Open Access Article
Open Access ArticleElectrochemical detection of FTO with N3-kethoxal labeling and MazF cleavage†
Chen Chen,
Mei Zhao,
Jingyi Guo,
Xia Kuang,
Zilin Chen and
Fang Wang
and
Fang Wang *
*
School of Pharmaceutical Sciences, Key Laboratory of Combinatorial Biosynthesis and Drug Discovery (MOE), Wuhan University, Wuhan, 430071, China. E-mail: fwang@whu.edu.cn; Fax: +86-27-68759850; Tel: +86-27-68759829
First published on 13th August 2024
Abstract
N6-Methyladenosine (m6A) is a prevalent modification in eukaryotic mRNAs and is linked to various human cancers. The fat mass and obesity-associated protein (FTO), a key m6A demethylase, is crucial in m6A regulation, affecting many biological processes and diseases. Detecting FTO is vital for clinical and research applications. Our study leverages the specific cleavage properties of the MazF endoribonuclease to design an electrochemical method with signal amplification guided by streptavidin-horseradish peroxidase (SA-HRP), intended for FTO detection. Initially, the compound N3-kethoxal is employed for its reversible tagging ability, selectively attaching to guanine (G) bases. Subsequently, dibenzocyclooctyne polyethylene glycol biotin (DBCO-PEG4-Biotin), is introduced through a reaction with N3-kethoxal. HRP is then employed to catalyze the redox system to enhance the current response further. A promising linear correlation between the peak current and the FTO concentration was observed within the range of 7.90 × 10−8 to 3.50 × 10−7 M, with a detection limit of 5.80 × 10−8 M. Moreover, this method assessed the FTO inhibitor FB23's inhibitory effect, revealing a final IC50 value of 54.73 nM. This result aligns with the IC50 value of 60 nM obtained through alternative methods and is very close to the values reported in the literature. The study provides reference value for research into obesity, diabetes, cancer, and other FTO-related diseases, as well as for the screening of potential therapeutic drugs.
1. Introduction
N6-Methyladenosine (m6A) is recognized as one of the most common epigenetic modifications.1,2 Numerous studies have linked anomalies in m6A methylation to the development of various cancers.3,4 The fat mass and obesity-associated protein (FTO), known for its demethylase activity on m6A, can remove the methyl group from m6A-modified mRNA, thereby rendering the m6A modifications reversible and dynamic.5,6 This regulatory mechanism influences a wide range of biological processes and diseases. Evidence has shown that FTO can impact mRNA metabolism and various cellular processes, contributing to conditions like obesity7 and cancer.8 Particularly, FTO dysregulation and subsequent m6A modification can lead to abnormal gene expression, affecting disease outcomes.9 Abnormal expression of FTO often co-occurs with related diseases, including type II diabetes,10 arthritis,11 leukemia,12 lung cancer,13 rectal cancer,14 breast cancer,15 liver cancer,16 and others.17 Moreover, recent findings indicate that abnormal expression of FTO may contribute to a spectrum of mental illness.18 Consequently, monitoring FTO activity is of paramount importance. Furthermore, the identification and screening of FTO protein inhibitors carry significant implications for the diagnosis and treatment of diseases associated with FTO.Currently, methods such as liquid chromatography-mass spectrometry (HPLC-MS),19 fluorescence,20 photoelectrochemistry,21 and CRISPR/Cas techniques22 have been used to detect FTO and find potential new treatments for clinically related diseases. Electrochemical methods have gained prominence in detecting both small molecules and biomacromolecules due to their remarkable sensitivity, selectivity, low energy consumption, simplicity, efficiency, and convenience.23–30 The ability to easily miniaturize and make electrochemical sensors portable provides significant advantages for tumor marker detection in clinical settings.31,32 These methods are widely used for detecting nucleic acid methylation modifications, offering highly selective and sensitive identification of methylation sites in DNA or proteins. This capability facilitates a deeper understanding of methylation's role in biological processes.33–35 Although electrochemical techniques can quickly and effectively assess changes in FTO enzyme activity and its substrate-binding affinity,36,37 highlighting their vital applications in studying nucleic acid methylation and FTO activity, there are relatively few reports on related methods.
Our research focuses on developing an innovative electrochemical method for detecting FTO enzyme activity. This method harnesses the specific recognition and cleavage capabilities of Escherichia coli MazF,38 combined with horseradish peroxidase (HRP) labelling for enhanced signal amplification. Building on the ground-breaking work of Prof. Weng et al.,39 who discovered that N3-kethoxal rapidly and efficiently targets and reacts with guanine residues in single-stranded DNA (ssDNA) and single-stranded RNA (ssRNA) for effective guanine labelling, our approach further advances this concept avoiding the commonly used electrochemical signal probes in nucleic acid electrochemical sensors such as Ru compounds and methylene. Our approach further advances this concept by replacing the commonly used Ru compounds and MB as electrochemical signal probes in nucleic acid electrochemical sensors. We utilize the copper-free click reaction between azide groups and dibenzocyclooctyne-R (DBCO-R), a widely-used technique for introducing molecular probes, enabling us to apply dibenzocyclooctyne polyethylene glycol biotin (DBCO-PEG4-Biotin) labeling.40,41 Additionally, our method incorporates the assessment of the inhibitory effects of FB23 on FTO activity, determining an IC50 of 54.73 nM, which aligns with results from other methodologies. These results are crucial as understanding and influencing FTO activity is key in tackling diseases related to this enzyme. Our findings have significant implications for research into conditions like obesity, diabetes, and cancer that are associated with FTO, and they could aid in the screening of new therapeutic agents.
2. Materials and methods
2.1 Reagents and apparatus
N3-kethoxal was a gift from Prof. Xiaocheng Weng, College of Chemistry and Molecular Sciences of Wuhan University. DBCO-PEG4-Biotin was purchased from Click Chemistry Tools (Beijing, China). Tris(2-carboxyethyl)-phosphine hydrochloride (TCEP), 6-mercapto-1-hexanol (MCH), NaCl, bovine serum albumin (BSA), and hydrogen tetrachloroaurate trihydrate (HAuCl4·3H2O) were purchased from Aladdin (Shanghai, China). Streptavidin-horseradish peroxidase (SA-HRP) was obtained from Sangon Biotech (Shanghai, China). Recombinant FTO (ALKBH9) protein was purchased from Active Motif (California, CA, USA) and mRNA Interferase-MazF was obtained from TARAKA Biomedical Technology Co., Ltd (Dalian, China). All aqueous solutions were prepared with ultrapure water purified by a Milli-Q water purification system (MA, USA). All HPLC-purified oligonucleotides were supplied by TAKARA Biomedical Technology Co., Ltd (Dalian, China), and the sequences are listed in Table 1.| Name | Sequence (5′-3′) |
|---|---|
| ssRNA | SH-(CH2)6-UUGGUUUUUUUUGGACAUGUAUAUAGU |
| m6A ssRNA | SH-(CH2)6-UUGGUUUUUUUUGGm6ACAUGUAUAUAGU |
RNA was quantified by Nanodrop 2000c (Thermo Scientific, USA). Gel images were recorded by Pharos FX molecular imager (Bio-Rad, USA). RNA after reaction was purified by oligo concentration clean kits (Zymo Research, USA).
Electrochemical experiments were performed on the CHI842B electrochemical workstation (Shanghai Chenhua Instruments, China) accompanied by a conventional three-electrode system consisting of an AuNPs-modified glassy carbon electrode (AuNPs/GCE) which served as working electrode, platinum wire and saturated calomel electrode served as auxiliary electrode and reference electrode, respectively. Electrochemical impedance spectroscopy (EIS) experiments were performed on the VersaSTAT 3F electrochemical workstation (Ametek, USA).
2.2 Gold nanoparticles deposition
The bare glassy carbon electrode (GCE, A = 0.071 cm2) was mechanically polished with polishing micro-cloth containing 0.05 μm Al2O3 slurry to a mirror finish, which was carefully cleaned in deionized water, ethanol, and deionized water in turn via ultra-sonication each for 40 seconds. After being dried with N2, GCE was immersed in 3 mM HAuCl4 solution containing 0.1 M KNO3 for AuNPs deposition at −0.2 V for 200 s. The prepared electrode is denoted as AuNPs/GCE.2.3 Denaturing polyacrylamide gel electrophoresis (PAGE) analysis
Before proceeding with denaturing PAGE, m6A ssRNAs were reacted as follows: (1) with 5 μL of 0.3 μg μL−1 FTO; (2) with 5 μL of 0.3 μg μL−1 FTO followed by 5 μL of 1 U mL−1 MazF; (3) with 5 μL of 10 mM N3-kethoxal (in DMSO); (4) with 5 μL of 10 mM N3-kethoxal (in DMSO) followed by 5 μL of 10 mM DBCO-PEG4-Biotin (in DMSO); and (5) with 5 μL of 10 mM DBCO-PEG4-Biotin (in DMSO), then mixed with deionized formamide to a final volume of 15 μL (V/V, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8). Next, a 20% denaturing PAGE was prepared according to a previous reference, using 1× TBE buffer.42 The PAGE process was conducted at a continuous voltage of 340 V for about 2 hours at room temperature. The final PAGE products were stained with GelRed for approximately 1 hour and then imaged on a Pharos FX molecular imager operated in fluorescence mode (λex = 532 nm).
8). Next, a 20% denaturing PAGE was prepared according to a previous reference, using 1× TBE buffer.42 The PAGE process was conducted at a continuous voltage of 340 V for about 2 hours at room temperature. The final PAGE products were stained with GelRed for approximately 1 hour and then imaged on a Pharos FX molecular imager operated in fluorescence mode (λex = 532 nm).
2.4 Fabrication of the electrochemical biosensor
5 μL of m6A ssRNA (0.1 μM) was dropped on the surface of the AuNPs/GCE electrode overnight at 37 °C and then the electrode surface was carefully rinsed with 1 mL of RNase-free water to remove the unabsorbed m6A RNA. 5 μL of 1 mM MCH solution and 5 μL of 2.5% BSA solution were successively dropped onto the electrode surface for blocking of the nonspecific active sites and incubated for 1 hour in turn. The electrode, labeled as m6A ssRNA/BSA/MCH/AuNPs/GCE, was incubated with different concentrations of FTO in FTO reaction buffer at 37 °C for 4 hours. After that, 5 μL of 0.1 U μL−1 MazF (including 1× MazF buffer) was applied onto the electrode surface and left for 30 minutes. Subsequently, 5 μL of 1 mM N3-kethoxal and 1 mM DBCO-PEG4-Biotin (containing reaction buffer) were added to the electrode surface and allowed to react for 1.5 hours. Then, the electrode was marked with 5 μL of 1 U mL−1 SA-HRP and incubated at 37 °C for 30 minutes, after which it was left to await testing. After each step, the electrode was rinsed and dried with 1 mL of RNase-free water, using a pipette.2.5 Electrochemical detection
The electrochemical detection was performed at the prepared electrochemical biosensor in 5 mL of 0.1 M PB buffer solution (pH 7.4) containing 2.5 mM HQ and 2.5 mM H2O2. The responses were recorded by differential pulse voltammetry (DPV) (potential range: +0.3 to −0.3 V; amplitude: 0.05 V; pulse width: 0.05 s; pulse period: 0.2 s). EIS and cyclic voltammetry (CV) characterization were carried out in 5 mL of 0.1 M KCl solution containing 1 mM [Fe (CN)6]3−/4−.2.6 The inhibitory effect of FB23 on FTO
At a concentration of 0.06 μg μL−1 for the FTO protein, varying concentrations of FB23 (20, 40, 60, 80, 100, and 120 nM) were introduced during the demethylation phase of the FTO enzyme. Subsequent electrochemical signal readings were recorded and graphed for further analysis.3. Results and discussion
3.1 Principle of the electrochemical detection
The electrochemical assay for FTO detection is illustrated in Scheme 1. Initially, AuNPs are electrodeposited onto a GCE to enhance the electrode's active surface area and conductivity, and to provide binding sites for the subsequent nucleic acid strands. The designed m6A ssRNA is then assembled onto the AuNPs/GCE via Au–S bonds. The electrode surface is blocked with 1 mM MCH and 2.5% BSA for 1 hour to close off nonspecific active sites on the AuNPS/GCE respectively. MCH not only blocks the Au sites that haven't bonded with the m6A ssRNA but also ensures that the nucleic acid strand is oriented perpendicularly to the electrode surface, facilitating subsequent reactions. To avoid protein adsorption and prevent false positives from SA-HRP adsorption, BSA is employed for additional blocking. BSA can adsorb to the AuNPS/GCE surface through nonspecific adsorption and mechanical accumulation, further shielding more nonspecific sites. The free thiol groups in BSA can also enhance their bond with the electrode through Au–S, thus extensively covering the nonspecific active sites and minimizing errors in subsequent protein adsorption.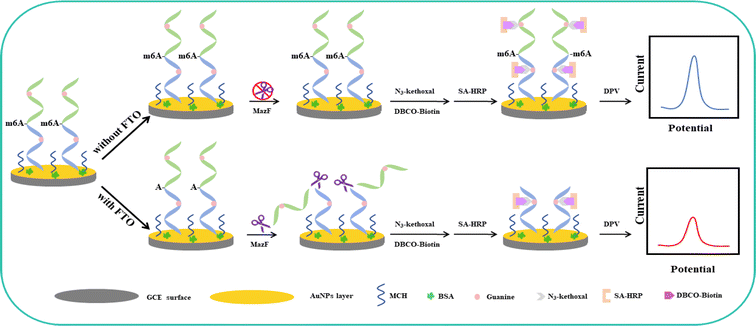 | ||
| Scheme 1 Electrochemical detection schematic for FTO based on the specific cleavage action of MazF and signal amplification of HRP. | ||
After the blocking process, the electrode surface was exposed to FTO protein, followed by treatment with MazF. MazF is an endoribonuclease that specifically recognizes and cleaves the ACA sequence, which is exposed following the demethylation of m6ACA by FTO, resulting in the truncation of the RNA strand containing G. In contrast, the RNA in the control group, which was not treated with FTO, retains its original length and G content due to the absence of the ACA cleavage site necessary for MazF recognition and action. Following this, reactions with N3-kethoxal and DBCO-PEG4-Biotin occurred. After labeling with SA-HRP, the strands that were not subjected to FTO demethylation and subsequent MazF cleavage retained a higher G content, which, in turn, provided more binding sites for HRP, resulting in increased labeling. Contrarily, RNA strands that experienced FTO demethylation and MazF cleavage displayed reduced G content, leading to fewer binding sites and consequently less HRP labelling. The reaction mechanism of the electrochemical biosensor can be summarized as follows: Initially, hydrogen peroxide in the solution is reduced by the immobilized Horseradish Peroxidase (HRP) in its reduced state HRP(Red), forming water and oxidizing the HRP to its oxidized state HRP(Ox) according to the reaction: H2O2 + HRP(Red) → H2O + HRP(Ox). Subsequently, the oxidized HRP (HRP(Ox)) is regenerated with the assistance of a mediator, which itself is oxidized from its reduced state (HQ(Red)) to its oxidized state (HQ(Ox)) during this enzymatic reaction: HRP(Ox) + HQ(Red) → HRP(Red) + HQ(Ox) + H2O. Finally, the produced HQ(Ox) is electrochemically reduced on the electrode, regenerating HQ(Red) and resulting in a significant increase in the reduction current: HQ(Ox) + 2H+ + 2e− → HQ(Red).43,44 To conclude, an amplification of the electrochemical signal was achieved using HRP's catalysis involving the HQ and H2O2 redox system. By evaluating the disparities in the electrochemical signals, the precise quantity of FTO protein present was ascertainable.
3.2 PAGE analysis
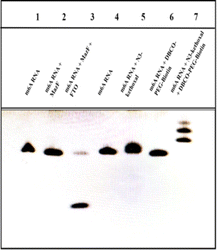
PAGE is utilized to corroborate the demethylation effectuated by MazF, MazF's specific recognition and cleavage, as well as the reactions involving N3-kethoxal, guanine (G), and DBCO-PEG4-Biotin on the ssRNA, as depicted in Fig. 1. Bands on the gel are positioned relative to their molecular weights. The larger the molecular weight post-treatment, the slower the movement through the gel and the higher its position. Bands 1 to 3 represent different stages of m6A ssRNA treatment: untreated, treated with MazF, and treated with FTO and MazF, respectively. The results elucidate that m6A ssRNA, treated with both FTO and MazF, migrates faster compared to untreated m6A ssRNA or m6A ssRNA solely treated with MazF, indicating successful oxidation of m6A by FTO and subsequent recognition and cleavage by MazF. This results in a reduced molecular weight due to shorter RNA strands, facilitating faster electrophoresis. Bands 4 to 7 delineate the progression post various treatments. Comparing the bands illustrates that the N3-kethoxal treated m6A ssRNA (band 5) migrates slower than the untreated m6A ssRNA (band 4), and when treated with both N3-kethoxal and DBCO-PEG4-Biotin (band 7), it moves even slower compared to band 5. However, m6A ssRNA treated solely with DBCO-PEG4-Biotin (band 6) exhibits a similar migration pattern to the untreated sample (band 4). These observations signify that N3-kethoxal effectively reacts with G on the m6A ssRNA, increasing the molecular weight and subsequently reducing the electrophoresis speed. Conversely, DBCO-PEG4-Biotin, when used alone, doesn't react significantly with the m6A ssRNA, maintaining the original molecular weight and electrophoresis speed. Band 7, post its exposure to a copper-free click chemistry reaction involving azide groups and DBCO-PEG4-Biotin, experienced an introduction of biotin, incrementing its molecular weight further, thereby slowing its electrophoretic speed relative to band 4.3.3 Feasibility of the electrochemical detection
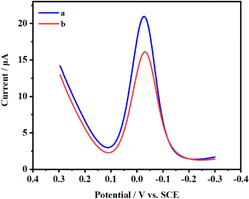
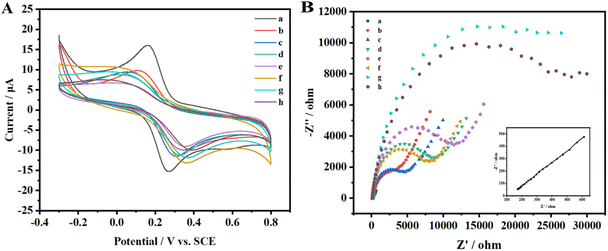
The differential pulse voltammograms were captured, as depicted in Fig. 2, at FTO concentrations of 0 and 0.08 μg μL−1, respectively. Curve a demonstrates that without FTO's demethylation, MazF is unable to interact with the m6ACA sequence. This leads to a substantial number of SA-HRP labels on the ssRNA present on the sensor, catalyzing the HQ-H2O2 redox reaction and consequently producing a prominent peak current. On the contrary, curve b reveals that in the presence of the FTO protein, there is a notable decline in the peak current. This indicates that FTO facilitates the conversion of m6ACA into ACA, enabling MazF to recognize and cleave it. During the rinsing process, the G residues on the cleaved RNA strands are removed by RNase free water. This results in a diminished residue containing G, reduced HRP labeling, and subsequently, a decrease in the electrochemical signals observed. Such observations underscore that the peak current exhibited by this method correlates with the FTO concentration to a certain degree. This provides a foundation for further research and analysis to be conducted on this relationship.Cyclic voltammetry is employed to characterize each phase of the modified electrode's assembly process, as illustrated in Fig. 3A. Initially, one pair of well-defined redox peaks for [Fe(CN)6]3−/4− are observed at the AuNPs/GCE (curve a). However, upon affixing the m6A ssRNA onto the AuNPs/GCE, a decline in the peak current (Ip) of [Fe(CN)6]3−/4− is found (curve b), coupled with an increase in peak potential differences (ΔEp). The m6A ssRNA, possessing a negatively charged phosphate backbone, assembles on the electrode, creating electrostatic repulsion with similarly negatively charged [Fe(CN)6]3−/4− ions. This effectively impedes the electron transfer of [Fe(CN)6]3−/4− on the AuNPs/GCE surface. MCH is subsequently utilized for initial blocking to eliminate non-specific active sites and to orient the m6A ssRNA upright on the electrode. When m6A RNA/AuNPs/GCE is immersed in MCH, a significant drop in the Ip of [Fe(CN)6]3−/4− is noted (curve c). A multitude of MCH molecules occupies the active sites on the AuNPs/GCE through Au–S bonds, reducing the chance of interaction between [Fe(CN)6]3−/4− and the sensing interface, which signifies the successful assembly of MCH on the electrode. In a further step to avoid non-specific adsorption of the macromolecular protein complex, SA-HRP, BSA as an inert protein is employed for additional electrode blocking. Post-BSA incubation, a continuous and significant reduction in Ip and a rise in ΔEp are apparent (curve d), indicating the successful integration of BSA onto the electrode. Cyclic voltammograms (CVs) show that the curve post-treatment with the FTO protein (curve e) exhibits a significant deviation from the pre-treatment (curve d), attributable to the adsorption of FTO on the electrode surface. This adsorption hindered electron transfer, leading to a reduction in the peak current (Ip) and an increase in the peak-to-peak separation (ΔEp). Following MazF protein treatment, the CV plot (curve f) reveals a marginal Ip enhancement without any significant alteration in ΔEp. MazF cleavage reduces the steric hindrance of the modified m6A ssRNA on the electrode, subtly amplifying the peak potential. Subsequent reactions with N3-kethoxal and DBCO-PEG4-Biotin and CV scanning of the modified electrodes showcase a rise in ΔEp and a fall in Ip (curve g). The introduction of biotin, which has a larger molecular weight, further amplifies the impedance of electron flow on the electrode surface, indicating successful modification of the target strand with N3-kethoxal and DBCO-PEG4-Biotin. Lastly, SA-HRP is integrated into the electrode surface via efficient and specific biotin-streptavidin binding. A continued dip in Ip and elevation in ΔEp (curve h) are observed due to the substantial molecular weight of SA-HRP, confirming its successful assembly on the electrode. Besides CV, serves as another instrumental technique in characterizing the process of electrode surface modification. The relevant Nyquist curve is shown in Fig. 3B. In EIS analysis, the semicircle diameter of the Nyquist plot represents the electron transfer resistance (Ret). The larger the value, the greater the impedance of the electrode sensing interface. In the plot, curves a–h represent sequential steps of electrode assembly and modification. Initial observations reveal a subtle increase in impedance following the assembly of the target strand on the AuNPs/GCE surface (curve a). As molecular entities like MCH and BSA are incrementally assembled onto the m6A ssRNA/AuNPs/GCE surface (curve b–d), a pronounced and gradual augmentation in the electrode surface's impedance can be found. It is notable that the impedance continues to increase even after the FTO treatment (curve e). Conversely, a minor reduction in impedance is detected following MazF treatment (curve f), an implication of FTO's successful oxidation of m6A. Owing to its ACA sequence, the oxidized ssRNA undergoes MazF-induced cleavage. Concludingly, the impedance gradually increased with the successive modifications of N3-kethoxal, DBCO-PEG4-Biotin (curve g), and SA-HRP (curve h), and the results demonstrated by the EIS for each step are in consistent with the CV. This coherence substantiates the achievement of each articulated phase in the electrode assembly and modification process.
3.4 Condition optimization
To enhance the efficacy of the novel electrochemical detection method presented in this study, we strategically selected several pivotal factors that could substantially influence the outcomes. These factors include the reaction time of N3-kethoxal and DBCO-PEG4-Biotin on the electrode, the combined reaction concentration of N3-kethoxal and DBCO-PEG4-Biotin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on the electrode, and the incubation duration of FTO on the electrode. By meticulously analyzing these variables, we aim to refine and optimize the detection performance of the designed electrochemical method to ensure it yields accurate and reliable results.
1) on the electrode, and the incubation duration of FTO on the electrode. By meticulously analyzing these variables, we aim to refine and optimize the detection performance of the designed electrochemical method to ensure it yields accurate and reliable results.
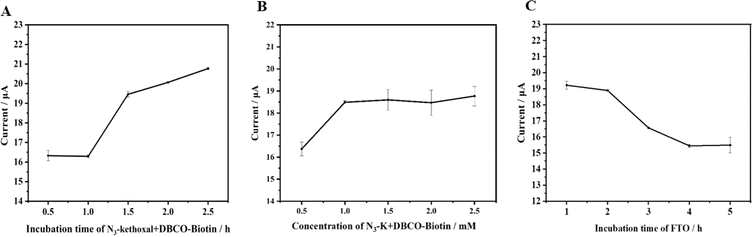
Fig. 4A illustrates that the DPV response intensifies in correspondence with the reaction time of N3-kethoxal and DBCO-PEG4-Biotin on the electrode. However, the enhancement is marginal, and the trend begins to plateau after 1.5 hours. It is hypothesized that beyond 1.5 hours, owing to the confined volume of fluid on the electrode and the reversible nature of the reaction between N3-kethoxal and guanine (G), the signal ceases to escalate. In contemplation of the sensor's overarching stability and sensitivity, 1.5 hours has been decisively identified as the optimum reaction time.
Fig. 4B demonstrates that the DPV response amplifies with the increasing reaction concentration of N3-kethoxal and DBCO-PEG4-Biotin on the electrode, levelling off beyond 1 mM. This plateau arises due to the finite quantity of RNA initially bonded to the electrode via the Au–S linkage. Despite the escalating concentrations of N3-kethoxal and DBCO-PEG4-Biotin, the reaction involving N3-kethoxal and guanine (G) on the ssRNA reaches a saturation point, preventing further enhancement of the electrochemical signal in response to increased N3-kethoxal concentration. Consequently, 1 mM has been determined as the optimal reaction concentration for subsequent experiments.
At an FTO concentration of 1.58 × 10−7 M, the impact of varying FTO incubation times was examined. Fig. 4C illustrates that the DPV response diminishes as FTO incubation time extends, signifying an increased action of FTO on the target methylated RNA strands over time, resulting in a higher quantity of demethylated RNA. Subsequently, MazF truncates these strands, reducing the number of N3-kethoxal labels on the strands and decreasing the subsequent introduction of SA-HRP. This increases the electrochemical signal difference between the control and experimental groups. However, beyond an incubation period of 4 hours, the diminishing signal becomes marginal and tends to stabilize. To optimize time efficiency and ensure maximum sensor stability, an incubation time of 4 hours was deemed optimal for the FTO protein on the electrode.
3.5 Analysis of FTO enzyme activity
To assess the detection efficiency of the method proposed in this study for FTO protein, we employed this approach to analyze standard FTO protein samples across varying concentrations. The outcomes are delineated in Fig. 5A. Under the optimized experimental conditions, a discernible decrement in the current signal of the HQ-H2O2 redox system is observed commensurate with the escalating concentration of FTO protein. The results show that there is a good linear correlation between peak current and FTO protein concentration within the concentration range of 7.90 × 10−8 to 3.50 × 10−7 M (Fig. S1†). This relationship is meticulously characterized by the fitting equation y = −32.88x + 17.24, boasting an R2 value of 0.94. The method exhibits a detection limit of 5.80 × 10−8 M, underscoring its sensitivity and precision in the quantification of FTO protein.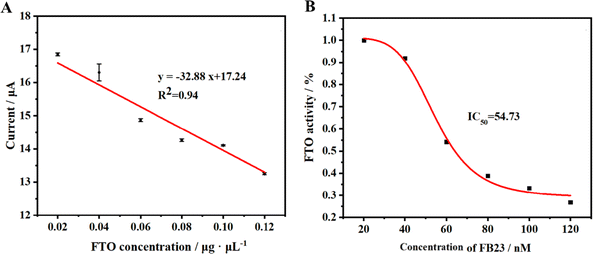 | ||
| Fig. 5 (A) The linear relationship between current variation value and FTO concentration. (B) Inhibition effect of FB23 on FTO in vitro. | ||
3.6 Verification of inhibitor screening by this method
The FTO protein plays a significant role in the onset and progression of certain diseases within the body. Thus, inhibiting its functions opens a potential pathway for treating related diseases. Our research introduces a novel electrochemical method, allowing for a detailed investigation of the inhibitory actions against the FTO protein. In our experiments, we selected FB23, a recognized inhibitor, to evaluate the effectiveness of our proposed method. We exposed the FTO protein to varying concentrations of FB23 and monitored the electrochemical DPV responses. The results, depicted in Fig. 5B, confirm the capability of our method to validate the inhibitory effects of FB23 on the FTO protein. Based on the data, it can be calculated that the IC50 of FB23 for FTO protein inhibition using our method is 54.73 nM, which is close to the reported value of 60 nM,45 substantiating the reliability of our method for screening potential inhibitors of the FTO enzyme. Our approach holds promise as a useful tool in the ongoing exploration of treatments for diseases associated with the FTO protein.3.7 Stability and selectivity
The selectivity of this method was thoroughly examined. In this section, DPV was utilized to assess the selectivity of the electrochemical sensor. We studied the potential interference of various substances such as rabbit IgG (Immunoglobulin G), human IgM (Immunoglobulin M), TET1 (Ten-Eleven Translocation), Ab (Albumin from chicken egg white), and UDG (Uracil DNA Glycosylase) on the detection of FTO. As depicted in Fig. 6(A) and S2,† the results indicate that under the utilized method, these common proteins did not influence the experimental outcomes. Furthermore, to enhance the practicality of the electrochemical sensor, the stability of the sensor during storage was meticulously evaluated, which is crucial for its reliability and consistency. With an FTO concentration of 0.02 μg μL−1, the detection outcomes are illustrated in Fig. 6(B). Curve a represents the detection outcomes on the initial day, while curve b displays the results after five days. It was observed that the Ip detected after five days exhibited a slight reduction of about 0.5% compared to the Ip detected on the initial day. This indicates that the constructed sensor possesses commendable stability over a brief storage period. However, due to factors such as RNA instability at the electrodes, it is advisable to avoid using the pre-prepared sensor for extended testing periods to ensure the accuracy and reliability of the results.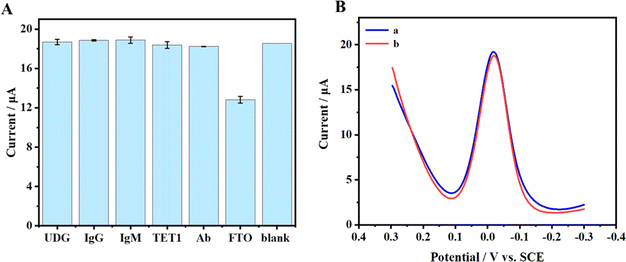 | ||
| Fig. 6 (A) The effects of five proteins on FTO detection. (B) DPVs (a) before and (b) after storage at 4 °C for 5 days with the FTO concentration of 7.90 × 10−8 M. | ||
3.8 FTO detection methods
The culmination of our method was realized following a comprehensive series of optimization and research endeavors. A notable linear correlation has been observed between the peak current and the FTO protein concentration, particularly within a concentration spectrum of 7.90 × 10−8 to 3.50 × 10−7 M. Impressively, this method achieves a detection limit as profound as 5.80 × 10−8 M. Unfortunately, compared to other methods, our approach exhibits somewhat limited detection sensitivity and range.22,36,46 Efforts to enhance signal amplification could significantly address this issue. Nonetheless, the innovative improvements and solid research behind our method continue to bolster its practicality and effectiveness in real-world applications (Table 2).Based on the comparative results, our method has significant potential for further improvement. Our future efforts will concentrate on enhancing the sensor's performance. Notably, the incorporation of the N3-kethoxal small molecule introduces a novel electrochemical probe that can precisely interact with guanine in single-stranded nucleic acids. This interaction generates electrochemical signals, providing an innovative approach for electrochemical sensors.
4. Conclusions
Our novel approach combines the RNA-specific cleavage protein MazF with the unique binding of N3-kethoxal to guanine (G), creating an effective platform for FTO detection, without using conventional direct electrochemical signal molecular probes. We successfully demonstrated this method's ability to detect the inhibitory effects of FB23 on FTO, offering a new way to screen for inhibitors of the FTO enzyme. The comparative analysis indicates substantial potential for advancing our method. Future endeavours will focus on optimizing the sensor's performance. Significantly, the integration of the N3-kethoxal small molecule introduces an advanced electrochemical probe capable of specific interaction with guanine residues in single-stranded nucleic acids. This interaction facilitates the generation of electrochemical signals, presenting a novel and innovative approach for electrochemical sensor development. Looking forward, we envision integrating our method with advanced microelectrode systems, such as laser-induced graphene electrodes, to meet specific user requirements. Furthermore, the potential applications extend to the identification of new FTO inhibitors, a crucial step in addressing diseases associated with FTO. This underscores the importance and impact of our approach in the broader context of biomedical research and therapeutic development.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 22277094). We would like to extend my sincere gratitude to Prof. Xiaocheng Weng from the College of Chemistry and Molecular Sciences at Wuhan University for generously providing the N3-kethoxal.References
- X. Wang, B. S. Zhao, I. A. Roundtree, Z. Lu, D. Han, H. Ma, X. Weng, K. Chen, H. Shi and C. He, N(6)-methyladenosine modulates messenger RNA translation efficiency, Cell, 2015, 161(6), 1388–1399, DOI:10.1016/j.cell.2015.05.014.
- I. A. Roundtree, M. E. Evans, T. Pan and C. He, Dynamic RNA modifications in gene expression regulation, Cell, 2017, 169(7), 1187–1200, DOI:10.1016/j.cell.2017.05.045.
- P. A. Boriack-Sjodin, S. Ribich and R. A. Copeland, RNA-modifying proteins as anticancer drug targets, Nat. Rev. Drug Discov., 2018, 17(6), 435–453, DOI:10.1038/nrd.2018.71.
- H. Huang, H. Weng and J. Chen, M(6)A modification in coding and non-coding RNAs: roles and therapeutic implications in cancer, Cancer Cell, 2020, 37(3), 270–288, DOI:10.1016/j.ccell.2020.02.004.
- G. Jia, Y. Fu, X. Zhao, Q. Dai, G. Zheng, Y. Yang, C. Yi, T. Lindahl, T. Pan, Y. Yang and C. He, N6-Methyladenosine in nuclear RNA is a major substrate of the obesity-associated FTO, Nat. Chem. Biol., 2011, 7(12), 885–887, DOI:10.1038/NCHEMBIO.687.
- C. He, Grand challenge commentary: RNA epigenetics?, Nat. Chem. Biol., 2010, 6(12), 863–865, DOI:10.1038/nchembio.482.
- T. M. Frayling, N. J. Timpson, M. N. Weedon, E. Zeggini, R. M. Freathy, C. M. Lindgren, J. R. Perry, K. S. Elliott, H. Lango, N. W. Rayner, B. Shields, L. W. Harries, J. C. Barrett, S. Ellard, C. J. Groves, B. Knight, A. M. Patch, A. R. Ness, S. Ebrahim, D. A. Lawlor, S. M. Ring, Y. Ben-Shlomo, M. R. Jarvelin, U. Sovio, A. J. Bennett, D. Melzer, L. Ferrucci, R. J. Loos, I. Barroso, N. J. Wareham, F. Karpe, K. R. Owen, L. R. Cardon, M. Walker, G. A. Hitman, C. N. Palmer, A. S. Doney, A. D. Morris, G. D. Smith, A. T. Hattersley and M. I. McCarthy, A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity, Science, 2007, 316(5826), 889–894, DOI:10.1126/science.1141634.
- Y. Li, R. Su, X. Deng, Y. Chen and J. Chen, FTO in cancer: functions, molecular mechanisms, and therapeutic implications, Trends Cancer, 2022, 8(7), 598–614, DOI:10.1016/j.trecan.2022.02.010.
- D. Xu, W. Shao, Y. Jiang, X. Wang, Y. Liu and X. Liu, FTO expression is associated with the occurrence of gastric cancer and prognosis, Oncol. Rep., 2017, 38(4), 2285–2292, DOI:10.3892/or.2017.5904.
- Q. Sun, H. Geng, M. Zhao, Y. Li, X. Chen, Q. Sha, P. Lai, D. Tang, D. Yang, J. Liang and M. Guo, FTO-mediated m(6)A modification of SOCS1 mRNA promotes the progression of diabetic kidney disease, Clin. Transl. Med., 2022, 12(6), e942, DOI:10.1002/ctm2.942.
- J. Yang, M. Zhang, D. Yang, Y. Ma, Y. Tang, M. Xing, L. Li, L. Chen, Y. Jin and C. Ma, m6A-mediated upregulation of AC008 promotes osteoarthritis progression through the miR-328-3p-AQP1/ANKH axis, Exp. Mol. Med., 2021, 53(11), 1723–1734, DOI:10.1038/s12276-021-00696-7.
- F. Camera, I. Romero-Camarero, B. H. Revell, F. Amaral, O. J. Sinclair, F. Simeoni, D. H. Wiseman, L. Stojic and T. Somervaille, Differentiation block in acute myeloid leukemia regulated by intronic sequences of FTO, Iscience, 2023, 26(8), 107319, DOI:10.1016/j.isci.2023.107319.
- Q. Wang, L. Zhang, Z. Su, W. Li, Y. Jia and J. Zhang, Serum exosomal m6A demethylase FTO promotes gefitinib resistance in non-small cell lung cancer by up-regulating FLRT3, PTGIS and SIRPalpha expression, Pulm. Pharmacol. Ther., 2023, 82, 102227, DOI:10.1016/j.pupt.2023.102227.
- D. Y. Ruan, T. Li, Y. N. Wang, Q. Meng, Y. Li, K. Yu, M. Wang, J. F. Lin, L. Z. Luo, D. S. Wang, J. Z. Lin, L. Bai, Z. X. Liu, Q. Zhao, X. Y. Wu, H. Q. Ju and R. H. Xu, FTO downregulation mediated by hypoxia facilitates colorectal cancer metastasis, Oncogene, 2021, 40(33), 5168–5181, DOI:10.1038/s41388-021-01916-0.
- S. Keelan, S. Charmsaz, S. Purcell, D. Vareslija, S. Cocchiglia, F. Bane, A. Hill and L. Young, M6A demethylase FTO a potential target in brain metastatic breast cancer, Br. J. Surg., 2021, 108 DOI:10.1093/bjs/znab117.028.
- M. J. Mittenbuhler, K. Saedler, H. Nolte, L. Kern, J. Zhou, S. B. Qian, L. Meder, R. T. Ullrich, J. C. Bruning and F. T. Wunderlich, Hepatic FTO is dispensable for the regulation of metabolism but counteracts HCC development in vivo, Mol. Metab., 2020, 42, 101085, DOI:10.1016/j.molmet.2020.101085.
- X. Yang, F. Shao, D. Guo, W. Wang, J. Wang, R. Zhu, Y. Gao, J. He and Z. Lu, WNT/beta-catenin-suppressed FTO expression increases m(6)A of c-Myc mRNA to promote tumor cell glycolysis and tumorigenesis, Cell Death Dis., 2021, 12(5), 462, DOI:10.1038/s41419-021-03739-z.
- R. Chang, Z. Huang, S. Zhao, J. Zou, Y. Li and S. Tan, Emerging roles of FTO in neuropsychiatric disorders, BioMed Res. Int., 2022, 2022, 2677312, DOI:10.1155/2022/2677312.
- J. Wei, F. Liu, Z. Lu, Q. Fei, Y. Ai, P. C. He, H. Shi, X. Cui, R. Su, A. Klungland, G. Jia, J. Chen and C. He, Differential m6A, m6Am, and m1A demethylation mediated by FTO in the cell nucleus and cytoplasm, Mol. Cell, 2018, 71(6), 973–985, DOI:10.1016/j.molcel.2018.08.011.
- X. Han, Y. Li, Z. Y. Wang, L. Z. Liu, J. G. Qiu, B. J. Liu and C. Y. Zhang, Label-free and sensitive detection of RNA demethylase FTO with primer generation rolling circle amplification, Chem. Commun., 2022, 58(10), 1565–1568, 10.1039/d1cc06493b.
- X. Cui, Y. Zhou, Y. Zheng, L. Cao, L. Gao, J. Duan, H. Yin and S. Ai, Investigation of the enhanced photoactivity of CdS/Bi2MoO6/MoSe2 and its application in antibody-free enzyme-assisted photoelectrochemical strategy for detection of N6-methyladenosine and FTO protein, Mater. Today Nano, 2022, 20 DOI:10.1016/j.mtnano.2022.100269.
- Z. Y. Wang, D. L. Li, X. Tian, Y. Li and C. Y. Zhang, Single-molecule counting of FTO in human breast tissues based on a rolling circle transcription amplification-driven clustered regularly interspaced short palindromic repeat horizontal line Cas12a, Anal. Chem., 2022, 94(32), 11425–11432, DOI:10.1021/acs.analchem.2c02578.
- C. Kalinke, A. P. Zanicoski-Moscardi, P. R. de Oliveira, A. S. Mangrich, L. H. Marcolino-Junior and M. F. Bergamini, Simple and low-cost sensor based on activated biochar for the stripping voltammetric detection of caffeic acid, Microchem. J., 2020, 159 DOI:10.1016/j.microc.2020.105380.
- M. David, M. Florescu and C. Bala, Biosensors for antioxidants detection: trends and perspectives, Biosensors, 2020, 10(9) DOI:10.3390/bios10090112.
- A. V. Bounegru and C. Apetrei, Voltamperometric sensors and biosensors based on carbon nanomaterials used for detecting caffeic acid-a review, Int. J. Mol. Sci., 2020, 21(23) DOI:10.3390/ijms21239275.
- P. C. Pwavodi, V. H. Ozyurt, S. Asir and M. Ozsoz, Electrochemical sensor for determination of various phenolic compounds in wine samples using Fe3O4 nanoparticles modified carbon paste electrode, Micromachines, 2021, 12(3) DOI:10.3390/mi12030312.
- C. Sainz-Urruela, S. Vera-Lopez, A. M. San and A. M. Diez-Pascual, Graphene-based sensors for the detection of bioactive compounds: a review, Int. J. Mol. Sci., 2021, 22(7) DOI:10.3390/ijms22073316.
- M. Qi, D. Lv, Y. Zhang, D. Wang, X. Chen, Z. Zhu, Z. Hong, Y. Chai, H. Zhang and Y. Cao, Development of a surface plasmon resonance biosensor for accurate and sensitive quantitation of small molecules in blood samples, J. Pharm. Anal., 2022, 12(6), 929–936, DOI:10.1016/j.jpha.2022.06.003.
- G. Feng, Y. Yang, J. Zeng, J. Zhu, J. Liu, L. Wu, Z. Yang, G. Yang, Q. Mei, Q. Chen and F. Ran, Highly sensitive electrochemical determination of rutin based on the synergistic effect of 3D porous carbon and cobalt tungstate nanosheets, J. Pharm. Anal., 2022, 12(3), 453–459, DOI:10.1016/j.jpha.2021.09.007.
- L. Qi, J. Zhang, Y. Gao, P. Gong, C. Liang, Y. Su, Q. Zeng and Y. Zhang, Peptide-RNA complexation-induced fluorescence "turn on" displacement assay for the recognition of small ligands targeting HIV-1 RNA, J. Pharm. Anal., 2022, 12(6), 923–928, DOI:10.1016/j.jpha.2022.07.003.
- L. Cao, Y. Zhou, L. Gao, Y. Zheng, X. Cui, H. Yin, S. Wang, M. Zhang, H. Zhang and S. Ai, Photoelectrochemical biosensor for DNA demethylase detection based on enzymatically induced double-stranded DNA digestion by endonuclease-exonuclease system and Bi4O5Br2-Au/CdS photoactive material, Talanta, 2023, 262, 124670, DOI:10.1016/j.talanta.2023.124670.
- D. Chen, Y. Wu, R. D. Tilley and J. J. Gooding, Rapid and ultrasensitive electrochemical detection of DNA methylation for ovarian cancer diagnosis, Biosens. Bioelectron., 2022, 206, 114126, DOI:10.1016/j.bios.2022.114126.
- Y. Zhou, H. Yin, W. Zhao and S. Ai, Electrochemical, electrochemiluminescent and photoelectrochemical bioanalysis of epigenetic modifiers: a comprehensive review, Coord. Chem. Rev., 2020, 424, 213519, DOI:10.1016/j.ccr.2020.213519.
- H. Yang, Y. Wang, J. Tang, F. Wang and Z. Chen, End-labeling-based electrochemical strategy for detection of adenine methylation in nucleic acid by differential pulse voltammetry, Microchim. Acta, 2021, 188(8), 250, DOI:10.1007/s00604-021-04898-8.
- T. Dai, Q. Pu, Y. Guo, C. Zuo, S. Bai, Y. Yang, D. Yin, Y. Li, S. Sheng, Y. Tao, J. Fang, W. Yu and G. Xie, Analogous modified DNA probe and immune competition method-based electrochemical biosensor for RNA modification, Biosens. Bioelectron., 2018, 114, 72–77, DOI:10.1016/j.bios.2018.05.018.
- X. Yang, S. Qiao, W. Zhao, S. Li, Y. Qiao, Y. Jiang, Y. Zhou and Y. Li, Homogeneous Electrochemiluminescence for highly sensitive determination of demethylase FTO based on target-regulated DNAzyme cleavage and host-guest interaction, Anal. Chem., 2023, 95(30), 11420–11428, DOI:10.1021/acs.analchem.3c01661.
- H. Gao, X. Song, Q. Chen, R. Yuan and Y. Xiang, Target-promoted specific activation of m6A-DNAzyme for SPEXPAR-amplified and highly sensitive non-label electrochemical assay of FTO demethylase, Anal. Chim. Acta, 2023, 1247, 340902, DOI:10.1016/j.aca.2023.340902.
- Y. Zhang, J. Zhang, K. P. Hoeflich, M. Ikura, G. Qing and M. Inouye, MazF cleaves cellular mRNAs specifically at ACA to block protein synthesis in Escherichia coli, Mol. Cell, 2003, 12(4), 913–923, DOI:10.1016/s1097-2765(03)00402-7.
- X. Weng, J. Gong, Y. Chen, T. Wu, F. Wang, S. Yang, Y. Yuan, G. Luo, K. Chen, L. Hu, H. Ma, P. Wang, Q. C. Zhang, X. Zhou and C. He, Keth-seq for transcriptome-wide RNA structure mapping, Nat. Chem. Biol., 2020, 16(5), 489–492, DOI:10.1038/s41589-019-0459-3.
- T. J. Desai, B. Habulihaz, J. R. Cannon, A. Chandramohan, H. Kaan, A. Sadruddin, T. Y. Yuen, C. Johannes, D. Thean, C. J. Brown, D. P. Lane, A. W. Partridge, R. Evers, T. K. Sawyer and J. Hochman, Liposome click membrane permeability assay for identifying permeable peptides, Pharm. Res., 2021, 38(5), 843–850, DOI:10.1007/s11095-021-03005-z.
- J. Tang, G. Zou, C. Chen, J. Ren, F. Wang and Z. Chen, Highly Selective electrochemical detection of 5-formyluracil relying on (2-benzimidazolyl) acetonitrile labeling, Anal. Chem., 2021, 93(49), 16439–16446, DOI:10.1021/acs.analchem.1c03389.
- S. Lim, H. Y. Yoon, S. J. Park, S. Song, M. K. Shim, S. Yang, S. W. Kang, D. K. Lim, B. S. Kim, S. H. Moon and K. Kim, Predicting in vivo therapeutic efficacy of bioorthogonally labeled endothelial progenitor cells in hind limb ischemia models via non-invasive fluorescence molecular tomography, Biomaterials, 2021, 266, 120472, DOI:10.1016/j.biomaterials.2020.120472.
- Y. R. Chen, Y. Chai, L. Xu, N. Wang, X. Li and L. Zhang, Amperometric bydrogen peroxide biosensor based on the immobilization of horseradish peroxidase (HRP) on the layer-by-layer assembly films of gold colloidal nanoparticles and toluidine blue, Electroanalysis, 2006, 18, 471–477, DOI:10.1002/ELAN.200503424.
- Y. Xiao, H. Ju and H. Chen, Hydrogen peroxide sensor based on horseradish peroxidase-labeled Au colloids immobilized on gold electrode surface by cysteamine monolayer, Anal. Chim. Acta, 1999, 391, 73–82, DOI:10.1016/S0003-2670(99)00196-8.
- Y. Huang, R. Su, Y. Sheng, L. Dong, Z. Dong, H. Xu, T. Ni, Z. S. Zhang, T. Zhang, C. Li, L. Han, Z. Zhu, F. Lian, J. Wei, Q. Deng, Y. Wang, M. Wunderlich, Z. Gao, G. Pan, D. Zhong, H. Zhou, N. Zhang, J. Gan, H. Jiang, J. C. Mulloy, Z. Qian, J. Chen and C. G. Yang, Small-Molecule Targeting of Oncogenic FTO Demethylase in Acute Myeloid Leukemia, Cancer Cell, 2019, 35(4), 677–691, DOI:10.1016/j.ccell.2019.03.006.
- L. Zhao, T. Fan, Y. Han, Y. Wang, Y. Jiang and F. Liu, Demethylase FTO activity analysis based on methyl sensitive enzyme MazF and hybridization chain reaction, Sensor. Actuator. B Chem., 2021, 341, 129983, DOI:10.1016/j.snb.2021.129983.
- B. Ge, Z. Li, L. Yang, R. Wang and J. Chang, Characterization of the interaction of FTO protein with thioglycolic acid capped CdTe quantum dots and its analytical application, Spectrochim. Acta, Part A, 2015, 149, 667–673, DOI:10.1016/j.saa.2015.04.106.
- Z. Y. Wang, D. L. Li, X. Tian, Y. Li and C. Y. Zhang, Single-Molecule Counting of FTO in Human Breast Tissues Based on a Rolling Circle Transcription Amplification-Driven Clustered Regularly Interspaced Short Palindromic Repeat-Cas12a, Anal. Chem., 2022, 94(32), 11425–11432, DOI:10.1021/acs.analchem.2c02578.
Footnote |
| † Electronic supplementary information (ESI) available: The current variation values with different FTO concentrations are shown in Fig. S1. The DPV of five proteins (UDG, IgG, IgM, TET, Ab) are shown in Fig. S2. See DOI: https://doi.org/10.1039/d4ra03989k |
| This journal is © The Royal Society of Chemistry 2024 |