Open Access Article
Open Access ArticleStructural characterization of PHOX2B and its DNA interaction shed light on the molecular basis of the +7Ala variant pathogenicity in CCHS†
Donatella
Diana‡
a,
Luciano
Pirone‡
a,
Luigi
Russo‡
b,
Gianluca
D’Abrosca
c,
Manoj
Madheswaran
 b,
Roberta
Benfante
def,
Simona
Di Lascio
d,
Laura
Caldinelli
g,
Diego
Fornasari
d,
Clementina
Acconcia
b,
Andrea
Corvino
b,
Nataliia
Ventserova
b,
Loredano
Pollegioni
g,
Carla
Isernia
b,
Roberta
Benfante
def,
Simona
Di Lascio
d,
Laura
Caldinelli
g,
Diego
Fornasari
d,
Clementina
Acconcia
b,
Andrea
Corvino
b,
Nataliia
Ventserova
b,
Loredano
Pollegioni
g,
Carla
Isernia
 b,
Sonia
Di Gaetano
a,
Gaetano
Malgieri
*b,
Emilia M.
Pedone
b,
Sonia
Di Gaetano
a,
Gaetano
Malgieri
*b,
Emilia M.
Pedone
 *a and
Roberto
Fattorusso
*a and
Roberto
Fattorusso
 *b
*b
aCNR – Institute of Biostructures and Bioimaging, Via Pietro Castellino 111, 80131 Naples, Italy. E-mail: emilia.pedone@cnr.it
bDepartment of Environmental, Biological and Pharmaceutical Sciences and Technologies – University of Campania “Luigi Vanvitelli”, Via Vivaldi 43, 81100 Caserta, Italy. E-mail: gaetano.malgieri@unicampania.it; roberto.fattorusso@unicampania.it
cDepartment of Clinical and Experimental Medicine – University of Foggia, Viale Luigi Pinto, 71122 Foggia, Italy
dDepartment of Medical Biotechnology and Translational Medicine (BIOMETRA), Università degli Studi di Milano, Milan, Italy
eCNR – Institute of Neuroscience, Vedano Al Lambro (MB), Italy
fNeuroMi – Milan Center for Neuroscience, University of Milano Bicocca, Milan, Italy
gDepartment of Biotechnology and Life Sciences, University of Insubria, Via J.H. Dunant 3, 21100 Varese, Italy
First published on 1st May 2024
Abstract
An expansion of poly-alanine up to +13 residues in the C-terminus of the transcription factor PHOX2B underlies the onset of congenital central hypoventilation syndrome (CCHS). Recent studies demonstrated that the alanine tract expansion influences PHOX2B folding and activity. Therefore, structural information on PHOX2B is an important target for obtaining clues to elucidate the insurgence of the alanine expansion-related syndrome and also for defining a viable therapy. Here we report by NMR spectroscopy the structural characterization of the homeodomain (HD) of PHOX2B and HD + C-terminus PHOX2B protein, free and in the presence of the target DNA. The obtained structural data are then exploited to obtain a structural model of the PHOX2B–DNA interaction. In addition, the variant +7Ala, responsible for one of the most frequent forms of the syndrome, was analysed, showing different conformational proprieties in solution and a strong propensity to aggregation. Our data suggest that the elongated poly-alanine tract would be related to disease onset through a loss-of-function mechanism. Overall, this study paves the way for the future rational design of therapeutic drugs, suggesting as a possible therapeutic route the use of specific anti-aggregating molecules capable of preventing variant aggregation and possibly restoring the DNA-binding activity of PHOX2B.
Introduction
Nearly 500 human proteins contain polyAla tracts. In particular, alanine tracts are encoded by imperfect trinucleotide repeats (GCNs), and polyAla expansions are reported to cause at least nine different diseases including developmental defects and neurological disorders.1,2In general, little is known about the effect of polyAla expansions on disease-associated proteins that natively contain polyAla sequences. Until now, there have been no detailed structural studies of protein structures in the field of polyA, and furthermore, experimental structures have been reported for only two related proteins, HOXA13 (ref. 3) and PABPN1.4 Because of the apparent similarities in mutation patterns with polyGln expansion diseases, polyAla, too, might promote misfolding and aggregation as observed in the study carried out by Hernandez and Facelli in 2020 by structural prediction analyses; indeed, this mechanism has been proposed suggesting the formation of amyloid fibrils.5 Differently, other studies indicated that polyAla aggregates into α-helical structures of an amorphous nature.6
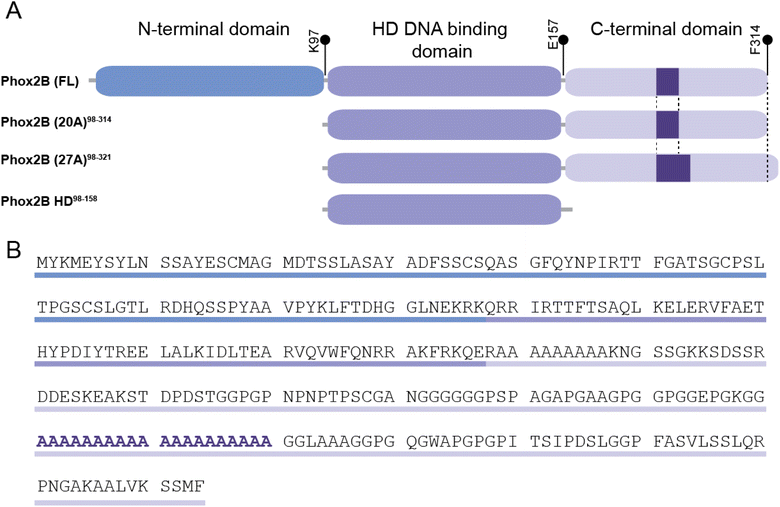
PHOX2B is a transcription factor that plays a key role in the development of the autonomic nervous system and neural structures involved in the control of respiration.7–9 It is a protein of 314 amino acids that can be subdivided into an N-terminal domain of 97 residues, a central domain (the DNA binding motif) containing a conserved homeodomain of 60 residues, and, within the C-terminal domain, two poly-alanine (polyAla) tracts of 9 and 20 residues, respectively (Fig. 1 A and B). About 90% of congenital central hypoventilation syndrome (CCHS) patients, characterized by defective autonomic control of breathing, show polyalanine triplet expansions, ranging from +5 to +13 alanine residues, of the 20 alanine stretch region of transcription factor PHOX2B,10,11 rendering this protein an intriguing target to understand the insurgence of this syndrome and for the design of a novel therapeutic approach.12 Indeed, CCHS is considered a life-long disease and nowadays there is no pharmacological intervention, so there is a compelling need for ventilatory support such as tracheostomy or a nasal mask. A correlation between expanded alanine tract length and severity of the phenotype has been reported.11 Moreover, functional studies have correlated expansions of polyAla with a decreased activation of dopamine-beta-hydroxylase (DBH) and PHOX2A promoters by PHOX2B, due to polyAla length-dependent formation of cytoplasmic aggregates that prevent the protein from performing its activities as a transcription factor such as entering the nucleus, binding DNA and activating transcription.13–15
In such a context, our previous studies, which focused on the biochemical characterization of different PHOX2B variants, highlighted the propensity of the pathogenic PHOX2B variant containing the polyalanine expansion (+7 alanines) to aggregate, especially in the presence of DNA and, unexpectedly, the formation of fibrils which possibly play a role in the insurgence of CCHS.16 The entire wild-type PHOX2B could not be produced in a recombinant form as a folded protein able to bind DNA if not under denaturing conditions. Therefore, our study was focused on the full-length protein lacking the 97 unstructured N-terminal amino acids (PHOX2B-20A).
Absolute limitations to the comprehension of the pathogenesis of CCHS, and the development of new and effective treatments for this disease are particularly correlated with the substantial lack of information on the structure–function relationships on wild-type and variant PHOX2B proteins.
Therefore, the aim of this study is to elucidate the pathogenesis of CCHS by a structural biology approach carrying out a characterization by NMR spectroscopy of PHOX2B-20A, its homeodomain and the pathological variant (+7 alanines). Within this frame, we here report the structural characterization by solution NMR spectroscopy of the wild-type PHOX2B-20A and its homeodomain and of their interaction with the DNA target, providing novel insights for the elucidation of the molecular mechanisms involved in the insurgence of the pathology.
Experimental section
NMR spectroscopy
All NMR experiments were carried out at 25 °C using a Bruker Avance III HD 600 MHz spectrometer, equipped with a triple resonance Prodigy N2 cryoprobe with a z-axis pulse field gradient, at the NMR laboratory of the Department of Environmental, Biological and Pharmaceutical Sciences and Technologies of the University of Campania “Luigi Vanvitelli” (Caserta, Italy).Recombinant 15N/13C- and 15N-labeled samples, were prepared through a procedure similar to that previously described,16 but cells were grown in M9 medium with the addition of 13C-labeled glucose and 15N-labeled ammonium chloride.
NMR structural studies were conducted on samples by dissolving proteins in 20 mM sodium phosphate pH 7.4, 200 mM NaCl, 0.02% sodium azide and 10% 2H2O to a final concentration of ∼100 μM for PHOX2B-HD and ∼80 μM for PHOX2B-20A and PHOX2B-27A.
NMR chemical shift assignment
Backbone resonances Cα, Cβ, C′, N, HN and Hα of PHOX2B-HD were assigned by analysing the standard triple resonance experiments as 3D HNCA, 3D CBCANH, 3D CBCA(CO)NH, 3D HNCO and 3D HNHα.17 In the case of PHOX2B-20A, considering the lack of chemical shift dispersion and severe signal-overlap for the C-terminal domain resonances, we analysed the conventional triple resonance spectra together with advanced NMR experiments suitable for intrinsically disordered proteins (IDPs) such as 3D HNN18 and 1H,15N T2-filter HSQC based experiments.19 These latter were conducted by using a relaxation-compensated Carr–Purcell–Meiboom–Gill sequence period of 100 ms.All NMR data were processed with the software NMRpipe.20 2D and 3D NMR spectra were analysed using CARA21 and SPARKY software.22
NMR structure and dynamics of PHOX2B-HD
Secondary structural elements of both PHOX2B-HD were initially identified by evaluation of the deviation of the observed Cα and Hα chemical shifts from the random coil values calculated as proposed by Kjaergaard et al.23,24 Dihedral angle restraints were calculated from HN, Cα, Hα and N chemical shifts with the software TALOS+.25 The structure calculation was performed with the program CS-Rosetta26 using as structural restraints the torsion angles ϕ/ψ derived from TALOS+ database and the HN, Cα, Hα, Cβ, C′ and N chemical shifts of those residues indicated by TALOS+ to be rigid in the pico-second timescale with an order parameter S2 > 0.7. A set of 200 fragment candidates matching the experimental chemical shifts was used to calculate 3000 conformers. After this, the energy of the calculated Rosetta conformers was rescored against the observed chemical shifts and the 20 structures with the lowest energy were selected to define the final structural ensemble.NMR structures were visualized and evaluated by using the program MOLMOL,27 CHIMERA28 and PROCHECK-NMR.29 Backbone dynamics of PHOX2B-HD were investigated by analyzing the model-free order parameters (S2) and R2 relaxation rates. The per-residue S2 values for the backbone amide groups were predicted from the backbone and Cβ chemical shifts using the random coil index approach.30 The relaxation rates R2 were estimated by analysing in the 1H,15N-HSQC spectrum the 15N linewidth (λ) that is related to R2, and this shows the dynamic features of molecules in solution. Moreover, hydrodynamic proprieties and NMR relaxation parameters of PHOX2B-HD were predicted from the representative NMR ensemble by HYDRONMR using an AER value of 3.3 Å which is the average atomic element radius for most proteins which ranges between 2.8 and 3.8 Å.31
NMR analysis of the PHOX2B–DNA interaction
Secondary structure elements of the free PHOX2B-20A were identified by analysing the Cα secondary chemical shifts (Δδ) which are the difference between the observed chemical shift (δobs) and the residue-specific random coil shift (δcoil) predicted as reported above. Chemical shift perturbation (CSP) studies of PHOX2B-HD with its 25-bp DNA target sequence named ATTA2 (5′-TAGTGTGATTGAATTAAAGGGCAGG-3′)32 (purchased from Sigma-Aldrich, St. Louis, Missouri, USA) were carried out using 15N-labeled PHOX2B dissolved in 300 μL at 100 μM concentration in 20 mM sodium phosphate pH 7.4, 200 mM NaCl, 0.02% sodium azide and 10% 2H2O. The DNA fragment was first dissolved at 300 μM concentration in 20 mM sodium phosphate pH 7.4 buffer, 50 mM NaCl, and then carefully mixed with PHOX2B-HD at a final concentration of 10 and 50 μM, as reported in Omichinsky et al.33 2D 1H,15N-HSQC spectra were acquired in the absence and presence of DNA. Starting from the amide resonances for PHOX2B-HD free, average combined chemical shift changes for PHOX2B-HD bound were determined using the following equation: ΔδHNav = [((ΔδH)2+(ΔδN/5)2)/2]1/2, where ΔδH and ΔδN are the chemical shift variations of the amide proton and nitrogen resonances,34,35 respectively. Moreover, intensity reduction of the amide cross-peaks over one standard deviation was also taken into account to define the DNA-binding site. The same procedure was employed for CSP studies of PHOX2B-20A with DNA.Homology modelling and dynamics studies of the PHOX2B-20A homodimer
The 3D structure of the homeodomain within PHOX2B-20A was built based on the primary sequence by the Alphafold methodology using as a template, for the region Gln98 to Glu157, the representative NMR structure obtained as reported above. In particular, the HD domain of PHOX2B-structural prediction was obtained using Alphafold v2.1 with default parameters.36 Thus, the calculation was launched with one homo-oligomer, MMseqs2 option for multiple sequence alignment (MSA) searching, unpaired mode for generating separate MSA for each protein and no filter options for pair_cov (minimum coverage with query (%)) and pair_qid (minimum sequence identity with query (%)). The structural models were generated using the following setting parameters: number of models = 5; max recycles = 3. The calculated conformers were very similar and the one with the highest rank based on pLDDT was selected as a reference structure and was used for the molecular docking as reported in the next paragraph. The HD domain of PHOX2B-20A was visualized and analysed using PyMOL 2.1 (https://pymol.org/2/) and CHIMERA.28 In all cases the identification of the secondary structure elements was performed using the DSSP software.37Building of the PHOX2B-20A homodimer/DNA complex by molecular docking
The DNA three-dimensional structure for the docking investigation was built using the software package 3DNA38 from the 25 bp double-stranded oligonucleotide corresponding to the ATTA2 site of the PHOX2B promoter32 used to explore the DNA binding abilities of PHOX2B-20A in a previous publication.16 To reproduce the DNA binding mechanism of the PHOX2B-20A dimer, according to the correlation plots of the NMR chemical shifts, we treated the two homeodomains as independent units excluding the contribution of the C-terminal tail.The software HADDOCK39 was used to dock the PHOX2B-20A homodimer to DNA using the ambiguous interaction restraints (AIRs) identified by evaluating the chemical shift variations observed for PHOX2B-HD upon addition of DNA, by DISPLAR analysis40 and by considering the solvent-accessible surface area (ASA) of PHOX2B-20A residues defined using the ProtSA approach.41 In particular, nine residues (Arg100, Tyr122, Asp124, Arg141, Asn148, Arg150, Ala151, Arg154, and Arg158) of each PHOX2B-20A chain, showing significant chemical shift perturbations (CSP > CSPaverage) upon binding to DNA, with >50% solvent accessibility were defined as “active” residues. In addition, a further ten residues (Gln143, Val144, Trp145, Gln147, Arg149, Lys152, Phe153, Lys155, Gln156, and Glu157) were considered “active” as revealed by DISPLAR and ASA analysis. In the case of the DNA, we considered as “active” the nucleotides encompassing the 5′-ATTA-3′ recognition site and the incomplete ATT sequence located 5′ to the ATTA motif. In both cases, the “passive” amino acids and nucleotides were automatically defined by the HADDOCK software. The structural model of the PHOX2B-20A homodimer/DNA complex was obtained following the HADDOCK 2.4 protocol using optimal settings for protein/DNA docking. In particular, during rigid-body energy minimization, 2000 docking conformers were calculated. The best 200 structures in terms of intermolecular energies were then used for the semi-flexible simulating annealing, followed by explicit water refinement. Then, the obtained structures were clustered using FCC (fraction of common contacts) method with a cut-off of 0.60. The representative structure from the lowest energy cluster having the lowest HADDOCK-score and Z-score values was accepted as the reference structural model of the complex.
Results
Structure and dynamics of the PHOX2B homeodomain
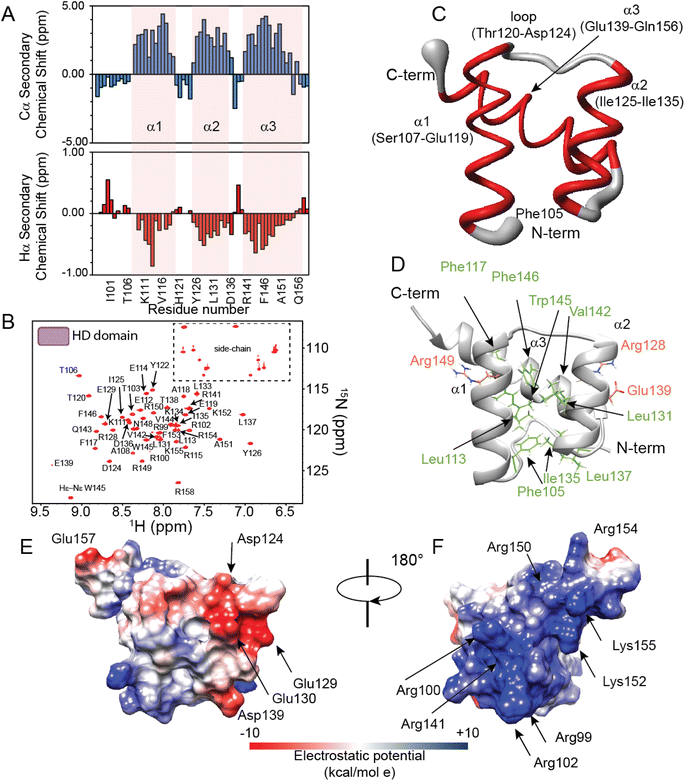
To elucidate the structural and dynamical features of the DNA recognition mechanism by PHOX2B-20A, we first investigated by NMR spectroscopy the PHOX2B-HD98–158 fragment (hereinafter called PHOX2B-HD), obtained from the wild-type protein upon deletion of the N- and C-terminal domains, that includes the homeodomain encompassing the region from Gln98 to Glu157 (Fig. 1A and B). Its 1H,15N-HSQC spectrum shows good cross-peak dispersion in both nitrogen and proton dimensions, revealing a well-defined native structure in aqueous solution for PHOX2B-HD (Fig. 2A and B), obtaining good quality triple resonance spectra that allowed a nearly complete backbone assignment (1HN, 1Hα, 15N, 13Cα, 13Cβ and 13CO) (ESI Table S1†). After this, secondary structure elements of PHOX2B-HD were identified by analysing the backbone chemical shifts that are sensitive reporters of the secondary structure content.42,43 As illustrated in Fig. 2A, the observed Hα and Cα chemical shifts (δobs) significantly deviate from the random coil value (δcoil) predicted as reported in the Materials and methods section. Specifically, the positive deviations of Cα and the negative Hα secondary chemical shifts (δobs − δcoil) strongly suggest that PHOX2B-HD presents, as reported for other homeodomains,44–46 three α-helical regions with residues Ser107–Glu119, Ile125–Ile135 and Ala140–Phe153 forming α1, α2 and α3 helices, respectively. Then, to provide a rigorous high-resolution description of the conformational properties of the homeodomain, we determined the 3D structure of PHOX2B-HD using as structural constraints the assigned backbone chemical shifts and torsion angles φ/ψ derived from the TALOS+ database of those residues reported by TALOS+ to be rigid in the picosecond timescale with an order parameter S2 > 0.7 (for details see the Materials and methods section).A high-quality structure (Table S1†) was obtained for PHOX2B-HD (Fig. 2C and D) consisting of a very well-defined globular domain in the region encompassing residues from Ser107 to Lys152 (Table S1†), as indicated by the root-mean-square deviation (rmsd) of the backbone of 0.527 Å. The NMR structure reveals that PHOX2B-HD presents the typical topology of the homeodomain with three α-helices and an N-terminal arm (residues Gln98–Thr106) lacking any preferential conformation. In detail, helices α1 (Ser107–Glu119) and α2 (Ile125–Ile135) are antiparallel, while helix α3 (Ala140–Gln156), also called the recognition helix, perpendicularly faces them (Fig. 2C and D). α1 and α2 are connected by a loop of five residues (Thr120, His121, Tyr122, Pro123, and Asp124) (Fig. 2C and D) which are in extended conformation with alternating residues either exposed or buried in the core domain. Helices α2 and α3 are connected by a turn constituting a so-called helix-turn-helix (HTH) motif.47 The conserved residues Phe105, Leu113, Phe117, Leu131, Ile135, Leu137, Val142, Trp145 and Phe146 play the primary role in defining a hydrophobic core in which the aliphatic arms of the charged and polar residues Arg128, Glu139 and Arg149 are buried and contribute to the core (Fig. 2D). This fold is further stabilized by several long-range hydrogen bonds seen in the ensemble of structures, including bonds between the Gln109 side chain and Ile135 backbone carbonyl, Glu112 and Lys134, and Glu114 and Arg149 side chains. Analysis of the electrostatic surface of PHOX2B-HD indicates that α3 and the N-terminal arm are rich in positively charged residues while α1 and α2 helices are negatively charged. Overall, the domain shows few small hydrophobic pockets (Fig. 2E and F).
Then, we investigated protein motions of PHOX2B-HD in the nanosecond-to-picosecond and millisecond-microsecond timescales by analysing the model-free order parameters (S2) estimated for the backbone amide group from the assigned chemical shifts (for details see the Materials and methods section, please). We also compared the 15N transverse relaxation rates (R2) obtained from 15N linewidths with the relaxation parameters predicted from the representative PHOX2B-HD NMR ensemble (Fig. S1A and B†) using HYDRONMR software.48–50S2 is a measure of the partial restriction in the ns–ps timescale of the H–N bond in a molecule-fixed frame that assumes values from 0 (a large amount of motion) to 1 (no motion). As reported in Fig. S1A,† PHOX2B-HD presents S2 values in the range of 0.725–0.926 indicating that the homeodomain is characterized by a reduced internal mobility in the ns–ps timescale. Moreover, the comparison of the 15N R2 rates estimated from the 15N linewidths (R2 average value of 8.13 ± 0.5 s−1) with the values back-calculated from the NMR structure (R2 average value 8.09 ± 0.24 s−1) (Fig. S1B†) demonstrated that PHOX2B-HD adopts a compact globular fold with a remarkable conformational rigidity in the ms–μs timescale.
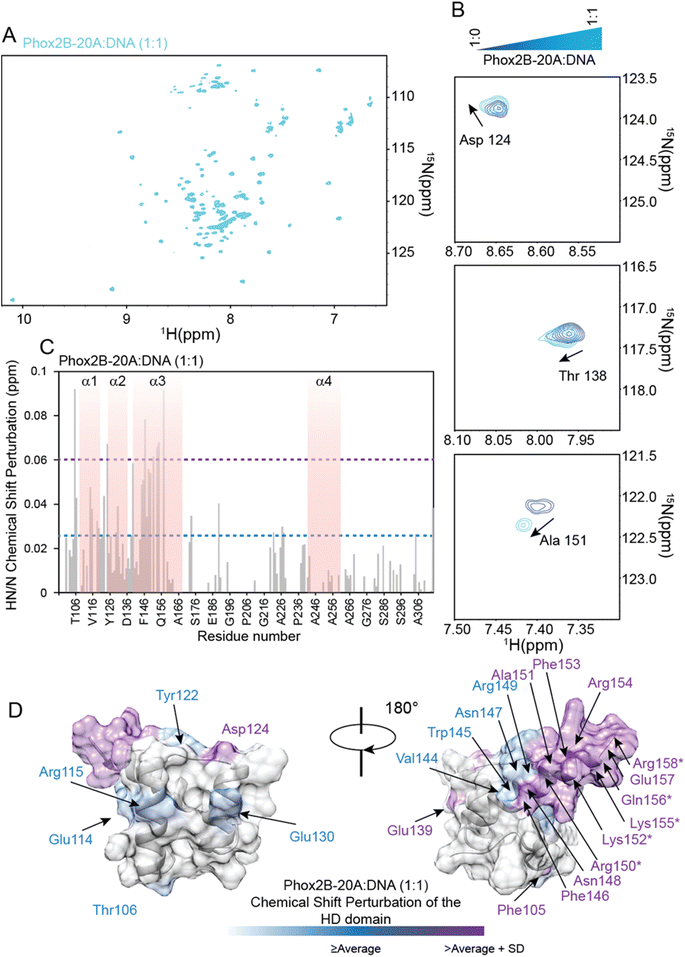
PHOX2B homeodomain–DNA interaction
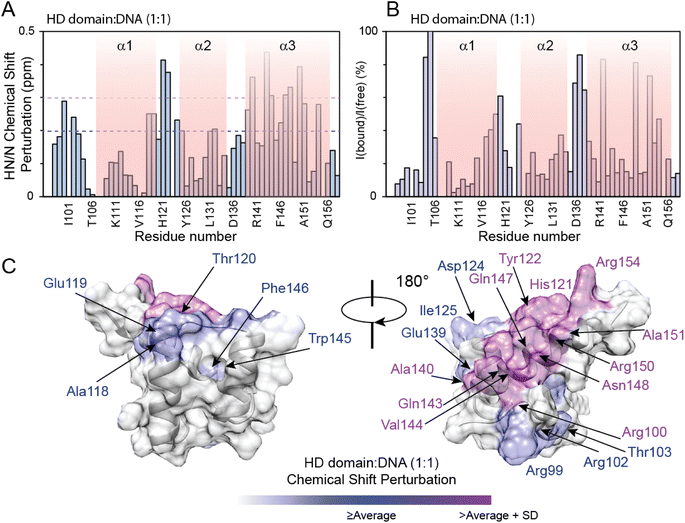
The PHOX2B-HD/DNA interaction was investigated by evaluating the chemical shift perturbation (CSP) and intensity ratio of the homeodomain resonances upon DNA binding (Fig. 3). In particular, the 1H,15N-HSQC spectrum of 15N–13C labeled PHOX2B-HD in the presence of its DNA target sequence (named ATTA2) that has been shown to be bound by PHOX2B with high affinity,14,32 was acquired (molar ratio PHOX2B-HD/DNA 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Notably, a subset of PHOX2B-HD resonances in the 11H,15N-HSQC spectrum showed significant variations upon DNA addition, indicating that several residues of the homeodomain were involved in the DNA binding mechanism. The 1H and 15N chemical shift changes were evaluated by applying for each residue a combined weighted chemical shift perturbation (see the Materials and methods section). Significant perturbations were observed for Arg100 (N-terminal arm); His121 and Tyr122 (loop between helices α1 and α2); Ala140, Gln143, Val144, Gln147, Asn148, Arg150, Ala151 and Arg154 (helix α3) (Fig. 3A–C). Moreover, small significant differences were observed for Arg99, Arg102 and Thr103 (N-terminal arm); Ala118 and Glu119 (edge of helix α1); Thr120 and Asp124 (turn between helices α1 and α2); Ile125, Leu131, and Ala132 (helix α2); Leu137 (turn between helices α2 and α3); Glu139, Trp145 and Phe146 (helix α3) (Fig. 3A and C). In addition, the comparison of the HN/N cross-peak intensities of PHOX2B-HD in the absence and presence of DNA showed a significant reduction (>average value) mainly for the residues located in the: (i) N-terminal arm (i.e. Gln98, Arg99, Arg100, Arg102 and Thr103); (ii) loop connecting the helices α1 to α2 (i.e. Tyr126, Arg128, Glu129, Glu130 and Leu131); (iii) α3-helix (i.e. Ala140, Arg141, Gln143, Trp145, Phe146, Gln147, Asn148, Arg150, Ala151 and Gln156) (Fig. 3B). Taken together, CSP data and intensity profile analysis (Fig. 3A–C) demonstrate that the DNA recognition mechanism by PHOX2B-HD is principally driven by the α3-helix of the HTH motif that, as observed for other members of the homeodomain family, has a crucial role in the DNA binding process by interacting with the DNA major groove. Moreover, NMR structural data indicate that the PHOX2B-HD/DNA complex is further stabilized by the positively charged region of the N-terminal arm preceding the first helix. This latter observation is in agreement with previously published data indicating that the N-terminal region of the homeodomain participates in DNA binding by making specific interactions with the DNA minor groove.44,45
1). Notably, a subset of PHOX2B-HD resonances in the 11H,15N-HSQC spectrum showed significant variations upon DNA addition, indicating that several residues of the homeodomain were involved in the DNA binding mechanism. The 1H and 15N chemical shift changes were evaluated by applying for each residue a combined weighted chemical shift perturbation (see the Materials and methods section). Significant perturbations were observed for Arg100 (N-terminal arm); His121 and Tyr122 (loop between helices α1 and α2); Ala140, Gln143, Val144, Gln147, Asn148, Arg150, Ala151 and Arg154 (helix α3) (Fig. 3A–C). Moreover, small significant differences were observed for Arg99, Arg102 and Thr103 (N-terminal arm); Ala118 and Glu119 (edge of helix α1); Thr120 and Asp124 (turn between helices α1 and α2); Ile125, Leu131, and Ala132 (helix α2); Leu137 (turn between helices α2 and α3); Glu139, Trp145 and Phe146 (helix α3) (Fig. 3A and C). In addition, the comparison of the HN/N cross-peak intensities of PHOX2B-HD in the absence and presence of DNA showed a significant reduction (>average value) mainly for the residues located in the: (i) N-terminal arm (i.e. Gln98, Arg99, Arg100, Arg102 and Thr103); (ii) loop connecting the helices α1 to α2 (i.e. Tyr126, Arg128, Glu129, Glu130 and Leu131); (iii) α3-helix (i.e. Ala140, Arg141, Gln143, Trp145, Phe146, Gln147, Asn148, Arg150, Ala151 and Gln156) (Fig. 3B). Taken together, CSP data and intensity profile analysis (Fig. 3A–C) demonstrate that the DNA recognition mechanism by PHOX2B-HD is principally driven by the α3-helix of the HTH motif that, as observed for other members of the homeodomain family, has a crucial role in the DNA binding process by interacting with the DNA major groove. Moreover, NMR structural data indicate that the PHOX2B-HD/DNA complex is further stabilized by the positively charged region of the N-terminal arm preceding the first helix. This latter observation is in agreement with previously published data indicating that the N-terminal region of the homeodomain participates in DNA binding by making specific interactions with the DNA minor groove.44,45
PHOX2B-20A structural and dynamical characterization
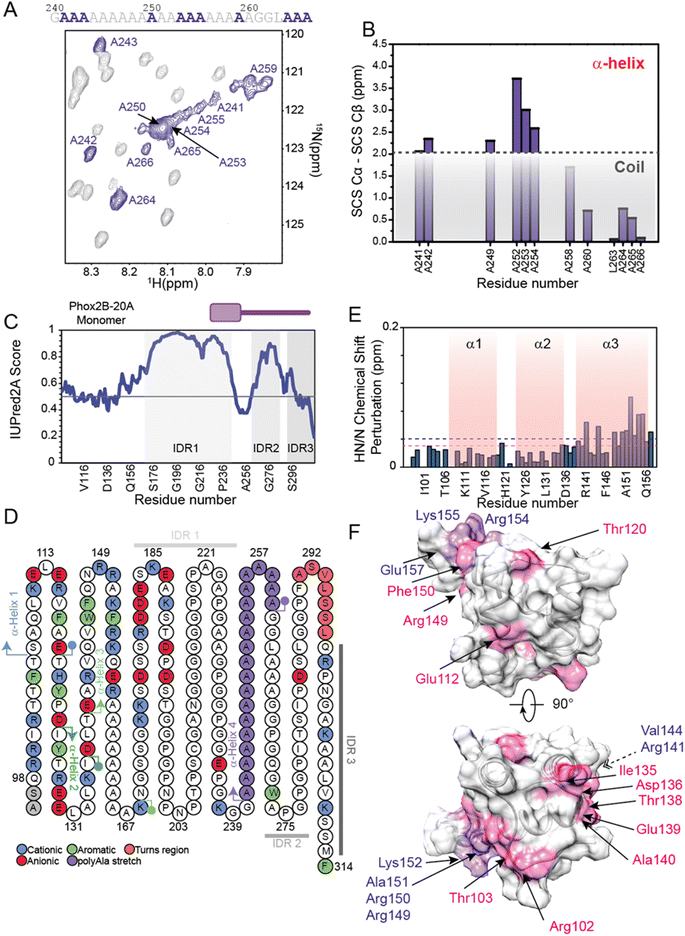
The 1H,15N-HSQC spectrum of PHOX2B-20A shows a well-defined set of signals, well distributed and dispersed, indicating how PHOX2B-20A contains a well-structured domain, and strong HN/N cross-peak signals condensed at 8 ppm due to a poorly structured part of the protein (Fig. S2†). For this reason, the backbone chemical shift assignment for this protein, especially for this second set of resonances, resulted in extremely challenging51 with several highly overlapped signals arising from the homorepeat regions (Fig. S2 and S3†). To overcome the problem of signal overlap, we conducted a series of multi-dimensional experiments suitable for IDPs including 1H,15N T2-filter HSQC-based NMR techniques (Fig. S4†). By using an ad hoc assignment strategy, we assigned 96 and 69% of the HN, N, Cα, and Cβ chemical shifts for HD and PHOX2B-20A respectively. Particularly, the HD of PHOX2B-20A region resonances was assigned based on PHOX2B-HD assignments (Fig. S2†). In agreement with the previously described light scattering data,16 which indicate a monomeric nature of PHOX2B-20A in solution, the 15N R2 rates of the homeodomain portion, estimated by analysing the 15N linewidth in the 1H,15N–HSQC spectrum, give an average value of 9.11 ± 2.04 s−1, similar to the values back-calculated for PHOX2B-HD from the NMR structure, showing that the PHOX2B-20A protein exists as a monomer. Interestingly, R2 values higher than the average have been found for residues at the N-terminal region of the globular domain, likely due to chemical exchange processes.The obtained assignment allowed us to explore the secondary structure organization of PHOX2B-20A (Fig. 4A, B, S5 and S6†). PHOX2B-20A consists of a well-defined N-terminal domain, the HD, followed by a mostly disordered C-terminus (Fig. 4C and S6†). Accordingly, most of the signals in the C-terminal region of PHOX2B-20A show chemical shifts typically found in residues in largely unfolded proteins that are exposed to the solvent and therefore experience a non-unique local chemical environment (Fig. S5 and S6†).
Cα and ΔδCα − ΔδCβ secondary chemical shifts (Fig. 4B, S5A and B†) clearly indicate that PHOX2B-20A presents in addition to the three helices, forming the HD domain, a fourth α-helix (α4) encompassing the poly-Ala region from Ala241 to Ala260 (Fig. 4B). Note that, while the residues Ala259–Ala266 at the C-terminus of α4 show a helical propensity, Cα chemical shifts of most of the residues flanking this polyAla region only slightly deviate from the random coil value confirming that the α4 helix is located in the middle of a largely unfolded C-terminal domain (Fig. 4B and S5B†).
In detail, according to the IUPred2A score, Cα secondary chemical shifts analysis indicates that the C-terminal domain of PHOX2B-20A presents three intrinsically disordered regions (Fig. 4C, D and S6†). Particularly, the highly dynamic C-terminal domain presents a first intrinsically disordered region (IDR1) from Gly170 to Gly240 that is connected, through the stable poly-Ala α-helical segment, to a second IDR (IDR2) encompassing the region from Ala260 to Pro290. IDR2 is linked by a short turn region (residues Phe291, Ala292, Ser293, Val294, Leu295, Ser296 and Ser297) to IDR3 comprising the portion from Leu298 to Phe314 (Fig. 4C and D).
Moreover, the comparison of the length and distribution of the secondary structure within PHOX2B-20A with the PHOX2B-HD NMR structure suggests that in the PHOX2B-20A the third α-helix (α3) of the homeodomain is three turns longer including residues (Fig. S5A†).
The C-terminal part of α3 connects the homeodomain to the disordered C-terminal tail. Interestingly, by comparing the 1H,15N-HSQC spectrum measured for PHOX2B-20A with that acquired for the PHOX2B-HD (Fig. S2†), significant chemical shift perturbations were observed for most of the residues forming the homeodomain, also within the N-terminus. In particular, chemical shift variations greater than the standard deviation were observed for Arg141, Val144, Arg149, Arg150, Ala151, Lys152, Arg154, Lys155 and Glu157 belonging to the α3 helix (Fig. 4E, F and S7†).Yet, significant chemical shift perturbations (CSPs > CSPavg) were observed for Arg102 and Thr103 (N-terminal tail); Glu112 located inside the first α-helix (α1); Thr120 within the loop connecting α1 to α2; Ile135, Asp136, Thr138, and Glu139 located in the region linking the α2 to α3; Ala140, Arg149 and Phe150 within α3.
As chemical shifts are extremely sensitive to changes in the local chemical environment of a residue, this indicates that the regions of the two proteins with these super-imposable resonances adopt a highly similar structure.52 However, some signals experience a shift in their position in the spectrum of PHOX2B-20A with respect to their equivalent position in the HD spectrum. These chemical shift variations reflect a perturbation in the local environment of the corresponding residues caused by changes in the conformation of either these residues themselves or spatially proximal residues. Although it is quite plausible that the addition of the C-terminal region to PHOX2B-HD perturbs the chemical environment of contiguous portions of the HD domain, the overall chemical shifts perturbations observed (Fig. 4E) require an additional molecular interpretation. The chemical shift perturbations observed for a large number of residues located in regions far from the HD-domain C-term (Fig. 4E) suggest that in PHOX2B-20A the homeodomain cross-talks with the unstructured C-terminal domain that bears the three IDRs.
While IDRs are generally characterized by a high primary sequence variability, charge distribution confers specific properties to the IDRs of the transcription factors.53–55 Therefore, we analysed the charge characteristics of PHOX2B-20A IDRs in order to understand their functional roles. IDR1 presents a positively charged stretch interrupted by acidic and hydrophobic residues; IDR2 is rich in hydrophobic residues: Ala 16.1%, Gly 32.3% and Pro 19.4%; IDR3 contains Ala (17.6%), Leu (11.8%) and three positively charged residues (17.7%). Overall, our analysis indicates that the IDRs of PHOX2B-20A, considering the great number of hydrophobic amino-acids rather than charged residues, have a marginal role in the DNA recognition process but they may play an important role in mediating protein–protein and dimerization mechanisms.
Structural insights into the PHOX2B-27A variant
The conformational characterization of the PHOX2B variant containing an elongated 27 alanine stretch, named PHOX2B-27A, was also carried out by means of NMR methodologies. Interestingly, the comparison of the 15N,1H-HSQC spectrum of PHOX2B-27A (Fig. S8†) with that acquired for PHOX2B-20A indicates that the two proteins present different conformational properties in solution. Indeed, PHOX2B-27A HSQC shows a general line broadening of the resonances and the disappearance of most HSQC signals. This behavior is again in agreement with our previously reported study16 which shows how the insertion of the seven alanine residues at position 260 leads to a strong aggregation propensity for PHOX2B-27A.PHOX2B-20A interaction with DNA
The identification of the key residues involved in the molecular mechanism by which PHOX2B-20A recognizes its cognate DNA was assessed by means of backbone 1H/15N CSP mapping. We analyzed 15N,1H-HSQC spectra of PHOX2B-20A acquired before and upon the addition of 1 equivalent of the ATTA2 DNA sequence (Fig. 5A and B). This choice was made on the basis of previous studies that have documented the formation of a homodimer upon PHOX2B-20A interaction with DNA.16 Chemical cross-linking experiments have shown the formation of mixed oligomeric species that disappear when the protein: DNA ratio is 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.16 Upon addition of an equimolar DNA aliquot, the 1H/15N correlation spectrum of PHOX2B-20A exhibits substantial chemical shift variations for a large number of residues along with signal loss and broadening for the C-terminal residues of the homeodomain (Arg150, Lys152, Lys155, Gln156 and Arg158) (Fig. 5B and C). Larger CSPs were observed for Phe105 (N-term arm); Asp124 (loop between α1 and α2); Glu139, Phe146, Asn148, Ala151, Phe153, Arg154 and Glu157 (α3 helix). Small chemical shift perturbations were detected for Thr106 (N-term arm); Glu114 and Arg115 (α1 helix); Tyr122 (loop between α1 and α2); Glu130 (α2 helix); Val144, Trp145, Asn147 and Arg149 (α3 helix) (Fig. 5C and D). Moreover, most of the residues of the C-terminal domain show chemical shift variations (CSPs < CSPavg), that although smaller than those shown by the HD (Fig. 5C) demonstrate that, as previously reported,15 the C-terminal domain is essential for PHOX2B-20A/DNA recognition. Overall, CSP data allowed the identification of the DNA binding surface of PHOX2B-20A (Fig. 5D). Interestingly, this surface largely matches the surface that we have identified as responsible for the DNA binding activity of PHOX2B-HD. In accordance with the literature data on the homeodomain-containing proteins,47 also in the interaction of PHOX2B-20A with its target DNA, helix α3 and the N-terminal portion of the HD are confirmed as the key structural elements. In addition, significant perturbations were found also for the signals arising from residues included in the helix α1 in which, in particular, Leu110, Glu112, Ala118 and Glu119 give rise to two signals each. As helix α1 remains essentially unperturbed upon the DNA interaction of PHOX2B-HD, these data strongly indicate α1 to be a mediator for the formation of the homo-dimeric structure that binds the DNA. Indeed, the presence of a double set of helix 1 chemical shifts suggests a slight asymmetry of the homodimer, where the two α1 helices experience different chemical environments; the two sets of signals of this secondary structure are likely due to its involvement in the interaction with the C-terminal residues to form the dimer in a head to tail fashion (Fig. S11†). In addition, the good correlation of the HN and N chemical shifts observed for PHOX2B-20A with the values obtained for PHOX2B-HD indicates that the PHOX2B-20A DNA binding occurs by a recognition mechanism in which the homeodomain acts as a (partial) independent unit (Fig. S9A and B†).
1.16 Upon addition of an equimolar DNA aliquot, the 1H/15N correlation spectrum of PHOX2B-20A exhibits substantial chemical shift variations for a large number of residues along with signal loss and broadening for the C-terminal residues of the homeodomain (Arg150, Lys152, Lys155, Gln156 and Arg158) (Fig. 5B and C). Larger CSPs were observed for Phe105 (N-term arm); Asp124 (loop between α1 and α2); Glu139, Phe146, Asn148, Ala151, Phe153, Arg154 and Glu157 (α3 helix). Small chemical shift perturbations were detected for Thr106 (N-term arm); Glu114 and Arg115 (α1 helix); Tyr122 (loop between α1 and α2); Glu130 (α2 helix); Val144, Trp145, Asn147 and Arg149 (α3 helix) (Fig. 5C and D). Moreover, most of the residues of the C-terminal domain show chemical shift variations (CSPs < CSPavg), that although smaller than those shown by the HD (Fig. 5C) demonstrate that, as previously reported,15 the C-terminal domain is essential for PHOX2B-20A/DNA recognition. Overall, CSP data allowed the identification of the DNA binding surface of PHOX2B-20A (Fig. 5D). Interestingly, this surface largely matches the surface that we have identified as responsible for the DNA binding activity of PHOX2B-HD. In accordance with the literature data on the homeodomain-containing proteins,47 also in the interaction of PHOX2B-20A with its target DNA, helix α3 and the N-terminal portion of the HD are confirmed as the key structural elements. In addition, significant perturbations were found also for the signals arising from residues included in the helix α1 in which, in particular, Leu110, Glu112, Ala118 and Glu119 give rise to two signals each. As helix α1 remains essentially unperturbed upon the DNA interaction of PHOX2B-HD, these data strongly indicate α1 to be a mediator for the formation of the homo-dimeric structure that binds the DNA. Indeed, the presence of a double set of helix 1 chemical shifts suggests a slight asymmetry of the homodimer, where the two α1 helices experience different chemical environments; the two sets of signals of this secondary structure are likely due to its involvement in the interaction with the C-terminal residues to form the dimer in a head to tail fashion (Fig. S11†). In addition, the good correlation of the HN and N chemical shifts observed for PHOX2B-20A with the values obtained for PHOX2B-HD indicates that the PHOX2B-20A DNA binding occurs by a recognition mechanism in which the homeodomain acts as a (partial) independent unit (Fig. S9A and B†).
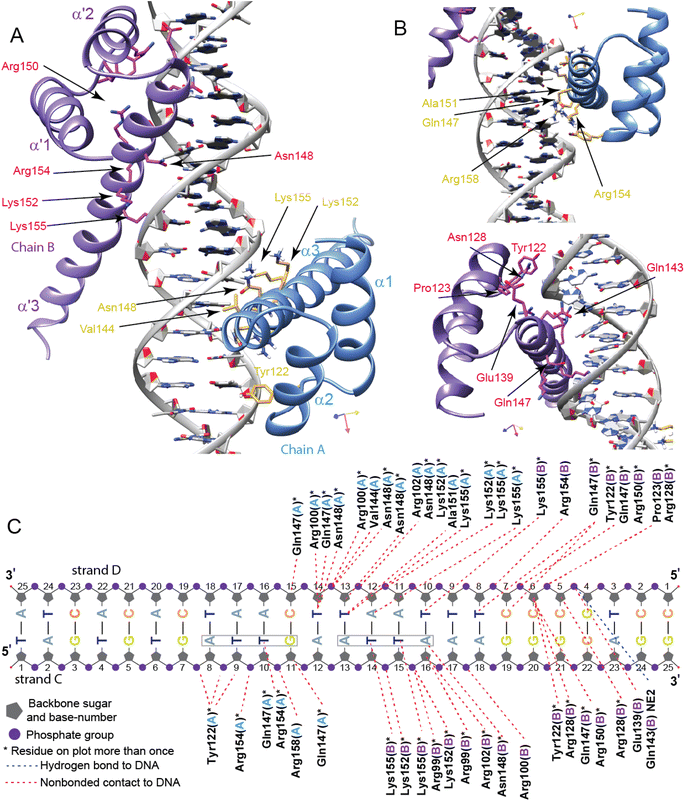
Structure of the PHOX2B-20A/DNA complex
To further describe the molecular machinery governing the DNA recognition mechanism by PHOX2B-20A, we calculated a structural model of the PHOX2B-20A homodimer/DNA complex using an ad hoc strategy (for details see the Materials and methods section and Fig. S12†) based on the data-driven docking program HADDOCK. The structural model of the PHOX2B-20A homodimer/DNA complex clearly indicates that the DNA-binding surface of each PHOX2B-20A monomer is mainly composed of the HTH recognition motif of the homeodomain, with helix α3 responsible for the majority of DNA contacts (Fig. 6A and B).The two HD domains (HD-A and HD-B) of the PHOX2B-20A homodimer wrap the DNA regions surrounding the single 5′-ATTA-3′ recognition site. A second incomplete ATT sequence is present 5′ to the ATTA motif (Fig. 6C). Of note there are no experimental data supporting the binding of PHOX2B to this motif. Helix α3 (HD-A) and the counterpart α3′ (HD-B) mainly recognize the major groove by making contacts with bases and the phosphate backbone of the DNA. In particular, α3 helix residues (i.e. Val144, Glu147, Asp148, Ala151, Lys152, Arg154, and Lys155) display multiple non-bonded contacts with T14′ and T13′ nucleotides of the DNA strand D and with T10 and G11 of the DNA strand C. Moreover, the interaction between the HD-A and DNA is further stabilized by the N-term arm preceding the α1 helix. In detail, Arg100 and Arg102 show multiple non-bonded contacts with the T13′ nucleotide of the DNA strand D. In addition, Tyr122 forms contacts with the phosphate backbone of G7 and A8 (DNA strand C) nucleotides, whereas Arg158 interacts with the phosphate backbone of T10. For HD-B, the side chain of Gln143′ in the α3′ helix is hydrogen bonding with the phosphate backbone of G4′; Arg154′ displays non-bonded contacts with T8′ nucleotide (DNA strand D); Gln147′ and Arg150′ forms non-bonded contacts with T8′ and C6′; Asn148′ forms non-bonded contact with A16; Lys152′ shows multiple interactions with the phosphate backbone of T14 and T15; Lys155′ interacts with the T10′ nucleotide of the DNA strand D and with T14 and T15 nucleotides located inside the ATTA recognition site. To note, different to HD-A, DNA binding is mediated in HD-B through non-bonded contacts made by Pro123′ located within the loop α1′– α2′ and Arg128′ of the α2′ helix belonging to the HTH recognition motif. In addition, as observed for the monomer A, the N-term arm of chain B contributes to stabilizing the PHOX2B-20A dimer/DNA complex. In particular, Arg100′ makes contact with the phosphate backbone of A17 (DNA strand C) whereas Arg99′ and Arg102′ show multiple non-bonded interactions with the A16 nucleotide.
Discussion
PHOX2B is a key target to understand the insurgence of the CCHS and for the design of therapy capable of tackling such a syndrome. This prompted us to realize the present study whose rationale has been the research of molecular details capable of explaining why the expansion of the poly-alanine tract prevents the protein from performing its activities and leads to the development of the disease. For this reason, we have performed a comparative study of PHOX2B-20A, PHOX2B-HD, and PHOX2B-27A, which are respectively the wild-type protein (20-Ala) lacking the N-terminal hypothetical unstructured 97 residues, its isolated homeodomain and the most common pathological variant (+7 alanines). The 60 amino acids long PHOX2B-HD shows, as expected, the classical structure of an HTH motif typical of the homeobox domains.47 The homeobox domain or ‘homeodomain’ is a highly conserved eukaryotic DNA-binding domain that can both bind DNA and mediate protein–protein interactions, well known for its role in transcription regulation during vertebrate development. We show that PHOX2B-HD binds DNA, as already reported for the other HDs, exploiting the recognition code44–46 contained in its helix 3. The insertion of this helix in the DNA major groove is then stabilized by the interaction of the unstructured N-terminal tail with the minor groove.PHOX2B-20A contains a compact globular HD domain and a highly dynamic C-terminal domain that bears a polyAla sequence forming a helical structure. The PHOX2B-20A structural and dynamical behaviour described here indicates that the two domains very likely cross-talk through transient interactions:56 the C-terminal tail transiently interacts with the HD without altering its tertiary organization.
This finding is of utmost importance also in consideration of the fact that the HD domain plays a major role in protein stabilization and solubilisation.47 Indeed, the deletion of the HD leads to a highly unstable and little soluble protein as observed in previous CD studies.16 Moreover, the expansions of the 20 alanine tract, a feature that is in clear relation with the insurgence of the CCHS, leads to a destabilization of the protein, decreases its DNA binding capability and its transcriptional activity, and influences interactions with other partner proteins.57 Alanine expansions, as seen also here in the case of PHOX2B-27A, have been also shown to induce protein oligomerization and destabilization in vitro and in cell overexpression assays.57 Interestingly, while this paper was under revision a new study has been published,58 describing in detail the behavior of the PHOX2B C-terminal domain, using mostly NMR spectroscopy, and its main results well complement our findings.
Thus, in light of this evidence, the interaction documented by our data allows the protein to populate transiently a more compact structure that protects the entire protein from the aggregation propensity of the polyAla sequences.59 The proper polyAla tract, generally proposed as a flexible spacer element between protein domains,60 likely allows the appropriate HD/C-term interaction. Any modification of this spacer could in principle interfere with this interaction and with the formation of the transient compact structure, allowing the lengthened polyAla tract to form aggregates that impair the protein function.
This hypothesis is further supported by our study of PHOX2B-20A-DNA interaction. We show, by means of an integrated approach that combines CSPs with in silico data, that the same recognition mode of the isolated HD is used by the full-length protein, PHOX2B-20A, to contact its cognate DNA: the interaction with the ATTA2 sequence is essentially mediated by the third helix of the HD and the N-terminal tail. Indeed, the CSP analysis indicates that upon DNA addition the perturbed surface of the HD contained in the PHOX2B-20A protein is not exactly the same as the isolated HD. Interestingly, our data outline perturbations for residues belonging to the first helix that, in contrast, remain unperturbed in the PHOX2B-HD protein. In particular, the 15N–1H cross peaks of these residues are doubled in the presence of DNA indicating that the α1 possibly experiences two different chemical environments, due to the involvement of this secondary structural element in the interaction with the C-terminal residues to form the dimer in a head to tail fashion, in a slight asymmetry. The C-terminal tail likely recruits a second protein first helix to form a dimeric structure that binds the sequence of the ATTA2 oligonucleotide well. The two molecules insert their helix 3 in the DNA major grove while the N-terminal portion comes into contact with the minor groove and further stabilizes the entire complex. These data agree with previously reported studies,15,16 which show that PHOX2B-20A binding to the DNA is severely affected upon deletion of the C-terminal domain or by expansion of the polyalanine tract, indicating the C-terminal tail to be a key structural element to form the functional complex.
The same studies have also shown how a protein variant, namely PHOX2B-0Ala, in which the polyAla tract is missing, which shares with PHOX2B-20A the same stability and the same capability to bind the cognate DNA. Our findings can be interpreted in light of these literature data16 and indicate that the polyAla tract within the long C-terminal domain does not play a fundamental role in the DNA binding activity of PHOX2B. On the other hand, our data can lead to the speculation that the polyAla tract, as already documented for other proteins,60 is a fundamental spacer that allows the C-terminal domain of PHOX2B-20A to interact with other proteins such as transcriptional coactivators or corepressors.16 Expansions of amino acid repeats occur in IDRs of transcription factors leading to diseases associated with protein aggregation.61–64 In particular, alanine repeats have been recently shown64 to alter, both in vitro and in vivo, the phase separation capacity in the HOXD13 transcription factor impairing its ability to co-condense with transcriptional co-activators leading to the alteration of the transcriptional program in a cell-specific manner. As the contribution of aggregates to “repeat expansion pathologies” has been controversial, the distinct behavior of the liquid–liquid phase separation observed suggests that the separation of transcriptional condensates may be at the basis of the observed pathologies.
In PHOX2B, the polyAla expansions decrease its DNA binding and transcriptional activity, likely impairing the formation of the stabilizing transient interaction between the HD and the C-terminal tail, leading to the formation of non-functional oligomers, not necessarily toxic, mediated by the elongated poly-alanine tract and to the possible insurgence of the disease through a loss-of-function mechanism.
Overall, our study contributes to shed light on the molecular mechanism underlying the CCHS syndrome and suggests as a possible therapeutic route the use of specific anti-aggregating molecules capable of preventing variant aggregation and possibly restoring the PHOX2B DNA binding activity.
Data availability
PDB ID 8P7G, BMRB ID 34822.Author contributions
Donatella Diana, Luciano Pirone, Luigi Russo, Gianluca D'Abrosca, Manoj Madheswaran, Clementina Acconcia, Andrea Corvino, Nataliia Ventserova, Carla Isernia and Sonia Di Gaetano produced and purified the samples, performed all the experiments and analyzed the data. Donatella Diana, Luciano Pirone, Luigi Russo, Gianluca D'Abrosca, Carla Isernia and Sonia Di Gaetano designed the experiments and supervised the study. Roberta Benfante, Simona Di Lascio, Laura Caldinelli, Diego Fornasari and Loredano Pollegioni supervised the study. Gaetano Malgieri, Emilia M. Pedone and Roberto Fattorusso conceived and designed the study and wrote the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This study was funded by grants PRIN 2022 PNRR P2022AW2H9 to GM and PRIN 2022 2022PAAYZE to RF, both from Ministero dell'Università e della Ricerca. Emilia M. Pedone thanks AISICC OdV (Associazione italiana per la Sindrome da Ipoventilazione Centrale Congenita - Sindrome di Ondine) for the support.References
- A. Albrecht and S. Mundlos, The other trinucleotide repeat: polyalanine expansion disorders, Curr. Opin. Genet. Dev., 2005, 15, 285–293 CrossRef CAS PubMed.
- C. Shoubridge and J. Gecz, Polyalanine tract disorders and neurocognitive phenotypes, Adv. Exp. Med. Biol., 2012, 769, 185–203 CrossRef CAS PubMed.
- Y. Zhang, C. A. Larsen, H. S. Stadler and J. B. Ames, Structural basis for sequence specific DNA binding and protein dimerization of HOXA13, PLoS One, 2011, 6, e23069 CrossRef CAS PubMed.
- H. Ge, D. Zhou, S. Tong, Y. Gao, M. Teng and L. Niu, Crystal structure and possible dimerization of the single RRM of human PABPN1, Proteins, 2008, 71, 1539–1545 CrossRef CAS PubMed.
- R. Hernandez and J. C. Facelli, Understanding protein structural changes for oncogenic missense variants, Heliyon, 2021, 7, e06013 CrossRef CAS PubMed.
- S. Polling, A. R. Ormsby, R. J. Wood, K. Lee, C. Shoubridge, J. N. Hughes, P. Q. Thomas, M. D. Griffin, A. F. Hill, Q. Bowden, T. Böcking and D. M. Hatters, Polyalanine expansions drive a shift into α-helical clusters without amyloid-fibril formation, Nat. Struct. Mol. Biol., 2015, 22, 1008–1015 CrossRef CAS PubMed.
- A. Pattyn, X. Morin, H. Cremer, C. Goridis and J. F. Brunet, The homeobox gene Phox2b is essential for the development of autonomic neural crest derivatives, Nature, 1999, 399, 366–370 CrossRef CAS PubMed.
- A. Pattyn, C. Goridis and J. F. Brunet, Specification of the central noradrenergic phenotype by the homeobox gene Phox2b, Mol. Cell. Neurosci., 2000, 15, 235–243 CrossRef CAS PubMed.
- R. L. Stornetta, T. S. Moreira, A. C. Takakura, B. J. Kang, D. A. Chang, G. H. West, J. F. Brunet, D. K. Mulkey, D. A. Bayliss and P. G. Guyenet, Expression of Phox2b by brainstem neurons involved in chemosensory integration in the adult rat, J. Neurosci., 2006, 26, 10305–10314 CrossRef CAS PubMed.
- J. Amiel, B. Laudier, T. Attié-Bitach, H. Trang, L. de Pontual, B. Gener, D. Trochet, H. Etchevers, P. Ray, M. Simonneau, M. Vekemans, A. Munnich, C. Gaultier and S. Lyonnet, Polyalanine expansion and frameshift mutations of the paired-like homeobox gene PHOX2B in congenital central hypoventilation syndrome, Nat. Genet., 2003, 33, 459–461 CrossRef CAS PubMed.
- I. Matera, T. Bachetti, F. Puppo, M. Di Duca, F. Morandi, G. M. Casiraghi, M. R. Cilio, R. Hennekam, R. Hofstra, J. G. Schöber, R. Ravazzolo, G. Ottonello and I. Ceccherini, PHOX2B mutations and polyalanine expansions correlate with the severity of the respiratory phenotype and associated symptoms in both congenital and late onset Central Hypoventilation syndrome, J. Med. Genet., 2004, 41, 373–380 CrossRef CAS PubMed.
- S. Di Lascio, R. Benfante, S. Cardani and D. Fornasari, Research Advances on Therapeutic Approaches to Congenital Central Hypoventilation Syndrome (CCHS), Front. Neurosci., 2020, 14, 615666 CrossRef PubMed.
- T. Bachetti, I. Matera, S. Borghini, M. Di Duca, R. Ravazzolo and I. Ceccherini, Distinct pathogenetic mechanisms for PHOX2B associated polyalanine expansions and frameshift mutations in congenital central hypoventilation syndrome, Hum. Mol. Genet., 2005, 14, 1815–1824 CrossRef CAS PubMed.
- S. Di Lascio, T. Bachetti, E. Saba, I. Ceccherini, R. Benfante and D. Fornasari, Transcriptional dysregulation and impairment of PHOX2B auto-regulatory mechanism induced by polyalanine expansion mutations associated with congenital central hypoventilation syndrome, Neurobiol. Dis., 2013, 50, 187–200 CrossRef CAS PubMed.
- S. Di Lascio, D. Belperio, R. Benfante and D. Fornasari, Alanine Expansions Associated with Congenital Central Hypoventilation Syndrome Impair PHOX2B Homeodomain-mediated Dimerization and Nuclear Import, J. Biol. Chem., 2016, 291, 13375–13393 CrossRef CAS PubMed.
- L. Pirone, L. Caldinelli, S. Di Lascio, R. Di Girolamo, S. Di Gaetano, D. Fornasari, L. Pollegioni, R. Benfante and E. Pedone, Molecular insights into the role of the polyalanine region in mediating PHOX2B aggregation, FEBS J., 2019, 286, 2505–2521 CrossRef CAS PubMed.
- S. Grzesiek, H. Döbeli, R. Gentz, G. Garotta, A. M. Labhardt and A. Bax, 1H, 13C, and 15N NMR backbone assignments and secondary structure of human interferon-gamma, Biochemistry, 1992, 31, 8180–8190 CrossRef CAS PubMed.
- S. C. Panchal, N. S. Bhavesh and R. V. Hosur, Improved 3D triple resonance experiments, HNN and HN(C)N, for HN and 15N sequential correlations in (13C, 15N) labeled proteins: application to unfolded proteins, J. Biomol. NMR, 2001, 20, 135–147 CrossRef CAS PubMed.
- B. Farina, A. Del Gatto, D. Comegna, S. Di Gaetano, D. Capasso, C. Isernia, M. Saviano, R. Fattorusso, L. Zaccaro and L. Russo, Conformational studies of RGDechi peptide by natural-abundance NMR spectroscopy, J. Pept. Sci., 2019, 25, e3166 CrossRef PubMed.
- F. Delaglio, S. Grzesiek, G. W. Vuister, G. Zhu, J. Pfeifer and A. Bax, NMRPipe: a multidimensional spectral processing system based on UNIX pipes, J. Biomol. NMR, 1995, 6, 277–293 CrossRef CAS PubMed.
- R. Keller, The Computer Aided Resonance Assignment Tutorial, Cantina-Verlag, Switzerland, 2004 Search PubMed.
- W. Lee, M. Tonelli and J. L. Markley, NMRFAM-SPARKY: enhanced software for biomolecular NMR spectroscopy, Bioinformatics, 2015, 31, 1325–1327 CrossRef PubMed.
- M. Kjaergaard, S. Brander and F. M. Poulsen, Random coil chemical shift for intrinsically disordered proteins: effects of temperature and pH, J. Biomol. NMR, 2011, 49, 139–149 CrossRef CAS PubMed.
- M. Kjaergaard and F. M. Poulsen, Sequence correction of random coil chemical shifts: correlation between neighbor correction factors and changes in the Ramachandran distribution, J. Biomol. NMR, 2011, 50, 157–165 CrossRef CAS PubMed.
- Y. Shen, F. Delaglio, G. Cornilescu and A. Bax, TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts, J. Biomol. NMR, 2009, 44, 213–223 CrossRef CAS PubMed.
- Y. Shen, O. Lange, F. Delaglio, P. Rossi, J. M. Aramini, G. Liu, A. Eletsky, Y. Wu, K. K. Singarapu, A. Lemak, A. Ignatchenko, C. H. Arrowsmith, T. Szyperski, G. T. Montelione, D. Baker and A. Bax, Consistent blind protein structure generation from NMR chemical shift data, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4685–4690 CrossRef CAS PubMed.
- R. Koradi, M. Billeter and K. Wüthrich, MOLMOL: a program for display and analysis of macromolecular structures, J. Mol. Graphics, 1996, 14(51–55), 29–32 Search PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, UCSF Chimera-a visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- R. A. Laskowski, J. A. Rullmannn, M. W. MacArthur, R. Kaptein and J. M. Thornton, AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR, J. Biomol. NMR, 1996, 8, 477–486 CrossRef CAS PubMed.
- M. V. Berjanskii and D. S. Wishart, The RCI server: rapid and accurate calculation of protein flexibility using chemical shifts, Nucleic Acids Res., 2007, 35, W531–W537 CrossRef PubMed.
- P. Bernadó, J. García de la Torre and M. Pons, Interpretation of 15N NMR relaxation data of globular proteins using hydrodynamic calculations with HYDRONMR, J. Biomol. NMR, 2002, 23, 139–150 CrossRef PubMed.
- F. Cargnin, A. Flora, S. Di Lascio, E. Battaglioli, R. Longhi, F. Clementi and D. Fornasari, PHOX2B regulates its own expression by a transcriptional auto-regulatory mechanism, J. Biol. Chem., 2005, 280, 37439–37448 CrossRef CAS PubMed.
- J. G. Omichinski, G. M. Clore, O. Schaad, G. Felsenfeld, C. Trainor, E. Appella, S. J. Stahl and A. M. Gronenborn, NMR structure of a specific DNA complex of Zn-containing DNA binding domain of GATA-1, Science, 1993, 261, 438–446 CrossRef CAS PubMed.
- D. S. Garrett, Y. J. Seok, A. Peterkofsky, G. M. Clore and A. M. Gronenborn, Identification by NMR of the binding surface for the histidine-containing phosphocarrier protein HPr on the N-terminal domain of enzyme I of the Escherichia coli phosphotransferase system, Biochemistry, 1997, 36, 4393–4398 CrossRef CAS PubMed.
- M. P. Foster, D. S. Wuttke, K. R. Clemens, W. Jahnke, I. Radhakrishnan, L. Tennant, M. Reymond, J. Chung and P. E. Wright, Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA, J. Biomol. NMR, 1998, 12, 51–71 CrossRef CAS PubMed.
- J. Jumper, R. Evans, A. Pritzel, T. Green, M. Figurnov, O. Ronneberger, K. Tunyasuvunakool, R. Bates, A. Žídek, A. Potapenko, A. Bridgland, C. Meyer, S. A. A. Kohl, A. J. Ballard, A. Cowie, B. Romera-Paredes, S. Nikolov, R. Jain, J. Adler, T. Back, S. Petersen, D. Reiman, E. Clancy, M. Zielinski, M. Steinegger, M. Pacholska, T. Berghammer, S. Bodenstein, D. Silver, O. Vinyals, A. W. Senior, K. Kavukcuoglu, P. Kohli and D. Hassabis, Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- W. Kabsch and C. Sander, Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features, Biopolymers, 1983, 22, 2577–2637 CrossRef CAS PubMed.
- A. V. Colasanti, X. J. Lu and W. K. Olson, Analyzing and building nucleic acid structures with 3DNA, J. Visualized Exp., 2013, e4401 Search PubMed.
- S. J. de Vries, M. van Dijk and A. M. Bonvin, The HADDOCK web server for data-driven biomolecular docking, Nat. Protoc., 2010, 5, 883–897 CrossRef CAS PubMed.
- H. Tjong and H. X. Zhou, DISPLAR: an accurate method for predicting DNA-binding sites on protein surfaces, Nucleic Acids Res., 2007, 35, 1465–1477 CrossRef CAS PubMed.
- J. Estrada, P. Bernadó, M. Blackledge and J. Sancho, ProtSA: a web application for calculating sequence specific protein solvent accessibilities in the unfolded ensemble, BMC Bioinf., 2009, 10, 104 CrossRef PubMed.
- D. S. Wishart, B. D. Sykes and F. M. Richards, The chemical shift index: a fast and simple method for the assignment of protein secondary structure through NMR spectroscopy, Biochemistry, 1992, 31, 1647–1651 CrossRef CAS PubMed.
- J. A. Marsh, V. K. Singh, Z. Jia and J. D. Forman-Kay, Sensitivity of secondary structure propensities to sequence differences between alpha- and gamma-synuclein: implications for fibrillation, Protein Sci., 2006, 15, 2795–2804 CrossRef CAS PubMed.
- M. Billeter, Y. Q. Qian, G. Otting, M. Müller, W. Gehring and K. Wüthrich, Determination of the nuclear magnetic resonance solution structure of an Antennapedia homeodomain-DNA complex, J. Mol. Biol., 1993, 234, 1084–1093 CrossRef CAS PubMed.
- M. Billeter, P. Güntert, P. Luginbühl and K. Wüthrich, Hydration and DNA recognition by homeodomains, Cell, 1996, 85, 1057–1065 CrossRef CAS PubMed.
- J. S. Anderson, M. D. Forman, S. Modleski, F. W. Dahlquist and S. M. Baxter, Cooperative ordering in homeodomain-DNA recognition: solution structure and dynamics of the MATa1 homeodomain, Biochemistry, 2000, 39, 10045–10054 CrossRef CAS PubMed.
- S. C. Harrison, A structural taxonomy of DNA-binding domains, Nature, 1991, 353, 715–719 CrossRef CAS PubMed.
- J. García de la Torre, M. L. Huertas and B. Carrasco, HYDRONMR: prediction of NMR relaxation of globular proteins from atomic-level structures and hydrodynamic calculations, J. Magn. Reson., 2000, 147, 138–146 CrossRef PubMed.
- A. Ortega, D. Amorós and J. García de la Torre, Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models, Biophys. J., 2011, 101, 892–898 CrossRef CAS PubMed.
- Y. Zhang, K. A. Edmonds, D. J. Raines, B. A. Murphy, H. Wu, C. Guo, E. M. Nolan, M. S. VanNieuwenhze, A. K. Duhme-Klair and D. P. Giedroc, The Pneumococcal Iron Uptake Protein A (PiuA) Specifically Recognizes Tetradentate Fe, J. Mol. Biol., 2020, 432, 5390–5410 CrossRef CAS PubMed.
- C. A. Elena-Real, A. Urbanek, L. Imbert, A. Morató, A. Fournet, F. Allemand, N. Sibille, J. Boisbouvier and P. Bernadó, Site-Specific Introduction of Alanines for the Nuclear Magnetic Resonance Investigation of Low-Complexity Regions and Large Biomolecular Assemblies, ACS Chem. Biol., 2023, 18, 2039–2049 CrossRef CAS PubMed.
- G. D'Abrosca, A. Paladino, I. Baglivo, L. Russo, M. Sassano, R. Grazioso, R. Iacovino, L. Pirone, E. M. Pedone, P. V. Pedone, C. Isernia, R. Fattorusso and G. Malgieri, Structural Insight of the Full-Length Ros Protein: A Prototype of the Prokaryotic Zinc-Finger Family, Sci. Rep., 2020, 10, 9283 CrossRef PubMed.
- A. Vovk and A. Zilman, Effects of Sequence Composition, Patterning and Hydrodynamics on the Conformation and Dynamics of Intrinsically Disordered Proteins, Int. J. Mol. Sci., 2023, 24, 1444 CrossRef CAS PubMed.
- D. Vuzman, A. Azia and Y. Levy, Searching DNA via a “Monkey Bar” mechanism: the significance of disordered tails, J. Mol. Biol., 2010, 396, 674–684 CrossRef CAS PubMed.
- M. A. Lohrum, D. B. Woods, R. L. Ludwig, E. Bálint and K. H. Vousden, C-terminal ubiquitination of p53 contributes to nuclear export, Mol. Cell. Biol., 2001, 21, 8521–8532 CrossRef CAS PubMed.
- L. Russo, G. Salzano, A. Corvino, E. Bistaffa, F. Moda, L. Celauro, G. D'Abrosca, C. Isernia, D. Milardi, G. Giachin, G. Malgieri, G. Legname and R. Fattorusso, Structural and dynamical determinants of a β-sheet-enriched intermediate involved in amyloid fibrillar assembly of human prion protein, Chem. Sci., 2022, 13, 10406–10427 RSC.
- S. Di Lascio, R. Benfante, S. Cardani and D. Fornasari, Advances in the molecular biology and pathogenesis of congenital central hypoventilation syndrome - implications for new therapeutic targets, Expert Opin. Orphan Drugs, 2018, 6, 719–731 CrossRef CAS.
- R. Antón, M. Treviño, D. Pantoja-Uceda, S. Félix, M. Babu, E. J. Cabrita, M. Zweckstetter, P. Tinnefeld, A. M. Vera and J. Oroz, Alternative low-populated conformations prompt phase transitions in polyalanine repeat expansions, Nat. Commun., 2024, 15, 1925 CrossRef PubMed.
- T. C. Chen and J. R. Huang, Musashi-1: An Example of How Polyalanine Tracts Contribute to Self-Association in the Intrinsically Disordered Regions of RNA-Binding Proteins, Int. J. Mol. Sci., 2020, 21, 2289 CrossRef CAS PubMed.
- R. Hernandez and J. C. Facelli, Structure analysis of the proteins associated with polyA repeat expansion disorders, J. Biomol. Struct. Dyn., 2022, 40, 5556–5565 CrossRef CAS PubMed.
- A. L. Darling and V. N. Uversky, Intrinsic Disorder in Proteins with Pathogenic Repeat Expansions, Molecules, 2017, 22, 2027 CrossRef PubMed.
- A. R. La Spada and J. P. Taylor, Repeat expansion disease: progress and puzzles in disease pathogenesis, Nat. Rev. Genet., 2010, 11, 247–258 CrossRef CAS PubMed.
- H. T. Orr and H. Y. Zoghbi, Trinucleotide repeat disorders, Annu. Rev. Neurosci., 2007, 30, 575–621 CrossRef CAS PubMed.
- S. Basu, S. D. Mackowiak, H. Niskanen, D. Knezevic, V. Asimi, S. Grosswendt, H. Geertsema, S. Ali, I. Jerković, H. Ewers, S. Mundlos, A. Meissner, D. M. Ibrahim and D. Hnisz, Unblending of Transcriptional Condensates in Human Repeat Expansion Disease, Cell, 2020, 181, 1062–1079 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc06427a |
| ‡ These authors equally contributed to the work. |
| This journal is © The Royal Society of Chemistry 2024 |