 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of ultrasonic-assisted extraction of total flavonoids from Oxalis corniculata by a hybrid response surface methodology-artificial neural network-genetic algorithm (RSM-ANN-GA) approach, coupled with an assessment of antioxidant activities†
Deng-Zhao Jiang *ab,
Dan-Ping Yua,
Ming Zenga,
Wen-Bo Liu
*ab,
Dan-Ping Yua,
Ming Zenga,
Wen-Bo Liu a,
Dong-Lin Lic and
Ke-Yue Liuab
a,
Dong-Lin Lic and
Ke-Yue Liuab
aSchool of Pharmacy and Life Science, Jiujiang University, Jiujiang 332005, China. E-mail: jiangdengzhao@126.com
bJiujiang Key Laboratory for the Development and Utilization of Traditional Chinese Medicine Resources in Northwest Jiangxi, Jiujiang 332005, China
cAnalytical and Testing Center, Jiujiang University, Jiujiang 332005, China
First published on 10th December 2024
Abstract
The objective of this research endeavor is to refine the ultrasonic-assisted extraction technique for total flavonoids from Oxalis corniculata (TFO), utilizing a synergistic approach combining response surface methodology (RSM) and artificial neural network integrated with genetic algorithm (RSM-ANN-GA). The optimized extraction parameters determined through RSM yielded a TFO concentration of 13.538 mg g−1 under the following conditions: an ethanol concentration of 61.95%, a liquid–solid ratio of 41.06 mL g−1, an ultrasonic power setting of 351.57 W, and an ultrasonic exposure duration of 58.95 minutes. Conversely, the RSM-ANN-GA approach identified an even more refined set of conditions, achieving a TFO concentration of 13.7844 mg g−1, with an ethanol concentration of 58.93%, a liquid–solid ratio of 41.16 mL g−1, an ultrasonic power of 350.22 W, and an ultrasonic exposure time of 58.18 minutes. These findings underscore the superior predictive accuracy and enhanced extraction efficiency offered by the RSM-ANN-GA model over the conventional RSM method. Furthermore, the study demonstrated that TFO possesses a potent antioxidant effect, as evidenced by its ability to scavenge DPPH, hydroxyl, and superoxide anion free radicals in vitro, highlighting its potential as a valuable source of natural antioxidants.
1 Introduction
Oxalis corniculata, a globally widespread member of the Oxalidaceae family, is prevalent in Asia, South Africa, and South America. Known as an invasive yet medicinally potent weed, it has been valued in traditional medicine in China, Pakistan, and India.1 Recent studies have reported various pharmacological activities, including anticancer, antibacterial, antifungal, insecticidal, and antioxidant effects, etc.2–12 Phytochemical analysis reveals the presence of alkaloids, flavonoids, terpenoids, glycosides, and saponin, etc.6–8,12–14 Among these, flavonoids stand out for their strong antioxidant and antimicrobial properties. These bioactive compounds make O. corniculata a valuable resource for advancing modern pharmacology, nutraceutical development, and cosmeceutical formulations.5,13Conventional extraction methods for flavonoids, such as percolation and decoction, require significant solvent and energy. Ultrasound-assisted extraction (UAE) is a more efficient and sustainable alternative. UAE uses ultrasonic waves to create acoustic cavitation, which disrupts cellular structures and enhances the release and solubilization of target compounds. This improves extraction efficiency and reduces environmental impact. UAE accelerates the leaching of bioactive compounds, boosting extraction efficacy while minimizing resource consumption. Therefore, UAE has become widely adopted for extracting bioactive plant components.15–19
The efficacy of component extraction depends on factors such as solvent concentration, extraction duration, ultrasonic power, pH levels, and temperature, etc. The complex nature of botanical constituents often results in a non-linear extraction process. Despite its inefficiency and resource wastage, the one-variable-at-a-time (OVAT) method is still widely used for its simplicity and intuitiveness. However, OVAT fails to detect interactions between factors and lacks a comprehensive understanding of all determinants.20
In contrast, the design of experiments (DoE) approach offers significant advantages by requiring fewer experiments and lower costs.21 Orthogonal design, a traditional DoE method, investigates the impact of different parameters through a limited number of experimental runs. When used correctly, orthogonal design effectively determines the optimal combination of factor levels, making it a valuable tool for optimizing extraction processes.21
Novel optimization methodologies, such as Response Surface Methodology (RSM) and Artificial Neural Networks (ANN), have gained attention for extracting plant-based medicinal ingredients. RSM, a statistical technique, models complex processes and optimizes parameter interactions.22,23 ANNs add computational intelligence by recognizing intricate patterns and making sophisticated predictions.24–27 Compared to RSM, the integration of RSM, ANN, and Genetic Algorithms (GA)—known as RSM-ANN-GA—demonstrates superior proficiency in analyzing observational data and creating predictive models for complex, nonlinear processes. This combination has evolved into a more powerful and accurate optimization tool.25,27–31
Current literature shows a notable lack of research on enhancing the extraction of TFO, particularly using ANN methodologies. To address this, we first employed RSM to optimize the TFO extraction process. Next, an ANN model was developed using the experimental data from RSM design points. Finally, the optimal extraction conditions were determined using a synergistic RSM-ANN-GA approach, followed by in vitro assessments of the antioxidant activities of TFO.
2 Results and discussion
2.1. Standard curve
The deduced linear regression equation manifests as: A = 9.8182C + 0.0264, complemented by a correlation coefficient R = 0.9995. Such results unequivocally validate that, within the concentration domain spanning from 0.03 to 0.06 mg mL−1, the concentration of rutin bears an exceptional linear relationship with absorbance. This finding attests to the precision and steadfast reliability of the adopted measurement method (Fig. 1).2.2. Identification and characterization of TFO
Following the interaction between the extraction solution and magnesium hydrochloride powder, a notable change occurred, manifesting as a deep purple coloration, which unequivocally signals the inclusion of flavonoids in the extracted mixture. The FTIR spectral profile for the extract derived from UAE is delineated in Fig. 2, revealing the molecular vibrations characteristic of the constituents within the extract.The broad and intense absorption band observed around 37.70 cm−1 is likely attributable to the stretching vibrations of the O–H group, which typically occur within the range of 3600–3200 cm−1.
An absorption peak at 2921.69 cm−1 could be attributed to the stretching vibrations of both aliphatic and aromatic C–H groups, which are commonly observed in the range of 2950–2850 cm−1. Meanwhile, the peak at approximately 1655.33 cm−1 may be indicative of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations, typically found within the spectral window of 1680–1620 cm−1. Peaks located at 1606.38 cm−1 and 1449.25 cm−1 might be associated with the skeletal vibrations of the benzene ring, while the feature at 1067.09 cm−1 is likely due to the deformation vibrations of C–C bonds. All the identified absorptions, when considered in concert, point unmistakably toward the existence of flavonoids, their distinctive spectral signatures weaving a tale of the sample's chemical composition.15,32
O stretching vibrations, typically found within the spectral window of 1680–1620 cm−1. Peaks located at 1606.38 cm−1 and 1449.25 cm−1 might be associated with the skeletal vibrations of the benzene ring, while the feature at 1067.09 cm−1 is likely due to the deformation vibrations of C–C bonds. All the identified absorptions, when considered in concert, point unmistakably toward the existence of flavonoids, their distinctive spectral signatures weaving a tale of the sample's chemical composition.15,32
2.3. Single factor experimental results and analysis
In accordance with the empirical findings delineated herein, the factorial design encompasses three hierarchical levels for each of the four investigated parameters. Specifically, the ethanol concentration (X1) spans a spectrum from 50% to 60% and 75%; the ultrasonic power (X2) is delineated at 300 W, 350 W and 400 W; the liquid–solid ratio (X3) is calibrated at 35, 40 and 45 mL g−1; and the extraction time (X4) is set at intervals of 50, 60 and 70 minutes, as meticulously tabulated in Table S1.†
2.4. Optimization of extraction parameters utilizing RSM
The Box–Behnken design with RSM and ANN results were shown in Table S2.† Utilizing the sophisticated capabilities of Design-Expert 10.0 software, an exhaustive analysis of variance (ANOVA) was conducted on the dataset amalgamating both empirical and projected values, as outlined in Table 1. Adopting the extraction yield of TFO (Y) as the focal response parameter, the implementation of a quadratic regression analytical framework resulted in the derivation of a comprehensive quadratic multinomial regression equation. This equation elucidates the intricate interplay between the extraction yield of TFO (Y) and the quartet of independent variables: ethanol concentration (X1), liquid–solid ratio (X2), ultrasonic power (X3), and ultrasonic time (X4):| Y = +13.41 + 0.65X1 + 0.53X2 + 0.094X3 − 0.096X4 + 0.22X1X2 − 0.098X1X3 + 0.24X1X4 + 0.19X2X3 − 0.50X2X4 − 0.030X3X4 − 1.84X12 − 1.48X22 − 1.90X32 − 1.19X42 |
| Source | Sum of squares | df | Mean square | F value | p-Value Prob > F | |
|---|---|---|---|---|---|---|
| Model | 55.59 | 14 | 3.97 | 42.60 | <0.0001 | Significant |
| X1-ethanol concentration | 5.10 | 1 | 5.10 | 54.66 | <0.0001 | Significant |
| X2-liquid–solid ratio | 3.31 | 1 | 3.31 | 35.48 | <0.0001 | Significant |
| X3-ultrasonic power | 0.11 | 1 | 0.11 | 1.14 | 0.3034 | |
| X4-ultrasonic time | 0.11 | 1 | 0.11 | 1.18 | 0.2953 | |
| X1X2 | 0.20 | 1 | 0.20 | 2.12 | 0.1671 | |
| X1X3 | 0.04 | 1 | 0.04 | 0.41 | 0.5334 | |
| X1X4 | 0.24 | 1 | 0.24 | 2.58 | 0.1308 | |
| X2X3 | 0.14 | 1 | 0.14 | 1.55 | 0.2337 | |
| X2X4 | 1.01 | 1 | 1.01 | 10.83 | 0.0054 | Significant |
| X3X4 | 0.0036 | 1 | 0.0036 | 0.04 | 0.8470 | |
| X12 | 22.03 | 1 | 22.03 | 236.36 | <0.0001 | Significant |
| X22 | 14.27 | 1 | 14.27 | 153.04 | <0.0001 | Significant |
| X32 | 23.52 | 1 | 23.52 | 252.33 | <0.0001 | Significant |
| X42 | 9.25 | 1 | 9.25 | 99.25 | <0.0001 | Significant |
| Residual | 1.31 | 14 | 0.09 | |||
| Lack of fit | 1.08 | 10 | 0.11 | 1.91 | 0.2783 | Not significant |
| Pure error | 0.23 | 4 | 0.06 | |||
| R2 | 0.9771 | |||||
| Adj. R2 | 0.9541 | |||||
| Pred. R2 | 0.8845 |
An inspection of Table 1 reveals that the regression model, marked by a p-value less than 0.001, underscores the exceptional significance of the regression equation model, signifying an optimally efficacious fit. The insignificance of the lack of fit test (p > 0.05) corroborates the veracity of the quadratic polynomial regression model, substantiating its appropriateness for the dataset at hand. The concordance between the coefficient of determination (R2 = 0.9771) and the adjusted determination coefficient (adj. R2 = 0.9541) is indicative of a well-calibrated model, underscoring the reliability of predictions derived from this regression equation.27 An F-value of 1.91 for the lack of fit suggests that any discrepancy between the model's predictions and the actual observations is not statistically significant relative to the intrinsic variability (pure error). This finding implies that there is a 27.83% probability that an F-value of this magnitude could arise solely from random fluctuations or noise.
In the significance testing of the coefficients for the quadratic model regression equation, the Prob > F values for X1 (ethanol concentration) and X2 (liquid-to-solid ratio) were both lower than 0.01, indicating a highly significant linear effect on the TFO ultrasonic extraction efficiency. The hierarchy of factors impacting the extraction yield of TFO is delineated as follows: X1 (ethanol concentration) > X2 (liquid–solid ratio) > X4 (ultrasonic time) > X3 (ultrasonic power). The Prob > F values for X12, X22, X32 and X42 were less than 0.01, indicating a highly significant curvilinear effect on the TFO ultrasonic extraction efficiency.
The 3D graphical representations (illustrated in Fig. 4) elucidate the interactive influences exerted by the independent variables upon the yields of TFO. A steeper incline within these visual depictions signifies a more pronounced interaction between the paired factors, thereby highlighting their collective potency in modulating the response variable.39 Analysis revealed that the Prob > F values for the interactions involving X1X2, X1X3, X1X4, X2X3 and X3X4 were all above 0.05, which implies insignificant interaction effects on the ultrasonic extraction rate; on the other hand, the interaction term X2X4 had a Prob > F value lower than 0.01, indicating a highly significant interaction effect. The optimal extraction parameters ascertained through RSM were delineated as follows: an ethanol concentration of 61.95%, a liquid–solid ratio of 41.06 mL g−1, an ultrasonic power setting of 351.57 W, and an ultrasonic exposure duration of 58.95 minutes, yielding a TFO concentration of 13.538 mg g−1. In consideration of practical experimental constraints, these conditions were judiciously adjusted for the validation experiment, resulting in an ethanol concentration of 62%, a liquid–solid ratio of 41 mL g−1, an ultrasonic system power of 350 W, and an ultrasonic treatment interval of 59 minutes.
2.5. Optimization of extraction parameters utilizing RSM-ANN-GA
Utilizing the dataset derived from RSM analyses, a Back Propagation (BP) Artificial Neural Network (ANN) was trained, incorporating four pivotal factors-ethanol concentration, liquid–solid ratio, ultrasonic power and ultrasonic time-as input parameters, with the extraction yield of TFO serving as the output metric. The architectural topology of this artificial neural network is visually represented in Fig. S1.† The hyperbolic tangent sigmoid (“tansig”) function was utilized as the activation function bridging the input layer and the hidden layer, whereas the linear (“purelin”) function served as the transfer mechanism linking the hidden layer to the output layer. Employing the Levenberg–Marquardt backpropagation (“trainlm”) algorithm as the training function, the network was meticulously calibrated with a regimen encompassing 1000 epochs, 25 iterations per epoch, and a training goal error of 0.0001. Consequently, a network topology featuring 4 input nodes, 10 hidden nodes, and a solitary output node was architecturally realized, as depicted in Fig. S1.† As exemplified in Fig. S2† and 5, the best validation performance was 0.32438 at epoch 6 and the correlation coefficients pertaining to the training data, validation data, test data, and the aggregate dataset were recorded at 0.99916, 0.96762, 0.94672, and 0.93688, respectively. Notably, the correlation coefficients across all four sample categories surpassed the benchmark of 0.90, a testament to the commendable fitting capacity of the BP-ANN model for training, validation, and testing samples alike. In essence, the constructed BP-ANN model manifests potent predictive potential with regard to the test outcomes.To optimize the output maximization, the network data emanating from the RSM-ANN-GA integration was adopted as the objective function. The quartet of independent variables was conceptualized as matrix variables, subject to boundary constraints, as delineated hereafter:
| [50; 35; 300; 50] ≤ [X1; X2; X3; X4] ≤ [70; 45; 400; 70] |
The optimal extraction parameters, as determined through the sophisticated amalgamation of RSM-ANN-GA, were delineated as follows: an ethanol concentration of 58.93%, a liquid–solid ratio of 41.16 mL g−1, an ultrasonic power setting of 350.22 W, and an ultrasonic treatment duration of 58.18 minutes, culminating in a TFO concentration of 13.7844 mg g−1. In deference to the practicalities of experimental execution, these conditions were judiciously refined for the validation phase, resulting in an ethanol concentration of 59%, a liquid–solid ratio of 41 mL g−1, an ultrasonic power output of 350 W, and an ultrasonic treatment interval of 58 minutes.
2.6. Validation experiment
In accordance with the optimal extraction parameters ascertained via RSM and the integrated RSM-ANN-GA approach, a trio of replicate experiments was meticulously conducted for each method. Subsequently, the relative errors were computed to critically assess the predictive efficacy of the respective methodologies. As evidenced by the tabulated data in Table 2, the prediction outcome of the RSM-ANN-GA integration exhibited a notably reduced relative error, accompanied by a superior extraction yield compared to the standalone RSM technique.| Method | The optimal extraction conditions | Result (TFO) | |||||
|---|---|---|---|---|---|---|---|
| Ethanol concentration (X1) | Liquid–solid ratio (X2) | Ultrasonic power (X3) | Ultrasonic time (X4) | Experiment | Predicted | RE (%) | |
| RSM | 62 | 41 | 350 | 59 | 13.3533 ± 0.0241 | 13.538 | −1.36 ± 0.18 |
| RSM-ANN-GA | 59 | 41 | 350 | 58 | 13.6608 ± 0.0251 | 13.7844 | −0.90 ± 0.18 |
2.7. Antioxidant activity of TFO
Under physiological homeostasis, the generation and scavenging of free radicals within the body maintain a delicate equilibrium. However, an overabundance of free radicals can precipitate a redox imbalance, disrupting the organism's metabolic functions and culminating in the manifestation of disease states. This perturbation of the body's natural redox equilibrium, triggered by an excess of free radicals, leads to oxidative stress, which is implicated in the pathogenesis of numerous maladies.Extensive scholarly investigations have unequivocally demonstrated that an array of antioxidant enzymes inherent in the human body possess the capability to efficaciously neutralize free radicals.40–42 However, concomitant with the progression of chronological age, there ensues a decrement in the activity of these antioxidant enzymes, thereby attenuating the organism's proficiency in scavenging free radicals. This decline precipitates an accumulation of free radicals, a concomitant elevation in the concentration of malondialdehyde-a marker of oxidative stress-and an exacerbation of the extent of oxidative damage inflicted upon the body. Such a cascade of events culminates in the onset and progression of senescence.43
To ascertain the antioxidant efficacy of TFO, a series of in vitro assays were meticulously conducted within the ambit of this investigation. As illustrated in Fig. 6, TFO demonstrated a pronounced propensity to scavenge DPPH, hydroxyl, and superoxide anion radicals (DPPH˙, ˙OH and ˙O2−, respectively), albeit exhibiting a less pronounced effect compared to vitamin C (VC) when administered at equivalent concentrations.
3 Experimental section
3.1. Materials
Oxalis corniculata, sourced from the Lushan Mountain Nature Reserve (situated between 115°52′ and 116°08′ east longitude and 29°26′ to 29°41′ north latitude) in Jiangxi Province, was taxonomically authenticated by Dr Keyue Liu from the School of Pharmacy and Life Sciences at Jiujiang University, Jiujiang, China. The plant material underwent a controlled drying process at 60 °C for a duration of 12 hours (water content less than 5%), post which it was finely ground and subsequently sieved through a 40 mesh sieve. Thereafter, the processed herbs were meticulously stored under cool conditions to preserve their integrity and pharmacological properties.3.2. Chemicals and reagents
The 1,1-diphenyl-2-picrylhydrazyl (DPPH) reagent (lot: 8R7RL-JT, purity > 97%) was acquired from Tokyo Chemical Industry Development Co., Ltd., based in Shanghai, China. Rutin (lot: M1013A, purity > 97%) was obtained from Dalian Meilun Biotech Co., Ltd., headquartered in Dalian, China. For Fourier transform infrared (FTIR) spectroscopic analysis, potassium bromide of spectral purity grade was sourced from Tianjin Kemiou Chemical Reagent Co., Ltd., situated in Tianjin, China. Ultra-pure water utilized in the experimental procedures was produced using a Milli-Q purification system. All other chemical reagents and solvents employed throughout this investigation were of analytical grade, ensuring the highest quality standards for accurate and reliable results.3.3. Experimental design
With meticulous precision, volumes of 0.8, 1.0, 1.2, 1.4, 1.6, 1.8, and 2.0 milliliters of the rutin standard solution were aliquoted. Each aliquot was then supplemented with 0.3 mL of a 5% sodium nitrite solution, vigorously shaken, and allowed to rest for a period of 6 minutes. Subsequently, 0.3 mL of a 10% aluminum nitrate solution was added to each sample, followed by another round of thorough mixing and a subsequent resting phase of 6 minutes. Thereafter, 4 mL of a 0.1 mol per L sodium hydroxide solution was introduced. The solutions were adjusted to a final volume of 10 mL with 60% ethanol, shaken rigorously, and left to equilibrate for a 10 minute interval. Spectral scans in the UV-VIS range from 400 to 600 nm were performed on the rutin standard solutions. The absorption peak at 505.2 nm was selected for quantitative analysis. A linear regression equation was established by plotting the concentration of the standard solution (mg mL−1) against the absorbance readings.
The extraction of TFO was facilitated through an ultrasonic-assisted extraction (UAE) technique. Precisely 1.00 g of the powdered plant material was weighed and subjected to extraction with a predetermined volume of ethanol. The mixture was exposed to ultrasonic agitation for a specified duration, followed by filtration to separate the solid residue from the liquid extract. Ethanol was then added to the filtrate to achieve a fixed volume of 50.00 mL. The total flavonoid content of the extract was assessed following the NaNO2–Al(NO3)3–NaOH colorimetric assay protocol. The concentration of TFO was subsequently quantified by applying the linear regression equation derived from the calibration curve.
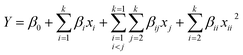 | (1) |
Within the quadratic model equation, the parameters β0, βi, βii, and βij represent the regression coefficients, where β0 signifies the intercept, βi denotes the linear effect coefficients, βii corresponds to the squared term coefficients indicating the curvature of the response surface, and βij represents the interaction effect coefficients. Here, xi and xj denote the coded levels of the independent variables that exert influence over the dependent response variable Y, while k signifies the total number of variables under consideration.
Analysis of Variance (ANOVA) was employed to scrutinize the RSM fitting model, thereby evaluating the statistical significance of each constituent term within the model. This rigorous statistical test facilitated the determination of whether the variations in the response variable could be attributed to the independent variables or were simply due to random error.
Guided by the minimization of the Mean Square Error (MSE) function, a 4-10-1 neural network model architecture was meticulously designed, as depicted in Fig. 7. In the construction of this network, the Levenberg–Marquardt algorithm was selected for its efficiency in minimizing the error. Of the total RSM samples, 70% (comprising 21 data points) were allocated for the training phase to teach the network the underlying patterns in the data. An additional 15% (equivalent to 4 points) were reserved for validation purposes, ensuring the network's generalizability beyond the training dataset. Similarly, another 15% (also 4 points) were set aside for testing the network's performance on unseen data. The optimal ANN model was identified based on the lowest MSE value and the highest correlation coefficient (R value). GA optimization, encompassing operations like reproduction, crossover, and mutation, was subsequently employed to fine-tune the model parameters for enhanced performance.25
With meticulous precision, the samples were weighed, and a series of triplicate experiments were meticulously executed in strict adherence to the optimal extraction parameters forecasted by both the RSM and the RSM-ANN-GA models. The discrepancies between the actual extraction yields of TFO and those predicted by the models were quantified as relative errors, offering a measure of the models' predictive accuracy.
3.4. Antioxidative study of TFO in vitro
Pursuant to the delineated optimal extraction protocols, the isolation of TFO was successfully executed. Upon completion of the evaporation process, the concentration of flavonoids was quantified at an impressive 76.8%. The antioxidant potential of the extracted TFO was subsequently appraised through the measurement of its ability to scavenge DPPH radicals (DPPH˙), hydroxyl radicals (˙OH), and superoxide anions (˙O2−). This evaluation was contextualized against vitamin C (VC), a widely recognized antioxidant, which served as the benchmark standard in assessing the comparative efficacy of TFO's free-radical scavenging capabilities.
 | (2) |
 | (3) |
 | (4) |
3.5. Statistical analysis
The outcomes of experiments were articulated as the mean and standard deviation (mean ± SD). The Design-Expert Software 13.0 and Neural Network Toolbox™ in MATLAB R2019a were used for RSM and RSM-ANN-GA analysis, respectively.4 Conclusions
This comprehensive study aims to optimize the UAE process for the efficient extraction of TFO. The optimization strategy builds upon the RSM by further employing ANN and GA to obtain the optimal process conditions. This method combines the advantages of RSM, ANN, and GA, not only enhancing the precision of the extraction parameters but also highlighting the superiority of the RSM-ANN-GA hybrid model in terms of predictive accuracy and extraction efficiency.Additionally, the study delves into the antioxidant properties of the extracted total flavonoids, providing critical insights into their potential health benefits. Through rigorous in vitro testing, the antioxidant activities of the flavonoids were assessed, revealing their effectiveness in scavenging reactive oxygen species such as DPPH, hydroxyl, and superoxide anions. This research not only contributes to the advancement of extraction techniques but also highlights the therapeutic potential of O. corniculata as a rich source of natural antioxidants.
In general, in consonance with a plethora of preceding investigations, the ANN emerges as a remarkably potent mathematical construct, adept at both optimizing and predicting the intricacies of the extraction process.25,27,30 This finding underscores the considerable potential of this methodology for broader application within the domain of traditional Chinese medicine, particularly concerning the extraction of its pharmacologically active constituents. Given its effectiveness in enhancing process efficiency and predictive accuracy, utilizing artificial neural networks to optimize extraction methods such as enzyme-assisted extraction (EAE), microwave-assisted extraction (MAE) and deep eutectic solvent extraction (DESE) represents a promising avenue for future research and practical applications.
In recent years, alongside ultrasonic-assisted extraction (UAE), a variety of innovative extraction techniques including supercritical fluid extraction (SCFE),49 subcritical water extraction (SWE),50 hot pressurized liquid extraction (HPLE),51 microwave-assisted extraction (MAE),52 high hydrostatic pressure extraction (HHPE),53 pulsed electric field (PEF) extraction,54 and high-voltage electrical discharge (HVED)55 extraction have gained prominence in the field of plant component extraction. Additionally, hybrid methodologies such as SCFE coupled with pressurized liquid extraction (SCFE-PLE),56 integration of HHPE with UAE (HHPE-UAE),57 synergistic application of PEF with UAE (PEF-UAE),58 and the combination of HVED with UAE (HVED-UAE)57 have emerged. Therefore, adopting innovative extraction techniques, particularly utilizing the synergistic effects of combined methods, and integrating ANN for process optimization, can help enhance extraction efficiency and improve the quality of extracted products, making it worthy of further exploration in future research.
Abbreviations
| TFO | Total flavonoids from Oxalis corniculata |
| RSM | Response surface methodology |
| ANN | Artificial neural networks |
| GA | Genetic algorithms |
| UAE | Ultrasound-assisted extraction |
| OVAT | One-variable-at-a-time |
| DoE | Design of experiments |
| FTIR | Fourier-transform infrared |
| BP-ANN | Back propagation artificial neural network |
| DPPH | 1,1-Diphenyl-2-picrylhydrazyl |
| BBD | Box–Behnken design |
| ANOVA | Analysis of variance |
| MSE | Mean square error |
| DPPH˙ | DPPH radicals |
| ˙OH | Hydroxyl radicals |
| ˙O2− | Superoxide anions |
| EAE | Enzyme-assisted extraction |
| DESE | Deep eutectic solvent extraction |
| SCFE | Supercritical fluid extraction |
| SWE | Subcritical water extraction |
| HPLE | Hot pressurized liquid extraction |
| MAE | Microwave-assisted extraction |
| HHPE | High hydrostatic pressure extraction |
| PEF | Pulsed electric field |
Data availability
Data are contained within the article or ESI.†Author contributions
Conceptualization, D.-Z. J. and K.-Y. L.; methodology, D.-P. Y.; data analysis, M. Z. and W.-B. L.; resources, K.-Y. L.; FTIR spectrum analysis, D.-L. L. and M. Z.; investigation, M. Z. and D.-Z. J.; writing—original draft preparation, D.-P. Y.; writing—review and editing, supervision, D.-Z. J.; funding acquisition, D.-Z. J. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This research was funded by Natural Science Foundation of Jiangxi Province of China, grant number 20192BAB205106. Our heartfelt gratitude extends to the Jiujiang Key Laboratory for the Development and Utilization of Traditional Chinese Medicine Resources in Northwest Jiangxi, as well as the Analytical and Testing Center at Jiujiang University, for their invaluable support throughout our research endeavors. Their contributions have significantly enriched our work and facilitated our progress in this field. Additionally, we are profoundly appreciative of the generous MATLAB R2019a software support provided by Dr Xiao-gang Dong from the School of Computer and Big Data Science at Jiujiang University. His expertise and assistance have been instrumental in our computational analyses, enabling us to achieve greater accuracy and efficiency in our research.References
- I. Sarfraz, A. Rasul, G. Hussain, M. A. Shah, B. Nageen, F. Jabeen, Z. Selamoglu, I. Ucak, M. Asrar and S. Adem, Comb. Chem. High Throughput Screening, 2022, 25, 1181–1186 CrossRef CAS.
- H. Salahuddin, Q. Mansoor, R. Batool, A. A. Farooqi, T. Mahmood and M. Ismail, Cell. Mol. Biol., 2016, 62, 60–63 CAS.
- A. R. Gholipour, L. Jafari, M. Ramezanpour, M. Evazalipour, M. Chavoshi, F. Yousefbeyk, S. J. Kargar Moghaddam, M. H. Yekta Kooshali, N. Ramezanpour, P. Daei, S. Ghasemi and M. Hamidi, Galen Med. J., 2022, 11, e2484 Search PubMed.
- P. Das, K. Kumar, A. Nambiraj, R. Awasthi, K. Dua and H. Malipeddi, Recent Pat. Drug Delivery Formulation, 2018, 12, 170–178 CrossRef CAS PubMed.
- H. Golbarg and M. J. Mehdipour Moghaddam, BioMed Res. Int., 2021, 2021, 9981915 CrossRef.
- A. Rehman, A. Rehman and I. Ahmad, Int. J. Anal. Chem., 2015, 2015, 842468 CrossRef.
- T. Gao, W. Hu, Z. Zhang, Z. Tang, Y. Chen, Z. Zhang, S. Yuan, T. Chen, Y. Huang, S. Feng, L. Zhou, C. Ding and M. Yuan, J. Food Biochem., 2022, 46, e14235 CAS.
- M. R. Khan and H. Zehra, Exp. Toxicol. Pathol., 2013, 65, 327–334 CrossRef CAS PubMed.
- G. Sreejith, M. Jayasree, P. G. Latha, S. R. Suja, S. Shyamal, V. J. Shine, G. I. Anuja, S. Sini, P. Shikha, N. M. Krishnakumar, V. Vilash, S. Shoumya and S. Rajasekharan, Indian J. Exp. Biol., 2014, 52, 147–152 CAS.
- P. A. Abhilash, P. Nisha, A. Prathapan, S. V. Nampoothiri, O. Lijo Cherian, T. K. Sunitha and K. G. Raghu, Exp. Toxicol. Pathol., 2011, 63, 535–540 CrossRef CAS.
- S. S. Sakat, P. Tupe and A. Juvekar, Indian J. Pharm. Sci., 2012, 74, 48–53 CrossRef CAS PubMed.
- D. Manna, P. K. Dutta, B. Achari and A. Lohia, Antimicrob. Agents Chemother., 2010, 54, 4825–4832 CrossRef CAS PubMed.
- J. G. Handique, M. P. Boruah and D. Kalita, Nat. Prod. Commun., 2012, 7, 1021–1023 CAS.
- H. Mizokami, K. Tomita-Yokotani and K. Yoshitama, J. Plant Res., 2008, 121, 133–136 CrossRef CAS PubMed.
- G. Chu, R. Liang, C. Wan, J. Yang, J. Li, R. Wang, L. Du and R. Lin, Molecules, 2022, 27, 4837 CrossRef CAS PubMed.
- X. Li, S. Chen, J. Zhang, L. Yu, W. Chen and Y. Zhang, Molecules, 2022, 27, 3069 CrossRef CAS.
- J. Liu, Y. Zhang, M. Zhang, Q. Wang and J. Xie, J. Food Biochem., 2022, 46, e14433 CAS.
- Y. Liu, W. Zhe, R. Zhang, Z. Peng, Y. Wang, H. Gao, Z. Guo and J. Xiao, Ultrason. Sonochem., 2022, 86, 106005 CrossRef CAS.
- D. Rosarina, D. R. Narawangsa, N. S. R. Chandra, E. Sari and H. Hermansyah, Molecules, 2022, 27, 6080 CrossRef CAS.
- G. Zhou, J. Ma, Y. Tang, X. Wang, J. Zhang, X. Yao, W. Jiang and J. A. Duan, BioMed Res. Int., 2018, 2018, 4598067 Search PubMed.
- I. D. Boateng, D. A. Soetanto, F. N. Li, X. M. Yang and Y. Y. Li, Ind. Crops Prod., 2021, 170, 113828 CrossRef CAS.
- D. Tramontin, S. E. Cadena-Carrera, J. Assreuy, R. Nunes, J. R. Santin, A. Bolzan and M. Quadri, J. Food Sci. Technol., 2021, 58, 3303–3313 CrossRef CAS.
- P. Alam, O. M. Noman, R. N. Herqash, O. M. Almarfadi, A. Akhtar and A. S. Alqahtani, Molecules, 2022, 27, 298 CrossRef CAS.
- K. Wang, Y. Mao, C. Wang, Q. Ke, M. Zhao and Q. Wang, Environ. Sci. Pollut. Res. Int., 2022, 29, 36075–36087 CrossRef CAS.
- K. Ameer, S. Ameer, Y. M. Kim, M. Nadeem, M. K. Park, M. A. Murtaza, M. A. Khan, M. A. Nasir, G. Mueen-Ud-Din, S. Mahmood, T. Kausar and M. Abubakar, Foods, 2022, 11, 883 CrossRef CAS PubMed.
- M. C. da Silva Sauthier, E. G. P. da Silva, B. R. da Silva Santos, E. F. R. Silva, J. da Cruz Caldas, L. A. Cavalcante Minho, A. M. P. Dos Santos and W. N. L. Dos Santos, Food Chem., 2019, 273, 115–123 Search PubMed.
- C. E. Onu, J. T. Nwabanne, P. E. Ohale and C. O. Asadu, S. Afr. J. Chem. Eng., 2021, 36, 24–42 Search PubMed.
- T. Aung, S. J. Kim and J. B. Eun, Food Chem., 2022, 366, 130689 CAS.
- H. Chen, B. Wang, J. Li, J. Xu, J. Zeng, W. Gao and K. Chen, Int. J. Biol. Macromol., 2023, 226, 982–995 CrossRef CAS PubMed.
- C. M. Agu, M. C. Menkiti, E. B. Ekwe and A. C. Agulanna, Artif. Intell. Agric., 2020, 4, 1–11 Search PubMed.
- S. Chen, H. Zhang, L. Yang, S. Zhang and H. Jiang, Foods, 2023, 12, 619 CrossRef CAS PubMed.
- O. C. Edward, S. S. Thomas, K. O. Cha, H. A. Jung, A. Han and Y. S. Cha, Nutr. Res. Pract., 2022, 16, 549–564 CrossRef CAS.
- D. B. Muniz-Marquez, G. C. Martinez-Avila, J. E. Wong-Paz, R. Belmares-Cerda, R. Rodriguez-Herrera and C. N. Aguilar, Ultrason. Sonochem., 2013, 20, 1149–1154 CrossRef CAS PubMed.
- X. Yang, Y. Li, S. Li, A. O. Oladejo, S. Ruan, Y. Wang, S. Huang and H. Ma, Ultrason. Sonochem., 2017, 38, 19–28 CrossRef CAS.
- C. D. Fernando and P. Soysa, Nutr. J., 2015, 14, 74 CrossRef PubMed.
- C. C. Coussios, C. H. Farny, G. T. Haar and R. A. Roy, Int. J. Hyperthermia, 2007, 23, 105–120 CrossRef CAS PubMed.
- K. V. Mahindrakar and V. K. Rathod, Chem. Eng. Process., 2020, 149, 107841 CrossRef CAS.
- T. P. Vo, L. N. H. Nguyen, N. P. T. Le, T. P. Mai and D. Q. Nguyen, Curr. Res. Food Sci., 2022, 5, 2013–2021 CrossRef CAS PubMed.
- M. Hadidi, A. Ibarz and J. Pagan, Food Chem., 2020, 309, 125786 CrossRef CAS PubMed.
- V. Galvan-Alvarez, A. Gallego-Selles, M. Martinez-Canton, E. Garcia-Gonzalez, M. Gelabert-Rebato, J. G. Ponce-Gonzalez, S. Larsen, D. Morales-Alamo, J. Losa-Reyna, I. Perez-Suarez, C. Dorado, M. Perez-Valera, H. C. Holmberg, R. Boushel, P. de Pablos Velasco, J. W. Helge, M. Martin-Rincon and J. A. L. Calbet, Free Radical Biol. Med., 2023, 209, 282–291 CrossRef CAS.
- P. K. R. Thavanati, K. R. Kanala, A. E. de Dios and J. M. C. Garzat, J. Gerontol., Ser. A, 2008, 63, 360–364 CrossRef.
- F. Pohl and P. Kong Thoo Lin, Molecules, 2018, 23, 3283 CrossRef.
- R. Chen, X. Zhou, Q. Deng, M. Yang, S. Li, Q. Zhang, Y. Sun and H. Chen, Int. J. Biol. Macromol., 2024, 257, 128669 CrossRef CAS.
- A. Pandey, T. Belwal, K. C. Sekar, I. D. Bhatt and R. S. Rawal, Ind. Crops Prod., 2018, 119, 218–225 CrossRef CAS.
- A. Weremfo, F. Adulley, K. Dabie, S. Abassah-Oppong and E. Peprah-Yamoah, J. Appl. Res. Med. Aromat. Plants, 2022, 30, 100387 CAS.
- L. Zhao, Z. Ji, K. Li, B. Wang, Y. Zeng and S. Tian, BMC Complementary Med. Ther., 2020, 20, 228 CrossRef CAS.
- J. Liu, D. Chen, Z. Wang, C. Chen, D. Ning and S. Zhao, Food Sci. Nutr., 2019, 7, 969–976 CrossRef CAS.
- C. Fu, T. Wang, Y. Wang, X. Chen, J. Jiao, F. Ma, M. Zhong and K. Bi, J. Pharm. Biomed. Anal., 2011, 55, 1067–1074 CrossRef CAS PubMed.
- L. Campone, R. Celano, A. Lisa Piccinelli, I. Pagano, S. Carabetta, R. D. Sanzo, M. Russo, E. Ibañez, A. Cifuentes and L. Rastrelli, Food Chem., 2018, 269, 495–502 CrossRef CAS.
- J. Vladić, M. Jakovljević, M. Molnar, S. Vidović, M. Tomić, Z. Drinić and S. Jokić, Molecules, 2020, 25, 1878 CrossRef.
- Y. F. Shang, J. L. Xu, W. J. Lee and B. H. Um, S. Afr. J. Bot., 2017, 109, 75–80 CrossRef CAS.
- P. Skenderidis, S. Leontopoulos, K. Petrotos and I. Giavasis, Foods, 2020, 9, 1055 CrossRef.
- L. Fernandes, S. I. P. Casal, J. A. Pereira, E. Ramalhosa and J. A. Saraiva, High Pressure Res., 2017, 37, 415–429 CrossRef CAS.
- V. M. Pappas, A. Lakka, D. Palaiogiannis, V. Athanasiadis, E. Bozinou, G. Ntourtoglou, D. P. Makris, V. G. Dourtoglou and S. I. Lalas, Antioxidants, 2021, 10, 1554 CrossRef CAS PubMed.
- J. Xi, L. He and L.-g. Yan, Food Chem., 2017, 230, 354–361 CAS.
- D. M. Ferro, S. Mazzutti, L. Vitali, C. M. Oliveira Müller and S. R. S. Ferreira, J. Supercrit. Fluids, 2019, 149, 10–19 CAS.
- T. M. C. Nguyen, M. Gavahian and P.-J. Tsai, LWT, 2021, 143, 111131 CAS.
- A. Görgüç, C. Bircan and F. M. Yılmaz, Food Chem., 2019, 283, 637–645 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05077k |
| This journal is © The Royal Society of Chemistry 2024 |







