Open Access Article
Open Access ArticleFormation of giant plasma membrane vesicles for biological and medical applications: a review
Yang
Li
,
Songyang
Liu
,
Wanyu
Xu
,
Kemin
Wang
 ,
Fengjiao
He
* and
Jianbo
Liu
,
Fengjiao
He
* and
Jianbo
Liu
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, College of Biology, Key Laboratory for Bionanotechnology and Molecular Engineering of Hunan Province, Hunan University, Changsha 410082, People's Republic of China. E-mail: fengjiaohe@hnu.edu.cn; liujianbo@hnu.edu.cn
First published on 2nd May 2023
Abstract
Giant plasma membrane vesicles (GPMVs) are micron-sized biomembrane vesicles that are isolated directly from living cells. They retain the lipid and protein complexity of the plasma membrane of the parent cell while lack the energy-mediated pathway of the cell; therefore, PMVs provide an alternative to vesicles constructed from synthetic or purified lipids as an experimental model system for use in a wide range of biomedical applications. In this review, we provide an overview of recent research on the formation of PMVs and on their unique structure and functional properties. We then discuss efforts to exploit them in biological and biomedical applications. For example, PMVs have been developed as cell mimics and plasma models to study the structure and composition of the plasma membrane and its interactions with other molecules, as drug carriers for targeted delivery and as probes for disease diagnostics. Finally, we provide some perspectives on the challenges and future developments of PMV research in biomimetic science and biomedical applications. GPMVs discussed in this review are restricted to those produced from mammalian cells.
1. Introduction
A cell membrane plays an indispensable and vital role in cells.1 Specifically, cells maintain their structural stability due to the rigidity of the cell membrane. In addition, the cell membrane not only acts as a barrier to separate the internal and external spaces of the cell but also plays an important role in cell-to-cell communication.2,3 The cell membrane is an inextricable topic in the mystery of life because the cell membrane is such a loyal guardian of the cell. Studies involving cells always have some degree of connection with the cell membrane. However, it is not easy to study the cell membrane in living cells directly because of its compositional and topographical complexity.4“Giant plasma membrane vesicles” (GPMVs) were first proposed in the 1970s by Scott, who provided a new technique for the isolation of plasma membranes.5 GPMVs are cell-derived vesicles that largely retain the compositional lipid and protein complexity of the parent cell plasma membrane but lack cortical actin cytoskeletons and intracellular processes.4 Thus, a substitute research object for living cells has been found. GPMVs are perfect cell mimics for studying the membrane organization and biophysical properties of the cell membrane without disturbances from intracellular dynamics. In addition, GPMVs are important biomimetic models for the study of the interactions between substances and plasma membranes and can be exploited as drug carriers for disease treatment.
GPMVs are emerging as potential and promising experimental tools, but related reviews are limited. Existing reviews have focused on the use of GPMVs for understanding plasma membrane structures and dynamics, while lack a more comprehensive overview of GPMVs.4 In this review, we provide an overview of GPMVs, which includes their formation methods, their unique structural and functional properties, and their biomedical applications (Fig. 1). We discuss efforts to exploit them as cell mimics, plasma membrane models and drug carriers for biomedical applications in particular. It's worth noting that the GPMVs discussed in this review are restricted to those produced from mammalian cells.
2. Formation of GPMVs and experimental factors
GPMVs are micrometre-scale spherical protrusions from the surface of parent cells (Fig. 2A). Protocols have been established to chemically induce the shedding of a substantial proportion of the plasma membrane in the form of GPMVs.6 Recently, a series of new simulation approaches have been reported. For example, Liu et al. observed plasma membrane vesiculation under light irradiation in their research and subsequently developed a nanomaterial-assisted strategy to generate GPMVs (Fig. 2B).7 This method has the advantage of involving no toxic chemical agents. A pulse laser irradiation strategy was found to induce the generation of GPMVs due to intracellular cavitation.8,9 A method of producing GPMVs using a hypertonic vesiculation buffer containing chloride salts was also described.10,11 This method can vesiculate Chinese hamster ovary cells.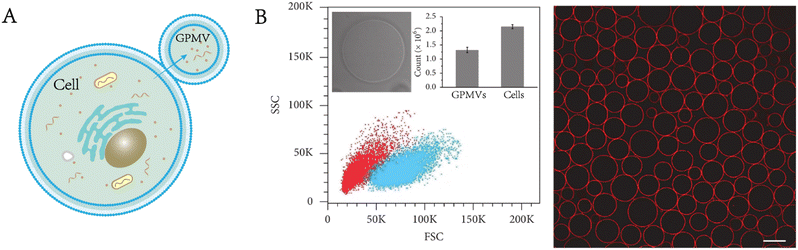 | ||
| Fig. 2 Formation and characterization of GPMVs. A) Diagram of GPMV formation. B) Flow cytometry analysis and morphological characterization of GPMVs acquired by the nanomaterial-assisted strategy. Scale bar, 20 μm (adapted with permission from ref. 7. Copyright 2019, AAAS). | ||
However, at present, the most widely used method of GPMV preparation is to expose cells to aldehydes and disulfide blocking agents (vesiculation agents). Vesiculation agent-mediated GPMV formation includes several different procedures. After removing the culture medium and washing, a buffer solution containing vesiculation (see below for specific components) agents is added to cells. The cells are cultured at 37 °C for several hours, during which GPMVs are automatically produced and shed into the supernatant. The production of GPMVs may be due to the combination of vesiculation agents with free amino or sulfhydryl groups in cells, weakening the binding between the cytoskeleton and plasma membrane, followed by expansion of the plasma membrane caused by intracellular pressure.5,12 To summarize the above methods of GPMV formation, we present Table 1 to show the details.
| Method | Vesiculation procedure | Ingredients | Size of GPMVs | Parent cells | Ref. |
|---|---|---|---|---|---|
| Nanomaterial-assisted strategy | Cells are incubated with carboxylfullerenes then irradiated with white light for 30 min | 0.5 mg mL−1 carboxylfullerenes | 5–22 μm | HeLa, HepG2, MCF-7, HEK-293, DC2.4, and CCRF-CEM cells | 7, 57 |
| Pulsed-laser irradiation strategy | Femtosecond laser pulses are used to stimulate GPMV formation on individual living cells | 3 nJ per pulse | 0.5–5 μm | KB, FaDu, A431, MCF-7, MCA207, jar, and Jeg-3, UMSCC-22A, Rat2, and mouse red blood cells | 8, 9 |
| 90 MHz | |||||
| 60 fs duration | |||||
| Osmotic buffer strategy | Osmotic buffer is added to cells and the cells are incubated for 13 h at 37 °C | 200 mM NaCl | 17.2 ± 4.4 μm | CHO cells | 10, 11 |
| 5 mM KCl | |||||
| 0.5 mM MgCl2 | |||||
| 0.75 mM CaCl2 | |||||
| 100 mM bicine | |||||
| pH = 8.5 | |||||
| Vesiculation agent-mediated strategy | A buffer solution containing vesiculation agents is added to cells. The cells are cultured at 37 °C for 2 h | 10 mM HEPES | 13.8 ± 3.7 μm | HEK-293 cells, HepG2 cells, CHO-K1 cells and HeLa cells | 15, 16, 20, 21, 38, 45, 63 |
| 150 mM NaCl | |||||
| 2 mM CaCl2 | |||||
| 2 mM dithiothreitol | |||||
| 25 mM polyformaldehyde | |||||
| pH = 7.4 |
A variety of mammalian cell types have been developed to produce GPMVs.6 However, adherent cells are often better choices since the generation of GPMVs involves the detachment of vesicles from cells and separation from parent cells. If suspended cells are used, this process is not easy to achieve. Many factors affect the preparation efficiency of GPMVs.
Vesiculation agents
Vesiculation agents play important roles in the formation of GPMVs. In addition to low concentrations of formaldehyde (or polyformaldehyde), other chemical agents can also induce vesiculation, such as N-ethylmaleimide, iodoacetate, cacodylate and p-chloromercuribenzoate.13 Dithiothreitol has been proven to be an effective auxiliary agent to promote vesiculation. Vesiculation agent concentrations will also affect the vesiculation efficiency of GPMVs, as proven by relevant studies.14 Other researchers have compared the difference in GPMVs when polyformaldehyde/dithiothreitol and calmidazolium are used as vesiculation agents.15 Different vesiculation agents produce different particle size distributions of GPMVs produced.Cations
The presence of cations is necessary for vesiculation, including both divalent cations and monovalent cations; calcium ions are the best divalent cations to promote vesiculation.13Temperature and pH
The influence of temperature and pH is very easy to understand. GPMVs are produced from parent cells, and factors affecting the conditions of parent cells will certainly affect the formation of GPMVs. Maximum vesiculation occurs at an optimum temperature of 37 °C and pH of 7–7.5. When the pH is above 8 or below 6, vesiculation is hardly observed.13Time
There has been much discussion about vesiculation time. The number and size of GPMVs increase with time in a certain range, but the optimal vesiculation time should be adjusted according to the specific research situation.Cell density
Cell density also affects the vesiculation of GPMVs, and the recommended cell density is 70%.63. Structural and functional properties of GPMVs
These prepared GPMVs are filled with cytoplasm associated with the parent cells but lack large organelles (e.g., nuclei, Golgi apparatus) and other intracellular structures. Their membrane is rich in lipids and proteins from the plasma membrane of the parent cells, and exactly mirrors the composition of the cells from which they originate while providing the experimental flexibility of synthetic vesicles.16,17 The proteomic analysis carried out by Bauer et al. identified 313 proteins from the GPMVs of NG108-15 cells.12 Their research results showed that 43 proteins were anchored on the membrane, of which 40 proteins were unique to the plasma membrane, whereas 221 proteins were from the cytoplasm.GPMVs have been used to evaluate the permeability of proteins, small molecules and drug-like molecules through the plasma membrane. It is noteworthy that the vesicles are not completely sealed. According to the research of Skinkle et al., GPMVs show passive permeability to hydrophobic molecules as large as 40 kDa, in contrast to synthesized unilamellar lipid vesicles, probably as a result of the rupture of GPMVs forming large and stable holes (Fig. 3A).17 GPMVs demonstrate cell-like mechanical properties. Dimova found that the mechanical properties of GPMVs correlate with lipid order, viscosity and cell density.18 When exposed to lipopolysaccharides, the membrane of GPMVs began to wrinkle unevenly and generated buds.19 The shape change of GPMVs is irreversible, in contrast to that of giant unilamellar vesicles.
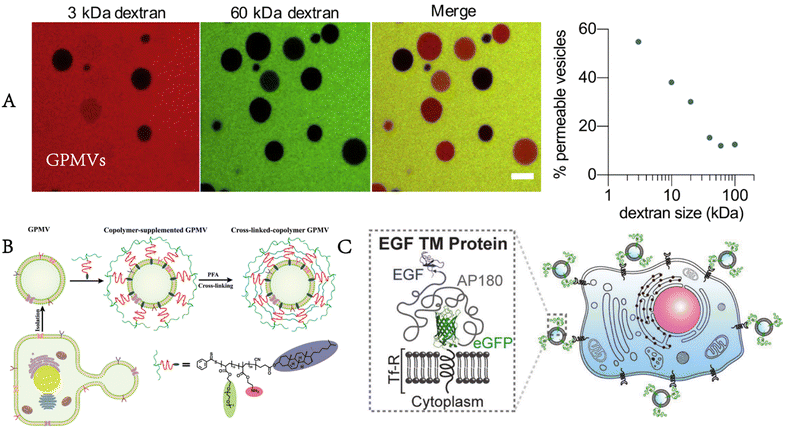 | ||
| Fig. 3 Properties and functionalities of GPMVs. A) GPMVs show passive permeability to dextran as large as 40 kDa (adapted with permission from ref. 17. Copyright 2020, Elsevier). B) After the preparation and separation of GPMVs, the copolymer is inserted into their membrane, and finally, a cross-linked polymer shell is formed around them to enhance their stability and functionality (reproduced with permission from ref. 20. Copyright 2022, Wiley). C) Targeted proteins are also present on the membrane of GPMVs produced by cells expressing targeted proteins (EGF). Adapted from Fig. 3 in ref. 17. Fig. 1 in ref. 20 and Fig. 1 in ref. 21. Scale bar, 10 μm (adapted with permission from ref. 21. Copyright 2016, Wiley). | ||
Some changes or enhancements can be made to GPMVs to make them more suitable for the expected applications. Palivan enhanced the mechanical properties and stability of GPMVs by modifying them with a specially designed diblock copolymer, cholesteryl-poly[2-aminoethyl methacrylate-b-poly(ethylene glycol) methyl ether acrylate].20 In addition, the pH responsiveness of the copolymer layer allows controlled cargo loading and release (Fig. 3B). This approach can be used for drug release in slightly acidic environments for cancer therapy. The transformation of GPMVs can also start with parent cells because GPMVs retain the membrane composition of the parent cells. When GPMVs are separated from parent cells expressing specific membrane proteins, these proteins are also present on the membrane of GPMVs (Fig. 3C).21
4. Biological and biomedical applications of GPMVs
4.1. GPMVs as cell mimics
In recent years, cell mimics have attracted increasing attention (terms such as “artificial cell”, “synthesized cell” and “protocell” are often mentioned). Scientists are trying to achieve the complexity of living cells by using cell mimics, although there is still a long way to go. Phospholipid vesicles,22 polymersomes,23 proteinosomes,24 colloidosomes,25 and coacervate droplets26 are widely used as cell mimics. For example, Chen et al. constructed multicompartment protocells for the spatial organization of enzymatic reactions using coacervate microdroplets.27 Zhang et al. designed a new cytomimetic model by integrating phospholipid membranes and coacervate microdroplets.28 Liu et al. utilized vasoactive erythrocyte membrane-enclosed coacervate protocells to produce nitric oxide for vasodilation by an enzymatic cascade reaction.29 However, there is still a large discrepancy between these mimics and living cells. As we mentioned before, GPMVs retain considerable aspects of the composition and structure of living cells, giving them innate advantages as cell mimics with appropriate modifications.Einfalt et al. designed GPMV-based artificial cells containing nanosized polymersomes as intracellular artificial organelles.16 On the one hand, GPMVs retain the components of the cytoplasm and plasma membrane of donor cells. On the other hand, the adjustable components of artificial organelles (enzymes, proteins, etc.) allow the controllability of intracellular reactions, thus realizing customizable cell mimics. The adjustable artificial organelles provide the possibility of diverse artificial cells, enabling more extensive applications in the field of cell mimics (Fig. 4A). Furthermore, researchers transferred polystyrene carboxylated nanoparticles into GPMVs, laying a foundation for further application of GPMVs in the field of cell mimics (Fig. 4B).30 However, it should be noted that although they have potential to be excellent models, GPMVs have not been studied extensively as cell mimics, leaving much room for further exploration. The reason for this may be that GPMVs retain the relative integrity of the membrane when they are prepared, requiring additional effort to achieve combination with other new components, which can be completed during the preparation process if synthesized phospholipid vesicles are used.
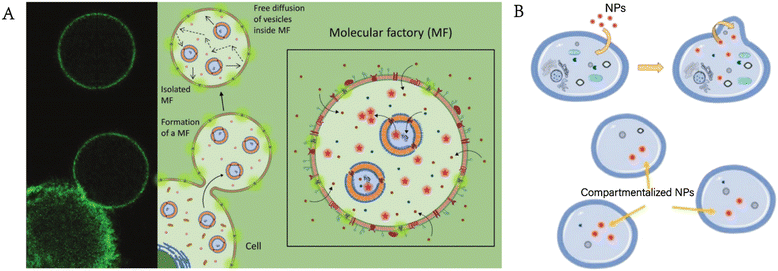 | ||
| Fig. 4 GPMVs as cell mimics. A) Cell mimics consisting of GPMVs produced from parent cells with internalized polymer artificial organelles also contain artificial organelles (reproduced with permission from ref. 16. Copyright 2020, Wiley). B) A similar method can be used to internalize nanoparticles into GPMVs (adapted with permission from ref. 30. Copyright 2021, American Chemical Society). | ||
Sedgwick et al. revealed the relationship between the production of GPMVs and the efflux of cholesterol from cells.31 Specifically, the molecules that promote the efflux of cholesterol can promote the formation of GPMVs, while those that inhibit the efflux of cholesterol hinder GPMV formation. Thus, the formation of GPMVs is a means to monitor the cholesterol content of cells to better understand the pathology of some diseases related to the abnormal accumulation of cholesterol in cells, such as Niemann–Pick type C and Alzheimer's disease.
4.2. GPMVs as plasma membrane models
Baumgart et al. proved in their research that the vast majority of GPMVs can form optically distinguishable domains when the temperature is below approximately 25 °C. They found that the lipid distribution in GPMVs was similar to that in model membrane systems with the coexistence of liquid-order and liquid-disorder fluid phases and imaged GPMVs to characterize the distribution of proteins in different membrane phases.33 For example, the transmembrane IgE receptor FcεRI was preferentially separated into a liquid-disordered-like phase, and the separation of other common membrane-related receptors was also reported. Here, GPMVs provided an effective method to characterize the heterogeneity of biomembranes. At the same time, this discovery verified the central principle of the membrane raft hypothesis, that is, the ability of eukaryotic membranes to form coexisting liquid domains, thus proving the rationality of the membrane raft hypothesis. In addition, nanodomains were observed at higher temperatures (physiological temperatures), and lipid replacement in the plasma membrane can regulate the tendency of the plasma membrane to form ordered domains.35
Levental et al. further clarified that the properties of the coexisting phases formed in GPMVs depended on the conditions for separating GPMVs, especially the vesiculation agents.36 Different separation conditions affected the phase separation temperature, the protein distribution among phases and the relative order between coexisting phases. The cholesterol-rich domain in the plasma membrane is considered to exclude many transmembrane proteins. GPMVs were used to quantitatively prove that the helical tetraspan peripheral myelin protein22 (PMP22) showed an obvious preference for the ordered membrane domain, which promoted the formation of the ordered membrane domains and stabilized the ordered membrane domains.37 Furthermore, Fricke et al. built a high-content platform that can be used to discover the chemical substances regulating plasma membrane rafts.38 The platform proved its reliability by verifying existing valve regulators, such as C6- and C8-ceramides, miltefosine, and epigallocatechin gallate (Fig. 5A).
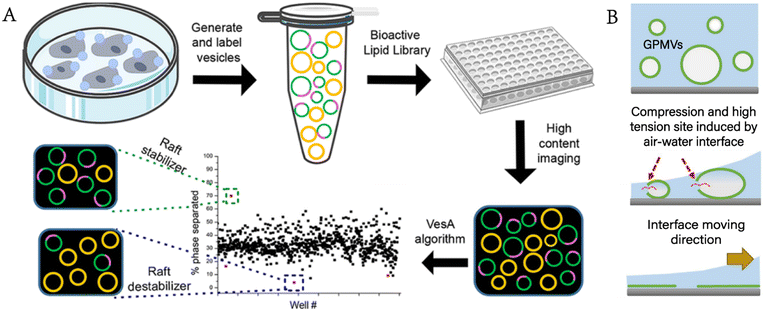 | ||
| Fig. 5 GPMVs as plasma membrane models for the study of the plasma membrane. A) Construction of a high-content imaging platform to discover chemical modulators of plasma membrane rafts (reproduced with permission from ref. 38. Copyright 2022, American Chemical Society). B) Rupturing GPMVs by air–water interface compression to form a microscale support membrane for plasma membrane protein study (adapted with permission from ref. 41. Copyright 2017, Springer). | ||
Moreover, Levental et al. used GPMVs to evaluate the plasma membrane structure and protein distribution.39 Cells realize their biological functions by regulating their shape and the lateral organization of membrane-related proteins, and the recruitment of proteins to specific areas of the membrane can promote regulation. Moreno-Pescador et al. used GPMVs derived from transfected cells to study the response of different types of membrane proteins to curvature changes and lipid phases. Nanoscale membrane curvature was generated by extracting nanotubes from these vesicles with optical traps.40 Chiang et al. developed a method for studying plasma membrane proteins using the air–water interface to rupture GPMVs and deposited them on a planar support to form a nanoscale supported membrane with natural plasma membrane proteins (Fig. 5B).41
GPMVs serve as tools to study the factors affecting membrane fusion.42 In the research of Kalyana et al., GPMVs were used to form cell membranes in a developed suspension lipid bilayer system to study the assembly of lipids and proteins.43 GPMVs are also used as sources of membrane and protein materials for building vesicle–nanotube networks. The vesicle–nanotube network system constructed by Bauer et al. requires obtaining membrane proteins and lipids from natural sources with high yield and functionality retention, making GPMVs become perfect precursors.44
 | ||
| Fig. 6 GPMVs as plasma membrane models for the study of interactions with the plasma membrane. A) GPMVs are used to study the interaction between amphiphilic CdTe@mPEG-SH nanoparticles and the plasma membrane. The nanoparticles enter the cell by the endocytic-escape pathway, thus filling the whole cell. B) Various characterization methods show that low-density lipoproteins can interact directly with the plasma membrane without receptor mediation (reproduced with permission from ref. 46. Copyright 2019, American Chemical Society). | ||
The interaction of the cell membrane and lipoprotein particles, whose task is to exchange cell membrane components and participate in cell lipid metabolism, maintaining lipid balance, is generally considered to be related to specific receptors on the cell membrane. The receptor-independent interaction pathway has been noted and confirmed in some studies through the interaction between lipoprotein particles and GPMVs, where lipoprotein particles can still transfer cargoes to the plasma membrane (Fig. 6B).46,47 These findings may help to better understand the biology of lipoprotein granules and to guide the treatment of dyslipidaemia diseases.
By adjusting the composition of the membrane, GPMVs can also be used to clarify the mechanism and behaviour of different kinds of cell-penetrating peptides in crossing the plasma membrane.48 For example, arginine-rich cell-penetrating peptides require nucleolin- and cholesterol-poor subdomains for translocation across membranes,49 glycosaminoglycans are required for the translocation of amphipathic cell-penetrating peptides across membranes,50 and the translocation of cell-penetrating peptides across the plasma membrane is controlled by cholesterol and the microenvironment created by membranous proteins.51
With insights into GPMVs, Steinkuhler et al. found that the interaction between lipid domains and membrane curvature produced a “superelastic” response, which represents the large deformation ability of the living cells' plasma membrane.52 Applying the curvature parameters that allow the superelastic behaviour, a vesicle analogue containing only lipids was designed through the simulation of GPMVs, with the denaturation ability three times that of traditional lipid vesicles, which made the bottom-up designed artificial cells more likely to approach the properties of living cells.
4.3. GPMVs as drug carriers
To date, liposomes,53,54 nanoparticles55,56 and exosomes57,58 have been widely studied and discussed as drug carriers. Compared with the above traditional drug carriers, the advantages of better biocompatibility, lower toxicity, accessible preparation and low immunogenicity imply that GPMVs also have potential as drug carriers.The research of Zemljic et al. demonstrated that calcein AM and dextran (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MW) can penetrate the membrane of GPMVs and remain in the membrane after washing, which is different from their behaviour in synthetic vesicles, and further, GPMVs can deliver dextran to cells grown in vitro.19 Their research shows that GPMVs are potentially useful as drug delivery systems. How to target is an important issue when using any material as a drug carrier. The conjugation of ligands to traditional drug carriers (as we mentioned before) is not easy to control, which often limits the consistency and complexity of ligands and thus limits the application of such drug carriers. This problem can be solved by extracting GPMVs from donor cells expressing engineered transmembrane targeting proteins because GPMVs retain the plasma membrane composition of their donor cells, as confirmed in the research of Zhao et al.21 This is undoubtedly another advantage of GPMVs as drug carriers.
000 MW) can penetrate the membrane of GPMVs and remain in the membrane after washing, which is different from their behaviour in synthetic vesicles, and further, GPMVs can deliver dextran to cells grown in vitro.19 Their research shows that GPMVs are potentially useful as drug delivery systems. How to target is an important issue when using any material as a drug carrier. The conjugation of ligands to traditional drug carriers (as we mentioned before) is not easy to control, which often limits the consistency and complexity of ligands and thus limits the application of such drug carriers. This problem can be solved by extracting GPMVs from donor cells expressing engineered transmembrane targeting proteins because GPMVs retain the plasma membrane composition of their donor cells, as confirmed in the research of Zhao et al.21 This is undoubtedly another advantage of GPMVs as drug carriers.
Luo et al. encapsulated photosensitizers and photothermal therapeutic agents in GPMVs modified with cholesterol-labelled aptamers on the surface for targeted drug delivery and synergistic photodynamic/photothermal therapy.59 GPMVs release therapeutic drugs by membrane fusion-mediated therapeutic transportation to kill targeted cancer cells with no obvious toxicity to normal cells (Fig. 7A). Nanoparticle or liposome drug carriers enter cells through endocytosis and are often trapped in the endosomal lumen.60,61 Ineffective endoplasmic escape limits the efficiency of cytoplasmic delivery and increases the concentration of drugs needed to kill cancer cells.62 When two neighbouring cells meet, they can exchange cargoes (metabolites, second messengers, peptides and siRNA) through the gap junctions on their membrane, which provides a way to avoid endocytosis. Gadok et al. extracted GPMVs from donor cells that overexpressed gap junctions and developed the so-called connectosomes for directly delivering drugs to the cytoplasm of cancer cells without endocytosis.63 The results showed that compared with either free or liposomal doxorubicin, this method reduced the therapeutic effective dose (LD50) by more than one order of magnitude. The drugs entered the connectosomes by the pretreatment of donor cells with a drug-containing medium (Fig. 7B).
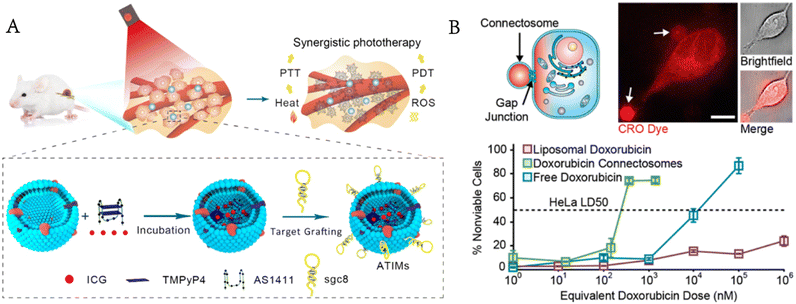 | ||
| Fig. 7 GPMVs for drug delivery. A) Photosensitizers and photothermal agents are simultaneously encapsulated in GPMVs for synergistic photodynamic/photothermal therapy of cancer (reproduced with permission from ref. 59. Copyright 2019, American Chemical Society). B) GPMVs act as connectosomes to deliver drugs to cancer cells (reproduced with permission from ref. 63. Copyright 2016, American Chemical Society). | ||
5. Challenges and prospects
This review has summarized the formation methods, properties, and biomedical applications of GPMVs. We emphatically discuss the biological and medical applications of GPMVs as cell mimics, plasma membrane models and drug carriers. Currently, the most widely used method to produce GPMVs is the vesiculation agent-mediated strategy, which can produce a large number of GPMVs in a matter of hours, but also causes the cross-linking of the plasma membrane proteins and destroys the asymmetry of the membrane, interfering with the study of the plasma membrane.4,6 Compared with synthetic phospholipid vesicles, GPMVs show better stability and biocompatibility. Compared with extracellular vesicles or cell membrane-derived vesicles, GPMVs require no complex procedures for preparation or isolation, making GPMVs promising drug carriers. Besides, the ability to retain proteins on the parent cell membrane makes GPMVs worth exploring as targeted drug carriers. But there are also some problems that need to be solved, such as leakage when loading small molecules.20 Programmed methods for loading cargoes also need to be developed.The potential of GPMVs as cell mimics has not yet been fully explored, probably as a result of difficulty of loading biomacromolecules into GPMVs. Cell communication, one of whose ways is with the help of small molecules, is one of the key focuses in the field of cell mimicking. In previous studies, giant monolayer vesicles have been used more frequently for biomimetic cell communication.64 GPMVs are well worth exploring as their alternatives. There have been a lot of research studies on GPMVs as plasma membrane models. But an underappreciated fact is that GPMVs can also be used as sources of plasma membranes; after all, GPMV vesiculation was originally proposed as a method of plasma membrane isolation. For example, plasma membrane vesicles are widely used for the coating of nanoparticles, where the plasma membranes are obtained through a series of procedures starting with cell lysis.65,66 GPMVs, which exclude structures such as large organelles of cells, can be used as an alternative method of obtaining plasma membranes.
In short, GPMVs are experimental tools with unique, even irreplaceable, advantages in certain fields. The ability of GPMVs to display a diverse range of unique structural and functional properties will enable the development of these biomimetic systems for a broad range of biological and biomedical applications.
Author contributions
Y. Li wrote the draft of the manuscript. F. He and J. Liu proposed the conception and critically reviewed the manuscript. All the authors thoroughly read the final manuscript draft and gave permission for its submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the National Natural Science Foundation of China (21190044, 22177032, and 32101082) and the Natural Science Foundation in Hunan Province (2022RC3047, 2021JJ10013, and 2022JJ40037) for financial support.References
- D. Casares, P. V. Escriba and C. A. Rossello, Int. J. Mol. Sci., 2019, 20, 2167 CrossRef CAS PubMed.
- E. Sackmann, Can. J. Phys., 1990, 68, 999–1012 CrossRef CAS.
- P. V. Escriba, Biochim. Biophys. Acta, Biomembr., 2017, 1859, 1493–1506 CrossRef CAS PubMed.
- E. Sezgin, Biochim. Biophys. Acta, Biomembr., 2022, 1864, 183857 CrossRef CAS PubMed.
- R. E. Scott, Science, 1976, 194, 743–745 CrossRef CAS PubMed.
- E. Sezgin, H. J. Kaiser, T. Baumgart, P. Schwille, K. Simons and I. Levental, Nat. Protoc., 2012, 7, 1042–1051 CrossRef CAS PubMed.
- Q. Liu, C. Bi, J. Li, X. Liu, R. Peng, C. Jin, Y. Sun, Y. Lyu, H. Liu, H. Wang, C. Luo and W. Tan, Research, 2019, 2019, 6523970 CAS.
- C. V. Kelly, M. M. Kober, P. Kinnunen, D. A. Reis, B. G. Orr and M. M. Banaszak Holl, J. Biol. Phys., 2009, 35, 279–295 CrossRef CAS PubMed.
- J. Y. Tinevez, U. Schulze, G. Salbreux, J. Roensch, J. F. Joanny and E. Paluch, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 18581–18586 CrossRef CAS PubMed.
- N. Del Piccolo, J. Placone, L. He, S. C. Agudelo and K. Hristova, Anal. Chem., 2012, 84, 8650–8655 CrossRef CAS PubMed.
- S. Sarabipour, N. D. Piccolo and K. Hristova, Acc. Chem. Res., 2015, 48, 2262–2269 CrossRef CAS PubMed.
- B. Bauer, M. Davidson and O. Orwar, Angew. Chem., Int. Ed., 2009, 48, 1656–1659 CrossRef CAS PubMed.
- R. E. Scott and P. B. Maercklein, J. Cell Sci., 1979, 35, 245–252 CrossRef CAS PubMed.
- Z. Gerstle, R. Desai and S. L. Veatch, Methods Enzymol., 2018, 603, 129–150 CAS.
- N. K. Teiwes, I. Mey, P. C. Baumann, L. Strieker, U. Unkelbach and C. Steinem, ACS Appl. Mater. Interfaces, 2021, 13, 25805–25812 CrossRef CAS PubMed.
- T. Einfalt, M. Garni, D. Witzigmann, S. Sieber, N. Baltisberger, J. Huwyler, W. Meier and C. G. Palivan, Adv. Sci., 2020, 7, 1901923 CrossRef CAS PubMed.
- A. D. Skinkle, K. R. Levental and I. Levental, Biophys. J., 2020, 118, 1292–1300 CrossRef CAS PubMed.
- J. Steinkuhler, E. Sezgin, I. Urbancic, C. Eggeling and R. Dimova, Commun. Biol., 2019, 2, 337 CrossRef PubMed.
- S. Zemljic Jokhadar, U. Klancnik, M. Grundner, T. Svelc Kebe, S. Vrhovec Hartman, M. Liovic and J. Derganc, BMC Biophys., 2018, 11, 1 CrossRef PubMed.
- X. Huang, D. Hurlimann, H. T. Spanke, D. Wu, M. Skowicki, I. A. Dinu, E. R. Dufresne and C. G. Palivan, Adv. Healthcare Mater., 2022, 11, e2202100 CrossRef PubMed.
- C. Zhao, D. J. Busch, C. P. Vershel and J. C. Stachowiak, Small, 2016, 12, 3837–3848 CrossRef CAS PubMed.
- A. Chen and P. Walde, Cold Spring Harbor Perspect. Biol., 2010, 2, a002170 Search PubMed.
- F. Mason and P. Thordarson, J. Appl. Polym. Sci., 2017, 55, 3817–3825 Search PubMed.
- X. Huang, A. J. Patil, M. Li and S. Mann, J. Am. Chem. Soc., 2014, 136, 9225–9234 CrossRef CAS PubMed.
- M. Li, R. L. Harbron, J. V. Weaver, B. P. Binks and S. Mann, Nat. Chem., 2013, 5, 529–536 CrossRef CAS PubMed.
- S. Koga, D. S. Williams, A. W. Perriman and S. Mann, Nat. Chem., 2011, 3, 720–724 CrossRef CAS PubMed.
- Y. Chen, M. Yuan, Y. Zhang, S. Liu, X. Yang, K. Wang and J. Liu, Chem. Sci., 2020, 11, 8617–8625 RSC.
- Y. Zhang, Y. Chen, X. Yang, X. He, M. Li, S. Liu, K. Wang, J. Liu and S. Mann, J. Am. Chem. Soc., 2021, 143, 2866–2874 CrossRef CAS PubMed.
- S. Liu, Y. Zhang, M. Li, L. Xiong, Z. Zhang, X. Yang, X. He, K. Wang, J. Liu and S. Mann, Nat. Chem., 2020, 12, 1165–1173 CrossRef CAS PubMed.
- L. Zartner, M. Garni, I. Craciun, T. Einfalt and C. G. Palivan, Biomacromolecules, 2021, 22, 106–115 CrossRef CAS PubMed.
- A. Sedgwick, M. Olivia Balmert and C. D'Souza-Schorey, Exp. Cell Res., 2018, 365, 194–207 CrossRef CAS PubMed.
- K. Simons and E. Ikonen, Nature, 1997, 387, 569–572 CrossRef CAS PubMed.
- T. Baumgart, A. T. Hammond, P. Sengupta, S. T. Hess, D. A. Holowka, B. A. Baird and W. W. Webb, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 3165–3170 CrossRef CAS PubMed.
- G. I. Mashanov, T. A. Nenasheva, A. Mashanova, R. Lape, N. J. M. Birdsall, L. Sivilotti and J. E. Molloy, Faraday Discuss., 2021, 232, 358–374 RSC.
- G. Li, Q. Wang, S. Kakuda and E. London, J. Lipid Res., 2020, 61, 758–766 CrossRef CAS PubMed.
- I. Levental, M. Grzybek and K. Simons, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 11411–11416 CrossRef CAS PubMed.
- T. Marinko, A. K. Kenworthy and C. R. Sanders, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 14168–14177 CrossRef PubMed.
- N. Fricke, K. Raghunathan, A. Tiwari, K. M. Stefanski, M. Balakrishnan, A. G. Waterson, R. Capone, H. Huang, C. R. Sanders, J. A. Bauer and A. K. Kenworthy, ACS Cent. Sci., 2022, 8, 370–378 CrossRef CAS PubMed.
- R. Levental and I. Levental, Methods Mol. Biol., 2015, 1232, 65–77 CrossRef PubMed.
- G. Moreno-Pescador, C. D. Florentsen, H. Ostbye, S. L. Sonder, T. L. Boye, E. L. Veje, A. K. Sonne, S. Semsey, J. Nylandsted, R. Daniels and P. M. Bendix, ACS Nano, 2019, 13, 6689–6701 CrossRef CAS PubMed.
- P. C. Chiang, K. Tanady, L. T. Huang and L. Chao, Sci. Rep., 2017, 7, 15139 CrossRef PubMed.
- H. Xu, M. Cai, J. Gao, Y. Shi, J. Chen, Q. Wu, J. Zhang, J. Jiang and H. Wang, Talanta, 2021, 226, 122091 CrossRef CAS PubMed.
- R. V. Kalyana Sundaram, M. Bera, J. Coleman, J. S. Weerakkody, S. S. Krishnakumar and S. Ramakrishnan, Small, 2022, 18, e2205567 CrossRef PubMed.
- B. Bauer, M. Davidson and O. Orwar, Langmuir, 2006, 22, 9329–9332 CrossRef CAS PubMed.
- A. Dubavik, E. Sezgin, V. Lesnyak, N. Gaponik, P. Schwille and A. Eychmuller, ACS Nano, 2012, 6, 2150–2156 CrossRef CAS PubMed.
- M. Axmann, E. Sezgin, A. Karner, J. Novacek, M. D. Brodesser, C. Rohrl, J. Preiner, H. Stangl and B. Plochberger, Nano Lett., 2019, 19, 2562–2567 CrossRef CAS PubMed.
- A. Plochberger, T. Sych, F. Weber, J. Novacek, M. Axmann, H. Stangl and E. Sezgin, Biochemistry, 2020, 59, 4421–4428 CrossRef PubMed.
- P. Saalik, A. Niinep, J. Pae, M. Hansen, D. Lubenets, U. Langel and M. Pooga, J. Controlled Release, 2011, 153, 117–125 CrossRef PubMed.
- A. Lorents, P. Saalik, U. Langel and M. Pooga, Bioconjugate Chem., 2018, 29, 1168–1177 CrossRef CAS PubMed.
- J. Pae, L. Liivamagi, D. Lubenets, P. Arukuusk, U. Langel and M. Pooga, Biochim. Biophys. Acta, 2016, 1858, 1860–1867 CrossRef CAS PubMed.
- J. Pae, P. Saalik, L. Liivamagi, D. Lubenets, P. Arukuusk, U. Langel and M. Pooga, J. Controlled Release, 2014, 192, 103–113 CrossRef CAS PubMed.
- J. Steinkuhler, P. Fonda, T. Bhatia, Z. Zhao, F. S. C. Leomil, R. Lipowsky and R. Dimova, Adv. Sci., 2021, 8, e2102109 CrossRef PubMed.
- T. Li, D. Cipolla, T. Rades and B. J. Boyd, J. Controlled Release, 2018, 288, 96–110 CrossRef CAS PubMed.
- T. M. Allen and P. R. Cullis, Adv. Drug Delivery Rev., 2013, 65, 36–48 CrossRef CAS PubMed.
- Y. Liu, G. Yang, Y. Hui, S. Ranaweera and C. X. Zhao, Small, 2022, 18, e2106580 CrossRef PubMed.
- J. Mitchell, M. M. Billingsley, R. M. Haley, M. E. Wechsler, N. A. Peppas and R. Langer, Nat. Rev. Drug Discovery, 2021, 20, 101–124 CrossRef PubMed.
- X. Zhao, D. Wu, X. Ma, J. Wang, W. Hou and W. Zhang, Biomed. Pharmacother., 2020, 128, 110237 CrossRef CAS PubMed.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed.
- C. Luo, X. Hu, R. Peng, H. Huang, Q. Liu and W. Tan, ACS Appl. Mater. Interfaces, 2019, 11, 43811–43819 CrossRef CAS PubMed.
- G. Sahay, D. Y. Alakhova and A. V. Kabanov, J. Controlled Release, 2010, 145, 182–195 CrossRef CAS PubMed.
- D. Thevenin, M. An and D. M. Engelman, Chem. Biol., 2009, 16, 754–762 CrossRef CAS PubMed.
- M. Bareford and P. W. Swaan, Adv. Drug Delivery Rev., 2007, 59, 748–758 CrossRef PubMed.
- A. K. Gadok, D. J. Busch, S. Ferrati, B. Li, H. D. Smyth and J. C. Stachowiak, J. Am. Chem. Soc., 2016, 138, 12833–12840 CrossRef CAS PubMed.
- H. Chen, W. Xu, H. Shi, Y. Qiao, X. He, J. Zheng, S. Zhou, X. Yang, K. Wang and J. Liu, Angew. Chem., Int. Ed., 2023, 62, e202301559 CrossRef PubMed.
- Q. V. Le, J. Lee, G. Shim and Y. K. Oh, Acta Pharm. Sin. B, 2021, 11, 2096–2113 CrossRef CAS PubMed.
- S. Zhou, B. Wang, C. Wang, Q. Wang and L. Zhang, Nanomedicine, 2020, 15, 625–641 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2023 |