Open Access Article
Open Access ArticlePaper/GO/e-Au flexible SERS sensors for in situ detection of tricyclazole in orange juice and on cucumber skin at the sub-ppb level: machine learning-assisted data analysis†
Ha Anh
Nguyen‡
 *a,
Quan Doan
Mai‡
*a,
Quan Doan
Mai‡
 a,
Dao Thi
Nguyet Nga
a,
Dao Thi
Nguyet Nga
 a,
Minh Khanh
Pham
a,
Quoc Khanh
Nguyen
b,
Trong Hiep
Do
b,
Van Thien
Luong
b,
Vu Dinh
Lam
c and
Anh-Tuan
Le
a,
Minh Khanh
Pham
a,
Quoc Khanh
Nguyen
b,
Trong Hiep
Do
b,
Van Thien
Luong
b,
Vu Dinh
Lam
c and
Anh-Tuan
Le
 *ad
*ad
aPhenikaa University Nano Institute (PHENA), Phenikaa University, Hanoi 12116, Vietnam. E-mail: anh.nguyenha@phenikaa-uni.edu.vn; tuan.leanh@phenikaa-uni.edu.vn
bFaculty of Computer Science, Phenikaa University, Hanoi 12116, Vietnam
cInstitute of Materials Science (IMS), Graduate University of Science and Technology (GUST), Vietnam Academy of Science and Technology, 18 Hoang Quoc Viet, Hanoi 10000, Vietnam
dFaculty of Materials Science and Engineering (MSE), Phenikaa University, Hanoi 12116, Vietnam
First published on 24th April 2024
Abstract
Despite being an excellent surface enhanced Raman scattering (SERS) active material, gold nanoparticles were difficult to be loaded onto the surface of filter paper to fabricate flexible SERS substrates. In this study, electrochemically synthesized gold nanoparticles (e-AuNPs) were deposited on graphene oxide (GO) nanosheets in solution by ultrasonication, resulting in the formation of a GO/Au hybrid material. Thanks to the support of GO, the hybrid material could adhere onto the surface of filter paper, which was immersed into a GO/Au solution for 24 h and dried naturally at room temperature. The paper-based materials were then employed as substrates for a surface enhanced Raman scattering (SERS) sensing platform to detect tricyclazole (TCZ), a widely used pesticide, resulting in better sensitivity compared to the use of paper/Au SERS sensors. With the most optimal GO content of 4%, paper/GO/Au SERS sensors could achieve a limit of detection of 1.32 × 10−10 M in standard solutions. Furthermore, the filter paper-based SERS sensors also exhibited significant advantages in sample collection in real samples. On one hand, the sensors were dipped into orange juice, allowing TCZ molecules in this real sample to be adsorbed onto their SERS active surface. On the other hand, they were pasted onto cucumber skin to collect the analytes. As a result, the paper/GO/Au SERS sensors could sense TCZ in orange juice and on cucumber skin at concentrations as low as 10−9 M (∼2 ppb). In addition, a machine learning model was designed and developed, allowing the sensing system to discriminate TCZ from nine other organic compounds and predict the presence of TCZ on cucumber skin at concentrations down to 10−9 M.
1. Introduction
Raman spectroscopy is a non-destructive analytical tool to study molecular structures. It provides detailed information about the unique vibration modes of samples, allowing the identification of chemical compounds.1 Since its discovery in 1977, surface-enhanced Raman spectroscopy (SERS) has gained increasing attention as it inherits from Raman the most important characteristic of chemical fingerprinting while improving its sensitivity. In the SERS phenomenon, Raman scattering of analytes is significantly enhanced when they are adsorbed onto the surface of plasmonic materials, allowing the detection of analytes at trace and even single-molecule levels.2,3 Noble metal nanoparticles (NPs), such as Au, Ag and Cu, have been employed as active substrates for SERS-based applications for environmental,4–6 food7,8 and health9,10 safety monitoring.In order to improve the performance of the SERS-based sensing systems, on one hand, many research groups have focused on modification of active materials to improve the sensitivity and reliability of the sensors. Plasmonic NPs with sharp tips and corners, such as nanostars,11,12 nanoflowers,13 nanocubes,14,15etc., were prepared as SERS substrates to sense various chemical and biological analytes. Besides, SERS active materials have been grown directly on rigid substrates to form nanogaps as well as generate high uniformity of the substrates.16,17 These strategies aimed to create hot-spots, where the electromagnetic field is particularly intense, allowing the Raman signal to be enhanced in the electromagnetic mechanism (EM). Several other groups modified the active substrates to promote substrate-to-analyte charge transfer to enhance the Raman signal in the chemical mechanism (CM). Different semiconductor–metal composites were fabricated and employed as SERS substrates, showing an enhancement in the SERS signal of analytes, compared to using pure plasmonic nanomaterials.18–20 Besides, with large surface area and excellent adsorption ability, graphene has been considered as the star of the two-dimensional (2D) materials family.21 Many SERS sensing systems have been designed and developed based on plasmonic materials and graphene or its derivatives.22,23
On the other hand, researchers have been more concerned about the practicability of the SERS sensors in real samples. Real applications are challenging for the development of the SERS sensing platform because real samples are much more complicated than standard ones in the laboratory. To be specific, standard samples are usually prepared in solution, which makes it convenient for them to be drop-cast on the surface of widely used rigid substrates, such as silicon wafers and glass slides. However, in real samples, it requires SERS sensors to detect analytes not only in solution but also on different surfaces, either planar or non-planar ones. Thus, flexible substrates, which can be pasted and/or wrapped onto different surfaces, have been designed for both material deposition and sample collection. For example, polydimethylsiloxane (PDMS) films loaded with Au nanostars and nanorods have been fabricated to detect thiabendazole and thiram, respectively, on fruit skin.24,25 In another approach, commercial adhesive tape was employed to fabricate “sticky” substrates.26–29 Although the use of adhesive tape is cost- and time-effective, this sticky substrate is only suitable to collect materials and samples on dry surfaces, which would limit the variety of materials and samples. Being hydrophilic but insoluble in water, filter paper is more versatile. SERS active materials have been introduced onto in-paper substrates by drop-casting,30 dip-coating,31 spray coating32 and thermal inkjet printing.33 Subsequently, the as-prepared substrates can be dipped into aqueous media, or be attached to wet surfaces via van der Waals forces to absorb analytes. In a recent study, we fabricated an in-paper SERS sensor by dip-coating filter paper into a silver nanoparticle (AgNP) solution for 24 h and then drying naturally at room temperature. The versatile AgNP-based SERS sensor could detect methylene blue in river water and thiram on apple skin at concentrations down to 1.0 × 10−10 M.34 In another study, Oliveira et al. prepared an Ag-based paper SERS sensor by a drop-casting method to detect rhodamine-6G with detection limit (LOD) as low as 11.4 ± 0.2 pg.35 These one-step substrate preparation methods were expected to be utilized to fabricate other paper-based substrates using other SERS active nanomaterials. Nevertheless, poor adsorption of gold nanoparticles (AuNPs) on the cellulose fiber network of filter paper, resulting in low SERS sensing performance of the SERS substrate, was reported in several studies.36,37 However, it did not prevent researchers from developing the idea of combining the excellent SERS performance of AuNPs and the versatility of filter paper. Jang et al. suggested performing the spray coating technique to load AuNPs onto filter paper, leading to enhancements in SERS signals over 2–5 times.36 However, it required the utilization of a spray gun tool purged with N2 gas, which should be performed in a laboratory by skilled labor. A simpler approach was proposed by Moram et al., in which aggregation of AuNPs was induced by NaCl before the dip coating step.38 The addition of NaCl solution (50 mM) led to an intermediate state of aggregation of AuNPs, reducing the distance between them and forming more hot spots. As a result, the SERS signal of MB on an aggregated-AuNP-based substrate was significantly improved. Nevertheless, the aggregation could not be well-controlled, resulting in poor uniformity of the in-paper substrate. Moreover, larger concentrations of NaCl promote the over-aggregation of AuNPs, and in contrast, reduce the number of hot spots, which was reflected by the rapid decrease in the SERS signal of MB on those substrates.
Another issue for the application of SERS sensors is complicated data analysis with the spectral overlapping of various molecules preventing sample detection and identification.39,40 Recently, machine learning has been developed as a solution to discriminate chemical and biological compounds with similar Raman spectra.41–43
In this study, we utilized graphene oxide (GO) nanosheets as the support for grafting electrochemically synthesized AuNPs (e-AuNPs). Subsequently, the composite material was loaded onto filter paper by dip-coating to fabricate a paper/GO/e-Au SERS sensor for detection of tricyclazole (TCZ), a commonly used pesticide. With the most optimal GO content of 4%, it could detect TCZ at concentrations down to 1.32 × 10−10 M in standard solutions and 10−9 M in real samples of orange juice and cucumber skin. Thus, we have fabricated a versatile in-paper substrate. Moreover, we also developed a machine learning model, allowing the SERS signal of TCZ to be distinguished from the signals of nine other organic compounds. On real samples of cucumber skin, the artificial neural network could detect and identify TCZ at concentrations down to 10−9 M, which was in agreement with the practicability result of the sensing system. Hence, with the machine learning-assisted SERS sensing system, we have targeted all three approaches as discussed above, including: (i) developing a versatile flexible in-paper substrate for detection of an analyte in solution and non-planar surface; (ii) improving the SERS sensing system by material modification; and (iii) discriminating the desired analyte from other organic compounds via machine learning.
2. Materials and methods
2.1. Materials
Sodium citrate (Na3C6H5O7, 99.9%), tricyclazole (C9H7N3S, 97.0%), ethanol (C2H5OH, 98%), graphite flakes, nitric acid (HNO3, 90%), potassium permanganate (KMnO4, 99%), sodium nitrate (NaNO3, 99%) and sulfuric acid (H2SO4, 98%) were purchased from Shanghai Chemical Reagent and used directly without further purification. Two gold plates (purity: 99.99%) were prepared with dimensions of (100 mm × 5 mm × 0.5 mm). Filter paper was supplied from Hangzhou Special Paper Industry Co., Ltd (NEWSTAR INDUSTRY). Double distilled water was used throughout the experiments.2.2. Electrochemically synthesized gold nanoparticles and their characterization
e-AuNPs were synthesized electrochemically in a 200 mL beaker containing 0.1% Na3C6H5O7 in distilled water. The electrodes were polished and washed to remove surface oxides before the synthesis. Electrolysis was carried out at room temperature with magnetic stirring at 200 rpm for 2 hours. This resulted in a wine-red colloidal e-AuNP solution. The shapes and sizes of e-AuNPs were recorded by scanning electron microscopy (SEM, Hitachi S-4800) operated at an acceleration voltage of 5 kV, which revealed the spherical shape of the NPs with an average diameter of 19 nm (Fig. S1a†). UV-Vis absorption spectra were recorded using a JENWAY 6850 spectrophotometer. The as-prepared e-AuNPs exhibited a plasmonic band at ∼532 nm (Fig. S1b†). Full information about NP characterization is reported in our previous study.442.3. Preparation of graphene oxide sheets
GO was synthesized using the Hummers' method as described in a previous study of our group.45 A mixture of graphite flakes, HNO3 and KMnO4 was generated with a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5 wt. The mixture was transferred to a Teflon autoclave, sealed, and heated in a microwave reactor (UWave-2000 Multifunctional Microwave, MW-2000/SINEO) with a power of 800 W for 1 min to obtained layered graphite. 2.0 g of layered graphite, 8.0 g of KMnO4 and 1.0 g of NaNO3 were added gradually into 160 mL of H2SO4 at 5 °C, followed by constant stirring for 30 min. The temperature of the solution was gradually increased up to 45 °C. The solution was stirred constantly for 2 hours. GO was generated when deionized water was dropped into the solution and the temperature was raised to 95 °C for 1 hour. Subsequently, H2O2 (30%) and HCl (10%) were added into GO solution to eliminate excessive KMnO4 and metal ions, and dissolve MnO2. Finally, GO was washed with distilled water using centrifugation.
1.5 wt. The mixture was transferred to a Teflon autoclave, sealed, and heated in a microwave reactor (UWave-2000 Multifunctional Microwave, MW-2000/SINEO) with a power of 800 W for 1 min to obtained layered graphite. 2.0 g of layered graphite, 8.0 g of KMnO4 and 1.0 g of NaNO3 were added gradually into 160 mL of H2SO4 at 5 °C, followed by constant stirring for 30 min. The temperature of the solution was gradually increased up to 45 °C. The solution was stirred constantly for 2 hours. GO was generated when deionized water was dropped into the solution and the temperature was raised to 95 °C for 1 hour. Subsequently, H2O2 (30%) and HCl (10%) were added into GO solution to eliminate excessive KMnO4 and metal ions, and dissolve MnO2. Finally, GO was washed with distilled water using centrifugation.
2.4. Fabrication of paper/e-Au and paper/GO/e-Au flexible SERS sensors
A mixture of e-AuNPs and GO in solution underwent 30 minutes of ultrasonication for deposition of e-AuNPs onto GO nanosheets, resulting in a GO/e-Au solution. Filter paper was cut into 1 cm × 1 cm pieces and immersed in the solution for 24 hours and dried naturally at room temperature. The structure of cellulose fibers within filter paper before and after the loading of e-AuNPs and GO/e-Au was observed under a DN-117M digital microscope using a 40× objective lens. The arrangement of e-AuNPs and GO/e-Au on the cellulose fiber surface in paper/e-Au and paper/GO/e-Au was analyzed by scanning electron microscopy (SEM, Hitachi S-4800) operated at an acceleration voltage of 5 kV.2.5. Standard samples: sample collection and SERS measurements
Distilled water was used to prepare solutions with various concentrations of TCZ ranging from 10−11 M to 10−3 M. In each SERS measurement, 5 μL of TCZ solution was drop-cast onto paper/e-Au and paper/GO/e-Au flexible SERS sensors and dried naturally at room temperature. Subsequently, SERS spectra were recorded from the substrate using a MacroRaman™ Raman spectrometer (Horiba) with 785 nm laser excitation. Raman measurements were taken using a 100× objective with a numerical aperture of 0.90. The laser power was set at 45 mW, with a 45° contact angle, resulting in a diffraction-limited laser spot diameter of 1.1 μm (1.22λ/NA) and a focal length of 115 nm. Each measurement lasted for 10 seconds with three accumulations. Finally, the spectrum was obtained after baseline calibration.2.6. Real samples: sample collection and SERS measurements
Cucumbers were obtained from a local supermarket in Hanoi, Vietnam, and thoroughly washed. For each measurement, 10 μL of TCZ solution at various concentrations (ranging from 10−10 M to 10−6 M) was directly dropped onto the surface of the cucumber skin and allowed to dry at room temperature. Following this, 10 μL of 98% ethanol was dropped onto the spiked cucumber skin, and a paper/GO/e-Au SERS sensor was immediately attached to the peel. The SERS paper was pressed onto the surface for 3 minutes and then carefully peeled off, following a procedure referred to as the “paste and peel off” method. The SERS paper was subsequently dried at room temperature. SERS measurements were conducted according to the procedure outlined in the previous section.Orange juice was also bought from a supermarket in Hanoi, Vietnam, and used directly for analysis. To achieve different concentrations (ranging from 10−10 M to 10−6 M), TCZ was spiked into the samples. The SERS paper was immersed in 10 mL of the aqueous TCZ solution for 30 minutes. Afterward, they were removed and allowed to naturally dry at room temperature, following a procedure referred to as the “dip and dry” method.
The recovery rate of each measurement is calculated using the formula:
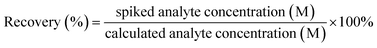 | (1) |
2.7. Machine learning-assisted data analysis
In order to speed up the process of classifying various pesticides acquired through the SERS sensors we leverage machine learning methodologies. In particular, our dataset consists of 50 spectral samples of 10 different compounds, each having 5 samples. Each data specimen encompasses intensity measurements at 2047 discrete Raman shift values, where only peak values, which retain the most salient information, are selected to feed the ML model. Depending on the compound's nature, the number of retained peaks varies from 2 to 4, resulting in a total of 34 Raman shift values per sample in the ultimate dataset. This dataset is partitioned into two subsets, namely, a training set and a testing set, comprising 30 and 20 samples, respectively. In order to mitigate the occurrence of data samples exclusively appearing in either the training or test set, we deliberately select the training set and testing set to ensure each compound had three samples in the training set and two samples in the test set.A wide range of multi-class classification ML models such as Logistic Regression, K-nearest Neighbor, Support Vector Machine, Decision Tree, Random Forest, and Naive Bayes are employed for classifying pesticides. The details of these models can be found in a textbook.46 We first train these models using the training set and subsequently make predictions on the trained model to measure the accuracy of the substance classification within the testing set. The model achieving the highest classification accuracy will be selected for realistic substance classifications.
All aforementioned data analytics, dimensionality reduction, and model training are executed utilizing the Python programming framework, where Pandas, Scikit-learn, and Matplotlib are the main Python packages used in this contribution. Herein, note that we implement the grid search method in Python to find the best hyperparameters for each model, namely, Logistic Regression (C=10, solver=‘liblinear’), K-nearest Neighbor (metric=‘cosine’, n_neighbors=1), Support Vector Machine (C=10, degree=2, kernel=‘linear’), Decision Tree (criterion=‘entropy’, max_depth=6, min_samples_split=2), Random Forest (criterion=‘gini’, max_depth=4, min_samples_split=5), and Naive Bayes (alpha=0.01).
All aforementioned data analytics, dimensionality reduction, and model training are executed utilizing the Python programming framework, where Pandas, Scikit-learn, and Matplotlib are the main Python packages used in this contribution.
3. Results and discussion
3.1. Fabrication of paper/GO/e-Au flexible SERS sensors
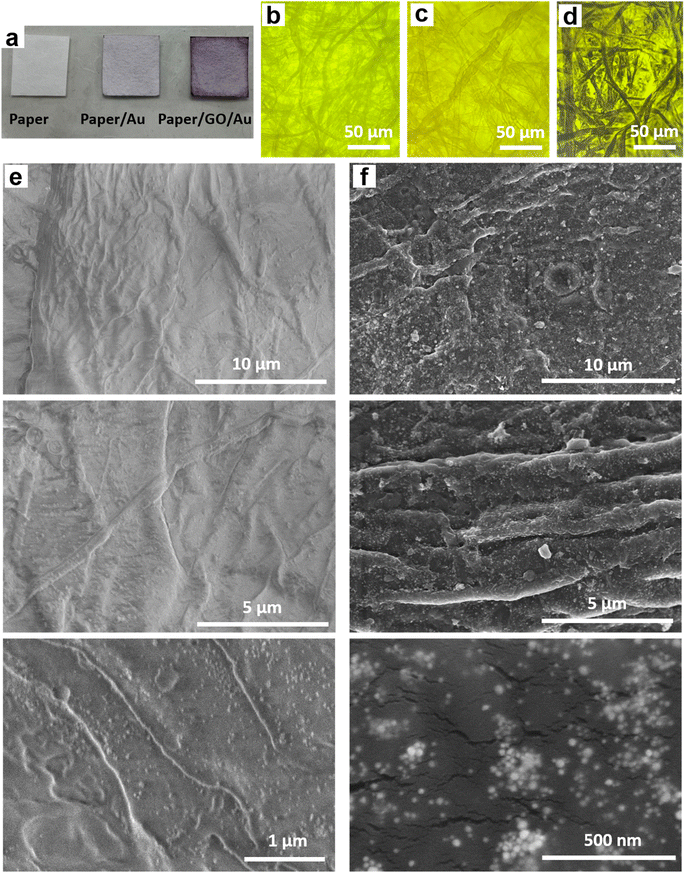
Paper/e-Au flexible SERS sensors were fabricated as described in our previous study,34 in which the e-AgNP solution was replaced by an e-AuNP one. As e-AuNPs were also coated with citrate, it was expected that e-AuNPs could be attached to the filter paper via hydrogen bonds between their coating molecules and –OH groups on the cellulose chains of the paper. More importantly, Au–O could also be formed thanks to the presence of –OH groups on the surface of cellulose, leading to direct grafting of e-AuNPs onto the paper surface. However, after 24 h of soaking in the e-AuNP solution the filter paper only showed a slight change in color, which can be observed with the bare eye (Fig. 1a) as well as under a microscope (Fig. 1b and c). The deposition of e-AuNPs on the filter paper was confirmed by a SEM image (Fig. 1e), and is represented by the presence of bright grains on the surface of the paper. However, NP density was much lower than that when using e-AgNP solutions.34 Thus, despite using the same method of substrate preparation, fewer e-AuNPs were anchored on the cellulose matrix of the filter paper. This was an unexpected result as Au–O bonding was reported to be a strong covalent one,47 even stronger than Ag–O bonding.48 However, in a 2015 study, Sun et al. calculated the binding energies of O in Mx–O (M = Ag or Au) models. The authors claimed the binding energy of the Au–O or Ag–O bond could be suppressed by the neighboring Au or Ag atoms. This suppressing effect of the neighboring Au atoms is stronger than that by the Ag ones. Hence, despite the intrinsically stronger Au–O bond, the Au–O bond in Aux–O usually appeared to be weaker than the Ag–O bond in Agx–O.49 In this case, formation of the Au–O bond between the e-AuNPs and cellulose fibers of the filter paper could have been suppressed by many neighboring atoms, and therefore, it was weaker than the Ag–O bond. Hence, there were only a few e-AuNPs attached onto the surface of the filter paper. Furthermore, in our previous study, the size of e-AuNPs was reported to be 19 nm in diameter (Fig. S1a†).44 The small size and weak Au–O bond would have prevented them from being anchored on the filter paper. Instead, they tended to be washed out through it. Therefore, this simple dip-and-dry method was not suitable to load e-AuNPs onto the paper substrate. Therefore, to combine the advantages of e-AuNPs and filter paper to fabricate a flexible SERS substrate, it required some modifications, which would allow the material to effectively adsorb onto the cellulose fibers. With a large surface, sheet-like GO was selected to combine with e-AuNPs to prepare a SERS active material which would not be easily filtered through the filter paper.Pristine GO was synthesized using a modified Hummers' method, resulting in a sheet-like structure, which was report in our 2021 study.50 GO and e-AuNPs were mixed together at different ratios, including 1, 2, 3, 4 and 5 wt% of GO. Subsequently, the mixture underwent sonication for 30 min, allowing the anchoring of e-AuNPs onto the GO surface. It was reported that the carbon to oxygen ratio in GO could reach approximately three to one,51 thanks to the presence of oxygenated functional groups such as ketone, hydroxyl, carboxyl and epoxy, exposed on both its edges and basal plane.52,53 Except for the epoxy group, in which the O atom shares two stable covalent bonds with two C atoms, other functional group can be available for the attachment of e-AuNPs via Au–O bonds (Fig. 2 – left side). Although cellulose fibers are also rich in oxygen, the oxygen-containing groups on the surface of cellulose chains need to interact with each other to maintain the stability of the fibers. Therefore, compared to cellulose fibers, oxygen-containing functional groups on the surface of GO sheets could be more available to form Au–O bonds. Moreover, in a 2015 study, Wang and Liu performed a force analysis at the AuNP/GO interface, revealing that the attachment of AuNPs on the GO surface promoted wrinkle formation and the wrinkling in turn strengthened the binding of AuNPs.54
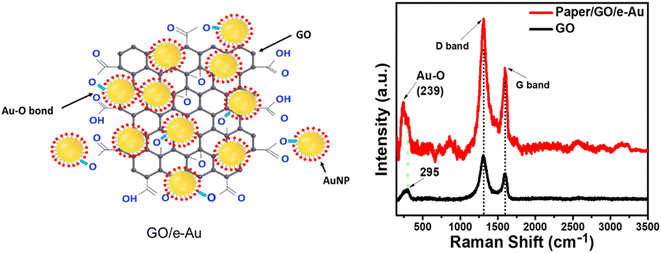 | ||
| Fig. 2 (Left) Formation of Au–O bonds between e-AuNPs and GO; (right) Raman spectra of GO (black) and paper/GO/e-Au with GO content of 4% (red). | ||
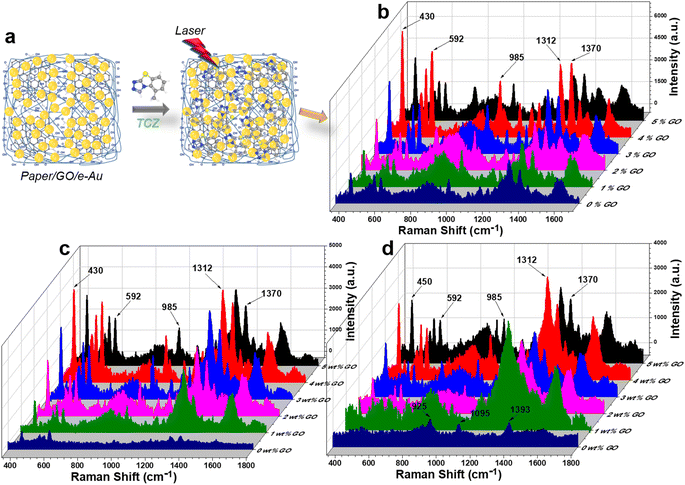
Paper/GO/e-Au SERS sensors were fabricated using filter paper and GO/Au solutions with different GO ratios by a dip-and-dry method. Thanks to the presence of GO, we could observe a significant change in the color of the filter paper by the bare eye (Fig. 1a). A microphotograph of the paper/GO/e-Au material with GO content of 4% (Fig. 1d) shows that the cellulose fibers of the filter paper were covered by a black material, which is the characteristic color of GO. This was confirmed by Raman spectroscopy. Fig. 2 shows the Raman spectrum of paper/GO/e-Au, in which the characteristic D- and G-bands of GO are obviously detected. In comparison to that of GO, the Raman spectrum of paper/GO/e-Au is noisier, which could be due to the presence of cellulose. However, it does not exhibit the characteristic peaks of cellulose such as those at 1093 cm−1, 1127 cm−1 and 1382 cm−1. In fact, these bands usually show low Raman intensity.34,55 Moreover, they are overlapped with the D- and G-bands of GO, which exhibit extremely high intensity in the Raman spectra. In the SEM image (Fig. 1f), the bright particles representing the presence of e-AuNPs were also revealed to be denser on the scaffold of GO on the filter paper. Therefore, with the assistance of GO during the fabrication of the in-paper SERS sensors, we enriched the density of the SERS active materials. As a result, the peak at 239 cm−1 was detected at high intensity in the Raman spectrum of paper/GO/e-Au, corresponding to the Au–O stretching mode,56 which was schematically described in Fig. 2. This peak differs from the band in the Raman spectrum of GO (290 cm−1), which could be due to excessive chemicals which were not eliminated properly during GO synthesis.
3.2. SERS sensing performance: a comparison of paper/e-Au and paper/GO/e-Au flexible SERS sensors
For standard samples, TCZ solution prepared in distilled water was drop-cast onto in-paper substrates as demonstrated in Fig. 3a. On filter paper, in the absence of the active SERS substrate, there was no peak of TCZ detected (Fig. S2†). Fig. 3c compares the SERS spectra of TCZ (10−5 M) on different e-AuNP-based in-paper substrates without and with GO at different GO contents (i.e., 1, 2, 3, 4 and 5% wt). Without GO, the SERS intensity of TCZ on the paper/e-Au substrate was low with characteristic peaks at 430 cm−1, 592 cm−1, 985 cm−1, 1312 cm−1 and 1372 cm−1. However, the intensities of the peaks at 592 cm−1 and 985 cm−1 are relatively low, so they could not be clearly detected as Raman peaks (Fig. S2†). In the presence of GO in the in-paper substrate, we could observe those characteristic bands of TCZ with obvious improvement in intensity (Fig. 3b and S2†). The band at 430 cm−1 represents C–N–C deformation vibration. The band at 592 cm−1 is assigned to C–S–C deformation vibration. The band at 985 cm−1 is associated with C–C symmetric stretching vibration. The bands at 1312 cm−1 and 1372 cm−1 represent C–N stretching vibration.8,57,58 In a recent study concerning the use of AgNPs to detect TCZ, Su et al. proposed that Ag–S bonds would have formed, allowing TCZ to replace citrate and fluorescein on the surface of AgNPs. In this case, Au–S bond formation would have also occurred. The sulfur atom in the TCZ molecule, which already shares two covalent bonds with two carbon atoms, could have been linked to Au via a dative bond with two shared electrons (Fig. S4†).59,60 Therefore, the band of the C–S–C bond remained even when TCZ was linked to AuNPs. Besides, the formation of the dative bond also reduced the distance between TCZ and AuNPs, allowing TCZ to better experience the SERS effect via EM enhancement. In addition, this chemical linkage was convenient for charge transfer.With the addition of GO, SERS enhancement of TCZ has been improved. It is clear that at low content of GO (1–4%), the SERS intensity of TCZ increases with the increase of GO content. Higher content of GO could have improved the adsorption of TCZ onto paper/GO/e-Au. In addition, the increase in GO content might have provided more carboxyl groups for the attachment of AuNPs, leading to the enrichment of AuNPs on the cellulose fibers. However, when GO content reaches 5%, the TCZ intensity decreases compared to the use of paper/4% GO/e-Au. It is worth mentioning that the decrease is the most significant at the band of 592 cm−1 (63%) while the decreases in the levels of TCZ intensity are 45%, 50%, 47%, and 45% at 430 cm−1, 985 cm−1, 1312 cm−1 and 1372 cm−1, respectively. Fig. S3† shows the intensity of the band at 592 cm−1 of TCZ (10−5 M) on paper/GO/e-Au with GO content ranging from 0% to 5%. A similar trend could be observed with the concentrations of 10−4 M and 10−6 M TCZ (Fig. 3b and d). As the band at 592 cm−1 was assigned to C–S–C deformation vibration, which is closest to the Au surface due to Au–S bonding, the decrease at this band might be related to the decrease of Au surface area on the substrate. Hence, a high GO content of 5% could have allowed GO nanosheets to envelop AuNPs, preventing TCZ to bind directly to AuNPs via Au–S linkage. Moreover, it was reported that the encapsulation of AuNPs by GO could reduce the SERS effect of the plasmonic material because it could cover AuNPs and block the nanogaps between them, preventing analytes from experiencing EM enhancements.21,61 Hence, with a GO content of 5%, we also observed the decrease in SERS intensity at other characteristic bands of TCZ due to partial encapsulation of AuNPs by GO. To overcome this, Xu et al. had to perform thermal annealing at 400 °C for 2 h on their graphene-encapsulated SERS substrate to activate its SERS effect.61 Nevertheless, in the case of depositing GO/e-Au composites on paper substrates, that thermal treatment was not a suitable approach for our system. Therefore, we optimized the performance of the paper/GO/e-Au substrate by selecting the most appropriate content of GO sheets to prevent them from covering the e-AuNPs. Therefore, the GO content of 4% was selected to further develop TCZ sensors.
The presence of GO nanosheets not only enriched e-AuNPs on the in-paper SERS sensors, but also improved the adsorption of analytes onto the substrate. Kim et al. stated that molecules containing a large number of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds were more adhesive to the GO surface.62 Meanwhile, TCZ contains an aromatic ring with 3 C
C bonds were more adhesive to the GO surface.62 Meanwhile, TCZ contains an aromatic ring with 3 C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds. It allowed TCZ to be adsorbed onto the GO surface. As a result, in addition to the TCZ molecules that bind directly to the Au surface, in the presence of the GO scaffold, more TCZ molecules would locate near AuNPs to experience the resonance effect around the SERS active materials. This might also explain the appearance of the band at 985 cm−1, which represents the C–C symmetric stretching vibration, in the SERS spectra of TCZ on all GO-containing in-paper substrates.
C bonds. It allowed TCZ to be adsorbed onto the GO surface. As a result, in addition to the TCZ molecules that bind directly to the Au surface, in the presence of the GO scaffold, more TCZ molecules would locate near AuNPs to experience the resonance effect around the SERS active materials. This might also explain the appearance of the band at 985 cm−1, which represents the C–C symmetric stretching vibration, in the SERS spectra of TCZ on all GO-containing in-paper substrates.
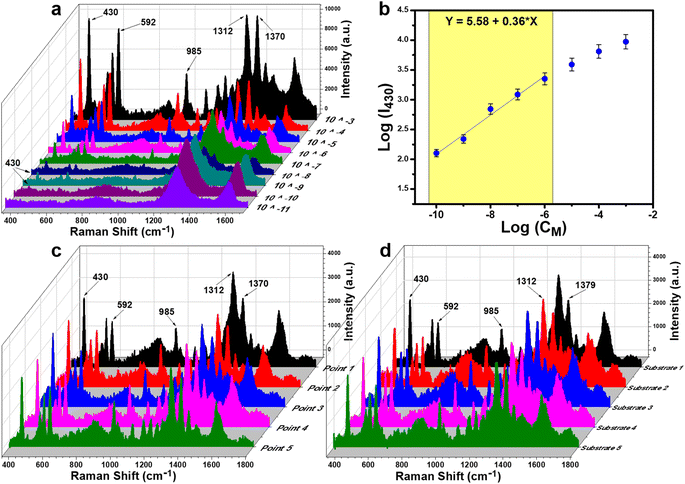
Paper/4% GO/e-Au was selected to be the substrate for the TCZ sensors. Nine TCZ solutions at different concentrations (10−3 M to 10−11 M) were prepared in distilled water. Subsequently, these samples were drop-cast on the prepared substrate. Fig. 4a demonstrates the SERS spectra of TCZ at these concentrations on the paper/4% GO/e-Au substrate. It is obvious that the SERS intensity of TCZ increases with the increase of its concentration. Down to the concentration of 10−10 M, the characteristic bands of TCZ are still able to be detected, and then disappear at the concentration of 10−11 M. The plot of logarithmic SERS intensity at 430 cm−1 against the logarithmic concentration of TCZ shows a good linear relationship in the region from 10−6 M to 10−10 M with a linear regression of R2 = 0.98 (Fig. 4b). For each point of the calibration curve, SERS signals were recorded three times. Based on the linear equation in Fig. 4b, the LOD was calculated to be 1.32 × 10−10 M, which is lower than that of many established SERS-based sensors for TCZ detection using noble metal NPs as shown in Table 1. Moreover, thanks to the assistance of GO nanosheets and cellulose fibers, e-AuNPs were spread quite evenly on the SERS paper. Uniformity of the SERS sensors was evaluated by measuring five random points on one substrate (Fig. 4c), revealing an RSD of 9.5%. Besides, five different paper/4% GO/e-Au substrates were prepared independently using the method as described in Section 2.4. The SERS signals of TCZ (10−5 M) on these substrates were recorded, resulting in five spectra as shown in Fig. 4d. The RSD value for the reproducibility of the method was calculated to be 12.1%. LOD and RSD values were calculated as described in the ESI.†
| Material | Measuring support | LOD | Linear range | Real sample | Sample collection method | Ref. |
|---|---|---|---|---|---|---|
| Au@AgNPs | Capillary tube | 2.75 × 10−7 M | 2.75 × 10−5 to 2.75 × 10−7 M | Pear | Homogenizing and centrifuging samples for extraction; dipping SERS substrates into sample solutions | 8 |
| rGO–Ag nanocomposite | Glass slide | 10−6 M | 10−3 to 10−6 M | — | — | 63 |
| AgNPs | Si substrate | 5.28 × 10−9 M | 5.28 × 10−4 to 5.28 × 10−9 M | Lettuce | Homogenizing and centrifuging samples for extraction; drop-casting the sample solution onto the SERS substrate | 64 |
| Multi-stacked Au–Ag bimetallic nanowires | Microwell (active substrates fixed within the wells) | 5.5 × 10−8 M | 5.5 × 10−4 to 5.5 × 10−8 M | Whole milk | Directly drop-casting milk samples onto SERS substrates/dipping SERS substrates into milk samples | 65 |
| GO/e-Au | Filter paper | 1.32 × 10−10 M | 10−6 to 10−11 M | Cucumber | “Paste and peel off” for cucumber skin | This work |
| Orange juice | “Dip and dry” for orange juice |
3.3. Practicability of paper/GO/e-Au sensors to detect TCZ in real samples
As discussed in our previous study,34 in-paper SERS sensors were convenient to detect analytes using two sample collection methods: “paste and peel off” and “dip and dry”. Using both methods, the practicability of paper/GO/e-Au sensors to detect TCZ was evaluated on cucumber skin and orange juice, respectively.The “paste and peel off” method was employed to collect TCZ on cucumber peel as described in Section 2.5 and Fig. 5a. To disassociate TCZ from the peel, alcohol solution was sprayed on the cucumber skin. This simple extraction allowed TCZ to be exposed on the cucumber peel. Moreover, water in the alcohol solution slowed the evaporation of the solution, and therefore, the in-paper sensors could adhere on it to adsorb the analyte extracted from the cucumber skin. Fig. 5b shows the SERS spectra of TCZ at different concentrations (10−6 M to 10−10 M) on cucumber peel collected on the SERS sensors. The characteristic peaks of TCZ can be detected in the spectra at concentrations down to 10−9 M. The recovery rates range from 78 to 93% (Table 2). A part of spiked TCZ might have been absorbed into deeper layers of the peel matrix, which could not be extracted by alcohol solution, and therefore, the recovery rates were lower than 100%. Moreover, the paper/GO/Ag SERS sensors allowed the detection of TCZ at concentrations as low as 10−9 M (∼2 ppb), which is much lower than the maximal residue limit (MRL) of 2 ppm in vegetables in China and the MRL of 0.01 ppm in vegetables in the European Union (EU) and Japan. In addition, with the use of this in-paper SERS sensor, we could avoid a step of sample preparation including homogenizing and multiple rounds of centrifuging, which was usually performed for sample collection in fruit and vegetable samples (Table 1).
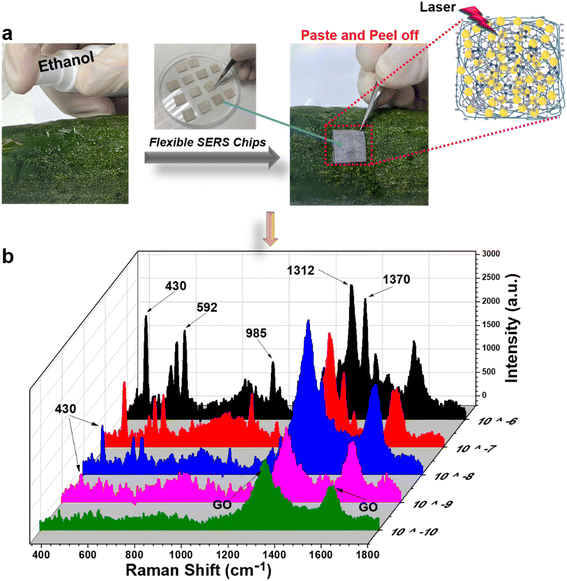 | ||
| Fig. 5 (a) “Paste and peel off” method to collect TCZ on the surface of a cucumber; (b) SERS spectra of thiram (10−10 M to 10−4 M) on the surface of a cucumber using the paper/GO/e-Au SERS substrate. | ||
| Real sample | Spiked concentration (M) | Detected concentration (M) | Recovery (%) |
|---|---|---|---|
| Cucumber skin | 10−6 | 9.32 × 10−7 | 93 |
| 10−7 | 8.21 × 10−8 | 82 | |
| 10−8 | 8.54 × 10−9 | 85 | |
| 10−9 | 7.82 × 10−10 | 78 | |
| Orange juice | 10−6 | 8.72 × 10−7 | 87 |
| 10−7 | 9.11 × 10−8 | 91 | |
| 10−8 | 7.58 × 10−9 | 76 | |
| 10−9 | 8.59 × 10−10 | 86 |
The “dip and dry” method was utilized to collect TCZ spiked in orange juice at different concentrations from 10−6 M to 10−10 M as described in Section 2.5 and Fig. 6a. Fig. 6b shows the SERS spectra of TCZ at these concentrations on paper/GO/e-Au SERS sensors. Similar to the sample on cucumber skin, characteristic peaks can be detected in the spectra of TCZ at concentrations down to 10−9 M. The recovery rates range from 76 to 91% (Table 2). Once again, the recovery rates are all lower than 100%. It is worth mentioning that for real samples, both cucumber skin and orange juice, sample collection methods were different from that used to collect TCZ in standard samples, for which, we drop-cast TCZ solution directly onto the flexible SERS substrate. As a result, the in-paper substrate could adsorb more TCZ molecules, leading to a higher SERS signal. Therefore, the recovery rates of the real sample are all lower than 100%. However, in this study, we aim to develop a flexible SERS substrate that can collect TCZ directly from the real samples without complicated pre-preparation, and the concentration of TCZ collected from different samples with different natures using different sample collection methods can be calculated using the same calibration curve with acceptable recovery rates. Paper/GO/e-Au SERS sensors were still able to detect TCZ at concentrations as low as 10−9 M in orange juice and on cucumber skin using a fast and simple procedure; therefore, the SERS papers are promising to be employed in other real applications.
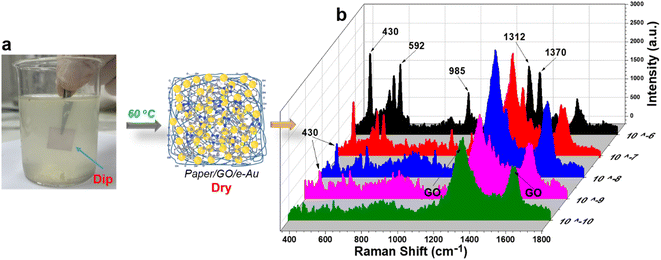 | ||
| Fig. 6 (a) “Dip and dry” method to collect TCZ in orange juice; (b) SERS spectra of TCZ (10−10 M to 10−4 M) in orange juice using paper/GO/e-Au SERS substrates. | ||
3.4. Machine learning-assisted data analysis
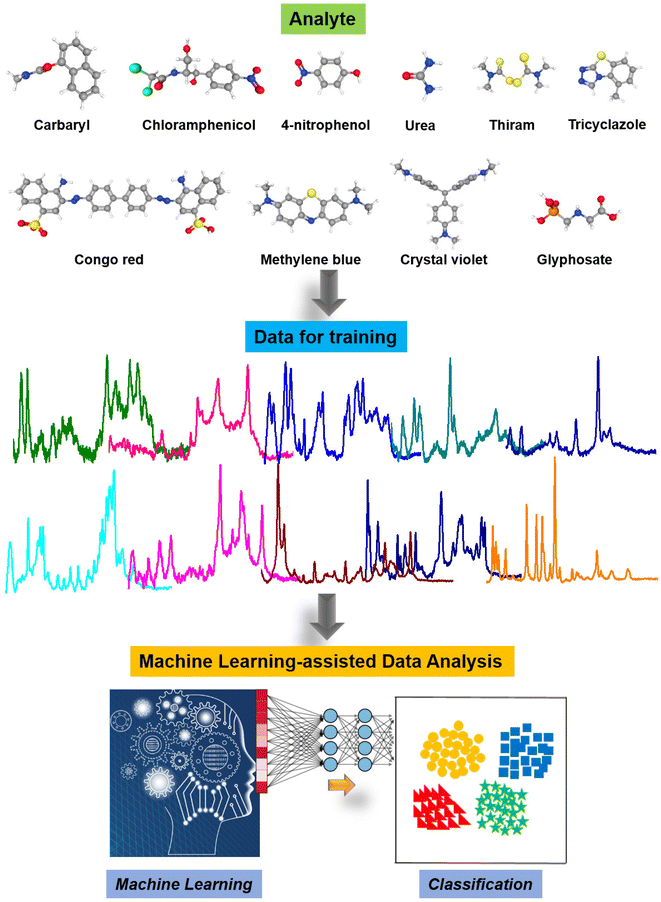
Fig. 7 illustrates the machine learning-assisted system we designed for discriminating TCZ from nine other organic compounds, including carbaryl, chloramphenicol, 4-nitrophenol, urea, thiram, Congo red, methylene blue, crystal violet and glyphosate. In addition to those of TCZ, the SERS spectra of these compounds were recorded and employed as training data, enriching the neural network. For each compound, we selected several characteristic peaks as shown in Table S1.† Training and testing were performed as described in Section 2.6. The mean normalized number of spectra sorted in the validation set is presented as a confusion matrix (Fig. 8A), showing excellent capability for discriminating TCZ from other compounds with an accuracy of 100%. The network could predict the presence of TCZ not only in standard samples, but also in real samples at different concentrations. Fig. 8B demonstrates the prediction of the network using the SERS spectra of TCZ collected on cucumber skin as described in Section 3.4 at the concentration range of 10−6 M to 10−10 M. From 10−6 M to 10−9 M, the network could predict the presence of TCZ with accuracies higher than 50%. Nevertheless, at a concentration of 10−10 M, the prediction was incorrect. This result was in agreement with the SERS spectrum of TCZ (10−10 M) in Fig. 5b, in which we hardly detected the characteristic bands of TCZ. This preliminary result is encouraging as the system could predict the presence of TCZ on real samples at concentrations down to 10−9 M (∼2 ppb), which is lower than the MRL of TCZ established in various countries and regions around the world.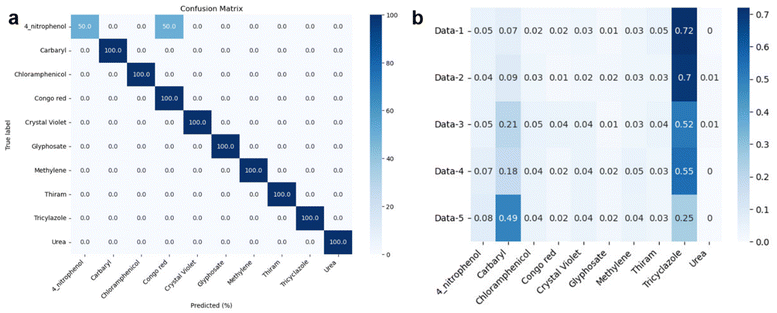 | ||
| Fig. 8 (a) Confusion matrix and (b) prediction of TCZ on cucumber skin (data-1: TCZ (10−6 M); data-2: TCZ (10−7 M); data-3: TCZ (10−8 M); data-4: TCZ (10−9 M); data-5: TCZ (10−10 M)). | ||
4. Conclusions
In this study, we designed and developed a flexible sensing platform based on paper/GO/e-Au SERS sensors and an artificial neural network to detect and identify TCZ in real samples. With the most optimal content of 4%, GO not only played an important role in material fabrication as a platform for the anchoring of e-AuNPs, but also promoted the adsorption of the analyte, resulting in an impressive LOD of 1.32 × 10−10 M in standard samples. Moreover, the flexible substrate could detect TCZ in real samples of cucumber skin and orange juice at concentrations down to 10−9 M (∼2 ppb). Furthermore, we developed a machine learning model, which could discriminate TCZ from nine other organic compounds. Also, it could predict the presence of TCZ on cucumber skin at concentrations as low as 10−9 M. The study confirms the potential of integrating machine learning and flexible substrates for fast determination of chemicals in real samples.Author contributions
H. A. Nguyen: conceptualization, methodology, formal analysis, writing – original draft; Q. D. Mai: conceptualization, validation, investigation, writing – review & editing; D. T. N. Nga: conceptualization, validation, investigation; M. K. Pham: validation, investigation; Q. K. Nguyen: methodology, validation, AI-data analysis; T. H. Do: methodology, validation, AI-data analysis; V. T. Luong: methodology, validation, AI-data analysis, writing – review & editing; V. D. Lam: methodology, validation, formal analysis; A. T. Le: conceptualization, methodology, supervision, project administration, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This research was financially supported by Phenikaa University & A&A Green Phoenix Group JSC for the Key Research Group (NEB Lab). The authors would like to acknowledge the support for Raman & microscopy measurements from NEB Lab (Phenikaa University) and SEM characterization from GUST-VAST.References
- D. R. Walt, I. Biran and T. K. Mandal, in Encyclopedia of Physical Science and Technology, ed. R. A. Meyers, Academic Press, New York, 3rd edn, 2003, pp. 803–829, DOI:10.1016/B0-12-227410-5/00954-6.
- X. X. Han, R. S. Rodriguez, C. L. Haynes, Y. Ozaki and B. Zhao, Nat. Rev. Methods Primers, 2022, 1, 87 CrossRef.
- K. Chen, Y. H. Ong, C. Yuen and Q. Liu, in Imaging in Dermatology, ed. M. R. Hamblin, P. Avci and G. K. Gupta, Academic Press, Boston, 2016, pp. 141–154, DOI:10.1016/B978-0-12-802838-4.00013-3.
- J. F. Li, Y. F. Huang, Y. Ding, Z. L. Yang, S. B. Li, X. S. Zhou, F. R. Fan, W. Zhang, Z. Y. Zhou, D. Y. Wu, B. Ren, Z. L. Wang and Z. Q. Tian, Nature, 2010, 464, 392–395 CrossRef CAS PubMed.
- R. A. Halvorson and P. J. Vikesland, Environ. Sci. Technol., 2010, 44, 7749–7755 CrossRef CAS PubMed.
- W. Liao, Q. Lin, S. Xie, Y. He, Y. Tian and Y. Duan, Anal. Chim. Acta, 2018, 1043, 64–71 CrossRef CAS PubMed.
- L. Wu, W. Zhang, C. Liu, M. F. Foda and Y. Zhu, Food Chem., 2020, 328, 127106 CrossRef CAS PubMed.
- N. Hussain, H. Pu and D.-W. Sun, Food Chem., 2021, 350, 129025 CrossRef CAS PubMed.
- Y. Peng, C. Lin, Y. Li, Y. Gao, J. Wang, J. He, Z. Huang, J. Liu, X. Luo and Y. Yang, Matter, 2022, 5, 694–709 CrossRef CAS PubMed.
- R. Haldavnekar, K. Venkatakrishnan and B. Tan, Nat. Commun., 2018, 9, 3065 CrossRef PubMed.
- A. S. D. S. Indrasekara, S. Meyers, S. Shubeita, L. C. Feldman, T. Gustafsson and L. Fabris, Nanoscale, 2014, 6, 8891–8899 RSC.
- X. Qi, X. Wang, Y. Dong, J. Xie, X. Gui, J. Bai, J. Duan, J. Liu and H. Yao, Spectrochim. Acta, Part A, 2022, 272, 120955 CrossRef CAS PubMed.
- Y. Xu, F. Y. H. Kutsanedzie, M. M. Hassan, J. Zhu, H. Li and Q. Chen, Sens. Actuators, B, 2020, 324, 128718 CrossRef CAS.
- H.-C. Hu, S.-H. Wu, L.-X. Jin and J.-J. Sun, Biosens. Bioelectron., 2022, 210, 114283 CrossRef CAS PubMed.
- J. M. McLellan, Z.-Y. Li, A. R. Siekkinen and Y. Xia, Nano Lett., 2007, 7, 1013–1017 CrossRef CAS PubMed.
- Y. Shan, Y. Yang, Y. Cao, C. Fu and Z. Huang, Nanotechnology, 2016, 27, 145502 CrossRef PubMed.
- T. Xu, J. Fu, X. Wang, G. Lu and B. Liu, Front. Chem., 2022, 10, 887900 CrossRef CAS PubMed.
- S. Cong, X. Liu, Y. Jiang, W. Zhang and Z. Zhao, Innovation, 2020, 1, 100051 CAS.
- J. Zhou, J. Zhang, H. Yang, Z. Wang, J.-a. Shi, W. Zhou, N. Jiang, G. Xian, Q. Qi, Y. Weng, C. Shen, Z. Cheng and S. He, Nanoscale, 2019, 11, 11782–11788 RSC.
- L.-J. Gu, C.-L. Ma, X.-H. Zhang, W. Zhang, S. Cong and Z.-G. Zhao, Chem. Commun., 2018, 54, 6332–6335 RSC.
- N. Zhang, L. Tong and J. Zhang, Chem. Mater., 2016, 28, 6426–6435 CrossRef CAS.
- Y. Long, W. Wang, W. Xiong and H. Li, J. Alloys Compd., 2022, 901, 163660 CrossRef CAS.
- C. Zhu, Q. Zhao, X. Wang, Z. Li and X. Hu, Microchem. J., 2021, 165, 106090 CrossRef CAS.
- B. Fortuni, Y. Fujita, M. Ricci, T. Inose, R. Aubert, G. Lu, J. A. Hutchison, J. Hofkens, L. Latterini and H. Uji-i, Chem. Commun., 2017, 53, 5121–5124 RSC.
- S. Kumar, P. Goel and J. P. Singh, Sens. Actuators, B, 2017, 241, 577–583 CrossRef CAS.
- J. Jiang, S. Zou, Y. Li, F. Zhao, J. Chen, S. Wang, H. Wu, J. Xu, M. Chu, J. Liao and Z. Zhang, Microchim. Acta, 2019, 186, 603 CrossRef PubMed.
- J. Chen, Y. Huang, P. Kannan, L. Zhang, Z. Lin, J. Zhang, T. Chen and L. Guo, Anal. Chem., 2016, 88, 2149–2155 CrossRef CAS PubMed.
- J. Jiang, S. Zou, L. Ma, S. Wang, J. Liao and Z. Zhang, ACS Appl. Mater. Interfaces, 2018, 10, 9129–9135 CrossRef CAS PubMed.
- J. Sitjar, J.-D. Liao, H. Lee, L. P. Pan, B. H. Liu, W.-e. Fu and G. D. Chen, Nanomaterials, 2019, 9, 1750 CrossRef CAS PubMed.
- Y. Hong, Y. Li, L. Huang, W. He, S. Wang, C. Wang, G. Zhou, Y. Chen, X. Zhou, Y. Huang, W. Huang, T. Gong and Z. Zhou, J. Biophotonics, 2020, 13, e201960176 CrossRef CAS PubMed.
- C. Wang, B. Liu and X. Dou, Sens. Actuators, B, 2016, 231, 357–364 CrossRef CAS.
- H. S. Siebe, Q. Chen, X. Li, Y. Xu, W. R. Browne and S. E. J. Bell, Analyst, 2021, 146, 1281–1288 RSC.
- P. M. Fierro-Mercado and S. P. Hernández-Rivera, Int. J. Spectrosc., 2012, 2012, 716527 Search PubMed.
- Q. D. Mai, H. A. Nguyen, N. X. Dinh, N. T. Thu Thuy, Q. H. Tran, P. C. Thanh, A.-T. Pham and A.-T. Le, Talanta, 2023, 253, 124114 CrossRef CAS.
- M. J. Oliveira, P. Quaresma, M. Peixoto de Almeida, A. Araújo, E. Pereira, E. Fortunato, R. Martins, R. Franco and H. Águas, Sci. Rep., 2017, 7, 2480 CrossRef PubMed.
- W. Jang, H. Byun and J.-H. Kim, Mater. Chem. Phys., 2020, 240, 122124 CrossRef CAS.
- S. S. B. Moram, C. Byram, S. N. Shibu, B. M. Chilukamarri and V. R. Soma, ACS Omega, 2018, 3, 8190–8201 CrossRef CAS PubMed.
- S. S. B. Moram, C. Byram and V. R. Soma, Bull. Mater. Sci., 2020, 43, 53 CrossRef CAS.
- N. Kim, M. R. Thomas, M. S. Bergholt, I. J. Pence, H. Seong, P. Charchar, N. Todorova, A. Nagelkerke, A. Belessiotis-Richards, D. J. Payne, A. Gelmi, I. Yarovsky and M. M. Stevens, Nat. Commun., 2020, 11, 207 CrossRef CAS PubMed.
- H. Zhou, L. Xu, Z. Ren, J. Zhu and C. Lee, Nanoscale Adv., 2023, 5, 538–570 RSC.
- F. U. Ciloglu, M. Hora, A. Gundogdu, M. Kahraman, M. Tokmakci and O. Aydin, Anal. Chim. Acta, 2022, 1221, 340094 CrossRef CAS PubMed.
- J. Q. Li, P. V. Dukes, W. Lee, M. Sarkis and T. Vo-Dinh, J. Raman Spectrosc., 2022, 53, 2044–2057 CrossRef CAS PubMed.
- Y. X. Leong, Y. H. Lee, C. S. L. Koh, G. C. Phan-Quang, X. Han, I. Y. Phang and X. Y. Ling, Nano Lett., 2021, 21, 2642–2649 CrossRef CAS PubMed.
- M. Q. Doan, N. H. Anh, N. X. Quang, N. X. Dinh, D. Q. Tri, T. Q. Huy and A.-T. Le, J. Electron. Mater., 2022, 51, 150–162 CrossRef CAS.
- P. N. Minh, V.-T. Hoang, N. X. Dinh, O. Van Hoang, N. Van Cuong, D. Thi Bich Hop, T. Q. Tuan, N. T. Khi, T. Q. Huy and A.-T. Le, New J. Chem., 2020, 44, 7611–7620 RSC.
- S. Ben-David and S. Shalev-Shwartz, Understanding Machine Learning: from Theory to Algorithms, Cambridge University Press, Cambridge, 2014 Search PubMed.
- H.-J. Zhai, C. Bürgel, V. Bonacic-Koutecky and L.-S. Wang, J. Am. Chem. Soc., 2008, 130, 9156–9167 CrossRef CAS PubMed.
- K. A. Moltved and K. P. Kepp, J. Phys. Chem. C, 2019, 123, 18432–18444 CrossRef CAS.
- K. Sun, M. Kohyama, S. Tanaka and S. Takeda, J. Energy Chem., 2015, 24, 485–489 CrossRef.
- M. Q. Doan, N. H. Anh, H. Van Tuan, N. C. Tu, N. H. Lam, N. T. Khi, V. N. Phan, P. D. Thang and A.-T. Le, Adsorpt. Sci. Technol., 2021, 2021, 1169599 CrossRef.
- I. Tiginyanu, V. Ursaki and V. Popa, in Nanocoatings and Ultra-Thin Films, ed. A. S. H. Makhlouf and I. Tiginyanu, Woodhead Publishing, 2011, pp. 330–354, DOI:10.1533/9780857094902.2.330.
- S. Azizighannad and S. Mitra, Sci. Rep., 2018, 8, 10083 CrossRef PubMed.
- A. Abdelkhalek, M. A. El-Latif, H. Ibrahim, H. Hamad and M. Showman, Sci. Rep., 2022, 12, 7060 CrossRef CAS PubMed.
- F. Wang and J. Liu, Nanoscale, 2015, 7, 919–923 RSC.
- J. H. Wiley and R. H. Atalla, Carbohydr. Res., 1987, 160, 113–129 CrossRef CAS.
- A. A. Bessonov, T. V. Basova, V. G. Kiselev, L. A. Sheludyakova, N. B. Morozova and I. K. Igumenov, Vib. Spectrosc., 2009, 51, 283–288 CrossRef CAS.
- H. Tang, D. Fang, Q. Li, P. Cao, J. Geng, T. Sui, X. Wang, J. Iqbal and Y. Du, J. Food Sci., 2012, 77, T105–T109 CAS.
- Q.-Q. Li, Y.-P. Du, Y. Xu, X. Wang, S.-Q. Ma, J.-P. Geng, P. Cao and T. Sui, Chin. Chem. Lett., 2013, 24, 332–334 CrossRef CAS.
- H. Häkkinen, Nat. Chem., 2012, 4, 443–455 CrossRef PubMed.
- D. Santamaría-Pérez, D. Daisenberger, J. Ruiz-Fuertes, T. Marqueño, R. Chulia-Jordan, C. Muehle, M. Jansen, P. Rodriguez-Hernandez, A. Muñoz, E. R. Johnson and A. Otero-de-la-Roza, Chem. Sci., 2019, 10, 6467–6475 RSC.
- W. Xu, J. Xiao, Y. Chen, Y. Chen, X. Ling and J. Zhang, Adv. Mater., 2013, 25, 928–933 CrossRef CAS PubMed.
- T.-H. Kim, K.-B. Lee and J.-W. Choi, Biomaterials, 2013, 34, 8660–8670 CrossRef CAS PubMed.
- V. Van Cat, N. X. Dinh, L. T. Tam, N. V. Quy, V. N. Phan and A.-T. Le, Mater. Today Commun., 2019, 21, 100639 CrossRef CAS.
- W. Chen, F. Long, G. Song, J. Chen, S. Peng and P. Li, J. Raman Spectrosc., 2020, 51, 611–618 CrossRef CAS.
- S. Kim, W. Choi, D. J. Kim, H. S. Jung, D.-H. Kim, S.-H. Kim and S.-G. Park, Nanoscale, 2020, 12, 12942–12949 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3na01113e |
| ‡ H. A. Nguyen and Q. D. Mai contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |