Open Access Article
Open Access ArticleThe role of pH-induced tautomerism of polyvinylpyrrolidone on the size, stability, and antioxidant and antibacterial activities of silver nanoparticles synthesized using microwave radiation
Nurul Ismillayli ab,
Suprapto Suprapto
ab,
Suprapto Suprapto a,
Eko Santoso
a,
Eko Santoso a,
Reva Edra Nugraha
a,
Reva Edra Nugraha c,
Holilah Holilah
c,
Holilah Holilah d,
Hasliza Bahruji
d,
Hasliza Bahruji e,
Aishah Abdul Jalil
e,
Aishah Abdul Jalil fg,
Dhony Hermanto
fg,
Dhony Hermanto b and
Didik Prasetyoko
b and
Didik Prasetyoko *a
*a
aDepartment of Chemistry, Faculty of Science and Data Analytics, Institut Teknologi Sepuluh Nopember, Keputih, Sukolilo, Surabaya 60111, Indonesia. E-mail: didikp@chem.its.ac.id
bDepartment of Chemistry, Faculty of Mathematics and Natural Sciences, University of Mataram, Mataram, 83125, Indonesia
cDepartment of Chemical Engineering, Faculty of Engineering, Universitas Pembangunan Nasional “Veteran” Jawa Timur, Surabaya, East Java 60294, Indonesia
dResearch Center for Biomass and Bioproducts, National Research and Innovation Agency of Indonesia (BRIN), Cibinong, 16911, Indonesia
eCentre of Advanced Material and Energy Sciences, Universiti Brunei Darussalam, Jalan Tungku Link, BE 1410, Brunei
fCentre of Hydrogen Energy, Institute of Future Energy, Universiti Teknologi Malaysia, 81310, Skudai, Johor Bahru, Johor, Malaysia
gDepartment of Chemical Engineering, Faculty of Chemical and Energy Engineering, Universiti Teknologi Malaysia, 81310, Skudai, Johor Bahru, Johor, Malaysia
First published on 2nd February 2024
Abstract
Tautomerism alters the structure and properties of materials, which can be exploited to control their chemical and biological activities. The role of pH-induced tautomerism of polyvinylpyrrolidone (PVP) was determined by measuring the size, stability, and antioxidant and antibacterial properties of microwave synthesized-silver nanoparticles (AgNPs). TEM and XRD analyses confirmed the formation of face-centered cubic silver nanoparticles. PVP stabilized the AgNPs by interaction with the carbonyl or hydroxyl groups depending on the tautomerization under different pH conditions. At pH 4, PVP was stable in the keto tautomer, stabilizing Ag through electron donation of oxygen atoms in the carbonyl group, producing smaller AgNPs with a higher zeta potential. At pH 7 and 9, the enol tautomer PVP stabilized the AgNPs via oxygen atoms in the hydroxyl group, forming large nanoparticles. The keto form of PVP improved the stability and antioxidant and antibacterial properties of AgNPs compared with the enol form. This study also excluded the antioxidant contribution of PVP via hydrogen donation to free radicals. A facile method for controlling the size of AgNPs by adapting the pH-induced tautomerism of PVP that affects their stability and antioxidant and antibacterial activities is thus reported.
Introduction
The role of silver nanoparticles in pharmaceutical and biomedical applications has increased in the last decade, including in antibacterial, antioxidant, anticancer applications,1,2 drug-delivery systems,3,4 diagnostics,5 and sensor devices.6 Their vast application has encouraged the development of facile and fast methods to produce a high yield of biocompatible and nontoxic AgNPs. The antibacterial and oxidative activities of AgNPs depend on their intrinsic characteristics, such as size, shape, composition, stability, and surrounding media.7 As nanosensors, the performance of AgNPs relies on the uniformity and size of the nanoparticles.8,9 Therefore, synthesis methods for AgNPs to produce nanoparticles with a uniform structure, narrow particle size, and high stability have been investigated.Silver nitrate reduction is widely employed for fabricating AgNPs because of its cost efficiency for large-scale production and ease of controlling the shape and size of AgNPs.10 A mixture of PVP as a capping and reducing agent was applied to produce AgNPs with flower-shaped nanostructures and uniform nanoparticles.11,12 Microwave irradiation provides homogeneous heating for the fast nucleation of Ag to form uniform structures.13,14 The utilization of a capping agent can help control AgNP crystal growth and enable them to maintain their stability against aggregation in solution.15,16 In particular, PVP saturates the silver surface, preventing agglomeration and maintaining long-term stability due to the steric hindrance of its hydrophobic carbon chain.17 Furthermore, PVP controls the shape of nanoparticles by promoting anisotropic growth into various morphologies.18 Several parameters can also affect the shape and size of the synthesized AgNPs, such as the molecular weight, PVP concentration, and solvent.19–21
PVP is stable in the keto form at acidic pH, whereas the enol tautomer exists at pH 7 and is predominant at pH > 10.22 The keto–enol tautomerism of PVP transforms its functional group to give it different properties when it interacts with precursors. For example, in the synthesis of silica-coated graphite, the enol tautomer of PVP rich in hydroxyl groups enhanced the interaction with the silicic acid precursor.23 In the synthesis of Au nanoparticles, the enol PVP readily donated the lone pair of electrons on the O-atom of the hydroxyl group to form Au–O–C bonds.24 Because the shape and size of AgNPs are affected by the characteristics and functional groups of the capping agent,25 the tautomerism of PVP may transform the properties of AgNPs due to the different interactions with the functional groups on the Ag+ precursor. Herein, the effect of the pH-induced tautomerism of PVP on the size, stability, antioxidant, and antibacterial properties of silver nanoparticles (AgNPs) was investigated. Also, the antioxidant activity of PVP-stabilized AgNPs was compared with that of PVP, AgNO3, and ascorbic acid solutions to exclude the antioxidant contribution of PVP via hydrogen donation to free radicals.26,27
Experimental
Materials
Analytical grade silver nitrate (AgNO3), ascorbic acid (AA, C6H8O6), polyvinyl pyrrolidone, (PVP, (C6H9NO)n, 30.000 kDa), sodium hydroxide, methanol, and Mueller–Hinton agar were purchased from Merck (Darmstadt, Germany). 1,1-Diphenyl-2-picrylhydrazyl (DPPH) was purchased from Sigma-Aldrich (St. Louis, MO, USA). Deionized water was used in all the preparations.Synthesis of AgNPs and their characterization
First, 0.01 M AgNO3, PVP, and 0.025 M ascorbic acid (AA) solutions were freshly prepared before the synthesis of AgNPs. Herein, 0.4% PVP aqueous solution at pH 4 was prepared by dissolving 0.4 g of PVP in deionized water up to 100 mL. PVP solutions at pH 7 and 9 were obtained by adding 0.5% NaOH solution under stirring until the desired pH was reached. The PVP solutions at pH 4, 7, and 9 are referred to as PVP4, PVP7, and PVP9, respectively. The PVP (100 mL) solution was treated under microwave radiation for 3 min at 80 W. Then, 60 mL of AgNO3 solution was added to the PVP solution and stirred for 10 min. As a reducing agent, AA solution was added dropwise to the Ag+–PVP solution while stirring vigorously, and then the mixture was treated under microwave irradiation for 3 min at 80 W. A domestic microwave in an open air system (AQUA AEM-S18125) was used without modification. The solution was centrifuged two times at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 45 min. Then, the separated AgNPs were freeze-dried for further analysis and application. The AgNPs produced using PVP solutions at pH 4, 7, and 9 were labeled as AgNP4, AgNP7, and AgNP9, respectively.
000 rpm for 45 min. Then, the separated AgNPs were freeze-dried for further analysis and application. The AgNPs produced using PVP solutions at pH 4, 7, and 9 were labeled as AgNP4, AgNP7, and AgNP9, respectively.
The keto–enol tautomerism of PVP was confirmed by scanning the absorbance of PVP solution at pH 4, 7, and 9 at a wavelength range of 200–700 nm using a 7809 UV-vis spectrophotometer (Labo-hub (China)). The analysis was also carried out on a mixture of PVP and Ag+ solution to identify the possibility of AgNP formation without adding AA, and the mixture of Ag+ with AA only. The solution was placed in a closed container and incubated at room temperature in the dark until the third day. The concentration of AgNPs was determined using an atomic absorption spectrometer (Thermo Scientific iCE 3000, USA). The stability of the AgNPs was monitored by comparing their intensity before and after storage for five months. The functional groups in AgNPs were identified using FTIR spectroscopy (PerkinElmer, USA) with the KBr pellet method. The size distribution (size and polydispersity index, PDI), zeta potential, and conductance of AgNPs were analyzed using a Malvern Zetasizer (Malvern Instrument, UK). A Hitachi H9500 TEM system (Hitachi, Japan) was employed to describe the morphology and size of AgNP4. The TEM images were analyzed using ImageJ to obtain the particle size distribution and the descriptive statistics of the particles (including mean diameter, standard deviation, maximum, and minimum size). The crystallinity of particles was determined using XRD (Philips X'pert PW 3050, the Netherlands) with Cu Kα radiation (λ = 1.54060 Å) at 30 mA and 40 kV. The XRD patterns were recorded at 2θ from 5° to 90°, and the average crystallite of AgNPs was calculated using the Debye–Scherrer equation:
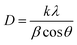 | (1) |
Antioxidant assay
The antioxidant activity of AgNPs was measured following a previously reported method29 with slight modification. Different concentrations of AgNPs and ascorbic acid solutions (as standard) were added to 1 mL of freshly prepared methanolic solution containing DPPH (2 mM) and shaken vigorously to form a homogeneous mixture. The mixture was incubated in the dark at room temperature for 30 min. Sample absorbance was measured at 517 nm wavelength using methanol as a blank. The same treatment was applied to the PVP solution at pH 4. The inhibition percentage was calculated using the following equation:
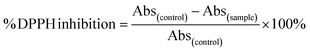 | (2) |
Antioxidant activity was expressed as the number of molecules required to reduce the initial DPPH absorbance by 50% (IC50), which was determined graphically by plotting the percentage loss of DPPH as a function of concentration. The standard deviation was calculated based on three replicate experiments.
Antibacterial assay
Antibacterial activity was determined using the well-diffusion method against Gram-negative (E. coli) and Gram-positive (S. aureus) bacteria based on a previously reported method.30 Bacterial cultures were incubated at 37 °C for 24 hours. The optical density of the culture was adjusted to 0.5 McFarland standard [1.5 × 108 colony forming units (CFU) mL−1] and diluted to give a final working concentration of 1 × 106 CFU mL−1. Bacteria were inoculated in an agar plate, and each sample (AgNP and PVP solutions with a concentration of 10 mg L−1) was added to the wells in the agar plate. Ciprofloxacin (Cip) was selected as the positive control for the study of the antibacterial activity of the AgNPs and PVPs. The samples were incubated overnight in Petri dishes at 37 °C, and the diameter of the inhibition zone was measured to evaluate the antibacterial activity of each sample. All the experiments were performed in triplicate.Results and discussion
Synthesis and characterization of the AgNPs
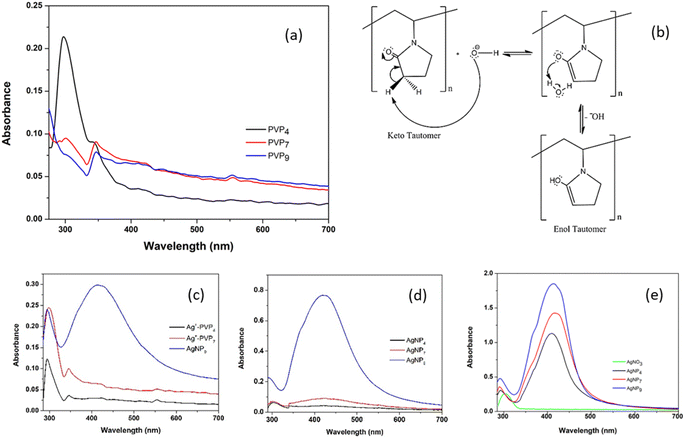
The keto–enol tautomerism of PVP at pH 4, 7, and 9 was monitored using UV-vis analysis (Fig. 1a). The adsorption peak at ∼300 nm indicated the n → π* transition of the carbonyl band (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) of the pyrrolidine ring.31 At pH 4, the PVP absorbance was higher than the absorbance at neutral and alkaline pH, implying that the keto tautomer was stable under acidic conditions. PVP was stable as an enol tautomer at pH 7 and 9. The increase in hydroxyl OH− from NaOH solution led to the formation of an enol tautomer following the reaction with the alpha hydrogen on the pyrrolidone ring, as shown in Fig. 1b. This confirmed the formation of the PVP enol tautomer under alkaline conditions, as reported previously.22,23
O) of the pyrrolidine ring.31 At pH 4, the PVP absorbance was higher than the absorbance at neutral and alkaline pH, implying that the keto tautomer was stable under acidic conditions. PVP was stable as an enol tautomer at pH 7 and 9. The increase in hydroxyl OH− from NaOH solution led to the formation of an enol tautomer following the reaction with the alpha hydrogen on the pyrrolidone ring, as shown in Fig. 1b. This confirmed the formation of the PVP enol tautomer under alkaline conditions, as reported previously.22,23
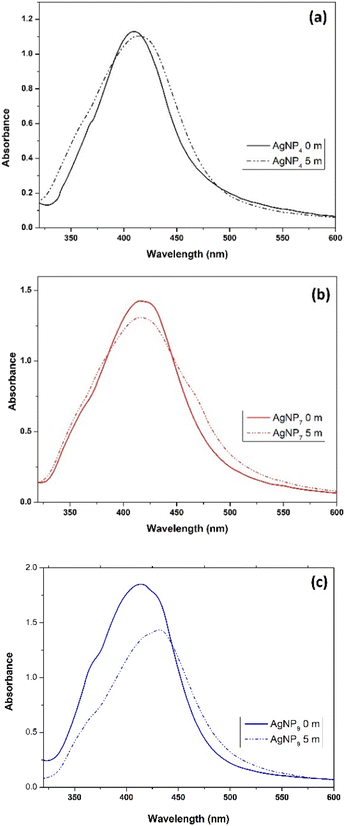
The enol tautomer has hydroxyl groups attached to the pyrrolidone ring, simultaneously acting as a reducing agent during AgNP formation without an external reducing agent (AA). Fig. 1c shows the UV-vis spectra of Ag–PVP mixtures at different pH values that were subsequently analyzed after the synthesis, which was denoted as day 0. A broad adsorption peak at 420 nm was observed, which was ascribed to the surface plasmon resonance of AgNPs, which confirmed the formation of AgNPs at pH 9. Meanwhile, the surface plasmon resonance of AgNP was negligible at pH 4, as the presence of PVP in its keto tautomer prevented it from reducing Ag+ to Ag0. At pH 7, the number of OH− groups was insufficient to induce AgNP formation. However, when the Ag–PVP mixtures were incubated for three days, AgNPs began to form on the third day of incubation, as shown in Fig. 1d. No plasmonic resonance was observed at pH 4. Meanwhile, the intensity was higher at pH 9, indicating the continuous growth of AgNPs.
The synthesis of AgNPs was also conducted in the presence of ascorbic acid (AA) as a reducing agent to accelerate the formation of AgNPs at different pH values. AgNP formation was visually monitored from the changes of colorless solution into brownish-yellow solution. Fig. 1e shows the absorption peak at 300 nm, indicating Ag+ in the solution.28 Broad peaks centered at ∼410, 414, and 415 nm were observed for AgNP4, AgNP7, and AgNP9, respectively. The SPR intensity of the AgNPs increased with increasing pH, suggesting that alkaline conditions produced a high yield of AgNPs (Fig. 1e). The effect of the PVP enol tautomer at pH 9 was also observed in the presence of AA. The redshift at higher pH implied the formation of large Ag nanoparticles.
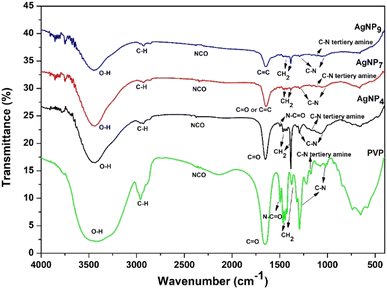
FTIR analysis of AgNPs/PVP was performed to investigate the keto–enol phenomenon at different pH (Fig. 2). The –CH2 groups in the skeleton chain of PVP were observed at 1465–1385 cm−1.32 The bands at 1295, 1078, and 1021 cm−1 indicated the C–N groups in PVP,33 and specifically, the tertiary amine C–N absorption appeared at 1172 cm−1. The bands were shifted to 1294, 1162, 1069, and 1065 cm−1 in the keto form (AgNP4), whereas in the enol form (AgNP7 and AgNP9), they were shifted to 1292, 1166, 1058, and 1029 cm−1, respectively. The absorption at 2364 cm−1 indicated the NCO functional group in PVP, which was shifted to 2378 cm−1 in both the keto and enol forms. The appearance of the OH group in PVP, especially the keto tautomer, was possible because of the hygroscopic nature of PVP in binding to water. The peak for the vibration of –OH was shifted to 3437 and 3444 cm−1 for AgNP7 and AgNP9, respectively. The peak at 1654 cm−1 was attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibration of PVP,32,33 and was shifted to 1650 cm−1 in AgNP4 (keto form) and to 1647 cm−1, indicating the C
O vibration of PVP,32,33 and was shifted to 1650 cm−1 in AgNP4 (keto form) and to 1647 cm−1, indicating the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C vibration of the enol form in AgNP7 and AgNP9.31,34 The N–C
C vibration of the enol form in AgNP7 and AgNP9.31,34 The N–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O band at 1495 cm−1 for PVP and AgNP4 also evidenced the keto tautomer formation. In contrast, this band was almost absent in AgNP7 and AgNP9. In addition, keto tautomer formation was dominant at acidic pH, as indicated by the sharp peak for the pyrrolidone ring at 1385 cm−1 (–CH2 bending), while in neutral and alkaline conditions this had much lower intensity due to the change of –CH2 in the keto form to
O band at 1495 cm−1 for PVP and AgNP4 also evidenced the keto tautomer formation. In contrast, this band was almost absent in AgNP7 and AgNP9. In addition, keto tautomer formation was dominant at acidic pH, as indicated by the sharp peak for the pyrrolidone ring at 1385 cm−1 (–CH2 bending), while in neutral and alkaline conditions this had much lower intensity due to the change of –CH2 in the keto form to ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH in the enol form. FTIR data showed shifts in the C
CH in the enol form. FTIR data showed shifts in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–N, and NCO bands in keto form, and –OH, C–N, and NCO bands in the enol form. However, the shifts were more obvious in the C
O, C–N, and NCO bands in keto form, and –OH, C–N, and NCO bands in the enol form. However, the shifts were more obvious in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and –OH bands than in the C–N and NCO bands, suggesting the role of oxygen groups in the capping action of PVP.
O and –OH bands than in the C–N and NCO bands, suggesting the role of oxygen groups in the capping action of PVP.
The mechanism for AgNP synthesis is proposed in Fig. 3. The oxygen and nitrogen atoms in the PVP may have a role in forming the PVP complex with Ag+ ions. However, the lone pair of electrons on the tertiary amines is from a highly soft ligand with poor complexing with Ag+ ions. Therefore, PVP coordinates with the silver ion through the lone pair electrons of the oxygen atoms, anchoring the PVP polymer chain to the Ag surfaces.18 The complex facilitates silver ion reduction and silver nucleation to form stable AgNPs.35 Then, ascorbic acid, as a reducing agent, donates electrons by turning them into semidehydroascorbic acid and dehydroascorbic acid radicals. Semidehydroascorbic acid is a potent reducing agent that reduces Ag+ ions to Ag0 nanoparticles.36 In the keto form, the silver ion coordinates with the oxygen atoms in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group (Fig. 3a), whereas the C
O group (Fig. 3a), whereas the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group is replaced by the –OH group in the enol form (Fig. 3b). The PVP surrounds the silver surface through physical and chemical bonding, preventing contact between particles that can result in particle agglomeration.
O group is replaced by the –OH group in the enol form (Fig. 3b). The PVP surrounds the silver surface through physical and chemical bonding, preventing contact between particles that can result in particle agglomeration.
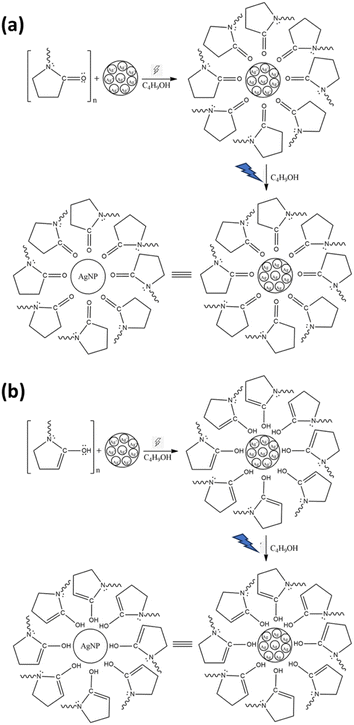 | ||
| Fig. 3 Proposed mechanism of AgNP formation using PVP in the keto form (a) and PVP in the enol form (b). | ||
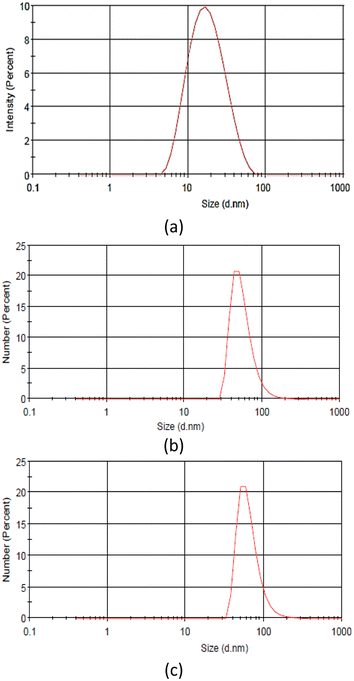
Due to tautomerization, the different functional groups in PVP affect the nucleation and nanoparticle growth. The PVP enol tautomer produced AgNPs with a broader particle size distribution than the keto tautomer, as indicated by the redshift and peak broadening of the AgNP spectra synthesized at a high pH.37 The particle size determined using the particle size analyzer in Fig. 4 and Table 1 also aligned with the UV-visible spectra. The zeta potential of AgNPs showed a negatively charged surface, resulting in electrostatic repulsion between the particles. The high negative value obtained at pH 4 indicated the stability of the AgNPs. Generally, synthesis under alkaline conditions decreases the particle size of AgNPs but increases the negative charge zeta potential of AgNPs.9,37,38
| AgNPs | Size (nm) | Zeta potential (mV) |
|---|---|---|
| AgNP4 | 27.50 ± 5.68 | −26.1 |
| AgNP7 | 56.83 ± 22.26 | −22.1 |
| AgNP9 | 65.64 ± 25.37 | −21.5 |
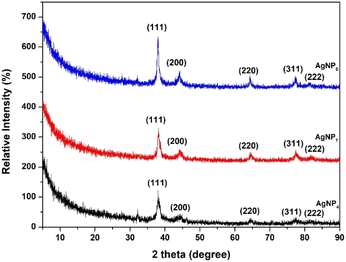
The XRD patterns of the AgNPs showed four diffraction peaks between 2θ = 5–80° (Fig. 5). The peaks were indexed to the (111), (200), (220), (311), and (222) planes, which confirmed the formation of a face-centered cubic (fcc) structure of pure silver.28,39 The crystal sizes of AgNPs calculated using the Scherrer equation based on the broadening of the (111) reflection were determined at 32.7, 45.8, and 76.3 nm for AgNP4, AgNP7, and AgNP9, respectively. This confirmed that higher pH induces the formation of larger AgNP crystals.
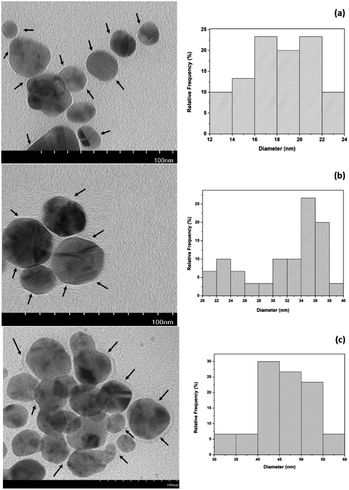
TEM analysis was used to determine the shape of the AgNPs, showing they were spherical and surrounded by a thin PVP layer, as shown in Fig. 6. The size ranged from 12.29 to 23.78 nm, giving an average diameter of 18.18 ± 2.99 nm for AgNP4. AgNP7 and AgNP9 have average diameters of 31.64 ± 5.47 nm and 46.15 ± 6.05 nm, respectively. The PVP in the keto form produced relatively smaller and uniform AgNPs than the enol form. The AgNPs obtained in this study were significantly smaller than those synthesized at room temperature using PVP and ascorbic acid.11 Another study using PVP in an ultrasonic-assisted synthesis with a double reducing agent of ascorbic acid and glucose produced AgNPs sized 16–34 nm.12 The homogeneous and fast microwave heating increases the reduction and stabilization of AgNPs, resulting in short reaction times, low energy consumption, and uniform-sized AgNPs.13,14,40
The effect of the AgNP size on their stability and antioxidant and antibacterial properties was investigated. The stability of AgNPs after five months of storage is shown in Fig. 7. The C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional group in PVP provided a more effective capping action for AgNPs than the –OH groups due to the π bond of the carbonyl group and silver.41 Hence, the keto form of PVP provided better stability for the AgNPs than the enol form.
O functional group in PVP provided a more effective capping action for AgNPs than the –OH groups due to the π bond of the carbonyl group and silver.41 Hence, the keto form of PVP provided better stability for the AgNPs than the enol form.
Antioxidant activity of the AgNPs
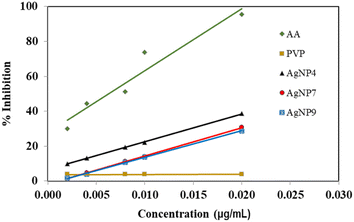
The antioxidant activity of the AgNPs was measured by monitoring the absorption of DPPH solution at 517 nm. The antioxidant activity is generally measured based on the amount needed to reduce 50% of DPPH radicals via proton or electron transfer. Compounds with antioxidant properties donate hydrogen atoms or transfer one electron to reduce DPPH, thereby decreasing the absorption. The decrease in absorption is proportional to the number of electrons received by DPPH.29 The antioxidant activity of AgNPs was compared with that of AA as the standard solution and PVP as the capping agent. Ascorbic acid, AA displayed a high antioxidant activity with an IC50 value of 6.3 μg mL−1 (Fig. 8). The AgNPs showed IC50 values of 27.3, 32.0, and 33.9 μg mL−1 for AgNP4, AgNP7, and AgNP9, respectively. The AgNPs obtained at pH 4 exhibited a higher percentage of radical inhibition than AgNP7 and AgNP9, suggesting the importance of a small nanoparticle size in exhibiting high antioxidant activity.27 The antioxidant activity of AgNPs obtained in this study was higher than that produced using PVP and Pandanus atrocarpus extract, despite the antioxidant activity being elevated by the polyphenol and flavonoid compounds in the Pandanus atrocarpus extract.42 The antioxidant activity of PVP in Fig. 8 showed a substantial IC50 value of 5076.5 μg mL−1, implying there was no antioxidant activity from PVP. Therefore, the antioxidant activity originated solely from the AgNPs due to single electron transfer (SET) from the Ag nanoparticles.43 AgNPs released electrons to form Ag+ ions, which occurred either on the surface of nanoparticles or via the release from nanoparticles.44,45 The effective PVP capping action on AgNP4 formed well-dispersed small nanoparticles that could effectively transfer electrons to DPPH. This also aligned with the negative surface charge of AgNP4, as shown in Table 1.Antibacterial activity of the AgNPs
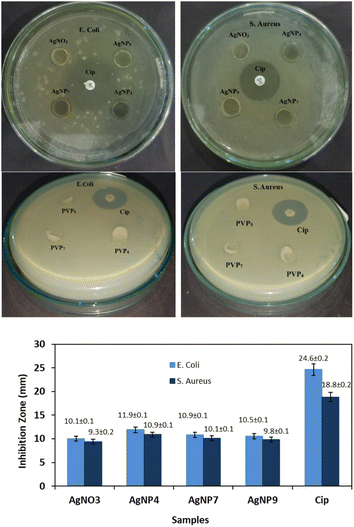
The antibacterial activity of the AgNPs was evaluated against E. coli and S. aureus, as shown in Fig. 9. Note that PVP4, PVP7, and PVP9 solutions showed negligible antibacterial activity, as indicated by the absence of an inhibition zone. The antibacterial activity of AgNO3 investigated for comparison showed lower activity than the AgNP samples. AgNP4 provided the broadest inhibition zone, indicating it has the most potent antibacterial activity among the three nanoparticles tested. Previous research has proposed several mechanisms regarding the antibacterial properties of silver nanoparticles. Small Ag nanoparticles enhanced antibacterial activity by providing a large surface area for efficient contact and penetration of AgNPs on the bacterial membrane.7,46 The penetration causes a leakage of cellular contents, allowing AgNPs to enter the cytoplasm and interact with biomolecules, such as proteins, DNA, and enzymes. The damage in the intracellular structure eventually kills the bacteria. AgNPs also inhibit the activity of antioxidant enzymes, such as glutathione, causing the accumulation of reactive oxygen species (ROS), which has implications for decreased respiration, ATP, and DNA damage.2,47 Living cells exposed to AgNPs for 5 min increased the formation of mitochondrial ROS, with the quantity increasing as the AgNP size reduced.48 The release of Ag+ ions from AgNPs has a high affinity for the thiol groups of many vital enzymes and proteins, leading to their deactivation and ultimately causing cell death.2,30 AgNPs were observed to be more effective against Gram-negative bacteria (E. coli) than against Gram-positive bacteria (S. aureus). The rigid and thick peptidoglycan layer on the surface prevents the entry of AgNPs.2,49Manipulating the keto–enol tautomerization of PVP allows controlling the AgNPs size through different stabilization mechanisms of the PVP functional group. Different sizes of AgNps affect the stability and antioxidant and antibacterial properties of the particles. PVP in the keto form produced smaller-sized AgNPs that were stable for long-term storage and exhibited higher antioxidant and antibacterial properties than AgNPs capped by the enol form of PVP.
Conclusions
This study characterized the impact of the pH-induced tautomerism of PVP on the size, stability, and antioxidant and antibacterial properties of microwave-synthesized silver nanoparticles (AgNPs). The acidic pH of the PVP solution induced the formation of a keto tautomer, whereas alkaline pH led to the formation of the enol tautomer PVP. The presence of hydroxyl groups in the enol tautomer allowed PVP to play a dual role as a capping agent and a reducing agent. The different functional groups in PVP affected the size of the nanoparticles. TEM, PSA, and XRD analyses confirmed the formation of large spherical AgNPs at high pH. The zeta potential values also became more negative for the smaller-sized AgNPs, indicating that high-stability nanoparticles were obtained. This was also supported by the stability of AgNP4, which was higher than that of AgNP7 and AgNP9 after five months of storage. This indicated that the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O functional group in PVP provided a more effective capping action for AgNPs than the –OH groups. The antioxidant and antibacterial activities were shown to be solely from the AgNPs, in which the keto form of PVP provided better stability and antioxidant and antibacterial properties for AgNPs than the enol form. AgNP4 exhibited a high antioxidant activity with an IC50 value of 27.3 μg mL−1, followed by AgNP7 and AgNP9 with IC50 values of 32.0 μg mL−1 and 33.9 μg mL−1, respectively. Test of the antibacterial activity using the well-diffusion method showed the broadest inhibition zone was achieved when using the smallest AgNPs (AgNP4) on E. coli and S. aureus. Therefore, controlling the pH of PVP solution can be employed as a fast synthesis method to form different sized AgNPs, which can significantly affect their stability and antioxidant and antibacterial activities.
O functional group in PVP provided a more effective capping action for AgNPs than the –OH groups. The antioxidant and antibacterial activities were shown to be solely from the AgNPs, in which the keto form of PVP provided better stability and antioxidant and antibacterial properties for AgNPs than the enol form. AgNP4 exhibited a high antioxidant activity with an IC50 value of 27.3 μg mL−1, followed by AgNP7 and AgNP9 with IC50 values of 32.0 μg mL−1 and 33.9 μg mL−1, respectively. Test of the antibacterial activity using the well-diffusion method showed the broadest inhibition zone was achieved when using the smallest AgNPs (AgNP4) on E. coli and S. aureus. Therefore, controlling the pH of PVP solution can be employed as a fast synthesis method to form different sized AgNPs, which can significantly affect their stability and antioxidant and antibacterial activities.
Author contributions
Nurul Ismillayli: conceptualization, methodology, data curation, writing – original draft. Suprapto Suprapto: validation, supervision, writing – review & editing. Eko Santoso: writing – review & editing. Holilah Holilah: formal analysis. Hasliza Bahruji: visualization, writing – review & editing. Reva Endra Nugraha: formal analysis. Aishah Abdul Jalil: writing – review & editing. Dhony Hermanto: writing – review & editing. Didik Prasetyoko: conceptualization validation, supervision, writing – review & editing.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
The authors thank the Ministry of Education, Culture, Research, and Technology (Center for Higher Education Funding-BPPT), Lembaga Pengelola Dana Pendidikan (LPDP), and Institut Teknologi Sepuluh Nopember grant (no. 2267/PKS/ITS/2023 and ORM 2024) for supporting this study.Notes and references
- J. Mishra, A. Kour, S. D. Amin and J. J. Panda, Colloid Interface Sci. Commun., 2021, 44, 100459 CrossRef CAS.
- F. Jalilian, A. Chahardoli, K. Sadrjavadi, A. Fattahi and Y. Shokoohinia, Adv. Powder Technol., 2020, 31, 1323–1332 CrossRef CAS.
- H. A. Hussein and M. A. Abdullah, Appl. Nanosci., 2022, 12, 3071–3096 CrossRef CAS.
- L. Qiu, J. W. Li, C. Y. Hong and C. Y. Pan, ACS Appl. Mater. Interfaces, 2017, 9, 40887–40897 CrossRef CAS PubMed.
- C.-W. Yen, H. de Puig, J. Tam, J. Gómez-Márquez, I. Bosch, K. Hamad-Schifferli and L. Gehrke, Lab Chip, 2015, 15, 1638–1641 RSC.
- N. D. Nguyen, T. Van Nguyen, A. D. Chu, H. V. Tran, L. T. Tran and C. D. Huynh, Arabian J. Chem., 2018, 11, 1134–1143 CrossRef CAS.
- I. X. Yin, J. Zhang, I. S. Zhao, M. L. Mei, Q. Li and C. H. Chu, Int. J. Nanomed., 2020, 15, 2555–2562 CrossRef CAS PubMed.
- B. Ajitha, Y. A. Kumar Reddy, P. S. Reddy, H. J. Jeon and C. W. Ahn, RSC Adv., 2016, 6, 36171–36179 RSC.
- L. A. P. Gontijo, E. Raphael, D. P. S. Ferrari, J. L. Ferrari, J. P. Lyon and M. A. Schiavon, Rev. Mater., 2020, 25, 1–10 Search PubMed.
- C. Quintero-quiroz, N. Acevedo, J. Zapata-giraldo, L. E. Botero, J. Quintero, D. Zárate-triviño, J. Saldarriaga and V. Z. Pérez, Biomater. Res., 2019, 23, 1–15 CrossRef PubMed.
- Y. Wu, P. Liang, Q. min Dong, Y. Bai, Z. Yu, J. Huang, Y. Zhong, Y. C. Dai, D. Ni, H. bo Shu and C. U. Pittman, Food Chem., 2017, 237, 974–980 CrossRef CAS PubMed.
- W. Li, X. Xu, W. Li, P. Liu, Y. Zhao, Q. Cen and M. Chen, J. Mater. Res. Technol., 2020, 9, 142–151 CrossRef CAS.
- H. Wang, C. G. Yuan, C. Liu, X. Duan, Q. Guo, Y. Shen, J. Liu and Y. Chen, J. Environ. Sci., 2022, 115, 286–293 CrossRef CAS PubMed.
- S. Mukherji, S. Bharti, G. Shukla and S. Mukherji, Phys. Sci. Rev., 2019, 4, 1–73 Search PubMed.
- A. M. Ferreira, A. Vikulina, M. Loughlin and D. Volodkin, RSC Adv., 2023, 13, 10542–10555 RSC.
- T. H. Yang, Y. Shi, A. Janssen and Y. Xia, Angew. Chem. Int. Ed., 2020, 59, 15378–15401 CrossRef CAS PubMed.
- R. Andrea, B. Péter, E. Boka, D. Zakupszky, N. Igaz, B. Szerencsés, I. Pfeiffer, Z. Kónya and M. Kiricsi, Int. J. Mol. Sci., 2021, 22, 1–21 Search PubMed.
- K. M. Koczkur, S. Mourdikoudis, L. Polavarapu and S. E. Skrabalak, Dalton Trans., 2015, 44, 17883–17905 RSC.
- B. Khodashenas and H. R. Ghorbani, Arabian J. Chem., 2019, 12, 1823–1838 CrossRef CAS.
- J. Li, K. Inukai, Y. Takahashi, A. Tsuruta and W. Shin, Integr. Med. Res., 2017, 1–10 Search PubMed.
- C. Wu, B. P. Mosher, K. Lyons and T. Zeng, J. Nanosci. Nanotechnol., 2010, 10, 2342–2347 CrossRef CAS PubMed.
- T. Guaragnone, M. Rossi, D. Chelazzi, R. Mastrangelo, M. Severi, E. Fratini and P. Baglioni, ACS Appl. Mater. Interfaces, 2022, 14, 7471–7485 CrossRef CAS PubMed.
- S. Choi, K. Kim, J. Nam and S. E. Shim, Carbon, 2013, 60, 254–265 CrossRef CAS.
- M. Behera and S. Ram, Appl. Nanosci., 2014, 4, 247–254 CrossRef CAS.
- C. V. Restrepo and C. C. Villa, Environ. Nanotechnol., Monit. Manage., 2021, 15, 100428 CAS.
- S. Bhakya, S. Muthukrishnan, M. Sukumaran and M. Muthukumar, Appl. Nanosci., 2016, 6, 755–766 CrossRef CAS.
- J. Flieger, W. Franus, R. Panek, M. Szymańska-Chargot, W. Flieger, M. Flieger and P. Kołodziej, Molecules, 2021, 26(16), 1–21 CrossRef PubMed.
- A. Mirzaei, K. Janghorban, B. Hashemi, M. Bonyani, S. G. Leonardi and G. Neri, J. Nanostruct. Chem., 2017, 7, 37–46 CrossRef CAS.
- A. U. Khan, Q. Yuan, Z. U. H. Khan, A. Ahmad, F. U. Khan, K. Tahir, M. Shakeel and S. Ullah, J. Photochem. Photobiol., B, 2018, 183, 367–373 CrossRef PubMed.
- E. Urnukhsaikhan, B. E. Bold, A. Gunbileg and N. Sukhbaatar, Sci. Rep., 2021, 11, 1–12 CrossRef PubMed.
- H. M. Zidan, E. M. Abdelrazek, A. M. Abdelghany and A. E. Tarabiah, J. Mater. Res. Technol., 2019, 8, 904–913 CrossRef CAS.
- R. Zein, I. Alghoraibi, C. Soukkarieh, M. T. Ismail and A. Alahmad, Micromachines, 2022, 13(5), 777 CrossRef.
- R. Bryaskova, D. Pencheva, S. Nikolov and T. Kantardjiev, J. Chem. Biol., 2011, 4, 185–191 CrossRef PubMed.
- A. B. D. Nandiyanto, R. Oktiani and R. Ragadhita, Indones. J. Sci. Technol., 2019, 4, 97–118 CrossRef.
- W. A. Shaikh, S. Chakraborty, G. Owens and R. U. Islam, Appl. Nanosci., 2021, 11, 2625–2660 CrossRef CAS.
- N. Zhang, X. Yu, J. Hu, F. Xue and E. Ding, RSC Adv., 2013, 3, 13740–13747 RSC.
- M. K. Alqadi, O. A. Abo Noqtah, F. Y. Alzoubi, J. Alzouby and K. Aljarrah, Mater. Sci.-Pol., 2014, 32, 107–111 CrossRef CAS.
- I. Fernando and Y. Zhou, Chemosphere, 2019, 216, 297–305 CrossRef CAS.
- P. K. Sahoo, S. S. Kalyan Kamal, T. Jagadeesh Kumar, B. Sreedhar, A. K. Singh and S. K. Srivastava, Def. Sci. J., 2009, 59, 447–455 CrossRef CAS.
- S. Joseph and B. Mathew, J. Mol. Liq., 2015, 204, 184–191 CrossRef CAS.
- S. Muthaiah, A. Bhatia and M. Kannan, Stab. Appl. Coord. Compd., 2020, 1–18 CAS.
- A. H. Jabbar, H. S. O. Al-janabi, M. Q. Hamzah, S. O. Mesan, A. N. Tuama, A. S. Binti Ameruddin and M. A. Agam, J. Xi'an Univ. Archit. amp; Technol., 2020, 12, 235–251 Search PubMed.
- T. Kalaiyarasan, V. K. Bharti and O. P. Chaurasia, RSC Adv., 2017, 7, 51130–51141 RSC.
- A. O. Docea, D. Calina, A. M. Buga, O. Zlatian, M. M. B. Paoliello, G. D. Mogosanu, C. T. Streba, E. L. Popescu, A. E. Stoica, A. C. Bîrcă, B. Ştefan Vasile, A. M. Grumezescu and L. Mogoanta, Int. J. Mol. Sci., 2020, 21, 1233 CrossRef CAS PubMed.
- D. Hermanto, N. Ismillayli, I. Irmawanti, I. Fathulloh, D. N. Cahyani, U. K. Zuryati, H. Muliasari and R. Wirawan, Karbala Int. J. Mod. Sci., 2023, 9, 80–88 CrossRef.
- Z. Lu, K. Rong, J. Li, H. Yang and R. Chen, J. Mater. Sci.: Mater., 2013, 24, 1465–1471 CAS.
- J. Jin, X. Wu, J. Xu, B. Wang, F. Jiang and Y. Liu, Biomater. Sci., 2017, 5, 247–257 RSC.
- A. Onodera, F. Nishiumi, K. Kakiguchi, A. Tanaka, N. Tanabe, A. Honma, K. Yayama, Y. Yoshioka, K. Nakahira, S. Yonemura, I. Yanagihara, Y. Tsutsumi and Y. Kawai, Toxicol Rep, 2015, 2, 574–579 CrossRef CAS PubMed.
- D. Hermanto, N. Ismillayli, D. H. Fatwa, U. K. Zuryati, H. Muliasari, R. Wirawan, D. Prasetyoko and S. Suprapto, S. Afr. J. Chem. Eng., 2024, 47, 136–141 Search PubMed.
| This journal is © The Royal Society of Chemistry 2024 |