Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Analytical method for the determination of the absorption coefficient of DNA-stabilized silver nanoclusters†
Giacomo
Romolini
 a,
Cecilia
Cerretani
a,
Cecilia
Cerretani
 *a,
Vanessa
Rück
*a,
Vanessa
Rück
 a,
Mikkel Baldtzer
Liisberg
a,
Mikkel Baldtzer
Liisberg
 a,
Christian Brinch
Mollerup
a,
Christian Brinch
Mollerup
 b and
Tom
Vosch
b and
Tom
Vosch
 *a
*a
aDepartment of Chemistry, University of Copenhagen, Universitetsparken 5, DK-2100 Copenhagen, Denmark. E-mail: cece@chem.ku.dk; tom@chem.ku.dk
bDepartment of Forensic Medicine, University of Copenhagen, Frederik V's Vej 11, DK-2100 Copenhagen, Denmark
First published on 18th June 2024
Abstract
DNA-stabilized silver nanoclusters (DNA-AgNCs) are biocompatible emitters formed by silver atoms and cations encapsulated in DNA oligomers. Here, we present an analytical approach to calculate the molar absorption coefficient (ε) of these systems, which consists of combining UV-Vis spectroscopy, electrospray ionization-mass spectrometry (ESI-MS), and inductively coupled plasma-optical emission spectrometry (ICP-OES). ESI-MS enables the determination of the number of silvers bound to the DNA strands, whereas ICP-OES allows measurement of the total amount of silver in solution. The data is used to calculate the concentration of DNA-AgNCs and together with UV-Vis absorbance, allows for the calculation of ε. We compare the obtained ε with the experimental values previously determined through fluorescence correlation spectroscopy (FCS) and theoretical estimates based on the ε of the DNA itself. Finally, the experimental radiative decay rates (kf) and ε values are evaluated and compared to those typically found for organic fluorophores, highlighting the molecular-like nature of the DNA-AgNC emission.
Introduction
The use of single-stranded DNA oligomers offers the possibility to easily synthesize and tune the optical properties of silver nanoclusters. Particularly, the length and specific motifs of the oligomers define the number of silver atoms and cations in the resulting silver nanocluster (AgNC).1,2To get a better insight into the relationship between the structure and the photophysical properties, it is fundamental to measure spectroscopic characteristics such as absorption and emission spectra, fluorescence decay times and quantum yields from purified samples. All these features can be obtained without knowing the accurate concentration of DNA-AgNCs, which is instead relevant if one wants to compare experimental molar absorption coefficients with theoretical values3 or calculate two-photon absorption cross-sections.4 The concentration of DNA-AgNCs is usually estimated based on the theoretical absorption coefficient of the DNA at 260 nm (εth260) and the number of DNA strands encapsulating the AgNC. However, it is important to realize that this εth260 is an estimate,5–7 and it is unknown how much the AgNC itself contributes or affects the overall absorption in the UV region.8,9 For example, zeolite-stabilized AgNCs often show strong optical transitions in the UV range,10,11 which are attributed to excitations into higher excited states centered on the AgNCs.
Classically, ε can be determined by weighing a certain amount of compound with known molecular weight and dissolving it in a known volume of solvent. However, this approach has never been reported for DNA-AgNCs because it relies on the capability of preparing enough material and the knowledge of the exact composition of the DNA-AgNCs, including the number of structurally bound water molecules. Single crystal X-ray diffraction showed that a large number of water molecules are indeed bound to DNA-AgNCs, and its removal could lead to changes in the optical properties.3,12,13 Therefore, alternative approaches to determine ε have been proposed in the past.
One of these is based on fluorescence correlation spectroscopy (FCS), which provided estimates for the molar absorption coefficient (εexpFCS,NC) of some NIR14,15 and visible emissive DNA-AgNCs.16–19 FCS allows one to obtain the number of diffusing molecules in a reference-calibrated volume, which can be used to determine the concentration of the investigated compound. However, not all DNA-AgNCs are bright enough for FCS, especially if they have significant probabilities of evolving into long-lived (dark) states.15,17,20
Another method that has been employed is based on fluorescence saturation spectroscopy.8,21 It relies on the deviation of the linear fluorescence response as a function of excitation intensity, using a reference with a known ε. This approach requires intense fs-laser pulses to saturate the optical transition, and while elegant, might not be directly accessible to everyone. Additionally, photobleaching under these intense conditions and the buildup of long-lived states could affect the outcome.
Here, we present an analytical approach to calculate the molar absorption coefficient (εexpNC) of DNA-AgNCs. This method consists of combining electrospray ionization-mass spectrometry (ESI-MS), inductively coupled plasma-optical emission spectrometry (ICP-OES) and UV-Vis absorption spectroscopy. The first technique allows for the determination of the number of silvers within the DNA-AgNCs and the number of DNA strands, while the second is employed to measure the total amount of silver in solution (after digestion of DNA-AgNCs in HNO3). The εexpNC values obtained with our method are in good agreement with εexpFCS,NC values reported in the literature. We also discuss the limitations of the presented method and why estimates using εth260 can lead to significant deviations.
Results and discussion
Synthesis and purification
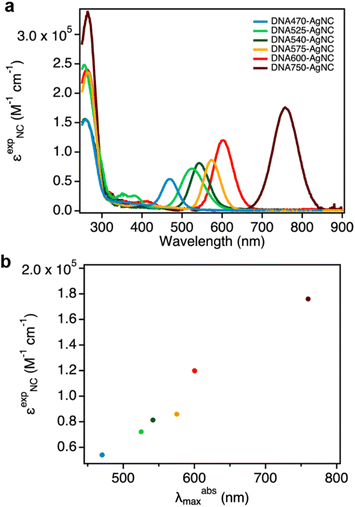
In this work, we determined the εexpNC of six DNA-AgNCs with AgNC-related absorption bands that span the visible and NIR-I range (Fig. 1a).13,15,17,22–24 The DNA-AgNCs are named after the AgNC absorption maximum, and hence they will be referred to as: DNA470-AgNC, DNA525-AgNC, DNA540-AgNC, DNA575-AgNC, DNA600-AgNC and DNA750-AgNC (see Experimental section for the DNA sequences).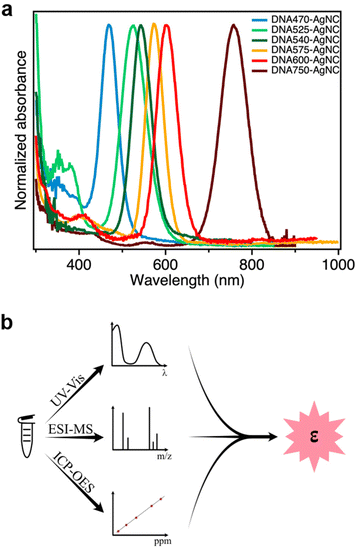 | ||
| Fig. 1 (a) Normalized absorption spectra of the six DNA-AgNCs used in this study; (b) Outline of the analytical approach to determine εexpNC. | ||
DNA-AgNCs are synthesized using the standard method of Ag+ reduction by NaBH4 after a 15-minute incubation in the presence of DNA. It is known that this reduction leads to a distribution of DNA-AgNCs and silver nanoparticles (AgNPs), thus a subsequent purification step is required. To achieve this, high-performance liquid chromatography (HPLC) is employed, and the purity is evaluated with absorption and fluorescence spectroscopy. The synthesis and purification conditions are reported in the Experimental section below and the detailed protocols are reported elsewhere.13,15,17,22–24
Quantification of the molar absorption coefficient
In order to determine εexpNC of the DNA-AgNCs, we developed an analytical methodology consisting of three steps, as outlined in Fig. 1b.Step 1. A known volume of the concentrated stock solution was diluted in 10 mM ammonium acetate, and the absorption spectrum of the diluted solution was then measured (Fig. 1a). In this method we assume that all features in the absorption spectrum are due to absorption and that loss in transmitted light caused by scattering is negligible.
Step 2. The number of silver atoms and stabilizing DNA strands per DNA-AgNC was determined by ESI-MS (see ESI for details†). The DNA-AgNC compositions are reported in Table 1, and the mass spectra for DNA575-AgNC, DNA600-AgNC and DNA750-AgNC can be found in Fig. S3, S4 and S5,† respectively, whereas the remaining mass spectra have been published recently.13,23,25
| Determination of DNA-AgNC concentration | ||||
|---|---|---|---|---|
| Sample | ICP-OES | ESI-MS | DNA-AgNC conc. [μM] | |
| Ag content [ppm] | Ag atoms/cluster | # DNA strands | ||
| DNA470-AgNC | 25.7 ± 0.2 | 11 (ref. 13) | 2 | 21.7 ± 0.6 |
| DNA525-AgNC | 34.7 ± 0.3 | 16 (ref. 25) | 2 | 20.1 ± 0.4 |
| DNA540-AgNC | 23.0 ± 0.1 | 18 (ref. 23) | 2 | 11.8 ± 0.2 |
| DNA575-AgNC | 46.0 ± 0.1 | 14 | 1 | 30.5 ± 0.3 |
| DNA600-AgNC | 27.4 ± 0.2 | 15 | 1 | 16.9 ± 0.4 |
| DNA750-AgNC | 46.8 ± 0.2 | 21 | 2 | 20.7 ± 0.3 |
Step 3. To quantify the total silver content from purified DNA-AgNCs, the stock solutions used in Step 1 were dissolved in HNO3, and the amount of Ag was determined by ICP-OES (see Experimental section for details). The obtained values ranged from 23 to 47 ppm (see Table 1 and Fig. S9–S14† for calibration curves).
The concentration of DNA-AgNCs can be then calculated by combining the data obtained from ICP-OES and ESI-MS (eqn (2)). The εexpNC values are finally determined with the help of UV-Vis absorption spectra and the Lambert–Beer law (eqn (3)). The detailed calculations can be found in the Experimental section, and the six εexpNC values are reported in Table 2. The εexpNC values span from 5.40 × 104 to 1.76 × 105 M−1 cm−1, and are similar to absorption coefficients of organic fluorophores.26Fig. 2a shows the absorption spectra in units of εexpNC for the six DNA-AgNCs analyzed in this study. It is worth noticing that for the investigated DNA-AgNCs a fairly linear correlation between εexpNC and the maximum absorption wavelength was found (Fig. 2b). This rough trend can be also observed for organic chromophores26 and can be tied back to the probability of spontaneous emission.27
| Determination of molar absorption coefficients | ||||
|---|---|---|---|---|
| Sample | ε th260 [M−1 cm−1] | ε thNC [M−1 cm−1] | ε expNC [M−1 cm−1] | ε expFCS,NC [M−1 cm−1] |
| DNA470-AgNC | 182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/198 000a/198![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800b 800b |
63![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/68 000a/68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 642b 642b |
54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— |
| DNA525-AgNC | 196![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 226a/213 226a/213![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000b 000b |
57![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/62 000a/62![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 640b 640b |
72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— |
| DNA540-AgNC | 192![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200a/221 200a/221![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800b 800b |
100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/115 000a/115![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 790b 790b |
81![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— |
| DNA575-AgNC | 182![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300a/193 300a/193![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200b 200b |
68![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/72 000a/72![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 460b 460b |
86![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
— |
| DNA600-AgNC | 255![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 300a/284 300a/284![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 420b 420b |
128![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/142 000a/142![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 760b 760b |
120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
| DNA750-AgNC | 266![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600a/277 600a/277![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 880b 880b |
143![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000a/150 000a/150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 470b 470b |
176![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
180![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 000 |
Comparing experimental and theoretical molar absorption coefficients of DNA-AgNCs
The εexpFCS,NC values of DNA600-AgNC and DNA750-AgNC have been previously reported in the literature to be 1.5 × 105 and 1.8 × 105 M−1 cm−1, respectively.15,17 The εexpNC values determined with our new method for DNA600-AgNC and DNA750-AgNC are in line with these reported εexpFCS,NC values, with the largest relative deviation of 25% observed for DNA600-AgNC. It is worth noting that in FCS, significant errors in the determination of the number of molecules can occur (especially below one molecule on average) due to contributions from uncorrelated background signals.28We also compared εexpNC with estimated absorption coefficients of the AgNC (εthNC) based on the theoretical DNA absorption coefficients at 260 nm (εth260). These εthNC values were calculated using eqn (4) (see Experimental section). Depending on which method was used to calculate the theoretical εth260, both positive and negative relative deviations up to 42% were observed between εthNC and εexpNC.
There are several approaches to calculate these theoretical values. In Table 2, the εth260 values on the left side of the first column are calculated using the nearest-neighbor method and are the values provided by the manufacturer (or twice those values if there are 2 DNA strands per AgNC).29
Alternatively, an estimate for εth260 can be obtained by summing up the ε values for the individual nucleotides.7 This second method was employed to calculate the εth260 values on the right side of the first column in Table 2, which are approximately 4–15% higher than those determined with the nearest-neighbor method. Cavaluzzi et al. showed that this second method provided more accurate estimates for small oligonucleotides.7
Additionally, the binding of silver to the nucleobases, particularly when the DNA-AgNCs are formed, could influence the absorption coefficients similarly to the hyperchromicity effect observed in melting curves of double-stranded DNA and DNA-metal base pairs.30–33 So far, the region below 300 nm in the excitation spectra of DNA-AgNCs has been interpreted as the result of a direct energy transfer from the DNA nucleobases to the silver core.8,9 However, while often assumed to be solely related to the DNA absorption, the AgNCs can exhibit absorption to higher excited states in this UV region. Careful analysis of the absorption spectra reveals indeed that some DNA-AgNCs present a shoulder around 300 nm (e.g., DNA525-AgNC, see Fig. 2a) that is unlikely to originate purely from DNA absorption. Since all the samples were HPLC purified, we believe that the absorption below 300 nm is partly due to an AgNC-related transition, resulting either from a direct excitation into higher-lying electronic states or mixed DNA/AgNC states.
Interestingly, when DNA-AgNC solutions age and decompose, the ratio of DNA to AgNC-related peak in the Vis/NIR range can change dramatically. For DNA750-AgNC, we indeed noticed that the ratio in the absorption spectrum changes over time (Fig. S15†), hence εth260-based values become increasingly unreliable for calculating the concentration of the remaining DNA-AgNCs.
It is also equally important to discuss limitations and potential sources of error in our new method. First, the purity of the samples needs to be assessed by comparing excitation spectra with absorption spectra. It is indeed paramount that DNA-AgNCs are pure and do not have Ag-containing byproducts. For example, in the case of DNA470-AgNC, we can see an additional small absorption band at 400 nm (Fig. 1) that is absent in the excitation spectrum (Fig. S6a†), indicating the presence of another species. Hence, HPLC purification was not completely successful in this case, and ICP-OES might have detected extra silvers not related to the cluster of interest, resulting most likely in an underestimation of εexpNC. Additionally, if the ionization process in the ESI-MS promotes the dissociation of Ag+ attached to either the DNA strands or the AgNC itself, we might also underestimate the number of silvers associated with the DNA-AgNC, which also leads to an underestimation of εexpNC. See ESI† for further discussion of ESI-MS limitations.
Molar absorption coefficients and radiative rate constants of DNA-AgNCs
The quantum yields and decay times of DNA-AgNCs allowed us to calculate the radiative rate constants (kf), which are reported in Table 3.20 We note that this is under the assumption that the non-radiative pathway is in direct competition with the radiative one. This might not always be true, as we have demonstrated in the past for red and NIR emissive DNA-AgNCs.34–36 Additionally, in the case of DNA525-AgNC and DNA750-AgNC, femtosecond transient absorption spectroscopy by Chen et al. and Thyrhaug et al.37,38 demonstrated that the Franck–Condon state quickly deactivates to a lower-energy excited state, the fluorescent state. As a result, the absorbing and emitting states are not the same and hence correlations between kf and ∈, as predicted by the Strickler–Berg relationship, might not hold.27In Fig. 3, the kf values are plotted against εexpNC and the maximum absorption wavelengths for the six measured DNA-AgNCs. We note that these values are in line with typical values obtained for organic fluorophores, supporting the molecular-like emission of DNA-AgNCs.26
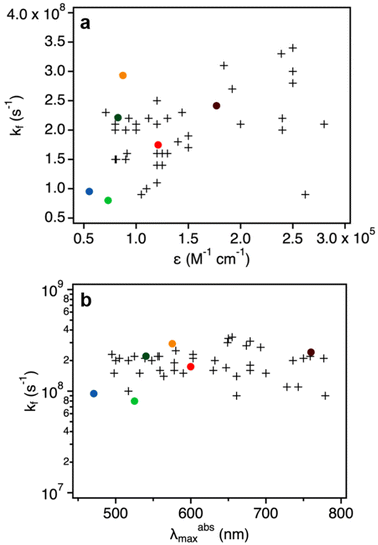 | ||
| Fig. 3 (a) Radiative rate constants (kf, Table 3) as a function of molar absorption coefficients. The colored dots refer to DNA-AgNCs (εexpNC, Table 2), while the crosses represent organic fluorophores reported by Maillard et al.26 (b) Radiative rate constants (kf) of DNA-AgNCs (colored dots) and organic fluorophores (crosses, data by Maillard et al.26) as a function of their absorption maximum in the Vis/NIR range (λabsmax). | ||
Experimental
Reagents
The oligonucleotides and the nuclease-free water were purchased from Integrated DNA Technologies (IDT). All DNA sequences are listed below, together with their corresponding names. AgNO3 (≥99.998%), NaBH4 (≥99.99%), and ammonium acetate (NH4OAc, ≥98%) were purchased from Sigma Aldrich. HNO3 (>68%) trace analysis-grade was purchased from Thermo Fisher. Alexa 750 Fluor™ Carboxylic Acid, tris(triethylammonium) salt was purchased from Thermo Fisher Scientific. All chemicals were used as received and dissolved in nuclease-free H2O.DNA sequences:
• DNA470: 5′-ATCCACGAG-3′
• DNA525: 5′-CACCTAGCGA-3′
• DNA540: 5′-TGGACGGCGG-3′
• DNA575: 5′-TTCCCACCCACCCCGGCCCGTT-3′
• DNA600: 5′-CACCGCTTTTGCCTTTTGGGGACGGATA-3′
• DNA750: 5′-CCCACCCACCCTCCCA-3′
Synthesis and purification of DNA-AgNCs
All DNA-AgNCs were synthesized by mixing the hydrated DNA with AgNO3 in a 10 mM NH4OAc solution (pH 7). After 15 min, the reaction mixture was reduced with NaBH4. The final ratio of the components is reported in the table below:The samples were stored in the fridge or at room temperature in the dark for 1 to 6 days before purification, as previously reported. For DNA750-AgNCs, the sample was instead kept in the fridge for 3 days prior to HPLC.
The HPLC purification was performed using a preparative HPLC system from Agilent Technologies (1100 series) with an Agilent Technologies 1260 Infinity fluorescence detector, an Agilent Technologies 1100 Series UV-Vis detector, and a Kinetex C18 column (5 μm, 100 Å, 250 × 4.6 mm; Phenomenex), equipped with a fraction collector. The mobile phase was a gradient mixture of 35 mM triethylammonium acetate (TEAA) buffer in water (A) and methanol (B). The flow rate was set to 1 mL min−1, whereas the specific elution gradient differs for each DNA-AgNC, and it is reported elsewhere.13,22–24 The only exceptions are DNA600-AgNC and DNA750-AgNC, which were purified with Luna C18(2) column (5 μm, 100 Å, 250 × 10 mm; Phenomenex), and the flow rate set to 4.7 mL min−1.
For DNA600-AgNC, the gradient was varied from 15% to 95% B as follows: 0–10 min 15% B, 10–61.7 min linear increase of B until 27%, 61.7–64 min constant at 27% B, and finally from 27% to 95% B in 4 min (total run time = 68 min). The fraction was collected based on the absorption signal at 600 nm.
For DNA750-AgNC, the following gradient was instead used: 0–10 min 15% B, 10–75.8 min increase of B until 29%, 75.8–82 min from 29% to 95% B. The fraction collection was based on the absorption signal at 730 nm.
Both runs were followed by 5 min of washing with 95% B to remove any traces of the samples from the column. The HPLC chromatograms of DNA600-AgNC and DNA750-AgNC are shown in Fig. S1 and S2†, respectively.
Electrospray ionization-mass spectrometry
ESI-MS measurements were performed with a Xevo G2-XS QTof (Waters Corporation), using negative ion mode with a 2 kV capillary voltage, 30 V cone voltage and no collision energy. Spectra were collected from 750 to 4000 m/z, with a scan time of 1 s. For DNA575-AgNC, the source temperature was 80 °C with a cone gas flow of 45 L h−1, and the desolvation temperature and gas flow were 150 °C and 450 L h−1, respectively. For DNA600-AgNC and DNA750-AgNC, the source temperature was instead set to 150 °C with a cone gas flow of 150 L h−1, and the desolvation temperature and gas flow were 600 °C and 1000 L h−1, respectively. The QTOF was calibrated using ESI-L Low Tune Mix (Agilent Technologies, Santa Clara, CA, USA), which contained compounds in the mass range of 1034 to 2834 m/z. All samples were injected using an Acquity I-Class Plus system (Waters) with a flow-through needle autosampler, with a flow of 0.1 mL min−1 50 mM NH4OAc buffer at pH 7–MeOH (80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) and using 2–5 μL injection volume. The system was operated using UNIFI v.1.9.4 (Waters), and the final spectra were generated by averaging multiple spectra surrounding the apex of the observed peak.
20) and using 2–5 μL injection volume. The system was operated using UNIFI v.1.9.4 (Waters), and the final spectra were generated by averaging multiple spectra surrounding the apex of the observed peak.
The recorded data were analyzed and fitted with the open-source software EnviPat Web (https://www.envipat.eawag.ch/index.php). ESI-MS of DNA470-AgNC, DNA525-AgNC, and DNA540-AgNC are reported elsewhere.13,23,25
Chemical composition
All the samples for the elemental analysis were prepared as follows. A certain volume of the DNA-AgNC stock solution was digested in 147 μL of HNO3 (>68%). The solution was mixed until it turned colorless and transparent, then it was brought to volume in a 2 mL volumetric flask with MilliQ water in order to obtain a 5% HNO3 solution.The volume of the stock solution was chosen in such a way that the concentration of Ag+ (ppm) in 5% HNO3 would be within and, possibly, in the middle of the calibration curves created with ICP-OES. The Ag+ concentration was estimated from the absorption band at 260 nm, assuming that all the absorption comes from the DNA strands:
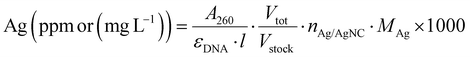 | (1) |
The calibration curve was built with the following silver standard concentrations: 1 ppm, 5 ppm, 10 ppm, 17 ppm, 30 ppm, 40 ppm, and 60 ppm, prepared by diluting a certified standard (Silver, plasma standard solution, Specpure™ Ag 1000 μg mL−1, Thermo Scientific Chemicals, 1000 μg mL−1 in 5% HNO3).
ICP-OES was conducted using a Thermofisher scientific iCAP PRO X DUO equipped with a photomultiplier tube detector. The Argon 99.999% gas was utilized for the plasma, and the operating pressure ranged from 5.5 to 6 bar. The instrument has a cooling system with pressure ranging from 2 to 6 bar. The employed purge gas is the same Argon 99.999% at the same pressure. The instrument is equipped with a CETAC ASX-280 autosampler from Teledyne.
To quantify the amount of silver, three different measurements were carried out, and each value was evaluated at three emission lines of silver: 243.8 nm, 328 nm, and 338 nm. The amount of silver obtained at each wavelength was then averaged and used to calculate the absorption coefficient.
The amount of silver in ppm obtained from ICP-OES was then converted to the molar concentration of DNA-AgNCs using the following formula:
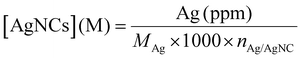 | (2) |
Optical spectroscopy
All absorption measurements were performed on a Cary 300 UV-Vis spectrophotometer from Agilent Technologies using a deuterium lamp for ultraviolet radiation and a halogen lamp for visible and near-infrared radiation. The measurements were carried out in a single-beam configuration with a 0/100% transmittance baseline correction. Every spectrum was subtracted by the absorption spectrum of the corresponding blank (10 mM NH4OAc).The εexpNC is calculated by applying the Lambert–Beer law:
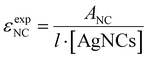 | (3) |
The εthNC value is estimated from εth260 in the following way:
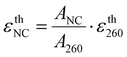 | (4) |
Steady-state fluorescence measurements were performed for DNA750-AgNC using a FluoTime300 instrument from PicoQuant. The emission spectrum was recorded by exciting with a 726 nm picosecond-pulsed laser from Picoquant (LDH-P-C-730).
The excitation spectra were measured using a QuantaMaster400 from PTI/HORIBA with a xenon arc lamp.
All spectra were corrected for the wavelength dependency of the detector and the excitation spectra were additionally corrected for the lamp power. For all fluorescence measurements, the absorbance of the investigated compound was kept below 0.1 at the excitation wavelength in order to avoid inner filter effects.
Time-resolved fluorescence measurements were carried out for DNA750-AgNC using a Fluotime300 instrument from PicoQuant with a 726 nm picosecond-pulsed laser (LDH-P-C-730) as excitation source. The intensity decay was monitored at 825 nm. The repetition rate was set to 25 MHz and the integration time was chosen to be 15 s in order to reach at least 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 counts in the maximum. The data was analyzed using Fluofit v.4.6 software from PicoQuant. The decay curve was fitted with a bi-exponential reconvolution model including the instrument response function (IRF), see Fig. S8.† The obtained amplitude (αi) and decay time (τi) components were used to calculate the intensity-weighted average decay time τexp at the selected emission wavelength.20
000 counts in the maximum. The data was analyzed using Fluofit v.4.6 software from PicoQuant. The decay curve was fitted with a bi-exponential reconvolution model including the instrument response function (IRF), see Fig. S8.† The obtained amplitude (αi) and decay time (τi) components were used to calculate the intensity-weighted average decay time τexp at the selected emission wavelength.20
Fluorescence quantum yield calculation
The quantum yield of DNA750-AgNC was determined in 10 mM NH4OAc aqueous solution at 25 °C, using Alexa 750 Fluor™ as reference dye in a PBS buffer (50 mM K3PO4 and 150 mM NaCl) with pH = 7.2. Absorption and emission spectra of the DNA750-AgNC and the reference compound were measured at five different concentrations, and the quantum yield was calculated according to the following formula:20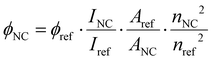 | (5) |
The emission spectra, fluorescence decays and quantum yield values for the other five DNA-AgNCs are reported elsewhere,13,17,22–24 while the spectra and data measured for DNA750-AgNC are reported in the Fig. S7 and S8.†
The experimental radiative rate constant (kf) is calculated from:
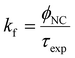 | (6) |
Conclusions
In this study, we introduced an analytical method for determining the absorption coefficient of DNA-AgNCs. This approach integrates UV-Vis absorption, ESI-MS, and ICP-OES measurements. The concentration of DNA-AgNCs was obtained from the ESI-MS and ICP-OES data, and it was combined with the corresponding absorbance to calculate εexpNC using the Lambert–Beer law. The values derived with this method span from 5.40 × 104 to 1.76 × 105 M−1 cm−1 for the six investigated DNA-AgNCs. For DNA600-AgNC and DNA750-AgNC, εexpNC values were in line with the FCS-based values reported in the literature. We discussed the limitations of using εthNC based on the DNA absorption peak at 260 nm and the possible sources of error in our new method. Finally, experimentally determined kf values were plotted as a function of the molar absorption coefficients of DNA-AgNCs. The εexpNC and kf values are in line with those of organic fluorophores, highlighting the molecular-like nature of DNA-AgNC emission.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
G. R., C. C., V. R., and T. V. acknowledge funding from the Villum Foundation (VKR023115), the Independent Research Fund Denmark (0136-00024B) and the Novo Nordisk Foundation (NNF22OC0073734). We thank Tatiana Yuli Morante Bligaard and the Center for High Entropy Alloy Catalysis (DNRF 149) for providing access to ICP-OES.References
- A. Gonzàlez-Rosell, C. Cerretani, P. Mastracco, T. Vosch and S. M. Copp, Nanoscale Adv., 2021, 3, 1230–1260 RSC.
- J. T. Petty, J. Zheng, N. V. Hud and R. M. Dickson, J. Am. Chem. Soc., 2004, 126, 5207–5212 CrossRef CAS PubMed.
- S. Malola, M. F. Matus and H. Häkkinen, J. Phys. Chem. C, 2023, 127, 16553–16559 CrossRef CAS.
- S. A. Patel, C. I. Richards, J.-C. Hsiang and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 11602–11603 CrossRef CAS PubMed.
- C. R. Cantor and I. Tinoco, J. Mol. Biol., 1965, 13, 65–77 CrossRef CAS PubMed.
- C. R. Cantor, M. M. Warshaw and H. Shapiro, Biopolymers, 1970, 9, 1059–1077 CrossRef CAS PubMed.
- M. J. Cavaluzzi and P. N. Borer, Nucleic Acids Res., 2004, 32, e13–e13 CrossRef PubMed.
- I. L. Volkov, Z. V. Reveguk, P. Y. Serdobintsev, R. R. Ramazanov and A. I. Kononov, Nucleic Acids Res., 2017, 46, 3543–3551 CrossRef PubMed.
- P. R. O'Neill, E. G. Gwinn and D. K. Fygenson, J. Phys. Chem. C, 2011, 115, 24061–24066 CrossRef.
- G. Romolini, J. A. Steele, J. Hofkens, M. B. J. Roeffaers and E. Coutino-Gonzalez, Adv. Opt. Mater., 2021, 9, 2170094 CrossRef.
- L. Sun, M. Keshavarz, G. Romolini, B. Dieu, J. Hofkens, F. de Jong, E. Fron, M. B. J. Roeffaers and M. Van der Auweraer, Chem. Sci., 2022, 13, 11560–11569 RSC.
- C. Cerretani, H. Kanazawa, T. Vosch and J. Kondo, Angew. Chem., Int. Ed., 2019, 58, 17153–17157 CrossRef CAS PubMed.
- V. Rück, V. A. Neacşu, M. B. Liisberg, C. B. Mollerup, P. H. Ju, T. Vosch, J. Kondo and C. Cerretani, Adv. Opt. Mater., 2024, 12, 2301928 CrossRef.
- T. Vosch, Y. Antoku, J.-C. Hsiang, C. I. Richards, J. I. Gonzalez and R. M. Dickson, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 12616–12621 CrossRef CAS PubMed.
- J. T. Petty, C. Fan, S. P. Story, B. Sengupta, A. St John Iyer, Z. Prudowsky and R. M. Dickson, J. Phys. Chem. Lett., 2010, 1, 2524–2529 CrossRef CAS PubMed.
- C. I. Richards, S. Choi, J.-C. Hsiang, Y. Antoku, T. Vosch, A. Bongiorno, Y.-L. Tzeng and R. M. Dickson, J. Am. Chem. Soc., 2008, 130, 5038–5039 CrossRef CAS PubMed.
- S. A. Bogh, C. Cerretani, L. Kacenauskaite, M. R. Carro-Temboury and T. Vosch, ACS Omega, 2017, 2, 4657–4664 CrossRef CAS PubMed.
- B. Sengupta, C. M. Ritchie, J. G. Buckman, K. R. Johnsen, P. M. Goodwin and J. T. Petty, J. Phys. Chem. C, 2008, 112, 18776–18782 CrossRef CAS PubMed.
- J. Sharma, H.-C. Yeh, H. Yoo, J. H. Werner and J. S. Martinez, Chem. Commun., 2010, 46, 3280–3282 RSC.
- J. R. Lakowicz, Principles of fluorescence spectroscopy, 2006 Search PubMed.
- I. Volkov, T. Sych, P. Serdobintsev, Z. Reveguk and A. Kononov, J. Lumin., 2016, 172, 175–179 CrossRef CAS.
- C. Cerretani, M. B. Liisberg, V. Rück, J. Kondo and T. Vosch, Nanoscale Adv., 2022, 4, 3212–3217 RSC.
- V. Rück, M. B. Liisberg, C. B. Mollerup, Y. He, J. Chen, C. Cerretani and T. Vosch, Angew. Chem., Int. Ed., 2023, 62, e202309760 CrossRef PubMed.
- C. Cerretani and T. Vosch, ACS Omega, 2019, 4, 7895–7902 CrossRef CAS PubMed.
- A. Gonzàlez-Rosell, S. Malola, R. Guha, N. R. Arevalos, M. F. Matus, M. E. Goulet, E. Haapaniemi, B. B. Katz, T. Vosch, J. Kondo, H. Häkkinen and S. M. Copp, J. Am. Chem. Soc., 2023, 145, 10721–10729 CrossRef PubMed.
- J. Maillard, K. Klehs, C. Rumble, E. Vauthey, M. Heilemann and A. Fürstenberg, Chem. Sci., 2021, 12, 1352–1362 RSC.
- S. J. Strickler and R. A. Berg, J. Chem. Phys., 1962, 37, 814–822 CrossRef CAS.
- https://www.picoquant.com/images/uploads/page/files/7351/appnote_quantfcs.pdf .
- https://eu.idtdna.com/pages/tools/oligoanalyzer .
- G. H. Clever, C. Kaul and T. Carell, Angew. Chem., Int. Ed., 2007, 46, 6226–6236 CrossRef CAS PubMed.
- M. Daune, C. A. Dekker and H. K. Schachman, Biopolymers, 1966, 4, 51–76 CrossRef CAS.
- T. Yamane and N. Davidson, Biochim. Biophys. Acta, 1962, 55, 609–621 CrossRef CAS PubMed.
- H. Mei, I. Röhl and F. Seela, J. Org. Chem., 2013, 78, 9457–9463 CrossRef CAS PubMed.
- V. A. Neacşu, C. Cerretani, M. B. Liisberg, S. M. Swasey, E. G. Gwinn, S. M. Copp and T. Vosch, Chem. Commun., 2020, 56, 6384–6387 RSC.
- S. Krause, M. R. Carro-Temboury, C. Cerretani and T. Vosch, Phys. Chem. Chem. Phys., 2018, 20, 16316–16319 RSC.
- C. Cerretani, G. Palm-Henriksen, M. B. Liisberg and T. Vosch, Chem. Sci., 2021, 12, 16100–16105 RSC.
- J. Chen, A. Kumar, C. Cerretani, T. Vosch, D. Zigmantas and E. Thyrhaug, J. Phys. Chem. Lett., 2023, 14, 4078–4083 CrossRef CAS PubMed.
- E. Thyrhaug, S. A. Bogh, M. R. Carro-Temboury, C. S. Madsen, T. Vosch and D. Zigmantas, Nat. Commun., 2017, 8, 15577 CrossRef CAS PubMed.
- https://www.thermofisher.com/uk/en/home/references/molecular-probes-the-handbook/tables/fluorescence-quantum-yields-and-lifetimes-for-alexa-fluor-dyes.html .
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr01765j |
| This journal is © The Royal Society of Chemistry 2024 |