Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A comprehensive investigation of the biophysical approach for aptamer functionalized nanoparticles in cancer therapy: a review
Alanthatta Govindan
Navaneeth
a and
Subramani
Karthikeyan
 *b
*b
aDepartment of Physics, School of Advanced Sciences, Vellore Institute of Technology (VIT), Chennai Campus, Vandalur-Kelambakkam Road, Chennai - 600127, India
bCenter for Health Care and Advanced Research, Vellore Institute of Technology (VIT), Chennai Campus, Vandalur-Kelambakkam Road, Chennai - 600127, India. E-mail: s.karthikeyan@vit.ac.in
First published on 12th September 2024
Abstract
Aptamers stand out for their remarkable specificity and versatility, making them an invaluable tool in cancer therapy. When combined with nanoparticles, they form a dynamic platform for targeted drug delivery and diagnostics, leveraging enhanced cellular take up and the enhanced permeability and retention effect. This review explores both experimental and computational studies that probe the intricate interactions between aptamers and nanoparticles. By combining theoretical insights with empirical studies, this approach deepens our understanding of aptamer–nanoparticle conjugation, opening new avenues to enhance therapeutic efficacy and reduce off-target effects. Recent advancements in the field are critically analysed, spotlighting transformative studies that highlight the potential of this approach. Offering a comprehensive overview of current achievements and future prospects, this article aims to establish the pivotal role of aptamer-functionalized nanoparticles in personalized cancer treatment strategies.
1. Introduction
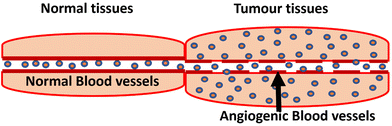
Cancer is currently one of the most serious threats to public health, causing numerous fatalities every year. It results from multiple factors, including genetic, lifestyle, and environmental factors. According to GLOBCAN, from 2020 to 2040, the number of cancer cases in India will increase by 57.5%, reaching 2.08 million. China has the highest number of cancer cases worldwide; India ranks third, behind China and the United States.1 Even though cancer is referred to as a single condition, it is a group of diseases characterised by the uncontrolled growth and spread of abnormal cells. These abnormal cells can invade and destroy neighbouring tissues, and they can also travel throughout the body via the bloodstream or lymphatic system.2 When cancer progresses to a metastatic state or begins to spread to nearby organs, a systemic chemotherapy approach is required for effective cancer management. In chemotherapy, the chemodrugs circulate in the bloodstream and kill the rapidly growing cancer cells. Normal cells in hair follicles and bone marrow also rapidly develop, since chemodrugs attack all rapidly growing cells, they are unable to differentiate these normal healthy cells from cancerous cells, which leads to numerous side effects, such as anaemia, gastrointestinal problems, vomiting, and nausea.3–5 In order to overcome this limitation, a distinct strategy with fewer off-target effects and high treatment efficacy is required. Targeted drug delivery is one of the possible solutions for meeting this need. In this particular treatment modality, a carrier will transport the drug molecule to the desired location, with nanomaterials serving as one such carrier. Nanomaterials are used to transport various types of molecules, such as nucleic acids, proteins, and microRNA.6 The major feature that makes nanoparticles appropriate for cancer therapeutic applications is their size, which ranges from 5 to 100 nm. Furthermore, it improves biodistribution and dosage tolerance for cancer cells, improves the solubility of poorly soluble medicines and slows down their metabolism.7 The physical phenomenon that promotes nanoparticle accumulation in tumour tissues is known as the EPR (enhanced permeability and retention) effect, which is illustrated in Fig. 1. The factor that facilitates the EPR effect is abnormal angiogenesis.8 This is one of the defining characteristics of malignancy, characterized by the uncontrolled growth of blood vessels. Due to their rapid growth, these blood vessels are porous and irregular. The size of these pores can range from a few hundred nanometers to micrometers.7 Nanoparticles with diameters between 10 nm and 100 nm are ideally sized to pass through the gaps in these blood vessels.9Active targeting increases the EPR effect by adding tumour-specific ligands to the surface of nanoparticles. To achieve this, a range of targeted ligands such as aptamers, antibodies, antibody fragments, human transferrin proteins, sugars, peptides, and vitamins such as folate, etc. are employed. The functionalized nanoparticles penetrate the tumour through the EPR effect, and then the targeting ligand binds to the cancer biomarker; this may increase the retention time of the nanocarrier–drug combination inside the tumour cell.10 Aptamer conjugated nanomaterials may offer a safer and more efficient way to fulfil the growing demand for new cancer treatments by combining the features of nanomaterials with the unique identification capacity of aptamers.11
In our comprehensive review, we embark on a journey through the intricate biophysical mechanisms that underpin the functionalization of nanoparticles with aptamers, specifically tailored for cancer therapy. Delving deep into the art of aptamer selection, the nuances of aptamer–nanoparticle design, and the critical factors influencing their interactions, we aim to provide a panoramic understanding of this cutting-edge approach. We explore the latest experimental and computational techniques employed to characterize these functionalized nanoparticles, illuminating their potential to redefine targeted cancer treatment. By unravelling both the promising advancements and challenges in this dynamic field, our insights aim to chart a course towards future innovations in cancer therapy.
2. Aptamers: history and background
Tuerk introduced the term “aptamer”, which is derived from the combination of the Latin word aptus (‘to fit’) and Greek word meros (‘part’), in 1990 to describe short single- or double-stranded DNA or RNA molecules.12 They are generated from a pool of random sequences, through a method known as SELEX. To date, thousands of aptamers have been selected against a wide range of targets, including metal ions, viruses, bacteria, peptides, proteins, cells, and even targets within live animals.13,14 Aptamers show great affinity for their target with a Kd value in the nanomolar to picomolar range.15 Also, the high selectivity of aptamers can differentiate closely related isoforms of the same target molecule. For example, theophylline, theobromine and caffeine have similar structures. Theobromine is an isomer of theophylline with a methyl group in a different position and caffeine differs from theophylline by a single methyl group. The anti-theophylline RNA aptamer shows high discrimination against both of these analogues, and it is proved that the binding affinity of the RNA aptamer with theophylline is 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 times higher than that with caffeine.16 The aptamers are capable of maintaining their binding and inhibitory activity even after being immobilised on carrier material, labelled with different functional groups, and expressed in living cells. Furthermore, their characteristics such as the ability to directly bind with targets, high resistance to various pH and organic solvent environments, low immunogenicity and cytotoxicity, strong chemical stability, molecular weight, and the potential to discover new signature molecules, make them promising candidates for different applications such as target validation, purification processes, drug discovery, therapy, and diagnostics.17–20
000 times higher than that with caffeine.16 The aptamers are capable of maintaining their binding and inhibitory activity even after being immobilised on carrier material, labelled with different functional groups, and expressed in living cells. Furthermore, their characteristics such as the ability to directly bind with targets, high resistance to various pH and organic solvent environments, low immunogenicity and cytotoxicity, strong chemical stability, molecular weight, and the potential to discover new signature molecules, make them promising candidates for different applications such as target validation, purification processes, drug discovery, therapy, and diagnostics.17–20
Nucleic acid aptamers, including DNA and RNA, exhibit similar properties but differ notably in chemical stability and target accessibility. RNA aptamers are chemically less stable due to the presence of a reactive hydroxyl group, whereas DNA aptamers, which lack this group, are more chemically stable. The functionality of both DNA and RNA aptamers relies on their stable three-dimensional structures, influenced by the primary nucleotide sequence, the length of the nucleic acid molecule, and environmental conditions. Typically, aptamers range in size from 25 to 100 nucleotides.21 Common structural motifs include internal loops, stems, hairpin structures, purine-rich bulges, pseudoknots, tetra loops, and G-quadruplexes.22 In 1993, Bock et al. reported the first protein-targeted DNA aptamer, a 15-nucleotide sequence against human thrombin.23 This DNA aptamer features a stacked G-quartet structure, similar to natural single-stranded DNAs, and was developed to inhibit thrombin-catalyzed fibrin clot formation with a binding constant (Kd) of 200 nmol L−1. Additionally, thrombin-specific RNA aptamers have been developed. In 1994, Kubik et al. introduced the first RNA aptamer that selectively bound to thrombin's heparin site, reporting a Kd value of 2–5 nmol L−1, indicating a tenfold higher affinity compared to the DNA aptamer targeting the same protein.24 This demonstrates that the RNA aptamer has a higher affinity for thrombin than its DNA counterpart. Subsequently, White et al. developed an RNA aptamer targeting the fibrinogen binding site of thrombin.25 Later, Gronewold et al. investigated the binding affinity of this RNA aptamer using surface plasmon resonance, binding assays, and acoustic methods, reporting Kd values of 113 nmol L−1, 294 nmol L−1, and 181 nmol L−1, respectively.26
Peptide aptamers, unlike nucleic acid aptamers, are artificial proteins engineered to bind specific target molecules. According to Colas et al., a peptide aptamer is a combinatorial protein molecule with a variable peptide sequence that shows affinity for a target protein and is displayed on an inert scaffold protein.27–29 Because both termini of the variable sequence are fused to the scaffold protein, peptide aptamers are doubly constrained.28,30 This double constraint sets them apart from other artificial combinatorial proteins, which typically consist of a peptide sequence terminally fused to the target protein. Peptide aptamers exhibit molecular recognition similar to antibodies but with enhanced characteristics such as small size, high-yield bacterial expression, rapid folding properties, high solubility, high stability, and the possibility of chemical synthesis. Interestingly, the binding affinity of these combinatorial proteins is increased by the constraints imposed by the scaffold31 (Fig. 2).
 | ||
| Fig. 2 Overall structures of PvLDH complexed with pL1 that fold into a hairpin–bulge contact, gold sphere represents a magnesium ion, (a) tetrameric PvLDH interacts with two DNA aptamers; PvLDH subunits A, B, C, and D are coloured orange, yellow, light green, and dark green, respectively. pL1 A and B are coloured blue and marine blue, respectively. (b) Dimeric PvLDH interacts with a DNA aptamer. PvLDH subunits A and B are coloured orange and yellow, respectively. (c) The structure and schematic of a DNA aptamer pL1. Ribbon depiction of pL1, with secondary structure. The strands comprising the bulge, hairpin loop, and stems are coloured cyan, purple, and black. (d) A comparison of RNA T-loop and DNA T-loop-like motifs that induce the hairpin–bulge interaction. N symbolises nucleotides, while R stands for purine. The nucleotides at the T and G turns are boxed in yellowish green. Adenine, guanine, thymine, and cytosine are coloured red, green, yellow, and blue, respectively.32 | ||
3. SELEX-aptamer generation
Craig Tuerk and Larry Gold first introduced the idea of SELEX (systematic evolution of ligands by exponential enrichment). They used this method to select an RNA sequence targeting T4 DNA polymerase from a combinatorial library of random sequences.12 Theoretically, this library contains 4N modified or unmodified single-stranded oligonucleotide sequences, created by standard organic synthesis. The SELEX process primarily involves three steps: (1) incubating a library of randomized sequences with a target molecule, (2) separating the bound sequences from the unbound ones, and (3) collecting and amplifying the bound sequences using PCR.33 This selection process is repeated until the sequences with the desired affinity for the target are enriched. The basic methods for aptamer selection have remained largely the same; there have been significant advancements in several areas, including characterisation, target types and modification (how aptamers can be modified for different applications).34Recent advancements in computerized analysis and sequencing technologies have enhanced our understanding of aptamer dynamics during the selection process. This progress will help identify effective sequences with high affinity in a shorter time.35 Additionally, new polymerases have been introduced to create sequence libraries with high nuclease hydrolysis stability, reducing the need for labour-intensive post-selection modifications. To further enhance serum stability for therapeutic applications, various derivatives with modifications on the sugar rings are used to incorporate unnatural nucleotides into oligonucleotides.36 These modifications include 2′-fluoro (2′-F) ribose, 2′-amino (2′-NH2) ribose, 2′-O-methyl (2′-OMe) ribose, and locked nucleic acids (LNAs), which bridge the 2′ and 4′ positions of ribose covalently.34
4. Aptamer–target interactions in SELEX
The interaction between the aptamer and its target is the primary factor that determines the effectiveness and quality of the SELEX process.37 The efficiency of this interaction is influenced by several variables, including the secondary structure of the aptamer, the diversity of aptamer sequences, the type and size of the target molecule, the immobilization matrix, physicochemical properties, and chemical modifications.38,39 It is critical to characterise this aptamer–target interaction for the tailoring of an aptamer with good affinity. Several biophysical instruments or techniques have been developed to measure aptamer–target binding affinity, including capillary electrophoresis (CE), isothermal titration calorimetry (ITC), surface plasmon resonance (SPR), circular dichroism (CD), microscale thermophoresis (MST), photonic crystal surface waves (PC SWs), and a quantum crystal microbalance (QCM). CD and CE are used to investigate the aptamer's conformational changes upon binding to its target. Similarly, SPR, PCSW, and QCM are used to monitor the affinity interaction and related kinetics in real time. ITC and MST are employed to investigate the thermodynamic properties of the aptamer–target interaction38,39 (Fig. 3).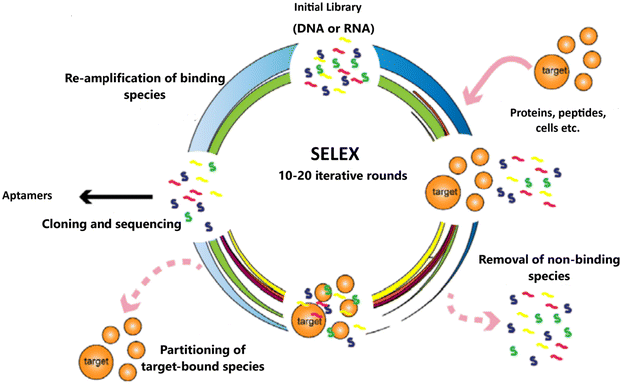 | ||
| Fig. 3 Schematic illustration of the SELEX process for the DNA and RNA library.40 | ||
5. The factors influencing aptamer–target interactions
5.1 Target molecule
The interaction between an aptamer and its target is influenced by the structure, shape, and dimensions of the target molecule. The target molecule can be either smaller or larger than the aptamer. When the target molecule is smaller, such as aromatic ligands, ions, oligosaccharides, or charged amino acids, it integrates into the aptamer structure. This interaction is typically facilitated by stacking interactions (with flat aromatic ligands and ions),38–40 electrostatic complementarity (with oligosaccharides and charged amino acids),41,42 or hydrogen bonding.43–46In contrast, if the target molecule is a larger protein, the aptamer integrates into the protein structure or attaches to its surface. Due to protein's complex structure, this interaction is more diverse.42,47,48 However, hydrogen bonding still plays a significant role in this interactions.
5.2 Aptamer structure complexity and stability
The next factor influencing aptamer–target interactions is the structural complexity of the aptamer. Due to sequence-dependent propensities, aptamers can assume a multimeric state by forming G-quadruplexes or i-motifs, both of which facilitate interactions between multiple oligonucleotides.41–43 According to the “induced fit” principle, the formation of an aptamer–target complex can result in conformational changes to the target, the aptamer, or both.44–46 This induced fit leads to structural complementarity, influencing short-range interactions such as hydrogen bonding and van der Waals contacts. Conformational changes can also lead to catalytic activity, where the aptamers are known as aptazymes. The negative charges on the surface of the target can negatively impact the aptamer–target interactions due to repulsive interactions with the highly electronegative phosphate groups in DNA and RNA.47,48 While positive charges can enhance the interaction strength, they may also increase the likelihood of non-specific binding. Generally, aptamers are hydrophilic, making them less likely to interact with hydrophobic targets.49,50In biological systems, the stability and functionality of unmodified aptamers are compromised by nuclease degradation.51,52 The stability of aptamers in intracellular environments, which is a major concern for therapeutic applications, can be enhanced from minutes to hours through chemical modifications or by selecting polyvalent aptamers.53 Additionally, pre-SELEX and post-SELEX chemical modifications play a crucial role in tailoring aptamers for various applications. This enhancement can be achieved through terminal modifications, such as adding modified nucleotides to the aptamer backbone, or by nucleotide modifications using different nucleic acids.54
In 2012, Li et al. introduced a polyvalent aptamer with multivalent interactions for increasing the nuclease stability. This is accomplished by covalently linking desired linkers to aptamers or by combining nanoparticles with aptamers.55 In addition, the polyvalent aptamers have a low dissociation constant, and thus, a higher binding affinity. Seferos et al. developed a gold nanoparticle–polyvalent oligonucleotide complex with enhanced nuclease stability.56 This complex features, a 13 nm gold nanoparticle (Au NP) functionalized with both antisense oligonucleotides and synthetic peptides. It exhibits perinuclear localization and enhanced gene regulation activity when tested in cellular models.
5.3 pH and temperature
The tertiary complex formed between an aptamer and its target is stabilized through a binding mechanism that relies on protonation.57 If the pH is greater than the critical pKa value of the target, the aptamer–target complex may form via hydrogen bonding. However, when the pH is lower than the pKa of the target, binding-linked protonation prevents the target from binding to the aptamer. Plich et al. found that the pH of an RNA–aminoglycoside complex affected its thermodynamic stability. They also observed that the protonation at acidic pH decreased aptamer–target coulombic repulsion, leading to prolonged retention times.58 The interaction between the aptamer and its target molecule involves thermodynamic processes such as entropy and enthalpy.59 Therefore, temperature significantly influences binding affinity. According to McKeague et al., the optimal temperature for achieving optimal binding kinetics and high affinity is approximately 37 °C.606. Types of SELEX
6.1 Capillary electrophoresis SELEX
Capillary electrophoresis (CE) serves not only as a selection tool but also as an analytical technique capable of determining binding affinity and monitoring the enrichment of ssDNA in each round of selection cycles, as well as assessing the quality of both the target and the ssDNA library. It offers advantages over other conventional SELEX methods, such as enhanced analysis capabilities, reduced sample and analytical reagent consumption, and improved screening efficiency.61–65 CE-SELEX typically requires only 1–4 rounds for aptamer selection, significantly reducing selection cycles and time.66,67 Furthermore, in CE-SELEX, aptamer binding and separation take place in free solution, eliminating steric hindrance of the interaction. Although all conventional SELEX methods, including CE-SELEX, rapid-SELEX, and microfluidic SELEX, necessitate prior knowledge of the target, they also require purified protein for aptamer selection. Consequently, aptamers selected in vitro using these methods often fail to recognize their target proteins in their native conformation. Traditionally, CE-SELEX was primarily associated with selecting aptamers for large protein targets. However, in recent times, this technique has been successfully employed to select aptamers that target small peptide molecules such as neuropeptide Y, which has a size of 4272 Da—smaller even than DNA. With small molecule targets, the mobility of the complex changes minimally compared to non-binding sequences, posing challenges in distinguishing between binding and non-binding sequences. Nonetheless, PCR can exponentially amplify nucleic acids even from small sample amounts, enabling sufficient enrichment. Bowser's team pioneered the concept of CE-SELEX, applying it to proteins, peptides, and small molecules.65,68 They utilized high-throughput sequencing techniques to analyze the evolution of sequence enrichment and affinity differences. Using this approach, Bowser and Mendonsa successfully selected an aptamer targeting IgE (with a Kd of less than 30 nM) after just four rounds of selection.68 These efforts represent significant advancements and innovations in SELEX. In 2005, Mosing et al. also employed this method to develop an aptamer for HIV reverse transcriptase following four rounds of selection.696.2 Toggle SELEX
Many potential therapeutic molecules show excellent in vitro activity but often fail to progress to clinical trials due to their lack of expected efficacy in vivo. This discrepancy may stem from molecules developed to target human proteins exhibiting lower affinity for analogous proteins in animal models.25 To address this issue, innovative models enable the substitution or insertion of human homologues into other species. For example, the in vivo efficacy of a human L-selectin aptamer was demonstrated in a xenogeneic model (SCID mouse/human lymphocyte),70 although such approaches are costly and less feasible. In response to these challenges, White et al. introduced a novel selection technique aimed at enhancing the development of therapeutic molecules that exhibit cross-reactivity between species.25 They introduced the concept of toggle SELEX to develop aptamers that were clinically relevant in humans and testable in animal models. Using this cross-reactive selection strategy, they successfully created a nuclease-resistant RNA ligand capable of binding to both human and porcine thrombin by alternating the protein target between these two species during selection rounds. Such aptamers demonstrate the ability to inhibit both porcine and human thrombin activity.256.3 Microfluidic SELEX
Serum comprises a diverse array of components, including proteins, enzymes, lipids, hormones, small molecules, and electrolytes. In this technique, aptamers are selected against a serum pool. Initially, the nucleotide library undergoes negative SELEX to eliminate sequences binding to normal serum components, thereby enhancing the affinity of the isolated aptamers.71 This refinement step improves the effectiveness of the selection process,72 enabling the creation of highly specific and selective aptamers suitable for various applications. Using this approach, aptamer apt-5 was isolated, which targeted human CLEC3b and demonstrated significant impacts on tumour immunity and infection.73,74 Additionally, Li et al. identified six distinct aptamers by utilizing serum samples from individuals with lung cancer.6.4 Cell-SELEX
The in vitro method used for selecting aptamers against living cells is known as cell-SELEX.75 This approach is effective at identifying high-affinity aptamers against various targets, such as the purified extracellular domain of prostate-specific membrane antigen (PSMA),76 pure MUC1 peptides,77 protein tyrosine phosphatase 1B (PTP1B),78 and the cell adhesion molecule P-selectin.79 The initial application of cell-SELEX involved targeting the pathogenic organism Trypanosoma brucei.80 The study led to the discovery of three RNA aptamers that specifically targeted the effective bloodstream life cycle stage of the parasite. These aptamers are directed towards the parasite's flagellar pocket; however, none of them exhibit binding to VSG proteins, which are highly prevalent polypeptides on the trypanosome's surface.The cell-SELEX technique, predominantly utilized in cancer therapy, generates a range of aptamers aimed at different types of cancer cells. These encompass cells originating from human hepatocarcinoma, mouse tumour endothelial cells, small cell lung cancer, glioblastoma multiforme, lung adenocarcinoma, colorectal cancer, and prostate cancer. Compared to conventional SELEX, cell-SELEX offers numerous advantages. For instance, this technology enables the identification of both established and novel biomarkers present on the cell membrane surface.81 Cell-SELEX specifically targets live cells with a multitude of surface molecules, facilitating the development of a diverse panel of aptamers. This capability is invaluable for precise disease diagnosis. Daniel et al. isolated a pool of highly specific DNA aptamers by targeting the glioblastoma-derived cell line U251. They subsequently identified and characterized tenascin-C as a target protein for one of the isolated oligonucleotides; this is an extracellular protein found in the tumour matrix.82 However, the selected aptamer exhibits reduced affinity at physiological pH and is susceptible to nuclease degradation. Subsequently, Hicke et al. described a modified DNA aptamer stabilized against nucleases, targeting tenascin-C. This aptamer binds specifically to the fibrinogen-like domain of tenascin-C.75 While this technique offers significant benefits, it also presents certain technical challenges. Specifically, selecting an aptamer against a target protein that is expressed at low levels on the cell membrane can be difficult. In cell-SELEX, it is critical to avoid erroneous binding caused by cell death and dying cells during the selection process, as this can diminish the overall quality of the selection.84 Therefore, high-speed fluorescence-activated sorting (FACS) can be employed to effectively separate live cells from dead ones.76
To date numerous aptamers have been developed by using cell-SELEX to diagnose different types of cancers, which includes glioblastoma, leukaemia, cholangiocarcinoma, liver cancer and breast cancer; these aptamers have also been applied in cancer classification and treatment. Zhao et al. explored the specificity of four aptamers (s1, s6, s11e, and s15) selected for adenocarcinoma A549, evaluating them across different cancer cell types.73 Their study revealed that while all four aptamers could detect adenocarcinoma cells, only s11e could identify both types of squamous carcinoma cells. This research highlights the potential of these aptamers in differentiating between distinct subtypes of large carcinoma cells. Moreover, the binding affinities of these four aptamers to different samples of large carcinoma cells vary, suggesting their capability to discern molecular distinctions. In a related study, Shangguan et al. isolated a DNA aptamer, sgc8c, specifically targeting the T cell acute lymphoblastic leukemia cell line CCRF-CEM, with a focus on diagnosing acute lymphoblastic leukemia.100 They demonstrated that sgc8c could detect leukemia cell lines in human bone marrow samples obtained from clinical specimens, highlighting its promise as a molecular probe for cancer detection. Suwussa et al. developed ACMNPs (aptamer-conjugated magnetic nanoparticles) capable of distinguishing between various types of cancer cells.91 This study involved selecting different aptamers—sgc8c targeting CCRF-CEM cells, KDED2a-3 targeting DLD-1 cells, and KCHA10 targeting HCT 116 cells—and coupling them with magnetic nanoparticles. These nanostructures exhibit enhanced stability and are employed for cancer detection. The specificity and selectivity of the aptamer–nanoparticle complexes were evaluated in diverse complex biological environments and cell mixtures, including fetal bovine serum albumin (BSA), whole blood, and human plasma. Notably, in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 mixture of target and non-target cells, the sgc8c aptamer–NP complex demonstrated high sensitivity. Similarly, the selectivity of the other two aptamer complexes was also found to be high. For further details, refer to Table 1, which lists the variety of aptamers selected through cell-SELEX, their targets, and the corresponding cancer types.
100 mixture of target and non-target cells, the sgc8c aptamer–NP complex demonstrated high sensitivity. Similarly, the selectivity of the other two aptamer complexes was also found to be high. For further details, refer to Table 1, which lists the variety of aptamers selected through cell-SELEX, their targets, and the corresponding cancer types.
| Aptamer | Target | Cancer type | Ref. |
|---|---|---|---|
| TTA1 | Matrix protein, tenascin-C | Breast, glioblastoma, lung and colon cancer | Hicke et al.77 |
| AS1411 | Nucleolin (plasma membrane of cancer cell) | Breast, prostate, lung, pancreatic cancer and renal carcinoma | Girvan et al.78 |
| MUC-1 | Mucin (expressed on human adenocarcinomas) | Lung cancer | Ferreria et al.79 |
| Apta-3, Apta-5 | Carcinoembryonic antigen (CEA) in T84 cells | Colorectal cancer | Melo et al.80 |
| Apc1 | Caco2 | Li et al.81 | |
| SKBR-3-R1 Tr | Luminal A breast cancer subtype SKBR3 cells | Breast cancer | Ferreira et al.82 |
| Ag-9, Ag-9g, A10, A10-3, A10 L, A10-3.2, A10-3-J | PSMA | Prostate cancer | Lupold,83 Leach et al.,84 Dassie et al.,85 Rockey et al.86 |
| HPV-07 | Type-16 virus-like-particle | Cervical cancer | Rockey et al.,86 Trausch et al.87 |
| c-9s | Ca ski cells, HeLa cells | Wang et al.71 | |
| Seq-3, seq-6, seq-19 and seq-54 | Circulating tumour cells | Gastric cancer | Li et al.88 |
| Spl3 EN-2 binding | Cyto-skeleton associated protein-4 | Bladder cancer | Kim et al.89 |
| ssDNA aptamer | Engrailed 2 (EN-2) | Sun et al.90 |
6.5 Tissue SELEX
Generally, for isolating aptamers against live cells, cell-SELEX is developed as a modified SELEX strategy that uses the living cell as the target. However, this process is time consuming and laborious, and the binding affinity and specificity of the selected aptamers are highly dependent on the selection process.92 In addition, the primary cultured cell line cannot reflect the actual in vivo tumour occurrence and development, which limits the clinical application of the cell-SELEX aptamers. Accordingly, a new variant of the selection technique known as tissue SELEX is introduced. Tissue SELEX enables the identification of aptamers against different target molecules, including membrane components and intracellular substances. As an example, Li et al. developed aptamer BC15 by targeting the intracellular protein hnRNP A1 through tissue-based SELEX.92 By using tissue SELEX, Zhong et al. isolated aptamer TC-16, which showed high affinity and specificity for papillary thyroid carcinoma (PTC).936.6 Bead based SELEX
Immobilizing target molecules on various solid supports, such as agarose, Sepharose, and Sephadex, is a commonly used approach in SELEX.94 Bead-based SELEX can be performed in two ways: first, by immobilizing the target on a solid support and then incubating it with an oligonucleotide library; second, by incubating the target with an oligonucleotide library and then selecting the best aptamer–target complex based on the highest affinity interaction.95,96 In both methods, steady-state equilibrium binding occurs, while non-equilibrium binding under flow conditions occurs only in the first method. This enables users to select aptamers based on various kinetic parameters. In the pre-immobilization scenario, the density of the target molecule on the solid support is crucial, as it influences the enrichment of aptamer sequences in each round of SELEX. An example of bead-based selection is the isolation of a DNA aptamer against staphylococcal enterotoxin B (SEB).97 Another example is the isolation of a PDL-1 (programmed death ligand 1) specific aptamer with a Kd value of 64.77 nM after 10 rounds of SELEX to serve as a cancer biomarker.286.7 SPR-SELEX
The primary reason that conventional SELEX is time-consuming is that selecting and evaluating the binding affinity of an aptamer is a two-step process.94 SPR-SELEX addresses this limitation by evaluating pool enrichment in real-time, round by round, during the selection process. This method monitors binding by tracking the SPR signal during pool injection and partitioning. If no further evolution occurs in the SPR signal, the selection procedure can be stopped. SPR-SELEX accelerates aptamer isolation by simultaneously selecting and evaluating binding sites. This approach is applicable to any type of target, provided that the oligonucleotide–target dissociation does not exceed the instrument's recovery time. Dausse et al. successfully employed SPR-SELEX to isolate a high-affinity RNA aptamer (Kd value = 8 nM) that formed stable loop–loop complexes with the target after six rounds of selection.987. Aptamer-conjugated nanoparticles in targeted drug delivery
Aptamers serve as ideal targeting ligands for functionalizing nanomaterials in various applications such as therapies, diagnostics, and drug delivery. Despite showing promising results in preclinical tests, aptamer–nanoparticle complexes have not yet progressed to the clinical stage of therapeutic development.11Table 2 details the aptamers utilized to functionalize nanoparticles for targeted cancer therapy, emphasizing their potential to enhance the precision and effectiveness of cancer treatments.| Aptamer | Nanomaterial | Drug | Target | Cancer cell line | Ref. |
|---|---|---|---|---|---|
| A10, RNA | PLA–PEG–COOH | Rho-labelled dextran | PSMA | Prostate cancer | 99 |
| A10, RNA | PLA–PEG–COOH | Docetaxel | PSMA | Prostate cancer | 100 |
| A10, RNA | PLA–PEG–COOH | Cisplatin | PSMA | Prostate cancer | 101 |
| A10, RNA | SPION | Doxorubicin | PSMA | LNCaP cell line | 102 |
| A10, RNA | H40–PLA–PEG | Doxorubicin | PSMA | CWR22Rγ1 cells | 103 |
| A10, RNA | PLGA | Triplex forming oligonucleotide | PSMA | LNCaP cells | 104 |
| A10-3.2, RNA | PEG–PAMAM | mRNA | PSMA | Prostate cancer | 105 |
| A10-3.2, RNA | Atelocollagen | mRNA | PSMA | Prostate cancer | 106 |
| A10, RNA and DUP-1 | PEG–gold nanostar | None (PTT) | Prostate cell | Prostate cancer | 107 |
| A9, RNA | SPION | Doxorubicin | PSMA | LNCaP cell line | 108 |
| A9, RNA | ONT–PAMAM dendrimer | Doxorubicin | PSMA | Prostate cancer | 109 |
| AS1411, DNA | Liposome | Cisplatin | Nucleolin | MCF-7 cells | 110 |
| AS1411, DNA | PEG–PLGA | Paclitaxel | Nucleolin | Glioma | 111 |
| AS1411, DNA | PLGA–lecithin–PGA | Paclitaxel | Nucleolin | GI-1 and MCF-7 cells | 112 |
| AS1411, DNA | PLGA | Paclitaxel | Nucleolin | GI-1 cells | 113 |
| AS1411, DNA | PEG–PCL | Docitaxel, DiR, coumarin-6 | Nucleolin | bEnd.3 and C6 cells | 114 |
| AS1411, DNA | Mesoporous silica | Gold nanorods | Nucleolin | MCF-7 cells | 115 |
| AS1411, DNA | Mesoporous silica | Fluorescein | Nucleolin | MDA-MB 231 | 116 |
| AS1411, DNA | DNA nanotrains | DOX, EPI and DAU | Nucleolin | HeLa cells | 117 |
| Sgc8c, DNA | Aptamer DNA | Antisense ONT to P-gp | CCRF-CEM cells | ALL | 118 |
| S2.2, DNA | ZnO nanoparticle | Doxorubicin | Mucin-1 | MCF-7 cells | 119 |
| 5TR1, DNA | PLGA–chitosan | Epirubicin | Mucin-1 | MCF-7 cells c26 cells | 120 |
| S15, DNA | PEG–PCL | Paclitaxel | NSCLC | A549 cells | 121 |
8. Aptamer–nanoparticle conjugation
The direct or indirect attachment of an aptamer to a nanoparticle is achieved using a linker molecule, often referred to as a spacer or bridge. Additionally, this conjugation can occur through covalent or non-covalent interactions.11 Typically, covalent conjugation occurs when functional groups at the end of an aptamer interact with functional groups on the nanoparticle's surface. Common functional groups on an aptamer include primary amino groups and thiol groups, while nanoparticles may have carboxylic acid groups, aldehyde groups, or maleimide groups on their surfaces.10,122,123 Moreover, the interaction of aptamer functional groups with linker molecules, metal ions, and inorganic molecules in nanoparticles also facilitates covalent conjugation. Common types of covalent bonds in aptamer–nanoparticle conjugation include amide linkages, thioester bonds formed from the interaction between carboxylic acid and thiol groups, ester bonds resulting from the interaction between primary amine groups and thiols, disulfide bonds created by the interaction of two thiol groups, and Au–S and Ag–S bonds where thiol groups interact with gold or silver. Noncovalent conjugation encompasses both electrostatic and affinity interactions.124 The latter includes streptavidin–biotin and avidin–biotin interactions, while the former involves a linker molecule facilitating the interaction between the aptamer and nanoparticle through opposing charges. Most reported aptamer–nanoparticle conjugates are direct and covalent.125 Farokhzad et al. suggested that covalently attached aptamer nanoparticles were extremely stable in physiological salts and pH.109. Aptamer–biological material conjugation for cancer therapy
Biological materials, which are biocompatible and naturally occurring, are designed to perform, enhance, or substitute natural functions, primarily for medicinal and pharmaceutical applications. Aptamers have been integrated with various biological materials, such as RNAi reagents, microsomes, antibodies, and DNA origami. These innovative conjugates are employed in the treatment of numerous diseases, including cancer, diabetes, and rheumatoid arthritis.1269.1 Aptamer–antibody and short peptide conjugation
A wide range of small peptides and antibodies are produced to treat various disorders. However, these protein drugs often lack cell selectivity and efficient tumour penetration. Conjugating aptamers with short peptides and antibodies may help overcome these limitations. In 2016, Heo et al. developed an aptamer–antibody complex, named an oligobody, for targeted cancer therapy.126 They conjugated an aptamer, t44-OME, to anti-cotinine antibodies. This t44-OME–anti-cotinine antibody complex penetrates cancer tissue more deeply than the antibody alone. Moreover, the anti-cotinine antibody significantly increases the pharmacokinetics of the t44-OME aptamer without affecting its affinity, indicating that the aptamer and antibody mutually enhance each other's pharmacokinetic properties. Similarly, Passariello et al. developed a bispecific aptamer–antibody complex.127 This complex includes an anti-epidermal growth factor receptor (EGFR) aptamer and an anti-epidermal growth factor receptor 2 (ErbB2) compact antibody. The aptamer–antibody complex enhances the cancer cell killing ability by redirecting T cells against cancer cells. They demonstrated that this complex not only increased cell specificity but also improved the pharmacokinetics of the aptamer.In addition to antibodies, short peptides are also combined with aptamers to enhance their specificity and bioactivity. Rajabnejad et al. developed an aptamer–peptide conjugate,128 comprising an anti-nucleolin aptamer (AS1411) and the short peptide metilin, which targeted cancer cells. Metilin, a 26-amino acid peptide, is renowned for its anticancer properties. However, many studies have found that, while metilin is effective against cancer, it also causes significant adverse effects such as hemolysis and liver injury. Pusuluri et al. introduced a new aptamer–peptide conjugate incorporating chemotherapy drugs to enhance potency.129 In this study, they conjugated a drug preloaded peptide (doxorubicin and camptothecin (CPT)) with an anti-nucleolin aptamer, aiming to avoid suboptimal exposure to individual drugs. Typically, chemotherapy drug combinations are administered at maximum tolerated doses, which can cause severe side effects and reduce therapeutic efficiency. However, the aptamer–peptide conjugate has the potential to mitigate these limitations. This conjugate achieved effective anti-tumor activity in vivo with an optimal drug concentration, approximately 500 μg kg−1 dose of doxorubicin and 350 μg kg−1 dose of CP.
9.2 Aptamer–nucleic acid drug conjugation
The emergence of RNA interference (RNAi) in 1990 paved the way for the use of nucleic acid drugs to treat a wide range of disorders.144 Today, a variety of nucleic acid drugs, including antisense nucleotides, miRNA, siRNA, mRNA, and aptamers, have been developed. Additionally, various delivery platforms are being designed to transport these drugs.145,146 Among them, viral vectors are the most common; however, they have limitations such as immunogenicity, biohazards, and mutagenesis.147 On the other hand, non-viral vectors like liposomes are safer but lack specificity and efficiency.148,149 Therefore, there is a need for a more specific and biologically secure delivery system for nucleic acid-based drugs. Consequently, the aptamer–siRNA complex has been extensively investigated over the past decade. The conjugation of aptamers and siRNA can be accomplished through covalent or non-covalent interactions. Additionally, a linker, U′–U′–U′, is often added between the aptamer and siRNA for effective delivery.130 Since both aptamers and siRNA are nucleic acid drugs, complementary annealing is also used for their conjugation. Zhou et al. introduced the concept of a sticky bridge to enhance aptamer–siRNA conjugation.131 This aptamer–siRNA complex has been shown to inhibit HIV-1 replication and infectivity in cultured T cells and primary blood mononuclear cells. Recently, Jeong et al. developed a multivalent aptamer–siRNA complex to overcome multiple drug resistance in various cancer cells.132 This conjugate is constructed using an aptamer against mucin-1, siRNA against bcl-2 genes, and the chemotherapeutic drug doxorubicin.9.3 Aptamer–DNA nanostructure conjugation
Due to its low immunogenicity and excellent biocompatibility, DNA is ideal for a variety of biological applications.154 Furthermore, the programmable structure of DNA can significantly contribute to DNA nanotechnology, enabling the design of smart drug delivery systems. Additionally, the three-dimensional structure and form of DNA are adjustable, with its size maintainable at around 50 nm, which is optimal for cell entry. Despite its high delivery rate, DNA alone lacks targeting and precision, necessitating the integration of aptamers to enhance specificity.133–137 Since both aptamers and DNA are derived from nucleic acid components, aptamers can be easily integrated into DNA structures. Zhu et al. constructed a DNA nanotrain by assembling aptamers onto DNA via complementary sequences.138 Similarly, Han et al. developed a DNA tetrahedron by conjugating anti-cancer aptamers.139,140 Chemotherapy drugs such as doxorubicin often cause significant side effects, reducing the quality of therapy. Doxorubicin, with its excellent binding affinity to DNA duplexes, can be integrated into aptamer–DNA complexes to minimize adverse effects on healthy cells and tissues. Zeng et al. demonstrated this by creating a DOX-integrated aptamer–DNA complex, which reduced the side effects typically associated with doxorubicin.141,142 In addition to enhancing targeted efficacy, aptamers facilitate regulated drug release in DNA–aptamer complexes. Church and colleagues initially developed an aptamer–DNA origami combination for the encapsulation and targeted delivery of thrombin to cancer blood vessels. The anti-nucleolin aptamer in this complex has molecular triggering activity, aiding in the release of the chemotherapeutic drug thrombin from the complex. This integration not only targets drug delivery but also ensures controlled release, maximizing therapeutic efficacy while minimizing side effects143 (Fig. 4).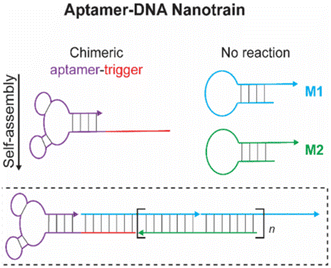 | ||
| Fig. 4 Schematic representation of DNA nanostructure assembly. DNA nanotrain containing aptamers assembled by complementarity of the sequence.138 | ||
10. Aptamer–non-biological material conjugation for cancer therapy
Beyond biological materials, significant advancements have been made in a diverse array of nanomaterials, including micelles, hydrogels, gold nanoparticles, silica nanoparticles, and liposomes, for biomedical applications. The development of new synthesis and characterization techniques has empowered researchers to meticulously design nanomaterials with precise control over their size, shape, and properties. These nanoparticles boast unique attributes, such as a large surface area, high loading capacity, flexibility, and the enhanced permeability and retention (EPR) effect, making them outstanding candidates for therapeutic use.154,155 Additionally, certain nanomaterials exhibit extraordinary features, like the surface plasmon resonance phenomenon. To further enhance their therapeutic potential, nanoparticles are often conjugated with targeted ligands, such as aptamers, improving their efficacy and specificity.10.1 Aptamer–polymeric nanoparticle conjugation
These particles, ranging in size from 1 to 1000 nm, have proved to be highly effective in the targeted delivery of drugs for various disorders. Numerous polymeric nanoparticles, including micelles, hydrogels, and branched polymeric structures, are currently being developed and investigated.154 Micelles, which are polymeric nanospheres composed of amphiphilic copolymer blocks formed through a thermodynamic process, offer numerous advantages as nanocarriers. These benefits include strong biodegradability, biocompatibility, a high drug payload capacity, and extended circulation and retention times.155 Additionally, the hydrophobic reservoir cores of micelles aid in the delivery of poorly soluble drug. To enhance their application, researchers have begun modifying the micelle nanostructure by incorporating targeting ligands such as aptamers. The idea of aptamer–micelle conjugation was first introduced by Miyamoto and colleagues in 2007.144 Following that, a number of aptamer–micelle conjugates were developed for medicinal purposes. In general, aptamers are conjugated to micelles using a variety of techniques, including click chemistry,157 base-pairing hybridization,158 and 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide (EDC)-mediated conjugation.159 In the aptamer–micelle complex, the aptamer acts as a probe for cancer-related markers such as EGFR,160 MUC1,161 PTK7 (protein tyrosine kinase),138 PSMA (prostate-specific membrane antigen),115 and HER2 (human epidermal growth factor 2).162 Xu et al. created a spherical-shaped aptamer–micelle conjugate with a diameter of 69 nm by coating a unimolecular micelle on the surface of the aptamer.103 In this case, the aptamer was chosen to target the prostate cancer biomarker PSMA. The DOX-loaded aptamer–micelle complex exhibited higher cytotoxic activity against PSMA-positive CWR22Rv1 cancer cells compared to the aptamer-free DOX–micelle complex.163 Subsequently, a number of aptamers targeting different cell lines, including MDA-MB-231 cells145 and Ramos cells,146 were incorporated into micelles. In 2009, Wu et al. developed a micelle structure functionalized with an aptamer targeting Ramos cells.146 The other benefits offered by aptamers to micelle nanostructures include increased cellular take up and reduced systemic toxicity. In 2015, Li et al. developed a micelle nanostructure consisting of AS1411 aptamers, the amphiphilic polymer Pluronic F127, and CD-PELA copolymers, employed to deliver DOX to human breast tumours.147 The results showed that aptamer AS1411 in the micelle nanostructure improved the cellular take up and cell specificity of doxorubicin via nucleolin-mediated endocytosis.For further applications, the aptamer–micelle nanostructure is conjugated to functional elements such as imaging agents and stimuli-responsive components. For example, in 2014, Tian et al. developed a multifunctional micelle–aptamer nanostructure by combining a NIR photosensitizer (R16FP) and a pH-active fluorescent probe (BDP-668).145 In this conjugate, the NIR photosensitizer mediated the lysosomal destruction of cancer cells by generating reactive oxygen species (ROS), while BDP-668 enhanced the visualization of therapeutic changes in real-time.
Hydrogels represent another category of polymeric nanomaterials, composed of hydrophilic and cross-linked polymeric networks. These materials are designed to respond to various stimuli, including pH,148 temperature,149 light exposure,150 magnetic fields,151 and ionic strength.152 The conjugation of aptamers with hydrogels has a range of applications in the biomedical field, such as targeted delivery, capture and release of molecules, enhanced control of the gel–sol transition, and volume changes.153 However, the high material permeability of hydrogels limits their application in controlled drug delivery, making it challenging to achieve precise drug release. Incorporating aptamers into hydrogels via an oligo-driven gel–sol transition may provide a solution to this limitation.154 In 2008, Yang et al. designed a hydrogel cross-linked with an adenosine-specific aptamer system.155 The gel–sol transition of this system, triggered by adenosine binding, was monitored by a gold nanoparticle, facilitating selective drug release. Following this, Wang et al. developed aptamer-conjugated hydrogel systems for the selective capture and release of individual proteins and cells.156–159 They also created an aptamer-incorporated hydrogel capable of releasing BB-PDGF (B chain platelet-derived growth factor) by controlling analyte–ligand binding. Additionally, they designed a capture-and-release aptamer–hydrogel complex for cancer cells.160 Future research may focus on the potential of these systems for tissue remodeling and regeneration.
10.2 Aptamer–gold nanoparticle conjugation
Gold nanoparticles are highly valued in biomedical applications due to their diverse physical and chemical properties. Advances in synthesis techniques further enhance their versatility across various applications.161 Their optical, electrical, magnetic, and biochemical properties make them desirable for diverse uses including bioimaging, drug delivery, radiation, and photothermal therapy.162–165 Recently, there has been increasing interest in functionalizing gold nanoparticles with aptamers. Numerous aptamer-functionalized gold nanomaterials have been developed, with a predominant focus on applications in cancer therapy. Wu et al. developed a nanosystem composed of Ag, Au, and aptamer S2.2, specifically targeting the MUC1 cell surface biomarker highly expressed in breast cancer MCF-7 cells.166 This system utilizes inherent surface-enhanced Raman scattering (SERS) for bioimaging and phototherapy applications. Aptamers with structure-switching capabilities enable controlled drug release, facilitating precise and selective drug delivery. In 2012, Wang et al. introduced a nanocomplex comprising gold nanorods (Au NRs), an aptamer switch probe (ASP), and the photosensitizer Ce6 (Chlorine6), utilized for targeted photodynamic therapy (PDT) and photothermal therapy (PTT).193 ASP undergoes structural changes in the presence of cancer cells, displacing Ce6 from the gold nanostructure's surface to induce singlet oxygen for PDT. The radiation absorbed by AuNRs further enhances cell death through PTT, resulting in a synergistic therapeutic effect. The inherent PTT property of gold nanoparticles offers potential for novel combination cancer therapies. Yang et al. then developed the Apt–AuNP–GO nanocomplex, combining aptamer-functionalized gold nanoparticles with graphene oxide, which exhibited therapeutic efficacy against cancer cells through near-infrared (NIR) light-activatable PTT.167 This nanocomplex demonstrates a therapeutic response in cultured human breast cancer cells by modulating heat shock proteins. Building upon this approach, Yang et al. engineered a therapeutic complex comprising aptamers, gold nanoparticles (NPs), graphene oxide (GO), and an HSP 70 inhibitor, showcasing beneficial therapeutic effects for breast cancer cells. Recent advancements also include image-guided therapies facilitated by aptamer–gold NP complexes. For instance, He and colleagues developed aptamers selected against A549 cells, labeling Raman-tagged gold nanoparticles in 2019 for combined chemo-PTT.168 Incorporating a Raman signal agent (4-MBA) into gold NPs, this conjugate demonstrates promising properties for targeting and eliminating cancer cells.This evolving field underscores the potential of aptamer-functionalized gold nanoparticles for enhancing targeted therapies and advancing the frontier of cancer treatment through innovative nanoengineering approaches.
10.3 Aptamer–magnetic nanoparticle conjugation
The inherent super-paramagnetism of magnetic nanoparticles has led to their use in a variety of applications, including drug administration, MRI, cancer therapy, and individual cell isolation.169–172 To enhance their properties for specific molecular capture and collection, these magnetic nanoparticles are often functionalized with aptamers. In 2013, Dai et al. introduced a novel nanosystem by incorporating an anti-multidrug-resistant-bacteria (MDRB) aptamer onto a magnetic core–plasmonic shell (MCPS).173 In this nanocarrier, aptamer S8-7 helps to hold methylene blue (MB), which is used for fluorescence imaging and photodynamic therapy (PDT). Additionally, the gold coating on this nanocarrier enables the combined photothermal destruction of target bacteria, demonstrating its potential as a multifunctional therapeutic tool for MDRB in clinical practice. Furthermore, the photothermal therapy (PTT) efficiency of magnetic nanoparticles can be enhanced by incorporating aptamers with exceptional tumour-targeting capabilities. Aravind et al. developed a new aptamer-loaded magnetic fluid nanoparticle system in 2013, which included a paclitaxel-loaded fluorescent label for targeted chemotherapy, optical imaging, and drug release.174 The aptamer in this complex targets nucleolin, a protein, the increased expression of which is linked to a poorer cancer prognosis. This system not only delivers paclitaxel via the nucleolin-specific aptamer (NSA) but also has the potential to magnetically direct the payload to tumour regions. In 2019, Zhao et al. designed a nanocomplex of aptamer-conjugated Fe3O4@carbon@DOX for combined chemophotothermal therapy (CPTT).175 Aptamer sgc8 in this nanocomplex targets the lung cancer cell line A549. This nanocomplex exhibits enhanced photothermal conversion efficiency and pH or heat-induced DOX release during PTT. Additionally, the system reduces the contrast enhancement of MRI signals, indicating its potential as a contrast agent for MRI.10.4 Aptamer–mesoporous silica nanoparticle conjugation
MSNs (mesoporous silica nanoparticles), a ceramic material, are potential candidates for targeted drug delivery.176,177 Advances in synthesis techniques have improved their pore size, surface area, and structure, resulting in a high drug loading capacity and facilitating the incorporation of various functional groups.178–181 Aptamers offer numerous advantages as targeting ligands for MSNs. In therapeutic applications, Ap–MSN (aptamer–mesoporous nanoparticle) complexes are primarily intended as drug release regulators, nanocarriers, and drug trackers. Gao et al. developed an aptamer–MSN complex consisting of thrombin-binding aptamers, lipid-coated MSNs, and the chemotherapeutic drug docetaxel (DTX).182,183 This aptamer@MSN@DTX complex exhibits excellent anticancer activity in thrombin-overexpressing cancer cells. Furthermore, the structure-switching properties of aptamers, when binding to target cells, enhance the drug-releasing mechanism of the aptamer–MSN complex.184,185 In 2012, He et al. incorporated an ATP-responsive aptamer onto MSNs. This nanosystem includes two single-stranded DNA molecules (ssDNA1 and ssDNA2) along with the aptamer and MSNs. The nanocomplex is designed so that the aptamer is initially hybridized with two DNA sequences, and the resulting complex is conjugated to the MSN via the click chemistry technique. This approach blocks the pores of MSNs, preventing loss of the loaded substance. The presence of ATP enhances the interaction between the aptamer and the target, facilitating their separation from the nanoparticle complex and resulting in the release of drug molecules. In 2017, Li et al. used the EpCAM aptamer, an overexpressed surface biomarker in human colon adenocarcinomas, to functionalize MSN carriers for transporting the anti-cancer drug DM1 to colorectal cancer cells.186 The incorporated aptamer not only facilitates drug delivery but also reduces systemic toxicity. Additionally, dopamine HCL (PDA), a pH-responsive agent, was conjugated to MSNs to regulate the release of DM1 in the EpCAM-positive SW480 colon cancer cell line in response to pH stimuli.10.5 Aptamer–carbon nanomaterial conjugation
Currently, one-dimensional carbon nanotubes (CNTs) are attracting significant scientific interest as drug carriers and biosensors. Their unique characteristics, such as large surface area, easy modifiability, and high in vivo stability, make them promising for biomedical applications. Incorporating aptamers and CNTs enhances their therapeutic efficacy. The strong affinity of these targeting ligands has led to notable advancements in the aptamer–CNT field within a short period.187–191This section discusses recent efforts made to develop carbon nanomaterial–aptamer conjugates that have been found to enhance therapeutic delivery. In 2015, Mohammadi et al. designed an RNA aptamer-incorporated single-walled carbon nanotube (SWNT) complex that significantly improved cancer therapy efficacy.187 The nanosystem consists of EpCAM-targeting RNA aptamers, SWNTs, and a piperazine–PEA derivative (polyethyleneimine), specifically targeting EpCAM-expressing cancer cells. The aptamer@SWNT@PPEA combination effectively targets EpCAM-positive cancer cells and promotes apoptosis by inhibiting BCL91. Aptamer-incorporated SWNTs are not only effective drug carriers but also promising agents for photodynamic therapy (PDT). Following this concept, Zhu et al. developed a novel nanosystem consisting of a DNA aptamer, SWNTs, and a photosensitizer for controllable singlet oxygen generation (SOG).189 In the absence of the target α-thrombin, the photosensitizer quenches SOG. However, when the target is present, the aptamer detaches from the SWNT surface and binds to the target. Additionally, a chemotherapy drug, daunorubicin, was linked to a leukemia-targeting aptamer–SWNT to enhance pH-dependent drug delivery into T-cells.192 Recently, an aptamer-incorporated SWNT was developed for NIR laser-controlled cancer treatment. The SWNT is conjugated with an aptamer specific for sgc8, controlled by complementary DNA strands. NIR laser exposure causes the dehybridization of the DNA strands, allowing the aptamer to recognize the target and release the drug DOX.190
11. Biophysical approach for aptamer-based targeted drug delivery
The biophysical approach for drug delivery involves applying physics and biology principles to the design and optimisation of drug delivery systems for improved therapeutic outcomes. The objective is to comprehend the physical interactions between drug carriers, therapeutic agents, and biological systems in order to develop efficient and targeted drug delivery strategies.193 Utilising the biophysical properties of drug carriers and their interactions with the biological environment, this strategy enhances the specificity, efficacy, and safety of drug delivery.19411.1 Spectra assisted methods
The UV-Vis spectroscopic method is employed to analyse the specific interaction between an aptamer and its target. The dissociation constant (Kd) is calculated by measuring the changes in absorbance as the target concentration varies, while the aptamer concentration remains constant.195 Generally, this method is used in drug delivery, particularly to confirm the conjugation of drugs, aptamers and nanoparticles. Zhao et al. developed aptamer-functionalized Fe3O4@carbon@doxorubicin nanoparticles for chemophotothermal therapy.175 The UV-Vis spectrum of this aptamer-functionalized nanosystem shows a significant distinction from that of free DOX (Fig. 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 175). The UV-Vis spectra are measured before and after aptamer conjugation to evaluate the effectiveness of aptamer coupling with the nanocomplex. After aptamer conjugation, the peak shifts to 260 nm compared to the nanocomplex without the aptamer, indicating successful conjugation. Tan et al. developed a novel theragnostic system using endoglin aptamer-conjugated fluorescent nanoparticles. The UV-Vis spectra of YQ26-FSiNPs and Lib-FSiNPs showed a DNA absorbance peak at 260 nm, which was absent in the nanoparticle complex without the aptamer (FSiNPs), further confirming the successful conjugation of the aptamer to the nanoparticle complex (illustrated in Fig. 5b
175). The UV-Vis spectra are measured before and after aptamer conjugation to evaluate the effectiveness of aptamer coupling with the nanocomplex. After aptamer conjugation, the peak shifts to 260 nm compared to the nanocomplex without the aptamer, indicating successful conjugation. Tan et al. developed a novel theragnostic system using endoglin aptamer-conjugated fluorescent nanoparticles. The UV-Vis spectra of YQ26-FSiNPs and Lib-FSiNPs showed a DNA absorbance peak at 260 nm, which was absent in the nanoparticle complex without the aptamer (FSiNPs), further confirming the successful conjugation of the aptamer to the nanoparticle complex (illustrated in Fig. 5b![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 196).
196).
 | ||
| Fig. 5 UV-Vis spectras of (a) DOX and Fe3O4@C@DOX; the red line represents the Fe3O4 @C@DOX complex while the black line represents free DOX. (b) YQ26-FSiNPs, Lib-FSiNPs, and FSiNPs the blue lines represents FSiNPs, Lib-FSiNPs, and YQ26-FSiNPs, respectively. All three show an absorption peak at 260 nm.196 | ||
FT-IR is another spectroscopic technique used to characterize aptamer–nanoparticle conjugation by identifying the functional groups involved. Tan et al. performed FT-IR spectral analysis on the YQ26-FSiNPs nanocomplex to verify the presence of functional groups in the aptamer–nanocomplex.196 The results indicated that both the complex with and without the aptamer exhibited absorption peaks at 1100 cm−1 and 1660 cm−1, corresponding to Si–O stretching vibrations and carbonyl stretching (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) vibrations, respectively. Additionally, the YQ26-FSiNPs complex showed an extra peak at 1550 cm−1 due to amide II stretching, confirming the successful conjugation of the YQ26 aptamer to the nanocomplex.
O) vibrations, respectively. Additionally, the YQ26-FSiNPs complex showed an extra peak at 1550 cm−1 due to amide II stretching, confirming the successful conjugation of the YQ26 aptamer to the nanocomplex.
Circular dichroism (CD) is a spectrum-assisted technique used to analyze conformational changes in aptamer structures during the formation of secondary structures.197,198 In 2011, Lin et al. identified the secondary structure of an aptamer using CD spectra,198 while Bowser et al. reported that different secondary structures exhibited distinct CD spectra patterns. For example, a G-quadruplex shows a negative band at 255 nm and positive bands at 245 and 290 nm.195 Furthermore, this spectroscopic method is useful for understanding structural changes in aptamers during their binding interactions with targets.
11.2 Surface morphological studies
There are numerous techniques for investigating the morphological properties of aptamers and aptamer–NP complexes, including XPS (X-ray photoelectron spectroscopy) and TEM (transmission electron microscopy). Generally, XPS is used to determine the elemental composition of a surface.197,199,200 Guo et al. developed aptamer-functionalized PEG–PLGA nanoparticles for improved anti-glioma drug delivery.111 In their study, XPS was employed to identify the elements present in the aptamer–nanoparticle complex. The investigation revealed that the C1s spectrum contained four peaks at around 285.0 eV, 286.8 eV, 287.6 eV, and 289.4 eV, with the 286.8 eV peak representing C–O–C bonds of the PEG component. Additionally, O1s decomposition confirmed the existence of two forms of oxygen: O–C and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, at 532.7 eV and 533.6 eV, respectively. The peak at 132 eV corresponded to P2p3, which was identified at 0.1% of the total number of C, O, N, and P atoms in the aptamer–nanocomplex. Based on the chemical composition, they concluded that phosphorus could only be attributed to the aptamer, confirming the incorporation of the aptamer onto the nanoparticle surface.
C, at 532.7 eV and 533.6 eV, respectively. The peak at 132 eV corresponded to P2p3, which was identified at 0.1% of the total number of C, O, N, and P atoms in the aptamer–nanocomplex. Based on the chemical composition, they concluded that phosphorus could only be attributed to the aptamer, confirming the incorporation of the aptamer onto the nanoparticle surface.
TEM is another technique used to study surface morphology. In the same study, Guo et al. also employed TEM to examine the surface morphology of the aptamer–nanoparticle complex. The study revealed that both types of nanoparticles had a spherical shape, smooth surface, and moderate uniformity. Additionally, TEM results indicated that Ap–PTX–NP had a slightly larger spherical volume compared to PTX–NP111 (Fig. 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 111). In a separate study, Xu et al. developed aptamer-incorporated doxorubicin–unimolecular micelles for targeted therapy of prostate cancer. TEM analysis revealed that the micelles were spherical with diameters ranging from 20 to 37 nm.103 Later, in 2019, Wen et al. created core–shell bifunctional nanoprobes and investigated morphological changes during the formation of the core–shell structure through TEM analysis. These observations confirmed the formation of AuNC/SiO2/Apt nanoprobes.201 Additionally, Tao et al. developed aptamer–nanoparticle conjugates for breast cancer therapy, utilizing TEM to examine the morphology of the aptamer–nanoparticle complex. The TEM images indicated that the complex exhibited a spherical shape with an average particle size of 90 nm.202
111). In a separate study, Xu et al. developed aptamer-incorporated doxorubicin–unimolecular micelles for targeted therapy of prostate cancer. TEM analysis revealed that the micelles were spherical with diameters ranging from 20 to 37 nm.103 Later, in 2019, Wen et al. created core–shell bifunctional nanoprobes and investigated morphological changes during the formation of the core–shell structure through TEM analysis. These observations confirmed the formation of AuNC/SiO2/Apt nanoprobes.201 Additionally, Tao et al. developed aptamer–nanoparticle conjugates for breast cancer therapy, utilizing TEM to examine the morphology of the aptamer–nanoparticle complex. The TEM images indicated that the complex exhibited a spherical shape with an average particle size of 90 nm.202
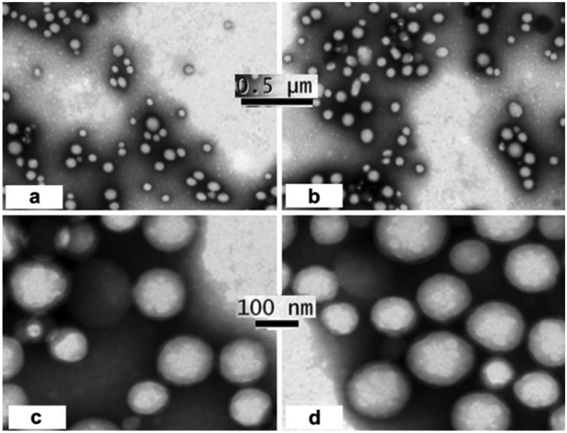 | ||
| Fig. 6 TEM images of PTX-NPs (a & c) and AP-PTX NPs (b & d).111 | ||
11.3 Affinity chromatography
HPLC (high performance liquid chromatography) techniques are used to analyse aptamer–target binding both qualitatively and quantitatively. This involves the pressurized passage of a liquid solvent through an aptamer-immobilized adsorbent column. The interaction is evaluated by plotting analyte concentrations against retention time on a chromatogram. This method provides insights into the equilibrium distribution of aptamer, target, and aptamer–target complexes, which can be utilized to estimate the Kd value.203 Furthermore, HPLC can assess the binding affinity of aptamers. Deng et al. employed affinity chromatography techniques to study the binding interaction of a DNA aptamer to adenosine. Specifically, frontal chromatography was used to quantitatively evaluate column loading and the dissociation constant (Kd) of the immobilized aptamer.20311.4 Electrophoresis
Electrophoresis, employed to analyze aptamer–target interactions, encompasses various forms such as GE (gel electrophoresis), ACE (affinity capillary electrophoresis), and ME (microchip electrophoresis). GE is a common technique for examining aptamer–target affinity interactions. Aptamers, being negatively charged, exhibit negative electrophoretic mobility, while target proteins typically show minimal polar mobility. The resulting aptamer–protein tertiary complex displays moderate electrophoretic mobility.195 Techniques like P-labelling of nucleic acids (NAs), UV-absorbance spectra, and staining of aptamers and proteins are available for analysing aptamer concentrations in aptamer–target complexes. Despite being sensitive and cost-effective, these methods are time-consuming.Affinity capillary electrophoresis (ACE) serves as an alternative to gel electrophoresis, focusing on the charge and size of aptamers and targets in free solution under an electric field.60,195 ACE is categorized as competitive and noncompetitive types. Noncompetitive ACE utilizes capillary electrophoresis (CE) assisted by laser-induced fluorescence (LIF) to analyse aptamer and protein concentrations through peak analysis.204,205 In competitive electrophoresis, two aptamers with the same target are used, one of which is labelled with a fluorescent probe (APF). The fluorescence intensity of the APF-T complex decreases due to the competitive binding of the unlabelled aptamer (AP) with APF for the binding site, generating data with two peaks that quantify binding affinity.205 Li and colleagues applied these concepts to investigate the thrombin affinity of 15-mer and 29-mer aptamers.206
Microchip electrophoresis (ME) and automated microchip electrophoresis (AME) are also utilized for electrophoresis binding studies, utilizing an intercalating fluorescent dye for molecular detection. Compared to GE and ACE, microchip electrophoresis offers advantages such as avoiding gel casting, quicker sensing times, reduced sample and reagent volumes, and the ability to characterize the binding of small molecules.207,208 However, their applicability in basic laboratories is limited due to the complexity of developing specialized platforms for electrophoresis.
11.5 Atomic force microscopy (AFM)
AFM (atomic force microscopy) is a scanning probe microscopy method that offers high-resolution imaging of nanoscale structures. Utilizing a cantilever, AFM can examine sample surfaces with precision and measure the affinity force between the sample and the cantilever. This capability makes AFM suitable for studying aptamer–biomolecule binding affinities.209 Notably, AFM does not require sample fixation or dehydration, allowing the examination of cells under aqueous conditions and in their natural state.210,211 This feature makes AFM well-suited for investigating protein surfaces and the topology of living cells.AFM data typically include a force histogram (FH) and a topology image. The affinity force of aptamer–target binding is determined by the peak location in the adhesion measurement histogram (in pN). Miyachi et al. indicated that higher pN values correlated with stronger affinity forces (AF) and, consequently, higher binding affinity (BA). Topology imaging provides insights into how aptamer binding modifies the surface conformation of the target. For instance, Mosafer et al. used AFM to explore the morphological properties of magnetic PLGA nanoparticles in their study.212
11.6 Thermodynamic characterization
Microscale thermophoresis (MST) and isothermal titration calorimetry (ITC) are thermodynamically related techniques used to measure the affinity between aptamers and their targets.213–216 In ITC, the concentration of the analyte (complementary molecule) is varied while keeping the concentration of the titrant (aptamer) constant.217 The formation of the tertiary complex is the preferred exothermic reaction, and the cell temperature is carefully controlled to eliminate other potential exothermic sources. In aptamer–target interactions, the change in energy is directly proportional to the binding stoichiometry. Pei et al. developed an aptamer-tethered nanostructure and conducted an ITC study to elucidate the thermodynamic binding characteristics of DOX, EPI, or DAU to AS1411. The resulting thermodynamic parameters from this study provide insights into the intermolecular interactions between aptamer AS1411 and the loaded anthracyclines.117 MST (microscale thermophoresis) is a relatively new biophysical technique that utilizes thermal gradient generation with an infrared laser.218,219 The thermophoretic mobility of biomolecules, influenced by factors such as charge, molecular size, solvation shell, and conformation, determines their behavior in MST. The formation of aptamer–target complexes alters these characteristics, resulting in a measurable change in mobility that quantifies their binding affinity. Due to its speed, specificity, and versatility, MST is expected to compete effectively with other biophysical technologies in the near future.11.7 Computational techniques (in silico)
| Docking software | Source | Ref. |
|---|---|---|
| GRAMM | https://vakser.bioinformatics.ku.edu/resources/gramm/gramm1 | 232 and 233 |
| Hex | https://hexserver.loria.fr/ | 234 and 235 |
| ZDOCK | https://zdock.umassmed.edu | 236 and 237 |
| HADDOCK | https://haddock.chem.uu.nl/Haddock | 238 and 239 |
| NPDock | https://genesilico.pl/NPDock | 240 |
| AutoDockTools | https://mgltools.scripps.edu | 241 |
| AutoDock | https://autodock.scripps.edu/resources/references | 242 |
In summary, MD simulations provide deeper insights into aptamer–target interactions, crucial for optimizing and rationally designing DNA aptamers.229 For instance, Zhang et al. developed the ssDNA aptamer Z100 against zearalenone (ZEN) and employed in silico docking analysis using AutoDock Vina version 1.1.2 to investigate their interaction, resulting in a binding free energy of approximately −9.6 kcal mol−1.230 Similarly, Farahbakhsh et al. conducted an in silico study on aptamer AS1411 targeting nucleolin in cancer cells, identifying the SS2-55 and SS4-54 loops as active sites through molecular docking simulations.231
12. Conclusion
This review delves into the specifics of aptamer-functionalized nanoparticles in cancer therapy, highlighting their potential as a targeted treatment option. Aptamers, short single-stranded oligonucleotides, possess unique properties that make them highly suitable for targeted therapy. Given the off-target effects associated with conventional chemotherapy and other treatment modalities, there is an urgent need for innovative therapeutic strategies in cancer treatment. This landscape is conducive to the advancement of targeted drug delivery systems. Aptamer-functionalized nanoparticles emerge as promising candidates for anticancer therapy. Traditional nano-mediated drug delivery primarily relies on the enhanced permeability and retention (EPR) effect, which has inherent limitations. In contrast, receptor-mediated targeting using aptamer-functionalized nanoparticles can circumvent these drawbacks. Despite growing interest in these active targeting ligands, their clinical application remains limited. Therefore, addressing the current state of aptamers is crucial for optimizing their efficacy and minimizing side effects.This review traces the journey from the discovery of the first aptamer in 1990 to the latest advancements in SELEX, various in silico approaches, and the development of diverse aptamer-functionalized nanoparticles. By revisiting the literature and examining current innovations, we aim to chart a course for future research into the potential of these active targeting ligands in cancer treatment. Adopting a biophysical approach, we explore a range of quantitative and qualitative methods to analyse molecular interactions, physical properties, and behaviours of aptamer–nanoparticle conjugates. This understanding is vital for the effective development of these conjugates for applications in medicine, diagnostics, and beyond. By gaining deeper insights into these interactions, we can better harness the potential of aptamer-functionalized nanoparticles in targeted cancer therapy.
13. Challenges and future perspective
Aptamer-incorporated nanoparticles hold immense promise for advancing anticancer drug development and applications. Despite growing interest in these active targeting ligands, their clinical and preclinical use remains limited. To fully leverage the potential of aptamers while minimizing side effects, it is essential to address their current state.In 1996, the United States FDA approved the first nanotechnology-based anticancer drug, which encapsulated doxorubicin in PEGylated liposomes. Today, approximately ten nanoparticle-based cancer drugs have received approval from the FDA or other agencies and are commercially available. These drugs are either untargeted or passively targeted, extending the drug's half-life and reducing side effects to some extent. However, the therapeutic improvements have been marginal, and off-target effects remain a significant issue, highlighting the need for actively targeted nanoparticles. Currently, over a dozen nanoparticles for cancer therapy are in clinical trials, with many featuring active targeting. However, none of these employ aptamer functionalization. Actively targeted nanoparticles, particularly those functionalized with aptamers, possess tremendous potential for future nanodrug development and applications. Therefore, intensified research efforts, improved experimental designs, and overcoming barriers to clinical translation are imperative. A major obstacle to the clinical application of aptamer-conjugated nanoparticles is limited information on their target effects and toxicity in animals or humans. Additionally, the availability of aptamers for clinical use is restricted. Aptamers, as short single-stranded nucleic acids, are prone to degradation in nuclease-rich physiological environments. Most aptamers reported in the scientific literature are obtained through in vitro selection, raising concerns about their in vivo affinity under physiological conditions. Thus, innovative aptamer stabilization techniques and SELEX (systematic evolution of ligands by exponential enrichment) strategies are urgently needed. Before clinical trials can commence, the biosafety of aptamer-based nanomedicines must be thoroughly evaluated. Aptamers contain foreign nucleic acids, posing risks of genomic insertion and immune responses. Additionally, some aptamer-functionalized materials exhibit inherent cytotoxicity. For example, cadmium-containing quantum dots (QDs) have been shown to induce DNA damage in cells and exhibit high toxicity. Systematic toxicity evaluations of candidate aptamer-integrated remedies are essential to ensure biosafety in clinical trials.
In summary, this innovative approach to aptamer–nanoparticle conjugation offers profound insights that are crucial for developing a highly effective and specific aptamer-functionalized nanoparticle system for cancer therapy. Addressing the current limitations and challenges is imperative to fully harness the potential of these advanced systems. By overcoming these hurdles, the door to the successful clinical application of aptamer-functionalized nanoparticles in targeted cancer therapy can be opened, promising a new era in cancer treatment.
Author contributions
Alanthatta Govindan Navaneeth: first author, literature survey, main script writing. Subramani Karthikeyan: corresponding author, main script correction.Data availability
No primary research results, software or code have been included and no new data were generated or analysed as part of this review.Conflicts of interest
The authors report there are no competing interests to declare.Acknowledgements
The authors thank VIT University for providing a VIT RGMES SEED GRANT for carrying out this research work.References
- J. Ferlay, M. Colombet and I. Soerjomataram, et al., Cancer statistics for the year 2020: An overview, Int. J. Cancer, 2021, 149, 778–789 CrossRef CAS PubMed.
- P. A. Jones and S. B. Baylin, The epigenomics of cancer, Cell, 2007, 128, 683–692 CrossRef CAS PubMed.
- T. N. Seyfried and L. C. Huysentruyt, On the origin of cancer metastasis, Crit. Rev. Oncog., 2013, 18, 43–73 CrossRef PubMed.
- I. J. Fidler, Cancer metastasis, Br. Med. Bull., 1991, 47, 157–177 CrossRef CAS PubMed.
- N. Behranvand, F. Nasri and R. Zolfaghari Emameh, et al., Chemotherapy: a double-edged sword in cancer treatment, Cancer Immunol. Immunother., 2022, 71, 507–526, DOI:10.1007/s00262-021-03013-3.
- R. Liu, C. Luo and Z. Pang, et al., Advances of nanoparticles as drug delivery systems for disease diagnosis and treatment, Chin. Chem. Lett., 2023, 34, 107518 CrossRef CAS.
- M. A. Subhan, F. Parveen and N. Filipczak, et al., Approaches to Improve EPR-Based Drug Delivery for Cancer Therapy and Diagnosis, J. Pers. Med., 2023, 13, 389 CrossRef PubMed.
- J. A. Stephenson, J. C. Goddard and O. Al-Taan, et al., Tumour Angiogenesis: A Growth Area—From John Hunter to Judah Folkman and Beyond, J. Cancer Res., 2013, 2013, 1–6 CrossRef.
- S. Gavas, S. Quazi and T. M. Karpiński, Nanoparticles for Cancer Therapy: Current Progress and Challenges, Nanoscale Res. Lett., 2021, 16(1), 173 CrossRef CAS PubMed.
- O. C. Farokhzad, J. M. Karp and R. Langer, Nanoparticle-aptamer bioconjugates for cancer targeting, Expert Opin. Drug Delivery, 2006, 3, 311–324 CrossRef CAS PubMed.
- Z. Fu and J. Xiang, Aptamer-functionalized nanoparticles in targeted delivery and cancer therapy, Int. J. Mol. Sci., 2020, 21, 1–39 Search PubMed.
- C. Tuerk and L. Gold, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 1990, 505–510 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, SELEX – A (r)evolutionary method to generate high-affinity nucleic acid ligands, Biomol. Eng., 2007, 24, 381–403 CrossRef CAS PubMed.
- J. Zhou and J. Rossi, Aptamers as targeted therapeutics: Current potential and challenges, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- D. Proske, M. Blank and R. Buhmann, et al., Aptamers – Basic research, drug development, and clinical applications, Appl. Microbiol. Biotechnol., 2005, 69, 367–374 CrossRef CAS PubMed.
- R. D. Jenison, S. C. Gill and A. Pardi, et al., High-resolution molecular discrimination by RNA, Science, 1994, 263, 1425–1429 CrossRef CAS PubMed.
- M. M. Soldevilla, H. Villanueva and F. Pastor, Aptamers: a feasible technology in cancer immunotherapy, J. Immunol. Res., 2016, 2016, 1083738 CAS.
- K. Ghasemii, M. Darroudi and I. Rahimmanesh, et al., Advances in aptamer-based drug delivery vehicles for cancer therapy, Biomater. Adv., 2022, 140, 213077 CrossRef CAS PubMed.
- A. Wochner, M. Menger and M. Rimmele, Characterisation of aptamers for therapeutic studies, Expert Opin. Drug Discovery, 2007, 2, 1205–1224 CrossRef CAS PubMed.
- C. Chandola and M. Neerathilingam, Aptamers for targeted delivery: current challenges and future opportunities, Role Nov. Drug Deliv. Veh. Nanobiomedicine, 2019, pp. 1–22 Search PubMed.
- A. V. Lakhin, V. Z. Tarantul and L. V. Gening, Aptamers: problems, solutions and prospects, Acta Nat., 2013, 5, 34–43 CrossRef CAS.
- J. B.-H. Tok, J. Cho and R. R. Rando, RNA aptamers that specifically bind to a 16S ribosomal RNA decoding region construct, Nucleic Acids Res., 2000, 28, 2902–2910 CrossRef CAS PubMed.
- L. C. Bock, L. C. Griffin and J. A. Latham, et al., Selection of single-stranded DNA molecules that bind and inhibit human thrombin, Nature, 1992, 355, 564–566, DOI:10.1038/355564a0.
- M. F. Kubik, A. W. Stephens and D. Schneider, et al., High-affinity RNA ligands to human alpha-thrombin, Nucleic Acids Res., 1994, 22, 2619–2626 CrossRef CAS PubMed.
- R. White, C. Rusconi and E. Scardino, et al., Generation of species cross-reactive aptamers using “toggle” SELEX, Mol. Ther., 2001, 4, 567–573 CrossRef CAS PubMed.
- T. M. A. Gronewold, S. Glass and E. Quandt, et al., Monitoring complex formation in the blood-coagulation cascade using aptamer-coated SAW sensors, Biosens. Bioelectron., 2005, 20, 2044–2052 CrossRef CAS PubMed.
- P. Colas, B. Cohen and T. Jessen, et al., Genetic selection of peptide aptamers that recognize and inhibit cyclin-dependent kinase 2, Nature, 1996, 380, 548–550 CrossRef CAS PubMed.
- R. Yazdian-Robati, M. Ramezani and M. Khedri, et al., An aptamer for recognizing the transmembrane protein PDL-1 (programmed death-ligand 1), and its application to fluorometric single cell detection of human ovarian carcinoma cells, Microchim. Acta, 2017, 184, 4029–4035 CrossRef CAS . Available from: https://api.semanticscholar.org/CorpusID:102924897.
- P. Colas, The eleven-year switch of peptide aptamers, J. Biol., 2008, 7, 2 CrossRef PubMed.
- I. C. Baines and P. Colas, Peptide aptamers as guides for small-molecule drug discovery, Drug Discovery Today, 2006, 11, 334–341 CrossRef CAS PubMed.
- F. Hoppe-Seyler, I. Crnkovic-Mertens and C. Denk, et al., Peptide aptamers: new tools to study protein interactions, J. Steroid Biochem. Mol. Biol., 2001, 78, 105–111 CrossRef CAS PubMed.
- S. J. Choi and C. Ban, Crystal structure of a DNA aptamer bound to PvLDH elucidates novel single-stranded DNA structural elements for folding and recognition, Sci. Rep., 2016, 6, 1–13 CrossRef PubMed.
- D. O'Connell, A. Koenig and S. Jennings, et al., Calcium-dependent oligonucleotide antagonists specific for L-selectin, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 5883–5887 CrossRef PubMed.
- K. E. Maier and M. Levy, From selection hits to clinical leads: progress in aptamer discovery, Mol. Ther. – Methods Clin. Dev., 2016, 3, 16014, DOI:10.1038/mtm.2016.14.
- S. Ni, Z. Zhuo and Y. Pan, et al., Recent Progress in Aptamer Discoveries and Modifications for Therapeutic Applications, ACS Appl. Mater. Interfaces, 2021, 13, 9500–9519 CrossRef CAS PubMed.
- K. Li, J. Deng and H. Jin, et al., Chemical modification improves the stability of the DNA aptamer GBI-10 and its affinity towards tenascin-C, Org. Biomol. Chem., 2017, 15, 1174–1182, 10.1039/C6OB02577C.
- A. L. Chang, M. McKeague and C. D. Smolke, Facile characterization of aptamer kinetic and equilibrium binding properties using surface plasmon resonance, Methods Enzymol, Elsevier, 2014, pp. 451–466 Search PubMed.
- A. Abe, S. Kosugi and K. Yoshida, et al., Genome sequencing reveals agronomically important loci in rice using MutMap, Nat. Biotechnol., 2012, 30, 174–178, DOI:10.1038/nbt.2095.
- B. Santosh and P. K. Yadava, Nucleic acid aptamers: research tools in disease diagnostics and therapeutics, BioMed Res. Int., 2014, 2014, 540451 Search PubMed.
- Z. Zhuo, Y. Yu and M. Wang, et al., Recent advances in SELEX technology and aptamer applications in biomedicine, Int. J. Mol. Sci., 2017, 18, 1–19 Search PubMed.
- Y. Lei, X. He and J. Tang, et al., Ultra-pH-responsive split i-motif based aptamer anchoring strategy for specific activatable imaging of acidic tumor microenvironment, Chem. Commun., 2018, 54, 10288–10291 RSC.
- S. Kolesnikova and E. A. Curtis, Structure and Function of Multimeric G-Quadruplexes, Molecules, 2019, 24, 3074 CrossRef CAS PubMed.
- A. A. Novoseltseva, N. M. Ivanov and R. A. Novikov, et al., Structural and Functional Aspects of G-Quadruplex Aptamers Which Bind a Broad Range of Influenza A Viruses, Biomolecules, 2020, 10(1), 119 CrossRef CAS PubMed.
- T. Sakamoto, E. Ennifar and Y. Nakamura, Thermodynamic study of aptamers binding to their target proteins, Biochimie, 2018, 145, 91–97 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0300908417302626.
- J. R. Williamson, Induced fit in RNA–protein recognition, Nat. Struct. Biol., 2000, 7, 834–837, DOI:10.1038/79575.
- S. Jones, D. T. Daley and N. M. Luscombe, et al., Protein-RNA interactions: a structural analysis, Nucleic Acids Res., 2001, 29, 943–954 CrossRef CAS PubMed.
- M. Rimmele, Nucleic acid aptamers as tools and drugs: recent developments, ChemBioChem, 2003, 4, 963–971 CrossRef CAS PubMed.
- Y. Feng, T. T. Baig and R. P. Love, et al., Suppression of APOBEC3-mediated restriction of HIV-1 by Vif, Front. Microbiol., 2014, 5, 450 Search PubMed.
- R. D. Jenison, S. C. Gill and A. Pardi, et al., High-resolution molecular discrimination by RNA, Science, 1994, 263(5152), 1425–1429 CrossRef CAS PubMed.
- M. Michaud, E. Jourdan and A. Villet, et al., A DNA aptamer as a new target-specific chiral selector for HPLC, J. Am. Chem. Soc., 2003, 125, 8672–8679 CrossRef CAS PubMed.
- P. R. Bouchard, R. M. Hutabarat and K. M. Thompson, Discovery and development of therapeutic aptamers, Annu. Rev. Pharmacol. Toxicol., 2010, 50, 237–257 CrossRef CAS PubMed.
- T. Wang, C. Chen and L. M. Larcher, et al., Three decades of nucleic acid aptamer technologies: Lessons learned, progress and opportunities on aptamer development, Biotechnol. Adv., 2019, 37, 28–50 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S0734975018301770.
- D. Musumeci and D. Montesarchio, Polyvalent nucleic acid aptamers and modulation of their activity: a focus on the thrombin binding aptamer, Pharmacol. Ther., 2012, 136, 202–215 CrossRef CAS PubMed.
- C. Platella, C. Riccardi and D. Montesarchio, et al., G-quadruplex-based aptamers against protein targets in therapy and diagnostics, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1429–1447 CrossRef CAS PubMed.
- L. Li, Q. Yin and J. Cheng, et al., Polyvalent mesoporous silica nanoparticle-aptamer bioconjugates target breast cancer cells, Adv. Healthc. Mater., 2012, 1, 567–572 CrossRef CAS PubMed.
- P. C. Patel, D. A. Giljohann and D. S. Seferos, et al., Peptide antisense nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17222–17226 CrossRef CAS PubMed.
- X. Lin, Q. Chen and W. Liu, et al., Assay of multiplex proteins from cell metabolism based on tunable aptamer and microchip electrophoresis, Biosens. Bioelectron., 2015, 63, 105–111 CrossRef CAS PubMed.
- C. M. Barbieri, T.-K. Li and S. Guo, et al., Aminoglycoside Complexation with a DNA·RNA Hybrid Duplex: The Thermodynamics of Recognition and Inhibition of RNA Processing Enzymes, J. Am. Chem. Soc., 2003, 125, 6469–6477 CrossRef CAS PubMed.
- R. Wang, J. Zhao and T. Jiang, et al., Selection and characterization of DNA aptamers for use in detection of avian influenza virus H5N1, J. Virol. Methods, 2013, 189, 362–369 CrossRef CAS PubMed.
- M. McKeague, E. M. McConnell and J. Cruz-Toledo, et al., Analysis of in vitro aptamer selection parameters, J. Mol. Evol., 2015, 81, 150–161 CrossRef CAS PubMed.
- F. Almasi, S. L. Mousavi Gargari and F. Bitaraf, et al., Development of a Single Stranded DNA Aptamer as a Molecular Probe for LNCap Cells Using Cell-SELEX, Avicenna J. Med. Biotechnol., 2016, 8, 104–111 Search PubMed.
- K. Setlem, B. Mondal and S. Ramlal, et al., Immuno Affinity SELEX for Simple, Rapid, and Cost-Effective Aptamer Enrichment and Identification against Aflatoxin B1, Front. Microbiol., 2016, 7, 1909 Search PubMed.
- L. Wang, R. Wang and F. Chen, et al., QCM-based aptamer selection and detection of Salmonella typhimurium, Food Chem., 2017, 221, 776–782 CrossRef CAS PubMed.
- S. D. Mendonsa and M. T. Bowser, In Vitro Selection of Aptamers with Affinity for Neuropeptide Y Using Capillary Electrophoresis, J. Am. Chem. Soc., 2005, 127, 9382–9383, DOI:10.1021/ja052406n.
- S. D. Mendonsa and M. T. Bowser, In Vitro Evolution of Functional DNA Using Capillary Electrophoresis, J. Am. Chem. Soc., 2004, 126, 20–21, DOI:10.1021/ja037832s.
- L. Dong, Q. Tan and W. Ye, et al., Screening and Identifying a Novel ssDNA Aptamer against Alpha-fetoprotein Using CE-SELEX, Sci. Rep., 2015, 5, 15552, DOI:10.1038/srep15552.
- N. S. Hamedani and J. Müller, Capillary Electrophoresis for the Selection of DNA Aptamers Recognizing Activated Protein C, Methods Mol. Biol., 2016, 1380, 61–75 CrossRef CAS PubMed.
- S. D. Mendonsa and M. T. Bowser, In vitro selection of high-affinity DNA ligands for human IgE using capillary electrophoresis, Anal. Chem., 2004, 76, 5387–5392 CrossRef CAS PubMed.
- R. K. Mosing and M. T. Bowser, Isolating Aptamers Using Capillary Electrophoresis–SELEX (CE–SELEX), in Nucleic Acid and Peptide Aptamers, Methods and Protocols, ed. G. Mayer, Humana Press, Totowa, NJ, 2009, pp. 33–43. DOI:10.1007/978-1-59745-557-2_3.
- S. R. Watson, Y. F. Chang and D. O'Connell, et al., Anti-L-selectin aptamers: binding characteristics, pharmacokinetic parameters, and activity against an intravascular target in vivo, Antisense Nucleic Acid Drug Dev., 2000, 10, 63–75 CrossRef CAS PubMed.
- J. Wang, T. Gao and Y. Luo, et al., In vitro selection of a DNA aptamer by cell-SELEX as a molecular probe for cervical cancer recognition and imaging, J. Mol. Evol., 2019, 87, 72–82 CrossRef CAS PubMed.
- Y. Zheng, Y. Zhao and Y. Di, et al., DNA aptamers from whole-serum SELEX as new diagnostic agents against gastric cancer, RSC Adv., 2019, 9, 950–957 RSC.
- Y. Zhao, L. He and B. Huang, et al., Identification of a novel DNA aptamer that selectively targets lung cancer serum, RSC Adv., 2021, 11, 33759–33769 RSC.
- Y. Guo, K. Li and Y. Gao, et al., CLEC3B Identified as a Potential Lung Cancer Biomarker in Serum by Aptamer-Capture Technology, ChemistrySelect, 2021, 6, 5640–5645 CrossRef CAS.
- B. J. Hicke, C. Marion and Y. F. Chang, et al., Tenascin-C aptamers are generated using tumor cells and purified protein, J. Biol. Chem., 2001, 276, 48644–48654 CrossRef CAS PubMed.
- K. Germer, M. Leonard and X. Zhang, RNA aptamers and their therapeutic and diagnostic applications, Int. J. Biochem. Mol. Biol., 2013, 4, 27 CAS.
- B. J. Hicke, A. W. Stephens and T. Gould, et al., Tumor targeting by an aptamer, J. Nucl. Med., 2006, 47, 668–678 CAS.
- A. C. Girvan, Y. Teng and L. K. Casson, et al., AGRO100 inhibits activation of nuclear factor-kappaB (NF-kappaB) by forming a complex with NF-kappaB essential modulator (NEMO) and nucleolin, Mol. Cancer Ther., 2006, 5, 1790–1799 CrossRef CAS PubMed.
- C. S. M. Ferreira, C. S. Matthews and S. Missailidis, DNA aptamers that bind to MUC1 tumour marker: design and characterization of MUC1-binding single-stranded DNA aptamers, Tumour Biol., 2006, 27, 289–301 CrossRef CAS PubMed.
- M. I. A. de Melo, C. R. Correa and P. da Silva Cunha, et al., DNA aptamers selection for carcinoembryonic antigen (CEA), Bioorg. Med. Chem. Lett., 2020, 30, 127278 CrossRef PubMed.
- K. Li, L. Qi and L. Gao, et al., Selection and preliminary application of a single stranded DNA aptamer targeting colorectal cancer serum, RSC Adv., 2019, 9, 38867–38876 RSC.
- D. Ferreira, J. Barbosa and D. A. Sousa, et al., Selection of aptamers against triple negative breast cancer cells using high throughput sequencing, Sci. Rep., 2021, 11, 8614, DOI:10.1038/s41598-021-87998-y.
- S. E. Lupold, Aptamers and apple pies: a mini-review of PSMA aptamers and lessons from Donald S. Coffey, Am. J. Clin. Exp. Urol., 2018, 6, 78 Search PubMed.
- J. C. Leach, A. Wang and K. Ye, et al., A RNA-DNA hybrid aptamer for nanoparticle-based prostate tumor targeted drug delivery, Int. J. Mol. Sci., 2016, 17, 380 CrossRef PubMed.
- J. P. Dassie, X. Liu and G. S. Thomas, et al., Systemic administration of optimized aptamer-siRNA chimeras promotes regression of PSMA-expressing tumors, Nat. Biotechnol., 2009, 27, 839–846 CrossRef CAS PubMed.
- W. M. Rockey, F. J. Hernandez and S.-Y. Huang, et al., Rational truncation of an RNA aptamer to prostate-specific membrane antigen using computational structural modeling, Nucleic Acid Ther., 2011, 21, 299–314 CrossRef CAS PubMed.
- J. J. Trausch, M. Shank-Retzlaff and T. Verch, Development and characterization of an HPV Type-16 specific modified DNA aptamer for the improvement of potency assays, Anal. Chem., 2017, 89, 3554–3561 CrossRef CAS PubMed.
- C. Li, S. Yang and R. Li, et al., Dual-Aptamer-Targeted Immunomagnetic Nanoparticles to Accurately Explore the Correlations between Circulating Tumor Cells and Gastric Cancer, ACS Appl. Mater. Interfaces, 2022, 14, 7646–7658, DOI:10.1021/acsami.1c22720.
- E. Kim, M. Kang and C. Ban, Aptamer-antibody hybrid ELONA that uses hybridization chain reaction to detect a urinary biomarker EN2 for bladder and prostate cancer, Sci. Rep., 2022, 12, 1–14 CrossRef PubMed.
- X. Sun, L. Xie and S. Qiu, et al., Elucidation of CKAP4-remodeled cell mechanics in driving metastasis of bladder cancer through aptamer-based target discovery, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, e2110500119 CrossRef CAS PubMed.
- B. Suwussa, C. Tao and S. Mohammed Ibrahim, et al., Pattern Recognition of Cancer Cells Using Aptamer-Conjugated Magnetic Nanoparticles, ACS Nano, 2012, 6(5), 3974–3981 CrossRef PubMed.
- S. Li, H. Xu and H. Ding, et al., Identification of an aptamer targeting hnRNP A1 by tissue slide-based SELEX, J. Pathol., 2009, 218, 327–336 CrossRef CAS PubMed.
- W. Zhong, Y. Pu and W. Tan, et al., Identification and Application of an Aptamer Targeting Papillary Thyroid Carcinoma Using Tissue-SELEX, Anal. Chem., 2019, 91, 8289–8297 CrossRef CAS PubMed.
- P. Bayat, R. Nosrati and M. Alibolandi, et al., SELEX methods on the road to protein targeting with nucleic acid aptamers, Biochimie, 2018, 154, 132–155, DOI:10.1016/j.biochi.2018.09.001.
- L. Zhou, P. Li and M. Yang, et al., Generation and characterization of novel DNA aptamers against coat protein of grouper nervous necrosis virus (GNNV) with antiviral activities and delivery potential in grouper cells, Antiviral Res., 2016, 129, 104–114 CrossRef CAS PubMed.
- A. Ozer, B. S. White and J. T. Lis, et al., Density-dependent cooperative non-specific binding in solid-phase SELEX affinity selection, Nucleic Acids Res., 2013, 41, 7167–7175 CrossRef CAS PubMed.
- J. G. Bruno and J. L. Kiel, Use of magnetic beads in selection and detection of biotoxin aptamers by electrochemiluminescence and enzymatic methods, BioTechniques, 2002, 32, 178–180 CrossRef CAS PubMed , 182–183.
- E. Dausse, A. Barré and A. Aimé, et al., Aptamer selection by direct microfluidic recovery and surface plasmon resonance evaluation, Biosens. Bioelectron., 2016, 80, 418–425 CrossRef CAS PubMed.
- P. Magala, W. E. Bocik and A. Majumdar, et al., Conformational Dynamics Modulate Activation of the Ubiquitin Conjugating Enzyme Ube2g2, ACS Omega, 2017, 2, 4581–4592 CrossRef CAS PubMed.
- D Shangguan, Y Li and Z Tang, et al., Aptamers Evolved from Live Cells as Effective Molecular Probes for Cancer Study, Proc. Natl. Acad. Sci. U. S. A., 2006, 103(32), 11838–11843, DOI:10.1073/pnas.0602615103.
- S. Dhar, F. X. Gu and R. Langer, et al., Targeted delivery of cisplatin to prostate cancer cells by aptamer functionalized Pt(IV) prodrug-PLGA-PEG nanoparticles, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 17356–17361 CrossRef CAS PubMed.
- A. Z. Wang, V. Bagalkot and C. C. Vasilliou, et al., Superparamagnetic iron oxide nanoparticle-aptamer bioconjugates for combined prostate cancer imaging and therapy, ChemMedChem, 2008, 3, 1311–1315 CrossRef CAS PubMed.
- W. Xu, I. A. Siddiqui and M. Nihal, et al., Aptamer-conjugated and doxorubicin-loaded unimolecular micelles for targeted therapy of prostate cancer, Biomaterials, 2013, 34, 5244–5253 CrossRef CAS PubMed.
- Y. H. Lee and G. G. Song, Un co r re c te d Un co r re c te d, Neoplasma, 2013, 60, 607–616 Search PubMed.
- X. Wu, Z. Tai and Q. Zhu, et al., Study on the prostate cancer-targeting mechanism of aptamer-modified nanoparticles and their potential anticancer effect in vivo, Int. J. Nanomed., 2014, 9, 5431–5440 Search PubMed.
- Z. Hao, W. Fan and J. Hao, et al., Efficient delivery of micro RNA to bone-metastatic prostate tumors by using aptamer-conjugated atelocollagen in vitro and in vivo, Drug Delivery, 2016, 23, 874–881 CrossRef PubMed.
- H. Jo, H. Youn and S. Lee, et al., Ultra-effective photothermal therapy for prostate cancer cells using dual aptamer-modified gold nanostars, J. Mater. Chem. B, 2014, 2, 4862–4867, 10.1039/C4TB00643G.
- I.-H. Lee, S. An and M. K. Yu, et al., Targeted chemoimmunotherapy using drug-loaded aptamer-dendrimer bioconjugates, J. Controlled Release, 2011, 155, 435–441 CrossRef CAS PubMed.
- M. K. Yu, D. Kim and I.-H. Lee, et al., Image-guided prostate cancer therapy using aptamer-functionalized thermally cross-linked superparamagnetic iron oxide nanoparticles, Small, 2011, 7, 2241–2249 CrossRef CAS PubMed.
- Z. Cao, R. Tong and A. Mishra, et al., Reversible cell-specific drug delivery with aptamer-functionalized liposomes, Angew. Chem., Int. Ed., 2009, 48, 6494–6498 CrossRef CAS PubMed.
- J. Guo, X. Gao and L. Su, et al., Aptamer-functionalized PEG-PLGA nanoparticles for enhanced anti-glioma drug delivery, Biomaterials, 2011, 32, 8010–8020, DOI:10.1016/j.biomaterials.2011.07.004.
- A. Aravind, P. Jeyamohan and R. Nair, et al., AS1411 aptamer tagged PLGA-lecithin-PEG nanoparticles for tumor cell targeting and drug delivery, Biotechnol. Bioeng., 2012, 109, 2920–2931 CrossRef CAS PubMed.
- A. Aravind, S. H. Varghese and S. Veeranarayanan, et al., Aptamer-labeled PLGA nanoparticles for targeting cancer cells, Cancer Nanotechnol., 2012, 3, 1–12 CrossRef CAS PubMed.
- H. Gao, J. Qian and S. Cao, et al., Precise glioma targeting of and penetration by aptamer and peptide dual-functioned nanoparticles, Biomaterials, 2012, 33, 5115–5123 CrossRef CAS PubMed.
- X. Yang, X. Liu and Z. Liu, et al., Near-infrared light-triggered, targeted drug delivery to cancer cells by aptamer gated nanovehicles, Adv. Mater., 2012, 24, 2890–2895 CrossRef CAS PubMed.
- F. J. Hernandez, L. I. Hernandez and A. Pinto, et al., Targeting cancer cells with controlled release nanocapsules based on a single aptamer, Chem. Commun., 2013, 49, 1285–1287 RSC.
- W. Pei, M. Liu and Y. Wu, et al., High payload and targeted release of anthracyclines by aptamer-tethered DNA nanotrains – Thermodynamic and release kinetic study, Eur. J. Pharm. Sci., 2020, 148, 105319, DOI:10.1016/j.ejps.2020.105319.
- C. Wu, D. Han and T. Chen, et al., Building a multifunctional aptamer-based DNA nanoassembly for targeted cancer therapy, J. Am. Chem. Soc., 2013, 135, 18644–18650 CrossRef CAS PubMed.
- Z. Han, X. Wang and C. Heng, et al., Synergistically enhanced photocatalytic and chemotherapeutic effects of aptamer-functionalized ZnO nanoparticles towards cancer cells, Phys. Chem. Chem. Phys., 2015, 17, 21576–21582, 10.1039/C5CP02139A.
- S. Taghavi, M. Ramezani and M. Alibolandi, et al., Chitosan-modified PLGA nanoparticles tagged with 5TR1 aptamer for in vivo tumor-targeted drug delivery, Cancer Lett., 2017, 400, 1–8 CrossRef CAS PubMed.
- D. Barak, S. Engelberg and Y. G. Assaraf, et al., Selective Targeting and Eradication of Various Human Non-Small Cell Lung Cancer Cell Lines Using Self-Assembled Aptamer-Decorated Nanoparticles, Pharmaceutics, 2022, 14, 1650 CrossRef CAS PubMed.
- O. C. Farokhzad, A. Khademhosseini and S. Jon, et al., Microfluidic system for studying the interaction of nanoparticles and microparticles with cells, Anal. Chem., 2005, 77, 5453–5459 CrossRef CAS PubMed.
- D. Sehgal and I. K. Vijay, A method for the high efficiency of water-soluble carbodiimide-mediated amidation, Anal. Biochem., 1994, 218, 87–91 CrossRef CAS PubMed.
- O. C. Farokhzad, S. Jon and A. Khademhosseini, et al., Nanoparticle-aptamer bioconjugates: a new approach for targeting prostate cancer cells, Cancer Res., 2004, 64, 7668–7672 CrossRef CAS PubMed.
- P. Ray and R. R. White, Aptamers for targeted drug delivery, Pharmaceuticals, 2010, 3, 1761–1778 CrossRef CAS PubMed.
- K. Heo, S.-W. Min and H. J. Sung, et al., An aptamer-antibody complex (oligobody) as a novel delivery platform for targeted cancer therapies, J. Controlled Release, 2016, 229, 1–9 CrossRef CAS PubMed.
- M. Passariello, S. Camorani and C. Vetrei, et al., Novel Human Bispecific Aptamer-Antibody Conjugates for Efficient Cancer Cell Killing, Cancers, 2019, 11, 1268 CrossRef CAS PubMed.
- S. H. Rajabnejad, A. Mokhtarzadeh and K. Abnous, et al., Targeted delivery of melittin to cancer cells by AS1411 anti-nucleolin aptamer, Drug Dev. Ind. Pharm., 2018, 44, 982–987 CrossRef CAS PubMed.
- A. Pusuluri, V. Krishnan and V. Lensch, et al., Treating Tumors at Low Drug Doses Using an Aptamer-Peptide Synergistic Drug Conjugate, Angew. Chem., Int. Ed., 2019, 58, 1437–1441 CrossRef CAS PubMed.
- A. Gilboa-Geffen, P. Hamar and M. T. N. Le, et al., Gene Knockdown by EpCAM Aptamer-siRNA Chimeras Suppresses Epithelial Breast Cancers and Their Tumor-Initiating Cells, Mol. Cancer Ther., 2015, 14, 2279–2291 CrossRef CAS PubMed.
- J. Zhou, P. Swiderski and H. Li, et al., Selection, characterization and application of new RNA HIV gp 120 aptamers for facile delivery of Dicer substrate siRNAs into HIV infected cells, Nucleic Acids Res., 2009, 37, 3094–3109 CrossRef CAS PubMed.
- H. Jeong, S. H. Lee and Y. Hwang, et al., Multivalent Aptamer-RNA Conjugates for Simple and Efficient Delivery of Doxorubicin/siRNA into Multidrug-Resistant Cells, Macromol. Biosci., 2017, 17(4) DOI:10.1002/mabi.201600343.
- R. M. Zadegan, M. D. E. Jepsen and L. L. Hildebrandt, et al., Construction of a fuzzy and Boolean logic gates based on DNA, Small, 2015, 11, 1811–1817 CrossRef CAS PubMed.
- E. S. Andersen, M. Dong and M. M. Nielsen, et al., Self-assembly of a nanoscale DNA box with a controllable lid, Nature, 2009, 459, 73–76 CrossRef CAS PubMed.
- V. S. Chambers, G. Marsico and J. M. Boutell, et al., High-throughput sequencing of DNA G-quadruplex structures in the human genome, Nat. Biotechnol., 2015, 33, 877–881 CrossRef CAS PubMed.
- R. Hänsel-Hertsch, D. Beraldi and S. V. Lensing, et al., G-quadruplex structures mark human regulatory chromatin, Nat. Genet., 2016, 48, 1267–1272 CrossRef PubMed.
- A. Malugin and H. Ghandehari, Cellular uptake and toxicity of gold nanoparticles in prostate cancer cells: a comparative study of rods and spheres, J. Appl. Toxicol., 2010, 30, 212–217 CrossRef PubMed.
- G. Zhu, J. Zheng and E. Song, et al., Self-assembled, aptamer-tethered DNA nanotrains for targeted transport of molecular drugs in cancer theranostics, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 7998–8003 CrossRef CAS PubMed.
- Q. Li, D. Zhao and X. Shao, et al., Aptamer-Modified Tetrahedral DNA Nanostructure for Tumor-Targeted Drug Delivery, ACS Appl. Mater. Interfaces, 2017, 9, 36695–36701 CrossRef CAS PubMed.
- X. Han, Y. Jiang and S. Li, et al., Multivalent aptamer-modified tetrahedral DNA nanocage demonstrates high selectivity and safety for anti-tumor therapy, Nanoscale, 2018, 11, 339–347 RSC.
- Y. Zeng, J. Liu and S. Yang, et al., Time-lapse live cell imaging to monitor doxorubicin release from DNA origami nanostructures, J. Mater. Chem. B, 2018, 6, 1605–1612 RSC.
- C. Wiraja, Y. Zhu and D. C. S. Lio, et al., Framework nucleic acids as programmable carrier for transdermal drug delivery, Nat. Commun., 2019, 10, 1147 CrossRef PubMed.
- R. Jahanban-Esfahlan, K. Seidi and A. Jahanban-Esfahlan, et al., Static DNA Nanostructures For Cancer Theranostics: Recent Progress In Design And Applications, Nanotechnol., Sci. Appl., 2019, 12, 25–46 CrossRef CAS PubMed.
- D. Miyamoto, Z. Tang and T. Takarada, et al., Turbidimetric detection of ATP using polymeric micelles and DNA aptamers, Chem. Commun., 2007, 4743–4745 RSC.
- J. Tian, L. Ding and H. Ju, et al., A multifunctional nanomicelle for real-time targeted imaging and precise near-infrared cancer therapy, Angew. Chem., Int. Ed., 2014, 53, 9544–9549 CrossRef CAS PubMed.
- Y. Wu, K. Sefah and H. Liu, et al., DNA aptamer-micelle as an efficient detection/delivery vehicle toward cancer cells, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 5–10 CrossRef CAS PubMed.
- X. Li, Y. Yu and Q. Ji, et al., Targeted delivery of anticancer drugs by aptamer AS1411 mediated Pluronic F127/cyclodextrin-linked polymer composite micelles, Nanomedicine, 2015, 11, 175–184 CrossRef CAS PubMed.
- B. Jeong, S. W. Kim and Y. H. Bae, Thermosensitive sol-gel reversible hydrogels, Adv. Drug Delivery Rev., 2002, 54, 37–51 CrossRef CAS PubMed.
- A. Richter, G. Paschew and S. Klatt, et al., Review on Hydrogel-based pH Sensors and Microsensors, Sensors, 2008, 8, 561–581 CrossRef CAS PubMed.
- Y.-L. Zhao and J. F. Stoddart, Azobenzene-based light-responsive hydrogel system, Langmuir, 2009, 25, 8442–8446 CrossRef CAS PubMed.
- T.-Y. Liu, S.-H. Hu and T.-Y. Liu, et al., Magnetic-sensitive behavior of intelligent ferrogels for controlled release of drug, Langmuir, 2006, 22, 5974–5978 CrossRef CAS PubMed.
- R. Rodríguez, C. Alvarez-Lorenzo and A. Concheiro, Cationic cellulose hydrogels: kinetics of the cross-linking process and characterization as pH-/ion-sensitive drug delivery systems, J. Controlled Release, 2003, 86, 253–265 CrossRef PubMed.
- N. Zhao, A. Suzuki and X. Zhang, et al., Dual Aptamer-Functionalized in Situ Injectable Fibrin Hydrogel for Promotion of Angiogenesis via Codelivery of Vascular Endothelial Growth Factor and Platelet-Derived Growth Factor-BB, ACS Appl. Mater. Interfaces, 2019, 11, 18123–18132 CrossRef CAS PubMed.
- M. Liu, L. Wang and Y. Lo, et al., Aptamer-Enabled Nanomaterials for Therapeutics, Targeting and Imaging, Cells, 2022, 11, 159 CrossRef CAS PubMed.
- H. Yang, H. Liu and H. Kang, et al., Engineering target-responsive hydrogels based on aptamer-target interactions, J. Am. Chem. Soc., 2008, 130, 6320–6321 CrossRef CAS PubMed.
- B. Soontornworajit, J. Zhou and M. P. Snipes, et al., Affinity hydrogels for controlled protein release using nucleic acid aptamers and complementary oligonucleotides, Biomaterials, 2011, 32, 6839–6849 CrossRef CAS PubMed.
- M. R. Battig, B. Soontornworajit and Y. Wang, Programmable release of multiple protein drugs from aptamer-functionalized hydrogels via nucleic acid hybridization, J. Am. Chem. Soc., 2012, 134, 12410–12413 CrossRef CAS PubMed.
- Z. Zhang, N. Chen and S. Li, et al., Programmable hydrogels for controlled cell catch and release using hybridized aptamers and complementary sequences, J. Am. Chem. Soc., 2012, 134, 15716–15719 CrossRef CAS PubMed.
- Z. Zhang, S. Li and N. Chen, et al., Programmable display of DNA-protein chimeras for controlling cell-hydrogel interactions via reversible intermolecular hybridization, Biomacromolecules, 2013, 14, 1174–1180 CrossRef CAS PubMed.
- N. Zhao, M. R. Battig and M. Xu, et al., Development of a Dual-Functional Hydrogel Using RGD and Anti-VEGF Aptamer, Macromol. Biosci., 2017, 17, 1700201 CrossRef PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, A study of the nucleation and growth processes in the synthesis of colloidal gold, Discuss. Faraday Soc., 1951, 11, 55–75, 10.1039/DF9511100055.
- L. Cole, R. Ross and J. Tilley, et al., Gold nanoparticles as contrast agents in X-ray imaging and computed tomography, Nanomedicine, 2015, 10, 321–341 CrossRef CAS PubMed.
- P. Ghosh, G. Han and M. De, et al., Gold nanoparticles in delivery applications, Adv. Drug Delivery Rev., 2008, 60, 1307–1315 CrossRef CAS PubMed.
- A. Nicolás-Boluda, J. Vaquero and G. Laurent, et al., Photothermal Depletion of Cancer-Associated Fibroblasts Normalizes Tumor Stiffness in Desmoplastic Cholangiocarcinoma, ACS Nano, 2020, 14, 5738–5753 CrossRef PubMed.
- X.-D. Zhang, D. Wu and X. Shen, et al., Size-dependent radiosensitization of PEG-coated gold nanoparticles for cancer radiation therapy, Biomaterials, 2012, 33, 6408–6419 CrossRef CAS PubMed.
- P. Wu, Y. Gao and H. Zhang, et al., Aptamer-guided silver-gold bimetallic nanostructures with highly active surface-enhanced Raman scattering for specific detection and near-infrared photothermal therapy of human breast cancer cells, Anal. Chem., 2012, 84, 7692–7699 CrossRef CAS PubMed.
- L. Yang, Y.-T. Tseng and G. Suo, et al., Photothermal therapeutic response of cancer cells to aptamer-gold nanoparticle-hybridized graphene oxide under NIR illumination, ACS Appl. Mater. Interfaces, 2015, 7, 5097–5106 CrossRef CAS PubMed.
- J. He, J. Dong and Y. Hu, et al., Design of Raman tag-bridged core–shell Au@Cu3(BTC)2 nanoparticles for Raman imaging and synergistic chemo-photothermal therapy, Nanoscale, 2019, 11, 6089–6100, 10.1039/C9NR00041K.
- E. Taboada, E. Rodríguez and A. Roig, et al., Relaxometric and magnetic characterization of ultrasmall iron oxide nanoparticles with high magnetization. Evaluation as potential T1 magnetic resonance imaging contrast agents for molecular imaging, Langmuir, 2007, 23, 4583–4588 CrossRef CAS PubMed.
- D.-L. Zhao, X.-X. Wang and X.-W. Zeng, et al., Preparation and inductive heating property of Fe3O4–chitosan composite nanoparticles in an AC magnetic field for localized hyperthermia, J. Alloys Compd., 2009, 477, 739–743 CrossRef CAS.
- T. Vangijzegem, D. Stanicki and S. Laurent, Magnetic iron oxide nanoparticles for drug delivery: applications and characteristics, Expert Opin. Drug Delivery, 2019, 16, 69–78 CrossRef CAS PubMed.
- J. Gao, H. Gu and B. Xu, Multifunctional magnetic nanoparticles: design, synthesis, and biomedical applications, Acc. Chem. Res., 2009, 42, 1097–1107 CrossRef CAS PubMed.
- X. Dai, Z. Fan and Y. Lu, et al., Multifunctional nanoplatforms for targeted multidrug-resistant-bacteria theranostic applications, ACS Appl. Mater. Interfaces, 2013, 5, 11348–11354 CrossRef CAS PubMed.
- A. Aravind, R. Nair and S. Raveendran, et al., Aptamer conjugated paclitaxel and magnetic fluid loaded fluorescently tagged PLGA nanoparticles for targeted cancer therapy, J. Magn. Magn. Mater., 2013, 344, 116–123 CrossRef CAS . Available from: https://www.sciencedirect.com/science/article/pii/S030488531300365X.
- C. Zhao, X. Song and W. Jin, et al., Image-guided cancer therapy using aptamer-functionalized cross-linked magnetic-responsive Fe3O4@carbon nanoparticles, Anal. Chim. Acta, 2019, 1056, 108–116 CrossRef CAS PubMed.
- T. Yanagisawa, T. Shimizu and K. Kuroda, et al., The Preparation of Alkyltrimethylammonium–Kanemite Complexes and Their Conversion to Microporous Materials, Bull. Chem. Soc. Jpn., 1990, 63, 988–992, DOI:10.1246/bcsj.63.988.
- C. T. Kresge, M. E. Leonowicz and W. J. Roth, et al., Ordered mesoporous molecular sieves synthesized by a liquid-crystal template mechanism, Nature, 1992, 359, 710–712, DOI:10.1038/359710a0.
- M. Xu, Y. Wen and Y. Liu, et al., Hollow mesoporous ruthenium nanoparticles conjugated bispecific antibody for targeted anti-colorectal cancer response of combination therapy, Nanoscale, 2019, 11, 9661–9678, 10.1039/C9NR01904A.
- Y.-P. Chen, C.-T. Chen and Y. Hung, et al., A new strategy for intracellular delivery of enzyme using mesoporous silica nanoparticles: superoxide dismutase, J. Am. Chem. Soc., 2013, 135, 1516–1523 CrossRef CAS PubMed.
- Y. Hoshikawa, H. Yabe and A. Nomura, et al., Mesoporous Silica Nanoparticles with Remarkable Stability and Dispersibility for Antireflective Coatings, Chem. Mater., 2009, 22(1), 12–14 CrossRef.
- L. L. Hench, The story of Bioglass, J. Mater. Sci. Mater. Med., 2006, 17, 967–978 CrossRef CAS PubMed.
- L. Gao, Y. Cui and Q. He, et al., Selective recognition of co-assembled thrombin aptamer and docetaxel on mesoporous silica nanoparticles against tumor cell proliferation, Chem. – Eur. J., 2011, 17, 13170–13174 CrossRef CAS PubMed.
- M. Alibolandi, F. Hoseini and M. Mohammadi, et al., Curcumin-entrapped MUC-1 aptamer targeted dendrimer-gold hybrid nanostructure as a theranostic system for colon adenocarcinoma, Int. J. Pharm., 2018, 549, 67–75 CrossRef CAS PubMed.
- F.-F. Zheng, P.-H. Zhang and Y. Xi, et al., Aptamer/Graphene Quantum Dots Nanocomposite Capped Fluorescent Mesoporous Silica Nanoparticles for Intracellular Drug Delivery and Real-Time Monitoring of Drug Release, Anal. Chem., 2015, 87, 11739–11745 CrossRef CAS PubMed.
- X. He, Y. Zhao and D. He, et al., ATP-responsive controlled release system using aptamer-functionalized mesoporous silica nanoparticles, Langmuir, 2012, 28, 12909–12915 CrossRef CAS PubMed.
- Y. Li, Y. Duo and S. Bao, et al., EpCAM aptamer-functionalized polydopamine-coated mesoporous silica nanoparticles loaded with DM1 for targeted therapy in colorectal cancer, Int. J. Nanomed., 2017, 12, 6239–6257 CrossRef CAS PubMed.
- M. Mohammadi, Z. Salmasi and M. Hashemi, et al., Single-walled carbon nanotubes functionalized with aptamer and piperazine-polyethylenimine derivative for targeted siRNA delivery into breast cancer cells, Int. J. Pharm., 2015, 485, 50–60 CrossRef CAS PubMed.
- S. Liu, L. Wei and L. Hao, et al., Sharper and faster “nano darts” kill more bacteria: a study of antibacterial activity of individually dispersed pristine single-walled carbon nanotube, ACS Nano, 2009, 3, 3891–3902 CrossRef CAS PubMed.
- Z. Zhu, Z. Tang and J. A. Phillips, et al., Regulation of singlet oxygen generation using single-walled carbon nanotubes, J. Am. Chem. Soc., 2008, 130, 10856–10857 CrossRef CAS PubMed.
- Y. Yang, J. Liu and X. Sun, et al., Near-infrared light-activated cancer cell targeting and drug delivery with aptamer-modified nanostructures, Nano Res., 2016, 9, 139–148, DOI:10.1007/s12274-015-0898-4.
- M. Pourmadadi, A. Moammeri and A. Shamsabadipour, et al., Application of Various Optical and Electrochemical Nanobiosensors for Detecting Cancer Antigen 125 (CA-125): A Review, Biosensors, 2023, 13(1), 99 CrossRef CAS PubMed.
- S. M. Taghdisi, P. Lavaee and M. Ramezani, et al., Reversible targeting and controlled release delivery of daunorubicin to cancer cells by aptamer-wrapped carbon nanotubes, Eur. J. Pharm. Biopharm., 2011, 77, 200–206 CrossRef CAS PubMed.
- J. Wang, G. Zhu and M. You, et al., Assembly of Aptamer Switch Probes and Photosensitizer on Gold Nanorods for Targeted Photothermal and Photodynamic Cancer Therapy, ACS Nano., 2012, 6(6), 5070–5077 CrossRef CAS PubMed.
- C. Peetla, A. Stine and V. Labhasetwar, Biophysical interactions with model lipid membranes: Applications in drug discovery and drug delivery, Mol. Pharm., 2009, 6, 1264–1276 CrossRef CAS PubMed.
- M. Jing and M. T. Bowser, Methods for measuring aptamer-protein equilibria: a review, Anal. Chim. Acta, 2011, 686, 9–18 CrossRef CAS PubMed.
- J. Tan, N. Yang and L. Zhong, et al., A New Theranostic System Based on Endoglin Aptamer Conjugated Fluorescent Silica Nanoparticles, Theranostics, 2017, 7, 4862–4876 CrossRef CAS PubMed.
- M. McKeague and M. C. Derosa, Challenges and opportunities for small molecule aptamer development, J. Nucleic Acids, 2012, 2012, 748913 Search PubMed.
- P.-H. Lin, R.-H. Chen and C.-H. Lee, et al., Studies of the binding mechanism between aptamers and thrombin by circular dichroism, surface plasmon resonance and isothermal titration calorimetry, Colloids Surf., B, 2011, 88, 552–558 CrossRef CAS PubMed.
- R. F. Macaya, P. Schultze and F. W. Smith, et al., Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 3745–3749 CrossRef CAS PubMed.
- V. J. B. Ruigrok, M. Levisson and J. Hekelaar, et al., Characterization of aptamer-protein complexes by X-ray crystallography and alternative approaches, Int. J. Mol. Sci., 2012, 13, 10537–10552 CrossRef CAS PubMed.
- S. Wen and X. Miao, et al., Aptamer-Conjugated Au Nanocage/SiO2 Core-Shell Bifunctional Nanoprobes with High Stability and Biocompatibility for Cellular SERS Imaging and Near-Infrared Photothermal Therapy, ACS Sens., 2019, 4(2), 301–308, DOI:10.1021/acssensors.8b00682.
- W. Tao, X. Zeng and J. Wu, et al., Polydopamine-based surface modification of novel nanoparticle-aptamer bioconjugates for in vivo breast cancer targeting and enhanced therapeutic effects, Theranostics, 2016, 6, 470–484 CrossRef CAS PubMed.
- Q. Deng, I. German and D. Buchanan, et al., Retention and separation of adenosine and analogues by affinity chromatography with an aptamer stationary phase, Anal. Chem., 2001, 73, 5415–5421 CrossRef CAS PubMed.
- D. N. Stratis-Cullum, S. McMasters and P. M. Pellegrino, Evaluation of relative aptamer binding to Campylobacter jejuni bacteria using affinity probe capillary electrophoresis, Anal. Lett., 2009, 42, 2389–2402 CrossRef CAS.
- X. C. Le, Q.-H. Wan and M. T. Lam, Fluorescence polarization detection for affinity capillary electrophoresis, Electrophoresis, 2002, 23, 903–908 CrossRef CAS PubMed.
- Y. Li, L. Guo and F. Zhang, et al., High-sensitive determination of human α-thrombin by its 29-mer aptamer in affinity probe capillary electrophoresis, Electrophoresis, 2008, 29, 2570–2577 CrossRef CAS PubMed.
- J. Hu and C. J. Easley, A simple and rapid approach for measurement of dissociation constants of DNA aptamers against proteins and small molecules via automated microchip electrophoresis, Analyst, 2011, 136, 3461–3468 RSC.
- F. Nishikawa, H. Arakawa and S. Nishikawa, Application of microchip electrophoresis in the analysis of RNA aptamer-protein interactions, Nucleosides, Nucleotides Nucleic Acids, 2006, 25, 369–382 CrossRef CAS PubMed.
- Y. Miyachi, N. Shimizu and C. Ogino, et al., Selection of DNA aptamers using atomic force microscopy, Nucleic Acids Res., 2010, 38, e21 CrossRef PubMed.
- L. Kailas, E. C. Ratcliffe and E. J. Hayhurst, et al., Immobilizing live bacteria for AFM imaging of cellular processes, Ultramicroscopy, 2009, 109, 775–780 CrossRef CAS PubMed.
- N. E. Lonergan, L. D. Britt and C. J. Sullivan, Immobilizing live Escherichia coli for AFM studies of surface dynamics, Ultramicroscopy, 2014, 137, 30–39 CrossRef CAS PubMed.
- J. Mosafer, K. Abnous and M. Tafaghodi, et al., In vitro and in vivo evaluation of anti-nucleolin-targeted magnetic PLGA nanoparticles loaded with doxorubicin as a theranostic agent for enhanced targeted cancer imaging and therapy, Eur. J. Pharm. Biopharm., 2017, 113, 60–74, DOI:10.1016/j.ejpb.2016.12.009.
- M. Kwon, S. M. Chun and S. Jeong, et al., In vitro selection of RNA against kanamycin B, Mol. Cells, 2001, 11, 303–311 CrossRef CAS PubMed.
- M. N. Win, J. S. Klein and C. D. Smolke, Codeine-binding RNA aptamers and rapid determination of their binding constants using a direct coupling surface plasmon resonance assay, Nucleic Acids Res., 2006, 34, 5670–5682 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, FluMag-SELEX as an advantageous method for DNA aptamer selection, Anal. Bioanal. Chem., 2005, 383, 83–91 CrossRef CAS PubMed.
- R. Stoltenburg, C. Reinemann and B. Strehlitz, SELEX – A (r)evolutionary method to generate high-affinity nucleic acid ligands, Biomol. Eng., 2007, 24, 381–403 CrossRef CAS PubMed.
- R. Stoltenburg, T. Schubert and B. Strehlitz, In vitro Selection and Interaction Studies of a DNA Aptamer Targeting Protein A, PLoS One, 2015, 10, e0134403 CrossRef PubMed.
- P. Amero, C. L. Esposito and A. Rienzo, et al., Identification of an Interfering Ligand Aptamer for EphB2/3 Receptors, Nucleic Acid Ther., 2016, 26, 102–110 CrossRef CAS PubMed.
- C. Entzian and T. Schubert, Studying small molecule-aptamer interactions using MicroScale Thermophoresis (MST), Methods, 2016, 97, 27–34 CrossRef CAS PubMed.
- M. Asmari, M. Waqas and A. E. Ibrahim, et al., Microscale Thermophoresis and Molecular Modelling to Explore the Chelating Drug Transportation in the Milk to Infant, Molecules, 2022, 27(14), 4604 CrossRef CAS PubMed.
- D. Sun, M. Sun and J. Zhang, et al., Computational tools for aptamer identification and optimization, TrAC, Trends Anal. Chem., 2022, 157, 116767 CrossRef CAS.
- M. Sherman and L. Contreras, Computational approaches in design of nucleic acid-based therapeutics, Curr. Opin. Biotechnol, 2018, 53, 232–239 CrossRef CAS PubMed . Available from: https://www.sciencedirect.com/science/article/pii/S095816691730188X.
- J. Caroli, C. Taccioli and A. De La Fuente, et al., APTANI: A computational tool to select aptamers through sequence-structure motif analysis of HT-SELEX data, Bioinformatics, 2016, 32, 161–164 CrossRef CAS PubMed.
- T. N. Navien, R. Thevendran and H. Y. Hamdani, et al., In silico molecular docking in DNA aptamer development, Biochimie, 2021, 180, 54–67, DOI:10.1016/j.biochi.2020.10.005.
- J. Si, J. Cui and J. Cheng, et al., Computational Prediction of RNA-Binding Proteins and Binding Sites, Int. J. Mol. Sci., 2015, 16, 26303–26317 CrossRef CAS PubMed.
- B. Deng, Y. Lin and C. Wang, et al., Aptamer binding assays for proteins: the thrombin example – a review, Anal. Chim. Acta, 2014, 837, 1–15 CrossRef CAS PubMed.
- S. Y. Tan, C. Acquah and A. Sidhu, et al., SELEX Modifications and Bioanalytical Techniques for Aptamer–Target Binding Characterization, Crit. Rev. Anal. Chem., 2016, 46, 521–537 CrossRef CAS PubMed.
- T. Hayashi, H. Oshima and T. Mashima, et al., Binding of an RNA aptamer and a partial peptide of a prion protein: crucial importance of water entropy in molecular recognition, Nucleic Acids Res., 2014, 42, 6861–6875 CrossRef CAS PubMed.
- V. Tereshko, E. Skripkin and D. J. Patel, Encapsulating streptomycin within a small 40-mer RNA, Chem. Biol., 2003, 10, 175–187 CrossRef CAS PubMed.
- Y. Zhang, T. Lu and Y. Wang, et al., Selection of a DNA Aptamer against Zearalenone and Docking Analysis for Highly Sensitive Rapid Visual Detection with Label-Free Aptasensor, J. Agric. Food Chem., 2018, 66, 12102–12110 CrossRef CAS PubMed.
- Z. Farahbakhsh, M. R. Zamani and M. Rafienia, et al., In silico activity of AS1411 aptamer against nucleolin of cancer cells, Iran J. Blood Cancer, 2020, 12, 95–100 Search PubMed.
- E. Katchalski-Katzir, I. Shariv and M. Eisenstein, et al., Molecular surface recognition: determination of geometric fit between proteins and their ligands by correlation techniques, Proc. Natl. Acad. Sci. U. S. A., 1992, 89, 2195–2199 CrossRef CAS PubMed.
- A. Tovchigrechko and I. A. Vakser, GRAMM-X public web server for protein-protein docking, Nucleic Acids Res., 2006, 34, W310–W314 CrossRef CAS PubMed.
- D. W. Ritchie and G. J. Kemp, Protein docking using spherical polar Fourier correlations, Proteins, 2000, 39, 178–194 CrossRef CAS.
- D. W. Ritchie and V. Venkatraman, Ultra-fast FFT protein docking on graphics processors, Bioinformatics, 2010, 26, 2398–2405 CrossRef CAS PubMed.
- B. G. Pierce, K. Wiehe and H. Hwang, et al., ZDOCK server: interactive docking prediction of protein-protein complexes and symmetric multimers, Bioinformatics, 2014, 30, 1771–1773 CrossRef CAS PubMed.
- Q.-L. Wang, H.-F. Cui and J.-F. Du, et al., In silico post-SELEX screening and experimental characterizations for acquisition of high affinity DNA aptamers against carcinoembryonic antigen, RSC Adv., 2019, 9, 6328–6334 RSC.
- S. J. de Vries, M. van Dijk and A. M. J. J. Bonvin, The HADDOCK web server for data-driven biomolecular docking, Nat. Protoc., 2010, 5, 883–897 CrossRef CAS PubMed.
- M. van Dijk, A. D. J. van Dijk and V. Hsu, et al., Information-driven protein-DNA docking using HADDOCK: it is a matter of flexibility, Nucleic Acids Res., 2006, 34, 3317–3325 CrossRef CAS PubMed.
- I. Tuszynska, M. Magnus and K. Jonak, et al., NPDock: a web server for protein-nucleic acid docking, Nucleic Acids Res., 2015, 43, W425–W430 CrossRef CAS PubMed.
- S. Forli, R. Huey and M. E. Pique, et al., Computational protein-ligand docking and virtual drug screening with the AutoDock suite, Nat. Protoc., 2016, 11, 905–919 CrossRef CAS PubMed.
- V. Y. Tanchuk, V. O. Tanin and A. I. Vovk, et al., A New, Improved Hybrid Scoring Function for Molecular Docking and Scoring Based on AutoDock and AutoDock Vina, Chem. Biol. Drug Des., 2016, 87, 618–625 CrossRef CAS PubMed.
- J. Xiao and F. R. Salsbury, Molecular dynamics simulations of aptamer-binding reveal generalized allostery in thrombin, J. Biomol. Struct. Dyn., 2017, 35, 3354–3369, DOI:10.1080/07391102.2016.1254682.
- C. Q. Vu, P. Rotkrua and B. Soontornworajit, et al., Effect of PDGF-B aptamer on PDGFRβ/PDGF-B interaction: Molecular dynamics study, J. Mol. Graphics Modell., 2018, 82, 145–156 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2024 |