Open Access Article
Open Access ArticleFluorescence nanoprobes bearing low temperature-derived biochar nanoparticles as efficient quenchers for the detection of single-stranded DNA and 17β-estradiol and their analytical potential†
Xiaoli Qi‡
,
Hui Hu‡,
Lina Liang,
Yuqing Lin,
Yudan Liu,
Haifeng Sun and
Yunxian Piao *
*
Key Laboratory of Groundwater Resources and Environment (Jilin University), Ministry of Education, Jilin Provincial Key Laboratory of Water Resources and Environment, College of New Energy and Environment, Jilin University, Changchun 130021, China. E-mail: yxpiao@jlu.edu.cn
First published on 3rd September 2024
Abstract
Bagasse-derived biochar nanoparticles obtained under a low pyrolysis condition (400 °C) were first revealed to be capable of highly efficiently quenching the fluorescence of 6-carboxyfluorescein, with a significantly improved quenching rate constant over that of other quenchers and high-temperature prepared ones, and were designated as bagasse-derived quencher nanoparticles (BQNPs). The BQNPs are suitable for the construction of fluorescence nanoprobes, taking advantage of their various beneficial properties, including low cost, environmental friendliness, high dispersibility, and rich functional groups that allow their easy and versatile molecular modification. They were demonstrated to be capable of stably binding single-stranded oligonucleotides through both adsorption and covalent interactions and were utilized for the construction of both BQNPs/DNA and BQNPs/aptamer probes. The BQNPs/DNA probe had strong resistance against degradation by deoxyribonuclease I and showed high precision and selectivity for the detection of single-stranded DNA, with a limit of detection of 1.04 nM. Moreover, the BQNPs/aptamer probe demonstrated the rapid and sensitive detection of 17β-estradiol (E2) with a limit of detection of 0.4 ng mL−1 with no cross-reactivity with the analogues, and it was also applied for real environmental sample detection and demonstrated reasonable signal recoveries. Benefiting from their strong quenching ability, low cost, and great dispersibility, the BQNPs show great potential for the development of cost-effective and sensitive fluorescence sensors.
Introduction
Nucleic acid fluorescent probes are known to be sensitive and reliable and are widely used in biological and environmental analyses, as well as in clinical and medical diagnostics.1–3 These sensors typically use fluorescence resonance energy transfer (FRET) or quenching mechanisms to detect signals.4 They consist of a DNA probe molecule with a fluorophore and quencher molecule.5 Nowadays, various kinds of nanomaterials have been employed to serve as quenchers, including carbon nanomaterials, gold nanoparticles, metal–organic frameworks, upconversion nanoparticles, and polymer nanostructures.3,6–8 Nevertheless, it would still be worthwhile to explore novel fluorescence nanoquenchers, featuring excellent quenching ability, good biological stability, easy preparation, and low cost, for constructing high-performance and cost-effective fluorescence sensing platforms.As an emerging green and environmentally friendly carbonaceous material, biochar obtained from the pyrolysis of waste biomass has the properties of good source abundance from a wide range of sources, extremely low cost, easy synthesis, large specific surface area, porous structure, rich functional groups, and good stability. Recently, some additional promising characteristics of biochar have improved its attractiveness, such as high level sp2 carbon structure representative of a good graphite structure, great defect structure, and electron transfer ability, which have broadened its application potential to not only soil amendment and chemical adsorption but also to advanced oxidative catalytic processes, electrocatalytic reactions, and capacitor and electrode constructions.9,10 Based on its physical and chemical properties, it is likely that rationally synthesized biochar may present fluorescence-quenching capabilities, yet, to the best of our knowledge, this has been rarely examined to date. Accordingly, it would be constructive and meaningful to explore the fluorescence-quenching properties of biochar to develop low-cost and alternative fluorescence nanoquencher probes.
In the present study, bagasse-derived quencher nanoparticles (BQNPs) obtained from the pyrolysis of bagasse with a lower temperature setting (400 °C) were first demonstrated to have an outstanding fluorescence-quenching ability toward 6-carboxyl fluorescein (FAM). The quenching efficiency of the BQNPs was evaluated by the quenching rate constant, and this was also compared to other well-known carbonyl nanoquenchers. Since BQNPs are greatly dispersible, have a large surface area, and rich functional groups, they are ideally suitable for various modifications with biomolecules. As a proof of concept, the BQNPs were utilized to construct nucleic acid-based fluorescence nanoprobes and their detection performances toward single-stranded target DNA (tDNA) and 17β-Estradiol (E2) were assessed, as shown in Scheme 1. It is considered that the precise analysis of nucleic acids is significant for the diagnosis of cancer, infections, and genetic abnormalities,11 and E2 is the most toxic estrogenic compound that is frequently found in varying water bodies, which can cause serious health problems.12,13 The single-stranded oligonucleotides could be attached on the BQNPs by either adsorption or covalent interactions. The fabricated hybridization probe and aptamer probe were demonstrated to be capable of the sensitive detection of single-stranded DNA and E2 with a low detection limit and high selectivity. Hence, the BQNPs have great potential for developing low-cost quencher sensors.
Materials and methods
Chemicals and reagents
17β-Estradiol (E2, ≥98%), bisphenol A (BPA, ≥99%), Trizma hydrochloride (Tris–HCl), sodium chloride (NaCl), potassium chloride (KCl), magnesium chloride hexahydrate (MgCl2·6H2O), fluorescein, DNase I (deoxyribonuclease I), and N-(3-dimethylaminopropyl)-N′-ethylcarbodiimide hydrochloride (EDC, ≥99%) were obtained from Sigma-Aldrich (Shanghai, China). The other endocrine disrupter chemicals (EDCs), including ethinyl estradiol (EE, ≥98%), were purchased from Aladdin Industrial (Shanghai, China). Estrone (≥99%) was obtained from Bailingwei (Beijing, China). Testosterone (≥98%) was purchased from Yuanye Biology (Shanghai, China).All the DNA oligonucleotides labelled with or without 6-carboxyl fluorescein (FAM) were synthesized by Sangon Biotechnology Co. Ltd (Shanghai, China). The sequences of oligonucleotides used in this work were:
| DNA probe: 5′-(FAM)-AAGGGATGCCGTTTGGGCCCAAGTTCGGCATAGTG-3′ |
| Target DNA: 5′-CACTATGCCGAACTTGGGCCCAAACGGCATCCCTT-3′ |
One-base mismatched target DNA:
T1: 5′-CACTATGCCGAACTTGG![[C with combining low line]](https://www.rsc.org/images/entities/char_0043_0332.gif) CCCAAACGGCATCCCTT-3′ CCCAAACGGCATCCCTT-3′ |
T2: 5′-CACTATGCCGAACTTGG![[T with combining low line]](https://www.rsc.org/images/entities/char_0054_0332.gif) CCCAAACGGCATCCCTT-3′ CCCAAACGGCATCCCTT-3′ |
T3: 5′-CACTATGCCGAACTTGG![[A with combining low line]](https://www.rsc.org/images/entities/char_0041_0332.gif) CCCAAACGGCATCCCTT-3′ CCCAAACGGCATCCCTT-3′ |
Non-complementary target DNA:
| T4: 5′-TAATACTGCCACTTCAAGACCTGGCGGCCACCCGT-3′ |
| E2 aptamer probe: 5′-(FAM)-AAGGGATGCCGTTTGGGCCCAAGTTCGGCATAGTG-(NH2)-3′ (35 mer) |
The stock solution of 100 μM oligonucleotides was dissolved in deionized water and stored at −20 °C before use. All the reagents were of analytical grade and used without further treatment. Deionized water, obtained from a Milli-Q water purification system (Millipore), was used in all the experiments. All the measurements were performed at room temperature in 100 mM Tris–HCl buffer (pH 8.0) containing 200 mM NaCl, 25 mM KCl, and 10 mM MgCl2.
Characterization measurements
Fluorescent emission spectra were recorded using an RF-5301PC luminescence spectrometer (Shimadzu, Kyoto, Japan) with 1.0 cm micro quartz cells at room temperature. Absorption spectra were recorded on a 2600 UV-vis spectroscopy instrument (Shimadzu, Tokyo, Japan). Biochar samples were prepared in a tubular furnace (OTF-1200X, Hefeikejing Materials Technology Co., Ltd China). A planetary ball mill (Nanchang City Hengshun Chemical Equipment Manufacturing Co., Ltd) was used to grind the biochar. The zeta potential and size of the biochar were measured using a NANO ZS90 Zetasizer (Malvern Panalytical, UK). Transmission electron microscopy (TEM) images and high-resolution transmission electron microscopy images of the biochar were obtained by field emission transmission electron microscopy (FESEM, JEM-2200FS, Japan) on a super-thin carbon support film at an acceleration voltage of 200 kV. Fourier transform infrared spectroscopy (FTIR) was performed on an FTIR spectrometer (VERTEX-70, USA). Circular dichroism (CD) spectroscopy was performed using a J-810 circular dichroism system (JASCO, Japan), and the Raman spectra were recorded using a confocal microscope Raman spectrometer (LabRAM HR Evolution, HORIBA, France). The powder X-ray diffraction (XRD) patterns were recorded on a D8 advance diffractometer (Brucker, Germany) equipped with Cu Kα radiation (λ = 1.54 Å) over the 2θ range of 10–80°.Preparation of the bagasse-derived quencher nanoparticles (BQNPs)
The raw biomass from agricultural waste bagasse was used to prepare the biochar nanoparticles. The bagasse was washed with deionized water three times to remove residual sugar and then exsiccated for 12 h at 80 °C in a vacuum drying oven. The dried bagasse was pyrolyzed at 400 °C in a tubular furnace with a flow of N2 gas to prevent organic materials from igniting. 14 The heating rate for pyrolysis was 7 °C min−1, and the residence time was 2 h. The produced rough carbon material (biochar) was ground into fine powder using a planetary ball mill for 2 h at 300 rpm, and then dispersed in deionized water, followed by ultrasonication for 10 min. Afterwards, the mixture was centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 3 min, and the nanoparticles in the liquid supernatant were collected by filtration through a membrane (cut-off size of 0.22 μm). After drying in a vacuum oven, these nanoparticles were ready for usage and denoted as bagasse-derived quencher nanoparticles (BQNPs).
000 rpm for 3 min, and the nanoparticles in the liquid supernatant were collected by filtration through a membrane (cut-off size of 0.22 μm). After drying in a vacuum oven, these nanoparticles were ready for usage and denoted as bagasse-derived quencher nanoparticles (BQNPs).
Preparation of the BQNPs/DNA probe and tDNA detection
A solution of BQNPs (100 μg mL−1) prepared in 100 mM Tris–HCl buffer (pH 8) was incubated with 30 nM of DNA probe for 5 min at room temperature. After thoroughly washing off the unbound DNA with Tris–HCl buffer, the produced composites of BQNPs and DNA probe (BQNPs/DNA probe) were ready for tDNA detection.For DNA sensing, the BQNPs/DNA probe was incubated with various concentrations of tDNA ranging from 2 to 400 nM in 100 mM Tris–HCl buffer (pH 8.0) for 60 min at room temperature. The fluorescence intensities were measured with the excitation and emission wavelength of 492 nm and 518 nm, respectively.
The stability of the sensing system was tested by treating the BQNPs/DNA probe with DNase I (2 U mL−1) for 10 min at room temperature, and after inactivating the activity of the DNase for 10 min at 80 °C, the fluorescence profile and the target analyte detection ability were evaluated.
Preparation of the BQNPs/aptamer probe and E2 detection
Conjugates of the BQNPs/aptamer probe were synthesized by coupling the amino group (–NH2) at the 3′ termini of the FAM-labelled E2 aptamer probe to the carboxyl group (–COOH) of BQNPs for E2 detection. Briefly, the carboxyl groups were first activated by EDC (10 mg mL−1) in MES buffer (100 mM, pH 5) for 20 min at room temperature. Upon activation, the solution was washed three times with deionized water to remove excess EDC. Subsequently, the activated BQNPs (200 μg mL−1) were reacted with FAM-labelled E2 aptamer probe (200 nM) in deionized water for 2 h at room temperature with gentle shaking (200 rpm). This was followed by centrifugation (10 min, 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) and thoroughly washing with deionized water to remove free aptamers. Then the obtained BQNPs/aptamer probe was stored at 4 °C for further use.
000 rpm) and thoroughly washing with deionized water to remove free aptamers. Then the obtained BQNPs/aptamer probe was stored at 4 °C for further use.
For E2 detection, a stock solution of E2 was prepared in ethanol and diluted with the binding buffer (100 mM Tris–HCl buffer) to various concentrations. Next, 50 μL of BQNPs/aptamer probe (10 μg mL−1) dispersed in the binding buffer was incubated with specific concentrations of E2 ranging from 1 to 500 ng mL−1 with shaking (200 rpm) for 15 min at room temperature. Then the fluorescence intensities of the solution were measured in a micro quartz cuvette cell with the excitation and emission wavelengths of 492 and 518 nm, respectively.
For real-sample environmental water detection, the sensor was applied to the analysis of E2 levels in three real water samples (tap water, lake water, and ground water). The tap water was obtained from the laboratory water tap. The lake water was obtained from Nanhu Lake, Changchun, China. The groundwater sample was obtained from the Changbaishan Hotel, Changchun, China. Before detection, the real water samples were filtered with a 0.22 μm filter membrane to remove the suspended microorganisms and solid matter. Then the water samples were spiked to attain specific concentrations of E2 (5, 8, 10, 20 ng mL−1), followed by quantitative sensing with the proposed method. All the measurements were performed in triplicate.
Results and discussion
Structural properties of the BQNPs
BQNPs were prepared from the biomass resources by the pyrolysis of bagasse at 400 °C under nitrogen protection, followed by grinding and size screening. The BQNPs were highly dispersive and presented nanosized and irregular structures (see TEM images in Fig. 1A). From the TEM image analysis, the average size of the BQNPs was 117.1 ± 38.4 nm (Fig. 1E). The particles from two further random batches exhibited a similar size distribution and average size value (Fig. S1†), indicating that the particle-generating approach was dependable and could be replicated. Upon further magnification under HR-TEM, many black dots smaller than 3 nm with an average size of 2.2 nm could be observed on the surfaces of the BQNPs (Fig. 1B), and almost each of them showed crystalline stripes representing a graphene-like nanosheet. Also, on the surfaces of BQNPs, aromatic cluster-like structures were visible (Fig. 1C and D), which was in accordance with the findings in rice-straw-derived biochar.15 These structural characteristics would contribute to an easy and strong attachment of single-stranded DNA to the BQNPs.The structural properties of the BQNPs were also characterized by means of Raman spectroscopy, FTIR, and XRD. The Raman spectra of the BQNPs (Fig. 1F) displayed D (∼1343.2 cm−1) and G (∼1595.4 cm−1) bands, which were assigned to local defects/disorders and stretching vibrations of the graphitized structure of the carbon atoms, indicating the crystallinity of the graphene nanosheets. 16 The intensity ratio of the D and G bands (R = ID/IG) was calculated as 0.57. The FTIR spectra of the BQNPs were obtained in the range of 400–4000 cm−1 (Fig. 1G). The broad band at 3405 cm−1 represented the band stretching of the hydrogen-bonded hydroxyl group (–OH), and the peaks at 1704 and 1600 cm−1 were characteristic peaks of the aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups, respectively,17,18 which contribute to the excellent water solubility and further modification capability. The XRD profile of the BQNPs exhibited two broad diffraction peaks around 2θ = 27° and 2θ = 43° (Fig. 1H) attributed to the crystal planes of parallel stacked graphene sheets and the presence of a honeycomb structure formed by sp2 hybridized carbons,19,20 consistent with the results from the HR-TEM analysis, and which could contribute to π–π stacking interactions of single-stranded DNA. After DNA adsorption on the BQNPs, there was a negligible change in the corresponding diffraction peaks (Fig. S2B†), indicating there was no significant structural alteration cause by the DNA attaching.
O groups, respectively,17,18 which contribute to the excellent water solubility and further modification capability. The XRD profile of the BQNPs exhibited two broad diffraction peaks around 2θ = 27° and 2θ = 43° (Fig. 1H) attributed to the crystal planes of parallel stacked graphene sheets and the presence of a honeycomb structure formed by sp2 hybridized carbons,19,20 consistent with the results from the HR-TEM analysis, and which could contribute to π–π stacking interactions of single-stranded DNA. After DNA adsorption on the BQNPs, there was a negligible change in the corresponding diffraction peaks (Fig. S2B†), indicating there was no significant structural alteration cause by the DNA attaching.
Fluorescence-quenching ability of the BQNPs
Fluorescence quenching denotes the process of decreasing the fluorescence intensity of a fluorophore by a quencher,21,22 and it requires molecular contact between them. To investigate the quenching ability of the BQNPs, the fluorescence intensities of FAM after incubating in varying concentrations of BQNPs solution were examined. As shown in Fig. 2B, the fluorescence of FAM gradually decreased with the increase in BQNPs concentration. This observation demonstrates that the BQNPs could act as an efficient fluorescence quencher of FAM. Fluorescence quenching can occur through a static process, dynamic process, or a combination of both. Static quenching creates a non-emissive ground-state complex between the fluorophore and quencher, while dynamic quenching occurs when the quencher deactivates the excited state of the fluorophore, often through a collisional mechanism. 23The Stern–Volmer plot is a graphical representation used to study the quenching of fluorescence and evaluate the fluorescence-quenching mechanism. The Stern–Volmer equation is given by: 21
If fluorescence quenching is due to a purely static or purely dynamic mechanism, the Stern–Volmer plot of I0/I vs. Q will be linear. Based on the nonlinear nature of Stern–Volmer plot, it is possible that both dynamic and static quenching may be involved. 23 An upward curve in the Stern–Volmer plot of the system, as shown here in Fig. 2C, illustrates a complex fluorescence-quenching process, which consists of both static and dynamic processes for quenching fluorescence.
FRET is a non-radiative process in which energy is transferred from the excited state of a donor to an acceptor through dipole–dipole interactions.25 The prerequisites for its occurrence are that the distance between the donor and acceptor is within the nanometre scale (smaller than 10 nm), and there is spectral overlap between the donors and acceptors. 26,27
To investigate whether FRET could occur between BQNPs and FAM, the UV-vis absorption spectrum of the BQNPs and the fluorescence emission spectrum of the fluorescein FAM were obtained. As shown in Fig. 2A, the BQNPs absorption spectrum (blue line) exhibited a wide absorption spectrum from 200 to 700 nm, which indicates a good spectral overlap with the fluorescence emission spectrum (orange line) of FAM, which has an emission peak at about 518 nm. This suggests that FRET could occur between the BQNPs and FAM, in which the BQNP could be a fluorescence acceptor and FAM could be a fluorescence donor.
In addition, the quenching efficiencies of BQNPs prepared at varying pyrolysis temperatures (400 °C, 600 °C, and 800 °C) were investigated by Stern–Volmer analysis.
According to the Stern–Volmer plots (Fig. 2C), the kq of BQNPs prepared at 400 °C (33.08 × 109 L (g s)−1) was higher than that prepared at 600 °C (12.14 × 109 L (g s)−1) and 800 °C (10.31 × 109 L (g s)−1), indicating that BQNPs obtained at lower temperature present the highest quenching ability towards FAM. This was supposed to be related to the greater amounts of carboxyl groups distributed on the surface of BQNPs obtained at lower temperature (see Fig. 1G and S2A†), since the carboxyl groups are generally considered good electron acceptors in the non-resonance energy-transfer process by deprotonating and negatively charging. Moreover, it was found that the kq of BQNP was also higher than that of other reported quenchers, such as graphene sheets (1.25 × 109 L (g s)−1), single walled carbon nanotubes (0.233 × 109 L (g s)−1), and graphene oxide (0.03 × 109 L (g s)−1).28 This was a possibility related to the high dispersibility and large surface area of the BQNPs, which enable more sites for access to FAM and fluorescence quenching.29,30 This result highlights that the BQNPs would be a good choice as an alternative nanoquencher, and their great quenching ability would be significant for enhancing the fluorescence sensing signal through the increment of the signal to background ratio. Here, BQNPs usually refer to those prepared at 400 °C, unless otherwise explained.
Feasibility of the BQNPs/DNA probe for DNA detection
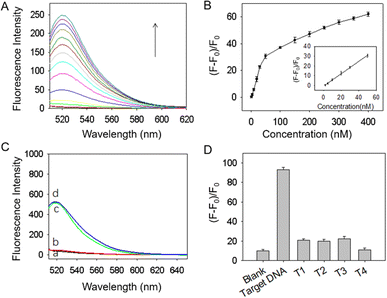
The feasibility of BQNPs as nanoquenchers for the fluorescence detection of nucleic acid was thus firstly demonstrated. As indicated in Fig. 3A, a strong fluorescence emission was presented at around 518 nm in the case of the FAM-labelled DNA probe (curve a), and the intensity did not change with the addition of the tDNA (curve b). It was found that when adding BQNPs to the DNA probe, more than 98% of the fluorescence was reduced (curve c), and when the tDNA was incorporated, approximately 63% of the fluorescence signal was recovered (curve d). This suggested that the DNA probe could attach to the surface of BQNPs, which brought the FAM molecules close to BQNPs, causing fluorescence quenching, and with the presence of the tDNA, the double-stranded DNA formed by the hybridization should be detached due to the lower binding force, resulting in the recovery of the fluorescence signal. The incomplete signal recovery may be related to the binding of a certain part of the tDNA to unoccupied surface areas on the BQNPs resulting in a decreased hybridization and recovery efficiency.
The possible mechanism for the binding of DNA probe to the BQNPs was assessed by isoelectric point (pI) analysis (Fig. S3†). By the pH analysis method, the pI of BQNPs was measured to be 7.2, suggesting that the BQNPs should be negatively charged in the detection solution (pH 8), which means that the BQNPs should not be electrostatically attracted to the DNA probe due to its negatively charged backbone. Instead, considering the presence of sp2 hybridized carbon in the BQNPs (XRD in Fig. 1H), the π–π interaction between the aromatic ring in the nucleotide bases of DNA and the BQNPs would be primarily responsible for the attachment of the DNA probe.
In order to more in-depth confirm the binding of the DNA probe to the BQNPs, the structural change of DNA was investigated using circular dichroism analysis. As shown in Fig. 3B, the probe DNA presented characteristic ellipticity positive peaks at 267 and 289 nm, and a negative peak at 246 nm (curve b). Also in case of the BQNPs/DNA probe, the positive peaks were reduced, while the negative peak disappeared, suggesting a structural alteration of the DNA probe, which occurred due to the attaching of the molecular backbone to the BQNPs (curve c). Moreover, after addition of the tDNA, there was a slight conformational change of the DNA probe caused by the hybridization (curve d).
The quenching efficiency was determined as (F0 − F)/F0, where F0 and F represent the fluorescence intensities of the DNA probe in the absence and presence of BQNPs, respectively. Fig. S4A† shows the fluorescence intensity changes of the DNA probe (30 nM) when reacted with different concentrations of BQNPs. The fluorescence intensity decreased as the concentration of BQNPs increased from 0 to 500 μg mL−1, and the highest quenching efficiency (99%) was achieved at 100 μg mL−1 of BQNPs (Fig. S4B†).
A study of the fluorescence-quenching kinetics was next conducted to determine the time required to completely quench the fluorescence of the DNA probe by the BQNPs. The quenching kinetics was assessed by monitoring the changes of the fluorescence intensities immediately after the addition of the BQNPs. As shown in Fig. S4C,† the fluorescence decrease was swift within the first 1 min (quenching efficiency of 95%), and it gradually reached equilibrium within 5 min (quenching efficiency of 99%). Apparently, the BQNPs were very effective at quenching FAM labelled on the DNA probe, which was attributed to the rapid and strong attachment of the DNA probe. Therefore, to obtain the optimum sensing performance, 5 min was used for quenching the fluorescence of the DNA probe.
The effect of the incubation time for the hybridization of tDNA in the detection process was evaluated, and 60 min was found to be optimal to achieve the highest signal recovery (Fig. S4D†). The effect of the incubation temperature during tDNA detection was also investigated, since temperature changes may affect the buffer conditions and the stability of DNA. As shown in Fig. S4E,† the fluorescence sensing signal was extremely weak when the incubation temperature was 4 °C. In contrast, the response signals increased significantly to nearly the same levels in the cases of 25 °C and 40 °C, showing that the proposed probe has a wide range of working temperatures. In the subsequent investigation, the incubation temperature was set at 25 °C.
The stability of the BQNPs/DNA probe was evaluated by treating it with DNase I. According to Fig. 4C, the BQNPs/DNA probe showed negligible changes in fluorescence signal after treatment with DNase I (see curves a and b), while after adding tDNA, the fluorescence signal recovered to almost its original level (see curves c and d). Clearly, the DNA probe on the BQNPs was resistant to DNase I cleavage and even remained normally functional with its adsorption to the BQNPs.
This sensing platform is based on the high selectivity of the single nucleotide chain, which contributes to its specificity. Therefore, we also evaluated the selectivity of the designed BQNPs/DNA probe in response to irrelevant DNA, such as mismatched single-base DNA (T1, T2, and T3) and non-complementary DNA (T4). As shown in Fig. 4D, the fluorescence signal increased significantly after incubation with tDNA, whereas the responses to the non-complementary DNA and one-base mismatched DNA were negligible. Based on these results, the proposed BQNPs/DNA probe demonstrated sufficient selectivity towards tDNA and potentially could be used for single nucleotide polymorphism analysis.
Feasibility of the BQNP/aptamer probe for E2 detection
To investigate whether the E2 aptamer probe was covalently bonded to BQNPs, FTIR spectral profiles of the original BQNPs, E2 aptamer probe, and BQNPs/aptamer probe after removing the unbound aptamers were analyzed. As shown in Fig. 5B, the characteristic peaks of BQNPs were observed at 1600 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1700 cm−1 (C
C), 1700 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and 3370 cm−1 (–OH).17,18 The characteristic peaks of the E2 aptamer were observed at 965, 1084, and 1229 cm−1, corresponding to phosphate asymmetric and symmetric vibrations, while the vibration at 1649 cm−1 was due to the presence of both C
O), and 3370 cm−1 (–OH).17,18 The characteristic peaks of the E2 aptamer were observed at 965, 1084, and 1229 cm−1, corresponding to phosphate asymmetric and symmetric vibrations, while the vibration at 1649 cm−1 was due to the presence of both C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N double bonds and N
N double bonds and N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N double bonds.31 In the BQNPs/aptamer probe, there was another characteristic peak of the E2 aptamer at 3416 cm−1, which was ascribed to the nucleic acid bases of NH stretching,17,18 indicating that the E2 aptamer was successfully combined with BQNPs.
N double bonds.31 In the BQNPs/aptamer probe, there was another characteristic peak of the E2 aptamer at 3416 cm−1, which was ascribed to the nucleic acid bases of NH stretching,17,18 indicating that the E2 aptamer was successfully combined with BQNPs.
The viability of a fluorescent probe-constructed using BQNPs and E2 aptamers was first established for E2 detection. As shown in Fig. 5A (curve a), the free FAM-labelled E2 aptamers exhibited a strong fluorescence emission at 518 nm, and after adding the target E2, the fluorescence signal was not changed (curve b), demonstrating that the fluorescence intensity of free aptamers was not affected by E2. In contrast, the BQNPs effectively quenched FAM fluorescence by more than 98% in the case of the BQNPs/aptamer probe (curve c). Furthermore, the quenching efficiency was higher than that of most reported nanoquenchers.32,33 In the presence of the target E2, the aptamer changes its secondary structure upon the selective capture of the target E2, leading to the formation of aptamer/E2 complexes with weak affinities to BQNPs, which allows the E2 aptamers to move away from the BQNPs surface, hence allowing the FAM fluorescence to be recovered (curve d). The achievement of a partial signal recovery was possibly due to the binding of some E2 to unoccupied surface areas of BQNPs, thereby the amount of E2 captured by the aptamer was reduced. In addition, it may possibly be related to incomplete detachment of the aptamers from the BQNPs following E2 capture.
Several irrelevant endocrine disrupting chemicals, including estrone (E1), ethinyl estradiol (EE), testosterone, and bisphenol A (BPA), were considered in order to test the specificity and selectivity of the proposed fluorescence probe. In Fig. 6C, it can be seen that the irrelevant chemicals exhibited negligible fluorescence responses despite their concentrations being 2-fold higher than the target molecules (E2). Consequently, the BQNPs/aptamer probe demonstrated excellent specificity for detecting the target molecules due to their high affinity for the target molecules (E2).
| Sample sources | Spiked E2 concentration (ng mL−1) | Mean found (ng mL−1) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Tap water | 5 | 4.86 | 97.2 | 2.8 |
| 8 | 8.51 | 106.4 | 6.4 | |
| 10 | 10.38 | 103.8 | 3.8 | |
| 20 | 19.36 | 96.8 | 3.2 | |
| Lake water | 5 | 4.76 | 95.3 | 4.7 |
| 8 | 8.5 | 106.1 | 6.1 | |
| 10 | 10.37 | 103.7 | 3.7 | |
| 20 | 19.3 | 96.4 | 3.6 | |
| Underground water | 5 | 4.76 | 95.2 | 4.8 |
| 8 | 8.46 | 105.7 | 5.7 | |
| 10 | 10.4 | 104.1 | 4.1 | |
| 20 | 19.4 | 96.9 | 3.1 |
Conclusions
Bagasse-derived quencher nanoparticles (BQNPs) obtained via low-temperature (400 °C) pyrolysis were demonstrated to possess a remarkable quenching ability towards the fluorescence of 6-carboxyfluorescein. These nanoparticles exhibited a significantly higher quenching rate constant compared to those prepared at high temperatures and also compared to other typical quenchers. Taking advantages of the distinctive characteristics of BQNPs, including their high quenching efficiency, rich functional groups, and excellent dispersibility, as a proof of concept, BQNP/DNA and BQNP/aptamer probes were successfully constructed by adsorption and covalent interactions of single-stranded oligonucleotides. Both probes were capable of the selective and sensitive detection of single-stranded DNA and 17β-estradiol, with satisfactory detection limits of 1.04 nM and 0.4 ng mL−1, respectively. Because of the outstanding quenching performance, extremely good cost-effectiveness, great dispersibility, and environmental friendliness, the BQNPs have significant potential for the development of low-cost and sensitive fluorescence sensors.Data availability
The authors confirm that the data supporting the findings of this study are available within the article and its ESI.†Author contributions
Xiaoli Qi was responsible for conceptualization, data curation, formal analysis, investigation, methodology, validation, writing – original draft, writing – review & editing. Hui Hu was responsible for investigation, data curation, formal analysis, methodology, validation, visualization, writing – original draft. Yunxian Piao was responsible for funding acquisition, conceptualization, project administration, supervision, writing – original draft, writing – review & editing. Lina Liang and Yuqing Lin, were responsible for conceptualization, resources, writing – original draft. Yudan Liu and Haifeng Sun were responsible for methodology, formal analysis.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Science and Technology Development Program of Jilin Province, China (20220101062JC) and the National Key Research and Development Program of China (2019YFC1804800).References
- X. Qi, H. Hu, Y. Yang and Y. Piao, Analyst, 2018, 143, 4163–4170 RSC.
- T. Ueno and T. Nagano, Nat. Methods, 2011, 8, 642–645 CrossRef CAS PubMed.
- G. Chen, W. Bai, Y. Jin and J. Zheng, Talanta, 2021, 232, 122405 CrossRef CAS PubMed.
- F. Zu, F. Yan, Z. Bai, J. Xu, Y. Wang, Y. Huang and X. Zhou, Microchim. Acta, 2017, 184, 1899–1914 CrossRef CAS.
- K. Quan, C. Yi, X. Yang, X. He, J. Huang and K. Wang, TrAC, Trends Anal. Chem., 2020, 124, 115784 CrossRef CAS.
- Y.-D. Ye, L. Xia, D.-D. Xu, X.-J. Xing, D.-W. Pang and H.-W. Tang, Biosens. Bioelectron., 2016, 85, 837–843 CrossRef CAS PubMed.
- B. Heli and A. Ajji, Sci. Rep., 2020, 10, 14367 CrossRef CAS PubMed.
- Q. Li, J. Bai, S. Ren, J. Wang, Y. Gao, S. Li, Y. Peng, B. Ning and Z. Gao, Anal. Bioanal. Chem., 2019, 411, 171–179 CrossRef CAS PubMed.
- D. Akhil, D. Lakshmi, A. Kartik, D.-V. N. Vo, J. Arun and K. P. Gopinath, Environ. Chem. Lett., 2021, 19, 2261–2297 CrossRef CAS.
- Y. Yang, Y. Piao, R. Wang, Y. Su, N. Liu and Y. Lei, J. Hazard. Mater. Adv., 2022, 8, 100171 CrossRef CAS.
- S. He, B. Song, D. Li, C. Zhu, W. Qi, Y. Wen, L. Wang, S. Song, H. Fang and C. Fan, Adv. Funct. Mater., 2010, 20, 453–459 CrossRef CAS.
- Q. Han, X. Shen, W. Zhu, C. Zhu, X. Zhou and H. Jiang, Biosens. Bioelectron., 2016, 79, 180–186 CrossRef CAS PubMed.
- Y. Tan and T. Wei, Talanta, 2015, 141, 279–287 CrossRef CAS PubMed.
- E.-B. Son, K.-M. Poo, J.-S. Chang and K.-J. Chae, Sci. Total Environ., 2018, 615, 161–168 CrossRef CAS PubMed.
- X. Xiao and B. Chen, Environ. Sci. Technol., 2017, 51, 5473–5482 CrossRef CAS PubMed.
- M. Wang, B. Hu, C. Yang, Z. Zhang, L. He, S. Fang, X. Qu and Q. Zhang, Biosens. Bioelectron., 2018, 99, 176–185 CrossRef CAS PubMed.
- K. Arora, A. Chaubey, R. Singhal, R. P. Singh, M. K. Pandey, S. B. Samanta, B. D. Malhotra and S. Chand, Biosens. Bioelectron., 2006, 21, 1777–1783 CrossRef CAS PubMed.
- Y. Gao, A. Pramanik, S. Begum, C. Sweet, S. Jones, A. Alamgir and P. C. Ray, ACS Omega, 2017, 2, 7730–7738 CrossRef CAS PubMed.
- G. Fey, Synth. Met., 2003, 139, 71–80 CrossRef CAS.
- S.-W. Han, D.-W. Jung, J.-H. Jeong and E.-S. Oh, Chem. Eng. J., 2014, 254, 597–604 CrossRef CAS.
- M. Acar, E. Bozkurt, K. Meral, M. Arık and Y. Onganer, J. Lumin., 2015, 157, 10–15 CrossRef CAS.
- Y. Chang, Z. Zhang, H. Liu, N. Wang and J. Tang, Analyst, 2016, 141, 4719–4724 RSC.
- Principles of Fluorescence Spectroscopy, ed. J. R. Lakowicz, Springer US, Boston, MA, 2006 Search PubMed.
- K. P. L. Kuijpers, C. Bottecchia, D. Cambié, K. Drummen, N. J. König and T. Noël, Angew Chem. Int. Ed. Engl., 2018, 57, 11278–11282 CrossRef CAS PubMed.
- W. Li, Z. Liang, P. Wang and Q. Ma, Biosens. Bioelectron., 2024, 249, 116008 CrossRef CAS PubMed.
- Z. S. Pehlivan, M. Torabfam, H. Kurt, C. Ow-Yang, N. Hildebrandt and M. Yüce, Microchim. Acta, 2019, 186, 563 CrossRef CAS PubMed.
- J. Shi, F. Tian, J. Lyu and M. Yang, J. Mater. Chem. B, 2015, 3, 6989–7005 RSC.
- X.-F. Zhang and X. Shao, J. Photochem. Photobiol., A, 2014, 278, 69–74 CrossRef CAS.
- D. F. Becheru, G. M. Vlăsceanu, A. Banciu, E. Vasile, M. Ioniţă and J. S. Burns, Int. J. Mol. Sci., 2018, 19, 3230 CrossRef PubMed.
- E. Morales-Narváez, B. Pérez-López, L. B. Pires and A. Merkoçi, Carbon, 2012, 50, 2987–2993 CrossRef.
- A. J. Patil, J. L. Vickery, T. B. Scott and S. Mann, Adv. Mater., 2009, 21, 3159–3164 CrossRef CAS.
- T. S. Bronder, M. P. Jessing, A. Poghossian, M. Keusgen and M. J. Schöning, Anal. Chem., 2018, 90, 7747–7753 CrossRef CAS PubMed.
- X. Zhang, K. Jiao, S. Liu and Y. Hu, Anal. Chem., 2009, 81, 6006–6012 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03168g |
| ‡ Xiaoli Qi and Hui Hu contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |