Open Access Article
Open Access ArticleIn vitro selection of N1-methyladenosine-sensitive RNA-cleaving deoxyribozymes with 105-fold selectivity over unmethylated RNA†
Jiarong
Shi
a,
Qiang
Zhang
b,
Yunping
Wu
a,
Yangyang
Chang
a and
Meng
Liu
 *a
*a
aSchool of Environmental Science and Technology, Key Laboratory of Industrial Ecology and Environmental Engineering (Ministry of Education), Dalian University of Technology, Dalian POCT Laboratory, Dalian, 116024, China. E-mail: mliu@dlut.edu.cn
bMOE Key Laboratory of Bio-Intelligent Manufacturing, School of Bioengineering, Dalian University of Technology, Dalian 116024, China
First published on 24th July 2024
Abstract
RNA-cleaving DNAzymes (RCDs) are catalytically active DNA molecules that cleave a wide range of RNA targets with extremely high sequence-selectivity, but none is able to faithfully discriminate methylated from unmethylated RNA (typically <30-fold). We report the first efforts to isolate RCDs from a random-sequence DNA pool by in vitro selection that cleave RNA/DNA chimera containing N1-methyladenosine (m1A), one of the most prevalent RNA modifications that plays important regulatory roles in gene expression and human cancers. A cis-acting deoxyribozyme, RCD1-S2m1A, exhibits an observed rate constant (kobs) of 5.3 × 10−2 min−1, resulting in up to 105-fold faster cleavage of the m1A-modified versus unmethylated RNA. Furthermore, a trans-acting fluorogenic deoxyribozyme was constructed by labeling a fluorophore and a quencher at the 5′ and 3′ ends of the chimeric substrate, respectively. It permits the synchronization of RNA-cleaving with real-time fluorescence signaling, thus allowing the selective monitoring of ALKBH3-mediated demethylation and inhibitor screening in living cells.
Introduction
Enzyme-mediated chemical modification of RNA bases is one of the factors that endow RNA with multiple biological activities among the more than 170 types of modified nucleotides present in natural RNA.1–3N1-Methyladenosine (m1A), as the post-transcriptional methylation modification, was first documented in yeast tRNAPhe in 1961.4,5 The methyl group in m1A disrupts the Watson–Crick base pairing and results in a positive charge to the nucleobase; thus m1A may form alternative secondary structures and block reverse transcription (RT).6–8 In human cells, m1A at positions 9 (m1A9) and 58 (m1A58) of tRNAs can be catalyzed by TRMT10C, TRMT61B, or the methyltransferase TRMT6/TRMT61A complex7 and erased by the demethylase AlkB homologue 1 (ALKBH1) or AlkB homologue 3 (ALKBH3).9–11 By participating in various physiological processes, m1A has regulatory roles in the pathogenesis of tumor and non-tumor diseases.12ALKBH3, also referred to as prostate cancer antigen-1 (PCA-1), is a 37.9 kDa enzyme composed of 286 amino acids and requiring 2-oxoglutarate (2OG) and Fe(II) for its activity.13,14 Although it has a conserved amino acid sequence and a tertiary structure similar to other AlkB homologous proteins, ALKBH3 has unique recognition and removal functions for m1A and m3C damage to ssDNA/RNA.15 Recent study has linked the expression of ALKBH3 to tumor growth, suggesting its biological and clinical significance.16,17 Increased levels of ALKBH3 have been detected in prostate tumor sections.18 ALKBH3 may also serve as a new diagnostic biomarker as well as a promising therapeutic target in various human diseases, including head and neck squamous cell carcinoma, breast cancer, and lung cancer.19 These studies also suggest the possibility of inhibiting ALKBH3 as a therapeutic approach for solid tumors.17 Despite this progress, further gaining a thorough insight into the functions of ALKBH3 needs a robust biosensing tool capable of measuring its activity in living cells.
RNA-cleaving DNAzymes (RCDs) are catalytically active DNA molecules that cleave RNA substrates.20–22 To date, many RCDs have been identified through an in vitro selection technique from random-sequence DNA pools.23–26 RCDs possess several physical and chemical properties that make them attractive in biosensing, diagnosis, and cancer therapy.27–31 Very recently, Höbartner and coworkers have evolved RCDs for the site-specific interrogation of RNA modifications, including N6-methyladenosine (m6A), 3-methylcytidine (m3C), N4-methylcytidine (m4C), and 5-methylcytidine (m5C).32–34 However, these RCDs only displayed 5- to 30-fold accelerated cleavage rates for methylated versus unmodified RNA. Furthermore, they have hypothesized and confirmed that RCDs effectively discriminate larger RNA modifications (e.g., N6-isopentenyladenosine),35 rather than small chemical modifications (e.g., m6A, m3C, m4C, and m5C). Therefore, it remains a great challenge to create RCDs that distinguish the methylation site of occurrence in the target RNA. Furthermore, improved site-specific cleavage would benefit applications in characterizing RNA-modification-associated proteins (e.g., ALKBH3).
Toward this end, we set out to use novel in vitro selection to generate RCDs that could robustly differentiate m1A and unmethylated RNA. Instead of all-RNA substrates, m1A-modified chimeric (RNA/DNA) substrates were purposely used so that specific m1A sites could be unambiguously targeted. To further ensure the desired specificity, counter selection is carried out to eliminate sequences with cross-activities for natural unmethylated RNA. Our work led to the isolation of an RCD, termed RCD1-S2m1A. Further truncation of the original RCD1-S2m1A produced a shortened version, RCD1S-S3m1A, that had a kobs of 6.2 × 10−2 min−1, a cleavage rate that is ∼105-fold faster for methylated versus unmethylated RNA. We have also created a related trans-acting version, RCD1T/Sm1A, that has uniquely synchronized RNA cleaving with fluorescence-signaling ability. This enables the development of fluorogenic RCD sensors for the monitoring of ALKBH3-catalyzed demethylation and ALKBH3 inhibitor screening in living cells.
Results and discussion
In vitro selection and characterization of m1A-sensitive RCDs
The in vitro selection strategy included six key steps, as illustrated in Fig. 1a. A pool of single-stranded 76 nt (nt: nucleotide) DNA containing 40 random-sequence nucleotides, named DL1, was used for the selection (see Table S1 in the ESI† for the sequences of all the DNA oligonucleotides used in this work). In step I, DL1 was first phosphorylated and ligated to a chimeric RNA/DNA substrate S1 (33 nt) that contains a single adenine ribonucleotide (rA) as the cleavage site (named S1A). The ligated DL1-S1A (109 nt) was purified by denaturing gel electrophoresis (dPAGE), followed by 24-h incubation with 1 × selection buffer (1 × SB; 50 mM HEPES, 150 mM NaCl, 50 mM KCl, 15 mM MgCl2, 5 mM MnCl2, and 0.01% Tween 20) in step II. This step represents the counter selection step to remove any self-cleaving and nonspecific DNAzymes. The uncleaved DL1-S1A molecules were purified by dPAGE and then digested with EcoRV in the presence of a splint DNA strand to regenerate the DNA pool in step III. The purified DL1 was then ligated to the chimeric substrate S2 containing N1-methylated ribonucleotide adenosine (m1A) as the cleavage site (named S2m1A, step IV). Upon purification using dPAGE, the ligated construct DL1-S2m1A was incubated for 12 h in 1 × SB: this procedure was the positive selection step aimed at isolating m1A-sensitive DNAzymes (step V). The cleaved DNA molecules were purified by dPAGE, amplified by the polymerase chain reaction (PCR) in step VI, and used for the next round of selection (detailed experimental protocols are provided in the ESI†).The selection progress is summarized in Fig. 1b. A detectable cleavage activity was observed after 4 rounds. By round 5, ∼5% of the DL1-S2m1A was cleaved after a 12-h incubation, compared to <1% for DL1-S1A following a 24-h incubation. The reaction time was then progressively reduced to isolate the most efficient DNAzymes. The positive selection was then allowed to proceed for 3 h in round 7 and 1 h in rounds 8 and 9, and the reaction time was further reduced to 30 min in rounds 10 and 11, to 20 min for rounds 12 and 13, to 5 min for rounds 14 and 15, and finally to 1 min for round 16. The enriched sequences from round 16 were subjected to high-throughput DNA sequencing using our previously published protocol.36 The top 5 sequences were chemically synthesized (Table S2†) and tested for cleavage activity (Fig. S1†). A DNAzyme with the highest cleavage activity, named RCD1-S2m1A, was internally labeled with a fluorophore (F) and chosen for further investigation.
RCD1-S2m1A was first assessed for metal dependence. Seven different divalent metal ions, including Mg2+, Hg2+, Zn2+, Ni2+, Ca2+, Cd2+, and Ba2+, were tested (Fig. S2†). RCD1-S2m1A was extremely specific for Mn2+ (causing 27% of cleavage). Furthermore, its cleavage activity increased as the Mn2+ concentration was increased from 1 mM to 10 mM (Fig. S3†). None of the other metal ions can induce cleavage. We next examined its cleavage activity at different reaction temperatures. A strong cleavage activity was found at 37 °C (36%, Fig. S4†). In contrast, reduced activity was observed when the reaction temperature was decreased to 25 °C or increased to 65 °C. We also examined the activity of RCD1-S2m1A when the reaction pH was varied between 3.5 and 8.5; the highest activity was obtained at pH 7.5–8.5 (Fig. S5†). Note that the spontaneous degradation of S2m1A at pH 8.5 was ∼2%. The kinetic analysis revealed the observed rate constants (kobs) of 5.3 × 10−2 min−1 and a final cleavage yield of 31% (Fig. 2a). For comparison, RCD1 did not show distinct cleavage activity towards S1A within 60 min (0%, Fig. 2b). Therefore, the cleavage rate was assumed to be a background rate of ∼10−7 min−1.37 This represents ∼105-fold faster cleavage of the m1A-modified versus unmethylated RNA. Furthermore, RCD1 cleaved only m1A-modified RNA and was completely inhibited by m6A- and N6-isopentenyladenosine (i6A)-containing substrates (Fig. S6†).
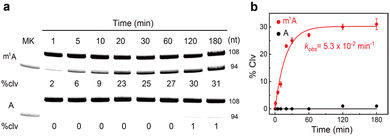 | ||
| Fig. 2 (a) 10% dPAGE analysis and (b) the kinetic responses of RCD1 to substrates S1A and S2m1A. The kobs is given in the graph. The error bars represent standard deviations of three independent experiments. Reaction conditions: 1 × selection buffer (50 mM HEPES, 150 mM NaCl, 50 mM KCl, 15 mM MgCl2, 5 mM MnCl2, and 0.01% Tween 20). An uncropped gel image is provided in the ESI.† MK, marker (94 nt); % clv, % cleavage. | ||
Sequence optimization by nucleotide truncation
We investigated whether the sequence of RCD1-S2m1A could be minimized. Its putative secondary structure was obtained using the M-fold program. As shown in Fig. 3a, it comprises six short duplexes (P1–P6), five hairpin loops (L1–L5), five inter-helical unpaired elements (J1/2, J2/1, J3/4, J4/5 and J5/6), and two single-stranded regions (SS1 and SS2). The effects of deleting nucleotides from the 3′ end were then assessed (Fig. 3b). The first 9 nucleotides in SS2 were completely dispensable, as the deletion of the entire sequence slightly enhanced the activity (34% vs. 32%). Interestingly, removing the L5 and P6 resulted in a very significant improvement in activity (63% vs. 32%). However, the truncated construct also exhibited high activity against S1A (41%). This finding reveals that L5 and P6 appear to play a structural role that is important for catalytic selectivity. Deleting the next J5/6, L4 and P5 elements resulted in a complete loss of activity (0%), suggesting the significant roles of these nucleotides in the catalytic function. L3 and P4 can be completely removed without affecting the cleavage activity (32%). In contrast, L2 and P3 are catalytically essential since their removal also caused complete loss of activity (0%). | ||
| Fig. 3 (a) Proposed secondary structure model of cis-acting RCD1-S2m1A. Individual elements are marked as SS (single-stranded region), P (pairing region), L (loop), and J (junction between two pairing regions). F = fluorescein-dT. (b) 10% dPAGE analysis of various truncated RCD1 constructs on substrates S1A (bottom) and S2m1A (top). MK, marker (108 nt); % clv, % cleavage. An uncropped gel image is provided in the ESI.† (c) Secondary structural model for a shortened cis-acting DNAzyme, RCD1S-S3m1A. (d) Kinetic analysis of RCD1S to substrates S3A and S3m1A. The kobs is given in the graph. The error bars represent standard deviations of three independent experiments. (e) Designing a trans-acting DNAzyme system, RCD1T/Sm1A. F = fluorescein-dC; Q = BHQ1-dT. (f) Examination of the real-time signaling capability of RCD1T/Sm1A and RCD1T/SA. | ||
On the basis of this finding, there is a significantly shortened cis-acting version, denoted as RCD1S-S3m1A (Fig. 3c), in which SS2, L3 and P4 were removed along with the deletion of SS1. Note that SS1 can be completely removed without affecting the cleavage activity (32%) (Fig. S7†). We determined the kobs value for RCD1S-S3m1A to be 6.2 × 10−2 min−1 (Fig. 3d), and only 2% cleavage product was observed in 180 min against S3A (Fig. S8†). One point merits further attention. Many well-known RCDs, such as 10-23 and 8-17 deoxyribozymes,20–22 have two binding arms to interact with their substrates while placing the catalytic core across the cleavage site. Although the M-fold program suggests five pairing regions within RCD1S-S3m1A, one of which links the substrate S3m1A to RCD1S, it does not predict any non-Watson–Crick interactions that involve the nucleotides between RCD1S and S3m1A. It is possible that a large number of nucleotides are engaged in tertiary interactions for structure folding or catalytic function.
According to the proposed secondary structure of RCD1S-S3m1A, we designed a trans-acting DNA enzyme system, denoted as RCD1T/Sm1A, by replacing P2 and L1 existing in RCD1S-S3m1A with a stem made of seven base-pairs (Fig. 3e). Sm1A was modified with a fluorophore (F) at the 5′-end and a quencher (Q) at the 3′-end. Initially, the short distance between F and Q results in maximal fluorescence quenching. Following cleavage by RCD1T, the free 5′ cleavage fragment of Sm1A was expected to be released, thus leading to increased fluorescence. This hypothesis was supported by the real-time fluorescence signaling results: RCD1T/Sm1A exhibited a very large signaling magnitude (i.e., F/F0, defined as the fluorescence response in the presence of RCD1T over that in the absence of RCD1T). In contrast, RCD1T/SA was not able to produce a change in the fluorescence signal.
RCD1T system for in vitro analysis of ALKBH3 inhibitors
ALKBH3-mediated m1A demethylation is essential for cell proliferation and tumor progression.16,17 Discovering small molecules that specifically modulate the activity of ALKBH3 is of significant biological and clinical interest.19 It is known that ALKBH3 strongly prefers single-stranded RNA (ssRNA) and single-stranded DNA (ssDNA) substrates with site-specifically incorporated m1A.13,14 We first investigated the demethylation activity of ALKBH3 toward m1A-containing chimeric RNA/DNA substrate Sm1A. As shown in Fig. 4a, we treated Sm1A with ALKBH3 for 2 h (pH 7.0 at 37 °C). It is expected that ALKBH3 catalyzed the oxidative demethylation of m1A to adenine ribonucleotide, thereby inhibiting the catalytic activity of RCD1T. This was confirmed experimentally (Fig. 4b): while Sm1A alone was cleaved by RCD1T (24%, lane 3), treatment with ALKBH3 indeed inhibited the cleavage of Sm1A, as evidenced by the reduction of small cleavage fragments (2%, lane 4).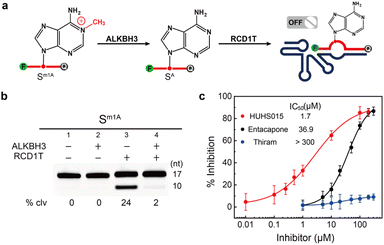 | ||
| Fig. 4 (a) Proposed oxidative demethylation of m1A to adenosine in Sm1A by ALKBH3, which will efficiently inhibit the cleavage reaction by RCD1T. (b) 10% dPAGE analysis of the cleavage reaction mixtures containing various combinations of Sm1A, ALKBH3 and RCD1T. An uncropped gel image is provided in the ESI.† (c) Inhibition of ALKBH3 by various concentrations of HUHS015, entacapone and thiram. % Inhibition was defined as (FInhibitor – Fno Inhibitor)/(Fno ALKBH3 – Fno Inhibitor), where FInhibitor and Fno Inhibitor refer to the fluorescence intensity with and without an inhibitor and Fno ALKBH3 is the initial reading in the absence of ALKBH3. | ||
The real-time signaling ability of the RCD1T/Sm1A system was also assessed following ALKBH3 treatment for different periods of time (Fig. S9†). Rapid signal generation was observed in the absence of ALKBH3. Upon introduction of ALKBH3, the signaling rate and the fluorescence enhancement were significantly decreased. Longer incubation times (up to 1 h) resulted in no obvious change in the fluorescence signal because more Sm1A molecules were converted into SA. RCD1T/Sm1A was also examined for fluorescence signaling with an increase in the ALKBH3 concentration in real time. Its signaling intensity decreased as the ALKBH3 concentration was increased between 0.01 and 500 nM (Fig. S10†). In addition, other unintended proteins and small molecules were also tested for their ability to inhibit the DNAzyme activity. No decrease in fluorescence was observed when the RCD1T/Sm1A system was treated with bovine serum albumin (BSA), polynucleotide kinase (PNK), alkaline phosphatase (ALP), uracil DNA glycosylase (UDG), thrombin, adenosine 5′-triphosphate (ATP), glutathione (GSH) and glucose (Fig. S11†).
We next examined the possibility of exploiting the RCD1T/Sm1A system for ALKBH3 inhibitor screening. We measured the inhibition of ALKBH3 by HUHS015, a potent prostate cancer antigen-1 PCA-1/ALKBH3 inhibitor.38 As expected, we discovered that HUHS015 inhibits ALKBH3 with an IC50 of 1.7 μM (Fig. 4c), comparable to the previously reported value (IC50 = 0.67 μM). Control experiments using entacapone (a potential inhibitor for obesity-related protein FTO)39 and thiram (a potent inhibitor against the TRMT6/TRMT61A complex)40 indicated that they exhibited >20-fold weaker inhibition of ALKBH3. Taken together, these results suggested that the proposed RCD1T1 system can be used for the screening of ALKBH3 inhibitors in vitro.
RCD1T system for cell imaging of ALKBH3 inhibitors
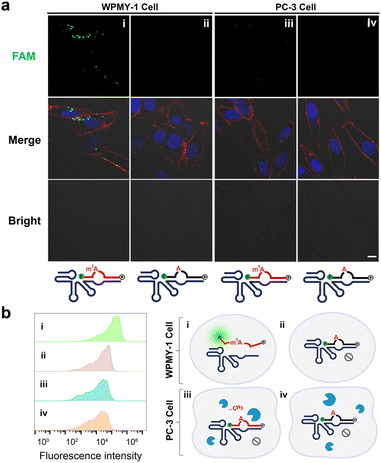
Finally, we investigated the feasibility of performing intracellular ALKBH3 inhibitor imaging in living cells (Fig. 5a). The PC-3 cell, known for its high expression of ALKBH3,41 was used for the assay. RCD1T was delivered into live cells along with the dually labelled substrate Sm1A using the standard Lipofectamine transfection procedure. The fluorescence signaling of the RCD1T/Sm1A system inside cells was measured by confocal microscopy imaging. In the absence of the ALKBH3 inhibitor, ALKBH3 catalyzes the removal of the m1A modification on Sm1A in live cells, thereby inactivating the catalytic activity of RCD1T. However, the introduction of the inhibitor should efficiently inhibit the demethylation activity of ALKBH3. This action should restore the catalytic activity of RCD1T and result in the generation of a fluorescence signal.All the PC-3 cell groups were first pre-treated with HUHS015 for 12 h to suppress the demethylation activity of ALKBH3. As shown in Fig. 5b and S12,† Sm1A was stable under intracellular conditions, as revealed by the intact fluorescence in Sm1A-transfected PC-3 cells (sample i). By carrying out the RNA-cleaving reaction, an intense fluorescence signal was produced in HUHS015-pretreated cells (sample iii). In contrast, cells not incubated with Mn2+ displayed a minimal signal under the same conditions (sample ii). One mutant RCD1T (RCD1TM) was also examined. Since several nucleotides crucial for the cleavage reaction were mutated in RCD1TM, it was expected that the fluorescence signal would be eliminated in HUHS015-pretreated cells. This was confirmed by the experimental observations: no obvious signal was observed in the RCD1TM/Sm1A-treated cells even in the presence of Mn2+ (sample iv). As a control, RCD1T/SA-treated cells were also tested (sample v). We observed a very low fluorescence signal in these treated cells.
We also investigated fluorescence responses of RCD1T/Sm1A-transfected PC-3 cells treated with varying concentrations of HUHS015 (Fig. S13†). HUHS015 induced a fluorescence increase in a dose-dependent manner. These results strongly suggest that our RCD1T system is effective in monitoring intracellular ALKBH3 inhibitors.
We also introduced the same RCD1T system into WPMY-1 cells, which are human normal prostate matrix immortalized cells lacking ALKBH3.42Fig. 6a indicates that a significant increase in fluorescence was observed in RCD1T/Sm1A-transfected WPMY-1 cells (sample i) but not in control RCD1T/SA-transfected cells (sample ii). For comparison, no obvious fluorescence signal was observed in PC-3 cells transfected with RCD1T/Sm1A (sample iii) or RCD1T/SA (sample iv). These results were consistent with those of flow cytometry assay (Fig. 6b and S14†): RCD1T/Sm1A-transfected WPMY-1 cells display large signaling. Taken together, we can conclude that (Fig. 6b): (1) cleavage of Sm1A by RCD1T occurs in WPMY-1 cells lacking ALKBH3, thus producing a strong fluorescence signal; (2) overexpressing ALKBH3 has robust demethylation activity against the Sm1A substrate in the PC-3 cell line, thus inhibiting the RNA-cleaving activity of RCD1T; (3) RCD1T exhibits extremely high specificity for Sm1A over SA in living cells.
Conclusions
In summary, we address the question of whether RNA-cleaving DNAzymes can be created that faithfully discriminate methylated from unmodified RNA. We have created one remarkable DNAzyme therefrom, a cis-acting deoxyribozyme RCD1, which exhibits a ∼105-fold higher kobs value for m1A-modified versus unmethylated RNA. To the best of our knowledge, no comparable deoxyribozymes have been reported that are sensitive to the presence of RNA modifications. A trans-acting DNA molecule, RCD1T, was also developed that cleaves the substrate Sm1A labeled with a FAM fluorophore at the 5′-end and a quencher at the 3′-end. This produces a very large fluorescence enhancement upon RNA cleavage.We finally describe the development and characterization of a fluorescent RCD1T system that can be used for the imaging of ALKBH3-mediated RNA demethylation and inhibitor screening in living cells. It was demonstrated that RCD1T can efficiently cleave an m1A-containing RNA substrate and record robust fluorescence signals in the presence of ALKBH3 inhibitors. In the future, we will continue to pursue the precise structure of RCD1T using high-resolution techniques, including nuclear magnetic resonance and X-ray crystallography. We envision that the strategies demonstrated in this work can expand the practical utilities of DNAzymes in RNA modification studies, clinical diagnosis and drug discovery.
Data availability
The data supporting this article have been included in the ESI.†Author contributions
M. L. applied for the funding. M. L. conceived the idea and supervised the project. J. R. S. performed the experiments and data collection. J. R. S., Q. Z., Y. Y. C., Y. P. W., and M. L. analyzed the data. J. R. S., Q. Z., Y. Y. C., Y. P. W., and M. L. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC; Grant No. 22176024) and the National Key R&D Program of China (No. 2023YFC3205804).Notes and references
- P. Boccaletto, M. A. Machnicka, E. Purta, P. Piatkowski, B. Baginski, T. K. Wirecki, V. Crecy-Lagard, R. Ross, P. A. Limbach, A. Kotter, M. Helm and J. M. Bujnicki, Nucleic Acids Res., 2018, 46, D303–D307 CrossRef CAS PubMed.
- I. A. Roundtree, M. E. Evans, T. Pan and C. He, Cell, 2017, 169, 1187–1200 CrossRef CAS PubMed.
- M. Frye, B. T. Harada, M. Behm and C. He, Science, 2018, 361, 1346–1349 CrossRef CAS PubMed.
- D. B. Dunn, Biochim. Biophys. Acta, 1961, 46, 198–200 CrossRef CAS PubMed.
- Y. Chen, S. Yang, S. Peng, W. Li, F. Wu, Q. Yao, F. Wang, X. Weng and X. Zhou, Chem. Sci., 2019, 10, 2975–2979 RSC.
- M. Safra, A. Sas-Chen, R. Nir, R. Winkler, A. Nachshon, D. Bar-Yaacov, M. Erlacher, W. Rossmanith, N. Stern-Ginossar and S. Schwartz, Nature, 2017, 551, 251–255 CrossRef CAS PubMed.
- P. Boccaletto, F. Stefaniak, A. Ray, A. Cappannini, S. Mukherjee, E. Purta, M. Kurkowska, N. Shirvanizadeh, E. Destefanis, P. Groza, G. Avşar, A. Romitelli, P. Pir, E. Dassi, S. G. Conticello, F. Aguilo and J. M. Bujnicki, Nucleic Acids Res., 2022, 50, D231–D235 CrossRef CAS PubMed.
- H. Zhou, I. J. Kimsey, E. N. Nikolova, B. Sathyamoorthy, G. Grazioli, J. McSally, T. Bai, C. H. Wunderlich, C. Kreutz, I. Andricioaei and H. M. AI-Hashimi, Nat. Struct. Mol. Biol., 2016, 23, 803–810 CrossRef CAS PubMed.
- E. Vilardo, C. Nachbagauer, A. Buzet, A. Taschner, J. Holzmann and W. Rossmanith, Nucleic Acids Res., 2012, 40, 11583–11593 CrossRef CAS PubMed.
- F. Liu, W. Clark, G. Luo, X. Wang, Y. Fu, J. Wei, X. Wang, Z. Hao, Q. Dai, G. Zheng, H. Ma, D. Han, M. Evans, A. Klungland, T. Pan and C. He, Cell, 2016, 167, 816–828 CrossRef CAS PubMed.
- Z. Hu, A. Sun, J. Yang, G. Naz, G. Sun, Z. Li, J. Jie, G. Liu, S. Zhang and X. Zhang, Chem. Sci., 2023, 14, 5945–5955 RSC.
- Z. Chen, M. Qi, B. Shen, G. Luo, Y. Wu, J. Li, Z. Lu, Z. Zheng, Q. Dai and H. Wang, Nucleic Acids Res., 2019, 47, 2533–2545 CrossRef CAS PubMed.
- P. A. Aas, M. Otterlei, P. O. Falnes, C. B. Vågbø, F. Skorpen, M. Akbari, O. Sundheim, M. Bjørås, G. Slupphaug, E. Seeberg and H. E. Krokan, Nature, 2003, 421, 859–863 CrossRef CAS PubMed.
- R. Ougland, C.-M. Zhang, A. Liiv, R. F. Johansen, E. Seeberg, Y.-M. Hou, J. Remme and P. O. Falnes, Mol. Cell, 2004, 16, 107–116 CrossRef CAS PubMed.
- Z. Chen, M. Qi, B. Shen, G. Luo, Y. Wu, J. Li, Z. Lu, Z. Zheng, Q. Dai and H. Wang, Nucleic Acids Res., 2019, 47, 2533–2545 CrossRef CAS PubMed.
- I. Barbieri and T. Kouzarides, Nat. Rev. Cancer, 2020, 20, 303–322 CrossRef CAS PubMed.
- I. Yamato, M. Sho, K. Shimada, K. Hotta, Y. Ueda, S. Yasuda, N. Shigi, N. Konishi, K. Tsujikawa and Y. Nakajima, Cancer Res., 2012, 72, 4829–4839 CrossRef CAS PubMed.
- K. Bian, S. Lenz, Q. Tang, F. Chen, R. Qi, M. Jost, C. L. Drennan, J. M. Essigmann, S. D. Wetmore and D. Li, Nucleic Acids Res., 2019, 47, 5522–5529 CrossRef CAS PubMed.
- M. Tasaki, K. Shimada, H. Kimura, K. Tsujikawa and N. Konishi, British J Cancer., 2011, 104, 700–706 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- S. W. Santoro and G. F. Joyce, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 4262–4266 CrossRef CAS PubMed.
- S. K. Silverman, Nucleic Acids Res., 2005, 33, 6151–6163 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- D. L. Robertson and G. F. Joyce, Nature, 1990, 344, 467–468 CrossRef CAS PubMed.
- Y. Wang, E. Liu, C. H. Lam and D. M. Perrin, Chem. Sci., 2018, 9, 1813–1821 RSC.
- E. M. McConnell, I. Cozma, Q. Mou, J. D. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC.
- Y. Wang, K. Nguyen, R. C. Spitale and J. C. Chaput, Nat. Chem., 2021, 13, 319–326 CrossRef CAS PubMed.
- M. Liu, D. Chang and Y. Li, Acc. Chem. Res., 2017, 50, 2273–2283 CrossRef CAS PubMed.
- Q. Hu, Z. Tong, A. Yalikong, L.-P. Ge, Q. Shi, X. Du, P. Wang, X.-Y. Liu, W. Zhan, X. Gao, D. Sun, T. Fu, D. Ye, C. Fan, J. Liu, Y.-S. Zhong, Y.-Z. Jiang and H. Gu, Nat. Chem., 2024, 16, 122–131 CrossRef CAS PubMed.
- K. Chiba, T. Yamaguchi and S. Obika, Chem. Sci., 2023, 14, 7620–7629 RSC.
- M. V. Sednev, V. Mykhailiuk, P. Choudhury, J. Halang, K. E. Sloan, M. T. Bohnsack and C. Höbartner, Angew. Chem., Int. Ed., 2018, 57, 15117–15121 CrossRef CAS PubMed.
- A. Liaqat, M. V. Sednev, C. Stiller and C. Höbartner, Angew. Chem., Int. Ed., 2021, 60, 19058–19062 CrossRef CAS PubMed.
- R. Micura and C. Höbartner, Chem. Soc. Rev., 2020, 49, 7331–7353 RSC.
- A. Liaqat, C. Stiller, M. Michel, M. V. Sednev and C. Höbartner, Angew. Chem., Int. Ed., 2020, 59, 18627–18631 CrossRef CAS PubMed.
- Q. Zhou, G. Zhang, Y. Wu, Q. Zhang, Y. Liu, Y. Chang and M. Liu, J. Am. Chem. Soc., 2023, 145, 21370–21377 CrossRef CAS PubMed.
- Y. Li and R. R. Breaker, J. Am. Chem. Soc., 1999, 121, 5364–5372 CrossRef CAS.
- S. Nakao, M. Mabuchi, T. Shimizu, Y. Itoh, Y. Takeuchi, M. Ueda, H. Mizuno, N. Shigi, I. Ohshio, K. Jinguji, Y. Ueda, M. Yamamoto, T. Furukawa, S. Aoki, K. Tsujikawa and A. Tanaka, Bioorg. Med. Chem. Lett., 2014, 24, 1071–1074 CrossRef CAS PubMed.
- L. Xie, L. Liu and L. Cheng, Biochemistry, 2020, 59, 230–239 CrossRef CAS PubMed.
- Y. Wang, J. Wang, X. Li, X. Xiong, J. Wang, Z. Zhou, X. Zhu, Y. Gu, D. Dominissini, L. He, Y. Tian, C. Yi and Z. Fan, Nat. Commun., 2021, 12, 6314 CrossRef CAS PubMed.
- A. A. Beharry, S. Lacoste, T. R. O'Connor and E. T. Kool, J. Am. Chem. Soc., 2016, 138, 3647–3650 CrossRef CAS PubMed.
- X. Li, X. Xiong, K. Wang, L. Wang, X. Shu, S. Ma and C. Yi, Nat. Chem. Biol., 2016, 12, 311–316 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc02943g |
| This journal is © The Royal Society of Chemistry 2024 |