showed a LOD of 200 copies per μL.197 Apart from SARS-CoV-2, pathogens such as HIV, HPV, Pseudomonas aeruginosa and Staphylococcus aureus have also been detected using CRISPR-based LFAs.198–201The techniques discussed above have provided scientific evidence for their application in routine use. Table 2 gives a summary of the techniques discussed above, their working principle and LODs. Even though they have enormous potential, these techniques have their limitations as well. The molecular diagnostic platforms provide highly sensitive and specific detection but require skilled professionals and a well-equipped lab. The hurdles presented by molecular diagnostic techniques have mostly been resolved by the use of biosensor-based diagnostic platforms and LFA devices as these require no or very minimal sample preparation and can be performed in a POC setting. Nonetheless, they have their drawbacks like the requirement for specific analyzers, due to less stringent performing conditions they are more prone to errors and are mostly considered as presumptive tests, results of the visual qualitative test may differ from person to person and since these tests do not require skilled individuals, there may be variations in results due to human error. The advantages and limitations of each of the discussed diagnostic platforms are described in Table 3.
| Technique | Principle | Target organism/molecule | LOD | Time | Ref. |
|---|---|---|---|---|---|
| NA – not available. | |||||
| ELISA | |||||
| Invader assisted ELISA coupled with oligonucleotide probes and AuNPs | Detection of antigens followed by oligonucleotide addition and quantification using the signal produced by amplification of oligonucleotide | HBV | 24 pg mL−1 | 210–420 min | 55 |
| Au/Ag nanohybrid based ELISA | Detection of antigens followed by quantification using colorimetric measurement | Norovirus | 10.8 pg mL−1 | 135 min | 57 |
| Fluorescence AuNPs-LISA | Detection of antigens and quantification using fluorescence measurement | Opisthorchis viverrini | 36.97 ng mL−1 | 280 min | 58 |
| Fluorescence ELISA | Detection of antigens and quantification using fluorescence measurement | HBV | 5 × 10−4 IU mL−1 | 90 min | 60 |
| Ultrasensitive ELISA with thio-NAD cycling | Detection of antigens and quantification using thio-NADH signal measurement | SARS-CoV-2 | 2.62 × 10−19 moles per assay | 160 min | 63 |
| Ultrasensitive ELISA with thio-NAD cycling | Detection of antigens and quantification using thio-NADH signal measurement | SARS-CoV-2 | 2.95 × 10−17 moles per assay | 180 min | 64 |
| Ultrasensitive ELISA with thio-NAD cycling | Detection of antigens and quantification using thio-NADH signal measurement | Mycobacterium tuberculosis | 0.15 pg mL−1 | 300 min | 65 |
| Ultrasensitive ELISA with thio-NAD cycling | Detection of antigens and quantification using thio-NADH signal measurement | Mycobacterium tuberculosis | 0.10 pg mL−1 | 300 min | 66 |
| PCR | |||||
| Droplet magnetofluidic based PCR | Detection, amplification and quantification of nucleic acids using functionalized magnetic particles | HCV | 4500 IU mL−1 | 60 min | 81 |
| LIGHT | Au film based photonic thermal cycler for rapid detection, amplification and quantification of nucleic acids | Escherichia coli | 103 CFU mL−1 | 13 min | 83 |
| Isothermal amplification | |||||
| RT-LAMP | Detection, amplification and quantification of nucleic acids | WNV | 0.1 PFU | 60 min | 92 |
| LAMP | Detection, amplification and quantification of nucleic acids | NDV | 0.5 pg plasmid per reaction | 120 min | 93 |
| LAMP | Detection, amplification and quantification of nucleic acids | Leishmania donovani | 100 fg | 60 min | 94 |
| NASBA | Detection and amplification of nucleic acids and quantification using ECL signal count | Dengue virus | 1 PFU mL−1 | 120 min | 95 |
| NASBA | Detection and amplification of nucleic acids and quantification using ECL signal count | LACV | 0.0175 PFU | 240 min | 96 |
| RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | Mycobacterium tuberculosis (IS6110) | 6.25 fg DNA per reaction | 20 min | 97 |
| RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | Mycobacterium tuberculosis (IS1081) | 20 fg DNA per reaction | 20 min | 97 |
| RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | HIV-1 | 3 copies per reaction | 20 min | 98 |
| RPA based lateral flow assay | Detection and amplification of nucleic acids and visual interpretation | Plasmodium falciparum | 100 fg DNA per μL | 15 min | 99 |
| Real time RT-LAMP | Detection, amplification and quantification of nucleic acids | SARS-CoV-2 | 1 RNA copy per reaction | 30 min | 100 |
| Real time NASBA | Detection, amplification and quantification of nucleic acids | SARS-CoV-2 | 200 RNA copies per mL | 90 min | 104 |
| Reverse transcription RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | SARS-CoV-2 (RNA-dependent RNA polymerase) | 2 RNA molecules per reaction | 15 min | 105 |
| Reverse transcription RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | SARS-CoV-2 (envelope protein) | 15 RNA molecules per reaction | 15 min | 105 |
| Reverse transcription RPA | Detection, amplification of nucleic acids and quantification using fluorescence measurement | SARS-CoV-2 (nucleocapsid protein) | 15 RNA molecules per reaction | 15 min | 105 |
| CRISPR | |||||
| DETECTR | Cas12a ssDNase activation with isothermal amplification of the target sequence | HPV 16 | NA | 1 h | 112 |
| DETECTR | Cas12a ssDNase activation with isothermal amplification of the target sequence | HPV 18 | NA | 1 h | 112 |
| ctPCR | Amplification of target DNA using universal primers followed by Cas9 cutting (C), A tailing (A) and T adaptor ligation (T) followed by amplification of CAT treated DNA using general-specific primers | HPV 16 | 400 copies per μL | 3 to 4 h | 113 |
| ctPCR | Amplification of target DNA using universal primers followed by Cas9 cutting (C), A tailing (A) and T adaptor ligation (T) followed by amplification of CAT treated DNA using general-specific primers | HPV 18 | 400 copies per μL | 3 to 4 h | 113 |
| CARP | Cleavage of target DNA using the sgRNA–Cas9 complex followed by ligation and amplification by PCR | HPV 16 | 0.002 ng of genomic DNA (cervical carcinoma cells) | 3 h | 114 |
| 0.02 ng DNA (clinical samples) | |||||
| CARP | Cleavage of target DNA using the sgRNA–Cas9 complex followed by ligation and amplification by PCR | HPV 18 | 0.002 ng of genomic DNA (cervical carcinoma cells) | 3 h | 114 |
| 0.02 ng DNA (clinical samples) | |||||
| SHERLOCK | Cas13 guided target recognition followed by isothermal amplification | Zika virus | 2 aM | NA | 115 |
| SHERLOCK | Cas13 guided target recognition followed by isothermal amplification | Dengue virus | 2 aM | NA | 115 |
| SHERLOCKv2 | Cas 12, Csm6 and Cas13 guided target recognition and signal enhancement followed by isothermal amplification | Zika virus | 8 zM | <90 min | 116 |
| SHERLOCKv2 | Cas 12, Csm6 and Cas13 guided target recognition and signal enhancement followed by isothermal amplification | Dengue virus | 8 zM | <90 min | 116 |
| HUDSON combined SHERLOCK | Lysis of viral particles and inactivation of ribonucleases found in bodily fluids using heat and chemical reduction followed by RPA | Ebola virus | 10 copies per μL for Ebola virus | 1 h | 118 |
| HUDSON combined SHERLOCK | Lysis of viral particles and inactivation of ribonucleases found in bodily fluids using heat and chemical reduction followed by RPA | Lassa virus | 10 copies per μL for Lassa virus clade II | 3 h | 118 |
| 100 copies per μL for Lassa virus | |||||
| HUDSON combined SHERLOCK | Lysis of viral particles and inactivation of ribonucleases found in bodily fluids using heat and chemical reduction followed by RPA | Zika virus | 1 copy per μL | <2 h | 117 |
| HUDSON combined SHERLOCK | Lysis of viral particles and inactivation of ribonucleases found in bodily fluids using heat and chemical reduction followed by RPA | Dengue virus | 1 copy per μL | <1 h | 117 |
| NASBA-CRISPR cleavage | Cas9 mediated cleavage followed by NASBA | Zika virus | 2.8 fM | NA | 119 |
| Automated microfluidic device | Cas13a mediated target detection followed by fluorometric measurement of nonspecific cleavage products of Cas13a | Ebola virus | 20 PFU mL−1 | 5 min | 120 |
| CARMEN | Microfluidic chip with CRISPR-based nucleic acid detection reagents in the wells to identify amplified samples, allowing for accurate detection of the crRNA–target complex. | 169 human associated viruses, HIV drug-resistance mutations, influenza A virus subtyping, dengue virus, Zika virus, HCV | NA | 1–3 hours | 121 |
| PCR-CRISPR method | PCR in combination with CRISPR–Cas13a, in which crRNAs recognize DNA targets | HBV | 1 copy per test | 15 min after the PCR amplification step | 122 |
| Type III CRISPR-based method | Without RNA extraction or preamplification, the type III-A CRISPR complex is able to detect RNA directly with the help of Can2 nuclease | SARS-CoV-2 | 15 fM | <20 min | 123 |
| Electrochemical biosensors | |||||
| Carbon electrode-based biosensor | Detection of change in electrical conductivity | SARS-CoV-2 | 90 fM | 30 s | 45 |
| Organic electrochemical transistor | Detection of change in current | Human influenza A virus | 0.025 hemagglutination units | 10 min | 129 |
| Graphene-based field-effect transistors | Detection of change in resistance of the electrode surface | HIV (p24 protein) | 100 fg mL−1 | NA | 130 |
| SPR-based biosensors | |||||
| SPR biosensor | LSPR signal detection | HIV | 48 fM | 60 min | 134 |
| AuNP sandwich-immunoassay LSPR chip | LSPR signal detection | HBV | 100 fg mL−1 | 15 min | 135 |
| LSPR biosensor | LSPR signal detection | Mycobacterium tuberculosis | NA | NA | 44 |
| Plasmonic NP-Qdot-molecular beacon biosensor | LSPR-mediated fluorescence | Zika virus | 1.7 RNA copies per mL | 3 min | 136 |
| SPR biosensor | SPR shift | Dengue virus | 0.08 pM | 8 min | 137 |
| SERS-based biosensors | |||||
| SERS-based colloidal aptasensors | SERS spectra and signal intensity | Influenza A viruses | 2 × 105 viral particles per mL | 15 min | 40 |
| SERS-based aptasensors | SERS signal intensity | SARS-CoV-2 | 5.5 × 104 TCID50 mL−1 | 7 min | 142 |
| SERS-based biosensor | SERS spectra and signal intensity | SARS-CoV-2 | 80 copies per mL | 5 min | 143 |
| Fluorescence-based biosensors | |||||
| Fluorescent paper-based DNA biosensor | Fluorescence detection | HCV | 5 pmol | 20 min | 41 |
| Metasurface fluorescent biosensor | Fluorescence detection | SARS-CoV-2 | 100 fM | 30 min | 147 |
| G-quadruplex-based fluorescent aptasensor | Fluorescence quenching detection | Dengue virus | 2.51 nM (buffer) 8.13 nM (serum) | 30 min | 148 |
| Chemiluminescence-based biosensors | |||||
| ECL-based immunoassay | ECL signal detection | Zika virus | 0.01 PFU μL−1 | NA | 43 |
| Dual wavelength ratiometric (ECL) biosensor | Electrochemiluminescence resonance energy transfer (ECL-RET) detection | SARS-CoV-2 | 7.8 aM | NA | 156 |
| ECL nanostructured DNA biosensor | ECL signal detection | SARS-CoV-2 | 514 aM | NA | 157 |
| AuNP-based LFAs | |||||
| AuNP-based LFA | Detection of antigens based on color development | Brucella spp. | 107 CFU mL−1 | 20 min | 47 |
| SERS-based LFA | Detection of antigens by colorimetric and SERS signal measurement | Zika virus | 0.7 ng mL−1 | NA | 168 |
| SERS-based LFA | Detection of antigens by colorimetric and SERS signal measurement | Dengue virus | 7.67 ng mL−1 | NA | 168 |
| AuNP-based LFA | Detection of multiple targets based on specificity of capture antibodies and resultant color development | CD81 | 3.4 × 106 EVs per μL | 15 min | 169 |
| AuNP-based LFA | Detection of multiple targets based on specificity of capture antibodies and resultant color development | CD9 | 3.4 × 106 EVs per μL | 15 min | 169 |
| Fluorescent NP-based LFAs | |||||
| Europium(III) microparticle-based LFA | Detection of antigens coupled with measurement of the fluorescence signal | HBV | 0.31 IU mL−1 | 15 min | 171 |
| Europium chelate based LFA | Detection of antigens coupled with measurement of the fluorescence signal | HBV | 0.05 IU mL−1 | 20 min | 172 |
| Europium chelate based LFA | Detection of antibodies coupled with measurement of the fluorescence signal | HBV | 5.78 IU mL−1 | 20 min | 172 |
| Europium chelate based LFA | Detection of antibodies coupled with measurement of fluorescence | HCV | NA | 20 min | 172 |
| Fluorescent dye-based LFAs | |||||
| Cy5 doped silica NP-based LFA | Detection of antigens coupled with measurement of the fluorescence signal | Influenza A virus | 250 ng mL−1 | 30 min | 177 |
| QD-based LFAs | |||||
| QD-based fluorescence LFA | Detection of antigens coupled with measurement of the fluorescence signal | Zika virus | 0.045 ng mL−1 (buffer) | 20 min | 46 |
| 0.15 ng mL−1 (serum) | |||||
| QD-based fluorescent nanobead LFA | Detection of antibodies coupled with measurement of the fluorescence signal | SARS-CoV-2 | NA | 15 min | 181 |
| QD-based fluorescent nanobead LFA | Detection of antigens coupled with measurement of the fluorescence signal | Avian influenza virus H5N1 | 25 PFU mL−1 | 15 min | 181 |
| QD-based LFA | Detection of DNA based on strand displacement amplification and quantification using measurement of the fluorescence signal | HIV | 0.76 pM | 15 min | 182 |
| Carbon nanoparticle-based LFAs | |||||
| Carbon nanotag based LFA | Detection of antigens by visual observation | Influenza A virus | 350 TCID50 mL−1 | 15 min | 187 |
| CSN-based LFA | Detection of antigens coupled with measurement of fluorescence | Severe fever with thrombocytopenia syndrome virus | 10 pg mL−1 | 15 min | 188 |
| MNP-based LFAs | |||||
| Amp-MNP-based LFA | Detection and enrichment of bacteria using Amp-MNPs | Salmonella enteritidis | 102 CFU mL−1 | 40 min | 191 |
| Multichannel magnetic SERS-based LFA | Detection of antigens and enrichment using MNPs followed by measurement of SERS spectra | SARS-CoV-2 | 8 pg mL−1 | 30 min | 192 |
| Multichannel magnetic SERS-based LFA | Detection of antigens and enrichment using MNPs followed by measurement of SERS spectra | RSV | 8 pg mL−1 | 30 min | 192 |
| Multichannel magnetic SERS-based LFA | Detection of antigens and enrichment using MNPs followed by measurement of SERS spectra | Influenza A virus | 85 copies per mL | 30 min | 192 |
| CRISPR-based LFAs | |||||
| CRISPR–Cas12 based DETECTR LFA | Cas12a ssDNase activation with isothermal amplification of the target sequence and visualization of the Cas12 detection reaction using a FAM-biotin reporter molecule and lateral flow strips designed to capture labeled nucleic acids | SARS-CoV-2 | 10 copies per μL | <40 min | 193 |
| CRISPR/Cas9-mediated LFA | RT-RPA followed by recognition of biotinylated amplicons by Cas9/sgRNA, binding of AuNP–DNA probes to sgRNA and accumulation on the test line | SARS-CoV-2 (envelope gene) | 10 copies per reaction | <60 min | 194 |
| CRISPR/Cas9-mediated LFA | RT-RPA followed by recognition of biotinylated amplicons by Cas9/sgRNA, binding of AuNP–DNA probes to sgRNA and accumulation on the test line | SARS-CoV-2 (Orf1ab gene) | 10 copies per reaction | <60 min | 194 |
| Bio-SCAN | Isothermal amplification with FAM-labeled primers followed by recognition by bio-dCas9, which immobilizes the AuNP–anti-FAM antibody complex at the test band for visual detection | SARS-CoV-2 | 4 copies per μL | <60 min | 195 |
| SHINEv2 | RPA-based amplification and Cas13-based detection followed by visualization using a biotinylated FAM reporter | SARS-CoV-2 | 200 copies per μL | <90 min | 197 |
| CRISPR–Cas12a mediated SERS LFA | CRISPR mediated signal amplification combined with SERS signal detection | HIV-1 | 0.3 fM | <60 min | 198 |
| CRISPR/Cas-isothermal amplification based lateral flow biosensor | Cas12a-mediated trans-cleavage of the reporter ssDNA upon target recognition and result visualization using a DNA probe-based lateral flow biosensor | HPV 16 | 3.1 attomoles | 50 min | 199 |
| CRISPR/Cas-isothermal amplification based lateral flow biosensor | Cas12a-mediated trans-cleavage of the reporter ssDNA upon target recognition and result visualization using a DNA probe-based lateral flow biosensor | HPV 18 | 3.1 attomoles | 50 min | 199 |
| CRISPR/Cas-isothermal amplification based lateral flow biosensor | Cas12a-mediated trans-cleavage of the reporter ssDNA upon target recognition and result visualization using a DNA probe-based lateral flow biosensor | Pseudomonas aeruginosa | 1 aM | 50 min | 200 |
| CRISPR/Cas-recombinase-assisted amplification based lateral flow biosensor | Recombinase assisted amplification of target DNA followed by trans-cleavage activity of Cas12a/crRNA and visualization using fluorescence strip reader or naked eye | Staphylococcus aureus | 75 aM | 70 min | 201 |
| Technique | Advantages | Limitations |
|---|---|---|
| Molecular diagnostics | Highly sensitive and specific; can detect multiple pathogens as required based on the use of multiple primers (PCR) and antibodies (ELISA) at a time; can be paired with multiple detection platforms, for example: isothermal amplification based LFA | Laborious procedure; requires skilled professionals; expensive instrumentation required; highly expensive test; non-portable; generally requires large sample sizes |
| Biosensors | No-to-minimal sample preparation required; rapid test; allow testing with various sample types; are able to detect small changes and hence are highly sensitive and specific; can be quantitative and qualitative; portable; compact in size; require less sample; easy to use | Specific readers or analyzers are required; stable for less time; high cost of development; require regular calibration; changing environmental factors such as temperature and humidity might affect test performance |
| Lateral flow assay | Low-cost assay; requires less sample volume; various sample types can be used; generally one-step assay; can be qualitative, semi-quantitative and quantitative depending on detection labels used; no-to-minimal sample preparation is required; long shelf life of test devices; portable and can be used at point-of-care; easy to use; sensitivity and specificity in concordance with molecular diagnostic techniques such as RT-PCR and ELISA | May require specific readers or analyzers depending upon the detection label used; presumptive tests and requires confirmatory tests; change in sample volume other than mentioned leads to result variation; is prone to interfering molecules present in various samples; result interpretation of colorimetric LFA can vary from person-to-person |
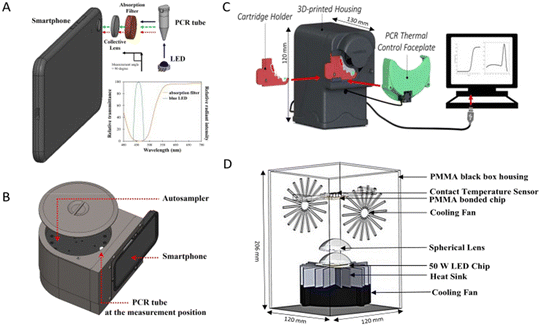
Advancements in the field of biological science have led to massive improvements in diagnostic platforms. Along with these improvements, the need for rapid and POC diagnosis have led researchers to innovate technologies to solve these problems. In this regard, attempts have been made to miniaturise the detection platforms and make them portable. A portable fluorometer which could read fluorescence with the help of a smartphone (Fig. 8A and B) was developed which had similar efficacy as compared to the conventional diagnostic instrument.59 Similarly, a portable PCR instrument (Fig. 8C) was fabricated by Shin et al., 2018.81 An enhanced technique of photonic PCR which was low-cost, simple, robust and without the need for a heating block was developed by Son et al., 2015.82 Another group of researchers developed a portable photonic PCR system with its main characteristics of rapid and POC application (Fig. 8D).84 Making progress in the technique of photonic PCR, a platform named LIGHT was developed which could yield results by utilising the sample directly without any need for pretreatment.83 A portable LAMP device which could be used with the help of a smartphone app along with a portable fluorescence reader was developed by Puri et al., 2021 with POC application as its main target.94 An optimized RT-LAMP based testing method was developed by Wei et al., 2021. The method did not need specialized or proprietary equipment or reagents and did not require any pretreatment of the sample,103 thus enhancing its capability to be used with multiple compatible instruments. And with the advent of portable isothermal platforms, such techniques display huge potential for POC application, especially in regions where resources are limited. Myhrvold et al., 2018 developed a sample preparation protocol named HUDSON117 to be paired with SHERLOCK which was a nucleic acid detection technology based on the distinct properties of CRISPR enzymes.115 SHERLOCK was capable of multiplexed quantitative and highly sensitive detection, combined with lateral flow for visual readout. By combining SHERLOCK and HUDSON, a detection system that could identify viruses directly from body fluids without the need for instruments was developed.117 The biosensor and LFA-based diagnostic platforms have an edge over molecular diagnostic techniques as they are highly portable and have been developed with the aim of POC application. These innovations in technology present promising prospects to overcome the current challenges and provide solutions for rapid and POC diagnostics without compromising the sensitivity and specificity of the process.
 | ||
| Fig. 8 Portable diagnostic platforms. A) Components of a smartphone-based fluorometer (reproduced from ref. 59 with permission from Elsevier, copyright 2021); B) a smartphone-based fluorometer along with an autosampler for simultaneously measuring multiple samples (reproduced from ref. 59 with permission from Elsevier, copyright 2021); C) a portable PCR instrument along with a USB readout unit (reproduced from ref. 81 with permission from Springer Nature, copyright 2018); D) schematic diagram for the design of a photonic PCR system (reproduced from ref. 84 with permission from Elsevier, copyright 2023). | ||
Conclusion
The massive threat posed by infectious diseases is not unknown. Traditional detection platforms have served as reliable tools for in vitro diagnosis. They have been a major aid in improving the scenario of the global health community. But with the increasing population, the need for early and accurate diagnosis has increased manifold. To fulfill current needs, the conventional platforms are insufficient, which thus has led to the development of newer technologies.The field of infectious disease diagnostics has undergone a revolution in recent years, with the development of new diagnostic techniques along with improvements in traditional detection methods that offer significant advantages over existing techniques. Nanoparticles, magnetic particles, specific nucleic acid probes, enzyme cycling and photonic thermocycling platforms have been merged with the conventional systems to improve their detection limits and reduce the turnaround time.55,57,58,63,81,83 While being independent of thermocycling, exhibiting equal or better accuracy and reducing the time required for results, isothermal amplification-based nucleic acid detection platforms have been developed.92–94,97–100,105,112,115–117,120 Even though the traditional techniques have evolved, they are still not able to reach the most vulnerable populations due to their stringent requirement of working conditions and need for advanced infrastructure. To resolve these problems, advancements in the field of POC diagnostics have been emerging recently in the form of technologies such as biosensors, lateral flow assays and point-of-care molecular techniques.40–43,81–84,171,172,193–197 These techniques have been proven to have comparable sensitivity, specificity, and accuracy. Moreover, the flexibility of these platforms enables them to be modified and used for detection of a wider range of disease-causing pathogens. As a result, they have the potential to transform the way infectious diseases are diagnosed and managed. Platforms which can detect multiple targets using a single test can reduce the cost of testing and increase screening at the grassroot level.
The development of technologies which have reduced the need for sample pretreatment or require no sample pretreatment and are available on portable systems has displayed true POC potential.81,83,103,197 The integration of smartphones with diagnostic systems for the purpose of performing analysis and obtaining results has increased the user-friendliness of these platforms and opened a new channel for communication and sharing of data. Currently, most of these platforms have been tested in research settings. Thus, the main hurdles that are faced by them are their capability of production at a large scale, actual POC usability, stability, storage and most importantly cost-effectiveness. Also, much research needs to be done to reduce the errors that might occur during tests and proper training and education need to be imparted among healthcare providers so that they can use these tests and interpret results effectively. Prospective future developments can focus on developing self-test kits and miniaturizing these platforms such that non-experts could use the test at the ease of their homes. Further research needs to be done to utilize more such techniques which use advanced detection systems and to reduce the paucity in information that currently exists regarding them.
The current innovations have numerously increased the potential for reach of diagnostic platforms. Therefore, a future is not unimaginable where diagnostic systems are compact, utilize samples without pretreatment, give accurate results and are operable using smartphones. These techniques have the potential to save lives and improve the quality of life for millions of people. As the world becomes increasingly interconnected, the need for effective diagnostic tools is more important than ever.
Author contributions
Shrikant P. Pawar and Shraddha Shaligram conceptualized the idea and designed the outline of the manuscript. Sahil Syed, Alisha Rahaman, Abhijit Mondal and Shraddha Shaligram were involved in writing the preliminary draft of the manuscript and designing all the tables and figures. Sahil Syed edited and finalized the manuscript for submission. All authors made substantial contributions to the acquisition, analysis and interpretation of data. All authors have carefully read and approved the final version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Mylab Discovery Solutions, Baner, India for kindly providing us with the facility.Notes and references
- P. T. J. Johnson, J. C. De Roode and A. Fenton, Why infectious disease research needs community ecology, Science, 2015, 349(6252), 1259504 CrossRef PubMed.
- H. W. Boucher, G. H. Talbot, J. S. Bradley, J. E. Edwards, D. Gilbert and L. B. Rice, et al., Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America, Clin. Infect. Dis., 2009, 48(1), 1–12 CrossRef PubMed.
- L. C. McDonald, D. N. Gerding, S. Johnson, J. S. Bakken, K. C. Carroll and S. E. Coffin, et al., Clinical Practice Guidelines for Clostridium difficile Infection in Adults and Children: 2017 Update by the Infectious Diseases Society of America (IDSA) and Society for Healthcare Epidemiology of America (SHEA), Clin. Infect. Dis., 2018, 66(7), e1–e48 CrossRef CAS PubMed.
- A. Zumla, M. Rao, R. S. Wallis, S. H. E. Kaufmann, R. Rustomjee and P. Mwaba, et al., Host-directed therapies for infectious diseases: Current status, recent progress, and future prospects, Lancet Infect. Dis., 2016, 16(4), e47–e63 CrossRef PubMed.
- J. Libertucci and V. B. Young, The role of the microbiota in infectious diseases, Nat. Microbiol., 2018, 4(1), 35–45 CrossRef PubMed.
- GBD Mortality and Causes of Death Collaborators, Global, regional, and national age–sex specific all-cause and cause-specific mortality for 240 causes of death, 1990–2013: a systematic analysis for the Global Burden of Disease Study 2013, Lancet, 2015, 385(9963), 117–171 CrossRef PubMed.
- C. Abat, H. Chaudet, J. M. Rolain, P. Colson and D. Raoult, Traditional and syndromic surveillance of infectious diseases and pathogens, Int. J. Infect. Dis. Infect. Dis., 2016, 48, 22–28 CrossRef PubMed.
- World Health Organization, The World health report: 2004: changing history, World Health Organization, Geneva, 2004 Search PubMed.
- Y. C. Wu, C. S. Chen and Y. J. Chan, The outbreak of COVID-19: An overview, J. Chin. Med. Assoc., 2020, 83(3), 217–220 CrossRef CAS PubMed.
- A. Remuzzi and G. Remuzzi, COVID-19 and Italy: what next?, Lancet., 2020, 395(10231), 1225–1228 CrossRef CAS PubMed.
- C. Sohrabi, Z. Alsafi, N. O'Neill, M. Khan, A. Kerwan and A. Al-Jabir, et al., World Health Organization declares global emergency: A review of the 2019 novel coronavirus (COVID-19), Int. J. Surg., 2020, 76, 71–76 CrossRef PubMed.
- World Health Organization, WHO Coronavirus (COVID-19) Dashboard, 2023, [cited 2023 Jun 27], Available from: https://covid19.who.int/ Search PubMed.
- L. Jones, D. Palumbo and D. Brown, Coronavirus: How the pandemic has changed the world economy, BBC News, 2021, [cited 2023 Jun 27]; Available from: https://www.bbc.com/news/business-51706225 Search PubMed.
- D. M. Morens and A. S. Fauci, The 1918 Influenza Pandemic: Insights for the 21st Century, J. Infect. Dis., 2007, 195(7), 1018–1028 CrossRef PubMed.
- M. E. Nickol and J. Kindrachuk, A year of terror and a century of reflection: perspectives on the great influenza pandemic of 1918–1919, BMC Infect. Dis., 2019, 19, 117 CrossRef PubMed.
- T. G. Ksiazek, D. Erdman, C. S. Goldsmith, S. R. Zaki, T. Peret and S. Emery, et al., A Novel Coronavirus Associated with Severe Acute Respiratory Syndrome, N. Engl. J. Med., 2003, 348(20), 1953–1966 CrossRef CAS PubMed.
- P. A. Rota, M. S. Oberste, S. S. Monroe, W. A. Nix, R. Campagnoli and J. P. Icenogle, et al., Characterization of a Novel Coronavirus Associated with Severe Acute Respiratory Syndrome, Science, 2003, 300(5624), 1394–1399 CrossRef CAS PubMed.
- World Health Organization, Summary table of SARS cases by country, 1 November 2002-7 August 2003, Weekly Epidemiological Record = Relevé épidémiologique hebdomadaire, 2003, 78, 35, pp. 310–311, Available from: https://apps.who.int/iris/handle/10665/232252.
- A. Assiri, A. McGeer, T. M. Perl, C. S. Price, A. A. Al Rabeeah and D. A. T. Cummings, et al., Hospital Outbreak of Middle East Respiratory Syndrome Coronavirus, N. Engl. J. Med., 2013, 369(5), 407–416 CrossRef CAS PubMed.
- K. Mobaraki and J. Ahmadzadeh, Current epidemiological status of Middle East respiratory syndrome coronavirus in the world from 1.1.2017 to 17.1.2018: a cross-sectional study, BMC Infect. Dis., 2019, 19, 351 CrossRef PubMed.
- C. E. M. Coltart, B. Lindsey, I. Ghinai, A. M. Johnson and D. L. Heymann, The Ebola outbreak, 2013–2016: old lessons for new epidemics, Philos. Trans. R. Soc., B, 2017, 372(1721), 20160297 CrossRef PubMed.
- B. Kreuels, D. Wichmann, P. Emmerich, J. Schmidt-Chanasit, G. de Heer and S. Kluge, et al., A Case of Severe Ebola Virus Infection Complicated by Gram-Negative Septicemia, N. Engl. J. Med., 2014, 371(25), 2394–2401 CrossRef CAS PubMed.
- D. M. Morens, G. K. Folkers and A. S. Fauci, The challenge of emerging and re-emerging infectious diseases, Nature, 2004, 430(6996), 242–249 CrossRef CAS PubMed.
- Institute of Medicine, Board on Global Health, Committee on Emerging Microbial Threats to Health in the 21st Century, Microbial Threats to Health: Emergence, Detection, and Response, ed. M. S. Smolinski, M. A. Hamburg and J. Lederberg, The National Academies Press, Washington D.C., 2003 Search PubMed.
- S. Binder, A. M. Levitt, J. J. Sacks and J. M. Hughes, Emerging Infectious Diseases: Public Health Issues for the 21st Century, Science, 1999, 284(5418), 1311–1313 CrossRef CAS PubMed.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Point-of-care diagnostics for infectious diseases: From methods to devices, Nano Today, 2021, 37, 101092 CrossRef CAS PubMed.
- J. Weigl, Challenges in infectious disease control and the current pandemic by skewed distributions, Prävention und Gesundheitsförderung, 2020, 15(2), 97–101 CrossRef.
- J. A. I. Weigl, T. Werlang, M. Wessendorf and H. Helbing, Efficient measures to control an outbreak of SARS-CoV2 in an elderly care home and to prevent a spill over into the general population, Res Sq., 2021 DOI:10.21203/rs.3.rs-484166/v1.
- Y. Wang, Z. Wang, J. Wang, M. Li, S. Wang and X. He, et al., Evolution and control of the COVID-19 pandemic: A global perspective, Cities, 2022, 130, 103907 CrossRef PubMed.
- A. Wilder-Smith and D. O. Freedman, Isolation, quarantine, social distancing and community containment: pivotal role for old-style public health measures in the novel coronavirus (2019-nCoV) outbreak, J. Travel Med., 2020, 27(2), taaa020 CrossRef PubMed.
- J. M. van Seventer and N. S. Hochberg, Principles of Infectious Diseases: Transmission, Diagnosis, Prevention, and Control, in International Encyclopedia of Public Health, ed. S. R. Quah, Elsevier, 2017, pp. 22–39 Search PubMed.
- M. Miller and J. Dillon, Early diagnosis improves outcomes in hepatitis C, Practitioner, 2015, 259(1787), 25–27 Search PubMed.
- P. Rajapaksha, A. Elbourne, S. Gangadoo, R. Brown, D. Cozzolino and J. Chapman, A review of methods for the detection of pathogenic microorganisms, Analyst, 2019, 144(2), 396–411 RSC.
- E. Gupta, P. Bhalla, N. Khurana and T. Singh, Histopathology For The Diagnosis Of Infectious Diseases, Indian J Med Microbiol., 2009, 27(2), 100–106 CrossRef CAS PubMed.
- S. P. Naber, Molecular Pathology -- Diagnosis of Infectious Disease, N. Engl. J. Med., 1994, 331(18), 1212–1215 CrossRef CAS PubMed.
- U. Reischl, Application of molecular biology-based methods to the diagnosis of infectious diseases, Front. Biosci.-Landmark, 1996, 1(5), 72–77 CrossRef PubMed.
- T. S. Hauck, S. Giri, Y. Gao and W. C. W. Chan, Nanotechnology diagnostics for infectious diseases prevalent in developing countries, Adv. Drug Delivery Rev., 2010, 62(4–5), 438–448 CrossRef CAS PubMed.
- I. M. Mackay, Real-time PCR in the microbiology laboratory, Clin. Microbiol. Infect., 2004, 10(3), 190–212 CrossRef CAS PubMed.
- J. R. Strich and D. S. Chertow, CRISPR-Cas Biology and Its Application to Infectious Diseases, J. Clin. Microbiol., 2019, 57(4), 1307–1318 CrossRef PubMed.
- D. Gribanyov, G. Zhdanov, A. Olenin, G. Lisichkin, A. Gambaryan and V. Kukushkin, et al., SERS-Based Colloidal Aptasensors for Quantitative Determination of Influenza Virus, Int. J. Mol. Sci., 2021, 22(4), 1842 CrossRef CAS PubMed.
- P. Teengam, N. Nisab, N. Chuaypen, P. Tangkijvanich, T. Vilaivan and O. Chailapakul, Fluorescent paper-based DNA sensor using pyrrolidinyl peptide nucleic acids for hepatitis C virus detection, Biosens. Bioelectron., 2021, 189, 113381 CrossRef CAS PubMed.
- D. Wang, S. He, X. Wang, Y. Yan, J. Liu and S. Wu, et al., Rapid lateral flow immunoassay for the fluorescence detection of SARS-CoV-2 RNA, Nat. Biomed. Eng., 2020, 4(12), 1150–1158 CrossRef CAS PubMed.
- D. Acharya, P. Bastola, L. Le, A. M. Paul, E. Fernandez and M. S. Diamond, et al., An ultrasensitive electrogenerated chemiluminescence-based immunoassay for specific detection of Zika virus, Sci. Rep., 2016, 6(1), 32227 CrossRef CAS PubMed.
- W. Sun, S. Yuan, H. Huang, N. Liu and Y. Tan, A label-free biosensor based on localized surface plasmon resonance for diagnosis of tuberculosis, J. Microbiol. Methods, 2017, 142, 41–45 CrossRef CAS PubMed.
- S. Mahari, A. Roberts, D. Shahdeo and S. Gandhi, eCovSens-Ultrasensitive Novel In-House Built Printed Circuit Board Based Electrochemical Device for Rapid Detection of nCovid-19 antigen, a spike protein domain 1 of SARS-CoV-2, bioRxiv, 2020, preprint, DOI:10.1101/2020.04.24.059204.
- Z. Rong, Q. Wang, N. Sun, X. Jia, K. Wang and R. Xiao, et al., Smartphone-based fluorescent lateral flow immunoassay platform for highly sensitive point-of-care detection of Zika virus nonstructural protein 1, Anal. Chim. Acta, 2019, 1055, 140–147 CrossRef CAS PubMed.
- C. Prakash, B. Kumar, R. P. Singh, P. Singh, G. Shrinet and A. Das, et al., Development and evaluation of a gold nanoparticle based Lateral Flow assay (LFA) strip test for detection of Brucella spp, J. Microbiol. Methods, 2021, 184, 106185 CrossRef CAS PubMed.
- E. M. Santos, W. Sheng, R. Esmatpour Salmani, S. Tahmasebi Nick, A. Ghanbarpour and H. Gholami, et al., Design of Large Stokes Shift Fluorescent Proteins Based on Excited State Proton Transfer of an Engineered Photobase, J. Am. Chem. Soc., 2021, 143(37), 15091–15102 CrossRef CAS PubMed.
- J. Bogdanovic, M. Koets, I. Sander, I. Wouters, T. Meijster and D. Heederik, et al., Rapid detection of fungal α-amylase in the work environment with a lateral flow immunoassay, J. Allergy Clin. Immunol., 2006, 118(5), 1157–1163 CrossRef CAS PubMed.
- X. Chi, D. Huang, Z. Zhao, Z. Zhou, Z. Yin and J. Gao, Nanoprobes for in vitro diagnostics of cancer and infectious diseases, Biomaterials, 2012, 33(1), 189–206 CrossRef CAS PubMed.
- A. Rostami, P. Karanis and S. Fallahi, Advances in serological, imaging techniques and molecular diagnosis of Toxoplasma gondii infection, Infection, 2018, 46(3), 303–315 CrossRef CAS PubMed.
- S. Kurkela and D. W. G. Brown, Molecular diagnostic techniques, Medicine, 2009, 37(10), 535–540 CrossRef PubMed.
- S. Aydin, A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA, Peptides, 2015, 72, 4–15 CrossRef CAS PubMed.
- T. Bhardwaj and T. K. Sharma, Enzyme-Linked Immunosorbent Assay-Based Nanosensors for the Detection of Pathogenic Bacteria, in Nanosensors for Point-of-Care Diagnostics of Pathogenic Bacteria, ed. A. Acharya and N. K. Singhal, Springer Nature Singapore, Singapore, 2023, pp. 59–83 Search PubMed.
- Q. Song, X. Qi, H. Jia, L. He, S. Kumar and J. L. Pitman, et al., Invader Assisted Enzyme-Linked Immunosorbent Assay for Colorimetric Detection of Disease Biomarkers Using Oligonucleotide Probe-Modified Gold Nanoparticles, J. Biomed. Nanotechnol., 2016, 12(4), 831–839 CrossRef CAS PubMed.
- N. Tsurusawa, J. Chang, M. Namba, D. Makioka, S. Yamura and K. Iha, et al., Modified ELISA for Ultrasensitive Diagnosis, J. Clin. Med., 2021, 10(21), 5197 CrossRef CAS PubMed.
- I. M. Khoris, K. Takemura, J. Lee, T. Hara, F. Abe and T. Suzuki, et al., Enhanced colorimetric detection of norovirus using in-situ growth of Ag shell on Au NPs, Biosens. Bioelectron., 2019, 126, 425–432 CrossRef CAS PubMed.
- W. Taron, W. Jamnongkan, J. Phetcharaburanin, P. Klanrit, N. Namwat and A. Techasen, et al., A fluorescence AuNPs-LISA: A new approach for Opisthorchis viverrini (Ov) antigen detection with a simple fluorescent enhancement strategy by surfactant micelle in urine samples, Spectrochim. Acta, Part A, 2021, 254, 119633 CrossRef CAS PubMed.
- W. Taron, K. Phooplub, S. Sanchimplee, K. Piyanamvanich, W. Jamnongkan and A. Techasen, et al., Smartphone-based fluorescent ELISA with simple fluorescent enhancement strategy for Opisthorchis viverrini (Ov) antigen detection in urine samples, Sens. Actuators, B, 2021, 348, 130705 CrossRef CAS.
- Y. Wu, W. Guo, W. Peng, Q. Zhao, J. Piao and B. Zhang, et al., Enhanced Fluorescence ELISA Based on HAT Triggering Fluorescence “Turn-on” with Enzyme–Antibody Dual Labeled AuNP Probes for Ultrasensitive Detection of AFP and HBsAg, ACS Appl. Mater. Interfaces, 2017, 9(11), 9369–9377 CrossRef CAS PubMed.
- S. Watabe, H. Kodama, M. Kaneda, M. Morikawa, K. Nakaishi and T. Yoshimura, et al., Ultrasensitive enzyme-linked immunosorbent assay (ELISA) of proteins by combination with the thio-NAD cycling method, Biophysics, 2014, 10, 49–54 CrossRef PubMed.
- A. Iwai, T. Yoshimura, K. Wada, S. Watabe, Y. Sakamoto and E. Ito, et al., Spectrophotometric Method for the Assay of Steroid 5α-Reductase Activity of Rat Liver and Prostate Microsomes, Anal. Sci., 2013, 29(4), 455–459 CrossRef CAS PubMed.
- Y. Kyosei, M. Namba, D. Makioka, A. Kokubun, S. Watabe and T. Yoshimura, et al., Ultrasensitive Detection of SARS-CoV-2 Spike Proteins Using the Thio-NAD Cycling Reaction: A Preliminary Study before Clinical Trials, Microorganisms, 2021, 9(11), 2214 CrossRef CAS PubMed.
- Y. Kyosei, M. Namba, S. Yamura, S. Watabe, T. Yoshimura and T. Sasaki, et al., Improved Detection Sensitivity of an Antigen Test for SARS-CoV-2 Nucleocapsid Proteins with Thio-NAD Cycling, Biol. Pharm. Bull., 2021, 44(9), 1332–1336 CrossRef CAS PubMed.
- K. Sakashita, R. Takeuchi, K. Takeda, M. Takamori, K. Ito and Y. Igarashi, et al., Ultrasensitive enzyme-linked immunosorbent assay for the detection of MPT64 secretory antigen to evaluate Mycobacterium tuberculosis viability in sputum, Int. J. Infect. Dis., 2020, 96, 244–253 CrossRef CAS PubMed.
- W. H. Wang, R. Takeuchi, S. H. Jain, Y. H. Jiang, S. Watanuki and Y. Ohtaki, et al., A novel, rapid (within hours) culture-free diagnostic method for detecting live Mycobacterium tuberculosis with high sensitivity, EBioMedicine, 2020, 60, 103007 CrossRef PubMed.
- W. Zheng, S. M. LaCourse, B. Song, D. K. Singh, M. Khanna and J. Olivo, et al., Diagnosis of paediatric tuberculosis by optically detecting two virulence factors on extracellular vesicles in blood samples, Nat. Biomed. Eng., 2022, 6(8), 979–991 CrossRef CAS PubMed.
- J. S. Schorey and C. V. Harding, Extracellular vesicles and infectious diseases: new complexity to an old story, J. Clin. Invest., 2016, 126(4), 1181–1189 CrossRef PubMed.
- E. Hosseini-Beheshti and G. E. R. Grau, Extracellular vesicles as mediators of immunopathology in infectious diseases, Immunol. Cell Biol., 2018, 96(7), 694–703 CrossRef PubMed.
- H. Zhu, H. Zhang, Y. Xu, S. Laššáková, M. Korabečná and P. Neužil, PCR past, present and future, BioTechniques, 2020, 69(4), 317–325 CrossRef CAS PubMed.
- A. J. Jeffreys, V. Wilson, R. Neumann and J. Keyte, Amplification of human minisatellites by the polymerase chain reaction: towards DNA fingerprinting of single cells, Nucleic Acids Res., 1988, 16(23), 10953–10971 CrossRef CAS PubMed.
- P. Markoulatos, N. Siafakas and M. Moncany, Multiplex polymerase chain reaction: A practical approach, J. Clin. Lab. Anal., 2002, 16(1), 47–51 CrossRef CAS PubMed.
- R. Higuchi, C. Fockler, G. Dollinger and R. Watson, Kinetic PCR Analysis: Real-time Monitoring of DNA Amplification Reactions, Nat. Biotechnol., 1993, 11(9), 1026–1030 CrossRef CAS PubMed.
- J. Mackay and O. Landt, Real-Time PCR Fluorescent Chemistries, in Protocols for Nucleic Acid Analysis by Nonradioactive Probes, ed. J. Mackay and E. Hilario, Humana Press, New Jersey, 2007, pp. 237–262 Search PubMed.
- M. Becker-André and K. Hahlbrock, Absolute mRNA quantification using the polymerase chain reaction (PCR). A novel approach by a PCR aided transcipt titration assay (PATTY), Nucleic Acids Res., 1989, 17(22), 9437–9446 CrossRef PubMed.
- B. Vogelstein and K. W. Kinzler, Digital PCR, Proc. Natl. Acad. Sci. U. S. A., 1999, 96(16), 9236–9241 CrossRef CAS PubMed.
- A. M. Streets and Y. Huang, Microfluidics for biological measurements with single-molecule resolution, Curr. Opin. Biotechnol., 2014, 25, 69–77 CrossRef CAS PubMed.
- A. Rival, D. Jary, C. Delattre, Y. Fouillet, G. Castellan and A. Bellemin-Comte, et al., An EWOD-based microfluidic chip for single-cell isolation, mRNA purification and subsequent multiplex qPCR, Lab Chip, 2014, 14(19), 3739–3749 RSC.
- B. Hwang, J. H. Lee and D. Bang, Single-cell RNA sequencing technologies and bioinformatics pipelines, Exp. Mol. Med., 2018, 50(8), 1–14 CrossRef CAS PubMed.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf and B. J. Hindson, et al., Absolute quantification by droplet digital PCR versus analog real-time PCR, Nat. Methods, 2013, 10(10), 1003–1005 CrossRef CAS PubMed.
- D. J. Shin, A. Y. Trick, Y. H. Hsieh, D. L. Thomas and T. H. Wang, Sample-to-Answer Droplet Magnetofluidic Platform for Point-of-Care Hepatitis C Viral Load Quantitation, Sci. Rep., 2018, 8(1), 9793 CrossRef PubMed.
- J. H. Son, B. Cho, S. Hong, S. H. Lee, O. Hoxha and A. J. Haack, et al., Ultrafast photonic PCR, Light: Sci. Appl., 2015, 4(7), e280 CrossRef CAS.
- B. Cho, S. H. Lee, J. Song, S. Bhattacharjee, J. Feng and S. Hong, et al., Nanophotonic Cell Lysis and Polymerase Chain Reaction with Gravity-Driven Cell Enrichment for Rapid Detection of Pathogens, ACS Nano, 2019, 13(12), 13866–13874 CrossRef CAS PubMed.
- J. Nabuti, A. R. Fath Elbab, A. Abdel-Mawgood, M. Yoshihisa and H. M. H. Shalaby, Highly efficient photonic PCR system based on plasmonic heating of gold nanofilms, Biosens. Bioelectron.: X, 2023, 14, 100346 CAS.
- W. W. Bazié, J. Boucher, B. Goyer, D. Kania, I. T. Traoré and D. Y. Somé, et al., HIV Replication Increases the Mitochondrial DNA Content of Plasma Extracellular Vesicles, Int. J. Mol. Sci., 2023, 24(3), 1924 CrossRef PubMed.
- F. Huang, J. Bai, J. Zhang, D. Yang, H. Fan and L. Huang, et al., Identification of potential diagnostic biomarkers for pneumonia caused by adenovirus infection in children by screening serum exosomal microRNAs, Mol. Med. Rep., 2019, 19(5), 4306–4314 CAS.
- T. Notomi, H. Okayama, H. Masubuchi, T. Yonekawa, K. Watanabe and N. Amino, et al., Loop-mediated isothermal amplification of DNA, Nucleic Acids Res., 2000, 28(12), 63–63 CrossRef PubMed.
- I. M. Lobato and C. K. O'Sullivan, Recombinase polymerase amplification: Basics, applications and recent advances, TrAC, Trends Anal. Chem., 2018, 98, 19–35 CrossRef CAS PubMed.
- M. M. Ali, F. Li, Z. Zhang, K. Zhang, D. K. Kang and J. A. Ankrum, et al., Rolling circle amplification: a versatile tool for chemical biology, materials science and medicine, Chem. Soc. Rev., 2014, 43(10), 3324 RSC.
- M. Fakruddin, R. M. Mazumdar, A. Chowdhury and M. K. S. Bin, Nucleic acid sequence based amplification (NASBA)-prospects and applications. International Journal of Life Science and Pharm, Research, 2012, 2(1), 106–121 CAS.
- Y. Zhao, F. Chen, Q. Li, L. Wang and C. Fan, Isothermal Amplification of Nucleic Acids, Chem. Rev., 2015, 115(22), 12491–12545 CrossRef CAS PubMed.
- M. Parida, G. Posadas, S. Inoue, F. Hasebe and K. Morita, Real-Time Reverse Transcription Loop-Mediated Isothermal Amplification for Rapid Detection of West Nile Virus, J. Clin. Microbiol., 2004, 42(1), 257–263 CrossRef CAS PubMed.
- H. M. Pham, C. Nakajima, K. Ohashi and M. Onuma, Loop-Mediated Isothermal Amplification for Rapid Detection of Newcastle Disease Virus, J. Clin. Microbiol., 2005, 43(4), 1646–1650 CrossRef CAS PubMed.
- M. Puri, H. K. Brar, N. Mittal, E. Madan, R. Srinivasan and K. Rawat, et al., Rapid diagnosis of Leishmania infection with a portable loop-mediated isothermal amplification device, J. Biosci., 2021, 46(4), 92 CrossRef CAS PubMed.
- W. Usawattanakul, A. Jittmittraphap, T. P. Endy, A. Nisalak, P. Tapchaisri and S. Looareesuwan, Rapid Detection of Dengue Viral RNA by Nucleic Acid Sequence-Based Amplification (NASBA), Dengue Bull., 2002, 26, 125–130 Search PubMed , Available from: https://apps.who.int/iris/bitstream/handle/10665/163750/dbv26p125.pdf.
- A. J. Lambert, R. S. Nasci, B. C. Cropp, D. A. Martin, B. C. Rose and B. J. Russell, et al., Nucleic Acid Amplification Assays for Detection of La Crosse Virus RNA, J. Clin. Microbiol., 2005, 43(4), 1885–1889 CrossRef CAS PubMed.
- D. S. Boyle, R. McNerney, H. Teng Low, B. T. Leader, A. C. Pérez-Osorio and J. C. Meyer, et al., Rapid Detection of Mycobacterium tuberculosis by Recombinase Polymerase Amplification, PLoS One, 2014, 9(8), e103091 CrossRef PubMed.
- D. S. Boyle, D. A. Lehman, L. Lillis, D. Peterson, M. Singhal and N. Armes, et al., Rapid Detection of HIV-1 Proviral DNA for Early Infant Diagnosis Using Recombinase Polymerase Amplification, MBio, 2013, 4(2), 10–128 CrossRef PubMed.
- S. Kersting, V. Rausch, F. F. Bier and M. von Nickisch-Rosenegk, Rapid detection of Plasmodium falciparum with isothermal recombinase polymerase amplification and lateral flow analysis, Malar. J., 2014, 13(1), 99 CrossRef PubMed.
- Y. L. Lau, I. Ismail, N. I. Mustapa, M. Y. Lai, T. S. Tuan Soh and A. Hassan, et al., Real-time reverse transcription loop-mediated isothermal amplification for rapid detection of SARS-CoV-2, PeerJ, 2020, 8, e9278 CrossRef PubMed.
- V. L. Fowler, B. Armson, J. L. Gonzales, E. L. Wise, E. L. A. Howson and Z. Vincent-Mistiaen, et al., A highly effective reverse-transcription loop-mediated isothermal amplification (RT-LAMP) assay for the rapid detection of SARS-CoV-2 infection, J. Infect., 2021, 82(1), 117–125 CrossRef CAS PubMed.
- L. Mautner, C. K. Baillie, H. M. Herold, W. Volkwein, P. Guertler and U. Eberle, et al., Rapid point-of-care detection of SARS-CoV-2 using reverse transcription loop-mediated isothermal amplification (RT-LAMP), Virol. J., 2020, 17, 160 CrossRef CAS PubMed.
- S. Wei, E. Kohl, A. Djandji, S. Morgan, S. Whittier and M. Mansukhani, et al., Direct diagnostic testing of SARS-CoV-2 without the need for prior RNA extraction, Sci. Rep., 2021, 11(1), 2402 CrossRef CAS PubMed.
- V. Kia, A. Tafti, M. Paryan and S. Mohammadi-Yeganeh, Evaluation of real-time NASBA assay for the detection of SARS-CoV-2 compared with real-time PCR, Ir. J. Med. Sci., 2023, 192(2), 723–729 CrossRef CAS PubMed.
- A. A. El Wahed, P. Patel, M. Maier, C. Pietsch, D. Rüster and S. Böhlken-Fascher, et al., Suitcase Lab for Rapid Detection of SARS-CoV-2 Based on Recombinase Polymerase Amplification Assay, Anal. Chem., 2021, 93(4), 2627–2634 CrossRef CAS PubMed.
- R. Barrangou, C. Fremaux, H. Deveau, M. Richards, P. Boyaval and S. Moineau, et al., CRISPR Provides Acquired Resistance Against Viruses in Prokaryotes, Science, 2007, 315(5819), 1709–1712 CrossRef CAS PubMed.
- R. Barrangou, The roles of CRISPR–Cas systems in adaptive immunity and beyond, Curr. Opin. Immunol., 2015, 32, 36–41 CrossRef CAS PubMed.
- J. E. Garneau, M. È. Dupuis, M. Villion, D. A. Romero, R. Barrangou and P. Boyaval, et al., The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA, Nature, 2010, 468(7320), 67–71 CrossRef CAS PubMed.
- E. V. Koonin, K. S. Makarova and F. Zhang, Diversity, classification and evolution of CRISPR-Cas systems, Curr. Opin. Microbiol., 2017, 37, 67–78 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf and E. V. Koonin, The basic building blocks and evolution of CRISPR–Cas systems, Biochem. Soc. Trans., 2013, 41(6), 1392–1400 CrossRef CAS PubMed.
- K. S. Makarova, Y. I. Wolf, O. S. Alkhnbashi, F. Costa, S. A. Shah and S. J. Saunders, et al., An updated evolutionary classification of CRISPR–Cas systems, Nat. Rev. Microbiol., 2015, 13(11), 722–736 CrossRef CAS PubMed.
- J. S. Chen, E. Ma, L. B. Harrington, M. Da Costa, X. Tian and J. M. Palefsky, et al., CRISPR-Cas12a target binding unleashes indiscriminate single-stranded DNase activity, Science, 2018, 360(6387), 436–439 CrossRef CAS PubMed.
- Q. Wang, B. Zhang, X. Xu, F. Long and J. Wang, CRISPR-typing PCR (ctPCR), a new Cas9-based DNA detection method, Sci. Rep., 2018, 8(1), 14126 CrossRef PubMed.
- B. Zhang, Q. Wang, X. Xu, Q. Xia, F. Long and W. Li, et al., Detection of target DNA with a novel Cas9/sgRNAs-associated reverse PCR (CARP) technique, Anal. Bioanal. Chem., 2018, 410(12), 2889–2900 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, J. W. Lee, P. Essletzbichler, A. J. Dy and J. Joung, et al., Nucleic acid detection with CRISPR-Cas13a/C2c2, Science, 2017, 356(6336), 438–442 CrossRef CAS PubMed.
- J. S. Gootenberg, O. O. Abudayyeh, M. J. Kellner, J. Joung, J. J. Collins and F. Zhang, Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6, Science, 1979, 360(6387), 439–444 CrossRef PubMed.
- C. Myhrvold, C. A. Freije, J. S. Gootenberg, O. O. Abudayyeh, H. C. Metsky and A. F. Durbin, et al., Field-deployable viral diagnostics using CRISPR-Cas13, Science, 2018, 360(6387), 444–448 CrossRef CAS PubMed.
- K. G. Barnes, A. E. Lachenauer, A. Nitido, S. Siddiqui, R. Gross and B. Beitzel, et al., Deployable CRISPR-Cas13a diagnostic tools to detect and report Ebola and Lassa virus cases in real-time, Nat. Commun., 2020, 11(1), 4131 CrossRef CAS PubMed.
- K. Pardee, A. A. Green, M. K. Takahashi, D. Braff, G. Lambert and J. W. Lee, et al., Rapid, Low-Cost Detection of Zika Virus Using Programmable Biomolecular Components, Cell, 2016, 165(5), 1255–1266 CrossRef CAS PubMed.
- P. Qin, M. Park, K. J. Alfson, M. Tamhankar, R. Carrion and J. L. Patterson, et al., Rapid and Fully Microfluidic Ebola Virus Detection with CRISPR-Cas13a, ACS Sens., 2019, 4(4), 1048–1054 CrossRef CAS PubMed.
- C. M. Ackerman, C. Myhrvold, S. G. Thakku, C. A. Freije, H. C. Metsky and D. K. Yang, et al., Massively multiplexed nucleic acid detection with Cas13, Nature, 2020, 582(7811), 277–282 CrossRef CAS PubMed.
- S. Wang, H. Li, Z. Kou, F. Ren, Y. Jin and L. Yang, et al., Highly sensitive and specific detection of hepatitis B virus DNA and drug resistance mutations utilizing the PCR-based CRISPR-Cas13a system, Clin. Microbiol. Infect., 2021, 27(3), 443–450 CrossRef CAS PubMed.
- A. Nemudraia, A. Nemudryi, M. Buyukyoruk, A. M. Scherffius, T. Zahl and T. Wiegand, et al., Sequence-specific capture and concentration of viral RNA by type III CRISPR system enhances diagnostic, Nat. Commun., 2022, 13(1), 7762 CrossRef CAS PubMed.
- B. Ning, Z. Huang, B. M. Youngquist, J. W. Scott, A. Niu and C. M. Bojanowski, et al., Liposome-mediated detection of SARS-CoV-2 RNA-positive extracellular vesicles in plasma, Nat. Nanotechnol., 2021, 16(9), 1039–1044 CrossRef CAS PubMed.
- J. Vidic, M. Manzano, C. M. Chang and N. Jaffrezic-Renault, Advanced biosensors for detection of pathogens related to livestock and poultry, Vet. Res., 2017, 48(1), 11 CrossRef PubMed.
- D. Grieshaber, R. MacKenzie, J. Vörös and E. Reimhult, Electrochemical Biosensors - Sensor Principles and Architectures, Sensors, 2008, 8(3), 1400–1458 CrossRef CAS PubMed.
- I. H. Cho, D. H. Kim and S. Park, Electrochemical biosensors: Perspective on functional nanomaterials for on-site analysis, Biomater. Res., 2020, 24(1), 6 CrossRef CAS PubMed.
- S. Imran, S. Ahmadi and K. Kerman, Electrochemical Biosensors for the Detection of SARS-CoV-2 and Other Viruses, Micromachines, 2021, 12(2), 174 CrossRef PubMed.
- W. Hai, T. Goda, H. Takeuchi, S. Yamaoka, Y. Horiguchi and A. Matsumoto, et al., Human influenza virus detection using sialyllactose-functionalized organic electrochemical transistors, Sens. Actuators, B, 2018, 260, 635–641 CrossRef CAS.
- S. Islam, S. Shukla, V. K. Bajpai, Y. K. Han, Y. S. Huh and A. Kumar, et al., A smart nanosensor for the detection of human immunodeficiency virus and associated cardiovascular and arthritis diseases using functionalized graphene-based transistors, Biosens. Bioelectron., 2019, 126, 792–799 CrossRef CAS PubMed.
- H. Nguyen, J. Park, S. Kang and M. Kim, Surface Plasmon Resonance: A Versatile Technique for Biosensor Applications, Sensors, 2015, 15(5), 10481–10510 CrossRef CAS PubMed.
- V. Shpacovitch and R. Hergenröder, Surface Plasmon Resonance (SPR)-Based Biosensors as Instruments with High Versatility and Sensitivity, Sensors, 2020, 20(11), 3010 CrossRef PubMed.
- H. Šípová and J. Homola, Surface plasmon resonance sensing of nucleic acids: A review, Anal. Chim. Acta, 2013, 773, 9–23 CrossRef PubMed.
- W. Diao, M. Tang, S. Ding, X. Li, W. Cheng and F. Mo, et al., Highly sensitive surface plasmon resonance biosensor for the detection of HIV-related DNA based on dynamic and structural DNA nanodevices, Biosens. Bioelectron., 2018, 100, 228–234 CrossRef CAS PubMed.
- J. Kim, S. Y. Oh, S. Shukla, S. B. Hong, N. S. Heo and V. Bajpai, et al., Heteroassembled gold nanoparticles with sandwich-immunoassay LSPR chip format for rapid and sensitive detection of hepatitis B virus surface antigen (HBsAg), Biosens. Bioelectron., 2018, 107, 118–122 CrossRef CAS PubMed.
- O. Adegoke, M. Morita, T. Kato, M. Ito, T. Suzuki and E. Y. Park, Localized surface plasmon resonance-mediated fluorescence signals in plasmonic nanoparticle-quantum dot hybrids for ultrasensitive Zika virus RNA detection via hairpin hybridization assays, Biosens. Bioelectron., 2017, 94, 513–522 CrossRef CAS PubMed.
- N. A. S. Omar, Y. W. Fen, J. Abdullah, A. R. Sadrolhosseini, Y. Mustapha Kamil and N. 'I. M. Fauzi, et al., Quantitative and Selective Surface Plasmon Resonance Response Based on a Reduced Graphene Oxide–Polyamidoamine Nanocomposite for Detection of Dengue Virus E-Proteins, Nanomaterials, 2020, 10(3), 569 CrossRef CAS PubMed.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguié and J. J. Baumberg, et al., Present and Future of Surface-Enhanced Raman Scattering, ACS Nano, 2020, 14(1), 28–117 CrossRef CAS PubMed.
- X. Liang, N. Li, R. Zhang, P. Yin, C. Zhang and N. Yang, et al., Carbon-based SERS biosensor: from substrate design to sensing and bioapplication, NPG Asia Mater., 2021, 13(1), 8 CrossRef CAS.
- L. Hamm, A. Gee and A. S. De Silva Indrasekara, Recent Advancement in the Surface-Enhanced Raman Spectroscopy-Based Biosensors for Infectious Disease Diagnosis, Appl. Sci., 2019, 9(7), 1448 CrossRef CAS.
- L. Ouyang, Y. Hu, L. Zhu, G. J. Cheng and J. Irudayaraj, A reusable laser wrapped graphene-Ag array based SERS sensor for trace detection of genomic DNA methylation, Biosens. Bioelectron., 2017, 92, 755–762 CrossRef CAS PubMed.
- E. Zavyalova, O. Ambartsumyan, G. Zhdanov, D. Gribanyov, V. Gushchin and A. Tkachuk, et al., SERS-Based Aptasensor for Rapid Quantitative Detection of SARS-CoV-2, Nanomaterials, 2021, 11(6), 1394 CrossRef CAS PubMed.
- Y. Yang, Y. Peng, C. Lin, L. Long, J. Hu and J. He, et al., Human ACE2-Functionalized Gold “Virus-Trap” Nanostructures for Accurate Capture of SARS-CoV-2 and Single-Virus SERS Detection, Nanomicro Lett., 2021, 13(1), 109 CAS.
- Y. T. Chen, Y. C. Lee, Y. H. Lai, J. C. Lim, N. T. Huang and C. T. Lin, et al., Review of Integrated Optical Biosensors for Point-of-Care Applications, Biosensors, 2020, 10(12), 209 CrossRef CAS PubMed.
- S. Kakkar, P. Gupta, N. Kumar and K. Kant, Progress in Fluorescence Biosensing and Food Safety towards Point-of-Detection (PoD) System, Biosensors, 2023, 13(2), 249 CrossRef CAS PubMed.
- K. Girigoswami and N. Akhtar, Nanobiosensors and fluorescence based biosensors: An overview, Int. J. Nano Dimens., 2019, 10, 1–17 CAS.
- M. Iwanaga, High-Sensitivity High-Throughput Detection of Nucleic Acid Targets on Metasurface Fluorescence Biosensors, Biosensors, 2021, 11(2), 33 CrossRef CAS PubMed.
- J. Mok, J. Jeon, J. Jo, E. Kim and C. Ban, Novel one-shot fluorescent aptasensor for dengue fever diagnosis using NS1-induced structural change of G-quadruplex aptamer, Sens. Actuators, B, 2021, 343, 130077 CrossRef CAS.
- A. M. García-Campaña and W. R. G. Baeyens, Principles and recent analytical applications of chemiluminescence, Analusis, 2000, 28(8), 686–698 CrossRef.
- K. Deshpande and L. Kanungo, Chemiluminescence and fluorescence biosensors for food application: A review, Sens. Actuators Rep., 2022, 5, 100137 CrossRef.
- A. P. Osipov, N. V. Zaitseva and A. M. Egorov, Chemiluminescent immunoenzyme biosensor with a thin-layer flow-through cell. Application for study of a real-time bimolecular antigen-antibody interaction, Biosens. Bioelectron., 1996, 11(9), 881–887 CrossRef CAS PubMed.
- A. B. Dippel, W. A. Anderson, R. S. Evans, S. Deutsch and M. C. Hammond, Chemiluminescent Biosensors for Detection of Second Messenger Cyclic di-GMP, ACS Chem. Biol., 2018, 13(7), 1872–1879 CrossRef CAS PubMed.
- Q. Zhu, Y. Chen, W. Wang, H. Zhang, C. Ren and H. Chen, et al., A sensitive biosensor for dopamine determination based on the unique catalytic chemiluminescence of metal–organic framework HKUST-1, Sens. Actuators, B, 2015, 210, 500–507 CrossRef CAS.
- L. Gutiérrez-Gálvez, T. García-Mendiola, C. Gutiérrez-Sánchez, T. Guerrero-Esteban, C. García-Diego and I. Buendía, et al., Carbon nanodot–based electrogenerated chemiluminescence biosensor for miRNA-21 detection, Microchim. Acta, 2021, 188(11), 398 CrossRef PubMed.
- G. Niu, L. Zhu, D. N. Ho, F. Zhang, H. Gao and Q. Quan, et al., Longitudinal Bioluminescence Imaging of the Dynamics of Doxorubicin Induced Apoptosis, Theranostics, 2013, 3(3), 190–200 CrossRef CAS PubMed.
- Z. Fan, B. Yao, Y. Ding, D. Xu, J. Zhao and K. Zhang, Rational engineering the DNA tetrahedrons of dual wavelength ratiometric electrochemiluminescence biosensor for high efficient detection of SARS-CoV-2 RdRp gene by using entropy-driven and bipedal DNA walker amplification strategy, Chem. Eng. J., 2022, 427, 131686 CrossRef CAS PubMed.
- L. Gutiérrez-Gálvez, R. del Caño, I. Menéndez-Luque, D. García-Nieto, M. Rodríguez-Peña and M. Luna, et al., Electrochemiluminescent nanostructured DNA biosensor for SARS-CoV-2 detection, Talanta, 2022, 240, 123203 CrossRef PubMed.
- K. M. Koczula and A. Gallotta, Lateral flow assays, Essays Biochem., 2016, 60(1), 111–120 CrossRef PubMed.
- E. B. Bahadır and M. K. Sezgintürk, Lateral flow assays: Principles, designs and labels, TrAC, Trends Anal. Chem., 2016, 82, 286–306 CrossRef.
- M. Sajid, A. N. Kawde and M. Daud, Designs, formats and applications of lateral flow assay: A literature review, J. Saudi Chem. Soc., 2015, 19(6), 689–705 CrossRef.
- T. Wang, L. Chen, A. Chikkanna, S. Chen, I. Brusius and N. Sbuh, et al., Development of nucleic acid aptamer-based lateral flow assays: A robust platform for cost-effective point-of-care diagnosis, Theranostics, 2021, 11(11), 5174–5196 CrossRef PubMed.
- Z. Li, Y. Yi, X. Luo, N. Xiong, Y. Liu and S. Li, et al., Development and clinical application of a rapid IgM-IgG combined antibody test for SARS-CoV-2 infection diagnosis, J. Med. Virol., 2020, 92(9), 1518–1524 CrossRef CAS PubMed.
- T. Xiang, Z. Jiang, J. Zheng, C. Lo, H. Tsou and G. Ren, et al., A novel double antibody sandwich-lateral flow immunoassay for the rapid and simple detection of hepatitis C virus, Int. J. Mol. Med., 2012, 30(5), 1041–1047 CrossRef CAS PubMed.
- F. M. Yrad, J. M. Castañares and E. C. Alocilja, Visual Detection of Dengue-1 RNA Using Gold Nanoparticle-Based Lateral Flow Biosensor, Diagnostics, 2019, 9(3), 74 CrossRef CAS PubMed.
- T. T. Le, P. Chang, D. J. Benton, J. W. McCauley, M. Iqbal and A. E. G. Cass, Dual Recognition Element Lateral Flow Assay Toward Multiplex Strain Specific Influenza Virus Detection, Anal. Chem., 2017, 89(12), 6781–6786 CrossRef CAS PubMed.
- B. B. Tasbasi, B. C. Guner, M. Sudagidan, S. Ucak, M. Kavruk and V. C. Ozalp, Label-free lateral flow assay for Listeria monocytogenes by aptamer-gated release of signal molecules, Anal. Biochem., 2019, 587, 113449 CrossRef CAS PubMed.
- S. Sloan-Dennison, E. O'Connor, J. W. Dear, D. Graham and K. Faulds, Towards quantitative point of care detection using SERS lateral flow immunoassays, Anal. Bioanal. Chem., 2022, 414(16), 4541–4549 CrossRef CAS PubMed.
- M. Sánchez-Purrà, M. Carré-Camps, H. de Puig, I. Bosch, L. Gehrke and K. Hamad-Schifferli, Surface-Enhanced Raman Spectroscopy-Based Sandwich Immunoassays for Multiplexed Detection of Zika and Dengue Viral Biomarkers, ACS Infect. Dis., 2017, 3(10), 767–776 CrossRef PubMed.
- M. Oliveira-Rodríguez, E. Serrano-Pertierra, A. C. García, S. López-Martín, M. Yañez-Mo and E. Cernuda-Morollón, et al., Point-of-care detection of extracellular vesicles: Sensitivity optimization and multiple-target detection, Biosens. Bioelectron., 2017, 87, 38–45 CrossRef PubMed.
- Z. Gao, Y. Hao, M. Zheng and Y. Chen, A fluorescent dye with large Stokes shift and high stability: synthesis and application to live cell imaging, RSC Adv., 2017, 7(13), 7604–7609 RSC.
- R. L. Liang, Q. T. Deng, Z. H. Chen, X. P. Xu, J. W. Zhou and J. Y. Liang, et al., Europium (III) chelate microparticle-based lateral flow immunoassay strips for rapid and quantitative detection of antibody to hepatitis B core antigen, Sci. Rep., 2017, 7(1), 14093 CrossRef PubMed.
- J. H. Ryu, M. Kwon, J. D. Moon, M. W. Hwang, J. M. Lee and K. H. Park, et al., Development of a Rapid Automated Fluorescent Lateral Flow Immunoassay to Detect Hepatitis B Surface Antigen (HBsAg), Antibody to HBsAg, and Antibody to Hepatitis C, Ann. Lab. Med., 2018, 38(6), 578–584 CrossRef CAS PubMed.
- X. E. Cao, J. Kim, S. Mehta and D. Erickson, Two-Color Duplex Platform for Point-of-Care Differential Detection of Malaria and Typhoid Fever, Anal. Chem., 2021, 93(36), 12175–12180 CrossRef CAS PubMed.
- Q. Y. Xie, Y. H. Wu, Q. R. Xiong, H. Y. Xu, Y. H. Xiong and K. Liu, et al., Advantages of fluorescent microspheres compared with colloidal gold as a label in immunochromatographic lateral flow assays, Biosens. Bioelectron., 2014, 54, 262–265 CrossRef CAS PubMed.
- E. Eltzov, S. Guttel, A. Low Yuen Kei, P. D. Sinawang, R. E. Ionescu and R. S. Marks, Lateral Flow Immunoassays - from Paper Strip to Smartphone Technology, Electroanalysis, 2015, 27(9), 2116–2130 CrossRef CAS.
- X. Jiao, T. Peng, Z. Liang, Y. Hu, B. Meng and Y. Zhao, et al., Lateral Flow Immunoassay Based on Time-Resolved Fluorescence Microspheres for Rapid and Quantitative Screening CA199 in Human Serum, Int. J. Mol. Sci., 2022, 23(17), 9991 CrossRef CAS PubMed.
- S. Bamrungsap, C. Apiwat, W. Chantima, T. Dharakul and N. Wiriyachaiporn, Rapid and sensitive lateral flow immunoassay for influenza antigen using fluorescently-doped silica nanoparticles, Microchim. Acta, 2014, 181(1–2), 223–230 CrossRef CAS.
- S. T. Selvan, P. K. Patra, C. Y. Ang and J. Y. Ying, Synthesis of Silica-Coated Semiconductor and Magnetic Quantum Dots and Their Use in the Imaging of Live Cells, Angew. Chem., 2007, 119(14), 2500–2504 CrossRef.
- L. Ma, C. Tu, P. Le, S. Chitoor, S. J. Lim and M. U. Zahid, et al., Multidentate Polymer Coatings for Compact and Homogeneous Quantum Dots with Efficient Bioconjugation, J. Am. Chem. Soc., 2016, 138(10), 3382–3394 CrossRef CAS PubMed.
- Y. Ma, Y. Li, S. Ma and X. Zhong, Highly bright water-soluble silica coated quantum dots with excellent stability, J. Mater. Chem. B, 2014, 2(31), 5043–5051 RSC.
- C. Li, Z. Zou, H. Liu, Y. Jin, G. Li and C. Yuan, et al., Synthesis of polystyrene-based fluorescent quantum dots nanolabel and its performance in H5N1 virus and SARS-CoV-2 antibody sensing, Talanta, 2021, 225, 122064 CrossRef CAS PubMed.
- X. Deng, C. Wang, Y. Gao, J. Li, W. Wen and X. Zhang, et al., Applying strand displacement amplification to quantum dots-based fluorescent lateral flow assay strips for HIV-DNA detection, Biosens. Bioelectron., 2018, 105, 211–217 CrossRef PubMed.
- W. Qiu, H. Xu, S. Takalkar, A. S. Gurung, B. Liu and Y. Zheng, et al., Carbon nanotube-based lateral flow biosensor for sensitive and rapid detection of DNA sequence, Biosens. Bioelectron., 2015, 64, 367–372 CrossRef CAS PubMed.
- N. Wiriyachaiporn, H. Sirikett and T. Dharakul, Rapid influenza a antigen detection using carbon nanostrings as label for lateral flow immunochromatographic assay, in 2013 13th IEEE International Conference on Nanotechnology (IEEE-NANO 2013), IEEE, 2013, pp. 166–169 Search PubMed.
- M. Lönnberg, M. Drevin and J. Carlsson, Ultra-sensitive immunochromatographic assay for quantitative determination of erythropoietin, J. Immunol. Methods, 2008, 339(2), 236–244 CrossRef PubMed.
- J. Liu, R. Li and B. Yang, Carbon Dots: A New Type of Carbon-Based Nanomaterial with Wide Applications, ACS Cent. Sci., 2020, 6(12), 2179–2195 CrossRef CAS PubMed.
- N. Wiriyachaiporn, H. Sirikett, W. Maneeprakorn and T. Dharakul, Carbon nanotag based visual detection of influenza A virus by a lateral flow immunoassay, Microchim. Acta, 2017, 184(6), 1827–1835 CrossRef CAS.
- L. D. Xu, Q. Zhang, S. N. Ding, J. J. Xu and H. Y. Chen, Ultrasensitive Detection of Severe Fever with Thrombocytopenia Syndrome Virus Based on Immunofluorescent Carbon Dots/SiO 2 Nanosphere-Based Lateral Flow Assay, ACS Omega, 2019, 4(25), 21431–21438 CrossRef CAS PubMed.
- V. G. Panferov, I. V. Safenkova, A. V. Zherdev and B. B. Dzantiev, Setting up the cut-off level of a sensitive barcode lateral flow assay with magnetic nanoparticles, Talanta, 2017, 164, 69–76 CrossRef CAS PubMed.
- A. Moyano, E. Serrano-Pertierra, M. Salvador, J. C. Martínez-García, M. Rivas and M. C. Blanco-López, Magnetic Lateral Flow Immunoassays, Diagnostics, 2020, 10(5), 288 CrossRef CAS PubMed.
- T. Bu, X. Yao, L. Huang, L. Dou, B. Zhao and B. Yang, et al., Dual recognition strategy and magnetic enrichment based lateral flow assay toward Salmonella enteritidis detection, Talanta, 2020, 206, 120204 CrossRef CAS PubMed.
- Z. Liu, C. Wang, S. Zheng, X. Yang, H. Han and Y. Dai, et al., Simultaneously ultrasensitive and quantitative detection of influenza A virus, SARS-CoV-2, and respiratory syncytial virus via multichannel magnetic SERS-based lateral flow immunoassay, Nanomedicine, 2023, 47, 102624 CrossRef CAS PubMed.
- J. P. Broughton, X. Deng, G. Yu, C. L. Fasching, V. Servellita and J. Singh, et al., CRISPR–Cas12-based detection of SARS-CoV-2, Nat. Biotechnol., 2020, 38(7), 870–874 CrossRef CAS PubMed.
- E. Xiong, L. Jiang, T. Tian, M. Hu, H. Yue and M. Huang, et al., Simultaneous Dual-Gene Diagnosis of SARS-CoV-2 Based on CRISPR/Cas9-Mediated Lateral Flow Assay, Angew. Chem., Int. Ed., 2021, 60(10), 5307–5315 CrossRef CAS PubMed.
- Z. Ali, E. Sánchez, M. Tehseen, A. Mahas, T. Marsic and R. Aman, et al., Bio-SCAN: A CRISPR/dCas9-Based Lateral Flow Assay for Rapid, Specific, and Sensitive Detection of SARS-CoV-2, ACS Synth. Biol., 2022, 11(1), 406–419 CrossRef CAS PubMed.
- J. Arizti-Sanz, C. A. Freije, A. C. Stanton, B. A. Petros, C. K. Boehm and S. Siddiqui, et al., Streamlined inactivation, amplification, and Cas13-based detection of SARS-CoV-2, Nat. Commun., 2020, 11(1), 5921 CrossRef CAS PubMed.
- J. Arizti-Sanz, A. Bradley, Y. B. Zhang, C. K. Boehm, C. A. Freije and M. E. Grunberg, et al., Simplified Cas13-based assays for the fast identification of SARS-CoV-2 and its variants, Nat. Biomed. Eng., 2022, 6(8), 932–943 CrossRef CAS PubMed.
- Y. Pang, Q. Li, C. Wang, S. zhen, Z. Sun and R. Xiao, CRISPR-cas12a mediated SERS lateral flow assay for amplification-free detection of double-stranded DNA and single-base mutation, Chem. Eng. J., 2022, 429, 132109 CrossRef CAS.
- O. Mukama, T. Yuan, Z. He, Z. Li, D. Habimana J. de and M. Hussain, et al., A high fidelity CRISPR/Cas12a based lateral flow biosensor for the detection of HPV16 and HPV18, Sens. Actuators, B, 2020, 316, 128119 CrossRef CAS.
- O. Mukama, J. Wu, Z. Li, Q. Liang, Z. Yi and X. Lu, et al., An ultrasensitive and specific point-of-care CRISPR/Cas12 based lateral flow biosensor for the rapid detection of nucleic acids, Biosens. Bioelectron., 2020, 159, 112143 CrossRef CAS PubMed.
- B. Zhou, Q. Ye, F. Li, X. Xiang, Y. Shang and C. Wang, et al., CRISPR/Cas12a based fluorescence-enhanced lateral flow biosensor for detection of Staphylococcus aureus, Sens. Actuators, B, 2022, 351, 130906 CrossRef CAS.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |