Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supramolecular complexation of phenylephrine by cucurbit[7]uril in aqueous solution
Khaleel I. Assaf *a,
Ayah N. Faraja,
Eyad S. M. Abu-Nameha and
Mohammad A. Alnajjarb
*a,
Ayah N. Faraja,
Eyad S. M. Abu-Nameha and
Mohammad A. Alnajjarb
aDepartment of Chemistry, Al-Balqa Applied University, Al-Salt 19117, Jordan. E-mail: khaleel.assaf@bau.edu.jo
bDepartment of Biology and Chemistry, Center for Cellular Nanoanalytics, Universität Osnabrück, 49069 Osnabrück, Germany
First published on 23rd April 2024
Abstract
Cucurbiturils (CBn) are known to establish stable host–guest complexes with a variety of drug molecules. Herein, the supramolecular complexation between cucurbit[7]uril (CB7) and phenylephrine hydrochloride is reported in aqueous solution. Phenylephrine forms inclusion complex with CB7 with high binding affinity (Kaffinity = 4.0 × 106 M−1), which allows for the development of a fluorescence-based sensing assay applying the dye displacement strategy. The structure of the host–guest inclusion complex is investigated by 1H NMR spectroscopy, in which complexation-induced chemical shifts indicate the immersion of the aromatic ring inside the hydrophobic cavity of CB7. Density functional theory (DFT) calculations support the 1H NMR results, and reveal that the complex is stabilized through intermolecular interactions between the polar groups on the phenylephrine and the carbonyl rims of CB7, as well as the hydrophobic effect. Moreover, preferential binding of phenylephrine in its protonated over the neutral form results in a complexation-induced pKa shift.
Introduction
Cucurbit[n]urils, CBn, are a family of interesting macrocyclic host molecules that are water-soluble.1,2 CBn and other homologues are synthesized by the condensation of glycoluril and formaldehyde in strongly acidic media,3,4 where the reaction composition representing the molecules was first identified by crystallography.5 CBn are composed of n (5–8, 10 and 14) repeating glycoluril units linked via two methylene bridges on each side, which results in a barrel-shaped molecules with two identical carbonyl rims and lipophilic cavity with low polarity and polarizability.1 CBn have found great interest due to their ability to encapsulate a wide spectrum of guest molecules within the hydrophobic cavity with high association constants.1,2 The host–guest complexation of CBn is driven by the hydrophobic effect and non-covalent interactions.6 The relocation from water into the hydrophobic cavity of a macrocycle changes the physical and chemical properties of the encapsulated guests, such as solubility and stability.7–10 Therefore, CBn and their derivatives are suitable for several lines of applications including pharmaceutical, agriculture and chemical industries.11–15 Several examples on the complexation of drug molecules with different CBn hosts in aqueous solution are documented in the literature.16 For example, CB7 forms host–guest complexes with a variety of bioactive analytes such as amino acids, steroids, and drug molecules.7,17–19In this work, we study the molecular recognition of phenylephrine by CB7 through host–guest complexation (Fig. 1). Indeed, CB7 was selected due to its moderate cavity size (242 Å) that fits small organic molecules, as well as its relatively high water solubility.20 Phenylephrine is used effectively to treat congestion, vasomotor rhinitis, hemorrhoids and hypotension, and commonly prescribed an alternative to pseudoephedrine.21,22 The supramolecular complexation is investigated by using different spectroscopic techniques and the structure of the host–guest complex is elucidated by nuclear magnetic resonance (NMR) spectroscopy and verified by quantum-chemical calculations.
Results and discussion
CBn have been widely used to complex drug molecules and other biologically relevant analytes, due to their low toxicity.23–25 Phenylephrine is structurally similar to catecholamines, a class of important pharmacological molecules that act as neurotransmitters and hormones.26 The supramolecular complexation of catecholamines, such as dopamine, norepinephrine, and epinephrine with different macrocyclic hosts has been subject for several studies.27–29 The encapsulation of a guest molecule (e.g., a dye) inside the hydrophobic cavity alerts its photophysical properties, including the absorption and fluorescence emission.30–32 This has extensively been applied to probe the host–guest complexation and allowed for the determination of the binding affinity of optically transparent guest molecules.7,33–35 Phenylephrine showed an absorption maximum in UV-range (λmax = 275 nm, in acidic solution and λmax = 290 nm, in basic solution). The addition of CB7 to a phenylephrine solution resulted in a hyperchromic shift, which in part a consequence of the CB7 absorption in the same region. Therefore, indicator displacement assay was chosen to determine the binding affinity of phenylephrine to CB7.33,36 Berberine chloride (Fig. 1) was selected as a sensitive dye, which is known to form stable inclusion complex with CB7.37 In aqueous solution, berberine showed a weak fluorescence emission (λem = 495 nm), which is significantly enhanced upon complexation with CB7, due to the relocation into the hydrophobic cavity and the rigid confinement.37 The binding constant of berberine was determined as 1.6 × 107 M−1, in a good agreement with reported values.37–39 The preformed CB7·berberine complex can potentially be used as a reporter pair for the sensing of optically transparent guest molecules.34 The addition of a competitive binder (e.g., phenylephrine) results in the displacement of the dye (i.e., berberine) and the restoration of the original fluorescence emission. Fig. 2 illustrates the principle of fluorescence-based dye displacement.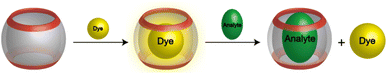 | ||
| Fig. 2 Schematic representation of the fluorescent-dye displacement assay principle for analyte binding using CBn/dye reporter pair. | ||
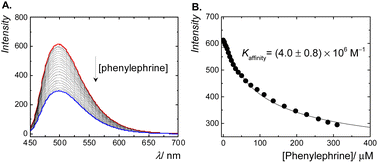
The fluorescence displacement titration of phenylephrine is shown in Fig. 3A. The fluorescence intensity of complexed berberine decreased upon the addition of phenylephrine. This indicates the displacement of berberine by phenylephrine. The emission intensity was then monitored as a function of the phenylephrine concentration and fitted according to a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding model, which resulted in a binding constant of 4.0 × 106 M−1 (Fig. 3B). The binding affinity of phenylephrine to CB7 is found to be higher than that of dopamine40 (a structurally similar catecholamine), which might be attributed to the position of a hydroxyl group at the side chain of phenylephrine rather than at the phenyl ring in dopamine.
1 binding model, which resulted in a binding constant of 4.0 × 106 M−1 (Fig. 3B). The binding affinity of phenylephrine to CB7 is found to be higher than that of dopamine40 (a structurally similar catecholamine), which might be attributed to the position of a hydroxyl group at the side chain of phenylephrine rather than at the phenyl ring in dopamine.
The sensitive fluorescence response of CB7·berberine to the phenylephrine could potentially be used for sensing purposes in aqueous solutions. A linear fluorescence response was observed in the concentration range of 0 to 30 μM of phenylephrine (Fig. 4, R2 = 0.997). Accordingly, the limits of detection (LOD) and quantification (LOQ) were estimated as 0.58 and 1.77 mg L−1. This represents a new fluorescence-based sensing method for phenylephrine.
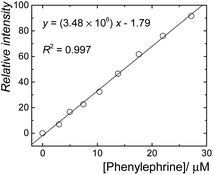 | ||
| Fig. 4 Linear fluorescence response for the detection of phenylephrine in aqueous solution using CB7·berberine reporter pair. | ||
CBn are known as strong binders for a variety of neutral and cationic guest molecules in aqueous solution.2 The preferential binding of the cationic form of guest molecules (e.g., ammonium-based) over their neutral form, modulates their acid dissociation constants (pKa), leading to a complexation-induced pKa shifts.9,15,41,42 The pKa values of free and complexed phenylephrine were determined by monitoring the absorbance as a function of pH (Fig. 5). The UV-spectra of free phenylephrine showed an absorption band with λmax of 275 nm at low pH (<7), which gradually shifts to a longer wavelength (bathochromic shift) as a function of increasing the pH (λmax of 290 nm), with two isosbestic points at 259 and 278 nm. The pKa of phenylephrine was determined as 9.35. Similarly, the pKa of complexed phenylephrine was measured in the presence of 1.0 mM CB7 to ensure a high degree of complexation at elevated pH values. No significant change in the λmax of phenylephrine was observed in the presence of CB7 in acidic solution, while a slight bathochromic shift (5 nm) was obtained in basic solution. CB7-complexed phenylephrine showed an increase in the pKa value to 10.52 (ΔpKa of 1.17). This shift to a higher pKa value indicated an increased basicity of phenylephrine in the presence of CB7 as consequence of the stabilization of the protonated form due to the ion–dipole interactions between the ammonium group in the phenylephrine and the carbonyl rim of CB7 (see below).
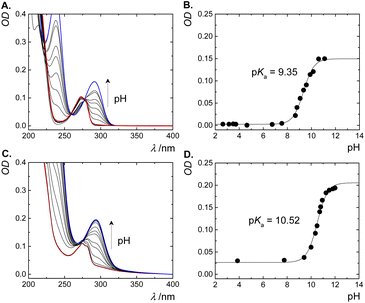 | ||
| Fig. 5 UV-absorption spectra of free phenylephrine and CB7·phenylephrine inclusion complex at different pH values (A and C, respectively). Fitted pH-dependent absorbance data at 295 nm (B and D). | ||
The formation of host–guest complexes between phenylephrine and CB7, in acidic aqueous solution, was investigated using 1H NMR spectroscopy, which is a powerful technique used to extract structural information. The formation of inclusion complexes in aqueous solution is usually associated with complexation-induced chemical shifts.43 In detail, we rely on chemical shift values that indicate whether the guest molecule or part of it is located inside the hydrophobic cavity or closer to the portals of the macrocyclic host. This is usually obtained by measuring the difference in 1H NMR chemical shifts of the bound (δbound) and free guest (δfree), which can be expressed as follows Δδ = δbound − δfree. In the case of CBn, the host protons are less informative, due to the absence of protons inside the cavity. Changes in the chemical shift of the guest protons can give a clear evidence for complexation. When, Δδ > 0, this means that the protons of the guest are located near to the carbonyl groups of CB7 and appear downfield shifted. When Δδ < 0, the protons of the guest are positioned inside the lipophilic cavity of CB7 and appear upfield shifted. Fig. 6 displays the 1H NMR spectra of free phenylephrine, free CB7, and their complex, dissolved in D2O. The phenylephrine signals were assigned as shown in the chemical structure Fig. 6B. The formation of the inclusion complex between CB7 and phenylephrine affects the proton chemical shifts based on their location within the cavity of CB7. The relocation of the guest into the hydrophobic cavity of CB7 induced significant upfield shifts due to the shielding effect. In contrast, protons that are located at the carbonyl rims are usually downfield shifted compared to the uncomplexed ones. The aromatic protons 4–7 were upfield shifted upon complexation (Δδ = −0.6 to −1 ppm), which indicated that the phenyl ring is positioned inside the cavity of CB7. Similarly, proton 3 experienced an upfield shift. In contrast, the methyl protons (1) were downfield shifted as expected from their location near the CB7 portal. The methylene protons (2) split upon complexation, which revealed different environments within the complex. These data revealed that the protonated ammonium group will be docked at the portal to enable ion–dipole interactions.
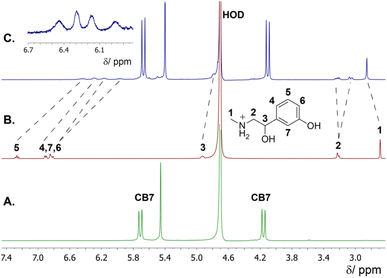 | ||
Fig. 6 1H NMR spectra of (A) free CB7 (2.0 mM), (B) free phenylephrine (2.0 mM), (C) CB7·phenylephrine complex (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in D2O. 1) in D2O. | ||
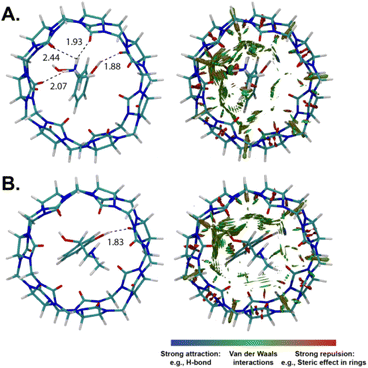
Density functional theory (DFT) calculations were performed to get additional structural information on the host–guest complexation. The optimized structures for the protonated/neutral phenylephrine as well as the non-covalent interactions (NCI) analysis are shown in Fig. 7. NCI analysis provide the visual illustration of the non-covalent interactions (Fig. 7B), such as hydrogen bonds (depicted with blue color), van der Waals interactions (depicted with green color), and repulsive steric interactions (depicted with red color).44 The optimized structure of CB7·phenylephrine in the protonated form indicated the formation of inclusion complex, in which the phenyl ring is encapsulated inside the cavity. In addition, the complex is stabilized by ion–dipole interaction between the ammonium group and the carbonyl rim from the upper side, as well as hydrogen bonding between the hydroxyl group and the lower rim (Fig. 7A). This result is in agreement with the 1H NMR data. For the unprotonated form of phenylephrine, the inclusion complex is stabilized by hydrogen bonding between the amine/hydroxyl groups and the carbonyl rims (Fig. 7B). It can be seen from the NCI analysis that the protonated form of phenylephrine involved more attraction interactions compared to the neutral one. The calculated interaction energy revealed a stabilization of the protonated complex by 63 kcal mol−1 relative to the neutral one.
Phenylephrine hydrochloride extended stability at low and ready-to-use concentrations has been recognized problematic.22,45,46 Consequently, there is a need to improve stability phenylephrine hydrochloride. In this regard, supramolecular complexation provides a versatile solution for drug stability.47 The host–guest complexation between CBn and several drug molecules has shown positive impact on their stability.16 Therefore, the complexation of phenylephrine hydrochloride by CB7 may serve as a potential strategy to enhance its stability and bioavailability.
Conclusions
In summary, we report on the host–guest complex between CB7 and phenylephrine in aqueous solution. CB7 forms inclusion complex with protonated phenylephrine with a high binding affinity, which is attributed to the combination of ion–dipole and hydrogen bonding interactions, as well as the hydrophobic effect. The supramolecular host–guest assembly resulted in a complexation-induced pKa shift, revealing an increase in the basicity of phenylephrine. The recognition of phenylephrine by CB7 enables the development of fluorescence-based sensing assay applying the indicator displacement strategy.Experimental details
CB7 was synthesized as previously reported.48 Phenylephrine hydrochloride and berberine chloride were purchased as a solid powder from Sigma-Aldrich (Germany). Hydrochloric acid and sodium hydroxide were used to adjust the pH. UV-Visible absorption measurements were performed with a Varian Cary 4000 spectrophotometer and the fluorescence spectra were recorded on a Varian Cary Eclipse. All optical measurements were performed at ambient temperature, using a rectangular quartz cuvette with 1 cm optical path length. Proton nuclear magnetic resonance (1H NMR) spectra were measured by using a 400 MHz FTNMR NanoBay spectrometer (Bruker, Switzerland) in D2O.Solutions were prepared in Milli-Q water and left to equilibrate before the measurements. The binding affinity of berberine dye to CB7 was measured by using fluorescence titration (pH = 3.0). The fluorescence emission (λex = 345 nm and λem = 495 nm) of berberine (20 μM, kept fixed during the titration) was measured as a function of CB7 concentration and the binding constant (Kaffinity) was fitted according to the non-linear curve fit for a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 binding stoichiometry.49 Indicator displacement experiment was performed to determine the binding constant of phenylephrine with CB7. In detail, phenylephrine was added to a solution of preformed CB7·berberine complex (using 35 μM of CB7 and 20 μM of berberine, both kept fixed during the titration), and the fluorescence emission (λex = 345 nm and λem = 495 nm) was recorded upon each addition. The data were fitted according to the non-linear curve fit.50 For the pKa determination, the absorption spectra of phenylephrine (50 μM) were recorded as a function of pH in the absence and presence of 1.0 mM CB7 (to ensure a high degree of complexation).51 1H NMR spectra for free CB7 (2.0 mM) and phenylephrine (2.0 mM), as well as the CB7·phenylephrine complex were measured in D2O.
1 binding stoichiometry.49 Indicator displacement experiment was performed to determine the binding constant of phenylephrine with CB7. In detail, phenylephrine was added to a solution of preformed CB7·berberine complex (using 35 μM of CB7 and 20 μM of berberine, both kept fixed during the titration), and the fluorescence emission (λex = 345 nm and λem = 495 nm) was recorded upon each addition. The data were fitted according to the non-linear curve fit.50 For the pKa determination, the absorption spectra of phenylephrine (50 μM) were recorded as a function of pH in the absence and presence of 1.0 mM CB7 (to ensure a high degree of complexation).51 1H NMR spectra for free CB7 (2.0 mM) and phenylephrine (2.0 mM), as well as the CB7·phenylephrine complex were measured in D2O.
Quantum-chemical calculations were performed using Gaussian 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 at the M06-2X/6-31+G** level of theory in the water applying the implicit solvation model based on density (smd) method.53 The non-covalent interactions (NCI) have been performed by using MULTIWFN v3.7.44
52 at the M06-2X/6-31+G** level of theory in the water applying the implicit solvation model based on density (smd) method.53 The non-covalent interactions (NCI) have been performed by using MULTIWFN v3.7.44
Author contributions
K. I. A. conceptualization, methodology, supervision, writing – review and editing. A. N. F. data curation, formal analysis, visualization, E. S. M. A. supervision, writing – review and editing. M. A. A. data curation, formal analysis, visualization, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Notes and references
- K. I. Assaf and W. M. Nau, Chem. Soc. Rev., 2015, 44, 394–418 RSC.
- S. J. Barrow, S. Kasera, M. J. Rowland, J. del Barrio and O. A. Scherman, Chem. Rev., 2015, 115, 12320–12406 CrossRef CAS PubMed.
- R. Behrend, E. Meyer and F. Rusche, Justus Liebigs Ann. Chem., 1905, 339, 1 CrossRef.
- W. A. Freeman, W. L. Mock and N.-Y. Shih, J. Am. Chem. Soc., 1981, 103, 7367–7368 CrossRef CAS.
- J. Kim, I.-S. Jung, S.-Y. Kim, E. Lee, J.-K. Kang, S. Sakamoto, K. Yamaguchi and K. Kim, J. Am. Chem. Soc., 2000, 122, 540–541 CrossRef CAS.
- F. Biedermann, W. M. Nau and H.-J. Schneider, Angew. Chem., Int. Ed., 2014, 53, 11158–11171 CrossRef CAS PubMed.
- A. I. Lazar, F. Biedermann, K. R. Mustafina, K. I. Assaf, A. Hennig and W. M. Nau, J. Am. Chem. Soc., 2016, 138, 13022–13029 CrossRef CAS PubMed.
- N. J. Wheate and C. Limantoro, Supramol. Chem., 2016, 28, 849–856 CrossRef CAS.
- N. Saleh, A. L. Koner and W. M. Nau, Angew. Chem., Int. Ed., 2008, 47, 5398–5401 CrossRef CAS PubMed.
- D. R. Boraste, G. Chakraborty, A. K. Ray, G. S. Shankarling and H. Pal, J. Photochem. Photobiol., A, 2018, 358, 26–37 CrossRef CAS.
- G. Cheng, J. Luo, Y. Liu, X. Chen, Z. Wu and T. Chen, ACS Appl. Bio Mater., 2020, 3, 8211–8240 CrossRef CAS PubMed.
- K. Kim, N. Selvapalam, Y. H. Ko, K. M. Park, D. Kim and J. Kim, Chem. Soc. Rev., 2007, 36, 267–279 RSC.
- H. Yin, Q. Cheng, D. Bardelang and R. Wang, JACS Au, 2023, 3, 2356–2377 CrossRef CAS PubMed.
- Z. Wang, C. Sun, K. Yang, X. Chen and R. Wang, Angew. Chem., Int. Ed., 2022, 61, e202206763 CrossRef CAS PubMed.
- F. J. R. Mejías, S. He, R. M. Varela, J. M. G. Molinillo, A. Barba-Bon, W. M. Nau and F. A. Macías, J. Agric. Food Chem., 2023, 71, 480–487 CrossRef PubMed.
- D. Das, K. I. Assaf and W. M. Nau, Front. Chem., 2019, 7, 619 CrossRef CAS PubMed.
- M. E. Bush, N. D. Bouley and A. R. Urbach, J. Am. Chem. Soc., 2005, 127, 14511–14517 CrossRef CAS PubMed.
- Z. Miskolczy, M. Megyesi, G. Tárkányi, R. Mizsei and L. Biczók, Org. Biomol. Chem., 2011, 9, 1061–1070 RSC.
- N. Saleh, I. Ghosh and W. M. Nau, in Supramolecular Systems in Biomedical Fields, ed. H.-J. Schneider, The Royal Society of Chemistry, 2013, p. 164 Search PubMed.
- E. Masson, X. Ling, R. Joseph, L. Kyeremeh-Mensah and X. Lu, RSC Adv., 2012, 2, 1213–1247 RSC.
- C. A. Gaglia, in Analytical Profiles of Drug Substances, ed. K. Florey, Academic Press, 1974, vol. 3, pp. 483–512 Search PubMed.
- J. A. Patel, V. D. Gupta and R. L. Mosier, Am. J. Hosp. Pharm., 1972, 29, 870–873 Search PubMed.
- V. D. Uzunova, C. Cullinane, K. Brix, W. M. Nau and A. I. Day, Org. Biomol. Chem., 2010, 8, 2037–2042 RSC.
- X. Zhang, X. Xu, S. Li, L.-H. Wang, J. Zhang and R. Wang, Sci. Rep., 2018, 8, 8819 CrossRef PubMed.
- E. Pashkina, A. Aktanova, O. Boeva, M. Bykova, E. Gavrilova, E. Goiman, E. Kovalenko, N. Saleh, L. Grishina and V. Kozlov, Pharmaceutics, 2024, 16, 127 CrossRef CAS PubMed.
- a. P. B. Molinoff and J. Axelrod, Annu. Rev. Biochem., 1971, 40, 465–500 CrossRef PubMed.
- S. K. S. Al-Burtomani and F. O. Suliman, RSC Adv., 2017, 7, 9888–9901 RSC.
- V. Sindelar, M. A. Cejas, F. M. Raymo, W. Chen, S. E. Parker and A. E. Kaifer, Chem.–Eur. J., 2005, 11, 7054–7059 CrossRef CAS PubMed.
- J. B. Pandya, S. M. Shinde and P. K. Jha, Struct. Chem., 2022, 33, 1321–1330 CrossRef CAS.
- A. L. Koner and W. M. Nau, Supramol. Chem., 2007, 19, 55–66 CrossRef CAS.
- W. M. Nau and J. Mohanty, Int. J. Photoenergy, 2005, 7, 568352 CrossRef.
- R. N. Dsouza, U. Pischel and W. M. Nau, Chem. Rev., 2011, 111, 7941–7980 CrossRef CAS PubMed.
- M. Florea and W. M. Nau, Angew. Chem., Int. Ed., 2011, 50, 9338–9342 CrossRef CAS PubMed.
- K. I. Assaf, M. Florea, J. Antony, N. M. Henriksen, J. Yin, A. Hansen, Z.-W. Qu, R. Sure, D. Klapstein, M. K. Gilson, S. Grimme and W. M. Nau, J. Phys. Chem. B, 2017, 121, 11144–11162 CrossRef CAS PubMed.
- Q. Duan, R. Chen, S. Deng, C. Yang, X. Ji, G. Qi, H. Li, X. Li, S. Chen, M. Lou and K. Lu, Front. Chem., 2023, 11, 1124705 CrossRef CAS PubMed.
- A. Praetorius, D. M. Bailey, T. Schwarzlose and W. M. Nau, Org. Lett., 2008, 10, 4089–4092 CrossRef CAS PubMed.
- M. Megyesi, L. Biczók and I. Jablonkai, J. Phys. Chem. C, 2008, 112, 3410–3416 CrossRef CAS.
- Z. Miskolczy and L. Biczók, J. Phys. Chem. B, 2014, 118, 2499–2505 CrossRef CAS PubMed.
- M. A. Alnajjar, W. M. Nau and A. Hennig, Org. Biomol. Chem., 2021, 19, 8521–8529 RSC.
- J. Chen-Wu, P. Máximo, P. Remón, A. J. Parola, N. Basílio and U. Pischel, Chem. Commun., 2023, 59, 3431–3434 RSC.
- I. Ghosh and W. M. Nau, Adv. Drug Delivery Rev., 2012, 64, 764–783 CrossRef CAS PubMed.
- N. Basílio, S. Gago, A. J. Parola and F. Pina, ACS Omega, 2017, 2, 70–75 CrossRef PubMed.
- V. V. Rudiger and H. J. Schneider, Eur. J. Chem., 2000, 6, 3771–3776 CrossRef CAS.
- T. Lu and F. Chen, J. Comput. Chem., 2012, 33, 580–592 CrossRef CAS PubMed.
- V. D. Gupta, Int. J. Pharm. Compd., 2004, 8, 153 Search PubMed.
- J. J. Jansen, A. R. Oldland and T. H. Kiser, Hosp. Pharm., 2014, 49, 455–457 CrossRef CAS PubMed.
- V. Aiassa, C. Garnero, A. Zoppi and M. R. Longhi, Pharmaceuticals, 2023, 16, 1074 CrossRef CAS PubMed.
- C. Marquez, H. Fang and W. M. Nau, IEEE Trans. NanoBiosci., 2004, 3, 39–45 CrossRef PubMed.
- H. Bakirci, X. Zhang and W. M. Nau, J. Org. Chem., 2005, 70, 39–46 CrossRef CAS PubMed.
- A. Hennig, H. Bakirci and W. M. Nau, Nat. Methods, 2007, 4, 629–632 CrossRef CAS PubMed.
- H. Bakirci, T. Schwarzlose, A. L. Koner and W. M. Nau, Chem.–Eur. J., 2006, 12, 4799–4807 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman, and D. J. Fox, Gaussian 16, Revision C.02, Gaussian, Inc., Wallingford CT, 2019 Search PubMed.
- A. V. Marenich, C. J. Cramer and D. G. Truhlar, J. Phys. Chem. B, 2009, 113, 6378–6396 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |