Open Access Article
Open Access ArticleDeep learning unlocks label-free viability assessment of cancer spheroids in microfluidics†
Chun-Cheng
Chiang‡
ab,
Rajiv
Anne‡
ac,
Pooja
Chawla
c,
Rachel M.
Shaw
c,
Sarah
He
ad,
Edwin C.
Rock
c,
Mengli
Zhou
abe,
Jinxiong
Cheng
ac,
Yi-Nan
Gong
af and
Yu-Chih
Chen
 *abcg
*abcg
aUPMC Hillman Cancer Center, University of Pittsburgh, 5115 Centre Ave, Pittsburgh, PA 15232, USA. E-mail: cheny25@upmc.edu; Tel: +1 412 623 7701
bDepartment of Computational and Systems Biology, University of Pittsburgh, 3420 Forbes Avenue, Pittsburgh, PA 15260, USA
cDepartment of Bioengineering, Swanson School of Engineering, University of Pittsburgh, 3700 O'Hara Street, Pittsburgh, PA 15260, USA
dCarnegie Mellon University, Department of Biological Sciences, 5000 Forbes Avenue, Pittsburgh, PA 15213, USA
eXiangya Hospital, Central South University, Changsha, Hunan 410008, China
fDepartment of Immunology, University of Pittsburgh School of Medicine, 3420 Forbes Avenue, Pittsburgh, PA 15260, USA
gCMU-Pitt Ph.D. Program in Computational Biology, University of Pittsburgh, 3420 Forbes Avenue, Pittsburgh, PA 15260, USA
First published on 22nd May 2024
Abstract
Despite recent advances in cancer treatment, refining therapeutic agents remains a critical task for oncologists. Precise evaluation of drug effectiveness necessitates the use of 3D cell culture instead of traditional 2D monolayers. Microfluidic platforms have enabled high-throughput drug screening with 3D models, but current viability assays for 3D cancer spheroids have limitations in reliability and cytotoxicity. This study introduces a deep learning model for non-destructive, label-free viability estimation based on phase-contrast images, providing a cost-effective, high-throughput solution for continuous spheroid monitoring in microfluidics. Microfluidic technology facilitated the creation of a high-throughput cancer spheroid platform with approximately 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 spheroids per chip for drug screening. Validation involved tests with eight conventional chemotherapeutic drugs, revealing a strong correlation between viability assessed via LIVE/DEAD staining and phase-contrast morphology. Extending the model's application to novel compounds and cell lines not in the training dataset yielded promising results, implying the potential for a universal viability estimation model. Experiments with an alternative microscopy setup supported the model's transferability across different laboratories. Using this method, we also tracked the dynamic changes in spheroid viability during the course of drug administration. In summary, this research integrates a robust platform with high-throughput microfluidic cancer spheroid assays and deep learning-based viability estimation, with broad applicability to various cell lines, compounds, and research settings.
000 spheroids per chip for drug screening. Validation involved tests with eight conventional chemotherapeutic drugs, revealing a strong correlation between viability assessed via LIVE/DEAD staining and phase-contrast morphology. Extending the model's application to novel compounds and cell lines not in the training dataset yielded promising results, implying the potential for a universal viability estimation model. Experiments with an alternative microscopy setup supported the model's transferability across different laboratories. Using this method, we also tracked the dynamic changes in spheroid viability during the course of drug administration. In summary, this research integrates a robust platform with high-throughput microfluidic cancer spheroid assays and deep learning-based viability estimation, with broad applicability to various cell lines, compounds, and research settings.
Introduction
Breast cancer is the leading type of cancer among women in the USA, and the second leading cause of cancer-related deaths among women.1 While surgical removal is beneficial, adjuvant chemotherapy is crucial in achieving cancer-free status and improving survival rates for breast cancer patients. However, breast cancer is notorious for its ability to become resistant to drugs.2 Therefore, identifying new compounds for either single or combined use has become a prominent focus in breast cancer research.3 As tumor cells in human bodies develop into three-dimensional (3D) structures, the conventional cell culture methods in dishes or plates are inadequate to accurately recapitulate the complex conditions of real tumors, due to the presence of tissue hypoxia, nutrient deprivation, and acidic environment in the central regions of tumors.4–7 The physical barrier in such environments results in a concentration gradient, leading to poor drug penetration during anti-cancer drug administration. Moreover, research has indicated that cancer cells tend to remain dormant in the core of the tumor and exhibit stem cell-like properties such as the ability to resist treatment and relapse.8–11 Hence, resistance to drugs can occur in animal or human models, despite demonstrating inhibitory effects on tumor cells in two-dimensional (2D) cell cultures.There are various 3D cell culture technologies that are currently available in laboratories and on the market. The hanging drop culture method suspends cells in small droplets that are hung upside down from a culture dish lid.12–15 The droplets are formed by carefully pipetting a small volume of cell suspension onto the lid of the culture dish. The surface tension of the liquid in the droplet allows it to hang in place, with the cells settling at the bottom of the droplet due to gravity and aggregating to spheroids. Despite its effectiveness, the hanging drop method can be limited by the need for careful handling and evaporation-induced osmotic pressure on the spheroids. One alternative approach to 3D culture is to embed cells in a hydrogel matrix, which can provide a more physiologically relevant environment that mimics the extracellular matrix in vivo.16,17 However, this method can also have some limitations. Nutrient exchange through the hydrogel can be slow, which can limit the size of the resulting spheroids and potentially affect their reproducibility. Another commonly used approach for 3D culture involves utilizing a low-attachment surface. Such a surface may be treated with a hydrophilic substance to decrease cell adhesion. One way to achieve this is by coating the surface with materials like poly(ethylene glycol) (PEG),18 poly(hydroxyethyl methacrylate) (pHEMA),11,19,20 or poly(oxyethylene) (POE) triblock polymer (Pluronic F108).21–23 By preventing cell adhesion on the coated substrate, cells will aggregate and form 3D spheroids, which can be used for applications such as drug screening and tissue engineering.
In addition to creating a non-adherent environment for cell aggregation, the precise formation of uniform spheroids from a small number of cells is another challenge. The conventional low-attachment plate method leads to the random generation of spheroids with different sizes throughout the well, resulting in unreliable results due to the significant impact of spheroid size on drug response.24,25 Microfluidics is an innovative and potent tool for high-throughput drug screening as it enables precise manipulation of cells in a controlled 3D environment.26–35 Thousands of micro-chambers on a single chip can host spheroids simultaneously for high-throughput drug screening.36 Additionally, the spontaneous loading of cells into all micro-chambers with good uniformity significantly reduces labor intensity and improves efficiency.
Although many challenges of high-throughput 3D cancer spheroid culture for drug screening have been addressed, there is still a significant issue with reliable and effective readout of the outcomes. Commonly used assays to measure cell viability and proliferation after treatment with chemical or physical stimuli can carry potential cytotoxicity and may lead to inconsistent and inaccurate results. Specifically, the BrdU assay can cause DNA damage,37 live/dead staining is cytotoxic,38,39 and the firefly luciferase assay requires cell lysis, which may lead to confounding experimental results.40 Moreover, though staining like trypan blue has low cost and rapid protocol, the results could be inconsistent and inaccurate, especially for 3D spheroid cultures.41
Recently, deep learning has made it possible to analyze cellular images with greater accuracy and efficiency. This has led to the development of high-throughput methods for classifying the status of cell cycle and identifying subcellular features using deep learning toolkits.42–47 Even unlabeled transmitted-light images can be used to predict different fluorescence labels like cell nuclei and cell type.48 Phase-contrast imaging can also be used computationally to perform live-dead assays on unlabeled cells.38,47,49 However, there has been relatively little research on detecting the viability of cancer spheroids without the use of labeling techniques. The existing works rely on specialized optical coherence tomography techniques.50 To overcome these limitations, we previously tested a deep learning model to estimate spheroid viability.51 However, this earlier investigation was confined to a single breast cancer cell line and three drugs, thus offering limited insights into the model's broader applicability. Therefore, in this study, we aim to take a more comprehensive strategy by training a versatile model using eight conventional chemotherapeutic compounds and subsequently validate the model's versatility by assessing its performance with novel compounds, additional cell lines, primary cells not utilized in the training dataset, and images sourced from external laboratories to demonstrate the generalizability of the approach. By combining microfluidics with a generic drug efficacy estimation approach, our method provides a cost-effective, high-throughput, and non-destructive way to assess tumor spheroid viability in real-time that can be widely adapted to many cell lines, compounds, and laboratories.
Methods
Microfluidic chip design and fabrication
A layer of micro-patterned PDMS (polydimethylsiloxane, Sylgard 184, Dow Corning) was bonded to a bottomless 6-channel slide (Sticky-Slide VI 0.4, 80608, ibidi) to create cancer spheroid devices. Each device, measuring 75.5 mm by 25.5 mm and following the 96-well plate format with a pitch of 9 mm between wells, can accommodate approximately 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 micro-chambers for spheroid culture and test 6 drug treatment conditions on a device.20 Sanitization to maintain a sterile condition. Pluronic® F-108 (BASF, CAS 9003-11-6) solution (5% in DI water) was applied to the device 12 hours before cell loading to create a non-adherent PDMS substrate for cell aggregation.23,52,53 Prior to experimentation, the devices were washed with PBS (Gibco 10010) for one hour to eliminate residual F-108 solution.
000 micro-chambers for spheroid culture and test 6 drug treatment conditions on a device.20 Sanitization to maintain a sterile condition. Pluronic® F-108 (BASF, CAS 9003-11-6) solution (5% in DI water) was applied to the device 12 hours before cell loading to create a non-adherent PDMS substrate for cell aggregation.23,52,53 Prior to experimentation, the devices were washed with PBS (Gibco 10010) for one hour to eliminate residual F-108 solution.
Cell culture
We cultured SUM159 and SUM149 cells in F-12 (Gibco 11765) media supplemented with 5% FBS (Gibco 16000), 1% pen/strep (Gibco 15070), 1% GlutaMax (Gibco 35050), 1 μg mL−1 hydrocortisone (Sigma H4001), and 5 μg mL−1 insulin (Sigma I6634), and 0.1% of plasmocin (InvivoGen, ant-mpp). We cultured T47D cells in RPMI 1640 medium (Gibco 1875) supplemented with 10% fetal bovine serum (FBS, Gibco 16000), 1% GlutaMax (Gibco 35050), 1% penicillin/streptomycin (pen/strep, Gibco 15070), and 0.1% of plasmocin (InvivoGen ant-mpp). Vari068 is a patient-derived cell line (originally derived from an ER-/PR-/Her2− breast cancer patient who had signed informed consent) adapted to the standard two-dimensional culture environment.11,54,55 We cultured Vari068 cells in DMEM Medium (Gibco 11995) supplemented with 10% fetal bovine serum (FBS, Gibco 16000), 1% GlutaMax (Gibco 35050), 1% penicillin/streptomycin (pen/strep, Gibco 15070), and 0.1% of plasmocin (InvivoGen ant-mpp). All cells were cultured in regular polystyrene culture dishes and passaged at or before cells reached 80% confluency. We maintained all cells at 37 °C in a humidified incubator with 5% CO2.Cell loading on microfluidic cancer spheroid chip
Breast cancer cells were harvested from a petri dish using a 0.05% trypsin/EDTA solution (Gibco, 25300) for 4–6 minutes depending on the cell lines. They were then centrifuged at 100 × g for 4 minutes and re-suspended to a concentration of 4 × 106 cells per mL in regular cell culture media. The microfluidic device was washed with cell culture media twice, and subsequently, 100 μL of the cell suspension was introduced into each inlet of the chip. Cells flowed into the spheroid microwells through designated inflow channels. Subsequently, we gently agitated the device to facilitate the settling of cells located on the sidewalls of microwells into the microwells. After a 5 minute cell loading period, the media in the outlet was removed, and 100 μL of fresh culture media was pipetted into the inlets to wash away excess cells for three times. Since the cells did not adhere to the PDMS substrate, they aggregated with each other within 24 hours.20Drug treatment and drug efficacy readout
We treated spheroids with conventional chemotherapeutics and novel inhibitors. Detailed list and targets are presented in Table S1.† All drugs were initially reconstituted in dimethylsulfoxide (DMSO) to create 10 mM stock solutions. Subsequently, a tenfold serial dilution was performed, ranging from 1 mM to 1 nM, with culture media for treatment purposes. These various drug concentrations were introduced into corresponding inlets on the microfluidic chip. We aspirated outlets and added 100 μL of the drug solution to the inlets. To minimize disruption of aggregated tumor spheroids, 20 μL of media was removed or added each time. Following a 72 hour drug treatment period, cancer spheroids were stained using the LIVE/DEAD Viability/Cytotoxicity Kit for mammalian cells (Life Technologies, L3224). This entailed employing 2 μM of LIVE (calcein-AM) and 4 μM of DEAD (ethidium homodimer-1) staining reagents diluted in media. Imaging experiments were conducted to validate that the fluorescence staining reagents do not alter the phase-contrast morphology of spheroids. To avoid disrupting the spheroids, the drug solution was aspirated from the devices and replaced with 100 μL staining solution, administered in 20 μL increments. An incubation period of 1 hour was employed in an incubator set at 37 °C within a 5% CO2 environment. Subsequently, the microfluidic device was subjected to microscopy for imaging.Image acquisition
Stained cells in the cancer spheroid devices were imaged using an inverted microscope (Nikon Ti2E). The phase-contrast and fluorescence images were taken with a 4× objective lens (1.6 μm per pixel) and a monochrome CMOS camera (Hamamatsu ORCA-Fusion Gen-III SCMOS Camera). The field of view is around 14 mm2, which can cover more than 600 spheroids per micro-chambers in an image. Live staining was imaged by a FITC filter set, and dead staining was imaged by a TRITC filter set. Focusing was performed to ensure the image remained in focus throughout the imaging experiments. To assess transferability across diverse imaging environments, a Lionheart FX automated microscope with Gen 5 software was employed.Image processing program
For image processing, we employed a MATLAB program using the circular Hough transform to accurately extract individual circular microwells containing tumor spheroids. This method detected circular microwells with a predetermined radius, which was effective due to the distinct circular edges and fixed size of each microwell. Phase-contrast images captured the tumor spheroid morphology within these circular microwells. The green and red fluorescence intensities were used to represent LIVE and DEAD signals, respectively. Background fluorescence for LIVE and DEAD staining was determined based on areas outside the circular microwells. To filter out empty microwells, we trained a Convolutional Neural Network (CNN) classification model using 1000 microwells with spheroids and 1000 without.Training of neural network
After image processing, a database with 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 845 spheroids was established for Convolutional Neural Network (CNN) training. The CNN architecture comprises an input layer, multiple hidden convolution layers, and an output layer. Input images undergo convolutional processing to generate classification or prediction outputs. The neural network's structures are available in Fig. S1.† In this investigation, phase-contrast images of tumor spheroids served as input predictor variables. For classification, each spheroid was attributed a categorical label based on its drug treatment concentration, forming the expected output. In cases of regression tasks, anticipated numeric output was assigned to each spheroid, representing its viability score as measured through LIVE/DEAD staining. Given that LIVE/DEAD staining induces background fluorescence, we conducted background subtraction from the signal. The viability score was derived using the formula:
845 spheroids was established for Convolutional Neural Network (CNN) training. The CNN architecture comprises an input layer, multiple hidden convolution layers, and an output layer. Input images undergo convolutional processing to generate classification or prediction outputs. The neural network's structures are available in Fig. S1.† In this investigation, phase-contrast images of tumor spheroids served as input predictor variables. For classification, each spheroid was attributed a categorical label based on its drug treatment concentration, forming the expected output. In cases of regression tasks, anticipated numeric output was assigned to each spheroid, representing its viability score as measured through LIVE/DEAD staining. Given that LIVE/DEAD staining induces background fluorescence, we conducted background subtraction from the signal. The viability score was derived using the formula: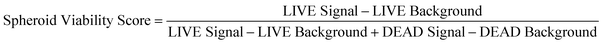 | (1) |
Results and discussion
High-throughput cancer spheroid formation on-chip
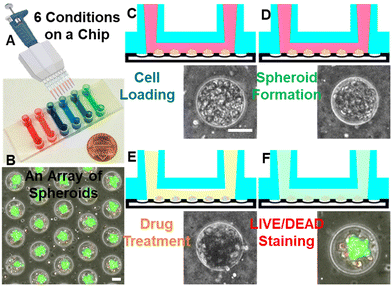
We developed a microfluidic high-throughput cancer spheroid formation platform. This platform leverages the favorable properties of PDMS as a substrate, characterized by its transparency, biocompatibility, gas permeability, and resistance to harsh chemicals, such as chemotherapeutics. The microfluidic substrate is patterned with a compact array of microwells. Each chip contains approximately 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 microwells, six channels for testing six conditions, each equipped with around 2000 spheroid-forming microwells (Fig. 1A and B). This high capacity enables us to conduct efficient and high-throughput cancer drug screening. Media exchange is streamlined, as our design facilitates easy refreshment of the culture medium without disturbing the isolated cancer spheroids in the microwells. This prevents undesired spheroid aggregation during media exchange, a common challenge in conventional non-adherent culture methods. The compact microwell layout with around 51 microwells per square millimeter permits the simultaneous imaging of over 600 microwells. This technology holds great potential for accelerating the identification of novel cancer therapies through high-throughput cancer spheroid testings.
000 microwells, six channels for testing six conditions, each equipped with around 2000 spheroid-forming microwells (Fig. 1A and B). This high capacity enables us to conduct efficient and high-throughput cancer drug screening. Media exchange is streamlined, as our design facilitates easy refreshment of the culture medium without disturbing the isolated cancer spheroids in the microwells. This prevents undesired spheroid aggregation during media exchange, a common challenge in conventional non-adherent culture methods. The compact microwell layout with around 51 microwells per square millimeter permits the simultaneous imaging of over 600 microwells. This technology holds great potential for accelerating the identification of novel cancer therapies through high-throughput cancer spheroid testings.
Cancer cells were retrieved from petri dishes and loaded into the microfluidic device via pipetting through designated inlets. Media flow directed the cells into the channels, and following gentle agitation to ensure that gravity guided the cells into the microwells (Fig. 1C). Surface modification with Pluronic F-108 prevented cell adhesion to the PDMS substrate, allowing cancer cells to aggregate as spheroids through interactions mediated by cell surface adhesion molecules. After a day of cultivation, the cancer spheroids compactly aggregated, becoming suitable for compound treatment and finally LIVE/DEAD staining (Fig. 1D–F). Thanks to the consistent size of the microwells and a uniform cell density per channel, the formation of uniform cancer spheroids (diameter: 67.9 ± 3.9 μm, N = 45, ± indicates standard deviation (S.D.)) ensured the reliability and reproducibility of the experimental results. To handle a large number of images generated by our platform, a customized MATLAB program was developed. This program includes an automated cell cropping function that accurately identifies circular chambers, cropping them for subsequent data analysis and machine learning applications. To eliminate empty microwells, we trained a CNN classification model employing 1000 microwells containing spheroids and 1000 without. This model exhibited an impressive 99% accuracy (Fig. S2†).
Morphological changes of spheroids after drug treatment
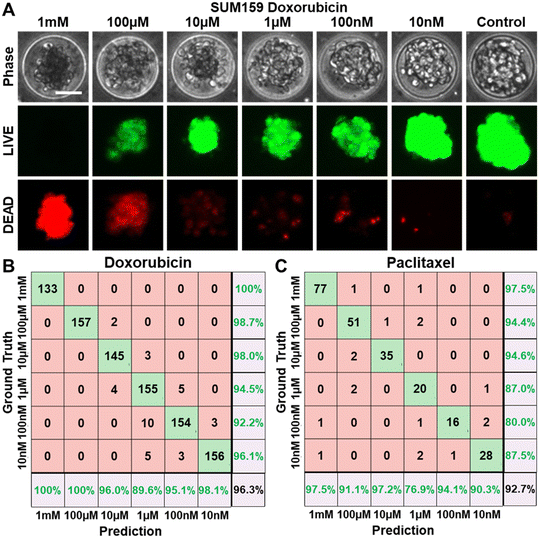
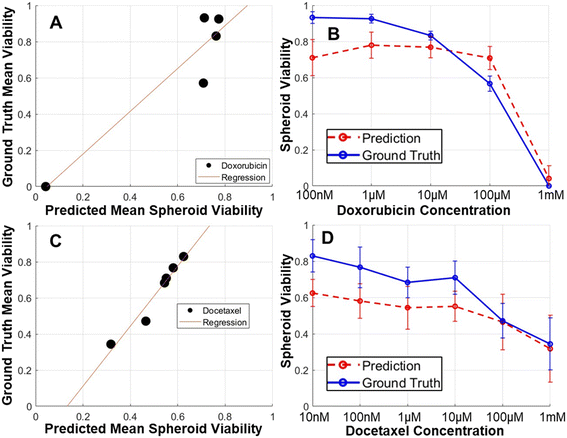
Using our cancer spheroid platform, we tested eight conventional chemotherapeutic drugs to a wide range of six concentrations over three days, based on established protocols. Take doxorubicin as an example: spheroids treated with low concentrations (e.g., 0.01 μM and 0.1 μM) displayed predominantly live cells (green fluorescence via LIVE staining) with few dead cells indicated by sporadic red fluorescence (DEAD staining) (Fig. 2A). The phase-contrast images also revealed a healthy spheroid morphology, featuring sharp boundaries and clear centers. In contrast, high concentrations (e.g., 100 μM, and 1000 μM) resulted in numerous dead cells at the spheroid core indicated by red fluorescence and a prevalence of dark, non-viable cells throughout the spheroid in phase-contrast imaging. These results underline the correlation between spheroid viability determined by fluorescence-based LIVE/DEAD staining and the phase-contrast morphology of spheroids. This correlation supports the feasibility of estimating viability via phase-contrast imaging. To further validate this approach, we employed a CNN model for image classification based on treatment doses. For doxorubicin, the model achieved an impressive 96.3% accuracy in distinguishing between the six concentrations, even though distinguishing low-dose treatments presented some challenges, which were caused by the minimal discernible morphological differences between low-dose treatments (Fig. 2B). Remarkably, high-dose versus low-dose separation yielded consistently high prediction accuracy. The classification of Paclitaxel treatment concentrations also yielded a high accuracy of 92.7% (Fig. 2C).Estimation of cancer spheroid viability using phase-contrast microscopy
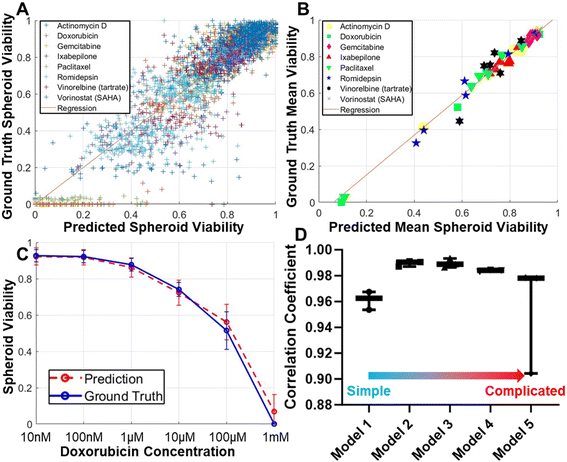
While phase-contrast morphology reveals substantial differences between healthy and compromised tumor spheroids, relying solely on qualitative morphological observations cannot offer a quantitative assessment of spheroid viability. To bridge this gap, we introduced a CNN model designed to quantitatively estimate viability scores. Our CNN model was rigorously trained, using phase-contrast images of spheroids as inputs, with the desired outputs being viability scores based on LIVE/DEAD staining. Our training dataset comprised spheroid images subjected to eight different conventional drugs, each administered at six different concentrations, encompassing a diverse range of treatment conditions. Within this dataset, 80% of the images were designated for training purposes, with the remaining 20% allocated for testing. We initiated our evaluation by comparing the viability scores of individual spheroids, as determined by LIVE/DEAD staining and as predicted by our CNN model from phase-contrast images. We achieved a remarkable high correlation coefficient of 0.908 (Fig. 3A). Given the high correlation, we observed a greater dispersion of data points among spheroids with low viability. This phenomenon can be attributed to two primary factors. Firstly, healthy spheroids typically exhibit a more uniform morphology compared to those that have been compromised. Additionally, our dataset contains a larger proportion of healthy spheroids compared to those significantly inhibited by treatment. Despite efforts to increase the representation of compromised spheroids by employing high drug concentrations of 100 μM or even 1 mM, they remain underrepresented. Further escalation of the drug concentration has practical limitations related to solubility and potential DMSO solvent toxicity. Both inherent morphological heterogeneity and dataset composition contribute to this issue. Given our high throughput to promptly analyze many spheroids, we averaged the spheroids treated under the same conditions to compute average viability scores, resulting in an even higher correlation coefficient of 0.989 (Fig. 3B). These encouraging findings from eight different compounds strongly suggest the feasibility of establishing a generic model for estimating spheroid viability from phase-contrast images. Furthermore, we demonstrated the efficacy of our method by generating treatment response curve based on both LIVE/DEAD staining (ground truth) and CNN predictions (Fig. 3C). In this comparison, the treatment response curve obtained from both approaches demonstrated remarkable consistency, providing validation for our label-free spheroid viability estimation method.Optimization of the convolutional neural network models
While our initial experiments yielded promising outcomes using a 7-layer CNN model, we conducted additional tests to explore optimization possibilities. We compared four possible alternative network structures (Fig. S1†), to investigate the potential enhancements achievable by modifying convolutional layer count and structure. The results, depicted in Fig. 3D, indicated that reducing the number of convolutional layers notably compromised prediction accuracy. Conversely, increasing the number of convolutional layers also yielded a reduction in accuracy due to potential model overfitting. These findings underscore the appropriateness of the originally selected model. Any further complexity would primarily extend computation times and reduce accuracy.Inter-drug validation of the CNN model
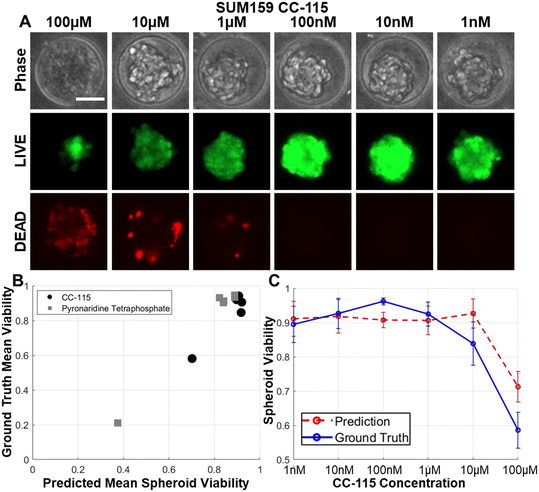
Having effectively predicted outcomes for eight drugs within the training dataset, we proceeded to assess the model's capability in predicting the effects of new compounds outside the training dataset. Given the dataset's diverse cell killing mechanisms, we expected that responses induced by other compounds share notable similarities in resulting spheroid morphology.59,60 To explore this, we tested novel compounds, namely CC-115, and pyronaridine tetraphosphate. CC-115, recognized as a dual inhibitor targeting mTOR kinase and DNA-dependent protein kinase, has demonstrated effectiveness in eliminating cancer cells with heightened oxidative stress levels, as evidenced in renal cell carcinoma, melanoma, and non-small cell lung cancer studies.61–65 Furthermore, its well-tolerated nature in the initial phase I trial has been established.62 Pyronaridine, an aminoquinoline-based antimalarial drug,66 has shown potential in overcoming cancer treatment resistance but remains relatively unexplored in breast cancer.67,68 As anticipated, the application of these compounds effectively inhibited breast cancer spheroids and resulted in consistent morphological changes similar to conventional chemotherapeutic drugs (Fig. 4A). Utilizing the model trained by eight conventional chemotherapeutic drugs, we achieved a high correlation coefficient of 0.961 when predicting the efficacy of the novel compounds (Fig. 4B). Notably, the treatment response curve derived from LIVE/DEAD staining and CNN predictions aligned well (Fig. 4C). This underscores the presence of shared morphological features among spheroids influenced by various drugs, affirming the model's adaptability.Inter-cell line validation of the CNN model
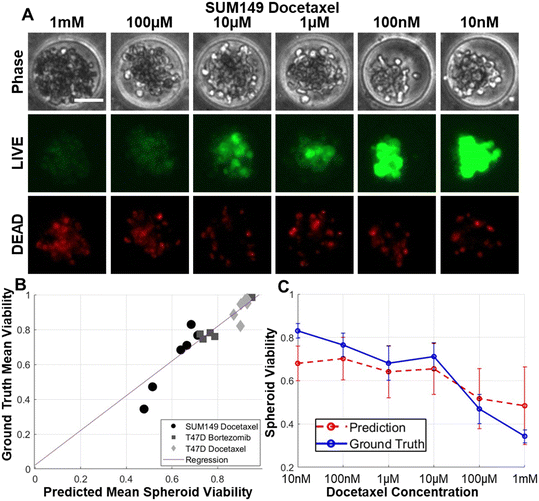
Broadening our investigation, we evaluated various breast cancer cell lines using the model initially trained by a triple-negative breast cancer (TNBC) cell line SUM159. This model was utilized to assess spheroid viability for both TNBC SUM149 and hormone receptor-positive (ER-positive and PR-positive) T47D cell lines. Upon subjecting SUM149 to docetaxel treatment, we observed comparable cell morphology in treated spheroids as SUM159 (Fig. 5A). Additionally, we assessed the effects of docetaxel on SUM149 and bortezomib and docetaxel on T47D (Fig. 5B). The correlation coefficient between the observed and predicted outcomes was 0.945 for SUM149 and T47D. Notably, despite both drugs (bortezomib and docetaxel) and cell lines (SUM149 and T47D) were not being part of the training dataset, the correlation coefficients remained high, only marginally lower than that of SUM159. Moreover, the treatment response curve exhibited consistency between the observed and predicted values (Fig. 5C). In order to enhance the clinical significance of our discoveries, we included breast cancer cells derived from a TNBC patient. The patient derived Vari068 cells successfully formed coherent cancer spheroids within our microfluidic platform. We treated doxorubicin to inhibit Vari068 spheroids and used the CNN model trained with SUM159 cells for predicting the viability of spheroids. Encouragingly, the ground truth viability aligns well with our prediction, displaying a robust correlation coefficient of 0.942 alongside consistent treatment response curves (Fig. 6A and B). This exploration of multiple cell lines yielded positive outcomes. The shared morphological features across diverse cells imply potential applicability of the model for wide-ranging drug screening initiatives.Validation of the model by different imaging environments
To assess the CNN model's transferability to analyze images acquired in diverse imaging settings, we conducted a side-by-side experiment involving two distinct imaging setups. Setup 1 featured a Nikon Ti2E microscope and Hamamatsu ORCA-Fusion Gen-III SCMOS camera, while setup 2 employed a BioTek Lionheart FX automated digital microscope. These variations in microscope types, objective lenses, and cameras created distinct imaging environments, providing a robust test for model transferability. To standardize for different magnifications, we resized the collected images before analysis. Notably, our model was exclusively trained using images from setup 1. As illustrated in Fig. 6C and D, our results demonstrated consistent spheroid viability assessments for images obtained from both setups with a high correlation coefficient of 0.975. These experiments unequivocally establish the method's transferability across different laboratory settings.Dynamic monitoring of tumor spheroid viability
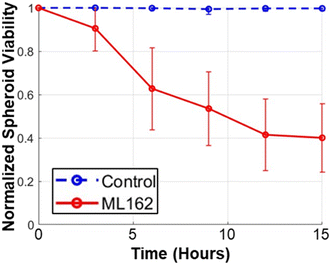
The label-free viability assessment method we employ eliminates the need for toxic LIVE/DEAD staining, allowing continuous and non-invasive monitoring of tumor spheroid viability in real-time. In our study, SUM159 spheroids were treated with ML162, a GPX4 inhibitor known to induce ferroptosis (Fig. 7).69,70 Using our model, we observed a rapid reduction in viability with 10 μM of ML162 treatment compared to the control group. This demonstrates the effectiveness of our approach in dynamic viability estimation without destructive methods, offering potential for accelerated drug discovery and efficacy testing.Conclusion
In cancer research, the search for improved therapeutic agents is a protracted endeavor. Conventional cell culture techniques in 2D dishes or plates fail to faithfully replicate tumor conditions. Various 3D cell culture methods, such as hanging drops, hydrogel embedding, and low-attachment surfaces, address some of these limitations. Microfluidics offers precise cell manipulation for high-throughput drug screening. However, existing viability assays for 3D cancer spheroids can be unreliable and cytotoxic. This study presents a deep learning model for non-invasive, label-free viability estimation of tumor spheroids, providing a cost-effective and high-throughput solution for continuous monitoring of spheroids over time. A high-throughput microfluidic cancer spheroid formation platform was developed. This device features around 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 microwells per chip across six channels for efficient high-throughput drug screening. It facilitates media exchange without disrupting the spheroids, ensuring reliable results. A custom MATLAB program was employed for precise image analysis to support machine learning applications. Eight conventional chemotherapeutic drugs were tested on cancer spheroids at various concentrations. Visual observations suggested a link between spheroid viability and their phase-contrast morphology, supporting viability estimation through phase-contrast imaging. A CNN classification model achieved remarkable accuracy in classifying images based on treatment dosages, even for the challenging task of distinguishing low-dose treatments. A CNN regression model was trained to quantitatively estimate viability scores based on phase-contrast images, showing a strong correlation with LIVE/DEAD staining. Further validation generated treatment response curves that closely aligned with ground truth values. Additional experiments explored alternative CNN models, affirming the appropriateness of the selected CNN model. The model exhibited robustness when tested with different drugs those were not in the training dataset, suggesting shared morphological features among spheroids affected by various compounds. Furthermore, the model effectively predicted viability for different breast cancer cell lines and patient derived cells. This research holds the potential for a universal model for spheroid viability estimation across compounds and cell lines. The study also assessed the CNN model's adaptability by comparing images from two different microscopy setups, demonstrating consistent spheroid viability assessments, and highlighting the method's transferability across diverse laboratory environments. In conclusion, the label-free approach offers several key advantages, including: (1) cost and time reduction by eliminating the need for fluorescent staining, (2) increased throughput by circumventing the time-consuming process of fluorescent microscopy, and (3) non-destructive measurement, enabling the tracking of spheroid changes throughout drug administration. The successful validation of our method across novel compounds, additional cell lines, primary cells not in the training dataset, and images sourced from external laboratories sets the stage for high-throughput screening of 3D spheroids with numerous compounds within microfluidics.
000 microwells per chip across six channels for efficient high-throughput drug screening. It facilitates media exchange without disrupting the spheroids, ensuring reliable results. A custom MATLAB program was employed for precise image analysis to support machine learning applications. Eight conventional chemotherapeutic drugs were tested on cancer spheroids at various concentrations. Visual observations suggested a link between spheroid viability and their phase-contrast morphology, supporting viability estimation through phase-contrast imaging. A CNN classification model achieved remarkable accuracy in classifying images based on treatment dosages, even for the challenging task of distinguishing low-dose treatments. A CNN regression model was trained to quantitatively estimate viability scores based on phase-contrast images, showing a strong correlation with LIVE/DEAD staining. Further validation generated treatment response curves that closely aligned with ground truth values. Additional experiments explored alternative CNN models, affirming the appropriateness of the selected CNN model. The model exhibited robustness when tested with different drugs those were not in the training dataset, suggesting shared morphological features among spheroids affected by various compounds. Furthermore, the model effectively predicted viability for different breast cancer cell lines and patient derived cells. This research holds the potential for a universal model for spheroid viability estimation across compounds and cell lines. The study also assessed the CNN model's adaptability by comparing images from two different microscopy setups, demonstrating consistent spheroid viability assessments, and highlighting the method's transferability across diverse laboratory environments. In conclusion, the label-free approach offers several key advantages, including: (1) cost and time reduction by eliminating the need for fluorescent staining, (2) increased throughput by circumventing the time-consuming process of fluorescent microscopy, and (3) non-destructive measurement, enabling the tracking of spheroid changes throughout drug administration. The successful validation of our method across novel compounds, additional cell lines, primary cells not in the training dataset, and images sourced from external laboratories sets the stage for high-throughput screening of 3D spheroids with numerous compounds within microfluidics.
Declaration of generative AI in scientific writing
During the preparation of this work the authors used ChatGPT 3.5 in order to improve readability and language. After using it, the authors reviewed and edited the content as needed and take full responsibility for the content of the publication.Author contributions
C.-C. C., E. C. R., and Y.-C. C. performed the fabrication of microfluidic devices. C.-C. C., S. H., M. Z., J. C., and Y.-C. C. performed cancer spheroid experiments. R. A., P. C., R. M. S., and Y.-C. C. developed the MATLAB software and deep learning models for image processing. Y. G. and Y.-C. C. supervised the study. C.-C. C., R. A., and Y.-C. C. wrote the manuscript. All authors discussed the results and commented on the manuscript.Conflicts of interest
The authors declare no competing financial interests.Acknowledgements
This study was generously supported by start-up funding from the UPMC Hillman Cancer Center, the Women's Cancer Research Center (WCRC) at Magee Women's Research Institute, the Pitt CTSI Pilot project (This project was supported by the National Institutes of Health through Grant Number UL1TR001857.), and the US National Institutes of Health grants P30 CA047904, P50 CA272218, and R35 GM150509 and the UPMC Competitive Medical Research Fund (CMRF) to Y.-C. C. We thank the Nanoscale Fabrication Characterization Facility of the University of Pittsburgh (Pittsburgh, PA) for device fabrication and Drs. Adrian Lee and Steffi Oesterreich (University of Pittsburgh) for insightful discussion.References
- R. L. Siegel, K. D. Miller, H. E. Fuchs and A. Jemal, Cancer Statistics, 2021, Ca-Cancer J. Clin., 2021, 71(1), 7–33, DOI:10.3322/caac.21654 , Epub 2021/01/13, PubMed PMID: 33433946.
- G. Cosentino, I. Plantamura, E. Tagliabue, M. V. Iorio and A. Cataldo, Breast Cancer Drug Resistance: Overcoming the Challenge by Capitalizing on MicroRNA and Tumor Microenvironment Interplay, Cancers, 2021, 13(15), 3691, DOI:10.3390/cancers13153691 , Epub 2021/08/08, PubMed PMID: 34359591, PMCID: PMC8345203.
- K. V. Kitaeva, C. S. Rutland, A. A. Rizvanov and V. V. Solovyeva, Cell Culture Based in vitro Test Systems for Anticancer Drug Screening, Front. Bioeng. Biotechnol., 2020, 8, 322, DOI:10.3389/fbioe.2020.00322 , Epub 2020/04/25, PubMed PMID: 32328489, PMCID: PMC7160228.
- B. Pinto, A. C. Henriques, P. M. A. Silva and H. Bousbaa, Three-Dimensional Spheroids as In Vitro Preclinical Models for Cancer Research, Pharmaceutics, 2020, 12(12), 1186, DOI:10.3390/pharmaceutics12121186 , Epub 2020/12/10, PubMed PMID: 33291351, PMCID: PMC7762220.
- S. Nath and G. R. Devi, Three-dimensional culture systems in cancer research: Focus on tumor spheroid model, Pharmacol. Ther., 2016, 163, 94–108, DOI:10.1016/j.pharmthera.2016.03.013 , Epub 2016/04/12, PubMed PMID: 27063403, PMCID: PMC4961208.
- S. J. Han, S. Kwon and K. S. Kim, Challenges of applying multicellular tumor spheroids in preclinical phase, Cancer Cell Int., 2021, 21(1), 152, DOI:10.1186/s12935-021-01853-8 , Epub 2021/03/06, PubMed PMID: 33663530, PMCID: PMC7934264.
- F. Perche and V. P. Torchilin, Cancer cell spheroids as a model to evaluate chemotherapy protocols, Cancer Biol. Ther., 2012, 13(12), 1205–1213, DOI:10.4161/cbt.21353 , Epub 2012/08/16, PubMed PMID: 22892843, PMCID: PMC3469478.
- T. Ishiguro, H. Ohata, A. Sato, K. Yamawaki, T. Enomoto and K. Okamoto, Tumor-derived spheroids: Relevance to cancer stem cells and clinical applications, Cancer Sci., 2017, 108(3), 283–289, DOI:10.1111/cas.13155 , Epub 2017/01/09, PubMed PMID: 28064442, PMCID: PMC5378268.
- J. Liao, F. Qian, N. Tchabo, P. Mhawech-Fauceglia, A. Beck, Z. Qian, X. Wang, W. J. Huss, S. B. Lele, C. D. Morrison and K. Odunsi, Ovarian cancer spheroid cells with stem cell-like properties contribute to tumor generation, metastasis and chemotherapy resistance through hypoxia-resistant metabolism, PLoS One, 2014, 9(1), e84941, DOI:10.1371/journal.pone.0084941 , Epub 2014/01/11, PubMed PMID: 24409314; PMCID: PMC3883678.
- C. Ginestier, M. H. Hur, E. Charafe-Jauffret, F. Monville, J. Dutcher, M. Brown, J. Jacquemier, P. Viens, C. G. Kleer, S. Liu, A. Schott, D. Hayes, D. Birnbaum, M. S. Wicha and G. Dontu, ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome, Cell Stem Cell, 2007, 1(5), 555–567, DOI:10.1016/j.stem.2007.08.014 , Epub 2008/03/29, PubMed PMID: 18371393, PMCID: PMC2423808.
- Y. C. Chen, P. N. Ingram, S. Fouladdel, S. P. McDermott, E. Azizi, M. S. Wicha and E. Yoon, High-Throughput Single-Cell Derived Sphere Formation for Cancer Stem-Like Cell Identification and Analysis, Sci. Rep., 2016, 6, 27301, DOI:10.1038/srep27301 , Epub 2016/06/14, PubMed PMID: 27292795, PMCID: PMC4904376.
- Y. C. Tung, A. Y. Hsiao, S. G. Allen, Y. S. Torisawa, M. Ho and S. Takayama, High-throughput 3D spheroid culture and drug testing using a 384 hanging drop array, Analyst, 2011, 136(3), 473–478, 10.1039/c0an00609b , Epub 2010/10/23, PubMed PMID: 20967331, PMCID: PMC7454010.
- D. Rodoplu, J. S. Matahum and C. H. Hsu, A microfluidic hanging drop-based spheroid co-culture platform for probing tumor angiogenesis, Lab Chip, 2022, 22(7), 1275–1285, 10.1039/d1lc01177d , Epub 2022/02/23, PubMed PMID: 35191460.
- R. Foty, A simple hanging drop cell culture protocol for generation of 3D spheroids, J. Visualized Exp., 2011,(51), 2720, DOI:10.3791/2720 , Epub 2011/05/19, PubMed PMID: 21587162, PMCID: PMC3197119.
- O. Frey, P. M. Misun, D. A. Fluri, J. G. Hengstler and A. Hierlemann, Reconfigurable microfluidic hanging drop network for multi-tissue interaction and analysis, Nat. Commun., 2014, 5, 4250, DOI:10.1038/ncomms5250 , Epub 2014/07/01, PubMed PMID: 24977495.
- Y. Li and E. Kumacheva, Hydrogel microenvironments for cancer spheroid growth and drug screening, Sci. Adv., 2018, 4(4), eaas8998, DOI:10.1126/sciadv.aas8998 , Epub 2018/05/03, PubMed PMID: 29719868, PMCID: PMC5922799.
- A. E. G. Baker, R. Y. Tam and M. S. Shoichet, Independently Tuning the Biochemical and Mechanical Properties of 3D Hyaluronan-Based Hydrogels with Oxime and Diels-Alder Chemistry to Culture Breast Cancer Spheroids, Biomacromolecules, 2017, 18(12), 4373–4384, DOI:10.1021/acs.biomac.7b01422 , Epub 2017/10/19, PubMed PMID: 29040808.
- S. Nam, R. Stowers, J. Lou, Y. Xia and O. Chaudhuri, Varying PEG density to control stress relaxation in alginate-PEG hydrogels for 3D cell culture studies, Biomaterials, 2019, 200, 15–24, DOI:10.1016/j.biomaterials.2019.02.004 , Epub 2019/02/12, PubMed PMID: 30743050, PMCID: PMC6463514.
- S. E. Lehman, J. M. McCracken, L. A. Miller, S. Jayalath and R. G. Nuzzo, Biocompliant Composite Au/pHEMA Plasmonic Scaffolds for 3D Cell Culture and Noninvasive Sensing of Cellular Metabolites, Adv. Healthcare Mater., 2021, 10(4), e2001040, DOI:10.1002/adhm.202001040 , Epub 2020/09/10, PubMed PMID: 32902201.
- Y. C. Chen, X. Lou, Z. Zhang, P. Ingram and E. Yoon, High-Throughput Cancer Cell Sphere Formation for Characterizing the Efficacy of Photo Dynamic Therapy in 3D Cell Cultures, Sci. Rep., 2015, 5, 12175, DOI:10.1038/srep12175 , Epub 2015/07/15, PubMed PMID: 26153550, PMCID: PMC4495468.
- V. A. Liu, W. E. Jastromb and S. N. Bhatia, Engineering protein and cell adhesivity using PEO-terminated triblock polymers, J. Biomed. Mater. Res., 2002, 60(1), 126–134, DOI:10.1002/jbm.10005 , Epub 2002/02/09, PubMed PMID: 11835168.
- W. Hellmich, J. Regtmeier, T. T. Duong, R. Ros, D. Anselmetti and A. Ros, Poly(oxyethylene) based surface coatings for poly(dimethylsiloxane) microchannels, Langmuir, 2005, 21(16), 7551–7557, DOI:10.1021/la0510432 , Epub 2005/07/27, PubMed PMID: 16042494.
- Y. H. Cheng, Y. C. Chen, R. Brien and E. Yoon, Scaling and automation of a high-throughput single-cell-derived tumor sphere assay chip, Lab Chip, 2016, 16(19), 3708–3717, 10.1039/c6lc00778c , Epub 2016/08/12, PubMed PMID: 27510097, PMCID: PMC6559352.
- A. R. Holub, A. Huo, K. Patel, V. Thakore, P. Chhibber and F. Erogbogbo, Assessing Advantages and Drawbacks of Rapidly Generated Ultra-Large 3D Breast Cancer Spheroids: Studies with Chemotherapeutics and Nanoparticles, Int. J. Mol. Sci., 2020, 21(12), 4413, DOI:10.3390/ijms21124413 , Epub 2020/06/25, PubMed PMID: 32575896, PMCID: PMC7352930.
- C. Y. Cho, T. H. Chiang, L. H. Hsieh, W. Y. Yang, H. H. Hsu, C. K. Yeh, C. C. Huang and J. H. Huang, Development of a Novel Hanging Drop Platform for Engineering Controllable 3D Microenvironments, Front. Cell Dev. Biol., 2020, 8, 327, DOI:10.3389/fcell.2020.00327 , Epub 2020/05/28, PubMed PMID: 32457907, PMCID: PMC7221142.
- A. Sontheimer-Phelps, B. A. Hassell and D. E. Ingber, Modelling cancer in microfluidic human organs-on-chips, Nat. Rev. Cancer, 2019, 19(2), 65–81, DOI:10.1038/s41568-018-0104-6 , Epub 2019/01/17, PubMed PMID: 30647431.
- Y. Lee, Z. Chen, W. Lim, H. Cho and S. Park, High-Throughput Screening of Anti-cancer Drugs Using a Microfluidic Spheroid Culture Device with a Concentration Gradient Generator, Curr. Protoc., 2022, 2(9), e529, DOI:10.1002/cpz1.529 , Epub 2022/09/07, PubMed PMID: 36066205.
- J. Ro, J. Kim and Y. K. Cho, Recent advances in spheroid-based microfluidic models to mimic the tumour microenvironment, Analyst, 2022, 147(10), 2023–2034, 10.1039/d2an00172a , Epub 2022/04/30, PubMed PMID: 35485712.
- B. Patra, C. C. Peng, W. H. Liao, C. H. Lee and Y. C. Tung, Drug testing and flow cytometry analysis on a large number of uniform sized tumor spheroids using a microfluidic device, Sci. Rep., 2016, 6, 21061, DOI:10.1038/srep21061 , Epub 2016/02/16, PubMed PMID: 26877244, PMCID: PMC4753452.
- S. Y. Jeong, J. H. Lee, Y. Shin, S. Chung and H. J. Kuh, Co-Culture of Tumor Spheroids and Fibroblasts in a Collagen Matrix-Incorporated Microfluidic Chip Mimics Reciprocal Activation in Solid Tumor Microenvironment, PLoS One, 2016, 11(7), e0159013, DOI:10.1371/journal.pone.0159013 , Epub 2016/07/09, PubMed PMID: 27391808, PMCID: PMC4938568.
- S. Gunti, A. T. K. Hoke, K. P. Vu and N. R. London Jr., Organoid and Spheroid Tumor Models: Techniques and Applications, Cancers, 2021, 13(4), 874, DOI:10.3390/cancers13040874 , Epub 2021/03/07, PubMed PMID: 33669619, PMCID: PMC7922036.
- M. D. Bourn, D. V. B. Batchelor, N. Ingram, J. R. McLaughlan, P. L. Coletta, S. D. Evans and S. A. Peyman, High-throughput microfluidics for evaluating microbubble enhanced delivery of cancer therapeutics in spheroid cultures, J. Controlled Release, 2020, 326, 13–24, DOI:10.1016/j.jconrel.2020.06.011 , Epub 2020/06/21, PubMed PMID: 32562855.
- A. Aung, S. K. Davey, J. Theprungsirikul, V. Kumar and S. Varghese, Deciphering the Mechanics of Cancer Spheroid Growth in 3D Environments through Microfluidics Driven Mechanical Actuation, Adv. Healthcare Mater., 2023, 12(14), e2201842, DOI:10.1002/adhm.202201842 , Epub 2022/11/16, PubMed PMID: 36377350, PMCID: PMC10183055.
- B. F. Bender, A. P. Aijian and R. L. Garrell, Digital microfluidics for spheroid-based invasion assays, Lab Chip, 2016, 16(8), 1505–1513, 10.1039/c5lc01569c , Epub 2016/03/30, PubMed PMID: 27020962.
- X. Y. Rima, J. Zhang, L. T. H. Nguyen, A. Rajasuriyar, M. J. Yoon, C. L. Chiang, N. Walters, K. J. Kwak, L. J. Lee and E. Reategui, Microfluidic harvesting of breast cancer tumor spheroid-derived extracellular vesicles from immobilized microgels for single-vesicle analysis, Lab Chip, 2022, 22(13), 2502–2518, 10.1039/d1lc01053k , Epub 2022/05/18, PubMed PMID: 35579189, PMCID: PMC9383696.
- Z. Zhang, Y. C. Chen, S. Urs, L. Chen, D. M. Simeone and E. Yoon, Scalable Multiplexed Drug-Combination Screening Platforms Using 3D Microtumor Model for Precision Medicine, Small, 2018, 14(42), e1703617, DOI:10.1002/smll.201703617 , Epub 2018/09/22, PubMed PMID: 30239130.
- L. H. Levkoff, G. P. Marshall 2nd, H. H. Ross, M. Caldeira, B. A. Reynolds, M. Cakiroglu, C. L. Mariani, W. J. Streit and E. D. Laywell, Bromodeoxyuridine inhibits cancer cell proliferation in vitro and in vivo, Neoplasia, 2008, 10(8), 804–816, DOI:10.1593/neo.08382 , Epub 2008/08/07, PubMed PMID: 18680882, PMCID: PMC2504767.
- C. Hu, S. He, Y. J. Lee, Y. He, E. M. Kong, H. Li, M. A. Anastasio and G. Popescu, Live-dead assay on unlabeled cells using phase imaging with computational specificity, Nat. Commun., 2022, 13(1), 713, DOI:10.1038/s41467-022-28214-x , Epub 2022/02/09, PubMed PMID: 35132059.
- L. Netuschil, T. M. Auschill, A. Sculean and N. B. Arweiler, Confusion over live/dead stainings for the detection of vital microorganisms in oral biofilms--which stain is suitable?, BMC Oral Health, 2014, 14, 2, DOI:10.1186/1472-6831-14-2 , Epub 2014/01/15, PubMed PMID: 24410850, PMCID: PMC3898065.
- S. T. Smale, Luciferase assay, Cold Spring Harb. Protoc., 2010, 2010(5), pdb prot5421, DOI:10.1101/pdb.prot5421 , Epub 2010/05/05, PubMed PMID: 20439408.
- T. L. Riss, R. A. Moravec, A. L. Niles, S. Duellman, H. A. Benink, T. J. Worzella and L. Minor, S. Markossian, A. Grossman, K. Brimacombe, M. Arkin, D. Auld, C. Austin, J. Baell, T. D. Y. Chung, N. P. Coussens, J. L. Dahlin, V. Devanarayan, T. L. Foley, M. Glicksman, K. Gorshkov, J. V. Haas, M. D. Hall, S. Hoare, J. Inglese, P. W. Iversen, S. C. Kales, M. Lal-Nag, Z. Li, J. McGee, O. McManus, T. Riss, P. Saradjian, G. S. Sittampalam, M. Tarselli, O. J. Trask Jr., Y. Wang, J. R. Weidner, M. J. Wildey, K. Wilson, M. Xia and X. Xu, Assay Guidance Manual, Bethesda (MD), 2004, PubMed PMID: 23805433 Search PubMed.
- P. Eulenberg, N. Kohler, T. Blasi, A. Filby, A. E. Carpenter, P. Rees, F. J. Theis and F. A. Wolf, Reconstructing cell cycle and disease progression using deep learning, Nat. Commun., 2017, 8(1), 463, DOI:10.1038/s41467-017-00623-3 , Epub 2017/09/08, PubMed PMID: 28878212, PMCID: PMC5587733.
- T. Blasi, H. Hennig, H. D. Summers, F. J. Theis, J. Cerveira, J. O. Patterson, D. Davies, A. Filby, A. E. Carpenter and P. Rees, Label-free cell cycle analysis for high-throughput imaging flow cytometry, Nat. Commun., 2016, 7, 10256, DOI:10.1038/ncomms10256 , Epub 2016/01/08, PubMed PMID: 26739115; PMCID: PMC4729834.
- N. U. Din and J. Yu, Training a deep learning model for single-cell segmentation without manual annotation, Sci. Rep., 2021, 11(1), 23995, DOI:10.1038/s41598-021-03299-4 , Epub 2021/12/16, PubMed PMID: 34907213, PMCID: PMC8671438.
- Y. Nagao, M. Sakamoto, T. Chinen, Y. Okada and D. Takao, Robust classification of cell cycle phase and biological feature extraction by image-based deep learning, Mol. Biol. Cell, 2020, 31(13), 1346–1354, DOI:10.1091/mbc.E20-03-0187 , Epub 2020/04/23, PubMed PMID: 32320349, PMCID: PMC7353138.
- N. Korber, MIA is an open-source standalone deep learning application for microscopic image analysis, Cells Rep. Methods, 2023, 3(7), 100517, DOI:10.1016/j.crmeth.2023.100517 , Epub 2023/08/03, PubMed PMID: 37533647, PMCID: PMC10391334.
- E. B. Hartnett, M. Zhou, Y. N. Gong and Y. C. Chen, LANCE: a Label-Free Live Apoptotic and Necrotic Cell Explorer Using Convolutional Neural Network Image Analysis, Anal. Chem., 2022, 94(43), 14827–14834, DOI:10.1021/acs.analchem.2c00878 , Epub 2022/10/18, PubMed PMID: 36251981.
- T. Parnamaa and L. Parts, Accurate Classification of Protein Subcellular Localization from High-Throughput Microscopy Images Using Deep Learning, G3: Genes, Genomes, Genet., 2017, 7(5), 1385–1392, DOI:10.1534/g3.116.033654 , Epub 2017/04/10, PubMed PMID: 28391243, PMCID: PMC5427497.
- H. Kobayashi, C. Lei, Y. Wu, A. Mao, Y. Jiang, B. Guo, Y. Ozeki and K. Goda, Label-free detection of cellular drug responses by high-throughput bright-field imaging and machine learning, Sci. Rep., 2017, 7(1), 12454, DOI:10.1038/s41598-017-12378-4 , Epub 2017/10/01, PubMed PMID: 28963483, PMCID: PMC5622112.
- I. Abd El-Sadek, L. T. Shen, T. Mori, S. Makita, P. Mukherjee, A. Lichtenegger, S. Matsusaka and Y. Yasuno, Label-free drug response evaluation of human derived tumor spheroids using three-dimensional dynamic optical coherence tomography, Sci. Rep., 2023, 13(1), 15377, DOI:10.1038/s41598-023-41846-3 , Epub 2023/09/17, PubMed PMID: 37717067, PMCID: PMC10505213.
- Z. Zhang, L. Chen, Y. Wang, T. Zhang, Y. C. Chen and E. Yoon, Label-Free Estimation of Therapeutic Efficacy on 3D Cancer Spheres Using Convolutional Neural Network Image Analysis, Anal. Chem., 2019, 91(21), 14093–14100, DOI:10.1021/acs.analchem.9b03896 , Epub 2019/10/12, PubMed PMID: 31601098.
- J. M. Corey, C. C. Gertz, T. J. Sutton, Q. Chen, K. B. Mycek, B. S. Wang, A. A. Martin, S. L. Johnson and E. L. Feldman, Patterning N-type and S-type neuroblastoma cells with Pluronic F108 and ECM proteins, J. Biomed. Mater. Res., Part A, 2010, 93(2), 673–686, DOI:10.1002/jbm.a.32485 , Epub 2009/07/18, PubMed PMID: 19609877, PMCID: PMC2845720.
- Z. F. Bielecka, K. Maliszewska-Olejniczak, I. J. Safir, C. Szczylik and A. M. Czarnecka, Three-dimensional cell culture model utilization in cancer stem cell research, Biol. Rev. Cambridge Philos. Soc., 2017, 92(3), 1505–1520, DOI:10.1111/brv.12293 , Epub 2016/08/23, PubMed PMID: 27545872.
- M. Liu, Y. Liu, L. Deng, D. Wang, X. He, L. Zhou, M. S. Wicha, F. Bai and S. Liu, Transcriptional profiles of different states of cancer stem cells in triple-negative breast cancer, Mol. Cancer, 2018, 17(1), 65, DOI:10.1186/s12943-018-0809-x , Epub 2018/02/24, PubMed PMID: 29471829, PMCID: PMC5824475.
- K. M. Aw Yong, P. J. Ulintz, S. Caceres, X. Cheng, L. Bao, Z. Wu, E. M. Jiagge and S. D. Merajver, Heterogeneity at the invasion front of triple negative breast cancer cells, Sci. Rep., 2020, 10(1), 5781, DOI:10.1038/s41598-020-62516-8 , Epub 2020/04/03, PubMed PMID: 32238832, PMCID: PMC7113246.
- C. Sumi, T. Ou, J. Takishima and S. Shirafuji, Considerations about L2- and L1-norm regularizations for ultrasound reverberation characteristics imaging and vectoral Doppler measurement, Annu. Int. Conf. IEEE Eng. Med. Biol. Soc., 2022, 2022, 3882–3886, DOI:10.1109/EMBC48229.2022.9870991 , Epub 2022/09/11, PubMed PMID: 36085805.
- B. Bilgic, I. Chatnuntawech, A. P. Fan, K. Setsompop, S. F. Cauley, L. L. Wald and E. Adalsteinsson, Fast image reconstruction with L2-regularization, J. Magn. Reson. Imaging, 2014, 40(1), 181–191, DOI:10.1002/jmri.24365 , Epub 2014/01/08, PubMed PMID: 24395184, PMCID: PMC4106040.
- Y. C. Chen, Z. Zhang and E. Yoon, Early Prediction of Single-Cell Derived Sphere Formation Rate Using Convolutional Neural Network Image Analysis, Anal. Chem., 2020, 92(11), 7717–7724, DOI:10.1021/acs.analchem.0c00710 , Epub 2020/05/20, PubMed PMID: 32427465, PMCID: PMC9552208.
- X. Tong, R. Tang, M. Xiao, J. Xu, W. Wang, B. Zhang, J. Liu, X. Yu and S. Shi, Targeting cell death pathways for cancer therapy: recent developments in necroptosis, pyroptosis, ferroptosis, and cuproptosis research, J. Hematol. Oncol., 2022, 15(1), 174, DOI:10.1186/s13045-022-01392-3 , Epub 2022/12/10, PubMed PMID: 36482419, PMCID: PMC9733270.
- B. A. Carneiro and W. S. El-Deiry, Targeting apoptosis in cancer therapy, Nat. Rev. Clin. Oncol., 2020, 17(7), 395–417, DOI:10.1038/s41571-020-0341-y , Epub 2020/03/24, PubMed PMID: 32203277, PMCID: PMC8211386.
- T. Tsuji, L. M. Sapinoso, T. Tran, B. Gaffney, L. Wong, S. Sankar, H. K. Raymon, D. S. Mortensen and S. Xu, CC-115, a dual inhibitor of mTOR kinase and DNA-PK, blocks DNA damage repair pathways and selectively inhibits ATM-deficient cell growth in vitro, Onco Targets Ther, 2017, 8(43), 74688–74702, DOI:10.18632/oncotarget.20342 , Epub 2017/11/02, PubMed PMID: 29088817, PMCID: PMC5650372.
- P. Munster, M. Mita, A. Mahipal, J. Nemunaitis, C. Massard, T. Mikkelsen, C. Cruz, L. Paz-Ares, M. Hidalgo, D. Rathkopf, G. Blumenschein , Jr., D. C. Smith, B. Eichhorst, T. Cloughesy, E. H. Filvaroff, S. Li, H. Raymon, H. de Haan, K. Hege and J. C. Bendell, First-In-Human Phase I Study Of A Dual mTOR Kinase And DNA-PK Inhibitor (CC-115) In Advanced Malignancy, Cancer Manage. Res., 2019, 11, 10463–10476, DOI:10.2147/CMAR.S208720 , Epub 2019/12/20, PubMed PMID: 31853198, PMCID: PMC6916675.
- F. Burkel, T. Jost, M. Hecht, L. Heinzerling, R. Fietkau and L. Distel, Dual mTOR/DNA-PK Inhibitor CC-115 Induces Cell Death in Melanoma Cells and Has Radiosensitizing Potential, Int. J. Mol. Sci., 2020, 21(23), 9321, DOI:10.3390/ijms21239321 , Epub 2020/12/11, PubMed PMID: 33297429, PMCID: PMC7730287.
- B. Zheng, X. Sun, X. F. Chen, Z. Chen, W. L. Zhu, H. Zhu and D. H. Gu, Dual inhibition of DNA-PKcs and mTOR by CC-115 potently inhibits human renal cell carcinoma cell growth, Aging, 2020, 12(20), 20445–20456, DOI:10.18632/aging.103847 , Epub 2020/10/29, PubMed PMID: 33109772, PMCID: PMC7655216.
- F. Chen, H. Zhao, C. Li, P. Li and Q. Zhang, An mTOR and DNA-PK dual inhibitor CC-115 hinders non-small cell lung cancer cell growth, Cell Death Discovery, 2022, 8(1), 293, DOI:10.1038/s41420-022-01082-6 , Epub 2022/06/19, PubMed PMID: 35717530, PMCID: PMC9206683.
- C. Bailly, Pyronaridine: An update of its pharmacological activities and mechanisms of action, Biopolymers, 2021, 112(4), e23398, DOI:10.1002/bip.23398 , Epub 2020/12/07, PubMed PMID: 33280083.
- J. Qi, C. Z. Yang, C. Y. Wang, S. B. Wang, M. Yang and J. H. Wang, Function and mechanism of pyronaridine: a new inhibitor of P-glycoprotein-mediated multidrug resistance, Acta Pharmacol. Sin., 2002, 23(6), 544–550 CAS , Epub 2002/06/13, PubMed PMID: 12060530.
- D. Duarte, I. Guerreiro and N. Vale, Novel Strategies for Cancer Combat: Drug Combination Using Repurposed Drugs Induces Synergistic Growth Inhibition of MCF-7 Breast and HT-29 Colon Cancer Cells, Curr. Issues Mol. Biol., 2022, 44(10), 4930–4949, DOI:10.3390/cimb44100335 , Epub 2022/10/27, PubMed PMID: 36286050, PMCID: PMC9601176.
- L. Wang, X. Chen and C. Yan, Ferroptosis: An emerging therapeutic opportunity for cancer, Genes Dis., 2022, 9(2), 334–346, DOI:10.1016/j.gendis.2020.09.005 , Epub 2020/09/29, PubMed PMID: 35224150, PMCID: PMC8843872.
- M. Zhou, Y. Ma, C. C. Chiang, E. C. Rock, S. C. Butler, R. Anne, S. Yatsenko, Y. Gong and Y. C. Chen, Single-cell morphological and transcriptome analysis unveil inhibitors of polyploid giant breast cancer cells in vitro, Commun. Biol., 2023, 6(1), 1301, DOI:10.1038/s42003-023-05674-5 , Epub 2023/12/22, PubMed PMID: 38129519, PMCID: PMC10739852.
Footnotes |
| † Electronic supplementary information (ESI) available: Supporting information includes: (1) detailed neural network structure, (2) prediction accuracy between chambers with and without spheroids, and (3) the list of compounds used in this study. See DOI: https://doi.org/10.1039/d4lc00197d |
| ‡ Equal contributions. |
| This journal is © The Royal Society of Chemistry 2024 |