Open Access Article
Open Access ArticleCharacterization of lactic acid bacteria isolated from human breast milk and their bioactive metabolites with potential application as a probiotic food supplement
Abel
Navarré†
,
Tiago
Nazareth†
,
Carlos
Luz
,
Giuseppe
Meca
and
Laura
Escrivá
 *
*
Laboratory of Food Chemistry and Toxicology, Faculty of Pharmacy, University of Valencia, Av/Vicent A. Estellés, s/n 46100 Burjassot, Valencia, Spain. E-mail: laura.escriva@uv.es
First published on 20th June 2024
Abstract
The probiotic properties of twenty-five lactic acid bacteria (LAB) isolated from human breast milk were investigated considering their resistance to gastrointestinal conditions and proteolytic activity. Seven LAB were identified and assessed for auto- and co-aggregation capacity, antibiotic resistance, and behavior during in vitro gastrointestinal digestion. Three Lacticaseibacillus strains were further evaluated for antifungal activity, metabolite production (HPLC-Q-TOF-MS/MS and GC-MS/MS) and proteolytic profiles (SDS-PAGE and HPLC-DAD) in fermented milk, whey, and soy beverage. All strains resisted in vitro gastrointestinal digestion with viable counts higher than 7.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1 after the colonic phase. Remarkable proteolytic activity was observed for 18/25 strains. Bacterial auto- and co-aggregation of 7 selected strains reached values up to 23 and 20%, respectively. L. rhamnosus B5H2, L. rhamnosus B9H2 and L. paracasei B10L2 inhibited P. verrucosum, F. verticillioides and F. graminearum fungal growth, highlighting L. rhamnosus B5H2. Several metabolites were identified, including antifungal compounds such as phenylacetic acid and 3-phenyllactic acid, and volatile organic compounds produced in fermented milk, whey, and soy beverage. SDS-PAGE demonstrated bacterial hydrolysis of the main milk (caseins) and soy (glycines and beta-conglycines) proteins, with no apparent hydrolysis of whey proteins. However, HPLC-DAD revealed alpha-lactoglobulin reduction up to 82% and 54% in milk and whey, respectively, with L. rhamnosus B5H2 showing the highest proteolytic activity. Overall, the three selected Lacticaseibacillus strains demonstrated probiotic capacity highlighting L. rhamnosus B5H2 with remarkable potential for generating bioactive metabolites and peptides which are capable of promoting human health.
log10 CFU mL−1 after the colonic phase. Remarkable proteolytic activity was observed for 18/25 strains. Bacterial auto- and co-aggregation of 7 selected strains reached values up to 23 and 20%, respectively. L. rhamnosus B5H2, L. rhamnosus B9H2 and L. paracasei B10L2 inhibited P. verrucosum, F. verticillioides and F. graminearum fungal growth, highlighting L. rhamnosus B5H2. Several metabolites were identified, including antifungal compounds such as phenylacetic acid and 3-phenyllactic acid, and volatile organic compounds produced in fermented milk, whey, and soy beverage. SDS-PAGE demonstrated bacterial hydrolysis of the main milk (caseins) and soy (glycines and beta-conglycines) proteins, with no apparent hydrolysis of whey proteins. However, HPLC-DAD revealed alpha-lactoglobulin reduction up to 82% and 54% in milk and whey, respectively, with L. rhamnosus B5H2 showing the highest proteolytic activity. Overall, the three selected Lacticaseibacillus strains demonstrated probiotic capacity highlighting L. rhamnosus B5H2 with remarkable potential for generating bioactive metabolites and peptides which are capable of promoting human health.
1. Introduction
Lactic acid bacteria (LAB) are a heterogeneous group of Gram-positive and catalase-negative bacteria that produce organic acids, mainly lactic acid, after glucose fermentation. LAB are naturally found in several foods, as well as in human and animal cavities, and many strains have been described as probiotics.1 Human breast milk is an important source of lactic acid bacteria, contains more than 200 strains, the most important of which are Lactobacilli, Bacteroides and Bifidobacterium. Among these populations, probiotic bacteria are present in an amount of 101–107 colony forming units (CFU) per mL.2The food fermentation and probiotic industry is rising nowadays for both their organoleptic properties and health benefits. In fact, the fermented food and beverage market is expected to grow by $533 million through 2026.3 These foods are increasingly in demand since probiotics have been associated with a healthy immune and digestive system, among other effects.4 An adequate supply of probiotic microorganisms with food supports appropriate formation of the microbiological profile, provides maximum benefits from microbiological homeostasis in the gastrointestinal tract, affects the maturation and development of the immune system, the integrity of the gastrointestinal mucosa and the production of secretory IgA antibodies, contributes to the formation of the immune system associated with the gastrointestinal mucosa, and prevents gastrointestinal infections by eliminating or reducing the number of pathogenic microflora.2
To use a microorganism in food within the European Union it needs to comply with specific parameters established by the European Food Safety Authority (EFSA) including strain identification, absence of antibiotic resistance, and production of antimicrobial substances, among others.5 The ability of LAB to produce antibacterial6 and antifungal substances7 is known and should be evaluated. According to the International Scientific Association for Probiotics and Prebiotics (ISAPP), a probiotic is a live microorganism that, when administered in adequate amounts, confers a health benefit to the host.4 The probiotic must be identified at the species level (preferably at the strain level, as the beneficial effects are shown to be strain-dependent). It must have a proven beneficial effect, like antagonism of pathogenic microorganisms or the production of bioactive metabolites such as organic acids or short-chain fatty acids.8 These effects must occur with a functional dose; therefore probiotics must be present at 8–9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU g−1 in the product before ingestion to ensure that a sufficient therapeutic minimum of 6–7
log10 CFU g−1 in the product before ingestion to ensure that a sufficient therapeutic minimum of 6–7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU g−1 can reach the colon.9 Moreover, resistance to stomach acid and tolerance to bile salts are two fundamental properties that allow probiotics to survive during passage through the gastrointestinal tract.9 Finally, microorganisms must have auto-aggregation capacity, a requirement to adhere to the intestinal epithelium and perform their function,10 as well as co-aggregation with other pathogenic microorganisms, this being directly related to their antimicrobial activity.
log10 CFU g−1 can reach the colon.9 Moreover, resistance to stomach acid and tolerance to bile salts are two fundamental properties that allow probiotics to survive during passage through the gastrointestinal tract.9 Finally, microorganisms must have auto-aggregation capacity, a requirement to adhere to the intestinal epithelium and perform their function,10 as well as co-aggregation with other pathogenic microorganisms, this being directly related to their antimicrobial activity.
LAB generate several bioactive compounds during fermentation and proteolytic hydrolysis of foods, including peptides, amino acids, organic acids, bacteriocin, vitamins, exopolysaccharides, and flavour substances.11 LAB bioactive peptides with antioxidant and antimicrobial activity, or inhibitory activity of Angiotensin I-Converting Enzyme (ACE) have been described.12 Knowing the proteolytic capacity of LAB strains is essential to determine the possible production of bioactive peptides, as well as other substances such as volatile organic compounds (VOCs) with functional capacity.
The isolation and characterization of LAB with probiotic properties is important to develop fermented products and functional foods with health-promoting properties. Certain probiotic strains such as Lacticaseibacillus spp. are commonly used as starters for the production of fermented foods, mainly dairy products. Fermented milk is one of the main forms of probiotic consumption worldwide; however, there are other food matrices recently introduced to the market, such as vegetable beverages, which could be susceptible to fermentation and production of probiotic foods.13
The objective of the present study was to isolate, characterize and select the most relevant LAB based on their probiotic capacity and proteolytic activity to find probiotic candidates to produce bioactive metabolites and peptides during fermentation of food matrices. Accordingly, after confirming their resistance during in vitro simulated gastrointestinal digestion the best three strains were deeply evaluated for their antifungal activity, metabolite and VOC production, as well as proteolytic capacity in three food matrices: cow milk, soy beverage and milk whey.
2. Materials and methods
2.1. Microorganisms and reagents
The LAB used were isolated from breast milk samples obtained from healthy volunteers from a partnership with La Fe Hospital (Valencia, Spain). Mycotoxigenic fungi Aspergillus flavus ISPA8111, Aspergillus niger CECT2088, Fusarium graminearum ITEM126, Fusarium verticillioides ITEM12043, Penicillium commune CECT20767 and Penicillium verrucosum VTTD-01847 were acquired from CECT. Milli-Q water (<18 MΩ cm−1 resistivity) was obtained from a Milli-Q purification system (Millipore, Bedford, MA, USA). Chromatographic solvents (Milli-Q water, acetonitrile >99.9% and trifluoroacetic acid 99%) were obtained from Thermo Fisher Scientific (Alcobendas, Madrid, Spain). Porcine bile salts, pepsin, pancreatin and antibiotics (ampicillin, vancomycin, gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline and chloramphenicol) were obtained from Sigma-Aldrich (Germany). Agela Technologies (Delaware, USA) supplied the reagents for the QuEChERS method. Microorganism culture media were provided by Thermo Fisher Scientific (Oxoid, UK).2.2. Bacterial isolation and phenotypic screening
The isolated bacteria were isolated from human breast milk from women donor volunteers. Women were selected based on specific characteristics including the absence of any pathology, the absence of recent hospital admission, and the absence of antibiotic treatment in the last three months. The drug research Ethics Committee of Hospital Universitario y Politécnico La Fe provided a favorable report for the project. Informed written consent was obtained from all participants prior to inclusion in the study. A total of ten breast milk samples were inoculated at 10% in modified MRS broth (Oxoid, Ireland) and incubated at 37 °C for 48 h under anaerobic conditions (Mikrobiologie Anaerocult A strips). After that, the inoculation of each bacterial solution (10 μL) on MRS agar plates was performed by the triple streak method and the plates were incubated (37 °C, 48 h) for single colony growth. To isolate bacterial strains, five single colonies of each plate were transferred and incubated again (37 °C, 48 h) in fresh MRS agar plates until being observed under the microscope; similar growth and morphology were observed for all colonies in a plate, obtaining pure bacterial cultures. Then, Gram differential staining was performed on the 50 isolated strains. Briefly, single colonies were spread on slides with 10 μL of distilled water, fixed by heat and subjected consecutively to gentian violet dye; lugol solution; alcohol–acetone (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v); and safranin (1 minute each) before observation under the microscope (100×). Moreover, colonies were spread on slides with 10 μL of hydrogen peroxide (30%) to assess catalase activity. Gram-positive catalase-negative strains were selected and kept at −80 °C in MRS broth-glycerol (70
1, v/v); and safranin (1 minute each) before observation under the microscope (100×). Moreover, colonies were spread on slides with 10 μL of hydrogen peroxide (30%) to assess catalase activity. Gram-positive catalase-negative strains were selected and kept at −80 °C in MRS broth-glycerol (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 v/v).
30 v/v).
2.3. In vitro proteolytic activity
To assess the proteolytic activity of the 25 Gram-positive catalase-negative selected strains, agar plates enriched with skim milk were prepared by mixing the minimal medium (10 g L−1 glucose, 2.5 g L−1 ammonium sulfate, 6.25 g L−1 sodium chloride, 0.25 g L−1 magnesium sulfate, 2.5 g L−1 potassium phosphate and 0.0625 g L−1 manganese sulfate) with 14 and 28 g L−1 of skim milk powder (Corporación Alimentaria Peñasanta SA, Granada, Spain). Then, 5 μL of each strain solution at exponentially growing culture (37 °C, 24 h) were inoculated into the center of the plates and incubated (37 °C for 48 h). Then, the size of the proteolysis halo was measured in triplicate and the most active strains (with the highest proteolysis halo) were selected for further analysis.2.4. Resistance to the acidic environment and bile salts
A total of 25 Gram-positive catalase-negative strains were tested for their resistance to the gastrointestinal environment by their incubation (4 and 6 h) under different conditions: (a) MRS broth (control), (b) MRS broth adjusted to pH 2 with 1 M HCl, (c) MRS broth with 0.3% of bile salts, and (d) MRS broth adjusted to pH 2 with 0.3% of bile salts. Strains at the exponential growth phase (20 μL) were incubated with 200 μL of each medium in 96-well plates (8 replicates/condition) at 37 °C under slight agitation. Control media without bacterial inoculation were also tested for each condition. The optical density (OD) at 600 nm was measured after 4 and 6 h incubation and bacterial growth (%) was calculated by comparing each modified medium with MRS control media, assuming 100% growth.2.5. Strain identification by MALDI-TOF/MS and 16S rRNA gene sequencing
On the one hand, seven bacterial strains were identified by matrix-assisted laser desorption/ionization with time-of-flight detection mass spectrometry (MALDI-TOF/MS) by the CECT (Universitat de València, Parque Científico, Spain) following the protocol recommended by Bruker Daltonics using the “extended direct transfer” method. The strains were analyzed from the original culture and 3 spectra per strain were obtained. MALDI-TOF/MS profiles were obtained using a Microflex L20 mass spectrometer equipped with an N2 laser and spectra were acquired in positive linear ion mode with an accelerating voltage of 20 kV. Each spectrum corresponds to the addition of 240 shots per target and the mass range used for the analysis was 2000–20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da. Identification was carried out following the MALDI Biotyper Realtime Classification (RTC) method with respect to the MBT 7854 and MBT 7311_RUO databases (Bruker Daltonics).
000 Da. Identification was carried out following the MALDI Biotyper Realtime Classification (RTC) method with respect to the MBT 7854 and MBT 7311_RUO databases (Bruker Daltonics).
On the other hand, selected strains were identified by 16S ribosomal (rRNA) gene sequencing by DNA extraction with a High Pure PCR Template Preparation Kit (Roche, Madrid, Spain). The 16S rRNA sequence was amplified and sequenced using the Applied Biosystems ABI PRISM BigDye Terminator Cycle Sequencing Ready Reaction Kit (Foster City, CA, USA). DNA amplification templates were obtained using polymerase chain reaction (PCR) with universal primers that amplify a 1000 bp region of the 16S rRNA gene: 616 V, 50-AGAGTTTGATYMTGGCTCAG-30; and 699R, 50-RGGGTTGCGCTCGTT-30. Primers (616V and 699R), Taq DNA polymerase, and deoxyribonucleotide triphosphate mix were obtained from Thermo Fisher Scientific (Waltham, MA, USA). DNA template amplification was performed by an initial denaturation (94 °C for 10 min), 40 cycles of denaturation (94 °C for 1 min), annealing (55 °C for 1 min), extension (72 °C for 1 min), and final extension (72 °C for 10 min). PCR products were evaluated for their integrity by single band development following electrophoresis (1 h at 100 V) in 2% (w/v) agarose gels in Tris–borate ethylenediaminetetraacetic acid buffer. A commercial mi-PCR Purification Kit (Metabion GmbH, Planegg, Germany) was used for amplicon purification, followed by sequencing reactions using the BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems), premixed format. The sequences obtained were aligned and compared with the on-line tool BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi), identifying the strains based on the highest scores.
2.6. Auto-aggregation and co-aggregation tests
For the aggregation and co-aggregation tests, the studied LAB strains and the pathogen microorganism Salmonella enterica 554 CECT (S. enterica) were grown in MRS broth and Tryptic Soy Broth (TSB) all under the same conditions (24 h, 37 °C). Firstly, for the auto-aggregation test LAB strains were washed twice with 0.01 M PBS buffer at pH 7.2. After that, LAB suspensions were diluted in PBS adjusting turbidity at 0.5 McFarland. The aggregation ability was then determined by measuring the absorbance at 600 nm at different time points, at 0 h and after 4 h incubation (37 °C). To calculate the auto-aggregation ratio, the following formula was applied with OD values at 0 and 4 h:| [Auto-aggregation (%) = (OD0h − OD4h)/OD0h × 100]. |
Secondly, the coaggregation ability was determined by washing and adjusting turbidity at 0.5 McFarland for both LAB and S. enterica. After 4 h of LAB and S. enterica co-incubation (37 °C), the absorbance at 600 nm was determined at 0 and 4 h incubation, calculating the co-aggregation rate based on the following formula:
| [Co-aggregation (%) = (OD0h − OD4h)/OD0h × 100]. |
Triplicate experiments were performed for each LAB and assay, and the L. plantarum ATCC 14917T probiotic strain was used as the reference control strain in both assays.
2.7. Antibiotic resistance
The antibiotic resistance of the studied LAB was evaluated based on the requirements of the European Food Safety Authority (EFSA) by testing their susceptibility against antibiotics of clinical importance, including ampicillin, vancomycin, gentamicin, kanamycin, streptomycin, erythromycin, clindamycin, tetracycline, and chloramphenicol. The Minimum Inhibitory Concentration (MIC) values were determined by the broth microdilution method, according to ISO standard 10932.14 A 96 well plate was prepared with two-fold dilutions in a range of 0.25–256 μg mL−1 of Mueller Hilton Broth (Oxoid, Ireland). The L. plantarum ATCC 14917T probiotic strain was used as the reference control. Ten microliters of LAB suspension adjusted to a standard turbidity of 0.5 McFarland was inoculated in 11 mL of Mueller Hilton Broth, and 100 μL of this suspension was inoculated in each well (8 replicates per condition). The plates were incubated at 37 °C for 18 h and the MIC of each antibiotic was established as the concentration where turbidity was not observed, considered as the lowest concentration of the antibiotic causing the absence of bacterial growth.2.8. In vitro digestion in MRS broth
To evaluate LAB strain viability during a simulated digestion process, an in vitro digestion was performed using the methodology described by Escrivá et al.15 with some modifications. Artificial saliva was prepared beforehand by mixing the inorganic phase [10 mL KCl (89.6 g L−1), 10 mL KSCN (20 g L−1), 10 mL NaH2PO4 (8.8 g L−1), 10 mL NaSO4 (57 g L−1), 1.7 mL NaCl (175.3 g L−1) and 20 mL NaHCO3 (84.7 g L−1)] with distilled water at a final volume of 490 mL. The pH was adjusted to 6.8 ± 0.2 with 0.1 M HCl and sterilized in an autoclave (15 min, 121 °C). After that, the organic phase (290 mg of α-amylase, 25 mg of mucin, and 8 mL of urea 20 g L−1) was added aseptically. The sterile pH adjustment solutions consisting of HCl 6 N, HCl 0.1 N, NaHCO3 1 N, NaHCO3 0.1 N and NaOH 0.5 N, as well as the enzyme mixture consisting of pepsin (1 g in 25 mL of sterile HCl 0.1 N) and pancreatin–bile salts (100 mg of pancreatin and 625 mg of bile salts in 25 mL of sterile NaHCO3 0.1 N) were also prepared. For the oral–gastric phase, 10 mL of MRS broth with the bacterial culture at exponential growth (24 h, 37 °C) were introduced in a sterile opaque flask, and 6 mL of artificial saliva and 84 mL of sterile distilled water were added. The pH was adjusted to 2 ± 0.2 with 6 N HCl, 0.5 mL of pepsin solution was added, and the samples were incubated for 2 h at 37 °C under slight agitation. Subsequently, for the duodenal phase, the pH was adjusted to 6.8 ± 0.2 with NaHCO3 1 N, 1.25 mL of the pancreatin–bile salt solution was added, and the samples were incubated again for 2 h at 37 °C with agitation. Finally, for the colonic phase the pH was adjusted to 7 ± 0.2 with NaOH 0.5 N and the samples were incubated for 48 h at 37 °C in anaerobiosis. Bacterial counting was performed in all phases, as well as initially prior to the simulated digestion, by seeding serial dilutions in PBS on MRS agar plates in duplicate and incubating at 37 °C for 48 h.2.9. Antifungal activity
Three LAB strains (Lacticaseibacillus rhamnosus B5H2, Lacticaseibacillus rhamnosus B9H2 and Lacticaseibacillus paracasei B10L2) were selected for further assays based on the results obtained for proteolytic activity and simulated gastrointestinal digestions in MRS broth. To evaluate the antifungal activity of the three selected LAB the overlay assay was performed. Ten microliters of an exponentially growing culture (MRS broth, 37 °C, 24 h) were placed at the center of MRS agar plates and incubated at 37 °C for 48 h. After that, the plates were covered with Potato Dextrose Agar (PDA) for antifungal activity determination against Aspergillus flavus ISPA8111, Aspergillus niger CECT2088, Fusarium graminearum ITEM126, Fusarium verticillioides ITEM12043, Penicillium commune CECT20767 and Penicillium verrucosum VTT D-01847. Fungi were incubated at 25 °C for 48 h, and the inhibition halo produced was measured in triplicate.2.10. LAB metabolite analysis by HPLC-Q-TOF-MS/MS
To analyze the metabolites produced by LAB fermentation, MRS broth was fermented (37 °C, 24 h) and the samples were diluted (1/4) with Milli-Q water and purified by the QuEChERS method. Briefly, 10 mL of sample was gently mixed with 4 g of MgSO4, 1 g f NaCl, and 10 mL of acetonitrile. Then, the samples were centrifuged (4000 rpm, 10 min, 4 °C), and the supernatant was collected and mixed with 150 mg C18 and 900 mg MgSO4. After that, the samples were centrifuged, the supernatant was dried in a Turbovap (LV, Zymark, Runcorn, UK), and the dry extract was reconstituted in 2 mL of Milli-Q water with 10% acetonitrile prior to filtration (0.22 μm) and injection into the HPLC-Q-TOF-MS/MS system. The chromatographic instrument consisted of an Agilent 1200 LC (Palo Alto, CA, USA) equipped with a vacuum degasser, a binary pump, an autosampler and a bioZen peptide C18 column (50 × 2.1 mm, 2.6 μm, Phenomenex, Madrid, Spain). Mobile phases were Milli-Q water–0.1% formic acid (A) and acetonitrile–0.1% formic acid (B), with a flow rate of 0.4 mL min−1 and an elution gradient (A![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) B) as 95
B) as 95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5% (0 min), 5
5% (0 min), 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95% (30 min) and 95–5% (37 min). 20 μL of sample was injected, performed in duplicate. The detection instrument consisted of an Agilent 6540 ultra-high definition accurate-mass Q-TOF mass spectrometer equipped with an Agilent dual jet stream ESI interface in positive and negative ionization modes. The mass spectrometer was operated in the scan range of 100–3000 m/z, with 13 L min−1 drying gas flow (N2), 35 psi nebulizer pressure, 325 °C gas drying temperature, 4 kV capillary voltage, 175 V fragment voltage and 10, 20 and 40 eV collision energy values. The spectrum generated was analyzed using a LAB metabolite personal database (METLIN PCDL B.08.00) for untargeted metabolome analysis considering compounds with scores >95% and delta errors <5 ppm. A heatmap of metabolites was obtained for each sample using the MetaboAnalyst software.
95% (30 min) and 95–5% (37 min). 20 μL of sample was injected, performed in duplicate. The detection instrument consisted of an Agilent 6540 ultra-high definition accurate-mass Q-TOF mass spectrometer equipped with an Agilent dual jet stream ESI interface in positive and negative ionization modes. The mass spectrometer was operated in the scan range of 100–3000 m/z, with 13 L min−1 drying gas flow (N2), 35 psi nebulizer pressure, 325 °C gas drying temperature, 4 kV capillary voltage, 175 V fragment voltage and 10, 20 and 40 eV collision energy values. The spectrum generated was analyzed using a LAB metabolite personal database (METLIN PCDL B.08.00) for untargeted metabolome analysis considering compounds with scores >95% and delta errors <5 ppm. A heatmap of metabolites was obtained for each sample using the MetaboAnalyst software.
2.11. Analysis of volatile organic compounds by GC-MS/MS
The analysis of the volatile organic compounds (VOCs) was performed using a gas chromatography system coupled to a triple quadrupole mass spectrometer (GC-MS/MS) following the methodology described by Lafuente et al.16 with some modifications adapted to the samples. Firstly, 10 mL of CFS were placed in a 20 mL glass vial and incubated for 45 min at 50 °C in a water bath under constant agitation. VOCs were extracted from the vial headspace by solid-phase microextraction (SPME) through a silica fiber (80 μm × 10 mm) coated with divinylbenzene/carbon-wide range/polydimethylsiloxane (DVB/C-WR/PDMS) (Supelco, Bellafonte, PA, USA). The fiber was introduced into the GC-MS/MS system in spitless mode for desorption at 250 °C for 10 minutes twice. The GC (Agilent 7890A) was equipped with an HP-5MS column (30 m × 0.25 mm, 0.25 μm 5% diphenyl/95% dimethylpolysiloxane) (J&W Scientific, Folsom, CA, USA), which was used for chromatographic separation. The temperature program started for 2 min at 40 °C and reached 160 °C at 6 °C min−1. Then, the temperature increased to 260 °C at 10 °C min−1 and was kept constant for 40 min. The collision and quenching gases were nitrogen (1.5 mL min−1) and helium (2.5 mL min−1), respectively, both at 99.999% purity (Carburos Metálicos S.L., Barcelona, Spain). Compounds were detected in a range of m/z of 40–50 Da in full scan mode. Data were acquired using Agilent Masshunter software (version B.04.00) and compounds were identified using NIST Atomic Spectra Database version 1.6 (Gaithersburg, MD, USA) with a spectral similarity of 80%. The linear retention index (LRI) was obtained from the time retention of an alkane solution (C8–C20) injected under the same conditions. A heatmap of VOCs was obtained for each sample using the MetaboAnalyst free software.2.12. LAB fermentation in food matrices
Three food products were selected to evaluate LAB proteolytic activity during the fermentation process in human eatable matrices, as well as to analyze the VOCs produced by the LAB strains; skimmed cow milk (Calidad Pascual SAU, Aranda de Duero, Spain), goat milk whey (ALCLIPOR Company SAL, Benassal, Spain) and soy drink (Esnelat SL, Gipuzkoa, Spain). Food matrices were prepared by adjusting the protein content at 0.325% (w/v) with minimal medium (10 g L−1 glucose, 2.5 g L−1 ammonium sulfate, 6.25 g L−1 sodium chloride, 0.25 g L−1 magnesium sulfate, 2.5 g L−1 potassium phosphate and 0.0625 g L−1 manganese sulfate). The food matrices were pasteurized (80 °C, 30 min) and inoculated individually with the three selected LAB at 5%, with a control sample without bacterial inoculation. The samples were incubated at 37 °C for 72 h under agitation, after which they were centrifuged (4000 rpm, 10 min) and the cell-free supernatant (CFS) was frozen (−18 °C) for further analysis.2.13. LAB proteolysis determined by SDS-PAGE and HPLC-DAD
The protein hydrolysis degree of food matrices after fermentation (37 °C, 72 h) with LAB strains was evaluated by sodium dodecyl sulphate-polyacrylamide gel electrophoresis – SDS-PAGE (Mini-Protean TGX Gels, Bio-Rad Laboratories Inc., USA). Briefly, 1 mL of CFS from each fermented food matrix was centrifuged (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min, 25 °C), and 200 μL of the supernatant were transferred to an Eppendorf tube, mixed with 800 μL of cold acetone, and kept at −20 °C for 24 h for protein precipitation. After that, samples were centrifuged (14
000 rpm, 10 min, 25 °C), and 200 μL of the supernatant were transferred to an Eppendorf tube, mixed with 800 μL of cold acetone, and kept at −20 °C for 24 h for protein precipitation. After that, samples were centrifuged (14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min) and the supernatant was removed. The resulting protein pellet was resuspended in 50 μL of Milli-Q water, mixed with 1.6% dithiothreitol in sample buffer at a 1
000 rpm, 10 min) and the supernatant was removed. The resulting protein pellet was resuspended in 50 μL of Milli-Q water, mixed with 1.6% dithiothreitol in sample buffer at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (2% SDS, 20% glycerol, 625 mM Tris-HCl and 0.01% bromophenol blue in Milli-Q water), and heated to 95 °C for 5 min. Then, 20 μL of each sample was loaded into each gel column, as well as 10 μL of a protein marker (Precision Plus Protein [All Blue], Bio-Rad Laboratories Inc., USA). After performing the electrophoresis (30 min at 80 V, and 50 min at 100 V) with the running buffer 10× (3% Trizma base, 14.4% glycine, and 1% SDS in Milli-Q water), gels were washed with a fixing solution (water–methanol–glacial acetic acid, 50
1 ratio (2% SDS, 20% glycerol, 625 mM Tris-HCl and 0.01% bromophenol blue in Milli-Q water), and heated to 95 °C for 5 min. Then, 20 μL of each sample was loaded into each gel column, as well as 10 μL of a protein marker (Precision Plus Protein [All Blue], Bio-Rad Laboratories Inc., USA). After performing the electrophoresis (30 min at 80 V, and 50 min at 100 V) with the running buffer 10× (3% Trizma base, 14.4% glycine, and 1% SDS in Milli-Q water), gels were washed with a fixing solution (water–methanol–glacial acetic acid, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) for 35 min under agitation, then with a staining solution (0.1% Brilliant Blue R-250, 50% water, 40% MeOH, and 10% acetic acid) for 35 min under agitation, and finally with a destaining solution (water–methanol–glacial acetic acid, 70
10) for 35 min under agitation, then with a staining solution (0.1% Brilliant Blue R-250, 50% water, 40% MeOH, and 10% acetic acid) for 35 min under agitation, and finally with a destaining solution (water–methanol–glacial acetic acid, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20
20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) for 24 h under agitation. Finally, protein bands were visualized and identified by comparison with the protein marker.
10) for 24 h under agitation. Finally, protein bands were visualized and identified by comparison with the protein marker.
On the other hand, CFSs from fermented food matrices were centrifuged (11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 10 min), filtered (0.22 μm) and vialized prior to chromatographic analysis in an Agilent 1100 chromatograph equipped with an LC-7100 pump and an autosampler L-2200 coupled to a DAD L-7455 detector (Hitachi, Tokyo, Japan) set at 214 nm. Mobile phases consisted of Milli-Q water–0.1% trifluoroacetic acid (A) and acetonitrile 0.1% trifluoroacetic acid (B) and the injection volume was 20 μL. The elution gradient was set at 5% at 0 min, 35% at 10 min, 100% at 30 min and finally 5% at 50 min, with the B phase. The column used for chromatographic separation was the Aeris peptide XB-C18 (100 × 4.6 mm, 3.6 μM ID, Phenomenex, Madrid, Spain) with a 1 mL min−1 flow rate. Standard calibration lines (25–200 μg mL−1) of alpha-lactoglobulin and beta-lactoglobulin (Sigma-Aldrich, Germany) were also injected to calculate sample protein concentration, as well as the percentage of reduction in fermented food matrices with respect to the non-fermented control.
000 rpm, 10 min), filtered (0.22 μm) and vialized prior to chromatographic analysis in an Agilent 1100 chromatograph equipped with an LC-7100 pump and an autosampler L-2200 coupled to a DAD L-7455 detector (Hitachi, Tokyo, Japan) set at 214 nm. Mobile phases consisted of Milli-Q water–0.1% trifluoroacetic acid (A) and acetonitrile 0.1% trifluoroacetic acid (B) and the injection volume was 20 μL. The elution gradient was set at 5% at 0 min, 35% at 10 min, 100% at 30 min and finally 5% at 50 min, with the B phase. The column used for chromatographic separation was the Aeris peptide XB-C18 (100 × 4.6 mm, 3.6 μM ID, Phenomenex, Madrid, Spain) with a 1 mL min−1 flow rate. Standard calibration lines (25–200 μg mL−1) of alpha-lactoglobulin and beta-lactoglobulin (Sigma-Aldrich, Germany) were also injected to calculate sample protein concentration, as well as the percentage of reduction in fermented food matrices with respect to the non-fermented control.
3. Results and discussion
3.1. In vitro proteolytic activity
From the 50 isolated strains, 25 were Gram-positive catalase-negative, so they were selected to continue with the proteolytic activity study. As shown in Table 1, all strains but two (B10H2 and B23L4) caused proteolysis to a greater or lesser degree, showing proteolytic halos from 0.93 to 1.40 cm, with 18 strains generating halos higher than 1 cm. The strains with higher proteolytic activity were B9H2 (1.40 cm), B5H2 (1.37 cm), and B10L2 (1.29 cm), followed by B2H2, B3H2, B8H2, B1H2 and B7L4, all with proteolytic halos higher than 1.25 cm (Table 1).| LAB strain | Halo (cm) |
|---|---|
| B1H2 | 1.22 ± 0.15 |
| B2H2 | 1.26 ± 0.06 |
| B3H2 | 1.26 ± 0.12 |
| B5H2 | 1.37 ± 0.06 |
| B7H2 | 1.19 ± 0.10 |
| B8H2 | 1.23 ± 0.06 |
| B9H2 | 1.40 ± 0.10 |
| B10H2 | 0.00 ± 0.00 |
| B1L2 | 1.03 ± 0.06 |
| B2L2 | 1.06 ± 0.06 |
| B3L2 | 0.99 ± 0.10 |
| B6L2 | 0.95 ± 0.15 |
| B7L2 | 0.96 ± 0.12 |
| B9L2 | 1.06 ± 0.06 |
| B10L2 | 1.29 ± 0.10 |
| B5L4 | 1.06 ± 0.06 |
| B6L4 | 1.09 ± 0.10 |
| B7L4 | 1.22 ± 0.15 |
| B8L4 | 1.16 ± 0.06 |
| B11L4 | 1.09 ± 0.10 |
| B13L4 | 0.99 ± 0.10 |
| B14L4 | 0.93 ± 0.06 |
| B15L4 | 1.02 ± 0.15 |
| B20L4 | 1.09 ± 0.10 |
| B23L4 | 0.00 ± 0.00 |
The proteolytic activity of LAB has garnered great interest due to its ability to enhance many desirable food qualities, and it has been extensively studied in several matrices for their industrial importance and essential role in ensuring bacterial survival.17 Many LAB are known to have proteolytic activity in milk and whey.17 Atanasova et al.18 reported the proteolytic activity of 58 LAB strains in goat milk, highlighting strains from Lactobacillus lactis, Lactococcus lactis, and Streptococcus thermophilus. The proteolytic activity of LAB (specifically Lactobacillus strains) in milk agar by proteolytic halo measurement has been previously reported,19 as well as LAB proteolytic activity in gelatin20 and vegetable proteins, such as legumes.21
3.2. Resistance to an acidic environment and bile salts
The 25 Gram-positive catalase-negative strains were assessed to evaluate their resistance to the gastrointestinal conditions including acidic pH and bile salts at two incubation times (4 and 6 h), simulating harsher conditions than those found in the gastrointestinal tract, where it is estimated that the ingested compounds start to leave the stomach (with acidic pH) after 1–3 h.22 LAB strain survival (% viability) was evaluated in modified MRS media including acidic conditions (pH = 2) and bile salts (0.3%) with acidic media (pH = 2), and it was compared to strain resistance in control MRS broth.As shown in Table 2, all strains were able to grow under strongly acidic conditions (pH = 2) as well as in the presence of bile salts (0.3%), after 4 and 6 h incubation. The survival rate, expressed as viability (%), varied considerably depending on the studied strain and as expected, it decreased as the incubation time increased in all strains. Viability after 4 h incubation under acidic conditions ranged between 50.8% and 112.7% compared to the control, with 16 strains showing viabilities higher than 75%. Interestingly, three strains (B2H2, B3H2 and B5H2) manage to slightly increase viability compared to the control (100%). Survival values decreased after 6 h incubation between 36.8 and 67.8%, where 17 strains demonstrated survival rates higher than 50%. The most resistant strains to acidic media at both incubation times were B5H2 and B3H2, followed by B2H2 and B9H2. When bile acids were added to the acidic media a slight increase was observed in strain survival. After 2 h incubation with bile acids at pH = 2 the strain viability ranged between 51.7 and 109.1%, with 20 strains reaching viabilities higher than 75%. When the exposure time increased up to 6 h survival rates decreased between 34.7 and 105.0%, with all strains but three reaching survival values higher than 50%. The slight increase observed in strain viability in the presence of bile salts may be explained to be due to the strains’ bile salt hydrolase activity that may influence the survival rate when using bile salts as metabolic substrates.23,24 Although all strains were found to be resistant to the studied gastrointestinal conditions including bile salts (0.3%) and/or the acid environment (pH = 2), a more complete in vitro simulated digestion assay should be performed to evaluate their behavior during the different digestion steps and to confirm strain resistance under the gastrointestinal digestion process.
| Strain | Viability (%) – 4 h incubation | Viability (%) – 6 h incubation | ||||
|---|---|---|---|---|---|---|
| Control | pH = 2 | pH = 2 – bile salts (0.3%) | Control | pH = 2 | pH = 2 – bile salts (0.3%) | |
| B1H2 | 100.0 ± 6.8 | 83.8 ± 6.5 | 94.4 ± 9.5 | 100.0 ± 5.6 | 50.8 ± 5.2 | 52.2 ± 5.9 |
| B2H2 | 100.0 ± 8.8 | 100.1 ± 6.9 | 103.5 ± 9.6 | 100.0 ± 6.2 | 65.0 ± 8.4 | 68.8 ± 6.6 |
| B3H2 | 100.0 ± 8.9 | 109.2 ± 13.7 | 109.1 ± 12.8 | 100.0 ± 6.3 | 65.2 ± 8.0 | 70.5 ± 10.5 |
| B5H2 | 100.0 ± 10.0 | 112.7 ± 12.3 | 93.9 ± 10.0 | 100.0 ± 7.0 | 67.8 ± 7.2 | 62.5 ± 9.7 |
| B7H2 | 100.0 ± 9.5 | 79.0 ± 8.7 | 106.4 ± 12.3 | 100.0 ± 10.5 | 49.7 ± 5.1 | 63.8 ± 7.3 |
| B8H2 | 100.0 ± 6.9 | 85.3 ± 13.5 | 101.4 ± 4.7 | 100.0 ± 4.0 | 51.9 ± 8.7 | 61.1 ± 4.4 |
| B9H2 | 100.0 ± 8.2 | 83.1 ± 7.0 | 104.6 ± 6.3 | 100.0 ± 9.0 | 65.2 ± 3.0 | 69.1 ± 2.9 |
| B10H2 | 100.0 ± 5.0 | 79.8 ± 6.3 | 107.2 ± 10.2 | 100.0 ± 5.0 | 45.4 ± 4.3 | 59.4 ± 6.8 |
| B1L2 | 100.0 ± 4.6 | 50.8 ± 3.1 | 58.7 ± 1.7 | 100.0 ± 3.5 | 38.2 ± 1.5 | 42.3 ± 1.7 |
| B2L2 | 100.0 ± 10.8 | 52.0 ± 4.3 | 60.0 ± 5.2 | 100.0 ± 6.9 | 38.2 ± 2.7 | 42.1 ± 3.5 |
| B3L2 | 100.0 ± 4.9 | 50.8 ± 1.6 | 51.7 ± 2.0 | 100.0 ± 5.5 | 36.8 ± 1.6 | 34.6 ± 1.6 |
| B6L2 | 100.0 ± 4.9 | 78.1 ± 3.6 | 88.3 ± 4.1 | 100.0 ± 4.0 | 54.6 ± 2.6 | 58.4 ± 2.8 |
| B7L2 | 100.0 ± 6.1 | 78.6 ± 3.6 | 89.4 ± 5.5 | 100.0 ± 4.8 | 57.0 ± 3.1 | 63.1 ± 3.9 |
| B9L2 | 100.0 ± 5.3 | 57.7 ± 3.2 | 74.5 ± 3.8 | 100.0 ± 4.4 | 42.5 ± 1.8 | 51.3 ± 2.9 |
| B10L2 | 100.0 ± 3.8 | 81.3 ± 4.4 | 87.0 ± 10.9 | 100.0 ± 3.5 | 53.4 ± 2.2 | 58.6 ± 4.8 |
| B5L4 | 100.0 ± 14.4 | 53.5 ± 9.9 | 71.0 ± 14.0 | 100.0 ± 12.3 | 37.2 ± 7.6 | 54.1 ± 11.7 |
| B6L4 | 100.0 ± 14.3 | 70.2 ± 9.0 | 106.5 ± 7.6 | 100.0 ± 11.4 | 50.1 ± 7.4 | 94.5 ± 8.0 |
| B7L4 | 100.0 ± 14.2 | 77.9 ± 13.5 | 102.8 ± 8.9 | 100.0 ± 9.4 | 56.8 ± 8.0 | 100.4 ± 10.5 |
| B8L4 | 100.0 ± 17.0 | 73.4 ± 16.4 | 104.2 ± 11.5 | 100.0 ± 13.4 | 55.0 ± 9.6 | 87.5 ± 12.3 |
| B11L4 | 100.0 ± 15.3 | 74.3 ± 14.9 | 103.6 ± 19.1 | 100.0 ± 11.1 | 57.3 ± 14.7 | 77.6 ± 11.7 |
| B13L4 | 100.0 ± 10.1 | 63.9 ± 14.7 | 106.6 ± 10.4 | 100.0 ± 14.6 | 51.5 ± 9.3 | 92.2 ± 9.4 |
| B14L4 | 100.0 ± 14.9 | 87.5 ± 14.7 | 107.5 ± 15.8 | 100.0 ± 12.8 | 61.4 ± 12.0 | 74.8 ± 11.3 |
| B15L4 | 100.0 ± 15.4 | 76.5 ± 10.6 | 103.3 ± 14.5 | 100.0 ± 10.2 | 54.5 ± 7.1 | 105.0 ± 12.0 |
| B20L4 | 100.0 ± 7.9 | 78.2 ± 29.5 | 100.1 ± 17.3 | 100.0 ± 14.4 | 47.8 ± 17.4 | 99.2 ± 9.5 |
| B23L4 | 100.0 ± 12.2 | 84.2 ± 12.8 | 103.7 ± 14.9 | 100.0 ± 14.6 | 57.3 ± 12.0 | 101.1 ± 12.1 |
LAB resistance to gastrointestinal conditions has been described in previous studies with related methodology reporting similar survival percentages, between 55 and 90%;25 however, resistance seems to directly depend on the specific type of strain, which could explain the high variability observed between strains (i.e. from 34.6% for B3L2 at pH = 2 and bile salts during 6 h of incubation, to 105% for B15L4 under the same conditions).
3.3. MALDI-TOF/MS and 16S rRNA gene sequencing identification
The strains that showed the highest proteolytic activity and high resistance under the studied gastrointestinal conditions were selected for identification. Seven strains namely B6L2, B7L2, B10L2, B2H2, B3H2, B5H2 and B9H2 were identified at the species level after MALDI-TOF analysis using the MBT database with the highest log (score) values. Moreover, strain identification at species level was confirmed by obtaining >99% of 16S rRNA sequence similarity after comparing the obtained full sequence with the on-line BLAST tool. The seven identified strains belonged to Lacticaseibacillus spp. (Table 3), specifically identifying two species, L. rhamnosus (B2H2, B3H2, B5H2 and B9H2) and L. paracasei (B6L2, B7L2 and B10L2).| LAB strain identification | |
|---|---|
| B6L2 | Lacticaseibacillus paracasei DSM 2649 |
| B7L2 | Lacticaseibacillus paracasei DSM 20020 |
| B10L2 | Lacticaseibacillus paracasei DSM 20244 |
| B2H2 | Lacticaseibacillus rhamnosus DSM 20711 |
| B3H2 | Lacticaseibacillus rhamnosus DSM 20021T |
| B5H2 | Lacticaseibacillus rhamnosus D155 ZZMK |
| B9H2 | Lacticaseibacillus rhamnosus DSM 20245 |
3.4. Auto-aggregation and co-aggregation assays
On the one hand, the auto-aggregation assay evidenced values between 10.79 ± 0.73 and 24.30 ± 1.60, highlighting four strains; L. rhamnosus B3H2, L. rhamnosus B5H2, L. paracasei B10L2 (with values >20%), and L. rhamnosus B2H2; that showed values above the auto-aggregation values obtained for L. plantarum ATCC 14917T, used as a probiotic control. On the other hand, values for co-aggregation with the pathogenic strain S. entrerica ranged between 9.15 ± 0.45 and 19.86 ± 1.70, with the highest levels for L. rhamnosus B9H2, L. paracasei B6L2, and L. rhamnosus B5H2, all higher than 19% and above the control strain co-aggregation levels (Table 4). The obtained values for auto-aggregation and co-aggregation to S. enterica were similar to those reported in other studies at the same incubation time.26 The higher auto-aggregation percentages obtained for the studied strains compared to the reference control may indicate greater adhesion capacity to the intestinal epithelium. Moreover, higher co-aggregation to S. enterica may lead to a higher competition capacity against pathogenic microorganisms of the digestive tract. Overall, all analyzed strains showed auto-aggregation and co-aggregation capacity against S. enterica, being a preliminary indication of adhesion to the intestinal epithelium and potential antimicrobial capacity, highlighting L. rhamnosus B3H2 as the one that showed the greatest auto-aggregation, although with scarce co-aggregation, and L. rhamnosus B9H2 as the one with the greatest co-aggregation and moderate auto-aggregation capacity. However, L. rhamnosus B5H2 was the only strain that reached higher values in both auto-aggregation and co-aggregation assays than the reference strain used as a probiotic control.| LAB strain | Auto-aggregation (%) | Co-aggregation (%) |
|---|---|---|
| L. paracasei B6L2 | 18.30 ± 1.41 | 19.68 ± 0.77 |
| L. paracasei B7L2 | 10.79 ± 0.73 | 15.06 ± 1.21 |
| L. paracasei B10L2 | 20.13 ± 0.93 | 13.74 ± 1.71 |
| L. rhamnosus B2H2 | 19.98 ± 0.71 | 16.80 ± 1.66 |
| L. rhamnosus B3H2 | 24.30 ± 1.60 | 9.15 ± 0.45 |
| L. rhamnosus B5H2 | 22.95 ± 1.69 | 19.43 ± 0.09 |
| L. rhamnosus B9H2 | 16.95 ± 1.25 | 19.86 ± 1.70 |
| L. plantarum ATCC 14917T (reference control) | 19.63 ± 1.38 | 19.01 ± 0.35 |
3.6. Antibiotic resistance
Resistance to antibiotic treatment of the seven identified strains and the reference control was evaluated according to EFSA recommendations.5 No growth of any tested LAB strain, as well as reference control strain, was observed above the minimum antibiotic concentration (0.25 mg L−1), so this concentration was established as the MIC. Thus, all seven strains showed MIC values below the cut-off level established by EFSA for the 9 tested antibiotics, confirming their potential safe use as probiotics.3.7. LAB in vitro digestion in MRS broth and LAB strain selection
To confirm the strain resistance to gastrointestinal digestion, the selected LAB were subjected to a simulated in vitro gastrointestinal digestion in MRS broth. All seven strains were able to withstand the conditions of the simulated digestion at all gastrointestinal steps; gastric, duodenal and colonic phases. Bacterial counts, expressed as log10 CFU mL−1, were determined after each digestion phase and immediately before the digestion process (initial count). As shown in Table 5, the initial bacterial count was similar for all strains ranging from 9.3 to 9.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1 except L. rhamnosus B2H2 with 8.8
log10 CFU mL−1 except L. rhamnosus B2H2 with 8.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1 as the initial count. Greater differences were observed after the gastric phase, where the bacterial count decreased for all strains to values between 0.2 and 5.9
log10 CFU mL−1 as the initial count. Greater differences were observed after the gastric phase, where the bacterial count decreased for all strains to values between 0.2 and 5.9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1. These indicate viability reduction from 3.5 (L. paracasei B10L2) up to 9.3
log10 CFU mL−1. These indicate viability reduction from 3.5 (L. paracasei B10L2) up to 9.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 units (L. paracasei B6L2). At this point, the most resistant strain was L. paracasei B10L2 followed by L. rhamnosus B5H2 and L. rhamnosus B9H2, all with reductions lower than 6
log10 units (L. paracasei B6L2). At this point, the most resistant strain was L. paracasei B10L2 followed by L. rhamnosus B5H2 and L. rhamnosus B9H2, all with reductions lower than 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 from the initial count. This considerable decrease is explained by the effect of pepsin and acidic pH, unfavorable conditions to which the strains are subjected during the 2 h of gastric digestion. It is well known that, throughout the passage through the stomach the viable cell count and the survival rate of probiotic microorganisms are reduced due to the extreme pH of stomach acid. The decrease of probiotic viability between 1 and 4
log10 from the initial count. This considerable decrease is explained by the effect of pepsin and acidic pH, unfavorable conditions to which the strains are subjected during the 2 h of gastric digestion. It is well known that, throughout the passage through the stomach the viable cell count and the survival rate of probiotic microorganisms are reduced due to the extreme pH of stomach acid. The decrease of probiotic viability between 1 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU g−1 during the passage through the gastrointestinal tract has been previously reported.9 The low acidity of the stomach is mainly the first barrier against microorganism survival in the gastrointestinal tract and many ingested bacteria die or show considerably reduced viable counts. In fact, when probiotics reach the stomach it is the point at which the greatest loss of bacterial viability is expected due to the acidic environment and the release of pepsin, a proteolytic enzyme which breaks down proteins.27
log10 CFU g−1 during the passage through the gastrointestinal tract has been previously reported.9 The low acidity of the stomach is mainly the first barrier against microorganism survival in the gastrointestinal tract and many ingested bacteria die or show considerably reduced viable counts. In fact, when probiotics reach the stomach it is the point at which the greatest loss of bacterial viability is expected due to the acidic environment and the release of pepsin, a proteolytic enzyme which breaks down proteins.27
| Strain | Concentration (log10 CFU mL−1) | |||
|---|---|---|---|---|
| Initial | Gastric | Duodenal | Colonic | |
| L. paracasei B6L2 | 9.5 ± 0.1 | 0.2 ± 0.1 | 1.5 ± 0.1 | 8.5 ± 0.2 |
| L. paracasei B7L2 | 9.3 ± 0.1 | 0.9 ± 0.1 | 2.6 ± 0.1 | 8.1 ± 0.1 |
| L. paracasei B10L2 | 9.4 ± 0.1 | 5.9 ± 0.1 | 6.1 ± 0.2 | 9.5 ± 0.2 |
| L. rhamnosus B2H2 | 8.8 ± 0.1 | 1.3 ± 0.1 | 3.0 ± 0.1 | 8.9 ± 0.1 |
| L. rhamnosus B3H2 | 9.5 ± 0.1 | 2.5 ± 0.1 | 3.8 ± 0.1 | 7.9 ± 0.1 |
| L. rhamnosus B5H2 | 9.5 ± 0.1 | 4.4 ± 0.1 | 4.9 ± 0.1 | 9.4 ± 0.1 |
| L. rhamnosus B9H2 | 9.8 ± 0.2 | 4.3 ± 0.1 | 4.7 ± 0.2 | 9.8 ± 0.2 |
After the duodenal digestion, all strains showed considerably increased counts reaching values from 1.5 to 6.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1, indicating that the viability increases between 0.2 and 1.7
log10 CFU mL−1, indicating that the viability increases between 0.2 and 1.7![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 units from the gastric phase, with again the most resistant strains being L. paracasei B10L2, L. rhamnosus B5H2 and L. rhamnosus B9H2 (Table 5). After passing through the stomach, probiotics reach the small intestine where abundant pancreatic juice and bile acids are present. Under the neutralizing effect of the intestinal fluid, the pH in the small intestine is about 6.0–7.0, much milder than that of the gastric fluid.27 The return to a pH close to neutral (pH = 6.8) during the two hours of the duodenal digestion step could explain the strain growth and the corresponding bacterial count increase. However, bile acids and digestive enzymes can also impact probiotic viability through cell membrane disruption and DNA damage,28 therefore, as it was expected, viable counts obtained after duodenal digestion were still far from the initial count, prior to the simulated gastrointestinal digestion.
log10 units from the gastric phase, with again the most resistant strains being L. paracasei B10L2, L. rhamnosus B5H2 and L. rhamnosus B9H2 (Table 5). After passing through the stomach, probiotics reach the small intestine where abundant pancreatic juice and bile acids are present. Under the neutralizing effect of the intestinal fluid, the pH in the small intestine is about 6.0–7.0, much milder than that of the gastric fluid.27 The return to a pH close to neutral (pH = 6.8) during the two hours of the duodenal digestion step could explain the strain growth and the corresponding bacterial count increase. However, bile acids and digestive enzymes can also impact probiotic viability through cell membrane disruption and DNA damage,28 therefore, as it was expected, viable counts obtained after duodenal digestion were still far from the initial count, prior to the simulated gastrointestinal digestion.
Finally, after the 48 h of colonic incubation under anaerobic conditions the bacterial count reached values close to the initial concentration, between 7.7 and 9.8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1 (Table 5). This indicated a huge viable count recovery, with an increase between 5 and 8.3
log10 CFU mL−1 (Table 5). This indicated a huge viable count recovery, with an increase between 5 and 8.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 from the gastric count. Moreover, some strains reached the same values as the initial count (L. rhamnosus B9H2) or even exceeded it as was the case of L. paracasei B10L2 and L. rhamnosus B2H2, with an increase of 0.1
log10 from the gastric count. Moreover, some strains reached the same values as the initial count (L. rhamnosus B9H2) or even exceeded it as was the case of L. paracasei B10L2 and L. rhamnosus B2H2, with an increase of 0.1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 units from the initial viable count. The favorable colonic conditions for LAB growth explained this remarkable count increase observed at the end of the simulated digestion. Other studies reported recoveries of up to 9
log10 units from the initial viable count. The favorable colonic conditions for LAB growth explained this remarkable count increase observed at the end of the simulated digestion. Other studies reported recoveries of up to 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 units in commercial Lactobacillus strains,9 although showing high variability among strains and higher initial concentrations, and in all cases there was a difference of at least 1
log10 units in commercial Lactobacillus strains,9 although showing high variability among strains and higher initial concentrations, and in all cases there was a difference of at least 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 between the initial and colonic counts. Similar reductions were observed for L. paracasei B6L2, L. paracasei B7L2 and L. rhamnosus B3H2, the only strains that had reduced viability at the end of simulated in vitro digestion (Table 5). However, since the colon is where the largest bacterial density is expected to be found (11–12
log10 between the initial and colonic counts. Similar reductions were observed for L. paracasei B6L2, L. paracasei B7L2 and L. rhamnosus B3H2, the only strains that had reduced viability at the end of simulated in vitro digestion (Table 5). However, since the colon is where the largest bacterial density is expected to be found (11–12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1) when considering the whole human organism, probiotics might face resistance to commensal bacterial colonization, competing for nutrients and adhesion sites with the host microbiota to successfully colonize the mucosa and proliferate;29 therefore these factors should be further considered when extrapolating data to in vivo conditions.
log10 CFU mL−1) when considering the whole human organism, probiotics might face resistance to commensal bacterial colonization, competing for nutrients and adhesion sites with the host microbiota to successfully colonize the mucosa and proliferate;29 therefore these factors should be further considered when extrapolating data to in vivo conditions.
Overall, all seven strains were found to be resistant to the in vitro gastrointestinal digestion process, highlighting L. rhamnosus B5H2, L. rhamnosus B9H2 and L. paracasei B10L2 reaching the highest viable count (higher than 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1) at the colonic phase; therefore they were selected to continue their characterization by evaluating their antifungal activity, and deeply analyzing the produced metabolites after fermentation in MRS broth, as well as in whole cow milk, soy drink and milk whey as representative food matrices.
log10 CFU mL−1) at the colonic phase; therefore they were selected to continue their characterization by evaluating their antifungal activity, and deeply analyzing the produced metabolites after fermentation in MRS broth, as well as in whole cow milk, soy drink and milk whey as representative food matrices.
3.8. Antifungal activity
To continue the study of the selected LAB strains, their antifungal activity against mycotoxigenic fungi was evaluated by the overlay assay. After fungal and LAB superimposed growth, the halos observed indicated fungal growth inhibition exerted by the studied LAB (Fig. 1).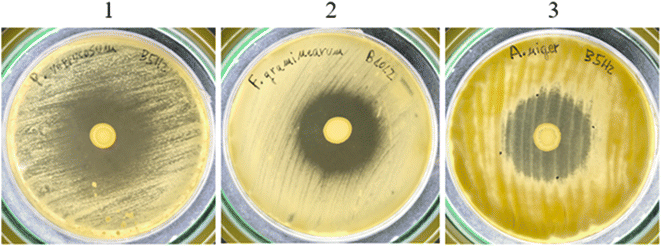 | ||
| Fig. 1 Overlay assay against mycotoxigenic fungi. 1: L. rhamnosus B5H2 inhibiting P. verrucosum; 2: L. paracasei B10L2 inhibiting F. graminearum; and 3: L. rhamnosus B5H2 inhibiting A. niger. | ||
As shown in Table 6, all three LAB were able to inhibit at least three different fungal strains. L. rhamnosus B9H2 and L. paracasei B10L2 showed growth inhibition of P. verrucosum, F. verticillioides and F. graminearum, while L. rhamnosus B5H2 also reduced A. niger growth. This means that P. verrucosum, and F. verticillioides were inhibited by all three LAB strains showing inhibition halos higher than 0.5 cm. Also F. verticillioides growth was inhibited by all three strains, with higher activity for L. rhamnosus B5H2 and L. paracasei B10L2 (halos > 0.5 cm), while A. niger growth was strongly inhibited (halo > 1 cm) by L. rhamnosus B5H2. On the other hand, two fungal strains (P. commune and A. flavus) were not inhibited by any of the studied LAB (Table 6).
| Mycotoxigenic fungi | L. rhamnosus B9H2 | L. rhamnosus B5H2 | L. paracasei B10L2 |
|---|---|---|---|
| − (no inhibition halo), + (<0.5 cm halo), ++ (0.5–1 cm halo), and +++ (>1 cm halo). | |||
| P. verrucosum | ++ | ++ | ++ |
| P. commune | − | − | − |
| F. verticillioides | + | ++ | ++ |
| F. graminearum | ++ | ++ | ++ |
| A. flavus | − | − | − |
| A. niger | − | +++ | − |
Such antifungal activity has been extensively described for many LAB strains and it is explained to be due to several metabolites produced by LAB, especially organic acids and phenolic acids, among others.30 Furthermore, the overlay technique has been widely used in numerous studies to qualitatively evaluate the direct antifungal capacity of Lacticaseibacillus spp. strains, reporting antifungal activity against Aspergillus and Penicillium genera as in the present work, showing greater resistance of Aspergillus species against LAB strains,31 in accordance with the observed results for A. flavus and A. niger.
3.9. Metabolite analysis by HPLC-Q-TOF-MS/MS
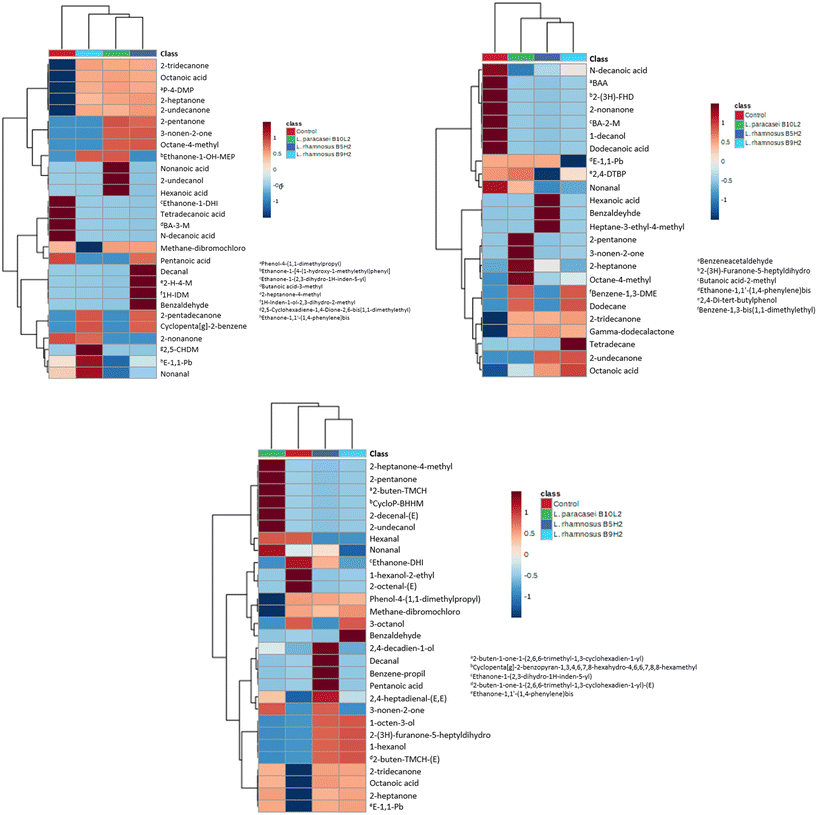
To study LAB metabolites, samples obtained from MRS broth fermentation were diluted and purified by the QuEChERS method for HPLC-Q-TOF-MS/MS analysis. A total of 25 metabolites were found in fermented samples. From them, 14 metabolites were not detected in control (non-fermented) samples, confirming that their production is due to LAB metabolism. Specifically, 13 compounds were produced by L. rhamnosus B5H2 fermentation, 11 by L. rhamnosus B9H2, and 12 by L. paracasei B10L2. From these LAB metabolites 6 were commonly produced by all three strains (phenylacetic acid, 4-hydroxyphenyllactic acid, benzoic acid, 3-phenyllactic acid, methyl nicotinate and 2-ethyl-2-hydroxybutyric acid), as shown in Fig. 2.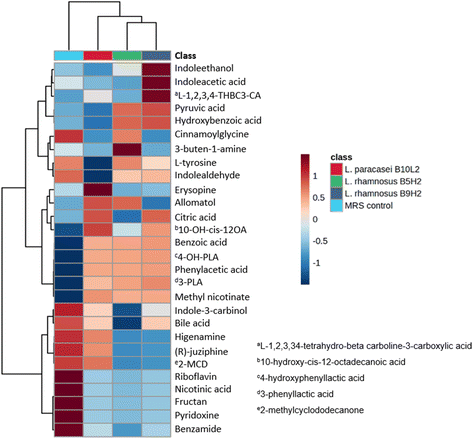 | ||
| Fig. 2 Heatmap of identified metabolites produced by L. rhamnosus B5H2, L. rhamnosus B9H2, L. paracasei B10L2 and in non-fermented MRS broth (control). | ||
Several bioactive metabolites, mainly organic and phenolic acids, were found in fermented samples. Citric acid, an organic acid frequently used as a food preservative due to its antimicrobial activity,32 was produced by L. paracasei B10L2 and L. rhamnosus B9H2. Five phenolic acids (phenylacetic acid, 4-hydroxyphenyllactic acid, benzoic acid, 3-phenyllactic acid and hydroxybenzoic acid) were found in LAB fermented samples. The highest abundance of benzoic acid, with described properties such as improving intestinal function and antimicrobial activity,33 was observed with L. rhamnosus B9H2, although it was also produced by all other strains. 3-Phenyllactic acid and phenylacetic acid, both exhibiting antifungal activity,34 were generated by all three strains. Although phenylacetic acid is a metabolite commonly produced by plants its production by microorganisms has been previously reported.35 Moreover, 10-hydroxy-cis-12-octadecenoic acid, with anti-inflammatory and antimicrobial effects,36 was found to be mainly produced by L. paracasei B10L2 and L. rhamnosus B9H2. In addition, it has been reported that this metabolite produced by microorganisms could improve the deterioration of the intestinal barrier.37
On the other hand, as shown in Fig. 2, a decrease in bile acid concentration compared to that of the MRS control was observed for all strains, highlighting L. rhamnosus B5H2. This decrease is in agreement with previous results on in vitro digestion, and may indicate a possible use of bile acid as a metabolic substrate by LAB.23 Moreover, there was a correlation between the direct antifungal activity observed (overlay assay) and the produced metabolites identified, since several of them, such as 3-phenyllactic acid and phenylacetic acid, have been found to exhibit antifungal activity.38 Indeed, other metabolites with functional capacity in the intestinal function, such as benzoic acid and 10-hydroxy-cis-12-octadecaenoic acid, have been detected pointing to the studied LAB strains as potential probiotics.
Fig. 2 shows a heatmap of the identified metabolites produced by L. rhamnosus B5H2, L. rhamnosus B9H2, and L. paracasei B10L2 and in non-fermented MRS broth (control).
3.10. Analysis of volatile organic compounds (VOCs) by GC-MS/MS
To continue evaluating LAB metabolites, the VOCs produced after food matrix (milk, whey and soy drink) fermentation were determined by GC-MS/MS. VOCs produced in fermented matrices were compared to their respective non-fermented foods, as controls. In general, several acids, alcohols, aldehydes and ketones were found in the fermented samples, identifying the metabolites specifically produced by LAB fermentation, as they were not found in control non-fermented samples.After milk fermentation a total of 28 VOCs were found in LAB fermented samples, of which 19 were not detected (or detected at a very low concentration) in control milk; therefore they were produced by LAB metabolism with milk as the substrate. The LAB strain that produced more VOCs was L. rhamnosus B5H2 (16 metabolites), followed by L. paracasei B10L2 (13 metabolites) and L. rhamnosus B9H2 (12 metabolites). Among these compounds 5 were commonly found in all LAB fermented milk samples, namely 2-heptanone, 2-tridecanone, 2-undecanone, octanoic acid, and phenol,4-(1,1-dimethylpropyl); so they were common metabolism products of all three studied LAB. Interestingly, other metabolites present in non-fermented milk were not detected in fermented samples (i.e. tetradecanoic acid, butanoic acid 3-methyl, n-decanoic acid, and ethanone 1-(2,3-dihydro-1H-inden-5-yl)); therefore they were possibly metabolized to other compounds during LAB fermentation. Fig. 3a shows a heatmap of the identified VOCs in fermented milk samples by L. rhamnosus B5H2, L. rhamnosus B9H2 and L. paracasei B10L2, as well as in non-fermented milk (control).
LAB fermentation of whey samples produced a total of 24 different VOCs, 14 of which were confirmed as produced by LAB metabolism. Each strain was able to produce 7 (L. rhamnosus B9H2), 8 (L. rhamnosus B5H2) and 11 (L. paracasei B10L2) metabolites, highlighting 2-tridecanone, acetoin, and γ-dodecalactone, produced by all three strains. As found in milk samples, some compounds present in non-fermented whey were not detected after LAB fermentation, such as dodecanoic acid, benzeneacetaldehyde, 1-decanol, 2-nonanone, butanoic acid 2-methyl, and 2(3H)-furanone-5-heptyldihydro; suggesting that these compounds may serve as substrates for LAB metabolism. Fig. 3b shows a heatmap of the identified VOCs in fermented whey samples by all three studied LAB, as well as in non-fermented whey (control).
VOCs produced after LAB fermentation in soy beverage were also evaluated as examples of vegetal origin food and non-dairy products. In total, 29 different VOCs were detected, with 22 identified as LAB metabolism products since they were not found in control soy beverage. As for milk and whey samples, L. rhamnosus B5H2 and L. paracasei B10L2 were the strains that produced the highest number of metabolites (17 and 14 VOCs, respectively), followed by L. rhamnosus B9H2 that produced 12 metabolites. The VOCs simultaneously produced by all three strains included octanoic acid, 2-heptanone, ethanone, 1,1′-(1,4-phenylene)-bis, and 2-tridecanone. Compounds such as 1-hexanol, 2-ethyl, and 2-octenal were exclusively produced in non-fermented soy samples. Fig. 3c shows a heatmap of the identified VOCs in fermented soy beverage samples by all three studied LAB, as well as in non-fermented soy drink (control).
Regarding the VOC profiles observed in dairy samples (milk and whey), similarities and disparities were observed with previous studies. As in the present work, Zhang et al.39 evaluated the metabolites produced in dairy samples fermented by Lacticaseibacillus spp. species, reporting an increase in benzaldehyde and organic acid (i.e. hexanoic acid, octanoic acid, etc.) production in fermented samples compared to the non-fermented control. However, other metabolites such as nonanal, 2-tridecanone or 2-undecanone that showed an increase in fermented samples of the present study were reported to decrease after fermentation. These differences may be explained by the specificity of VOC production by each single strain, as well as by the type of matrix used, since differences may be found despite being all dairy products, for example the production of hexanoic acid by L. rhamnosus B5H2 differs between milk and whey in the present work, as seen in Fig. 3a and b.
Several identified metabolites have been reported as bioactive compounds; 2-heptanone (produced by all strains in milk and soy samples, as well as by L. paracasei B10L2 and L. rhamnosus B5H2 in whey) showed an antifungal character against several species of the Fusarium genus after being produced by Bacillus strains;40 1-octen-3-ol (produced by L. rhamnosus B5H2 and L. rhamnosus B9H2 in soy beverage) showed antifungal activity against Fusarium spp. fungi and antimicrobial activity against foodborne pathogenic bacteria such as Staphylococcus aureus or Escherichia coli;41 and 2-tridecanone, produced by all strains in all samples, was described as a biostimulant in plants.42
Previous studies have reported a relationship between the production of LAB metabolites, mainly organic acids and VOCs, and antifungal activity by the strain. The interaction between fungi and LAB may lead to an increase in organic acids and other compounds that, associated with VOCs, can increase the antifungal capacity.43 In addition to the generated bioactivity, organic acids, alcohols, ketones, and esters are some of the flavoring compounds made by LAB with multiple metabolic pathways involved. The citric acid pathway (Krebs cycle) is one of the metabolic pathways that synthesize intermediate compounds such as citric acid and succinic acid, which contribute to flavor formation. In addition, sugar metabolism leads to the production of sugar alcohol, which contributes to the food sweet taste.11
As previously reported, production of LAB metabolites is closely related to the strain and the fermented matrix. In the present work, L. rhamnosus B5H2 was the strain that produced the highest number of VOCs during fermentation, especially in milk and soy matrices, where 15 and 14 VOCs were identified, respectively, and 8 VOCs were identified after whey fermentation. A similar trend was found for L. paracasei B10L2 with 12 VOCs produced in milk and soy beverages, while 8 VOCs were produced in whey; meanwhile L. rhamnosus B9H2 produced 9 VOCs in milk and in soy, but 7 VOCs in whey.This preference for milk as probiotic growth matrix and metabolites production could be explained since dairy products are considered primary dietary sources for LAB. Dairy products are highly suitable substrates for LAB growth and fermentation, where they can be naturally found or added afterwards.44 Interestingly, despite their similarities, the milk matrix proved to be more appropriate for VOC formation than whey, although many studies have successfully reported whey fermentation by LAB. Moreover, the soy beverage showed high potential as a fermentation substrate for VOC production.
In summary, the strains having potential for VOC production were L. rhamnosus B5H2 > L. paracasei B10L2 > L. rhamnosus B9H2 in all three studied matrices, while the more suitable matrices for all studied strains were milk > soy beverage > whey.
3.11. Proteolysis analysis by SDS-PAGE and HPLC-DAD
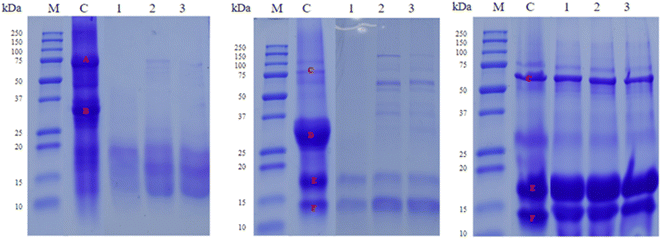
To evaluate the hydrolysis degree of food proteins after LAB fermentation SDS-PAGE and HPLC-DAD analysis were performed. Firstly, SDS-PAGE showed that all three strains had proteolytic activity over the main proteins of the tested food matrices, except for whey (Fig. 4). The main protein groups in the analyzed food samples were identified based on their molecular weight. As shown in Fig. 4, the ‘A’ band corresponds to the beta-conglycine region, and ‘B’ to the glycinine region in soy beverage;45 while in dairy matrices (milk and whey) the ‘C’ band was identified as the albumin region, ‘D’ as the casein region, ‘E’ as the beta-lactoglobulin region, and ‘F’ as the alpha-lactoglobulin region.46In soy beverage, similar proteolysis was observed between L. rhamnosus B9H2 and L. paracasei B10L2 strains, hydrolyzing glycinins more markedly, while the L. rhamnosus B5H2 strain produced higher proteolysis, hydrolyzing more noticeably the main proteins (‘A’ and ‘B’ bands). For dairy products (milk and whey), high hydrolysis of caseins (‘D’) was observed for all strains, especially in milk, also showing high proteolysis of albumins (‘C’) and moderate proteolysis of lactoglobulins (‘E’ and ‘F’), with L. rhamnosus B5H2 as the most proteolytic strain. However, no apparent hydrolysis of albumins and lactoglobulins was evidenced in whey by any of the studied strains (Fig. 4). Overall, the hydrolysis of the main milk proteins (caseins) and soy proteins (glycines and beta-conglycines) was observed; however, whey proteins (alpha-lactoglobulins and beta-lactoglobulins) did not show apparent protein hydrolysis (Fig. 4).
Other studies reported proteolytic activities of several LAB after 6 and 24 h incubation in whey by SDS-PAGE, where the majority digested protein fractions were 69 and 50 kDa fractions; however, some strains poorly hydrolyzed this fraction and preferentially hydrolyzed the 25 kDa or 80 kDa fraction, confirming different casein and whey protein degradation degrees for different strains or species; the variability of protein degradation was previously observed in milk proteins.47
In the present work, since electrophoresis does not allow the quantification of the protein hydrolysis degree, a peptide screening through the analysis of the hydrolyzed proteins by HPLC-DAD was performed to confirm the observed protein hydrolysis in milk and soy matrices as well as to verify whether there was hydrolysis of proteins other than milk caseins (lactoglobulins), soy glycines and soy beta-conglycines.
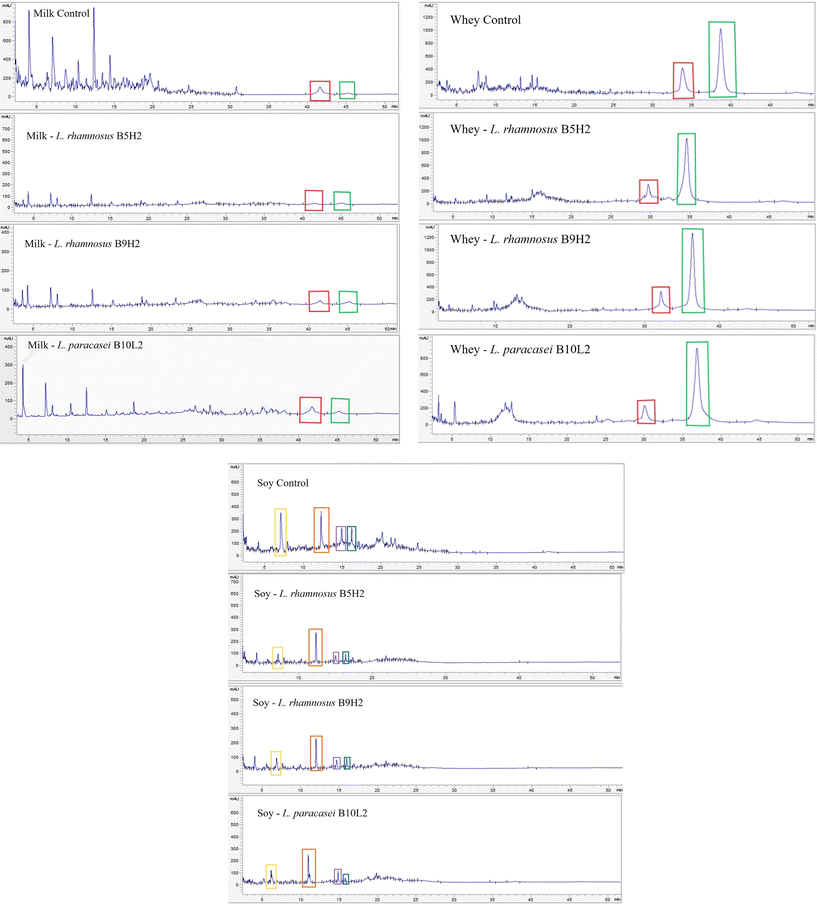
Fig. 5a and b show the HPLC-DAD chromatograms of milk and whey, respectively, where the peaks corresponding to alpha-lactoglobulin (red frame) and beta-lactoglobulin (green frame) were identified by their retention time after injecting the corresponding protein standards (Sigma-Aldrich, Germany). As shown in Fig. 5a, the alpha-lactoglobulin (red frame) control peak was clearly reduced in fermented samples, especially with L. rhamnosus B5H2 and L. rhamnosus B9H2, while the beta-lactoglobulin (green frame) protein peak did not show relevant changes. Protein concentration was calculated by area interpolation in a standard curve, confirming a reduction of alpha-lactoglobulin from 205.15 μg mL−1 in the control to 116.80 μg mL−1 (L. paracasei B10L2), 55.47 μg mL−1 (L. rhamnosus B9H2), and 37.67 μg mL−1 (L. rhamnosus B5H2) in fermented samples, which corresponded to a protein reduction from 100% (control) up to 57, 27 and 18%, respectively (Table 7).
| Samples | Alpha-lactoglobulin | Beta-lactoglobulin | ||
|---|---|---|---|---|
| Concentration (μg mL−1) | Variation (%) | Concentration (μg mL−1) | Variation (%) | |
| Milk | ||||
| Control | 205.15 | 100 | 51.40 | 100 |
| L. rhamnosus B5H2 | 37.67 | 18 | 52.01 | 101 |
| L. rhamnosus B9H2 | 55.47 | 27 | 53.28 | 106 |
| L. paracasei B10L2 | 116.80 | 57 | 53.81 | 127 |
| Whey | ||||
| Control | 929.35 | 100 | 2998.52 | 100 |
| L. rhamnosus B5H2 | 424.05 | 46 | 3029.06 | 101 |
| L. rhamnosus B9H2 | 762.95 | 82 | 3248.95 | 104 |
| L. paracasei B10L2 | 562.22 | 61 | 3131.59 | 105 |
A similar protein hydrolysis pattern was observed in whey samples, although less markedly and not evidenced after SDS-PAGE (Fig. 4). HPLC-DAD analysis showed a higher alpha-lactoglobulin peak in the control sample compared to those in LAB fermented samples, confirming bacterial protein hydrolysis to some extent for all three studied LAB in milk whey (Fig. 5b). Indeed, a reduction from 929.35 μg mL−1 in the control up to 762.95 μg mL−1 (L. rhamnosus B9H2), 562.22 μg mL−1 (L. paracasei B10L2), and 424.05 μg mL−1 (L. rhamnosus B5H2) was observed in LAB fermented whey samples, which represent reductions from 100% (control) up to 82% (L. rhamnosus B9H2), 61% (L. paracasei B10L2), and 46% (L. rhamnosus B5H2), as shown in Table 7. On the other side, beta-lactoglobulin did not show apparent reduction with any LAB strain, neither in milk nor whey samples.
With regard to soy beverage samples, four major peaks were observed in control samples with retention times: 7.1 min (peak 1; yellow frame); 12.3 min (peak 2; orange frame); 14.9 min (peak 3; purple frame); and 16.2 min (peak 4; blue frame), which were notably reduced in all LAB fermented soy samples, especially in the case of peaks 1, 3 and 4 (Fig. 5c), reaching reductions from 100% in the control up to 14.81% (peak 1, by L. rhamnosus B5H2), 56.58% (peak 2, by L. rhamnosus B9H2), 37.71% (peak 3, by L. rhamnosus B9H2), and 19.53% (peak 4, by L. paracasei B10L2).
Compared to the highly studied LAB proteolytic systems in dairy milk, less research has been focused on proteins from plant-based foods, where available research has been mainly conducted on soy as one of the most used plant-based dairy alternatives until recently.48 The benefits of peptides derived from soy beverage LAB fermentation have been previously described,49 and include increased antioxidant activity, increased bioavailability of amino acids, and increased inhibition of angiotensin-converting enzyme. However, the study of fermented non-dairy beverages is novel, giving rise to the use of LAB strains to ferment plant-based products with beneficial properties for health, since the study of strains with proteolytic capacity on these matrices could allow the creation of new probiotic food supplements. Thus, from previous studies it is known that LAB proteolytic systems can degrade the main soy proteins, especially beta-conglycins, and the proteolysis degree is related to the viability when fermenting the matrix,48 which agrees with the results shown in Fig. 5c, where intense hydrolysis was observed due to fermentation by all strains, especially of the protein framed in yellow.
The proteolytic activity of Lacticaseibacillus spp. is commonly used for dairy production to manufacture fermented dairy products such as cheese, yogurt, and kefir, among others. The intricate proteolysis mechanism enables these LAB to effectively break down casein into smaller peptides and free amino acids, making them a widely employed starter culture that enhances the flavor and texture of numerous dairy products.50 Moreover, many studies indicated that milk proteolysis by Lactobacillus spp. produces bioactive peptides, such as angiotensin-converting enzyme inhibitory peptides that can prevent cardiovascular disease related to hypertension.50 Other studies demonstrated the ability of LAB strains (specifically L. plantarum) to produce bioactive peptides in soy beverages.51 Moreover, the revalorization of industrial by-products, such as whey, and the production of new fermented foods with novel matrices, like soy, with a positive impact on consumer health are receiving increasing attention nowadays.15,51 However, due to the diversity of LAB strains and the complexity of the fermentation substrates,50 further studies should evaluate the bioactive peptides originating at each specific combination of the LAB strain and food substrate.
4. Conclusions
All the 25 isolated LAB showed resistance to pH 2 and bile salts, with 18 strains showing proteolytic activity. The seven most remarkable strains were identified and they showed the absence of antibiotic resistance, as well as auto-aggregation (up to 23%) and co-aggregation (up to 20%) capacity. Their resistance during in vitro simulated gastrointestinal digestion was confirmed, highlighting L. rhamnosus B5H2, L. rhamnosus B9H2 and L. paracasei B10L2, which reached bacterial viable counts higher than 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log10 CFU mL−1 after the colonic phase. All three LAB showed antifungal activity against P. verrucosum, F. verticillioides and F. graminearum, with L. rhamnosus B5H2 being the most active, and produced bioactive metabolites after MRS broth fermentation, including the antifungal compounds phenylacetic acid and 3-phenyllactic acid, as well as metabolites with antimicrobial and anti-inflammatory activities (10-hydroxy-cis-12-octadecenoic acid and benzoic acid). Moreover, all three strains produced VOCs in fermented milk, soy beverage and milk whey, highlighting the presence of L. rhamnosus B5H2 in all food matrices. The hydrolysis of the main milk (caseins) and soy proteins (glycines and beta-conglycines) by the studied LAB was evidenced, with alpha-lactoglobulin reduction in milk (82%) and whey (54%), highlighting L. rhamnosus B5H2 proteolytic activity. Overall, the three selected strains demonstrated probiotic capacity with L. rhamnosus B5H2 showing remarkable potential and this needs further investigation.
log10 CFU mL−1 after the colonic phase. All three LAB showed antifungal activity against P. verrucosum, F. verticillioides and F. graminearum, with L. rhamnosus B5H2 being the most active, and produced bioactive metabolites after MRS broth fermentation, including the antifungal compounds phenylacetic acid and 3-phenyllactic acid, as well as metabolites with antimicrobial and anti-inflammatory activities (10-hydroxy-cis-12-octadecenoic acid and benzoic acid). Moreover, all three strains produced VOCs in fermented milk, soy beverage and milk whey, highlighting the presence of L. rhamnosus B5H2 in all food matrices. The hydrolysis of the main milk (caseins) and soy proteins (glycines and beta-conglycines) by the studied LAB was evidenced, with alpha-lactoglobulin reduction in milk (82%) and whey (54%), highlighting L. rhamnosus B5H2 proteolytic activity. Overall, the three selected strains demonstrated probiotic capacity with L. rhamnosus B5H2 showing remarkable potential and this needs further investigation.
Data availability
In overall, the three selected strains demonstrated probiotic capacity with L. rhamnosus B5H2 as remarkable potential for further investigation.Author contributions
Abel Navarré: formal analysis, data curation, and writing – original draft. Tiago Nazareth: formal analysis, data curation, and writing – original draft. Carlos Luz: writing – review and editing. Giuseppe Meca: methodology and supervision. Laura Escrivá: methodology, supervision, and writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study was supported by the Ministry of Science and Innovation; projects PID2022-140722OB-I00/AEI/10.13039/501100011033/Unión Europea NextGenerationEU/PRTR and PCI2022-132937.References
- H. König and J. Fröhlich, Chapter one - Lactic acid bacteria, in Biology of Microorganisms on Grapes, in Must and in Wine, Springer International Publishing, 2017, pp. 3–41. DOI:10.1007/978-3-319-60021-5_1.
- K. Łubiech and M. Twarużek, Lactobacillus Bacteria in Breast Milk, Nutrients, 2020, 12(12), 3783 CrossRef PubMed , https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7764098/.
- A. F. El-Sheikha, Why Fermented Foods are the Promising Food Trends in the Future?, Curr. Res. Nutr. Food Sci., 2022, 10, 3, DOI:10.12944/CRNFSJ.10.3.1.
- C. Hill, F. Guarner, G. Reid, G. R. Gibson, D. J. Merenstein, B. Pot, L. Morelli, R. B. Canani, H. J. Flint, S. Salminen, P. C. Calder and M. E. Sanders, The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic, Nat. Rev. Gastroenterol. Hepatol., 2014, 11, 506–514, DOI:10.1038/nrgastro.2014.66.
- G. Rychen, G. Aquilina, G. Azimonti, V. Bampidis, M. d. L. Bastos, G. Bories, A. Chesson, P. S. Cocconcelli, G. Flachowsky, J. Gropp, B. Kolar, M. Kouba, M. López-Alonso, S. López, A. Mantovani, B. Mayo, F. Ramos, M. Saarela and J. Galobart, EFSA Panel on Additives and Products or Substances used in Animal Feed (FEEDAP), Guidance on the characterisation of microorganisms used as feed additives or as production organisms, EFSA J., 2018, 16(3), e05206, DOI:10.2903/j.efsa.2018.5206.
- P. Saranraj, M. A. Naidu and P. Sivasakthivelan, Lactic acid bacteria and its antimicrobial properties: a review, Int. J. Pharm. Biol. Arch., 2013, 4(6), 1124–1133 Search PubMed , https://api.semanticscholar.org/CorpusID:32008504.
- S. Crowley, J. Mahony and D. van Sinderen, Current perspectives on antifungal lactic acid bacteria as natural bio-preservatives, Trends Food Sci. Technol., 2013, 33(2), 93–109, DOI:10.1016/j.tifs.2013.07.004.
- R. Ballan, C. Battistini, D. Xavier-Santos and S. M. I. Saad, Chapter nine - Interactions of probiotics and prebiotics with the gut microbiota, Prog. Mol. Biol. Transl. Sci., 2020, 171, 265–300, DOI:10.1016/bs.pmbts.2020.03.008.
- M. da Silva, B. L. Tagliapietra, V. D. A. Flores and N. S. Richards, In vitro test to evaluate survival in the gastrointestinal tract of commercial probiotics, Curr. Res. Food Sci., 2021, 4, 320–325, DOI:10.1016/j.crfs.2021.04.006.
- G. V. De Melo, B. De Oliveira, A. I. Magalhães, V. Thomaz-Soccol and C. R. Soccol, How to select a probiotic? A review and update of methods and criteria, Biotechnol. Adv., 2018, 36(8), 2060–2076, DOI:10.1016/j.biotechadv.2018.09.003.
- B. N. Abdul Hakim, N. J. Xuan and S. N. H. Oslan, A Comprehensive Review of Bioactive Compounds from Lactic Acid Bacteria: Potential Functions as Functional Food in Dietetics and the Food Industry, Foods, 2023, 12(15), 2850, DOI:10.3390/foods12152850.
- A. Sánchez and A. Vázquez, Bioactive peptides: A review, Food Qual. Saf., 2017, 1(1), 29–46, DOI:10.1093/fqsafe/fyx006.
- C. S. Ranadheera, J. K. Vidanarachchi, R. S. Rocha, A. G. Cruz and S. Ajlouni, Probiotic Delivery through Fermentation: Dairy vs. Non-Dairy Beverages, Fermentation, 2017, 3(4), 67, DOI:10.3390/fermentation3040067.
- ISO 10932:2010. IDF International Organization for Standardization 10932:2010 (IDF 223:2010), milk and milk products—determination of the minimal inhibitory concentration (MIC) of antibiotics applicable to bifidobacteria and non-enterococcal lactic acid bacteria, 2010. https://www.iso.org/standard/46434.html.
- L. Escrivá, L. Manyes, P. Vila-Donat, G. Font, G. Meca and M. Lozano, Bioaccessibility and bioavailability of bioactive compounds from yellow mustard flour and milk whey fermented with lactic acid bacteria, Food Funct., 2021, 12(22), 11250–11261, 10.1039/D1FO02059E.
- C. Lafuente, J. Calpe, L. Musto, T. d. M. Nazareth, V. Dopazo, G. Meca and C. Luz, Preparation of Sourdoughs Fermented with Isolated Lactic Acid Bacteria and Characterization of Their Antifungal Properties, Foods, 2023, 12(4), 686, DOI:10.3390/foods12040686.
- Y. H. Lim, H. L. Foo, T. C. Loh, R. Mohamad and N. Abdullah, Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements, J. Anim. Sci. Biotechnol., 2019, 10, 15, DOI:10.1186/s40104-019-0323-z.
- J. Atanasova, P. Moncheva and I. Ivanova, Proteolytic and antimicrobial activity of lactic acid bacteria grown in goat milk, Biotechnol. Biotechnol. Equip., 2014, 28(6), 1073–1078, DOI:10.1080/13102818.2014.971487.
- C. Raveschot, B. Cudennec, B. Deracinois, M. Frémont, M. Vaeremans, J. Dugersuren, S. Demberel, D. Drider, P. Dhulster, F. Coutte and C. Flahaut, Proteolytic activity of Lactobacillus strains isolated from Mongolian traditional dairy products: A multiparametric analysis, Food Chem., 2020, 304, 125415, DOI:10.1016/j.foodchem.2019.125415.
- H. Wang, X. Zhang, Z. Chen, G. Hao and G. Li, Two Potential Probiotic Bacillus with Proteolytic Activity to Dietary Protein from Adult Feces, Biocontrol Sci., 2021, 26(4), 221–224, DOI:10.4265/bio.26.221.
- J. Kim, H. Kim, H. J. Jeon, Y. H. Jung and J. Yang, Lacticaseibacillus casei IDCC 3451 Strengthen Digestibility of Plant-based Proteins in Mice, Probiotics Antimicrob. Proteins, 2024, 16(3), 927–935, DOI:10.1007/s12602-023-10091-5.
- I. Sensoy, A review on the food digestion in the digestive tract and the used in vitro models, Curr. Res. Food Sci., 2021, 4, 308–319, DOI:10.1016/j.crfs.2021.04.004.
- Y. Yang, Y. Liu, S. Zhou, L. Huang, Y. Chen and H. Huan, Bile salt hydrolase can improve Lactobacillus plantarum survival in gastrointestinal tract by enhancing their adhesion ability, FEMS Microbiol. Lett., 2019, 366(8), fnz100, DOI:10.1093/femsle/fnz100.
- M. Begley, C. G. M. Gahan and C. Hill, The interaction between bacteria and bile, FEMS Microbiol. Rev., 2005, 29(4), 625–651, DOI:10.1016/j.femsre.2004.09.003.
- M. Li, Y. Wang, H. Cui, Y. Li, Y. Sun and H.-J. Qiu, Characterization of Lactic Acid Bacteria Isolated From the Gastrointestinal Tract of a Wild Boar as Potential Probiotics, Front. Vet. Sci., 2020, 7, 49 DOI:10.3389/fvets.2020.00049.
- A. Zawistowska-Rojek, A. Kośmider, K. Stępień and S. Tyski, Adhesion and aggregation properties of Lactobacillaceae strains as protection ways against enteropathogenic bacteria, Arch. Microbiol., 2022, 204(5), 285, DOI:10.1007/s00203-022-02889-8.
- M. T. Cook, G. Tzortzis, D. Charalampopoulos and V. V. Khutoryanskiy, Microencapsulation of probiotics for gastrointestinal delivery, J. Controlled Release, 2012, 162(1), 56–67, DOI:10.1016/j.jconrel.2012.06.003.
- S. Han, Y. Lu, J. Xie, Y. Fei, G. Zheng, Z. Wang, J. Liu, L. Lv, Z. Ling, B. Berglund, M. Yao and L. Li, Probiotic Gastrointestinal Transit and Colonization After Oral Administration A Long Journey, Front. Cell. Infect. Microbiol., 2021, 11, 609722, DOI:10.3389/fcimb.2021.609722.
- M. F. Yao, J. J. Xie, H. J. Du, D. J. McClements, H. Xiao and L. J. Li, (2020). Progress in microencapsulation of probiotics, Compr. Rev. Food Sci. Food Saf., 2020, 19(2), 857–874, DOI:10.1111/1541-4337.12532.
- J. I. I. Fugaban, E. S. Jung, S. D. Todorov and W. H. Holzapfel, Evaluation of Antifungal Metabolites Produced by Lactic Acid Bacteria, Probiotics Antimicrob. Proteins, 2023, 15(5), 1447–1463, DOI:10.1007/s12602-022-09995-5.
- T. d. M. Nazareth, J. Calpe, C. Luz, J. Mañes and G. Meca, Manufacture of a Potential Antifungal Ingredient Using Lactic Acid Bacteria from Dry-Cured Sausages, Foods, 2023, 12(7), 1427, DOI:10.3390/foods12071427.
- K.-S. Ng, M. F. Bambace and C. Schwab, Microbially produced short-chain carboxylic acids are ancient food biopreservatives with complex mode of action, Curr. Opin. Food Sci., 2023, 52, 101066, DOI:10.1016/j.cofs.2023.101066.
- X. Mao, Q. Yang, D. Chen, B. Yu and J. He, Benzoic Acid Used as Food and Feed Additives Can Regulate Gut Functions, BioMed Res. Int., 2019, 2019, e5721585, DOI:10.1155/2019/5721585.
- W. Mu, S. Yu, L. Zhu, T. Zhang and B. Jiang, Recent research on 3-phenyllactic acid, a broad-spectrum antimicrobial compound, Appl. Microbiol. Biotechnol., 2012, 95(5), 1155–1163, DOI:10.1007/s00253-012-4269-8.
- S. D. Cook, An Historical Review of Phenylacetic Acid, Plant Cell Physiol., 2019, 60(2), 243–254, DOI:10.1093/pcp/pcz004.
- B. Sulijaya, N. Takahashi and K. Yamazaki, Lactobacillus-Derived Bioactive Metabolites for the Regulation of Periodontal Health Evidences to Clinical Setting, Molecules, 2020, 25(9), 2088, DOI:10.3390/molecules25092088.
- J. Miyamoto, T. Mizukure, S.-B. Park, S. Kishino, I. Kimura, K. Hirano, P. Bergamo, M. Rossi, T. Suzuki, M. Arita, J. Ogawa and S. Tanabe, A Gut Microbial Metabolite of Linoleic Acid, 10-Hydroxy-cis-12-octadecenoic Acid, Ameliorates Intestinal Epithelial Barrier Impairment Partially via GPR40-MEK-ERK Pathway, J. Biol. Chem., 2015, 290(5), 2902–2918, DOI:10.1074/jbc.M114.610733.
- H. Abouloifa, I. Hasnaoui, Y. Rokni, R. Bellaouchi, N. Ghabbour, S. Karboune, M. Brasca, A. Abousalham, B. Jaouadi, E. Saalaoui and A. Asehraou, Chapter Two—Antifungal activity of lactic acid bacteria and their application in food biopreservation, in Advances in Applied Microbiology, ed. E. G. M. Gadd and S. Sariaslani, 2022, vol. 120, pp. 33–77. DOI:10.1016/bs.aambs.2022.07.001.
- L. Zhang, S. Mi, R. Liu, Y. Sang and X. Wang, Evaluation of Volatile Compounds in Milks Fermented Using Traditional Starter Cultures and Probiotics Based on Odor Activity Value and Chemometric Techniques, Molecules, 2020, 25(5), 1129 CrossRef PubMed.
- B. Piechulla, M. C. Lemfack and N. Magnus, Bioactive Bacterial Organic Volatiles An Overview and Critical Comments, in Bacterial Volatile Compounds as Mediators of Airborne Interactions, ed. E. C.-M. Ryu, L. Weisskopf and B. Piechulla, 2020, pp. 39–92. DOI:10.1007/978-981-15-7293-7_2.
- C. Xiong, Q. Li, S. Li, C. Chen, Z. Chen and W. Huang, In vitro Antimicrobial Activities and Mechanism of 1-Octen-3-ol against Food-related Bacteria and Pathogenic Fungi, J. Oleo Sci., 2017, 66(9), 1041–1049, DOI:10.5650/jos.ess16196.
- A. Rani, A. Rana, R. K. Dhaka, A. P. Singh, M. Chahar, S. Singh, L. Nain, K. P. Singh and D. Minz, Bacterial volatile organic compounds as biopesticides, growth promoters and plant-defense elicitors Current understanding and future scope, Biotechnol. Adv., 2023, 63, 108078, DOI:10.1016/j.biotechadv.2022.108078.
- A. Digaitiene, Å.S Hansen, G. Juodeikiene, D. Eidukonyte and J. Josephsen, Lactic acid bacteria isolated from rye sourdoughs produce bacteriocin-like inhibitory substances active against Bacillus subtilis and fungi, J. Appl. Microbiol., 2012, 112(4), 732–742, DOI:10.1111/j.1365-2672.2012.05249.x.
- D. Ağagündüz, B. Yılmaz, T.Ö Şahin, B. E. Güneşliol, Ş. Ayten, P. Russo, G. Spano, J. M. Rocha, E. Bartkiene and F. Özogul, Dairy Lactic Acid Bacteria and Their Potential Function in Dietetics The Food-Gut-Health Axis, Foods, 2021, 10(12), 3099, DOI:10.3390/foods10123099.
- Y.-H. Hsiao, C.-J. Yu, W.-T. Li and J.-F. Hsieh, Coagulation of β-conglycinin, glycinin and isoflavones induced by calcium chloride in soymilk, Sci. Rep., 2015, 5(1), 13018 CrossRef CAS PubMed.
- J. Čurlej, P. Zajác, J. Čapla, J. Golian, L. Benešová, A. Partika, A. Fehér and S. Jakabová, The Effect of Heat Treatment on Cow’s Milk Protein Profiles, Foods, 2022, 11(7), 1023 CrossRef.
- M. Pescuma, E. M. Héber, E. Bru, G. F. de Valdez and F. Mozzi, Diversity in growth and protein degradation by dairy relevant lactic acid bacteria species in reconstituted whey, J. Dairy Res., 2012, 79(2), 201–208, DOI:10.1017/S0022029912000040.
- A. R. Harper, R. C. J. Dobson, V. K. Morris and G.-J. Moggré, Fermentation of plant-based dairy alternatives by lactic acid bacteria, Microb. Biotechnol., 2022, 15(5), 1404–1421, DOI:10.1111/1751-7915.14008.
- M. Kumari, A. Kokkiligadda, V. Dasriya and H. Naithani, Functional relevance and health benefits of soymilk fermented by lactic acid bacteria, J. Appl. Microbiol., 2022, 133(1), 104–119, DOI:10.1111/jam.15342.
- N. Şanlier, B. B. Gökcen and A. C. Sezgin, Health benefits of fermented foods, Crit. Rev. Food Sci. Nutr., 2019, 59(3), 506–527, DOI:10.1080/10408398.2017.1383355.
- B. P. Singh and S. Vij, Growth and bioactive peptides production potential of Lactobacillus plantarum strain C2 in soy milk A LC-MS/MS based revelation for peptides biofunctionality, LWT–Food Sci. Technol., 2017, 86, 293–301, DOI:10.1016/j.lwt.2017.08.013.
Footnote |
| † Both authors contributed equally to the manuscript. |
| This journal is © The Royal Society of Chemistry 2024 |