Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Improving the colloidal stability of protein@ZIF-8 nanoparticles in biologically relevant buffers†
Justin
Van Houten‡
,
Ruben Castillo
Barberi‡
,
Jared
King
and
Alana F.
Ogata
 *
*
Department of Chemistry, University of Toronto, UTM 1867 Inner Circle Road, Mississauga, ON L5L 1C6, Canada. E-mail: Alana.ogata@utoronto.ca
First published on 10th June 2024
Abstract
ZIF-8 is one of the most common metal–organic frameworks used in protein encapsulation and is advantageous for the protection of protein function and delivery of protein cargo. However, protein@ZIF-8 composites typically degrade in aqueous solutions, which significantly limits their use in most biological applications that require stability in biologically relevant buffers. Here, we present a facile synthetic approach to produce reproducible and precise protein@ZIF-8 nanoparticles with bovine serum albumin (BSA) and demonstrate their colloidal stability in various buffers. We optimized the biomimetic mineralization of BSA-encapsulated ZIF-8 nanoparticles (BSA@c-ZIF-8) and demonstrated accurate synthesis of 107 ± 9 nm BSA@c-ZIF-8 nanoparticles for 36 independent batches. Dynamic light scattering and supplementary analytical techniques such as powder X-ray diffraction and scanning electron microscopy established BSA@c-ZIF-8's colloidal stability for 14 days in phosphate buffered saline, imidazole, and water. This stability enhancement is ascribed to the BSA-induced negative surface charge (−36 ± 10 mV), which repels chelating anions derived from buffers like phosphate buffered saline. To further improve the colloidal stability of BSA@c-ZIF-8 in buffers, we tested eight polymer coatings and show that poly(acrylic acid) (PAA), hexadecyltrimethylammonium bromide (CTAB), dextran and poly(vinyl alcohol) (PVA) improve colloidal stability and provide a route for further surface functionalization. This advancement can enable application of protein@ZIF-8 nanoparticles in analytical and biotechnological practices, overcoming previously described buffer instability.
Introduction
In recent years, zeolitic imidazolate framework 8 (ZIF-8) has garnered increasing attention for its ability to encapsulate proteins via biomimetic mineralization and produce colloidal protein@ZIF-8 nanoparticles for applications in therapeutics and biotechnologies.1–11 Protein encapsulation in ZIF-8 nanoparticles has been demonstrated to protect protein structure and function, and enhance protein activity.12–14 However, the application of protein@ZIF-8 materials in biological applications is limited by the degradation of ZIF-8 in aqueous buffered solutions.8,9,15 Vasquez-Hernandez et al. demonstrated that ZIF-8 degrades in phosphate buffered saline (PBS) due to the chelation of phosphate ions to Zn nodes ultimately causing the de-coordination of ZIF-8.8 Luzuriaga et al.'s work similarly demonstrates the instability of GFP@ZIF-8 in numerous buffer systems, including PBS, HCO3−, 2-[4-(2-hydroxyethyl)piperazin-1-yl]ethanesulfonic acid and Tris–HCl.9 Gao et al. showed pH-dependent degradation of protein@ZIF-8 nanoparticles and that the release behaviour of protein was significantly influenced by nanoparticle morphology.16 The buffer-induced degradation of ZIF-8 nanoparticles with encapsulated proteins is advantageous for drug delivery applications. However, this attribute poses a significant challenge in realizing protein@ZIF-8 nanomaterials in other biological applications where colloidal stability in aqueous solutions is necessary.17 Thus, the development of synthetic strategies to improve the stability of protein@ZIF-8 nanomaterials in relevant buffer systems is required.Previous studies on ZIF-8 without protein achieved improvement in stability in aqueous solutions using surfactants or capping agents. Chen et al., incubated ZIF-8 with various ligands/polymers and determined poly(acrylic acid) to be effective at limiting degradation in PBS and showed stability for 48 h. The authors ascribe the increase in stability to the COO− moieties on the polymer repelling the chelating PO43− ligands derived from the buffer.18 Other groups have found success in the stabilization of ZIF-8 structures with PEGs via in situ addition. Cui's group demonstrates the use of 8-arm PEGs for the mineralization of ZIF-8 and report good drug loading and stability in PBS and cell media.19–21 Tian and coworkers describe a similar improvement in stability with poly(dopamine) (PDA) coatings,22 which was attributed to a 18.7 nm thick layer of PDA on the surface of the ZIF-8, preventing recrystallization of the material under aqueous conditions.22 This was further demonstrated by Zhang et al., through the post synthetic modification of ZIF-8 with 5,6-dimethylbenzimidazole where steric hinderance provided by the bulkiness of this ligand over 2-methylimidazole (HmIm) improved the stability of ZIF-8.23 However, these strategies have not been used to improve the stability of protein@ZIF-8 nanomaterials.
Herein, we describe a synthetic approach for BSA@c-ZIF-8 nanoparticles that are 107 ± 9 nm in size and optimize their colloidal stability in biologically relevant buffers. BSA was used as an inexpensive model protein and is one of the most studied proteins in the biomimetic mineralization of protein@ZIF-8 materials.24,25 We evaluated the BSA@c-ZIF-8 with and without polymer coatings and assessed colloidal stability in water, phosphate buffered saline (PBS, pH 6.1–8.1), imidazole buffer (IM), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 2-morpholinoethanesulphonic acid (MES), and 3-(N-morpholino)propanesulfonic acid (MOPS) buffers. The temporal aggregation dynamics of BSA@c-ZIF-8 was assessed using dynamic light scattering (DLS).9,26,27 Orthogonal techniques, such as cryogenic electron microscopy (cryo-EM), Fourier transform infrared spectroscopy (FTIR), powder X-ray diffraction (pXRD), scanning electron microscopy (SEM) and ζ potential (ζp) were used to confirm ZIF-8 capping, distribution, and structure. From these studies, we produced BSA@c-ZIF-8 nanoparticles that show colloidal stability in water and PBS for 14 days and we provide synthetic routes for polymer-coated BSA@c-ZIF-8 that can serve as the foundation for customized functionalization.
Experimental section
Materials
MilliQ water (<18.2 Ω). For cell media studies the water used was 0.03 micron filtered, DNase, RNase and protease free, and purchased from Fisher Bioreagents. Zinc acetate dihydrate (99%) and 2-methylimidazole (99%) were purchased from Sigma Aldrich and used without further purification. Bovine serum albumin (BSA; lyophilized powder) was purchased from Sigma Aldrich. Dextran (40k Mn), polyethylenimine, branched (PEI; 10k Mn), poly(vinyl alcohol) (PVA; 13–23k Mn), poly(acrylic acid) (PAA; 30k Mn), hexadecyltrimethylammonium bromide (CTAB, ≥99.0%), 3,4-dihydroxyhydrocinnamic acid (CA, 98%), L-cystine (L-SH, ≥98.0%), and Tween-20 (viscous liquid) were purchased from Sigma Aldrich. Phosphate buffer saline (PBS), imidazole buffer (IM), 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) (acid free), 2-morpholinoethanesulphonic acid (MES) (acid free), and 3-(N-morpholino)propanesulfonic acid (MOPS) (acid free) were purchased from BioShop. HCl (conc., Fischer, ACS Plus grade) and NaOH (10 M, Sigma, >99%) were used for pH adjustments. For cell media studies Dulbecco's modified Eagle's medium (DMEM) and Grace's fortified media were purchased from ThermoFisher Scientific/Gibco. Lennox Broth (LB) and Gamborg's minimal organics powder were purchased from Sigma Aldrich.Methods
Synthesis of ZIF-8
Aqueous solutions of 4000 mM HmIm and 40 mM zinc acetate dihydrate (Zn(OAc)2) were prepared and were allowed to age for 15 minutes before use. Then, 10 mL of the 4000 mM HmIm solution was added to 10 mL of 40 mM Zn(OAc)2. The solution was allowed to react for 1 h before centrifugation at 5k rpm for 30 minutes, the supernatant was collected, and the pellet was discarded. The supernatant was then centrifuged at 14k rpm for 30 minutes. The pellet was re-dispersed in water and the second supernatant was discarded.Adsorption of BSA to ZIF-8
100 μL of the as-prepared ZIF-8 were incubated in 9.9 mL of 1 mg mL−1, 5 mg mL−1, and 10 mg mL−1 BSA and allowed to age for 1 day. Adsorption was confirmed using ζ potential.Synthesis of BSA@c-ZIF-8
Aqueous solutions of 1400 mM HmIm and 20 mM Zn(OAc)2 were prepared and were allowed to age for 15 minutes before use. Then, 10 mL of the 1400 mM HmIm solution was added to 200 mg of BSA slowly. The HmIm/BSA solution was allowed to age for approximately one hour before use. Then, 10 mL of Zn(OAc)2 solution was added to the HmIm/BSA solution for a final volume of 20 mL. The reaction was allowed to proceed for 16 h at room temperature, unstirred, before the product was isolated by centrifugation. First, the solution was centrifuged at 5k rpm for 30 minutes, the supernatant was collected, and then the pellet was discarded. The supernatant was then centrifuged at 14k rpm for 30 minutes. The pellet was redispersed in water and the second supernatant was discarded.Incubation of BSA@c-ZIF-8 in various cell media
1 mL of as-prepared BSA@c-ZIF-8 was added to 9 mL of cell media. The cell media used was either DMEM fortified with 10% FBS, LB broth, Gamborg's media or Grace's fortified media. The size and PDI were monitored over the course of 6 h, measured every 30 minutes.Synthesis of polymer-BSA@c-ZIF-8
100 μL of the prepared BSA@c-ZIF-8 solution was mixed with 900 μL of a surfactant solution in water, where the surfactant concentration was 1 mg mL−1 in all cases. This mixture is referred to as pre-wash polymer-BSA@c-ZIF-8. The samples were left to stand for 1 hour before use. To prepare the post-wash polymer-BSA@c-ZIF-8, the pre-wash mixture was centrifuged at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 minutes. After centrifugation, the supernatant was discarded and the remaining pellet was re-suspended in 1 mL of water.
000 rpm for 30 minutes. After centrifugation, the supernatant was discarded and the remaining pellet was re-suspended in 1 mL of water.
Incubation of ZIF-8, BSA adsorbed ZIF-8, BSA@c-ZIF-8, and polymer-BSA@c-ZIF-8 in buffers
100 μL of the prepared crystals was added to 9.9 mL of buffer and allowed to sit unstirred for 6 h–14 days. Buffers include 1× PBS (pH 6.1–8.1), 10 mM IM (pH 7.5), 10 mM HEPES (pH 7.4), 10 mM MES (pH 6.0), and 10 mM MOPS (pH 7.2).Powder X-ray diffraction measurements
20 mL of the as-prepared crystals were allowed to dry under vacuum for 4 hours. The sample was then powdered using a pestle and mortar. Approximately 15 mg of dry crystals were placed on an amorphous silicon wafer and X-ray diffractograms were acquired using a D2 phaser with a Cu K-α (1.5406 Å) X-ray source (Bruker, Germany).Fourier transform infrared spectroscopy
20 mL of as-prepared crystals were allowed to dry under vacuum for 4 hours. The sample was then powdered using a pestle and mortar. Approximately 1 mg of dry crystals was placed on the diamond and analysed. The FTIR used was a Nicolette IS-20 with ATR (ThermoFisher, USA).ζ potential measurements
Approximately 750 μL of the crystals in surfactant was placed in a DS1070 folded capillary ζ cell from Malvern Panalytical (Malvern Panalytical, USA). Each sample was allowed to equilibrate to 25 °C before measuring and measured thrice for three independent batches. All ζ potential measurements were acquired using a Zetasizer Nano series ZS with DTS 1070 disposable cells (Malvern Panalytical, USA).Dynamic light scattering measurements
The size and polydispersity index (PDI) of the crystals were monitored hourly and daily with DLS. Approximately 750 μL of the sample was placed in a quartz cuvette. All dynamic light scattering measurements were acquired using a Zetasizer Nano series ZS (Malvern Panalytical, USA).Scanning electron microscopy
Prepared crystals were diluted 1000-fold. Then, 40 μL of the diluted crystals was drop-casted onto gold coated glass (∼10 Å). The gold coated glass was adhered to an aluminium stub with carbon tape and carbon paint. The sample was allowed to dry on the glass and was coated with gold again (∼10 Å). The samples were imaged with an SU7000 scanning electron microscope from Hitachi (Japan). The working distance ranged from 2–6 mm. The accelerating voltage ranged from 1 kV to 15 kV.Cryogenic electron microscopy (CryoEM)
Prior to spotting the sample, the grids (Quantifoil R2,2 300 mesh, EMS) were charged for 30 seconds with a Pelco EasiGlow glow discharge cleaning system (Ted Pella). Then, 4 μL of sample was deposited onto the grid. Grids were blotted and plunge frozen using a Vitrobot IV (FEI, USA). The vitrobot temperature was set to 23 °C, humidity was set to 100%, blot time was set to 4 seconds, wait time was set to 5 seconds, blot force was set to 0 (AU), blot total was set to 1, and drain time was set to 0 seconds. Samples were loaded into the TEM using a Gatan 626 single tilt cryo-EM holder. All images were taken using a Talos L120C TEM (Thermo Scientific (USA)) equipped with a BM-Ceta metal-oxide semiconductor camera. Micrographs were collected at a defocus of −5.0 to −2.5 μm and dosing range of 2–4 electrons per Å2.Results and discussion
Biomimetic mineralization of protein@ZIF-8 is advantageous compared to post-synthetic modifications of ZIF-8 with proteins to enhance protein function and promote ZIF-8 crystallization. Here, we hypothesize that protein encapsulation in ZIF-8 by biomimetic mineralization is not only important for protein immobilization but can also be key in improving colloidal stability in biologically relevant buffers, compared to protein adsorbed to the surface of ZIF-8. We first assessed the colloidal stability of ZIF-8 nanoparticles without (hydrodynamic radius = 158 ± 5 nm, PDI = 0.11 ± 0.11) and with post-synthetic modification with BSA (hydrodynamic radius = 154 ± 2 nm, PDI = 0.16 ± 0.04, Fig. S1, ESI†). We assessed surface modification using ζ potential measurements and show that the surface charge of ZIF-8 nanoparticles changes from 39 ± 6 mV to −30 ± 6 mV upon incubation with 10 mg mL−1 of BSA, indicating that the BSA is adsorbed to the ZIF-8 surface (Fig. S3, ESI†). We then tested the stability of ZIF-8 with and without adsorbed BSA in PBS, IM, HEPES, MES and MOPS buffers over 24 hours (Fig. S4–S10, ESI†). ZIF-8 nanoparticles aggregate in all solutions except water and IM, resulting in micron-sized particles or PDI values above 0.2 (Fig. S4 and S5, ESI†). With adsorbed BSA, similar degradation behaviour is observed in all environments excluding water and PBS (pH 7.6) (Fig. S8 and S9, ESI†). In MES, PBS (pH 6.1), and PBS (pH 6.6), ZIF-8 nanoparticles show desorbed BSA, confirmed by a peak at Dh = 10 nm in the CONTIN plots (Fig. S10 and S11, ESI†). These results show that adsorption of BSA to the surface of ZIF-8 nanoparticles only improves colloidal stability in one of nine buffers, possibly due to the desorption of BSA, thus, supporting the need for encapsulated protein in ZIF-8.To optimize the biomimetic mineralization of colloidally stable ZIF-8 nanoparticles with encapsulated BSA, we tested various precursor and BSA concentrations based on previous reports,25 which show that BSA@ZIF8 materials can produce a range of polymorphs, sodalite (sod), diamondoid (dia), U13, ZIF-C, and amorphous (am).26,27 Our goal was to produce BSA@ZIF-8 nanoparticles with a sod crystal topology, due to its protective effects of encapsulated biomolecules.28 We also aimed to produce monodisperse particles with a polydispersity index below 0.2.
Synthesis with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 Zn
100 Zn![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) HmIm and 10 mg mL−1 BSA produced the am polymorph (Fig. S14, ESI†). Therefore, to obtain the sod BSA@ZIF-8 nanoparticles, we tested varying concentrations of the Zn(OAc)2, Hmlm precursors at 1 mg mL−1 BSA (Fig. S12 and Tables S1, S2, ESI†). A summary of the BSA@ZIF-8 materials produced is shown in Fig. S12 (ESI†). We found that at Zn(OAc)2 concentrations between 18–50 mM sod BSA@ZIF-8 was produced, while Zn(OAc)2 concentrations of 63 mM and 88 mM co-crystalize both sod and dia-ZIF-8 topologies. Varying the Hmlm concentration between 700–1225 mM produced sod BSA@ZIF-8, while HmIm concentrations below 700 mM produced dia BSA@ZIF-8 (Fig. S12i and j, ESI†).27 However, the sod BSA@ZIF-8 ratios synthesized produced PDI values above 0.2 (Tables S1 and S2, ESI†). We then tested BSA concentrations of 5 mg mL−1, 10 mg mL−1, and 20 mg mL−1 (Table 1 and Fig. S13, ESI†), which resulted in a final BSA@ZIF-8 with an optimal PDI of 0.06 ± 0.02 and size of 108 ± 2 nm with a coefficient of variation (CV) of 2% across 3 batches measured thrice (n = 9). This new material, BSA@c-ZIF-86 (hereafter, BSA@c-ZIF-8) was used for the remainder of the work.
HmIm and 10 mg mL−1 BSA produced the am polymorph (Fig. S14, ESI†). Therefore, to obtain the sod BSA@ZIF-8 nanoparticles, we tested varying concentrations of the Zn(OAc)2, Hmlm precursors at 1 mg mL−1 BSA (Fig. S12 and Tables S1, S2, ESI†). A summary of the BSA@ZIF-8 materials produced is shown in Fig. S12 (ESI†). We found that at Zn(OAc)2 concentrations between 18–50 mM sod BSA@ZIF-8 was produced, while Zn(OAc)2 concentrations of 63 mM and 88 mM co-crystalize both sod and dia-ZIF-8 topologies. Varying the Hmlm concentration between 700–1225 mM produced sod BSA@ZIF-8, while HmIm concentrations below 700 mM produced dia BSA@ZIF-8 (Fig. S12i and j, ESI†).27 However, the sod BSA@ZIF-8 ratios synthesized produced PDI values above 0.2 (Tables S1 and S2, ESI†). We then tested BSA concentrations of 5 mg mL−1, 10 mg mL−1, and 20 mg mL−1 (Table 1 and Fig. S13, ESI†), which resulted in a final BSA@ZIF-8 with an optimal PDI of 0.06 ± 0.02 and size of 108 ± 2 nm with a coefficient of variation (CV) of 2% across 3 batches measured thrice (n = 9). This new material, BSA@c-ZIF-86 (hereafter, BSA@c-ZIF-8) was used for the remainder of the work.
| Sample ID | Zn(OAc)2 (mM) | HMIM (mM) | BSA (mg mL−1) | Hydrodynamic radius (d nm) | Coefficient of variation HDR (CV%) | PDI | Coefficient of variation PDI (CV%) |
|---|---|---|---|---|---|---|---|
| a Did not redisperse. | |||||||
| BSA@c-ZIF-81a | 20 | 700 | 5 | N/A | N/A | N/A | N/A |
| BSA@c-ZIF-82 | 20 | 700 | 20 | 161 ± 21 | 13 | 0.30 ± 0.08 | 27 |
| BSA@c-ZIF-83 | 20 | 700 | 10 | 137 ± 7 | 5 | 0.15 ± 0.03 | 17 |
| BSA@c-ZIF-84 | 5 | 700 | 10 | 103 ± 8 | 8 | 0.13 ± 0.06 | 49 |
| BSA@c-ZIF-85 | 10 | 350 | 10 | 89 ± 43 | 48 | 0.40 ± 0.29 | 29 |
| BSA@c-ZIF-86 | 10 | 700 | 10 | 108 ± 2 | 2 | 0.06 ± 0.02 | 31 |
The size and morphology of BSA@c-ZIF-8 were characterized using SEM, which showed an even distribution of spherical nanoparticles (Fig. 1(a)), and were further validated using cryo-EM (Fig. 1(b)). We then tested the synthetic reproducibility of BSA@c-ZIF-8 across 36 batches using DLS and achieved a mean hydrodynamic radius of 107 ± 9 nm with a CV of 8% and a PDI of 0.08 ± 0.06 (Fig. 1(c)). Manual measurements of particle sizes from cryo-EM and SEM images confirmed sizes of 110 ± 30 nm (CV = 25%) and 110 ± 30 nm (CV = 22%), respectively (Fig. 1(d) and Fig. S15, ESI†). We note that in Fig. S12 (ESI†) the BSA@ZIF-8 structures with 1 mg mL−1 exhibit large dodecahedron structures, characteristic of sod-ZIF-8. However, at 10 mg mL−1 BSA concentrations, the dodecahedron morphology is lost (Fig. 1(a) and (b)). PXRD analysis of BSA@c-ZIF-8 confirmed characteristic peaks centred at 2θ = 7.3, 10.3, 12.7, 14.9, 17.7, 22.1, and 26.7 for ZIF-8 nanoparticles, corresponding to (011), (002), (112), (022), (222), (114), and (134) planes, aligning with previously published diffraction patterns for sod ZIF-8 (Fig. 1(e), and Fig. S16, ESI†).25 The presence of characteristic sod ZIF-8 diffraction patterns indicate that the crystal structure remains constant in BSA@ZIF-8 particles, while the morphology is spherical and uneven. We attribute these morphological changes to the incorporation of BSA in the crystallization of BSA@ZIF-8. High concentrations of BSA can show non-classical nucleation and growth via Zn2+ and BSA prenucleation complexes, rather than classical growth that results in facetted crystals.25,26 This can result in small particles that lack the visible dodecahedron topology.29,30 Previous reports also show that at high BSA concentrations a ZIF-8 core forms first followed by heterogenous nucleation of the BSA@ZIF-8 shell on the surface, resulting in a spherical or core–shell morphology.27,31 The surface charge of the BSA@c-ZIF-8 nanoparticles was −36 ± 10 mV, attributed to the presence of encapsulated BSA (Fig. 1(f)).32 FTIR confirmed the encapsulation of BSA within ZIF-8, as evidenced by a distinct amide shift at 1650 cm−1 and bands between 1300 and 1500 cm−1 that correspond to the imidazole ring structure, while stretches below 850 cm−1 represent the in and out of plane bending of the imidazole ring from the bidentate HmIm ligand (Fig. 1(g) and Fig. S17, ESI†).30,33,34
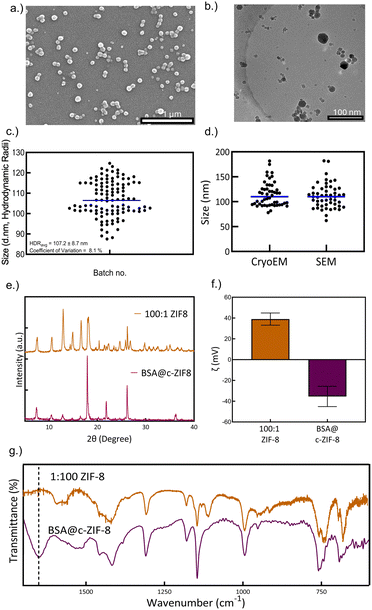 | ||
| Fig. 1 Synthesis and characterization of BSA@c-ZIF-8. (a) Scanning electron micrograph of BSA@c-ZIF-8, (b) cryogenic electron micrograph of BSA@c-ZIF-8, and (c) average size of the BSA@ZIF-8 particles by DLS. The average peak hydrodynamic radii and coefficient of variation of the hydrodynamic radii across 36 batches, each measured thrice, is provided in the inset. The error on the average size represents the standard deviation across 36 batches with 3 replicate measurements per batch. (d) The average size across 50 particles as manually measured on cryogenic and scanning electron micrographs. Measurements were completed with ImageJ scale-pixel matching. Fig. S15a and b (ESI†) show annotated SEM and Cryo-EM images, which the values were derived from. (e) The powder X-ray diffractograms of c-ZIF and BSA@c-ZIF-8 after drying. (f) The ζ potential of c-ZIF8 and BSA@c-ZIF-8, where the bars and error bars are the average and standard deviation of three independent batches, measured thrice (n = 9). (g) The FTIR spectrum of c-ZIF-8 and BSA@ZIF-8. The dotted line at the 1650 cm−1 peak denotes the characteristic proteinaceous peak. | ||
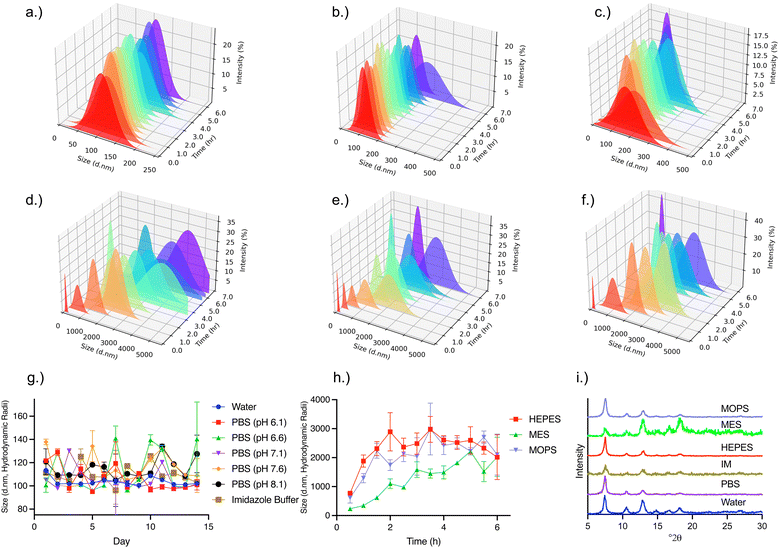
Previous reports of protein@ZIF-8 materials demonstrate instability in common buffers and water in 6 h.7,9,35–37 Here, the stability of the BSA@c-ZIF-8 in water, PBS, IM, HEPES, MES and MOPs buffers was assessed using DLS to monitor changes in the hydrodynamic radii over time (Fig. 2(a)–(f)). We established BSA@c-ZIF-8 colloidal stability based on the following parameters from DLS measurements:
1. The PDI remains monodisperse and unaggregated (<0.2 PDI).
2. The size distribution (from CONTIN plots of each sample must be monomodal, and show one particle population, absent of newly formed particles or precipitates.
3. The hydrodynamic radius has minimal shift (<100%) across the duration of the study.
As depicted in Fig. 2(a), the BSA@c-ZIF-8 exhibited stability in water over 6 h with a 3% variability in average hydrodynamic radius. The CONTIN plots show a consistent, unimodal, distribution with a CV of 1%. Stability was also observed in PBS (pH 7.1) and IM buffers (Fig. 2(b) and (c)), where the variability in hydrodynamic size was 3% and 7%, respectively, after 6 hours of incubation. This variability increased to just 6% and 9% after 14 days for IM and PBS, respectively (Fig. 2(g)). The SEM micrographs of the BSA@c-ZIF-8 sample after 14 days incubation in PBS is consistent with the micrograph of the as-prepared BSA@c-ZIF-8 samples (Fig. S18, ESI†). We attribute this stability to the negative ζ potential of BSA@c-ZIF-8, which prevents the interaction of the PO43− anions with the Zn2+ nodes. While the stability in imidazole can be ascribed to the imidazolium cations having a low affinity for Zn2+ ions, thus retaining stability. Given the importance of PBS for analytical and drug delivery fields, we evaluated the stability of BSA@c-ZIF-8 in PBS with a pH of 6.1, 6.6, 7.1, 7.6, or 8.1, and found no significant difference in size or PDI after 14 days for the pHs tested (Fig. S19, ESI†), suggesting that BSA@c-ZIF-8 does not have the same pH dependent stability seen in other ZIF-8 composites.9
In contrast, BSA@c-ZIF-8 in HEPES, MES, and MOPS, show an increase in size from an initial 107 ± 9 nm to 2100 ± 800 nm for HEPES, 2200 ± 700 nm for MES, and 2200 ± 800 nm for MOPS over 6 h (Fig. 2(d)–(f) and Fig. S19, ESI†). The corresponding CONTIN plots remained monomodal across the 6 h for these buffers, which suggest the size increase is due to aggregation of nanoparticles. The crystal structure of BSA@c-ZIF-8 is retained after incubation in all buffers, independent of aggregation or stability (Fig. 2(i)), and there is no evidence of precursor structures suggesting that unstable dispersions do not re-dissolve into their precursor components (Fig. S16, ESI†).
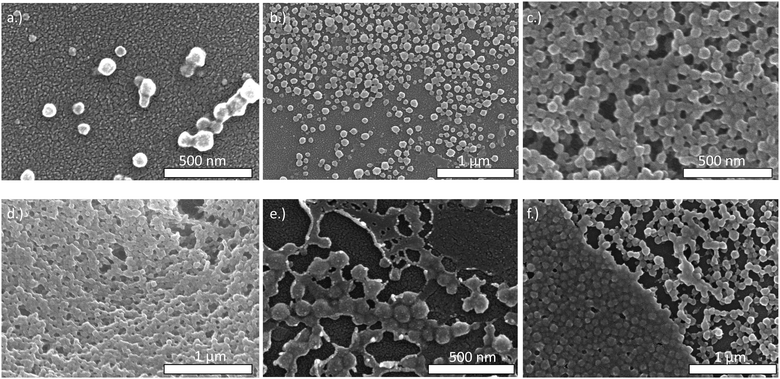
We assessed changes in BSA@c-ZIF-8 morphology after incubation in the buffers by SEM. We manually measured each particle, confirming the size after incubating in water was 110 ± 10 nm (n = 50), in IM was 100 ± 10 nm (n = 50) and in PBS was 110 ± 10 nm (n = 50), in good agreement with the DLS results (Fig. 3(a)–(c)). SEMs show aggregation of BSA@c-ZIF-8 after incubation in HEPES, MES, and MOPS (Fig. 3(d)–(f)). We hypothesized that the morpholine buffers can chelate Zn2+ from the ZIF-8 lattice, causing aggregation.
Protein@ZIF-8 nanoparticles can also be applied to cell culturing environments. Numerous applications have explored ZIF-8 biocomposites as plasmid and protein vectors for generous expression and drug delivery, but investigations into the stability within cell media have only been explored in a few studies with limited investigation into the temporal aggregation dynamics of ZIF-8.9,19,20 Thus, we evaluated the stability of our nanoparticles in various cell media including DMEM fortified with 10% FBS, LB broth, Gamborg's and Grace's media. We chose DMEM with 10% FBS due its application in the growth of mammalian cell lines,38 LB for its use in bacterial growth,39 and Gamborg's40 and Grace's media41 for representative use in plant and insect cell culture, respectively.
We show that BSA@c-ZIF-8 nanoparticles were stable in three of four cell media tested. We find that there is little deviation in the size of our nanoparticles across 6 h in all three batches tested (<10%, n = 9), while the PDI remains below 0.2 for DMEM 10% FBS, LB broth and Gamborg's media (Fig. 4(a), (b), (d), (e), (g) and (h)). However, when incubated in Grace's media, the nanoparticles precipitate instantly into micron-sized structures comprised of aggregated BSA@c-ZIF-8 (Fig. S21, ESI†). At t = 0 we find that the CONTIN plot is bifurcated into a peak at 200 ± 10 nm and a peak at 2000 ± 1000 nm (PDI > 0.9). At t = 15 min we find a monomodal peak at 5000 ± 2000 nm (PDI > 0.4), indicating full aggregation of BSA@c-ZIF-8 in this media. We attribute the aggregation in Grace's media to high concentrations of amino acids (AA) mediating degradation of our nanoparticles. The use of hydrophobic AA has been shown to elicit changes at Zn binding sites via AA–Zn linkages (AA = Val, Leu).42 AAs are present in LB as oligopeptides. We hypothesize the larger size of oligopeptides prevents interaction at the Zn2+ site and therefore prevents destabilization in LB broth.
Here, BSA@c-ZIF-8 nanoparticles show enhanced stability compared to other protein@ZIF-8 constructs presented in the literature. We attribute this difference to the high negative surface charge. Reported protein@ZIF-8 constructs range from −2.1 mV to −27 ± 5 mV,27,43,44 while the material produced in our work was −36 ± 10 mV. We hypothesize that our material's repulsive effect towards negatively charged chelators is higher than previously described protein@ZIF-8 due to this surface charge.
After we established the stability of our nanoparticles in buffer and cell media, we attempted to improve the stability of our particles in the selected buffer systems. Based on our literature review, previous enhancement of colloidal stability for ZIF-8 is based on two factors:
1. Minimizing the chemical interactions between ions such as H2PO4−, HPO42−, HCO− and the Zn2+ in the ZIF-8 lattice using chemical surfactants like PAA. The negatively charged COO− groups on the PAA and ligands like it, provide electro-repulsive forces to the anions, preventing dissolution of ZIF-8.
2. Preventing the re-crystallization of Zn2+ through surface functionalization or fortification using polymeric materials like PDA and dextran, which create a physical barrier between the ZIF-8 particle and solution, preventing aggregation.
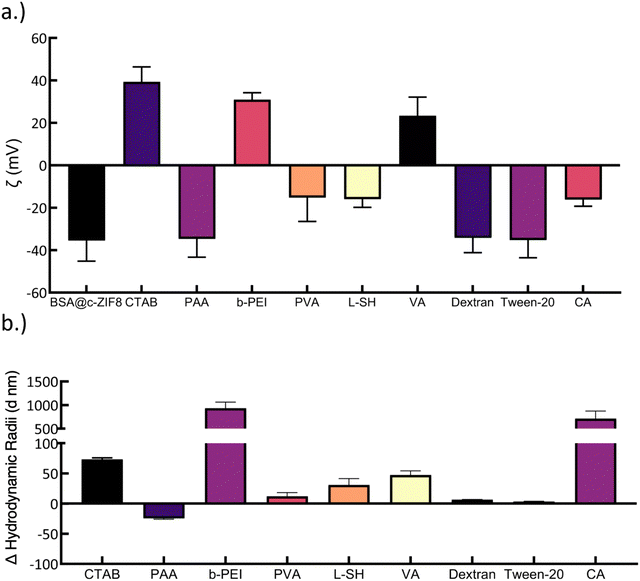
To assess improvements in the stability of BSA@c-ZIF-8 in all buffers and modify the surface for functionalization, we incubated the BSA@c-ZIF-8 nanoparticles in 1 mg mL−1 of surfactants for 24 h. We tested eight surfactants to analyse the impact of surface passivation on stability, a strategy previously employed for ZIF-8 stability by Li et al., with various surfactants.18 The authors of this work show that PAA enhances the stability on non-proteinaceous ZIF-8 to 48 h, up from 0 h.18 In addition to PAA, PVA and dextran have been used to stabilize and further modify nanoparticles via 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide/N-hydroxysuccinimide (EDC/NHS) chemistry.45–48 CTAB is also a common surfactant that has been used for templated synthesis, but we hypothesized that its specific binding to ZIF-8 would enhance the stability of the nanoparticles.49 Tween-20, a non-ionic surfactant, has been shown to limit non-specific fouling interactions and was investigated as a result.50 CA was also tested, as we predicted that small molecules would bind to the negatively charged surface, leaving carboxylic acid groups free in solution, allowing for EDC/NHS conjugation. Branched-PEI (b-PEI) is another polymer that has been employed to improve the thermal and mechanical stability of MIL-101(Cr) and ZIF-8.51,52 Branched PEI was used in this work over linear PEI as it is more easily functionalized.53
To assess polymer coating onto BSA@c-ZIF-8, we compared the ζ potential of BSA@c-ZIF-8 with and without surfactants and confirmed significant changes in the surface ζ potential for seven of ten surfactants tested (Fig. 5(a)). In brief, BSA@c-ZIF-8 begins at −36 ± 10 mV and changes to a positive surface charge of 39 ± 7 mV for CTAB, 31 ± 3 mV for b-PEI, −15 ± 11 mV for PVA, −16 ± 4 mV, and −16 ± 3 for CA. We did not find statistically significant shifts in the ζ potential for PAA, Tween-20 and dextran. We ascribe this lack of change in ζ potential to the inherent negative charge of these surfactants.18 We monitored the average hydrodynamic radius of BSA@c-ZIF-8 (107 ± 9 nm) and showed colloidal stability was retained for polymer-BSA@c-ZIF-8 in all surfactants except for b-PEI (HDR = 1100 ± 10 nm; PDI = 0.58 ± 0.1), CA (HDR = 80 ± 200 nm; PDI = 0.1 ± 0.1), L-SH (HDR = 660 ± 60; PDI = 0.35 ± 0.05), and Tween-20 (HDR = 180 ± 20 nm; PDI = 0.07 ± 0.02) (Fig. 3(b) and Fig. S22, ESI†). Polymer-BSA@c-ZIF-8 nanoparticles with CTAB, PAA, PVA, and dextran remain colloidally stable and have changes in hydrodynamic radius of 74 ± 3 nm, −25 ± 2 nm, 12 ± 6 nm, and 6 ± 1 nm, compared to BSA@c-ZIF-8. We ascribe the changes in size to polymers/small molecules assembling on the surface of BSA@c-ZIF-8.54 We verified the DLS results for the coating experiments with SEM (Fig. S22 and S23, ESI†) and showed colloidal particles, however Tween-20-BSA@c-ZIF-8 showed aggregation and amorphous material and therefore was excluded from further analysis.19–21,55–57
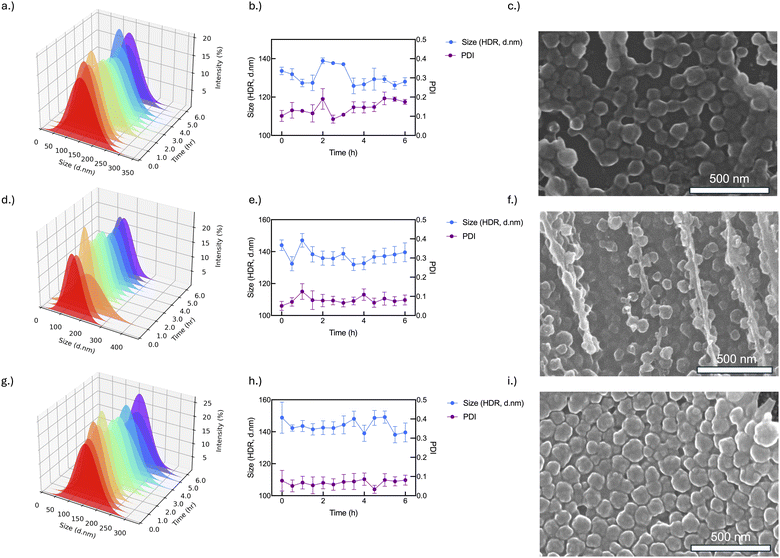
Studies have shown that nanoparticle stability can be enhanced with surface coated polymers or with excess polymer in solution. Here, we tested both polymer-BSA@c-ZIF8 nanoparticles in excess surfactant (pre-wash polymer-BSA@c-ZIF-8) and polymer-BSA@c-ZIF-8 nanoparticles (post-wash polymer-BSA@c-ZIF-8). We evaluated the stability and ζ potential of pre-wash polymer-BSA@c-ZIF-8 (Fig. 6(a)–(d) and Fig. S24, ESI†) and post-wash polymer-BSA@c-ZIF-8 (Fig. 6(e)–(h) and Fig. S24, ESI†). We provide a schematic illustration of the creation of “pre-wash” and “post-wash” samples (Scheme 1). We rationalized that some long-term storage conditions would allow for excess surfactant to remain in solution, while other applications would not benefit from additional surfactant in solution.
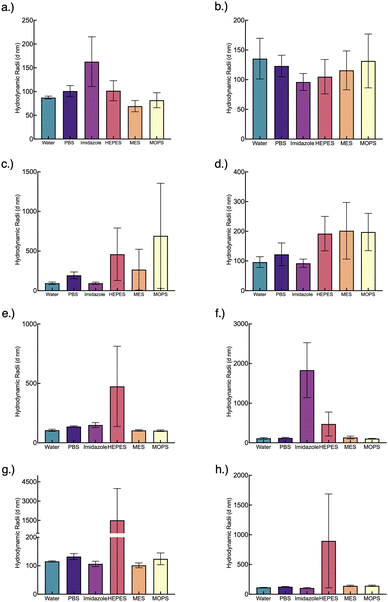 | ||
| Fig. 6 Polymer-BSA@c-ZIF-8 size change after conjugation (a)–(d) pre-wash and (e)–(h) post-wash. (a) and (e) show PAA-BSA@c-ZIF-8 size pre- and post-wash, respectively. (b) and (f) show CTAB-BSA@c-ZIF-8 size pre- and post-wash, respectively. (c) and (g) show Dextran-BSA@c-ZIF-8 size pre- and post-wash, respectively. (d) and (e) show PVA-BSA@c-ZIF-8 size pre- and post-wash, respectively. The associated PDIs can be found in Table S3 (ESI†). Each bar and error bar represents the average and standard deviation of 3 batches measured thrice (n = 9). | ||
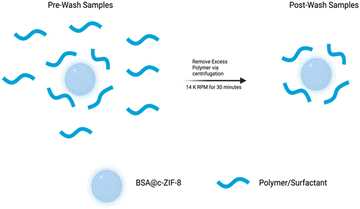 | ||
| Scheme 1 Illustration of the preparation of “Post-Wash” BSA@c-ZIF-8 samples from “Pre-Wash” BSA@c-ZIF-8 samples. | ||
Stability criteria based on a PDI < 0.2, a monomodal CONTIN plot, and <100% change in size were used to determine stable polymer-BSA@c-ZIF-8 nanoparticles in buffers for three batches. A summary is found in Table S4 (ESI†). We show that pre-wash CTAB-BSA@c-ZIF-8 and PAA-BSA@c-ZIF-8 were stable in PBS, IM MES, MOPS and HEPES for 24 hours (Fig. 5(a)–(d) and Fig. S25–S32 discussion, ESI†). These results agree with previously published work, PAA has been shown previously to enhance the stability of ZIF-8 by Chen et al., extending the stability of ZIF-8 in PBS from hours to days (48 h).18 Pre-wash Dextran-BSA@c-ZIF-8 and PVA-BSA@c-ZIF-8 were only stable in water, imidazole, and PBS, while stability in MES and MOPS was improved using post-wash Dextran-BSA@c-ZIF-8 and PVA-BSA@c-ZIF-8 (Fig. 6(e)–(h) and Fig. S33–S36, ESI†). A summary of colloidally stable polymer-BSA-@c-ZIF-8 is provided in Table 2.
| Surfactant | Pre-wash | Post-wash | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PBS (pH 7.1) | Imidazole (pH 7.5) | HEPES (pH 7.4) | MES (pH 6.0) | MOPS (pH 7.2) | PBS (pH 7.1) | Imidazole (pH 7.5) | HEPES (pH 7.4) | MES (pH 6.0) | MOPS (pH 7.2) | |
| PAA | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✓ | ✓ |
| CTAB | ✓ | ✓ | ✓ | ✓ | ✓ | ✓ | ✗ | ✗ | ✓ | ✓ |
| Dextran | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ | ✓ | ✓ |
| PVA | ✓ | ✓ | ✗ | ✗ | ✗ | ✓ | ✓ | ✗ | ✓ | ✓ |
Conclusion
This work optimized the colloidal stability of BSA@c-ZIF-8 nanoparticles in biologically relevant buffer systems. We developed BSA@c-ZIF-8 nanoparticles of 107 ± 9 nm and evaluated their colloidal stability in buffers and show enhanced colloidal stability compared to ZIF-8. Our study shows that the BSA@c-ZIF-8 nanoparticles maintain their colloidal integrity up to 14 days in PBS, IM, and water. The stability of BSA@c-ZIF-8 in PBS is contrasted by the instability in HEPES, MES, and MOPS buffers, which promoted particle aggregation. The application of surfactants such as PAA, CTAB, dextran, and PVA mitigated this instability, preserving colloidal integrity across all buffers except HEPES over 24 h. We attribute the reported stability of BSA@c-ZIF-8 to the highly negative surface charge on the nanoparticle repelling negatively charged chelating agents in buffer, such as HPO42−. We hypothesize that the high negative surface charge is due to BSA molecules concentrated at the surface of the BSA@c-ZIF-8 nanoparticles. The negative surface charge is maintained after washing, compared to BSA adsorbed onto the surface of ZIF-8, which we attribute to BSA crystallized in ZIF-8 on the surface of the particle. Previous reports showed that BSA@ZIF-8 can form core–shell structures, with a ZIF-8 core and a BSA@ZIF-8 shell.26,27 Future studies will include probing the spatial distribution of protein molecules in protein@c-ZIF-8 nanoparticles and formation mechanisms that result in these particle morphologies. Additional studies will investigate the optimization of colloidally stable protein@c-ZIF-8 nanoparticles with other biomolecules such as enzymes for applications in catalysis or drug delivery. Additionally, identification of polymer-BSA@c-ZIF-8 coatings provides a basis to explore functionalization of these BSA@c-ZIF-8 nanoparticles via common EDC–NHS or tetrameric antibody complexes for applications in analytical assays. These findings demonstrate improved colloidal stability of BSA@c-ZIF-8 nanoparticles in biologically relevant buffers that can enable new implementations in biological applications.Author contributions
Justin Van Houten: conceptualization, methodology, validation, investigation, data curation, writing – original draft, writing – review & editing, and visualization. Ruben Castillo Barberi: conceptualization, methodology, validation, investigation, data curation, writing – review & editing, and visualization. Jared King: methodology and investigation. Alana F. Ogata: conceptualization, resources, writing – review & editing, supervision, project administration, and funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This material is primarly funded by the Natural Sciences and Engineering Research Council of Canada (NSERC – 05329). The authors thank Peter Mitrakos from the UTM Core Facilities for their guidance DLS. The authors thank Sal Boccia from the Open Centre for the Characterization of Advanced Materials (OCCAM) for their guidance on scanning electron microscopy experiments. The authors thank Lindsey Fiddes of the Temetry Microscopy Facility for their guidance with cryogenic electron microscopy experiments. The authors thanks Kezia Suryoraharjo for helpful discussions and Justin Sung-Ho Kim for guidance in generating graphical representations. The authors thank the Shin Lab and Raneem Akel for their guidance and assistance in the preparation of DMEM. The authors wish to acknowledge this land on which the University of Toronto operates. For thousands of years it has been the traditional land of the Huron-Wendat, the Seneca, and the Mississaugas of the Credit. Today, this meeting place is still the home to many Indigenous people from across Turtle Island and we are grateful to have the opportunity to work on this land.Notes and references
- B. P. Carpenter, A. R. Talosig, J. T. Mulvey, J. G. Merham, J. Esquivel, B. Rose, A. F. Ogata, D. A. Fishman and J. P. Patterson, Role of Molecular Modification and Protein Folding in the Nucleation and Growth of Protein–Metal–Organic Frameworks, Chem. Mater., 2022, 34(18), 8336–8344, DOI:10.1021/acs.chemmater.2c01903
.
- N. K. Maddigan, A. Tarzia, D. M. Huang, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Protein Surface Functionalisation as a General Strategy for Facilitating Biomimetic Mineralisation of ZIF-8, Chem. Sci., 2018, 9(18), 4217–4223, 10.1039/C8SC00825F
.
- X. Gu and A. F. Palmer, ZIF-8 Metal–Organic Framework Nanoparticles Loaded with Hemoglobin as a Potential Red Blood Cell Substitute, ACS Appl. Nano Mater., 2022, 5(4), 5670–5679, DOI:10.1021/acsanm.2c00608
.
- L. Wang, W. Zhi, D. Lian, Y. Wang, J. Han and Y. Wang, HRP@ZIF-8/DNA Hybrids: Functionality Integration of ZIF-8 via Biomineralization and Surface Absorption, ACS Sustainable Chem. Eng., 2019, 7(17), 14611–14620, DOI:10.1021/acssuschemeng.9b02348
.
- C. Wang, G. Sudlow, Z. Wang, S. Cao, Q. Jiang, A. Neiner, J. J. Morrissey, E. D. Kharasch, S. Achilefu and S. Singamaneni, Metal–Organic Framework Encapsulation Preserves the Bioactivity of Protein Therapeutics, Adv. Healthcare Mater., 2018, 7(22), 1800950, DOI:10.1002/adhm.201800950
.
- S. Kumari, T. S. Howlett, R. N. Ehrman, S. Koirala, O. Trashi, I. Trashi, Y. H. Wijesundara and J. J. Gassensmith,
In Vivo Biocompatibility of ZIF-8 for Slow Release via Intranasal Administration, Chem. Sci., 2023, 14(21), 5774–5782, 10.1039/D3SC00500C
.
- S. Kumari, Y. H. Wijesundara, T. S. Howlett, M. Waliullah, F. C. Herbert, A. Raja, I. Trashi, R. A. Bernal and J. J. Gassensmith, Biolistic Delivery of Liposomes Protected in Metal-Organic Frameworks, Proc. Natl. Acad. Sci. U. S. A., 2023, 120(11), e2218247120, DOI:10.1073/pnas.2218247120
.
- M. Velásquez-Hernández, J. de, R. Ricco, F. Carraro, F. T. Limpoco, M. Linares-Moreau, E. Leitner, H. Wiltsche, J. Rattenberger, H. Schröttner, P. Frühwirt, E. M. Stadler, G. Gescheidt, H. Amenitsch, C. J. Doonan and P. Falcaro, Degradation of ZIF-8 in Phosphate Buffered Saline Media, CrystEngComm, 2019, 21(31), 4538–4544, 10.1039/C9CE00757A
.
- M. A. Luzuriaga, C. E. Benjamin, M. W. Gaertner, H. Lee, F. C. Herbert, S. Mallick and J. J. Gassensmith, ZIF-8 Degrades in Cell Media, Serum, and Some—but Not All—Common Laboratory Buffers, Supramol. Chem., 2019, 31(8), 485–490, DOI:10.1080/10610278.2019.1616089
.
- O. R. Brohlin, R. N. Ehrman, F. C. Herbert, Y. H. Wijesundara, A. Raja, A. Shahrivarkevishahi, S. D. Diwakara, R. A. Smaldone and J. J. Gassensmith, Zeolitic Imidazolate Framework Nanoencapsulation of CpG for Stabilization and Enhancement of Immunoadjuvancy, ACS Appl. Nano Mater., 2022, 5(10), 13697–13704, DOI:10.1021/acsanm.1c03555
.
- E. Astria, M. Thonhofer, R. Ricco, W. Liang, A. Chemelli, A. Tarzia, K. Alt, C. E. Hagemeyer, J. Rattenberger, H. Schroettner, T. Wrodnigg, H. Amenitsch, D. M. Huang, C. J. Doonan and P. Falcaro, Carbohydrates@MOFs, Mater. Horiz., 2019, 6(5), 969–977, 10.1039/C8MH01611A
.
- T. Knedel, E. Ricklefs, C. Schlüsener, V. B. Urlacher and C. Janiak, Laccase Encapsulation in ZIF-8 Metal–Organic Framework Shows Stability Enhancement and Substrate Selectivity, ChemistryOpen, 2019, 8(11), 1337–1344, DOI:10.1002/open.201900146
.
- M. Hartmann and D. Jung, Biocatalysis with Enzymes Immobilized on Mesoporous Hosts: The Status Quo and Future Trends, J. Mater. Chem., 2010, 20(5), 844–857, 10.1039/B907869J
.
- D.-G. Zhang, Y.-J. Pan, B.-Q. Chen, X.-C. Lu, Q.-X. Xu, P. Wang, R. K. Kankala, N.-N. Jiang, S.-B. Wang and A.-Z. Chen, Protein-Guided Biomimetic Nanomaterials: A Versatile Theranostic Nanoplatform for Biomedical Applications, Nanoscale, 2024, 16(4), 1633–1649, 10.1039/D3NR05495K
.
- H. Zhang, M. Zhao and Y. S. Lin, Stability of ZIF-8 in Water under Ambient Conditions, Microporous Mesoporous Mater., 2019, 279, 201–210, DOI:10.1016/j.micromeso.2018.12.035
.
- J. Gao, W. Chu, X. Ding, L. Ding, Q. Guo and Y. Fu, Degradation Kinetic Studies of BSA@ZIF-8 Nanoparticles with Various Zinc Precursors, Metal-to-Ligand Ratios, and PH Conditions, ACS Omega, 2023, 8(47), 44601–44610, DOI:10.1021/acsomega.3c04973
.
- A. Huang, L. Tong, X. Kou, R. Gao, Z.-W. Li, S. Huang, F. Zhu, G. Chen and G. Ouyang, Structural and Functional Insights into the Biomineralized Zeolite Imidazole Frameworks, ACS Nano, 2023, 17(23), 24130–24140, DOI:10.1021/acsnano.3c09118
.
- W. Chen, W. Cai, H. Liu, L. Fu, W. Lu, C. Zhang, M. K. ADale, C. Da, H. Pan, S. Kong, J. Wang, L. Du and C. Wu, Facially- Controllable Synthesis of Zeolitic Imidezolate Framework-8 Nanocrystal and Its Colloidal Stability in Phosphate Buffered Saline, Mater. Chem. Phys., 2020, 245, 122576, DOI:10.1016/J.MATCHEMPHYS.2019.122576
.
- N. Wang, G. Zhang, P. Zhang, K. Zhao, Y. Tian and J. Cui, Vaccination of TLR7/8 Agonist-Conjugated Antigen Nanoparticles for Cancer Immunotherapy, Adv. Healthcare Mater., 2023, 12(22), 2300249, DOI:10.1002/adhm.202300249
.
- Y. Tian, Z. Gao, M. Li, M. Hu, J. Hao and J. Cui, Interface Assembly of Polymer Networks on Metal–Organic Frameworks for the Engineering of Functional Nanoparticles, Chem. Mater., 2023, 35(14), 5593–5601, DOI:10.1021/acs.chemmater.3c01094
.
- Q. Yu, Y. Tian, M. Li, Y. Jiang, H. Sun, G. Zhang, Z. Gao, W. Zhang, J. Hao, M. Hu and J. Cui, Poly(Ethylene Glycol)-Mediated Mineralization of Metal–Organic Frameworks, Chem. Commun., 2020, 56(75), 11078–11081, 10.1039/D0CC03734F
.
- Q. Tian, X. Jia, J. Yang, S. Wang, Y. Li, D. Shao and H. Song, Polydopamine-Stabilized ZIF-8: Improved Water Stability and Lubrication Performance, Appl. Surf. Sci., 2022, 578, 152120, DOI:10.1016/j.apsusc.2021.152120
.
- H. Zhang, J. James, M. Zhao, Y. Yao, Y. Zhang, B. Zhang and Y. S. Lin, Improving Hydrostability of ZIF-8 Membranes via Surface Ligand Exchange, J. Membr. Sci., 2017, 532, 1–8, DOI:10.1016/j.memsci.2017.01.065
.
- R. N. Ehrman, O. R. Brohlin, Y. H. Wijesundara, S. Kumari, O. Trashi, T. S. Howlett, I. Trashi, F. C. Herbert, A. Raja, S. Koirala, N. Tran, N. M. Al-Kharji, W. Tang, M. C. Senarathna, L. M. Hagge, R. A. Smaldone and J. J. Gassensmith, A Scalable Synthesis of Adjuvanting Antigen Depots Based on Metal–Organic Frameworks, Chem. Sci., 2024, 15(8), 2731–2744, 10.1039/D3SC06734C
.
- F. Carraro, M. Velásquez-Hernández, J. de, E. Astria, W. Liang, L. Twight, C. Parise, M. Ge, Z. Huang, R. Ricco, X. Zou, L. Villanova, C. O. Kappe, C. Doonan and P. Falcaro, Phase Dependent Encapsulation and Release Profile of ZIF-Based Biocomposites, Chem. Sci., 2020, 11(13), 3397–3404, 10.1039/C9SC05433B
.
- W. Liang, R. Ricco, N. K. Maddigan, R. P. Dickinson, H. Xu, Q. Li, C. J. Sumby, S. G. Bell, P. Falcaro and C. J. Doonan, Control of Structure Topology and Spatial Distribution of Biomacromolecules in Protein@ZIF-8 Biocomposites, Chem. Mater., 2018, 30(3), 1069–1077, DOI:10.1021/acs.chemmater.7b04977
.
- A. F. Ogata, A. M. Rakowski, B. P. Carpenter, D. A. Fishman, J. G. Merham, P. J. Hurst and J. P. Patterson, Direct Observation of Amorphous Precursor Phases in the Nucleation of Protein–Metal–Organic Frameworks, J. Am. Chem. Soc., 2020, 142(3), 1433–1442, DOI:10.1021/jacs.9b11371
.
- C. Wang, G. Sudlow, Z. Wang, S. Cao, Q. Jiang, A. Neiner, J. J. Morrissey, E. D. Kharasch, S. Achilefu and S. Singamaneni, Metal–Organic Framework Encapsulation Preserves the Bioactivity of Protein Therapeutics, Adv. Healthcare Mater., 2018, 7(22), 1800950, DOI:10.1002/adhm.201800950
.
- J. J. Beh, J. K. Lim, E. P. Ng and B. S. Ooi, Synthesis and Size Control of Zeolitic Imidazolate Framework-8 (ZIF-8): From the Perspective of Reaction Kinetics and Thermodynamics of Nucleation, Mater. Chem. Phys., 2018, 216, 393–401, DOI:10.1016/j.matchemphys.2018.06.022
.
- A. Mittal, S. Gandhi and I. Roy, Mechanistic Interaction Studies of Synthesized ZIF-8 Nanoparticles with Bovine Serum Albumin Using Spectroscopic and Molecular Docking Approaches, Sci. Rep., 2022, 12(1), 10331, DOI:10.1038/s41598-022-14630-y
.
- G. Mo, X. He, D. Qin, S. Meng, Y. Wu and B. Deng, Spatially-Resolved Dual-Potential Sandwich Electrochemiluminescence Immunosensor for the Simultaneous Determination of Carbohydrate Antigen 19–9 and Carbohydrate Antigen 24-2, Biosens. Bioelectron., 2021, 178, 113024, DOI:10.1016/J.BIOS.2021.113024
.
- M. Bukackova, P. Rusnok and R. Marsalek, Mathematical Methods in the Calculation of the Zeta Potential of BSA, J. Solution Chem., 2018, 47(12), 1942–1952, DOI:10.1007/s10953-018-0830-0
.
- Y. Zhang, Y. Jia, M. Li and L. Hou, Influence of the 2-Methylimidazole/Zinc Nitrate Hexahydrate Molar Ratio on the Synthesis of Zeolitic Imidazolate Framework-8 Crystals at Room Temperature, Sci. Rep., 2018, 8(1), 9597, DOI:10.1038/s41598-018-28015-7
.
- J. Gao, W. Chu, X. Ding, L. Ding, Q. Guo and Y. Fu, Degradation Kinetic Studies of BSA@ZIF-8 Nanoparticles with Various Zinc Precursors, Metal-to-Ligand Ratios, and PH Conditions, ACS Omega, 2023, 8, 44610, DOI:10.1021/ACSOMEGA.3C04973
.
- H. Zhang, M. Zhao and Y. S. Lin, Stability of ZIF-8 in Water under Ambient Conditions, Microporous Mesoporous Mater., 2019, 279, 201–210, DOI:10.1016/j.micromeso.2018.12.035
.
- H. Zhang, D. Liu, Y. Yao, B. Zhang and Y. S. Lin, Stability of ZIF-8 Membranes and Crystalline Powders in Water at Room Temperature, J. Membr. Sci., 2015, 485, 103–111, DOI:10.1016/j.memsci.2015.03.023
.
- S. A. Butonova, E. V. Ikonnikova, A. Sharsheeva, I. Yu Chernyshov, O. A. Kuchur, I. S. Mukhin, E. Hey-Hawkins, A. V. Vinogradov and M. I. Morozov, Degradation Kinetic Study of ZIF-8 Microcrystals with and without the Presence of Lactic Acid, RSC Adv., 2021, 11(62), 39169–39176, 10.1039/D1RA07089D
.
- M. I. Saleh, B. Rühle, S. Wang, J. Radnik, Y. You and U. Resch-Genger, Assessing the Protective Effects of Different Surface Coatings on NaYF4:Yb3+, Er3+ Upconverting Nanoparticles in Buffer and DMEM, Sci. Rep., 2020, 10(1), 19318, DOI:10.1038/s41598-020-76116-z
.
- A. R. Tuttle, N. D. Trahan and M. S. Son, Growth and Maintenance of Escherichia Coli Laboratory Strains, Curr. Protoc., 2021, 1(1), e20, DOI:10.1002/cpz1.20
.
- M. Hashemi, A. Moieni, M. S. Sabet, A. Mokhtassi-Bidgoli and S. Mojarrad Nanas, Introducing Gamborg's B5, a High-Potential Medium for Isolated Microspore Culture, and Presenting a New MS Medium-Based Protocol for Androgenic Plant Regeneration in Eggplant (Solanum Melongena L.), Plant Cell, Tissue Organ Cult., 2024, 156(3), 79, DOI:10.1007/s11240-024-02706-8
.
- F. R. X. Batista, C. A. Pereira, R. Z. Mendonça and A. M. Moraes, Enhancement of Sf9 Cells and Baculovirus Production Employing Grace's Medium Supplemented with Milk Whey Ultrafiltrate, Cytotechnology, 2005, 49(1), 1–9, DOI:10.1007/s10616-005-4206-0
.
- S. Sun, Z. Zhang, Y. Xiang, M. Cao and D. Yu, Amino Acid-Mediated Synthesis of the ZIF-8 Nanozyme That Reproduces Both the Zinc-Coordinated Active Center and Hydrophobic Pocket of Natural Carbonic Anhydrase, Langmuir, 2022, 38(4), 1621–1630, DOI:10.1021/acs.langmuir.1c03118
.
- S. Peng, J. Liu, Y. Qin, H. Wang, B. Cao, L. Lu and X. Yu, Metal–Organic Framework Encapsulating Hemoglobin as a High-Stable and Long-Circulating Oxygen Carriers to Treat Hemorrhagic Shock, ACS Appl. Mater. Interfaces, 2019, 11(39), 35604–35612, DOI:10.1021/acsami.9b15037
.
- X. Gu and A. F. Palmer, ZIF-8 Metal–Organic Framework Nanoparticles Loaded with Hemoglobin as a Potential Red Blood Cell Substitute, ACS Appl. Nano Mater., 2022, 5(4), 5670–5679, DOI:10.1021/acsanm.2c00608
.
- S.-H. Jung, J.-W. Jung, I.-B. Suh, J. S. Yuk, W.-J. Kim, E. Y. Choi, Y.-M. Kim and K.-S. Ha, Analysis of C-Reactive Protein on Amide-Linked N-Hydroxysuccinimide−Dextran Arrays with a Spectral Surface Plasmon Resonance Biosensor for Serodiagnosis, Anal. Chem., 2007, 79(15), 5703–5710, DOI:10.1021/ac070433l
.
- S. Tarvirdipour, E. Vasheghani-Farahani, M. Soleimani and H. Bardania, Functionalized Magnetic Dextran-Spermine Nanocarriers for Targeted Delivery of Doxorubicin to Breast Cancer Cells, Int. J. Pharm., 2016, 501(1–2), 331–341, DOI:10.1016/j.ijpharm.2016.02.012
.
- S. B. Makhluf, R. Abu-Mukh, S. Rubinstein, H. Breitbart and A. Gedanken, Modified PVA–Fe3O4 Nanoparticles as Protein Carriers into Sperm Cells, Small, 2008, 4(9), 1453–1458, DOI:10.1002/smll.200701308
.
- C.-H. Kuo, Y.-C. Liu, C.-M. J. Chang, J.-H. Chen, C. Chang and C.-J. Shieh, Optimum Conditions for Lipase Immobilization on Chitosan-Coated Fe3O4 Nanoparticles, Carbohydr. Polym., 2012, 87(4), 2538–2545, DOI:10.1016/j.carbpol.2011.11.026
.
- G. Zheng, Z. Chen, K. Sentosun, I. Pérez-Juste, S. Bals, L. M. Liz-Marzán, I. Pastoriza-Santos, J. Pérez-Juste and M. Hong, Shape Control in ZIF-8 Nanocrystals and Metal Nanoparticles@ZIF-8 Heterostructures, Nanoscale, 2017, 9(43), 16645–16651, 10.1039/C7NR03739B
.
- K. L. Brogan, J. H. Shin and M. H. Schoenfisch, Influence of Surfactants and Antibody Immobilization Strategy on Reducing Nonspecific
Protein Interactions for Molecular Recognition Force Microscopy, Langmuir, 2004, 20(22), 9729–9735, DOI:10.1021/la048437y
.
- Z. Hu, H. Zhang, X.-F. Zhang, M. Jia and J. Yao, Polyethylenimine Grafted ZIF-8@cellulose Acetate Membrane for Enhanced Gas Separation, J. Membr. Sci., 2022, 662, 120996, DOI:10.1016/j.memsci.2022.120996
.
- Q. Xin, J. Ouyang, T. Liu, Z. Li, Z. Li, Y. Liu, S. Wang, H. Wu, Z. Jiang and X. Cao, Enhanced Interfacial Interaction and CO 2 Separation Performance of Mixed Matrix Membrane by Incorporating Polyethylenimine-Decorated Metal–Organic Frameworks, ACS Appl. Mater. Interfaces, 2015, 7(2), 1065–1077, DOI:10.1021/am504742q
.
- M. Jäger, S. Schubert, S. Ochrimenko, D. Fischer and U. S. Schubert, Branched and Linear Poly(Ethylene Imine)-Based Conjugates: Synthetic Modification, Characterization, and Application, Chem. Soc. Rev., 2012, 41(13), 4755, 10.1039/c2cs35146c
.
- P. Wu, T. Sun, X. Jiang and S. Kondrat, Hydrodynamic Properties of Polymers Screening the Electrokinetic Flow: Insights from a Computational Study, Polymers, 2019, 11(6), 1038, DOI:10.3390/polym11061038
.
- F. Asadi, S. N. Azizi and M. J. Chaichi, Green Synthesis of Fluorescent PEG-ZnS QDs Encapsulated into Co-MOFs as an Effective Sensor for Ultrasensitive Detection of Copper Ions in Tap Water, Mater. Sci. Eng., C, 2019, 105, 110058, DOI:10.1016/j.msec.2019.110058
.
- A. N. Generalova, V. V. Rocheva, A. V. Nechaev, D. A. Khochenkov, N. V. Sholina, V. A. Semchishen, V. P. Zubov, A. V. Koroleva, B. N. Chichkov and E. V. Khaydukov, PEG-Modified Upconversion Nanoparticles for in Vivo Optical Imaging of Tumors, RSC Adv., 2016, 6(36), 30089–30097, 10.1039/C5RA25304G
.
- C. Fang, N. Bhattarai, C. Sun and M. Zhang, Functionalized Nanoparticles with Long-Term Stability in Biological Media, Small, 2009, 5(14), 1637–1641, DOI:10.1002/smll.200801647
.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00377b |
| ‡ These authors have contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |