Open Access Article
Open Access ArticleArtificial tongue based on carbon dots and a porphyrin derivative for pattern recognition of metal ions†
Xiaowei
Liu
a,
Zhiwei
Zhang
a,
Xuetao
Yan
b,
Xinying
Zhang
a,
Francois
Amblard
c,
Yingying
Chen
 *a and
Lingyan
Feng
*a and
Lingyan
Feng
 *a
*a
aMaterials Genome Institute, Shanghai University, 333 Nanchen Road, Shanghai 200444, China. E-mail: chenyy08@t.shu.edu.cn; lingyan.feng@t.shu.edu.cn
bCollege of Qianweichang, Shanghai University, 99 Shangda Road, Shanghai 200444, China
cDepartment of Physics and Life Sciences, Ulsan National Institute of Science and Technology, South Korea
First published on 22nd November 2022
Abstract
An artificial tongue for the discrimination of different metal ions based on fluorescent carbon dots (C-dots) and meso-tetrakis(4-(N-methylpyridiumyl))porphyrin (TMPyP) was developed in this manuscript. Fluorescence resonance energy transfer (FRET) between C-dots and TMPyP was applied for the sensor array consisting of C-dots, TMPyP and a C-dot–TMPyP complex. Fifteen metal ions have been effectively identified by the sensor array with different fingerprint-like patterns which were analyzed from the distinct fluorescence responses through principal component analysis (PCA) and hierarchical cluster analysis (HCA), providing a fast and efficient method for the detection of metal ions.
Introduction
Inspired by the mammalian taste and olfactory system, an “artificial tongue” based on sensor arrays has been employed to detect analytes such as proteins,1,2 bacteria3 and metal ions4,5 by generating a unique fingerprint-like pattern. Different from the traditional “lock and key” principle, the sensing array in this strategy needs not to be highly specific to the analytes. Varieties of analytes are identified by a distinct fingerprint-like pattern assigned to each analyte through a plurality of cross reaction sensing elements. Compared to other methods in detecting metal ions, the sensor array provides the advantages of simple design, convenient operation and high efficiency.Fluorescence resonance energy transfer (FRET) is a nonradiative process whereby an excited state donor transfers energy to a proximal ground state acceptor, and the acceptor must absorb the energy at the emission wavelength of the donor.6 Fluorescent materials such as organic dyes,7 metal chelates,8 and nanoparticles9,10 have already been employed for FRET energy donors. Recently, fluorescent carbon dots (C-dots) have attracted intensive attention in FRET because of their high photostability, excellent biocompatibility and facile preparation.11–13 Pang and collaborators proposed a novel thrombin aptamer biosensor based on FRET from polyacrylic acid (PAA) functionalized up-conversion fluorescent molecules (UCPs) to C-dots, to monitor the human plasma thrombin level.14 Zheng et al. have developed a FRET drug delivery platform between C-dots and fluorescent drug molecules to enhance drug delivery and facilitate cell imaging and real-time monitoring of drug release.15 It has potential to fabricate sensors based on FRET between C-dots and other acceptors.
meso-Tetrakis(4-(N-methylpyridiumyl))porphyrin (TMPyP), a cationic porphyrin derivative, exhibits strong fluorescence and is widely used as an optical indicator ligand. Since TMPyP is a positive molecule, it can easily combine with negative substances on the surface. In this work, we constructed a FRET system between C-dots and TMPyP (in which C-dots are donors and TMPyP is an acceptor), and further utilized the system to construct a novel array-based sensor for metal ion detection. As shown in Scheme 1, the C-dots, TMPyP and C-Dot–TMPyP complex are designed as a sensor array; when different metal ion solutions are added to the C-dots and TMPyP solutions separately, the fluorescence hardly changes, while the fluorescence changes a lot as certain metal ions are added to the C-dot–TMPyP solution. With the increase of metal ion concentration, C-dots and TMPyP molecules are surrounded by metal ions, and the compounds are in the critical range of energy migration. The process of FRET is completely limited owing to the excited state energy transfer from TMPyP to metal ions, which affects the intrinsic fluorescence emission of TMPyP. Therefore, when different metal ions are added to the sensor array, they interact to a greater or lesser extent with the surface groups of the free C-dots, TMPyP and C-dot–TMPyP complex. The distinct responses to different metal ions of the sensor array result in a unique fingerprint-like pattern to discriminate them from each other.
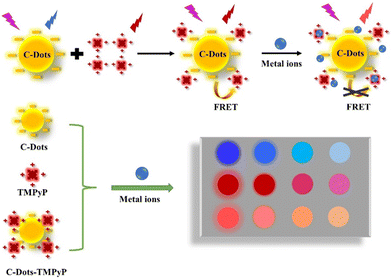 | ||
| Scheme 1 Schematic illustration of the array-based artificial tongue for discrimination of metal ions. | ||
Experimental
Reagents and materials
All the reagents were purchased from formal commercial suppliers and were used without any further purification. L-Cysteine (L-Cys) and meso-tetrakis(4-(N-methylpyridiumyl))porphyrin (TMPyP) were purchased from Sigma-Aldrich (Shanghai, China). Na2HPO4, NaH2PO4, KCl, MgCl2, NaCl, MnCl2, BaCl2, Ni(NO3)2, CuCl2, Al(NO3)3, CaCl2, FeCl2, ZnCl2, FeCl3, Cd(NO3)2, and PbCl2 were bought from Aladdin Chemistry Co. Ltd. (Shanghai, China). NaOH was obtained from Sinopharm Chemical Reagent Company (Shanghai, China).Characterization and instrument
Transmission electron microscopy (TEM) images were obtained on an FEI/Philips Tecnai F20 (200 kV) transmission electron microscope. Dynamic light scattering and zeta potential measurement were performed using a Litesizer 500 instrument. X-ray photoelectron spectroscopy (XPS) images were recorded on an Axis Ultra instrument. Fourier transform infrared (FT-IR) spectra were obtained on a Nicolet iS50 spectrometer. UV-vis absorption spectra were acquired on a Perkin-Elmer Lambda 750 UV-vis spectrometer. The fluorescence spectra were measured on an FS5 fluorescence spectrometer.Preparation of C-dots
The C-dots were synthesized by a hydrothermal method according to Kang et al.16 0.6 g L-Cys and 1.0 g NaOH were fully dissolved in 8.0 mL water under ultrasonication for 10 minutes. The above solution was transferred to a 25 mL Teflon-sealed autoclave and heated at 120 °C for 16 h to obtain a brown sample. The crude solution was dialyzed through a 1 kDa cellulose dialysis membrane for 72 hours to remove impurities. The aqueous solution was then lyophilized to gain a brown solid.17Preparation of C-dot–TMPyP
Porphyrin reserve solution was prepared by dissolving porphyrin solid in phosphate buffer (PB) (1 mM Na2HPO4 and 1 mM NaH2PO4, pH = 7.4). A Perkin-Elmer Lambda 750 UV-vis spectrophotometer was used (ε = 2.66 × 10−5 M−1 cm−1 at λ = 422 nm) to prepare the solution of TMPyP with a concentration of 100 μM based on Lambert–Beer Law (A = εcl). The C-dot–TMPyP solution was prepared by incubating TMPyP (10 μM, 1.8 mL) with carbon dots (50 μg mL−1, 200 μL) at pH = 7.4 for 24 h at room temperature (25 °C). Note that light should be avoided for all the steps.Cytotoxicity assay
The cytotoxicity was evaluated with HeLa cells by MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] assay according to Feng et al.18 The cells were cultured in 25 cm2 flasks for two days at 37 °C in a humidified atmosphere of 5% CO2 per 95% air, then attached to a 96-well plate and incubated with C-dots of different concentrations (0, 20, 40, 80, 100, 200, 300, 400 μg mL−1) for 24 hours. The viabilities of the HeLa cells were analyzed by a standard MTT assay.Preparation of the sensor array
Free C-dots, TMPyP and C-dot–TMPyP were constructed as a fluorescent sensor array. The first sensor was prepared by adding 200 μL C-dots (50 μg ml−1) into 1.8 mL aqueous solution at room temperature. The other two sensors, the TMPyP solution (10 μM) and C-dot–TMPyP complex solution, were prepared as mentioned before. Fifteen metal ions (Al3+, Ba2+, Ca2+, Cd2+, Co2+, Cu2+, Fe2+, Fe3+, K+, Mg2+, Mn2+, Na+, Ni2+, Pb2+, Zn2+) were added to the sensor array at the same concentration (16 μM). The fluorescence responses of the array were measured using a fluorescence spectrometer at emission wavelengths of 402 nm, 715 nm and 717 nm, respectively, and the corresponding excitation wavelengths were 320 nm, 420 nm and 320 nm.Statistical analysis methods
The fingerprint-like patterns were processed by principal component analysis (PCA) in SPSS Statistics (Version 21.0) software. PCA is a common statistical analysis method to convert a group of variables into some linearly unrelated components through orthogonal transformation.19 The fingerprint patterns of different specimens were exhibited with 2D plots by PCA mapping. Hierarchical cluster analysis (HCA) is a statistical classification method based on the relative distance between all data pairs in the full vector space, providing a straightforward way to analyze similar responses to different analytes. The Euclidean distance, which refers to the real distance between two points or the natural length of the vector in an n-dimensional space, is chosen to describe the similarity among distinct metal ions in our strategy.Results and discussion
Morphological and spectral characterization of C-dots
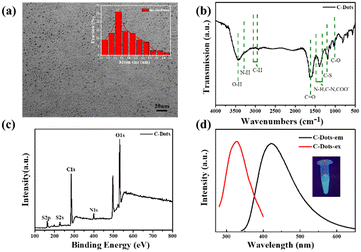
The C-dots were synthesized from L-Cys and NaOH by a hydrothermal method. Fig. 1a displays the typical TEM image of C-dots, which demonstrated that the as-prepared C-dots were well dispersed with a uniform size of about 1–3 nm (the average particle size is 1.7 nm according to dynamic light scattering). The functional groups on the surface of the C-dots were analyzed by FT-IR spectroscopy as shown in Fig. 1b. The intense peak at 3475 cm−1 and the small hump at 3218 cm−1 were caused by O–H and N–H vibration separately, which implied that there were lots of residual hydroxyl and amino groups on the surface of the C-dots. The peak value at 2950–3050 cm−1 was assigned to the C–H group. The two peaks centered at 1650 cm−1 and 1050 cm−1 correspond to the stretching vibration of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O. The peak positions of C–N, COO–, and N–H bonds were at 1320–1480 cm−1. These groups on the surface of the C-dots improved the hydrophilicity and stability in aqueous solution, which were confirmed in the TEM images. Fig. 1c depicts the full-scan XPS of C-dots, where S (2p), C (1s), N (1s) and O (1s) correspond to the four peaks located at 168.2, 286.85, 400.12 and 532.83 eV, respectively. The high-resolution scan expanded images are shown in Fig. S1† to further prove the chemical structure of the C-dots. The optimal excitation and emission wavelengths of the C-dots were located at 320 nm and 420 nm separately, and the inset photograph in Fig. 1d is the optical image of the C-dot aqueous solution under UV light at 365 nm, which indicated the excellent optical properties of the synthetic C-dots. The UV-vis absorption spectrum of C-dots (Fig. 2a) had an obvious peak centered at 260 nm, which was consistent with the previously reported carbon dots and attributed to the electron transitions from π–π* of C
O and C–O. The peak positions of C–N, COO–, and N–H bonds were at 1320–1480 cm−1. These groups on the surface of the C-dots improved the hydrophilicity and stability in aqueous solution, which were confirmed in the TEM images. Fig. 1c depicts the full-scan XPS of C-dots, where S (2p), C (1s), N (1s) and O (1s) correspond to the four peaks located at 168.2, 286.85, 400.12 and 532.83 eV, respectively. The high-resolution scan expanded images are shown in Fig. S1† to further prove the chemical structure of the C-dots. The optimal excitation and emission wavelengths of the C-dots were located at 320 nm and 420 nm separately, and the inset photograph in Fig. 1d is the optical image of the C-dot aqueous solution under UV light at 365 nm, which indicated the excellent optical properties of the synthetic C-dots. The UV-vis absorption spectrum of C-dots (Fig. 2a) had an obvious peak centered at 260 nm, which was consistent with the previously reported carbon dots and attributed to the electron transitions from π–π* of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C of the sp2-hybridized carbon network.
C of the sp2-hybridized carbon network.
The MTT method was employed to assess the cytotoxicity and biocompatibility of C-dots for potential application in intercellular detection. Fig. S2† suggests that more than 90% cell viability was observed after 24 hours of incubation with HeLa cells and C-dots (from 0 μg mL−1 to 400 μg mL−1), demonstrating a good biocompatibility of the C-dots with living cells.
Fluorescence resonance energy transfer between C-dots and TMPyP
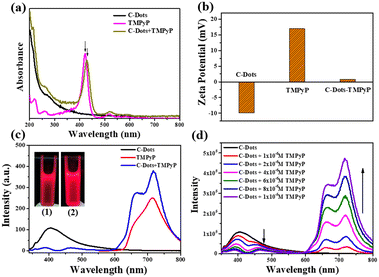
When the emission spectra of a receptor overlap the energy of a donor, it is possible to form a good donor–receptor pair. As a cationic porphyrin derivative, TMPyP exhibits prominent fluorescence properties with the excitation wavelength at 420 nm, and is normally applied as an optical indicator ligand. Since TMPyP is a positive molecule, it is feasible to combine it with negative C-dots on the surface.As observed in Fig. 2a, the UV-vis spectrum of TMPyP solution had a strong Soret band at 420 nm with four weak Q-band absorption peaks. After being mixed with the C-dot solution, the intensity of TMPyP at 420 nm decreased lightly. The Soret band had a red shift of 15 nm and a wider half bandwidth compared with free TMPyP, which was attributed to the electron transfer between the two delocalized π electronic systems of C-dots and TMPyP.20 When the C-dots solutions were titrated with TMPyP (0–10 μM) of different concentrations, the intensity of the Soret band at 435 nm increased gradually with the increase of the concentration (Fig. S3†). The net charge of the C-dot–TMPyP complex was further verified by zeta potentials in Fig. 2b. Due to the carboxyl and hydroxyl functional groups on the surface of the C-dots, the ξ-potential was −9.9 mV. TMPyP is a positive porphyrin, where the ξ-potential was +16.9 mV. It was feasible to form a C-dot–TMPyP complex through electrostatic interactions, not to mention a strong π–π stacking. Fig. S4† reveals that the emission spectrum of C-dots overlapped the absorption spectrum of TMPyP to some extent. It was easy to employ C-dots as energy donors and TMPyP as a fluorescence acceptor. Under the irradiation of a 365 nm UV lamp, the fluorescence intensity of C-dot–TMPyP seemed stronger than that of TMPyP (Fig. 2c inset), which was further confirmed in the fluorescence spectra (Fig. 2c). The emission intensity at 420 nm in the C-dot–TMPyP conjugated system was significantly lower than that in C-dots. In contrast, the emission intensity at 600–800 nm of C-dot–TMPyP was increased relative to free TMPyP, demonstrating that the FRET phenomenon occurred. According to the FRET mechanism, C-dots absorbed the energy from incident light, and the generated excited state energy was directly transferred to the nearby receptor TMPyP without emitting photons (Fig. S5†). The process of energy transfer resulted in the decrease of C-dot fluorescence (even quenching) and the enhancement of TMPyP fluorescence.21Fig. 2d shows that with the increase of TMPyP concentration (0–10 μM), the fluorescence of C-dots decreased gradually under the same excitation wavelength, especially at 420–440 nm (absorption peak of porphyrin), while the fluorescence intensity between 600 nm and 800 nm increased correspondingly. In addition, a shorter lifetime of C-dot–TMPyP compared with that of the only donor C-Dots further proved the FRET process (Fig. S6†).
Metal ion discrimination
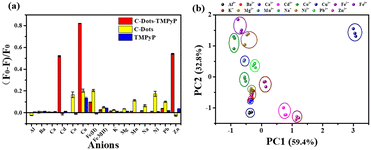
Despite recent advances in the detection of heavy metal ions, there remain numerous challenges in developing convenient, effective and low-cost technologies. Our array-based artificial tongue contains three fluorescent sensors: C-dots, TMPyP and a C-dot–TMPyP complex. When different metal ions are added to the sensor array, they would interact to a greater or lesser extent with the surface groups of C-dots, TMPyP and C-dot–TMPyP. The distinct fluorescence responses to different metal ions give rise to a unique fingerprint-like pattern for discrimination. Fifteen metal ions (Al3+, Ba2+, Ca2+, Cd2+, Co2+, Cu2+, Fe2+, Fe3+, K+, Mg2+, Mn2+, Na+, Ni2+, Pb2+, Zn2+) were chosen to verify the feasibility of the sensor array. Different metal ions were added to the array separately at the same concentration; the array under UV light is exhibited in Fig. S7.† While the fluorescence responses of each sensor also changed to different degrees, as shown in Fig. S8,† the fluorescence changes of different metal ions at 405, 715 and 717 nm in the three sensor elements were recorded respectively.The combinations of fluorescence responses from the three sensors are plotted in Fig. 3a; it was suggested that each sensor expressed unique patterns with different metal ions. Herein, each analyte was monitored with the sensor array three times, which resulted in a data matrix containing 135 data points (3 sensors × 15 analytes × 3 times). Principal component analysis (PCA) was used to process the multivariate data in SPSS Statistics. Three principal components were generated by PCA using the fingerprints of different samples and the first two principal components were selected to produce two-dimensional plots. Each cluster containing three points represented the response pattern of an individual analyte. PC1 and PC2 were used as the X and Y coordinates to draw the PCA scattering diagram of different metal ions (Fig. 3b). It was displayed that all 15 clusters were well separated from each other, which proved that the sensor array could accurately identify distinct metal ions. Among all the metal ions, Cd2+, Cu2+, Fe2+ and Zn2+ ions were more likely to interact with the three sensors and induced unique fluorescence responses. The more independent spatial positions of the four ions led to an easier discrimination. The distribution of another four metal ions (Ba2+, Ca2+, K+, Mg2+) in the figure was relatively close; it was better to distinguish them by amplifying their area separately (Fig. S9†).
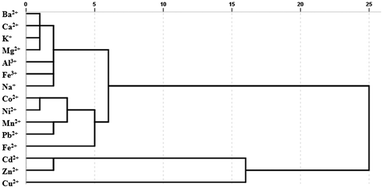
A statistical classification method, hierarchical clustering analysis (HCA), was applied to analyse similar responses to different metal ions. Similar responses to metal ions tended to form a cluster apart from less similar metal ions. According to the different interactions between metal ions and sensors, different classifications were determined. The HCA dendrogram described the precise classification of metal ions in Fig. 4. The length of the horizontal lines indicated the relative difference in the Euclidean distance of the metal ions. Fe2+ and Fe3+ were divided into two clusters to distinguish effectively. Cu2+, Cd2+ and Zn2+ are proved to interrupt the FRET process by breaking the C-dot–TMPyP complex to restore the fluorescence of the carbon dots.22 The flat porphyrin structure in the C-dot–TMPyP complex accelerated the binding of Cu2+, Cd2+, and Zn2+ to the porphyrin ring compared with free TMPyP according to Fig. 3a. On the other hand, the amino groups on the surface of C-dots could combine with Cu2+ to form cupric amine, which led to the selectivity and strong quenching of fluorescence through the inner filter effect.23
Further responses of the sensor array to different concentrations of single metal ions were investigated. Cu2+, Cd2+, Zn2+ and Fe2+ were chosen to be tested due to their unique fluorescence responses, despite the sensor array being available for all the 15 kinds of metal ions. Four different concentrations of each metal ion were titrated to the sensor array respectively, and the corresponding fluorescence spectra are shown in Fig. S10a–d.† As the four assays used the same analysis method, Cu2+ was taken for a detailed discussion. When Cu2+ was added to the C-dot–TMPyP solution, it caused a large fluorescence change compared with free C-dots and TMPyP. With the increase of Cu2+ concentration, C-dots and TMPyP molecules were surrounded by Cu2+, and the excited state energy of TMPyP transferred to Cu2+ as the concentration reached the critical of energy migration; the process of FRET was completely limited and affected the intrinsic fluorescence emission of the TMPyP molecules. PCA was used to analyse the obtained fluorescence data in Fig. S10e;† the response mode divided the Cu2+ samples of different concentrations into four clusters. There was a linear relationship between the concentration and the fluorescence intensity within the concentration range of 0–12 μM, and the limit of detection (LOD) for Cu2+ was calculated to be as low as 0.25 μM (Fig. S10a inset†). The information of all the calibration curves is displayed in Table S1.† Compared with the reported work for detecting metal ions, the sensor platform was more facile and effective.
Conclusions
In summary, we have constructed an artificial tongue based on a sensor array which consists of C-dots, TMPyP and C-dot–TMPyP for the discrimination of different metal ions. Fifteen kinds of metal ions can be well identified by PCA and HCA multivariable chemometric analysis technology. The sensor array possesses the advantages of convenient operation, low-cost and high efficiency without complex instruments, and is potentially considered as a green and user-friendly analytical tool. We are hopeful to extend the receptor to the detection of a broad spectrum of analytes, providing the possibility for the application in the biomedical field in the future.Author contributions
Yingying Chen: investigation and writing – review & editing. Xiaowei Liu: formal analysis and writing – original draft preparation. Xuetan Yan: investigation and validation. Xinying Zhang: characterization. Zhiwei Zhang: investigation. Francois Amblard: methodology. Lingyan Feng: project administration, conceptualization and supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors gratefully acknowledge the National Natural Science Foundation of China [Grant No. 22177067], the Shanghai Rising-Star Program [No. 20QA1403400] and the Shanghai Sailing Program [No. 20YF1413000].Notes and references
- C. Hou, J. Dong, G. Zhang, Y. Lei, M. Yang, Y. Zhang, Z. Liu, S. Zhang and D. Huo, Biosens. Bioelectron., 2011, 26, 3981–3986 CrossRef CAS PubMed.
- Y. Tao, X. Ran, J. Ren and X. Qu, Small, 2014, 10, 3667–3671 CrossRef CAS PubMed.
- G.-J. Liu, S.-N. Tian, C.-Y. Li, G.-W. Xing and L. Zhou, ACS Appl. Mater. Interfaces, 2017, 9, 28331–28338 CrossRef CAS PubMed.
- S. Abbasi-Moayed, H. Golmohammadi and M. R. Hormozi-Nezhad, Nanoscale, 2018, 10, 2492–2502 RSC.
- H. Qiu, F. Pu, X. Ran, J. Ren and X. Qu, Chem. – Eur. J., 2017, 23, 9258–9261 CrossRef CAS PubMed.
- G. Chen, F. Song, X. Xiong and X. Peng, Ind. Eng. Chem. Res., 2013, 52, 11228–11245 CrossRef CAS.
- Z. Lei, C. Sun, P. Pei, S. Wang, D. Li, X. Zhang and F. Zhang, Angew. Chem., Int. Ed., 2019, 58, 8166–8171 CrossRef CAS PubMed.
- L. Kokko, T. Lövgren and T. Soukka, Anal. Chim. Acta, 2007, 585, 17–23 CrossRef CAS PubMed.
- J. Shi, C. Chan, Y. Pang, W. Ye, F. Tian, J. Lyu, Y. Zhang and M. Yang, Biosens. Bioelectron., 2015, 67, 595–600 CrossRef CAS PubMed.
- Y. Liu, Q. Ouyang, H. Li, M. Chen, Z. Zhang and Q. Chen, J. Agric. Food Chem., 2018, 66, 6188–6195 CrossRef CAS PubMed.
- M. Han, S. Zhu, S. Lu, Y. Song, T. Feng, S. Tao, J. Liu and B. Yang, Nano Today, 2018, 19, 201–218 CrossRef CAS.
- K. A. S. Fernando, S. Sahu, Y. Liu, W. K. Lewis, E. A. Guliants, A. Jafariyan, P. Wang, C. E. Bunker and Y.-P. Sun, ACS Appl. Mater. Interfaces, 2015, 7, 8363–8376 CrossRef CAS PubMed.
- M. L. Liu, B. B. Chen, C. M. Li and C. Z. Huang, Green Chem., 2019, 21, 449–471 RSC.
- Y. Wang, L. Bao, Z. Liu and D.-W. Pang, Anal. Chem., 2011, 83, 8130–8137 CrossRef CAS PubMed.
- J. Tang, B. Kong, H. Wu, M. Xu, Y. Wang, Y. Wang, D. Zhao and G. Zheng, Adv. Mater., 2013, 25, 6569–6574 CrossRef CAS PubMed.
- L. Hu, Y. Sun, Y. Zhou, L. Bai, Y. Zhang, M. Han, H. Huang, Y. Liu and Z. Kang, Inorg. Chem. Front., 2017, 4, 946–953 RSC.
- X. Liu, J. Lu, J. Chen, M. Zhang, Y. Chen, F. Xing and L. Feng, Front. Chem., 2020, 8, 670 CrossRef CAS PubMed.
- J. Chen, X. Liu, X. Hou, Y. Chen, F. Xing and L. Feng, Anal. Bioanal. Chem., 2020, 412, 2893–2901 CrossRef CAS PubMed.
- Z. Li, J. R. Askim and K. S. Suslick, Chem. Rev., 2019, 119, 231–292 CrossRef CAS PubMed.
- X. Wang, K. Qu, B. Xu, J. Ren and X. Qu, Nano Res., 2011, 4, 908–920 CrossRef CAS.
- S. Hu, Q. Zhao, Q. Chang, J. Yang and J. Liu, RSC Adv., 2014, 4, 41069–41075 RSC.
- Y. Xu, L. Zhao, H. Bai, W. Hong, C. Li and G. Shi, J. Am. Chem. Soc., 2009, 131, 13490–13497 CrossRef CAS PubMed.
- S. Liu, J. Tian, L. Wang, Y. Zhang, X. Qin, Y. Luo, A. M. Asiri, A. O. Al-Youbi and X. Sun, Adv. Mater., 2012, 24, 2037–2041 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00155a |
| This journal is © The Royal Society of Chemistry 2023 |