Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Comparative metabolomics study on the secondary metabolites of the red alga, Corallina officinalis and its associated endosymbiotic fungi†
Sherif M. Shamaa,
Ahmed M. Elissawy bc,
Mohamed A. Salema,
Fadia S. Youssef
bc,
Mohamed A. Salema,
Fadia S. Youssef b,
Mohamed S. Elnaggar
b,
Mohamed S. Elnaggar b,
Hesham R. El-Seedid,
Shaden A. M. Khalifaef,
Khaled Brikig,
Dalia Ibrahim Hamdana and
Abdel Nasser B. Singab
b,
Hesham R. El-Seedid,
Shaden A. M. Khalifaef,
Khaled Brikig,
Dalia Ibrahim Hamdana and
Abdel Nasser B. Singab *bc
*bc
aDepartment of Pharmacognosy and Natural Products, Faculty of Pharmacy, Menoufia University, Shibin Elkom, 32511, Egypt. E-mail: dean@pharma.asu.edu.eg
bDepartment of Pharmacognosy, Faculty of Pharmacy, Ain-Shams University, Cairo 11566, Egypt
cCenter of Drug Discovery Research and Development, Ain-Shams University, Cairo 11566, Egypt
dChemistry Department, Faculty of Science, Islamic University of Madinah, P. O. Box: 170, Madinah 42351, Saudi Arabia
eInternational Research Center for Food Nutrition and Safety, Jiangsu University, Zhenjiang 212013, China
fPsychiatry and Neurology Department, Capio Saint Göran's Hospital, Sankt Göransplan 1, 112 19 Stockholm, Sweden
gLaboratory of Organic Chemistry and Natural Substance, University Ziane Achour, Djelfa, Algeria
First published on 20th June 2024
Abstract
Marine endosymbionts have gained remarkable interest in the last three decades in terms of natural products (NPs) isolated thereof, emphasizing the chemical correlations with those isolated from the host marine organism. The current study aimed to conduct comparative metabolic profiling of the marine red algae Corallina officinalis, and three fungal endosymbionts isolated from its inner tissues namely, Aspergillus nidulans, A. flavipes and A. flavus. The ethyl acetate (EtOAc) extracts of the host organism as well as the isolated endosymbionts were analyzed using ultra-high performance liquid chromatography coupled to high resolution tandem mass spectrometry (UHPLC-MS/MS)in both positive and negative ion modes, applying both full scan (FS) and all ion fragmentation (AIF) modes. Extensive interpretation of the LC-MS/MS spectra had led to the identification of 76 metabolites belonging to different phytochemical classes including alkaloids, polyketides, sesquiterpenes, butyrolactones, peptides, fatty acids, isocoumarins, quinones, among others. Metabolites were tentatively identified by comparing the accurate mass and fragmentation pattern with metabolites previously reported in the literature, as well as bioinformatics analysis using GNPS. A relationship between the host C. officinalis and its endophytes (A. flavus, A. nidulans, and A. flavipes) was discovered. C. officinalis shares common metabolites with at least one of the three endosymbiotic fungi. Some metabolites have been identified in endophytes and do not exist in their host. Multivariate analysis (MVA) revealed discrimination of A. flavipes from Corallina officinalis and other associated endophytic Aspergillus fungi (A. flavus and A. nidulans).
1 Introduction
The expanding number of marine-derived natural products described from endosymbiotic fungi encourages additional action and in-depth research for marine environment-derived natural bioactive compounds from a drug discovery perspective.1,2 Macroalgae are divided into three primary phyla based on their coloration, which includes red seaweed (Rhodophyta), brown seaweed (Phaeophyta), and green seaweed (Chlorophyta).3 Corallina officinalis, a red seaweed, has a long history of use in traditional Chinese medicine and is a well-known edible seaweed in China and many other nations.4Red algae are known to contain a variety of secondary metabolites, including sulfated sugars, halogenated mono- and diterpenes, sterols, alkaloids, and polyphenols. Many reports demonstrated the biological impact of the secondary metabolites isolated from genus Corallina and the endophytes isolated thereof; for example, The in vitro assay of total extract of C. officinalis exhibited antiprotozoal activity against Trypanosoma cruzi.5 The new cyclic depsipeptides isolated from culture broth of Staphylococcus sp. derived from C. officinalis, known as cyclo (2α, 3-diamino-propoincacid-L-Asn-3-β-hydroxy-5-methyl-tetradecanoicacid-L-Leu1-L-Asp-L-Val-L-Leu2-L-Leu3) and cyclo(L-pro-L-omet) exhibiting potent antibacterial and antifungal activities.6–8
Aspergillus is a large genus with more than 180 different anamorphic species distributed in various ecological niches. As a common fungal endosymbiont, it is regarded as a source of vast classes of chemical compounds with interesting biological functions. Diverse Aspergillus species have demonstrated their capacity to produce a wide range of secondary metabolites, including butenolides, alkaloids, terpenoids, cytochalasins, phenalenones, terphenyls, xanthones, sterols, and anthraquinone derivatives with a variety of biological activities, including anti-cancer, anti-fungal, anti-bacterial, anti-viral, anti-inflammatory, anti-trypanosomal and anti-leishmanial activities.8–14
Recently, metabolomic studies became an indispensable tool in natural products chemistry where it provides broad qualitative and quantitative profiles of metabolites in organisms under diverse environmental situations. Under stressful circumstances, both plants and microbes create a wide variety of metabolites with distinct chemistries and bioactivities. Through integrated softwares, platforms, libraries and databases it is possible to unveil the intricate interactions between endophytes and their host organisms.15
UPLC/MS/MS provides a highly sensitive and adaptable tool and represents the basic tool in metabolomic studies with the ability to analyze minor chemicals providing crucial structural information for identification. However, massive volumes of spectra may be produced using mass spectrometry, which increases the complexity of the analysis. Considering that, the online platform Global Natural Products Social Molecular Networking (GNPS) allows the use of several different mass spectrometry-based metabolomics tools to analyze large sets of data. Moreover, numerous tools offered by GNPS have the ability to automatically search for a spectral match, and the organization provides a public spectrum library.16,17
Finding a link between the secondary metabolites produced by the host organism and those produced by the endophytic community is essential to unveil the ecological significance of endophytes and to open new avenue to discover the need of such mutualistic relationship.
In the present study and in continuation of our ongoing research on marine endosymbiotic fungal products, we herein report a comparative LC-MS-MS metabolomics study on the ethyl acetate extracts of the red algae C. officinalis and three endosymbiotic fungi isolated from its inner tissue namely; A. nidulans, A. flavipes and A. flavus. The identified metabolites belonged to various chemical classes such as alkaloids, anthraquinones, polyketides, sesquiterpenes, butyrolactones, peptides, fatty acids, isocoumarins, quinones, among other miscellaneous compounds.
2 Experimental
2.1. Fungal material
The fungi A. nidulans, A. flavipes and A. flavus were isolated from the inner tissues of the red algae C. officinalis. The algae was collected in the Mediterranean Sea close to Alexanderia, Egypt, in September 2018. For isolation of the fungal strain, the Algae was rinsed with distilled water and then surface sterilization using 70% ethanol was performed for 2 min. Small samples from the inner tissues of the algae were aseptically cut using sterilized blade and pressed onto malt agar plate (15 g per L malt extract, 15 g per L agar, 0.2 g per L chloramphenicol to suppress bacterial growth, pH adjusted to 7.4–7.8 using 10% NaOH). After incubation at 25 °C the fungal strains under investigation were found to grow out of the algal tissue. Pure fungal strains were grown by repeated reinoculation on fresh culture media.2.2. Identification of the fungal strains
The isolated fungal strains were identified as A. nidulans, A. flavipes and A. flavus using a molecular biological protocol by DNA amplification and sequencing of the ITS region as previously reported.18 The obtained data of sequencing were submitted to GenBank with the accession number OQ930448 for A. nidulans, OQ930542 for A. flavipes and OR120990 for A. flavus.2.3. Cultivation and extraction
Small scale fermentation of the fungal strains was performed on solid rice culture media (100 g rice in 110 mL distilled water, autoclaved for 20 min at 121 °C) in 1 L Erlenmeyer flask (3 flasks) for 30 days at 25 °C under static conditions. After incubation, the fungal cultures were extracted with ethyl acetate, filtered, and evaporated with a rotary evaporator to yield the ethyl acetate extracts for A. nidulans (350 mg), A. flavipes (400 mg) and A. flavus (500 mg). Similarly, the algae for C. officinalis was extracted and evaporated to yield 400 mg total extract. Aliquots (10 mg) of each extraction was dissolved in 1 ml of 50% methanol per water, centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 5 min and filtered through nylon syringe filters (0.22 μm) before subjected to further analysis.
000 rpm for 5 min and filtered through nylon syringe filters (0.22 μm) before subjected to further analysis.
2.4. Ultra-performance liquid chromatography (UPLC) analysis
The metabolites of the extracts were analyzed on a reversed phase C18 column (High Strength Silica (HSS) T3, 100 mm × 2.1 mm, 1.7 μm diameter particles, Waters™, Waters Corporation, Milford, MA 01757, USA), connected to ultra-performance liquid chromatography (LC) system (Waters™ Acquity UPLC system, Waters Corporation, Milford, MA 01757, USA).16 The injection volume was four μL and the flow rate was adjusted to 400 μL min−1. The mobile phases used for chromatographic separation were water containing 0.1% formic acid (A) and acetonitrile containing 0.1% formic acid (B). The following gradient was applied: 1 min 99% A, 13 min linear gradient from 99% A to 45% A, 14.5 min linear gradient from 45% A to 30% A, 15.5 min linear gradient from 30% A to 1% A. The gradient was hold at 1% A from 15.5 to 17 min, followed by linear gradient from 1% A to 99% A to 17.5 min. Finally, the column was re-equilibrated for 2.5 min at 99% A.2.5. High-resolution electrospray ionisation orbitrap mass spectrometry (HR-ESI-orbitrap-MS) analysis
The mass spectra were acquired, covering a mass range 100–1500 m/z, by orbitrap-type high resolution MS and MS/MS (Thermo Scientific™ Exactive™, Thermo Fisher, Bremen, Germany).19 Collision Induced Dissociation (CID) was obtained using a normalized collision energy of 35 eV. Tandem mass spectrometry (MS/MS) data were acquired by using the data-Independent acquisition in both in negative and positive ion modes. Instrument control, data acquisition and processing were performed using Xcalibur software package (Thermo Fischer Scientific, San Jose, CA, USA).2.6. LC-MS-based data processing and multivariate statistical analysis
LC-MS/MS data processing was performed using Mass Spectrometry-Data Independent Analysis (MS-DIAL) software.20 The following parameters were used: MS and MS/MS tolerance of 0.01 and 0.05 Da, respectively, retention time = 2–17 min, MS mass range = 50–1500 Da, minimum peak height = 1 × 103 amplitude and retention time tolerance of 0.25 min. Post MS-DIAL data processing, the GNPS export files were imported into the GNPS platform using the WinSCP server.21 The GNPS feature processing was achieved following specific parameters such as a fragment ion mass tolerance of m/z (0.25 Da), a minimum number of common fragment ions (5), and a minimum cosine score (0.7). Subsequently, a search of the bronze spectral library was conducted, with the top 10 hits per spectrum. Metabolomic data analysis and interpretation were performed using MetaboAnalyst.22 The resulting data matrix (.csv file) was directly imported to the Metaboanalyst 5 platform (https://www.metaboanalyst.ca/). The dataset was then pareto-scaled and log2 transformed to standardize variables and minimize redundancy. Subsequently, the data was subjected to different statistical analysis methods, including principal component analysis (PCA) and hierarchical cluster analysis (HCA), as unsupervised methods, next to Partial Least Squares Discriminant Analysis (PLS-DA) as a supervised method.3 Results and discussion
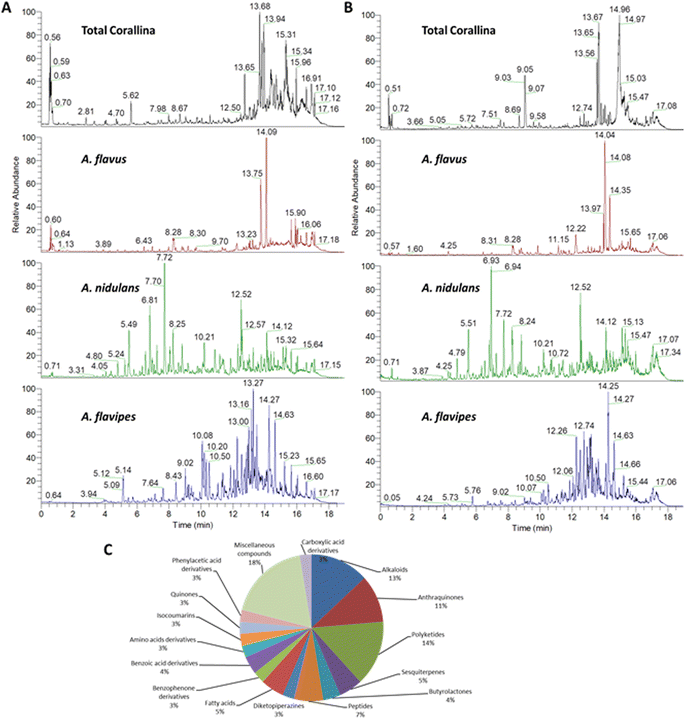
3.1. Chemical profiling of the red alga C. officinalis, and associated Aspergillus sp.
The chemical profiles of the ethyl acetate extracts from the red alga C. officinalis, and associated Aspergillus. species (A. flavipes, A. flavus and A. nidulans) were analyzed by LC-ESI-HRMS analysis which was achieved using alternating full scan (FS) and all ion fragmentation (AIF) modes in positive (+) and negative (−) modes. Representative chromatograms are shown in Fig. 1. In total 76 compounds were annotated based on retention times, accurate mass, fragmentation pattern using the available literature23–25 as well as the MS/MS databases and bioinformatics analysis using GNPS21 (Tables 1 and S1†). The annotated compounds included 11 polyketides, 8 anthraquinones, 12 alkaloids, 5 peptides, 4 sesquiterpenes, 3 butrylactone derivatives, 2 benzophenone derivatives, 4 fatty acids, 2 quinones, 3 amino acids, 7 carboxylic acids and 15 miscellaneous compounds (Fig. 1).| Peak no. | tR (min) | Annotated compound | Class | Relative abundancea | Ref. | |||
|---|---|---|---|---|---|---|---|---|
| Total Corallina | A. flavus | A. nidulans | A. flavipes | |||||
| a Symbols represent the relative abundance, (++++) for area ≥1 × 109, (+++) for area ≥1 × 107, (++) for area ≥1 × 105, (+) for area ≥1 × 103, (−) for not detected. | ||||||||
| 1 | 4.06 | Orsellinic acid | Benzoic acid derivatives | +++ | ++ | +++ | +++ | 9 |
| 2 | 4.14 | p-Hydroxyphenyl acetic acid | Phenylacetic acid derivatives | ++ | ++ | ++ | ++ | 45kra |
| 3 | 4.17 | 5-Hydroxyaverantin | Anthraquinones | − | ++ | ++ | + | GNPS |
| 4 | 5.09 | Terrein | Polyketides | ++ | ++ | ++ | +++ | GNPS |
| 5 | 5.13 | 3-Methylorsellinicacid | Benzoic acid derivatives | ++ | + | ++ | ++ | 46 |
| 6 | 5.20 | Kojic acid methyl ether | Gamma-pyrone derivatives | ++ | ++ | ++ | +++ | 47 |
| 7 | 5.33 | Insulicolide A | Sesquiterpenes | + | − | − | ++ | 48 |
| 8 | 5.45 | Orsellinaldehyde | Dihydroxybenzaldehydes | ++ | ++ | ++ | +++ | 49 |
| 9 | 5.63 | Sinapine | Alkaloids | ++ | +++ | ++ | ++ | GNPS |
| 10 | 6.18 | N-acetyl-L-leucine | Amino acids | +++ | +++ | +++ | +++ | GNPS |
| 11 | 6.43 | Penidiamide | Peptides | + | +++ | ++ | + | GNPS |
| 12 | 6.60 | Asperflavin | Anthraquinones | ++ | + | + | ++ | 50 |
| 13 | 6.78 | Phenylalanine | Amino acids | ++ | +++ | ++ | ++ | GNPS |
| 14 | 6.92 | Speradine D | Alkaloids | ++ | ++ | +++ | − | 51 |
| 15 | 6.92 | Cichorine | Alkaloids | ++ | ++ | ++++ | ++ | 52 |
| 16 | 7.71 | Ferulic acid | Cinnamic acid derivatives | ++ | ++ | ++++ | +++ | 53 |
| 17 | 7.71 | Nidulol | Furane derivatives | + | ++ | ++++ | ++ | 54 |
| 18 | 7.97 | 2-O-methyl-9-dehydroxyeurotinone | Anthraquinones | + | ++ | ++ | +++ | 55 |
| 19 | 8.36 | Asperfuranone | Polyketides | ++ | ++ | ++ | ++ | 56 |
| 20 | 8.40 | 4-Hydroxyphenylpyruvic acid | Phenylpyruvic acid derivatives | +++ | ++ | ++ | +++ | GNPS |
| 21 | 8.66 | Homogenentisic acid | Phenylacetic acid derivatives | + | ++ | ++ | ++ | GNPS |
| 22 | 8.69 | Alternariol monomethyl ether | Isocoumarins | − | + | +++ | ++ | 57 |
| 23 | 8.96 | Daidzein | Isoflavonoids | +++ | ++ | + | + | GNPS |
| 24 | 9.02 | Sterigmatocystin hemiacetal | Xanthone polyketides | + | ++ | ++ | ++++ | 58 |
| 25 | 9.20 | Erythroglaucin | Quinones | − | + | +++ | + | 59 |
| 26 | 9.22 | 1-Hydroxy-6-methyl-8-hydroxymethylxanthone | Xanthone polyketides | + | − | +++ | + | GNPS |
| 27 | 9.32 | 2,3,6,8,9-Pentahydroxy-1-oxo-3-(2-oxopropyl)-1,2,3,4-tetra-hydroanthracene-2-carboxylic acid | Carboxylic acid derivatives | − | ++ | ++ | +++ | GNPS |
| 28 | 9.39 | Sterigmatocystin | Xanthone polyketides | + | ++ | + | ++++ | 58, 60 and 61 |
| 29 | 9.45 | Atrochryson carboxylic acid | Carboxylic acid derivatives | + | ++ | ++ | ++++ | GNPS |
| 30 | 9.70 | Aspergilloid C | Sesquiterpenes | ++ | +++ | ++ | + | 62 |
| 31 | 9.77 | Violaceol I | Diphenyl- ethers | + | + | ++++ | ++ | GNPS |
| 32 | 9.80 | Versiquinazoline E | Alkaloids | ++ | − | ++ | + | 63 |
| 33 | 9.93 | Aspulvinone E | Butenolides | + | + | ++ | +++ | GNPS |
| 34 | 10.02 | Aspergiterpenoid A | Sesquiterpenes | ++ | ++ | ++ | ++ | 64 |
| 35 | 10.08 | Epicoccolide B | Polyketides | − | ++ | ++ | ++++ | 65 |
| 36 | 10.20 | 5-Methoxydihydrosterigmatocystin | Xanthone polyketides | ++ | ++ | ++ | ++++ | 58 |
| 37 | 10.87 | Alternariol | Isocoumarinss | − | ++ | ++++ | ++ | 57 |
| 38 | 11.33 | Curvularin | Polyketide | ++ | +++ | +++ | ++++ | GNPS |
| 39 | 11.96 | Aspoquinoline C | Alkaloids | + | + | + | ++ | 66 |
| 40 | 12.12 | Cytochalasin Z17 | Alkaloids | + | + | + | +++ | 67 |
| 41 | 12.35 | Dethiosecoemestrin | Diketopiperazines | − | − | +++ | − | GNPS |
| 42 | 12.58 | Emestrin | Alkaloids | − | + | +++ | − | 68 |
| 43 | 12.67 | Cytochalasin z8 | Alkaloids | + | + | + | +++ | 67 |
| 44 | 12.70 | Fellutamide A | Peptides | + | − | ++ | +++ | 69 |
| 45 | 12.74 | Acyl-hemiacetal sterigmatocystin | Xanthone polyketides | − | + | +++ | − | 58 |
| 46 | 12.80 | Butyrolactone I | Butyrolactone derivatives | ++ | ++ | +++ | ++++ | GNPS |
| 47 | 13.27 | Butyrolactone VII | Butyrolactone derivatives | + | ++ | ++ | ++++ | GNPS |
| 48 | 13.27 | 2-O-methylbutyrolactone II | Butyrolactone derivatives | − | + | + | +++ | GNPS |
| 49 | 13.32 | Aspergillide E | Macrolides | + | + | +++ | ++ | 70 |
| 50 | 13.40 | Emericellamide C | Peptides | + | + | +++ | − | 71 |
| 51 | 13.45 | Fellutamide D | Peptides | + | ++ | ++ | +++ | 72 |
| 52 | 13.49 | Emodin | Anthraquinones | ++ | ++ | +++ | ++ | GNPS |
| 53 | 13.49 | Eurotinone | Anthraquinoness | − | ++ | ++ | ++++ | 73 |
| 54 | 13.49 | Monodictyphenone | Polyketides | − | ++ | ++ | ++++ | 74 |
| 55 | 13.72 | Cyclopiazonic acid | Alkaloids | + | +++ | +++ | + | 75 |
| 56 | 13.76 | Sydonic acid | Sesquiterpenes | ++++ | ++++ | ++++ | ++++ | 76 |
| 57 | 13.92 | Emericellamide A | Polyketides | ++ | + | +++ | − | 71 |
| 58 | 13.99 | Asperphenamate | Phenylalanine derivatives | + | ++ | ++ | +++ | 77 |
| 59 | 13.99 | 2-(((2-Ethylhexyl)oxy)carbonyl)benzoic acid | Benzoic acid derivatives | +++ | +++ | ++ | +++ | GNPS |
| 60 | 14.08 | 2-Methyleurotinone | Anthraquinones | ++ | ++ | ++ | ++++ | 78 |
| 61 | 14.11 | Emericellamide E | Polyketides | +++ | − | ++++ | ++ | 71 |
| 62 | 14.15 | 1-Hexadecanoyl glycerophosphocholine | Lipids | +++ | +++ | ++ | ++ | GNPS |
| 63 | 14.22 | 8-Hydroxy-9,12-octadecadienoic acid | Fatty acids | +++ | +++ | +++ | +++ | GNPS |
| 64 | 14.25 | 6,8-O-dimethylaverantin | Anthraquinones | + | ++ | ++ | ++++ | GNPS |
| 65 | 14.25 | 5,2′-Dihydroxy-3,7,8-trimethoxyflavone | Flavonoids | − | − | + | +++ | GNPS |
| 66 | 14.26 | Hormonemate F | Polyketides | ++ | ++ | ++ | ++++ | GNPS |
| 67 | 14.35 | Flavoglaucin | Quinones | +++ | ++ | ++ | + | 59 |
| 68 | 14.37 | Scopularide E | Peptides | ++ | + | +++ | − | GNPS |
| 69 | 14.56 | Aversin | Anthraquinones | − | ++ | ++++ | ++ | 61 |
| 70 | 14.5 | 7-Hydroxy-8,14-dimethyl-9-hexadecanoic acid | Fatty acids | +++ | +++ | +++ | +++ | 79 |
| 71 | 14.69 | Isoechinulins C | Diketopiperazines | + | ++ | ++ | ++ | 80 |
| 72 | 14.91 | Wentinoid A | Diterpenoids | ++ | ++ | ++ | ++ | 81 |
| 73 | 15.62 | Glycerol linoleate | Fatty acids | +++ | +++ | +++ | +++ | 47 |
| 74 | 15.65 | Arugosin G | Benzophenone derivatives | + | + | + | +++ | 82 |
| 75 | 15.6 | Linoleic acid | Fatty acids | ++ | ++ | ++ | ++ | 47 |
| 76 | 16.22 | Versiquinazoline (J) | Alkaloids | ++ | +++ | ++ | ++ | 63 |
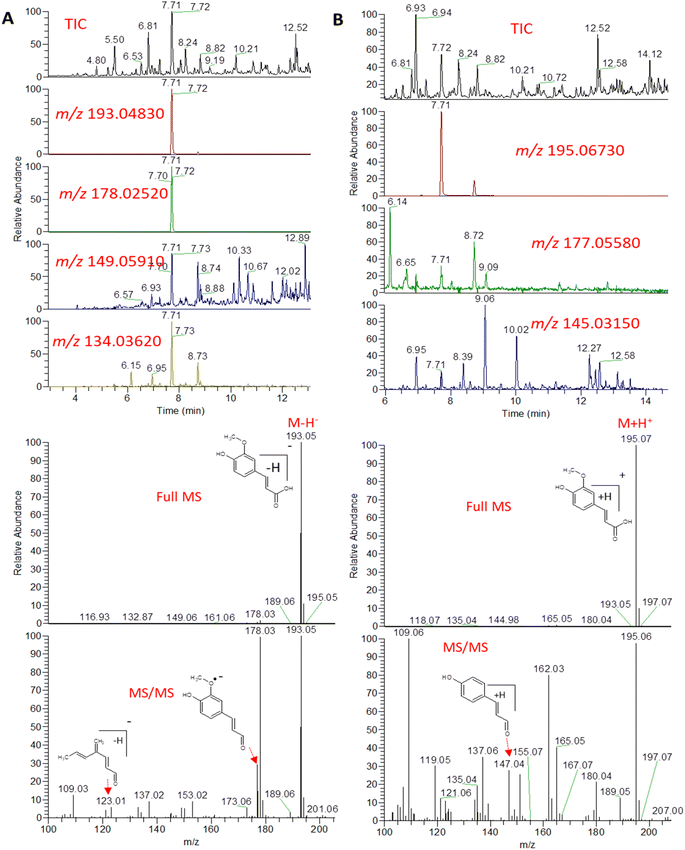
An example description of the workflow used for annotation of metabolites is described here showing ferulic acid. The identification of ferulic acid was achieved in both negative and positive electrospray ionization modes by the MS2 analysis (Fig. 2). In negative ionization mode, a molecular ion peak was observed at m/z 193.04830 equivalent to the deprotonated adduct [M − H]− with the chemical formula C10H9O4−. Characteristic product ion fragments were observed at m/z 178.02520 (loss of CH3 from the precursor ion), m/z 176.0450 (loss of OH from the precursor ion), m/z 149.05910 (loss of CO2 from the precursor ion) and m/z 134.03620 (loss of CH3 with CO2 from the precursor ion).26,27 In positive ionization mode, a molecular ion peak was observed at m/z 195.06730 equivalent to the protonated adduct [M + H]+with the chemical formula C10H11O4+. Characteristic product ion fragments were observed at m/z 180.04380 (loss of CH3 from the precursor ion), m/z 177.05580 (loss of water from the precursor ion), [M + H–H2O]+, and m/z 147.04610 (loss of OCH3 and OH from the precursor ion).28 Other example description for the annotated metabolites are described in the detailed classification in the following sections.
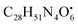 with [M + H]+ m/z 539.3798 followed by continuous cleavage of different amide bonds and loss of different amino acid fragments occurred to this moiety till it reaches the last fragment.30 Polyketides were the major secondary metabolites exhibited by the data extracted from LC-MS, where, terrein (4), epicoccolide B (35), curvularin (38), emericellamide A (57), emericellamide E (61) and hormonemate F (66) were detected at different retention times in both negative and positive ionization modes. Compounds 4 was identified as terrain,24 it showed [M − H]− at m/z 153.05495 with characteristic fragments through the loss of C2H6 unit ([M − H]−–30)123.0442 and loss of H2O ([M − H]−–18)135.0442. Polyketides were the major secondary metabolites exhibited by the data extracted from LC-MS, with total number of 11 where, 6 of which were found in the host algal sample and its three endosymbiotic fungi such as terrein (4) (tR 5.09 min), sterigmatocystin hemiacetal (24) (tR 9.02 min), sterigmatocystin (28) (tR 9.39 min), 5-methoxydihydrosterigmatocystin (36) (tR 10.20 min), curvularin (38) (tR 11.33 min) and hormonemate F (66) (tR 14.26 min). Epicoccolide B (35) (tR 10.08 min), was detected in all three Aspergillus extracts and totally absent in the host algal extract. Emericellamide E (61) (tR 14.11 min) and 1-hydroxy-6-methyl-8-hydroxymethylxanthone (26) (tR 9.22 min), were not detected in Aspergillus flavus extract, while they were detected in the other three extracts. Acyl-hemiacetal sterigmatocystin (45) (tR 12.74 min), was not identified in the host algal extract and its endosymbiotic A. flavipes, while was detected in A. flavus and A. nidulans extracts. Emericellamide A (57) (tR 13.92 min), was not detected in A. flavipes extract, while was detected in the remaining three sample extracts.
with [M + H]+ m/z 539.3798 followed by continuous cleavage of different amide bonds and loss of different amino acid fragments occurred to this moiety till it reaches the last fragment.30 Polyketides were the major secondary metabolites exhibited by the data extracted from LC-MS, where, terrein (4), epicoccolide B (35), curvularin (38), emericellamide A (57), emericellamide E (61) and hormonemate F (66) were detected at different retention times in both negative and positive ionization modes. Compounds 4 was identified as terrain,24 it showed [M − H]− at m/z 153.05495 with characteristic fragments through the loss of C2H6 unit ([M − H]−–30)123.0442 and loss of H2O ([M − H]−–18)135.0442. Polyketides were the major secondary metabolites exhibited by the data extracted from LC-MS, with total number of 11 where, 6 of which were found in the host algal sample and its three endosymbiotic fungi such as terrein (4) (tR 5.09 min), sterigmatocystin hemiacetal (24) (tR 9.02 min), sterigmatocystin (28) (tR 9.39 min), 5-methoxydihydrosterigmatocystin (36) (tR 10.20 min), curvularin (38) (tR 11.33 min) and hormonemate F (66) (tR 14.26 min). Epicoccolide B (35) (tR 10.08 min), was detected in all three Aspergillus extracts and totally absent in the host algal extract. Emericellamide E (61) (tR 14.11 min) and 1-hydroxy-6-methyl-8-hydroxymethylxanthone (26) (tR 9.22 min), were not detected in Aspergillus flavus extract, while they were detected in the other three extracts. Acyl-hemiacetal sterigmatocystin (45) (tR 12.74 min), was not identified in the host algal extract and its endosymbiotic A. flavipes, while was detected in A. flavus and A. nidulans extracts. Emericellamide A (57) (tR 13.92 min), was not detected in A. flavipes extract, while was detected in the remaining three sample extracts.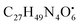 with [M + H]+ m/z 525.3638 witnessed continuous cleavage of different amide bonds and loss of different amino acid fragments to produce different product ions including 454.3265, 341.2424, and 244.1647.30 Emericellamide C (50) (tR 13.44 min) and scopularide E (68) (tR 14.37 min) (Fig. S5†) were not detected only in A. flavipes extract, while detected in the other three extracts. Fellutamide A (44) (tR 12.70 min), was detected in A. nidulans extract, A. flavipes extract and the host algal extract, whereas, not detected in A. flavus extract.
with [M + H]+ m/z 525.3638 witnessed continuous cleavage of different amide bonds and loss of different amino acid fragments to produce different product ions including 454.3265, 341.2424, and 244.1647.30 Emericellamide C (50) (tR 13.44 min) and scopularide E (68) (tR 14.37 min) (Fig. S5†) were not detected only in A. flavipes extract, while detected in the other three extracts. Fellutamide A (44) (tR 12.70 min), was detected in A. nidulans extract, A. flavipes extract and the host algal extract, whereas, not detected in A. flavus extract.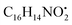 from the precursor ion), and m/z 238.1213 (loss of
from the precursor ion), and m/z 238.1213 (loss of 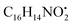 from the precursor ion).37 The identified amino acids, N-acetyl-L-leucine (10), (tR 6.18 min), phenylalanine (13), (tR 6.78 min) and asperphenamate (58) (tR 13.99 min) were observed in A. flavus, A. nidulans, A. flavipes and the algal extracts. Two identified carboxylic acid derivatives, 2, 3, 6, 8, 9-pentahydroxy-1-oxo-3-(2-oxopropyl)-1,2,3,4-tetra-hydroanthracene-2-carboxylic acid (27), (tR 9.32 min), was only not observed in the host algal extract. On the other hand, atrochryson carboxylic acid (29) (tR 9.45 min), was observed in all extracts. All three identified benzoic acid derivatives were observed in host algal extract and their three endosymbiotic fungal extracts, such as orsellinic acid (1) (tR 4.06 min), 3-methylorsellinic acid (5) (tR 5.13 min) and 2-(((2-ethylhexyl)oxy)carbonyl)benzoic acid (59) (tR 13.99 min) (Fig. S6†). Orsellinic acid (1) (benzoic acid derivative) showed molecular ion [M − H]− peak at m/z 167.03418 with the chemical formula C8H7O4− in negative electrospray ionization mode. Characteristic product ion fragments were observed at m/z 151.0393 (loss of OH from the precursor ion), and m/z 123.0443 (loss of carboxylic group from the precursor ion).38,39 Also two identified phenyl acetic acid derivatives were observed in all examined extracts, including p-hydroxyphenyl acetic acid (2) (tR 4.14 min) and homogenentisic acid (21) (tR 8.66 min).
from the precursor ion).37 The identified amino acids, N-acetyl-L-leucine (10), (tR 6.18 min), phenylalanine (13), (tR 6.78 min) and asperphenamate (58) (tR 13.99 min) were observed in A. flavus, A. nidulans, A. flavipes and the algal extracts. Two identified carboxylic acid derivatives, 2, 3, 6, 8, 9-pentahydroxy-1-oxo-3-(2-oxopropyl)-1,2,3,4-tetra-hydroanthracene-2-carboxylic acid (27), (tR 9.32 min), was only not observed in the host algal extract. On the other hand, atrochryson carboxylic acid (29) (tR 9.45 min), was observed in all extracts. All three identified benzoic acid derivatives were observed in host algal extract and their three endosymbiotic fungal extracts, such as orsellinic acid (1) (tR 4.06 min), 3-methylorsellinic acid (5) (tR 5.13 min) and 2-(((2-ethylhexyl)oxy)carbonyl)benzoic acid (59) (tR 13.99 min) (Fig. S6†). Orsellinic acid (1) (benzoic acid derivative) showed molecular ion [M − H]− peak at m/z 167.03418 with the chemical formula C8H7O4− in negative electrospray ionization mode. Characteristic product ion fragments were observed at m/z 151.0393 (loss of OH from the precursor ion), and m/z 123.0443 (loss of carboxylic group from the precursor ion).38,39 Also two identified phenyl acetic acid derivatives were observed in all examined extracts, including p-hydroxyphenyl acetic acid (2) (tR 4.14 min) and homogenentisic acid (21) (tR 8.66 min).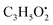 from the precursor ion), and m/z 175.1107 (loss of
from the precursor ion), and m/z 175.1107 (loss of 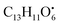 from the precursor ion)40 (Fig. S7†). Three butrylactone derivatives were identified in algal extract and there endosymbiotic fungal extracts. Butyrolactone I (46) (tR 12.80 min) (Fig. S8†) and butyrolactone VII (47) (tR 13.27) were observed in A. flavus, A. nidulans, A. flavipes and algal extracts. 2-O-methylbutyrolactone II (48) (tR 13.27 min) was totally not observed in the host algal extract, whereas observed in all three endosymbiotic fungal extracts (Fig. S9†).
from the precursor ion)40 (Fig. S7†). Three butrylactone derivatives were identified in algal extract and there endosymbiotic fungal extracts. Butyrolactone I (46) (tR 12.80 min) (Fig. S8†) and butyrolactone VII (47) (tR 13.27) were observed in A. flavus, A. nidulans, A. flavipes and algal extracts. 2-O-methylbutyrolactone II (48) (tR 13.27 min) was totally not observed in the host algal extract, whereas observed in all three endosymbiotic fungal extracts (Fig. S9†).Four sesquiterpenes were identified, three of them including aspergilloid C (30) (tR 9.70 min), aspergiterpenoid A (34) (tR 10.02 min) and sydonic acid (56) (tR 13.76 min) were detected in A. flavus, A. nidulans, A. flavipes and the algal extracts. On the other hand, insulicolide A (7) (tR 5.33 min) was not detected in A. flavus and A. nidulans extracts, while detected in A. flavipes and algal extracts.
Two quinone derivatives were identified. Erythroglaucin (25) (tR 9.20 min), was only not detected in the host algal extract, whereas detected in A. flavus, A. nidulans and A. flavipes extracts. On the other hand, flavoglaucin (67) (tR 14.35 min) was detected in all four sample extracts.
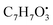 and
and 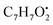 , respectively. Then the loss of CH3 from
, respectively. Then the loss of CH3 from 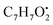 produced product ion at m/z 109.0343 with chemical formula
produced product ion at m/z 109.0343 with chemical formula 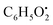 (Fig. S12†). Representative chemical structures of the annotated metabolites are shown in Fig. 3.
(Fig. S12†). Representative chemical structures of the annotated metabolites are shown in Fig. 3.
The comprehensive metabolomic study of the three different Aspergillus sp. revealed that polyketides, anthraquinoes and alkaloids are the major classes of the identified secondary metabolites. These results comply with the reported data in multiple reviews comprehensively describing hundreds of compounds belonging to the three mentioned classes of secondary metabolites from marine derived Aspergillus sp.42–44 Moreover, the presence of common compounds in the host algae C. officinalis as well as the three isolated Asprgillus endosymbionts unveils the presence mutualistic symbiotic relationship emphasizing the ecological significance the fungal endophytes, where they can provide bioactive metabolites as chemical defense strategy to the host organism and in turn secures a nutrient rich system to flourish.
3.2. Multivariate analysis of the LC-ESI-HRMS data
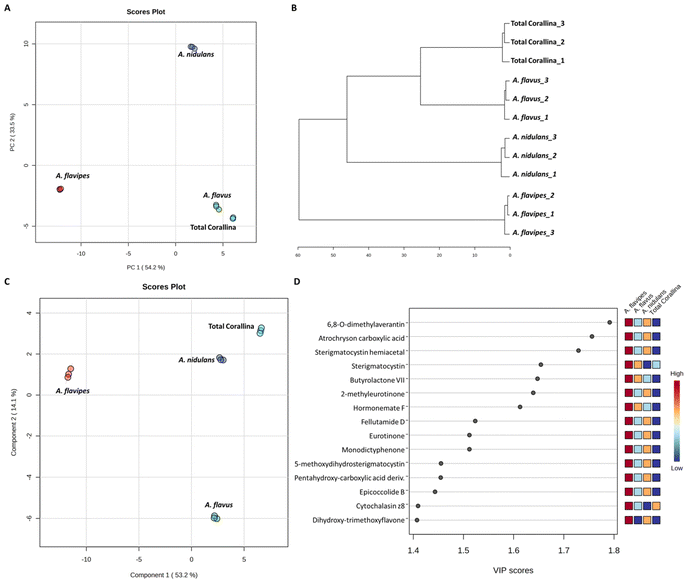
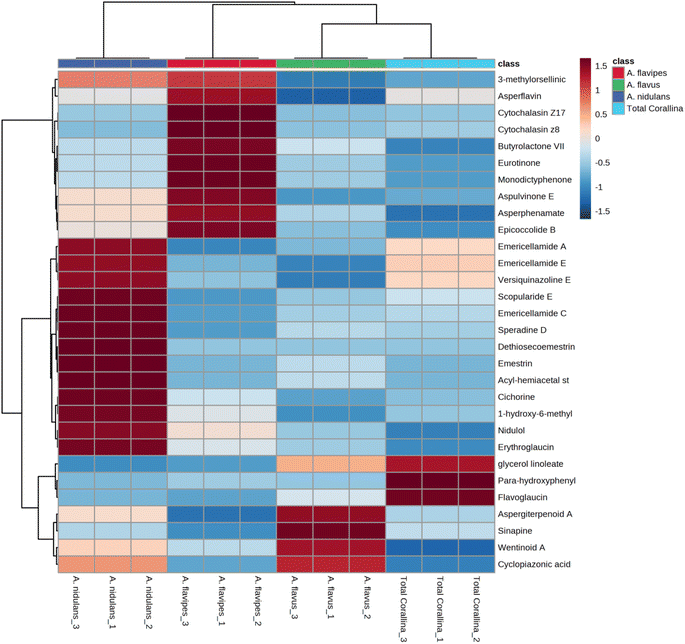
Non-targeted metabolomics approaches based on liquid chromatography-high resolution mass spectrometry (LC-HRMS) have been widely used for specific discrimination of different biological samples particularly from plants and fungi.83,84 Chemometrics tools such as principal component analysis (PCA), hierarchical clustering analysis (HCA) and partial least squares discriminant analysis (PLS-DA) provide a wealth of techniques for exploratory analysis and classification of multivariate data. The LC-MS-MS data of the total extract as well as the associated fungal extracts were subjected to multivariate analysis to evaluate their chemical diversity. Unsupervised classifications including principal component analysis (PCA) and hierarchical clustering analysis (HCA) were used to simplify a large amount of data without compromising the main information (Fig. 4A and B). PCA analysis revealed clustering of A. flavipes from C. officinalis and other associated endosymbiotic Aspergillus fungi (A. flavus and A. nidulans) among the first principal component (PC1) which accounted for 54.2% of the variance (Fig. 4A). On the other hand, the second principal component (PC2), which accounted for 33.5% of the variance, revealed a clear separation of A. nidulans from A. flavus and Corallina officinalis which were clustered together. HCA analysis revealed two main clusters; the first cluster included A. flavipes while the second cluster revealed separation of A. nidulans from A. flavus and C. officinalis, consistent with PCA results (Fig. 4B). Heatmap of the top 30 metabolites differentially changing between C. officinalis and associated endosymbiotic Aspergillus fungi is depicted in Fig. 5.Supervised classifications, on the other hand, including partial least squares discriminant analysis (PLS-DA) was used as multivariate dimensionality-reduction tool for discriminative variable selection (Fig. 4C). PLS-DA achieved the effective discrimination between the tested groups. A Variable Importance for Projection (VIP) score is a measure of a variable importance in the PLS-DA model. It summarizes the quantitative contribution of a variable to the model. The candidate metabolites with variable importance in the PLS-DA (VIP) responsible for the discrimination and with scores ≥1 were considered important in the PLS-DA model (Fig. 4D). Anthraquinones such as eurotinone, 2-methyleurotinone and 6,8-O-dimethylaverantin were found to be discriminatory metabolites for A. flavipes. Additionally, polyketides such as epicoccolide B, monodictyphenone and hormonemate F as well as other xanthones derivatives such as sterigmatocystin, sterigmatocystin hemiacetal and 5-methoxydihydrosterigmatocystin influence the metabolic discrimination of A. flavipes from Corallina officinalis and other associated endosymbiotic Aspergillus fungi (A. flavus and A. nidulans). Further, alkaloid derivatives such as sinapine, cyclopiazonic acid and versiquinazoline (J) together with fatty, benzoic and amino acids derivatives (e.g. N-acetyl-leucine, phenylalanine, 2-(((2-Ethylhexyl)oxy)carbonyl)benzoic acid, 8-Hydroxy-9,12-octadecadienoic acid, 7-hydroxy-8,14-dimethyl-9-hexadecanoic acid and glycerol linoleate) dominated A. flavus. Additionally, penidiamide, curvularin and aspergilloid C were found to discriminate this fungal species. Moreover, metabolites from diverse chemical classes such as cichorine, aversin, violaceol I, nidulol, alternariol and emericellamide E were found to be abundant in A. nidulans, discriminating it from C. officinalis and other associated endosymbiotic Aspergillus fungi. C. officinalis was found to accumulate flavoglaucin, emericellamide E, daidzein, orsellinic acid next to some fatty acids and organic acid derivatives.
4 Conclusions
In this study, we investigated the metabolic profile of C. officinalis and the three isolated endophytes A. nidulans, A. favipes, and A. flavus using LC-MS/MS. C. officinalis and its endosymbiotic fungi demonstrate a valuable source of various metabolites including alkaloids, polyketides, sesquiterpenes, butyrolactones, peptides, fatty acids, isocoumarins, and quinones. Based on LC-MS/MS, polyketides represented the highest percentage (14%) among other metabolite classes of C. officinalis and related endosymbiotic Aspergillus fungi, followed by alkaloids (13%), anthraquinones (11%), peptides (7%), fatty acids and sesquiterpenes (5% for each one), in addition to small amounts of other classes such as butyrolactones, diketopiperazines, isocoumarins, amino acid derivatives, benzoic acid derivatives, and other miscellaneous classes. All metabolites that were found in C. officinalis were also exist in its endophytes with different proportions. On the contrary, some metabolites have been identified in endosymbiotic fungi and did not exist in their host. The chemical profiling of many groups of algae and their endophytes is still unrevealed and needs to be explored. These findings will be of interest in pharmaceutical applications for drug discovery from natural resources. Further studies also should be pushed toward investigating the correlations between these endophytes and their hosts while predicting the viable applications and pharmacological effects.Author contributions
Sherif M. Shama: methodology, investigation, data curation, and writing – original draft, Ahmed M. Elissawy: writing – original draft, writing – review & editing, and supervision Mohamed A. Salem: methodology, data curation, formal analysis, writing – original draft, writing – review & editing, and supervision, Fadia S. Youssef: writing – review & editing and supervision, Mohamed S. Elnaggar: methodology, Hesham R. El-Seedi: formal analysis, writing – review & editing, Shaden A. M. Khalifa: formal analysis, writing – review & editing, Khaled Briki: writing – review, Dalia Ibrahim Hamdan: writing – review & editing and supervision, Abdel Nasser B. Singab: conceptualization, writing – review & editing, and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. N. S. & A. E. would like to acknowledge the Science, Technology and Innovation Fund Authority (STIFA) for funding through project number 46667 under the title “Sustainability of lab. Capacities of Center of Drug Discovery Research and Development”.References
- M. Rashmi, J. Kushveer and V. Sarma, Endophytes and Secondary Metabolites, Springer International Publishing, Cham, 2019, pp. 491–526 Search PubMed.
- C. Jimenez, ACS Med. Chem. Lett., 2018, 9, 959–961 CrossRef CAS PubMed.
- K. Sahayaraj, presented in part at the National Conference on Conservation and Sustainable Utilization of Marine Resources, Tamil Nadu, India, 2015, pp. 22–23.
- Y. Yang, D. Liu, J. Wu, Y. Chen and S. Wang, Int. J. Biol. Macromol., 2011, 49, 1031–1037 CrossRef CAS PubMed.
- A. Allmendinger, J. Spavieri, M. Kaiser, R. Casey, S. Hingley-Wilson, A. Lalvani, M. Guiry, G. Blunden and D. Tasdemir, Phytother. Res., 2010, 24, 1099–1103 CrossRef PubMed.
- R. F. A. Abdelhameed, S. S. Elhady, A. O. Noor, D. M. Almasri, A. A. Bagalagel, G. T. Maatooq, A. I. M. Khedr and K. Yamada, Metabolites, 2019, 9, 273 CrossRef CAS PubMed.
- R. R. Sun, F. P. Miao, J. Zhang, G. Wang, X. L. Yin and N. Y. Ji, Magn. Reson. Chem., 2013, 51, 65–68 CrossRef CAS PubMed.
- A. N. B. Singab, N. M. Mostafa, Y. A. Elkhawas, E. Al-Sayed, M. M. Bishr, A. M. Elissawy, M. S. Elnaggar, I. M. Fawzy, O. M. Salama, Y. H. Tsai and F. R. Chang, Mar. Drugs, 2022, 20, 331 CrossRef CAS PubMed.
- M. V. Nguyen, J. W. Han, H. Kim and G. J. Choi, ACS Omega, 2022, 7, 33273–33279 CrossRef CAS PubMed.
- J. Romsdahl and C. C. C. Wang, Medchemcomm, 2019, 10, 840–866 RSC.
- S. S. El-Hawary, A. S. Moawad, H. S. Bahr, U. R. Abdelmohsen and R. Mohammed, RSC Adv., 2020, 10, 22058–22079 RSC.
- M. S. Elnaggar, A. M. Elissawy, F. S. Youssef, M. Kicsak, T. Kurtan, A. N. B. Singab and R. Kalscheuer, RSC Adv., 2023, 13, 16480–16487 RSC.
- K. Amr, N. Ibrahim, A. M. Elissawy and A. N. B. Singab, Fungal Biol. Biotechnol., 2023, 10, 6 CrossRef PubMed.
- A. M. Elissawy, S. S. Ebada, M. L. Ashour, M. El-Neketi, W. Ebrahim and A. B. Singab, Phytochem. Lett., 2019, 29, 1–5 CrossRef CAS.
- A. F. Tawfike, C. Viegelmann and R. Edrada-Ebel, Methods Mol. Biol., 2013, 201(1055), 227–244 CrossRef PubMed.
- Y. Hou, D. Braun, C. Michel, J. Klassen, N. Adnani, T. Wyche and T. Bugni, Anal. Chem., 2012, 84(10), 4277–4283 CrossRef CAS PubMed.
- M. Tangerina, L. Furtado, V. Leite, A. Bauermeister, K. Velasco-Alzate, P. Jimenez, L. Garrido, G. Padilla, N. Lopes, L. Costa-Lotufo and M. Ferreira, PLoS One, 2020, 15(12), e0244385 CrossRef CAS PubMed.
- J. Kjer, A. Debbab, A. H. Aly and P. Proksch, Nat. Protoc., 2010, 5, 479–490 CrossRef CAS PubMed.
- M. A. Salem, M. M. Salama, S. M. Ezzat and Y. A. Hashem, Sci. Rep., 2022, 12, 20168 CrossRef CAS PubMed.
- H. Tsugawa, T. Cajka, T. Kind, Y. Ma, B. Higgins, K. Ikeda, M. Kanazawa, J. VanderGheynst, O. Fiehn and M. Arita, Nat. Methods, 2015, 12, 523–526 CrossRef CAS PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W. T. Liu, M. Crusemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderon, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C. C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C. C. Liaw, Y. L. Yang, H. U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O’Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. F. Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodriguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P. M. Allard, P. Phapale, L. F. Nothias, T. Alexandrov, M. Litaudon, J. L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D. T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Muller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. O. Palsson, K. Pogliano, R. G. Linington, M. Gutierrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- Z. Pang, J. Chong, G. Zhou, D. A. de Lima Morais, L. Chang, M. Barrette, C. Gauthier, P. E. Jacques, S. Li and J. Xia, Nucleic Acids Res., 2021, 49, W388–W396 CrossRef CAS PubMed.
- O. M. Khattab, D. M. El-Kersh, S. A. M. Khalifa, N. Yosri, H. R. El-Seedi and M. A. Farag, Plants, 2023, 12, 2078 CrossRef CAS PubMed.
- M. I. G. El-Din, N. M. Fahmy, F. L. Wu, M. M. Salem, O. M. Khattab, H. R. El-Seedi, M. Korinek, T. L. Hwang, A. K. Osman, M. El-Shazly and S. Fayez, Plants, 2022, 11, 1699 CrossRef CAS PubMed.
- I. M. El-Garawani, S. M. El-Sabbagh, N. H. Abbas, H. S. Ahmed, O. A. Eissa, D. M. Abo-Atya, S. A. M. Khalifa and H. R. El-Seedi, Sci. Rep., 2020, 10, 14076 CrossRef PubMed.
- Y. Hong, Z. Wang, C. J. Barrow, F. R. Dunshea and H. A. R. Suleria, Antioxidants, 2021, 10, 234 CrossRef CAS PubMed.
- K. Kramberger, D. Barlic-Maganja, D. Bandelj, A. Baruca Arbeiter, K. Peeters, A. Miklavcic Visnjevec and Z. Jenko Praznikar, Metabolites, 2020, 10, 403 CrossRef CAS PubMed.
- S. K. T. Seraglio, A. C. Valese, H. Daguer, G. Bergamo, M. S. Azevedo, L. V. Gonzaga, R. Fett and A. C. O. Costa, Food Res. Int., 2016, 87, 60–67 CrossRef CAS PubMed.
- N. K. Mund and E. Cellarova, Biotechnol. Adv., 2023, 63, 108104 CrossRef CAS PubMed.
- K. F. Nielsen and T. O. Larsen, Front. Microbiol., 2015, 6, 71 Search PubMed.
- M. Masi and A. Evidente, Toxins, 2020, 12, 714 CrossRef CAS PubMed.
- H. Li, C. Liang, Q. Chen and Z. Yang, Med. Hypotheses, 2011, 77, 1105–1107 CrossRef CAS PubMed.
- P. C. Rida, D. LiVecche, A. Ogden, J. Zhou and R. Aneja, Med. Res. Rev., 2015, 35, 1072–1096 CrossRef CAS PubMed.
- A. Pirillo and A. L. Catapano, Atherosclerosis, 2015, 243, 449–461 CrossRef CAS PubMed.
- J. Oszmiański, J. Kolniak-Ostek and A. Wojdyło, Eur. Food Res. Technol., 2013, 236, 699–706 CrossRef.
- L. Cui, Y. Liu, M. Liu, M. Ren, A. F. Ahmed and W. Kang, J. Future Foods, 2022, 2, 253–260 CrossRef.
- S. Kildgaard, M. Mansson, I. Dosen, A. Klitgaard, J. C. Frisvad, T. O. Larsen and K. F. Nielsen, Mar. Drugs, 2014, 12, 3681–3705 CrossRef PubMed.
- S. G. Musharraf, N. Kanwal, V. M. Thadhani and M. I. Choudhary, Anal. Methods UK, 2015, 7, 6066–6076 RSC.
- Y. Jin, Y. Ma, W. Xie, L. Hou, H. Xu, K. Zhang, L. Zhang and Y. Du, RSC Adv., 2018, 8, 15487–15500 RSC.
- L. Wu, C. L. Xie, X. W. Yang and G. Chen, Mar. Drugs, 2021, 20, 11 CrossRef PubMed.
- B. P. Lau, P. M. Scott, D. A. Lewis, S. R. Kanhere, C. Cleroux and V. A. Roscoe, J. Chromatogr. A, 2003, 998, 119–131 CrossRef CAS PubMed.
- F. youssef, E. Alshammari and M. Ashour, Int. J. Mol. Sci., 2021, 22, 1866 CrossRef CAS PubMed.
- S. Ghoran, F. Taktaz, S. Ayatollahi and A. Kijjoa, Mar. Drugs, 2022, 20, 474 CrossRef PubMed.
- X. Bai, Y. Sheng, Z. Tang, J. Pan, S. Wang, B. Tang, T. Zhou, L. Shi and H. Zhang, J. Fungi, 2023, 9, 261 CrossRef CAS PubMed.
- W. J. Zuo, P. F. Jin, W. H. Dong, H. F. Dai and W. L. Mei, Chin. J. Nat. Med., 2014, 12, 151–153 CAS.
- G. D. Chen, D. Hu, M. J. Huang, J. Tang, X. X. Wang, J. Zou, J. Xie, W. G. Zhang, L. D. Guo, X. S. Yao, I. Abe and H. Gao, Chem. Commun., 2020, 56, 4607–4610 RSC.
- E. M. Eliwa, M. M. El-Metwally, A. H. Halawa, A. M. El-Agrody, A. H. Bedair and M. Shaaban, J. At. Mol., 2017, 7, 1045 CAS.
- L. Rahbaek, C. Christophersen, J. Frisvad, H. S. Bengaard, S. Larsen and B. R. Rassing, J. Nat. Prod., 1997, 60, 811–813 CrossRef.
- H. Han, C. Yu, J. Qi, P. Wang, P. Zhao, W. Gong, C. Xie, X. Xia and C. Liu, Microb. Cell Fact., 2023, 22, 60 CrossRef CAS PubMed.
- X. Yang, M. C. Kang, Y. Li, E. A. Kim, S. M. Kang and Y. J. Jeon, Molecules, 2017, 22, 1823 CrossRef PubMed.
- V. Uka, G. G. Moore, N. Arroyo-Manzanares, D. Nebija, S. De Saeger and J. Diana Di Mavungu, Toxins, 2017, 9, 35 CrossRef PubMed.
- J. F. Sanchez, R. Entwistle, D. Corcoran, B. R. Oakley and C. C. Wang, Medchemcomm, 2012, 3, 997–1002 RSC.
- D. O. Abonyi, P. M. Eze, C. C. Abba, N. T. Ujam, P. Proksch, F. B. Okoye and C. O. Esimone, Eur. J. Biol. Res., 2018, 8, 157–167 Search PubMed.
- P. Aukamp and C. Holzapfel, S. Afr. J. Chem., 1968, 21, 26–32 Search PubMed.
- H. J. Yan, X. M. Li, C. S. Li and B. G. Wang, Helv. Chim. Acta, 2012, 95, 163–168 CrossRef CAS.
- C. C. Wang, Y. M. Chiang, M. B. Praseuth, P. L. Kuo, H. L. Liang and Y. L. Hsu, Basic Clin. Pharmacol. Toxicol., 2010, 107, 583–589 CrossRef CAS PubMed.
- K. Kong, Z. Huang, S. Shi, W. Pan and Y. Zhang, BMC Microbiol., 2023, 23, 30 CrossRef CAS PubMed.
- F. Song, B. Ren, C. Chen, K. Yu, X. Liu, Y. Zhang, N. Yang, H. He, X. Liu, H. Dai and L. Zhang, Appl. Microbiol. Biotechnol., 2014, 98, 3753–3758 CrossRef CAS PubMed.
- V. V. Volkov and C. C. Perry, Dyes Pigm., 2021, 195, 109719 CrossRef CAS.
- S. S. Zhang, A. O. Zhu, X. Bai, H. J. Zhu and F. Cao, Chem. Nat. Compd., 2020, 56, 964–967 CrossRef CAS.
- T. Q. Cao, Z. Liu, L. Dong, H. Lee, W. Ko, L. B. Vinh, N. Q. Tuan, Y. C. Kim, J. H. Sohn, J. H. Yim, D. S. Lee and H. Oh, Molecules, 2022, 27, 2851 CrossRef CAS PubMed.
- Z. Liu, J. Y. Zhao, S. F. Sun, Y. Li, J. Qu, H. T. Liu and Y. B. Liu, J. Nat. Prod., 2019, 82, 1063–1071 CrossRef CAS PubMed.
- Z. Cheng, L. Lou, D. Liu, X. Li, P. Proksch, S. Yin and W. Lin, J. Nat. Prod., 2016, 79, 2941–2952 CrossRef CAS PubMed.
- D. Li, Y. Xu, C. L. Shao, R. Y. Yang, C. J. Zheng, Y. Y. Chen, X. M. Fu, P. Y. Qian, Z. G. She, N. J. Voogd and C. Y. Wang, Mar. Drugs, 2012, 10, 234–241 CrossRef CAS PubMed.
- H. Q. Hu, Y. H. Li, Z. W. Fan, W. L. Yan, Z. H. He, T. H. Zhong, Y. B. Gai and X. W. Yang, Chem. Biodiversity, 2022, 19, e202100804 CrossRef CAS PubMed.
- K. Scherlach and C. Hertweck, Org. Biomol. Chem., 2006, 4, 3517–3520 RSC.
- Z. J. Lin, G. J. Zhang, T. J. Zhu, R. Liu, H. J. Wei and Q. Q. Gu, Helv. Chim. Acta, 2009, 92, 1538–1544 CrossRef CAS.
- F. Y. Lv, A. Mandi, X. M. Li, L. P. Chi, X. Li, B. G. Wang, T. Kurtan and L. H. Meng, Deep Sea Res., Part I, 2023, 195, 104004 CrossRef CAS.
- H. Shigemori, S. Wakuri, K. Yazawa, T. Nakamura, T. Sasaki and J. i. Kobayashi, Tetrahedron, 1991, 47, 8529–8534 CrossRef CAS.
- K. Kito, R. Ookura, S. Yoshida, M. Namikoshi, T. Ooi and T. Kusumi, Org. Lett., 2008, 10, 225–228 CrossRef CAS PubMed.
- M. T. Henke, PhD thesis, Northwestern University, 2016.
- D. Xu, J. Ondeyka, G. H. Harris, D. Zink, J. N. Kahn, H. Wang, G. Bills, G. Platas, W. Wang, A. A. Szewczak, P. Liberator, T. Roemer and S. B. Singh, J. Nat. Prod., 2011, 74, 1721–1730 CrossRef CAS PubMed.
- W. Wang, T. Zhu, H. Tao, Z. Lu, Y. Fang, Q. Gu and W. Zhu, J. Antibiot., 2007, 60, 603–607 CrossRef CAS PubMed.
- D. A. Sumilat, H. Yamazaki, K. Endo, H. Rotinsulu, D. S. Wewengkang, K. Ukai and M. Namikoshi, J. Nat. Med., 2017, 71, 776–779 CrossRef CAS PubMed.
- M. Shaaban, H. Nasr, T. K. Mohamed, S. F. Mahmoud, M. M. El-Metwally and A. B. Abdelwahab, Z. Naturforsch., C: J. Biosci., 2023, 78, 149–156 CrossRef CAS PubMed.
- Z. Y. Wu, F. Liu, S. Y. Ke, Z. G. Zhang, H. T. Hu, W. Fang, S. Y. J. Xiao, Y. N. Zhang, Y. Y. Wang and K. M. Wang, Plants, 2023, 12, 203 CrossRef CAS PubMed.
- N. Tibashailwa, F. Stephano, D. M. Shadrack, J. J. E. Munissi and S. S. Nyandoro, Neurotoxicology, 2023, 94, 147–157 CrossRef CAS PubMed.
- Y. Zhang, A. Jia, H. Chen, M. Wang, G. Ding, L. Sun, L. Li and M. Dai, J. Antibiot., 2017, 70, 1138–1141 CrossRef CAS PubMed.
- V. M. Dembitsky, T. Rezanka and E. E. Shubina, Phytochemistry, 1993, 34, 1057–1059 CrossRef CAS.
- H. Nagasawa, A. Isogai, A. Suzuki and S. Tamura, Agric. Biol. Chem., 1979, 43, 1759–1763 CAS.
- X. Li, X. D. Li, X. M. Li, G. M. Xu, Y. Liu and B. G. Wang, RSC Adv., 2017, 7, 4387–4394 RSC.
- A. Kralj, S. Kehraus, A. Krick, E. Eguereva, G. Kelter, M. Maurer, A. Wortmann, H. H. Fiebig and G. M. Konig, J. Nat. Prod., 2006, 69, 995–1000 CrossRef CAS PubMed.
- A. Serag, M. A. Salem, S. Gong, J. L. Wu and M. A. Farag, Metabolites, 2023, 13, 424 CrossRef CAS PubMed.
- M. A. Salem, L. Perez de Souza, A. Serag, A. R. Fernie, M. A. Farag, S. M. Ezzat and S. Alseekh, Metabolites, 2020, 10, 37 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra01055h |
| This journal is © The Royal Society of Chemistry 2024 |