Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Automated extraction of synthesis parameters of pulsed laser-deposited materials from scientific literature†
Rajan
Kumar
a,
Ablokit
Joshi
a,
Salman A.
Khan
 b and
Shikhar
Misra
b and
Shikhar
Misra
 *a
*a
aIndian Institute of Technology Kanpur, Kalyanpur, Kanpur, Uttar Pradesh 208016, India. E-mail: shikharm@iitk.ac.in
bDelaware Energy Institute, University of Delaware, 221 Academy Street, Newark, Delaware 19716, USA
First published on 5th April 2024
Abstract
The materials science literature contains a large amount of reliable and high-quality data and automatically extracting useful information, including processing parameters and materials property data from this scientific literature continues to be a challenge. The development of new materials is typically based on experimental trial and error approach to identify the optimized processing parameters. In this work, we present an approach at the intersection of Natural Language Processing (NLP) and Materials Science, focusing on the extraction and analysis of materials and processing parameters associated with Pulsed Laser Deposition (PLD). Using the MatSciBERT (Bidirectional Encoder Representations from Transformers)-based architecture, we achieved precise identification and categorization of different PLD synthesis parameters, including, deposition temperature and pressure, laser energy, laser wavelength, thin film material and substrate, using the Named Entity Recognition (NER) model. This involved meticulous data acquisition from over 6000 research articles, followed by pre-processing, feature extraction, and model training. The trained NER model showcased impressive micro and macro F1 scores of 80.2% and 81.4%, respectively. This highlights the potential of Literature-based Discovery (LBD) approaches in expediting material discovery processes. The insights gained from this study are expected to drive advancements in materials research, streamlining information extraction processes by building a searchable database, and accelerating discoveries in the domain of Pulsed Laser Deposition.
Introduction
The interdisciplinary field of materials science involves the study of materials, encompassing properties, compositions, behaviors, design, and characterization, which yields a plethora of high-quality and peer-reviewed published research works present in the scientific literature. These publications serve as reservoirs of invaluable knowledge essential for new discoveries and advancements in the field. However, accessing this wealth of knowledge demands an exhaustive exploration of literature, which is an extremely time-consuming and labor-intensive task, thus impeding the pace of new material discoveries. Therefore, to accelerate materials discovery, an efficient data-driven literature-based discovery (LBD) approach is needed, that is capable of extracting and harnessing knowledge from pre-existing literature.1–3 In this regard, Natural Language Processing (NLP), emerges as a powerful tool for extracting, storing, and analyzing information from a variety of written sources, including research articles, journal papers, review articles, and websites.4–8 Over time, advancements in NLP technology and computational power have enabled the pretraining of large language models (LLMs), enhancing the efficiency of entity/information extraction from scholarly literature and in their ability to capture contextual relationships among different tokens within a sentence or paragraph.9–16ChemDataExtractor, ChemicalTagger, and other NLP tools have been developed for materials science to extract data from chemical text.17 To increase the accuracy of chemical data extraction, sophisticated models based on deep convolutional and recurrent neural networks have recently been proposed.1,5,18,19 The development of Bidirectional Encoder Representations from Transformers (BERT) model, which is a neural network architecture, has led to its wide-scale adoption for training diverse Language Models tailored for domain-specific tasks. BERT can generate contextual embeddings and can be trained with vast training datasets. However, the standard BERT model, pretrained on English Wikipedia and BookCorpus, lacked the ability to comprehend domain-specific jargon prevalent in material science literature, including materials' properties and names.20 To bridge this gap, several domain-specific BERT models such as BioBERT, SciBERT, and clinical-BERT have emerged, which have been further trained on the respective literature and all of which exhibit superior performance for domain-specific tasks.18,21,22 Recently, a materials-aware language model was also developed, namely, MatSciBERT, which yielded state-of-the-art results for downstream tasks such as Named Entity Recognition (NER), Relation Classification, and Abstract Classification within the material science domain.19 While prior NER models like Matscholar and Materials Entity Recognition (MER) model exist, they lack support for domain-specific tasks.2,3,23,24 For example, Matscholar relies on Word2Vec embeddings, incapable of capturing token context, while existing (NER) models were pretrained on general text corpora rather than domain-specific material science corpora. Recently, LLMs have also been successfully applied to extract synthesis parameters from chemistry and materials science literature.25–29
In this work, a specialized MER model was developed to extract materials synthesized using the Pulsed Laser Deposition technique and the corresponding processing parameters from unstructured text. PLD is a Physical Vapor Deposition technique used in materials science and device fabrication, which uses a high-intensity laser to ablate a target material and then deposit the ablated material onto a substrate to form a thin film. Pre-trained weights of the MatSciBERT model were fine-tuned by training on text describing the synthesis of materials from PLD literature. The accuracy of the MER model was demonstrated through its high macro and micro F1 scores on a validation set, showcasing its reliable performance. Our work provides a platform for systematic analysis of unstructured PLD data and opens doors for future research to discover optimal processing parameters for the synthesis of various materials by PLD.
Data acquisition
The acquisition of data is a critical aspect of scientific research, providing the foundational basis upon which analyses, conclusions, and advancements are made. In this study, text data was mined from a vast corpus of scientific literature, specifically focused on research related to thin film deposition using Pulsed Laser Deposition (PLD). By employing web scraping, XML/HTML article extraction, and paragraph-level extraction and classification, a comprehensive dataset was compiled upon which further analysis was done.Digital object identifier (DOI)
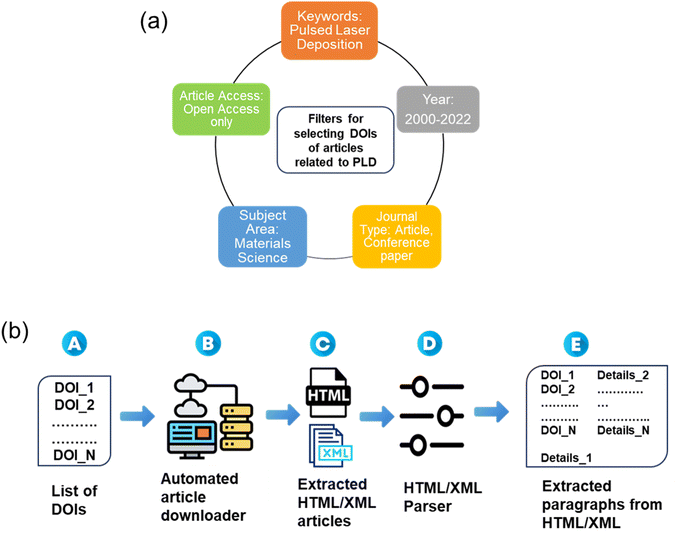
DOI acquisition was performed to procure research articles pertinent to PLD, by doing a Scopus search, coupled with precise filters (keywords, journal type, article access, subject area, and year), shown in Fig. 1a, yielding a total of about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 DOIs. The DOIs were segregated based on the respective publishers, enabling the development of publisher-specific Python scripts for subsequent download requests. An automated pipeline was established for efficient article downloading based on the curated DOIs list. This pipeline, employing publisher-specific approaches, successfully downloaded journal and conference articles by major publishers like Elsevier, Springer Nature, and other open-source articles in HTML and XML formats. The dataset thus obtained included a substantial collection of 5300 XML and 3677 HTML research articles, published between the years 2000–2022, as shown in Fig. 1a.
000 DOIs. The DOIs were segregated based on the respective publishers, enabling the development of publisher-specific Python scripts for subsequent download requests. An automated pipeline was established for efficient article downloading based on the curated DOIs list. This pipeline, employing publisher-specific approaches, successfully downloaded journal and conference articles by major publishers like Elsevier, Springer Nature, and other open-source articles in HTML and XML formats. The dataset thus obtained included a substantial collection of 5300 XML and 3677 HTML research articles, published between the years 2000–2022, as shown in Fig. 1a.
Web scraping and XML/HTML article extraction
Web scraping was utilized to retrieve the research articles based on the DOI list of the research articles relevant to PLD (Figure 1b A). By using customized web scrapers, relevant information including article titles, authors, abstracts, publication dates, in addition to the raw-text files were downloaded in HTML/XML format (Fig. 1b, B–C). The extracted web content, often in the form of HTML or XML, necessitates structured processing to further extract useful information using NER. Leveraging XML's hierarchical structure and HTML's markup tags, ElementTree Python library was used for effective parsing of the articles (Fig. 1b, D). This process involved extracting specific sections like metadata (comprising DOI, title, abstract, and authors' details), paragraphs, and object information (pertaining to figures and tables) in a systematic and efficient manner.Paragraph extraction
Further, the processed text was used to extract individual paragraphs in the research articles (Fig. 1b, E). The ‘para’ tag in XML served as a vital indicator for paragraph identification. Similarly, BeautifulSoup, another python library, which can extract specific data from HTML documents by searching for tags, attributes, or text content, was used for the extraction of all the paragraphs and metadata of HTML articles. In this step, the individual paragraphs were obtained and were associated with their respective DOI, title, and abstract. The result was a structured database of articles' textual contents, enhancing the granularity of our dataset and facilitating targeted analysis, as shown schematically in Fig. 1b. In addition, in the process of paragraph extraction, it was important to ensure data quality by identifying and addressing outliers. We encountered paragraphs that were either irrelevant or exceptionally small in size. Employing Python's regular expression library, we designed a robust mechanism to detect and remove these outlier paragraphs. Additionally, we addressed paragraphs riddled with unusual whitespaces, further refining our dataset and enhancing its quality and reducing the computational cost for the subsequent analysis. The code details are provided in the Data and Software Availability section.Methodology
Pre-processing
Text pre-processing was performed at various stages, including data cleaning, normalization, and entity categorization. In data cleaning, irrelevant paragraphs were removed, and consistency in formatting was ensured. Normalization encompassed standardizing units and formats of extracted parameters such as temperature, pressure, and energy. Entity categorization involves labeling the entities using an annotation tool by associating each entity with its respective entity, thereby making a training dataset for MER model training. The details of each of these steps are discussed later.Paragraph embeddings using BERT
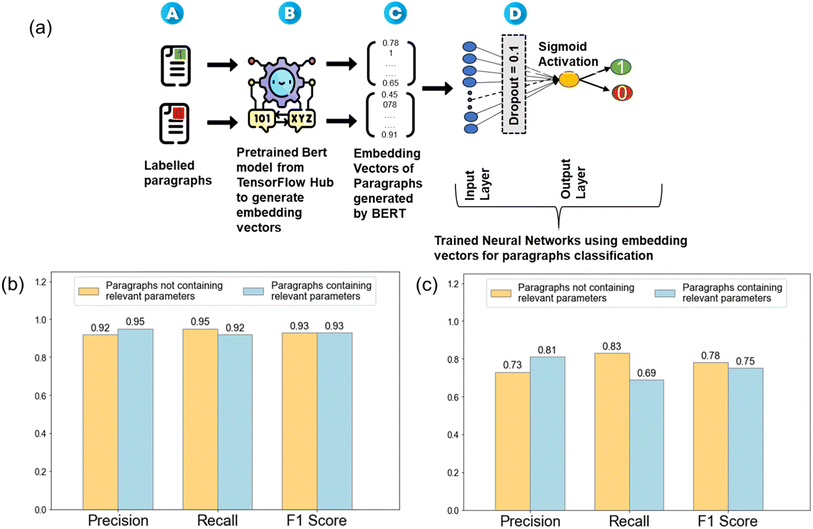
In Natural Language Processing (NLP), converting textual information into quantifiable features is essential for machine learning models. To achieve this, we utilized BERT-based embeddings to represent the contextualized features of each labeled paragraph.22 The BERT model used for generating embedding was a general pre-trained model from the TensorFlow hub. BERT embeddings have been demonstrated to encapsulate contextual understanding of the words, capturing the intricate relationships and meanings between them. The embeddings served as the feature vectors, which contain the contextual information of the paragraphs, that helps to train the subsequent classification model.Paragraph classification model
Further, a classification model was built to categorize paragraphs based on the presence or absence of processing parameters related to PLD. Typically, a manuscript contains only 1–2 synthesis paragraphs and not all papers follow the same format. Therefore, in order to generalize the process, we chose a binary classification approach, where a paragraph was labeled as a synthesis paragraph either containing relevant parameters (labelled as 1) or not containing them (labelled as 0). Hence, manual labelling of 5200 paragraphs as 0/1 was done for generating a training dataset. Out of 5200 labelled paragraphs, 577 paragraphs were labelled as 1 i.e., those paragraphs contained processing parameters and the remaining 4653 paragraphs were labelled as 0 i.e., paragraphs having no processing parameters. Additionally, the labeling was verified by another PLD domain expert to ensure consistency and accuracy. Therefore, this approach helped minimize error. The training of this classification model involved utilizing the embeddings generated earlier and designing a neural network-based classification model, using TensorFlow and Keras frameworks, Adam as the optimizer, and binary cross-entropy as the loss function during training. The NLP pipeline for paragraph classification is shown in Fig. 2a. The key evaluation metrics were precision, recall, and F1 score, providing a comprehensive understanding of the model's performance.Due to the imbalanced nature of the training dataset, both under-sampling and over-sampling techniques were explored to mitigate bias toward the majority class (label 0). Under-sampling involves reducing the number of instances of the over-represented class to balance the dataset, while over-sampling involves increasing the number of occurrences of the under-represented data to balance the dataset. Under-sampling, though simple, resulted in a significant loss of data and a lower average F1-score of 0.76. In contrast, over-sampling by replication of the minority class instances (label 1) proved to be a more effective approach. By ensuring an almost equal representation of both classes, the training dataset was balanced which enhanced the model's ability to generalize across the classes. The chosen model, trained on the balanced dataset, using the over-sampling technique, was then utilized for predicting paragraphs containing processing parameters of interest. Finally, 5918 paragraphs out of a vast database of 163![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 228 paragraphs were classified as paragraphs containing synthesis information with an average F1-score of 0.93, as shown in Fig. 2b and c.
228 paragraphs were classified as paragraphs containing synthesis information with an average F1-score of 0.93, as shown in Fig. 2b and c.
Materials entity recognition (MER)
MER involved the recognition and categorization of specific elements from unstructured textual data. In our context, these elements primarily consist of materials' formulae and names, and materials synthesis parameters associated with PLD experiments. By training a specialized NER model using MatSciBERT (discussed later), a BERT-based transformer model tailored for materials science, these entities were accurately identified and labelled from the text. The MatSciBERT model, pre-trained on a massive corpus of materials science-related text, provided a strong foundation for our NER task. MatSciBERT is a deep transformer encoder-based pre-trained language model. By fine-tuning these models with the processed text dataset, the model was able to recognize and classify material entities with high precision and recall.In order to streamline the process, reduce the computational cost, and increase the model efficiency, we took advantage of initially classifying the paragraphs followed by running the MER model on the extracted paragraphs. After extracting the paragraphs containing the processing parameters, material-specific entities and their respective processing parameters were identified using a MER model. To facilitate the creation of a labeled dataset for training the MER model, a labeling tool was utilized. In this case, an open-source tool, NER Annotator by Tecoholic, was employed to label entities.30 The resulting annotated dataset, formed the training data essential for the MER model.
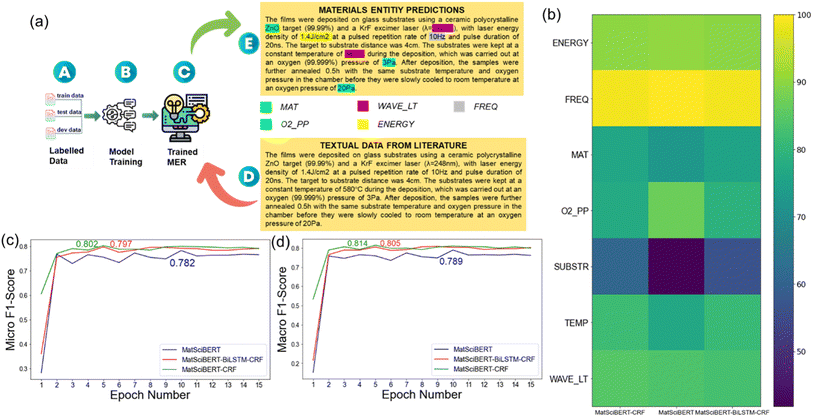
The training data for NER included domain-specific information related to PLD in materials science. Labels were classified as Deposited Material (MAT), Laser Fluence (ENERGY), Frequency (FREQ), O2 Partial Pressure (O2_PP), Substrate (SUBSTR), Temperature (TEMP), Wavelength (WAVE_LT), and Other (O). All these labels serve as key experimental parameters for thin film deposition using PLD. Fig. 3a shows an example of the predicted labels in a paragraph, after the MER model was trained on the labeled dataset. Further, three architectures were explored – MatSciBERT, MatSciBERT-CRF (Conditional Random Fields), and MatSciBERT-BiLSTM-CRF (Bidirectional Long Short-Term Memory with Conditional Random Fields), using the MatSciBERT embeddings. MatSciBERT-CRF, integrating MatSciBERT and a Conditional Random Field layer, demonstrated superior performance, compared to the other two architectures, in capturing label dependencies and making globally optimal predictions. With this model, the relevant materials entities were extracted from scientific literature, as shown in Fig. 3a. Note that in contrast to the MatSciBERT embeddings used for the MER model, we used BERT embeddings for paragraph classification as described above. Further refining of the obtained dataset was done by removing the outliers through a manual inspection to eliminate redundant or irrelevant entities, to ensure the accuracy and precision of our dataset. A few cases were encountered where there were multiple sets of synthesis parameters, for example, a paragraph having multiple temperature and pressure values. In such cases, our model returned a list of values, and then we used our human domain experts to further post-process those data points. For example, if a sample was deposited at multiple temperatures and pressures, then the mean value was taken. A systematic normalization approach (discussed later) was devised to standardize the units across the dataset, ensuring uniformity and facilitating meaningful comparisons and analyses.
Material entities extraction by MER model
The fine-tuned MatSciBERT model (PLD-BERT) was applied to extract processing parameters. These material and respective processing parameters were extracted and stored in an organized way from ∼6000 paragraphs.Unit normalization
The extracted data often comprises diverse units and inconsistent representations. Therefore, the normalization of units is a pivotal step in standardizing the representation of extracted processing parameters. It involves bringing all data into a consistent and uniform unit, facilitating meaningful comparisons and analyses. In the domain of material science, where units can vary widely, this process is particularly important. Therefore, normalization was performed for all the extracted entities, including material, wavelength, frequency, energy, temperature, and pressure. Following are the methods we used to normalize each parameter.Results
Trained MER Model's evaluation
Typical metrics, including accuracy, precision, recall (sensitivity), and F1 Score, were calculated to evaluate the accuracy of the trained model. In our case, there were 8 different labels, which is a multi-class classification. In the case of multi-class classification, one can calculate two versions of the F1 Score, i.e., micro and macro F1 Score. Both micro and macro F1 scores were calculated to decide the model performance. Micro F1 score, a composite metric, assesses the model's effectiveness by considering true positives, false positives, and false negatives across all classes. It treats the classification task as a single, multi-class problem, generating a singular F1 score that represents the overall model performance. This approach aggregates performance measures across all classes, making it suitable for scenarios with imbalanced class distributions, offering equal importance to each class. On the other hand, the macro F1 score calculates the F1 score for each class independently and then computes the average across all classes. Each class is given equal weight, irrespective of its prevalence in the dataset. This metric is beneficial when evaluating the model's performance in individual classes, providing a sense of its average performance across all classes.Fig. 3c and d shows the micro and macro F1 score for the three MatSciBERT architectures: MatSciBERT, MatSciBERT-BiLSTM-CRF, and MatSciBERT-CRF. MatSciBERT is combined with a layer of Conditional Random Field (CRF) in MatSciBERT-CRF. When tasked with assigning a label to each element in a series, sequence labeling jobs make use of CRF. MatSciBERT-BiLSTM-CRF combines MatSciBERT with layers of CRF and Bidirectional Long Short-Term Memory (BiLSTM). Recurrent neural networks (RNNs) of the BiLSTM type are capable of identifying sequential patterns in data.33 The best Micro F1 Scores were 78.2%, 80.2% and 79.7% for MatSciBERT, MatSciBERT-CRF and MatSciBERT-BiLSTM-CRF, respectively on the validation dataset, and was the metric based on which the final model was chosen (Fig. 3c). The Macro F1 Scores (for the corresponding best Micro F1 scores) were calculated as 78.9%, 81.4%, and 80.5% for MatSciBERT, MatSciBERT-CRF, and MatSciBERT-BiLSTM-CRF, respectively on the validation dataset (Fig. 3d). The detailed results of the F1 scores for the three models are included in the ESI (Tables 1–3†). Further, Fig. 3b shows a heatmap of the average F1 scores of the individual entities for each model. Table 1 summarizes the entity-wise average F1 scores for the three MatSciBERT architectures. MatSciBERT-CRF claims the best performance for 4 entities and slightly lags behind in two entities: ENERGY and FREQ. MatSciBERT-CRF lags behind in ‘O2_PP’ entity compared to MatSciBERT, however, it significantly outperforms in the ‘SUBSTR’ entity leading to overall higher Micro F1 score. The second-best performance is for MatSciBERT-BiLSTM-CRF, followed by the MatSciBERT model.
| Model | Energy | Freq | Mat | O2 PP | Substr | Temp | Wave Lt | Micro F1 score | Macro F1 score |
|---|---|---|---|---|---|---|---|---|---|
| MatSciBERT | 90.63 | 100 | 72.73 | 86.96 | 41.86 | 76.67 | 83.72 | 78.21 | 78.93 |
| MatSciBert-CRF | 90.09 | 98.90 | 76.29 | 77.42 | 60.98 | 81.63 | 84.78 | 80.25 | 81.44 |
| MatSciBert-BILSTM-CRF | 90 | 98.92 | 75.54 | 79.24 | 56.47 | 80.98 | 82.83 | 79.72 | 80.57 |
Synthesis insights
The distributions of normalized extracted parameters are shown in Fig. 4. The distribution plots provide useful insights on the key parameters in PLD experiments. Notably, temperature distribution indicated a prevalent range of 400 °C to 700 °C (65.53%) for thin film deposition, with the peaks usually at an interval of 50 °C in these experiments. Pressure distribution highlighted a common range below 100 mTorr, with a significant number of depositions occurring between 5 to 15 mTorr. These peaks in deposition temperature and pressure are typical of oxide thin film deposition, the reports of which are present in large numbers in the literature. Laser and laser fluence distribution is shown in Fig. 4c. Laser fluence energy distribution exhibited a prevalent range of 2 J cm−2 to 6 J cm−2. The laser wavelength distribution is shown in Fig. 4d and it predominately shows a wavelength of 248 nm, that is widely utilized for PLD, using KrF excimer laser. Additionally, frequency distribution revealed common usage at 5 Hz and 10 Hz. Further, the temperature and pressure distribution for some specific materials, namely ZnO, VO2, and BaTiO3 is also plotted and shown in Fig. 5. Clearly, the majority of the thin film depositions have been performed at 600 °C, 500 °C, and 700 °C for ZnO, VO2, and BaTiO3 respectively, which is typical for these materials.34–39 While the oxygen partial pressure indicate a vacuum deposition or very low oxygen partial pressures (0–10 mTorr) for most of these thin film depositions. For example, VO2 is a metastable phase deposited using the V2O5 target that shows a very narrow growth window near 10 mTorr oxygen partial pressure.40–42 The difference in growth temperature for the different materials can arise due to several factors, including bonding energy, substrate crystal structure, vapor pressure, diffusion coefficient, etc. Therefore, such a database can greatly help in quickly deciding the appropriate deposition parameters for a variety of materials.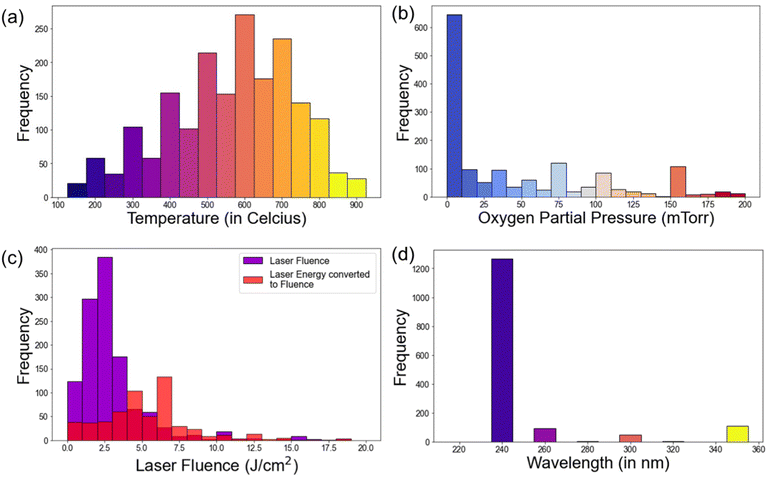 | ||
| Fig. 4 (a) Temperature, (b) pressure, (c) laser fluence, and (d) laser wavelength distribution of the thin films deposited using PLD. These data points are extracted using the PLD-BERT model. | ||
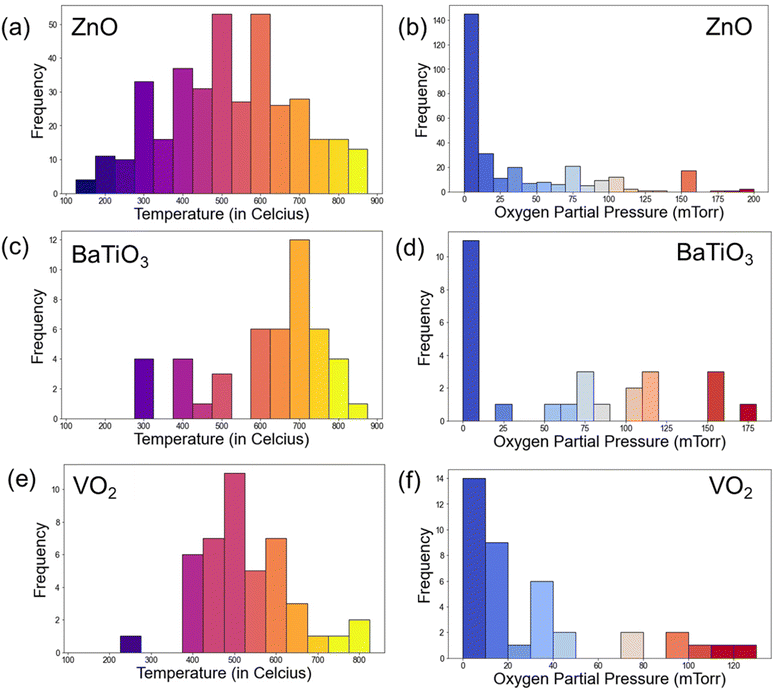 | ||
| Fig. 5 Temperature and pressure distribution of PLD deposited thin films of (a and b) ZnO, (c and d) BaTiO3, and (e and f) VO2. | ||
Conclusion
In this study, we successfully applied a large language model to extract experimental processing parameters for thin film deposition using PLD. We developed a MER model by fine-tuning the MatSciBERT model on PLD-specific literature. A meticulous approach to outlier removal and unit normalization ensured data quality and uniformity, essential for meaningful analysis. Model performance was assessed by calculating micro and macro F1 scores. The trained model achieved precise extraction and categorization of critical processing parameters such as deposition temperature, pressure, laser fluence and wavelength, and substrate from an extensive corpus of unstructured textual data. The insights derived from this work are anticipated to catalyze further advancements in materials research, streamlining knowledge acquisition and facilitating future discoveries in the domain of thin film deposition using PLD.Data and software availability
Code details for web scrapping and paragraph extraction with data cleaning is available as a jupyter notebook on Figshare at: 10.6084/m9.figshare.25265431. The fine-tuning code is available as a jupyter notebook on Figshare at: 10.6084/m9.figshare.24770952. The fine-tuned PLD-BERT model can be found on Figshare at: 10.6084/m9.figshare.24770895 and the annotated data sets used for fine-tuning can be found at: 10.6084/m9.figshare.24770916.Author contributions
S. M. proposed and initiated the study. R. K., and S. M. collected and analyzed the data, and built the ML models. S. A. K. assisted in the study design and in analyzing the results, while A. J. helped analyze the results. All authors contributed to writing and revising the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The work was partially supported by the IITK Start-up Fund and SERB SRG/2022/000580.References
- E. A. Olivetti, J. M. Cole, E. Kim, O. Kononova, G. Ceder and T. Y. J. Han, et al., Data-driven materials research enabled by natural language processing and information extraction, Appl. Phys. Rev., 2020, 7(4), 041317 CAS.
- L. Weston, V. Tshitoyan, J. Dagdelen, O. Kononova, A. Trewartha and K. A. Persson, et al., Named Entity Recognition and Normalization Applied to Large-Scale Information Extraction from the Materials Science Literature, J. Chem. Inf. Model., 2019, 59(9), 3692–3702 CrossRef CAS PubMed.
- V. Tshitoyan, J. Dagdelen, L. Weston, A. Dunn, Z. Rong and O. Kononova, et al., Unsupervised word embeddings capture latent knowledge from materials science literature, Nature, 2019, 571(7763), 95–98 CrossRef CAS PubMed.
- S. Huang and J. M. Cole, A database of battery materials auto-generated using ChemDataExtractor, Sci. Data, 2020, 7(1), 1–13 CrossRef PubMed.
- A. C. Vaucher, F. Zipoli, J. Geluykens, V. H. Nair, P. Schwaller and T. Laino, Automated extraction of chemical synthesis actions from experimental procedures, Nat. Commun., 2020, 11(1), 1–11 CrossRef PubMed.
- S. M. Azimi, D. Britz, M. Engstler, M. Fritz and F. Mücklich, Advanced steel microstructural classification by deep learning methods, Sci. Rep., 2018, 8(1), 1–14 CAS.
- K. T. Mukaddem, E. J. Beard, B. Yildirim and J. M. Cole, ImageDataExtractor: A Tool to Extract and Quantify Data from Microscopy Images, J. Chem. Inf. Model., 2020, 60(5), 2492–2509 CrossRef CAS PubMed.
- A. Friedrich, H. Adel, F. Tomazic, J. Hingerl, R. BenteauA. Maruscyk and et al., The SOFC-Exp corpus and neural approaches to information extraction in the materials science domain, Proceedings of the Annual Meeting of the Association for Computational Linguistics, 2020, 1255–1268 Search PubMed.
- E. A. Olivetti, J. M. Cole, E. Kim, O. Kononova, G. Ceder and T. Y. J. Han, et al., Data-driven materials research enabled by natural language processing and information extraction, Appl. Phys. Rev., 2020, 7(4), 041317 CAS.
- O. Kononova, T. He, H. Huo, A. Trewartha, E. A. Olivetti and G. Ceder, Opportunities and challenges of text mining in materials research, iScience, 2021, 24(3), 102155 CrossRef PubMed.
- A. Trewartha, N. Walker, H. Huo, S. Lee, K. Cruse and J. Dagdelen, et al., Quantifying the advantage of domain-specific pre-training on named entity recognition tasks in materials science, Patterns, 2022, 3(4), 100488 CrossRef PubMed.
- Z. Wang, O. Kononova, K. Cruse, T. He, H. Huo and Y. Fei, et al., Dataset of solution-based inorganic materials synthesis procedures extracted from the scientific literature, Sci. Data, 2022, 9(1), 1–11 CrossRef.
- W. Wang, X. Jiang, S. Tian, P. Liu, D. Dang and Y. Su, et al., Automated pipeline for superalloy data by text mining, npj Comput. Mater., 2022, 8(1), 1–12 CrossRef.
- E. Kim, Z. Jensen, A. Van Grootel, K. Huang, M. Staib and S. Mysore, et al., Inorganic Materials Synthesis Planning with Literature-Trained Neural Networks, J. Chem. Inf. Model., 2020, 60(3), 1194–1201 CrossRef CAS PubMed.
- H. Huo, Z. Rong, O. Kononova, W. Sun, T. Botari and T. He, et al., Semi-supervised machine-learning classification of materials synthesis procedures, npj Comput. Mater., 2019, 5(1), 1–7 CrossRef.
- E. Kim, K. Huang, A. Saunders, A. McCallum, G. Ceder and E. Olivetti, Materials Synthesis Insights from Scientific Literature via Text Extraction and Machine Learning, Chem. Mater., 2017, 29(21), 9436–9444 CrossRef CAS.
- M. C. Swain and J. M. Cole, ChemDataExtractor: A Toolkit for Automated Extraction of Chemical Information from the Scientific Literature, J. Chem. Inf. Model., 2016, 56(10), 1894–1904 CrossRef CAS PubMed.
- I. Beltagy, K. Lo and A. Cohan, SciBERT: Pretrained Contextualized Embeddings for Scientific Text, arXiv, 2019, preprint, arXiv:1903.10676, DOI:10.48550/arXiv.1903.10676.
- T. Gupta, M. Zaki and N. M. A. Krishnan, Mausam. MatSciBERT: A materials domain language model for text mining and information extraction, npj Comput. Mater., 2022, 8(1), 1–11 CrossRef.
- J. Devlin, M. W. Chang, K. Lee and K. Toutanova, BERT: Pre-training of deep bidirectional transformers for language understanding, NAACL, 2019, 4171–4186 Search PubMed.
- J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim and C. H. So, et al., BioBERT: A pre-trained biomedical language representation model for biomedical text mining, Bioinformatics, 2020, 36(4), 1234–1240 CrossRef CAS PubMed.
- E. Alsentzer, J. R. Murphy, W. Boag, W. H. Weng, D. Jin, T. Naumann and et al.Publicly Available Clinical BERT Embeddings. arXiv:1904.03323, 2019 Search PubMed.
- M. S. U. Miah, J. Sulaiman, T. B. Sarwar, N. Ibrahim, M. Masuduzzaman and R. Jose, An automated materials and processes identification tool for material informatics using deep learning approach, Heliyon, 2023, 9(9), e20003 CrossRef PubMed.
- X. Zhao, J. Greenberg, Y. An and X. T. HuFine-Tuning BERT Model for Materials Named Entity Recognition, Proceedings - 2021 IEEE International Conference on Big Data, Big Data 2021, 2021, 3717–3720 Search PubMed.
- A. Dunn, J. Dagdelen, N. Walker, S. Lee, A. S. Rosen, G. Ceder and et al., Structured information extraction from complex scientific text with fine-tuned large language models, arXiv:2212.05238, 2022 Search PubMed.
- N. Walker, S. Lee, J. Dagdelen, K. Cruse, S. Gleason and A. Dunn, et al., Extracting structured seed-mediated gold nanorod growth procedures from scientific text with LLMs, Digital Discovery, 2023, 2(6), 1768–1782 RSC.
- M. C. Ramos, S. S. Michtavy, M. D. Porosoff and A. D. White, Bayesian Optimization of Catalysts With In-context Learning, arXiv:2304.05341, 2023 Search PubMed.
- A. D. White, G. M. Hocky, H. A. Gandhi, M. Ansari, S. Cox and G. P. Wellawatte, et al., Assessment of chemistry knowledge in large language models that generate code, Digital Discovery, 2023, 2(2), 368–376 RSC.
- A. M. Bran, S. Cox, O. Schilter, C. Baldassari, A. D. White and P. Schwaller. ChemCrow: Augmenting large-language models with chemistry tools, arXiv:2304.05376, 2023 Search PubMed.
- M. Neves and J. Ševa, An extensive review of tools for manual annotation of documents, Briefings Bioinf., 2021, 22(1), 146–163 CrossRef PubMed.
- Z. Vakulov, D. Khakhulin, E. Zamburg, A. Mikhaylichenko, V. A. Smirnov and R. Tominov, et al., Towards scalable large-area pulsed laser deposition, Materials, 2021, 14(17), 4854 CrossRef CAS PubMed.
- R. Delmdahl and R. Pätzel, Pulsed laser deposition-UV laser sources and applications, Appl. Phys. A: Mater. Sci. Process., 2008, 93(3), 611–615 CrossRef CAS.
- Z. Huang, W. Xu and K. Yu, Bidirectional LSTM-CRF Models for Sequence Tagging, arXiv:1508.01991. 2015 Search PubMed.
- S. Misra, D. Zhang, Z. Qi, D. Li, J. Lu and H. T. Chen, et al., Morphology control of self-assembled three-phase Au–BaTiO3–ZnO hybrid metamaterial for tunable optical properties, Cryst. Growth Des., 2020, 20(9), 6101–6108 CrossRef CAS.
- S. Misra, L. Li, D. Zhang, J. Jian, Z. Qi and M. Fan, et al., Self-Assembled Ordered Three-Phase Au–BaTiO3–ZnO Vertically Aligned Nanocomposites Achieved by a Templating Method, Adv. Mater., 2019, 31(7), 1806529 CrossRef PubMed.
- B. Zhang, J. Huang, B. Rutherford, P. Lu, S. Misra and M. Kalaswad, et al., Tunable room temperature multiferroic Fe-BaTiO3 vertically aligned nanocomposite with perpendicular magnetic anisotropy, Mater. Today Nano, 2020, 11, 100083 CrossRef.
- D. Zhang, S. Misra, L. Li, X. Wang, J. Jian and P. Lu, et al., Tunable Optical Properties in Self-Assembled Oxide–Metal Hybrid Thin Films via Au-Phase Geometry Control: From Nanopillars to Nanodisks, Adv. Opt. Mater., 2020, 8(4), 1901359 CrossRef CAS.
- S. Misra, D. Zhang, P. Lu and H. Wang, Thermal stability of self-assembled ordered three-phase Au–BaTiO3–ZnO nanocomposite thin films: via in situ heating in TEM, Nanoscale, 2020, 12(46), 23673–23681 RSC.
- S. Misra, M. Kalaswad, D. Zhang and H. Wang, Dynamic tuning of dielectric permittivity in BaTiO3via electrical biasing, Mater. Res. Lett., 2020, 8(9), 321–327 CrossRef CAS.
- Y. Ji, Z. Qi, S. Misra, R. Jin, X. Ou and Y. Lin, et al., Breaking Lattice Symmetry in Highly Strained Epitaxial VO2 Films on Faceted Nanosurface, ACS Appl. Mater. Interfaces, 2019, 11(47), 44905–44912 CrossRef CAS PubMed.
- J. Jian, X. Wang, S. Misra, X. Sun, Z. Qi and X. Gao, et al., Broad Range Tuning of Phase Transition Property in VO2 Through Metal-Ceramic Nanocomposite Design, Adv. Funct. Mater., 2019, 29(36), 1903690 CrossRef.
- Z. He, J. Jian, S. Misra, X. Gao, X. Wang and Z. Qi, et al., Bidirectional tuning of phase transition properties in Pt : VO2 nanocomposite thin films, Nanoscale, 2020, 12(34), 17886–17894 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Tables for the F1 scores for the individual entities on the validation dataset for the individual epochs using the MatSciBERT, MatSciBERT-CRF, and MatSciBERT-BiLSTM-CRF architecture. See DOI: https://doi.org/10.1039/. |
| This journal is © The Royal Society of Chemistry 2024 |