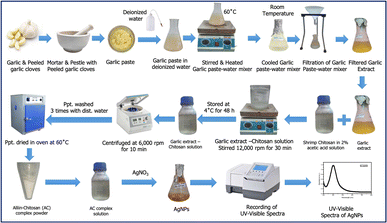
Open Access Article
Open Access ArticleUltrasensitive and selective colorimetric and smartphone-based detection of arsenic ions in aqueous solution using alliin–chitosan–AgNPs
Rintumoni Pawabc,
Ankur K. Guha d and
Chandan Tamuly
d and
Chandan Tamuly *ab
*ab
aNatural Product Chemistry Section, CSIR-North East Institute of Science and Technology, Itanagar, Arunachal Pradesh 791110, India. E-mail: c.tamuly@gmail.com
bAcademy of Scientific and Innovative Research (AcSIR), Ghaziabad 201002, India
cDept of Chemistry, Silapathar Science College, Silapathar, Assam 787059, India
dDept of Chemistry, Cotton University, Guwahati, Assam 781001, India
First published on 18th July 2024
Abstract
In this study, we developed a highly selective and sensitive colorimetric sensor for arsenic [As(III)] detection using alliin–chitosan-stabilized silver nanoparticles (AC–AgNPs). The AC–AgNPs were synthesized using a complex prepared by mixing aqueous garlic extract containing alliin and chitosan extracted from shrimp. The synthesis of AC–AgNPs was confirmed by UV-vis spectroscopy, which showed a surface plasmon resonance (SPR) band at 403 nm, and TEM analysis revealing spherical nanoparticles with a mean diameter of 7.57 ± 3.52 nm. Upon the addition of As3+ ions, the brownish-coloured solution of AC–AgNPs became colourless. Moreover, the computational study revealed that among all the metal ions, only As3+ was able to form a stable complex with AC–AgNPs, with a binding energy of 8.7 kcal mol−1. The sensor exhibited a linear response to As(III) concentrations ranging from 0.02 to 1.4 fM, with a detection limit of 0.023 fM. The highest activity was observed at pH 7 and temperature 25 °C. Interference studies demonstrated high selectivity against common metal ions. The study also demonstrated that the concentration of As3+ ions can be estimated by the decrease in red intensity and increase in green intensity in smartphone optical transduction signals. These results indicate the potential of the AC–AgNP-based sensor for reliable and efficient arsenic detection in environmental monitoring.
Introduction
The long-standing global issue of arsenic toxicity from contaminated water and food materials is a cause for serious concern, as it can increase the risk of cancer by altering DNA repair, DNA methylation, oxidative stress and genotoxicity. Among the four forms of arsenic, namely, metalloid (As0), arsenite (As3+), arsenate (As5+) and arsenic gas (AsH3), arsenite is considered the most toxic because it reacts with thiol and sulfhydryl groups present in proteins and enzymes, thereby disrupting their normal functioning.1Approximately 140 million individuals across more than 70 nations are consuming water with arsenic concentrations surpassing the provisional guideline set by the World Health Organization, which is 10 μg L−1.2,3 Accurate rapid detection of toxic arsenic ions is therefore indispensable to avoid toxicity. In addition, detecting arsenic at trace levels presents a significant scientific and technological challenge due to its chemical properties and the complex matrices in which it is found. Advancing detection technologies for arsenic can lead to improvements in analytical methods for other contaminants as well. Analytical methods such as hydride generation atomic fluorescence spectrometry, inductively coupled plasma atomic emission spectroscopy (ICP-AES), ICP mass spectrometry (ICP-MS), hydride generation atomic absorption spectroscopy, graphite furnace AAS and fluorescence spectrometry are highly sensitive and precise for the determination of arsenic ions.4 However, these typical analytical techniques are expensive, time-consuming and unsuitable for onsite analysis.5,6
Various nanomaterial based onsite detection techniques are explored in recent studies to detect analytes. The development of wearable sensors for real-time monitoring of biomarkers has also been explored in recent studies. Highly capable Al-doped NiO electrode has been developed for analyte like alcohol in sweat.7 Likewise, graphene nanoribbon/Co3O4-modified electrode has been explored for detection of H2O2, ZnO/NiO nanocomposite as a working electrode for sensing p-cresol, MnO2/rGO nanocomposites for the detection of the pollutant para-nitrophenol.8–10 Different biomaterial-based nanomaterials have also been explored in different studies. For example, cotton based colorimetric sensing has been explores for detection of ketones, gallic acid–AuNP@Tollens' complex as a highly sensitive plasmonic nanosensor for colorimetric and smartphone-based detection of formaldehyde and benzaldehyde in preserved food products, alliin–AgNPs for colorimetric detection of Hg2+ and Sn2+ ions in water.11–13
The detection of heavy metal ions in wastewater is a critical environmental challenge. Various techniques have been developed to address this issue, including electrochemical sensors, colorimetric sensors, and nanomaterial-based adsorbents. Recent advances have highlighted the efficiency of nanomaterials in enhancing the sensitivity and selectivity of these detection methods. Heavy metal detection technologies have followed the trend of autonomous, intelligent, comprehensive and accurate quick detection in recent decades. Spectroscopic detection, optical techniques and electrochemical methods are some of the most common techniques of heavy metal detection.14
Recent advancements in arsenic detection have centred around innovative colorimetric and nanomaterial-based methods. Researchers have developed several advanced methods including optical colorimetric platforms leveraging the redox chemistry of arsenic, biomolecule-modified nanomaterials, and arsenic-binding ligand-modified nanomaterials. These methods aim to enhance sensitivity, selectivity, and ease of use for on-site detection. Notable developments include enzyme-based sensors using gold nanoparticles for colorimetric detection and nanozymes that mimic enzymatic activity. Likewise, polymer hydrogel-based colorimetric strip sensors have shown promise, offering a simple and cost-effective solution for arsenic. Additionally, enzyme-inhibitory biosensors using gold nanoparticles and laccase-based systems have provided new avenues for arsenic detection in environmental samples. These advancements highlight the potential of integrating nanotechnology with traditional colorimetric approaches to create more efficient and reliable arsenic detection methods.15–17
Colorimetric nanosensors and biomolecule-modified nanomaterials represent significant advancements in arsenic detection, providing tools that are both sensitive and user-friendly. Colorimetric nanosensors, such as the paper-based sensor developed by Chanda's group, leverage gold nanoparticles (AuNPs) functionalized with ligands like thioguanine (TG) and meso-2,3-dimercaptosuccinic acid (DMSA) to visually indicate arsenic contamination in water through a color change. This simplicity and rapid response make them ideal for on-site applications, although there is a need for further development to enhance their specificity and selectivity in complex environmental samples.18
Similarly, biomolecule-modified nanomaterials use enzymes, aptamers, or peptides to achieve high specificity and sensitivity in arsenic detection. These nanosensors can translate the inhibition of enzyme activity by arsenic into measurable optical or electrochemical signals, offering rapid, on-site analysis capabilities. Despite their potential, these materials require extensive screening to validate their reliability, stability, and performance in various environmental conditions. Further exploration and development in this field are crucial to address existing challenges, such as cost, time efficiency, and stability in harsh environments. Advancing these technologies will ensure they are not only scientifically robust but also practically viable for real-world applications.17
Nanoscale materials have emerged as novel sensors due to their high surface area-to-volume ratio, which allows for a greater number of chemical and physical interactions to occur on the surface, as well as their unique chemical and physical properties, such as surface plasmon resonance (SPR). SPR is a collective oscillation of electrons at the interface of metallic structures that are produced through the electromagnetic interaction of the metal with incident light of a specific wavelength. In the case of nanoparticles, this process is generally referred to as localized surface plasmon resonance (LSPR) since the oscillation is localized in the region. LSPR occurs when the electron oscillations are in phase with the incident light frequency, resulting in an enhancement of the local electromagnetic field and a sharp spectral response (scattering and absorption). Moreover, for nanoparticles smaller than 15 nm, the spectral response is dominated by absorption, while for nanoparticles larger than 15 nm, the spectral response is dominated by scattering. Furthermore, the LSPR is different for different metals and depends on the shape, size, and dielectric environment of the surrounding medium.19 These variations can be used to develop a metal nanosensor using colorimetric and smartphone-based analysis. The search for such bioinorganic nanoscale sensors based on SPR is rapidly growing due to their ability to instantly detect toxic ions.20 These nanosensors rely on analyte-induced aggregation, which results in a visible colour change. Several advantageous nanosensors have been developed for the detection of As3+ ions, such as oligonucleotides (aptamers), glutathione, dithiothreitol and cysteine-functionalized AuNPs, citrate–AuNPs and polyethylene glycol-functionalized AgNPs.18,21–24 Likewise, smartphone-based methods based on the surface plasmon resonance of nanoparticles have also been used for the detection of As3+ ions.25
Noble metals such as gold and silver have an abundance of free electrons, resulting in strong surface plasmon resonance effects. Among these metals, silver (Ag) is the best material for plasmonics due to its low optical losses in the visible and near-infrared spectra and its resonance wavelength is situated in the visible and near-infrared regions, allowing colour changes to be detected by human eyes.26 Silver nanoparticles are generally nontoxic, making them safe. This makes them ideal for optical sensing applications. Despite these advantages and the high sensitivity of many AgNPs, the use of AgNPs for the detection of arsenic is less explored extensively.18,21–24,27
Similarly, biomaterials such as garlic and chitosan for the detection of As3+ ions are unexplored. Garlic cloves are known to contain a plethora of molecules, such as allicin, alliin, ajoene, diallyl sulfide, diallyl disulfide and vinyldithiins. Some of these molecules are known to possess antimicrobial and antioxidant properties, which can help protect the sensor from damage.28 Chitin is the second most abundant biopolymer on earth after cellulose and is found in the cell walls of some microorganisms and the exoskeletons of certain invertebrates. It is a homopolysaccharide derived from N-acetyl-D-glucosamine and is linked by β-1,4-glycosidic bonds. In most invertebrates, including crustaceans and insects, chitin is found in a partially deacetylated form, known as chitosan.29
The physicochemical and sensing properties of selected materials for arsenic detection are critical for ensuring effective and sensitive monitoring. Gold nanoparticles (AuNPs), functionalized with various ligands like thioguanine (TG) and meso-2,3-dimercaptosuccinic acid (DMSA), exhibit strong surface plasmon resonance, which is pivotal for colorimetric detection due to their visual colour change in the presence of arsenic ions. Silver nanoparticles (AgNPs) functionalized with multi-ligand systems such as asparagine, DTT, and GSH also show significant promise due to their excellent sensitivity and lower cost compared to gold. Another effective material is cerium oxide (CeO2) nanoparticles, which are notable for their stability and enzyme-mimicking catalytic properties that enhance their sensitivity in colorimetric assays. Copper nanoparticles (CuNPs), particularly when functionalized with organic molecules like ranolazine, provide an economical and stable option for arsenic detection, demonstrated by their colorimetric response and low detection limits. Additionally, calix[4]pyrrole tetrahydrazide (MCPTH) functionalized AuNPs and ferrihydrite-coated silica gel with silver nanoplates are utilized for both colorimetric and electrochemical sensing, leveraging their high surface area and reactivity to detect arsenic at low concentrations. Each of these materials brings unique advantages in terms of sensitivity, specificity, and cost-effectiveness, though ongoing research and development are essential to overcome challenges related to stability and reproducibility in complex environmental samples.17,30–32
Therefore, to develop a nanosensor for the detection of As3+ ions in aqueous solution considering matrices such as selectivity, accuracy, lower detection limit and an eco-friendly synthesis approach, the present study was designed to exploit the advantages of Ag-based nanoparticles and important biomaterials derived from garlic and chitosan. To minimize the cost, technical difficulties and ease of availability, colorimetric and smartphone-based methods based on surface plasmon resonance were chosen to develop the nanosensor. The synthesized nanoparticles were characterized using UV-visible spectroscopy, X-ray photon spectroscopy, X-ray diffraction (XRD), high-performance liquid chromatography, Fourier transform infrared spectroscopy, Raman spectroscopy and scanning electron microscopy. The theoretical structure of the nanoparticles and their interactions with As3+ ions were simulated using density functional theory (DFT) and the chemosensor was successfully applied for the determination of As3+ ions in groundwater samples.
Materials and methods
Materials
Garlic was procured from the Naharlagun market (27.1030°N, 93.7008°E, 155 m asl), Arunachal Pradesh, India. Analytical grade alliin (standard) and shrimp chitosan, acetonitrile, phosphoric acid, acetic acid, AsI3 (∼99%), AgNO3 (99.99%), BaCl2·2H2O (99%), CaCO3 (99%), CoCl2, CrCl2 (99.9%), CuSO4 (99.99), FeCl3 (96%), FeSO4·7H2O (≥99.0%), KI, SnCl2, NiCl2 (98%), PdCl2 (99%), Pb(NO3)2, ZnSO4·7H2O (99.99%) and Zn3(PO4)2 were purchased from Sigma-Aldrich.Instrumentation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (80
water (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) with 0.2% phosphoric acid. The mobile phase was sonicated for 15 min and degassed and the column temperature was maintained at 30 °C.
20) with 0.2% phosphoric acid. The mobile phase was sonicated for 15 min and degassed and the column temperature was maintained at 30 °C.The morphology of the nanoparticles was observed using a Sigma 300 VP Field Effect Scanning Electron Microscope (FE-SEM) (ZEISS, Germany) with a resolution of 1.2 nm at 15 kV. The elemental composition, empirical formula, chemical and electronic state of the elements within the nanoparticles were analysed using a ESCALAB Xi+ XPS (Thermo Fisher Scientific Inc., USA).
Techniques and procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes, after which it was left at 4 °C for 48 h. The solution was then centrifuged at 6000 rpm for 10 min. The precipitate was washed three times with distilled water and dried in an oven at 60 °C for 2 days, resulting in a white powder of 304.10 mg.33
000 rpm for 30 minutes, after which it was left at 4 °C for 48 h. The solution was then centrifuged at 6000 rpm for 10 min. The precipitate was washed three times with distilled water and dried in an oven at 60 °C for 2 days, resulting in a white powder of 304.10 mg.33Standard solutions of each metal ion were prepared in deionized water at a concentration of 1 mM. A solution of 5 mL of diluted AC–AgNPs in water was supplemented with 0.2 mL of 1 mM salt solution of each metal ion solution. The mixture was shaked using vortex for 2 minutes at room temperature to allow for interaction between the nanoparticles and the metal ions.
Changes in colour of the solution was recorded visually and the absorbance spectra of the mixtures were recorded using a UV-vis spectrophotometer across the wavelength range of 300–800 nm. The specific absorbance peak corresponding to the surface plasmon resonance (SPR) band of AC–AgNPs was monitored to observe any shifts or intensity changes indicative of metal ion interaction.
| Relative activity (%) = (M1/M2) × 100 | (1) |
For optimization of pH, the absorbance of 3 mL diluted AC–AgNPs were recorded at pH 2, 6, 7, 8 and 10 at room temperature (25 °C). For optimization of temperature, the absorbance of 3 mL diluted AC–AgNPs were recorded at 25 °C, 45 °C, 65 °C and 85 °C maintaining pH 7.
| Y = a + bX, | (2) |
The LoD and LoQ were calculated based on the standard deviation of the response and the slope using the following equations (eqn (3) and (4)):25,37
| LoD = 3.3 × SD/S | (3) |
| LoQ = 10 × SD/S | (4) |
| [At] = [A0] − k0t | (5) |
| Ln([At]/[A0]) = −k1t | (6) |
| 1/[At] = k2t + 1/[A0] | (7) |
Practical application and interference study
To assess the reliability of the developed method, the presence of As3+ ions in three ground water samples was tested. The samples were collected from different locations in the Dhemaji (Silapathar, 26°12′21.49′′N, 93°48′34.24′′E and Sripani, 27°34′4′′N 94°38′24′′E) and Lakhimpur (Boginadi, 27°36′08′′N, 94°18′26′′E) districts of Assam, India.A solution of 3 mL of diluted AC–AgNPs in water was supplemented with 0.2 mL of the sample water. The mixture was shaked using vortex for 2 minutes at room temperature to allow for interaction between the nanoparticles and the metal ions. The absorbance spectra of the mixtures were recorded using a UV-vis spectrophotometer across the wavelength range of 300–800 nm. The quantity of As3+ in the samples was estimated using the standard linear regression model derived based on the absorbance maxima and the concentration of As3+ solution in LoD calculation.
To study the interference of common anions and cations, a solution of 3 mL of diluted AC–AgNPs in water was supplemented with 0.2 mL 1 mM AsI3. To this solution 0.2 mL 1 mM salt solution of BaCl2, CaCO3, CoCl2, CrCl2, CuSO4, FeCl3, FeSO4, KI, SnCl2, NiCl2, PdCl2, Pb(NO3)2, ZnSO4 and Zn3(PO4)2 were added individually. The mixture was shaked using vortex for 2 minutes at room temperature to allow for interaction between the nanoparticles and the metal ions. The absorbance spectra of the mixtures were recorded using a UV-vis spectrophotometer across the wavelength range of 300–800 nm.
Five replications of control (without adding salts other than As3+) and of the mixture with added salts were used for recording absorbance data. Analysis of variance (ANOVA) was done using Origin 9 software to test where there is any variation in the absorbance after addition of other ions or not.
Smartphone integrated detection of As3+
A smartphone-based red-blue-green (RBG) colour analysis was tested for the detection of As3+ in addition to a colorimetric sensing method. For this study, 10–150 nM As3+ solution in water were added to 3 mL of the diluted AC–AgNPs. The mixture was shaked using vortex for 2 minutes at room temperature to allow for interaction between the nanoparticles and the metal ions. The chromogenic change associated with the variation in the concentration of As3+ was recorded using an inbuilt smartphone camera and a colour detector application ‘Color Meter’ (Fig. 2). This application displayed the percent intensity of the primary colours: red, blue and green. A regression model was then derived from plotting the red/green intensity ratio with the change in concentration, allowing for the estimation of the concentration of As3+ in unknown samples.13,25 | ||
| Fig. 2 Schematic diagram of the As3+ ion sensing technique using red-blue-green (RBG) colour detection application of smartphone. | ||
Results and discussion
Alliin–chitosan (AC) complex and alliin–chitosan–AgNPs (AC–AgNPs)
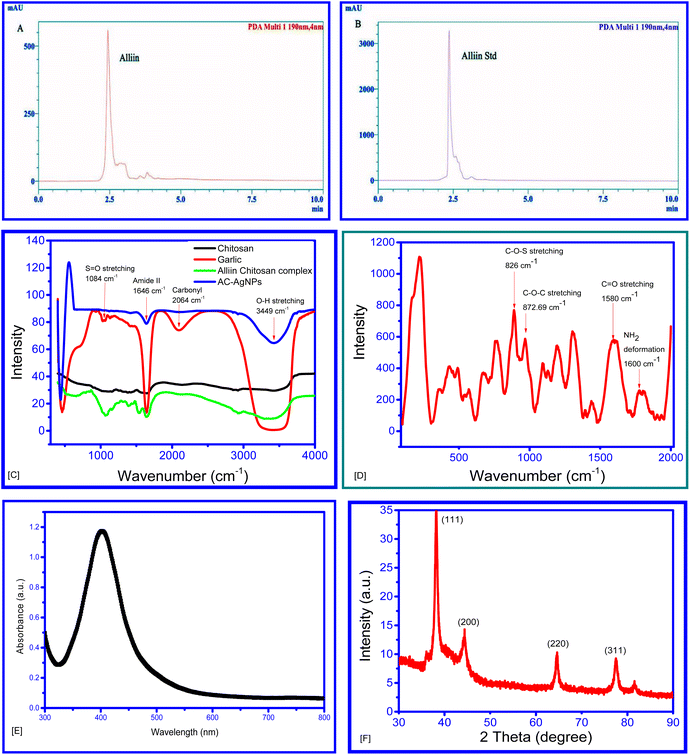
The garlic paste was immediately boiled after grinding to prevent the activity of the enzyme alliinase, which converts alliin to allicin.39,40 The HPLC chromatogram showed a peak similar to that of standard alliin. Consequently, alliin is likely to be the major phytochemical in garlic extract (Fig. 3A and B). The formation of the conjugated complex was confirmed through FTIR spectra of the garlic extract (GE), chitosan and GE–chitosan complex, hereafter called the alliin–chitosan (AC) complex (Fig. 3C). The FTIR spectrum of the complex differed from that of the aqueous extract of garlic clove, and chitosan. The presence of the absorbance peaks at 3449 (O–H stretching), 2064 (carbonyl group, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the carboxylic group), 1646 (amide II) and 1084 (S
O of the carboxylic group), 1646 (amide II) and 1084 (S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching) cm−1, similar to its precursor molecules, is indicative of the formation of the complex.41–43
O stretching) cm−1, similar to its precursor molecules, is indicative of the formation of the complex.41–43
Upon the addition of the alliin–chitosan complex, the colourless AgNO3 solution turned brown, indicating that the alliin–chitosan complex acts as a reducing agent to form AgNPs.44 The reduction of Ag+ to Ag0 and the formation of AgNPs were further validated by the UV-vis spectrum, which shows a characteristic surface plasmon resonance (SPR) band at λmax = 403 nm. This SPR band confirms the successful formation of AgNPs (Fig. 2E) (Paw et al. 2021).11,45–48
The band at 1600 cm−1 in the Raman spectrum of the AC–AgNPs is assigned to NH2 deformation (scissoring) (Fig. 2D), while the broad band at 1580 cm−1 is attributed to the asymmetric C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the carboxyl (O–C
O stretching vibrations of the carboxyl (O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) group. Moreover, the band at 872.69 cm−1 is likely due to the stretching vibration of the C–O–C of chitosan, and the band at 826 cm−1 is assigned to a C–O–S stretch.41,42
O) group. Moreover, the band at 872.69 cm−1 is likely due to the stretching vibration of the C–O–C of chitosan, and the band at 826 cm−1 is assigned to a C–O–S stretch.41,42
The amine group of chitosan is assumed to bind to the carboxy terminus of alliin, releasing a single hydroxyl ion. XPS for the N 1s scan also displays three peaks, demonstrating a shift in the bonding pattern of the N atom. Furthermore, this N atom of chitosan may be involved in binding with Ag+ during the formation of nanoparticles. Additionally, the oxygen of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of alliin may also be involved in binding with Ag+, thus leading to the formation of alliin–chitosan–silver nanoparticles (AC–AgNPs). This hypothesis is supported by the lack of an absorbance peak at 1084 cm−1 in the FTIR spectrum of the nanoparticles compared to that of the alliin–chitosan complex.
O of alliin may also be involved in binding with Ag+, thus leading to the formation of alliin–chitosan–silver nanoparticles (AC–AgNPs). This hypothesis is supported by the lack of an absorbance peak at 1084 cm−1 in the FTIR spectrum of the nanoparticles compared to that of the alliin–chitosan complex.
Fig. 4A and B shows TEM images of the AC–AgNPs, which confirmed their spherical structure. The size distribution of the AC–AgNPs varied from 7.61 to 21.46 nm, with a mean particle size of 7.57 ± 3.522 nm (Fig. 4C). AgNPs were reported to exhibit a similar size distribution depending on the synthesis method and morphology, further validating the successful synthesis of AC–AgNPs.11 The selected area diffraction (SAED) pattern of individual AC–AgNPs shows the lattice pattern structure of the AC–AgNP sample. SAED also showed a fcc crystal lattice with d-spacings of 0.29 nm, 0.17 nm, 0.16 nm and 0.11 nm, corresponding to diffraction angles of 30.2°, 50.2°, 56.7° and 83.9°, respectively (Fig. 4D). The interplanar distance from the TEM image was calculated to be 0.11 (311 plane, 2θ = 83.9), which is similar (0.11 ± 0.021 nm) to that calculated from the SAED image and XRD (0.12 nm).
The FESEM image of the AC–AgNPs showed that the nanoparticles were arranged like irregular masses of cotton (Fig. 4E and F). Energy-dispersive X-ray spectrometry (EDX) was used to determine the chemical composition of the nanoparticle surface. As shown in Fig. 4G and H the nanoparticle surface contained 12% C, 11% O and 77% Ag.
The XRD pattern of the synthesized AC–AgNPs is shown in (Fig. 3F). The diffraction peaks observed at 2θ values of 38.25°, 44.42°, 64.53°, and 77.59° correspond to the (111), (200), (220), and (311) planes of the face-centered cubic (FCC) structure of silver. This XRD data matches with standard data of the International Centre for Diffraction Data (ICDD) for Ag with PDF card no. 00-004-0783.49 The peaks are well-indexed and matched with the standard reference, confirming the crystalline nature of the silver nanoparticles.
The XPS survey scans at 369.79, 532.2, 163.2, 401 and 280 eV revealed the presence of Ag 3d, O 1s, S 2p, N 1s and C 1s (Fig. 5), respectively, with the twin peaks of Ag 3d suggesting that Ag0 was the major component (369.0 eV for Ag0 3d5/2 and 373.75 eV for Ag0 3d3/2), corresponding to the AgNPs (Fig. 5B).50,51 Moreover, the N 1s spectrum (Fig. 5E) showed three peaks at 399.65, 403.45 and 407.4 eV, suggesting the presence of amines/amides.52,53
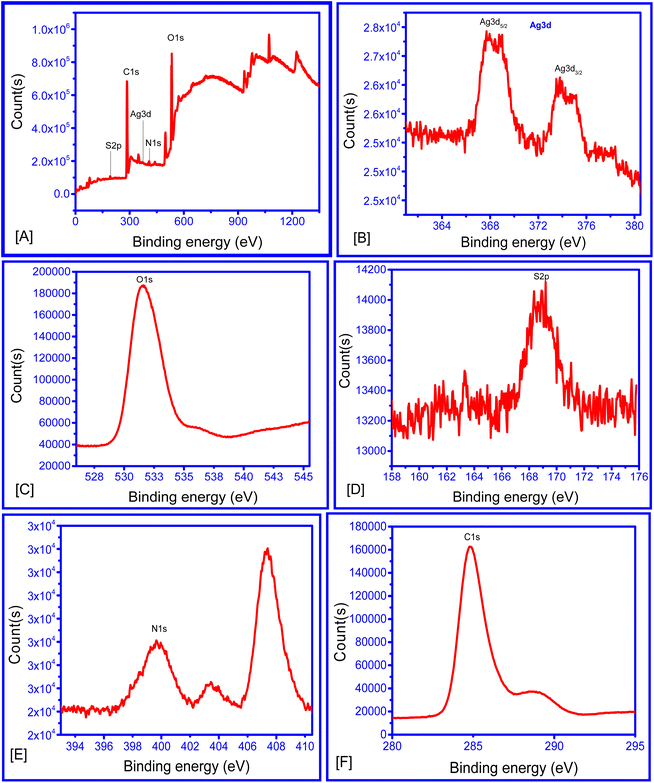 | ||
| Fig. 5 (A) XPS survey scan of AC–AgNPs, (B) XPS Ag 3d scan, (C) XPS (O 1s scan), (D) XPS S 2p scan, (E) XPS N 1s scan, (F) XPS C 1s scan. | ||
Alliin–chitosan–AgNPs (AC–AgNPs) as selective As3+ nanosensors
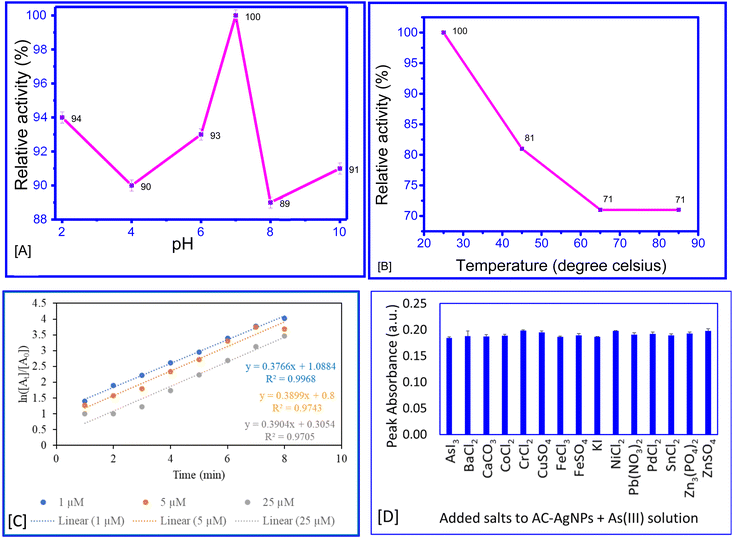
The sensitivity of AC–AgNPs was evaluated for the metal ions As3+, Sn2+, Co2+, Pb2+, Zn2+, Ni2+, Fe3+, Fe2+, Pd2+, Cu2+, Ca2+, Cr2+ and Ba2+. As3+ was found to be the only metal ion that caused a change in the colour of the AC–AgNP solution from brownish to colourless (Fig. 6A). The results showed that the presence of standard solution of the other ions did not cause apparent changes in the SPR band and colour of AC–AgNPs solution, in contrast to As3+, that turn the colour of the solution from brown to colourless.Chitosan has been widely recognized for its remarkable sorption capabilities, which are attributed to its replaceable functional groups, reusability, and environmentally friendly characteristics. The polysaccharide, derived from chitin through deacetylation, consists of 2-amino-2-deoxy-β-D-glucopyranose units linked by 1,4-glycosidic bonds. The extensive range of functional groups on chitosan, including amino, hydroxyl, and amide, contribute to its high affinity for arsenic adsorption through various mechanisms such as electrostatic interactions, ion-pair formation, ion exchange, diffusion, metal chelation, and complex formation.54 Likewise, alliin–AgNPs are known to be sensitive towards metal ions like Hg2+ and Sn2+.11 However, nanoparticle system prepared using alliin–chitosan complex, enhances the interaction with arsenic ions, leading to sensitive detection and selectivity. Thus, the inclusion of alliin provided sensitivity and a distinctive colorimetric response. This is particularly important for detecting low concentrations of arsenic in complex matrices. This unique interaction mechanism of alliin–chitosan complex and arsenic ions cannot be replicated by chitosan or alliin alone.
Thus, the synthesis and application of alliin–chitosan–AgNPs as a sensing material for arsenic detection is a novel approach. This combination leverages the unique properties of alliin and chitosan to enhance the sensitivity and selectivity of silver nanoparticles (AgNPs) towards arsenic ions. The synthesis process of alliin–chitosan–AgNPs is eco-friendly and cost-effective, utilizing readily available natural materials. This method offers a sustainable alternative to conventional sensing materials, contributing to green chemistry principles. The comprehensive characterization of the synthesized nanoparticles, combined with extensive validation through various analytical techniques, underscores the reliability and robustness of our sensor.
Possible mechanism of As3+ detection using AC–AgNPs
As indicated by HPLC, alliin is the major chemical constituent of hot aqueous garlic extract. So, it plays significant role in the synthesis of AC–AgNPs as reported in Fig. 6B. The garlic extract was mixed with chitosan to prepare the alliin–chitosan complex as the previous mention.The amine group of chitosan is assumed to bind with the hydroxyl group of the carboxy-terminal of alliin releasing one molecule of water. The amine group of chitosan is also reported to bind with the hydroxyl group of many other compounds in previous literature. XPS for the N 1s scan also shows three peaks indicating changes in the bonding pattern with the N atom. It also shows the involvement of this N atom of the chitosan in binding with Ag during the formation of the nanoparticle. In addition, the O of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of the alliin may also involve in binding with Ag forming the alliin–chitosan–silver nanoparticle (AC–AgNPs). It was indicated by the absence of an absorbance peak ∼ wavelength 1000 in FTIR spectra of the nanoparticle compared to the alliin–chitosan complex. Thus, the silver atoms may directly bind to N and O atoms of the alliin–chitosan moiety forming N⋯Ag and O⋯Ag bond. It may further form a cluster of three Ag0 ions with two alliin–chitosan moiety. After addition of As3+ ions, the solution becomes colourless Fig. 6B. This may be due to the analyte induced aggregation of AC–AgNPs in the solution.18,21–24
O of the alliin may also involve in binding with Ag forming the alliin–chitosan–silver nanoparticle (AC–AgNPs). It was indicated by the absence of an absorbance peak ∼ wavelength 1000 in FTIR spectra of the nanoparticle compared to the alliin–chitosan complex. Thus, the silver atoms may directly bind to N and O atoms of the alliin–chitosan moiety forming N⋯Ag and O⋯Ag bond. It may further form a cluster of three Ag0 ions with two alliin–chitosan moiety. After addition of As3+ ions, the solution becomes colourless Fig. 6B. This may be due to the analyte induced aggregation of AC–AgNPs in the solution.18,21–24
Computational analysis
Fig. 7 shows the computational analysis based optimized geometry of the AC–AgNPs, with Ag atoms bonded to the N and O atoms of the alliin–chitosan moiety and N⋯Ag and O⋯Ag distances of 3.45 and 3.21 Å, respectively. Moreover, the binding energy of the Ag3 cluster with the alliin–chitosan moiety was calculated to be 14.2 kcal mol−1, indicating the stable formation of Ag nanoparticles with the alliin–chitosan moiety. | ||
| Fig. 7 Optimized geometry of Ag(0) nanocluster stabilized by alliin–chitosan complex and its interactions with arsenic (As). | ||
To check the sensitivity of this AC–AgNP towards different metals, As3+, Fe3+, Fe2+, Ni2+, Pd2+, Pb2+, Na+, Zn2+, K+, Cu2+, Ca2+, Cr2+ and Ba2+ complexes with this AC–AgNP were tried to be optimized. However, except As3+, all other complexes were found to be broken after geometry optimization. This implies that only As3+ can form stable complex with AC–AgNP. The calculated binding energy of As33+ ring with AC–AgNP is 8.7 kcal mol−1. The optimized geometry of As3–AC–AgNP is shown in Fig. 2. As3 ring binds with Ag atoms of Ag3 ring and O and N atoms of the alliin–chitosan moiety as shown in Fig. 7.
Thus, this work provides detailed mechanistic insights into the interaction between alliin–chitosan–AgNPs and arsenic ions. The study elucidates the binding mechanism and the role of functional groups in the sensing process, offering valuable knowledge for the development of future sensors.
Limit of detection (LoD)
Fig. 6C shows that the absorption peak of the AC–AgNPs decreased with increasing concentrations of As3+ ions. A significant linear relationship existed between the changes in absorbance and the concentration of As3+ ions over the range of 0.02–1.40 fM at 403 nm. This regression model can be presented as Y = 0.3226 − 0.18217X, R2 = 94.23%, where Y = peak absorbance (nm) and X = concentration of arsenic (in femto molar) (Fig. 6D). The slope is significantly different from zero at the 95% significance level. The LoD and LoQ for the detection and estimation of As3+ ions were 0.021 fM and 0.063 fM (SD = 0.001155, slope = −0.18217), respectively. This LoD is lower than that of many other reported methods (Table 1). Thus, the presented sensor demonstrates significantly improved sensitivity and selectivity compared to existing methods.| Sl no. | Probe | Calibration range | LoD (references) |
|---|---|---|---|
| 1 | Inductively coupled plasma-atomic emission spectrometry with ultrasonic nebulization | 2.5–1000.8 μg L−1 | 0.8 μg L−1 (ref. 55) |
| 2 | Hydride generation with dielectric barrier discharge atomizer | 0.5–50 μg L−1 | 0.04 μg L−1 (ref. 56) |
| 3 | Citrate-AuNPs in presence of phytochelatin-like peptide | 0.04–1.2 μg L−1 | 0.02 μg L−1 (ref. 22) |
| 4 | Gold–thioguanine based nanosensor | 10–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg L−1 000 μg L−1 |
10 μg L−1 (ref. 23) |
| 5 | Glutathione–dithiothreitol–cysteine–2,6-pyridinedicarboxylic acid–AuNPs | 2–20 μg L−1 | 7 μg L−1 (ref. 57) |
| 6 | Hydride generation atomic absorption spectrometry | 0.5 to 8.0 μg L−1 | 0.07 μg L−1 (ref. 52) |
| 7 | Aptamer based AgNPs | 50–700 μg L−1 | 6 μg L−1 (ref. 53) |
| 8 | Polyethylene glycol functionalized AgNPs | 10–15 μg L−1 | 10 μg L−1 (ref. 24) |
| 9 | Cysteine-capped AgNPs, and methionine-capped AgNPs | 0.5–1000 μg L−1 | 0.5 μg L−1 (ref. 58) |
| 10 | Alliin–chitosan–AgNPs | 1.498–104.86 fg L−1 | 1.72 fg L−1 (present work) |
The highest activity of the AC–AgNPs for the detection of As3+ was recorded at pH 7 and 25 °C (Fig. 8A and B). A higher temperature may cause a breakdown of the nanoparticle, resulting in a decrease in the relative activity. Likewise, a change in pH may cause a change in the hydrogen ion concentration, influencing the activity of the nanoparticle.
A reaction kinetics study of the interaction between AC–AgNPs and As3+ revealed that the first-order model provided the best fit with the highest R2 value compared to the zero-order and second-order models (Fig. 8C). However, as the reaction involves two reactants, nanoparticles and As3+ ions, the kinetics seem to follow pseudo-first-order kinetics. This indicates that the rate is solely dependent on one specific reactant in solution.38
Practical application and interference study
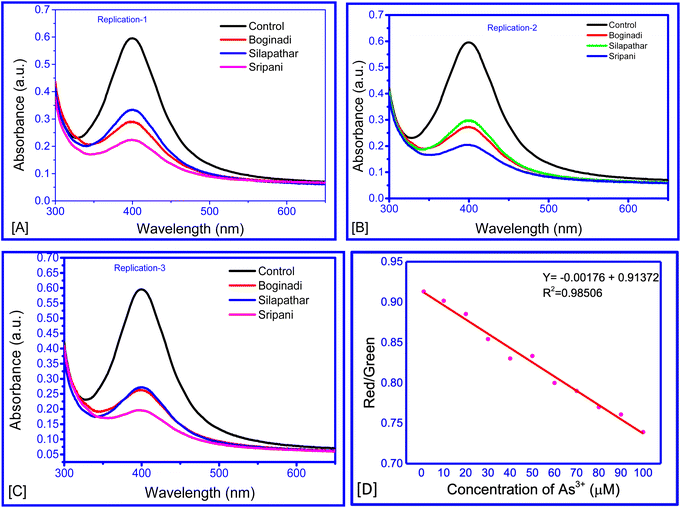
Four water samples were collected from tube wells in different locations of the Dhemaji and Lakhimpur district, Assam. India. The level of As3+ was calculated as 4.06–5.68 fM in untreated water samples (Fig. 9A–C and Table 2). These results show that the proposed method has practical applicability in the sensing of As3+ in environmental samples. Thus, this study introduces an innovative sensor based on alliin–chitosan–AgNPs for the detection of arsenic ions. The use of alliin and chitosan as stabilizing and functionalizing agents for AgNPs is novel, providing enhanced sensitivity and selectivity. This approach is not only cost-effective and eco-friendly but also offers significant improvements over existing detection methods. The mechanistic insights provided by this study further contribute to the understanding and development of advanced sensing materials.| Location of water sample source | Peak absorbance ± SD | Conc. of As3+ (femto molar) |
|---|---|---|
| Silapathar | 0.273 ± 0.014 | 0.271 ± 0.078 |
| Boginadi | 0.300 ± 0.030 | 0.126 ± 0.165 |
| Sripani | 0.206 ± 0.014 | 0.640 ± 0.079 |
Analysis of variance (ANOVA) revealed no significant difference between the absorbance of the AC–AgNP + As3+ solution and that of the AC–AgNP + As3+ solution with individually added metal ions (F = 1.962; P = 0.03694; df = 14, 60), indicating that these metal ions did not interfere significantly with the sensing of As3+ ions by the AC–AgNPs (Fig. 8D). This is likely due to the higher binding affinity of the sensor towards arsenite ions compared to other ions. The interference study demonstrates that the AC–AgNPs sensor is highly selective for As(III) ions, with minimal interference from other common anions. This high selectivity is crucial for accurate detection of As(III) in environmental water samples, ensuring reliable and precise measurements.
Smartphone-based detection of As3+ ions using AC–AgNPs
The optical transduction signals obtained using smartphones showed that with increasing concentrations of As3+ ions, the intensity percentage of the red colour decreased, whereas the percentage intensity of the green colour increased. This might be a result of the transformation to a lighter colour with increasing As3+ concentration. The ratio of red/green decreased with increasing As3+ concentration (Fig. 9D). A regression model of Y = −0.00176X + 0.91372 was derived for the range of 10–150 nM. This study demonstrated that the concentration of an analyte in any solution can be estimated from the change in the intensity of colours in terms of percentage.Conclusion
In this study, we successfully developed a highly selective and sensitive colorimetric sensor for the detection of arsenic (As(III)) ions using alliin–chitosan-stabilized silver nanoparticles (AC–AgNPs). The detection method demonstrated a significant colour change from brown to colorless upon the addition of As3+ ions, indicating the potential for visual detection. This change in color was quantitatively analyzed using a smartphone-based red-blue-green (RBG) color analysis, allowing for the estimation of As3+ concentration through a regression model derived from the red/green intensity ratio. The sensor exhibited a linear response to As(III) concentrations ranging from 0.02 to 1.4 fM with an impressive detection limit of 0.023 fM, making it suitable for detecting trace amounts of arsenic in environmental samples. Interference studies confirmed the high selectivity of the AC–AgNPs sensor against common metal ions such as Sn2+, Co2+, Pb2+, Zn2+, Ni2+, Fe3+, Fe2+, Pd2+, Cu2+, Ca2+, Cr2+, and Ba2+. Only As3+ ions caused a noticeable change in the colour of the AC–AgNPs solution, underscoring the sensor's specificity. Practical application of the developed method was demonstrated by testing ground water samples from Dhemaji and Lakhimpur districts of Assam, India. The samples were analyzed using the developed sensor, showing reliable detection and quantification of arsenic ions. These results indicate that the AC–AgNPs-based sensor is a promising tool for the reliable and efficient detection of arsenic in aqueous solutions, with potential applications in environmental monitoring and public health protection.Data availability
All the data are available in revised manuscript are original.Author contributions
RP: conceptualization, investigation, analysis, manuscript preparation. AKG: validation, software. CT: conceptualization, methodology, supervision, visualization, writing – review & editing. All read and approved the final version of the manuscript.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
The authors expressed their sincere thanks to Director, CSIR-North East Institute of Science and Technology Jorhat Assam for the continuous support and valuable suggestions provided throughout the research. The authors also thank SEED Division, DST, New Delhi (Ref No: DST/SEED/TSP/STI/2022/915) for providing financial support.References
- M. F. Hughes, Toxicol. Lett., 2002, 133, 1–16 CrossRef CAS PubMed.
- P. Ravenscroft, H. Brammer and K. Richards. Arsenic Pollution: A Global Synthesis, Wiley-Blackwell, 2009 Search PubMed.
- UNCF and WHO, Arsenic Primer: Guidance on the Investigation and Mitigation of Arsenic Contamination, United Nations Children's Fund and the World Health Organization, New York, 2018 Search PubMed.
- P. Niedzielski and M. Siepak, Pol. J. Environ. Stud., 2003, 12, 653–667 CAS.
- E. M. Jones, Arsenic 2000: An Overview of the Arsenic Issue in Bangladesh, Water Aid Bangladesh, Dhaka, Bangladesh, 2000 Search PubMed.
- S. Hu, J. Lu and C. Jing, J. Environ. Sci., 2012, 24, 1341–1346 CrossRef CAS.
- A. Singh, A. Sharma, A. K. Sundramoorthy and S. Arya, IEEE Sensor. J., 2023, 23, 22153–22160 CAS.
- P. Murugan, A. K. Sundramoorthy, R. D. Nagarajan, R. Atchudan, R. Shanmugam, D. Ganapathy, S. Arya, A. A. Alothman and M. Ouladsmane, J. Nanomater., 2022, 9866111 CAS.
- S. Dutt, A. Singh, R. Mahadeva, A. K. Sundramoorthy, V. Gupta and S. Arya, Diamond Relat. Mater., 2024, 141, 110554 CrossRef CAS.
- M. Thakur, A. Singh and A. Dubey, et al., Emergent Mater., 2024 DOI:10.1007/s42247-024-00729-7.
- R. Paw, M. Hazarika, P. K. Boruah, A. J. Kalita, A. K. Guha, M. R. Das and C. Tamuly, RSC Adv., 2021, 11, 14700–14709 RSC.
- A. Sharma, A. Singh, A. Khosla and S. Arya, J. Saudi Chem. Soc., 2021, 25, 101340 CrossRef CAS.
- N. Borah, D. Gogoi, N. C. Ghosh and C. Tamuly, Food Chem., 2023, 399, 133975 CrossRef CAS PubMed.
- A. Singh, S. S. Shah, C. Sharma, V. Gupta, A. K. Sundramoorthy, P. Kumar, S. Arya and J. Env, Chem. Eng., 2024, 12, 113032 CAS.
- T. Wang, R. D. Milton, S. Abdellaoui, D. P. Hickey and S. D. Minteer, Anal. Chem., 2016, 88, 3243–3248 CrossRef CAS.
- T. Wang, R. D. Milton, S. Abdellaoui, D. P. Hickey and S. D. Minteer, Anal. Chem., 2016, 88, 3243–3248 CrossRef CAS.
- K. H. Gebremedhin, M. H. Kahsay, N. K. Wegahita, T. Teklu, B. A. Berhe, A. G. Gebru, A. H. Tesfay and A. G. Asgedom, Discover Nano, 2024, 19(38) DOI:10.1186/s11671-024-03981-2.
- P. Nath, R. K. Arun and N. Chanda, RSC Adv., 2014, 4, 59558–59561 RSC.
- S. Kasani, K. Curtin and N. Wu, Nanophotonics, 2019, 8, 2065–2089 CrossRef CAS.
- L. L. Shen, G. R. Zhang, W. Li, M. Biesalski and B. J. M. Etzold, ACS Omega, 2017, 2, 4593–4603 CrossRef CAS.
- J. R. Kalluri, T. Arbneshi, S. A. Khan, A. Neely, P. Candice, B. Varisli, M. Washington, S. McAfee, B. Robinson, S. Banerjee, A. K. Singh, D. Senapati and P. C. Ray, Angew. Chem., 2009, 48, 9668–9671 CrossRef CAS.
- N. Xia, Y. Shi, R. Zhang, F. Zhao, F. Liu and L. Liu, Anal. Methods, 2012, 4, 3937–3941 RSC.
- S. Zhan, M. Yu, J. Lv, L. Wang and P. Zhou, Aust. J. Chem., 2014, 67, 813–818 CrossRef CAS.
- B. S. Boruah, N. K. Daimari and R. Biswas, Results Phys., 2019, 12, 2061–2065 CrossRef.
- S. Nag, A. Mondal, H. Hirani and P. Banerjee, Mater. Adv., 2022, 3, 4649–4658 RSC.
- H. Yu, Y. Peng, Y. Yang and Z.-Y. Li, npj Comput. Mater., 2019, 5(45) DOI:10.1038/s41524-019-0184-1.
- Silver Micro-Nanoparticles, ed. S. T. Galatage, A. S. Hebalkar, S. V. Dhobale, O. R. Mali, P. S. Kumbhar, S. V. Nikade, S. G. Killedar, S. Kumar, P. Kumar and C. S. Pathak, IntechOpen, 2021, pp. 1–19 Search PubMed.
- G. E.-S. Batiha, A. M. Beshbishy, L. G. Wasef, Y. H. A. Elewa, A. A. Al-Sagan, M. E. A. El-Hack, A. E. Taha, Y. M. Abd-Elhakim and H. P. Devkota, Nutrients, 2020, 12, 872 CrossRef CAS.
- A. Sharbidre, S. Sargar, H. Gogoi and R. Patil, Int. J. Trop. Insect Sci., 2021, 41, 1893–1900 CrossRef.
- N. Priyadarshni, P. Nath, Nagahanumaiah and N. Chanda, ACS Sustain. Chem. Eng., 2018, 6, 6264–6272 CrossRef CAS.
- A. Kongor, M. Panchal and M. Athar, et al., J. Inclusion Phenom. Macrocyclic Chem., 2020, 98, 29–41 CrossRef CAS.
- H. Kolya, K. Hashitsume and C. W. Kang, Toxics, 2021, 9, 143 CrossRef CAS PubMed.
- T. Pirak, A. Jangchud and P. Jantawat, Int. J. Food Sci. Technol., 2012, 47, 1339–1347 CrossRef CAS.
- A. Duru and C. E. Duru, Sci. Afr., 2020, 9, e00533 Search PubMed.
- Y. Zhao and D. G. Truhlar, Theor. Chem. Acc., 2008, 120, 215–241 Search PubMed.
- M. J. Frisch, et al., Gaussian 16, Revision A.03, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- ICH Q2, Validation of Analytical Procedures: Test and Methodology, 2005 Search PubMed.
- R. Painuli, S. Raghav and D. Kumar, ACS Omega, 2019, 4, 3635–3645 Search PubMed.
- T. Miron, T. Bercovici, A. Rabinkov, M. Wilchek and D. Mirelman, Anal. Biochem., 2004, 331, 364–369 CrossRef CAS.
- S. H. Omar and N. A. Al-Wabel, Saudi Pharm. J., 2010, 18, 51–58 CrossRef CAS.
- G. Cardenas and S. P. Miranda, J. Chil. Chem. Soc., 2004, 49, 291–295 CAS.
- V. Nikolić, M. Stanković, L. Nikolić, D. Cvetković, A. Kapor and M. Cakić, Chem. Ind. Chem. Eng. Q., 2005, 2, 69–73 CrossRef.
- T. Pirak, A. Jangchud and P. Jantawat, Int. J. Food Sci. Technol., 2012, 47, 1339–1347 CrossRef CAS.
- A. L. González, C. Noguez, J. Beránek and A. S. Barnard, J. Phys. Chem. C, 2014, 118, 9128–9136 CrossRef.
- A. A. Hebeish, M. H. El-Rafie, F. A. Abdel-Mohdy, E. S. Abdel Halim and H. E. Emam, Carbohydr. Polym., 2010, 82, 933–941 CrossRef CAS.
- I. Barwal, P. Ranjan, S. Kateriya and S. C. Yadav, J. Nanobiotechnol., 2011, 9, 56 CrossRef CAS PubMed.
- W. Yu and H. Xie, J. Nanomater., 2012, 435873 Search PubMed.
- Z. A. Ali, R. Yahya, S. D. Sekaran and R. Puteh, Adv. Mater. Sci. Eng., 2016, 4102196 Search PubMed.
- D. H. Wi, H. Yang, Y. Kim, H. Ahn, J. W. Hong and S. W. Han, J. Mater. Chem. A, 2023, 11, 1343–1350 RSC.
- S. Liang, G. Zhang, J. Min, J. Ding and X. Jiang, J. Nanomater., 2014, 684251 Search PubMed.
- S. Kumar-Krishnan, E. Prokhor dov, M. Hernández-Iturriaga, J. D. Mota-Morales, M. Vázquez-Lepe and Y. Kovalenko, et al., Eur. Polym. J., 2015, 67, 242–251 CrossRef CAS.
- C. Cerveira, D. Pozebon, D. P. de Moraes and J. C. S. de Fraga, Anal. Methods, 2015, 7, 4528–4534 RSC.
- F. Divsar, K. Habibzadeh, S. S. Shariati and M. Shahriarinour, Anal. Methods, 2015, 7, 4568–4576 RSC.
- A. Ayub and Z. A. Raza, Int. J. Biol. Macromol., 2021, 192, 1196–1216 CrossRef CAS PubMed.
- S. Karthikeyan and S. Hirata, Anal. Bioanal. Chem., 2003, 375, 139–144 CrossRef CAS.
- Z. Zhu, J. Liu, S. Zhang, X. Na and X. Zhang, Anal. Chim. Acta, 2008, 607, 136–141 CrossRef CAS PubMed.
- R. Domínguez-González, L. G. Varela and P. Bermejo-Barrera, Talanta, 2014, 118, 262–269 CrossRef PubMed.
- A. Saadati, F. Farshchi, M. Hasanzadeh, Y. Liu and F. Seidi, RSC Adv., 2022, 12, 21836–21850 RSC.
| This journal is © The Royal Society of Chemistry 2024 |