Open Access Article
Open Access ArticleSynergistic impact of nanoplastics and nanopesticides on Artemia salina and toxicity analysis†
Mahalakshmi
Kamalakannan
,
Durgalakshmi
Rajendran
,
John
Thomas
and
Natarajan
Chandrasekaran
 *
*
Centre for Nanobiotechnology, Vellore Institute of Technology, Vellore-632014, Tamil Nadu, India. E-mail: nchandrasekaran@vit.ac.in; nchandra40@hotmail.com; Fax: +91 416 2243092; Tel: +91 416 2202624
First published on 7th May 2024
Abstract
Polystyrene nanoplastics (PSNPs) when exposed to nanopermethrin (NPER) exacerbate toxicity on Artemia salina. In the environment, NPs act as a vector for other pollutants mainly heavy metals and pesticides. Nanopesticides are efficient compared to their bulk form. The adsorption of NPER on PSNPs was studied systematically and it was found that the binding of NPER is inversely proportional to its concentration. NPER adsorption on PSNPs followed pseudo-first-order kinetics with an adsorption percentage of 1.7%, 3.7%, 7.7%, 15.4%, and 30.8% when PSNPs were incubated with 2 mg L−1,4 mg L−1, 8 mg L−1, 16 mg L−1, and 32 mg L−1 of NPER. The adsorption followed the Langmuir isotherm. The increased hydrodynamic size of the NPER/PSNP complex was observed. Different characterization studies were performed for NPER, PSNPs, and their complex using Fourier transform infrared spectroscopy, field emission scanning electron microscopy, X-ray diffraction, and gas chromatography-mass spectrometry. The LC50 value for the NPER/PSNP complex treated with Artemia salina was 3.127 mg L−1, compared to LC50 NPER which was found to be 4.536 mg L−1. PSNPs had a lower mortality rate in Artemia salina, where 50% mortality (LC50) was not observed at their working concentration. Both the nanoforms led to morphological changes in Artemia salina. Reactive oxygen species increased to 87.94% for the NPER/PSNP complex, 78.93% for NPER, and 23.65% for PSNPs. Greater amounts of ROS in the cells may have led to SOD degradation. Superoxide dismutase activity for the NPER/PSNP complex was 1.2 U mg−1, NPER was 1.3 U mg−1, and PSNPs was 2.1 U mg−1. A lipid peroxidation study reveals that the melondialdehyde synthesis by NPER/PSNPs complex, NPER and PSNPs were found to be 2.21 nM mg−1, 1.59 nM mg−1, and 0.91 nM mg−1 respectively. Catalase activity in a complex of NPER/PSNPs, NPER, and PSNPs was found to be 1.25 U mg−1, 0.94 U mg−1, and 0.49 U mg−1. This study envisages the individual and combined toxicity of nanopesticides and PSNPs on aquatic organisms. Increased plastic usage and new-age chemicals for agriculture could result in the formation of a PSNPs–NPER complex potentially causing highly toxic effects on aquatic animals, compared to their pristine forms. Therefore, we should also consider the other side of nanotechnology in agriculture.
1. Introduction
Micronanoplastics (MNPs) are an emergent pollutant which cause environmental hazards to terrestrial and aquatic ecosystems.1 They enter the environment in two ways. One way is direct entry from consumer products like face cleansers, toothpaste, face creams, etc.,2 The wastewater from household drains and industries ends up in sewage treatment plants, where there is no proper treatment for MNPs, releasing them into the environment.3 Another way is that plastics continuously deteriorate into MNPs, enabling their penetration into all environmental niches.4 Trillions of floating MNPs exist in marine ecosystems.5 Furthermore, MNPs act as a vector to carry other pollutants (organic and inorganic chemicals) to non-source pollution points.6 MNPs along with other co-pollutants exhibit toxicity to aquatic organisms.7One of the co-pollutants is agricultural-based chemicals. Approximately, 2.5 million tonnes of bulk pesticides are used on agricultural lands annually.8 In addition, nanopesticides are synthesized with highly reactive surfaces for target specificity and high potency to control agricultural pests. Nanoforms of agricultural chemicals, like nanourea, nanodeltamethrin, nanopermethrin, micronutrient based nanopesticides, and many others are already in the market.9 Among the volume of these chemicals applied on land, only a small portion of them is effective, and the remaining wash away as residue, generating residual pollution. They cause toxicity to many aquatic invertebrates and fish since they are non-biodegradable.10 Most of the synthetic pyrethroids are neurotoxic and interfere with the function of nerve cells by interacting with ion channels and sodium channels, leading to paralysis.11 MNPs present in sewage sludge12 interact with these chemicals, when sludges are used as fertilizers. During surface runoff, these MNPs may transport the nanopesticides to nonpoint sources such a marine ecosystem, fresh water ecosystem and so many others.13 As a result, their combined toxicity on these ecosystems might be tremendous.
This study was undertaken to investigate such combined toxicity in a marine environment. Here, polystyrene nanoplastics and nanopermethrin were taken as representatives for MNPs and nanopesticides respectively. An aquatic invertebrate – Artemia salina was chosen as the model organism to study aquatic toxicity. These are zooplanktons, a nonselective filter feeder, which feed on debris in marine water bodies. These species are exposed to significant concentrations of plastics and co-contaminants in the natural environment viz., seawater. Zooplanktons which are feeding on these debris are an important source of nutrition for many secondary producers, including fish and crustaceans, and thus MNPs make their way into the food chain.14Artemia salina is used in aquaculture industries as a live feed by fish breeders,15 besides as a model organism to study toxicity of emergent pollutants.16 The genus Artemia have both reproducing and parthenogenic species. They are widely used in laboratory acute and chronic toxicity studies, because they are easy to culture owing to their small size and short life span of about 3–4 months.17,18
Ingestion of polystyrene nanoplastics causes several adverse effects on zooplanktons with increased oxidative stress.19 However, the combined effect of PSNPs and nanopermethrin (NPER) on the aquatic organisms is not yet investigated. This study offers the first evidence of the combined impact of nanoplastics and nanopesticides on a marine organism. The end points of the studies include effects of PSNPs, NPER and the PSNPs/NPER complex on Artemia salina's hatching rate, swimming behaviour, life cycle changes, and morphological changes, and biochemical analysis.
2. Materials and methods
Monodisperse polystyrene nanoplastics of 100 nm size (catalog no. 00876-15) were purchased from Polyscience Inc., USA; these polystyrene nanoplastics are not secondary as they are commercially purchased and do not contain any additives. Permethrin was purchased from Tagros Chemical India Ltd. Ammonium glycyrrhizinate (AG), sec-butyl alcohol (sec-BuOH), and n-butyl acetate (nBuAc) were procured from Sigma Aldrich India. Dimethyl sulphoxide (DMSO) was obtained from Hi-media laboratories, in India. Soybean lecithin with 92% soybean phosphatidylcholine (SbPC) was procured from Lipoid, Switzerland. Artemia salina cysts were acquired from Ocean Star International, Inc. USA. Natural seawater was collected from the Central Institute of Brackish Aquaculture (CIBA), Chennai, Tamil Nadu, India (13.0207° N, 80.2729° E).2.1 Nanopermethrin formulation and characterization
Formulation and characterization of nanopermethrin (NPER) was performed using our previous experimental method (Anjali et al., 2010).37 An o/w microemulsion containing permethrin was prepared by mixing the necessary amount of the aqueous and volatile organic phases with co-surfactants such as soybean lecithin, 92% soybean phosphatidylcholine and surfactant – ammonium glycyrrhizinate. Solvents from the resulting o/w microemulsion were quickly evaporated to produce nanopermethrin. The resulting sample was lyophilized at 95 °C for 24 hours to produce the water-dispersible powder and stored at 4 °C for further use. The NPER synthesized was characterized as given in Sections 2.5 and 2.6.2.2 Preparation of the sample
PSNPs (100 nm) were dispersed in filtered Milli-Q water and natural seawater at a stock concentration of 1000 mg L−1. Working concentrations of 5, 25, 50, 75, and 100 mg L−1 were set for experimental purposes. Formulated nanopermethrin was dispersed in natural seawater and Milli-Q water at a stock concentration of 100 mg L−1. Working concentrations of 2, 4, 8, 16, and 32 mg L−1 were prepared for experimental purposes. A complex of PSNPs and NPER was prepared to analyze their combinatorial effect. For the complexes, the concentration of polystyrene was kept constant (100 mg L−1) and nanopermethrin concentrations were varied as 2, 4, 8, 16, and 32 mg L−1.2.3 Effect of time on adsorption
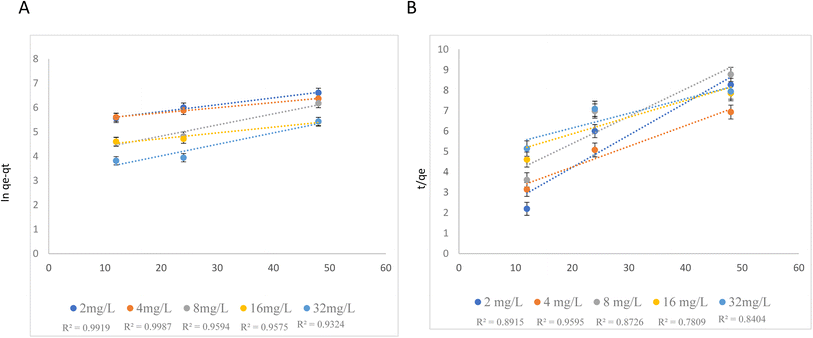
NPER of different concentrations viz., 2, 4, 8, 16, and 32 mg L−1 was mixed with a fixed concentration of PSNPs (100 mg L−1) and kept for interaction. The samples were taken at regular intervals (till the adsorption reaches the saturation point) and filtered through a 0.1 μm syringe filter. The filtrates were measured for unadsorbed NPER using a UV-spectrophotometer (Bio Spectrometer, Eppendorf India) at a wavelength of 230 nm (λmax). The adsorption kinetics of NPER on the PSNPs are predicted using pseudo-first-order (PFO) and pseudo-second-order (PSO) models. While the PSO model assumes that chemical surface interactions would determine how quickly the adsorbate accumulates on the adsorbent surface, the PFO model predicts that the adsorbate will accumulate on the adsorbent surface over time.One of the models that is most frequently used to calculate the adsorption rate constant is the PFO kinetic model, which uses the following equation20
ln(qe − qt) = ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe − K1t qe − K1t | (1) |
The slope and intercept of ln(qe − qt) against t plots were used to calculate the values of K1 and qe.
The PSO kinetic model is another widely used model which asserts that chemisorption is the adsorption mechanism and is represented as follows,21
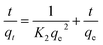 | (2) |
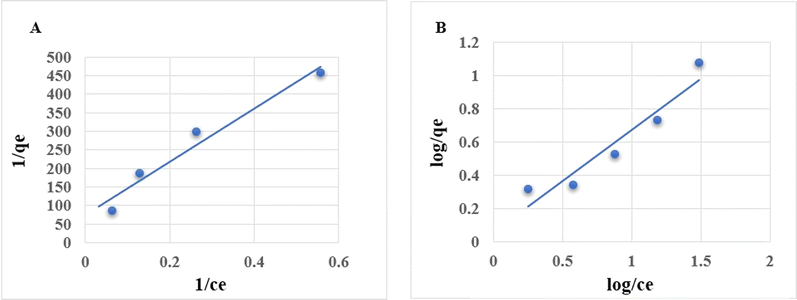
2.4 Adsorption isotherm
For isotherm analysis, initial concentrations of 2, 4, 8, 16, and 32 mg L−1 of NPER were mixed with a fixed concentration of 100 mg L−1 PSNPs and kept for equilibrium adsorption time optimised from the previous experiment, after which the samples were filtered and measured for absorbance.The Langmuir and Freundlich isotherm models were used to simulate the experimental results.
The uniform surface of the adsorbent and the formation of a monolayer during adsorption are presumptions made by the Langmuir isotherm.
The formula for the equation's linear form is:22
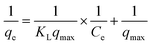 | (3) |
According to the Freundlich isotherm, the adsorbent's surface is heterogeneous, the sorption energy distribution is uniform, and sorption progresses in several layers, since there are an infinite number of centres that may be accessed.
Here is the equation's linear representation:23
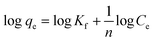 | (4) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) qe against log
qe against log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ce.
Ce.
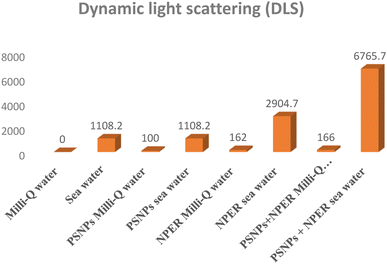
2.5 Characterization of nanoparticles and their complexes
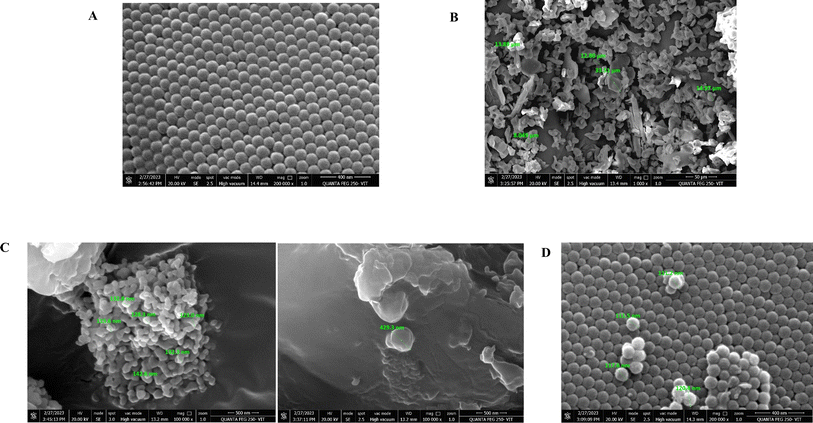
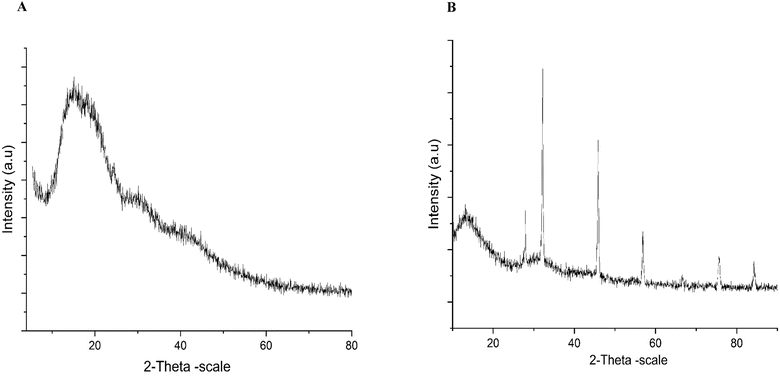
The prepared PSNPs and NPER dispersions were examined for particle size, and their stability was assessed by measuring the zeta potential using a nanoparticle analyzer (SZ100, Horiba Scientific, Japan). The adsorption of NPER on PSNPs was characterized by functional group analysis of polystyrene, permethrin and their complexes by Fourier transform infrared spectroscopy (FTIR-JASCO 6800 FT-IR spectrometer) in the wavenumber range of 400 cm−1 to 4000 cm−1. The morphology of PSNPs, NPER, and their complexes was observed by field emission scanning electron microscopy (FE-SEM) (Thermo Fisher FEI-Quanta 250 FEG, USA). The nanoparticles were subjected to gold sputtering and then the samples were observed under high magnification, and the elements were analyzed by EDX. The crystal structure of NPER and its complex with PSNPs was analyzed by X-ray diffraction analysis (XRD) (Bruker, D-8 Advance P-XRD).2.6 Gas chromatography-mass spectrometry
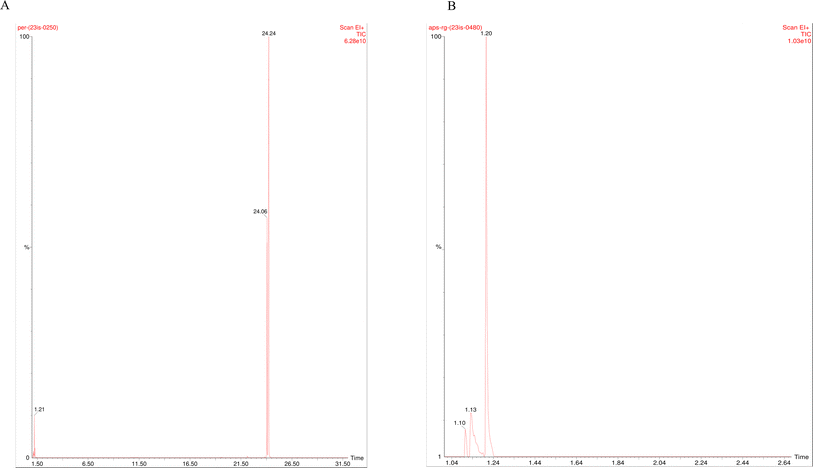
GC-MS was performed for permethrin to study the components present in the sample of NPER. A fused silica column filled with Elite 5MS (5% biphenyl, 95% dimethylpolysiloxane, 30 m, 0.25 mm ID, 250 m df) was utilized in the analysis, and helium carrier gas was used to separate the components at a constant flow rate of 1 mL min−1. During the chromatographic run, the injector temperature was set to 260 °C and the oven temperature for the 1 liter of extract sample was as follows: after reaching 60 °C for 2 minutes, 300 °C was reached at a rate of 10 °C per minute, and 300 °C was maintained for 6 minutes. Conditions for the mass detector were 240 °C transfer line and ion source, an electron impact in the ionization mode at 70 eV, a scan time of 0.2 s, and a scanning interval of 0.1 s. The component spectra were compared to a database of spectra for well-known components kept in the GC-MS NIST (2008) library.2.7 Toxicity assessment of pristine and complex particles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 01×. Whatman 0.45 μm filter paper was used to filter the natural seawater which was sterilized to eliminate any biological contaminants. The culture medium was consistently maintained at 28 °C (room temperature). The nauplii of Artemia salina were used for further experiments.
01×. Whatman 0.45 μm filter paper was used to filter the natural seawater which was sterilized to eliminate any biological contaminants. The culture medium was consistently maintained at 28 °C (room temperature). The nauplii of Artemia salina were used for further experiments.
The hatching percentage was calculated using the following formula25
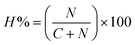 | (5) |
NPER at a sublethal concentration was made to interact with the Artemia salina cysts and was determined at a concentration of 0.1, 0.25, 0.5, and 1 mg L−1.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm and the following bioassays were carried out using the recovered supernatant.
000 rpm and the following bioassays were carried out using the recovered supernatant.
Protein content was estimated by the Bradford method for the treated and control groups.26 Absorbance was measured at 595 nm and bovine serum albumin was used as a standard.
DCFH-DA (dichloro-dihydro-fluorescein diacetate) was used to measure reactive oxygen species (ROS). The measurement of DCFH directly relates to the quantity of intracellular ROS generated. This technique involves mixing 80 μL of supernatant with 20 μL of DCFH-DA solution and then incubating for 30 min at room temperature (28 °C) in the dark. After incubation, ROS generation was measured at 485 nm excitation wavelength and in the emission range of 510–560 nm using a JASCO fluorescence (Japan) spectrophotometer FP8300.27
All aerobic species have the essential enzyme catalase, which hastens the conversion of hydrogen peroxide to water and oxygen. The CAT activity under oxidative stress was measured. 800 μL of the hydrogen peroxide solution were combined with 200 μL samples of supernatants. PBS was used as the control and the absorbance was immediately measured at 240 nm for 3 min27 The data were recorded using a UV-visible spectrophotometer – EVALUATION 220 (Thermo Scientific).
Measurement of SOD activity is crucial for understanding a biological system's antioxidant capabilities. A 24-well plate was used for the experiment, and the following chemicals were added sequentially: 70 μL of the recovered supernatant, 50 mM of Na2CO3 buffer (pH 10), 96 mM of NBT, 0.6% of Triton X-100, and 20 mM of hydroxylamine hydrochloride. The generated reaction mixture was left in the presence of fixed-wavelength visible light for 20 minutes. The absorbance of the reaction mixture at 560 nm was measured using a microplate reader – BIORAD.27
The most widely used technique for studying lipid peroxidation in biological systems is the malondialdehyde (MDA) assay, which measures thiobarbituric acid-reactive compounds (TBARS) by the thiobarbituric acid (TBA) test.28 As a gauge of oxidative stress, TBARS were evaluated to ascertain the lipid peroxidation products. One of the several end products produced by the formation of lipid hydroperoxides is MDA, which is now recognized as a trustworthy indicator of lipid peroxidation.29 100 μl of the supernatant was mixed thoroughly with 400 μl of a 0.25% mixture of TBA/TCA and incubated at 95 °C for 60 min. Following incubation, the tubes were placed in an ice breaker for a short period. The sample tube was then spun at 3000 rpm for 15 minutes to cool it, the supernatant was collected and the absorbance was measured at 532–600 nm.
The nature of the interaction between PSNPs and NPER has been identified. Eqn (6) was used to calculate the mixture's expected toxicity (Cexp) based on the toxicity (%) caused by pristine PSNPs and NPER.
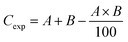 | (6) |
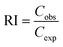 | (7) |
The interaction between the two pollutants is antagonistic when RI < 1, additive when RI = 1, and synergistic when RI > 1. Using the two-way ANOVA approach, a statistical analysis was conducted to compare the measured and shown levels of toxicity.27 After the RI value is established, every combination takes into account the kind of interaction between the pollutants.
3. Results and discussion
3.1 Adsorption kinetics
Kinetic experiments were performed by varying the initial concentrations of NPER from 2 to 32 mg L−1 and the amount of adsorption was measured at regular time intervals of 12, 24, and 48 hours. Pseudo-first-order and pseudo-second-order kinetics were plotted using the Qe (amount of NPER adsorbed on PSNPs (mg g−1)) values obtained from the experiment. Based on the correlation coefficient (R2) from the two plots (Fig. 1), it was found that the PFO model fits better for the adsorption of NPER on PSNPs than the PSO model. The maximum adsorption percentage for varied initial concentrations of 2, 4, 8, 16, and 32 mg L−1 was found to be 1.7%,3.7%,7.7%,15.4%, and 30.8% at 48 h. After 48 hours, aggregation was found in the experiment setup, and hence 48 h was considered as the saturation time. Pseudo-first-order kinetics sufficiently explains the adsorption data, and hence physisorption may be the main adsorption mechanism and the physical process may be the rate-limiting phase of adsorption.30 Adsorption of other co-pollutants on microplastics also involves physisorption31 Increasing concentration can cause variations in the rate constant K1 possibly due to saturation effects. At lower concentrations of NPER, the adsorption rate may be primarily determined by factors like heightened collision frequency. However, at higher concentrations, saturation effects may occur, causing active sites or reactants to become saturated and thereby altering the reaction rate. This can lead to a non-monotonic pattern in the concentration-rate constant relationship.323.2 Adsorption isotherm studies
Isotherm models such as Langmuir and Freundlich are employed to simulate the adsorption process.33 In Fig. 2, adsorption isotherm plots are presented. The results show that the Langmuir isotherm model (R2 = 0.9594), rather than the Freundlich isotherm model (R2 = 0.9101), may be more adequate to describe the behaviour of NPER adsorption on the PSNPs. The Langmuir model states that the sorption process is monolayer, and it occurs on a homogeneous surface. According to the results, the primary events are the creation of a monolayer on the homogenous adsorbent surface. Similar sorption processes were observed in the previous studies.34 Heavy metals, for example, lead, adsorb onto microplastics by monolayer adsorption.35 The adsorption process of pollutants onto other polymers such as polyvinyl chloride (PVC) microplastics followed the monolayer adsorption processes, based on the Langmuir model.363.3 Characterization of nanoparticles
| Sample | Zeta (mV) |
|---|---|
| Milli-Q water | 0.0 mV |
| Sea water | 0.6 mV |
| NPER + Milli-Q water | −80.3 mV |
| NPER + seawater | 0.3 mV |
| PSNPs + Milli-Q water | −31.2 mV |
| PSNPs + seawater | −3.8 mV |
| NPER + PSNPs + Milli-Q water complex | −68.1 mV |
| NPER + PSNPs + Seawater complex | 30.6 mV |
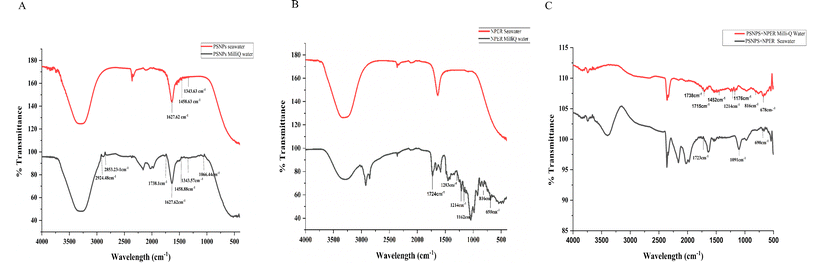
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond stretching), 1550–1750 cm−1 (C
C bond stretching), 1550–1750 cm−1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 2800–3000 cm−1 (C–H stretch aliphatic), 2800–3060 cm−1 (C–H), and aromatic 3610–3645 cm−1 (hydroxyl), and a similar significant polystyrene peak was reported in ref. 41. FTIR spectra of NPER had sharp characteristic peaks of nanopermethrin C
O), 2800–3000 cm−1 (C–H stretch aliphatic), 2800–3060 cm−1 (C–H), and aromatic 3610–3645 cm−1 (hydroxyl), and a similar significant polystyrene peak was reported in ref. 41. FTIR spectra of NPER had sharp characteristic peaks of nanopermethrin C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibration of carbonyl groups at 1724 cm−1, asymmetric stretching of C–O–C at 1283 cm−1, C–Cl stretch at 816 cm−1 and C–H bend at 690 cm−1,42 and similar significant permethrin peaks are reported in ref. 43. In the complex, significant peaks have exhibited some shift in the (C–H) bend at 670 cm−1, at 810 cm−1 (C–Cl stretch), asymmetric stretching of C–O–C at 1283 cm−1 and C
O stretching vibration of carbonyl groups at 1724 cm−1, asymmetric stretching of C–O–C at 1283 cm−1, C–Cl stretch at 816 cm−1 and C–H bend at 690 cm−1,42 and similar significant permethrin peaks are reported in ref. 43. In the complex, significant peaks have exhibited some shift in the (C–H) bend at 670 cm−1, at 810 cm−1 (C–Cl stretch), asymmetric stretching of C–O–C at 1283 cm−1 and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups at 1214 cm−1 The results demonstrate that NPER interacts with PSNPs in Milli-Q water and sea water and the adsorption of the complex shown the major nanopermethrin peak on the surface of the PSNPs and a peak shift is demonstrated as shown in Fig. 4.
O groups at 1214 cm−1 The results demonstrate that NPER interacts with PSNPs in Milli-Q water and sea water and the adsorption of the complex shown the major nanopermethrin peak on the surface of the PSNPs and a peak shift is demonstrated as shown in Fig. 4.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 625 spectra presented in Fig. S2.† Most hydrophobic pesticides such as HCB and bromophos-ethyl, were found to be absorbed in the soil at a relatively high level, despite being less than what had previously been reported for topsoil. The adsorption of the pesticides was demonstrated to have reached an apparent equilibrium in the soil using three model pesticides with high (desethylatrazine), medium (terbutryn), and low (bromophos-ethyl) polarity.47
625 spectra presented in Fig. S2.† Most hydrophobic pesticides such as HCB and bromophos-ethyl, were found to be absorbed in the soil at a relatively high level, despite being less than what had previously been reported for topsoil. The adsorption of the pesticides was demonstrated to have reached an apparent equilibrium in the soil using three model pesticides with high (desethylatrazine), medium (terbutryn), and low (bromophos-ethyl) polarity.47
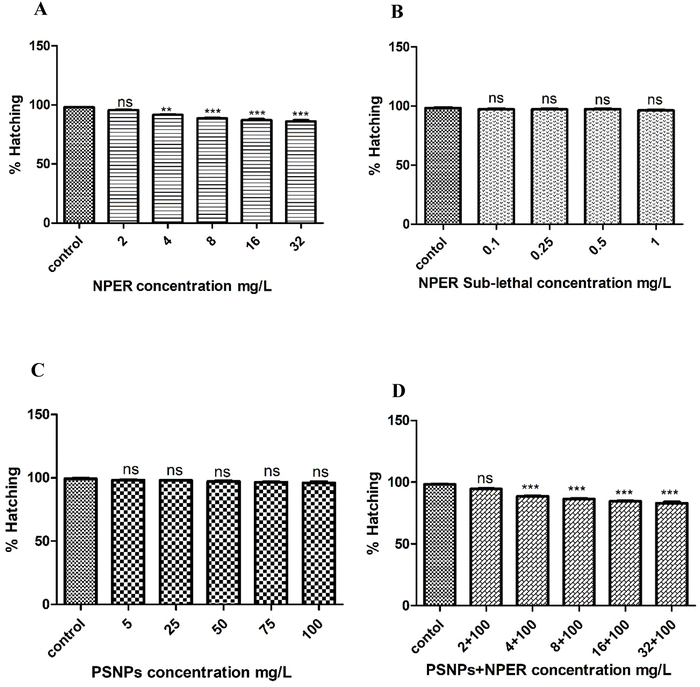
3.3.7.1 Effect on hatching. The hatching percentage of Artemia salina in the control was found to be 98% in seawater. When treated with NPER at different concentrations such as 2, 4, 8, 16, and 32 mg L−1, the hatching percentage of Artemia salina was decreased to 96, 91, 89, 88, and 87%, respectively. All the concentrations were statistically significant (p < 0.001) with the control except 2, 4 mg L−1. The hatching percentage of Artemia salina after interaction with pristine PSNPs at different concentrations such as 5, 25, 50, 75, and 100 mg L−1, was found to be 98.66, 98, 97.66, 97, and 97% respectively compared with that of the control, with no significant difference. But when the cysts were treated with the NPER + PSNP complex (2 + 100, 4 + 100, 8 + 100, 16 + 100, and 32 + 100 mg L−1), the hatching percentage was found to be 95, 89, 87, 85, and 84%, which are statistically significant when compared with the control (P < 0.001) except 2 + 100 mg L−1. This significant decrease in hatching percentage revealed the synergistic toxicity of PSNPs and NPER on Artemia salina. When subjected to lower sublethal permethrin concentrations (0.1, 0.25, 0.5, and 1 mg L−1), the hatching of decapsulated Artemia salina occurred in sublethal concentrations of 98, 97.66, 97.66, and 97.66%, respectively, compared with the control, and there is no significant difference as presented in Fig. 8. There is no significant difference between the control and pristine particles of PSNPs. Similar results were observed by Madhav et al., where the hatching ability of Artemia was not affected when cysts were exposed to lower exposure dosages of nanoparticles.25 The dormant cysts' metabolic activity is inhibited when nanoparticles enter the decapsulated cysts through their smooth outer layer.24 Thus, compared to pristine, the mixture of NPER and PSNPs significantly affects the hatching rate of Artemia salina.
3.3.7.2 Swimming performance. Organisms were taken at regular time intervals of 3, 6, 12, 24, and 48 h after being treated with pristine PSNPs, NPER and their complexes and their corresponding swimming behaviours were observed. It was noticed that the changes in the swimming performance of Artemia salina were not observed for control and PSNPs. But organisms treated with NPER and complex particles exhibited reduced swimming ability because of the morphological damage after particle intake as shown in Fig. 9. Neurotoxic compounds can cause abnormal swimming behavior or impair swimming capability in fish and other aquatic animals. This is due to the fact that pyrethroids, which are potent neurotoxins, can have a significant impact on the nervous system of these animals and permethrin is a neurotoxic chemical which impairs or causes erratic swimming behaviour in fish and other aquatic species.11
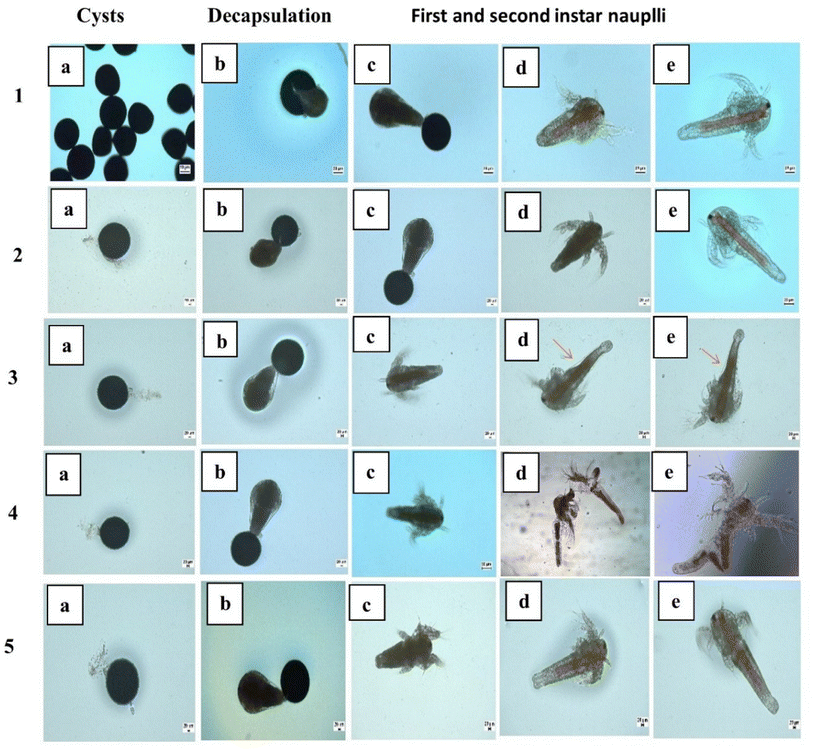
3.3.7.3 Artemia salina morphological changes. Developmental growth and morphology of Artemia salina, when exposed to nanopollutants were examined from cysts to 2nd instar nauplii (Fig. 9). Artemia salina nauplii were exposed to various concentrations of nanoparticles including pristine nanopermethrin (2, 4, 8, 16, and 32 mg L−1), polystyrene (5, 25, 50, 75, and 100 mg L−1), and a combinatorial mixture of NPER and PSNPs (2 + 100, 4 + 100, 8 + 100, 16 + 100, and 32 + 100 mg L−1) and the latter was also used at sub-lethal concentrations (0.1, 0.25, 0.5, and 1 mg L−1). After 48 hours, changes in morphology were observed under a microscope. The results showed that the animals in the control group and PSNP group did not show any signs of trauma or seem to have any evidence of damage, while pristine ermethrin damages Artemia salina at greater doses. Exposure of the cysts to the lower concentration of nanopermethrin did not affect Artemia salina growth, but it affected the morphological alterations in the gut region at the highest concentration of 32 mg L−1 as shown in Fig. 9 (3 d and e), and the magnified gut region is shown in Fig. S3.† The combination particles have a stronger impact on Artemia salina, injure the Artemia salina tissues in the gut region and lead to collapse of the system as shown in Fig. 9 (4 d and e). At the sublethal concentration there was no effect in morphological alterations. Intestinal alterations caused by the nanoparticles could be observed clearly under a phase contrast microscope. According to research, the NPER + PSNP complex led to a high death rate in second instar nauplii by developing more morphological damage in the gut region. Early studies report that the meta nauplii and Artemia salina's first and second instars can consume NPs. The accumulation of NPs in various physiological tissues caused the particles to travel up the food chain from Artemia salina to fish in aquatic environments.48 Intake of aggregated particles is the primary factor impacting the many toxicological reactions that are shown in the organism.49 A study on the ability of PSNPs to translocate through the intestine of Artemia salina was reported by by Albano et al. in 2021.50 The microscopy images of Artemia franciscana larvae treated with various concentrations of PS-COOH (5–100 μg mL−1) revealed the presence of aggregate particles, which were absent in the control group. A study conducted by Bergami et al. (2016) also reported a significant sequestration of PS NPs within the digestive tract.51
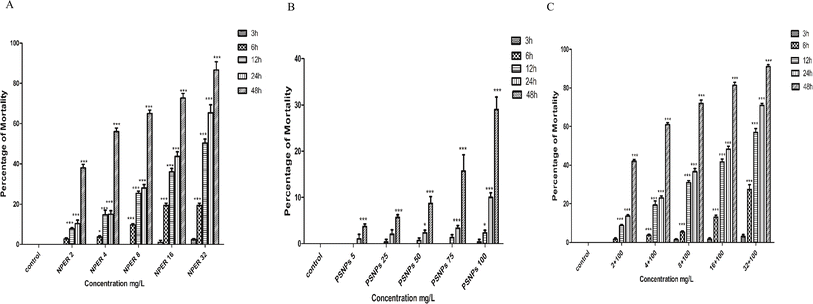
3.3.7.4 Assessment of acute toxicity of nanopermethrin and polystyrene. Compared to the control groups, all the treatment concentrations of NPER used in the experiment showed no significant difference until 3 h of treatment after which a significant difference was observed at all the concentrations (Fig. 10). In the case of PSNP treated Artemia salina, no significant difference was found until 12 h of treatment at all the concentrations when compared to the control. In the combination study, no significant difference was found until 3 h of treatment when compared to the control. This trend was observed at all concentrations. NPER treated Artemia salina was found to have an increased mortality at all the time intervals with increasing concentrations. Mortality was found to be time-dependent and it was found to increase with interaction time. PSNP's increasing concentration was found to lead to an increase in mortality at 24 and 48 h time intervals. Complex particles increased the mortality rate compared to pristine forms of nanoparticles. When compared with pristine NPER, the complex exhibits a significant difference after an interaction period of 12 h at 8 mg L−1. It was observed that the significant difference between the pristine and the combination treatment increased with the increase in the concentrations of the contaminants. Permethrin and other pyrethroids are found to be extremely dangerous for aquatic organisms. Permethrin was shown to have fatal concentration values (LC50) of 12.4 and 8.7 μg L−1 for the species Ceriodaphnia dubia and Daphnia magna, respectively.52 Permethrin was discovered to be more dangerous to sea urchin embryos than their sperm.53 The larvicidal activities of nanopermethrin and permethrin were studied comparatively against A. aegypti. The results showed that bulk permethrin-treatment leads to 100% mortality after 24 h, but nanopermethrin has a 100% mortality rate after 4 h37. Short-term exposure to PS-NH2 altered development and reduced nauplii growth in a concentration-dependent way, whereas chronic exposure reduced survival but not growth or feeding behaviour. A decrease in the antioxidant enzyme activity was the first sign of oxidative stress following brief exposure.5 Polystyrene has a lower mortality rate over the course of 48 hours, as evidenced by the less than 50% mortality rate. The LC50 values for each duplicate were calculated as the mean using the Probit approach,54 The LC50 value for PSNP treated Artemia salina is not shown here, since they have a lower percentage of Artemia salina mortality. Here, the complex particle toxicity was evaluated, and the precise value of the LC50, was calculated to be 3.127 mg L−1 which is lower than the LC50 value of pristine NPER. Hence, combined particles exhibit synergistic toxicity to Artemia salina.
The combined toxicity of NPER and PSNPs was validated by using an independent action model (Table 2). When a PSNP concentration of 100 mg L−1 and NPER concentration of 2 mg L−1 were combined, the RI values was observed to be decreased below 1,27 proving their antagonistic effects on Artemia salina. However, when the concentration of NPER was increased from 4 to 32 mg L−1, the complex exhibited synergistic effects on the organisms. Previous studies have reported that, the mode of action among mixed groups was antagonistic, which was observed in the mortality rate.
| Concentration of PSNPs (mg L−1) | Concentration of NPER (mg L−1) | Observed toxicity (%) | Expected toxicity (%) | Ratio of inhibition (RI) | P-Values | Mode of action |
|---|---|---|---|---|---|---|
| 100 | 2 | 38 | 55.98 | 0.75 | P < 0.05 | Antagonistic |
| 100 | 4 | 56 | 60.35 | 1.10 | P < 0.001 | Synergistic |
| 100 | 8 | 65 | 75.15 | 1.52 | P < 0.001 | Synergistic |
| 100 | 16 | 72.66 | 80.58 | 1.93 | P < 0.001 | Synergistic |
| 100 | 32 | 86.66 | 90.52 | 1.53 | P < 0.001 | Synergistic |
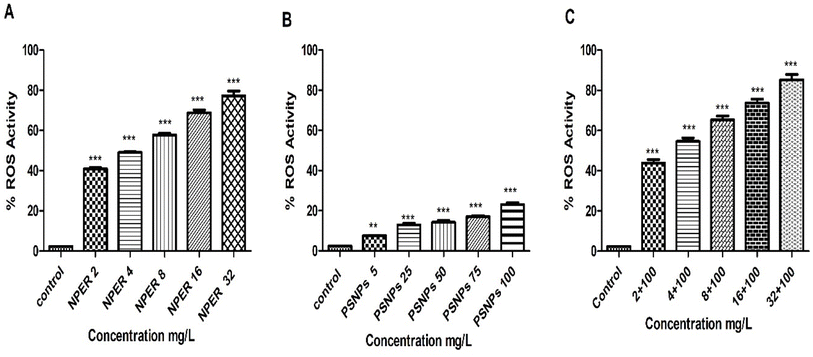
3.3.7.5 Reactive oxygen species. The detection of oxidative stress with the ROS assay is a crucial biological test. Based on the mechanism, oxidative stress releases free radicals, thus increasing the production of Reactive oxygen species (ROS). Artemia salina was treated with experimental particles to analyze the changes in the ROS levels, and the results are presented in Fig. 11. Compared to the control, the test groups (except for 5 mg L−1 PSNPs) experienced a statistically significant increase in ROS levels (p < 0.001). At all of the corresponding concentrations, the NPER–PSNP complex generated abundant ROS compared to pristine NPER and PSNPs, which was statistically significant (p < 0.001). Intriguingly, the significant difference between pristine and combination treatment groups increased with the increase in NPER concentration. When compared to the control group, the treated nanoparticles produce changes in Artemia salina's detrimental behaviour that leads to the generation of ROS, which is regarded as an indicator of toxic effects.18,48
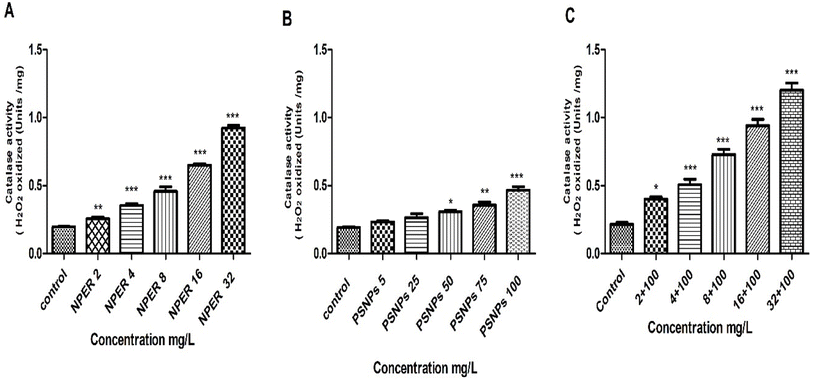
3.3.7.6 Catalase activity. Catalase is a marker for oxidative pressure in cells.55 The catalase (CAT) enzyme played a major role in mediating the antioxidant defense against ROS.56 The main cellular precursor of the most dangerous ROS, H2O2, is reduced by CAT, which is known to protect cells by lowering toxicity.57 The generation of oxygen and H2O2 from the disproportionation of superoxide also results in the activation of the CAT enzyme.58 When Artemia salina was treated with nanopollutants, CAT activity increased with increasing concentration as presented in Fig. 12. Regarding PSNPs, this increase was only highly significant at a concentration of 100 mg L−1 of PSNPs (p < 0.001) and less significant at the concentrations of 50 mg L−1 (p < 0.05) and 75 mg L−1 (p < 0.01). Treatment with pure NPER also increased CAT activity which is statistically significant at all concentrations, when compared to the control. CAT activity in the NPER–PSNP mixture also increased and becomes statistically significant (p < 0.001) on comparison to the control. Overall, the NPER–PSNP complex exhibits strong significance in stress responses (p < 0.001) compared to pristine PSNPs and NPER, revealing their synergistic toxicity.
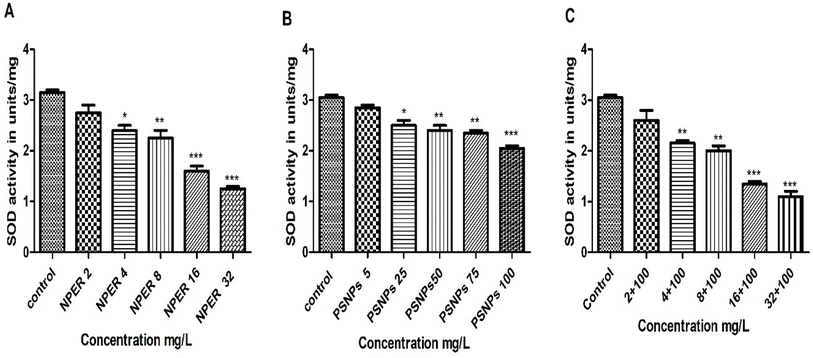
3.3.7.7 Superoxide dismutase. When Artemia salina was treated with PSNPs, SOD activity decreased with increasing concentration. This decrease in SOD activity was highly significant only at a concentration of PSNPs of 100 mg L−1 (p < 0.01) as shown in Fig. 13 and was noticed to be less significant at the concentrations of 50 mg L−1 and 75 mg L−1 (p < 0.01). The lowest concentrations of 5 mg L−1 and 25 mg L−1 were statistically insignificant on comparing with the control. Similarly, treatment with NPER also led to reduced SOD activity, when compared to the control, with the exception of 2 mg L−1, which is insignificant. All treated concentrations were extremely significant (especially 16 mg L−1 and 32 mg L−1 (p < 0.001)). The SOD activity after the NPER–PSNP complex treatment significantly decreased when compared to the control, and this result was highly significant at all treated concentrations (p < 0.001), with the exception of the concentrations below 2 + 100 mg L−1, which had an inconsequential effect. The reduction in SOD activity was higher in complex treated animals than pristine particle treatment, confirming their synergistic toxicity in the environment. Additionally, the greater levels of ROS in the cells may have caused SOD to degrade, which would explain why SOD activity decreased at greater concentrations of the complex.59 In general, nanoparticle toxicity was usually assessed by using the decrease in SOD activity in Artemia salina nauplii.60
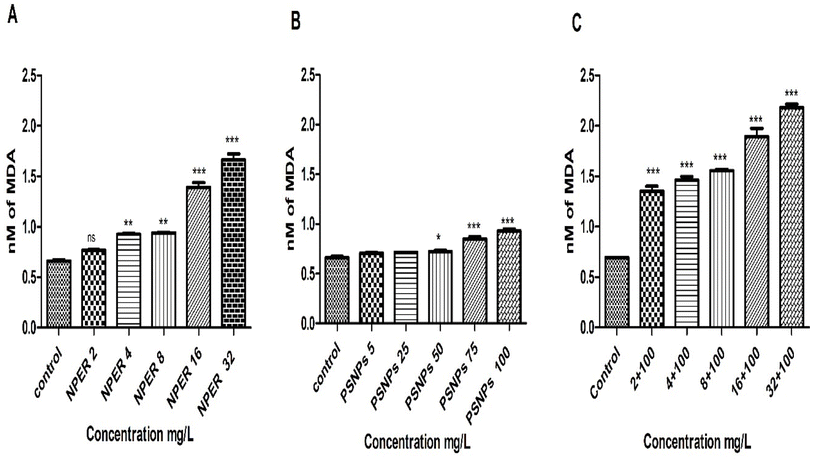
3.3.7.8 Lipid peroxidation. The generation of lipid peroxides in the host species cytoplasm as a result of lipid membrane damage has the potential to act as a key signal for the relationship between the induced toxicity and membrane integrity.61 MDA is a powerful sign of oxidative stress and a naturally occurring by-product of lipid peroxidation.62 MDA assay demonstrated a significant substantial connection between MDA levels and oxidative stress.63 A substantial difference (p < 0.001) at a high concentration of 100 mg L−1 PSNP treated Artemia salina is shown by the quantifiable MDA levels presented in Fig. 14. The significance reduced when concentration is lowered to 75 mg L−1 (p < 0.01). At a lower dosage of 2 mg L−1, pristine NPER is unremarkable, but at a higher concentration, it displays a very high MDA level (p < 0.001). Comparing control and pristine particles, the NPER–PSNP mixture exhibits a significant difference in the MDA level (p < 0.001) at all concentrations, further evidencing the impact of additive toxicity.
4. Conclusion
Environmental nanoplastics are a complex mixture of contaminants, and their toxicity when mixed with other pollutants can magnify the potential harm to ecosystems. Nanopesticides, which are designed to decrease the hazardous effects of conventional pesticides, are also accumulating in the environment, forming a monolayer adsorption over PSNPs by physisorption. The toxicity was determined using second instar nauplii, which are particularly vulnerable to the particles, and morphological harm was assessed using microscopic examinations. Toxicity assessment demonstrates that the combined particles are more hazardous than pristine PSNPs and NPER. All of the particles induced oxidative stress (ROS and LPO) in Artemia salina, although it is more intense in complex treated organisms. As a result, their influence on Artemia salina's antioxidant response system (SOD and catalase) is quite significant. Synergistic toxicity is observed when nanoplastics interact with other co-pollutant particles (nanopesticides), which produces higher oxidative stress to Artemia salina. However, further research is needed to better understand the extent and consequences of nanopesticide adsorption on nanoplastics. Efforts should be made to reduce the discharge of nanoplastics and other pollutants into the environment in order to counteract their combined harmful effects and safeguard the health of species and ecosystems.Abbreviations
| NPER | Nanopermethrin |
| MNPs | Micronanoplastics |
| PSNPs | Polystyrene nanoplastics |
| PFO | Pseudo-first-order |
| PSO | Pseudo-second-order |
| ROS | Reactive oxygen species |
| SOD | Superoxide dismutase |
| LPO | Lipid peroxidation |
| XRD | X-ray diffraction |
| GC-MS | Gas chromatography-mass spectrometry |
| FE-SEM | Field emission scanning electron microscopy |
Author contributions
Mahalakshmi Kamalakannan: investigation, methodology, data curation, writing – original draft, writing – review & editing; Durgalakshmi Rajendran: writing – review & editing; John Thomas: writing – review & editing; natarajan Chandrasekaran: conceptualization, supervision, resources, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors declare that there is no conflict of interest.Acknowledgements
The authors acknowledge the funding provided by the Indian Council of Medical Research (ICMR) vide Research Grant-F No. 36/2/2020/Toxi/BMS.References
- M. F. Cárdenas-Alcaide, J. A. Godínez-Alemán, R. B. González-González, H. M. N. Iqbal and R. Parra-Saldívar, Green Anal. Chem., 2022, 3, 100031 CrossRef.
- V. Gupta, S. Mohapatra, H. Mishra, U. Farooq, K. Kumar, M. Ansari, M. Aldawsari, A. Alalaiwe, M. Mirza and Z. Iqbal, Gels, 2022, 8, 173 CrossRef CAS PubMed.
- C. Moeck, G. Davies, S. Krause and U. Schneidewind, Grundwasser, 2023, 28, 23–35 CAS.
- D. Huang, H. Chen, M. Shen, J. Tao, S. Chen, L. Yin, W. Zhou, X. Wang, R. Xiao and R. Li, J. Hazard. Mater., 2022, 438, 129515 CrossRef CAS PubMed.
- I. Varó, A. Perini, A. Torreblanca, Y. Garcia, E. Bergami, M. L. Vannuccini and I. Corsi, Sci. Total Environ., 2019, 675, 570–580 CrossRef PubMed.
- C. Fajardo, C. Martín, G. Costa, S. Sánchez-Fortún, C. Rodríguez, J. J. de Lucas Burneo, M. Nande, G. Mengs and M. Martín, Chemosphere, 2022, 288, 132460 CrossRef CAS PubMed.
- T. Schell, R. Hurley, N. T. Buenaventura, P. V. Mauri, L. Nizzetto, A. Rico and M. Vighi, Environ. Pollut., 2022, 293, 118520 CrossRef CAS PubMed.
- P. Mishra, A. P. B. Balaji, J. S. Swathy, A. L. Paari, M. Kezhiah, B. K. Tyagi, A. Mukherjee and N. Chandrasekaran, Environ. Sci. Pollut. Res., 2016, 23, 24970–24982 CrossRef CAS PubMed.
- C. An, C. Sun, N. Li, B. Huang, J. Jiang, Y. Shen, C. Wang, X. Zhao, B. Cui, C. Wang, X. Li, S. Zhan, F. Gao, Z. Zeng, H. Cui and Y. Wang, J. Nanobiotechnol., 2022, 20, 11 CrossRef CAS PubMed.
- V. M. Pathak, V. K. Verma, B. S. Rawat, B. Kaur, N. Babu, A. Sharma, S. Dewali, M. Yadav, R. Kumari, S. Singh, A. Mohapatra, V. Pandey, N. Rana and J. M. Cunill, Front. Microbiol., 2022, 13, 962619 CrossRef PubMed.
- I. Werner and K. Moran, in ACS Symposium Series, American Chemical Society, 2008, vol. 991, pp. 310–334 Search PubMed.
- E. Dube and G. E. Okuthe, Int. J. Environ. Res. Public Health, 2023, 20, 6667 CrossRef CAS PubMed.
- F. Wang, J. Gao, W. Zhai, D. Liu, Z. Zhou and P. Wang, J. Hazard. Mater., 2020, 394, 122517 CrossRef CAS PubMed.
- M. Cole, P. K. Lindeque, E. Fileman, J. Clark, C. Lewis, C. Halsband and T. S. Galloway, Environ. Sci. Technol., 2016, 50, 3239–3246 CrossRef CAS PubMed.
- K. Madkour, M. A. O. Dawood and H. Sewilam, Ann. Anim. Sci., 2023, 23, 3–10 CrossRef.
- S. Rajabi, A. Ramazani, M. Hamidi and T. Naji, Daru, J. Pharm. Sci., 2015, 23, 20 CrossRef PubMed.
- G. K. Ñañez Pacheco, N. S. Sanabio Maldonado, R. Y. Pastrana Alta and H. Aguilar Vitorino, Ecotoxicol. Environ. Saf., 2021, 213, 112052 CrossRef PubMed.
- S. Zhu, M. Xue, F. Luo, W. Chen, B. Zhu and G. Wang, Environ. Pollut., 2017, 230, 683–691 CrossRef CAS PubMed.
- E. Bergami, E. Bocci, M. L. Vannuccini, M. Monopoli, A. Salvati, K. A. Dawson and I. Corsi, Ecotoxicol. Environ. Saf., 2016, 123, 18–25 CrossRef CAS PubMed.
- E. D. Revellame, D. L. Fortela, W. Sharp, R. Hernandez and M. E. Zappi, Clean. Eng. Technol., 2020, 1, 100032 CrossRef.
- K. D. Kowanga, E. Gatebe, G. O. Mauti and E. M. Mauti, J. Phytopharm., 2016, 5, 71–78 CrossRef.
- I. Langmuir, J. Am. Chem. Soc., 1918, 40, 1361–1403 CrossRef CAS.
- H. M. F. Freundlich, J. Phys. Chem., 1906, 57, 1100–1107 Search PubMed.
- C. Arulvasu, S. M. Jennifer, D. Prabhu and D. Chandhirasekar, Sci. World J., 2014, 2014, 256919 Search PubMed.
- M. R. Madhav, S. E. M. David, R. S. S. Kumar, J. S. Swathy, M. Bhuvaneshwari, A. Mukherjee and N. Chandrasekaran, Environ. Toxicol. Pharmacol., 2017, 52, 227–238 CrossRef CAS PubMed.
- M. M. Bradford, Anal. Biochem., 1976, 72, 248–254 CrossRef CAS PubMed.
- V. Thiagarajan, N. Seenivasan, D. Jenkins, N. Chandrasekaran and A. Mukherjee, Aquat. Toxicol., 2020, 225, 105541 CrossRef CAS PubMed.
- J. M. C. Gutteridge and B. Halliwell, Trends Biochem. Sci., 1990, 15, 129–135 CrossRef CAS PubMed.
- H. Ohkawa, N. Ohishi and K. Yagi, Anal. Biochem., 1979, 95, 351–358 CrossRef CAS PubMed.
- K. Wu, D. Su, J. Liu, R. Saha and J.-P. Wang, Nanotechnology, 2019, 30, 502003 CrossRef CAS PubMed.
- D. J. Sarkar, S. Das Sarkar, S. Mukherjee and B. K. Das, Contaminants in Drinking and Wastewater Sources, 2021, pp. 95–115 Search PubMed.
- A. Hubert, T. Aquino, H. Tabuteau, Y. Méheust and T. Le Borgne, Adv. Water Resour., 2020, 146, 103739 CrossRef CAS.
- W. K. Ho, J. C. F. Law, T. Zhang and K. S. Y. Leung, Water Res., 2020, 187, 116419 CrossRef CAS PubMed.
- S. Sharifi, R. Nabizadeh, B. Akbarpour, A. Azari, H. R. Ghaffari, S. Nazmara, B. Mahmoudi, L. Shiri and M. Yousefi, J. Environ. Health Sci. Eng., 2019, 17, 873–888 CrossRef CAS PubMed.
- T.-B. Nguyen, T.-B.-C. Ho, C. P. Huang, C.-W. Chen, W.-H. Chen, S. Hsieh, S.-L. Hsieh and C.-D. Dong, Chemosphere, 2022, 304, 135276 CrossRef CAS PubMed.
- P. Wu, Z. Cai, H. Jin and Y Tang, Sci. Total Environ., 2019, 650(1), 671–678 CrossRef CAS PubMed.
- C. H. Anjali, S. Sudheer Khan, K. Margulis-Goshen, S. Magdassi, A. Mukherjee and N. Chandrasekaran, Ecotoxicol. Environ. Saf., 2010, 73, 1932–1936 CrossRef CAS PubMed.
- M. L. Wells and E. D. Goldberg, Mar. Chem., 1993, 41, 353–358 CrossRef CAS.
- L. Manfra, A. Rotini, E. Bergami, G. Grassi, C. Faleri and I. Corsi, Ecotoxicol. Environ. Saf., 2017, 145, 557–563 CrossRef CAS PubMed.
- P. Mishra, A. P. B. Balaji, P. K. Dhal, R. S. Suresh Kumar, S. Magdassi, K. Margulis, B. K. Tyagi, A. Mukherjee and N. Chandrasekaran, Bull. Entomol. Res., 2017, 1–13 Search PubMed.
- M. R. Jung, F. D. Horgen, S. V. Orski, V. Rodriguez, K. L. Beers, G. H. Balazs, T. T. Jones, T. M. Work, K. C. Brignac, S. Royer, K. D. Hyrenbach, B. A. Jensen and J. M. Lynch, Mar. Pollut. Bull., 2018, 127, 704–716 CrossRef CAS PubMed.
- FAO, FAO Specifications and Evaluations for Agricultural Pesticides, Food and Agriculture Organisation of United Nations, 1994, pp. 1–46 Search PubMed.
- Z. Uslinawaty, M. Muin and Suhasman, IOP Conf. Ser.: Mater. Sci. Eng., 2020, 935, 012003 CAS.
- X. Tian, Q. Yu, X. Kong and M. Zhang, Front. Chem., 2022, 10, 847203 CrossRef CAS PubMed.
- J. Zhang, S. Zhan, L.-B. Zhong, X. Wang, Z. Qiu and Y.-M. Zheng, J. Hazard. Mater., 2023, 443, 130130 CrossRef CAS PubMed.
- M. A. Amin, M. A. El-degwy and B. A. Fayed, J. Pharm. Biol. Sci., 2017, 12, 42–45 Search PubMed.
- J. Tian and A. Rustum, RSC Adv., 2016, 6, 83020–83024 RSC.
- M. Ates, J. Daniels, Z. Arslan and I. O. Farah, Environ. Monit. Assess., 2013, 3339–3348 CrossRef CAS PubMed.
- M. R. Madhav, S. E. M. David, R. S. S. Kumar, J. S. Swathy, M. Bhuvaneshwari, A. Mukherjee and N. Chandrasekaran, Environ. Toxicol. Pharmacol., 2017, 52, 227–238 CrossRef CAS PubMed.
- G. Panarello, D. Di Paola, F. Capparucci, R. Crupi, E. Gugliandolo, N. Spanò, G. Capillo and S. Savoca, Appl. Sci., 2021, 11(8), 3352 CrossRef.
- E. Bergami, E. Bocci, M. L. Vannuccini, M. Monopoli, A. Salvati, K. A. Dawson and I. Corsi, Ecotoxicol. Environ. Saf., 2016, 123, 18–25 CrossRef CAS PubMed.
- A. Ç. Günal, S. K. Tunca, P. Arslan, G. Gül and A. S. Dinçel, Environ. Sci. Pollut. Res., 2021, 28, 52405–52417 CrossRef PubMed.
- B. Erkmen, Bull. Environ. Contam. Toxicol., 2015, 94, 419–424 CrossRef CAS PubMed.
- B. Nunes, F. Carvalho and L. Guilhermino, Ecotoxicol. Environ. Saf., 2005, 61, 413–419 CrossRef CAS PubMed.
- A. Nandi, L.-J. Yan, C. K. Jana and N. Das, Oxid. Med. Cell. Longevity, 2019, 2019, 1–19 CrossRef PubMed.
- O. M. Ighodaro and O. A. Akinloye, Alexandria J. Med., 2018, 54, 287–293 CrossRef.
- T. Konno, E. P. Melo, J. E. Chambers and E. Avezov, Cells, 2021, 10, 233 CrossRef CAS PubMed.
- Y. Wang, R. Branicky, A. Noë and S. Hekimi, J. Cell Biol., 2018, 217, 1915–1928 CrossRef CAS PubMed.
- R. H. Gottfredsen, U. G. Larsen, J. J. Enghild and S. V. Petersen, Redox Biol., 2013, 1, 24–31 CrossRef CAS PubMed.
- H. J. An, M. Sarkheil, H. S. Park, I. J. Yu and S. A. Johari, Comp. Biochem. Physiol., Part C: Toxicol. Pharmacol., 2019, 218, 62–69 CAS.
- M. P. Cajaraville, M. J. Bebianno, J. Blasco, C. Porte, C. Sarasquete and A. Viarengo, Sci. Total Environ., 2000, 247, 295–311 CrossRef CAS PubMed.
- C. H. Cordilea Hannah, IOSR J. Pharm. Biol. Sci., 2013, 6, 47–51 Search PubMed.
- M. Ates, V. Demir, Z. Arslan, J. Daniels, I. O. Farah and C. Bogatu, Environ. Toxicol., 2015, 30, 109–118 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4na00013g |
| This journal is © The Royal Society of Chemistry 2024 |