Rapid identification of novel indolylarylsulfone derivatives as potent HIV-1 NNRTIs via miniaturized CuAAC click-chemistry-based combinatorial libraries
Ping
Gao
 ab,
Shu
Song
a,
Christophe
Pannecouque
c,
Erik
De Clercq
c,
Peng
Zhan
ab,
Shu
Song
a,
Christophe
Pannecouque
c,
Erik
De Clercq
c,
Peng
Zhan
 *ad and
Xinyong
Liu
*ad and
Xinyong
Liu
 *ad
*ad
aDepartment of Medicinal Chemistry, Key Laboratory of Chemical Biology, Ministry of Education, School of Pharmaceutical Sciences, Shandong University, Ji'nan, 250012, China. E-mail: zhanpeng1982@sdu.edu.cn; xinyongl@sdu.edu.cn
bDepartment of Pharmacy, Shandong Provincial Hospital Affiliated to Shandong First Medical University, Jinan, Shandong 250021, China
cRega Institute for Medical Research, K. U. Leuven, Minderbroedersstraat 10, B-3000 Leuven, Belgium
dChina-Belgium Collaborative Research Center for Innovative Antiviral Drugs of Shandong Province, 44 West Culture Road, 250012 Jinan, Shandong, PR China
First published on 15th October 2024
Abstract
This article presents the rapid identification of novel indolylarylsulfone (IAS) derivatives as potent non-nucleoside reverse transcriptase inhibitors (NNRTIs) for HIV-1 through a miniaturized click-chemistry-based combinatorial library approach. Utilizing copper(I)-catalyzed azide–alkyne cycloaddition (CuAAC), a reliable and biocompatible click chemistry technique, the researchers synthesized and characterized a series of IAS derivatives. Several compounds selected through the in situ enzyme inhibition assay demonstrated promising activity in subsequent cellular level tests. Notably, compound C1N4 displayed the most potent anti-HIV-1 IIIB activity with an EC50 of 0.024 μM and low cytotoxicity (CC50 > 215.88 μM). Molecular docking studies provided insights into the binding mode of these novel compounds within the NNIBP, aiding in the structure-based design of future NNRTIs. The findings underscore the potential of click chemistry in the discovery of new anti-HIV agents with improved efficacy and safety profiles.
1. Introduction
The global pandemic of human immunodeficiency virus type 1 (HIV-1) remains as a major threat to human health worldwide.1 Since the beginning of the epidemic, 85.6 million [65.0–113.0 million] people have been infected with the HIV virus and about 40.4 million [32.9–51.3 million] people have died of HIV.2 Standard treatments for HIV-1 infection are represented by highly active antiretroviral therapies (HAART), which combine various anti-HIV drugs that target different stages of the virus's life cycle, and have achieved notable success in controlling HIV infection.3,4 Among the antiretroviral drugs approved by the U.S. Food and Drug Administration (FDA), non-nucleoside reverse transcriptase inhibitors (NNRTIs) have received a lot of attention due to their relatively favorable potency and low cytotoxicity. However, the rapid emergence of drug-resistant mutants with reduced susceptibility and adverse effects, such as drug–drug interactions and toxicities of long-term use, has seriously impaired the efficacy of current drugs.5–7 Therefore, there is a continuous need for the discovery of additional NNRTI families with a broad spectrum of activity against clinically relevant HIV-1 mutant strains and lower adverse effects.Indolylarylsulfones (IASs) have been considered as a class of highly active NNRTIs developed from the Merck prototype compound L-737,126 which possessed nanomolar inhibitory activity towards HIV-1 wild-type (WT) and several mutant strains. Derivatives of this scaffold have contributed several promising hit compounds.8–15 Crystallography based on the complex of HIV-1 RT and the indolylarylsulfone derivative 7e (Fig. 1) showed that the 2-position of the indole nucleus was located in the entrance channel (the protein/solvent interface between the p66 and p51 RT subunits) of the NNRTI binding pocket (NNIBP), mainly formed by Leu100, Glu138 and Val179. Additionally, the groups at this position could build potential hydrogen bonds with surrounding amino acids such as Glu138.11
 | ||
| Fig. 1 Indolylarylsulfone derivatives and schematic diagram showing the location of compound 7e within the NNRTI binding pocket of HIV-1 RT. Compound 7e (yellow) and amino acid side chains in the binding pocket (blue) are shown using a stick representation. The figure was generated with the PyMol software (http://www.pymol.org) (PDB code: 2RF2). | ||
Click chemistry, especially copper(I)-catalyzed azide–alkyne (3 + 2) dipolar cycloaddition (CuAAC), has drawn great attention and becomes a powerful tool for the generation of privileged medicinal skeletons in the discovery of bioactive molecules due to its high degree of reliability, complete specificity (chemoselectivity and regioselectivity), mild conditions, and the biocompatibility of the reactants.16–19 Therefore, in this study, a rapid method to discover hit compounds which combined parallel click chemistry synthesis via the CuAAC reaction with in situ biological screening was reported to identify novel HIV-1 NNRTIs through in-depth exploration of untapped chemical space around the entrance channel through chemical modifications at the 2-position of indole.
2. Results and discussion
2.1 Building and characterizing the libraries essential for click chemistry
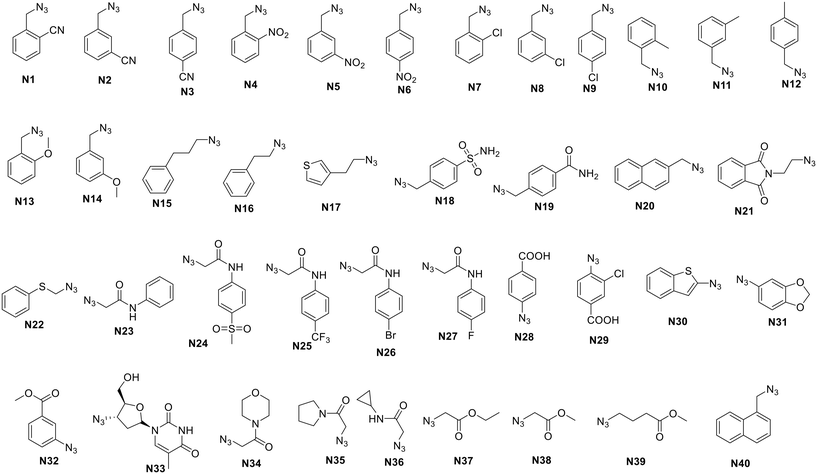
The synthetic routes of the two intermediates containing terminal alkynes are outlined in Scheme 1. Intermediate 4 was synthesized from the commercially available starting material ethyl 5-chloro-1H-indole-2-carboxylate (1) as previously described,12–15 which underwent the acylation reaction with prop-2-yn-1-amine or 2-methylbut-3-yn-2-amine to generate intermediates C1 and C2.The azide substituents (as shown in Fig. 2) were prepared as previously reported.18
2.2 Generation of the combinatorial library
The indolylarylsulfone derivative library was established in 96-well microtiter plates on a millimolar scale following the previously explored reaction conditions illustrated in Table 1.18 The prepared azide substituents, alkyne fragments, tris[(1-benzyl-1H-1,2,3-triazol-4-yl)methyl]amine (TBTA, the addition of TBTA could form complexes with copper ions to effectively increase the stability of monovalent copper and accelerate reaction rates20), CuSO4·5H2O and sodium ascorbate were added in sequence to a 96-well microtiter plate, and then the reaction mixtures were stirred in a constant temperature oscillation incubator at room temperature for 24 h. The synthetic process was detected by thin-layer chromatography (TLC) under UV light (λ = 254, 365 nm). After completion, the reaction mixtures were diluted with 150 μL DMSO and the final concentration of target compounds in each well was 2 mM. Finally, apart from 2 wells whose reaction didn't complete (C2N25, C2N26), 78 indolylarylsulfone derivatives were successfully obtained, which were used to perform enzymatic assays against HIV-1 RT directly.| Reagents | Concentration | Volume | Final concentration |
|---|---|---|---|
| Azide-unit | 35 mM DMSO | 20 μL | 7 mM |
| Alkyne-unit | 25 mM DMSO | 20 μL | 5 mM |
| TBTA | 10 mM DMSO | 10 μL | 20 mol% |
| CuSO4·5H2O | 4 mM MilliQ | 25 μL | 20 mol% |
| Sodium ascorbate | 20 mM MilliQ | 25 μL | 100 mol% |
| Total volume | 100 μL per well |
2.3 In situ screening against the RT
To rapidly identify potent RT inhibitors, all library compounds were subjected to enzymatic assays against HIV-1 RT. 0.66 mM was finally chosen as the inhibitor concentration after the pre-test. The biological results are demonstrated as the inhibition ratio in Fig. 3. L-737,126 was selected as the positive control. Blank controls were set up to eliminate the influence of DMSO, TBTA, CuSO4·5H2O and sodium ascorbate on reverse transcriptase activity experiments. As indicated, C1NX sublibraries showed significant inhibitory activity against HIV-1 RT, with some compounds exceeding the positive control. The percentages of inhibition of all of the compounds of sublibraries C2NX to RT were lower than L-737,126, indicating their weaker inhibitory activities. In addition, the overall reverse transcriptase inhibition rate of the C2 series is lower than that of the C1 series, possibly due to steric hindrance caused by the dimethyl spatial group in the linker impeding the binding between introduced substituents and the entrance channel in the NNIBP. Compounds with an inhibition rate of over 85% (C1N2, C1N4, C1N5, C1N15, C1N16, C1N22, C1N25, C1N26, C1N27, C1N37, C1N38, C1N39, C1N40) were selected for subsequent synthesis and activity screening.2.4 Anti-HIV biological evaluation
13 compounds in the HIV-1 RT inhibition assay were subsequently synthesized and evaluated for their activity against the WT HIV-1 strain (IIIB) and HIV-2 strain (ROD) in MT-4 cell cultures using the MTT method, respectively. What's more, the test compounds were also assayed against a panel of frequently encountered single (L100I, K103N, Y181C, Y188L, E138K) or double (F227L/V106A, K103N/Y181C (RES056)) mutant HIV-1 strains. The FDA-approved drugs nevirapine (NVP) and etravirine (ETV) were used as reference drugs. EC50 values (anti-HIV activity) and CC50 values (cytotoxicity) are illustrated in Table 2.| Compd. | Structure | EC50a | CC50b (μM) | SIc | |
|---|---|---|---|---|---|
| IIIB (μM) | ROD (μM) | IIIB | |||
| a EC50: concentration of the compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method. b CC50: concentration required to reduce the viability of mock-infected cell cultures by 50%, as determined by the MTT method. c SI: selectivity index, the ratio of CC50/EC50. | |||||
| C1N4 | 2-NO2–Bn | 0.024 ± 0.0065 | >215.88 | >215.88 | >9115 |
| C1N5 | 3-NO2–Bn | 0.23 ± 0.034 | >215.88 | >215.88 | >939 |
| C1N16 | (CH2)2Ph | 0.15 ± 0.052 | >4.69 | 161.65 ± 16.73 | 1080 |
| C1N22 | CH2S–Ph | 0.040 ± 0.0099 | >220.81 | >220.81 | >5507 |
| C1N25 | CH2CONH–4-CF3–Ph | 0.11 ± 0.038 | >16.99 | 16.99 ± 2.89 | 148 |
| C1N26 | CH2CONH–4-Br–Ph | 0.068 ± 0.018 | >15.77 | 15.77 ± 4.81 | 231 |
| C1N27 | CH2CONH–4-F–Ph | 0.087 ± 0.066 | >16.24 | 16.24 ± 3.55 | 188 |
| C1N37 | CH2COOEt | 0.032 ± 0.0034 | 225.55 ± 4.86 | >235.85 | >7328 |
| C1N38 | CH2COOCH3 | 0.045 ± 0.016 | >6.46 | 6.46 ± 0.70 | 143 |
| C1N39 | (CH2)3COOCH3 | 0.035 ± 0.0072 | >205.60 | 205.60 ± 15.96 | 5808 |
| C1N40 | 1-Naphthyl | 0.061 ± 0.018 | >214.01 | >214.01 s | >3513 |
| C1N2 | 3-CN–Bn | 0.062 ± 0.033 | >166.13 | 166.13 ± 39.16 | 2680 |
| C1N15 | (CH2)3Ph | 0.39 ± 0.16 | >116.89 | 116.89 ± 31.19 | 302 |
| L-737,126 |

|
0.0028 ± 0.00056 | >7.08 | 7.08 ± 1.38 | 2568 |
| Lamivudine | — | 5.02 ± 1.93 | — | >87.24 | >17 |
| Zidovudine | — | 0.019 ± 0.006 | — | >7.48 | >397 |
| Nevirapine | — | 0.16 ± 0.10 | — | >15.02 | >92 |
| Etravirine | — | 0.003 ± 0.001 | — | >6.33 | >1638 |
The biological testing results clearly demonstrated that all novel IASs displayed moderate to excellent potency against the wild-type HIV-1, with EC50 values ranging from 0.02 to 0.39 μM. Totally, 11 compounds were much better than that of NVP. Among them, compound C1N4 (EC50 = 0.024 μM) showed the best anti-HIV-1 IIIB activity with low cytotoxicity (CC50 > 215.88 μM).
An analysis of the aforementioned data allows us to delineate preliminary structure–activity relationships (SARs) with greater precision and academic rigor:
These refined SAR observations provide a more nuanced understanding of the molecular features that contribute to potent and selective inhibition of HIV-1 reverse transcriptase. They lay a solid foundation for the rational design of subsequent generations of NNRTIs with improved therapeutic potential.
These compounds were then evaluated for their inhibitory activities against a panel of frequently encountered single (L100I, K103N, Y181C, Y188L, E138K) or double (F227L/V106A, K103N/Y181C (RES056)) mutant HIV-1 strains. The results (Table 3) showed that all compounds exhibited moderate inhibitory activity against HIV-1 mutant strains, with inhibition levels ranging from submicromolar to micromolar. C1N25 exhibited a higher activity towards the Y188L mutant than L-737,126, indicating that the introduction of the trifluoromethyl group might form extra interactions with Y188L-RT.
| Compd. | EC50a (μM) | ||||||
|---|---|---|---|---|---|---|---|
| L100I | K103N | Y181C | Y188L | E138K | F227L + V106A | RES056 | |
| a EC50: concentration of the compound required to achieve 50% protection of MT-4 cell cultures against HIV-1-induced cytotoxicity, as determined by the MTT method. | |||||||
| C1N2 | 0.34 ± 0.03 | 1.24 ± 0.46 | 1.10 ± 0.38 | >35.33 | 0.35 ± 0.11 | 2.96 ± 0.98 | >166.13 |
| C1N4 | 0.15 ± 0.06 | 0.51 ± 0.21 | 0.29 ± 0.07 | 20.83 ± 10.83 | 0.15 ± 0.04 | 1.56 ± 0.30 | >194.24 |
| C1N5 | 1.90 ± 0.46 | 2.84 ± 0.30 | 3.04 ± 0.98 | 151.88 ± 7.97 | 1.30 ± 0.29 | 9.41 ± 3.24 | >215.88 |
| C1N15 | 1.87 ± 0.21 | 3.25 ± 1.01 | 2.71 ± 0.89 | >116.89 | 1.39 ± 0.19 | 9.70 ± 0.56 | >116.89 |
| C1N16 | 1.00 ± 0.68 | 0.54 ± 0.13 | 2.51 ± 0.54 | >161.65 | 0.87 ± 0.27 | 4.33 ± 0.72 | >4.69 |
| C1N22 | 0.15 ± 0.07 | 0.24 ± 0.10 | 0.67 ± 0.19 | 15.84 ± 7.02 | 0.18 ± 0.05 | 0.81 ± 0.12 | >220.81 |
| C1N25 | 0.59 ± 0.23 | 1.72 ± 0.70 | >7.41 | 0.40 ± 0.12 | 2.22 ± 0.38 | >16.99 | >16.99 |
| C1N26 | 0.38 ± 0.15 | 1.50 ± 0.42 | 0.54 ± 0.15 | 4.08 ± 1.47 | 0.15 ± 0.04 | 1.87 ± 0.34 | >15.77 |
| C1N27 | 0.53 ± 0.05 | 2.78 ± 0.66 | 1.07 ± 0.19 | 14.10 ± 3.23 | 0.56 ± 0.23 | 5.09 ± 1.84 | >16.24 |
| C1N37 | 0.17 ± 0.05 | 0.34 ± 0.06 | 0.63 ± 0.16 | 33.41 ± 7.84 | 0.10 ± 0.02 | 1.70 ± 0.45 | 204.41 ± 4.71 |
| C1N38 | 0.22 ± 0.06 | 0.14 ± 0.05 | 0.80 ± 0.16 | 11.67 ± 1.78 | 0.17 ± 0.05 | 1.07 ± 0.46 | >6.46 |
| C1N39 | 0.27 ± 0.15 | 0.51 ± 0.11 | 0.58 ± 0.11 | 9.04 ± 1.55 | 0.05 ± 0.01 | 1.66 ± 0.77 | >205.60 |
| C1N40 | 0.30 ± 0.06 | 0.70 ± 0.21 | 0.62 ± 0.11 | 24.97 ± 4.88 | 0.24 ± 0.10 | 1.99 ± 0.23 | 113.03 ± 22.93 |
| L-737,126 | 0.01 ± 0.00 | 0.02 ± 0.01 | 0.03 ± 0.01 | 1.17 ± 0.36 | 0.01 ± 0.00 | 0.08 ± 0.06 | >7.08 |
| Lamivudine | 1.62 ± 0.56 | 3.75 ± 1.13 | 4.79 ± 1.73 | 3.56 ± 0.93 | 6.64 ± 1.83 | 2.77 ± 0.75 | 8.35 ± 2.88 |
| Zidovudine | 0.006 ± 0.002 | 0.019 ± 0.005 | 0.016 ± 0.005 | 0.009 ± 0.003 | 0.027 ± 0.010 | 0.009 ± 0.003 | 0.002 ± 0.001 |
| Nevirapine | 1.03 ± 0.54 | 5.10 ± 1.00 | 6.69 ± 1.89 | ≥7.70 | 0.19 ± 0.06 | ≥5.41 | >15.02 |
| Etravirine | 0.005 ± 0.002 | 0.003 ± 0.001 | 0.014 ± 0.003 | 0.014 ± 0.005 | 0.007 ± 0.003 | 0.018 ± 0.005 | 0.038 ± 0.021 |
2.5 Molecular modeling analysis
With the aim of obtaining insight into the allosteric binding of the novel designed derivatives to the NNIBP, representative compounds C1N4 and C1N39 were selected for molecular docking studies using software Surflex-Dock module Sybyl-X 2.1, to predict the mode of action in wild-type HIV-1RT (PDB code: 2FR2) and the docking results were shown using PyMOL 1.5.As depicted in Fig. 4, both compounds C1N4 and C1N39 exhibit the archetypal horseshoe conformation within the non-nucleoside reverse transcriptase inhibitor binding pocket (NNIBP). This conformation is stabilized by an intramolecular hydrogen bond between the sulfone group and the amide nitrogen, which is essential for their biological activity. These compounds also engage in the canonical interactions characteristic of indolylarylsulfone (IAS) analogs with the reverse transcriptase enzyme. Specifically, the 3,5-dimethylbenzene moiety partakes in π–π stacking with the hydrophobic amino acids Tyr181, Tyr188, Phe227, and Trp229, while the indole parent ring nitrogen forms hydrogen bonds with Lys101. Moreover, the newly introduced triazolyl substituent adeptly occupies the tolerant region II of the NNIBP, enhancing the interaction profile of these compounds.
 | ||
| Fig. 4 Predicted binding modes of C1N4 (blue, left) and C1N39 (blue, right) in the HIV-1 RT NNIBP (PDB code: 2RF2). Amino acid residues important for ligand binding are represented in lines. H-bond interactions are indicated by red dashed lines. Hydrogens (nonpolar) are not shown. | ||
For compound C1N4, the carbonyl group of the amide forms an additional hydrogen bond with Lys101, and the terminal nitro group engages in bidentate hydrogen bonding with the conserved amino acids Ile180 and Arg172. These interactions significantly augment the affinity of C1N4 for the reverse transcriptase. In the case of C1N39, the carbonyl group of the amide is predicted to form an extra hydrogen bond with Lys103. The molecular docking simulations have been instrumental in elucidating the binding modes of these novel compounds, providing valuable insights that will inform the structure-based design of the next generation of potent NNRTIs.
It's worth mentioning that analogue C1N25, with the trifluoromethyl group, showed detrimental activity against the WT HIV-1 compared to the Br- and F- analogues (C1N26 and C1N27). However, C1N25 exhibited superior activity against the Y188L mutated strains. The free energy perturbation (FEP) calculation was performed for investigation. Firstly, three analogues were docked into the WT and Y188L mutated RT structure models, respectively. Then, the relative binding energy differences of these analogues were estimated. As illustrated in Table 4, the Br- and F-analogues (C1N26 and C1N27) showed favorable binding affinity compared to the CF3-analogue (C1N25) against the WT HIV-1, while the calculation showed the distinct result for the Y188L mutated strains. This result is consistent with the antiviral activity result.
| Compd. | ΔΔG WT (kcal mol−1) | ΔΔG Y188L (kcal mol−1) |
|---|---|---|
| C1N25 | 0.0 | 0.0 |
| C1N26 | −0.7 ± 0.41 | 0.6 ± 0.42 |
| C1N27 | −0.5 ± 0.43 | 0.8 ± 0.41 |
The relative binding affinity differences (ΔΔG, kcal mol−1) of C1N25, C1N26, and C1N27 were estimated using FEP. The binding affinity of C1N25 was set as the reference (ΔG = 0), and the difference between C1N26/C1N27 and C1N25 was calculated and the negative value means the superior binding affinity and the positive values suggests the weaker binding affinity.
3. Conclusion
In conclusion, this study reports the identification of highly potent HIV-1 indolylarylsulfone NNRTIs through the use of miniaturized CuAAC click-chemistry-based combinatorial libraries. 13 hits emerging from the enzyme-inhibitory screening were synthesized and evaluated for their anti-HIV-1 activity in vitro, which showed strong inhibitory activity against wild strains of HIV-1, with EC50 values in the range of 0.024–0.23 μM, most of which were much better than the positive control nevirapine (EC50 = 0.16 μM). Among them, C1N4 (EC50 = 0.024 μM) showed the best anti-HIV-1 IIIB activity with low cytotoxicity, high selectivity and safety (CC50 > 215.88 μM). Some of the compounds also showed high inhibitory activity against HIV-1 single mutant strains such as Y188L, E138K, etc., which is valuable for further research and development. Additionally, the binding dynamics are often altered by these multi-site occupying molecules,21 representing a pivotal area of focus for future research endeavors. All in all, the findings from this research not only validate the utility of click chemistry in the rapid discovery of potential therapeutic agents but also highlight the importance of structural innovation in improving drug efficacy and overcoming resistance challenges. The introduction of the 1,2,3-triazole ring has emerged as a key structural element, offering a platform for further optimization and the exploration of its role in enhancing inhibitory activity.4. Experimental section
4.1 Synthetic procedures and analytical data
Mass spectrometry was performed on an API 4000 triple quadrupole mass spectrometer (Applied Biosystems/MDS Sciex, Concord, ON, Canada). 1H NMR and 13C NMR spectra were recorded on a Bruker AV-400 spectrometer (Bruker BioSpin, Switzerland), using solvents as indicated. Chemical shifts were reported in δ values (ppm) with tetramethylsilane as the internal reference, and J values were reported in hertz (Hz). Melting points (mp) were determined on a micromelting point apparatus (Tian Jin Analytical Instrument Factory, Nankai, Tianjin, China). Flash column chromatography was performed on columns packed with silica gel 60 (200–300 mesh) (Qingdao waves silica gel desiccant Co., Ltd, Qingdao, China). Thin layer chromatography was performed on pre-coated HUANGHAI® HSGF254, 0.15–0.2 mm TLC-plates (Yantai Jiangyou Silica Gel Development Co., Ltd., Yantai, Shandong, China).5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-(prop-2-yn-1-yl)-1H-indole-2-carboxamide (C1). White solid, yield: 52%. 1H NMR (400 MHz, DMSO-d6) δ 13.08 (s, 1H), 9.34 (t, J = 5.4 Hz, 1H), 7.98 (d, J = 2.1 Hz, 1H), 7.65 (s, 2H), 7.54 (d, J = 8.7 Hz, 1H), 7.35 (dd, J = 8.7, 2.1 Hz, 1H), 7.27 (s, 1H), 4.21 (dd, J = 5.4, 2.6 Hz, 2H), 2.32 (s, 6H).
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-(2-methylbut-3-yn-2-yl)-1H-indole-2-carboxamide (C2). White solid, yield: 60%. 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H), 9.13 (s, 1H), 8.00 (d, J = 2.0 Hz, 1H), 7.62 (s, 2H), 7.53 (d, J = 8.7 Hz, 1H), 7.36 (dd, J = 8.8, 2.1 Hz, 1H), 7.28 (s, 1H), 3.35 (s, 1H), 2.32 (s, 6H), 1.68 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 158.46, 142.94, 139.57, 137.23, 135.28, 133.22, 127.81, 125.73, 125.29, 123.99, 119.49, 115.35, 111.91, 87.42, 72.48, 47.76, 28.96, 21.24.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 10 mL). Then a solution of 1 M freshly prepared sodiumascorbate (0.195 mmol, 0.04 g) and 7.5% CuSO4·5H2O (0.065 mmol, 0.017 g) in water were added to the mixture. The heterogeneous mixture was stirred vigorously at 50 °C for 4–12 h (monitored by TLC). Then 20 mL water was added to the mixture and extracted with ethyl acetate (3 × 10 mL). The organic phase was dried over anhydrous Na2SO4, filtered, and purified by flash column chromatography. The product was recrystallized from ethyl acetate/petroleum ether to afford the target compound CXNX.
1, 10 mL). Then a solution of 1 M freshly prepared sodiumascorbate (0.195 mmol, 0.04 g) and 7.5% CuSO4·5H2O (0.065 mmol, 0.017 g) in water were added to the mixture. The heterogeneous mixture was stirred vigorously at 50 °C for 4–12 h (monitored by TLC). Then 20 mL water was added to the mixture and extracted with ethyl acetate (3 × 10 mL). The organic phase was dried over anhydrous Na2SO4, filtered, and purified by flash column chromatography. The product was recrystallized from ethyl acetate/petroleum ether to afford the target compound CXNX.
5-Chloro-N-((1-(3-cyanobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-3-((3,5-dimethylphenyl)sulfonyl)-1H-indole-2-carboxamide (C1N2). White solid, yield: 77%. mp: 198–200 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H), 9.87 (s, 1H), 9.46 (t, J = 5.5 Hz, 1H), 7.96 (s, 1H), 7.93 (d, J = 2.0 Hz, 1H), 7.66 (s, 2H), 7.56–7.52 (m, 5H), 7.42 (m, 1H), 7.25 (s, 1H), 5.36 (s, 2H), 4.37 (d, J = 5.6 Hz, 2H), 2.30 (s, 6H). HRMS: m/z C28H23ClN6O3S: calcd. 558.1241, found 557.1176 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(2-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N4). White solid, yield: 89%. mp: 225–227 °C. 1H NMR (400 MHz, DMSO-d6) δ13.08 (s, 1H), 9.53 (t, J = 5.6 Hz, 1H), 8.17 (s, 1H), 8.13 (dd, J = 8.0, 1.5 Hz, 1H), 7.94 (d, J = 2.0 Hz, 1H), 7.63 (m, 4H), 7.53 (d, J = 8.7 Hz, 1H), 7.35 (dd, J = 8.8, 2.1 Hz, 1H), 7.25 (s, 1H), 7.05 (dd, J = 7.5, 1.5 Hz, 1H), 6.00 (s, 2H), 4.66 (d, J = 5.6 Hz, 2H), 2.28 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 159.77, 147.94, 144.53, 142.81, 139.45, 137.14, 135.18, 134.75, 133.31, 131.43, 130.44, 130.06, 127.74, 125.53, 124.70, 124.22, 119.35, 115.38, 112.19, 50.43, 35.53, 21.18. HRMS: m/z C27H23ClN6O5S: calcd. 578.1139, found 577.1063 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(3-nitrobenzyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N5). White solid, yield: 91%. mp: 230–232 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.06 (s, 1H), 9.52 (d, J = 5.7 Hz, 1H), 8.25 (d, J = 12.4 Hz, 2H), 8.17 (d, J = 8.3 Hz, 1H), 7.95 (d, J = 6.9 Hz, 1H), 7.77 (d, J = 7.6 Hz, 1H), 7.64 (d, J = 7.9 Hz, 1H), 7.59 (s, 2H), 7.53 (d, J = 8.7 Hz, 1H), 7.35 (d, J = 8.9 Hz, 1H), 7.23 (s, 1H), 5.81 (s, 2H), 4.65 (d, J = 5.5 Hz, 2H), 2.89 (s, 1H), 2.73 (s, 1H), 2.26 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 162.76, 159.76, 148.26, 144.67, 142.78, 139.41, 138.53, 137.08, 135.23, 135.14, 133.29, 130.81, 127.74, 125.58, 125.23, 124.18, 124.13, 123.58, 123.39, 119.37, 115.37, 112.22, 52.22, 35.52, 21.16. HRMS: m/z C27H23ClN6O5S: calcd. 578.1139, found 577.1049 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(3-phenylpropyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N15). White solid, yield: 79%. mp: 145–147 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.07 (s, 1H), 9.51 (t, J = 5.5 Hz, 1H), 8.14 (s, 1H), 7.95 (d, J = 1.9 Hz, 1H), 7.64 (s, 2H), 7.55 (d, J = 8.8 Hz, 1H), 7.37–7.33 (m, 1H), 7.29 (d, J = 7.1 Hz, 2H), 7.20 (td, J = 7.9, 3.3 Hz, 4H), 4.65 (d, J = 5.4 Hz, 2H), 4.38 (q, J = 6.2, 5.5 Hz, 2H), 2.29 (s, 6H), 2.21–2.01 (m, 4H). HRMS: m/z C29H28ClN5O3S: calcd. 561.1601, found 560.1543 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-phenethyl-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N16). White solid, yield: 82%. mp: 255–257 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.06 (s, 1H), 9.49 (s, 1H), 8.06 (s, 1H), 7.96 (s, 1H), 7.64 (s, 1H), 7.54 (d, J = 8.7 Hz, 1H), 7.39–7.32 (m, 2H), 7.28–7.22 (m, 3H), 7.20 (d, J = 7.8 Hz, 3H), 4.68–4.54 (m, 4H), 3.15 (t, J = 7.4 Hz, 2H), 2.30 (s, 6H). HRMS: m/z C28H26ClN5O3S: Calcd. 547.1445, Found 546.1382 [M-H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-((phenylthio)methyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N22). White solid, yield: 78%. mp: 180–182 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.09 (s, 1H), 9.51 (t, J = 5.7 Hz, 1H), 8.11 (s, 1H), 7.96 (d, J = 2.0 Hz, 1H), 7.65 (s, 2H), 7.55 (d, J = 8.8 Hz, 1H), 7.42 (dd, J = 8.1, 1.4 Hz, 2H), 7.37–7.22 (m, 5H), 5.98 (s, 2H), 4.62 (d, J = 5.5 Hz, 2H), 2.29 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 159.72, 144.72, 142.84, 139.48, 135.20, 133.36, 133.03, 130.88, 129.71, 128.07, 127.75, 125.25, 124.22, 123.37, 119.40, 115.44, 112.21, 52.17, 35.51, 21.21. HRMS: m/z C27H24ClN5O3S2: calcd. 565.1009, found 564.0940 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(2-oxo-2-((4-(trifluoromethyl)phenyl)amino)ethyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N25). White solid, yield: 70%. mp: 220–222 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H), 10.87 (s, 1H), 9.58 (t, J = 5.6 Hz, 1H), 8.16 (s, 1H), 7.97 (d, J = 2.0 Hz, 1H), 7.80 (d, J = 8.5 Hz, 2H), 7.71 (d, J = 8.5 Hz, 2H), 7.66 (s, 2H), 7.55 (d, J = 8.7 Hz, 1H), 7.36 (dd, J = 8.7, 2.1 Hz, 1H), 7.26 (s, 1H), 5.42 (s, 2H), 4.69 (d, J = 5.6 Hz, 2H), 2.30 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 165.40, 159.69, 144.12, 142.80, 142.41, 139.51, 136.90, 135.23, 133.30, 127.79, 126.76, 125.67, 125.29, 124.21, 119.62, 119.42, 115.43, 112.22, 52.75, 35.55, 21.18. HRMS: m/z C29H24ClF3N6O4S: calcd. 644.1220, found 643.1146 [M − H]−.
N-((1-(2-((4-Bromophenyl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl)-5-chloro-3-((3,5-dimethylphenyl)sulfonyl)-1H-indole-2-carboxamide (C1N26). White solid, yield: 88%. mp: 231–233 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H), 10.63 (s, 1H), 9.57 (t, J = 5.6 Hz, 1H), 8.14 (s, 1H), 7.97 (d, J = 2.0 Hz, 1H), 7.66 (s, 2H), 7.57–7.50 (m, 5H), 7.36 (dd, J = 8.7, 2.1 Hz, 1H), 7.25 (s, 1H), 5.36 (s, 2H), 4.69 (d, J = 5.6 Hz, 2H), 2.30 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 164.87, 159.68, 144.09, 142.83, 139.50, 138.23, 136.90, 135.23, 133.32, 132.22, 127.79, 125.70, 125.29, 125.23, 124.22, 121.59, 119.43, 115.87, 115.42, 112.23, 52.74, 35.56, 21.24, 21.19. HRMS: m/z C28H24BrClN6O4S: calcd. 654.0452, found 655.0363, 653.0398 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(2-((4-fluorophenyl)amino)-2-oxoethyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N27). White solid, yield: 93%. 1H NMR (400 MHz, DMSO-d6) δ 13.10 (s, 1H), 10.55 (s, 1H), 9.56 (s, 1H), 8.14 (s, 1H), 7.97 (d, J = 1.8 Hz, 1H), 7.66 (s, 2H), 7.63–7.45 (m, 3H), 7.40–7.33 (m, 1H), 7.27 (s, 1H), 7.23–7.13 (m, 2H), 5.35 (s, 2H), 2.39 (s, 2H), 2.30 (s, 6H). HRMS: m/z C28H24ClFN6O4S: calcd. 594.1252, found 593.1186 [M − H]−.
Ethyl-2-(4-((5-chloro-3-((3,5-dimethylphenyl)sulfonyl)-1H-indole-2-carboxamido)methyl)-1H-1,2,3-triazol-1-yl) acetate (C1N37). White solid, yield: 85%. mp: 204–207 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.11 (s, 1H), 9.56 (s, 1H), 8.15 (s, 1H), 7.96 (s, 1H), 7.66 (s, 2H), 7.54 (d, J = 8.5 Hz, 1H), 7.36 (d, J = 8.4 Hz, 1H), 7.26 (s, 1H), 5.43 (s, 2H), 4.67 (s, 2H), 4.18 (q, J = 6.9 Hz, 2H), 2.30 (s, 6H), 1.22 (t, J = 6.8 Hz, 3H). 13C NMR (100 MHz, DMSO-d6) δ 167.66, 159.65, 142.84, 139.50, 136.86, 135.23, 133.31, 127.78, 125.69, 125.30, 124.20, 119.43, 115.43, 61.96, 50.93, 35.53, 21.19, 14.45. HRMS: m/z C24H24ClN5O5S: calcd. 529.1187, found 528.1118 [M − H]−.
Methyl-2-(4-((5-chloro-3-((3,5-dimethylphenyl)sulfonyl)-1H-indole-2-carboxamido)methyl)-1H-1,2,3-triazol-1-yl)acetate (C1N38). White solid, yield: 84%. mp: 236–238 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.09 (s, 1H), 9.54 (d, J = 5.8 Hz, 1H), 8.12 (s, 1H), 7.96 (s, 1H), 7.65 (s, 2H), 7.54 (d, J = 8.7 Hz, 1H), 7.35 (d, J = 8.6 Hz, 1H), 7.26 (s, 1H), 5.44 (s, 2H), 4.67 (d, J = 5.3 Hz, 2H), 3.71 (s, 3H), 2.30 (s, 6H). HRMS: m/z C23H22ClN5O5S: calcd. 515.1030, found 514.0957 [M − H]−.
Methyl-4-(4-((5-chloro-3-((3,5-dimethylphenyl)sulfonyl)-1H-indole-2-carboxamido)methyl)-1H-1,2,3-triazol-1-yl)butanoate (C1N39). White solid, yield: 89%. mp: 168–170 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.08 (s, 1H), 9.50 (s, 1H), 8.12 (s, 1H), 7.95 (s, 1H), 7.64 (s, 2H), 7.54 (d, J = 8.3 Hz, 1H), 7.35 (d, J = 8.4 Hz, 1H), 7.26 (s, 1H), 4.64 (s, 2H), 4.41 (t, J = 6.7 Hz, 2H), 3.57 (s, 2H), 2.31 (d, J = 11.4 Hz, 9H), 2.06 (t, J = 6.7 Hz, 2H). HRMS: m/z C25H26ClN5O5S: calcd. 543.1343, found 542.1278 [M − H]−.
5-Chloro-3-((3,5-dimethylphenyl)sulfonyl)-N-((1-(naphthalen-1-ylmethyl)-1H-1,2,3-triazol-4-yl)methyl)-1H-indole-2-carboxamide (C1N40). White solid, yield: 91%. mp: 140–142 °C. 1H NMR (400 MHz, DMSO-d6) δ 13.05 (s, 1H), 9.48 (t, J = 5.6 Hz, 1H), 8.24–8.19 (m, 1H), 8.12 (s, 1H), 7.95 (dt, J = 9.1, 7.3 Hz, 3H), 7.62 (s, 2H), 7.59–7.50 (m, 3H), 7.44 (dd, J = 13.2, 7.2 Hz, 2H), 7.34 (dd, J = 8.7, 2.1 Hz, 1H), 7.23 (s, 1H), 6.11 (s, 2H), 4.61 (d, J = 5.5 Hz, 2H), 2.25 (s, 6H). 13C NMR (100 MHz, DMSO-d6) δ 159.71, 142.82, 139.43, 137.04, 135.15, 133.29, 131.97, 131.10, 129.47, 129.10, 127.74, 127.68, 127.24, 126.61, 125.96, 125.22, 124.21, 123.90, 123.77, 119.38, 115.37, 51.26, 35.58, 21.16. HRMS: m/z C31H26ClN5O3S: calcd. 583.1445, found 582.1378 [M − H]−.
4.2 In vitro anti-HIV assay and HIV-1 RT inhibition assay
Evaluation of the antiviral activity of the compounds against HIV in MT-4 cells (the HTLV-I infected MT-4 cell line was established as described by Miyoshi et al. 1981 (ref. 22) and was kindly provided by Dr. N. Yamamoto, Yamaguchi University, Yamaguchi, Japan) was performed using the MTT assay as previously described.23,24 A reverse transcriptase (RT) assay kit produced by Roche was selected for the RT inhibition assay, and the ELISA procedure for the RT inhibition assay was carried out following the description in the kit protocol.25,264.3 Molecular simulation experiment
The molecules for docking were optimized for 2000 generations until the maximum derivative of energy became 0.005 kcal mol−1 A−1, using the Tripos force field. Charges were computed and added according to Gasteiger–Hückel parameters. The published three dimensional crystal structures of RT complexes (PDB code: 2RF2)11 were retrieved from the Protein Data Bank and were used for the docking experiment by means of the surflex-docking module of Sybyl-X 2.1. The protein was prepared by using the biopolymer application accompanying Sybyl: the bound ligand was extracted from the complexes, water molecules were removed, hydrogen atoms were added, side chain amides and side chains bumps were fixed, and charges and atom types were assigned according to AMBER 99. After the protomol was generated, the optimized molecules were surflex-docked into the binding pocket of NNRTIs, with the relevant parameters set as defaults. The top-scoring pose was shown using the software PyMOL version 1.5 http://www.pymol.org). The secondary structure of the RT is shown in cartoons, and only the key residues for interactions with the inhibitor were shown in sticks and labeled. The potential hydrogen bonds were presented by dashed lines.The FEP calculations were carried out using the ligand FEP workflow of the academic Desmond module. The docking poses were used as the input structures. The calculation was performed using the OPLS_2005 force field and the other parameters were set as default.
Data availability
The data that support the findings of this study are available on request from the first author, [Ping Gao, E-mail: gaoping_candice@163.com], upon reasonable request.Author contributions
Ping Gao: synthesis, in situ screening against the RT, molecular modeling analysis, writing – original draft. Shu Song: synthesis, in situ screening against the RT. Christophe Pannecouque and Erik De Clercq: anti-HIV biological evaluation. Peng Zhan and Xinyong Liu: design, guidance, writing – review & editing.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We gratefully acknowledge financial support from the Key Research and Development Program, Ministry of Science and Technology of the People's Republic of China (Grant No. 2023YFC2606500; 2023YFE0206500), the Shandong Laboratory Program (SYS202205) and the Natural Science Foundation of Shandong Province (No. ZR2021QH324).References
- A. Mody, A. H. Sohn, C. Iwuji, R. K. J. Tan, F. Venter and E. H. Geng, HIV epidemiology, prevention, treatment, and implementation strategies for public health, Lancet, 2024, 403(10425), 471–492 CrossRef.
- https://www.who.int/data/gho/data/themes/theme-details/GHO/hiv-aids .
- S. I. Gubernick, N. Félix, D. Lee, J. J. Xu and B. Hamad, The HIV therapy market, Nat. Rev. Drug Discovery, 2016, 15(7), 451–452 CrossRef CAS PubMed.
- M. S. Cohen, Y. Q. Chen, M. McCauley, T. Gamble, M. C. Hosseinipour, N. Kumarasamy, J. G. Hakim, J. Kumwenda, B. Grinsztejn, J. H. Pilotto, S. V. Godbole, S. Mehendale, S. Chariyalertsak, B. R. Santos, K. H. Mayer, I. F. Hoffman, S. H. Eshleman, E. Piwowar-Manning, L. Wang, J. Makhema, L. A. Mills, G. de Bruyn, I. Sanne, J. Eron, J. Gallant, D. Havlir, S. Swindells, H. Ribaudo, V. Elharrar, D. Burns, T. E. Taha, K. Nielsen-Saines, D. Celentano, M. Essex and T. R. Fleming, HPTN 052 Study Team. Prevention of HIV-1 infection with early antiretroviral therapy, N. Engl. J. Med., 2011, 365(6), 493–505 CrossRef CAS PubMed.
- P. Zhan, C. Pannecouque, E. De Clercq and X. Liu, Anti-HIV drug discovery and development: current innovations and future trends, J. Med. Chem., 2016, 59(7), 2849–2878 CrossRef CAS.
- M. E. Cilento, K. A. Kirby and S. G. Sarafianos, Avoiding drug resistance in HIV reverse transcriptase, Chem. Rev., 2021, 121, 3271e96 CrossRef.
- A. M. Benedicto and Fuster-Martı'nez I, Tosca J, Esplugues JV, Blas-Garcı'a A, Apostolova N., NNRTI and liver damage: evidence of their association and the mechanisms involved, Cell, 2021, 10, 1687 CrossRef CAS.
- T. M. Williams, T. M. Ciccarone, S. C. MacTough, C. S. Rooney, S. K. Balani, J. H. Condra, E. A. Emini, M. E. Goldman, W. J. Greenlee and L. R. Kauffman, et al., 5-chloro-3-(phenylsulfonyl)indole-2-carboxamide: a novel, non-nucleoside inhibitor of HIV-1 reverse transcriptase, J. Med. Chem., 1993, 36(9), 1291–1294 CrossRef CAS.
- V. Famiglini and R. Silvestri, Indolylarylsulfones, a fascinating story of highly potent human immunodeficiency virus type 1 non-nucleoside reverse transcriptase inhibitors, Antiviral Chem. Chemother., 2018, 26, 2040206617753443 CrossRef CAS PubMed.
- R. Silvestri, G. De Martino, G. La Regina, M. Artico, S. Massa, L. Vargiu, M. Mura, A. G. Loi, T. Marceddu and P. La Colla, Novel indolyl aryl sulfones active against HIV-1 carrying NNRTI resistance mutations: synthesis and SAR studies, J. Med. Chem., 2003, 46, 2482e93 Search PubMed.
- Z. Zhao, S. E. Wolkenberg, P. E. Sanderson, M. Lu, V. Munshi, G. Moyer, M. Feng, A. V. Carella, L. T. Ecto, L. J. Gabryelski, M. T. Lai, S. G. Prasad, Y. Yan, G. B. McGaughey, M. D. Miller, C. W. Lindsley, G. D. Hartman, J. P. Vacca and T. M. Williams, Novel indole-3-sulfonamides as potent HIV non-nucleoside reverse transcriptase inhibitors (NNRTIs), Bioorg. Med. Chem. Lett., 2008, 18(2), 554–559 CrossRef CAS PubMed.
- X. Li, P. Gao, B. Huang, Z. Zhou, Z. Yu, Z. Yuan, H. Liu, C. Pannecouque, D. Daelemans, E. De Clercq, P. Zhan and X. Liu, Discovery of novel piperidine-substituted indolylarylsulfones as potent HIV NNRTIs via structure-guided scaffold morphing and fragment rearrangement, Eur. J. Med. Chem., 2017, 126, 190–201 CrossRef CAS.
- T. Zhao, Q. Meng, D. Kang, J. Ji, E. De Clercq, C. Pannecouque, X. Liu and P. Zhan, Discovery of novel indolylarylsulfones as potent HIV-1 NNRTIs via structure-guided scaffold morphing, Eur. J. Med. Chem., 2019, 182, 111619 CrossRef CAS.
- S. Gao, Y. Cheng, S. Song, L. Song, F. Zhao, S. Xu, D. Kang, L. Sun, P. Gao, E. De Clercq, C. Pannecouque, X. Liu and P. Zhan, Chemical space exploration around indolylarylsulfone scaffold led to a novel class of highly active HIV-1 NNRTIs with spiro structural features, Eur. J. Med. Chem., 2022, 238, 114471 CrossRef CAS PubMed.
- S. Gao, L. Song, Y. Cheng, F. Zhao, D. Kang, S. Song, M. Yang, B. Ye, W. Zhao, Y. Tang, E. De Clercq, C. Pannecouque, P. Zhan and X. Liu, Discovery of novel sulfonamide substituted indolylarylsulfones as potent HIV-1 inhibitors with better safety profiles, Acta Pharm. Sin. B, 2023, 13(6), 2747–2764 CrossRef CAS.
- X. Wang, B. Huang, X. Liu and P. Zhan, Discovery of bioactive molecules from CuAAC click-chemistry-based combinatorial libraries, Drug Discovery Today, 2016, 21(1), 118–132 CrossRef CAS.
- P. Gao, L. Sun, J. Zhou, X. Li, P. Zhan and X. Liu, Discovery of novel anti-HIV agents via Cu(I)-catalyzed azide-alkyne cycloaddition (CuAAC) click chemistry-based approach, Expert Opin. Drug Discovery, 2016, 11(9), 857–871 CrossRef CAS.
- D. Kang, D. Feng, L. Jing, Y. Sun, F. Wei, X. Jiang, G. Wu, E. De Clercq, C. Pannecouque, P. Zhan and X. Liu, In situ click chemistry-based rapid discovery of novel HIV-1 NNRTIs by exploiting the hydrophobic channel and tolerant regions of NNIBP, Eur. J. Med. Chem., 2020, 193, 112237 CrossRef CAS.
- Y. Sun, D. Feng, Z. Zhou, T. Zhang, E. De Clercq, C. Pannecouque, D. Kang, P. Zhan and X. Liu, In situ click chemistry-based discovery of 1,2,3-triazole-derived diarylpyrimidines as novel HIV-1 NNRTIs by exploiting the tolerant region I in binding pocket, Bioorg. Med. Chem., 2023, 96, 117484 CrossRef CAS PubMed.
- T. R. Chan, R. Hilgraf, K. B. Sharpless and V. V. Fokin, Polytriazoles as copper(I)-stabilizing ligands in catalysis, Org. Lett., 2004, 6(17), 2853–2855 CrossRef CAS.
- Y. Itoh, P. Zhan, T. Tojo, P. Jaikhan, Y. Ota, M. Suzuki, Y. Li, Z. Hui, Y. Moriyama, Y. Takada, Y. Yamashita, M. Oba, S. Uchida, M. Masuda, S. Ito, Y. Sowa, T. Sakai and T. Suzuki, Discovery of selective histone deacetylase 1 and 2 inhibitors: screening of a focused library constructed by click chemistry, kinetic binding analysis, and biological evaluation, J. Med. Chem., 2023, 66(22), 15171–15188 CrossRef CAS.
- I. Miyoshi, I. Kubonishi, S. Yoshimoto, T. Akagi, Y. Ohtsuki, Y. Shiraishi, K. Nagata and Y. Hinuma, Type-C virus particles in a cord T-cell line derived by co-cultivating normal human cord leukocytes and human leukaemic T-cells, Nature, 1981, 294, 770–771 CrossRef CAS.
- R. Pauwels, J. Balzarini, M. Baba, R. Snoeck, D. Schols, P. Herdewijn, J. Desmyter and E. De Clercq, Rapid and automated tetrazolium-based colorimetric assay for the detection of anti-HIV compounds, J. Virol. Methods, 1988, 20(4), 309–321 CrossRef CAS PubMed.
- C. Pannecouque, D. Daelemans and E. De Clercq, Tetrazolium-based colorimetric assay for the detection of HIV replication inhibitors: revisited 20 years later, Nat. Protoc., 2008, 3(3), 427–434 CrossRef CAS.
- Reverse Transcriptase Assay, Colorimetric kit, SandhoferStrasse 116, D-68305 Mannheim, Germany, Roche Diagnostics GmbH, Roche, Applied Science Search PubMed.
- K. Suzuki, B. P. Craddock, N. Okamoto, T. Kano and R. T. Steigbigel, Poly A-linked colorimetric microtiter plate assay for HIV reverse transcriptase, J. Virol. Methods, 1993, 44(2–3), 189–198 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2025 |