Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Computational investigation of copper-mediated conformational changes in α-synuclein dimer†
Loizos
Savva
 and
James A.
Platts
and
James A.
Platts
 *
*
School of Chemistry, Cardiff University, Park Place, Cardiff CF10 3AT, UK. E-mail: platts@cardiff.ac.uk; Tel: +44-2920-874950
First published on 4th January 2024
Abstract
We report molecular dynamics simulation of dimers of α-synuclein, the peptide closely associated with onset of Parkinson's disease, both as metal-free dimer and with inter-chain bridging provided by Cu(II) ions. Our investigation reveals that the presence of copper-induced inter-chain bridging not only stabilizes α-synuclein dimers, but also leads to enhanced β-sheet formation at critical regions within the N-terminal and NAC regions of the protein. These contacts are larger and longer-lived in the presence of copper, and as a result each peptide chain is more extended and less flexible than in the metal-free dimer. The persistence of these inter-peptide contacts underscores their significance in stabilising the dimers, potentially influencing the aggregation pathway. Moreover, the increased flexibility in the two termini, as well as the absence of persistent contacts in the metal-free dimer, correlates with the presence of amorphous aggregates. This phenomenon is known to mitigate fibrillation, while their absence in the metal-bound dimer suggests an increased propensity to form fibrils in the presence of copper ions.
Introduction
Aggregation of the α-synuclein (αS) protein has long been implicated in the development of Parkinson's disease (PD) – one of the most prevalent neuropathologies. The formation of fibril deposits of αS in Lewy bodies affects the neuronal concentration in the midbrain, and results in dopamine depletion associated with motor behaviour deficits. This 140-residue intrinsically disordered protein (IDP) is primarily found in presynaptic terminals of neurons,1 and is generally partitioned into three main regions: the N-terminal (residues M1-K60), characterised by a high content of amphipathic helices; the non-amyloid-β component (NAC – residues E61-V95), largely populated with hydrophobic residues; and the C-terminal (residues K96-A140), which contains most of the acidic residues in the protein.2,3 The N-terminal is distinguished by the four 11-mer [EQS]-K-T-K-[EQ]-[GQ]-V-X4 imperfect repeats, which constitute the region where the peptide assumes the most stable order of secondary characteristics. Within these repeats, β-hairpins have been found to form between residues L38-A53 and V63-T72,4–7 as well as α-helices, either in solution or as a response to lipid membrane interactions.2,8–13 This region has also been characterised by the facilitation of interactions with Cu(II) (amongst other metal ions), with studies using small-angle X-ray scattering (SAXS), nuclear magnetic resonance (NMR), circular dichroism (CD) and electron paramagnetic resonance (EPR) spectroscopy, indicating Cu(II) to bind to regions encompassing residues 1MDVFMKGLS9 and 48VAHGV52, but more specifically by coordination with residues M1-D2-H50.14–17 Prior work from our group, using molecular dynamics (MD) simulations, has identified the stabilisation of long-lasting β-hairpins as a result of Cu(II) coordination with the monomer of αS,7 believed to increase the propensity of inter-peptide bridging, and by extension increasing its ability to form aggregates.The work presented here follows these previous assessments on the effects of copper ions on the αS monomer,7 now examining the effect of Cu(II) on a dimer of αS, in an effort to elucidate the structural alterations upon dimerization. This should shed light on the possible fibrillation mechanism this protein undergoes, and provide further evidence on the aggregation-promoting features we have observed in the monomer, upon coordination of Cu(II). To study this dimer, we look at the cross-bridged αS, with copper ions interacting with residues M1 and D2 from one chain and residue H50 from the other.18,19
Under physiological conditions, αS normally exists in an equilibrium between the monomeric and membrane-bound forms,20 or in a tetrameric form which resists the formation of the aberrant aggregates.21 Concerning the aggregation of biomolecular systems, one usually looks for a nucleation–elongation process,22 with the off-pathway formation of amorphous aggregates, found to decrease the development of fibrils.23,24 This is in line with the mechanism seen in other systems, such as amyloid-β, where similar aggregates have also been suggested to modulate the formation of fibrils.25
Computational methods
The α-synuclein dimer systems were modelled in their extended conformation in DommiMOE, where the ligand-field molecular mechanics (LFMM) force field was used for an initial minimisation of the systems, both with and without Cu(II). For the metal-free system, the two chains were placed at a ca. 12° angle to each other, crossing at H50, as it has been found to be the region populated with folded secondary structural elements, and the likely area for inter-peptide interactions.26 The angle between the two chains was chosen after considering the increasing distance between residues with wider angles, where the system did not have enough time to form interactions, with the two monomers instead drifting away from each other. In the case of the metal-bound dimer, the two chains were placed in anti-parallel direction to each other, permitting bridging interactions from the two metal ions and residues M1 and D2 of one chain, and H50 from the other, Fig. 1.18 We note that the NMR structure of αS deposited as PDB entry 1XQ8 is not suitable for our purposes since it is micelle-bound,27,28 whereas all simulations here are in aqueous environment. | ||
| Fig. 1 Minimised structure of the copper-coordinated αS dimer, with CaDAM (blue-pink) bridging residues M1 and D2 from one chain with H50 from the other. | ||
Lack of detailed LFMM parameters for this specific system meant that an alternative model of Cu coordination was required. For the modelling of the Cu(II) metal ion, a cationic dummy atom model (CaDAM) was therefore implemented,29 to permit the flexible formation of metal-residue interactions within binding pockets identified in literature. In order to construct the model, four dummy atoms, each carrying +0.5 charge, were placed in a square planar arrangement around the uncharged metal centre.30 The MD simulations were performed using the AMBER16 suite,31 with the ff03ws force field,32 and Onufriev, Bashford, Case (OBC) continuum model.33–35 These were selected after an assessment of this combination for the MD simulations of the metal-free and bound αS monomers, reported in prior publications by our group.7,36 Having constructed and minimised the two systems, five independent MD simulations of 400 ns (200 million steps at 2 fs time step, with SHAKE holonomic restraints on bonds to hydrogen) were performed in the NVT ensemble (Langevin thermostat,37 310 K), with the first 100 ns of each run disregarded for equilibration, resulting in 1.5 μs data for analysis for each system. Most analysis was carried out using cpptraj,38 along with carma for clustering.39,40
Results and discussion
In order to confirm the equilibrated state of the proteins studied here, the root-mean-square-deviation (RMSD) of the two systems was examined, Fig. S1 and S2 (ESI†), where it was found that in all five runs, in both the metal-free and metal-bound dimers, the proteins settled to stable fluctuation around a mean within 100 ns. Hence, it was decided to consider the final 300 ns from each of the runs in the analysis.‡ Accelerated molecular dynamics simulations were also assessed, although the bias on the potential energy pushed the two chains apart within the first 100 ns, Fig. S3 (ESI†), and no meaningful analysis can be presented from these; the discussion is therefore focused on the 1.5 μs combined data from the conventional MD simulations.It is notable that the metal-free dimer adopts a broad range of relative chain orientations, whereas the Cu-bound system maintains the approximately anti-parallel arrangement that is required for M1-D2-H50 bridging coordination to be possible. This orientation, defined by the vectors between the terminal residues, remains approximately the same for the Cu-bound dimer, with a narrow distribution centred around 140°. In contrast, the chains in the metal-free system preferentially populate a perpendicular arrangement of chains, peaking between 60°–100°, but with appreciable density down to 20° and up to 160° Fig. 2. This is in accordance with the description of amorphous aggregates, hinting towards the formation of fibrillation-inhibitory structures in the case of the metal-free dimer.41,42
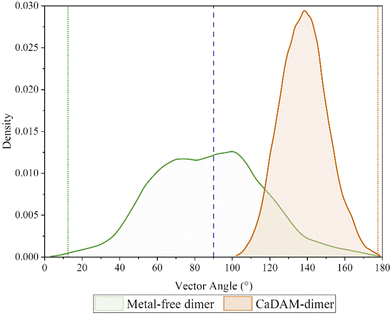 | ||
| Fig. 2 Density plot of the angle between the two vectors, corresponding to each of the chains in the dimers. The green/orange dotted lines indicate the angle of the starting conformation. | ||
Fig. 3, shows the average distance between the two Cu(II) ions and the backbone-C of each residue in the system, where it is clear that these remain within a close proximity to the residues they were first placed near, especially in the case of the Cu-facilitated bridging between M1-D2 from Chain A and H50 from Chain B. The reproduction of known binding modes in this analysis confirms the suitability of the CaDAM approach to Cu-coordination, as observed in numerous other examples of the biological chemistry of Cu(II).18,43
 | ||
| Fig. 3 Contact map of the average distance between the backbone-C and the CaDAM in the dimeric system. | ||
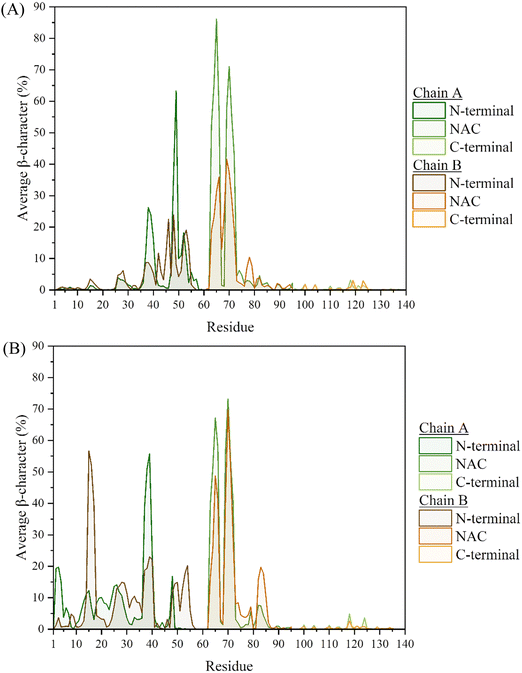
The secondary structural elements of each of the chains in the two systems, were also examined, with a breakdown of the mean percentage in each of the regions given in Tables S1 and S2 (ESI†). Considering the Cu(II)-bound monomer from our previous work,7 a clear increase in the β-characteristics can be seen throughout. In particular, the N-terminal of both chains exhibit ca. 3 times greater β-content, compared to the Cu(II)-bound monomer. The NAC-region of both chains also displays a similarly inreased β-content, from 5% in the monomer to 13–14% in the dimer (Table 1). In the case of the metal-free system, the β-content most influenced by the dimer formation appears to be in the NAC region, where Chain A is twice as populated with strands compared to the monomer. Of note is the fact that the characteristics between the two chains here, are not as alike as those seen in the Cu-bound dimer. This is also reflected in the average secondary structure plots, Fig. 4 and 5, where despite the characteristics following the same trend, they are generally more pronounced in Chain A, especially where the β-content is concerned.
 | ||
| Fig. 4 Average helical characteristics of the residues in the (A) metal-free and (B) CaDAM-bound dimeric system. | ||
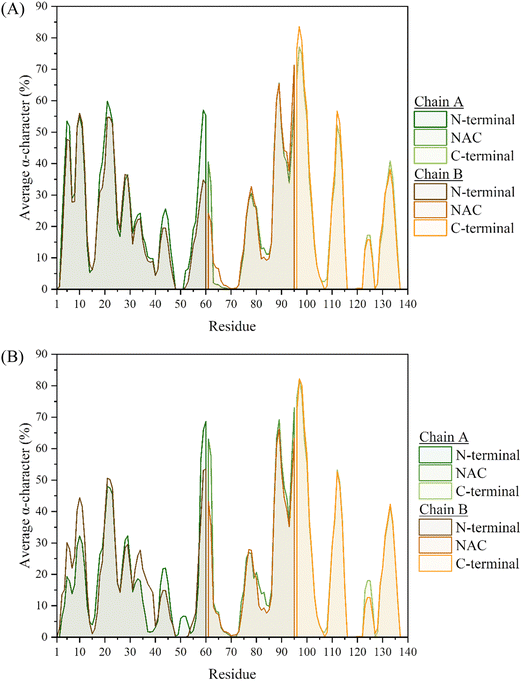
Interestingly, many similarities can be identified between the metal-free and metal-bound systems, with regards to the residues that are part of the most populated characteristics. In particular, the helical regions remain almost identical between the two systems, with the greatest content found between residues S87-K102. This region was also found to be the most populated in the monomeric system, and resulted in the formation of an amphipathic helix. The replication of this characteristic in the dimeric system, suggests that the presence of the metal ion, or additional chains, does not adversely affect the folding of this relatively stable region of secondary structure. In fact, the only major difference in helical content of the two dimer systems appears to be in the persistence of the region between residues V55-V63 (where one of the KTKEQV repeats is contained), which is about twice more pronounced in the metal-bound system. This region was also found to be populated with α-helices in the monomer systems,7 although at ca. 40–42% occupancy in both the metal-free and Cu-bound monomers. The inter-chain interactions could aid in that regard, with the stabilisation of these helices from hydrogen bonds formed between the two chains, especially in the CaDAM-bound dimer where these are more populated (vide infraFig. 10).
The greatest differences in the secondary characteristics arise from the β-strand content of the two systems. The most populated region involved residues V63-T72, which was also identified in the monomeric systems as one of the locations exhibiting long-lasting β-hairpin character, through the formation of anti-parallel β-strands. This characteristic is replicated here in the two chains of both systems through intra-chain anti-parallel β-strands, Fig. 6 and Fig. S4 (ESI†). Intra-chain interactions are also formed between frames 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–120
000–120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, through the formation of a parallel β-sheet. Upon assessing the per-residue secondary characteristics of the other regions in the two systems, with respect to the frames in the trajectories, the β-hairpin that was present between residues L38-A53 in the monomers is now repurposed towards the formation of inter-peptide β-sheets, bridging the two chains and by extension aiding in the conservation of the dimer, Fig. S6 (ESI†). While this region displays a high propensity for this inter-peptide interaction in the metal-free system, the same is not observed in the Cu-bound dimer. Upon a closer inspection of the per-residue characteristics, this bridging appears to be instead facilitated by the residues in the extended N-terminal region (between residues V15-V40), Fig. S4(B) and S5 (ESI†). From the former figure, it is clear that this β-sheet is not maintained in all five repeats, only appearing in three of them – in the rest (between frames 60
000, through the formation of a parallel β-sheet. Upon assessing the per-residue secondary characteristics of the other regions in the two systems, with respect to the frames in the trajectories, the β-hairpin that was present between residues L38-A53 in the monomers is now repurposed towards the formation of inter-peptide β-sheets, bridging the two chains and by extension aiding in the conservation of the dimer, Fig. S6 (ESI†). While this region displays a high propensity for this inter-peptide interaction in the metal-free system, the same is not observed in the Cu-bound dimer. Upon a closer inspection of the per-residue characteristics, this bridging appears to be instead facilitated by the residues in the extended N-terminal region (between residues V15-V40), Fig. S4(B) and S5 (ESI†). From the former figure, it is clear that this β-sheet is not maintained in all five repeats, only appearing in three of them – in the rest (between frames 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–120
000–120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000), the two chains instead appear to only be held together by the two Cu ions. This is in line with the short-lived nature of secondary characteristics seen in the monomer systems, owing to the intrinsically disordered nature of the protein. Overall, the N-terminal region of both systems give rise to the anchoring characteristics that help sustain the dimer.
000), the two chains instead appear to only be held together by the two Cu ions. This is in line with the short-lived nature of secondary characteristics seen in the monomer systems, owing to the intrinsically disordered nature of the protein. Overall, the N-terminal region of both systems give rise to the anchoring characteristics that help sustain the dimer.
 | ||
| Fig. 6 Evolution of secondary structural elements of each of residues V63–T72 in the two chains of the (A) metal-free and (B) Cu-bound dimers. The pink line separates the two chains. | ||
The snapshots of the systems given in Fig. S4 (ESI†), illustrate the areas where β-sheets can form, although they do not represent the average conformation in these systems. Therefore, clusters were created using principal component analysis (PCA) of the Cα atoms. For this, the carma software was used to execute PCA on the cartesian coordinates of backbone carbons.39 The results from this analysis are shown in Table S3 (ESI†), with the average cluster structure for each of the systems given in Fig. 7. As expected for IDPs, many frames do not fall into any cluster but in both metal-free and Cu-bound dimers we find one cluster with significant population (ca. 30% of frames), with others covering no more than 1 or 2% of frames. The interactions in the metal-free dimer are mainly focused on the intrachain V63-T72 β-hairpins, with the inter-chain interactions formed between the two hairpins. This is in accordance with the observations made above, for frames 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–120
000–120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, suggesting cluster 1 may be populated with frames from this region; confirmed from secondary structure analysis on these structures, Fig. 8, with ca., 16
000, suggesting cluster 1 may be populated with frames from this region; confirmed from secondary structure analysis on these structures, Fig. 8, with ca., 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (36% of the whole) structures exhibiting this character. The metal-bound system's average cluster structure is in accordance with the observations made in Fig. S5 (ESI†), discussed above, with an inter-peptide β-sheet between residues V37-Y39 from Chain A, and V15-A17 from Chain B, along with the interactions enforced by bridging Cu-coordination.
000 (36% of the whole) structures exhibiting this character. The metal-bound system's average cluster structure is in accordance with the observations made in Fig. S5 (ESI†), discussed above, with an inter-peptide β-sheet between residues V37-Y39 from Chain A, and V15-A17 from Chain B, along with the interactions enforced by bridging Cu-coordination.
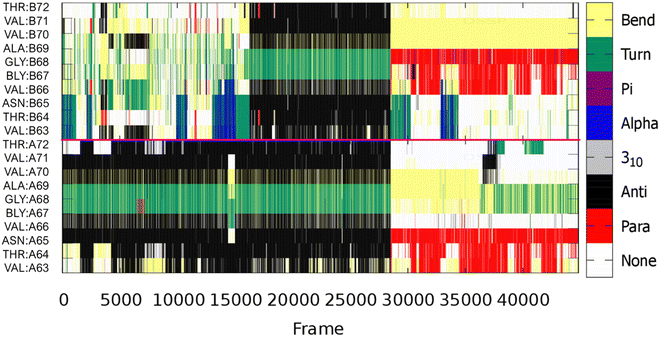 | ||
| Fig. 8 Evolution of secondary structural elements of each of residues V63-T72, from the most populated cluster, in the two chains of the metal-free dimer. The pink line separates the two chains. | ||
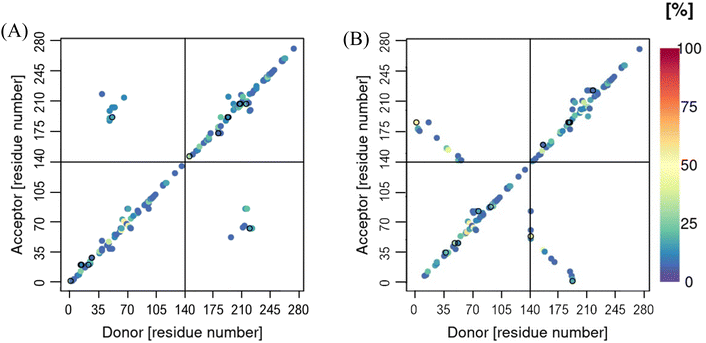
Cα-contact maps for the two systems are shown in Fig. 9, where the chains in both systems are found to interact through the N-terminal region. This interaction is more pronounced in the metal-bound system, with the metal ions bridging the two chains both at the beginning and the end of the N-termini. In the case of the metal-free system, the inter-peptide interactions are concentrated around residues V40-K60. These regions of close proximity between the two chains, unsuprisingly follow the areas of inter-peptide β-sheet formation, discussed above, as well as hydrogen bonds between the two chains, Fig. 10. These are also both more populated and longer-lasting in the Cu-αS dimer, decreasing flexibility. This is reflected in the RMSF plots, Fig. 11, where the metal-bound system is overall at least half as mobile as the metal-free system, especially in the chain ends. The shortest motions in the metal-free dimer are exhibited in the region between residues K45-T75, where the β-content is also the highest, Fig. 5(A). The flexibility of residues in the metal-bound system is much more evenly distributed, such that RMSF values for the Cu-bound dimer are as large as those in the metal-free dimer only at the very end of the C-terminus.
 | ||
| Fig. 9 Contact maps of the distance between the backbone-C from the dynamics of the (A) metal-free and (B) metal-bound dimeric systems. | ||
 | ||
| Fig. 11 Root mean square fluctuation, of the individual residues in the (A) metal-free and (B) metal-bound dimeric system. | ||
Despite the reduced RMSF of the metal-bound dimer, the size of the two chains in the metal-bound dimer is notably more extended than in the metal-free system, Table 2, with the Rg 5 Å higher in both chains. This is attributed to the extended intermolecular interactions in the N-terminal and NAC regions of the metal-bound system, which are more constrained in the metal-free dimer. This is especially evident from the total Rg of the systems, where the metal-bound system is ca. 13 Å larger than the metal-free system. It is also notable that each chain in the metal-free dimer is slightly larger than that found in the monomer under identical conditions (mean = 38.80 Å, sd = 3.96 Å),36 which we again attribute to inter-chain interactions acting to hinder folding of individual chains.
| Chain | Avg, Rg (Å) | SD (Å) | Max (Å) | Min (Å) |
|---|---|---|---|---|
| Free-dimer | ||||
| Chain A | 41.96 | 5.35 | 61.65 | 26.47 |
| Chain B | 42.27 | 5.76 | 63.27 | 26.00 |
| Total | 50.26 | 4.75 | 66.12 | 36.17 |
| Metal-dimer | ||||
| Chain A | 46.76 | 5.47 | 64.37 | 31.62 |
| Chain B | 47.85 | 7.77 | 77.51 | 28.26 |
| Total | 63.53 | 6.65 | 90.03 | 45.63 |
Conclusions
From the results presented here, we conclude that the cationic dummy atom model provides a sound description of Cu(II) binding to αS as a bridge between chains, maintaining interaction with residues M1-D2 of one chain and H50 of the other, as described in EPR experiments.18,19 Stable secondary characteristics were identified for both the free and CaDAM-bound systems, specifically intramolecular β-hairpin structures between residues V63-T72. This area was also identified in the metal-free and Cu(II)-bound monomeric systems, to exhibit a long-lasting β-hairpin. Furthermore, inter-peptide bridging was found to proceed through β-sheets formed between these β-hairpins, as well as between residues L38-A53, in the metal-free; while in the Cu-bound system, these interactions were maintained only between residues M1-V40. In the case of the metal-free dimer, the L38-A53 β-sheet has been repurposed from the β-hairpin structure observed in the monomeric system, while in the metal-bound system, a number of β-sheets are formed and maintained throughout the N-terminal, between the two binding sites. These bridging structures have been important in maintaining interactions between the two chains, specifically in the metal-free system, where in the absence of β-sheets shared between the two monomers, the dimer dissociated. These findings are in accord with those from Zhang et al.,44 who used discrete MD to examine αS dimer formation and showed dimers to be mostly dynamic with limited duration times, with unstructured formations dominating the ensemble but significant β-sheet formations between chains around the NAC region also evident.In the case of the Cu-bound dimer, however, even in the absence of these β-sheets, the dimer was able to maintain its inter-peptide linkage, through the metal-mediated bridges formed between the two chains. This speaks towards the stability of the multimers formed in the presence of the metal ion, which are less prone to dissociation and increase the persistence of interactions between chains. Specifically, the results obtained point towards the formation of amorphous aggregates in the case of the metal-free systems. These off-pathway aggregates have been described in the past as fibrillation-inhibitory structures,23,24 affirming the role of the metal ion towards the formation of fibril deposits in Lewy bodies.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the role of Advanced Research Computing @ Cardiff (ARCCA) in providing computational resources for simulations.References
- M. Goedert, R. Jakes and M. G. Spillantini, The Synucleinopathies: Twenty Years on, J. Parkinsons. Dis., 2017, 7, S53–S71 Search PubMed.
- K. Uéda, H. Fukushima, E. Masliah, Y. U. Xia, A. Iwai, M. Yoshimoto, D. A. C. Otero, J. Kondo, Y. Ihara and T. Saitoh, Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 11282–11286 CrossRef PubMed.
- A. D. Stephens, M. Zacharopoulou, R. Moons, G. Fusco, N. Seetaloo, A. Chiki, P. J. Woodhams, I. Mela, H. A. Lashuel, J. J. Phillips, A. De Simone, F. Sobott and G. S. K. Schierle, Extent of N-terminus exposure of monomeric alpha-synuclein determines its aggregation propensity, Nat. Commun., 2020, 11, 2820 CrossRef CAS PubMed.
- J. Li, V. N. Uversky and A. L. Fink, Effect of familial Parkinson's disease point mutations A30P and A53T on the structural properties, aggregation, and fibrillation of human α-synuclein, Biochemistry, 2001, 40, 11604–11613 CrossRef CAS PubMed.
- S. B. T. A. Amos, T. C. Schwarz, J. Shi, B. P. Cossins, T. S. Baker, R. J. Taylor, R. Konrat and M. S. P. Sansom, Membrane Interactions of α-Synuclein Revealed by Multiscale Molecular Dynamics Simulations, Markov State Models, and NMR, J. Phys. Chem. B, 2021, 125, 2929–2941 CrossRef CAS PubMed.
- H. Yu, W. Han, W. Ma and K. Schulten, Transient β-hairpin formation in α-synuclein monomer revealed by coarse-grained molecular dynamics simulation, J. Chem. Phys., 2015, 143, 243142 CrossRef PubMed.
- L. Savva and J. A. Platts, How Cu(II) binding affects structure and dynamics of α-synuclein revealed by molecular dynamics simulations, J. Inorg. Biochem., 2023, 239, 112068 CrossRef CAS PubMed.
- J. P. Segrest, H. De Loof, J. G. Dohlman, C. G. Brouillette and G. M. Anantharamaiah, Amphipathic helix motif: classes and properties, Proteins Struct. Funct. Bioinforma., 1990, 8, 103–117 CrossRef CAS PubMed.
- R. Bussell and D. Eliezer, A structural and functional role for 11-mer repeats in α-synuclein and other exchangeable lipid binding proteins, J. Mol. Biol., 2003, 329, 763–778 CrossRef CAS PubMed.
- C. C. Jao, B. G. Hegde, J. Chenb, I. S. Haworth and R. Langen, Structure of membrane-bound α-synuclein from site-directed spin labeling and computational refinement, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 19666–19671 CrossRef CAS PubMed.
- L. Xu, R. Nussinov and B. Ma, Coupling of the non-amyloid-component (NAC) domain and the KTK(E/Q)GV repeats stabilize the α-synuclein fibrils, Eur. J. Med. Chem., 2016, 121, 841–850 CrossRef CAS PubMed.
- M. D. Tuttle, G. Comellas, A. J. Nieuwkoop, D. J. Covell, D. A. Berthold, K. D. Kloepper, J. M. Courtney, J. K. Kim, A. M. Barclay, A. Kendall, W. Wan, G. Stubbs, C. D. Schwieters, V. M. Y. Lee, J. M. George and C. M. Rienstra, Solid-state NMR structure of a pathogenic fibril of full-length human α-synuclein, Nat. Struct. Mol. Biol., 2016, 23, 409–415 CrossRef CAS PubMed.
- D. E. Mor, S. E. Ugras, M. J. Daniels and H. Ischiropoulos, Dynamic structural flexibility of α-synuclein, Neurobiol. Dis., 2016, 88, 66–74 CrossRef CAS PubMed.
- A. Binolfi, R. M. Rasia, C. W. Bertoncini, M. Ceolin, M. Zweckstetter, C. Griesinger, T. M. Jovin and C. O. Fernández, Interaction of α-synuclein with divalent metal ions reveals key differences: a link between structure, binding specificity and fibrillation enhancement, J. Am. Chem. Soc., 2006, 128, 9893–9901 CrossRef CAS PubMed.
- R. M. Rasia, C. W. Bertoncini, D. Marsh, W. Hoyer, D. Cherny, M. Zweckstetter, C. Griesinger, T. M. Jovin and C. O. Fernández, Structural characterization of copper(II) binding to α-synuclein: insights into the bioinorganic chemistry of Parkinson's disease, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 4294–4299 CrossRef CAS.
- F. Camponeschi, D. Valensin, I. Tessari, L. Bubacco, S. Dell’Acqua, L. Casella, E. Monzani, E. Gaggelli and G. Valensin, Copper(I)-α-Synuclein Interaction: Structural Description of Two Independent and Competing Metal Binding Sites, Inorg. Chem., 2013, 52, 1358–1367 CrossRef CAS.
- S. C. Drew, L. L. Su, C. L. L. Pham, D. J. Tew, C. L. Masters, L. A. Miles, R. Cappai and K. J. Barnham, Cu2+ binding modes of recombinant α-synuclein – Insights from EPR spectroscopy, J. Am. Chem. Soc., 2008, 130, 7766–7773 CrossRef.
- S. C. Drew, The N Terminus of α-Synuclein Forms Cu II-Bridged Oligomers, Chem. – Eur. J., 2015, 21, 7111–7118 CrossRef CAS.
- S. C. Drew, Probing the quaternary structure of metal-bridged peptide oligomers, J. Inorg. Biochem., 2016, 158, 30–34 CrossRef CAS.
- J. T. Bendor, T. P. Logan and R. H. Edwards, The function of α-synuclein, Neuron, 2013, 79, 1044–1066 CrossRef CAS PubMed.
- T. Bartels, J. G. Choi and D. J. Selkoe, α-Synuclein occurs physiologically as a helically folded tetramer that resists aggregation, Nature, 2011, 477, 107–111 CrossRef CAS PubMed.
- C. Frieden, Protein aggregation processes: in search of the mechanism, Protein Sci., 2007, 16, 2334–2344 CrossRef CAS PubMed.
- L. Breydo, J. W. Wu and V. N. Uversky, α-Synuclein misfolding and Parkinson's disease, Biochim. Biophys. Acta, Mol. Basis Dis., 2012, 1822, 261–285 CrossRef CAS PubMed.
- G. Verma and R. Bhat, The Anthocyanidin Peonidin Interferes with an Early Step in the Fibrillation Pathway of α-Synuclein and Modulates It toward Amorphous Aggregates, ACS Chem. Neurosci., 2023, 14, 1424–1438 CAS.
- J. Wu, N. Österlund, H. Wang, R. Sternke-Hoffmann, H. Pupart, L. L. Ilag, A. Gräslund and J. Luo, Identifying the role of co-aggregation of Alzheimer’s amyloid-β with amorphous protein aggregates of non-amyloid proteins, Cell Rep. Phys. Sci., 2022, 3, 101028 CrossRef CAS.
- D. L. Abeyawardhane, R. D. Fernández, D. R. Heitger, M. K. Crozier, J. C. Wolver and H. R. Lucas, Copper Induced Radical Dimerization of α-Synuclein Requires Histidine, J. Am. Chem. Soc., 2018, 140, 17086–17094 CrossRef CAS PubMed.
- T. S. Ulmer, A. Bax, N. B. Cole and R. L. Nussbaum, Human micelle-bound alpha-synuclein (PDB: 1XQ8) DOI:10.2210/pdbixq8/pdb.
- T. S. Ulmer, A. Bax, N. B. Cole and R. L. Nussbaum, Structure and dynamics of micelle-bound human α-synuclein, J. Biol. Chem., 2005, 280, 9595–9603 CrossRef CAS PubMed.
- F. Duarte, P. Bauer, A. Barrozo, B. A. Amrein, M. Purg, J. Åqvist and S. C. L. Kamerlin, Force Field Independent Metal Parameters Using a Nonbonded Dummy Model, J. Phys. Chem. B, 2014, 118, 4351–4362 CrossRef CAS PubMed.
- Y. P. Pang, Successful molecular dynamics simulation of two zinc complexes bridged by a hydroxide in phosphotriesterase using the cationic dummy atom method, Proteins: Struct., Funct., Genet., 2001, 45, 183–189 CrossRef CAS PubMed.
- D. A. Case, R. M. Betz, D. S. Cerutti, T. E. Cheatham, T. A. Darden, R. E. Duke, T. J. Giese, H. Gohlke, A. W. Goetz, N. Homeyer, S. Izadi, P. Janowski, J. Kaus, A. Kovalenko, T. S. Lee, S. LeGrand, P. Li, C. Lin, T. Luchko, R. Luo, B. Madej, D. Mermelstein, K. M. Merz, G. Monard, H. Nguyen, H. T. Nguyen, I. Omelyan, A. Onufriev, D. R. Roe, A. Roitberg, C. Sagui, C. L. Simmerling, W. M. Botello-Smith, J. Swails, R. C. Walker, J. Wang, R. M. Wolf, X. Wu, L. Xiao and P. A. Kollman, AMBER 2016, San Francisco, 2016 Search PubMed.
- R. B. Best, W. Zheng and J. Mittal, Balanced protein-water interactions improve properties of disordered proteins and non-specific protein association, J. Chem. Theory Comput., 2014, 10, 5113–5124 CrossRef CAS PubMed.
- G. D. Hawkins, C. J. Cramer and D. G. Truhlar, Pairwise solute descreening of solute charges from a dielectric medium, Chem. Phys. Lett., 1995, 246, 122–129 CrossRef CAS.
- G. D. Hawkins, C. J. Cramer and D. G. Truhlar, Parametrized Models of Aqueous Free Energies of Solvation Based on Pairwise Descreening of Solute Atomic Charges from a Dielectric Medium, J. Phys. Chem., 1996, 100, 19824–19839 CrossRef CAS.
- A. Onufriev, D. Bashford and D. A. Case, Modification of the generalized born model suitable for macromolecules, J. Phys. Chem. B, 2000, 104, 3712–3720 CrossRef CAS.
- L. Savva and J. A. Platts, Evaluation of implicit solvent models in molecular dynamics simulation of α-Synuclein, J. Biomol. Struct. Dyn., 2022, 41, 1–16 CrossRef PubMed.
- J. A. Izaguirre, D. P. Catarello, J. M. Wozniak and R. D. Skeel, Langevin stabilization of molecular dynamics, J. Chem. Phys., 2001, 114, 2090–2098 CrossRef CAS.
- D. R. Roe and T. E. Cheatham, PTRAJ and CPPTRAJ: software for processing and analysis of molecular dynamics trajectory data, J. Chem. Theory Comput., 2013, 9, 3084–3095 CrossRef CAS PubMed.
- N. M. Glykos, Carma: a molecular dynamics analysis program, J. Comput. Chem., 2006, 27, 1765–1768 CrossRef CAS PubMed.
- A. S. Baltzis, P. I. Koukos and N. M. Glykos, Clustering of molecular dynamics trajectories via peak-picking in multidimensional PCA-derived distributions, arXiv, 2015, preprint, arXiv:1512.04024 DOI:10.48550/arXiv.1512.04024.
- J. Li, V. N. Uversky and A. L. Fink, Conformational Behavior of Human α-Synuclein is Modulated by Familial Parkinson's Disease Point Mutations A30P and A53T, Neurotoxicology, 2002, 23, 553–567 CrossRef CAS PubMed.
- L. R. Lemkau, G. Comellas, K. D. Kloepper, W. S. Woods, J. M. George and C. M. Rienstra, Mutant protein A30P α-synuclein adopts wild-type fibril structure, despite slower fibrillation kinetics, J. Biol. Chem., 2012, 287, 11526–11532 CrossRef CAS PubMed.
- A. Binolfi, L. Quintanar, C. W. Bertoncini, C. Griesinger and C. O. Fernández, Bioinorganic chemistry of copper coordination to alpha-synuclein: relevance to Parkinson's disease, Coord. Chem. Rev., 2012, 256, 2188–2201 CrossRef CAS.
- Y. Zhang, Y. Wang, Y. Liu, G. Wei, F. Ding and Y. Sun, Molecular Insights into the Misfolding and Dimerization Dynamics of the Full-Length α-Synuclein from Atomistic Discrete Molecular Dynamics Simulations, ACS Chem. Neurosci., 2022, 13, 3126–3137 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cp04697d |
| ‡ The 1.5 μs combined trajectories for the two systems can be viewed (in 50-frame increments) at: Cubound dimer: https://doi.org/10.6084/m9.figshare.23552151; metal-free dimer: https://doi.org/10.6084/m9.figshare.23552475. |
| This journal is © the Owner Societies 2024 |