Open Access Article
Open Access ArticleFine-tuning large language models for chemical text mining†
Wei
Zhang‡
ab,
Qinggong
Wang‡
c,
Xiangtai
Kong
ab,
Jiacheng
Xiong
ab,
Shengkun
Ni
ab,
Duanhua
Cao
ad,
Buying
Niu
ab,
Mingan
Chen
aef,
Yameng
Li
g,
Runze
Zhang
ab,
Yitian
Wang
ab,
Lehan
Zhang
ab,
Xutong
Li
ab,
Zhaoping
Xiong
g,
Qian
Shi
f,
Ziming
Huang
h,
Zunyun
Fu
*a and
Mingyue
Zheng
 *abc
*abc
aDrug Discovery and Design Center, State Key Laboratory of Drug Research, Shanghai Institute of Materia Medica, Chinese Academy of Sciences, 555 Zuchongzhi Road, Shanghai 201203, China. E-mail: myzheng@simm.ac.cn; fuzunyun@simm.ac.cn
bUniversity of Chinese Academy of Sciences, No. 19A Yuquan Road, Beijing 100049, China
cNanjing University of Chinese Medicine, 138 Xianlin Road, Nanjing 210023, China
dInnovation Institute for Artificial Intelligence in Medicine of Zhejiang University, College of Pharmaceutical Sciences, Zhejiang University, Hangzhou, Zhejiang 310058, China
eSchool of Physical Science and Technology, ShanghaiTech University, Shanghai 201210, China
fLingang Laboratory, Shanghai 200031, China
gProtonUnfold Technology Co., Ltd, Suzhou, China
hMedizinische Klinik und Poliklinik I, Klinikum der Universität München, Ludwig-Maximilians-Universität, Munich, Germany
First published on 7th June 2024
Abstract
Extracting knowledge from complex and diverse chemical texts is a pivotal task for both experimental and computational chemists. The task is still considered to be extremely challenging due to the complexity of the chemical language and scientific literature. This study explored the power of fine-tuned large language models (LLMs) on five intricate chemical text mining tasks: compound entity recognition, reaction role labelling, metal–organic framework (MOF) synthesis information extraction, nuclear magnetic resonance spectroscopy (NMR) data extraction, and the conversion of reaction paragraphs to action sequences. The fine-tuned LLMs demonstrated impressive performance, significantly reducing the need for repetitive and extensive prompt engineering experiments. For comparison, we guided ChatGPT (GPT-3.5-turbo) and GPT-4 with prompt engineering and fine-tuned GPT-3.5-turbo as well as other open-source LLMs such as Mistral, Llama3, Llama2, T5, and BART. The results showed that the fine-tuned ChatGPT models excelled in all tasks. They achieved exact accuracy levels ranging from 69% to 95% on these tasks with minimal annotated data. They even outperformed those task-adaptive pre-training and fine-tuning models that were based on a significantly larger amount of in-domain data. Notably, fine-tuned Mistral and Llama3 show competitive abilities. Given their versatility, robustness, and low-code capability, leveraging fine-tuned LLMs as flexible and effective toolkits for automated data acquisition could revolutionize chemical knowledge extraction.
Introduction
Chemical text mining is a crucial foundation in chemical research. It creates extensive databases that provide access to physicochemical properties and synthetic routes for experimental chemists. Additionally, it accumulates rich data and insights for computational chemists to use for modelling and predicting. More than just extracting information from chemical texts, the rule-based transformation of chemical text is particularly interesting. For instance, synthetic procedures can be converted into action sequences1,2 or programming languages.3–5 This allows them to be understood and executed by robotics for automated syntheses.However, converting structured data from intricate scientific literature is a challenging task, especially due to the complexity and heterogeneity of chemical language. As a result, a number of text-mining tools have been developed. For instance, ChemDataExtractor6,7 was created to extract chemical entities and their associated properties, measurements and relationships from chemical documents, using unsupervised word clustering, conditional random fields, rule-based grammar and dictionary matching. ChemRxnExtractor,8 a BERT-like model, was designed to extract the product and label associated reaction roles such as the reactant, catalyst, solvent, and temperature from paragraphs of synthesis experiments. Vaucher et al.1,2 developed task-adaptive pre-trained transformers to convert the synthesis protocol paragraphs into action sequences. SynthReader3 was built to convert literature syntheses to executable XDL formats, containing a series of domain-specific algorithms with predefined rules. Historically, the focus has been on designing models and algorithms specific to certain tasks, requiring extensive domain knowledge and sophisticated data processing. These tools, challenging to adapt for diverse extraction tasks, often require complementary collaboration to manage complex information extraction tasks, thus limiting their versatility and practicality.
Recently, large language models (LLMs), represented by ChatGPT released in November 2022, have shown the potential for Artificial General Intelligence (AGI). LLMs, such as GPT-3.5 and GPT-4, can generate logical insights or content that meets requirements based on human instructions. We are entering a new era where AGI and medicinal chemists might work together. There have been some assessments of ChatGPT's chemistry capabilities, including tasks like synonym transformation, property prediction, retrosynthesis, and molecule design.9–11 However, LLMs tend to “hallucinate”, meaning they generate unintended text that misaligns with established facts and real-world knowledge.12,13 Moreover, objectively evaluating the results of open-ended questions remains a significant challenge.
At this juncture, LLMs may still find it difficult to accurately answer factual and knowledge-based questions. However, using LLMs for knowledge extraction tasks should greatly alleviate hallucination and fully leverage their powerful text comprehension and processing capabilities, making them promising universal tools for chemical text mining. For instance, Zheng et al.14 used prompt engineering to guide ChatGPT in extracting information about metal–organic framework (MOF) synthesis. Patiny et al.15 tried to use ChatGPT to extract FAIR (Findable, Accessible, Interoperable, Reusable) data from publications. However, their approach of using LLMs simply based on prompt engineering tends to achieve poor performance in exact accuracy. According to the biomedical benchmark study by Chen et al.,16 ChatGPT performed significantly worse on biomedical text mining compared to existing models. These findings seem to contradict the common belief in the LLMs' superior comprehension abilities. Either way, LLMs have limitations due to their model architecture and memory, including a maximum length of prompt tokens. Besides, human expressions can be ambiguous, incomplete, vague, and difficult to refine. Outputs may not strictly adhere to formatting requirements, leading to misunderstanding and poor performance in mining complex text, such as patents or scientific literature. Therefore, zero-shot or few-shot prompts are often insufficient to address the diversity of scenarios and cannot guarantee the quality of extracted data.
In this study, we extensively explored the effectiveness of fine-tuning LLMs on five challenging tasks in chemical text mining: compound entity recognition, reaction role annotation, metal–organic framework (MOF) synthesis information extraction, nuclear magnetic resonance spectroscopy (NMR) data extraction, and conversion reaction paragraphs into action sequences. We found that fine-tuning GPT models significantly enhances performance in text mining tasks, compared to prompt-only versions, while also reducing dependency on the repetitive and extensive prompt engineering experiments. Meanwhile, we also evaluated prevalent generative pre-trained language models, such as Mistral,17 Llama3,18 Llama2,19 T5,20 and BART.21 Among these, fine-tuned ChatGPT (GPT-3.5-turbo) models achieved state-of-the-art (SOTA) performance across all five tasks. Remarkably, it even outperformed models that have been trained specifically for each task and subsequently fine-tuned, based on a significantly larger amount of in-domain data. This study highlights the potential of fine-tuning LLMs to revolutionize complex knowledge extraction with their versatility, robustness, and low code capability. Fine-tuned LLMs can be easily generalizable and can optimize the labor-intensive and time-consuming data collection workflow, even with few data. This will accelerate the discovery and creation of novel substances, making them powerful tools for universal use.
Results and discussion
Overview of chemical text mining tasks
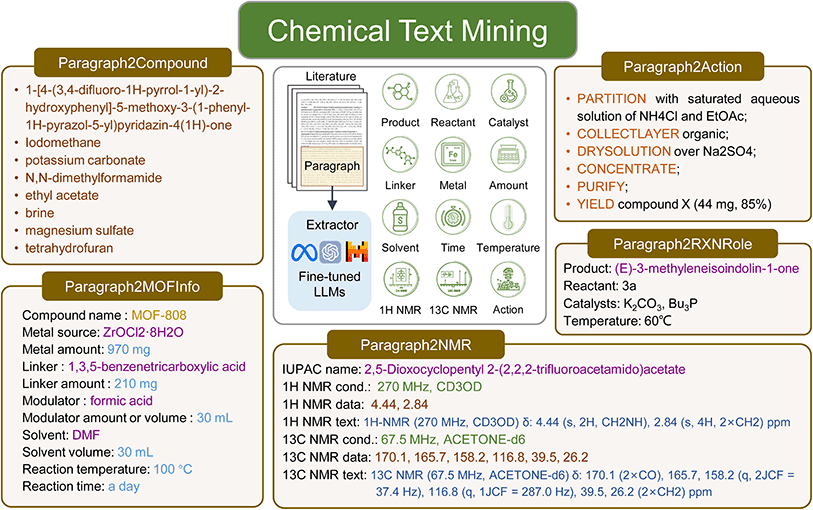
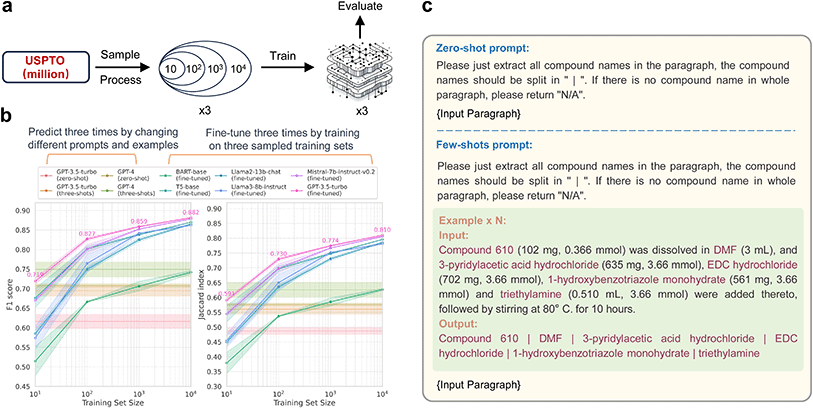
Given the complex and diverse information embedded in chemical literature, we designed five extraction tasks to demonstrate the potential and practicality of LLMs in chemical text mining (Fig. 1). The Paragraph2Compound task is a relatively simple task, aiming to extract all chemical compound entities from the given paragraph. The Paragraph2RXNRole task is to label the reaction roles including the product, reactant, catalyst, temperature, solvent, time, and yield in the paragraph. The Paragraph2MOFInfo task is to extract all MOF synthesis information including the compound name, metal source, metal amount, linker, linker amount, modulator, modulator amount or volume, solvent, solvent volume, reaction temperature and reaction time. The Paragraph2NMR task is designed to extract the IUPAC name, experimental conditions including frequency and solvent as well as chemical shift data for both 1H NMR and 13C NMR spectra. The Paragraph2Action task is to convert experimental procedures to structured synthetic steps (action sequences). The details of datasets used for the five chemical text mining tasks are listed in Table S1.† All tasks are unified to sequence-to-sequence formats to facilitate the use of LLMs. The details about using LLMs with prompt-engineering and fine-tuning can be found in the Methods section.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 samples, followed by randomly picking 1000, then 100, and finally 10. This sampling process ensures that each smaller subset is included in the larger one, with each subset used for individual training. Fig. 2b demonstrates the performance of prompt-only models and fine-tuned models, which are evaluated on a consistent evaluation set of 1000 samples across varying training data sizes. These results are obtained from three independent trials. In the case of prompt-only models, randomness is intentionally introduced by altering the prompt and examples (Fig. 2c and S2†). Given the task's straightforward nature and clear instructions, even the prompt-only language models achieved decent F1 scores over 0.6. For fine-tuned models, the sampling and training process for the training set is repeated three times, as depicted in Fig. 2a. As shown in Fig. 2b, all fine-tuned models demonstrate a performance improvement, especially in terms of the F1 score and Jaccard index, proportional to the increase in dataset size. These models outperform the prompt-only models designed for this task. When the training data size is substantial enough, the F1 scores of the fine-tuned models can reach close to 90%, and the Jaccard index can approach 80%. Notably, fine-tuned LLMs such as GPT-3.5-turbo showed minimal fluctuations and superior performance. However, it is essential to emphasize that the cost of fine-tuning GPT-3.5-turbo increased tenfold with each tenfold increase in data volume. Our experimentation was capped at 10
000 samples, followed by randomly picking 1000, then 100, and finally 10. This sampling process ensures that each smaller subset is included in the larger one, with each subset used for individual training. Fig. 2b demonstrates the performance of prompt-only models and fine-tuned models, which are evaluated on a consistent evaluation set of 1000 samples across varying training data sizes. These results are obtained from three independent trials. In the case of prompt-only models, randomness is intentionally introduced by altering the prompt and examples (Fig. 2c and S2†). Given the task's straightforward nature and clear instructions, even the prompt-only language models achieved decent F1 scores over 0.6. For fine-tuned models, the sampling and training process for the training set is repeated three times, as depicted in Fig. 2a. As shown in Fig. 2b, all fine-tuned models demonstrate a performance improvement, especially in terms of the F1 score and Jaccard index, proportional to the increase in dataset size. These models outperform the prompt-only models designed for this task. When the training data size is substantial enough, the F1 scores of the fine-tuned models can reach close to 90%, and the Jaccard index can approach 80%. Notably, fine-tuned LLMs such as GPT-3.5-turbo showed minimal fluctuations and superior performance. However, it is essential to emphasize that the cost of fine-tuning GPT-3.5-turbo increased tenfold with each tenfold increase in data volume. Our experimentation was capped at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 training samples for 3 epochs due to OpenAI's limitations, resulting in a nearly 90-dollar expense to fine-tune GPT-3.5-turbo—a low cost-effective investment in computational resources. In contrast, other fine-tuned language models have displayed notable cost advantages in this relatively simple compound name entity recognition task.
000 training samples for 3 epochs due to OpenAI's limitations, resulting in a nearly 90-dollar expense to fine-tune GPT-3.5-turbo—a low cost-effective investment in computational resources. In contrast, other fine-tuned language models have displayed notable cost advantages in this relatively simple compound name entity recognition task.
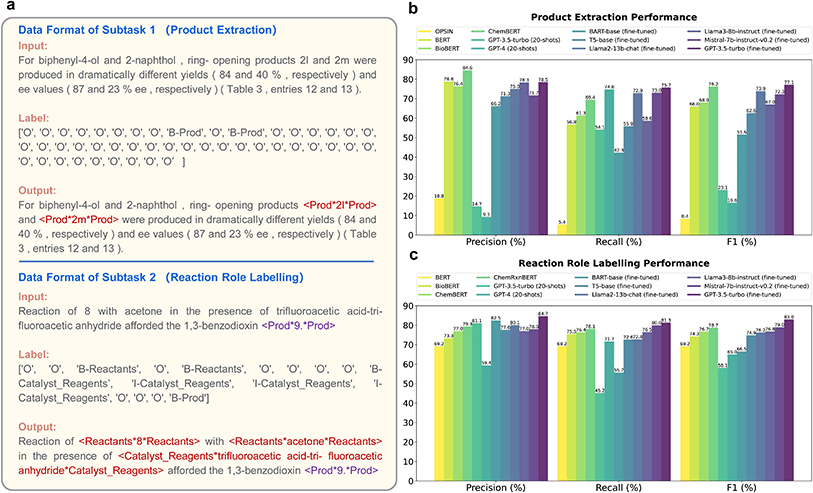 | ||
| Fig. 3 (a) Data formats of two subtasks in the Paragraph2RXNRole task. (b) Performance of product extraction. Concrete values can be found in Table S7.† (c) Performance of reaction role labelling. Concrete values can be found in Table S8.† | ||
For product extraction, the fine-tuned GPT-3.5-turbo (best over one epoch) achieved an F1 score of 77.1%, slightly surpassing the previous SOTA approach, ChemBERT, which scored 76.2% (Fig. 3b). For reaction role labelling, the fine-tuned GPT-3.5-turbo (best over five epochs) achieved an F1 score of 83.0%, significantly outperforming the previous SOTA approach, ChemRxnBERT, which scored 78.7% (Fig. 3c). It's notable that the fine-tuned GPT-3.5-turbo models, which cost only $1 and $5 respectively, demonstrated extremely high cost-effectiveness with small training datasets. In contrast, ChemBERT was domain-adaptive pre-trained on 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 478
478![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 043 sentences from 200
043 sentences from 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 journal articles, and ChemRxnBERT was further task-adaptive trained on 944
000 journal articles, and ChemRxnBERT was further task-adaptive trained on 944![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 733 reaction-inclusive sentences. We should also mention that the outputs of fine-tuned GPTs, Mistrals and Llamas align almost perfectly with the input text, with over 99% post-processing-free ratios. On the other hand, most outputs of fine-tuned T5 and BART require additional alignment due to their tokenization and vocabulary limitations, with a ratio of only 31% that does not require post-processing. Even after post-processing, the F1 scores of T5 and BART were significantly lower than those of token-classification BERT-like models or large language models.
733 reaction-inclusive sentences. We should also mention that the outputs of fine-tuned GPTs, Mistrals and Llamas align almost perfectly with the input text, with over 99% post-processing-free ratios. On the other hand, most outputs of fine-tuned T5 and BART require additional alignment due to their tokenization and vocabulary limitations, with a ratio of only 31% that does not require post-processing. Even after post-processing, the F1 scores of T5 and BART were significantly lower than those of token-classification BERT-like models or large language models.
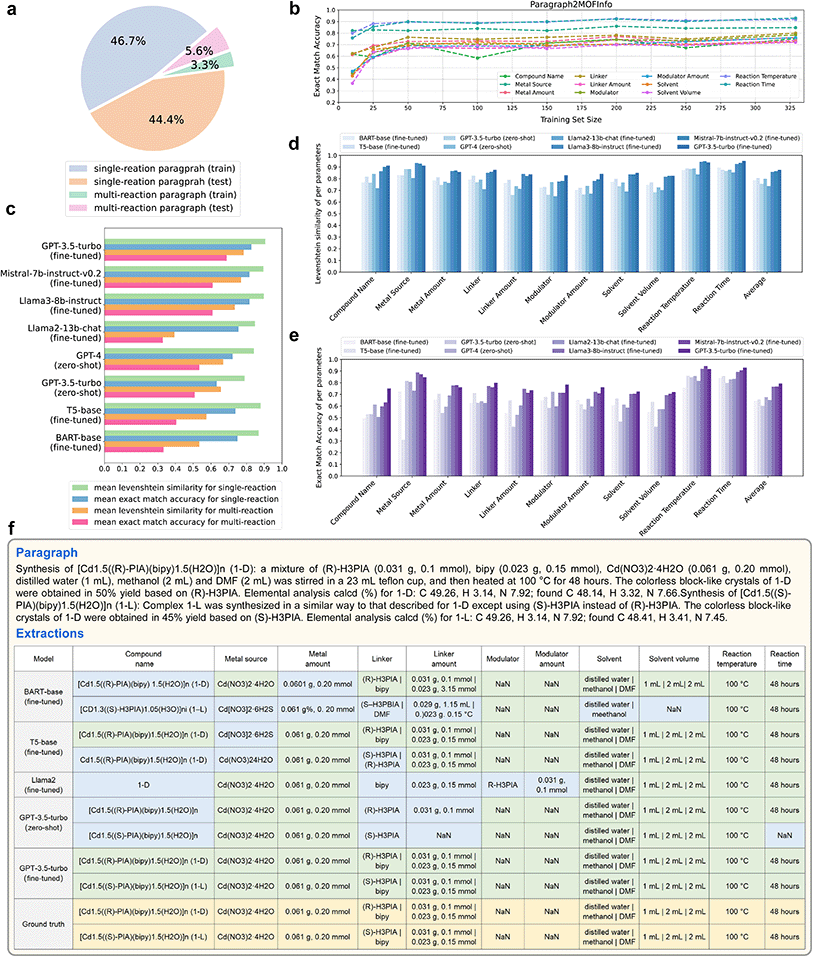 | ||
| Fig. 4 (a) A statistic of the Paragraph2MOFInfo dataset. (b) The performance of fine-tuned GPT-3.5-turbo across varying sizes of the training set. (c) Mean performance of Levenshtein similarity and exact match accuracy for extracting paragraphs containing single reactions and multiple reactions, respectively, by different models. Concrete values can be found in Table S9.† (d) Levenshtein similarity for 11 parameters in the Paragraph2MOFInfo task. Concrete values can be found in Table S10.† (e) Exact match accuracy for 11 parameters in the Paragraph2MOFInfo task. Concrete values can be found in Table S11.† (f) An example of extractions by different models from a multi-reaction MOF synthesis paragraph. The cells in yellow represented the ground truth. The cells in green represented the exact match predictions. The cells in blue represented the incorrect predictions. | ||
Exact accuracy rates for single and multiple reactions are 82.7% and 68.8%, respectively (Fig. 4c). As depicted in Fig. 4d and e, while most models achieve high Levenshtein similarity across the 11 parameters, only a few maintain high exact accuracy, which is the golden metric that we mainly focus on.
Considering that some MOF synthesis paragraphs may include multiple reactions, we provide an example of multi-reaction extraction by various models in Fig. 4f. The paragraph includes two reactions, the first with (R)-H3PIA and bipy as linkers, providing all reaction conditions explicitly, and the second with the substitution of (R)-H3PIA with (S)-H3PIA, keeping all other conditions unchanged. Most models successfully interpreted the semantics and extracted two reactions from the MOF synthesis paragraph. However, only the fine-tuned ChatGPT perfectly extracted information that matched our annotated ground truth. Other models showed varying degrees of incompleteness, particularly with items involving multiple components and their quantities.
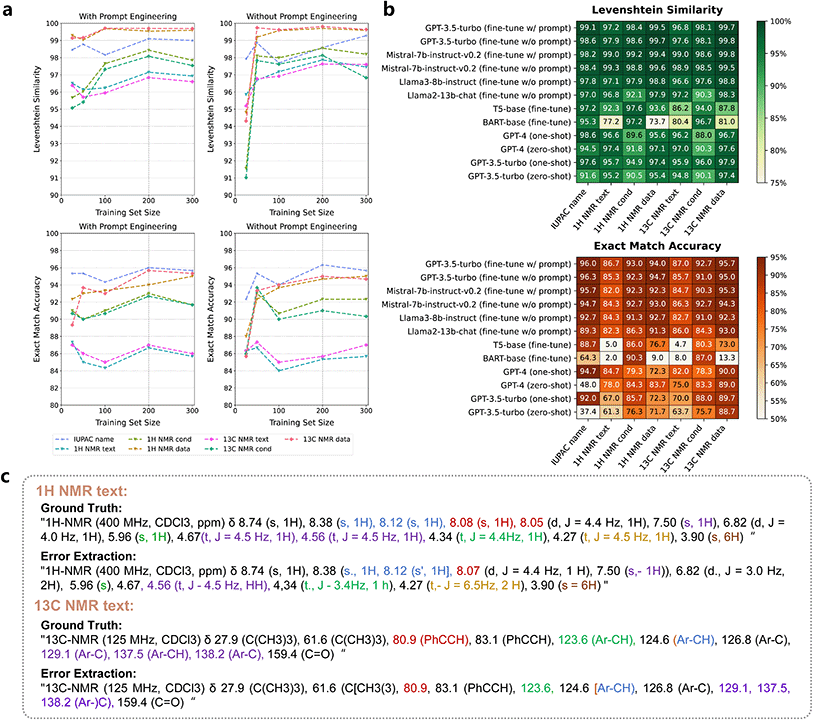 | ||
| Fig. 5 (a) The performance of fine-tuned GPT-3.5-turbo with and without prompt engineering as it varies with training data size in the Pargraph2NMR task. (b) Heat map illustrating Levenshtein similarity and exact match accuracy of various models in extracting NMR information. Concrete values can be found in Tables S12 and S13.† (c) Examples of error extractions by T5 and BART, compared with the ground truth. | ||
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168 augmented data. Interestingly, further improvement was achieved by augmenting the training data size to 14
168 augmented data. Interestingly, further improvement was achieved by augmenting the training data size to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168 when fine-tuning GPT-3.5-turbo. This resulted in 69.0% full sentence exact accuracy, an 86.4 modified BLEU score, and an 89.9% Levenshtein similarity (Table 1). For autonomous robots, it is challenging to generate instructions that follow strict syntax rules. Fine-tuning LLMs plays a crucial role in bridging the gap between fuzzy natural language and structured machine-executable programming languages, significantly improving the accuracy of customization with a small amount of annotated data. In similar tasks involving “fuzzy rules” or hard-to-define extraction, fine-tuning LLMs might offer considerable advantages in tailoring the transformation.
168 when fine-tuning GPT-3.5-turbo. This resulted in 69.0% full sentence exact accuracy, an 86.4 modified BLEU score, and an 89.9% Levenshtein similarity (Table 1). For autonomous robots, it is challenging to generate instructions that follow strict syntax rules. Fine-tuning LLMs plays a crucial role in bridging the gap between fuzzy natural language and structured machine-executable programming languages, significantly improving the accuracy of customization with a small amount of annotated data. In similar tasks involving “fuzzy rules” or hard-to-define extraction, fine-tuning LLMs might offer considerable advantages in tailoring the transformation.
| Model | Strategy | 100% acc. | 90% acc. | 75% acc. | Modified BLEU score | Levenshtein similarity | Cost |
|---|---|---|---|---|---|---|---|
| a The symbol “*” represented the results reported by Vaucher et al. The result in black bold is the best performance. The details of fine-tuning cost can be found in Table S3. | |||||||
| GPT-3.5-turbo (6-shot) | Prompt engineering without fine-tuning | 8.2 | 16.8 | 34.7 | 38.6 | 59.4 | 905 mean tokens |
| GPT-3.5-turbo (12-shot) | 8.8 | 19.3 | 42.3 | 43.1 | 62.3 | 1374 mean tokens | |
| GPT-3.5-turbo (18-shot) | 13.1 | 23.3 | 42.6 | 44.4 | 64.3 | 1670 mean tokens | |
| GPT-3.5-turbo (24-shot) | 14.8 | 25.9 | 45.5 | 47.0 | 65.8 | 2598 mean tokens | |
| GPT-3.5-turbo (30-shot) | 13.9 | 26.4 | 47.2 | 49.5 | 66.0 | 3610 mean tokens | |
| GPT-4 (6-shot) | Prompt engineering without fine-tuning | 13.4 | 23.3 | 44.9 | 44.7 | 54.5 | 861 mean tokens |
| GPT-4 (12-shot) | 20.7 | 30.7 | 51.1 | 51.4 | 69.2 | 1357 mean tokens | |
| GPT-4 (18-shot) | 21.9 | 33.0 | 56.5 | 53.8 | 63.0 | 1631 mean tokens | |
| GPT-4 (24-shot) | 22.7 | 35.8 | 58.2 | 56.7 | 65.1 | 2546 mean tokens | |
| GPT-4 (30-shot) | 26.1 | 40.0 | 61.6 | 59.8 | 67.7 | 3611 mean tokens | |
| GPT-4 (60-shot) | 32.7 | 43.8 | 63.3 | 65.0 | 72.8 | 7010 mean tokens, $ 41 | |
| Transformer (single model)* | No task-adaptive pretraining and fine-tuning on hand-annotated data (1060) | 13.1 | 15.1 | 21.9 | 22.5 | 45.9 | — |
| BART-base (fine-tuned) | 51.1 | 65.9 | 77.6 | 73.2 | 83.9 | 6 min on 1 × 40 GB A100 | |
| T5-base (fine-tuned) | 57.7 | 71.6 | 83.2 | 81.8 | 86.8 | 10 min on 1 × 40 GB A100 | |
| Lama2-13b-chat (qlora fine-tuned) | 56.8 | 66.8 | 80.7 | 80.3 | 86.0 | 40 min on 1 × 40 GB A100 | |
| Lama3-8b-instruct (fine-tuned) | 59.7 | 70.2 | 83.2 | 82.2 | 86.3 | 30 min on 4 × 40 GB A100 | |
| Mistral-7b-instruct-v0.2 (fine-tuned) | 64.8 | 73.6 | 85.5 | 85.9 | 88.7 | 8 min on 4 × 40 GB A100 | |
| GPT-3.5-turbo (fine-tuned) | 63.6 | 71.6 | 82.7 | 84.8 | 88.1 | 4 epochs, total 1 h, $ 4 | |
| Transformer (single model)* | No task-adaptive pretraining and fine-tuning on augmented data (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168) 168) |
37.8 | 47.7 | 62.8 | 64.7 | 76.4 | — |
| BART-base (fine-tuned) | 52.0 | 68.5 | 80.1 | 74.4 | 84.8 | 30 min on 1 × 40 GB A100 | |
| T5-base (fine-tuned) | 59.7 | 74.1 | 82.4 | 84.1 | 87.1 | 100 min on 1 × 40 GB A100 | |
| Llama2-13b-chat (qlora fine-tuned) | 62.2 | 71.6 | 84.1 | 84.3 | 87.5 | 5 hours on 1 × 40 GB A100 | |
| Lama3-8b-instruct (fine-tuned) | 56.0 | 67.0 | 80.4 | 81.4 | 84.8 | 100 min on 4 × 40 GB A100 | |
| Mistral-7b-instruct-v0.2 (fine-tuned) | 64.2 | 73.3 | 86.4 | 84.3 | 87.2 | 30 min on 4 × 40 GB A100 | |
| GPT-3.5-turbo (fine-tuned) | 69.0 | 78.1 | 86.9 | 86.4 | 89.9 | 5 epochs, total 1.5 h, $ 92 | |
| Transformer (single model)* | Task-adaptive pretraining (2 M) and fine-tuning on hand-annotate data (1060) | 56.8 | 67.3 | 80.4 | 81.5 | 85.7 | — |
| Transformer (single model)* | Task-adaptive pretraining (2 M) and fine-tuning on augmented data (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 168) 168) |
59.4 | 70.5 | 81.8 | 84.3 | 86.7 | — |
| Transformer (ensemble)* | 60.8 | 71.3 | 82.4 | 85.0 | 86.6 | — | |
Comparison of different methods for chemical text mining
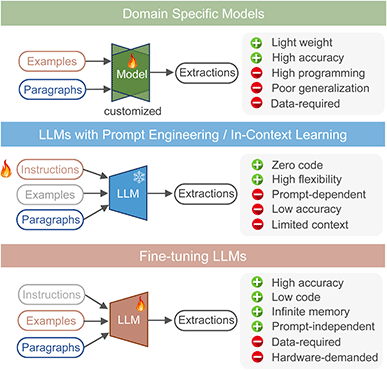
Chemical text mining expedites scientific discovery in chemistry. Previously, tasks involving complex chemical language and sophisticated processing depended on rule-based matching algorithms and custom-built domain-specific models. Now, leveraging universal LLMs' semantic understanding, long context window, and generation abilities offers promising and general approaches. These methods are illustrated in Fig. 6. In prompt engineering scenarios, LLMs' parameters remain fixed, solely relying on the provided examples to extract from new paragraphs. As for the training and fine-tuning process, a model learns the statistic extraction patterns from the training data by adjusting and optimizing the internal parameters.Undoubtedly, leveraging LLMs with prompt engineering is the most attractive approach because it does not require writing any code or retraining model parameters, only interacting with the large model through natural language instructions. However, relying solely on instructions without any examples (zero-shot) also makes it difficult to standardize the output of LLMs, which is crucial for formatting data extraction tasks. In the case of extracting NMR based solely on instructions (Fig. S8†), we repeatedly modify the instructions to ensure that the model can generate expected formatting results on a certain paragraph. However, when we used this carefully designed prompt for other paragraphs containing NMR, the extraction results did not meet the qualified formatting requirements again. This zero-shot approach resulted in poor performance across all five tasks, even using GPT-4.
Apart from instructions, providing few example pairs of paragraph-extraction as context can help LLMs learn the extraction patterns. In these few-shot sceneries (Fig. 2c, S2–S7†), as shown in Table 1, increasing the number of examples leads LLMs to extract more structured outputs. Ideally, the whole training set should serve as context. However, the upper limit of in-context learning is constrained by the maximum input length due to the memory limitation. The versions of GPT-3.5-Turbo-0613 and GPT-4-0613 we tested were limited to 4096 and 8192 tokens, respectively. Hence, comparing prompt engineering methods in zero-shot and few-shot sceneries to fine-tuned models trained with complete datasets can be somewhat unfair.
To compare the performance of in-context learning and fine-tuning approaches objectively, we should use an equal number of examples for both context and the fine-tuning data set. Here, we tested the latest version of GPT-3.5-turbo-0125, which expands the context length to 16 K and supports fine-tuning. We used a variety of action sequences during sampling to cover as many action types as possible. As the number of examples increased from 30 to 60, 90 and 120, both the performances of in-context learning and fine-tuning are increasing (Table S14†). Even when the same number of examples was provided for in-context learning as fine-tuning, the fine-tuned model typically outperforms by 10–20% on metrics like exact match accuracy and modified BLEU score. This could be attributed to information loss in in-context learning, while fine-tuning adjusts parameters to learn extraction patterns, thus maintaining higher accuracy.
In the test, we also find two features of fine-tuning LLMs: rapid performance convergence with small amounts of data and efficient training generalization. For the four tasks utilizing manually annotated data, the LLM's performance rapidly improved and converged with increasing sample sizes (Fig. S11†). This highlights that hundreds of high-quality data are enough to train an effective extractor, which is typically a manageable workload for manual annotation. Besides, LLMs can be easily adapted for specific text extraction tasks, requiring only a few epochs and a low learning rate for fine-tuning (Table S3†). However, they are also prone to overfitting if trained for an excessive number of epochs.
Starting with five chemical extraction tasks, we have proved the effectiveness of fine-tuning LLMs in the relatively small testing sets. This approach, when utilized for large-scale extraction in the future, promises to greatly improve data collection efficiency and accelerate scientific research and experimentation. For the Paragraph2MOFInfo task, we can document the synthesis conditions along with other key information such as MOF structures, pore characteristics, and functional performance. Using these data, we can develop machine learning models to optimize the synthesis novel MOF materials with functions such as new catalysts, gas storage and separation. For the Paragraph2NMR task, we can collect extensive NMR data with the corresponding compound names from millions of synthesis literature documents. This can help create an NMR database for retrieving similar spectra and structures, as well as constructing predictive models to identify molecules structures and analysing complex mixtures, which support drug development and quality control. For the action sequence transformation task, the extracted information is beneficial for automatic and robotic synthesis. It will improve reproducibility and minimize human errors, especially in high-throughput experiments.
Apart from the five mentioned extraction tasks, it can be easily extended to tasks related to extracting information from scientific literature and transforming data into a simple user-friendly reaction format22 that is both human- and machine-readable. This approach will significantly contribute to the development of extensive databases like the Open Reaction Database,23,24 SciFinder25 and Reaxys,26 which gather comprehensive synthesis data through automated curation and expert verification, to make data more findable, accessible, interoperable, and reusable (FAIR).
Nevertheless, leveraging fine-tuned LLMs is still insufficient to extract all synthesis information from chemical literature, which contains extensive complex figure and form contents. Recently, some tools have been developed to recognize molecular images27,28 and reaction diagrams29,30 from the literature. Integrating LLMs with these image recognition tools or developing advanced large multimodal models (LMMs) may be a promising unified solution for further chemical data mining. Notably, when extracting large amounts of data from copyrighted literature, it's essential to access the necessary permissions from scientific publications.
Herein, we have scratched the surface of the vast potential of LLMs in chemistry and materials science by fine-tuning LLMs for chemical text mining. We may notice that the gap between open-source language models and proprietary GPTs (GPT-3.5-turbo and GPT-4) has been narrowing from Llama2 to Llama3 and Mistral. This progress is due to the concerted efforts of researchers and communities in the direction of LLMs. Technically, advancements like more effective fine-tuning strategies, improved open-source model architectures, faster inference approaches, wider context windows, higher quality corpus, and lower computational costs in the era of LLMs are anticipated to further enhance text mining. Meanwhile, it's more essential to consider what else can be achieved with LLMs and how we can develop more effective LLMs for chemistry and materials science. For instance, LLMs have the potential to revolutionize predictive modelling by incorporating the extensive “fuzzy knowledge” encapsulated within scientific literature, especially in chemistry and drug discovery. By combining empirical results with documented knowledge, LLMs could assist chemists identify patterns in experiments that might otherwise be missed, predict properties of compounds and outcomes of reactions, and even generate new chemical hypotheses and theories. Furthermore, the integration of LLMs' comprehension with specialized tools could substantially lower the barrier of chemists to use these tools throughout the entire workflow, thanks to interactive interfaces in natural language. Future research could investigate how to merge formatted laboratory data with the wealth of information in scientific literature and develop the multimodal capability to enrich specific domain knowledge for LLMs. This endeavour will require a sustained, long-term effort.
Conclusions
In this work, we have demonstrated the effectiveness of fine-tuning LLMs in chemical text mining. We conducted five complex tasks: compound entity recognition, reaction role labelling, MOF synthesis information extraction, NMR data extraction, and the transformation of reaction procedures to action sequences. Chemical text mining remains a challenging professional domain when leveraging language model mining, even with prompt engineering. However, LLMs that are fine-tuned with appropriate annotations can produce structured outputs that perfectly fulfil human requirements not easily expressed in natural language. This feature fully utilizes their natural language understanding and formatting capability. Using chemical text mining as an example, this study provides guidance on fine-tuning of LLMs to serve as universal knowledge extraction toolkits. These toolkits can be easily extended for automated extraction from documents and rule-based formatted transformations. Our work lays the groundwork for the applications of LLMs in information extraction within the chemical domain, which will catalyse data-driven innovations in chemical and materials science.Methods
Dataset preparation
For the Paragraph2Compound task, we compiled an automatically annotated dataset. This dataset is based on the publicly accessed USPTO subset extracted by Lowe et al.,31,32 and includes millions of chemical reaction paragraphs from patents, each paired with compound tags. We used regular expressions to identify compound labels within each paragraph, separating them with the “|” symbol based on their sequential occurrence in the paragraph. For the Paragraph2RXNRole task, we used the manually annotated dataset by Guo et al.,8 following the same data partitioning strategy. We transformed the data from the BIO-token classification format into a sequence-to-sequence format using the annotation scheme “<Role*compound*Role>”. We processed paragraphs containing multiple central products and related reactions into several input and output pairs. For the Paragraph2MOFInfo task, we manually checked and re-annotated the raw data of Zheng et al.,14 transforming them into a sequence-to-sequence format. This dataset comprises MOF synthesis paragraphs, extraction by ChatGPT, and human-evaluated answers. For the Paragraph2NMR task, we manually curated a dataset of 600 high-quality annotations. These were mainly sourced from various literature studies on PubMed to ensure a wide diversity. The task aims to extract information such as the IUPAC name, experimental conditions, including the frequency and solvent, and chemical shift data from both 1H NMR and 13C NMR spectra. For the Paragraph2Action task, we utilized the hand-annotated dataset by Vaucher et al., employing the same data partitioning strategy. This dataset is derived from the Pistachio dataset by NextMove software.33 The details of datasets used for the five chemical text mining tasks are listed in Table S1.†Prompt-only ChatGPT
Prompt-only interaction enables users to efficiently communicate with large language models through simple prompts. This guides the model to produce relevant responses without further training. In a zero-shot scenario, the model generates responses using only a descriptive prompt and its pre-trained knowledge. However, in a few-shot approach, the model uses a small number of examples to improve its understanding and responses. To maximize the performance, we selected diverse examples and ensured a large number of tokens. We interacted with ChatGPT using API keys and employed model versions GPT-3.5-turbo-0613 and GPT-4-0613. The zero-shot and few-shot prompts for chemical text mining tasks can be found in Fig. S2–S8.†Fine-tuning ChatGPT
Since late August 2023, supervised fine-tuning capabilities have been available for the GPT-3.5-turbo model.34 The aim is to enhance performance in specific scenarios customized based on private data. In this study, we fine-tuned the GPT-3.5-turbo-0613 model for chemical text mining scenarios on five tasks. When discussing the performance in the Comparison of different methods for chemical text mining section, we fine-tuned the latest GPT-3.5-turbo-0125 model for fair comparison, which expanded the context length to 16 K and supported fine-tuning as well. We formatted the data into jsonl and uploaded them to OpenAI's cloud servers, and then initiated fine-tuning jobs. Once the training was complete, the fine-tuned GPT-3.5-turbo model was ready for inference. API keys were requisite throughout the training and inference procedures. Fine-tuning for the GPT-4-turbo model is not available now and is highly expected in the future.Fine-tuning open-source language models
We selected the most widely used and representative generative pre-trained language models such as Mistral,17 Llama3,18 Llama2,19 T5,20 and BART.21 These serve as baselines for a comprehensive comparison with the fine-tuned ChatGPT across five chemical text mining tasks. Considering performance, efficiency, and hardware resource constraints, we used full parameter fine-tuning for Mistral-7b-instruct-v0.2 and Llama3-8b-instruct on 4 × 40 GB A100, and full parameter fine-tuning for BART-base and T5-base on 1 × 40 GB A100. We applied multitask-learning to BART and T5 in the Paragraph2MOFInfo task and Paragraph2NMR task due to their limitations in generating multi-attribute long sentences (Fig. S9 and S10†), aiming to enhance their performance. This approach significantly improved their performance. For Llama2, we used Q-LoRA35 to efficiently fine-tune llama2-13b-chat on 1 × 40 GB A100. This method maintains most of the performance of full parameter fine-tuning while significantly reducing computational demands. We used vllm36 to speed up the inference of LLMs such as Mistral-7b-instruct-v0.2, Llama3-8b-instruct, and Llama2-13b-chat, which is tens of times faster than Hugging Face's pipeline. The inference of all fine-tuned models can run on 1 × 40 GB A100. To ensure optimal performance, we adjusted hyperparameters such as learning rates, lora_r, and lora_alpha during the fine-tuning process of baseline models (Table S2†). The hardware resources, memory cost, and runtimes of fine-tuning are provided for reference (Table S3†). More details of training, pre-processing, and post-processing can be found in the ESI.†Metrics for evaluation
Since fine-tuning ChatGPT does not allow for early stopping based on optimal validation loss, we report the performances of all models at the best epoch selected from the evaluation set for fair comparison. Given the task specifics, we use metrics including precision, recall, and F1 score for evaluating entity-level performance. For sentence-level performance assessment, we use Levenshtein similarity, exact match accuracy, partial accuracy, and a modified BLEU score.Data availability
All data and code of this work are available at GitHub: https://github.com/zw-SIMM/SFTLLMs_for_ChemText_Mining to allow replication of processing, fine-tuning and evaluation. All concrete values of performance in figures are listed in Section 5 of the ESI.†Author contributions
W. Z., J. C. X., Z. Y. F., and M. Y. Z. conceived the idea. M. Y. Z. and Z. Y. F. designed the research. W. Z., Q. G. W., and Z. M. H. implemented the codes. W. Z., Q. G. W., X. T. K., J. C. X., S. K. N., and Z. Y. F. collected, annotated, and processed the training data. D. H. C., B. Y. N., Q. S., and X. T. L. checked the data. Y. M. L., M. A. C., R. Z. Z., Y. T. W., and L. H. Z. benchmarked the models. W. Z. wrote the initial draft. M. Y. Z., Z. Y. F. and Z. P. X. reviewed and refined the article. All authors contributed to the analysis of the results. All authors read and approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank all contributions of the open-source community on LLMs. We appreciate Yaghi's group for guiding in ChatGPT prompt engineering for chemistry tasks. This work was supported by the National Natural Science Foundation of China (T2225002 and 82273855 to M. Y. Z. and 82204278 to X. T. L.), the National Key Research and Development Program of China (2022YFC3400504 to M. Y. Z.), the SIMM-SHUTCM Traditional Chinese Medicine Innovation Joint Research Program (E2G805H to M. Y. Z.), the Shanghai Post-doctoral Excellence Program (2023693 to Z. Y. F.) and the Shanghai Municipal Science and Technology Major Project.References
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Automated extraction of chemical synthesis actions from experimental procedures, Nat. Commun., 2020, 11, 3601 CrossRef PubMed.
- M. Suvarna, A. C. Vaucher, S. Mitchell, T. Laino and J. Pérez-Ramírez, Language models and protocol standardization guidelines for accelerating synthesis planning in heterogeneous catalysis, Nat. Commun., 2023, 14, 7964 CrossRef CAS PubMed.
- S. H. M. Mehr, M. Craven, A. I. Leonov, G. Keenan and L. Cronin, A universal system for digitization and automatic execution of the chemical synthesis literature, Science, 2020, 370, 101–108 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson and D. Angelone, Organic synthesis in a modular robotic system driven by a chemical programming language, Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- T. Ha, D. Lee, Y. Kwon, M. S. Park, S. Lee, J. Jang, B. Choi, H. Jeon, J. Kim and H. Choi, AI-driven robotic chemist for autonomous synthesis of organic molecules, Sci. Adv., 2023, 9, eadj0461 CrossRef CAS PubMed.
- M. C. Swain and J. M. Cole, ChemDataExtractor: a toolkit for automated extraction of chemical information from the scientific literature, J. Chem. Inf. Model., 2016, 56, 1894–1904 CrossRef CAS PubMed.
- J. Mavracic, C. J. Court, T. Isazawa, S. R. Elliott and J. M. Cole, ChemDataExtractor 2.0: autopopulated ontologies for materials science, J. Chem. Inf. Model., 2021, 61, 4280–4289 CrossRef CAS PubMed.
- J. Guo, A. S. Ibanez-Lopez, H. Gao, V. Quach, C. W. Coley, K. F. Jensen and R. Barzilay, Automated chemical reaction extraction from scientific literature, J. Chem. Inf. Model., 2021, 62, 2035–2045 CrossRef PubMed.
- C. M. Castro Nascimento and A. S. Pimentel, Do Large Language Models Understand Chemistry? A Conversation with ChatGPT, J. Chem. Inf. Model., 2023, 63, 1649–1655 CrossRef CAS PubMed.
- T. M. Clark, E. Anderson, N. M. Dickson-Karn, C. Soltanirad and N. Tafini, Comparing the Performance of College Chemistry Students with ChatGPT for Calculations Involving Acids and Bases, J. Chem. Educ., 2023, 100, 3934–3944 CrossRef CAS.
- T. Guo, K. Guo, Z. Liang, Z. Guo, N. V. Chawla, O. Wiest and X. Zhang, arXiv, 2023, preprint, arXiv:2305.18365, DOI:10.48550/arXiv.2108.09926.
- Z. Ji, N. Lee, R. Frieske, T. Yu, D. Su, Y. Xu, E. Ishii, Y. J. Bang, A. Madotto and P. Fung, Survey of hallucination in natural language generation, ACM Comput. Surv., 2023, 55, 1–38 CrossRef.
- Y. Zhang, Y. Li, L. Cui, D. Cai, L. Liu, T. Fu, X. Huang, E. Zhao, Y. Zhang and Y. Chen, arXiv, 2023, preprint, arXiv:2309.01219, DOI:10.48550/arXiv.2309.01219.
- Z. Zheng, O. Zhang, C. Borgs, J. T. Chayes and O. M. Yaghi, ChatGPT Chemistry Assistant for Text Mining and the Prediction of MOF Synthesis, J. Am. Chem. Soc., 2023, 145, 18048–18062 CrossRef CAS PubMed.
- L. Patiny and G. Godin, ChemRxiv, 2023 DOI:10.26434/chemrxiv-2023-05v1b-v2.
- Q. Chen, H. Sun, H. Liu, Y. Jiang, T. Ran, X. Jin, X. Xiao, Z. Lin, H. Chen and Z. Niu, An Extensive Benchmark Study on Biomedical Text Generation and Mining with ChatGPT, Bioinformatics, 2023, btad557 CrossRef CAS PubMed.
- A. Q. Jiang, A. Sablayrolles, A. Mensch, C. Bamford, D. S. Chaplot, D. d. l. Casas, F. Bressand, G. Lengyel, G. Lample and L. Saulnier, arXiv, 2023, preprint, arXiv:2310.06825, DOI:10.48550/arXiv.2310.06825.
- Llama3, https://llama.meta.com/llama3/, accessed April 26, 2024 Search PubMed.
- H. Touvron, L. Martin, K. Stone, P. Albert, A. Almahairi, Y. Babaei, N. Bashlykov, S. Batra, P. Bhargava and S. Bhosale, arXiv, 2023, preprint, arXiv:2307.09288, DOI:10.48550/arXiv.2307.09288.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, Exploring the limits of transfer learning with a unified text-to-text transformer, J. Mach. Learn. Res., 2020, 21, 5485–5551 Search PubMed.
- M. Lewis, Y. Liu, N. Goyal, M. Ghazvininejad, A. Mohamed, O. Levy, V. Stoyanov and L. Zettlemoyer, arXiv, 2019, preprint, arXiv:1910.1346, DOI:10.48550/arXiv.1910.13461.
- D. F. Nippa, A. T. Müller, K. Atz, D. B. Konrad, U. Grether, R. E. Martin and G. Schneider, ChemRxiv, 2024, preprint, DOI:10.26434/chemrxiv-2023-nfq7h-v2.
- S. M. Kearnes, M. R. Maser, M. Wleklinski, A. Kast, A. G. Doyle, S. D. Dreher, J. M. Hawkins, K. F. Jensen and C. W. Coley, The open reaction database, J. Am. Chem. Soc., 2021, 143, 18820–18826 CrossRef CAS PubMed.
- R. Mercado, S. M. Kearnes and C. W. Coley, Data sharing in chemistry: lessons learned and a case for mandating structured reaction data, J. Chem. Inf. Model., 2023, 63, 4253–4265 CrossRef CAS PubMed.
- SciFinder, https://scifinder-n.cas.org, accessed August 29, 2023 Search PubMed.
- Reaxys, https://www.reaxys.com, accessed August 29, 2023 Search PubMed.
- J. Xiong, X. Liu, Z. Li, H. Xiao, G. Wang, Z. Niu, C. Fei, F. Zhong, G. Wang and W. Zhang, αExtractor: a system for automatic extraction of chemical information from biomedical literature, Sci. China: Life Sci., 2023, 67, 618–621 CrossRef PubMed.
- Y. Qian, J. Guo, Z. Tu, Z. Li, C. W. Coley and R. Barzilay, MolScribe: Robust Molecular Structure Recognition with Image-to-Graph Generation, J. Chem. Inf. Model., 2023, 63, 1925–1934 CrossRef CAS PubMed.
- Y. Qian, J. Guo, Z. Tu, C. W. Coley and R. Barzilay, RxnScribe: A Sequence Generation Model for Reaction Diagram Parsing, J. Chem. Inf. Model., 2023, 63, 4030–4041 CrossRef CAS PubMed.
- D. M. Wilary and J. M. Cole, ReactionDataExtractor 2.0: a deep learning approach for data extraction from chemical reaction schemes, J. Chem. Inf. Model., 2023, 63, 6053–6067 CrossRef CAS PubMed.
- D. Lowe, Chemical reactions from US patents (1976-Sep2016), https://figshare.com/articles/dataset/Chemical_reactions_from_US_patents_1976-Sep2016_/5104873, accessed August 29, 2023, DOI:10.6084/m9.figshare.5104873.v1.
- D. M. Lowe, PhD thesis, University of Cambridge, 2012.
- Pistachio, https://www.nextmovesoftware.com/pistachio.html, accessed August 22, 2023 Search PubMed.
- A. Peng, M. Wu, J. Allard, L. Kilpatrick and S. Heidel, GPT-3.5 Turbo fine-tuning and API updates, https://openai.com/blog/gpt-3-5-turbo-fine-tuning-and-api-updates, accessed August 22, 2023 Search PubMed.
- T. Dettmers, A. Pagnoni, A. Holtzman and L. Zettlemoyer, arXiv, 2023, preprint, arXiv:2305.14314, DOI:10.48550/arXiv.2305.14314.
- W. Kwon, Z. Li, S. Zhuang, Y. Sheng, L. Zheng, C. H. Yu, J. Gonzalez, H. Zhang and I. Stoica, presented in part at the Proceedings of the 29th Symposium on Operating Systems Principles, Koblenz, Germany, 2023 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc00924j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |