Open Access Article
Open Access ArticlePreorganized cyclic modules facilitate the self-assembly of protein nanostructures†
Jaka
Snoj
 ab,
Fabio
Lapenta‡
a and
Roman
Jerala
ab,
Fabio
Lapenta‡
a and
Roman
Jerala
 *ac
*ac
aDepartment of Synthetic Biology and Immunology, National Institute of Chemistry, Hajdrihova 19, SI-1000 Ljubljana, Slovenia. E-mail: roman.jerala@ki.si
bInterdisciplinary Doctoral Program in Biomedicine, University of Ljubljana, Kongresni trg 12, SI-1000 Ljubljana, Slovenia
cEN-FIST Centre of Excellence, Trg OF 13, SI-1000 Ljubljana, Slovenia
First published on 5th February 2024
Abstract
The rational design of supramolecular assemblies aims to generate complex systems based on the simple information encoded in the chemical structure. Programmable molecules such as nucleic acids and polypeptides are particularly suitable for designing diverse assemblies and shapes not found in nature. Here, we describe a strategy for assembling modular architectures based on structurally and covalently preorganized subunits. Cyclization through spontaneous self-splicing of split intein and coiled-coil dimer-based interactions of polypeptide chains provide structural constraints, facilitating the desired assembly. We demonstrate the implementation of a strategy based on the preorganization of the subunits by designing a two-chain coiled-coil protein origami (CCPO) assembly that adopts a tetrahedral topology only when one or both subunit chains are covalently cyclized. Employing this strategy, we further design a 109 kDa trimeric CCPO assembly comprising 24 CC-forming segments. In this case, intein cyclization was crucial for the assembly of a concave octahedral scaffold, a newly designed protein fold. The study highlights the importance of preorganization of building modules to facilitate the self-assembly of higher-order supramolecular structures.
1. Introduction
Self-assembling supramolecular structures can be designed de novo from well-understood modules, where the information governing the assembly is encoded in the chemical structure of building molecules. This can be a formidable task, as the assembly has to overcome a substantial entropic barrier. Therefore, strategies to facilitate the assembly include the use of modules that reduce the degrees of freedom and facilitate the design. Among the polypeptide modules that can facilitate the assembly of novel complexes, coiled-coils (CC) stand out as they can be designed de novo and finely tuned, as the rules underlying the specificity of the CC-forming peptides are well-understood and many orthogonal CCs are already available.1–6 CCs are super-secondary structural elements composed of two or more α-helices intertwined in a supercoiled left-handed helix. Importantly, they display high specificity and known rules governing their oligomerization, enabling rational design.7–10 Structural motifs of CCs offer a relatively straightforward rational connection between sequence and structure, allowing to control oligomerization,11,12 peptide chain orientation,13,14 interaction specificity6,15,16 and stability5 making them suitable modular building blocks for building nanostructures de novo.12,17–22The use of CC modules enabled the design of large and complex nanostructures such as nanofibers,23 nanotubes,24 nanocages19,25,26 and nanomotors.27 Dimeric CC units, however, present some difficulties when used for the construction of large multimeric structures. Most notably, the flexibility imposed by the linkers between CC segments, while playing an important role in the assembly may lead to different and at times heterogeneous oligomerization states.28 This has been for instance shown on short CC peptides designed to assemble into a large variety of complexes, such as bundles, triangles and squares by the selection of linker length.29,30 Sets of coiled-coil dimer-forming peptides that interact specifically with each other (orthogonal sets) can be used for designing polyhedral protein nanostructures known as coiled-coil protein origami (CCPO).7 CC peptides in such structures are arranged in a defined order in the primary structure and guide the polypeptide to fold into a polyhedral topology, from either a single19,31–33 or multiple chains.34,35 Interestingly, in the wake of the spectacular success of machine learning-based protein structure prediction and design,36–38 the fold of coiled-coil-based protein nanostructures cannot yet be efficiently predicted by these methods, presumably due to the absence of such structures from the learning set underlying machine learning algorithms and their complex fold topology.
We have previously shown how CCPO polyhedral shapes can self-assemble in cells during protein translation19 and undergo structural rearrangement according to which CC modules are selected and how they are placed within the sequence.34,35 However, expanding this design strategy further, and applying it to larger CCPO architectures composed of multiple chains, requires a significant shift in the design rules, especially of the inter-connecting linkers between the building blocks, whose length could lead to the formation of alternative, heterogeneous tertiary and quaternary structures.30 Moreover, the overall topology of CC-based proteins and the positioning of the terminal CC segments have to be taken into consideration when constructing large CC assemblies. In this regard, we have previously noted that CC peptide pairs with low stability tend to destabilize the nanostructures19 and that specific arrangements of the building blocks allow the interface of oligomeric CC-based nanostructures to adopt the desired conformation.35
We reasoned that covalent cyclization could introduce an additional constraint for the preorganization of building modules to facilitate the assembly. Although rare, natural cyclized proteins have increased stability against thermal stress and proteolysis.39,40 Protein cyclization has been already used in bioengineering, mainly to increase stability.41 Covalent bonds within polypeptide chains can be introduced by enzymatic tools such as transglutaminases,42 tyrosinases,43 split cellular anchoring proteins (SpyCatcher/SpyTag)44 and backbone cyclization with inteins. Inteins catalyse a posttranslational reaction, analogous to RNA splicing, where as a result of their activity, they are ultimately excised from the flanking peptides followed by ligation of the remaining parts.45,46 Since their discovery in 1990 in yeast,47 additional inteins have been uncovered48 as well as split inteins, which consist of two inactive parts that regain the splicing capability after their spatial reconstitution.49,50 The important biotechnological potential of this class of proteins was soon recognized and applied to protein cyclization and other applications, such as tag removal, labelling and purification.51 Since the discovery of the first split intein, based on Npu DnaE found in the DNA polymerase III of cyanobacteria Nostoc punctiforme;52,53 more efficient and stable split inteins have been discovered54 or engineered.55,56 Inteins are used to enhance protein stability by decreasing the conformational entropy of the unfolded state by protein cyclization.57 However; despite their versatility, CC-based nanostructures stabilized by intein splicing or intein cyclization have been used only to construct planar triangles.58 On the other hand, cyclization has already been implemented in designed DNA nanostructures and has been shown to increase their stability against exonucleases.59,60 CCPO designs, which like DNA nanostructures, rely on the discrete interactions between building block segments, could equally benefit from cyclization, acquiring higher stability and more conformational homogeneity.
In this work we approached the design of supramolecular assemblies, stabilized by cyclization using the in vivo splicing activity of the naturally occurring split intein gp41.54 Here, we propose that polypeptide chains with several unpaired CC segments can be used as building blocks once the interface and the termini of the CC-based subunit are stabilized in a prearranged conformation. We used this approach to construct CCPO topologies including a tetrahedral fold, composed of two chains as well as an irregular concave octahedral architecture composed of three chains, thus demonstrating the versatility of this strategy for designing multiple-chain assemblies from preorganized subunits.
2. Experimental
2.1. Molecular cloning and expression gene preparation
Plasmid propagation and vector transfer were carried out using the E. coli strain DH5-α (F– φ80lacZΔM15 Δ(lacZYA-argF) U169 recA1 endA1 hsdR17(rK–, mK+) phoA supE44 λ– thi-1 gyrA96 relA1) (NEB, MA USA).Synthetic genes were purchased from Twist Bioscience (CA USA) or IDT (IA USA) and DNA oligonucleotides used in PCR reactions were purchased from IDT (IA USA). Genes coding for the proteins of interest were cloned in two different expression vectors pET41a+ (Genscript, NJ USA) and pCIRCgp41-1 a gift from Barbara Di Ventura & Roland Eils (Addgene plasmid # 74227; http://n2t.net/addgene:74227; RRID: Addgene_74227).57 Reading frames were optimized for E. coli codon usage using a software property of IDT (IA USA).
Gibson assembly61 was used to introduce, substitute or delete DNA segments in the genes. Amplification of DNA fragments was done using repliQa HiFi ToughMix® (Quantabio, Beverly, MA, USA) or Phusion® HotStart DNA polymerase (NEB, MA USA) in PCR reactions performed according to manufacturer instructions. Gibson assembly was performed with a mixture of the enzymes Taq Ligase (NEB, MA, USA), Phusion® Polymerase and T5 exonuclease (NEB, MA, USA) in reaction buffer (NEB, MA. USA) following the formulation provided by the manufacturer.61 The mixture was incubated for 1 h at 50 °C before transformation in competent E. coli cells. The plasmid transformation process was carried out utilizing a heat shock technique and following the manufacturer's protocol. Single colonies were then cultured in the presence of the antibiotic Kanamycin (Goldbio, MO, USA), at a final concentration of 50 μg ml−1, in Lysogeny broth (LB) media.
2.2. Protein production and lysis
In this study, protein production was carried out using the E. coli strain NiCO21(DE3) (can:CBD fhuA2 [lon] ompT gal (λ DE3) [dcm] arnA::CBD slyD::CBD glmS6Ala ΔhsdS λ DE3 = λ sBamHIo ΔEcoRI-B int:(lacI::PlacUV5:T7 gene1) i21 Δnin5) (NEB, MA USA) transformed with expression vectors containing the protein of interest. Overexpression of the protein was achieved through fermentation in Erlenmeyer flasks, using precultures inoculated in 100 ml LB media supplemented with Kanamycin (50 μg ml−1) and incubated at 37 °C, 160 RPM overnight. These precultures were then diluted to 0.1 OD in 5 L Erlenmeyer flasks filled with 1 L of LB media supplemented with antibiotics and grown at 37 °C until reaching the stationary phase. Upon reaching an OD value between 0.6 and 0.9, the cultures were induced with 1 mM IPTG (Goldbio, MO USA) and grown for four hours in agitation (180 RPM) at 30 °C. The bacteria were then harvested via centrifugation and frozen.The cellular pellets were resuspended in 10 ml of lysis buffer 50 mM Tris–HCl at pH 8.0, 150 mM NaCl, 10 mM imidazole, 18 U ml−1 Benzonase (Merck, Germany), 1 mM MgCl2, 2 μl ml−1 CPI (Protease Inhibitor Cocktails) (Millex Sigma-Aldrich, MO USA), 1 mM TCEP (Goldbio, MO USA) per liter of culture. Lysis was completed by conducting a thermal protocol, where the lysate was incubated for 10 min in boiling water, cooled in ice, and supplemented with an additional 0.06 μl ml−1 of Benzonase (250 U ml−1) (Merck, Germany) before centrifugation.
The cellular lysates were centrifuged at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g (4 °C) for 20 min and the soluble fraction was then filtered through 0.45 μm filter units (Sartorius Stedim, Germany) for further purification.
000×g (4 °C) for 20 min and the soluble fraction was then filtered through 0.45 μm filter units (Sartorius Stedim, Germany) for further purification.
2.3. Protein chromatography
A standard isolation protocol was composed of two chromatography steps: affinity (Ni-NTA) and size exclusion chromatography (SEC). All of the proteins that were cyclized via split gp41 intein (cySB6, cyRH1, cyRH2) contained cysteine residues (needed for intein splicing) and were therefore isolated in the presence of 1 mM TCEP (Goldbio, MO USA).In this study, for purifying proteins, we employed a combination of nickel-nitrilotriacetic acid (Ni-NTA) resin and size exclusion chromatography (SEC). The bacterial lysates were first filtered and then incubated with 5 ml of Ni-NTA resin (Goldbio, MO USA) in buffer A (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 10 mM imidazole) for 5 minutes. The resin was then washed with buffer A and buffer B (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 20 mM imidazole) and the bound proteins were eluted with buffer C (50 mM Tris–HCl pH 8.0, 150 mM NaCl, 300 mM imidazole).
The eluted proteins were then subjected to SEC using 320 ml of HiLoad Superdex™ 200 resin (GE Healthcare, IL USA) packed in a 26/600 XK column (GE Healthcare, IL USA) and the same amount of HiLoad Superdex™ 75 resin (GE Healthcare, IL USA) packed in a 26/600 XK column (GE Healthcare, IL USA) equilibrated with filtered and degassed SEC buffer (20 mM Tris–HCl pH 7.5, 150 mM NaCl, 10% v/v glycerol). The samples after NiNTA were concentrated using centrifugal filters (3k or 10k) (Amicon-ultra, Millex Sigma-Aldrich, MO USA) and filtered through 0.22 μm syringe filters (Millex Sigma-Aldrich, MO USA) before being injected into the column. The chromatography was run using an AKTA™ pure FPLC system (GE Healthcare, IL USA) in SEC buffer at a linear flow rate of 2.6 ml min−1 and the eluted protein fractions were collected separately.
The heterodimeric protein complexes (SB24-nnn, SB24-ncn) were obtained by combining the purified subunits SB9b and SB9c in an equimolar ratio at low concentrations (∼0.2 mg ml−1). Then component SB6/cySB6 was added in 20–30% molar excess. The mixture was then concentrated and purified via an additional SEC passage. The heterodimeric complex was collected after separation and further concentrated for additional characterization.
The heterodimeric protein complexes (DiTET nn, cc, cn) described in the article were obtained by combining the purified subunits in an equimolar ratio at low concentrations (∼0.2 mg ml−1) without additional chromatography passages. The complexes were afterwards concentrated using centrifugal filters (3k) (Amicon-ultra, Millex Sigma-Aldrich, MO USA) to a final concentration depending on the needs of the characterization methods and finally filtered through Durapore 0.1 μm centrifuge filters (Merck Millipore, MA, USA).
2.4. Elastase protease cleavage
A simple experiment was conducted to further confirm the success of intein cyclization. Either cyclic (cyRH1, cyRH2) or linear protein (RH1, RH2) variants were incubated for 10 min with 17 ng elastase peptidase per 1 μg of protein and then ran on tricine SDS-PAGE (Sup. Fig. 8†). Elastase cut proteins more or less specifically at positions V or A and as expected we observed the presence of a larger fragment in the case of the cut cyclic variant and no such fragment in the case of the linear variant indicating successful cyclization.2.5. Protein electrophoresis
In this study, we used SDS-PAGE to analyze protein samples. The SDS-PAGE was performed using a Bio-rad (CA, USA) 16.5% Tris/Tricine Precast Gels polyacrylamide gel in a Bio-rad (CA, USA) mini-PROTEAN™ apparatus at a constant voltage of 120 V. A pre-stained low-molecular weight ruler (Thermo Fisher Scientific, MA, USA) was used to calculate the molecular weight. The gels were then stained with InstantBlue™ (Millex Sigma-Aldrich, MO, USA).2.6. Circular dichroism
Circular dichroism (CD) spectra of protein samples were acquired using a Chirascan CD spectrometer (Applied Photophysics, UK) equipped with a Peltier temperature controller. The samples were prepared at a concentration of 0.3–0.5 mg ml−1 and were measured in a 1 mm cuvette (Hellma, Germany) in the far-UV range (200–280 nm) at a temperature of 5 °C. The spectra were recorded using 1 nm steps, 1 nm bandwidth, and 1 s sampling intervals. To investigate thermal denaturation, the samples were heated at a rate of 1 °C per minute from 5 °C to 92 °C and then quickly quenched. The CD signal was measured at 222 nm. When possible, the experimental data were analyzed using a two-state equilibrium model proposed by Drobnak et al.62 The helical content of the proteins was calculated according to the following equation:| Helical content (%) = MRE222/(MREH222 × (1–2.57/n)) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 deg cm2 dmol−1).63
500 deg cm2 dmol−1).63
2.7. Size exclusion chromatography coupled with multi-angle light scattering
Size exclusion chromatography multi-angle light scattering (SEC-MALS) measurements were conducted using an HPLC system (Waters, MA, USA). The system was coupled with a UV detector, a Dawn8+ multiple-angle light scattering detector (Wyatt, CA, USA) and a refractive index detector (RI500, Shodex, Japan). The protein samples were filtered through 0.1 μm Durapore centrifuge filters (Merck Millipore, MA, USA) before being injected onto a Superdex™ 200 increase 10/300 column (GE Healthcare, IL, USA), which had been previously equilibrated with SEC buffer B (20 mM Tris–HCl pH 7.5, 150 mM NaCl). The data analysis was performed using Astra 7.0 software (Wyatt, CA, USA) and the peaks of interest were analyzed.2.8. Modeling
Molecular models of designed oligomeric two-chain tetrahedra and a three-chain octahedron were built using Python module Modeller64,65 and Chimera.66 Briefly, the amino acid sequence is designed by selecting an appropriate CC peptide for each position in the polypeptide chain. The pdbs of individual CC pairs were generated using the online software CC builder 2.0.67 Next, the pdbs of individual segments were arranged in Chimera so their relative positions and orientations roughly corresponded to the positions of the predicted fold of the structure. Using Modeller,64 the segments were connected by specified linkers and pairs of corresponding CC pairs were formed. Slight randomization of spatial positioning of individual segments was performed to generate diversity between models. Additionally, Modeller performed convergence of N and C termini in the case of cyclized chains. For each protein, we generated a couple of thousand models.2.9. Small angle X-ray scattering
Scattering curves of complexes were measured at the P12 beamline of PETRA III – DESY (Hamburg, Germany),68 while batch measurements of single chain components were performed on a table-top instrument SAXSpoint 5.0 (Anton Paar, AT) with primux 100 micro Cu X-ray generator and 2D 1M EIGER2 R series detector located at the National Institute of Chemistry (NIC), Slovenia.SAXS experiments that were carried out using the advanced PETRA III facility utilized an X-ray wavelength of 1.24 Å and a Pilatus 6M detector that was positioned at a distance of 3 meters from the sample. The scattering vector was recorded in the range of 0.028–7.3 nm−1. The measurement of a single-chain protein SB6 was conducted utilizing a batch mode with the aid of a robotic sample changer operating in flow-through mode. A dilution series was used to evaluate the effects of concentration and included four concentrations ranging from 9.5 mg ml−1 to 1.2 mg ml−1. For each dilution sample (40 μl), data was obtained over 20 exposures with a duration of 0.05 s each. The frames that did not exhibit radiation damage were then averaged and integrated into the SASFLOW pipeline.69 Before and after each sample, buffer scattering data was collected for background subtraction purposes.
Size exclusion chromatography coupled SAXS (SEC-SAXS) was performed using a Superdex™ 200 increase 10/300 column (GE Healthcare, IL USA). The buffer used in the experiment was SEC buffer C (20 mM Tris–HCl (pH 7.5), 150 mM NaCl and 3% (v/v) glycerol) and the samples measured were DiTET-nn, DiTET-cc, DiTET-nc, SB24-nnn, SB24-ncn with their concentrations ranging from 11 to 18 mg ml−1. The mobile phase was run through the column at a 0.5 ml min−1 flow rate. During the experiments, 3000–3600 scattering frames were collected with an exposure time of 0.995 seconds.
The majority of batch measurements were performed at NIC on an Anton Paar instrument SAXSpoint 5.0 using a heated/cooled sample cell holder, low-volume ASX autosampler and the detector positioned 600 mm from the measuring capillary. Samples RH1, cyRH1, RH2, cyRH2 and cySB6 had a concentration of 5.4 mg ml−1, 14 mg ml−1, 22.7 mg ml−1, 8.96 mg ml−1, 8.74 mg ml−1, respectively. A dilution series consisting of four different concentrations was performed for each of the samples to assess concentration effects. Each sample (20 μl) and their matching buffer was loaded into the 1 mm flow-through quartz capillary with the ASX autosampler and the data were collected over 7 exposures each of 30 min. The temperature of the sample during measurements was kept at 10 °C via a Peltier unit, while the waiting samples were kept at 4 °C. Software SAXS Analysis by Anton Paar, (version 4.01) was used to transform and export the raw data: We determined the center of the beam, beamstop was masked, the intensity was normalized by transmittance, q transformation was performed, the data were transformed from 2d to 1d and finally exported in ATSAS format. The subsequent steps of data analysis were performed using the ATSAS suite.70 Frames not displaying any radiation damage were manually averaged and afterwards, buffer scattering was used for background subtraction. To assess concentration effects, a dilution series consisting of four concentrations was measured for the single-chain proteins. Analysis of scattering curves and ab initio modeling was performed using the ATSAS suite.70 Theoretical SAXS profiles were calculated from molecular models and compared to experimental data using Pepsi-SAXS.71 The agreement between theoretical and experimental curves was evaluated using the χ metric, with low values demonstrating a good fit. The models that agreed with the experimental curve best, were further refined using an online tool of ATSAS suite70 Sreflex,72 which performed flexible refinement of models to achieve a better fit.
Experimental scattering profiles and ideal theoretical scattering of the complexes SB24 were compared using the volatility ratio (VR) using a web application Sibyls (https://sibyls.als.lbl.gov/saxs-similarity/).73 VR was calculated by taking the ratio of two scattering profiles in the scattering vector range of 0.15–1.5 nm−1.
2.10. Isothermal titration calorimetry
An Isothermal Titration Calorimeter (ITC) manufactured by Malvern Panalytical (UK) was utilized in the experiments. Before the experiment, the titrant and the analyte samples were dialyzed overnight against phosphate buffer (20 mM NaH2PO4xH2O pH 6.8, 150 mM NaCl, 2 mM TCEP). The titrant solution was present in excess volume (300 ml) at a concentration ranging from 10–30 μM and was loaded into a stirring syringe. The analyte solution, with a volume of 1.4 ml and a concentration ranging from 1–3 μM, was placed in the isothermal cell. After an initial equilibration period of 3000 seconds, the analyte was titrated with 25–29 additions of 10 μl of titrant, with a time interval of 600 seconds between each addition. The first injection always consisted of 2 μl of the titrant. The heat effects were determined through data integration using Nitpic74 and fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 dissociation model with the software SedPhat.75 The data was visualized using Gussi software (Dr Chad Brautigam).
1 dissociation model with the software SedPhat.75 The data was visualized using Gussi software (Dr Chad Brautigam).
2.11. Software
Graphs were prepared with Matplotlib 3.6.2 (https://matplotlib.org/) and Gussi by Dr Chad Brautigam. Results from SEC-MALS and SAXS were analyzed as described in “Methods”. SAXS at EMBL-DESY data was acquired and initially analyzed with the SASFLOW pipeline.68 Cartoon schematics were generated with Inkscape (https://inkscape.org/). Images of molecular models were created using UCSF Chimera (https://www.cgl.ucsf.edu/chimera/) and ChimeraX (https://www.cgl.ucsf.edu/chimerax/). Figures were edited and generated with Microsoft PowerPoint (https://www.microsoft.com/en-ww/microsoft-365/powerpoint). ITC data were collected with the software VPViewer 1.4.12 (Malvern Panalytical, UK). The pdbs of individual CC pairs were generated using an online software CC builder 2.0.673. Results and discussion
3.1. Two-componential tetrahedra including cyclization
We set out to investigate how to assemble multiple-chain CCPOs using preorganized subunits stabilized by cyclization and intramolecular CC paring and how this could assist the assembly of oligomeric CC-based architectures. We combined three core design concepts to impose the correct assembly of the subunits according to the design.First, we paid attention to the arrangement of CC-forming peptides in individual subunits to minimize the number of undesired conformations of the final assembly. To prevent the undesired homodimerization of larger subunits, complementary CC pairs within a single subunit were strategically positioned at a minimal topological distance in the primary structure. In some subunits, only one intramolecular CC pair was introduced, and all other CC segments were left unpaired. These unpaired CC segments were designed to be complementary to the corresponding segments in the partner chain subunits.
Second, we designed subunits to have rigid geometrical faces, such as triangles, to avoid conformational variability such as in oblique shapes of four-sided faces. Third, we fixed the subunits in the preorganized shape to support the desired assembly. This was achieved by increasing the helical propensity of CC-forming modules by introducing salt bridges at positions b, c and f of the heptad repeats76 of a previously published orthogonal set of dimeric CC units5,15 (Fig. 1B).
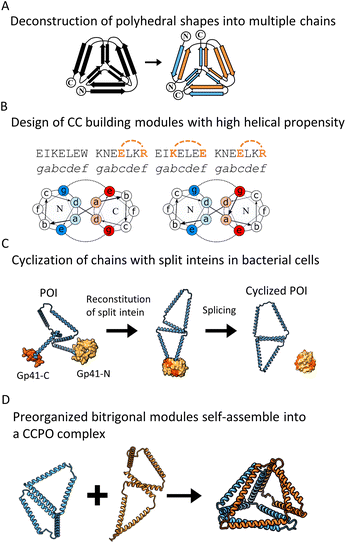 | ||
| Fig. 1 Design strategy for multi-chain CCPO structures and generation of preorganized building modules that include split intein cyclization and intrachain CC dimer. (A) Polyhedral shapes with edges formed by pairs of peptides were decomposed into multiple chains. (B) CC modules from the orthogonal library of dimeric CCs5,15 were selected to have a high helical propensity to increase their rigidity. This was achieved by the introduction of salt bridges between residues at the b, c and f positions of the CC dimer. (C) The splicing activity of split intein Gp41 was used to generate cyclic peptide chains in bacterial cells to preorganize the building modules to increase the predictability of the assembly. (D) Intramolecular CC dimer supported the preorganization of the cyclic chain to form merged bitrigonal modules. Molecular models were generated to predict the shape of the complexes. | ||
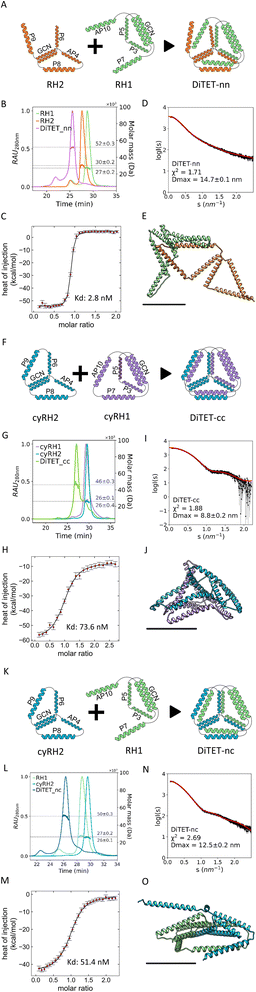
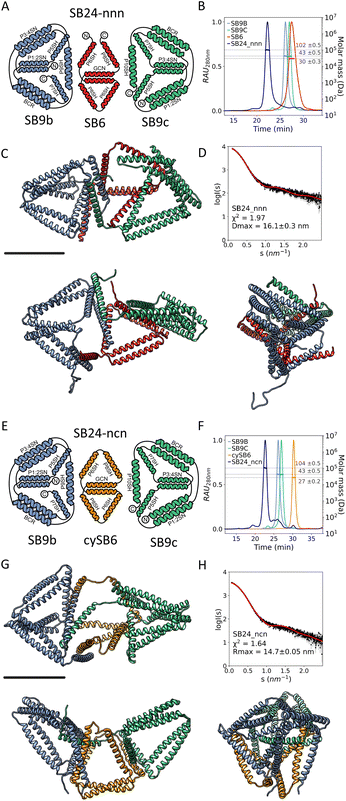
For our initial design, we selected a 12 CC-segment tetrahedral structure called TET12SN19 and decomposed it into a heterodimeric complex (Fig. 1A) with each unit composed of 6-segments (Fig. 1D left). Initially, we designed the subunits as two linear chains, each preorganized only by the presence of an intramolecular CC dimer, that self-assembles into a trigon with the N-terminus of the first and C-terminus of the sixth segment unconstrained. Additionally, we designed the alternative preorganized subunit variants, where the termini were constrained by an intein-mediated cyclization (Fig. 1C) shaping each of the subunits into a merged bitrigon. This was achieved by the genetic fusion of both parts of a highly efficient naturally occurring split intein Gp41 to the C- and N-termini of each subunit. Protein splicing occurred spontaneously during biosynthesis in bacterial cells, and those building subunits were isolated from the soluble fraction. By intein cyclization we were able to covalently link the N- and C-terminal segment, essentially creating a preorganized subunit comprising two trigons connected across one edge with an internal homodimeric CC with the remaining four unpaired CC dimer-forming segments (Fig. 1C right). In this manner, we aimed to self-assemble the tetrahedron from two bitrigon-forming subunits (Fig. 1D).
The linear (RH1 and RH2) and cyclized (cyRH1 and cyRH2) subunits for the two-chain tetrahedral CCPO assembly were produced separately in E. coli. SEC-MALS analysis indicated that the subunits were monodisperse with masses corresponding to the predicted values (RH1 = 27 kDa, RH2 = 30 kDa, cyRH1 = 26 kDa, cyRH2 = 26 kDa) (Fig. 2, B, G and L, and S1A to D, Table S1†). CD measurements confirmed the predicted high helicity of the individual subunits (Fig. S2B, S3B and Table S1†). The CD signal at 222 nm (Fig. S4A–D†) showed a gradual loss of helicity with the increased temperature, as expected for the nonglobular helical structure. After cooling, the polypeptide regained high helicity, similar to the measurements before the denaturation (Fig. S5A–D†), indicating a reversible unfolding-refolding. Each linear subunit and its cyclic counterpart, comprising identical amino acid sequences, exhibited different biophysical characteristics, indicating that the intein splicing reaction indeed successfully cyclized the proteins. Specifically, linear subunits had a lower elution time on SEC-MALS (RH1 = 28.7 min, RH2 = 27.6 min vs. cyRH1 = 29.3 min, cyRH2 = 29.6 min) (Fig. S2A and S3A†) and a larger Dmax measured by Small-Angle X-ray scattering (SAXS) in comparison to the cyclic polypeptide chains (RH1 = 32.5 ± 0.1 nm, RH2 = 17.1 ± 0.3 nm vs. cyRH1 = 16.6 ± 0.3 nm, cyRH2 = 10.5 ± 0.1 nm) (Fig. S2E and S3E†). By transforming SAXS curves into Kratky plots it is possible to better assess the degree of unfolding in the samples; unfolded proteins show a plateau in the Kratky plot at high q, while compact, globular proteins are distinguished by a bell-shaped peak. According to the Kratky plot of the SAXS curves, the linear subunits assumed a less ordered conformational state in the solution (Fig. S2D and S3D†) than cyclic subunits. Moreover, proteolytic treatment of the proteins with elastase further confirmed the success of intein cyclization (Fig. S6†). Comparing the cyclic subunits and noncyclic counterparts (Fig. S2C and S3C†) we observed no significant difference in the thermal denaturation of secondary structure elements indicating that the stability of the CC units is not affected by the cyclization.
Linear (RH1 and RH2) and cyclized (cyRH1 and cyRH2) subunits were mixed in all four different combinations to form dimeric complexes (DiTET-nn, DiTET-cc, DiTET-cn, DiTET-nc). Nomenclature of the complexes includes initials of the number of subunits constituting the complex (Di), initials of the intended polyhedron (TET), and letters at the end indicating the cyclic (c) or non-cyclic (i.e. linear) (n) nature of the first and second subunit. SEC-MALS of all complexes (Fig. 2B, G, L and S1K–N†) showed predominantly monodisperse dimers with masses around 50 kDa, corresponding to their theoretical size. Complex DiTET-nn had the lowest elution time (25.5 min), indicating a larger hydrodynamic diameter, while DiTET-cc had the highest elution time (27.1 min), pointing to a more compact shape. Both the monomeric TET12SN (26.6 min) and DiTET-nc (26.3 min) complexes had elution times between the DiTET-nn and DiTET-cc complexes (Fig. 3A). SEC-MALS chromatograms of DITET-cc and DiTET-nc showed a shoulder to the right of the main peak, corresponding to the monomeric components, which indicates these complexes' subunits may have slightly lower binding affinity compared to the rest. In the case of DiTET-cc the shoulder was reduced at high concentrations (S9). The formation of the complexes was monitored by ITC (Fig. 2C, H and M) and the data fitted to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 model, with Kd determined in the nanomolar range (Table 1). As indicated by SEC-MALS, the ITC confirmed that the binding affinity for the linear variants was higher compared to the cyclized versions.
1 model, with Kd determined in the nanomolar range (Table 1). As indicated by SEC-MALS, the ITC confirmed that the binding affinity for the linear variants was higher compared to the cyclized versions.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5. SAXS data was analyzed to determine Dmax from a pair-distance distribution function and to determine the radius of gyration (Rg)
1.5. SAXS data was analyzed to determine Dmax from a pair-distance distribution function and to determine the radius of gyration (Rg)
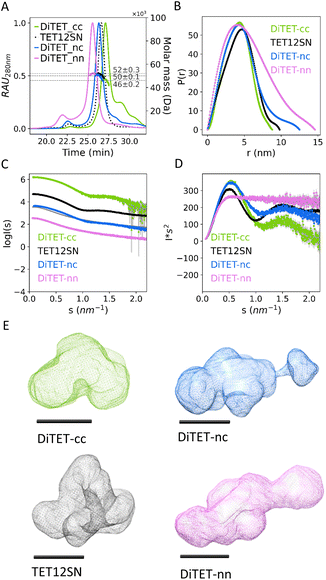
After proving the correct stoichiometry of the assembly, we then attempted to crystallize the complex and to obtain the structure using cryo-electron microscopy; however, the intrinsic flexibility of the loops on the vertices of the polyhedral structure prevented us from obtaining any high-resolution reconstruction of the complex. In this regard, we have already observed that CCPO structures seem inherently difficult to crystalize or vitrify; only recently an X-ray structure of the CCPO triangle has been determined based on the fortuitous packing of CC modules into a crystal lattice.77 We therefore resorted to using small-angle X-ray scattering (SAXS) (Fig. 2D, I and N, and Table 1) a method that allows determination of the molecular shape directly in solution. The experimental SAXS plot of the DiTET-nn complex matched the theoretical scattering of a molecular model with a flat and elongated conformation, suggesting misfolding. In contrast, assemblies DiTET-cc and DiTET-nc closely matched the theoretical scattering curve of the designed models with a tetrahedral shape (Fig. 2E, J and O). Both Dmax and radius of gyration (Rg) (Table 1) were in accordance with the SAXS fits, confirming that complexes containing cyclized subunits are more compact than the complexes without cyclized subunits.
SAXS curves of the three measured complexes were compared to the curve of the single-chain tetrahedral protein TET12SN (previously published data)19 (Fig. 3C and D). The scattering profile of DiTET-nn was the most divergent from the rest, lacking a scattering pattern characteristic for TET12SN tetrahedral topology, indicating a misfolded state. Dmax determined by SAXS (Fig. 3B and Table 1) was the highest for DiTET-nn (14.7 ± 0.1 nm), followed by DiTET-nc (12.5 ± 0.2 nm). The Dmax of DITET-cc (8.8 ± 0.2 nm) was similar to the diameter of the monomeric TET12SN (9.8 ± 0.1 nm), whose larger size, in comparison to DiTET-cc, might be an indication of some flexibility at the termini.
In addition, the ab initio SAXS reconstruction based on the pair distance distribution function (Fig. 3E) confirmed these results and, for complexes DiTET-cc, DiTET-nc, featured a concave depression indicating an internal cavity, which is characteristic of this type of de novo CCPO protein designs. Those results, therefore, demonstrate that the modular polypeptide assemblies can be achieved using preorganized cyclic subunits.
3.2. Three-componential asymmetric octahedra including cyclization
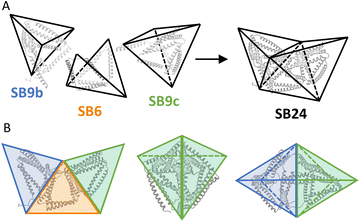
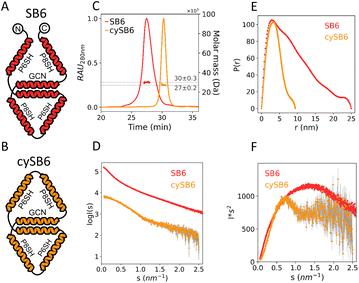
After establishing that chains with several unpaired CC-forming modules can be used as building blocks when stabilized by intein cyclization and pre-organized into trigonal shapes, we sought to implement these design principles for the assembly of more complex multi-chain design, advancing beyond the tetrahedral design, and two-chain assembly. Therefore, we selected to design an irregular octahedral CCPO shape from three polypeptide chains comprising 24 CC-forming segments. This fold entailed a particular concave shape, which would be difficult to achieve via the assembly of compact protein domain subunits or highly symmetric assemblies as in viral capsids, and here, we used it to showcase the use of the rational preorganized assembly of cyclized subunits.The assembly was divided into three pre-organized CC-based subunits to self-assemble into a concave irregular octahedral CCPO (Fig. 4). This heterotrimeric design consisted of two 9-segment pre-organized subunits (SB9b, SB9c) and a 6-segment polypeptide. We established the following nomenclature for the designed proteins: “SB” stood for “split boat” and the number denoted the number of CC-forming segments that were included in the protein/complex. The prefix “cy” described whether the protein was cyclized and letters at the end of the complex's name indicated cyclic (c) or non-cyclic (i.e. linear) (n) nature of the first (SB9b), second (SB6/cySB6) and third (SB9c) subunit. The two large subunits, composed of a 9 CC-forming segment contained three identical CC pairs (P3:4, P1:2, BCR:BCR) in a mirrored configuration. The three unpaired CC segments of each subunit constituted the binding interfaces between the SB9 subunits and a connecting smaller 6-CC-segment subunit. We were particularly interested to see how the short 6-CC peptide in either the linearized version or cyclized version (SB6 and cySB6, respectively) would affect the overall success of the assembly (SB24-nnn or cySB24-ncn, respectively) and therefore performed cyclization of the SB6 subunit similarly as for the tetrahedron as described above using split intein splicing (Fig. 1C).
Polypeptide subunits SB9b, SB9c, SB6 and cySB6 were expressed separately from E. coli and purified. SEC-MALS analysis (Fig. S1E–H†) showed that each component of the complex was monodisperse; while CD measurements confirmed that all the subunits assumed a highly helical secondary structure (SB6 α = 73.7%, cySB6 α = 77.8%, SB9b α = 62.2%, SB9c α = 67.1%) (Table S1†) and were able to refold after thermal denaturation (Fig. S4E–H and S5E–H†). The amino acid sequence of subunits cySB6 and SB6 was similar except for the intein splicing scar and some point mutations in the CC modules (Table S2†), however the biophysical characterization showed a difference in secondary and tertiary structure between the two (Fig. 5) suggesting that, cyclization with split inteins caused a more compact and less disordered fold. While SEC-MALS showed the proteins had a similar mass as theoretically predicted (SB6 = 30 kDa, cySB6 = 27 kDa) (Fig. S1E and F†), the larger elution time on SEC indicated a smaller hydrodynamic radius in the case of the cyclized variant (30.2 min) in comparison to the linear variant (27.4 min) (Fig. 5C). CD analysis showed a difference in thermal stability when comparing cyclic and linear subunits (Fig. S10†) which could be due to the point mutation differences in the sequence. SAXS analysis of the two proteins (Fig. 5D, S7A and B†) indicated SB6 had a hydrodynamic diameter larger than cySB6 (Dmax = 25 ± 0.1 nm and 9.4 ± 0.05 nm, respectively) (Fig. 5E, Table S1†). Kratky plot of SB6 showed a wide bell-shaped peak indicating a more unfolded conformation in solution than cySB6, shown by the narrower peak of the latter (Fig. 5F).
SEC-MALS experiments showed that a mixture of SB9b and SB9c had little to no tendency to form dimers (Fig. S1O†), even though they were complementary to each other across a single unpaired segment. On the other hand, mixing SB9b and SB9c with either SB6 or cySB6 in an equimolar ratio resulted in a predominantly monodisperse trimer (Fig. 6A and E) with an experimentally determined mass of 102 ± 0.5 kDa and 104 ± 0.6 kDa, respectively, corresponding to the theoretical size of the correctly assembled heterotrimers (SB24-nnn = 108.9 kDa, SB24-ncn = 108.6 kDa) (Fig. 6B and F, and S1I, J and Table S1†). The thermodynamics of the interaction between the subunits was analysed by ITC. Either SB6 or cySB6 was titrated into an equimolar mixture of SB9b and SB9c. The determined Kd pointed to a substantially higher affinity for the complex formed with the linear SB6 (Kd = 6.9 nM) compared to its cyclized counterpart cySB6 (Kd = 195.1 nM) (Fig. S8†). The linear subunit is more flexible in solution and has a higher conformational entropy than the cyclic subunit. This could contribute towards the higher affinity observed with the linear subunit, which however form a misfolded assembly upon binding to the other two proteins.
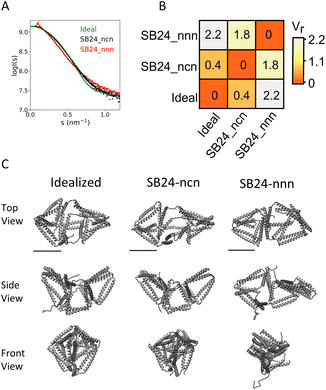
The size and shape of the complexes in the solution were assessed by SEC-SAXS. Experimental SAXS analysis of the complex SB24-nnn, comprising only linear chains, matched the theoretical scattering curve of a model with a collapsed subunit (χ2 = 1.97) (Fig. 6C and D). On the other hand, the complex SB24-ncn was shown to fit best to the scattering profile of models with a concave octahedral conformation (χ2 = 1.64) (Fig. 6G and H), with a Dmax (Dmax = 14.7 ± 0.05 nm), lower than SB24-nnn (Dmax = 16.1 ± 0.3 nm) (Table S1†). The difference between the experimental scattering profiles of the complexes and the theoretical scattering profile of an “idealized” model of an irregular octahedron were analysed with the volatility of ratio (Fig. 7), which is a metric that can identify local changes in otherwise structurally similar particles from their scattering profile.73 This revealed that the cyclized version of the complex, SB24ncn, assumed a conformation closer to the ideal model of a designed octahedron, suggesting that cyclization of the smaller subunit is sufficient to stabilize the complex and allow it to assume a conformation close to the original design.
4. Discussion
Rational design of designed supramolecular assemblies can be facilitated by modularity, as demonstrated before on organic molecules,78 DNA nanotechnology79 and polypeptide assemblies (e.g. using coiled coils,15,80 foldons28 and repeated protein domains81). Designed polypeptide assemblies are often characterized by a certain level of structural flexibility and heterogeneity (e.g. antigen presenting cages82,83 or multidomain structures26,28,84). Here, we have shown that the preorganization of subunits introduced through genetically encoded cyclization and intramolecular interactions allows greater control over structural flexibility and predictability in arranging tri-dimensional polypeptide assemblies, a principle that is likely to facilitate the self-assembly of modular protein complexes independently from the type of building blocks employed in the design.While particularly significant in the field of synthetic biology, where CC and CC-based structures have been widely employed, understanding the rules governing the folding and assembly of molecular architectures is relevant for the use of synthetic proteins in biomedicine and biotechnology. In this context, this study introduces a strategy that enhances the stability and predictability of CC-based assemblies.
The two-chain assembly of the tetrahedral topology demonstrated here clearly illustrates the advantages of structural preorganization via cyclization. While non-cyclized subunits assembled stoichiometrically, their interaction yielded a misfolded assembly as the interaction energy of the collapsed state of the complex was higher compared to when we employed cyclic subunits, which on the other hand, favored the correct folding of the heterodimeric complex. In this case, the assembly assumed the intended shape when at least one cyclized subunit was incorporated into the design, which was supported by an extensive biophysical characterization and SAXS analysis of the assembly in solution. Expanding the reach of the rational design we demonstrated the design and assembly of SB24, an irregular concave octahedral topology, so far not implemented at the molecular scale, crafted from 24 coiled-coil modules, which marks the first successful design from three pre-organized chains as well as the largest de novo 3D CC-based assembly to date.
In both cases (DiTET-cc and SB24-ncn) the molecular shape in agreement with the design was confirmed with a high degree of certainty by SAXS analysis. For the two-chain tetrahedron, the validity of its shape was further confirmed through ab initio reconstitution based on scattering profiles. Attempts to determine the high-resolution structure of both assemblies by either X-ray crystallography or cryo-electron microscopy have not been successful, due to the intrinsic flexibility of the assembly lacking a compact core. Notably, SAXS was able to identify the misfolded assemblies despite their correct stoichiometry. It helped us to understand how the flexibility within the subunit of the complex SB24-nnn caused a partial collapse, and how replacing the flexible polypeptide chain with a cyclic chain alleviated the flexibility of the CC segments at the termini. This approach enabled the construction of a CCPO architecture that assumes a defined asymmetric molecular envelope that would be otherwise unattainable.
Interestingly, ITC results revealed a slightly lower affinity between cyclized subunits compared to linear ones. This discrepancy may be due to strain in the interacting cyclized chains. A likely explanation is that the collapse of linear chains, characterized by a higher conformational entropy, added a non-specific contribution to the interaction energy, which however came at the expense of misfolding.
Intein-mediated cyclization enhanced the structural stability and definition of protein–protein interaction surfaces. This method of subunit stabilization, utilizing highly efficient split intein splicing during bacterial production, requires no additional steps in polypeptide subunit isolation.
Recently, computational techniques based on machine learning benefiting from the extensive 3D structural information of natural proteins collected to date36,38,85 have significantly improved the design and prediction of globular natural-like proteins. In contrast, the reliable design of modular assemblies, such as CCPO, remains challenging due to their reliance on long-range topological contacts and CC dimer orthogonality.5,10,86 Interestingly, algorithms, such as AlphaFold2, have not been able to predict the structure of CCPOs.77
The rigid nature of the used triangular building blocks, combined with the rational arrangement of interacting CC segments allows in principle for the design of larger multimeric CC-based architectures. Multimeric CCPO shapes could represent scaffolds for encapsulation of small cargo molecules, as the cavity of these structures is designable in shape and size, and most importantly, unlike cavities in virus-like particles, is not bound to the specific symmetry of the assembly and could harbor molecules with an asymmetric shape. Moreover, the cavity of SB24 is notably larger than previous CC-based designs e.g. monomeric tetrahedron,19 dimeric bipyramid.35
5. Conclusions
In this study, the preorganization of structural modules has been used as a powerful design principle for the macromolecular assembly, based on polypeptides in particular, although similar preorganization and cyclisation principles could likely also be applied to other types of molecules. Some supramolecular assemblies already used macrocycles as cyclic building units, where however the degree of oligomerization could not be precisely defined as in the case of CCPOs.87The preorganization strategy for macromolecular self-assembly can be used to assemble asymmetric, yet precisely defined nanostructures where each vertex or edge of the polyhedral structure could be individually addressed to provide the geometric arrangement of the selected functionalities, e.g. presentation of antigens for the stimulation of immune response, activation of cellular receptors, positioning of catalytic centers or binding sites. Additionally, such CC complexes could be engineered to undergo a reversible conformational change in response to chemical signals such as metal ions,34,88 pH89 or biological molecules,90 all cues already used to regulate CC assemblies.
Here, we demonstrated an innovative approach to protein assembly by integrating cyclic subunits with intramolecular CC complementarity to overcome flexibility challenges. This strategy enabled the design of larger CC-based structures, marking a substantial advancement in the field. Utilizing split intein cyclization, not only enhances the predictability in designing interaction interfaces but also stands out for its simplicity of implementation.
Data availability
The datasets supporting this article have been uploaded as part of the ESI. SAXS data and calculated structures are available at SASBDB (https://www.sasbdb.org/) under accession codes (SASDUN2, SASDUP2, SASDUQ2, SASDUR2, SASDUS2).Author contributions
Conceptualization: J. S., F. L., and R. J. Methodology: J. S., F. L. Investigation: J. S., F. L. Visualization: J. S. Supervision: R. J. Writing—original draft: J. S., F. L. and R. J. Writing—review and editing: J. S., F. L., and R. J.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The synchrotron SAXS data were collected at beamline P12 operated by EMBL Hamburg at the PETRA III storage ring (DESY, Hamburg, Germany). We wish to thank Tadej Satler for helping with matplolib scripts for graph generation and for helping with ITC measurements, Žiga Strmšek for helping with SAXS data collection and analysis, Jana Aupič for helping with molecular modeling, Klemen Mezgec for helping with visualization of the geometric models, Andreja Majerle for mentoring, Robert Bremšak and Tina Strmljan for technical support. This research was conducted with the support of the Slovenian Research Agency (P4-0176, J1-2481, J1-1711, Z1-3194), The European Research Council (ERC AdG MaCChines 787115), EU FET Open Project Virofight (#899619), Horizon EIC Pathfinder: project LoopOfFun 101070817.References
- J. M. Mason and K. M. Arndt, Coiled coil domains: stability, specificity, and biological implications, ChemBioChem, 2004, 5(2), 170–176 CrossRef CAS PubMed.
- P. Burkhard, J. Stetefeld and S. V. Strelkov, Coiled coils: a highly versatile protein folding motif, Trends Cell Biol., 2001, 11(2), 82–88 CrossRef CAS PubMed.
- M. Gruber and A. N. Lupas, Historical review: another 50th anniversary – new periodicities in coiled coils, Trends Biochem. Sci., 2003, 28(12), 679–685 CrossRef CAS PubMed.
- D. N. Woolfson, G. J. Bartlett, M. Bruning and A. R. Thomson, New currency for old rope: from coiled-coil assemblies to α-helical barrels, Curr. Opin. Struct. Biol., 2012, 22(4), 432–441, DOI:10.1016/j.sbi.2012.03.002.
- I. Drobnak, H. Gradišar, A. Ljubetič, E. Merljak and R. Jerala, Modulation of Coiled-Coil Dimer Stability through Surface Residues while Preserving Pairing Specificity, J. Am. Chem. Soc., 2017, 139(24), 8229–8236 CrossRef CAS PubMed.
- W. C. Boldridge, A. Ljubetič, H. Kim, N. Lubock, D. Szilágyi and J. Lee, et al., A multiplexed bacterial two-hybrid for rapid characterization of protein-protein interactions and iterative protein design, Nat. Commun., 2023, 14(1), 4636 CrossRef CAS PubMed.
- F. Lapenta, J. Aupič, Ž. Strmšek and R. Jerala, Coiled coil protein origami: from modular design principles towards biotechnological applications, Chem. Soc. Rev., 2018, 47(10), 3530–3542 RSC , available from: https://pubs.rsc.org/en/content/articlehtml/2018/cs/c7cs00822h.
- A. N. Lupas, J. Bassler and S. Dunin-Horkawicz, The Structure and Topology of α-Helical Coiled Coils, Subcell. Biochem., 2017, 82, 95–129 CAS.
- D. N. Woolfson, Coiled-coil design: updated and upgraded, Subcell. Biochem., 2017, 82, 35–61 CAS.
- D. N. Woolfson, The design of coiled-coil structures and assemblies, Adv. Protein Chem., 2005, 70, 79–112 CrossRef CAS PubMed.
- P. B. Harbury, T. Zhang, P. S. Kim and T. Alber, A switch between two-, three-, and four-stranded coiled coils in GCN4 leucine zipper mutants, Science, 1993, 262(5138), 1401–1407 CrossRef CAS PubMed.
- N. R. Zaccai, B. Chi, A. R. Thomson, A. L. Boyle, G. J. Bartlett and M. Bruning, et al., A de novo peptide hexamer with a mutable channel, Nat. Chem. Biol., 2011, 7(12), 935–941, DOI:10.1038/nchembio.692.
- M. J. Pandya, E. Cerasoli, A. Joseph, R. G. Stoneman, E. Waite and D. N. Woolfson, Sequence and structural duality: designing peptides to adopt two stable conformations, J. Am. Chem. Soc., 2004, 126(51), 17016–17024 CrossRef CAS PubMed.
- J. D. Steinkruger, G. J. Bartlett, E. B. Hadley, L. Fay, D. N. Woolfson and S. H. Gellman, The d′-D-D′ vertical triad is less discriminating than the a′-A-A′ vertical triad in the antiparallel coiled-coil dimer motif, J. Am. Chem. Soc., 2012, 134(5), 2626–2633 CrossRef CAS PubMed.
- H. Gradišar and R. Jerala, De novo design of orthogonal peptide pairs forming parallel coiled-coil heterodimers, J. Pept. Sci., 2011, 17(2), 100–106 CrossRef PubMed.
- K. E. Thompson, C. J. Bashor, W. A. Lim and A. E. Keating, SYNZIP protein interaction toolbox: in vitro and in vivo specifications of heterospecific coiled-coil interaction domains, ACS Synth. Biol., 2012, 1(4), 118–129 CrossRef CAS PubMed.
- P. S. Huang, S. E. Boyken and D. Baker, The coming of age of de novo protein design, Nature, 2016, 537(7620), 320–327 CrossRef CAS PubMed.
- J. Liu, Q. Zheng, Y. Deng, C. S. Cheng, N. R. Kallenbach and M. Lu, A seven-helix coiled coil, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(42), 15457–15462 CrossRef CAS PubMed.
- A. Ljubetič, F. Lapenta, H. Gradišar, I. Drobnak, J. Aupič and Ž. Strmšek, et al., Design of coiled-coil protein-origami cages that self-assemble in vitro and in vivo, Nat. Biotechnol., 2017, 35(11), 1094–1101 CrossRef PubMed.
- J. P. Schneider, A. Lombardi and W. F. DeGrado, Analysis and design of three-stranded coiled coils and three-helix bundles, Folding Des., 1998, 3(2), 29–40 CrossRef PubMed.
- A. R. Thomson, C. W. Wood, A. J. Burton, G. J. Bartlett, R. B. Sessions and R. L. Brady, et al., Computational design of water-soluble α-helical barrels, Science, 2014, 346(6208), 485–488 CrossRef CAS PubMed.
- E. A. Naudin, K. I. Albanese, A. J. Smith, B. Mylemans, E. G. Baker and O. D. Weiner, et al., From peptides to proteins: coiled-coil tetramers to single-chain 4-helix bundles, Chem. Sci., 2022, 13(38), 11330–11340 RSC.
- N. C. Burgess, T. H. Sharp, F. Thomas, C. W. Wood and R. Andrew, Modular Design of Self-assembling Peptide-based Nanotubes, J. Appl. Chem. Sci., 2015, 1–12 Search PubMed.
- C. Xu, R. Liu, A. K. Mehta, R. C. Guerrero-Ferreira, E. R. Wright and S. Dunin-Horkawicz, et al., Rational design of helical nanotubes from self-assembly of coiled-coil lock washers, J. Am. Chem. Soc., 2013, 135(41), 15565–15578 CrossRef CAS PubMed.
- J. M. Fletcher, R. L. Harniman, F. R. H. Barnes, A. L. Boyle, A. Collins and J. Mantell, et al., Self-assembling cages from coiled-coil peptide modules, Science, 2013, 340(6132), 595–599 CrossRef CAS PubMed.
- D. P. Patterson, M. Su, T. M. Franzmann, A. Sciore, G. Skiniotis and E. N. G. Marsh, Characterization of a highly flexible self-assembling protein system designed to form nanocages, Protein Sci., 2014, 23(2), 190–199 CrossRef CAS PubMed.
- H. R. Khataee and A. R. Khataee, Kinesin and dynein smart nanomotors: Towards bio-nanorobotic systems, Nano, 2010, 5(1), 13–23 CrossRef CAS.
- N. Kobayashi, K. Yanase, T. Sato, S. Unzai, M. H. Hecht and R. Arai, Self-Assembling Nano-Architectures Created from a Protein Nano-Building Block Using an Intermolecularly Folded Dimeric de Novo Protein, J. Am. Chem. Soc., 2015, 137(35), 11285–11293 CrossRef CAS PubMed.
- W. M. Park, M. Bedewy, K. K. Berggren and A. E. Keating, Modular assembly of a protein nanotriangle using orthogonally interacting coiled coils, Sci. Rep., 2017, 7(1), 10577 CrossRef PubMed.
- A. L. Boyle, E. H. C. Bromley, G. J. Bartlett, R. B. Sessions, T. H. Sharp and C. L. Williams, et al., Squaring the Circle in Peptide Assembly: From Fibers to Discrete Nanostructures by de Novo Design, J. Am. Chem. Soc., 2012, 134, 15457–15467 CrossRef CAS PubMed . Available from: https://pubs.acs.org/sharingguidelines.
- J. Aupič, Ž. Strmšek, F. Lapenta, A. Ljubetič, R. Jerala and D. Pahovnik, et al., Designed folding pathway of modular coiled-coil- based proteins, Nat. Commun., 2021, 12(1), 940 CrossRef PubMed.
- H. Gradišar, S. Božič, T. Doles, D. Vengust, I. Hafner-Bratkovič and A. Mertelj, et al., Design of a single-chain polypeptide tetrahedron assembled from coiled-coil segments, Nat. Chem. Biol., 2013, 9(6), 362–366 CrossRef PubMed.
- S. Božič Abram, H. Gradišar, J. Aupič, A. R. Round and R. Jerala, Triangular in Vivo Self-Assembling Coiled-Coil Protein Origami, ACS Chem. Biol., 2021, 16(2), 310–315 CrossRef PubMed.
- J. Aupič, F. Lapenta, Ž. Strmšek, E. Merljak, T. Plaper and R. Jerala, Metal ion-regulated assembly of designed modular protein cages, Sci. Adv., 2022, 8(24), eabm8243 CrossRef PubMed.
- F. Lapenta, J. Aupič, M. Vezzoli, Ž. Strmšek, S. Da Vela and D. I. Svergun, et al., Self-assembly and regulation of protein cages from pre-organised coiled-coil modules, Nat. Commun., 2021, 12(1), 939 CrossRef CAS PubMed.
- J. Dauparas, I. Anishchenko, N. Bennett, H. Bai, R. J. Ragotte and L. F. Milles, et al., Robust deep learning-based protein sequence design using ProteinMPNN, Science, 2022, 378(6615), 49–56 CrossRef CAS PubMed.
- Z. Yang, X. Zeng, Y. Zhao and R. Chen, AlphaFold2 and its applications in the fields of biology and medicine, Signal Transduction Targeted Ther., 2023, 8(1), 115 CrossRef PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov and O. Ronneberger, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596(7873), 583–589 CrossRef CAS PubMed.
- O. Saether, D. J. Craik, I. D. Campbell, K. Sletten, J. Juul and D. G. Norman, Elucidation of the Primary and Three-Dimensional Structure of the Uterotonic Polypeptide Kalata B1, Biochemistry, 1995, 34(13), 4147–4158 CrossRef CAS PubMed.
- B. Samyn, M. Martinez-Bueno, B. Devreese, M. Maqueda, A. Gálvez and E. Valdivia, et al., The cyclic structure of the enterococcal peptide antibiotic AS-48, FEBS Lett., 1994, 352(1), 87–90 CrossRef CAS PubMed.
- A. Purkayastha and T. J. Kang, Stabilization of Proteins by Covalent Cyclization, Biotechnol. Bioprocess. Eng, 2019, 24(5), 702–712 CrossRef CAS.
- J. Touati, A. Angelini, M. J. Hinner and C. Heinis, Enzymatic cyclisation of peptides with a transglutaminase, ChemBioChem, 2011, 12(1), 38–42 CrossRef CAS PubMed.
- M. J. Lobba, C. Fellmann, A. M. Marmelstein, J. C. Maza, E. N. Kissman and S. A. Robinson, et al., Site-Specific Bioconjugation through Enzyme-Catalyzed Tyrosine-Cysteine Bond Formation, ACS Cent. Sci., 2020, 6(9), 1564–1571 CrossRef CAS PubMed.
- C. Schoene, J. O. Fierer, S. P. Bennett and M. Howarth, SpyTag/SpyCatcher Cyclization Confers Resilience to Boiling on a Mesophilic Enzyme, Angew. Chem., 2014, 126(24), 6215–6218 CrossRef.
- H. Paulus, The chemical basis of protein splicing, Chem. Soc. Rev., 1998, 27(6), 375–386 RSC.
- N. I. Topilina and K. V. Mills, Recent advances in in vivo applications of intein-mediated protein splicing, Mobile DNA, 2014, 5(1), 1–14 CrossRef PubMed.
- R. Hirata, Y. Ohsumk, A. Nakano, H. Kawasaki, K. Suzuki and Y. Anraku, Molecular structure of a gene, VMA1, encoding the catalytic subunit of H(+)-translocating adenosine triphosphatase from vacuolar membranes of Saccharomyces cerevisiae, J. Biol. Chem., 1990, 265(12), 6726–6733 CrossRef CAS PubMed.
- F. B. Perler, G. J. Olsen and E. Adam, Compilation and analysis of intein sequences, Nucleic Acids Res., 1997, 25(6), 1087–1093 CrossRef CAS PubMed.
- N. H. Shah and T. W. Muir, Split inteins: Nature's protein ligases, Isr. J. Chem., 2011, 51(8–9), 854–861 CrossRef CAS PubMed.
- H. Wang, L. Wang, B. Zhong and Z. Dai, Protein Splicing of Inteins: A Powerful Tool in Synthetic Biology, Front. Bioeng. Biotechnol., 2022, 10, 1–9 Search PubMed.
- D. W. Wood and J. A. Camarero, Intein applications: From protein purification and labeling to metabolic control methods, J. Biol. Chem., 2014, 289(21), 14512–14519 CrossRef CAS PubMed.
- N. H. Shah, E. Eryilmaz, D. Cowburn and T. W. Muir, Naturally split inteins assemble through a “capture and collapse” mechanism, J. Am. Chem. Soc., 2013, 135(49), 18673–18681 CrossRef CAS PubMed.
- H. Wu, Z. Hu and X. Q. Liu, Protein frans-splicing by a split intein encoded in a split DnaE gene of Synechocystis sp. PCC6803, Proc. Natl. Acad. Sci. U. S. A., 1998, 95(16), 9226–9231 CrossRef CAS PubMed.
- P. Carvajal-Vallejos, R. Pallissé, H. D. Mootz and S. R. Schmidt, Unprecedented rates and efficiencies revealed for new natural split inteins from metagenomic sources, J. Biol. Chem., 2012, 287(34), 28686–28696 CrossRef CAS PubMed.
- S. Brenzel, T. Kurpiers and H. D. Mootz, Engineering artificially split inteins for applications in protein chemistry: biochemical characterization of the split Ssp DnaB intein and comparison to the split Sce VMA intein, Biochemistry, 2006, 45(6), 1571–1578 CrossRef CAS PubMed.
- A. J. Stevens, Z. Z. Brown, N. H. Shah, G. Sekar, D. Cowburn and T. W. Muir, Design of a Split Intein with Exceptional Protein Splicing Activity, J. Am. Chem. Soc., 2016, 138(7), 2162–2165 CrossRef CAS PubMed.
- M. C. Waldhauer, S. N. Schmitz, C. Ahlmann-Eltze, J. G. Gleixner, C. C. Schmelas and A. G. Huhn, et al., Backbone circularization of Bacillus subtilis family 11 xylanase increases its thermostability and its resistance against aggregation, Mol. Biosyst., 2015, 11(12), 3231–3243 RSC.
- W. Bai, C. J. Sargent, J. M. Choi, R. V. Pappu and F. Zhang, Covalently-assembled single-chain protein nanostructures with ultra-high stability, Nat. Commun., 2019, 10(1), 3317 CrossRef PubMed.
- S. Raniolo, S. Croce, R. P. Thomsen, A. H. Okholm, V. Unida and F. Iacovelli, et al., Cellular uptake of covalent and non-covalent DNA nanostructures with different sizes and geometries, Nanoscale, 2019, 11(22), 10808–10818 RSC.
- R. Li, H. Chen, H. Lee and J. H. Choi, Elucidating the mechanical energy for cyclization of a dna origami tile, Appl. Sci., 2021, 11(5), 1–15 CAS.
- D. G. Gibson, L. Young, R. Y. Chuang, J. C. Venter, C. A. Hutchison and H. O. Smith, Enzymatic assembly of DNA molecules up to several hundred kilobases, Nat. Methods, 2009, 6(5), 343–345 CrossRef CAS PubMed.
- I. Drobnak, G. Vesnaver and J. Lah, Model-Based Thermodynamic Analysis of Reversible Unfolding Processes, J. Phys. Chem. B, 2010, 114(26), 8713–8722 CrossRef CAS PubMed.
- Y. H. Chen, J. T. Yang and H. M. Martinez, Determination of the secondary structures of proteins by circular dichroism and optical rotatory dispersion, Biochemistry, 1972, 11(22), 4120–4131 CrossRef CAS PubMed.
- N. Eswar, B. Webb, M. A. Marti-Renom, M. S. Madhusudhan, D. Eramian and M. Shen, et al., Comparative Protein Structure Modeling Using MODELLER, Curr. Protoc. Protein Sci., 2007, 50(1) Search PubMed.
- B. Webb and A. Sali, Comparative protein structure modeling using MODELLER, Curr. Protoc. Bioinf., 2016, 2016 Search PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt and E. C. Meng, et al., UCSF Chimera - A visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25(13), 1605–1612 CrossRef CAS PubMed.
- C. W. Wood and D. N. Woolfson, CCBuilder 2.0: Powerful and accessible coiled-coil modeling, Protein Sci., 2018, 27(1), 103–111 CrossRef CAS PubMed.
- C. E. Blanchet, A. Spilotros, F. Schwemmer, M. A. Graewert, A. Kikhney and C. M. Jeffries, et al., Versatile sample environments and automation for biological solution X-ray scattering experiments at the P12 beamline (PETRA III, DESY), J. Appl. Crystallogr., 2015, 48, 431–443 CrossRef CAS PubMed.
- D. Franke, A. G. Kikhney and D. I. Svergun, Automated acquisition and analysis of small angle X-ray scattering data, Nucl. Instrum. Methods Phys. Res., Sect. A, 2012, 689, 52–59, DOI:10.1016/j.nima.2012.06.008.
- D. Franke, M. V. Petoukhov, P. V. Konarev, A. Panjkovich, A. Tuukkanen and H. D. T. Mertens, et al., ATSAS 2.8: A comprehensive data analysis suite for small-angle scattering from macromolecular solutions, J. Appl. Crystallogr., 2017, 50, 1212–1225 CrossRef CAS PubMed.
- S. Grudinin, M. Garkavenko and A. Kazennov, Pepsi-SAXS: An adaptive method for rapid and accurate computation of small-angle X-ray scattering profiles, Acta Crystallogr., Sect. D: Struct. Biol., 2017, 73(5), 449–464 CrossRef CAS PubMed.
- A. Panjkovich and D. I. Svergun, Deciphering conformational transitions of proteins by small angle X-ray scattering and normal mode analysis, Phys. Chem. Chem. Phys., 2016, 18(8), 5707–5719 RSC.
- G. L. Hura, H. Budworth, K. N. Dyer, R. P. Rambo, M. Hammel and C. T. McMurray, et al., Comprehensive macromolecular conformations mapped by quantitative SAXS analyses, Nat. Methods, 2013, 10(6), 453–454 CrossRef CAS PubMed . Available from: http://dx.doi.org/10.1038/nmeth.2453.
- C. S. Mocny and V. L. Pecoraro, De Novo Protein Design as a Methodology for Synthetic Bioinorganic Chemistry, Acc. Chem. Res., 2015, 48(8), 2388–2396 CrossRef CAS PubMed.
- J. C. D. Houtman, P. H. Brown, B. Bowden, H. Yamaguchi, E. Appella and L. E. Samelson, et al., Studying multisite binary and ternary protein interactions by global analysis of isothermal titration calorimetry data in SEDPHAT: Application to adaptor protein complexes in cell signaling, Protein Sci., 2007, 16(1), 30–42 CrossRef CAS PubMed.
- E. Lacroix, A. R. Viguera and L. Serrano, Elucidating the folding problem of alpha-helices: local motifs, long-range electrostatics, ionic-strength dependence and prediction of NMR parameters, J. Mol. Biol., 1998, 284(1), 173–191 CrossRef CAS PubMed.
- T. Satler, S. Hadži and R. Jerala, Crystal Structure of de Novo Designed Coiled-Coil Protein Origami Triangle, J. Am. Chem. Soc., 2023, 145(31), 16995–17000 CrossRef CAS PubMed.
- E. Mattia and S. Otto, Supramolecular systems chemistry, Nat. Nanotechnol., 2015, 10(2), 111–119 CrossRef CAS PubMed.
- Y. He, T. Ye, M. Su, C. Zhang, A. E. Ribbe and W. Jiang, et al., Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra, Nature, 2008, 452(7184), 198–201 CrossRef CAS PubMed.
- F. Thomas, A. L. Boyle, A. J. Burton and D. N. Woolfson, A set of de novo designed parallel heterodimeric coiled coils with quantified dissociation constants in the micromolar to sub-nanomolar regime, J. Am. Chem. Soc., 2013, 135(13), 5161–5166 CrossRef CAS PubMed.
- C. H. Bowen, C. J. Sargent, A. Wang, Y. Zhu, X. Chang and J. Li, et al., Microbial production of megadalton titin yields fibers with advantageous mechanical properties, Nat. Commun., 2021, 12(1), 1–12, DOI:10.1038/s41467-021-25360-6.
- K. El Bissati, Y. Zhou, S. M. Paulillo, S. K. Raman, C. P. Karch and S. Reed, et al., Engineering and characterization of a novel Self Assembling Protein for Toxoplasma peptide vaccine in HLA-A*11:01, HLA-A*02:01 and HLA-B*07:02 transgenic mice, Sci. Rep., 2020, 10(1), 16984 CrossRef CAS PubMed.
- Y. Yang, P. Ringler, S. A. Müller and P. Burkhard, Optimizing the refolding conditions of self-assembling polypeptide nanoparticles that serve as repetitive antigen display systems, J. Struct. Biol., 2012, 177(1), 168–176 CrossRef CAS PubMed.
- J. P. M. Vrancken, J. Aupič, C. Addy, R. Jerala, J. R. H. Tame and A. R. D. Voet, Molecular assemblies built with the artificial protein Pizza, J. Struct. Biol. X, 2020, 4, 100027 CAS.
- M. Mirdita, K. Schütze, Y. Moriwaki, L. Heo, S. Ovchinnikov and M. Steinegger, ColabFold: making protein folding accessible to all, Nat. Methods, 2022, 19(6), 679–682 CrossRef CAS PubMed.
- A. W. Reinke, J. Baek, O. Ashenberg and A. E. Keating, Networks of bZIP protein-protein interactions diversified over a billion years of evolution, Science, 2013, 340(6133), 730–734 CrossRef CAS PubMed.
- C. G. Pappas, P. K. Mandal, B. Liu, B. Kauffmann, X. Miao and D. Komáromy, et al., Emergence of low-symmetry foldamers from single monomers, Nat. Chem., 2020, 12(12), 1180–1186 CrossRef CAS PubMed.
- J. Aupič, F. Lapenta and R. Jerala, SwitCCh: Metal-Site Design for Controlling the Assembly of a Coiled-Coil Homodimer, ChemBioChem, 2018, 19(23), 2453–2457 CrossRef PubMed.
- R. M. Reja, M. Khan, S. K. Singh, R. Misra, A. Shiras and H. N. Gopi, pH sensitive coiled coils: a strategy for enhanced liposomal drug delivery, Nanoscale, 2016, 8(9), 5139–5145 RSC.
- C. Mueller and T. N. Grossmann, Coiled-Coil Peptide Beacon: A Tunable Conformational Switch for Protein Detection, Angew. Chem., Int. Ed., 2018, 57(52), 17079–17083 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06658d |
| ‡ University of Nova Gorica, Vipavska 13, SI5000 Nova Gorica, Slovenia |
| This journal is © The Royal Society of Chemistry 2024 |